Summary
Lungs undergo mechanical strain during breathing, but how these biophysical forces affect cell fate and tissue homeostasis are unclear. We show that biophysical forces through normal respiratory motion actively maintain alveolar type 1 (AT1) cell identity and restrict these cells from reprogramming into AT2 cells in the adult lung. AT1 cell fate is maintained at homeostasis by Cdc42 and Ptk2 mediated actin remodeling and cytoskeletal strain, and inactivation of these pathways causes a rapid reprogramming into the AT2 cell fate. This plasticity induces chromatin reorganization and changes in nuclear lamina-chromatin interactions, which can discriminate AT1 and AT2 cell identity. Unloading the biophysical forces of breathing movements leads to AT1-AT2 cell reprogramming, revealing that normal respiration is essential to maintain alveolar epithelial cell fate. These data demonstrate the integral function of mechanotransduction in maintaining lung cell fate and identifies the AT1 cell as an important mechanosensor in the alveolar niche.
Keywords: Biophysical forces, mechanotransduction, lung, alveolar epithelial cell, lamina-associated domain
Graphical Abstract
In Brief
Forces elicited by respiration actively induce changes in nuclear lamina-chromatin interactions to maintain alveolar type 1 (AT1) cell identity and restrict these cells from reprogramming into AT2 cells.
Introduction
Mammalian lungs have evolved to be flexible and elastic to undergo cyclical inflation and deflation of the alveoli for efficient gas exchange. Multiple cell lineages comprise the alveolar niche including alveolar type 1 (AT1) and alveolar type 2 (AT2) epithelial cells as well as various mesenchymal and endothelial cell types. Most studies on mechanotransduction in the lungs have focused on the response of how mesenchymal lineages respond to mechanical signals, and their ability to differentiate into myofibroblasts in diseases such as idiopathic pulmonary fibrosis (IPF).1 However, the ability of epithelial cells to respond to and transmit biophysical signals, in contrast to mesenchymal cells, remains poorly understood.
Previous studies have demonstrated a role for mechanotransduction in mesenchymal lineages in the lung in response to injury. Activation of fibroblasts by Yap/Taz through mechanotransduction drives lung fibrosis, whereas inhibition of Yap/Taz activity in fibroblasts prevents lung injury and fibrosis.2,3 In the lung alveolus, AT1 cells are required for efficient gas exchange across the endothelial capillary plexus while AT2 cells generate and recycle pulmonary surfactant and act as facultative progenitors that differentiate into AT1 cells.4,5 We previously showed that Yap/Taz are required to actively maintain the AT1 cell fate and loss of Yap/Taz leads to a rapid reprogramming of AT1 cells into AT2 cells.6 Moreover, nuclear Yap is found in AT1 and not AT2 cells, further suggesting a specific responsiveness to Hippo signaling in the AT1 lineage.6 While forced expression of an activated Yap protein in AT2 cells can lead to increased expression of some AT1 cell markers, these cells do not exhibit all traits of this lineage, including a consistently enlarged squamous shape.6 Since Yap/Taz can function as cytoplasmic mechanotransducers that translocate to the nucleus upon actin-regulated cell stretch and strain in vitro,7,8 these data raise the intriguing concept that mechanotransduction plays a specific role in AT1 rather than AT2 cells to maintain alveolar function in the homeostatic lung.
Using a multi-modal genetic and biophysical approach, we show that AT1 cell lineage is the mechanotransductive epithelial cell type in the lung alveolus. Physical constraint of AT2 cells suppresses their differentiation into AT1 cells. In contrast, loss of cytoskeletal-extracellular matrix interactions through Cdc42 and Ptk2 in AT1 cells leads to their rapid reprogramming into AT2 cells in vivo and is associated with loss of nuclear Yap expression. Single cell RNA-seq (scRNA-seq) analysis shows that these reprogrammed AT2 cells are highly similar to normal AT2 cells. Conversely, loss of Ptk2 in AT2 cells inhibits their differentiation into AT1 cells, demonstrating that integrin mediated mechanotransduction controls the bi-directional switch between these two alveolar cell fates. Loss of AT1 cell fate is accompanied by changes in lamina-associated chromatin domain (LAD) organization, causing sequestration or release of AT1 or AT2 specific loci at the nuclear periphery, inversely related to the changed fate. Importantly, unilateral unloading of tissue strain from breathing movements in vivo causes a spontaneous reprogramming of AT1 into AT2 cells, revealing that constant mechanical strain is required to maintain the AT1 fate in the adult lung. These studies reveal that AT1 cells are a nodal cell lineage in the lung for sensing and transmitting the normal mechanical forces due to breathing.
Results
Mechanical properties define AT1 cells in vivo
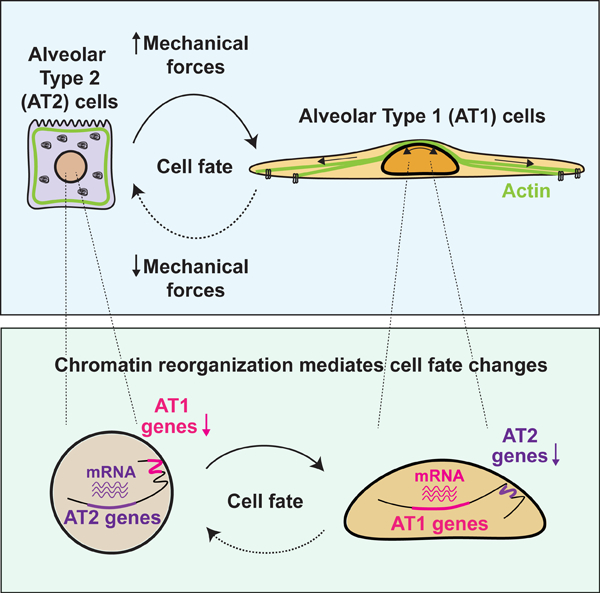
To gain insight into how biophysical forces affect alveolar epithelial cell fate in vivo, we examined actin cytoskeletal organization in AT1 and AT2 cells. We found that actin fibers and activated myosin contact the luminal side of the Hopx-EGFP+ AT1 nucleus, but not the Sftpc-EGFP+ AT2 nucleus (Fig. 1a, 1b and Fig. S1a). We also found that the AT1 nucleus is more elliptical in shape than the AT2 nucleus (Fig. 1c), which could reflect active cytoskeletal forces of AT1 cells resulting in changes in their nuclear shape.9,10 As we have previously reported, Yap localizes to the AT1 nucleus (Fig. 1d and Fig. S1b).6 Moreover, comparison of the AT1 and AT2 cell transcriptomes obtained from our previous study shows that the AT1 cell transcriptome is enriched for genes associated with actin cytoskeleton regulation, focal adhesion, and Hippo pathway (Fig. 1e).6 Examination of adult lung scRNA-seq data from a previous study reveals that expression of actin, non-muscle myosin, Cdc42-N-WASP-Arp2/3 complex, downstream target genes of Hippo pathway, and a set of AT1-specific integrins is enriched in AT1 cells (Fig. 1f–1h and S1c).11 On the other hand, AT2 transcriptome is enriched for genes associated with lipid metabolism (Fig. S1d). Of note, increased lipid metabolism utilization has recently been suggested as a general response to reduced actin contractility.12 Furthermore, we found that intermediate filaments, but not microtubules, are abundant in the perinuclear spaces of AT1 cells, suggesting that intermediate filaments could provide further structural support to AT1 cells (Fig. S1e). These findings suggest that biophysical forces preferentially control AT1 cell shape and function (Fig. 1i–j, Fig. S1f).
Figure 1. Actin-regulated biophysical forces define AT1 cells in vivo.
a. Phalloidin and activated myosin (phosphorylated myosin light chain; pMLC) staining of alveolar epithelial cells. F-actin and activated myosin contact luminal side of Hopx-EGFP+ AT1 nucleus, but not Sftpc-EGFP+ AT2 nucleus. Arrowheads indicate AT1 cells (top) and AT2 cells (bottom).
b. Quantification of perinuclear actin (n = 144 AT1 and n = 138 AT2 cells pooled from n = 5 mice) and perinuclear myosin (n = 130 AT1 and n = 153 AT2 cells pooled from n = 6 mice).
c. Representative 3D image of AT1 and AT2 nucleus (top). Quantification of nuclear sphericity and ellipticity for AT1 (n = 126) and AT2 (n = 185) cells pooled from n = 6 mice (bottom).
d. Yap localizes to AT1 nucleus, but not to AT2 cells.
e. Mechanotransduction-related GO terms and pathways are enriched in AT1 cells. RNA-seq data were obtained from Penkala et al.6
f-h. Actin, non-muscle myosin (f), Cdc42-N-WASP-Arp2/3 complex genes (g), and Hippo pathway downstream genes (h) are upregulated in AT1 cells. scRNA-seq data were obtained from Zepp et al.11
i. 3D images of AT1 and AT2 cell body. Imaged with lung strips from HopxCreERT2; R26RmTmG and SftpcCreERT2; R26RmTmG mice.
j. AT1 cells are subject to biophysical forces derived from tissue strain.
*** P < 0.001 by two-tailed t-test. Each dot represents an individual cell, and error bars indicate mean with s.d. Scale bars: a (left two panels), 10 μm; a (right three panels), d, 2.5 μm. See also Figure S1.
Actin mediated mechanotransduction maintains AT1 cell fate
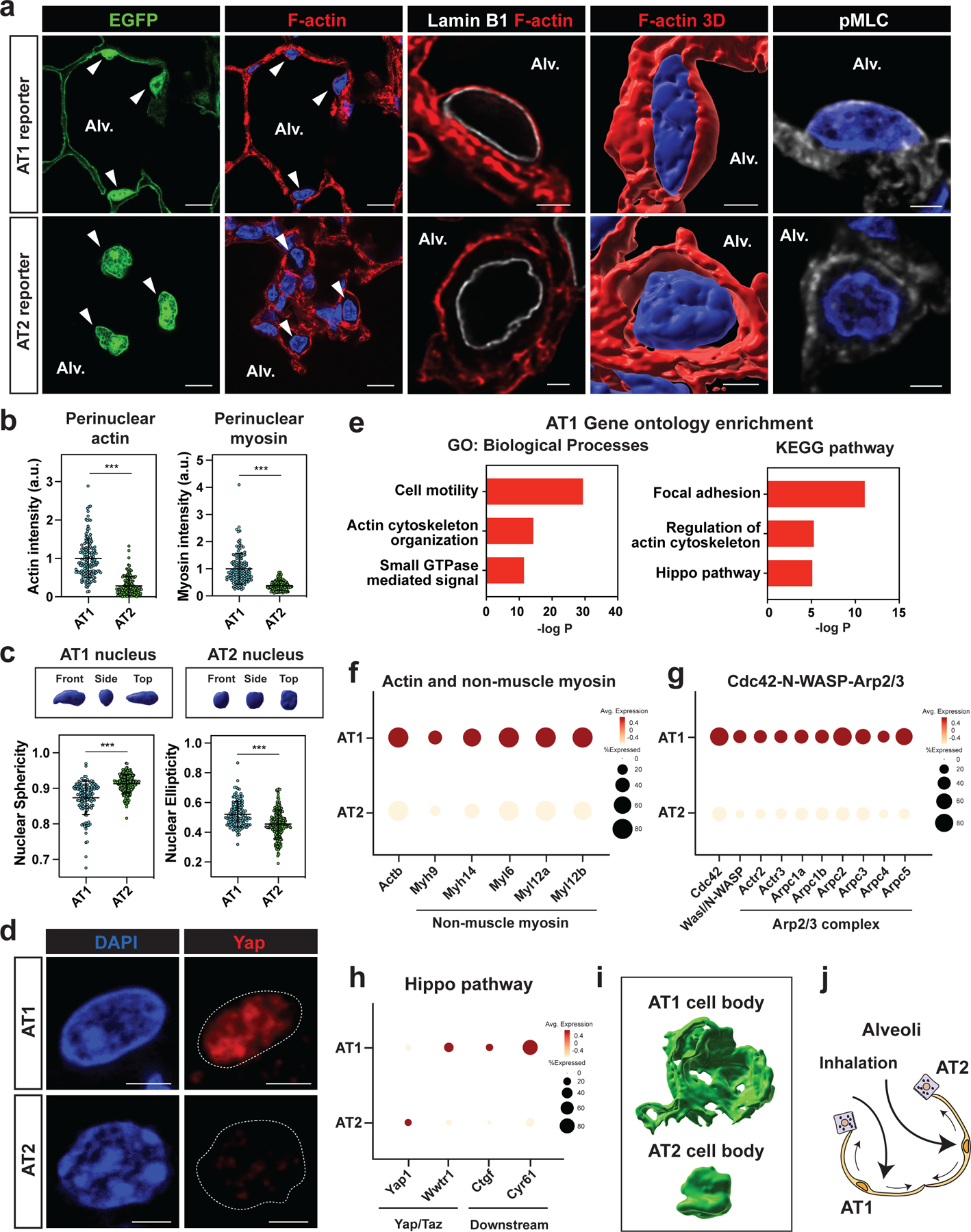
Small RhoGTPase Cdc42 is a critical regulator of actin cytoskeleton reorganization that is preferentially expressed in AT1 rather than AT2 cells in the adult mouse lung (Fig. 2a).11,13 To determine what role actin cytoskeleton reorganization plays in AT1 cells, we generated HopxCreERT2; Cdc42flox/flox; R26REYFP mice (hereafter called Cdc42AT1-KO) to inactivate Cdc42 specifically in AT1 cells. Lineage-traced cells were isolated (Fig. S2a), and Cdc42 deletion was confirmed by qPCR and immunoblotting (Fig. S2b, S2c). Loss of Cdc42 in AT1 cells led to increased expression of AT2 cell marker genes including Sftpc, suggesting that these cells re-programmed into AT2 cells (Fig. 2b–d). This loss of AT1 and gain of AT2 fate was rapid as it was observed as early as four days post-induction of cre recombinase (Fig. S2d). Cdc42 deletion did not affect the gross morphology of the lungs nor AT1 and AT2 cell numbers as assessed by histology (Fig. S2e–g), suggesting that AT2 cells differentiate into AT1 cells to compensate for the loss of AT1 cells in Cdc42AT1-KO mice.
Figure 2. Cdc42 deletion reprograms AT1 cells into AT2 cells.
a. AT1 cells express a higher level of Cdc42 than AT2 cells.
b-d. AT1 cell-specific Cdc42 deletion reprograms AT1 cells into AT2 cells. Cdc42 deletion and lineage-tracing were performed on HopxCreERT2; Cdc42flox/+; R26REYFP (control) and HopxCreERT2; Cdc42flox/flox; R26REYFP (Cdc42AT1-KO) mice and analyzed 14 days later (b). AT1 lineage cells express AT2 cell marker Sftpc in Cdc42AT1-KO mice (d). White arrowheads indicate AT1 cells and yellow arrowheads indicate AT2rep cells. Quantification of EYFP+ Sftpc+ AT2rep cells from n = 5 mice, 3 independent experiments (c).
e-f. scRNA-seq analysis of wildtype alveolar epithelial cells and lineage-traced cells from Cdc42AT1-KO mice. AT2rep cells co-cluster with wildtype AT2 cells (e). Correlation analysis shows high similarity between AT2rep and wildtype AT2 cells (f).
g. YAP does not localize to the EYFP+ Lamp3+ AT2rep nucleus in Cdc42AT1-KO mice, and F-actin does not contact AT2rep nucleus.
h. Quantification of Yap positivity for control AT1 cells and AT2rep cells (n = 5 mice).
i. Quantification of perinuclear actin for control AT1 cells, AT2 cells, and AT2rep cells (n = 5 mice). Control lung and Cdc42AT1-KO mouse lung were adhered to the same glass slide. Actin intensity for AT2 and AT2rep cells was standardized to that of control AT1 cells on the same glass slide. N.S., not significant by two-tailed t-test.
