Abstract
Nasopharyngeal carcinoma (NPC) is a common malignant carcinoma of the head and neck, and the biological mechanisms underlying the pathogenesis of NPC remain not fully understood. In the present study, we systematically analyzed four independent NPC transcriptomic datasets and focused on identifying the critical molecular networks and novel key hub genes implicated in NPC. We found totally 170 common overlapping differentially expressed genes (DEGs) in the four NPC datasets. GO and KEGG pathway analysis revealed that cell cycle dysregulation is a critical event in NPC. Protein–protein interaction (PPI) network analysis identified a 15 hub-gene core network with overexpressed kinesin family member 2C (KIF2C) as a central regulator. Loss-of-function study demonstrated that knockdown of KIF2C significantly inhibited cell growth and cell motility, and delayed cell cycle progression, accompanied with dramatic mitotic defects in spindle formation in NPC cells. RNA-seq analysis revealed that KIF2C knockdown led to deregulation of various downstream genes. KIF2C could also regulate the AKT/mTOR pathways, and enhance paclitaxel sensitivity in NPC cells. Taken together, our results suggest that cell cycle dysregulation is a critical event during NPC pathogenesis and KIF2C is a novel key mitotic hub gene with therapeutic potential in NPC.
Keywords: Bioinformatics, Cell cycle, KIF2C, Mitosis, Nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a common type of head and neck cancer arising in the nasopharyngeal mucosa, and shows a distinct geographical and racial distribution with particularly high incidence in South China and Southeast Asia.1, 2, 3, 4, 5, 6 With the advancement in diagnostic techniques, and individualized chemoradiotherapy, NPC incidence has declined progressively, accompanied by reduced mortality.7 However, some NPC patients still suffer from poor prognosis due to distant metastasis and/or recurrence.1,5 In addition, the biological mechanisms underlying the pathogenesis and development of nasopharyngeal carcinoma remain not fully understood.7 Thus, more studies are needed to identify novel critical molecular events and key hub genes to develop novel diagnostic and therapeutic strategies for the management and treatment of NPC.
Kinesin family member 2C (KIF2C), also known as mitotic centromere-associated kinesin (MCAK), belongs to the kinesin-13 family that has four members of KIF2A, KIF2B, KIF2C and KIF24.8,9 Like all other kinesin-13 family members, KIF2C contains a conserved kinesin motor domain in the middle of its amino acid sequence, and has the ability to depolymerize microtubules by removing tubulin subunits from the microtubule polymer end.8 KIF2C has been found to be predominantly localized at centrosomes, centromeres, astral microtubules and the plus ends of microtubules in several species.8 In accordance with these findings, functional studies revealed that KIF2C play multiple essential roles in spindle assembly, chromosome congression, kinetochore-microtubule attachment and chromosome segregation during mitosis.8 KIF2C also participates in cytoskeletal remodeling during migration and invasion.10, 11, 12 In addition, KIF2C has been also shown to play important roles in tumorigenesis and cancer development. Studies revealed that KIF2C is aberrantly overexpressed in several types of cancers including gastric cancer, breast cancer, colorectal cancer, glioma, lung cancer and liver cancer.13, 14, 15, 16, 17, 18 Limited functional studies demonstrated that KIF2C is implicated in the proliferation and migration of gastric, lung and liver cancer cells.9,13,18 However, the expression and the role of KIF2C as well as its underlying molecular mechanism in NPC remain elusive.
In this study, we exhaustively retrieved all the NPC gene expression profile datasets and selected four high quality NPC datasets. Our integrated analysis of the four independent NPC datasets revealed totally 170 common overlapping differentially expressed genes (DEGs). Further GO, KEGG pathway and PPI network analyses strongly suggested that cell cycle dysregulation is a critical event in NPC tumorigenesis. Among the key hub genes implicated in NPC, KIF2C is a central hub regulator. Our functional study indicated that KIF2C is an essential regulator for the malignant phenotype of NPC cells and thus might serve as a novel key therapeutic target in NPC.
Materials and methods
NPC expression profiling dataset selection
GEO database (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/gds/) was retrieved to select the NPC gene expression profile datasets. Datasets were eligible only if they fulfilled all the following criteria: (1) the dataset must contain both tumor and corresponding adjacent normal tissues; (2) the dataset must contain some previously well-validated NPC-related genes (e.g. FOXM1, SOX4, etc);6 (3) the dataset must contain at least three normal tissue samples and at least three tumor samples. The raw data of the selected datasets were then downloaded from the GEO database for further processing and analysis.
Dataset processing and differentially expressed gene (DEG) analysis
The raw data of the different datasets were independently processed. The gene expression profiling data were normalized by quantile method and analyzed by limma package in R software (Version 3.6.2). Differentially expressed genes (DEGs) were filtered by P value (<0.05) and gene expression fold change (FC, |FC|>1.5). The intersections of the DEGs in different datasets were then analyzed and combined to get the common DEGs. Finally, the heat maps of the common DEGs were established by pheatmap package in R.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis
ClusterProfiler package is an R package for comparing biological themes among gene clusters.19 The enrichment analysis of the common DEGs for the biological processes and pathways were analyzed by clusterProfiler in R software (Version 3.6.2). To screen out significant biological processes (BP) of GO and KEGG pathway enrichment terms, P < 0.05 was defined as the cut-off criterion. Some significant terms were presented as the bar chart which was produced by ggplot2 package in R.
Protein–protein interaction (PPI) network construction
STRING (Search Tool for the Retrieval of Interacting Genes) is an online tool for analyzing the protein–protein interaction (PPI) network (https://string-db.org/).20 Cytoscape is an open source software for integrating (https://cytoscape.org/).21 The PPI network for the overlapping DEGs was constructed by STRING online database with default parameters and then the relevant genes were imported into Cytoscape to build a summary network. Finally, Molecular Complex Detection (MCODE) plugin was used to screen out top key genes.
Cell culture, synchronization and paclitaxel treatment
Nasopharyngeal carcinoma (NPC) cell lines, HNE-1 and CNE-1, were both cultured in RPMI 1640 medium (Hyclone) containing 50 U/mL penicillin, 50 mg/mL streptomycin, and 10% (v/v) FBS (Invitrogen, Carlsbad, USA). The cells were routinely cultured at a humidified incubator containing 5% CO2 in a 37 °C environment. Cells were synchronized to the G2 phase with the Cdk1 inhibitor RO-3306 (9 μM) (Selleck Chemicals) for 18 h and released from the G2 block for 1 h in fresh media.22 Paclitaxel (Aladdin) was dissolved in DMSO and diluted to the concentration of 10 nM for treatment of NPC cells for 24 h.
Plasmid construction and lentiviral shRNA-mediated gene silencing
The double-stranded negative control short hairpin RNA (shRNA) oligonucleotides and shRNA targeting human KIF2C mRNA, designated as NCsh and KIF2Csh, were synthesized by Sangon (Sangon Biotech, Shanghai, China) and cloned into the AgeI/EcoRI sites of the pLKO.1-puro lentiviral vector with the T4 DNA Ligase Kit (Thermo Scientific). The shRNA sequences were provided in Table S1. The lentivirus particles were packaged by co-transfection of the vectors of pLKO.1-puro, pSPAx2 and pMD2.G into 293Ta cells by Neofect DNA® transfection reagent (Neofect, Beijing, China), followed by routine culture supernatant collection and concentration. The lentivirus particles were used to infect the HNE-1 and CNE-1 NPC cells with 5 μg/mL polybrene; 48 h later, cells were collected and subjected to subsequent analysis.
qRT-PCR and immunoblotting
Total RNA was isolated from cells using the Total RNA Kit I (Omega Bio-Tek) according to the manufacturer's instructions. Quantitative RT-PCR (qRT-PCR) was performed using the SYBR® Premix Ex Taq (Perfect Real Time, TAKARA) as described previously.23 GAPDH was used as an internal control. The information of the primers used was provided in Table S2. Immunoblotting analysis was conducted as described previously.24,25 Briefly, cells were collected and lysed with RIPA buffer supplemented with protease inhibitor Cocktail (Biotool, Houston, USA). The total proteins were determined using the Quick Start™ Bradford (Bio-Rad Laboratories, Hercules, USA) and then subjected to SDS-PAGE and immunoblotting. The blots were visualized by enhanced chemiluminescence (ECL, Bio-Rad Laboratories). The information of the antibodies used was provided in Table S3.
