Idiopathic hypogonadotropic hypogonadism (IHH) is a rare genetic disease with clinical and genetic heterogeneity. This study aimed to investigate a novel causal gene of IHH and a homozygous mutation (p. Ala515Val) in SEMA4D, and sought to determine the mechanism of SEMA4D promoting GnRH neurons migration. The detailed materials and methods were shown in Supplementary Methods. Combination of bioinformatics, in silico analysis and in vitro analysis indicated the homozygous mutation as a loss-of-function mutation. Functional experiments were conducted to explore SEMA4D modulating GN11 cells migration through SEMA4D/PlexinB1/Met/Rnd1/RhoA/Raf1/MAPK signaling pathway. The results of in vivo experiment demonstrated the reduced population of GnRH neurons at the hypothalamus in Sema4D−/− mice models with normal serum testosterone level, reproductive system, and quality of sperm, consistent with the oligogenic pathogenicity of IHH. In this study, we expanded the genetic spectrum of IHH, and provided theoretical basis for genetic diagnosis and personalized treatment of IHH patients.
IHH was characterized by absence or incomplete development of puberty and infertility caused by deficiency of gonadotropin-releasing hormone (GnRH). IHH patients are likely to recover reproduction through early and proper treatment. Genetic diagnosis is one of the important methods of early diagnosis of IHH. In addition to Mendelian modes of inheritance, more and more studies indicated an even more complex genetic architecture for IHH, often referred to as oligogenicity1,2. The top