Each dot represents an individual mouse, and error bars indicate mean with s.d. Scale bars: d, g (left), 25 μm; g (right 3 panels), 2.5 μm. See also Figures S2 and S3.
To assess how similar reprogrammed AT2 cells from Cdc42AT1-KO mice (hereafter called AT2rep) are to normal AT2 cells, we isolated lineage-traced cells from Cdc42AT1-KO mice (Fig. S3a), and performed scRNA-seq analysis and compared the data to wildtype AT1 and AT2 cells. We found that AT2rep cells co-cluster with wildtype AT2 cells (Fig. 2e, S3b), and expressed AT2 specific genes at a similar level as wildtype AT2 cells (Fig. S3c). Spearman rank-order correlation test between wildtype AT2 and AT2rep cells confirmed that gene expression in AT2rep cells correlates to a high degree with wildtype AT2 cells (Fig. 2f). The AT2rep cells express proteolytically processed mature Sftpc protein, suggesting that AT2rep cells are similar to wildtype AT2 cells (Fig. S3d).14 Moreover, Yap nuclear localization was not observed in Lamp3+ AT2rep cells and fewer actin fibers surrounded AT2rep nucleus in comparison to the AT1 nucleus (Fig. 2g–i and Fig. S3e). These results demonstrate that Cdc42-dependent rearrangement of the actin cytoskeleton contributes to the active maintenance of AT1 cell identity, prohibiting the reprogramming of these cells into AT2 cells.
Cell spreading determines AT2-AT1 cell differentiation
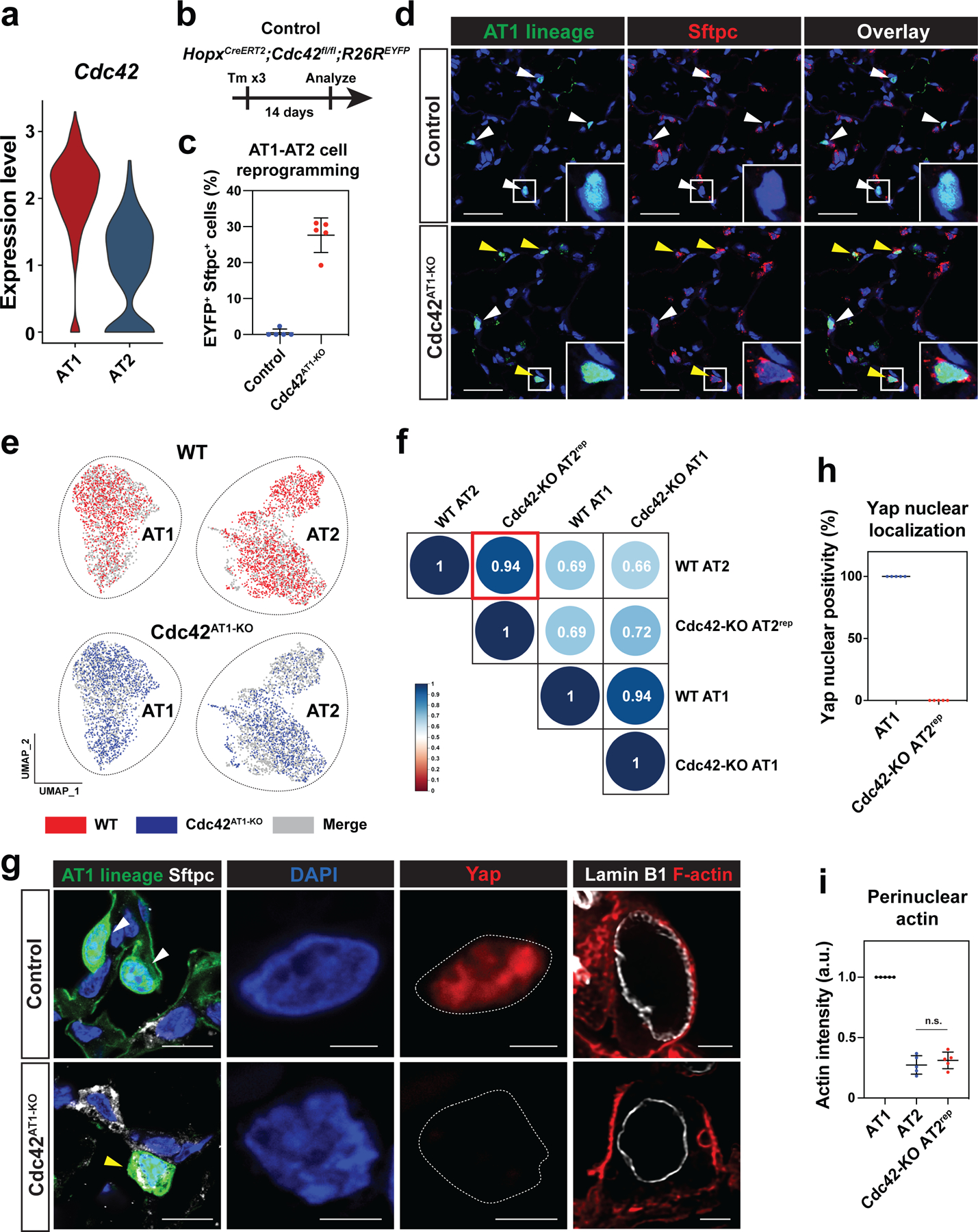
Primary AT2 cells autonomously spread and differentiate into AT1 cells between 24 and 48 hours after plating and acquire abundant cytoskeletal networks (Fig. S4a–d). To test our hypothesis that AT2 cells require cell spreading and actin-mediated cytoskeletal tension to differentiate into AT1 cells, we cultured primary mouse and human AT2 cells on micropatterned glass-bottom dishes to limit their ability to spread (Fig. 3a). The dishes are patterned with adherent circles ranging in size from 10 μm to 100 μm in diameter. AT2 cells adhering to circles of 10 μm in diameter were constrained in their ability to spread, whereas AT2 cells adhering to circles greater than 20 μm in diameter were not constrained and could spread freely. At 48 hours after plating, unconstrained mouse AT2 cells expressed the AT1 cell marker Ager and lost the AT2 cell marker Sftpc, while constrained mouse AT2 cells did not express Ager but retained Sftpc (Fig. 3b, 3d). AT2 cells cultured on elongated micropattern differentiated into AT1 cells at the same extent as AT2 cells cultured on a circular pattern, suggesting that cell spreading rather than cell shape determines the differentiation into AT1 cells (Fig. S4e, f). Similarly, unconstrained human AT2 cells acquired expression of the AT1 cell marker PDPN and lost SFTPC, while constrained human AT2 cells did not express PDPN but retained SFTPC (Fig. 3c, 3d and Fig. S5a–c). In mouse AT2 cells, Yap/Taz was found in the nucleus of unconstrained cells (Fig. 3e–f). Cytoplasmic actin fibers and activated myosin were abundant in unconstrained mouse AT2 cells and were associated with the nuclear periphery (Fig. 3g–h). YAP/TAZ translocated to the nucleus of unconstrained human AT2 cells, and cytoplasmic actin fibers were abundant in unconstrained human AT2 cells (Fig. S5d–f). These results suggest that AT2-AT1 cell differentiation requires cell spreading and cytoskeletal strain in vitro.
Figure 3. Cell spreading determines AT2-AT1 cell differentiation.
a. Mouse and human AT2 cells were cultured 48 h on micropatterned dishes with printed circles of different sizes.
b-c. Constrained AT2 cells do not differentiate into AT1 cells. Constrained mouse AT2 cells (10 μm circle) maintain Sftpc expression, while unconstrained mouse AT2 cells (> 20 μm circle) downregulate Sftpc and express AT1 cell marker Ager (b). CellMask is used to visualize the cell body. Constrained human AT2 cells maintain SFTPC expression, while unconstrained human AT2 cells downregulate SFTPC and express AT1 cell marker PDPN (c).
d. Ager/Sftpc ratio representing mouse AT2-AT1 cell differentiation was quantified with n = 145 constrained and n = 164 unconstrained AT2 cells pooled from 4 independent experiments (left). PDPN/SFTPC ratio representing human AT2-AT1 cell differentiation was quantified with n = 122 constrained, n = 106 unconstrained AT2 cells pooled from 3 independent experiments (right).
e-f. Yap/Taz localize to unconstrained mouse AT2 nucleus, but not to constrained AT2 nucleus (e). Yap/Taz nuclear to cytoplasm ratio was quantified with n = 131 constrained and n = 166 unconstrained mouse AT2 cells pooled from 4 independent experiments (f). g-h. Actin fibers and activated myosin are abundant in cytoplasm and perinuclear regions of unconstrained AT2 cells (g). Actin and myosin intensity in cytoplasmic and perinuclear regions were quantified with n = 33 constrained and n = 36 unconstrained mouse AT2 cells (h).
*** P < 0.001 by two-tailed t-test. For Box and whisker plots, bars represent min and max values. Scale bars: b, c, 10 μm; e, g, 2.5 μm. See also Figures S4 and S5.
AT2-AT1 cell differentiation and AT1 cell fate maintenance require Integrin-FAK signaling
Actin cytoskeleton transduces mechanical forces from the extracellular environment into the cell through integrin mediated connections and signaling, and Integrin-Focal Adhesion Kinase (FAK) pathway is the central integrin mediated signaling pathway implicated in cell spreading and actin cytoskeleton organization.15 Based on the results from micropattern experiments, we evaluated the role of Integrin-FAK pathway in AT2-AT1 cell differentiation and AT1 cell fate maintenance. AT1 and AT2 cells express cell-type specific integrin subunits (Fig. S1c). AT2 cells were cultured and treated with antibodies that block either AT1-specific integrins (anti-Itgb5 and anti-Itgb6) or AT2-specific integrin (anti-Itga6), FAK inhibitor PF573228, or actin polymerization inhibitor latrunculin B (Fig. 4a). Anti-Itgb5 and anti-Itgb6 antibodies, PF573228, and latrunculin B all attenuated AT2 cell spreading and AT2-AT1 cell differentiation (Fig. 4b, 4c) without affecting cell viability (Fig. S5g, S5h). However, the anti-Itga6 AT2-specific antibody did not have any effect on AT2 cell spreading or AT2-AT1 cell differentiation (Fig. 4b, 4c). These data suggest that the AT1-specific Integrin-FAK pathway and the acquisition of the cytoskeletal strain are essential for AT2-AT1 cell differentiation in vitro.
Figure 4. AT2-AT1 cell differentiation and AT1 cell fate maintenance require integrin signaling.
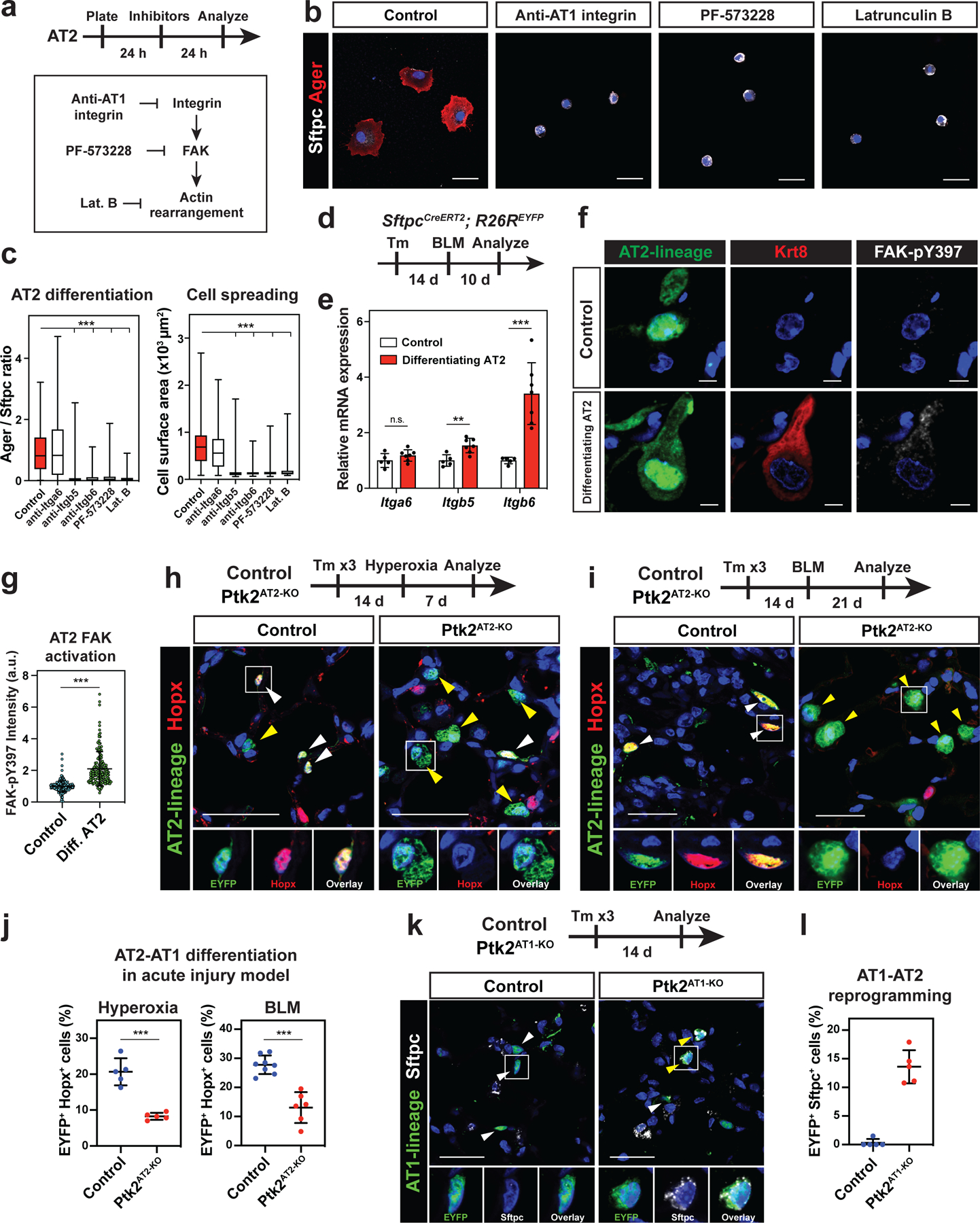
a-c. Integrin-FAK signaling is required for AT2 cell spreading and differentiation in vitro. Inhibitors were added 24 h after AT2 cell plating, and the cells were analyzed 24 h after the treatment (a). Anti-AT1 Integrin (Itgb6) antibody, FAK inhibitor PF 573228, or actin polymerization inhibitor Latrunculin B attenuated AT2 cell spreading and differentiation (b). The inhibitor-treated AT2 cells retained Sftpc+ Ager− AT2 cell state. Ager/Sftpc ratio and cell surface area were quantified for control (n = 135), anti-Itga6-treated (n = 69), anti-Itgb5-treated (n = 100), anti-Itgb6-treated (n = 126), PF 573228-treated (n = 123), or Latrunculin B-treated (n = 117) AT2 cells pooled from 3–6 independent experiments (c). d-g. Integrin-FAK signaling is activated during AT2-AT1 cell differentiation in vivo. Bleomycin (BLM) was intratracheally given to SftpcCreERT2; R26REYFP mice, and the mice were analyzed 10 days later (d). qPCR shows Itgb5, and Itgb6 are upregulated after acute lung injury, using sorted AT2 cells from PBS-treated control (n = 5 mice) and BLM-treated SftpcCreERT2; R26REYFP mice (n = 7 mice) from 3 independent experiments (e). FAK is activated (FAK-pY397+) in EYFP+ Krt8+ differentiating AT2 cells (f). FAK-pY397 intensity was quantified for n = 236 AT2 and n = 147 Krt8+ AT2 cells from n = 6 mice (g).