Cell growth and cell viability analysis
Cell growth was monitored by JULI Stage Real-time Cell History Recorder (NanoEntek, Seoul, South Korea). Briefly, cells were infected with NCsh and KIF2Csh lentiviral particles, respectively, and 48 h later, cells were collected and seeded into 6-well plates at the concentration of 5 × 105 cells/mL. Images were taken continuously by JULI Stage for 48 h with an interval of 2 h. The growth rate was quantified by JULI STAT. For cell viability analysis, trypan blue (Solarbio Life Sciences, Beijing, China) staining was performed by mixing 0.4% trypan blue solution with cell suspension at the ratio of 1:9 for 3 min. Cell number was counted by CellDrop FL Fluorescence Cell Counter (Devovix, USA).
Cell cycle analysis and apoptosis assay
Cell cycle analysis and apoptosis assay were performed as described previously.26 Briefly, cells were collected by trypsin digestion and low speed centrifugation, and washed with ice-cold phosphate-buffered saline (PBS). For cell cycle analysis, cells were fixed with 75% ethanol, and then subjected to FACS analysis. For apoptosis assay, cells were resuspended in binding buffer and stained with Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (BD Pharmingen, Franklin lake, USA) according to the manufacturer's instructions, and then subjected to FACS analysis.
Wound-healing assay and motility assay
Wound-healing assay was performed as described previously.23 Briefly, cells were infected with lentiviral particles of NCsh and KIF2Csh, and 48 h later, cells were scratched; images were taken at 0, 4, and 8 h under a Leica light microscope (DM4B, Leica Corporation, Wetzlar, Germany). The migration rate was quantified by Image J. For cell motility assay, cells were collected and resuspended at a concentration of 5 × 103 cells/mL in 6-well plates, and images were taken continuously by JULI Stage Real-time Cell History Recorder (NanoEntek, Seoul, South Korea) for 8 h with an interval of 15 min. The cell motility ability was quantified by Image-pro. Cells that were died, dividing or moved out of the field of view were excluded from the quantification criteria for motility assessment.
Indirect immunofluorescence assay
The indirect immunofluorescence assay was conducted as described previously.1 Briefly, cells were cultured on glass coverslips, fixed in ice-cold 4% formaldehyde for 20 min at room temperature, and permeabilized with 0.5% Triton X-100/PBS for 20 min. After being blocked with 1% bovine serum albumin (BSA) at room temperature for 2 h, the cells were incubated with the indicated primary antibodies at 4°C for overnight, and then incubated with the corresponding secondary antibodies at room temperature for 2 h. The nucleus was stained with 4′6-diamidino-2-phenylindole (DAPI, Vector Laboratories) for 20 min. Cells were visualized with Leica DM4000 versatile upright microscope and Leica SP8 confocal laser scanning microscope. The information of the antibodies used was provided in Table S3.
RNA-sequencing analysis
The KIF2C knockdown cells and its corresponding negative control cells were collected, total RNA was extracted, and cDNA libraries were then constructed and subjected to RNA-seq analysis as described previously.27 DEGs were subjected to GO and KEGG Pathway enrichment analyses. Gene Set Enrichment Analysis (GSEA; http://www.broad.mit.edu/gsea) was performed to identify the oncogenic signatures and significantly changed pathways.
Statistical analysis
Statistical analysis was conducted using the SPSS 16.0 statistical software package (SPSS Inc, Chicago, USA). Data were displayed as mean ± SD (standard deviation), and presented using GraphPad Prism (GraphPad Software Inc., San Diego, USA). The difference between different groups was evaluated by Student's t tests with a P value less than 0.05 indicating statistical significance.
Results
NPC expression profiling dataset selection
To screen and identify critical molecular events and novel hub cancer-associated genes implicated in NPC, we fully retrieved the public GEO database to select out NPC expression profiling datasets with high quality for deep data mining. Based on our selection criteria as described in Material and Methods, totally four NPC datasets were consequently chosen from the GEO database, including GSE12452, GSE53819, GSE13597, and GSE118719. The detailed information of the chosen datasets regarding tissue samples and platforms were summarized in Table 1.
Table 1.
The information of chosen NPC datasets with high quality.
Identification of DEGs in NPC
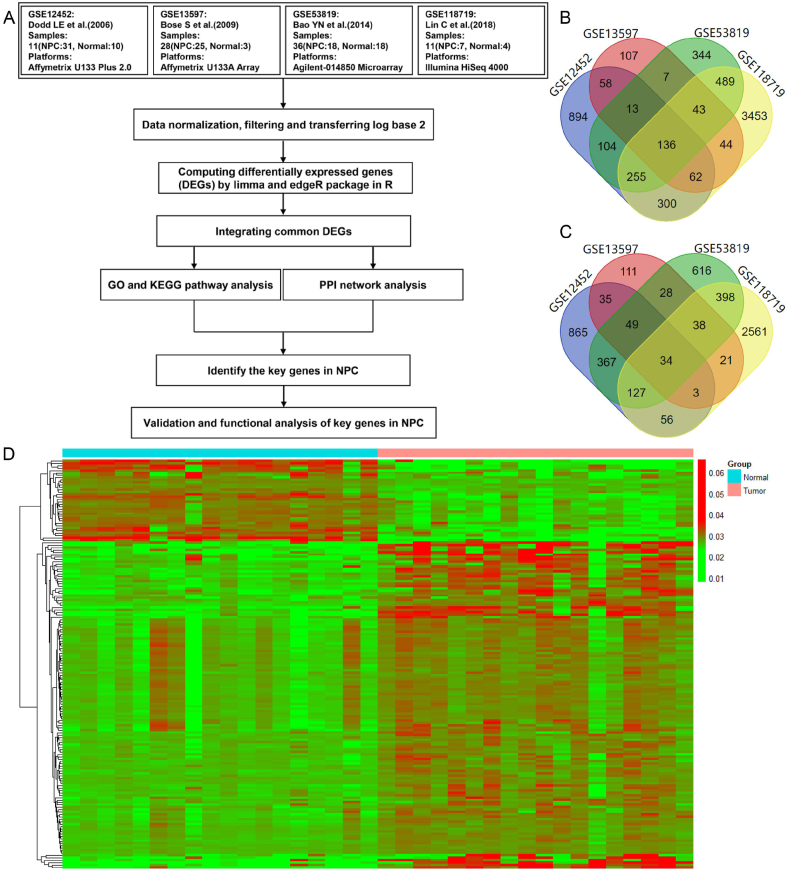
As described in Figure 1A, the four NPC expression datasets (GSE12452, GSE53819, GSE13597 and GSE118719) were independently analyzed. Using P < 0.05 and absolute gene expression fold change (FC) > 1.5 as cut-off criteria, a total of 170 overlapping common DEGs (Table 2), including 136 of upregulated genes (Fig. 1B) and 34 of downregulated genes (Fig. 1C), were identified from the four datasets. Of note, some of the common DEGs have been previously reported and validated in NPC, including FOXM1, SOX4, TYMS, MMP1 and CHEK1,32, 33, 34, 35, 36 whereas most of the common DEGs remain unexplored in NPC (Table 2). The heat map visualization of the common DEGs shows that the expression of downregulated genes in NPC tissues was significantly lower than that in normal tissues and the expression of upregulated genes in NPC tissues obviously higher than that in normal tissues (Fig. 1D; Fig. S1A–C). Taken together, these results suggest that the 170 common DEGs may play important roles in the tumorigenesis and development of NPC, which needs further investigation.
Figure 1.
Identification of differentially expressed genes in NPC. (A) Workflow of meta-analysis for identification of critical molecular networks and key hub genes in NPC. (B) Venn diagram of upregulated genes overlapped among the four GEO datasets (GSE12452, GSE53819, GSE13597 and GSE118719). (C) Venn diagram of downregulated genes overlapped among the four GEO datasets. (D) Heat map of differentially expressed genes in GSE53819. Gene expression levels were indicated by color intensities with red as highly expressed and green as lowly expressed.
Table 2.
