Abstract
Human hematopoietic stem cells (HSCs), which arise from aorta-gonad-mesonephros (AGM), are widely used to treat blood diseases and cancer. However, a technique for their robust generation in vitro is still missing. Here we show temporal manipulation of Wnt signaling is sufficient and essential to induce AGM-like hematopoiesis from human pluripotent stem cells. TGFβ inhibition at the stage of aorta-like SOX17+CD235a− hemogenic endothelium yielded AGM-like hematopoietic progenitors, which closely resembled primary cord blood HSCs at the transcriptional level and contained diverse lineage-primed progenitor populations via single cell RNA-sequencing analysis. Notably, the resulting definitive cells presented lymphoid and myeloid potential in vitro; and could home to a definitive hematopoietic site in zebrafish and rescue bloodless zebrafish after transplantation. Engraftment and multilineage repopulating activities were also observed in mouse recipients. Together, our work provided a chemically-defined and feeder-free culture platform for scalable generation of AGM-like hematopoietic progenitor cells, leading to enhanced production of functional blood and immune cells for various therapeutic applications.
Introduction
Hematopoietic stem cells (HSCs) lay the foundation of hematopoiesis and give rise to all functional myeloid and lymphoid cells, including erythrocytes, leukocytes, platelets, immune T and natural killer (NK) cells1,2. Perturbations in the hematopoietic system have caused numerous diseases such as anemia, thrombocytopenia and leukemia1,3. Hematopoietic cell transplantation (HCT) using HSCs and hematopoietic cell transfusion are widely used as primary treatments for many hematological diseases2,4 without alternative curative means. However, the lack of access to reliable cell sources of HSCs limits such therapeutic applications, since numbers of transplantable cells conventionally from bone marrow, cord blood, and mobilized peripheral blood are sometimes quite insufficient, and robust cell expansion strategies are still needed5,6. Other challenges, such as the risk of graft-versus-host diseases and shortage of human leukocyte antigen-matched cell donors, further hamper utility of these readily available HSCs2,7. Alternative cell sources of transplantable HSCs are thus needed to enhance healthcare efforts.
Human pluripotent stem cells (hPSCs) represent one potential source for transplantable HSCs and could serve as an in vitro model for elucidating underlying mechanisms of human hematopoiesis, due to their unique properties of capacity for self-renewal and pluripotency2,8. The past decade has witnessed rapid development of methodologies for de novo hematopoietic cell generation9, though most of them resemble yolk-sac-stage hematopoietic cells that lack long-term repopulating ability after transplantation10. This is partly due to the complex nature of the embryonic hematopoietic system composed of multiple stage-specific hematopoietic progenitor cells with distinct potential11. In mouse embryos, the earliest long-term repopulating HSCs arise from the aorta-gonad-mesonephros (AGM) region at embryonic day 119. Pre-HSCs produced in AGM home to fetal liver and mature to become repopulating HSCs12, highlighting the importance of AGM for definitive hematopoiesis and the need for reproducible methods to differentiate hPSCs into AGM-like HSCs. Recently, patterning of Wnt and Activin signaling was used to convert hPSCs into more aorta-like vasculature, promoting definitive AGM-like hematopoietic cells that could home to bone marrow10. However, a mixed culture of SOX17+ and SOX17- vasculature was induced respectively, promoting both definitive and primitive hematopoiesis. A platform for robust generation of homogenous SOX17+ vasculature and definitive hematopoietic cells is still missing.
Here, we sought to build a robust and straightforward differentiation platform to generate definitive AGM-like hematopoietic stem and progenitor cells (HSPCs) by recapitulating in vivo AGM hematopoiesis. It is well-established that repopulating HSCs develop from hemogenic endothelium (HE) in arterial vasculature through the endothelial-to-hematopoietic transition (EHT) process13–15. GSK3β inhibition of hPSCs induced robust generation of homogenous aorta-like CD31+CD34+ HE from hPSCs that are marked by SOX17, a transcription factor expressed in vascular structures of AGM and which is required for HSC generation from AGM10,16,17. We devised an all-in-one inducible Cas13d-mediated SOX17 knockdown platform, and found SOX17 knockdown significantly blocked formation of endothelial cells induced by Wnt activation. TGFβ inhibition significantly promoted the EHT process generating CD43+CD45+ hematopoietic cells that co-expressed CD44 and RUNX1, hallmarks of AGM-like hematopoietic cells10,18–20. Using an inducible shRNA CTNNB1 knockdown system, we demonstrated that Wnt inhibition was sufficient to induce hematopoiesis from HE, mimicking aspects of in vivo AGM hematopoiesis21. The resulting late-stage day 18 HSPCs resembled primary cord blood HSCs at global transcript levels, displayed lymphoid and myeloid potential in vitro, and homed to definitive caudal hematopoietic tissue (CHT) in zebrafish. Notably, our hPSC-derived hematopoietic cells also significantly rescued c-myb knockout bloodless zebrafish up to 4 days after transplantation, and presented engraftment and multilineage repopulating activities in mouse recipients. Single-cell RNA-sequencing (scRNA-seq) analysis identified similarities and differences between hPSC-derived and primary AGM hemato-endothelial progenitor cells. Discrete sub-populations, enriched for erythroid, myeloid, monocytic, granulocytic or megakaryocytic markers, and their developmental hierarchy were identified in the day 18 hPSC-derived HSPCs. Our findings provide fundamental advances in determining critical components for in vitro specification of more homogeneous definitive hematopoiesis. This simplified platform will offer a robust model for human hematopoiesis and pave the way to a scalable production of functional blood and immune cells for various therapeutic applications.
Results
Canonical Wnt signaling specifies homogenous aorta-like CD34+SOX17+ endothelium.
Producing hemogenic endothelium (HE) from hPSCs, marked by the expression of typical endothelial marker VE-cadherin (VE-cad), CD31, and CD34, is a vital step towards hematopoietic cell generation. Homogenous CD34+CD31+ HE was robustly induced from hPSCs via small-molecule activation of Wnt signaling (Fig. 1A–B)22,23. Since simultaneous modulation of Wnt and Activin signaling yields aorta-like SOX17+ vessels in AGM that give rise to hematopoietic cells10, we speculated that our small molecule-induced CD34+CD31+ cells are also SOX17+, which was confirmed by flow cytometry and immunostaining analysis of SOX17 expression (Fig. 1C–D, Supplementary Fig. 1A). Interestingly, GSK3β inhibition alone yielded a relatively homogenous CD34+SOX17+ population (Supplementary Fig. 1B–C), whereas both CD34+SOX17- and CD34+SOX17+ populations were observed in the day 6 embryoid body (EB) cultures with Activin A and Wnt modulation10,24. Under CHIR99021 (CHIR) treatment, CD34+SOX17+ HE emerged as early as day 4 (Supplementary Fig. 1D–E). Furthermore, resulting day 6 CD34+SOX17+ AGM-like endothelial cells did not express CD235a (Supplementary Fig. 1F), a well-known marker for primitive hematopoiesis25. Using an all-in-one inducible Cas13d-mediated gene knockdown system (Fig. 1E), we demonstrated that SOX17 knockdown (Fig. 1F) significantly blocked formation of endothelial cells (ECs) (Fig. 1G), consistent with previous studies26,27.
Figure 1.
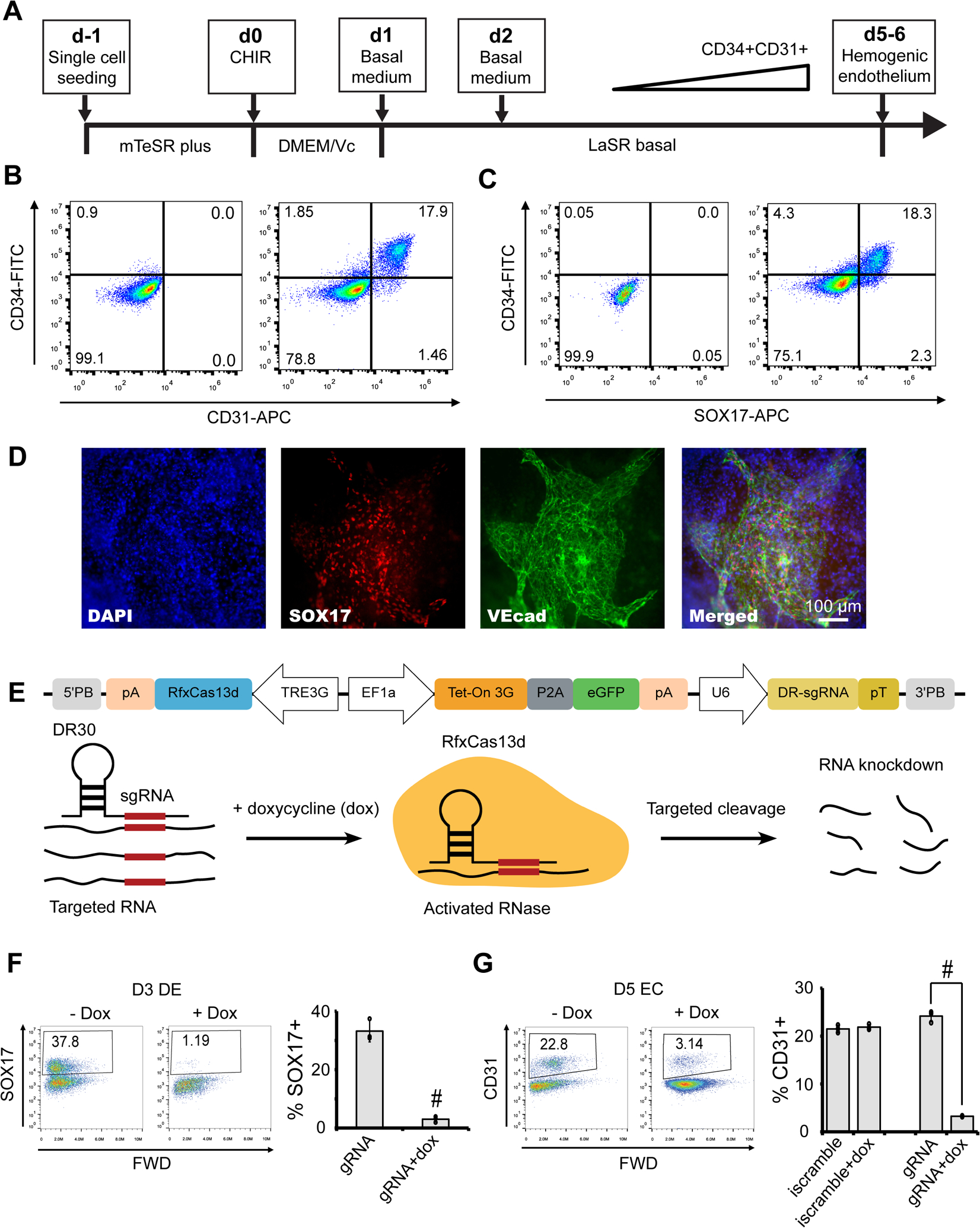
Canonical Wnt signaling specifies homogenous aorta-like CD34+ SOX17+ endothelium. (A) A schematic of the protocol used to differentiate hPSCs towards hemogenic endothelium. (B–D) 19-9-11 iPSC-derived day 5 cultures were subjected to flow cytometry analysis for CD34/CD31 (B) and CD34/SOX17 (C), and immunostaining analysis for SOX17 and CD34 (D). Scale bars, 200 μm. CHIR, CHIR99021. (E–G) Drug-resistant H9 hPSCs (E) were differentiated into definitive endoderm (F) and hemogenic endothelium (G) with or without doxycycline (dox) treatment, and day 3 definitive endoderm (DE) and day 5 endothelial cell (EC) cultures were subjected for flow cytometry analysis of SOX17 (F) and CD31 (G) expression. #P < 0.05, dox treatment versus no dox condition. Inducible Cas13d scramble gRNA (iscramble) line was used as negative control. Data are represented as mean ± s. e.m of three independent replicates.
Screening developmental signaling pathways reveals contribution from Wnt and TGFβ inhibitions to the hematopoiesis of hemogenic endothelium.
Many signaling pathways, including Wnt10,25, TGFβ28, BMP29,30, retinoic acid (RA)21, Notch31, etc., and their cross-talks, regulate hematopoietic cell specification at multiple stages. We next screened small molecules and cytokines regulating signaling pathways involved in the induction of hematopoiesis from hPSC-derived CD34+SOX17+ HE (Fig. 2A). SB431542 (SB), a TGFβ inhibitor, significantly enhanced generation of CD43+CD45+ hematopoietic cells (Fig. 2B–D), which was blocked by a GSK3β inhibitor, CHIR99021 (CHIR). Since SB may also block ALK5 receptor (also called TGFβ type I receptor) and BMP signaling, we treated HE with ALK5 inhibitor II and a BMP inhibitor (Dorsomorphin) and found that ALK5 inhibition could also efficiently induce hematopoiesis (Supplementary Fig. 2A), confirming the importance of TGFβ signaling in hematopoiesis. Wnt-C59, a Wnt inhibitor, also significantly induced emergence of hematopoietic cells (Fig. 2B–C). CD34+ HE and CD34- non-HE cells were sorted (Supplementary Fig. 2B) for hematopoietic cell differentiation and more CD45+ cells were produced from CD34+ cells (Supplementary Fig. 2C). Wnt-C59 and SB also induced hematopoiesis in H9 and 6-9-9 hPSCs with a similar efficiency (Supplementary Fig. 2D).
Figure 2.