h-j. AT2-specific FAK knockout attenuates AT2-AT1 cell differentiation in vivo. FAK (Ptk2) deletion and lineage-tracing was performed on SftpcCreERT2; Ptk2flox/+; R26REYFP (control) and SftpcCreERT2; Ptk2flox/flox; R26REYFP (Ptk2AT2-KO) mice. The mice were analyzed 7 days after hyperoxia lung injury (h) or 21 days after BLM injury (i). AT2 cells differentiate into Hopx+ AT1 cells in control (h, i left), but less so in the Ptk2AT2-KO mice (h, i right) after injury. Quantification of AT2-AT1 cell differentiation for hyperoxia (n = 5 control and n = 5 Ptk2AT2-KO mice from 2 independent experiments) and BLM (n = 8 control and n = 6 Ptk2AT2-KO mice from 3 independent experiments) acute lung injury model (j).
k-l. AT1 cell-specific FAK deletion reprograms AT1 cells into AT2 cells. FAK deletion and lineage-tracing were performed on HopxCreERT2; R26REYFP (control) and HopxCreERT2; Ptk2flox/flox; R26REYFP (AT1Ptk2-KO) mice and analyzed 14 days later. AT1 lineage cells in AT1Ptk2-KO mice express AT2 cell marker Sftpc (k). Quantification of EYFP+ Sftpc+ reprogrammed AT2 cells from n = 5 mice (l).
*** P < 0.001 and ** P < 0.01 by one-way ANOVA (c) and two-tailed t-test (e, g, j). Each dot represents an individual mouse or cell, and error bars indicate mean with s.d. For Box and whisker plots, bars represent min and max values. Scale bars: b, h, i, k, 25 μm; f, 10 μm. See also Figure S5.
Next, we assessed whether the Integrin-FAK pathway is essential for AT2-AT1 cell differentiation in vivo. We lineage labeled AT2 cells using SftpcCreERT2; R26REYFP mice and subjected them to acute lung injury (Fig. 4d). Ten days after the injury, the lineage labeled AT2 cells expressed higher levels of AT1-specific integrins (Itgb5, and Itgb6), but not the AT2-specific integrin Itga6, than uninjured control cells (Fig. 4e). Moreover, FAK phosphorylation was increased in Krt8+ transition state of AT2 cells differentiating into AT1 cells (Fig. 4f–g),16–18 suggesting that the Integrin-FAK pathway is activated during AT2-AT1 cell differentiation.19 To test whether FAK is required for AT2-AT1 cell differentiation, we inactivated FAK (also known as Ptk2) in AT2 cells using SftpcCreERT2; Ptk2flox/flox; R26REYFP (hereafter called Ptk2AT2-KO) mice. Ptk2 deletion was confirmed by qPCR and immunoblotting (Fig. S5i, S5j). Following acute lung injury induced by hyperoxia or bleomycin, lineage labeled AT2 cells exhibited a reduced ability to differentiate into AT1 cells in Ptk2AT2-KO mice compared to control mice (Fig. 4h–j). Loss of Ptk2 did not affect cell death or proliferation of AT2 cells (Fig. S5k–n). Importantly, loss of Ptk2 in AT1 cells led to their reprogramming into AT2 cells at homeostasis, suggesting that Ptk2 is required for AT1 cell fate maintenance (Fig. 4k, 4l). Collectively, these data demonstrate that transduction of extracellular mechanical forces into the cell through integrin signaling is required for AT2-AT1 cell differentiation and AT1 cell fate maintenance.
Nuclear lamina-chromatin interactions discriminate alveolar epithelial cell fates
External forces applied to the cell stretch the nuclear membrane and reorganize the genome, leading to alterations in gene expression.20,21 The precise molecular mechanisms underlying how nuclear sensing of mechanical forces results in gene expression changes are likely multifactorial. These include large scale physical changes in chromatin that involves reorganization and retention of specific domains at the nuclear periphery.22 To better understand genome organization and how it is affected by mechanical stress in AT1 and AT2 cells, we performed Lamin B1 ChIP-seq on AT1 and AT2 cells and analyzed the lamina-associated domains (LADs) (Fig. 5a).23,24 LADs are large domains of transcriptionally repressed chromatin located at the nuclear periphery that are associated with the nuclear lamina through interactions with the multiprotein linker of nucleoskeleton and cytoskeleton (LINC) complex.25 During cell state changes, certain loci are repositioned to and away from lamina, contributing to the expression patterns of cell lineage specific genes.26,27
Figure 5. Nuclear lamina-chromatin interactions discriminate alveolar epithelial cellular fate.
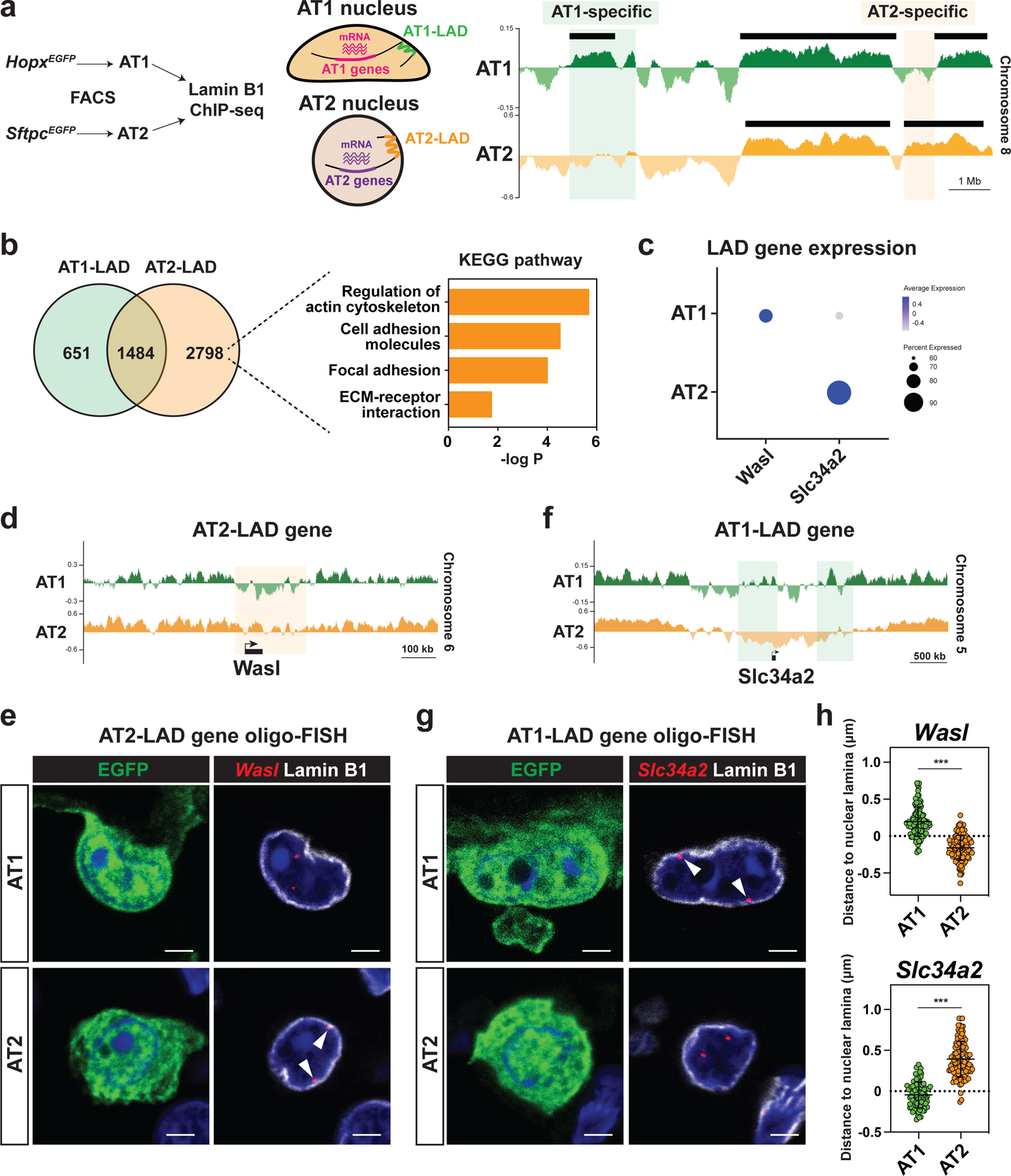
a. AT1 and AT2 cells were isolated from HopxEGFP (AT1) or SftpcEGFP (AT2) mice by fluorescence-activated cell sorting (FACS) and used for Lamin B1 ChIP-seq. Representative Lamin B1 ChIP-seq tracks (right) for AT1 and AT2 cells, showing AT1 and AT2-specific LADs. Black bars represent EDD-defined LADs.
b. Comparison of AT1 and AT2 LAD genes. Pathway enrichment analysis of AT2-LAD genes reveals an enrichment of genes associated with actin cytoskeleton and focal adhesion.
c. DotPlot showing LAD gene expression in AT1 and AT2 cells. scRNA-seq data obtained from this study were used.
d. Lamin B1 ChIP-seq tracks for AT1 and AT2 cells, showing that Wasl loses residence in AT1 cells.
e. Oligo-FISH of Wasl loci in lung sections from HopxEGFP AT1 and SftpcEGFP AT2 reporter mice. Arrowheads indicate Wasl locus.
f. Lamin B1 ChIP-seq tracks for AT1 and AT2 cells, showing that Slc34a2 loses residence in AT2 cells.
g. Oligo-FISH of Slc34a2 loci in lung sections from HopxEGFP AT1 and SftpcEGFP AT2 reporter mice. Arrowheads indicate Slc34a2 locus.
h. 3D Quantification of the distance between Wasl (n = 113 AT1 and n = 112 AT2 cells) or Slc34a2 (n = 101 AT1 and n = 112 AT2 cells) loci and Lamin B1. Imaris returned negative values when a FISH signal was embedded in the nuclear lamina.
*** P < 0.001 by two-tailed t-test. Each dot represents an individual cell, and error bars indicate mean with s.d. Scale bars: e, g, 2.5 μm. See also Figure S6.
Lamin B1 ChIP-seq identified 651 AT1-specific genes, 2798 AT2-specific genes, and 1484 shared genes located within LADs, by assessing the number of reads that mapped to the middle of the gene body ± 50 kb (Fig. 5b). The expression of AT1 and AT2 LAD genes was lower than that of non-LAD genes, confirming that genes found in LADs were repressed (Fig. S6a, S6b).26 Pathway enrichment analysis revealed that AT2 LAD genes were enriched for genes associated with actin cytoskeleton regulation and focal adhesion (Fig. 5b). Wasl, Rock1, and Ptk2 are genes enriched in pathways associated with regulation of actin cytoskeleton or focal adhesion. Wasl binds to Cdc42 to activate the Arp2/3 complex, leading to actin polymerization.28 Rock1 phosphorylates myosin and is a critical regulator of actomyosin contraction.29 We also found that the canonical AT1 cell marker Cav2 was found in an AT2 LAD and the canonical AT2 cell marker Slc34a2 was found in an AT1 LAD (Fig. 5c, 5f, S6f).
To validate our genomics data and confirm the nuclear locations of these LAD genes, we performed oligo-FISH to visualize the genomic loci of these genes. We reconstructed 3D images from confocal z stacks to precisely evaluate spatial positioning of each gene relative to the lamina (Fig. S6c). Consistent with the Lamin B1 ChIP-seq data (Fig. 5d, S6d–S6f), oligo-FISH confirmed that Wasl, Rock1, Ptk2, and Cav2 are located at the periphery of the AT2 but not in the AT1 nucleus (Fig. 5e, 5h, S6d–S6g). Conversely, we found that the AT2 cell marker gene Slc34a2 was enriched in an AT1 LAD (Fig. 5f). Oligo-FISH confirmed that Slc34a2 is located at the periphery of the AT1 nucleus, but not in the AT2 nucleus (Fig. 5g, 5h). These data suggest that nuclear lamina-chromatin interactions discriminate AT1 and AT2 cell-specific gene expression and that genes related to actin regulation and focal adhesion in AT2 cells are repressed by spatial repositioning to the nuclear periphery.
Alveolar epithelial fate changes involve changes in LAD organization
We next examined whether AT1-AT2 cell reprogramming and AT2-AT1 cell differentiation involved genome reorganization and changes in LADs given their ability to discriminate these two cell fates. We performed oligo-FISH on Cdc42AT1-KO and Ptk2AT1-KO mouse lungs and found that Wasl is peripherally located in Lamp3+ AT2rep cells (Fig. 6a, 6c), whereas Slc34a2 is not (Fig. 6b, 6c), suggesting that AT1-AT2 cell reprogramming involves chromatin reorganization and changes in LADs. Moreover, Wasl is peripherally located in constrained and FAK inhibitor-treated AT2 cells in vitro, whereas Slc34a2 is not (Fig. 6d, 6e). Additionally, in AT2 cells, we inhibited cytoplasm-to-nucleus mechanotransduction mediated by the LINC complex through expression of a dominant negative KASH protein and found that dominant negative KASH inhibits AT2-AT1 cell differentiation and spatial reorganization of relevant loci (Fig. 6f–i).30 These data indicate that biophysical strain maintains alveolar epithelial cell fate, in part, through control of nuclear lamina-chromatin interactions.
Figure 6. Alveolar epithelial fate changes involve LAD reorganization.
