Abstract
The mRNA polyadenylation plays essential function in regulation of mRNA metabolism. Mis-regulations of mRNA polyadenylation are frequently linked with aberrant gene expression and disease progression. Under the action of polyadenylate polymerase, poly(A) tail is synthesized after the polyadenylation signal (PAS) sites on the mRNAs. Alternative polyadenylation (APA) often occurs in mRNAs with multiple poly(A) sites, producing different 3′ ends for transcript variants, and therefore plays important functions in gene expression regulation. In this review, we first summarize the classical process of mRNA 3′-terminal formation and discuss the length control mechanisms of poly(A) in nucleus and cytoplasm. Then we review the research progress on alternative polyadenylation regulation and the APA site selection mechanism. Finally, we summarize the functional roles of APA in the regulation of gene expression and diseases including cancers.
Keywords: Alternative polyadenylation (APA), Cancer, Gene expression, mRNA polyadenylation, Poly(A)
Introduction
Gene expression is the process of synthesizing functional gene products from genetic information. This process is tightly regulated at various stages, including transcription, RNA processing, translation and post-translational modification. Splicing, capping and polyadenylation are the three main steps in processing precursor messenger RNA (pre-mRNA) into mature mRNA.1 Polyadenylation is the last key step in the maturation of mRNA, which involves cutting the 3′-terminal of pre-mRNA and adding poly(A) tail at cleavage site. This is important for the translation efficiency, stability and localization of mRNA.2 Almost all eukaryotic mRNA and many non-coding RNAs (ncRNAs) are polyadenylated. Many eukaryotic genes contain more than one poly(A) site, termed alternative polyadenylation (APA), leading to the production of different mRNA transcripts from the same gene.3,4 In recent years, the polyadenylation and APA have received considerable attention because several studies have revealed that this process plays a key role in a wide range of biological processes and the occurrence and development of diseases.5, 6, 7, 8 Given that previous reviews have provided detailed information on mRNA polyadenylation and poly(A) tail length control,5, 6, 7, 8, 9 here in this review we intend to summarize the recent progress on the functions and mechanisms underlying mRNA APA regulation and its roles in gene expression and diseases including cancers.
The process of polyadenylation
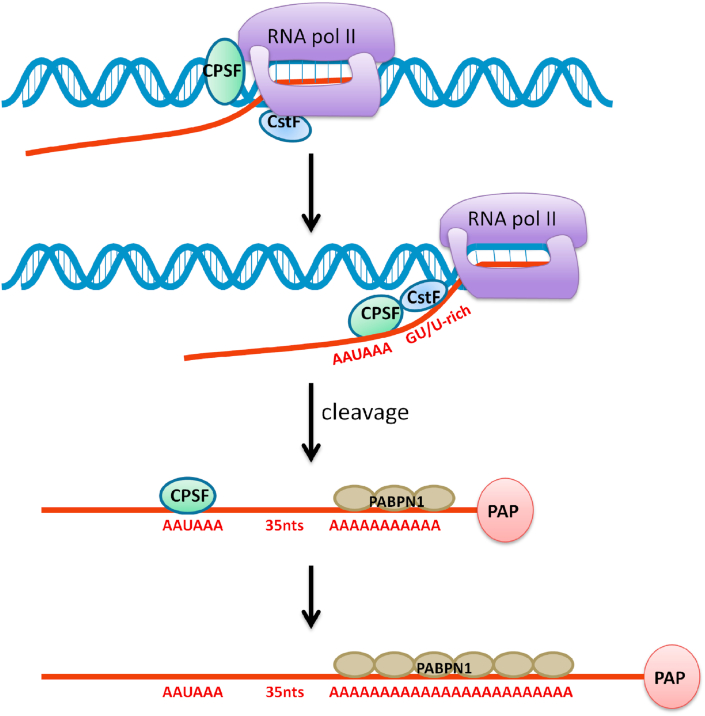
The formation of the mature mRNA 3′-terminal in eukaryotes involves two steps: cleavage and polyadenylation. Polyadenylation is a non-template addition of nucleotide. Generally, 250–300 nt long poly(A) tail is synthesized through the polyadenylation process under the action of polyadenylate polymerase. However, recent studies have shown that poly(A) tail length of most mRNAs in steady state is much shorter than 250 nt, as we would explain later.10 First, the cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF) protein complexes begin to bind at carboxy-terminal domain (CTD) of RNA polymerase II (RNAP II). When RNAP II progresses through the polyadenylation signal (PAS) and CstF is transferred to the new mRNA precursor, CPSF will bind to the PAS, and CstF will bind downstream of PAS. Meanwhile, CFIm complex binds to 20–30 nt upstream of PAS, and Pcf11(a subunit of CFIIm) binds to the CTD of RNAP II.11,12 Then CPSF and CstF initiate cleavage approximately 35 nt after the PAS. Immediately the polyadenylation polymerase (PAP) begins to synthesize poly(A) tails. Almost at the same time, the polyadenylation binding protein in the nucleus (PABPN1) binds to the newly-formed poly(A) sequence. CPSF begins to dissociate, while PAP continues to catalyze polyadenylation and synthesize poly(A) tails. PABPN1 will serve as a molecular ruler defining when polyadenylation stops. Then PAP began to dissociate, and PABPN1 continued to maintain the binding state13 (Fig. 1). In fact, the purified 3′ processing complex contains about 85 proteins, including known core 3′ processing factors and more than 50 proteins that may mediate cross-talk with other processes, but these proteins still need to be further studied.14
Figure 1.