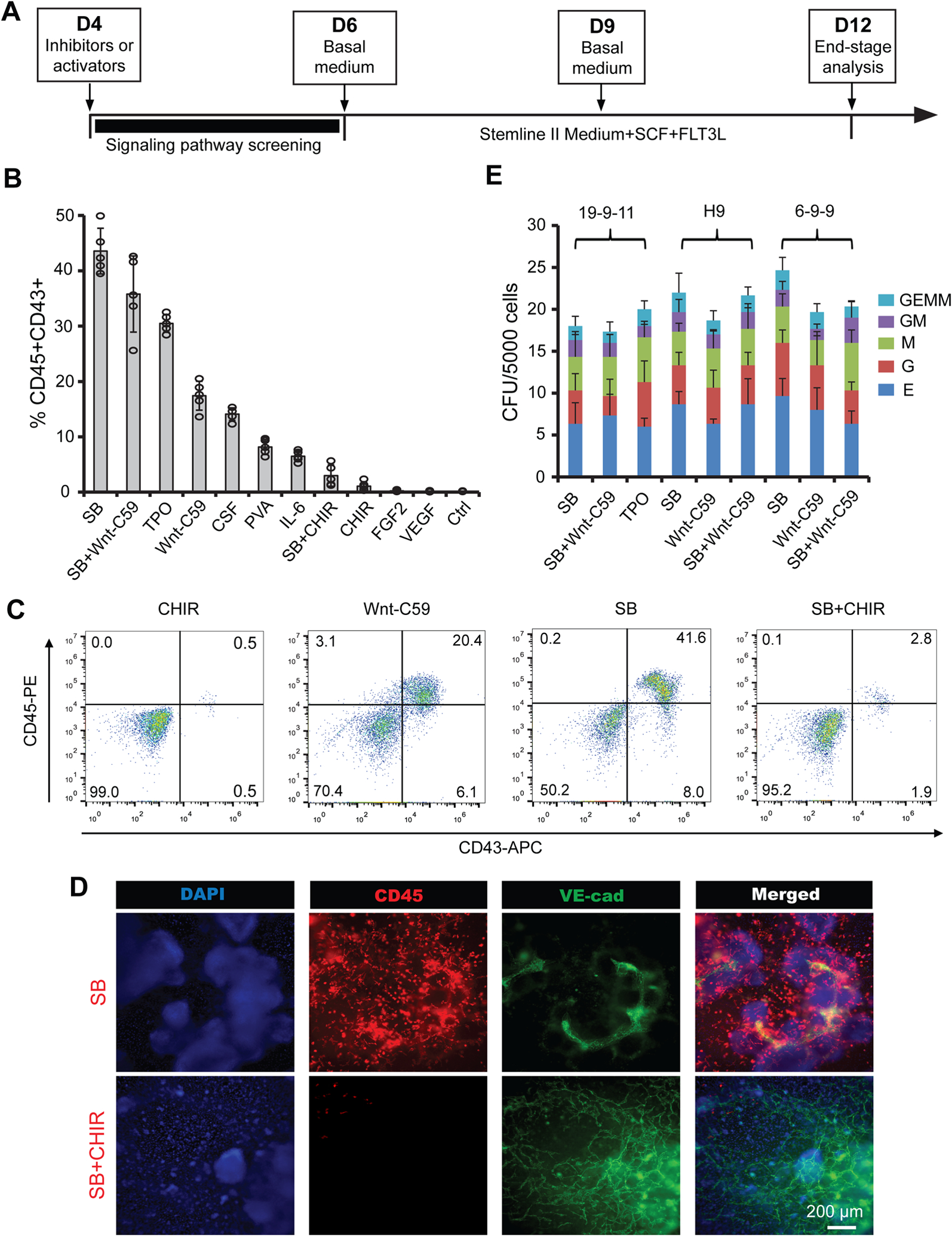
Wnt and TGFβ inhibitors significantly induce hematopoiesis of hemogenic endothelium. (A) A schematic of the protocol used to differentiate CD34 + SOX17+ hPSC-derived hemogenic endothelium towards hematopoietic cells. (B–D) 19-9-11 iPSC-derived cultures differentiated as shown in (A) with indicated molecular signaling regulators were subjected to flow cytometry analysis for CD45/CD43 (B), and representative flow plots were shown in (C) and immunostaining images were shown in (D). CHIR, CHIR99021; SB, SB431542; Ctrl, control; PVA, polyvinyl alcohol; TPO, thrombopoietin; CSF, colony-stimulating factor 3. (E) Colony-forming unit (CFU) assay was performed on day 12 differentiated cultures with indicated treatment using 19-9-11, H9 and 6-9-9 hPSC lines, and scored by cellular morphology: erythroid (E), granulocyte (G), granulocyte/macrophage (GM), macrophage (M), and multilineage progenitor (GEMM). Data are represented as mean ± s. e.m of three to five independent replicates. Scale bars, 200 μm.
Notably, a higher hematopoietic specifying efficiency was observed under SB treatment, suggesting potential cross-talk between TGFβ and other signaling pathways, such as RA signaling21, in addition to the Wnt inhibition, for the SB-induced robust hematopoiesis. Importantly, hematopoietic cells derived from hPSCs under different conditions could produce colony-forming units (CFUs) with various subtypes in methylcellulose (Fig. 2E, Supplementary Fig. 2E). Collectively, our results demonstrate that TGFβ and Wnt inhibition significantly enhances hematopoiesis of hPSC-derived HE, offering the potential for an accessible, simplified platform for further investigation of signaling pathways involved in human definitive hematopoiesis.
Wnt inhibition is sufficient to induce hematopoiesis of AGM-like hemogenic endothelium.
To further investigate the role of Wnt signaling during hematopoiesis, we performed RT-PCR analysis of the day 6 HE samples under different conditions from day 4 to 6. Low expression of WNT3A and AXIN2 (Fig. 3A), a downstream target of Wnt signaling, as well as low transcriptional activity of endogenous Wnt signaling in a 7TGP Wnt reporter line32 (Fig. 3B–C), were observed in both SB and Wnt-C59 cultures, indicating the critical role of Wnt inhibition at this stage. CTNNB1 knockdown via inducible beta-catenin shRNA (ishcat)32 further confirmed that Wnt inhibition is sufficient for hematopoiesis of HE (Fig. 3D–E). These findings are consistent with previous reports that genes antagonizing canonical Wnt signaling are enriched in human AGM cells10, and Wnt inhibition is required for hematopoietic transition from mouse AGM21.
Figure 3.
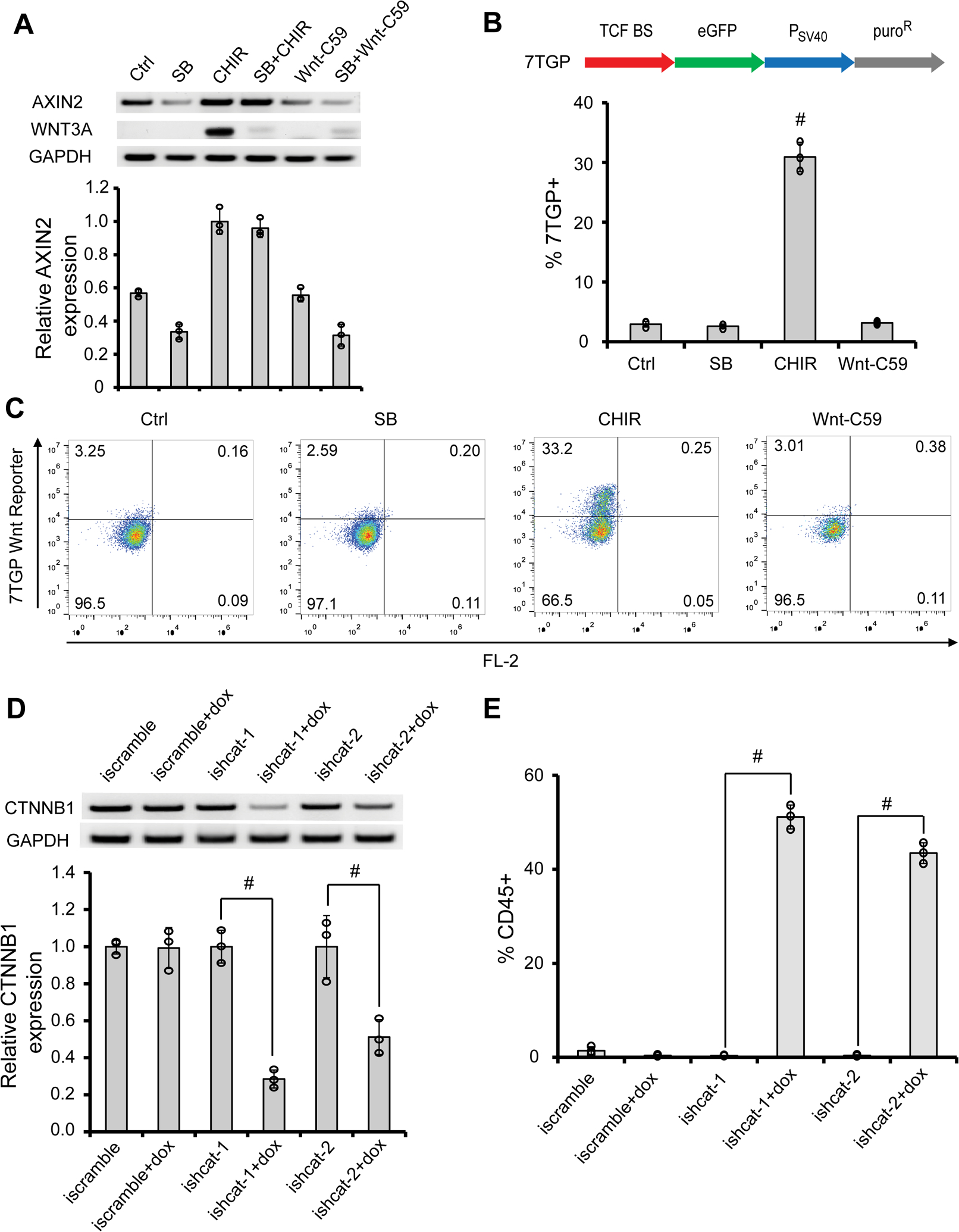
Wnt inhibition is sufficient for the AGM-like hematopoiesis. (A) RT-PCR analysis of 19-9-11 iPSC-derived day 6 cells for AXIN2, WNT3A and GAPDH expression was performed and quantified. (B–C) H9 7TGP Wnt reporter hPSCs (B) were differentiated as illustrated in Fig. 2A with indicated signaling modulators, and day 6 differentiation cultures were subjected to flow cytometry analysis for eGFP expression (B). #P < 0.05, CHIR versus other conditions. Representative flow plots were shown in (C). (D–E) 19-9-11 inducible shRNA CTNNB1 (beta-catenin) knockdown (ishcat) iPSCs were cultured as illustrated in Fig. 2A with or without doxycycline (dox) treatment from day 4 to day 6. At day 6, cells were subjected to RT-PCR analysis and quantified in (D). At day 12, cells were analyzed for CD45 expression by flow cytometry (E). Data are represented as mean ± s. e.m of three independent replicates. #P < 0.05, dox treatment versus no dox condition.
Chemically-defined conditions induce robust generation of AGM-like hematopoietic cells.
Since the window for SB treatment is vital for definitive hematopoiesis8, we optimized the culture conditions and identified day 4 to 6 as an optimal time to add SB (Supplementary Fig. 3A), which resulted in more than 40% CD45+ hematopoietic cells on day 12 (Supplementary Fig. 3B). We then screened different basal media, alone or in combinations, for conditions supporting robust hematopoietic cell generation (Supplementary Fig. 3C–E). While human serum (HS) significantly increased the purity of hematopoietic cells on day 12, the employment of undefined serum increases the complexity and reduces reproducibility. Thus, we employed optimal serum-free conditions, hereon referred to as the GiTi (Gsk3β inhibitor, TGFβ inhibitor) protocol, for subsequent experiments (Fig. 4A). Dynamic morphological changes (Supplementary Fig. 4A) and expressions of hematopoietic-specific markers (Fig. 4B–E, Supplementary Fig. 4B–C), including CD45, CD34, CD43, and CD4418,19, were observed along with the emergence of hematopoietic clusters from day 6. The cells were also positive for RUNX1 (Fig. 4F–G), which is expressed in AGM-derived repopulating HSCs33, confirming a definitive identity of our hematopoietic cells. Removal of SCF and FLT3L from the differentiation medium reduced both the yield and purity of CD45+ hematopoietic cells (Fig. 4H–I). HSPCs collected at day 10 and day 15 both formed CFUs with various subtypes (Supplementary Fig. 4D). Under current culture condition, the number of hPSC-derived CD43+CD45+ HSPCs significantly increased before day 20 and then decreased afterwards (Supplementary Fig. 4E), indicating a limitation of our recipe in supporting their long-term maintenance and expansion. Nevertheless, day 15 HSPCs maintained high viability (Supplementary Fig. 4F) and CFU forming ability (Supplementary Fig. 4G) after freeze-thaw, indicating the possibility for long-term storage and shipping. Collectively, we developed a chemically-defined, feeder-free monolayer culture platform for the generation of AGM-like hematopoietic cells from 11 (9 normal and 2 genetically-modified) hPSC lines (Supplementary Fig. 4H), highlighting its reproducibility and potential clinical application.
Figure 4.