five genes associated with IHH were FGFR1 (12%), KLA1 (7%), PROKR2 (6%), TACR3 (5%) and GNRHR (4.3%) all over the world 1. In our previous study, the top five genes related to IHH were PROKR2 (15.03%), ANOS1 (10.39%), CHD7 (8.5%), KISS1R (5.41%), FGFR1 (5.23%) and LHB (5.23%) in middle part of China 3. We need to expand the genetic spectrum of IHH and deepen the understanding of genotype-phenotype and genotype-prognosis relationship to perform more effective personalized treatment for IHH patients.
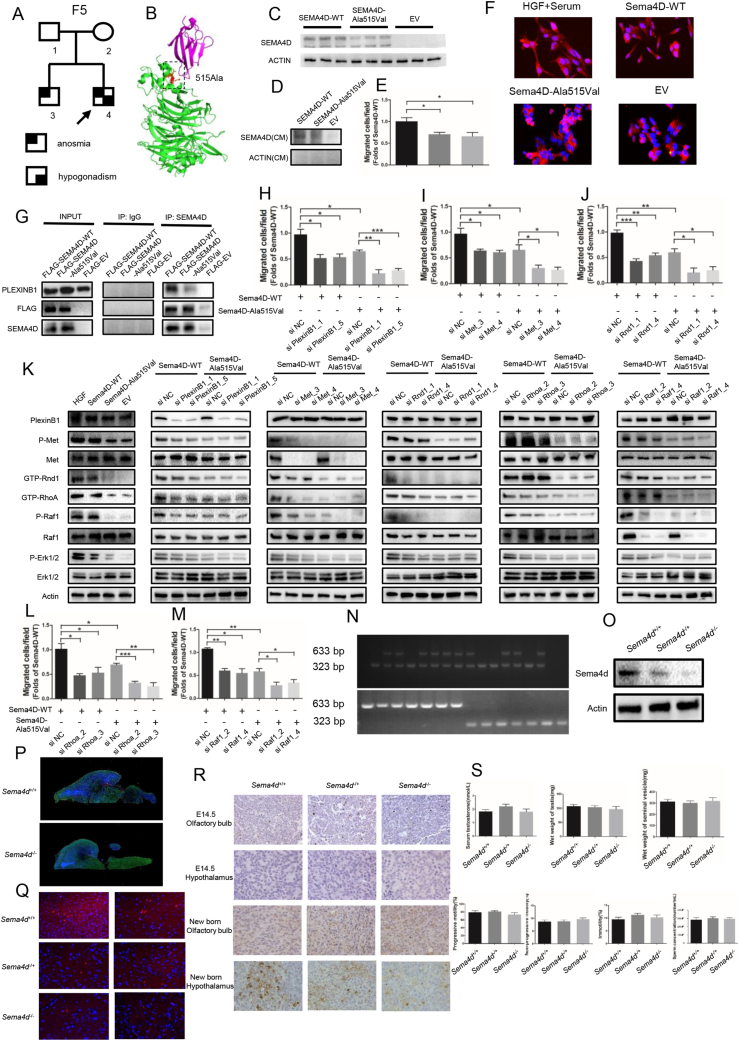
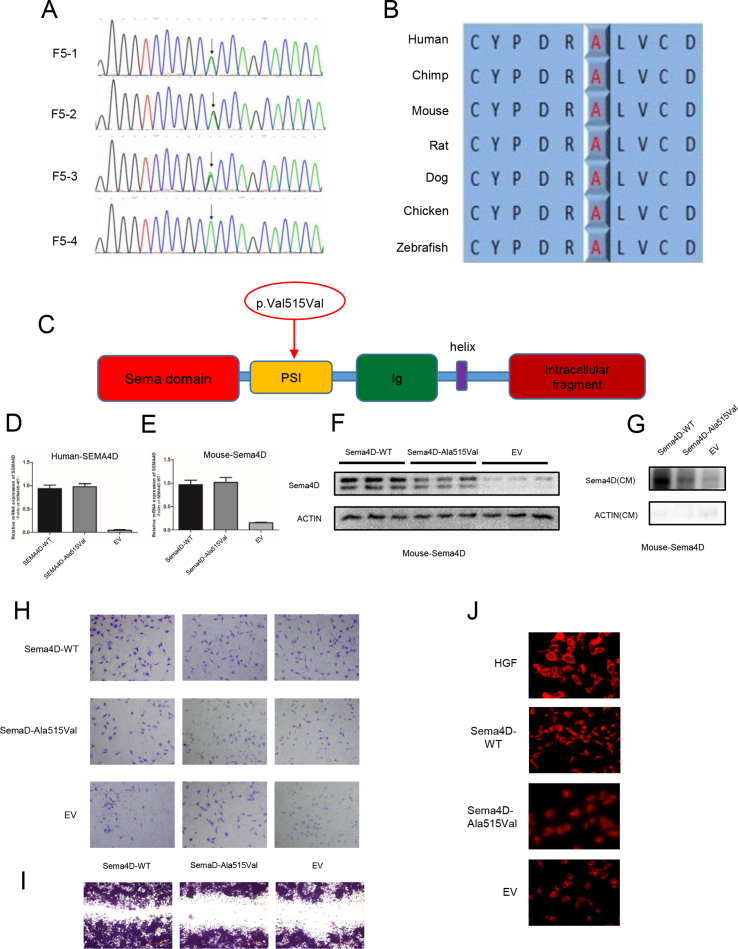
In our previous study, we identified three mutations in SEMA4D (NM_001371194.2, p. Ala515Val, p. Ser788Thr, p. Arg113Gln) from 153 IHH males (shown in Table S1) through target-gene sequence, especially one homozygous mutation (p. Ala515Val, rs75298730) in SEMA4D.3 The mutations were predicted as damaging or disease-causing in online scoring databases (shown in Table S2). The mutation frequency of SEMA4D was 1.96% (3/153), while there was no mutation identified in 100 normal people in our study. The allele frequency of the mutation (p. Ala515Val) was 0.022%–0.06% worldwide, and was 0.113%–0.3% in Asian people, according to these databases (shown in Table S3). The homozygous mutation was inherited from his parents (Fig. 1A; Fig. S1A). The mutant residue was highly conserved across various species, including human, chimpanzee, mouse, rat, dog, chicken and zebrafish (Fig. S1B), and was located at the start part of the α-helix in PSI domain of SEMA4D (Fig. 1B; Fig. S1C).
Figure 1.