The 170 common differentially expressed genes (DEGs) among four NPC datasets.
| DEGs | Genes Name |
|---|---|
| Up-regulated | ABCA3, COL4A5, BRCA1, LMNB2, PDCD5, FOXM1, HSPD1, CDK1, PLXNA1, FGF1, COL7A1, UNG, KIF14, MAD2L1, TNFAIP6, VRK2, IL23A, KIF4A, MCM10, TYMS, MELK, NDC80, OIP5, DUSP10, MMP1, TWSG1, KIF23, ASPN, PRC1, CDT1, CDC45, MCM4, CLEC5A, NREP, CCNF, NEK2, RCN2, CHST3, ESM1, ESPL1, SOX4, CHEK1, KIF18B, CST1, ATP2C1, AHCY, ARNTL2, HOXA10, ROBO1, PARPBP, INPP1, CENPN, PLK4, POLQ, KRMEN2, PUS7, TOP2A, FANCI, TNFRSF10B, GALNT11, CHAF1B, ERCC6L, PRAME, OLA1, LHX2, FJX1, FGD6, CENPF, ARNT2, TPX2, IGF2BP3, GINS1, BIRC5, RACGAP1, PSMC3IP, CDC6, FN1, NUAK1, LAMB3, WNT5A, PTGS2, STK3, PRKDC, DNA2, ZWINT, DOCK4, PTTG1, ECT2, VASH2, INSM1, GADD45A, POSTN, FSCN1, SRD5A1, CEP55, CTPS1, RBBP8, STAR, PMAIP1, RAD51AP1, PLAU, GJA1, MARK1, RAI14, DSG2, ZWILCH, HSPA4L, CKS1B, ITGAV, B4GALT6, KIF2C, NOX4, TFRC, PBK, TROAP, TK1, PLA2G3, ASPM, FAP, LAMB1, MYO19, STAP2, BUB1B, HJURP, RNASEH2A, DTL, TM7SF3, MEST, FERMT1, GMNN, KIF20A, HOXC6, NPL, TTK, NCAPG, NUSAP1 |
| Down-regulated | SLC15A2, CYP2F1, DLEC1, NDRG2, CR1, GNG7, CX3CR1, PAX5, ABLIM1, PLEKHB1, C7, CXCR5, PTGDS, CLU, DPEP2, MSLN, FAM107A, TFF3, SIDT1, ADRA2A, DGKD, ATP2A3, CD22, BACH2, RASGRP2, SERPINB7, VILL, LMO2, SERPINB6, AQP5, CR2, ADH1B, PIP, VIPR1 |
Functional enrichment analysis of DEGs in NPC
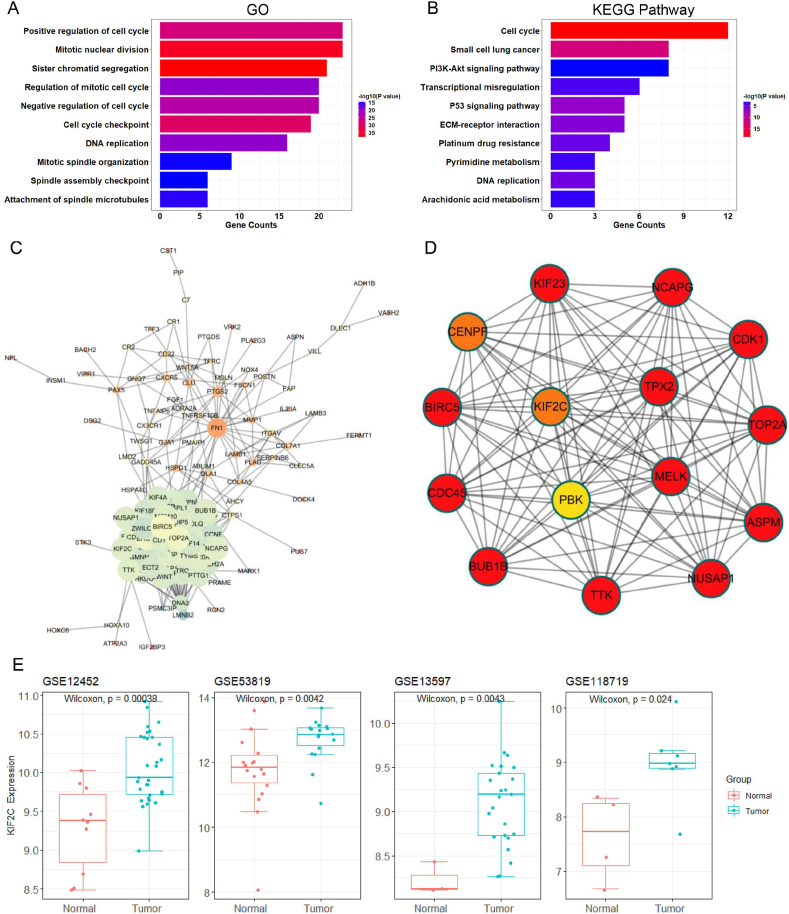
To gain insights into the molecular and cellular mechanisms underlying DEGs in NPC, we performed GO and pathway enrichment analysis. The GO biological processes (BP) analysis revealed that the common DEGs were enriched in cell cycle, DNA replication, mitotic division and organization, spindle assembly chromosome segregation and other cancer-associated BP terms (Fig. 2A; Table S4). The KEGG pathway analysis demonstrated that the common DEGs were not only associated with cell cycle and DNA replication, but also enriched in some cancer-related pathway such as PI3K-Akt and p53 signaling pathway, ECM-receptor interaction, small cell lung cancer and transcriptional misregulation (Fig. 2B). Of note, the KEGG pathway analysis also pointed to platinum drug resistance (Fig. 2B) and cell apoptosis (Table S5). Taken together, these data suggest that cell cycle dysregulation is a critical event implicated in NPC tumorigenesis.
Figure 2.
Functional enrichment analysis and PPI network of DEGs in NPC. (A) GO enrichment analysis of common DEGs. The abscissa represents the number of DEGs, and the ordinate represents the significantly enriched GO terms. The color intensity of the columns indicates the different -log10 of P value. (B) KEGG pathway enrichment analysis of common DEGs. The ordinate represents the significantly enriched KEGG pathways. (C) The concise complete PPI network of common DEGs. The size of circle indicates the number of edges, i.e., the larger the circle, the more the number of edges. (D) The core PPI network containing top 15 key hub DEGs constructed by MCODE plugin of Cytoscape. Assembly factor for spindle microtubules, ASPM; baculoviral IAP repeat containing 5, BIRC5; BUB1 mitotic checkpoint serine/threonine kinase B, BUB1B; cell division cycle 45, CDC45; cyclin dependent kinase 1, CDK1; centromere protein F, CENPF; kinesin family member 23, KIF23; maternal embryonic leucine zipper kinase, MRLK; non-SMC condensin I complex subunit G, NCAPG; nucleolar and spindle associated protein 1, NUSAP1; PDZ binding kinase, PBK; DNA topoisomerase II alpha, TOP2A; TPX2 microtubule nucleation factor, TPX2; TTK protein kinase, TTK. (E) The box–plot diagram of KIF2C expression in NPC datasets.
PPI network construction and identification of KI2C as a potential hub gene in NPC
The 170 overlapping common DEGs were then imported to STRING online database, and the interactions among these genes was searched using the default parameters. Consequently, we constructed a protein–protein network for the 170 common DEGs (Fig. S2). By removing some loosely connected or even totally isolated nodes, a more succinct PPI network was established, which contains 132 nodes and 1576 edges (Fig. 2C). The key modules were identified by MCODE plugin of Cytoscape with default parameters, and the top 15 key DEGs were identified as the potential hub genes in NPC, including ASPM, BIRC5, BUB1B, CDC45, CDK1, CENPF, KIF23, KIF2C, MRLK, NCAPG, NUSAP1, PBK, TOP2A, TPX2, TTK. The 15 hub genes are all cell cycle-related genes and show strong positive expression correlation with each other (Fig. S3), and more importantly they constitute a core interaction network (Fig. 2D). Of note, four genes, PBK, KIF2C, TPX2 and MELK, are located at the central of the module, strongly suggesting that they play important roles in NPC tumorigenesis. Through exhaustive literature review, we found that PBK, one of the four central hub genes, has been recently demonstrated as a novel theranostic target for NPC,37 which further supports the authenticity of our bioinformatic PPI analysis. Nevertheless, the role of KIF2C, TPX2 and MELK in NPC has not been investigated yet. Intriguingly, KIF2C has been shown to act as an important regulator of microtubule dynamics and also participate in some cancers such as hepatocellular carcinoma.8,9 We thus speculated that KIF2C may serve as a central hub regulator and a novel therapeutic target in NPC. In support of our speculation, detailed analysis revealed that KIF2C is indeed aberrantly overexpressed in all the four NPC datasets (Fig. 2E). Therefore, we decided to focus on investigating the functional implication of KIF2C in NPC.