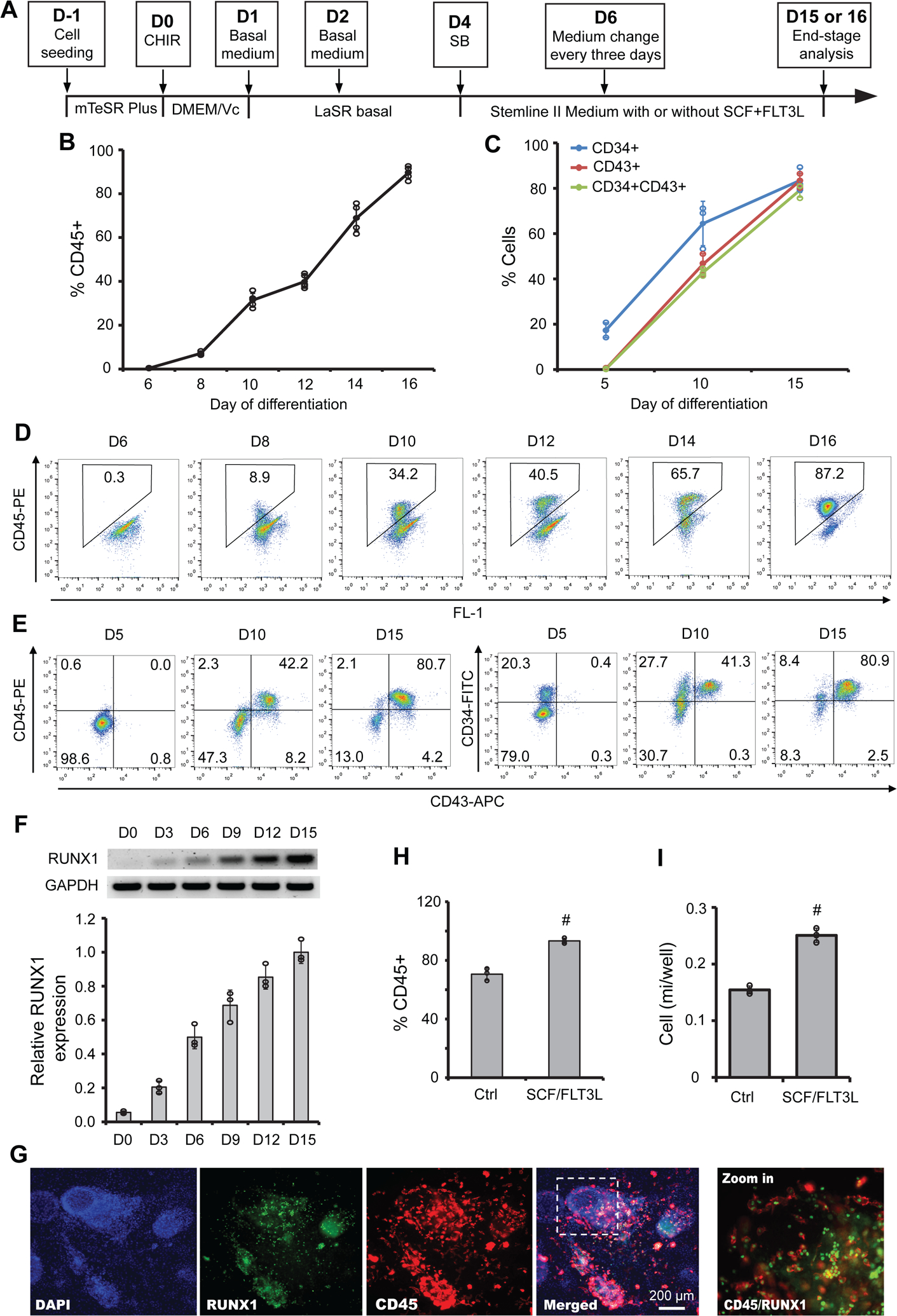
Chemically-defined conditions for robust AGM-like hematopoietic cell generation. (A) A schematic of the optimized protocol for differentiation of hPSCs to hematopoietic cells. (B–G) 19-9-11 iPSCs were differentiated as illustrated in (A). At different time points, CD45 (B) and CD34/CD43 (C) expression was assessed by flow cytometry. Representative flow plots of CD45, CD45/CD43, and CD34/CD43 expression were shown in (D) and (E). RT-PCR analysis of RUNX1 and GAPDH at indicated days was performed and quantified in (F). Representative immunostaining images of CD45 and RUNX1 expression were shown in (G). Scale bars, 200 μm. (H–I) 19-9-11 iPSCs were cultured as illustrated in (A) with or without the addition of SCF and FLT3L, and day 15 cultures were subjected for flow cytometry analysis of CD45 (H) and the yield of CD45+ cells were shown in (I). Data are represented as mean ± s. e.m of three to five independent replicates. #P < 0.05, SCF/FLT3L versus control condition (ctrl).
Single-cell RNA-sequencing analysis identifies similarities and differences between hPSC-derived and primary AGM endothelial cells.
To investigate the dynamics and heterogeneity of hemato-endothelial cells derived from hPSCs, scRNA-seq analysis was performed on day 5 adherent cells and day 8 suspension cells, and compared with that of primary AGM hemato-endothelial cells34. Unsupervised clustering and UMAP embedding of quality control (QC)-filtered scRNA-seq data (Supplementary Fig. 5A–C) revealed 3, 4, and 7 distinct clusters of cells in day 5 (Fig. 5A), day 8 (Fig. 5B) and AGM (Fig. 5C) samples, respectively. Cell identities were assigned to clusters based on their expression of key marker genes (Supplementary Fig. 5D–G). As expected, CDH5+SOX17+ endothelial cells and non-endothelial mesenchymal/stromal cells dominated day 5 and AGM cell clusters (Supplementary Fig. 5H–I), whereas fewer endothelial and mesenchymal/stromal cells were observed in day 8 cells, which mainly included clusters of endothelial (SOX17/CDH5), early (CAV1/RUNX1) hematopoietic, hematopoietic (MTTL3)10 and lineage-primed hematopoietic progenitors that were enriched in mitochondrial genes35 (Supplementary Fig. 5J). Importantly, high expression levels of definitive AGM hematopoiesis markers LMO4 and CD4418,19 and low expression levels of primitive marker GYPA (CD235a)25 were observed across all hemato-endothelial samples. The expression of CD44, recently identified as a marker and regulator of EHT19, was more active in day 5 and 8 cells, indicating these cells were undergoing EHT to achieve a hematopoietic fate. This observation was also confirmed by the lack of hematopoietic marker SPI1 and PTPRC (CD45) in day 5 and day 8 cells (Fig. 5A–B), while a small CD45+ cluster presented in the AGM (Fig. 5C).
Figure 5.
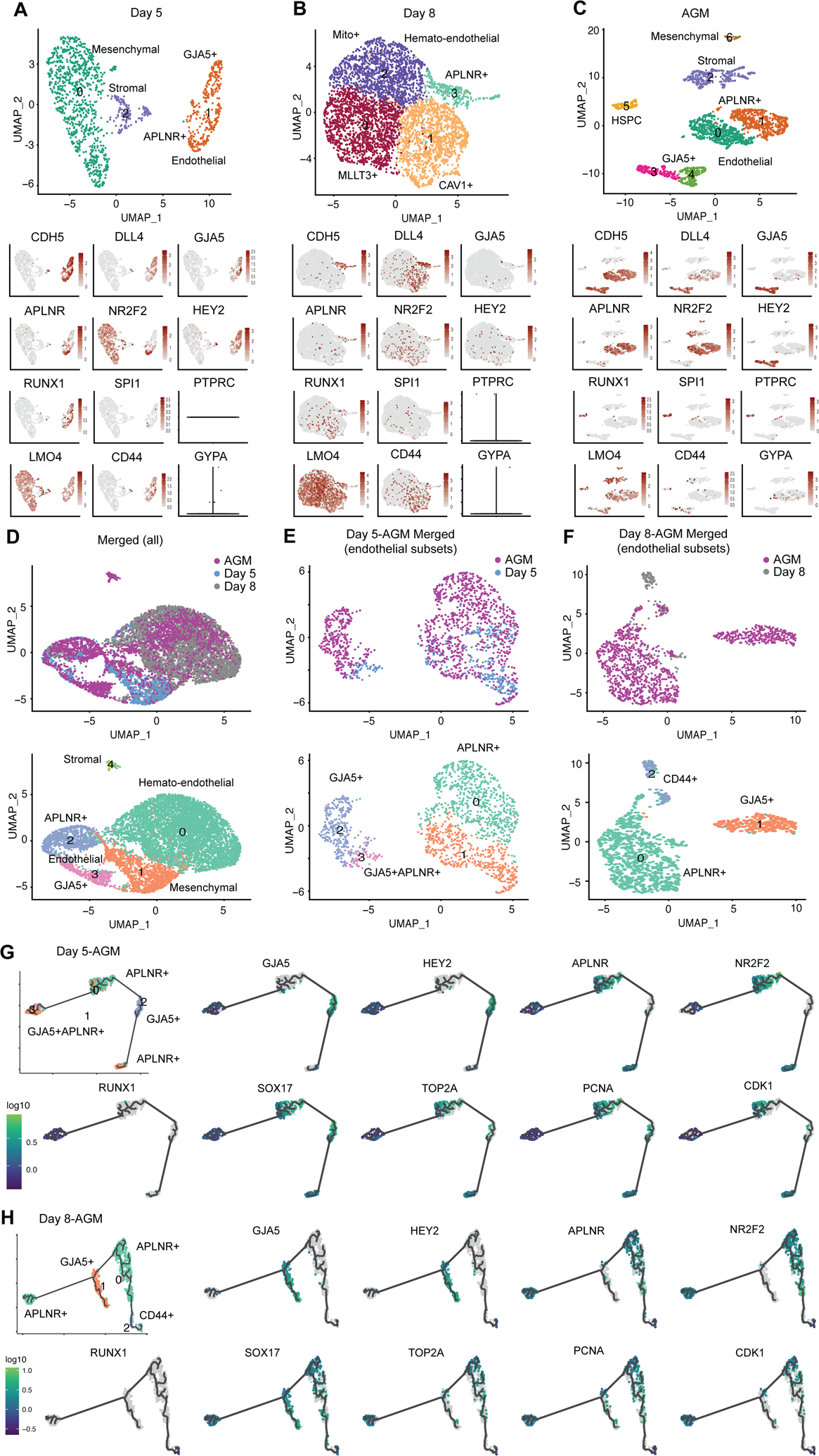
Comparative transcriptome analysis of hPSC-derived and primary aorta-gonad-mesonephros (AGM) hemato-endothelial cells. (A–C) Clustering and UMAP embedding of scRNA-seq data of day 5 (A), day 8 (B) and AGM [34] (C) samples were colored by meta-clusters to simplify visualization. Cell identities were assigned to the clusters based on their expression of key marker genes. Gene expression plots of endothelial and hematopoietic cell markers are also shown. Violin plots for PTPRC and GYPA at day 5 (A) and day 8 (B) were used due to their low expression. (D–F) Clustering and UMAP embedding of scRNA-seq data of merged day 5, day 8 and AGM samples (D), endothelial subsets of merged day 5/AGM (E) and day 8/AGM (F) were colored by meta-clusters to simplify visualization. The lower panels in (D) are cell clusters of merged scRNA-seq samples, in which cell identities were assigned based on their expression of key marker genes. (G–H) Pseudotime trajectory analysis using Monocle 3 of the merged day 5/AGM (G) and day 8/AGM (H) identifies the potential developmental trajectory within the heterogenous endothelial cells. Gene expression plots of hemato-endothelial and cell cycle markers for merged day 5/AGM and day 8/AGM samples were also shown.
To study the hierarchy of these hemato-endothelial cell populations, trajectory analysis was performed using the Monocle packages36. While a single developmental trajectory was not observed in the distinct cell clusters, day 5 and AGM cells arose from the same progenitor pool of MSX2+ mesenchymal (Supplementary Fig. 5H) and APLNR+ endothelial progenitor cells (Supplementary Fig. 5I), respectively. Trajectory analysis on day 8 cells demonstrated the emergence and development of hematopoietic progenitor cells from hemogenic endothelium (Supplementary Fig. 5J). To compare the similarities and differences between hPSC-derived and primary AGM cells, we directly merged day 5, day 8 and AGM cells in the UMAP, which displayed high degrees of overlap in terms of endothelial and hematopoietic cell clusters, respectively (Fig. 5D). Further analysis on the endothelial subsets in merged day 5/AGM and day 8/AGM cells identified two major distinct clusters that express venous (APLNR and NR2F2) and arterial (GJA5 and HEY2) markers34 (Fig. 5E–F). Four different endothelial cell clusters were identified in AGM cells, whereas most day 5 endothelial cells (clusters 1 and 3 in Fig. 5E) expressed both venous and arterial markers at lower expression levels than those of individual venous cluster 0 and arterial cluster 2, indicating a more homogenous population of our day 5 endothelial cells. Day 8 endothelial cells expressed predominantly venous markers APLNR and NR2F2 as well as EHT markers CD4418,19 and RUNX110 (Fig. 5F), suggestive of a more hematopoietic-primed progenitor population at day 8. A previous study using the force-directed graph has identified NR2F2+ venous endothelium enriched with cell cycle markers (TOP2A, PCNA, and CDK1) as the bridge hematogenic endothelium between GJA5+HEY2+ arterial endothelium and CD45+ hematopoietic cell populations in AGM34. Consistent with this finding, Monocle 3-based pseudotime trajectory analysis revealed a potential developmental trajectory of quiescent APLNR+ venous-to-GJA5+ arterial-to-proliferating APLNR+ venous-to-APLNR+GJA5+ endothelial populations in merged day 5-AGM cells (Fig. 5G), and a trajectory of quiescent APLNR+ venous-to-GJA5+ arterial-to-proliferating APLNR+ venous-to CD44+RUNX1+ hematopoietic populations in merged day 8-AGM cell (Fig. 5H, Supplementary Fig. 5K). Our scRNA-seq analysis collectively suggested a similar but more homogenous endothelium of our hPSC-derived cells as compared to the primary AGM cells at transcriptomic levels.
Transcriptome analysis reveals global similarity among hPSC-derived day 18 hematopoietic cells, human primary AGM hemato-endothelial cells and cord-blood HSCs.