The formation of 3′-end processing machinery. With the help of CPSF, CstF, RNAP II, PAP and other proteins, RNA completes 3′ end splicing and polyadenylation, forming poly(A) tail. PABPN1 binds to adenosine to control the length of the 3′ end.
The control of poly(A) tail length
Most eukaryotic mRNAs have a poly(A) tail generally limited to 250–300 nt. But recent studies have shown that most mRNA steady-state tails are much shorter than 250 nt, averaging between 50 and 100 nt, and the length and composition of tails vary in different creatures.15 During nuclear synthesis of poly(A) tails, CPSF binds to the PAS and recruits PAP, and the first PABPN1 molecule is added to the complex after its minimum binding site (12 nt) is formed. As the poly(A) tail synthesized by PAP is extended, it is covered by more PABPN1 molecules. However, the efficiency of PAP itself is very low, and it is unable to generate a specified length of tail. Only by affecting the activity of PAP through CPSF and PABPN1, can effective polyadenylation and length control be achieved. In the presence of these two factors, the chain length rate can be increased by 300–1000 times through the synergistic stimulation of CPSF and PABPN1. Then, the rapid elongation stops and the tail continues to grow at a low rate.16 When the poly(A) tail exceeds the critical length of 250 nt, the additional PABPN1 molecule can no longer be accommodated in the spherical RNA protein complex. Therefore, the contact between PAP and CPSF was interrupted. During the further elongation of poly(A) tail, PAP is stimulated only by PABPN1, and the synthesis rate is greatly reduced.17
The regulation and site selection mechanism of APA
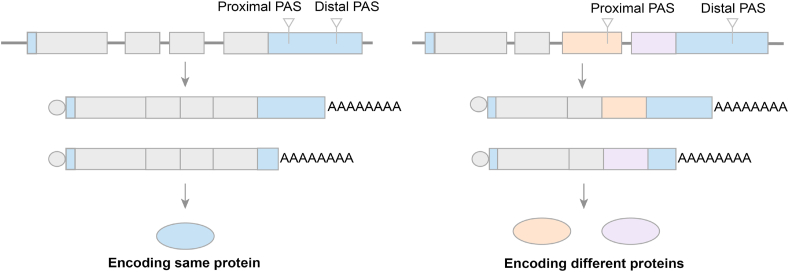
A gene can produce transcripts with multiple PASs, and the different usage of these sites can lead to the formation of different mRNA isoforms, which is called alternative polyadenylation.18 Transcriptome studies have shown that at least 70% of human genes contain multiple PASs, and alternative polyadenylation (APA) is far more common than recognized.19 In recent years, more and more studies have proved that APA plays an important role in regulating gene expression, and APA global deregulation has been found in various human diseases. According to the location of the APA site, APA is divided it into two categories (Fig. 2). Most APA sites are located in the 3′ untranslated region (3′ UTR) (UTR-APA), resulting in transcripts with different lengths of UTRs encoding the same protein. The feature of this type of APA is that it can be regulated globally and affect many transcripts in the cell, controlling the 3′-UTR length and potentially affecting protein abundance. UTR-APA regulates mRNA stability and translation, nuclear export and localization by regulating the targeting of microRNAs (miRNA) and binding to RNA binding proteins (RBPs). When the alternative poly(A) site is located in an internal intron or exon, it's defined as coding region APA (CR-APA). CR-APA controls the transcription of different mRNA isoforms in the coding region, leading to the expression of different protein isoforms. Similar to UTR-APA, CR-APA can also participate in gene regulation through various ways, including protein initiation and repression of gene expression.20
Figure 2.
Types and regulation of APA and mechanism of PAS selection. According to the location of APA sites, APA is divided into two categories. UTR-APA, located in the 3′ UTR, regulates mRNA and protein expression mainly by affecting the length of 3′ UTR. CR-APA, located in the CR, affects mRNA and cause the production of different proteins.