KIF2C silencing inhibits NPC cell growth
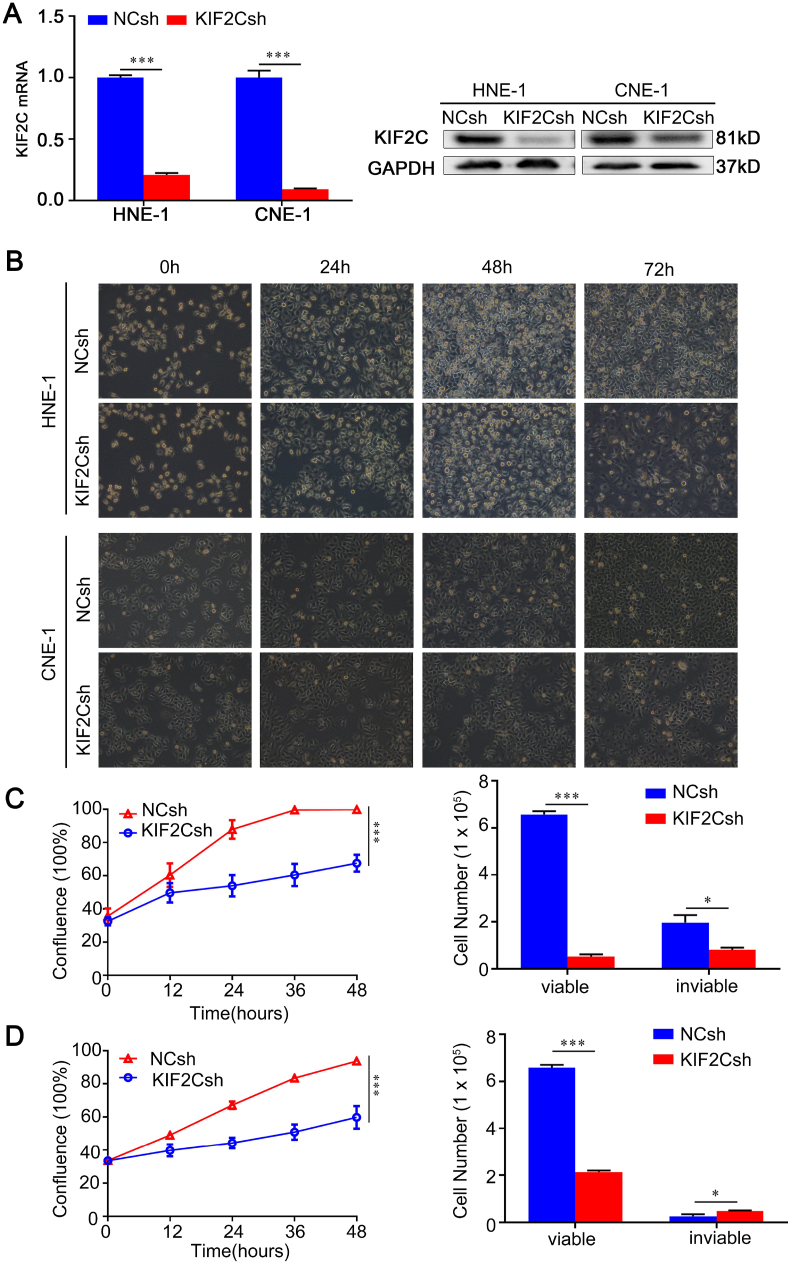
In the first step of the functional study regarding KIF2C in NPC, we examined the effect of a targeted depletion of KIF2C gene expression by lentivirus-mediated RNA interference (RNAi) on the cell growth of two different NPC cell lines, HNE-1 and CNE-1. Quantitative RT-PCR and immunoblotting analysis demonstrated that KIF2C expression was significantly silenced at both mRNA and protein levels, whereas the expression of internal control gene, GAPDH, remained unchanged (Fig. 3A). As shown in Figure 3B–D, knockdown of KIF2C expression caused obvious alterations in cell morphology and remarkable decrease in cell number in both HNE-1 and CNE-1 cells. In accordance with these observations, KIF2C knockdown inhibited the growth rate of both cell lines, and trypan blue exclusion assay revealed that KIF2C knockdown significantly reduced cell viability (Fig. 3C, D).
Figure 3.
Knockdown of KIF2C inhibits NPC cell growth. (A) RNAi-mediated KIF2C gene silencing. Human NPC cells (HNE-1 and CNE-1) were infected with lentivirus particles expressing negative control shRNA (NCsh) and shRNA against KIF2C (KIF2Csh). Forty-eight hours after infection, total RNA and whole cell lysates were prepared and subjected to RT-PCR and immunoblotting, respectively. (B) KIF2C knockdown causes changes in cell morphology. Cell morphology was monitored under phase-contrasted microscope at the indicated times after lentivirus infection (× 100). (C, D) KIF2C knockdown inhibits HNE-1 and CNE-1 cell growth, respectively. Cell growth was continuously monitored by JULI Stage Real-time Cell History Recorder, and cell viability was examined by trypan blue exclusion assay at the final time point.
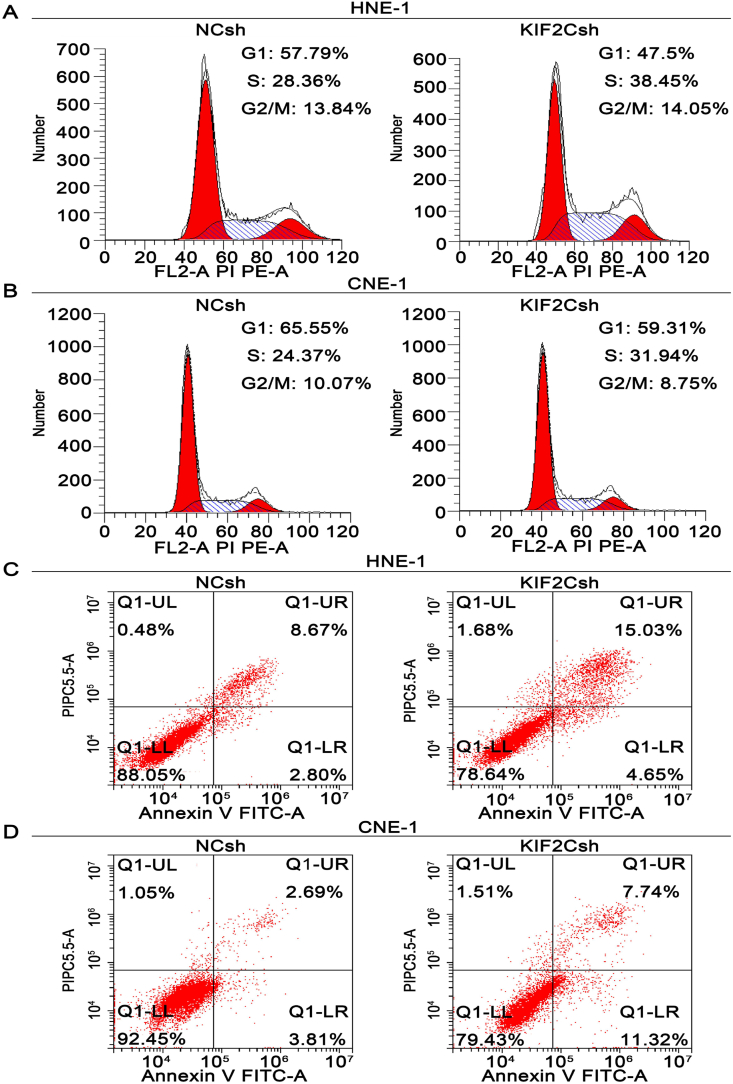
KIF2C silencing induces NPC cell cycle arrest and apoptosis
Next, we analyzed the effect of KIF2C knockdown on cell cycle progression and apoptosis. As shown in Figure 4A and B, KIF2C silencing resulted in a significant G2/M and S phase arrest in both CNE-1 and HNE-1 cells. About 38% of KIF2Csh-infected HNE-1 cells were arrested in S phase compared with about 28% in NCsh-infected control cells, and 32% of KIF2Csh-infected CNE-1 cells compared with 24% in control cells. To further determine whether KIF2C silencing induces apoptosis in NPC cells, Annexin V-FITC and PI double staining with flow cytometry was conducted to detect the apoptotic cells. The results showed that, 72 h after lentivirus infection, approximately 4.65% and 15.03% of KIF2C depleted HNE-1 cells displayed early apoptotic and late apoptotic/secondary necrotic phenotype, respectively, whereas only 2.80% and 8.67% of NCsh-infected cells had such respective presentations (Fig. 4C). Similarly, KIF2C knockdown in CNE-1 cells also caused an increase in the apoptotic population, approximately 11.32% and 7.74% in KIF2C knockdown group, but 3.81% and 2.69% in control group (Fig. 4D). These results suggested that KIF2C depletion induces cell cycle arrest and apoptotic cell death.