To further confirm the identity of hPSC-derived hematopoietic cells, we performed bulk RNA sequencing (RNA-seq) on 6-9-9 and H9-derived day 18 CD45+ hematopoietic cells. Hierarchical clustering analysis (Supplementary Fig. 6A) of RNA-seq expression data of hPSCs, hPSC-derived mesoderm (Mes), hematopoietic stem and progenitor cells (HSPCs), primary human neonatal cord-blood hematopoietic stem cells (CB-HSCs)37, 5-week AGM endothelial (AGM-En), stem/progenitor (AGM-S/P), and progenitor 1 (AGM-Pr1) cells10 demonstrated that our hPSC-derived hematopoietic cells were closely related to primary cord-blood HSCs, thus suggestive of a later hematopoietic stage of our day 18 cells as compared to the isolated early AGM hemato-endothelial progenitor cells10. The close relationship of transcriptional signatures between hPSC-derived hematopoietic cells and CB-HSCs was also confirmed by principal component analysis (PCA) on the RNA-seq data (Fig. 6A). In the 3D score plot of the first three principal components (PCs), hPSC-derived hematopoietic cells clustered with CB-HSCs, and were distinct from other cell populations, including hPSCs and hPSC-derived mesoderm, from which they originated.
Figure 6.
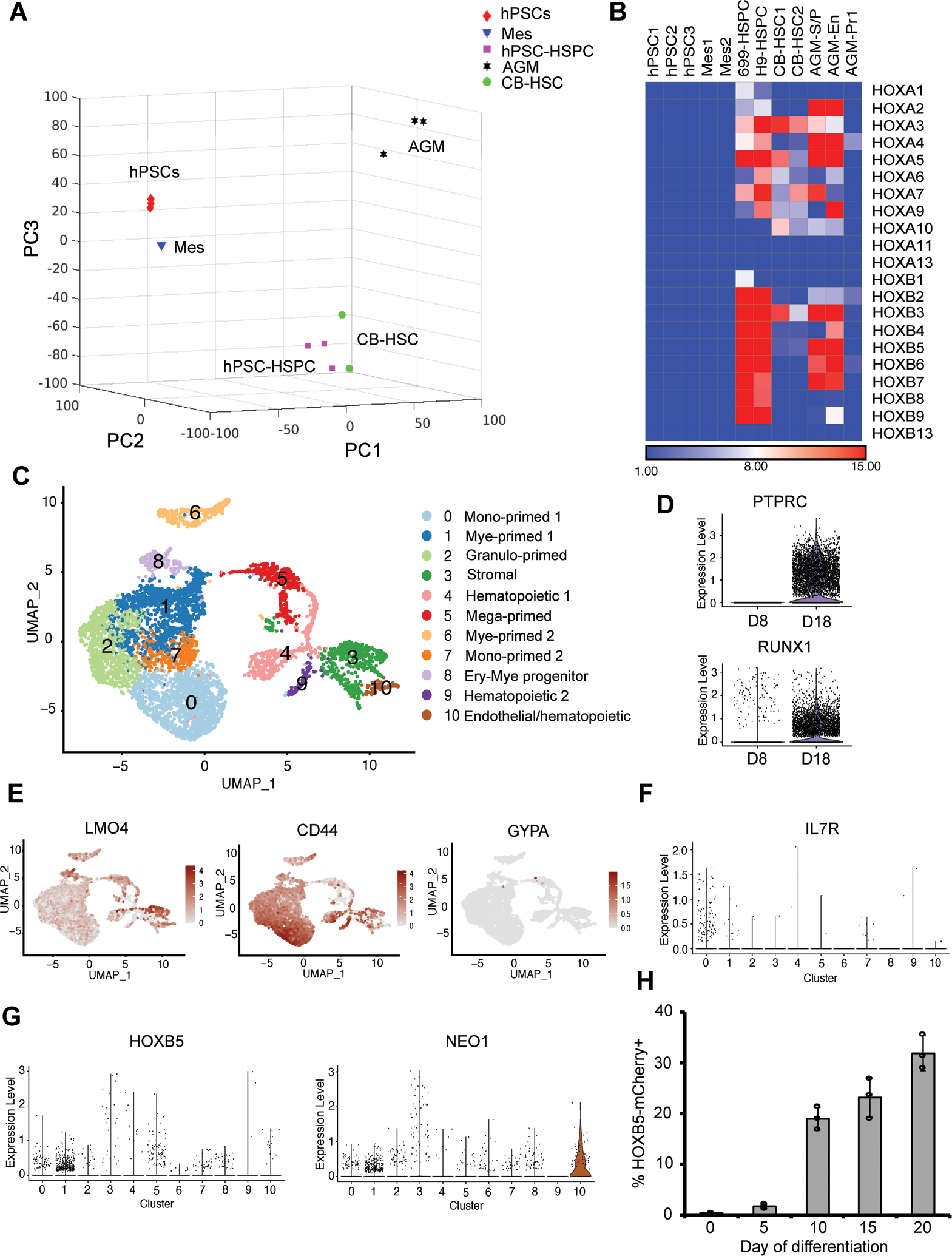
Transcriptome analysis of hPSC-derived definitive hematopoietic cells. (A) 3D scores plot of the first three principal components (PCs) from the principal component analysis on the RNA-sequencing data of hPSCs, hPSC-derived mesoderm (Mes), day 18 hematopoietic stem-like cells (hPSC-HSC), primary neonatal cord-blood HSCs (CB-HSC), 5-week aorta-gonad-mesonephros (AGM) endothelial, stem/progenitor and progenitor cells. Each data point corresponds to different biological samples. RNA-seq data of primary CB-HSC [37] and AGM cells [10] were obtained from previous publications. (B) Heatmap showing similar expression patterns of HOXA and HOXB gene clusters among hPSC-derived, CB and AGM hematopoietic cells. (C) UMAP embedding of day 18 scRNA-seq data colored by meta-clusters to simplify visualization. Mono: monocytes; Mye: myeloid cells; Granulo: granulocytes; Ery: erythroid cells; Mega: megakaryocytes. Violin plots of RNA counts of two hematopoietic progenitor markers PTPRC and RUNX1, and UMAP plots of two definitive and one primitive hematopoietic cell markers are shown in (D) and (E). (F–G) Violin plots of T-cell progenitor marker IL7R, and hematopoietic cell marker HOXB5 and NEO1 along different clusters are shown in (F) and (G). (H) VEcad-eGFP HOXB5-mCherry dual reporter H9 hPSCs were differentiated as illustrated in Fig. 4A. At different time points, HOXB5-mCherry expression was assessed by flow cytometry. Data are represented as mean ± s. e.m of three independent replicates.
Examination of specific hematopoietic genes revealed that hPSC-derived cells express many hematopoietic transcription factors (Supplementary Fig. 6B) and cell-surface markers (Supplementary Fig. 6C), though at lower expression levels than those in primary AGM and CB cells. Gene set enrichment analysis (GSEA) over hPSCs identified enriched hematopoiesis-related gene ontology (GO), including “aorta development”, “cell migration”, “hematopoietic stem cell proliferation”, “Notch signaling regulation”, which further confirmed the transcriptional similarity between hPSC-derived and AGM cells (Supplementary Fig. 6D). Bulk RNA-seq analysis also revealed similar expression patterns of HOXA, a landmark of AGM hematopoiesis10, and HOXB genes in the hPSC-derived and AGM hematopoietic cells (Fig. 6B). The hPSC-derived and CB cells also displayed similar HOXA expression. Collectively, our bulk RNA-seq data suggests the transcriptional similarity between our hPSC-derived cells and AGM as well as CB-HSCs, highlighting the definitive trajectory of our GiTi hematopoietic differentiation7.
Single-cell RNA-sequencing analysis identifies discrete sub-populations in hPSC-derived hematopoietic progenitor cells.
To investigate the dynamics and heterogeneity of hematopoietic cells emerged from hPSC-derived SOX17+CD34+ HE, scRNA-seq analysis was performed on both day 8 (Fig. 5B, Supplementary Fig. 5) and day 18 (Fig. 6C–G, Supplementary Fig. 7) suspension cells. Unsupervised clustering and UMAP embedding of quality control (QC)-filtered scRNA-seq data (Supplementary Fig. 7A) revealed 11 distinct clusters of cells on day 18 (Fig. 6C). As expected, hematopoietic markers PTPRC (CD45) and RUNX1 were highly expressed in most day 18 cells (Fig. 6D). Cell identities were assigned to clusters based on their expression of key marker genes (Supplementary Fig. 7B). Day 18 cells contained clusters of stromal (IGF2/COL1A1), endothelial, and hematopoietic cells, as well as clusters of progenitor cells primed towards megakaryocyte (GP9/PF4), monocyte (SPP1/CCL3 and CD74/MMP9), granulocyte (AZU1/PRTN3), myeloid (MPO/LYZ) and erythroid (KLF1/HBE1)10 (Supplementary Fig. 7B–F). Interestingly, hematopoietic cells under the current culture conditions were biased towards granulocyte fate, rather than erythroid lineages, as only a small fraction of cells expressed HBE1 or KLF138, indicating their definitive identity39. Both day 8 and 18 cells displayed high expression levels of definitive AGM hematopoiesis markers LMO4 and CD4418,19 (Fig. 5B, 6E). In contrast, only a few cells in both samples were positive for the primitive marker GYPA (CD235a)25 (Fig. 5B, 6E). The downregulation of LMO4 and upregulation of CD44 upon EHT was consistent with previous in vivo emergence of hematopoietic cells from aortic endothelium of mouse AGM19. In addition to its role in regulating EHT, CD44 is also a maker of adult HSCs40 and is involved in fetal HSC homing and long-term engraftment41, suggesting a potentially high homing ability of our hPSC-derived hematopoietic cells.
To study the hierarchy of our day 18 hematopoietic cell populations, pseudotime developmental trajectory analysis was performed using Monocle packages36. Trajectory analysis on day 8 cells demonstrated the emergence and development of hematopoietic progenitor cells from hemogenic endothelium (Supplementary Fig. 5J). For day 18 cells, a single developmental trajectory was not observed in Monocle 3-based pseudotime trajectory analysis (Supplementary Fig. 7B), indicating the high heterogeneity of our hPSC-derived hematopoietic cells. Monocle 2 pseudotime trajectory analysis revealed that day 18 hematopoietic progenitor cells branched from a central core to three distinct trajectories of monocyte-, granulocyte- and erythroid/megakaryocyte-primed lineages (Supplementary Fig. 7C–E). Additional endothelial (CAV1), hematopoietic (RUNX1), erythroid (KLF1/HBE1), and T-cell progenitor (IL7R) cells42 were also positioned to the trajectory map and represented by distinct branches (Fig. 6F, Supplementary Fig. 7F). Despite the lack of adult HBB expression in day 18 cells, high expression levels of HBA1 and HBG2, as well as low expression level of HBZ (Supplementary Fig. 7G), and the globin switches from ε to γ and ζ to α, suggested a fetal-like globin profiling. This observation coincides with the transition from the primitive yolk sac to definitive fetal liver and bone marrow hematopoiesis43, consistent with fetal-like globin switches and profiling observed in the CHIR99021/SB431542-induced AGM-like hematopoiesis from hPSCs10. HOXB cluster genes, such as HOXB544, are predominantly enriched in murine LT HSCs45. More recently, NEO146 was reported as a marker of more dormant subpopulations of HOXB5+ LT-HSCs. Consistent with bulk RNA-seq data (Fig. 6B), expression of HOXB5 and NEO1 was detected across different hematopoietic cell populations via scRNA-seq analysis (Fig. 6G). To monitor the dynamics of HOXB5 during hematopoietic cell cultures, mCherry fluorescent protein was knocked into the endogenous HOXB5 locus of H9 VE-cad-eGFP reporter line via CRISPR/Cas9-mediated homology-directed repair (HDR)47 (Supplementary Fig. 8A). PCR genotyping and sequencing identified 1 out of 17 picked clones successfully targeted in both alleles (Supplementary Fig. 8B). Immunostaining analysis confirmed the co-localization of HOXB5 and mCherry expression (Supplementary Fig. 8C). The mCherry signal was first detected at day 5 and elevated at day 20 (Fig. 6H, Supplementary Fig. 8D–E), indicating the potential presence of LT-HSCs. Collectively, our data revealed the heterogeneity and hierarchy of seemingly homogenous hPSC-derived HSPCs, highlighting the definitive trajectory of our GiTi hematopoietic differentiation7.
In vitro and in vivo characterization of hPSC-derived definitive hematopoietic cells.
To further assess their hematopoietic potential, we performed in vitro and in vivo functional assays on the hPSC-derived hematopoietic cells. In methylcellulose-based colony-forming unit assays, erythroid (E), granulocyte/macrophage (GM), macrophage (M), and multilineage progenitor (GEMM) colonies were generated (Supplementary Fig. 9A–C), confirming the multipotent potential of our hematopoietic cells. Lymphocytic T and natural killer (NK) cells, a hallmark of definitive hematopoiesis25, were also successfully generated on OP9-DLL4 feeder cells (Fig. 7A–B). Notably, the NK cells generated have presented anti-tumor cytotoxicity against various tumor cells (Fig. 7C) with a killing efficiency comparable to the reported numbers of primary NK and NK92 cells48. However, the tumor-killing efficiency of our hPSC-derived NK cells is lower than that of primitive yolk sac erythro-myeloid progenitor (EMP)-derived NK cells, which is likely due to their more granular phenotypes49. In summary, our results highlight that our hPSC-derived hematopoietic cells can produce off-the-shelf immune cells for both research and clinical applications.
Figure 7.