SEMA4D acts as a novel oligogenic pathogenic gene of IHH through the PlexinB1/MET/RND1/RHOA/RAF1/MAPK signaling axis. (A) The KS proband (F5-4) carried a homozygous mutation (p. Ala515Val) in SEMA4D inherited from his parents (F5-1, F5-2) who carried the heterozygous mutation (p. Ala515Val) in SEMA4D without clinical manifestations. His brother, F5-3, harbored the heterozygous mutation (p. Ala515Val) in SEMA4D with anosmia, but no hypogonadism. KS, Kallmann syndrome. (B) The mutation (p. Ala515Val) was mapped onto the ribbon diagram of the SEMA4D/PlexinB1 complex (PDB, 3OL2). The SEMA4D protein was colored as green, and the PlexinB1 was colored as magenta. The mutant residue and relevant residues were shown as stick in different color. (C) The protein expression of human wild type and mutant SEMA4D. (D) The protein expression of human wild type and mutant SEMA4D in conditioned medium from 293T cells transfected with Human-SEMA4D-WT and Human-SEMA4D-Ala515Val constructs. (E) Migrated cells in Transwell assay were presented through bar graph. Data are expressed as mean ± standard deviation (n = 3 per group). ∗P < 0.05. (F) Actin and DAPA staining showed the morphology of GN11 cells treated with HGF, Mouse-Sema4D-WT-CM, Mouse-Sema4D-Ala515Val-CM, and EV-CM after serum starve pretreatment. Scale bar = 20 μm. EV, empty vector; WT, wild type; CM, conditioned medium. (G) Pull-Down assay of SEMA4D and PlexinB1 with Input and IgG as control. (H–J) Migrated cells in Transwell assay of GN11 cells transfected with si PlexinB1, si Met and si Rnd1 were presented through bar graph respectively. Data are expressed as mean ± standard deviation (n = 3 per group). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001. (K) Representative Western blot results for PlexinB1, P-Met, Met, GTP-Rnd1, GTP-RhoA, P-Raf1, Raf1, P-Erk1/2 and Erk1/2 in GN11 cells treated with HGF, Mouse-Sema4D-WT-CM, Mouse-Sema4D-Ala515Val-CM, and EV-CM in the first column from left. Representative Western blot results for PlexinB1, P-Met, Met, GTP-Rnd1, GTP-RhoA, P-Raf1, Raf1, P-Erk1/2 and Erk1/2 in GN11 cells transfected with si PlexinB1, si Met, si Rnd1, si RhoA and si Raf1 in Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM treatment groups from the second column on the left to the right respectively. (L, M) Migrated cells in Transwell assay of GN11 cells transfected with si RhoA and si Raf1 were presented through bar graph. Data are expressed as mean ± standard deviation (n = 3 per group). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001. (N) The verification of genotype of experimental mice models. Only one band of 633 bp indicated Sema4D−/− genotype. Only one band of 323 bp indicated Sema4D+/+ genotype. Two bands of 633 bp and 323 bp indicated Sema4D−/+ genotype. (O) Representative Western blot results for SEMA4D in the experimental mice models. (P) The immunofluorescence staining of brain tissue in sagittal plane in new born Sema4D+/+ and Sema4D−/− mice models. DAPI, blue; Actin, Green; GnRH, Red. Scale bars = 100 μm. (Q) The immunofluorescence staining of hypothalamus tissue in new born experimental mice models. DAPI, blue; GnRH, Red. Scale bars = 20 μm. (R) The immunohistochemically staining result of GnRH in olfactory bulb and hypothalamus in E14.5 and new born experimental mice models. (S) The concentration of serum testosterone, wet weight of testis, wet weight of seminal vesicle, the progressive motility of sperm, the non-progressive motility of sperm, the immotility of sperm and the concentration of sperm in adult male mice models respectively (n = 3 per group). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001.
The functional studies showed that the protein expression of mutant SEMA4D (p. Ala515Val) was decreased with normal mRNA level (Fig. 1C; Fig. S1D). The protein level of mutant SEMA4D in conditioned medium was also decreased compared to wild-type SEMA4D (Fig. 1D). We found the same results in COS7 cells transfected with Mouse-Sema4D-WT or Mouse-Sema4D-Ala515Val constructs (Fig. S1E–G). The results of Transwell and scratch assay showed that the migrated GN11 cells were decreased by 40% in Sema4D-Ala515Val group compared to Sema4D-WT group (Fig. 1E; Fig. S1H, S1I). And the morphology of GN11 cells which were pretreated with serum-free medium could recover from shrinkage state in HGF and Sema4D-WT group, while the mutant Sema4D could not rescue the morphology changes of GN11 cells (Fig. 1F). These results indicated the mutation as a loss-of-function mutation.