Figure 4.
KIF2C silencing induces cell cycle arrest and apoptosis in NPC. HNE-1 and CNE-1 cells were infected with lentivirus particles expressing negative control shRNA (NCsh) and shRNA against KIF2C (KIF2Csh) as described in Fig. 3, and then subjected to cell cycle analysis (A, B) and apoptosis assay (C, D), respectively.
KIF2C silencing reduces NPC cell motility
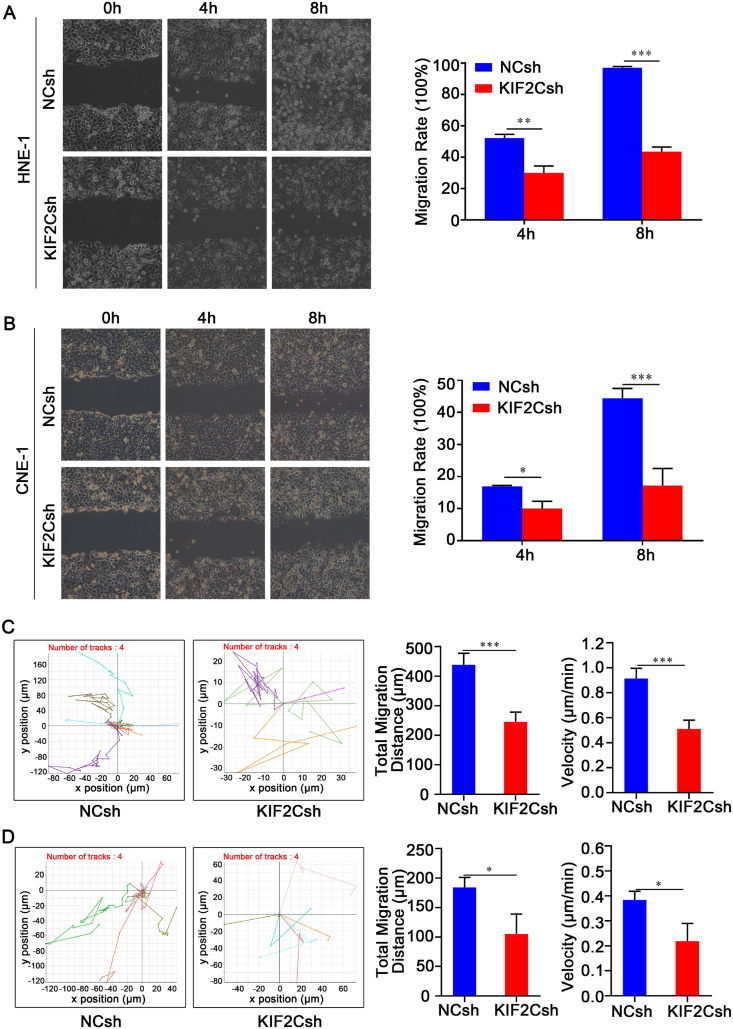
Furthermore, we assessed the effect of KIF2C knockdown on cell migration and motility. As shown in Figure 5A and B, wound healing assay revealed that in both HNE-1 and CNE-1 cells, knockdown of KIF2C significantly decreased the rate of lateral migration into a wound introduced in a confluent monolayer of cells compared with control cells. Additionally, cell motility assay by a live cell imaging system demonstrated that again in both HNE-1 and CNE-1 cells, the mean total migration distance of KIF2C knockdown cells was significantly greater than that of the control cells (Fig. 5C, D). Therefore, these results strongly indicate that KIF2C is essential for the migration and movement ability of NPC cells.
Figure 5.
KIF2C silencing reduces cell migration and motility in NPC. Cells were infected with lentivirus particles expressing negative control shRNA and KIF2C shRNA, and 48 h later, wound healing assay was conducted for HNE-1 (A) and CNE-1 (B) cells, respectively. Similarly, cell motilities were also monitored using a live cell imaging analyzer every 15 min for 8 h in both HNE-1 (C) and CNE-1 (D) cells. Motile trajectory of each cell for 8 h, mean total migration distances over 8 h and mean minutely migration speeds are presented.
KIF2C silencing causes mitotic defects in NPC cells
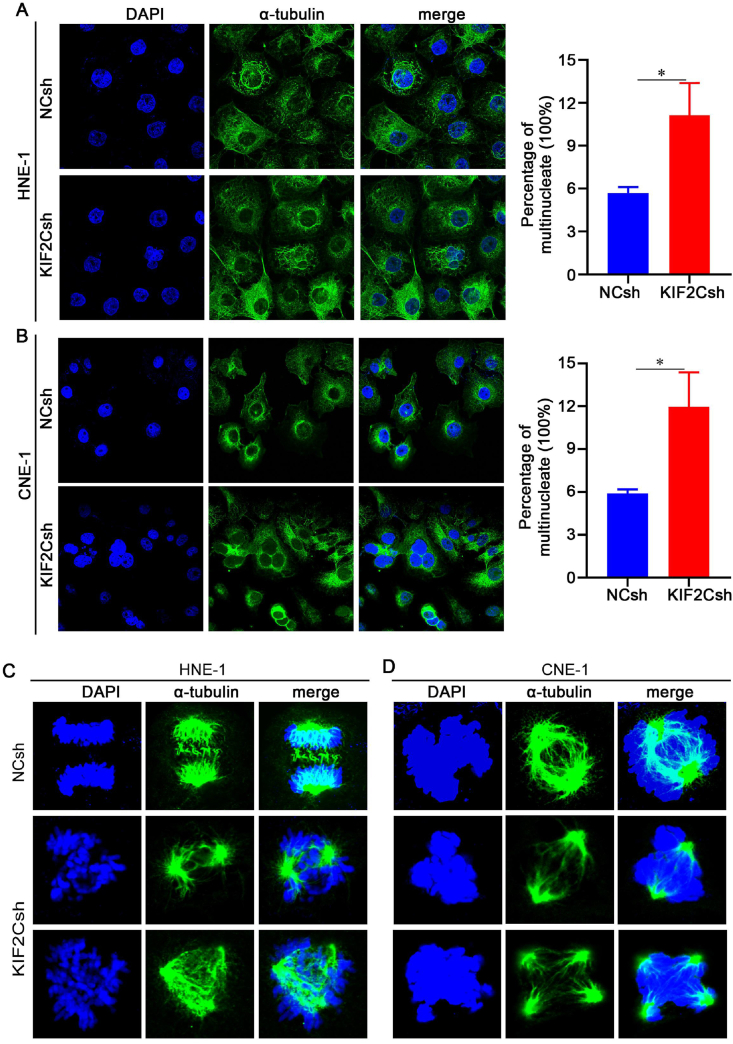
Moreover, we analyzed the effect of KIF2C knockdown on mitotic events in NPC cells by fluorescence microscopy. The results showed that KIF2C knockdown caused a significant increase in binulceate and multinucleate cells in both HNE-1 and CNE-1 cells (Fig. 6A, B). Remarkably, KIF2C knockdown also caused cells with mitotic defects in spindle structure and chromosome alignment (Fig. 6C, D). Lots of the KIF2C knockdown cells exhibited aberrant bipolar spindles with excessively long and disorganized astral microtubules extending toward the cell cortex (Fig. 6C, D). Meanwhile, the majority of KIF2C knockdown cells had disturbed prometaphase chromosome arrangements (Fig. 6C, D). These data are in accordance with the notion that one major function of KIF2C is to regulate spindle microtubule dynamics during mitosis to ensure proper spindle formation and proper attachment of chromosomes on the spindle.
Figure 6.