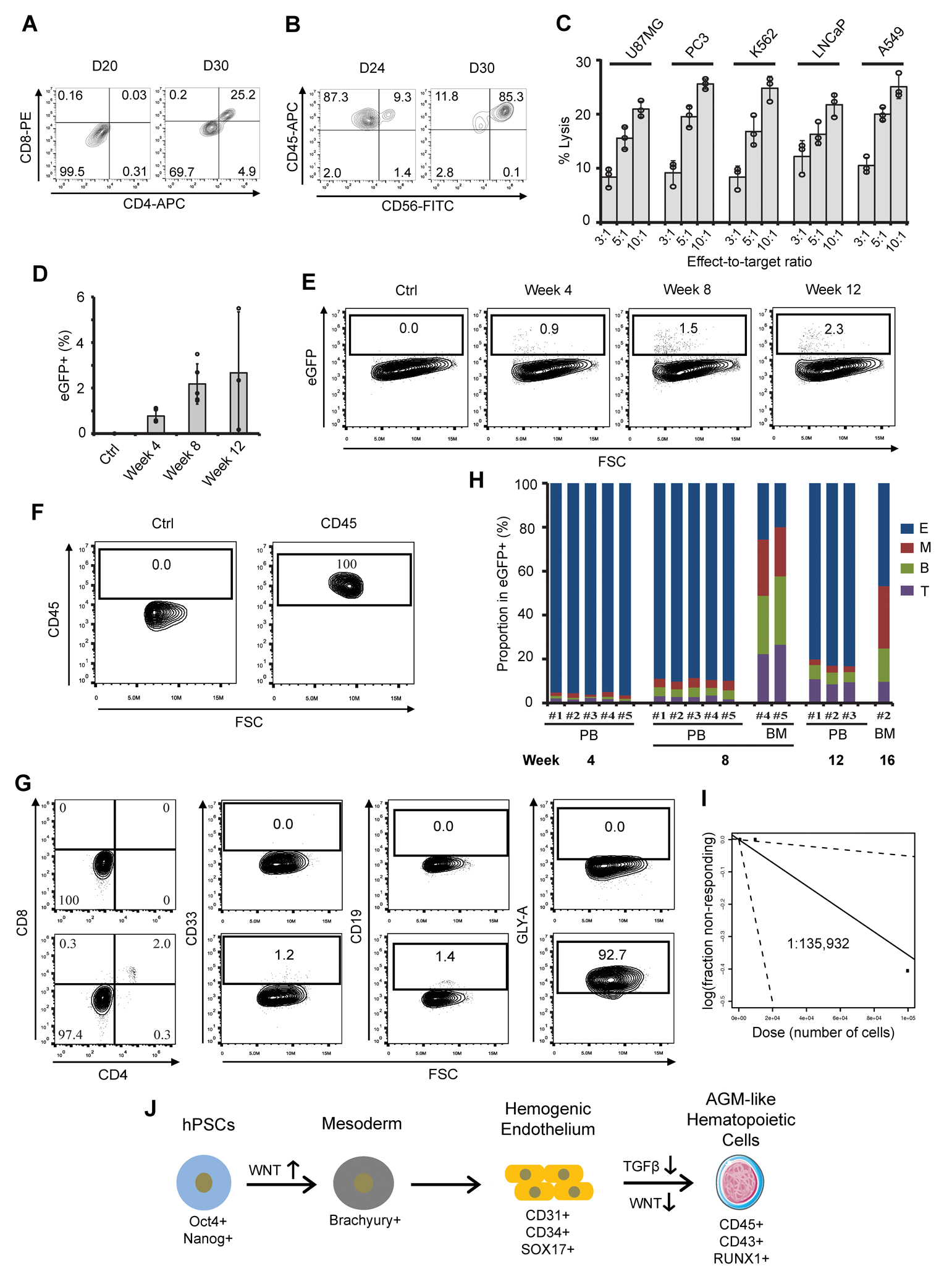
In vitro and in vivo characterization of hPSC-derived AGM-like hematopoietic cells. (A–C) Day 12 hPSC-derived hematopoietic cells were co-cultured with OP9-DLL4 for immune T and natural killer (NK) cell differentiation. At different time points, expression of CD4/CD8 (A) and CD45/CD56 (B) was assessed by flow cytometry. (C) hPSC-NK cells were co-cultured with different cancer cells at the indicated effector-to-target ratios, and the corresponding percentages of lysis were shown. (D–H) eGFP+ hPSC-derived hematopoietic stem and progenitor cells (HSPCs) were transplanted into femur of irradiated immunodeficient NRG mice for the assessment of engraftment and repopulation potentials. Engrafted human HSPCs were recorded at the indicated weeks (D) and representative flow plots of eGFP+ cells were shown in (E). The expressions of multi-lineage markers, including CD45 (F), CD4, CD8, CD33, GLY-A, and CD19 (G) in eGFP+ cells isolated from recipient peripheral blood (PB) and bone marrow (BM) were assessed by flow cytometry and quantified in (H). E: erythroid cells; M: myeloid cells; B: B cells; T: T cells. (I) Limiting dilution assay of hPSC-derived HSPCs during secondary transplantation. 103, 104, and 105 eGFP + cells isolated from bone marrow of primary recipients (#4 and #5) were transplanted to secondary recipients. Confidence intervals of 1/(stem cell frequency) were calculated by ELDA according to Poisson distribution. (J) A schematic model highlighting the specification of hPSCs to AGM-like hematopoietic cells by stage-specific modulation of Wnt with or without TGFβ signaling pathway. VEGF is required for endothelial specification of female hPSC lines.
LT-HSCs50 and hPSC-derived AGM-like cells10 are able to home to bone marrow after tail vein injection. In support of this, purified hPSC-derived mCherry+CD45+ hematopoietic cells were injected in the duct of Cuvier of 48–52 hours post-fertilization (hpf) zebrafish (Supplementary Fig. 9D), and mCherry+ cells were detected in the caudal hematopoietic tissue (CHT), a site of definitive hematopoiesis, within 1-hour post-transplantation (hpt) (Supplementary Fig. 9E). Compared to control neurons, more hematopoietic cells remained in the CHT at 5 hpt (Supplementary Fig. 9F–G). We also injected the mCherry+CD45+ hematopoietic cells directly into the circulation of c-myb knockout bloodless zebrafish embryos (Supplementary Fig. 9H) at 48–52 hpf, and mCherry+ hematopoietic cells were observed in the CHT up to 96 hpf (Supplementary Fig. 9I). Similar to cord blood CD34+ hematopoietic stem cells (CB-HSCs), hPSC-derived hematopoietic cells significantly reduced the death of bloodless zebrafish up to 4 days after transplantation (Supplementary Fig. 9J), highlighting their functional capacity. Collectively, our results demonstrated the multipotency and function of hPSC-derived definitive hematopoietic cells both in vitro and in vivo.
hPSC-derived HSPCs could engraft and repopulate in murine models.
To further confirm their in vivo homing and multi-lineage engraftment potentials, HSPCs derived from eGFP+ hPSCs (Supplementary Fig. 10A) were injected into the tail vein of irradiated NSG mice. After 15 hr, ~1.3% eGFP+ HSPCs homed to the femur bone marrow (BM) in all four recipient mice (Supplementary Fig. 10B–C). Similar to a previous report51, a significant number of eGFP+CD45+ HSPCs were detected in the mouse peripheral blood (PB) after one month (Supplementary Fig. 10D–E) but disappeared after three months (Supplementary Fig. 10F). Bone marrow cells were then harvested for chimerism rate analysis. Albeit at a relatively low rate (~0.1%), the homing and long-term (3-month) engraftment capacity of hPSC-derived HSPCs was confirmed by flow cytometry (Supplementary Fig. 10G) and PCR analysis (Supplementary Fig. 10H). The observed low-rate engraftment of our cells suggests potentially incorrect patterning of receptors required for their homing to BM, as exemplified by enhanced CXCR452–54 and reduced CD2655 expression on primary hematopoietic stem cells for improved homing and engraftment. To further determine the multi-lineage engraftment of our hPSC-derived HSPCs, we injected eGFP+ HSPCs intrafemorally into the bone marrow of primary recipients and harvested peripheral blood (PB) every four weeks. Increased chimaerism of human eGFP+ HSPCs in PB of primary recipients was observed for up to 12 weeks (Fig. 7D–F). Examination of PB and bone marrow revealed the multi-lineage engraftment of HSPCs with human GLY-A+ erythroid cells (E), CD33+ myeloid cells (M), CD19+ B cells (B), and CD4+CD8+ T cells (T) in 5 of 5 recipients (Fig. 7G–H, Supplementary Fig. 11A–E). To quantify the frequency of engraftable hematopoietic cells, we transplanted secondary recipients (3 mice/group) with 103, 104, and 105 eGFP+ HSPCs isolated from femur bone marrow of primary engrafted mice (#4 and #5 in Fig. 7H). One out of 3 secondary mice injected with 105 eGFP+ HSPCs from the primary recipient demonstrated multi-lineage engraftment (Supplementary Fig. 11F–H), indicating an estimated stem-cell frequency of 1 in 135,932 cells (Fig. 7I) for our hPSC-derived HSPCs, based on the calculation from ELDA software56 (http://bioinf.wehi.edu.au/software/elda/). Collectively, our mouse data provided preliminary evidence supporting the homing and multi-lineage engraftment activities of our hPSC-derived HSPCs. Further investigations and improvement of their long-term repopulating ability, such as the effects of hypoxia57–59 and HOXA genes10, are needed.
Discussion
While attempts were made to develop hematopoietic cell generation protocols from hPSCs with stage-specific employment of morphogens by recapitulating in vivo hematopoiesis, it remains unknown which developmental signaling pathways are sufficient and essential to specify human AGM-like cells, the first wave of LT-HSCs. In addition, efficient, cost-effective methods to generate homogenous AGM-like hemogenic and hematopoietic cells without primitive hematopoietic cells are lacking, limiting their large-scale production for clinical and research applications. This study demonstrates the robust and efficient generation of relatively homogeneous definitive AGM-like cells from various hPSC lines via sequential manipulation of Wnt and TGFβ signaling under chemically-defined and xeno-free conditions (Fig. 7J). Importantly, we also showed that stage-specific manipulation of Wnt signaling alone is sufficient to induce homogenous AGM-like SOX17+ hemogenic endothelium and hematopoiesis from hPSCs, further demonstrating the critical role of Wnt signaling during multiple stages of definitive hematopoiesis10,21,25.
This study also demonstrates transcriptional and functional similarity among hPSC-derived, primary AGM and cord blood (CB) hemato-endothelial cells. At global levels, hPSC-derived cells clustered closer to CB-HSCs than AGM hemato-endothelial progenitor cells, reflecting distinct developmental stages of the collected hPSC-derived and AGM cells. Further examination of specific hematopoietic genes and ontology confirmed the transcriptional similarity between hPSC-derived and AGM cells. Notably, bulk RNA-seq identified similar patterns of HOXA cluster gene patterns between these cells, highlighting their potential for repopulating HSC generation10. Comparative scRNA-seq analysis on day 5, day 8, and AGM cells revealed similarities and differences among the distinct endothelial subsets. In addition, our hPSC-derived hematopoietic cells presented lymphoid and myeloid potential in vivo. They were able to home to the definitive CHT site and partially rescued bloodless zebrafish after transplantation. Importantly, we also demonstrated that hPSC-derived hematopoietic cells could home to the bone marrow and repopulate in irradiated murine models. However, further investigations and improvement of their longer-term repopulating ability are still needed. For instance, it will be interesting to investigate the effects of HOXA genes on the engraftment of hPSC-derived hematopoietic cells, since expression of HOXA genes in our cells did not reach an AGM cell expression level10. Additional maturation strategies, such as co-culture with OP9 feeder cells60–62, hypoxic culture57–59, CXCR4 enhancement52–54, DPP4/CD26 inhibition55,63–65, and RA patterning10, may be required to improve the long-term repopulating capacity of hPSC-derived hematopoietic cells.
Collectively, our data demonstrated that stage-specific manipulation of Wnt and TGFβ signaling pathways (Fig. 7J) is sufficient to induce homogenous aorta-like SOX17+CD34+ hemogenic endothelium and definitive hematopoiesis from hPSCs, recapitulating many aspects of AGM hematopoiesis in human10 and mouse21 models. This finding is also consistent with a previous report that RA signaling-mediated Wnt inhibition is essential for HSC development from hemogenic endothelium in mice21. The simplified and scalable GiTi platform for robust definitive hematopoiesis will provide insights into molecular mechanisms of human hematopoietic development and has the potential to serve as an easily accessible cell source for treating various blood diseases and cancers.
Methods
Maintenance and differentiation of hPSCs.
19-9-11, 19-9-7, 6-9-9, IMR90C4, H1, H9 and H13 were obtained from WiCell and maintained on Matrigel- or iMatrix 511-coated plates in mTeSR plus or mTeSR1 medium according to a previously published method66. RUES2 were kindly provided by Dr. Ali H. Brivanlou at the Rockefeller University. The Kolf2 and CT2 data were acquired in Dr. Yang Yang’s lab and Dr. Ourania Andrisani’s lab at Purdue. H9 7TGFP Wnt reporter and 19-9-11 ischcat-1 as well as ischcat-2 lines32 were kindly provided by Dr. Sean Palecek at the University of Wisconsin-Madison. To make hematopoietic cells, hPSCs were dissociated with 0.5 mM EDTA and seeded onto iMatrix 511 or Matrigel-coated 6-, 12- or 24-well plate at a cell density between 10,000 and 80,000 cell/cm2 in mTeSR plus or mTeSR1 medium with 5 μM Y27632 for 24 hours (day −1). At day 0, cells were treated with 6 μM CHIR99021 (CHIR) in DMEM medium (ThermoFisher, 11965) supplemented with 100 μg/ml ascorbic acid (DMEM/Vc)22, followed by a medium change with LaSR basal medium at day 1, day 2 and day 3. Mesoderm differentiation could also be achieved by 2-day CHIR treatment in LaSR basal medium23. For female hPSC lines67, 50 ng/mL VEGF was added to the medium from day 2 to day 4. On day 4, medium was replaced by Stemline II medium (Sigma) supplemented 10 μM SB431542. After 2 days, SB431542-containing medium was aspirated and cells were maintained in Stemline II medium with or without 50 ng/mL SCF and 50 ng/mL FLT3L. On day 9 and every 2 to 3 days afterward, aspirate half medium and add fresh Stemline II medium with or without SCF/FLT3L until analysis. Other media used to induce hematopoietic cells were illustrated in Supplementary Fig. 3C.