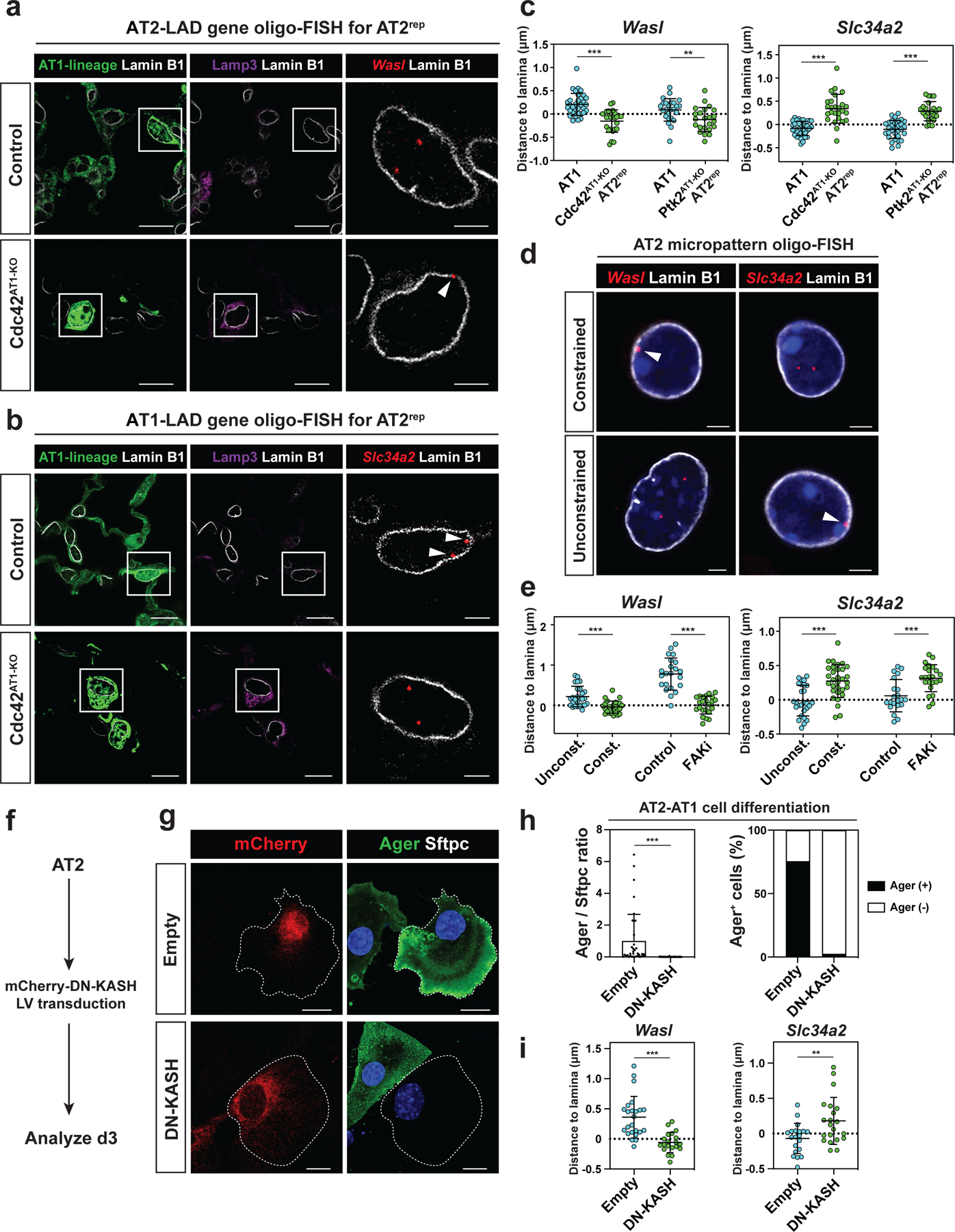
a-b. Oligo-FISH of Wasl (a) and Slc34a2 (b) loci in lung sections from control and Cdc42AT1-KO mice. Arrowheads indicate Wasl (a) or Slc34a2 (b).
c. Quantification of the distance between Wasl (n = 33 AT1 and n = 25 AT2rep cells for Cdc42AT1-KO, and n = 25 AT1 and n = 23 AT2rep cells for Ptk2AT1-KO) or Slc34a2 (n = 36 AT1 and n = 26 AT2rep cells for Cdc42AT1-KO, and n = 31 AT1 and n = 23 AT2rep cells for Ptk2AT1-KO) loci and Lamin B1 for control AT1 and AT2rep cells.
d. Oligo-FISH of Wasl and Slc34a2 loci for AT2 cells from micropattern experiments. Arrowheads indicate Wasl (left) or Slc34a2 (right).
e. Quantification of the distance between Wasl (n = 26 constrained, n = 26 unconstrained, n = 22 untreated, and n = 23 FAK inhibitor-treated cells) or Slc34a2 (n = 31 constrained, n = 27 unconstrained, n = 21 untreated, and n = 23 FAK inhibitor-treated cells) loci and Lamin B1.
f. Primary AT2 cells were transfected with lentivirus containing dominant-negative KASH and analyzed on day 3.
g-h. Dominant-negative KASH inhibits AT2 cell differentiation (g). Quantification of Ager/Sftpc ratio and Ager+ cell ratio from n = 37 control vector-transfected and n = 40 dominant-negative KASH-transfected cells from n = 3 experiments (h).
i. Quantification of the distance between Wasl (n = 24 control and n = 20 dominant-negative KASH-transfected cells) or Slc34a2 (n = 20 control and n = 21 dominant-negative KASH-transfected cells) loci and Lamin B1 from n = 3 experiments.
*** P < 0.001 and ** P < 0.01 by two-tailed t-test. Each dot represents an individual cell, and error bars indicate mean with s.d. Scale bars: a (left and middle), b (left and middle), g, 10 μm; a (right), b (right), d, 2.5 μm.
Alveolar cell fate is maintained by active breathing movements
The data presented thus far suggest that mechanotransductive pathways maintain AT1 cell fate in part through repositioning of LADs and alteration of cell type specific gene expression. However, whether actual changes in mechanical strain in vivo alters alveolar cell fate is unknown. To further understand the role of biophysical forces in the lungs, we developed an in vivo model that limits lung breathing movements we termed the unilateral lung ligation model. In this model, the left bronchus is clipped without damaging the blood vessels, thereby limiting left lung expansion and sparing right lung lobes (Fig. 7a). Assessment of apoptosis and the response to hypoxia by examination of Hif1a target gene expression showed that the unilateral lung ligation model does not lead to overt changes in cell death or induction of the transcriptional response to hypoxia, consistent with preservation of normal blood flow into the left lung (Fig. S7a–S7d). We lineage labeled AT1 cells using HopxCreERT2;R26REYFP mice and examined whether AT1 cells reprogram into AT2rep cells following the unilateral lung ligation (Fig. 7b). At 28 days following the unilateral lung ligation, nearly 30% of the lineage labeled cells expressed the AT2 cell marker Sftpc similar to the Cdc42AT1-KO mice (Fig. 7c, 7d). AT2rep cells composed 5.66 ± 1.50% (n = 5 mice) of all AT2 cell population and this AT1 reprogramming was observed as early as seven days after ligation (Fig. S7e). We performed scRNA-seq of lineage-traced AT1-derived cells from the unilateral lung ligation model and compared them to wildtype AT1 and AT2 cells. These AT2rep cells from the unilateral lung ligation model co-clustered with wildtype AT2 cells in UMAP space (Fig. 7e, S7f). Moreover, these AT2rep cells expressed AT2 specific genes to a similar extent as wildtype AT2 cells (Fig. S7g). Spearman rank-order correlation test confirmed that gene expression in AT2rep cells and wildtype AT2 cells are highly similar (Fig. 7f). In AT2rep cells, Yap nuclear localization was also not detected, and these cells had fewer actin fibers surrounding their nuclei compared to AT1 nuclei (Fig. 7g, 7h, S7h). Yap localization and actin fiber composition in AT1 and AT2 cells did not change after hyperventilation (Fig. S7i–j), suggesting that a constant actin-based cellular stretch from breathing movements rather than a cyclical nature of breathing likely plays a critical role in maintaining alveolar epithelial cell identity. These results, together with the results from Cdc42AT1-KO mice and Ptk2 deletion model, suggest that biophysical forces from breathing movements actively maintain AT1 cell fate (Fig. 7i).
Figure 7. Alveolar cell fate is maintained by active breathing movements.
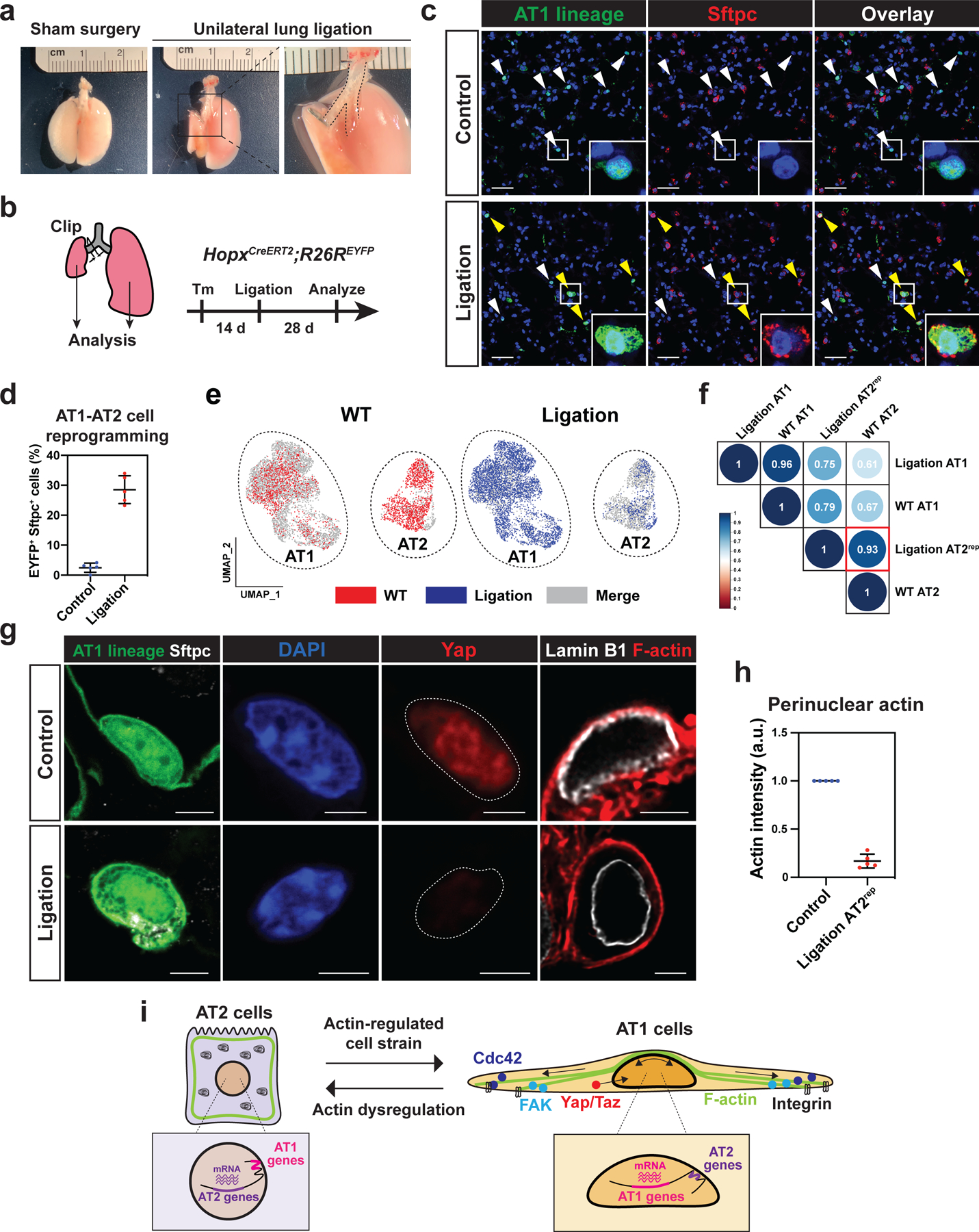
a-d. Unilateral lung ligation reprograms AT1 cells into AT2 cells. A gross morphological image of a mouse lung 28 days after sham surgery or unilateral lung ligation (a). The left main bronchus was ligated without affecting the left pulmonary artery or vein. Unilateral lung ligation and lineage-tracing were performed on HopxCreERT2; R26REYFP mice, and the mice were analyzed at 28 days later (b). Both the right (control) and left (ligated) lungs were analyzed. AT1 lineage cells express AT2 marker Sftpc in the ligated lung (c). White arrowheads indicate AT1 cells and yellow arrowheads indicate AT2rep cells. Quantification of EYFP+ Sftpc+ AT2rep cells from n = 5 mice from 3 independent experiments (d).
e-f. scRNA-seq analysis of wildtype alveolar epithelial cells and lineage-traced cells from HopxCreERT2; R26REYFP mice after unilateral lung ligation. AT2rep cells co-cluster with wildtype AT2 cells (e). Correlation analysis shows high similarity between AT2rep and wildtype AT2 cells (f).
g-h. YAP does not localize to the AT2rep nucleus and F-actin does not contact AT2rep nucleus. Representative images (g) and quantification (h) of perinuclear actin for control AT1 and AT2rep cells (n = 5 mice). Control lung and ligated left lung were adhered to the same glass slide. Actin intensity for AT2rep cells was standardized to that of control AT1 cells on the same glass slide.
i. Model of AT1 and AT2 cell regulation by biophysical forces.
Each dot represents an individual mouse, and error bars indicate mean with s.d. Scale bars: c, 25 μm; g, 2.5 μm. See also Figure S7.
Discussion
The lungs undergo constant cyclical stretch from normal breathing movements thereby applying mechanical forces to resident cells of the alveolus. In this study, we demonstrate that these breathing-induced biophysical forces maintain AT1 cell fate and control AT2-AT1 cell differentiation. Disruption of actin-regulated biophysical forces, cell adhesion, cell spreading, and unloading of the biophysical forces due to normal breathing movements, all lead to spontaneous and rapid reprogramming of AT1 cells into AT2rep cells that are transcriptionally highly similar to normal AT2 cells. This reprogramming is associated with dramatic changes in LAD organization and 3D spatial positioning of the genome, leading to lineage specific mechanosensitive gene expression changes in the lung alveolus. Together, these data reveal that the AT1 cell is a mechanosensor in the lung, requiring tissue strain from breathing movements to maintain its lineage fidelity during normal respiratory function. More broadly, our data reveal that epithelial cells play a critical role in mechanosensing to control cell identity in tissue homeostasis.
One of the enigmas of tissues such as the lungs is how they maintain cellular homeostasis when undergoing constant biophysical stresses due to rhythmic or cyclical forces. While mesenchymal cells are well known to sense and exert mechanical forces, the ability of epithelial cells to sense and respond to mechanotransduction is less well understood. During late stages of postnatal lung development called alveologenesis, a specialized type of mesenchymal cell called the secondary crest myofibroblast (SCMF) drives important aspects of alveolar remodeling through its ability to exert physical force and stretch the emerging alveolus to increase surface area.11 AT1 cells communicate with SCMFs, promoting their development and acquisition of the myofibroblast phenotype which increases its contractile abilities.11 Our current study indicates that AT1 cells also require a mechanotransduction feedback in the lung to maintain their fate during normal respiration.
Our study shows that AT1 cells, generally considered terminally differentiated, readily reprogram into AT2 cells when their cytoskeleton is perturbed, as demonstrated in the Cdc42 and Ptk2 loss of function studies as well as when normal breathing movements are ceased. A previous report showed that Cdc42 is essential for AT2-AT1 cell differentiation in a model of unilateral pneumonectomy.31 However, our data shows that Cdc42 expression is enriched in AT1 cells and is required to maintain AT1 cells fate during normal homeostasis in the absence of injury. The most harmonious manner in which to interpret these studies is that mechanotransduction plays a key role as AT2 cells differentiate into AT1 cells, rather than in the baseline AT2 cell homeostatic state. This is consistent with previous work showing that lipid metabolism utilization, which is a hallmark of AT2 and not AT1 cells, is associated with reduced extracellular matrix interactions associated mechanotransduction.12 This is also supported by the data presented here that integrin mediated signaling peaks during the differentiation of AT2 cells. In our models, AT1 cells that did not reprogram co-clustered with wildtype AT1 cells. However, not all AT1 cells reprogrammed suggesting that this lineage may respond heterogeneously to biophysical forces.