Mitotic defects in KIF2C knockdown NPC cells. (A, B) KIF2C knockdown causes bionucleate and multinucleate cells. HNE-1 and CNE-1 cells were infected with lentivirus particles expressing negative control (NC) shRNA and KIF2C shRNA, and 48 h later, cells were then fixed and nucleic DNA was stained with DAPI. Representative images are shown. The normal and defective cells in multiple fields were counted under a microscope, and the quantitative data (expressed as percentage of cells with defectives) are shown as a histogram. (C, D) KIF2C knockdown causes defective spindle formation. HNE-1 and CNE-1 cells were infected as in (A), and cells were then synchronized to the G2 phase using a specific and reversible Cdk1 inhibitor RO-3306 for 18 h and released for 1 h to allow cells to progress through mitosis. Cells were finally fixed and stained with the anti-α-tubulin antibody followed with FITC-conjugated secondary antibody.
Identification of oncogenic signatures and signaling pathways regulated by KIF2C
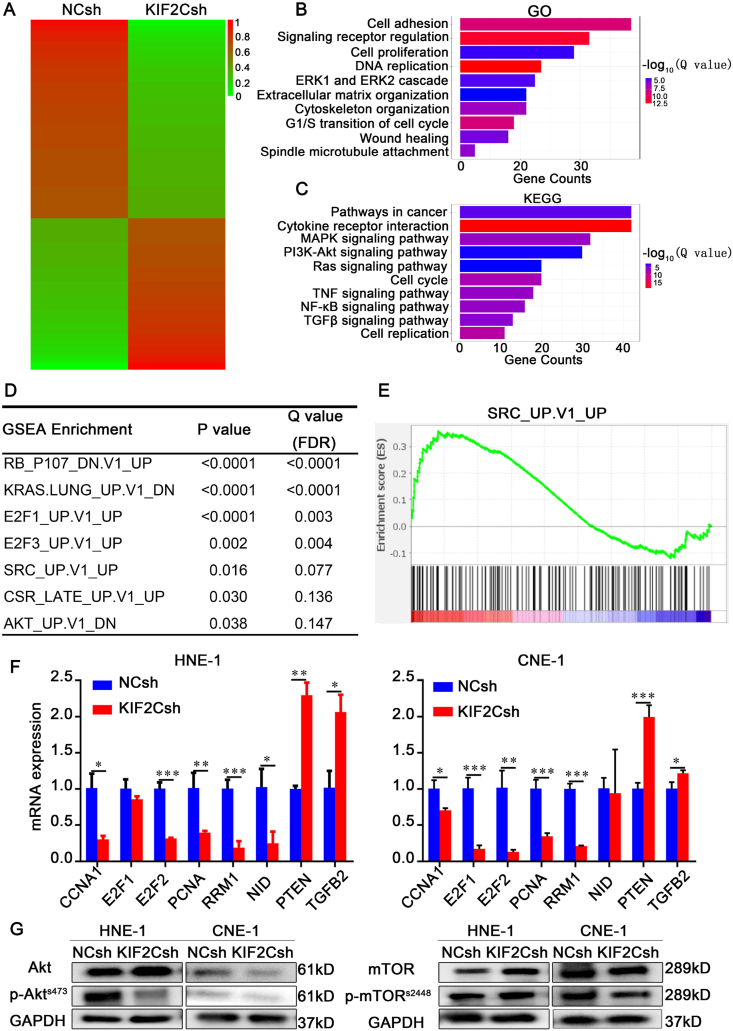
To further investigate the potential molecular mechanism behind KIF2C-related NPC tumorigenesis, we carried out RNA-seq analysis to examine the gene expression profile changes at whole genomic level in KIF2C knockdown cells. We found that KIF2C knockdown caused a significant gene expression change, with 1558 genes downregulated and 1385 genes upregulated after KIF2C knockdown (Fig. 7A). GO analysis revealed that the DEGs were mainly enriched in DNA replication, signaling receptor regulation, cell adhesion, cell proliferation, cytoskeleton organization, spindle microtubule attachment, and ERK1/2 cascade (Fig. 7B and Table S6). KEGG Pathway analysis demonstrated that DEGs were implicated in multiple tumor-associated pathways, such as MAPK, NF-κB, TNF, TGF-β, and PI3K-Akt signaling pathways (Fig. 7C and Table S7). In addition, GSEA analysis using Molecular Signature Database (MsigDB) revealed several cancer-associated gene sets, including E2F1 regulated gene signature (E2F1_UP.V1_UP), serum-stimulated gene signature (CSR_LATE_UP.V1_UP) and SRC regulated gene signature (SRC_UP.V1_UP) (Fig. 7D, E). Quantitative RT-PCR assay confirmed that a series of important genes contained in the affected signaling pathways and gene sets were indeed either downregulated or upregulated in KIF2C knockdowned HNE-1 and/or CNE-1 cells, including CCNA1, E2F1, E2F2, PCNA, RRM1, NID1, TGFB2, and PTEN (Fig. 7F). Moreover, immunoblotting verified that KIF2C knockdown reduced phosphorylation of AKT in both HNE-1 and CNE-1 cells. Of note, KIF2C knockdown resulted in a decrease of phosphorylated mTOR in CNE-1 cells but an increased mTOR phosphorylation in HNE-1 cells (Fig. 7G). These results together indicate that KIF2C might be an important regulator for multiple cancer-related signaling pathways in NPC cells.
Figure 7.
Downstream genes and pathways regulated by KIF2C. (A) RNA-seq analyses for KIF2C knockdown and its control HNE-1 cells. Heatmap shows the differentially expressed genes with fold change of expression more than 2.0. (B, C) GO enrichment and KEGG pathway analysis based on the differentially expressed genes between the KIF2C knockdown (KIF2Csh) and its control (NCsh) HNE-1 cells. (D) Top gene sets enriched by GSEA analysis. FDR, false discovery rate. (E) GSEA plot of the SRC oncogenic signature (SRC_UP.V1_UP) correlated with KIF2C depletion. (F) Verification of important KIF2C-regulated genes by qRT-PCR. (G) Detection of mTOR and AKT pathways in KIF2C knockdown HNE-1 and CNE-1 cells by immunoblotting.
KIF2C silencing enhances paclitaxel sensitivity in NPC cells
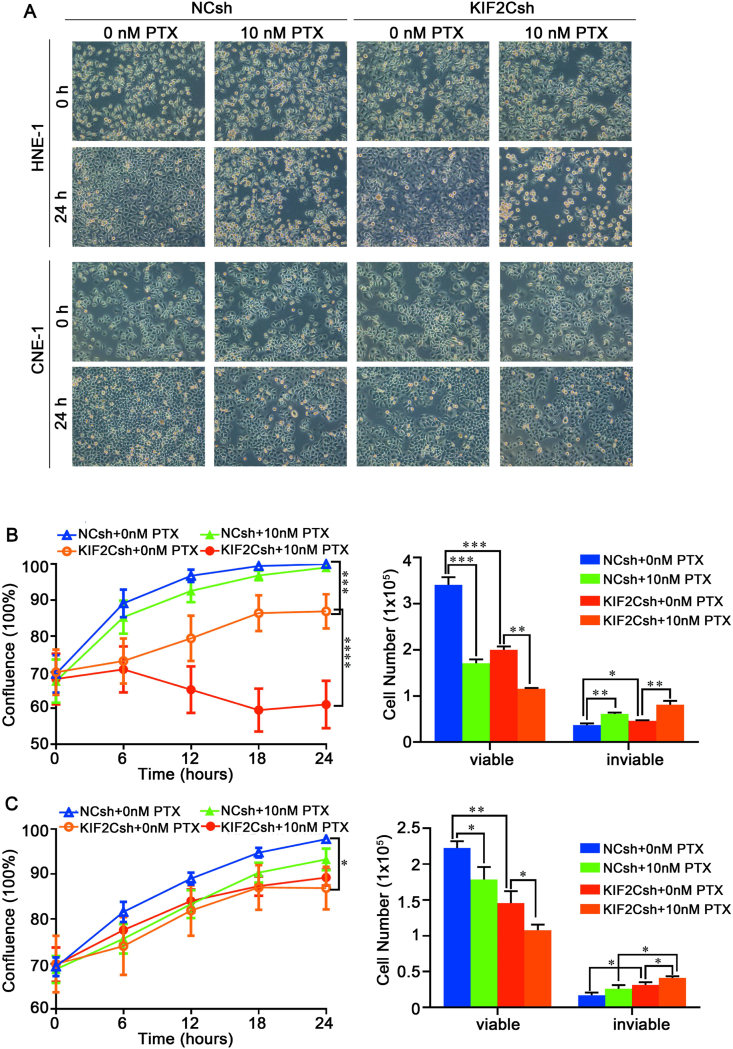
Finally, we sought to examine whether KIF2C knockdown could enhance paclitaxel sensitivity in NPC cells as KIF2C is a critical regulator of microtubule dynamics. The results showed that paclitaxel treatment could strongly inhibit cell growth and caused significant abnormal morphological changes in both HNE-1 and CNE-1 cells (Fig. 8A). After paclitaxel treatment, some cells were detached from the plates and floated on the media with round shape, indicating an obvious mitotic arrest. Subsequently, long-term paclitaxel treatment caused less confluent cell population in which some cells were completely detached from the plates and forms floating aggregates, indicating a significant cell death especially in HNE-1 cells. Notably, live cell growth monitoring and trypan blue exclusion assay revealed that KIF2C knockdown cells with 10 nM paclitaxel treatment had a dramatic decrease in cell growth and a significant increase in dead cells especially in HNE-1 cells as compared with KIF2C knockdown or paclitaxel treatment alone (Fig. 8B). Together, these data suggest that KIF2C knockdown can exacerbate the effect of paclitaxel to block mitotic progression and increase cell death.