Genome editing of hPSCs.
Two Cas9 sgRNAs targeting near the HOXB5 stop codon (1: GGCTCCTCTGGGCGGGCTCAGGG and 2: ATCGTAACACAAGGCGAGGCAGG with a G added at the beginning) were used. To generate the HOXB5–2A-mCherry donor plasmid, DNA fragments of about 800 bp in length were PCR amplified from genomic DNA before and after the stop codon of HOXB5 and were cloned into the VE‐cad-2A‐eGFP (Addgene #92309) and VE‐cad-2A‐mCherry (Addgene #31938) donor plasmids replacing the VEcad homologous arms. The resulting 3 μg gRNA1, 3 μg gRNA2, and 6 μg HOXB5‐2A‐mCherry donor plasmids were prepared in 100 μl stem cell nucleofection solution (Lonza, #VAPH-5012) and then co-nucleofected into 2.5–3 million singularized H9 hPSCs pretreated with 5 μM Y27632 overnight using program B-015 in a Nucleofector 2b. The nucleofected cells were subsequently plated onto one well of a Matrigel‐coated 6-well plate in 3 mL pre-warmed mTeSR plus with 10 μM Y27632. Twenty‐four hours later, and every day afterward, the medium was changed with fresh mTeSR plus. Once cells are confluent, 1 μg/ml puromycin was added to the mTeSR plus for selection for about 2 weeks. Single-cell clones were then picked into wells of a Matrigel‐coated 96‐well plate and subjected to PCR genotyping after 4–7 days. To generate an inducible gene knockdown system in hPSCs, RfxCas13d68,69 (Addgene #138147) was PCR amplified and cloned into our all-in-one PiggyBac (PB) backbone70 by replacing SARS-CoV2 N gene (Addgene #154399). The U6 driven construct, containing a 5’ direct repeat 30 (DR30) and a BbsI-based single guide RNA (sgRNA) cloning site, was then cloned right before the 3’ PB sequence (Fig. 1E), leading to an all-in-one PB inducible Cas13d-mediated gene knockdown plasmid (Addgene #155184). The SOX17 targeting sgRNA1 and sgRNA2 were designed using an online tool (https://cas13design.nygenome.org/ or https://www.rnatargeting.org/)and cloned into the Cas13d backbone to make SOX17 targeting plasmids (Addgene #155187 and #155188). The SOX17 knockdown plasmids were then used to transfect H9 hPSCs along with a hyPBase plasmid (kindly provided by Dr. Pentao Liu) via Lipofectamine Stem (ThermoFisher) according to the manufacturer’s instructions. Once transfected cells were confluent, 5 μg/ml puromycin or 20 μg/ml blasticidine (BSD) were used to select the drug-resistant hPSCs for one or two days, and the drugs were reapplied to the survived cells once they recovered and used consistently to maintain the engineered H9 hPSCs to avoid gene silencing during differentiation.
Hematopoietic colony-forming assay and Wright-Giemsa staining.
About 10,000 hPSC-derived hematopoietic cells on differentiation day 15 were grown in 1.5 mL cytokine containing MethoCult H4434 medium (StemCell Technologies, Vancouver) at 37 °C. After 14 days, the hematopoietic colonies were scored for colony-forming units (CFUs) according to cellular morphology. Hematopoietic cells were also seeded onto glass slides and stained with modified Wright-Giemsa stain solution.
NK and T cell differentiation from hematopoietic cells.
Both NK71 and T72 cell differentiations were performed on OP9-DLL4 feeder layer (kindly provided by Dr. Igor Slukvin at the University of Wisconsin Madison) in α-MEM medium supplemented with 20% FBS and 1% GlutaMAX. To initiate NK cell induction, day 12 hematopoietic cells were cultured on OP9-DLL4 with 100 ng/mL FLT3L, 5 ng/mL IL-7, 40 ng/mL SCF, and 35 nM UM171. After 7 days and every 7 days afterward, cells were transferred to fresh OP9-DLL4. After 14 to 21 days, floating cells were collected and subjected to flow cytometry analysis. A similar approach was used to induce T cell differentiation on OP9-DLL4, except different cytokines were used: 10 ng/mL SCF, 5 ng/mL IL-7 and 5 ng/mL FLT3L.
Flow cytometry and immunostaining analysis.
Floating hematopoietic cells were gently pipetted and filtered through a 70 or 100 μm strainer sitting on a 50 ml tube. The cells were then pelleted by centrifugation and washed once in PBS−/− solution with 1% bovine serum albumin (BSA). The cells were stained with appropriate conjugated antibodies (Supplementary Table 1) for 25 mins at room temperature in dark, and analyzed in Accuri C6 plus flow cytometer (Beckton Dickinson) after washing once with BSA-containing PBS−/− solution. FlowJo software was used to process collected flow data. Immunostaining assay was performed according to previous protocols73,74. Briefly, day 5 or day 15 cell cultures were fixed in PBS−/− with 4% formaldehyde for 15 mins at room temperature and stained with SOX17-APC75,76, CD34-FITC22,23, CD45-APC77,78 or RUNX1–48879,80 antibodies in 5% nonfat dry milk and 0.4% Triton X-100 solution for 30 mins in dark. After gentle washing and nuclei staining, the resulting cells were then imaged with a Leica DMi-8 fluorescent microscope and analyzed in ImageJ.
Bulk RNA sequencing and data analysis.
Total RNA of purified day 18 hPSC-derived CD45+ hematopoietic cells was prepared with the Direct-zol RNA MiniPrep Plus kit (Zymo Research) according to the manufacturer’s instructions. Samples were performed in Illumina HiSeq 2500 by GENEWIZ. HISAT2 program81 was employed to map the resulting 2×150 sequencing reads to the human genome (hg 19), and the python script rpkmforgenes.py82 was used to quantify the RefSeq transcript levels (RPKMs). The original fastq files and processed RPKM text files were submitted to NCBI GEO (GSE155196). RNA-seq data of human primary AGM and neonatal cord blood HSC samples were retrieved from NCBI (SRR3475781, 3475782, 347578310, 3039602, and 303960837). Hierarchical clustering of whole transcripts and heatmap of hematopoietic-specific genes were then plotted using Morpheus (Broad Institute). Principal component analysis (PCA) was processed in R program, and 3D score plot of the first three principal components (PCs) was plotted in MATLAB. GSEA software (Broad Institute) was used to perform gene ontology (GO) enrichment analysis and the values of normalized enrichment score (NES) were used to plot GO heatmap in Morpheus.
Single-cell RNA sequencing (scRNA-seq) analysis.
As previously described83, scRNA-seq was performed using the 10X Genomics 3’ v3 kit, following their protocol targeting recovery of 10,000 cells. Libraries were constructed per the manufacturer’s instructions and sequenced using Illumina’s NovaSeq 6000 platform in the Center for Medical Genomics at Indiana University. The average read depth across the samples was 92,000 reads/cell for the day 8 sample and 76,000 reads/cell for the day 18 sample. Reads were then aligned to the human genome GRCh37/hg19 using the CellRanger 2.1.0 software. Subsequent analysis was performed in R using the filtered barcode and count matrices produced by CellRanger. Seurat 3.1.0 was used to analyze the single-cell data84. Time points were processed individually and filtered for quality control (Supplementary Fig. 5A–C, 7A). Specifically, cells with fewer than 200 detected genes or more than 15% mitochondrial content were excluded from analysis, resulting in 4,676 cells for day 8 and 6,190 cells for day 18. The data was log-normalized, and the top 3000 variable genes were used for Principal Component Analysis (PCA) following Seurat’s tutorial as evaluated by elbow plots. The top 5 Principal Components (PCs) were used for the day 8 sample, and the top 15 PCs for the day 18 sample were used for downstream Louvain clustering and UMAP embedding to visualize the data. Seurat’s FindAllMarkers function was used to identify differentially expressed genes (DEGs) per cluster and then manually annotated based on enriched gene expression. All genes considered for cell-type classification had a P-value of less than 0.0001 using a Mann-Whitney Wilcoxon test. Monocle version 2 and 3 were used for pseudotime analysis and trajectory inference36. scRNA-seq data of day 5 and primary AGM samples were obtained from previous studies with accession numbers of GSE16140885 and GSE15187734. The resulting day 8 and 18 scRNA-seq raw and processed data can be accessed via GEO with accession number: GSE157944.
RT-PCR analysis.
Cells cultured on 24-well plate were collected and lysed in 500 μL TRIzol™ reagent (Invitrogen). Total RNA was then prepared with the Direct-zol RNA miniprep kit (Zymo) with in-column DNase treatment following the manufacture’s instruction. cDNA was reverse transcribed from 1 μg RNA with ProtoScript First Strand cDNA Synthesis Kit (NEB) and used for RT-PCR with GoTaq Green Master Mix (Promega). GAPDH was used as an endogenous housekeeping control, and the primer pairs for targeted genes were listed in Supplementary Table 2.
Transplantation of hPSC-derived hematopoietic cells into zebrafish.
About 200 purified mCherry+CD45+ of day 15 hPSC-derived hematopoietic cells were injected in the circulation of 48–52 hour old zebrafish via the duct of Cuvier86,87. After 1-, 3- and 5-hour post-transplantation (hpt), mCherry+ cells homed to CHT were recorded and quantified under a fluorescent microscope. PBS and hPSC-derived neurons66 were used as negative controls, and cord-blood (CB) HSCs were used as a positive control. Bloodless zebrafish were generated by injecting embryos at the 1-cell stage with Cas9 protein and two c-myb targeting sgRNAs88 or 0.2 mM c-myb–specific antisense morpholino89. ~200 cells were injected into the duct of Cuvier of 48–52 hour old bloodless zebrafish. At 1, 5, 48, 72, and 96 hpf, mCherry+ cells homed to CHT were recorded under a fluorescent microscope, and viable zebrafish were counted.
Transplantation of hPSC-derived hematopoietic cells into irradiated mice.
All animal experiments were performed in accordance with institutional guidelines approved by the Purdue Animal Care and Use Committee (PACUC) and Indiana University Institutional Animal Care and Use Committee (IACUC). NOD.Cg-RAG1tm1MomIL2rgtm1Wjl/SzJ (NRG) or NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NSG) mice were bred and housed at the animal care facility. 6- to 10-week-old female mice were irradiated (2.75 Gy) 24 hr before transplantation. Day 10 to day 15 eGFP+ HSPCs were purified via magnetic-activated cell sorting (MACS) using CD34 or CD44 microbeads. For the intravenous injection, PBS, 5×104, 105 or 5×105 sorted eGFP+ HSPCs were injected into the tail vein of sublethally irradiated NSG mice, and cells homed to the recipient bone marrow were analyzed after 15 hr. One or three months after transplantation, the percentage of human CD45+ cell chimerism was analyzed. For intrafemoral transplantation, a 28.5-gauge insulin needle was used to inject 25 μl volume of 105 cells into the femur bone marrow of five primary recipient NRG mice. Peripheral blood was collected every 4 weeks from the primary recipients for multilineage engraftment analysis. At week 8, eGFP+ HSPCs were isolated from the bone marrow of primary recipients and intrafemorally injected into the secondary recipient mice with different cell numbers of 103, 104, and 105 for limited dilution analysis. Peripheral blood was collected at week 4 and 6 for the multilineage engraftment analysis in secondary transplantation.
Statistical analysis.
Data are presented as mean ± standard error of the mean (s.e.m). Statistical significance was determined by Student’s t-test (two-tail) between two groups, and three or more groups were analyzed by one-way analysis of variance (ANOVA). P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank members of the Deng, Lian and Bao laboratories for technical assistance and critical reading of the manuscript, Tianxiao Han, Dr. Sean P. Palecek and Dr. David V. Schaffer for valuable discussion and advice. We also gratefully acknowledge the Purdue Flow Cytometry and Cell Separation Facility, Purdue Genomics Core Facility and the Center for Medical Genomics at Indiana University School of Medicine. This study was supported by startup funding from the Davidson School of Chemical Engineering and the College of Engineering at Purdue (X.B.), Showalter Research Trust (Young Investigator Award to X.B.), NIH NIGMS (grant no. R35GM119787 to Q.D.), NIH NHLBI (grant no. R35HL139599 to H.E.B.), and NIH NIDDK (grant no. U54DK106846).
Footnotes
Competing interests
A patent related to this manuscript is under application (Y.C. and X.B.).
Data availability
The final processed data and raw fastq files were submitted to Gene Expression Omnibus (GEO) with accession numbers: GSE155196 and GSE157944. Source data for the figures in this study are available in figshare with the identifier: http://doi.org/10.6084/m9.figshare.16556577. The authors declare that all other data supporting the findings of this study are available within the paper and its Supplementary Information.