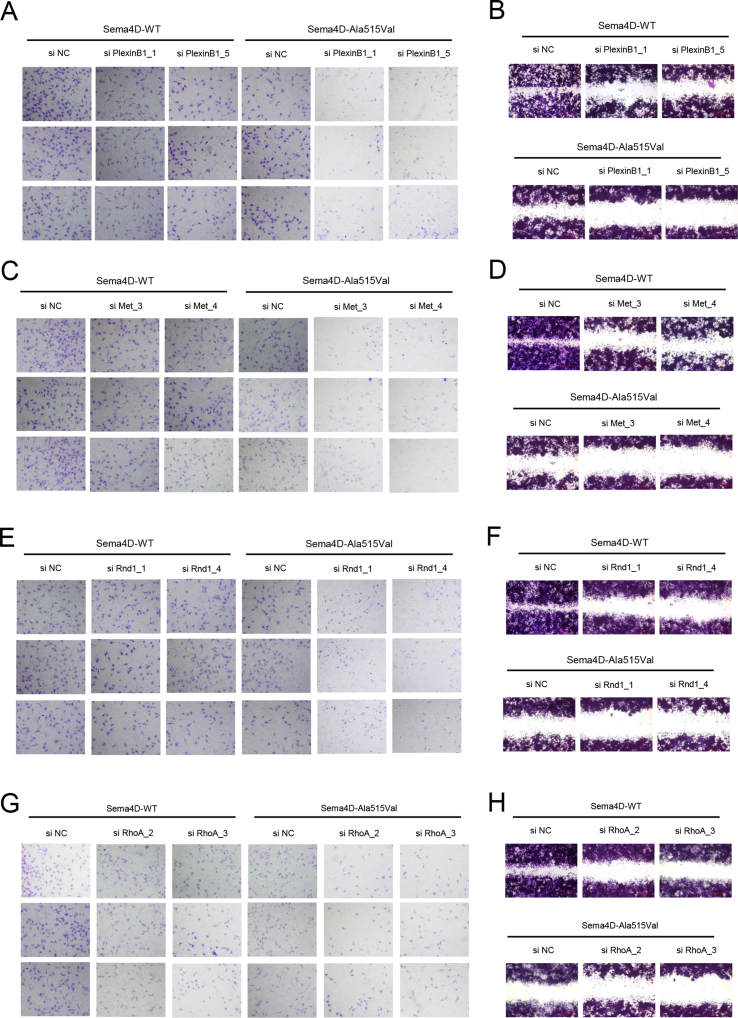
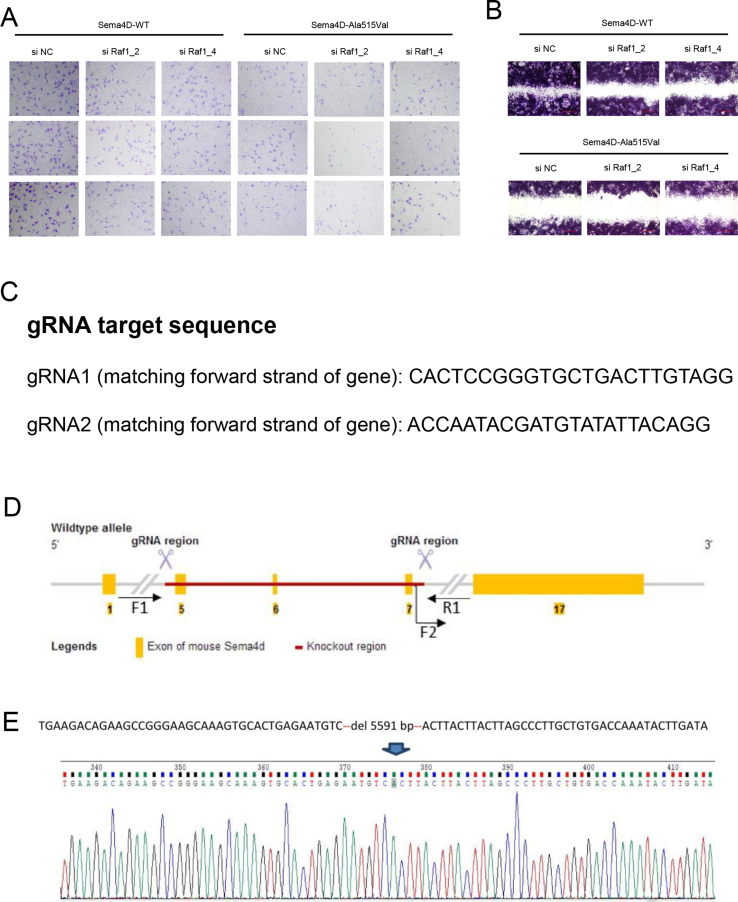
The results of pull-down assay indicated that SEMA4D could bind to PlexinB1, but the mutation damaged the binding capacity with less PlexinB1 pulled down (Fig. 1G). We also found that the phosphorylation of Met was decreased in GN11 cells treated with Mouse-Sema4D-Ala515Val-CM (Fig. S1J). The results of Western blot showed the down-regulated phosphorylation of Met, GTP-Rnd1, GTP-RhoA, phosphorylation of Raf1 and activity of MAPK signaling pathway in GN11 cells under Mouse-Sema4D-Ala515Val-CM treatment (Fig. S1K). Then the expression of PlexinB1, Met, Rnd1, RhoA and Raf1 in GN11 cells was down-regulated by si RNA respectively (shown in Table S4). The results of migration assay indicated the reduced migration of GN11 cells with decreased expression of PlexinB1, Met, Rnd1, RhoA or Raf1 in both Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM groups (Fig. 1H–J, L, M; Fig. S2, S3A, B). At the meantime, we found that the down-stream molecule was suppressed with the up-stream molecule unaffected when PlexinB1, Met, Rnd1, RhoA or Raf1 were down-regulated in Mouse-Sema4D-WT-CM treatment group. But all markers were at low levels in Mouse-Sema4D-Ala515Val-CM treatment group (Fig. 1K).
In in vitro experiment, we found that SEMA4D could bind to receptor PlexinB1, then recruited and phosphorylated Met to activate Rnd1 and RhoA by transferring GDP into GTP, then induced the phosphorylation of Raf1 and activated the MAPK signaling pathway to promote the migration of GN11 cells. And the mutation (p. Ala515Val) in SEMA4D could damage the biological function of SEMA4D in promoting GN11 cells migration.
The Sema4D knock-out mice models were established (Fig. 1N, O). The results of immunofluorescence showed the outline of the brain tissues from Sema4D−/− and Sema4D+/+ mice models (Fig. 1P). We could find that the migrated GnRH neurons at the hypothalamus in new born Sema4D−/− mice models were significantly decreased compared to Sema4D+/+ mice models (Fig. 1P, Q). We found most GnRH neurons were originated in olfactory bulbs of all