The regulation of APA
-
1
Regulation by 3′-end-processing factors
An important factor involved in APA cleavage is CstF64, which significantly increases the efficiency of proximal cleavage.21 For example, during the transition of B cells into plasma cells, IgM protein changes from a membrane-binding type to a secreted type. This switching is largely caused by the selection of the poly(A) site. Secretory type is produced at the proximal poly(A) site, while membrane-binding type is produced at the distal poly(A) site. Studies have shown that the transition from membrane-binding to secreted IgM is accompanied by a specific increase in CstF64 protein levels.22 Moreover, the upregulation of CstF64 and CstF complex can lead to the selection of poly(A) site from distal to proximal, causing the transition of IgM heavy chain from membrane-binding to secretory type.23
Another important APA regulatory factor is CFIm. Under normal circumstances, CFIm promotes the use of distal PAS, or may inhibit the use of proximal PAS.12 Distal site tends to be canonical PAS, while there are more PAS variants at proximal site. In the case of CFIm25 or CFIm68 depletion, the usage of proximal site increased and the 3′ UTR got shorter. It shows that although 3′-end-processing is completely dependent on CFIm at some canonical poly(A) sites, it is less dependent at some weak and noncanonical poly(A) sites. But CFIm59 knockdown did not induce the same results.12,24, 25, 26 This may be related to the interaction between CFIm59 and U2AF 65. The region 17–47 of U2AF 65 physically associates with CFIm59.27 Contrary to CFIm59 promoting the use of distal sites, U2AF 65 can activate proximal APA sites. When CFIm59 is knocked down, it may affect the promoting effect of U2AF 65 on the use of proximal sites.28
CPSF and CFII respectively participate in APA regulation through Fip1 and Pcf11.29 They both can promote the use of the proximal poly(A) site. Among them, Fip1 is particularly important for the self-renewal and somatic reprogramming of embryonic stem cells, and Pcf11 is also found to have an important influence on APA of metazoan cells.30 In addition, several poly(A) binding proteins also play an important role in APA regulation. For example, PABPN1 has been proved to be associated with the use of proximal sites. Knockout of PABPN1 promoted the use of proximal sites.30 PABPC1 plays a similar role in facilitating the use of distal site, with 3′ UTR shortened by knockout.31 CPEB1, in contrast, increases the use of proximal sites because it can assist CPSF to recruit to weaker DSEs.32
-
2
Regulation by transcription
The interaction between transcription and the 3′-end processing mechanism increases the poly(A) tail cutting efficiency and then promotes the use of proximal sites.33,34 Transcription elongation factor PAF1C and ELL2 can improve the utilization ratio of proximal site.35 And as confirmed by the analysis of the gene transcriptome, enhance of transcription activity can promote the cleavage of the proximal site. Because the proximal poly(A) sites are transcribed earlier than the distal sites, the 3′-processing machinery encounters them first, and they have an advantage over the distal poly(A) sites. Therefore, the use of proximal poly(A) sites should be positively correlated with the distance between adjacent poly(A) sites, and negatively correlated with transcription elongation.5 In addition, RNAP II can also influence the selection of poly(A) sites. Studies have shown that reducing the kinetics of RNAP II elongation can lead to increased use of the proximal poly(A) site in a large number of transcripts. This may be due to the fact that slower RNAP II will expose the proximal poly(A) signal to the 3′-processing complex for a longer time before transcription at the second poly(A) site. The phenomenon that RNAP II can also promote proximal PAS use if it is paused also supports this claim.36 Global analysis shows that, when genes are expressed at high level, short 3ʹUTR mRNA isoform tend to be richer, RNAP II will pause more frequently at the proximal PAS, therefore regulating the poly(A) site selection.
-
3
APA regulation by splicing
RNA splicing also plays important function in regulation of APA. In the case of intronic PASs, splicing and polyadenylation may compete with each other. In introns, the possibility of polyadenylation at intronic PAS is greatly increased if the 5′-cleavage site is weak.37 When the use of the 3′UTR proximal poly(A) site is increased, the cleavage at the intron poly(A) site is also increased. There are multiple protein interactions between core splicing factor and core polyadenylation factor, such as U1 snRNP and CPSF160, U2 snRNP and the CPSF complex, and U2AF and the CFI complex. Recent studies have shown that U1 snRNP, independent of its role in splicing, widely promoted the use of intron poly(A) sites and protected pre-mRNAs from premature cleavage.38 Moreover, the effect of U1 snRNP is dose-dependent. When the level of U1 snRNP decreases gently, the shift towards the proximal 3′UTR cleavage site will happen instead of using the upstream intron cleavage site. U2 snRNP mediates CPSF loading to pre-mRNA, and U2 snRNP cofactor (U2AF) interacts with CFIm59, both of which stimulated cleavage and polyadenylation.28