Figure 8.
KIF2C knockdown enhances paclitaxel sensitivity in NPC cells. HNE-1 and CNE-1 cells were infected with lentivirus particles expressing negative control shRNA (NCsh) and KIF2C shRNA (KIF2Csh), and 24 h later, cells were then treated with paclitaxel. Cell morphology (A), cell growth and viability of (B), HNE-1 (C) and CNE-1 were monitored as shown in Figure 3B and C.
Discussion
KIF2C is a novel key hub gene overexpressed in NPC
Numerous studies have previously demonstrated that KIF2C, one of the kinesin-13 family members, plays important roles in regulating cell cycle progression especially mitosis.8,9 KIF2C thus is supposed to be implicated in cancer development. However, the role of KIF2C in cancers has not been deeply and extensively explored. Nakamura et al13 demonstrated that elevated expression of KIF2C is associated with lymph node metastasis and poor prognosis in gastric cancer, and consistently, ectopic overexpression of KIF2C increases proliferation and migratory ability in gastric cancer cells. Wei et al9 reported that KIF2C is significantly up-regulated in hepatocellular carcinoma, and functional experiments revealed that KIF2C promotes hepatocellular carcinoma cell proliferation, migration, invasion, and metastasis. In the present study, we systematically analyzed multiple NPC gene expression datasets and found that KIF2C is a key hub gene up-regulated in several independent NPC cohorts. Notably, our results also revealed that KIF2C is a central hub gene of the core molecular network in NPC. Furthermore, our functional experiments demonstrated that KIF2C knockdown significantly inhibits NPC cell growth and motility accompanied by delayed cell cycle progression as well as remarkable mitotic defects. Our results indicate that KIF2C is a novel key cancerous gene in NPC by essentially regulating cell cycle progression, proliferation, and migration, and therefore might serve as a novel diagnostic and therapeutic target for the diagnosis and treatment of NPC.
KIF2C and mitotic dysfunction in NPC
According to our observations, mitotic arrest and mitotic defects were the prominent major phenotype in KIF2C-depleted NPC cells. In accordance with our observations, several previous studies also showed that knockdown of KIF2C causes remarkable spindle morphology and chromosome segregation defects in mitosis in normal and malignant cells, and the tested malignant cells are more sensitive to KIF2C knockdown than normal cells.8,12,38 Chemical compounds targeting microtubule dynamics such as paclitaxel of taxanes are routinely used as a first-line chemotherapeutic regimen for various types of cancer including NPC.39 Intriguingly, as a potent regulator of microtubule dynamics, overexpression of KIF2C has been shown to confer resistance to paclitaxel,40 and siRNA-mediated knockdown of KIF2C increases the cellular sensitivity to paclitaxel.41 Another study has found that downregulation of MCAK induced by cabazitaxel (a second-line taxane chemotherapy) sensitizes prostate cancer cells to docetaxel treatment.42 These studies indicate that KIF2C could antagonize tumor cell sensitivity to taxanes by modulating microtubule dynamics at least through its microtubule depolymerization. In addition, our present study also showed that KIF2C knockdown inhibits the motility of NPC cells. In consistence with our results, three studies have previously showed that elevated expression of KIF2C is positively correlated with the invasiveness and metastasis of cancer cells.13, 14, 15 Consistently, Braun et al10 reported that KIF2C modulates the microtubule dynamics, directional migration and invasion capacity of epithelial cells. Another study has showed that depletion of KIF2C by siRNA reduces the invasion of tumor cells.11,12 Of note, additional study showed that overexpression of MCAK can stimulate microtubule depolymerization and confer resistance to paclitaxel.41 Therefore, it warrants further in vivo systemic studies to investigate the therapeutic potential of KIF2C, used either as single therapy or in combination with other cancer drugs, for combating malignancy as well as taxane resistance in various types of cancer including NPC.
KIF2C and mTOR
One recent study has found that, by interacting with the component of TSC complex, Tre2-Bub2-Cdc16 (TBC) 1 domain family member 7 (TBC1D7), KIF2C can disrupt the formation of the TSC complex and enhance the mammalian target of rapamycin complex1 (mTORC1) signal transduction.9 Immunoblotting revealed that knockdown of KIF2C decreased the phosphorylation of mTOR whereas overexpression of KIF2C led to an enhancement of the phosphorylaed mTOR in two different hepatocellular carcinoma cell lines.9 However, the effects have not been quantitatively verified by ELISA assays. Of note, our results showed that knockdown of KIF2C led to decreased phosphorylation of mTOR in CNE-1 cells but increased phosphorylation of mTOR in HNE-1 cells, suggesting a complicated regulatory role of KIF2C in mTOR signaling pathway. Thus, further studies are needed to confirm and elucidate the role of KIF2C in mTOR as well as other potential signaling pathways.
In summary, our present study systematically analyzed the NPC transcriptomic datasets, and identified cell cycle dysregulation as a critical event in NPC, with KIF2C as a novel central hub gene. Our functional study indicated that KIF2C is an essential regulator for NPC cell cycle progression and cell motility via regulating various downstream genes and cancer-associated pathways, and thus might serve as a novel therapeutic target in NPC.
Author contributions
Conceptualization, Y.Bu. and J.Z.; investigation, X.Z., P.M., YX.B., C.T., Y.W., X.L., Y.B. and J.Z.; methodology, X.Z.; software, C.T.; validation, X.Z., P.M., YX.B. and C.T.; writing—original draft preparation, X.Z., P.M., YX.B. and C.T.; writing—review and editing, Y.B.; supervision, Y.B., X.L. and J.Z.; funding acquisition, Y.B and Y.W. All authors have read and agreed to the published version of the manuscript.
Conflict of interests
The authors declare no conflict of interests. The funders had no role in the design of the study, the collection, analyses, or interpretation of data, the writing of the manuscript, or the decision to publish the results.
Funding
This research was funded by the National Natural Science Foundation of China (No. 81902824) and The First Affiliated Hospital of Chongqing Medical University, Chongqing, China.
Acknowledgements
We are grateful to our lab members for valuable discussion.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2021.05.003.
Contributor Information
Youquan Bu, Email: buyqcn@cqmu.edu.cn, buyqcn@aliyun.com.
Jiang Zhu, Email: 201959@hospital.cqmu.edu.cn.