the mice, while there was nearly no GnRH neuron at the hypothalamus at E14.5 fetal mice. The GnRH neurons were migrated to the hypothalamus from olfactory bulbs in Sema4D+/+ mice models, while the migrated GnRH neurons to the hypothalamus were decreased in Sema4D−/− mice models at new born mice (Fig. S1R). However, we found the concentration of serum testosterone and the development of reproductive system were not different between Sema4D+/+ and Sema4D−/− mice models. Consistently, the quality of sperm was not affected in Sema4D−/− mouse models (Fig. 1S). The results were consistent with the oligogenic pathogenicity of IHH in clinical cohort. All three IHH patients carried other mutations in genes associated with IHH besides mutations in SEMA4D (shown in Table S5). In previous studies, IHH patients were often reported to carry more than one mutant gene. The common gene variants pattern included SEMA3E, SEMA3A, FGFR1, NELF, KAL1, PROKR2 and PROK2 genes 4. There were some other explanations of the results. Mason et al transplanted the tissue with normal GnRH neurons to hypogonadism mice, and found only one-third of neurons survival. But the surviving GnRH neurons were enough to improve the gonadal function of mice. GnRH neuron were powerful, and hundreds of GnRH neurons could maintain the development of reproductive system 5.
In the next phase of investigation, we are going to establish the mice model with both Sema4D and PlexinB1 knock-out, which could better verify the oligogenic pathogenicity of IHH. Furthermore, we will keep feeding and observing these Sema4D knock-out mice model until getting the results of the reproduction.
Author contributions
Daoqi Wang: Conceptualization, Data curation, Formal analysis and Roles/Writing - original draft. Yonghua Niu: Conceptualization, Data curation, and Roles/Writing - original draft. Jiahong Tan: Investigation and Methodology. Jiaxin Wang: Methodology. Le Ling: Validation. Yinwei Chen: Visualization. Jianan Gong: Methodology. Hao Xu: Funding acquisition. Qing Ling: Supervision. Jianhe Liu: Supervision. Jihong Liu: Supervision, Writing - review & editing.