-
4
The APA and chromatin
Previous research has found that, nucleosomes sharply decreased at PAS, while it increased significantly at the downstream of PAS, which may be due to the low intrinsic affinity of A-rich sequences for nucleosomes.39,40 This phenomenon is also observed in the cases of multiple poly(A) sites, in which the stronger poly(A) site shows more obvious depletion of PAS nucleosomes and enrichment of downstream nucleosomes, indicating a correlation between nucleosome localization and the use of PAS site.41 But the specific relationship remains to be further studied. In addition, chromatin may also affect APA through DNA methylation.42 For example, some genes are not methylated on paternally-derived chromosomes and are highly methylated on maternally-derived chromosomes, unmethylated genes use upstream poly(A) sites, and methylated genes use downstream poly(A) site,42 suggesting that DNA methylation may play a role in the regulation of APA.43
-
5
APA and tissue specificity
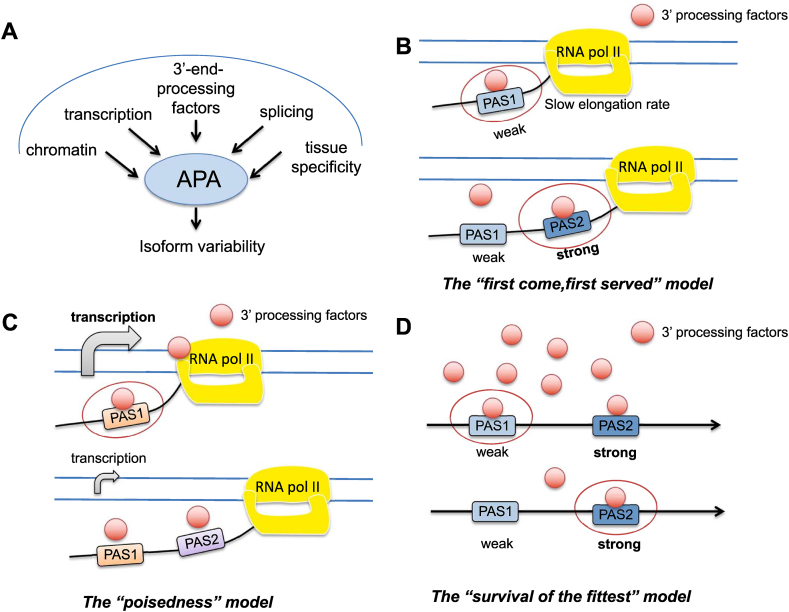
A large amount of evidence shows that APA exhibits certain tissue specificity. For example, transcripts in the nervous system and brain are characterized by the preferential use of the distal poly(A) site (which generates isoforms with a longer 3′UTRs), whereas transcripts tend to use more proximal sites in the placenta, ovary, and blood.44,45 The APA selection is much more similar in the same tissues of different species than between different tissues of the same species. Few tissue specific accessory proteins have been found capable of controlling APA. And changes in core polyadenylated protein levels seem to play a significant role. However, in metazoan or mammalian tissues, there are only a few physiological changes in the levels of core polyadenylated proteins, and the role of them may be limited. The underlying causes of APA tissue specificity remain to be further investigated46 (Fig. 3A).
Figure 3.
Regulation of APA and mechanism of PAS selection. (A) 3ʹ-end-processing factors, transcription, splicing and structure of chromatin can affect APA. And APA has tissue specificity. (B) When the elongation rate is slow, the proximal site has an obvious time advantage over the distal site because it is recognized by related factors first, so the possibility of selecting the proximal site increases. (C) APA tends to select proximal sites when transcriptional activity is high. Highly-expressed genes often own a shorter 3′UTR. (D) The distal site is stronger, while the proximal site is weaker. Only when 3ʹ-end-processing factor concentration is high, mRNA will choose the proximal site. The arrows represent transcription. The size of the arrow represents the rate of transcriptional elongation.
Mechanism of poly(A) site selection
There is still no definite conclusion about the selection mechanism of PASs in specific environments. But several models have been proposed. Here, we introduce three of them. The first is the “first come, first served” model (Fig. 3B). Because of its position, the proximal site is first transcribed, so there is more time for cleavage and polyadenylation, leading to an advantage over distal PASs. This proximal dominance is influenced by transcriptional elongation. When the elongation rate is slow, the time advantage is more obvious and the possibility of selecting proximal site is increased.51 It should be noted that there is no direct relationship between the transcriptional activity of genes and the transcriptional elongation. For transcriptional activity, there is a “poisedness” model (Fig. 3C). Transcription factors can regulate the efficiency of 3′-processing factor cotranscriptional re-cruitment, which in turn regulated APA.9 Numerous reports have shown that the expression of stronger promoters leads to a preference for proximal site, and the higher expression of genes tends to have shorter 3′UTRs.47
Another important model is the “survival of the fittest'' (Fig. 3D). In most cases, the level of core 3′-processing factors in proliferating cells is much higher than that in differentiated or resting cells. As mentioned above, distal poly(A) site preferentially uses the canonical PAS, whereas the prevalence of PAS variants increases at proximal poly(A) site. The distal site has stronger cis-elements than the proximal site and is more conserved. Therefore, the distal site is stronger than the proximal site. Only when 3′-end-processing factors are high, mRNA chooses proximal site. The actual sites of cleavage and polyadenylation selected are strongly influenced by nuclear cleavage and polyadenylation factors, as well as other protein concentrations.36 Therefore, in proliferating cells, it is more likely to recognize weaker proximal sites, while in differentiated or resting cells with insufficient 3′-end-processing factors, distal sites are preferentially selected. None of the current mechanisms can perfectively explain the poly(A) selection mechanism. Whether the selection of poly(A) site is mediated by multiple mechanisms or there is unknown mechanism still needs further study.37,48