References
- (1).Dzierzak E; Speck NA Of Lineage and Legacy: The Development of Mammalian Hematopoietic Stem Cells. Nature Immunology. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Takahashi K; Tanabe K; Ohnuki M; Narita M; Ichisaka T; Tomoda K; Yamanaka S Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131 (5), 861–872. [DOI] [PubMed] [Google Scholar]
- (3).Takahashi K; Yamanaka S Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126 (4), 663–676. [DOI] [PubMed] [Google Scholar]
- (4).Godin I; Dieterlen-Lièvre F; Cumano A Emergence of Multipotent Hemopoietic Cells in the Yolk Sac and Paraaortic Splanchnopleura in Mouse Embryos, Beginning at 8.5 Days Postcoitus. Proc. Natl. Acad. Sci. U. S. A 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ditadi A; Sturgeon CM; Tober J; Awong G; Kennedy M; Yzaguirre AD; Azzola L; Ng ES; Stanley EG; French DL; Cheng X; Gadue P; Speck NA; Elefanty AG; Keller G Human Definitive Haemogenic Endothelium and Arterial Vascular Endothelium Represent Distinct Lineages. Nat. Cell Biol 2015, 17 (5), 580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kaufman DS Toward Clinical Therapies Using Hematopoietic Cells Derived from Human Pluripotent Stem Cells. Blood. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).McKinney-Freeman S; Cahan P; Li H; Lacadie SA; Huang HT; Curran M; Loewer S; Naveiras O; Kathrein KL; Konantz M; Langdon EM; Lengerke C; Zon LI; Collins JJ; Daley GQ The Transcriptional Landscape of Hematopoietic Stem Cell Ontogeny. Cell Stem Cell 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kennedy M; Awong G; Sturgeon CM; Ditadi A; LaMotte-Mohs R; Zúñiga-Pflücker JC; Keller G T Lymphocyte Potential Marks the Emergence of Definitive Hematopoietic Progenitors in Human Pluripotent Stem Cell Differentiation Cultures. Cell Rep. 2012, 2 (6), 1722–1735. [DOI] [PubMed] [Google Scholar]
- (9).Ivanovs A; Rybtsov S; Ng ES; Stanley EG; Elefanty AG; Medvinsky A Human Haematopoietic Stem Cell Development: From the Embryo to the Dish. Development 2017, 144 (13), 2323–2337. [DOI] [PubMed] [Google Scholar]
- (10).Ng ES; Azzola L; Bruveris FF; Calvanese V; Phipson B; Vlahos K; Hirst C; Jokubaitis VJ; Yu QC; Maksimovic J; Liebscher S; Januar V; Zhang Z; Williams B; Conscience A; Durnall J; Jackson S; Costa M; Elliott D; Haylock DN; Nilsson SK; Saffery R; Schenke-Layland K; Oshlack A; Mikkola HKA; Stanley EG; Elefanty AG Differentiation of Human Embryonic Stem Cells to HOXA+ Hemogenic Vasculature That Resembles the Aorta-Gonad-Mesonephros. Nat. Biotechnol 2016, 34 (11), 1168–1179. [DOI] [PubMed] [Google Scholar]
- (11).Ivanovs A; Rybtsov S; Ng ES; Stanley EG; Elefanty AG; Medvinsky A Human Haematopoietic Stem Cell Development: From the Embryo to the Dish. Development 2017, 144 (13), 2323–2337. [DOI] [PubMed] [Google Scholar]
- (12).Rybtsov S; Ivanovs A; Zhao S; Medvinsky A Concealed Expansion of Immature Precursors Underpins Acute Burst of Adult HSC Activity in Foetal Liver. Development 2016, 143 (8), 1284–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Boisset J-C; van Cappellen W; Andrieu-Soler C; Galjart N; Dzierzak E; Robin C In Vivo Imaging of Haematopoietic Cells Emerging from the Mouse Aortic Endothelium. Nature 2010, 464 (7285), 116–120. [DOI] [PubMed] [Google Scholar]
- (14).Bertrand JY; Chi NC; Santoso B; Teng S; Stainier DYR; Traver D Haematopoietic Stem Cells Derive Directly from Aortic Endothelium during Development. Nature 2010, 464 (7285), 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kissa K; Herbomel P Blood Stem Cells Emerge from Aortic Endothelium by a Novel Type of Cell Transition. Nature 2010, 464 (7285), 112–115. [DOI] [PubMed] [Google Scholar]
- (16).Kim I; Saunders TL; Morrison SJ Sox17 Dependence Distinguishes the Transcriptional Regulation of Fetal from Adult Hematopoietic Stem Cells. Cell 2007, 130 (3), 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Clarke RL; Yzaguirre AD; Yashiro-Ohtani Y; Bondue A; Blanpain C; Pear WS; Speck NA; Keller G The Expression of Sox17 Identifies and Regulates Haemogenic Endothelium. Nat. Cell Biol 2013, 15 (5), 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fidanza A; Romanò N; Ramachandran P; Tamagno S; Lopez-Yrigoyen M; Taylor AH; Easterbrook J; Henderson B; Axton R; Henderson NC; Medvinsky A; Ottersbach K; Forrester LM LM Single Cell Transcriptome Analysis Reveals Markers of Naïve and Lineage-Primed Hematopoietic Progenitors Derived from Human Pluripotent Stem Cells. bioRxiv 2019, 602565. [Google Scholar]
- (19).Oatley M; Bölükbası ÖV; Svensson V; Shvartsman M; Ganter K; Zirngibl K; Pavlovich PV; Milchevskaya V; Foteva V; Natarajan KN; Baying B; Benes V; Patil KR; Teichmann SA; Lancrin C Single-Cell Transcriptomics Identifies CD44 as a Marker and Regulator of Endothelial to Haematopoietic Transition. Nat. Commun 2020, 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Motazedian A; Bruveris FF; Kumar SV; Schiesser JV; Chen T; Ng ES; Chidgey AP; Wells CA; Elefanty AG; Stanley EG Multipotent RAG1+ Progenitors Emerge Directly from Haemogenic Endothelium in Human Pluripotent Stem Cell-Derived Haematopoietic Organoids. Nat. Cell Biol 2020, 22 (1), 60–73. [DOI] [PubMed] [Google Scholar]
- (21).Chanda B; Ditadi A; Iscove NN; Keller G Retinoic Acid Signaling Is Essential for Embryonic Hematopoietic Stem Cell Development. Cell 2013, 155 (1), 215–227. [DOI] [PubMed] [Google Scholar]
- (22).Bao X; Lian X; Dunn KK; Shi M; Han T; Qian T; Bhute VJ; Canfield SG; Palecek SP Chemically-Defined Albumin-Free Differentiation of Human Pluripotent Stem Cells to Endothelial Progenitor Cells. Stem Cell Res 2015, 15 (1), 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lian X; Bao X; Al-Ahmad A; Liu J; Wu Y; Dong W; Dunn KK; Shusta EV; Palecek SP Efficient Differentiation of Human Pluripotent Stem Cells to Endothelial Progenitors via Small-Molecule Activation of WNT Signaling. Stem Cell Reports 2014, 3 (5), 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Palpant NJ; Pabon L; Friedman CE; Roberts M; Hadland B; Zaunbrecher RJ; Bernstein I; Zheng Y; Murry CE Generating High-Purity Cardiac and Endothelial Derivatives from Patterned Mesoderm Using Human Pluripotent Stem Cells. Nat. Protoc 2017, 12 (1), 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sturgeon CM; Ditadi A; Awong G; Kennedy M; Keller G Wnt Signaling Controls the Specification of Definitive and Primitive Hematopoiesis from Human Pluripotent Stem Cells. Nat. Biotechnol 2014, 32 (6), 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Jung HS; Uenishi GI; Park MA; Liu P; Raymond M; Thomson J; Ong I; Slukvin I SOX17 Is Essential for Integration of Arterial and HOXA Programs in Hemogenic Endothelium. Blood 2019, 134 (Supplement_1), 2476–2476. [Google Scholar]
- (27).Nakajima-Takagi Y; Osawa M; Oshima M; Takagi H; Miyagi S; Endoh M; Endo TA; Takayama N; Eto K; Toyoda T; Koseki H; Nakauchi H; Iwama A Role of SOX17 in Hematopoietic Development from Human Embryonic Stem Cells. Blood 2013, 121 (3), 447–458. [DOI] [PubMed] [Google Scholar]
- (28).Wang C; Tang X; Sun X; Miao Z; Lv Y; Yang Y; Zhang H; Zhang P; Liu Y; Du L; Gao Y; Yin M; Ding M; Deng H TGFβ Inhibition Enhances the Generation of Hematopoietic Progenitors from Human ES Cell-Derived Hemogenic Endothelial Cells Using a Stepwise Strategy. Cell Res. 2012, 22 (1), 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kirmizitas A; Meiklejohn S; Ciau-Uitz A; Stephenson R; Patient R Dissecting BMP Signaling Input into the Gene Regulatory Networks Driving Specification of the Blood Stem Cell Lineage. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (23), 5814–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Crisan M; Kartalaei PS; Vink C; Yamada-Inagawa T; Bollerot K; Van Ijcken W; Van Der Linden R; De Sousa Lopes SMC; Monteiro R; Mummery C; Dzierzak E BMP Signalling Differentially Regulates Distinct Haematopoietic Stem Cell Types. Nat. Commun 2015, 6 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Uenishi GI; Jung HS; Kumar A; Park MA; Hadland BK; McLeod E; Raymond M; Moskvin O; Zimmerman CE; Theisen DJ; Swanson S; Tamplin OJ; Zon LI; Thomson JA; Bernstein ID; Slukvin II NOTCH Signaling Specifies Arterial-Type Definitive Hemogenic Endothelium from Human Pluripotent Stem Cells. Nat. Commun 2018, 9 (1), 1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lian XJ; Hsiao C; Wilson G; Zhu KX; Hazeltine LB; Azarin SM; Raval KK; Zhang JH; Kamp TJ; Palecek SP Robust Cardiomyocyte Differentiation from Human Pluripotent Stem Cells via Temporal Modulation of Canonical Wnt Signaling. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (27), E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).North TE; De Bruijn MFTR; Stacy T; Talebian L; Lind E; Robin C; Binder M; Dzierzak E; Speck NA Runx1 Expression Marks Long-Term Repopulating Hematopoietic Stem Cells in the Midgestation Mouse Embryo. Immunity 2002, 16 (5), 661–672. [DOI] [PubMed] [Google Scholar]
- (34).Crosse EI; Gordon-Keylock S; Rybtsov S; Binagui-Casas A; Felchle H; Nnadi NC; Kirschner K; Chandra T; Tamagno S; Webb DJ; Rossi F; Anderson RA; Medvinsky A Multi-Layered Spatial Transcriptomics Identify Secretory Factors Promoting Human Hematopoietic Stem Cell Development. Cell Stem Cell 2020, 27 (5), 822–839.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Inoue SI; Noda S; Kashima K; Nakada K; Hayashi JI; Miyoshi H Mitochondrial Respiration Defects Modulate Differentiation but Not Proliferation of Hematopoietic Stem and Progenitor Cells. FEBS Lett 2010, 584 (15), 3402–3409. [DOI] [PubMed] [Google Scholar]
- (36).Trapnell C; Cacchiarelli D; Grimsby J; Pokharel P; Li S; Morse M; Lennon NJ; Livak KJ; Mikkelsen TS; Rinn JL The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat. Biotechnol 2014, 32 (4), 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Notta F; Zandi S; Takayama N; Dobson S; Gan OI; Wilson G; Kaufmann KB; McLeod J; Laurenti E; Dunant CF; McPherson JD; Stein LD; Dror Y; Dick JE Distinct Routes of Lineage Development Reshape the Human Blood Hierarchy across Ontogeny. Science (80-.). 2016, 351 (6269). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Siatecka M; Bieker JJ The Multifunctional Role of EKLF/KLF1 during Erythropoiesis. Blood. The American Society of Hematology; August 25, 2011, pp 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).McGrath KE; Frame JM; Fegan KH; Bowen JR; Conway SJ; Catherman SC; Kingsley PD; Koniski AD; Palis J Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep 2015, 11 (12), 1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zöller M CD44, Hyaluronan, the Hematopoietic Stem Cell, and Leukemia-Initiating Cells. Frontiers in Immunology. Frontiers Media S.A. 2015, p 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cao H; Heazlewood SY; Williams B; Cardozo D; Nigro J; Oteiza A; Nilsson SK The Role of CD44 in Fetal and Adult Hematopoietic Stem Cell Regulation. Haematologica 2016, 101 (1), 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Motazedian A; Bruveris FF; Kumar SV; Schiesser JV; Chen T; Ng ES; Chidgey AP; Wells CA; Elefanty AG; Stanley EG Multipotent RAG1+ Progenitors Emerge Directly from Haemogenic Endothelium in Human Pluripotent Stem Cell-Derived Haematopoietic Organoids. Nat. Cell Biol 2020, 22 (1), 60–73. [DOI] [PubMed] [Google Scholar]
- (43).Umeda K; Heike T; Yoshimoto M; Shiota M; Suemori H; Luo HY; Chui DHK; Torii R; Shibuya M; Nakatsuji N; Nakahata T Development of Primitive and Definitive Hematopoiesis from Non-Human Primate Embryonic Stem Cells in Vitro. Development 2004, 131 (8), 1869–1879. [DOI] [PubMed] [Google Scholar]
- (44).Chen JY; Miyanishi M; Wang SK; Yamazaki S; Sinha R; Kao KS; Seita J; Sahoo D; Nakauchi H; Weissman IL Hoxb5 Marks Long-Term Haematopoietic Stem Cells and Reveals a Homogenous Perivascular Niche. Nature 2016, 530 (7589), 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Qian P; De Kumar B; He XC; Nolte C; Gogol M; Ahn Y; Chen S; Li Z; Xu H; Perry JM; Hu D; Tao F; Zhao M; Han Y; Hall K; Peak A; Paulson A; Zhao C; Venkatraman A; Box A; Perera A; Haug JS; Parmely T; Li H; Krumlauf R; Li L Retinoid-Sensitive Epigenetic Regulation of the Hoxb Cluster Maintains Normal Hematopoiesis and Inhibits Leukemogenesis. Cell Stem Cell 2018, 22 (5), 740–754.e7. [DOI] [PubMed] [Google Scholar]