There is increasing interest in the effects of increased mechanical stress and strain on tissue homeostasis due to injury. However, it is less clear what roles such forces play in normal tissue homeostasis in tissues such as the lungs and heart, which undergo constant cyclical strain. Measuring and disrupting mechanotransduction in vivo in the lungs has been difficult to perform, but our unilateral lung ligation model allows the direct assessment of the effects of breathing cessation on cellular phenotypes. Our findings that breathing movements are required to maintain AT1 cell fate raise the possibility that other cell types in the lung or cells in other tissues that undergo constant cellular strain may also rely on mechanical strain to maintain their cell phenotype. Our studies show that chromatin association with the nuclear lamina underlies at least part of the mechanism maintaining AT1 cell fate in the lung. Sequestering regions of the genome to the nuclear periphery in LADs could be an important mechanism through which cells respond to altered mechanical forces.
Previous work has demonstrated that acute lung injury and regeneration results in a heterogeneous response leading to regions containing compacted tissue architecture and altered cell differentiation responses.5,32 Chronic lung diseases, such as IPF and COPD, also disrupt tissue architecture, with resulting changes in matrix stiffness and in the ability for proper inflation. Our study indicates that aberrant mechanical microenvironments could impact AT2-AT1 cell differentiation and AT1 cell fate maintenance, both of which are essential for lung regeneration. Future studies on how these injury and disease-associated changes in the biomechanics of lung tissue alter the trajectory of chronic disease or tissue regeneration after acute injury, could provide undiscovered avenues for the development of needed therapies for human lung disease.
Limitations of the Study
The primary lung AT2 epithelial cells used in our in vitro assays are isolated from harsh tissue digestions and are fragile. Moreover, AT1 cells are unable to be cultured in vitro likely due to their large extended shape and fragile nature. Future studies to examine how these cells respond to biophysical forces and the different molecular pathways involved in transmitting and receiving these forces in vitro and in vivo will provide further insight.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Edward E. Morrisey (emorrise@pennmedicine.upenn.edu).
Materials availability
All unique reagents generated in this study are available from the lead contact under material transfer request.
Data and code availability
ChIP-seq and scRNA-seq data generated during this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. RNA-seq data for AT1 and AT2 cells6 and scRNA-seq data for adult mouse lungs11 have been previously described and are publicly available (GSE148487 for RNA-seq and GSE149563 for scRNA-seq).
All original code has been deposited at Github (https://github.com/Morriseylab/LungBiophysical) and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Aves Labs | cat#: GFP-1020; RRID: AB_10000240 |
| Goat polyclonal anti-tdTomato | Origene | cat#: AB8181-200; RRID: AB_2722750 |
| Rabbit polyclonal anti-Phosphorylated Myosin Light Chain | Abcam | cat#: ab2480; RRID: AB_303094 |
| Rabbit polyclonal anti-Yap | Cell Signaling | cat#: 4912; RRID: AB_2218911 |
| Rabbit monoclonal anti-Yap/Taz | Cell Signaling | cat#: 8418; RRID: AB_10950494 |
| Rabbit polyclonal anti-Sftpc | Millipore | cat#: AB3786; RRID: AB_91588 |
| Rabbit polyclonal anti-Mature SFTPC | Seven Hills | cat#: WRAB-76694 |
| Rabbit polyclonal anti-SFTPC | Abcam | cat#: ab90716; RRID: AB_10674024 |
| Mouse monoclonal anti-HT2-280 | Terrace Biotech | cat#: TB-27AHT2-280; RRID: AB_2832931 |
| Rat monoclonal anti-Lamp3 | Novus | cat#: DDX0191P-100; RRID: AB_2827532 |
| Mouse monoclonal anti-Hopx | Santa Cruz | cat#: sc-398703; RRID: AB_2687966 |
| Rat monoclonal anti-Ager | R&D Systems | cat#: MAB1179; RRID: AB_2289349 |
| Rat monoclonal anti-PDPN | Thermo Fisher | cat#: 14–9381-82; RRID: AB_1603307 |
| Rat monoclonal anti-Krt8 | DSHB | cat#: TROMA-I; RRID: AB_2891089 |
| Rabbit monoclonal anti-FAK | Cell Signaling | cat#: 13009; RRID: AB_2798086 |
| Rabbit polyclonal anti-pFAK (Tyr397) | Thermo Fisher | cat#: 44–624G; RRID: AB_2533701 |
| Rabbit polyclonal anti-Lamin B1 | Abcam | cat#: ab16048; RRID: AB_443298 |
| Rabbit monoclonal anti-Cleaved Caspase-3 | R&D Systems | cat#: MAB835; RRID: AB_2243951 |
| Mouse monoclonal anti-Ki67 | BD Biosciences | cat#: 550609; RRID: AB_393778 |
| Rat monoclonal anti-α-Tubulin | Santa Cruz | cat#: sc-53029; RRID: AB_793541 |
| Mouse Pan-cytokeratin antibody cocktail | Invotrogen | cat#: MA5-13156; RRID: AB_10983023 |
| Mouse monoclonal anti-Cdc42 | Abcam | cat#: ab41429; RRID: AB_726768 |
| Rabbit monoclonal anti-β-actin | Cell Signaling | cat#: 4970; RRID: AB_2223172 |
| Mouse monoclonal anti-β-actin | Cell Signaling | cat#: 3700; RRID: AB_2242334 |
| Mouse monoclonal anti-β-actin, HRP conjugated | Proteintech | cat#: HRP-60008; RRID: AB_2819183 |
| Rat monoclonal anti-Integrin α6 | Invitrogen | cat#: 14–0495-82; RRID: AB_891480 |
| Mouse monoclonal anti-Integrin β5 | Invitrogen | cat#: 14–0497-82; RRID: AB_467288 |
| Mouse monoclonal anti-Integrin β6 | Sigma-Aldrich | cat#: MAB2077Z; RRID: AB_11210680 |
| Rat monoclonal anti-EpCAM, PE-Cy7 | eBioscience | cat#: 25–5791-80; RRID: AB_1724047 |
| Rat monoclonal anti-CD31, APC | eBioscience | cat#: 17–0311-82; RRID: AB_657735 |
| Rat monoclonal anti-CD45, APC | eBioscience | cat#: 17–0451-83; RRID: AB_469393 |
| Biological samples | ||
| Healthy human lung tissue | The University of Pennsylvania | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Bleomycin | Teva | N/A |
| Tamoxifen | Sigma-Aldrich | cat#: T5648 |
| SABM Basal Medium | Lonza | cat#: CC-3119 |
| SAGM SingleQuots Supplements and Growth Factors | Lonza | cat#: CC-4124 |
| EGF | PeproTech | cat#: AF-100-15 |
| Cholera toxin | Millipore | cat#: C9903 |
| Fibronectin | Sigma-Aldrich | cat#: F0895 |
| PF 573228 | Tocris | cat#: 3239 |
| Latrunculin B | Sigma-Aldrich | cat#: 428020 |
| Calcein, AM | Thermo Fisher | cat#: C1430 |
| Ethidium Homodimer-1 (EthD-1) | Thermo Fisher | cat#: E1169 |
| CellMask Green | Thermo Fisher | cat#: C37608 |
| Alexa Fluor Plus 647 Phalloidin | Thermo Fisher | cat#: A30107 |
| Mouse IgM microbeads | Miltenyi Biotec | cat#: 130–047-301 |
| CD31 microbeads, human | Miltenyi Biotec | cat#: 130–091-935 |
| CD45 microbeads, human | Miltenyi Biotec | cat#: 130–045-801 |
| Critical commercial assays | ||
| Micropatterned dishes | 4DCell | N/A |
| NEB Ultra II DNA Library Prep Kit | New England Biolabs | E7645 |
| Chromium Next GEM Single Cell 3’ Reagent Kits v3 | 10X Genomics | cat#: 1000121 |
| Deposited data | ||
| Single Cell RNA-sequencing | This paper | GEO: GSE202319 |
| Lamin B1 ChIP-seq | This paper | GEO: GSE202319 |
| Codes used in this study | This paper | DOI: 10.5281/zenodo.743 4692 |
| Experimental models: Organisms/strains | ||
| Mouse: SftpcCreERT2 | PMID: 21701069 | N/A |
| Mouse: HopxCreERT2, Hopxtm2.1(cre/ERT2)Joe/J | Jackson Laboratories | cat#: 017606 |
| Mouse: SftpcEGFP, B6N.Cg-Tg(Sftpc,-EGFP)1Dobb/J | Jackson Laboratories | cat#: 028356 |
| Mouse: HopxEGFP, Hopxtm3.1Joe/J | Jackson Laboratories | cat#: 029271 |
| Mouse: Ptk2Flox, B6.129P2(FVB)-Ptk2tm1.1Guan/J | Jackson Laboratories | cat#: 031956 |
| Mouse: Cdc42Flox, Cdc42tm1Yizh/J | Jackson Laboratories | cat#: 027576 |
| Mouse: Rosa26REYFP, B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J | Jackson Laboratories | cat#: 007903 |
| Mouse: Rosa26RmTmG, B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratories | cat#: 007676 |
| Recombinant DNA | ||
| Plasmid: mCherry-DN KASH | PMID: 21652697 | N/A |
| Software and algorithms | ||
| Prism 9.4.1 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Imaris | Oxford Instruments | https://imaris.oxinst.com/packages |
| Huygens | Scientific Volume Imaging | https://svi.nl/Huygens-Software |
| Fiji | NIH | https://fiji.sc |
| R 4.2.0 | R Project | https://www.r-project.org |
| Seurat V4.0.6 | Satija Lab | https://satijalab.org/seurat/ |
| corrplot | CRAN | https://cran.r-project.org/web/packages/corrplot/index.html |
| STARsolo v2.7.9a | PMID: 23104886 | https://github.com/alexdobin/STAR/blob/master/docs/STARsolo.md |
| bwa | PMID: 19451168 | https://bio-bwa.sourceforge.net |
| Picard Tools | Broad Institute | https://broadinstitute.github.io/picard/ |
| SAMtools | PMID: 19505943 | https://samtools.sourceforge.net |
| deepTools | PMID: 27079975 | https://pypi.org/project/deepTools/ |
| EDD | PMID: 24782521 | https://github.com/CollasLab/edd |
| DAVID | PMID: 35325185 | https://david.ncifcrf.gov |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
The normal human samples used in this study were from de-identified non-used lungs donated for organ transplantation following an established protocol (PROPEL, approved by University of Pennsylvania Institutional Review Board) with informed consent in accordance with institutional and NIH procedures. Consent was provided by next of kin or healthcare proxy. The institutional review board of the University of Pennsylvania approved this study, and all patient information was removed before use. This use does not meet the current NIH definition of human subject research, but all institutional procedures required for human subject research were followed throughout the reported experiments. The sample age and sex were: 33 years old male, 55 years old male, and 58 years old female.
Animals
All mouse experiments were performed under the protocols approved by the guidance of the University of Pennsylvania Institutional Animal Care and Use Committee. SftpcCreERT2, HopxCreERT2 (Stock No. 017606), SftpcEGFP (Stock No. 028356), HopxEGFP (Stock No. 029271), Ptk2Flox (Stock No: 031956), Cdc42Flox (Stock No: 027576), Rosa26REYFP (Stock No: 007903), and Rosa26RmTmG (Stock No: 007676) mice have been previously described.5,11,33–35 SftpcCreERT2 was generously provided by Chapman lab, and other strains were obtained from Jackson Laboratories (MA, USA). All experiments were performed on 6–12 week old mice that were maintained on a mixed C57BL/6 and CD1 background. Both male and female mice were used in all conditions. For Cre recombinase induction, tamoxifen (Sigma-Aldrich) was dissolved in corn oil to produce stock with a concentration of 20 mg/mL. For lineage tracing experiments, a single dose of 200 mg/kg tamoxifen was delivered by oral gavage except for HopxCreERT2; Rosa26RmTmG mice that were given 50 mg/kg tamoxifen. For gene knockout experiments, 200 mg/kg tamoxifen was given 3 consecutive days.
Bleomycin and hyperoxic lung injury
Two weeks after tamoxifen induction, 2U/kg Bleomycin (Teva) was intratracheally administered to anesthetized mice. Control animals received PBS intratracheally. A hyperoxic lung injury experiment was performed as previously described.6 Mice were placed in the hyperoxia chamber (BioSpherix) under 95% oxygen for 3 days.
Unilateral lung ligation
Mice were anesthetized with isoflurane and intubated for ventilation using a MiniVent ventilator (Harvard apparatus; tidal volume of 200 μL, respiratory rate of 150 breaths per minute). The thoracic cavity was exposed by incising 2 cm on the left lateral side of the skin, followed by a 1 cm incision at the fifth left intercostal space. The left main bronchus was clipped using a Microclip Applier and a Microclip (Horizon). Left lung collapse was visually confirmed after the clipping.
METHODS DETAILS
Histology
Mice were euthanized by CO2 inhalation, and the lungs were perfused with PBS via the right ventricle. Then, the lungs were inflated with 2% PFA at a constant pressure of 25 cm H2O and were fixed overnight at 4°C. Tissue was then dehydrated, paraffin embedded, and sectioned. Hematoxylin and eosin staining was performed as previously explained.5 For immunohistochemistry analysis, the following antibodies were used on paraffin sections: GFP (chicken, Aves, GFP-1020, 1:200), tdTomato (Goat, Origene, AB8181, 1:100), Sftpc (rabbit, Millipore, AB3786, 1:200), Hopx (mouse, Santa Cruz, sc-398703, 1:100), Ager (rat, R&D, MAB1179, 1:100), Krt8 (rat, DSHB, TROMA-1, 1:200), Phosphorylated Myosin Light Chain (rabbit, Abcam, ab2480, 1:100), FAK-pY397 (rabbit, Thermo, 44–624G, 1:100), Yap (rabbit, Cell Signaling, 4912S, 1:100), Lamp3 (rat, Novus, 1010E1.01, 1:100), Lamin B1 (rabbit, Abcam, ab16048, 1:50), Cleaved Caspase 3 (rabbit, R&D, MAB835, 1:100), Ki67 (mouse, BD Biosceinces, B56, 1:200), α-Tubulin (rat, Santa Cruz, sc-53029, 1:100), and pan-cytokeratin (mouse, Invitrogen, MA5–13156, 1:100). For phalloidin (Invitrogen, A30107, 1:100) staining of actin filaments, lungs were fixed with 2% PFA for 6 h at 4°C, incubated in 30% sucrose/PBS overnight, and embed in OCT Compound (Sakura FineTek) for preparation of frozen blocks and sections. Lung strip was prepared following protocol described in Yang et al.36
Flow cytometry and cell sorting
Flow cytometry and cell sorting experiments were performed by using cell analyzer CytoFLEX LX (Beckman Coulter) and cell sorter MoFlo Astrios (Beckman Coulter). Lungs were harvested and digested into single cell suspensions using collagenase I, dispase, and DNase, as previously explained.6,37 Red blood cells were removed with the ACK buffer and then stained with antibodies. The following antibodies were used for flow cytometry and cell sorting: EpCAM-PE-Cy7 (eBioscience, G8.8, 1:200), CD31-APC (eBioscience, 390, 1:200), and CD45-APC (eBioscience, 30-F11, 1:200). DAPI was used to gate out dead cells.