The functions of APA
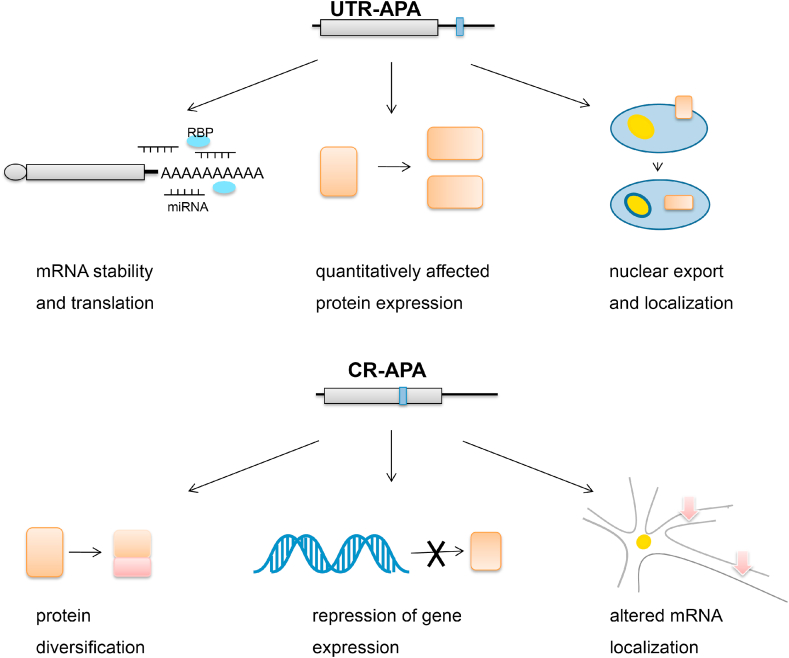
APA, as a post-transcriptional event, plays an important role in gene expression in eukaryotes. APA can influence the expression of genes containing multiple poly(A) sites by changing the length and composition of 3′UTR and affecting mRNA metabolism and protein expression, thereby affecting cell phenotype and even tissue and organ functions. The UTR-APA and CR-APA differ in terms of their functions in gene expression regulation. The UTR-APA mainly affects 3′UTR and quantitatively affects protein expression. The CR-APA not only influences the length and composition of 3′UTR, but also plays an important role in protein diversification and repression of gene expression. In addition, recent studies have found that UTR-APA can regulate protein localization independently of mRNA localization,49 while CR-APA changes mRNA localization due to the inclusion of different terminal exons50 (Fig. 4).
Figure 4.
Functions of different APA. APA can influence mRNA and protein through a variety of regulation in mRNA stability and localization, protein types and expression levels etc. The two types of APA are different in functions.
APA affects mRNA stability and translation mainly through miRNA and RBPs. miRNAs are small RNAs (22 nt) that regulate the stability and/or translation of their target complementary mRNAs. In mammals, more than half of the conserved miRNA targets are located in 3′UTRs. miRNA targeting ability is usually affected by the location of the targeted point in mRNA and surrounding sequence, and the targeted point that located at the ends of 3ʹUTR are more efficient than that in the middle. Therefore, in the case of short 3′UTRs, miRNA has a better working environment and stronger targeting efficiency.51 In addition to a large number of miRNA targets, there are also a lot of 3′UTR mRNA destabilization elements, such as AU-rich elements (AREs), GU-rich elements (GREs) and PUF protein-binding elements, etc. When the length and/or composition of the 3′UTR change, these mRNA destabilization elements are included or excluded, and then the combination of RBP will be affected, leading to the change of mRNA stability and translation.52
Some 3′UTR APA isoforms show differences in the abundance between nucleus and cytoplasm. There are more long 3′UTR isoforms enriched in the nucleus than in the cytoplasm.53,54 Although it has not been determined whether this is more influenced by mRNA stability or mainly because of the influence of APA on protein localization, APA plays a role in mRNA nuclear export and localization. In addition, even if the same protein is encoded, the mRNAs with different 3′UTR may lead to different protein subcellular localization. For example, there are two brain-derived neurotrophic factor (BDNF) transcripts in the brain that encode the same protein but have different lengths of 3′UTR. It was observed that BDNF mRNA with a long 3′UTR preferentially targets dendrites. And when three strong poly(A) sites are inserted behind the BDNF poly(A) site, which results in the shortening of its 3′UTR, the dendritic BDNF is significantly reduced.55 APA affects mRNA localization, which in turn can promote localized protein translation, thereby facilitating rapid local protein synthesis.48
UTR-APA can regulate protein localization independently of mRNA localization. At present, this mechanism has been found in CD47, CD44, ITGA1 and members of TNFRSF13C. In the case of CD47, the gene encoding the membrane protein CD47 has an alternative 3′UTR. CD47 from the longer isoform was highly expressed on the cell membrane, while CD47 from the shorter isoform was mainly localized in the endoplasmic reticulum. The reason is that 3′UTR of mRNA that encoding CD47 contains RBP HUR (Hu antigen R), and CD47 can be transferred to the cell membrane through the interaction between HUR and the SET of the translation site. The short mRNA isoforms lacked the sequences needed to assemble the HUR-SET complex, leading to CD47 localization primarily to the endoplasmic reticulum.56
In addition, CR-APA can affect protein expression and produce a variety of proteins through the use of different PAS. Previous studies have shown that protein truncation resulting from polyadenylation of protein introns can significantly affect full-length counterpart function. For example, retinoblastoma-binding protein 6 (RBBP6) can produce 4 isoforms via differential RNA processing.57 One of the isoforms come from the use of an intron PAS, which produces a truncated protein called Iso3. Iso3 will compete with RBBP6 and affect the function of RBBP6.58,59 Changes in mRNA localization by CR-APA can also be achieved by including different terminal exons. In Berkovits's study, the alternative last exons ALE isoform was located in neural projection in 80% of pairs with significant differential localization. Isomorphic analysis of different neuronal localization systems revealed associations between ALE isomorphic subcellular localization and APA.56
Abnormal APA regulation in tumors