Conflict of interests
The authors declared no conflict of interests.
Funding
The project was supported by the National Natural Science Foundation of China (No. 81671443 to J.L., No. 81601270 to H.X.) and Tongji Hospital Clinical Research Flash Ship Program (China) (No. 2019CR109).
Acknowledgements
We thank Prof. Chuanzhou Li for technical support in samples collection and mouse genotyping; the laboratory of Dr. Kun Qian for statistical analysis; Prof. Pamela Mellon for kindly providing us with GN11 cells; H. Nikki March, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/), for editing the English text of a draft of this manuscript; the Experimental Medicine Research Center of Tongji Hospital for microscopy and image analysis.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.05.030.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
The mutation (p. Ala515Val) in SEMA4D was a loss-of-function mutation. (A) The Sanger sequencing of SEMA4D gene in F5 family showed a homozygous mutation (p. Ala515Val) from the KS proband (F5-4). His parents (F5-1, F5-2) and brother carried the heterozygous mutation (p. Ala515Val) in SEMA4D. (B) Conservation analyses of the residue in SEMA4D protein were performed across different species, including human, chimpanzee, mouse, rat, dag, chicken and zebrafish. (C) The mutation (p. Ala515Val) was shown in the schematic diagram of SEMA4D. (D) The relative mRNA expression of human wild type and mutant SEMA4D. (E) The relative mRNA expression of mouse wild type and mutant SEMA4D. (F) The protein expression of mouse wild type and mutant SEMA4D. (G) The protein expression of mouse wild type and mutant SEMA4D in conditioned medium from COS7 cells transfected with Mouse-Sema4D-WT and Mouse-Sema4D-Ala515Val constructs. (H) Transwell assay of GN11 cells with Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM as lower chamber medium. (I) Scratch assay of GN11 cells treated with Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM. (J) P-Met staining of GN11 cells treated with HGF, Mouse-Sema4D-WT-CM, Mouse-Sema4D-Ala515Val-CM, and EV-CM.
Supplementary Fig. 2.
The migration of GN11 cells was modulated through the PlexinB1/MET/RND1/RHOA/RAF1/MAPK signaling axis. (A, C, E, G) Transwell assay of GN11 cells transfected with si PlexinB1, si Met, si Rnd1 and si RhoA in Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM treatment groups respectively. (B, D, F, H) Scratch assay of GN11 cells transfected with si PlexinB1 si Met, si Rnd1 and si RhoA in Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM treatment groups respectively.
Supplementary Fig. 3.
The gene editing strategy of Sema4D knock out mice model. (A) Transwell assay of GN11 cells transfected with si Raf1in Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM treatment groups. (B) Scratch assay of GN11 cells transfected with si Raf1in Mouse-Sema4D-WT-CM and Mouse-Sema4D-Ala515Val-CM treatment groups. (C) The gRNA target sequence used to knock out Sema4D in mice models. (D) The strategy of the establishment of Sema4D knock-out mice model. Finally, the axon 5, 6, 7 were deleted in Sema4D in genome DNA. (E) Verification of Sema4D knock-out mice model establishment. Total 5591bp nucleotides were deleted.
References
- 1.Balasubramanian R., Crowley W.F., Jr. Isolated GnRH deficiency: a disease model serving as a unique prism into the systems biology of the GnRH neuronal network. Mol Cell Endocrinol. 2011;346(1–2):4–12. doi: 10.1016/j.mce.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sykiotis G.P., Plummer L., Hughes V.A., et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci U S A. 2010;107(34):15140–15144. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou C., Niu Y., Xu H., et al. Mutation profiles and clinical characteristics of Chinese males with isolated hypogonadotropic hypogonadism. Fertil Steril. 2018;110(3):486–495. doi: 10.1016/j.fertnstert.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Cariboni A., André V., Chauvet S., et al. Dysfunctional SEMA3E signaling underlies gonadotropin-releasing hormone neuron deficiency in Kallmann syndrome. J Clin Invest. 2015;125(6):2413–2428. doi: 10.1172/JCI78448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mason A.J., Hayflick J.S., Zoeller R.T., et al. A deletion truncating the gonadotropin-releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science. 1986;234(4782):1366–1371. doi: 10.1126/science.3024317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.