Cell culture and immunocytochemistry
Mouse AT2 cells were sorted from the single cell suspension of SftpcEGFP mice lungs and plated in 4-well culture slide (Falcon) or micropatterned glass bottom dishes with MTEC/SAGM medium,4,32 containing SABM base media (Lonza), insulin, transferrin, bovine pituitary extract, gentamycin, retinoic acid, 0.1 μg/mL cholera toxin (Millipore), 25 ng/mL EGF (PeproTech), and 5% FBS. Human AT2 cells were obtained as previously explained,5 and were cultured on the micropatterned dishes with MTEC/SAGM medium supplemented with Antibiotic-Antimycotic (Thermo). Briefly, single-cell suspensions of human lung whole cells were incubated with human CD31 and CD45 microbeads (Miltenyi). After magnetic negative selection of CD31− CD45− population, cells were MACS purified using the mouse anti-HT2-280 IgM antibodies (Terrace, TB-27AHT2-280, 1:50) and anti-mouse IgM microbeads (Miltenyi, 1:20) using MACS LS Columns. Micropatterned dishes containing printed circles ranging in size from 10 μm to 100 μm or elongated shape 10 × 100 μm were custom-made by 4Dcell (Montreuil, France). Fibronectin (Sigma, 50 μg/mL) coating of the micropatterned dishes was performed according to the manufacturer’s protocol. Mouse AT2 cells were plated in micropatterned dishes at a density of 100,000 cells per dish in 0.4 mL medium. Human AT2 cells were plated at a density of 50,000–500,000 cells per dish in 0.4 mL medium. Then, 48 h after plating, the cells were fixed with 4% PFA for 15 min at RT.
For treatment with inhibitors, mouse AT2 cells were plated in fibronectin-coated (10 μg/mL, overnight) 4-well culture slide at a density of 100,000 cells per well in 0.5 mL medium. At 24 h after plating, the medium was replaced with fresh medium containing anti-Itgb6 antibody (Sigma, 10D5, 1:100), anti-Itgb5 antibody (Invitrogen, KN52, 1:100), anti-Itga6 antibody (Invitrogen, GoH3, 1:500), PF 573228 (Tocris, 10 μM), or Latrunculin B (Sigma, 0.5 μM). After 24 h incubation with the inhibitors, cells were fixed with 4% PFA for 15 min at RT. For immunocytochemistry analysis, fixed cells were permeabilized with 0.5% Triton X-100 for 10 min at RT. Actin was stained with Phalloidin (1:400), and cytoplasm was stained with CellMask (Thermo, 1:1,000). The following antibodies were used: Yap/Taz (rabbit, Cell Signaling, D24E4, 1:100), Phosphorylated Myosin Light Chain, α-Tubulin, Sftpc (for mouse, AB3786, 1:100), SFTPC (for human, rabbit, ab90716, 1:100), Ager, and PDPN (rat, Thermo, NZ-1.3, 1:100). For cell viability evaluation, the medium was replaced with HBSS containing 2 μM Calcein (Thermo) and 4 μM EthD-1 (Thermo) for 30 min at RT. Plasmid containing mCherry-DN KASH was generously provided by the Lammerding Lab.30 Lentivirus containing mcherry-DN KASH or mCherry was generated as previously explained.38 Sorted AT2 cells were centrifuged at 600g at 32°C for 60 min with lentivirus before plating.
DNA oligo FISH
DNA oligo FISH protocol was adopted from previous studies39 with modifications. In brief, after immunohistochemistry staining, sections were post-fixed with 4% PFA for 30 min at RT. Sections were then permeabilized with 0.7% Triton X-100 for 10 min at RT, incubated in 2X SSCT (2X SSC with 0.1% Tween) for 5 min at RT. Then sections were incubated in 2X SSCT with 50% Formamide for 5 min RT, 2.5 min 92°C, and 20 min 60°C. Hybridization was performed with ~50–100 pmol DNA primary oligo probes for 16 h 37°C. Sections were washed in 2X SSCT for 15 min at 60°C, 2X SSCT for 10min at RT, and 2X SSC for 10 min. Sections were then hybridized with secondary fluorescent DNA oligo probes for 2 h RT. Sections were washed in 2X SSCT for 5 min 60°C, 2X SSCT 5 min RT, and 2X SSC 5 min RT. DNA oligo libraries, with each oligo containing 42 nucleotides homologous to the region of interest in the mouse mm10 genome, were designed aiming for more than 5 probes/kb for 100 kb region using the OligoMiner.40 Single stranded probes were produced as previously described.41
Image analysis
All fluorescence images were acquired by a Leica TCS SP8 confocal microscope and processed with ImageJ or Imaris (Oxford Instruments). For perinuclear actin or myosin quantification, nuclear object demarcated by DAPI fluorescence was dilated by 0.17 μm. The perinuclear actin or myosin was quantified by subtracting fluorescence of the non-dilated nucleus from the dilated nucleus. For nuclear morphology analysis, z-stack images obtained with a 63x lens were processed by Imaris with default smoothening and thresholding. A larger value of prolate or oblate ellipticity was used as a representative value. Yap/Taz nuclear/cytoplasmic ratio was calculated by analyzing nuclear (DAPI fluorescence) and cytoplasmic (CellMask fluorescence) Yap/Taz intensity. Cell counting for lineage-tracing experiments was performed on at least five z-stack images with a 40x lens for each mouse, except for cCaspase 3+ cell counting, which was performed with a 20x lens. For FISH dots to nuclear lamina distance quantification and in vivo F-actin visualization, z-stack images were deconvoluted using Huygens software (Scientific Volume Imaging) and the distance was measured using Imaris as previously explained42. Representative 3D images were also generated using Imaris. Mean linear intercept was analyzed with MATLAB software on images acquired using EVOS FL Auto2 Imaging System as previously described.32
ChIP-seq
AT1 and AT2 cells were sorted by gating on DAPI− CD31/45− EpCAM+ EGFP+ poupulations from HopxEGFP (AT1) or SftpcEGFP (AT2) mice and were crosslinked by 1% formaldehyde (Thermo) for 10 min RT. Glycine (final 125 mM) was then added and incubated for 5 min RT. The cell pellets were flash frozen and stored at −80°C until ChIP and library preparation. ChIP, library preparation, and sequencing were performed as previously explained.26 Briefly, crude nuclei were isolated from the cell pellets and were sonicated with a Covaris S220 sonicator. Anti-Lamin B1 (Abcam, ab16048) bound to protein G magnetic beads (Thermo) were added to 500 μg input protein, and the mixture was incubated overnight at 4°C. Libraries were prepared using NEBNext Ultra II DNA library prep kit (NEB) and were sequenced on the Illumina NextSeq 500 instrument. Raw reads were aligned to mm10 genome using bwa, with parameters ‘-q 5 -l 32 -k 2’. Duplicates were removed using Picard tools, and all samples were merged and downsampled to 43 million (AT1) or 48.8 million (AT2) uniquely mapped reads using samtools. Input normalized bigwig coverage tracks for AT1 and AT2 cells were created using deeptools. To identify LAD-genes, the number of reads that mapped to the middle of the gene body ± 50kb were calculated for each gene by using GenomicRanges.43 For pathway enrichment analysis, genes were considered LAD-genes when the log2 fold change of the signal to input ratio was greater than 0.1. Genes with fewer than 100 reads were excluded from the analysis. LAD peaks were called using EDD.44 Olfr genes and Vmn1r/2r genes were excluded from the pathway enrichment analysis due to the species-specific nature of the mouse olfactory system. Gene ontology enrichment analysis was performed using DAVID.
Single cell RNA sequencing (scRNA-seq)
Lineage-traced cells were sorted by gating on DAPI− CD31/45-APC− EpCAM-PECy7+ EYFP+ populations. Cells were loaded onto the 10x Chromium (10X Genomics) aiming for 10,000 cells, and libraries were prepared according to the manufacturer’s protocol using Chromium Single Cell 3’ v3 chemistry. The libraries were sequenced on an Illumina Novaseq 6000 instrument. The data was processed by aligning the reads and obtaining unique molecular identifiers (UMIs) using STARsolo (v2.7.9a).45 The data was further processed and analyzed using the Seurat (V4.0.6; https://satijalab.org/seurat/) package. Cells with less than 1000 UMIs were removed. Also, cells with potential stress signals were removed if the percent mitochondrial reads were greater than 5%. Feature (gene) data was scaled in order to remove unwanted sources of variation using the Seurat SCTransform function based on percent mitochondrial reads, and the number of genes and reads. Libraries were integrated using the CCA method as implemented in Seurat. Non-linear dimension reduction was performed using uniform manifold projection (UMAP) and graph-based clustering was performed using the Louvain. Correlation analysis was performed in R and correlation matrix plots constructed using the corrplot R package.
Real-time qPCR analysis
FACS-isolated cells were pelleted, and RNA was extracted with Trizol Reagent (Invitrogen) according to the manufacturer’s protocol. SuperScript IV First-Strand synthesis system (Invitrogen) was used to generate cDNA. Real-time qPCR was performed with Power SYBR Green Master Mix (Thermo) and QuantStudio 7 PCR System (Applied Biosystems).
Immunoblotting
Cell lysates were separated by SDS-PAGE, transferred to PVDF membrane as previously described,6 and imaged using Odyssey Fc (LI-COR Biosciences) with IRDye 700 or 800-conjugated secondary antibodies (1:7,500) or ECL 2 Western Blotting Substrate (Thermo). The following primary antibodies were used: Cdc42 (mouse, ab41429, Abcam, 1:200), FAK (rabbit, D2R2E, Cell Signaling, 1:2,000), mature Sftpc (rabbit, WRAB-76694, 1:3,000, Seven Hills), β-Actin (rabbit, 13E5, Cell Signaling, 1:2,000; mouse, 8H10D10, Cell Signaling, 1:2,000; mouse, HRP-60008, 1:2,500, Protein Tech).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
All results are shown as mean ± s.d. A two-tailed t-test was used to compare two groups, and a one-way ANOVA was used to compare multiple groups. The difference was considered significant when p < 0.05. Statistical analysis was performed using Graphpad Prism 9.
Highlights
Mechanotransduction is a key feature of alveolar type 1 (AT1) cells in the lung
Actin remodeling and integrin signaling regulate AT1 and AT2 cell fate transitions
Physical blockade of respiratory movement leads to loss of AT1 cell identity
Nuclear lamina-chromatin interactions determine alveolar epithelial cell fate
Supplementary Material
Figure S1. Additional data on characteristics of AT1 and AT2 cells, related to Figure 1
a. The lungs of HopxEGFP and SftpcEGFP mice were stained for EGFP, Hopx, and Sftpc. EGFP+ Hopx+ AT1 cells from HopxEGFP mice are not Sftpc+ (left). EGFP+ Sftpc+ AT2 cells from SftpcEGFP mice are not Hopx+ (right).
b. Yap localizes to Hopx+ AT1 nucleus, but not to Lamp3+ AT2 cells. Arrowheads indicate AT1 cells.
c. DotPlot showing expression of integrins in AT1 and AT2 cells.
d. Lipid metabolism-related GO terms and pathways are enriched in AT2 cells.
e. Microtubules (α-Tubulin) and intermediate filaments (pan-cytokeratin) staining of AT1 and AT2 cells.
f. Representative 2D pictures from lung strips of HopxCreERT2; R26RmTmG mice, which were used to generate 3D images of AT1 cell body.
Scale bars: a, b, f, 10 μm; e, 2.5 μm.
Figure S2. Additional evaluation of Cdc42AT1-KO mice, related to Figure 2
a. A representative FACS gating strategy used in this study to isolate Hopx+ AT1 and Sftpc+ AT2 cells from SftpcEGFP, SftpcCreERT2; R26REYFP, HopxEGFP, and HopxCreERT2; R26REYFP mice. The EGFP channel voltage is lower for SftpcEGFP mice.
b-c. Gene knockout efficiency in Cdc42AT1-KO mice. qPCR of Cdc42 using sorted AT1 cells from control (n = 5 mice) and Cdc42AT1-KO (n = 5 mice) mice from two independent experiments (b). Immunoblot of Cdc42 using sorted AT1 cells from control and Cdc42AT1-KO mice from two independent experiments (c).
d. AT1 lineage cells express AT2 cell marker Sftpc in Cdc42AT1-KO mice four days after tamoxifen injection. Arrowheads indicate AT2rep cells.
e. H&E staining showed no gross changes in alveolar complexity between control and Cdc42AT1-KO mice.
f. Mean linear intercept (MLI) quantification for control (n = 5 mice) and Cdc42AT1-KO (n = 5 mice) mice 14 or 28 days after tamoxifen injection.
g. AT1 and AT2 cell numbers and AT1/AT2 cell ratio do not change in Cdc42AT1-KO mice (n = 5 mice).
*** P < 0.001 by two-tailed t-test, n.s. not significant. Each dot represents an individual mouse. Error bars indicate mean with s.d. Scale bars: d, 25 μm; e, 100 μm.
Figure S3. scRNA-seq and functional analysis of Cdc42AT1-KO mice, related to Figure 2
a. FACS gating strategy used to isolate EYFP+ lineage-traced cells from Cdc42AT1-KO mice or unilateral lung ligation model (Figure 7) for scRNA-seq.
b. Expression of Sftpc, Sftpb, Ager, and Hopx using UMAP plot from integrated data of control and Cdc42AT1-KO mice.
c. DotPlot showing AT1 and AT2-specific genes.
d. Immunoblot of mature Sftpc using EYFP+ lineage-traced cells from Cdc42AT1-KO mice, wildtype AT1 cells, and wildtype AT2 cells. For each cell type, n = 2 mice.
e. Low magnification pictures of YAP staining in Cdc42AT1-KO mice. Scale bar: 25 μm.
Figure S4. Additional data for primary AT2 cell culture experiment, related to Figure 3
a. Primary AT2 cells autonomously spread from day 1 to day 2 after plating.
b. Anti-Ager antibody did not label mouse Sftpc+ AT2 cells of the mouse lung section.
Arrowheads indicate AT2 cells.
c. Immunostaining of AT2 cells for α-Tubulin and Krt8.
d. Immunostaining of AT2 cells for F-actin and activated myosin.
e. Constrained AT2 cells compared to AT2 cells cultured on elongated micropattern.
Constrained mouse AT2 cells maintain Sftpc expression, while elongated mouse AT2 cells downregulate Sftpc and express AT1 cell marker Ager.
f. Ager/Sftpc ratio was quantified with n = 27 constrained, n = 23 unconstrained, and n = 29 elongated AT2 cells pooled from 2 independent experiments.
*** P < 0.001 by one-way ANOVA, n.s. not significant. For Box and whisker plots, bars represent min and max values. Scale bars: a, b (left), 25 μm; e, 10 μm; b (right) c, d, 2.5 μm.
Figure S5. Additional data for human AT2 cell culture and evaluation of Integrin-FAK signaling, related to Figures 3 and 4
a. A representative cytospin staining of MACS-purified human AT2 cells.
b. Anti-PDPN antibody did not label human SFTPC+ AT2 cells of the human lung section.