Although the extent and consequences of all APA events are unclear in both non-cancerous and cancerous cells, it is certain that APA events contribute to the development and/or maintenance of tumors. Justin Brumbaugh et al. found that there is a subset of APA genes regulated by CFIm25 in glioblastoma. After knocking down CFIm25, the 3′UTRs of at least 1450 genes were shortened. And with the downregulation of CFIm25 in glioblastoma cells, tumorigenicity enhanced and tumor size increased.60 In addition, a recent study focused on single-cell APA for different cell types in AML revealed that many leukemia cell marker genes such as NF-κB, GATA2 and IAP-family genes exhibit an APA variation, which affects the proliferation and differentiation of leukemia bone marrow mononuclear cells.61
About 70% miRNA targets and about 11% AREs of human protein-coding genes are located in 3′UTR.62 In a systematic survey of global APA landscape, overall APA was found to be shorter in tumor samples than in matched normal samples, and APA was more widely shortened in cancer cell lines than in tumor samples.63 The shortening of 3′UTR leads to decreasing of miRNA binding sites and disappearing of some mRNA destabilization elements. In normal cells, the proto-oncogene uses distal PAS in the protein coding region, and transcripts are normally regulated by miRNAs and/or RBPs.59 With the occurrence of APA events, once the cell selects proximal PAS to generate a short 3′UTR, it is possible to eliminate the binding sites of miRNA and/or RBP, resulting in the loss of normal control of the mRNA and inducing carcinogenesis.64 A study in breast cancer found that the number of miRNA targets in the alternative UTR (aUTR) region, which exists only in long isoforms, can affect the ratio of long and short isoforms (LSR) of target genes. The target sites of up-regulated miRNAs often appear in aUTR. And the genes whose aUTRs were targeted by up-regulated miRNAs in cancer cells had an overall lower LSR. The reduction of LSR of aUTR target genes of these abnormally up-regulated miRNAs is dose-dependent on the number of target sites.65 This result suggests that the main reason for LSR reduction may be degradation of long isoforms, and miRNAs may play a key role in the transition from normal to carcinogenic or proliferative status of APA patterns.
APA events can affect the expression of genes through protein expression and the production of truncated proteins, and promote the occurrence of tumors. For example, ACTH produced by non-pituitary tumors generally cannot be suppressed by exogenous glucocorticoids. Studies have shown that DMS-79 cells derived from an ectopic ACTH-producing tumor express abnormal glucocorticoid receptor (GR) mRNA. Due to the occurrence of intron APA, the protein encoded by this mRNA lacks a steroid-binding domain and cannot function as a transcription factor activated by a ligand. The APA event in GR gave DMS-79 cells the ability to resist glucocorticoids.66
Furthermore, Justin Brumbaugh et al. found that RNA-processing factor Nudt21 directs differential polyadenylation of over 1500 transcripts in cells acquiring pluripotency, which were strongly enriched for cancer-associated chromatin regulators. It supports a possible role of APA in driving tumorigenesis via affecting chromatin signaling.67 Besides, one of the characteristics of cancer cells is that they prefer aerobic glycolysis rather than the mitochondrial tricarboxylic acid cycle, which is commonly referred to as the Warburg effect. This effect can promote the uptake and incorporation of nutrients into the biomass.68 In some cancer cell lines, genes with shortened 3′UTRs were found to be involved in metabolism, glucose import and regulation of transport. The transfer of these genes to the proximal sites of APA may be related to the Warburg effect of tumor cells.69
However, not in all tumor cells, APA tends to select the proximal poly(A) site and cause the 3′UTR to shorten. For example, in a study of breast cancer APA sites, it was found that the estrogen-sensitive breast cancer cell line MCF7 tends to shorten 3′UTRs, while estrogen-independent and highly invasive MB231 prefers distal poly(A) sites. In the long 3′UTR gene list of MB231 cell line, 30 genes are related to apoptosis or programmed cell death. In addition, genes involved in the caspase pathway were found to be enriched in MB231, and four downstream genes (Caspase 6, DFFA, DFFB and PARP1) were switched to the distal APA site. The extension of these genes by 3′UTRs may help to avoid the apoptosis of MB231 cells.70 This finding indicated that the regulation and function of APA in tumors is very complicated. The transformation of normal cells into cancer cells could be an evolutionary dynamic process driven by mutations of some driving factors under the action of various environmental selection forces.71 Although APA plays a certain role in the occurrence and development of tumors, this effect on the overall environment is closely related. MCF7 is estrogen sensitive, and MB231 is estrogen independent and highly aggressive.72 The two are different at different states on the adaptive landscape, which may have caused the difference in APA events.