- (46).Gulati GS; Zukowska M; Noh J; Zhang A; Sinha R; George BM; Wesche DJ; Weissman IL; Szade K Neogenin-1 Distinguishes between Myeloid-Biased and Balanced Hoxb5+ Mouse Long-Term Hematopoietic Stem Cells. bioRxiv 2019, 608398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Bao X; Bhute VJ; Han T; Qian T; Lian X; Palecek SP Human Pluripotent Stem Cell‐derived Epicardial Progenitors Can Differentiate to Endocardial‐like Endothelial Cells. Bioeng. Transl. Med 2017, 2 (2), 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Li Y; Hermanson DL; Moriarity BS; Kaufman DS Human IPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-Tumor Activity. Cell Stem Cell 2018, 23 (2), 181–192.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Dege C; Fegan KH; Creamer JP; Berrien-Elliott MM; Luff SA; Kim D; Wagner JA; Kingsley PD; McGrath KE; Fehniger TA; Palis J; Sturgeon CM Potently Cytotoxic Natural Killer Cells Initially Emerge from Erythro-Myeloid Progenitors during Mammalian Development. Dev. Cell 2020, 53 (2), 229–239.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Heazlewood SY; Oteiza A; Cao H; Nilsson SK Analyzing Hematopoietic Stem Cell Homing, Lodgment, and Engraftment to Better Understand the Bone Marrow Niche. Ann. N. Y. Acad. Sci 2014, 1310 (1), 119–128. [DOI] [PubMed] [Google Scholar]
- (51).Zhu Y; Wang T; Gu J; Huang K; Zhang T; Zhang Z; Liu H; Tang J; Mai Y; Zhang Y; Li Y; Feng Y; Kang B; Li J; Shan Y; Chen Q; Zhang J; Long B; Wang J; Gao M; Zhang D; Zhou M; Zhong X; Chen J; Pei D; Nie J; Liu B; Pan G Characterization and Generation of Human Definitive Multipotent Hematopoietic Stem/Progenitor Cells. Cell Discov 2020, 6 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Huang X; Guo B; Liu S; Wan J; Broxmeyer HE Neutralizing Negative Epigenetic Regulation by HDAC5 Enhances Human Haematopoietic Stem Cell Homing and Engraftment. Nat. Commun 2018, 9 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Guo B; Huang X; Cooper S; Broxmeyer HE Glucocorticoid Hormone-Induced Chromatin Remodeling Enhances Human Hematopoietic Stem Cell Homing and Engraftment. Nat. Med 2017, 23 (4), 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Capitano ML; Hangoc G; Cooper S; Broxmeyer HE Mild Heat Treatment Primes Human CD34+ Cord Blood Cells for Migration toward SDF-1α and Enhances Engraftment in an NSG Mouse Model. Stem Cells 2015, 33 (6), 1975–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Christopherson KW; Hangoc G; Mantel CR; Broxmeyer HE Modulation of Hematopoietic Stem Cell Homing and Engraftment by CD26. Science (80-.). 2004, 305 (5686), 1000–1003. [DOI] [PubMed] [Google Scholar]
- (56).Sugimura R; Jha DK; Han A; Soria-Valles C; da Rocha EL; Lu Y-F; Goettel JA; Serrao E; Rowe RG; Malleshaiah M; Wong I; Sousa P; Zhu TN; Ditadi A; Keller G; Engelman AN; Snapper SB; Doulatov S; Daley GQ Haematopoietic Stem and Progenitor Cells from Human Pluripotent Stem Cells. Nature 2017, 545 (7655), 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Eliasson P; Rehn M; Hammar P; Larsson P; Sirenko O; Flippin LA; Cammenga J; Jönsson JI Hypoxia Mediates Low Cell-Cycle Activity and Increases the Proportion of Long-Term-Reconstituting Hematopoietic Stem Cells during in Vitro Culture. Exp. Hematol 2010, 38 (4), 301–310.e2. [DOI] [PubMed] [Google Scholar]
- (58).Ramírez-Bergeron DL; Runge A; Cowden Dahl KD; Fehling HJ; Keller G; Simon MC Hypoxia Affects Mesoderm and Enhances Hemangioblast Specification during Early Development. Development 2004, 131 (18), 4623–4634. [DOI] [PubMed] [Google Scholar]
- (59).Mantel CR; O’Leary HA; Chitteti BR; Huang X; Cooper S; Hangoc G; Brustovetsky N; Srour EF; Lee MR; Messina-Graham S; Haas DM; Falah N; Kapur R; Pelus LM; Bardeesy N; Fitamant J; Ivan M; Kim KS; Broxmeyer HE Enhancing Hematopoietic Stem Cell Transplantation Efficacy by Mitigating Oxygen Shock. Cell 2015, 161 (7), 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Taoudi S; Gonneau C; Moore K; Sheridan JM; Blackburn CC; Taylor E; Medvinsky A Extensive Hematopoietic Stem Cell Generation in the AGM Region via Maturation of VE-Cadherin+CD45+ Pre-Definitive HSCs. Cell Stem Cell 2008, 3 (1), 99–108. [DOI] [PubMed] [Google Scholar]
- (61).Rybtsov S; Sobiesiak M; Taoudi S; Souilhol C; Senserrich J; Liakhovitskaia A; Ivanovs A; Frampton J; Zhao S; Medvinsky A Hierarchical Organization and Early Hematopoietic Specification of the Developing HSC Lineage in the AGM Region. J. Exp. Med 2011, 208 (6), 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zhou F; Li X; Wang W; Zhu P; Zhou J; He W; Ding M; Xiong F; Zheng X; Li Z; Ni Y; Mu X; Wen L; Cheng T; Lan Y; Yuan W; Tang F; Liu B Tracing Haematopoietic Stem Cell Formation at Single-Cell Resolution. Nature 2016, 533 (7604), 487–492. [DOI] [PubMed] [Google Scholar]
- (63).Broxmeyer HE; Hoggatt J; O’leary HA; Mantel C; Chitteti BR; Cooper S; Messina-Graham S; Hangoc G; Farag S; Rohrabaugh SL; Ou X; Speth J; Pelus LM; Srour EF; Campbell TB Dipeptidylpeptidase 4 Negatively Regulates Colony-Stimulating Factor Activity and Stress Hematopoiesis. Nat. Med 2012, 18 (12), 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Farag SS; Srivastava S; Messina-Graham S; Schwartz J; Robertson MJ; Abonour R; Cornetta K; Wood L; Secrest A; Strother RM; Jones DR; Broxmeyer HE In Vivo DPP-4 Inhibition to Enhance Engraftment of Single-Unit Cord Blood Transplants in Adults with Hematological Malignancies. Stem Cells Dev 2013, 22 (7), 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Farag SS; Nelson R; Cairo MS; O’Leary HA; Zhang S; Huntley C; Delgado D; Schwartz J; Zaid MA; Abonour R; Robertson M; Broxmeyer H High-Dose Sitagliptin for Systemic Inhibition of Dipeptidylpeptidase-4 to Enhance Engraftment of Single Cord Umbilical Cord Blood Transplantation. Oncotarget 2017, 8 (66), 110350–110357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Chang Y; Hellwarth PB; Randolph LN; Sun Y; Xing Y; Zhu W; Lian XL; Bao X Fluorescent Indicators for Continuous and Lineage-Specific Reporting of Cell-Cycle Phases in Human Pluripotent Stem Cells. Biotechnol. Bioeng 2020, 117 (7), 2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Randolph LN; Bao X; Oddo M; Lian XL Sex-Dependent VEGF Expression Underlies Variations in Human Pluripotent Stem Cell to Endothelial Progenitor Differentiation. Sci. Rep 2019, 9 (1), 16696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Konermann S; Lotfy P; Brideau NJ; Oki J; Shokhirev MN; Hsu PD Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173 (3), 665–676.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Wessels HH; Méndez-Mancilla A; Guo X; Legut M; Daniloski Z; Sanjana NE Massively Parallel Cas13 Screens Reveal Principles for Guide RNA Design. Nat. Biotechnol 2020, 38 (6), 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Randolph LN; Bao X; Zhou C; Lian X An All-in-One, Tet-On 3G Inducible PiggyBac System for Human Pluripotent Stem Cells and Derivatives. Sci. Rep 2017, 7 (1), 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Mesquitta WT; Wandsnider M; Kang HJ; Thomson J; Moskvin O; Suknuntha K; Slukvin II UM171 Expands Distinct Types of Myeloid and NK Progenitors from Human Pluripotent Stem Cells. Sci. Rep 2019, 9 (1), 6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Kumar A; Lee JH; Suknuntha K; D’Souza SS; Thakur AS; Slukvin II NOTCH Activation at the Hematovascular Mesoderm Stage Facilitates Efficient Generation of T Cells with High Proliferation Potential from Human Pluripotent Stem Cells. J. Immunol 2019, 202 (3), 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Lian X; Zhang J; Azarin SM; Zhu K; Hazeltine LB; Bao X; Hsiao C; Kamp TJ; Palecek SP Directed Cardiomyocyte Differentiation from Human Pluripotent Stem Cells by Modulating Wnt/β-Catenin Signaling under Fully Defined Conditions. Nat. Protoc 2013, 8 (1), 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Bao X; Lian X; Qian T; Bhute VJ; Han T; Palecek SP Directed Differentiation and Long-Term Maintenance of Epicardial Cells Derived from Human Pluripotent Stem Cells under Fully Defined Conditions. Nat. Protoc 2017, 12 (9), 1890–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Jiang Y; Chen C; Randolph LN; Ye S; Zhang X; Bao X; Lian XL Generation of Pancreatic Progenitors from Human Pluripotent Stem Cells by Small Molecules. Stem Cell Reports 2021, 16 (9), 2395–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Jiang Y; Hoenisch RC; Chang Y; Bao X; Cameron CE; Lian XL Robust Genome and RNA Editing via CRISPR Nucleases in PiggyBac Systems. Bioact. Mater 2022, 14, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Raiter A; Zlotnik O; Lipovetsky J; Mugami S; Dar S; Lubin I; Sharon E; Cohen CJ; Yerushalmi R A Novel Role for an Old Target: CD45 for Breast Cancer Immunotherapy. Oncoimmunology 2021, 10 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Jung HS; Uenishi G; Park MA; Liu P; Suknuntha K; Raymond M; Choi YJ; Thomson JA; Ong IM; Slukvin II SOX17 Integrates HOXA and Arterial Programs in Hemogenic Endothelium to Drive Definitive Lympho-Myeloid Hematopoiesis. Cell Rep 2021, 34 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Hsu J; Huang HT; Lee CT; Choudhuri A; Wilson NK; Abraham BJ; Moignard V; Kucinski I; Yu S; Hyde RK; Tober J; Cai X; Li Y; Guo Y; Yang S; Superdock M; Trompouki E; Calero-Nieto FJ; Alireza G; Jiang J; Gao P; Gao L; Nguyen V; Robertson AL; Durand EM; Kathrein KL; Aifantis I; Gerber SA; Tong W; Tan K; Cantor AB; Zhou Y; Liu PP; Young RA; Göttgens B; Speck NA; Zon LI CHD7 and Runx1 Interaction Provides a Braking Mechanism for Hematopoietic Differentiation. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (38), 23626–23635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Peng S; Shi S; Tao G; Li Y; Xiao D; Wang L; He Q; Cai X; Xiao J JKAMP Inhibits the Osteogenic Capacity of Adipose-Derived Stem Cells in Diabetic Osteoporosis by Modulating the Wnt Signaling Pathway through Intragenic DNA Methylation. Stem Cell Res. Ther 2021, 12 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Kim D; Paggi JM; Park C; Bennett C; Salzberg SL Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol 2019, 37 (8), 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Ramsköld D; Wang ET; Burge CB; Sandberg R An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data. PLoS Comput. Biol 2009, 5 (12), e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Oprescu SN; Yue F; Qiu J; Brito LF; Kuang S Temporal Dynamics and Heterogeneity of Cell Populations during Skeletal Muscle Regeneration. iScience 2020, 23 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Stuart T; Butler A; Hoffman P; Hafemeister C; Papalexi E; Mauck WM; Hao Y; Stoeckius M; Smibert P; Satija R Comprehensive Integration of Single-Cell Data. Cell 2019, 177 (7), 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Randolph LN; Jiang Y; Chang Y; Bao X; Lian XL Direct Induction of Hemogenic Endothelial Progenitors from HPSCs by Defined Factors Revealed by Single-Cell Transcriptome Analysis. bioRxiv 2021, 2021.03.18.435636. [Google Scholar]
- (86).Staal FJT; Spaink HP; Fibbe WE Visualizing Human Hematopoietic Stem Cell Trafficking in Vivo Using a Zebrafish Xenograft Model. Stem Cells Dev 2016, 25 (4), 360–365. [DOI] [PubMed] [Google Scholar]
- (87).Hamilton N; Sabroe I; Renshaw SA A Method for Transplantation of Human HSCs into Zebrafish, to Replace Humanised Murine Transplantation Models. F1000Research 2018, 7, 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Zhou W; Pal AS; Hsu AYH; Gurol T; Zhu X; Wirbisky-Hershberger SE; Freeman JL; Kasinski AL; Deng Q MicroRNA-223 Suppresses the Canonical NF-KB Pathway in Basal Keratinocytes to Dampen Neutrophilic Inflammation. Cell Rep 2018, 22 (7), 1810–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Soza-Ried C; Hess I; Netuschil N; Schorpp M; Boehm T Essential Role of C-Myb in Definitive Hematopoiesis Is Evolutionarily Conserved. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (40), 17304–17308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final processed data and raw fastq files were submitted to Gene Expression Omnibus (GEO) with accession numbers: GSE155196 and GSE157944. Source data for the figures in this study are available in figshare with the identifier: http://doi.org/10.6084/m9.figshare.16556577. The authors declare that all other data supporting the findings of this study are available within the paper and its Supplementary Information.