Arrowheads indicate AT2 cells.
c. The average PDPN/SFTPC ratio in each human AT2 experiment. n = 3 human samples and experiments.
d-f. YAP/TAZ localize to unconstrained human AT2 nucleus, and actin fibers are more abundant in unconstrained human AT2 cells (d). YAP/TAZ localization was quantified with n = 101 constrained and n = 102 unconstrained human AT2 cells pooled from 3 independent experiments (e left). The average YAP/TAZ nuclear to cytoplasm ratio in each human AT2 experiment from n = 3 human samples and experiments (e right). Cytoplasmic actin intensity was quantified with n = 42 constrained and n = 52 unconstrained human AT2 cells (f).
g-h. Cell viability assay after inhibitor treatment of AT2 cells. The majority of AT2 cells were Calcein+ and viable 24 h after adding anti-Integrin antibodies, PF 573228, or Latrunculin B. Ethidium homodimer-1 (EthD-1) marked few non-viable cells (g). Quantification of viable cells from n = 3–6 individual experiments (h).
i-j. Gene knockout efficiency in Ptk2AT2-KO mice. qPCR of Ptk2 using the sorted AT2 cells from control (n = 5 mice) and Ptk2AT2-KO (n = 7 mice) mice from three independent experiments (i). Immunoblot of FAK (Ptk2) using the sorted AT2 cells from control and Ptk2AT2-KO mice from two independent experiments (j).
k-l. FAK deletion did not affect AT2 cell death. AT2-lineaged cells were stained for cCaspase 3 seven days after BLM injury (k). Quantification of cCaspase 3 positivity in control (n = 7) and Ptk2AT2-KO (n = 6) mice from two independent experiments (l).
m-n. FAK deletion did not affect AT2 cell proliferation. AT2-lineaged cells were stained for Ki67 seven days after BLM injury (m). Quantification of Ki67 positivity in control (n = 7) and Ptk2AT2-KO (n = 6) mice from two independent experiments (n).
*** P < 0.001, n.s. not significant. A two-tailed t-test was used for e, f, i, l, and n. A one-way ANOVA was used for h. Each dot represents an individual mouse (i, l, n) or an experiment (c, e right, h). Error bars indicate mean with s.d. Scale bars: g, 50 μm; a, b (left), k, m, 25 μm; b (right), d, 2.5 μm.
Figure S6. LAD gene expression and additional oligo-FISH analysis, related to Figure 5
a-b. Expression of LAD genes and non-LAD genes in AT1 (a) and AT2 (b) cells using the RNA-seq data.6 Bars represent min and max values.
c. Representative 3D reconstruction showing Wasl locus in relation to nuclear lamina.
d. Lamin B1 ChIP-seq tracks in AT1 and AT2 cells for Rock1 and oligo-FISH of Rock1 loci in lung sections from HopxEGFP AT1 and SftpcEGFP AT2 reporter mice. Arrowheads indicate each locus.
e. Lamin B1 ChIP-seq tracks and oligo-FISH for Ptk2 loci in AT1 and AT2 cells.
Arrowheads indicate each locus.
f. Lamin B1 ChIP-seq tracks and oligo-FISH for Cav1/ Cav2 loci in AT1 and AT2 cells.
Arrowheads indicate each locus.
g. 3D Quantification of the distance between Rock1 (n = 65 AT1 and n = 53 AT2 cells), Ptk2 (n = 76 AT1 and n = 47 AT2 cells), and Cav1/Cav2 (n = 40 AT1 and n = 62 AT2 cells) loci and Lamin B1. Each dot represents an individual cell, and error bars indicate mean with s.d.
*** P < 0.001 by two-tailed t-test. Scale bars: c, 1 μm; d, e, f, 2.5 μm.
Figure S7 Additional evaluation for ligation model, related to Figure 7
a, b. Apoptotic (cCaspase 3+) cells were not apparent in the ligation model (a). Quantification of cCaspase 3 positivity in control and ligated lung from n = 5 mice (b). Each dot represents an individual mouse. Error bars indicate mean with s.d. Statistical analysis was performed by one-way ANOVA, n.s. not significant.
c. Violin plots showing expression of Hif1a target genes.
d. Endomucin staining of the ligated lung.
e. AT1 lineage cells express AT2 cell marker Sftpc in ligated lung seven days after ligation. Arrowheads indicate AT2rep cells.
f. Expression of Lamp3, Sftpb, Ager, and Hopx using UMAP plot from integrated data of control and unilateral lung ligation model.
g. DotPlot showing AT1 and AT2-specific genes.
h. YAP does not localize to the EYFP+ Lamp3+ AT2rep nucleus after ligation.
i, j. Yap (i) and F-actin (j) staining after hyperventilation.
Scale bars: 25 μm.
Acknowledgements
We thank E. Joyce, S. Nguyen, D. Liberti, K. Shen, K. McDaid, T. Yamamoto, T. Sakamoto, B. Kim, the Flow Cytometry Core Laboratory of Children’s Hospital of Philadelphia, and CDB Microscopy Core at the University of Pennsylvania for technical assistance. We also thank Dr. Jan Lammerding for providing DN-KASH plasmids. This research was supported by the National Institutes of Health (HL152194, HL132999, HL134745, HL148857 to E.E.M.; DP2HL147123, HL148857 to R.J.; HL007586 to M.C.B., S.M.L., and J.D.P.; HL155821 to E.C.), Longfonds BREATH Consortium, Burroughs Wellcome Foundation (R.J.), the JSPS overseas research fellowship (K.S.), the fellowship of Uehara Memorial Foundation (K.S.), and the fellowship of Astellas Foundation for Research on Metabolic Disorders (K.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Authors declare no competing interests.
References
- 1.Tschumperlin DJ, Ligresti G, Hilscher MB, and Shah VH (2018). Mechanosensing and fibrosis. J Clin Invest 128, 74–84. 10.1172/JCI93561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haak AJ, Kostallari E, Sicard D, Ligresti G, Choi KM, Caporarello N, Jones DL, Tan Q, Meridew J, Diaz Espinosa AM, et al. (2019). Selective YAP/TAZ inhibition in fibroblasts via dopamine receptor D1 agonism reverses fibrosis. Sci Transl Med 11. 10.1126/scitranslmed.aau6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu F, Lagares D, Choi KM, Stopfer L, Marinkovic A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, et al. (2015). Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol 308, L344–357. 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BL (2013). Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036. 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penkala IJ, Liberti DC, Pankin J, Sivakumar A, Kremp MM, Jayachandran S, Katzen J, Leach JP, Windmueller R, Stolz K, et al. (2021). Age-dependent alveolar epithelial plasticity orchestrates lung homeostasis and regeneration. Cell Stem Cell 28, 1775–1789 e1775. 10.1016/j.stem.2021.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183. 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 8.Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, et al. (2017). Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171, 1397–1410 e1314. 10.1016/j.cell.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, and Wirtz D (2015). Cytoskeletal tension induces the polarized architecture of the nucleus. Biomaterials 48, 161–172. 10.1016/j.biomaterials.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazumder A, and Shivashankar GV (2010). Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development. J R Soc Interface 7 Suppl 3, S321–330. 10.1098/rsif.2010.0039.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zepp JA, Morley MP, Loebel C, Kremp MM, Chaudhry FN, Basil MC, Leach JP, Liberti DC, Niethamer TK, Ying Y, et al. (2021). Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science 371. 10.1126/science.abc3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romani P, Brian I, Santinon G, Pocaterra A, Audano M, Pedretti S, Mathieu S, Forcato M, Bicciato S, Manneville JB, et al. (2019). Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat Cell Biol 21, 338–347. 10.1038/s41556-018-0270-5. [DOI] [PubMed] [Google Scholar]
- 13.Hodge RG, and Ridley AJ (2016). Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol 17, 496–510. 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 14.Beers MF, and Lomax C (1995). Synthesis and processing of hydrophobic surfactant protein C by isolated rat type II cells. Am J Physiol 269, L744–753. 10.1152/ajplung.1995.269.6.L744. [DOI] [PubMed] [Google Scholar]
- 15.Geiger B, Spatz JP, and Bershadsky AD (2009). Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol 10, 21–33. 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 16.Choi J, Park JE, Tsagkogeorga G, Yanagita M, Koo BK, Han N, and Lee JH (2020). Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell 27, 366–382 e367. 10.1016/j.stem.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi Y, Tata A, Konkimalla A, Katsura H, Lee RF, Ou J, Banovich NE, Kropski JA, and Tata PR (2020). Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat Cell Biol 22, 934–946. 10.1038/s41556-020-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, Tsidiridis G, Lange M, Mattner LF, Yee M, et al. (2020). Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun 11, 3559. 10.1038/s41467-020-17358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra SK, Hanson DA, and Schlaepfer DD (2005). Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol 6, 56–68. 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 20.Le HQ, Ghatak S, Yeung CY, Tellkamp F, Gunschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, and Wickstrom SA (2016). Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol 18, 864–875. 10.1038/ncb3387. [DOI] [PubMed] [Google Scholar]
- 21.Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, and Wang N (2016). Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 15, 1287–1296. 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczesny SE, and Mauck RL (2017). The Nuclear Option: Evidence Implicating the Cell Nucleus in Mechanotransduction. J Biomech Eng 139. 10.1115/1.4035350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, and van Steensel B (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951. 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 24.Keough KC, Shah PP, Gjoni K, Santini GT, Wickramasinghe NM, Dundes CE, Karnay A, Chen A, Salomon REA, Walsh PJ, et al. (2021). An atlas of lamina-associated chromatin across twelve human cell types reveals an intermediate chromatin subtype. bioRxiv [DOI] [PMC free article] [PubMed]
- 25.Kirby TJ, and Lammerding J (2018). Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol 20, 373–381. 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah PP, Lv W, Rhoades JH, Poleshko A, Abbey D, Caporizzo MA, Linares-Saldana R, Heffler JG, Sayed N, Thomas D, et al. (2021). Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes. Cell Stem Cell 28, 938–954 e939. 10.1016/j.stem.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38, 603–613. 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takenawa T, and Suetsugu S (2007). The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 8, 37–48. 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 29.Julian L, and Olson MF (2014). Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases 5, e29846. 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, and Lammerding J (2011). The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem 286, 26743–26753. 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H, Yu Y, Huang H, Hu Y, Fu S, Wang Z, Shi M, Zhao X, Yuan J, Li J, et al. (2020). Progressive Pulmonary Fibrosis Is Caused by Elevated Mechanical Tension on Alveolar Stem Cells. Cell 180, 107–121 e117. 10.1016/j.cell.2019.11.027. [DOI] [PubMed] [Google Scholar]
- 32.Liberti DC, Kremp MM, Liberti WA 3rd, Penkala IJ, Li S, Zhou S, and Morrisey EE (2021). Alveolar epithelial cell fate is maintained in a spatially restricted manner to promote lung regeneration after acute injury. Cell Rep 35, 109092. 10.1016/j.celrep.2021.109092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, and Vu TH (2011). Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 121, 2855–2862. 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Liao G, Yang L, Campbell K, Nakafuku M, Kuan CY, and Zheng Y (2006). Cdc42 deficiency causes Sonic hedgehog-independent holoprosencephaly. Proc Natl Acad Sci U S A 103, 16520–16525. 10.1073/pnas.0603533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen TL, Park AY, Alcaraz A, Peng X, Jang I, Koni P, Flavell RA, Gu H, and Guan JL (2005). Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J Cell Biol 169, 941–952. 10.1083/jcb.200411155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Hernandez BJ, Martinez Alanis D, Narvaez del Pilar O, Vila-Ellis L, Akiyama H, Evans SE, Ostrin EJ, and Chen J (2016). The development and plasticity of alveolar type 1 cells. Development 143, 54–65. 10.1242/dev.130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, and Morrisey EE (2017). Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 170, 1134–1148 e1110. 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardenas-Diaz FL, Osorio-Quintero C, Diaz-Miranda MA, Kishore S, Leavens K, Jobaliya C, Stanescu D, Ortiz-Gonzalez X, Yoon C, Chen CS, et al. (2019). Modeling Monogenic Diabetes using Human ESCs Reveals Developmental and Metabolic Deficiencies Caused by Mutations in HNF1A. Cell Stem Cell 25, 273–289 e275. 10.1016/j.stem.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poleshko A, Smith CL, Nguyen SC, Sivaramakrishnan P, Wong KG, Murray JI, Lakadamyali M, Joyce EF, Jain R, and Epstein JA (2019). H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis. Elife 8. 10.7554/eLife.49278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beliveau BJ, Kishi JY, Nir G, Sasaki HM, Saka SK, Nguyen SC, Wu CT, and Yin P (2018). OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc Natl Acad Sci U S A 115, E2183–E2192. 10.1073/pnas.1714530115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosin LF, Nguyen SC, and Joyce EF (2018). Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLoS Genet 14, e1007393. 10.1371/journal.pgen.1007393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poleshko A, Shah PP, Gupta M, Babu A, Morley MP, Manderfield LJ, Ifkovits JL, Calderon D, Aghajanian H, Sierra-Pagan JE, et al. (2017). Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction. Cell 171, 573–587 e514. 10.1016/j.cell.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118. 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lund E, Oldenburg AR, and Collas P (2014). Enriched domain detector: a program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res 42, e92. 10.1093/nar/gku324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Additional data on characteristics of AT1 and AT2 cells, related to Figure 1
a. The lungs of HopxEGFP and SftpcEGFP mice were stained for EGFP, Hopx, and Sftpc. EGFP+ Hopx+ AT1 cells from HopxEGFP mice are not Sftpc+ (left). EGFP+ Sftpc+ AT2 cells from SftpcEGFP mice are not Hopx+ (right).
b. Yap localizes to Hopx+ AT1 nucleus, but not to Lamp3+ AT2 cells. Arrowheads indicate AT1 cells.
c. DotPlot showing expression of integrins in AT1 and AT2 cells.
d. Lipid metabolism-related GO terms and pathways are enriched in AT2 cells.
e. Microtubules (α-Tubulin) and intermediate filaments (pan-cytokeratin) staining of AT1 and AT2 cells.
f. Representative 2D pictures from lung strips of HopxCreERT2; R26RmTmG mice, which were used to generate 3D images of AT1 cell body.
Scale bars: a, b, f, 10 μm; e, 2.5 μm.
Figure S2. Additional evaluation of Cdc42AT1-KO mice, related to Figure 2
a. A representative FACS gating strategy used in this study to isolate Hopx+ AT1 and Sftpc+ AT2 cells from SftpcEGFP, SftpcCreERT2; R26REYFP, HopxEGFP, and HopxCreERT2; R26REYFP mice. The EGFP channel voltage is lower for SftpcEGFP mice.
b-c. Gene knockout efficiency in Cdc42AT1-KO mice. qPCR of Cdc42 using sorted AT1 cells from control (n = 5 mice) and Cdc42AT1-KO (n = 5 mice) mice from two independent experiments (b). Immunoblot of Cdc42 using sorted AT1 cells from control and Cdc42AT1-KO mice from two independent experiments (c).
d. AT1 lineage cells express AT2 cell marker Sftpc in Cdc42AT1-KO mice four days after tamoxifen injection. Arrowheads indicate AT2rep cells.
e. H&E staining showed no gross changes in alveolar complexity between control and Cdc42AT1-KO mice.
f. Mean linear intercept (MLI) quantification for control (n = 5 mice) and Cdc42AT1-KO (n = 5 mice) mice 14 or 28 days after tamoxifen injection.
g. AT1 and AT2 cell numbers and AT1/AT2 cell ratio do not change in Cdc42AT1-KO mice (n = 5 mice).