Besides, APA may have great potential in the treatment and clinical judgment of tumors.73 At least 214 antitumor drugs have been identified that are closely related to APA events, particularly pan-histone deacetylase (HDAC) inhibitors and DNA topoisomerase I inhibitors. APA events play a key role in the response to antitumor drugs. And 345 different APA events related to tumor isoforms, 66 related to tumor staging, and 1707 related to overall survival were identified in paired normal and tumor samples.74 For example, the shortening of CSNK1D transcript is associated with poor survival at KIRP,75 and CRTC1's shortening is associated with poor survival in LGG.76 These findings may provide an important clinical basis for diagnosis, treatment, and prognosis of cancer patients. The regulation and role of APA in tumor remains to be further studied in the future.
Concluding remarks
With the development of whole transcriptome sequencing, the regulatory mechanisms and functional effects of polyadenylation and APA are being revealed at an unprecedented rate. We began to gain a clearer understanding of the polyadenylation process and its key factors, poly(A) length control and influence, the way APA is regulated, APA function, and APA's role in tumors. The regulation of APA, in particular, is a complex process involving 3′-end-processing proteins, transcription, splicing, chromatin structure, tissue specificity and other factors. A deeper understanding of polyadenylation and APA can provide new information on the regulation of gene expression and achieve a more comprehensive and systematic understanding of gene expression. Abnormal APA isoforms detected in cancer without alteration of the proto-oncogene will contribute to the discovery of potential cancer-related genes and pathways. More importantly, a better understanding of polyadenylation and APA opens the possibility of developing new diagnostic and therapeutic approaches. Cancer type-specific APA may provide new targets for diagnosis and prognosis, which may help optimize patient outcomes using sophisticated medical methods.
Conflict of interests
All authors declare that they have no conflict of interests.
Funding
This research was supported by grants from National Natural Science Foundation of China (No. 81922052, 81974435, 81772999), the Natural Science Foundation of Guangdong Province, China (No. 2019B151502011), and the Guangzhou People's Livelihood Science and Technology Project, China (No. 201903010006).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Yeh H.S., Yong J. Alternative polyadenylation of mRNAs: 3'-untranslated region matters in gene expression. Mol Cell. 2016;39(4):281–285. doi: 10.14348/molcells.2016.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25(17):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian B., Hu J., Zhang H., et al. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Hu J., Recce M., et al. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res. 2005;33:D116–D120. doi: 10.1093/nar/gki055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Giammartino D.C., Nishida K., Manley J.L. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43(6):853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra M., Johnson E.L., Swamy V.S., et al. Alternative polyadenylation factors link cell cycle to migration. Genome Biol. 2018;19(1) doi: 10.1186/s13059-018-1551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner R.E., Pattison A.D., Beilharz T.H. Alternative polyadenylation in the regulation and dysregulation of gene expression. Semin Cell Dev Biol. 2018;75:61–69. doi: 10.1016/j.semcdb.2017.08.056. [DOI] [PubMed] [Google Scholar]
- 8.Stewart M. Polyadenylation and nuclear export of mRNAs. J Biol Chem. 2019;294(9):2977–2987. doi: 10.1074/jbc.REV118.005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18(12):2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandel C.R., Bai Y., Tong L. Protein factors in pre-mRNA 3'-end processing. Cell Mol Life Sci. 2008;65(7–8):1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner R.E., Henneken L.M., Liem-Weits M., et al. Requirement for cleavage factor IIm in the control of alternative polyadenylation in breast cancer cells. RNA. 2020;26(8):969–981. doi: 10.1261/rna.075226.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy J.G., Norbury C.J. Cleavage factor Im (CFIm) as a regulator of alternative polyadenylation. Biochem Soc Trans. 2016;44(4):1051–1057. doi: 10.1042/BST20160078. [DOI] [PubMed] [Google Scholar]
- 13.Chen W., Jia Q., Song Y., et al. Alternative polyadenylation: methods, findings, and impacts. Dev Reprod Biol. 2017;15(5):287–300. doi: 10.1016/j.gpb.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y., Di Giammartino D.C., Taylor D., et al. Molecular architecture of the human pre-mRNA 3' processing complex. Mol Cell. 2009;33(3):365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller R.W., Kühn U., Aragón M., et al. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J Mol Biol. 2000;297(3):569–583. doi: 10.1006/jmbi.2000.3572. [DOI] [PubMed] [Google Scholar]