Dataset availability
Datasets are available in a publicly accessible repository. Publicly available datasets were analyzed in this study. The data can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE12452; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE53819; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE13597; and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118719.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Feng X., Zhang C., Zhu L., et al. DEPDC1 is required for cell cycle progression and motility in nasopharyngeal carcinoma. Oncotarget. 2017;8(38):63605–63619. doi: 10.18632/oncotarget.18868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S.B., Liu Y.Y., Yuan L., et al. Autocrine INSL5 promotes tumor progression and glycolysis via activation of STAT5 signaling. EMBO Mol Med. 2020;12(9) doi: 10.15252/emmm.202012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bologna M., Corino V., Calareso G., et al. Baseline MRI-radiomics can predict overall survival in non-endemic EBV-related nasopharyngeal carcinoma patients. Cancers. 2020;12(10) doi: 10.3390/cancers12102958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsang C.M., Lui V.W.Y., Bruce J.P., et al. Translational genomics of nasopharyngeal cancer. Semin Cancer Biol. 2020;61:84–100. doi: 10.1016/j.semcancer.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Kang Y., He W., Ren C., et al. Advances in targeted therapy mainly based on signal pathways for nasopharyngeal carcinoma. Signal Transduct Target Ther. 2020;5(1) doi: 10.1038/s41392-020-00340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campion N.J., Ally M., Jank B.J., et al. The molecular march of primary and recurrent nasopharyngeal carcinoma. Oncogene. 2021;40(10):1757–1774. doi: 10.1038/s41388-020-01631-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y.P., Chan A.T.C., Le Q.T., et al. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 8.Ritter A., Kreis N.N., Louwen F., et al. Molecular insight into the regulation and function of MCAK. Crit Rev Biochem Mol Biol. 2015;51(4):228–245. doi: 10.1080/10409238.2016.1178705. [DOI] [PubMed] [Google Scholar]
- 9.Wei S., Dai M., Zhang C., et al. KIF2C: a novel link between Wnt/β-catenin and mTORC1 signaling in the pathogenesis of hepatocellular carcinoma. Protein Cell. 2020;12(10):788–809. doi: 10.1007/s13238-020-00766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun A., Dang K., Buslig F., et al. Rac1 and Aurora A regulate MCAK to polarize microtubule growth in migrating endothelial cells. J Cell Biol. 2014;206(1):97–112. doi: 10.1083/jcb.201401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichenlaub-Ritter U. Microtubule dynamics and tumor invasion involving MCAK. Cell Cycle. 2015;14(21) doi: 10.1080/15384101.2015.1093813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritter A., Sanhaji M., Friemel A., et al. Functional analysis of phosphorylation of the mitotic centromere-associated kinesin by Aurora B kinase in human tumor cells. Cell Cycle. 2015;14(23):3755–3767. doi: 10.1080/15384101.2015.1068481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura Y., Tanaka F., Haraguchi N., et al. Clinicopathological and biological significance of mitotic centromere-associated kinesin overexpression in human gastric cancer. Br J Cancer. 2007;97(4):543–549. doi: 10.1038/sj.bjc.6603905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimo A., Tanikawa C., Nishidate T., et al. Involvement of kinesin family member 2C/mitotic centromere-associated kinesin overexpression in mammary carcinogenesis. Cancer Sci. 2008;99(1):62–70. doi: 10.1111/j.1349-7006.2007.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa K., Kamohara Y., Tanaka F., et al. Mitotic centromere-associated kinesin is a novel marker for prognosis and lymph node metastasis in colorectal cancer. Br J Cancer. 2008;98(11):1824–1829. doi: 10.1038/sj.bjc.6604379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnjatic S., Cao Y., Reichelt U., et al. NY-CO-58/KIF2C is overexpressed in a variety of solid tumors and induces frequent T cell responses in patients with colorectal cancer. Int J Cancer. 2010;127(2):381–393. doi: 10.1002/ijc.25058. [DOI] [PubMed] [Google Scholar]
- 17.Bie L., Zhao G., Wang Y.P., et al. Kinesin family member 2C (KIF2C/MCAK) is a novel marker for prognosis in human gliomas. Clin Neurol Neurosurg. 2012;114(4):356–360. doi: 10.1016/j.clineuro.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Gan H., Lin L., Hu N., et al. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem Funct. 2019;37(6):424–431. doi: 10.1002/cbf.3420. [DOI] [PubMed] [Google Scholar]
- 19.Yu G., Wang L.G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Morris J.H., Cook H., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vassilev L.T., Tovar C., Chen S., et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103(28):10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Y., Xie M., Lan H., et al. PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int J Biochem Cell Biol. 2013;45(3):645–656. doi: 10.1016/j.biocel.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Bu Y., Suenaga Y., Okoshi R., et al. NFBD1/MDC1 participates in the regulation of G2/M transition in mammalian cells. Biochem Biophys Res Commun. 2010;397(2):157–162. doi: 10.1016/j.bbrc.2010.05.063. [DOI] [PubMed] [Google Scholar]
- 25.Hu L., Chen Q., Wang Y., et al. Sp1 Mediates the constitutive expression and repression of the PDSS2 gene in lung cancer cells. Genes. 2019;10(12) doi: 10.3390/genes10120977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang N., Jiang T., Wang Y., et al. BTG4 is a novel p53 target gene that inhibits cell growth and induces apoptosis. Genes. 2020;11(2) doi: 10.3390/genes11020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Y., Zhu H., Cai W., et al. B-Myb is up-regulated and promotes cell growth and motility in non-small cell lung cancer. Int J Mol Sci. 2017;18(6) doi: 10.3390/ijms18060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta S., den Boon J.A., Chen I.H., et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66(16):7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 29.Bao Y.N., Cao X., Luo D.H., et al. Urokinase-type plasminogen activator receptor signaling is critical in nasopharyngeal carcinoma cell growth and metastasis. Cell Cycle. 2014;13(12):1958–1969. doi: 10.4161/cc.28921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bose S., Yap L.F., Fung M., et al. The ATM tumour suppressor gene is down-regulated in EBV-associated nasopharyngeal carcinoma. J Pathol. 2009;217(3):345–352. doi: 10.1002/path.2487. [DOI] [PubMed] [Google Scholar]
- 31.Lin C., Zong J., Lin W., et al. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-κB and Erk1/2 pathways. J Exp Clin Cancer Res. 2018;37(1) doi: 10.1186/s13046-018-0953-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo W., Gao F., Li S., et al. FoxM1 promotes cell proliferation, invasion, and stem cell properties in nasopharyngeal carcinomsa. Front Oncol. 2018;8 doi: 10.3389/fonc.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi S., Cao X., Gu M., et al. Upregulated expression of SOX4 is associated with tumor growth and metastasis in nasopharyngeal carcinoma. Dis Markers. 2015;2015 doi: 10.1155/2015/658141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.W., Chen T.J., Lin L.C., et al. Overexpression of thymidylate synthetase confers an independent prognostic indicator in nasopharyngeal carcinoma. Exp Mol Pathol. 2013;95(1):83–90. doi: 10.1016/j.yexmp.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Pang J.S., Yen J.H., Wu H.T., et al. Gallic acid inhibited matrix invasion and AP-1/ETS-1-mediated MMP-1 transcription in human nasopharyngeal carcinoma cells. Int J Mol Sci. 2017;18(7) doi: 10.3390/ijms18071354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W.J., Wu S.P., Liu J.B., et al. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013;73(3):1219–1231. doi: 10.1158/0008-5472.CAN-12-1408. [DOI] [PubMed] [Google Scholar]
- 37.Wang M.Y., Lin Z.R., Cao Y., et al. PDZ binding kinase (PBK) is a theranostic target for nasopharyngeal carcinoma: driving tumor growth via ROS signaling and correlating with patient survival. Oncotarget. 2016;7(18):26604–26616. doi: 10.18632/oncotarget.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedrick D.G., Stout J.R., Walczak C.E. Effects of anti-microtubule agents on microtubule organization in cells lacking the kinesin-13 MCAK. Cell Cycle. 2008;7(14):2146–2156. doi: 10.4161/cc.7.14.6239. [DOI] [PubMed] [Google Scholar]
- 39.Hou Y., Zhu Q., Li Z., et al. The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis. 2017;8(3) doi: 10.1038/cddis.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganguly A., Yang H., Cabral F. Overexpression of mitotic centromere-associated Kinesin stimulates microtubule detachment and confers resistance to paclitaxel. Mol Cancer Ther. 2011;10(6):929–937. doi: 10.1158/1535-7163.MCT-10-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganguly A., Yang H., Pedroza M., et al. Mitotic centromere-associated kinesin (MCAK) mediates paclitaxel resistance. J Biol Chem. 2011;286(42):36378–36384. doi: 10.1074/jbc.M111.296483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin S.K., Kyprianou N. Exploitation of the androgen receptor to overcome taxane resistance in advanced prostate cancer. Adv Cancer Res. 2015;127:123–158. doi: 10.1016/bs.acr.2015.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.