*** P < 0.001 by two-tailed t-test, n.s. not significant. Each dot represents an individual mouse. Error bars indicate mean with s.d. Scale bars: d, 25 μm; e, 100 μm.
Figure S3. scRNA-seq and functional analysis of Cdc42AT1-KO mice, related to Figure 2
a. FACS gating strategy used to isolate EYFP+ lineage-traced cells from Cdc42AT1-KO mice or unilateral lung ligation model (Figure 7) for scRNA-seq.
b. Expression of Sftpc, Sftpb, Ager, and Hopx using UMAP plot from integrated data of control and Cdc42AT1-KO mice.
c. DotPlot showing AT1 and AT2-specific genes.
d. Immunoblot of mature Sftpc using EYFP+ lineage-traced cells from Cdc42AT1-KO mice, wildtype AT1 cells, and wildtype AT2 cells. For each cell type, n = 2 mice.
e. Low magnification pictures of YAP staining in Cdc42AT1-KO mice. Scale bar: 25 μm.
Figure S4. Additional data for primary AT2 cell culture experiment, related to Figure 3
a. Primary AT2 cells autonomously spread from day 1 to day 2 after plating.
b. Anti-Ager antibody did not label mouse Sftpc+ AT2 cells of the mouse lung section.
Arrowheads indicate AT2 cells.
c. Immunostaining of AT2 cells for α-Tubulin and Krt8.
d. Immunostaining of AT2 cells for F-actin and activated myosin.
e. Constrained AT2 cells compared to AT2 cells cultured on elongated micropattern.
Constrained mouse AT2 cells maintain Sftpc expression, while elongated mouse AT2 cells downregulate Sftpc and express AT1 cell marker Ager.
f. Ager/Sftpc ratio was quantified with n = 27 constrained, n = 23 unconstrained, and n = 29 elongated AT2 cells pooled from 2 independent experiments.
*** P < 0.001 by one-way ANOVA, n.s. not significant. For Box and whisker plots, bars represent min and max values. Scale bars: a, b (left), 25 μm; e, 10 μm; b (right) c, d, 2.5 μm.
Figure S5. Additional data for human AT2 cell culture and evaluation of Integrin-FAK signaling, related to Figures 3 and 4
a. A representative cytospin staining of MACS-purified human AT2 cells.
b. Anti-PDPN antibody did not label human SFTPC+ AT2 cells of the human lung section.
Arrowheads indicate AT2 cells.
c. The average PDPN/SFTPC ratio in each human AT2 experiment. n = 3 human samples and experiments.
d-f. YAP/TAZ localize to unconstrained human AT2 nucleus, and actin fibers are more abundant in unconstrained human AT2 cells (d). YAP/TAZ localization was quantified with n = 101 constrained and n = 102 unconstrained human AT2 cells pooled from 3 independent experiments (e left). The average YAP/TAZ nuclear to cytoplasm ratio in each human AT2 experiment from n = 3 human samples and experiments (e right). Cytoplasmic actin intensity was quantified with n = 42 constrained and n = 52 unconstrained human AT2 cells (f).
g-h. Cell viability assay after inhibitor treatment of AT2 cells. The majority of AT2 cells were Calcein+ and viable 24 h after adding anti-Integrin antibodies, PF 573228, or Latrunculin B. Ethidium homodimer-1 (EthD-1) marked few non-viable cells (g). Quantification of viable cells from n = 3–6 individual experiments (h).
i-j. Gene knockout efficiency in Ptk2AT2-KO mice. qPCR of Ptk2 using the sorted AT2 cells from control (n = 5 mice) and Ptk2AT2-KO (n = 7 mice) mice from three independent experiments (i). Immunoblot of FAK (Ptk2) using the sorted AT2 cells from control and Ptk2AT2-KO mice from two independent experiments (j).
k-l. FAK deletion did not affect AT2 cell death. AT2-lineaged cells were stained for cCaspase 3 seven days after BLM injury (k). Quantification of cCaspase 3 positivity in control (n = 7) and Ptk2AT2-KO (n = 6) mice from two independent experiments (l).
m-n. FAK deletion did not affect AT2 cell proliferation. AT2-lineaged cells were stained for Ki67 seven days after BLM injury (m). Quantification of Ki67 positivity in control (n = 7) and Ptk2AT2-KO (n = 6) mice from two independent experiments (n).
*** P < 0.001, n.s. not significant. A two-tailed t-test was used for e, f, i, l, and n. A one-way ANOVA was used for h. Each dot represents an individual mouse (i, l, n) or an experiment (c, e right, h). Error bars indicate mean with s.d. Scale bars: g, 50 μm; a, b (left), k, m, 25 μm; b (right), d, 2.5 μm.
Figure S6. LAD gene expression and additional oligo-FISH analysis, related to Figure 5
a-b. Expression of LAD genes and non-LAD genes in AT1 (a) and AT2 (b) cells using the RNA-seq data.6 Bars represent min and max values.
c. Representative 3D reconstruction showing Wasl locus in relation to nuclear lamina.
d. Lamin B1 ChIP-seq tracks in AT1 and AT2 cells for Rock1 and oligo-FISH of Rock1 loci in lung sections from HopxEGFP AT1 and SftpcEGFP AT2 reporter mice. Arrowheads indicate each locus.
e. Lamin B1 ChIP-seq tracks and oligo-FISH for Ptk2 loci in AT1 and AT2 cells.
Arrowheads indicate each locus.
f. Lamin B1 ChIP-seq tracks and oligo-FISH for Cav1/ Cav2 loci in AT1 and AT2 cells.
Arrowheads indicate each locus.
g. 3D Quantification of the distance between Rock1 (n = 65 AT1 and n = 53 AT2 cells), Ptk2 (n = 76 AT1 and n = 47 AT2 cells), and Cav1/Cav2 (n = 40 AT1 and n = 62 AT2 cells) loci and Lamin B1. Each dot represents an individual cell, and error bars indicate mean with s.d.
*** P < 0.001 by two-tailed t-test. Scale bars: c, 1 μm; d, e, f, 2.5 μm.
Figure S7 Additional evaluation for ligation model, related to Figure 7
a, b. Apoptotic (cCaspase 3+) cells were not apparent in the ligation model (a). Quantification of cCaspase 3 positivity in control and ligated lung from n = 5 mice (b). Each dot represents an individual mouse. Error bars indicate mean with s.d. Statistical analysis was performed by one-way ANOVA, n.s. not significant.
c. Violin plots showing expression of Hif1a target genes.
d. Endomucin staining of the ligated lung.
e. AT1 lineage cells express AT2 cell marker Sftpc in ligated lung seven days after ligation. Arrowheads indicate AT2rep cells.
f. Expression of Lamp3, Sftpb, Ager, and Hopx using UMAP plot from integrated data of control and unilateral lung ligation model.
g. DotPlot showing AT1 and AT2-specific genes.
h. YAP does not localize to the EYFP+ Lamp3+ AT2rep nucleus after ligation.
i, j. Yap (i) and F-actin (j) staining after hyperventilation.
Scale bars: 25 μm.
Data Availability Statement
ChIP-seq and scRNA-seq data generated during this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. RNA-seq data for AT1 and AT2 cells6 and scRNA-seq data for adult mouse lungs11 have been previously described and are publicly available (GSE148487 for RNA-seq and GSE149563 for scRNA-seq).
All original code has been deposited at Github (https://github.com/Morriseylab/LungBiophysical) and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Aves Labs | cat#: GFP-1020; RRID: AB_10000240 |
| Goat polyclonal anti-tdTomato | Origene | cat#: AB8181-200; RRID: AB_2722750 |
| Rabbit polyclonal anti-Phosphorylated Myosin Light Chain | Abcam | cat#: ab2480; RRID: AB_303094 |
| Rabbit polyclonal anti-Yap | Cell Signaling | cat#: 4912; RRID: AB_2218911 |
| Rabbit monoclonal anti-Yap/Taz | Cell Signaling | cat#: 8418; RRID: AB_10950494 |
| Rabbit polyclonal anti-Sftpc | Millipore | cat#: AB3786; RRID: AB_91588 |
| Rabbit polyclonal anti-Mature SFTPC | Seven Hills | cat#: WRAB-76694 |
| Rabbit polyclonal anti-SFTPC | Abcam | cat#: ab90716; RRID: AB_10674024 |
| Mouse monoclonal anti-HT2-280 | Terrace Biotech | cat#: TB-27AHT2-280; RRID: AB_2832931 |
| Rat monoclonal anti-Lamp3 | Novus | cat#: DDX0191P-100; RRID: AB_2827532 |
| Mouse monoclonal anti-Hopx | Santa Cruz | cat#: sc-398703; RRID: AB_2687966 |
| Rat monoclonal anti-Ager | R&D Systems | cat#: MAB1179; RRID: AB_2289349 |
| Rat monoclonal anti-PDPN | Thermo Fisher | cat#: 14–9381-82; RRID: AB_1603307 |
| Rat monoclonal anti-Krt8 | DSHB | cat#: TROMA-I; RRID: AB_2891089 |
| Rabbit monoclonal anti-FAK | Cell Signaling | cat#: 13009; RRID: AB_2798086 |
| Rabbit polyclonal anti-pFAK (Tyr397) | Thermo Fisher | cat#: 44–624G; RRID: AB_2533701 |
| Rabbit polyclonal anti-Lamin B1 | Abcam | cat#: ab16048; RRID: AB_443298 |
| Rabbit monoclonal anti-Cleaved Caspase-3 | R&D Systems | cat#: MAB835; RRID: AB_2243951 |
| Mouse monoclonal anti-Ki67 | BD Biosciences | cat#: 550609; RRID: AB_393778 |
| Rat monoclonal anti-α-Tubulin | Santa Cruz | cat#: sc-53029; RRID: AB_793541 |
| Mouse Pan-cytokeratin antibody cocktail | Invotrogen | cat#: MA5-13156; RRID: AB_10983023 |
| Mouse monoclonal anti-Cdc42 | Abcam | cat#: ab41429; RRID: AB_726768 |
| Rabbit monoclonal anti-β-actin | Cell Signaling | cat#: 4970; RRID: AB_2223172 |
| Mouse monoclonal anti-β-actin | Cell Signaling | cat#: 3700; RRID: AB_2242334 |
| Mouse monoclonal anti-β-actin, HRP conjugated | Proteintech | cat#: HRP-60008; RRID: AB_2819183 |
| Rat monoclonal anti-Integrin α6 | Invitrogen | cat#: 14–0495-82; RRID: AB_891480 |
| Mouse monoclonal anti-Integrin β5 | Invitrogen | cat#: 14–0497-82; RRID: AB_467288 |
| Mouse monoclonal anti-Integrin β6 | Sigma-Aldrich | cat#: MAB2077Z; RRID: AB_11210680 |
| Rat monoclonal anti-EpCAM, PE-Cy7 | eBioscience | cat#: 25–5791-80; RRID: AB_1724047 |
| Rat monoclonal anti-CD31, APC | eBioscience | cat#: 17–0311-82; RRID: AB_657735 |
| Rat monoclonal anti-CD45, APC | eBioscience | cat#: 17–0451-83; RRID: AB_469393 |
| Biological samples | ||
| Healthy human lung tissue | The University of Pennsylvania | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Bleomycin | Teva | N/A |
| Tamoxifen | Sigma-Aldrich | cat#: T5648 |
| SABM Basal Medium | Lonza | cat#: CC-3119 |
| SAGM SingleQuots Supplements and Growth Factors | Lonza | cat#: CC-4124 |
| EGF | PeproTech | cat#: AF-100-15 |
| Cholera toxin | Millipore | cat#: C9903 |
| Fibronectin | Sigma-Aldrich | cat#: F0895 |
| PF 573228 | Tocris | cat#: 3239 |
| Latrunculin B | Sigma-Aldrich | cat#: 428020 |
| Calcein, AM | Thermo Fisher | cat#: C1430 |
| Ethidium Homodimer-1 (EthD-1) | Thermo Fisher | cat#: E1169 |
| CellMask Green | Thermo Fisher | cat#: C37608 |
| Alexa Fluor Plus 647 Phalloidin | Thermo Fisher | cat#: A30107 |
| Mouse IgM microbeads | Miltenyi Biotec | cat#: 130–047-301 |
| CD31 microbeads, human | Miltenyi Biotec | cat#: 130–091-935 |
| CD45 microbeads, human | Miltenyi Biotec | cat#: 130–045-801 |
| Critical commercial assays | ||
| Micropatterned dishes | 4DCell | N/A |
| NEB Ultra II DNA Library Prep Kit | New England Biolabs | E7645 |
| Chromium Next GEM Single Cell 3’ Reagent Kits v3 | 10X Genomics | cat#: 1000121 |
| Deposited data | ||
| Single Cell RNA-sequencing | This paper | GEO: GSE202319 |
| Lamin B1 ChIP-seq | This paper | GEO: GSE202319 |
| Codes used in this study | This paper | DOI: 10.5281/zenodo.743 4692 |
| Experimental models: Organisms/strains | ||
| Mouse: SftpcCreERT2 | PMID: 21701069 | N/A |
| Mouse: HopxCreERT2, Hopxtm2.1(cre/ERT2)Joe/J | Jackson Laboratories | cat#: 017606 |
| Mouse: SftpcEGFP, B6N.Cg-Tg(Sftpc,-EGFP)1Dobb/J | Jackson Laboratories | cat#: 028356 |
| Mouse: HopxEGFP, Hopxtm3.1Joe/J | Jackson Laboratories | cat#: 029271 |
| Mouse: Ptk2Flox, B6.129P2(FVB)-Ptk2tm1.1Guan/J | Jackson Laboratories | cat#: 031956 |
| Mouse: Cdc42Flox, Cdc42tm1Yizh/J | Jackson Laboratories | cat#: 027576 |
| Mouse: Rosa26REYFP, B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J | Jackson Laboratories | cat#: 007903 |
| Mouse: Rosa26RmTmG, B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratories | cat#: 007676 |
| Recombinant DNA | ||
| Plasmid: mCherry-DN KASH | PMID: 21652697 | N/A |
| Software and algorithms | ||
| Prism 9.4.1 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Imaris | Oxford Instruments | https://imaris.oxinst.com/packages |
| Huygens | Scientific Volume Imaging | https://svi.nl/Huygens-Software |
| Fiji | NIH | https://fiji.sc |
| R 4.2.0 | R Project | https://www.r-project.org |
| Seurat V4.0.6 | Satija Lab | https://satijalab.org/seurat/ |
| corrplot | CRAN | https://cran.r-project.org/web/packages/corrplot/index.html |
| STARsolo v2.7.9a | PMID: 23104886 | https://github.com/alexdobin/STAR/blob/master/docs/STARsolo.md |
| bwa | PMID: 19451168 | https://bio-bwa.sourceforge.net |
| Picard Tools | Broad Institute | https://broadinstitute.github.io/picard/ |
| SAMtools | PMID: 19505943 | https://samtools.sourceforge.net |
| deepTools | PMID: 27079975 | https://pypi.org/project/deepTools/ |
| EDD | PMID: 24782521 | https://github.com/CollasLab/edd |
| DAVID | PMID: 35325185 | https://david.ncifcrf.gov |