- 16.Winter R., Kühn U., Hause G., et al. Polyalanine-independent conformational conversion of nuclear poly(A)-binding protein 1 (PABPN1) J Biol Chem. 2012;287(27):22662–22671. doi: 10.1074/jbc.M112.362327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangus D.A., Evans M.C., Agrin N.S., et al. Positive and negative regulation of poly(A) nuclease. Mol Cell Biol. 2004;24(12):5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beisang D., Bohjanen P.R. Perspectives on the ARE as it turns 25 years old. Wiley Interdiscip Rev RNA. 2012;3(5):719–731. doi: 10.1002/wrna.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoque M., Ji Z., Zheng D., et al. Analysis of alternative cleavage and polyadenylation by 3' region extraction and deep sequencing. Nat Methods. 2013;10(2):133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian B., Manley J.L. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18(1):18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao C., Biesinger J., Wan J., et al. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci U S A. 2012;109(46):18773–18778. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagaki Y., Seipelt R.L., Peterson M.L., et al. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87(5):941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 23.Takagaki Y., Manley J.L. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2(6):761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 24.Kubo T., Wada T., Yamaguchi Y., et al. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3'-UTRs. Nucleic Acids Res. 2006;34(21):6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruber A.R., Martin G., Keller W., et al. Cleavage factor Im is a key regulator of 3' UTR length. RNA Biol. 2012;9(12):1405–1412. doi: 10.4161/rna.22570. [DOI] [PubMed] [Google Scholar]
- 26.Kim S., Yamamoto J., Chen Y., et al. Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Gene Cell. 2010;15(9):1003–1013. doi: 10.1111/j.1365-2443.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 27.Millevoi S., Loulergue C., Dettwiler S., et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3' end processing machineries. EMBO J. 2006;25(20):4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kralovicova J., Knut M., Cross N.C., et al. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3' splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res. 2015;43(7):3747–3763. doi: 10.1093/nar/gkv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guéguéniat J., Dupin A.F., Stojko J., et al. Distinct roles of Pcf11 zinc-binding domains in pre-mRNA 3'-end processing. Nucleic Acids Res. 2017;45(17):10115–10131. doi: 10.1093/nar/gkx674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lackford B., Yao C., Charles G.M., et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33(8):878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., You B., Hoque M., et al. Systematic profiling of poly(A)+ transcripts modulated by core 3' end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 2015;11(4) doi: 10.1371/journal.pgen.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bava F.A., Eliscovich C., Ferreira P.G., et al. CPEB1 coordinates alternative 3'-UTR formation with translational regulation. Nature. 2013;495(7439):121–125. doi: 10.1038/nature11901. [DOI] [PubMed] [Google Scholar]
- 33.Gruber A.J., Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20(10):599–614. doi: 10.1038/s41576-019-0145-z. [DOI] [PubMed] [Google Scholar]
- 34.Chandrasekaran V., Juszkiewicz S., Choi J., et al. Mechanism of ribosome stalling during translation of a poly(A) tail. Nat Struct Mol Biol. 2019;26(12):1132–1140. doi: 10.1038/s41594-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martincic K., Alkan S.A., Cheatle A., et al. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10(10):1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Z., Luo W., Li W., et al. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elkon R., Drost J., van Haaften G., et al. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012;13(7) doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaida D., Berg M.G., Younis I., et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468(7324):664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal E., Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19(1):65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson H.C., Finch J.T., Luisi B.F., et al. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 41.Spies N., Nielsen C.B., Padgett R.A., et al. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36(2):245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood A.J., Roberts R.G., Monk D., et al. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3(2) doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowley M., Wood A.J., Böhm S., et al. Epigenetic control of alternative mRNA processing at the imprinted Herc3/Nap1l5 locus. Nucleic Acids Res. 2012;40(18):8917–8926. doi: 10.1093/nar/gks654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang E.T., Sandberg R., Luo S., et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H., Lee J.Y., Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6(12) doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacDonald C.C. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond (2018 update) Wiley Interdiscip Rev RNA. 2019;10(4) doi: 10.1002/wrna.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan S., Choi E.A., Shi Y. Pre-mRNA 3'-end processing complex assembly and function. Wiley Interdiscip Rev RNA. 2011;2(3):321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Liu L., Qiu Q., et al. Alternative polyadenylation: methods, mechanism, function, and role in cancer. J Exp Clin Cancer Res. 2021;40(1) doi: 10.1186/s13046-021-01852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereira-Castro I., Moreira A. On the function and relevance of alternative 3’-UTRs in gene expression regulation. Wiley Interdiscip Rev RNA. 2021;12(5) doi: 10.1002/wrna.1653. [DOI] [PubMed] [Google Scholar]
- 50.Taliaferro J.M., Vidaki M., Oliveira R., et al. Distal alternative last exons localize mRNAs to neural projections. Mol Cell. 2016;61(6):821–833. doi: 10.1016/j.molcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandberg R., Neilson J.R., Sarma A., et al. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 53.Neve J., Burger K., Li W., et al. Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 2016;26(1):24–35. doi: 10.1101/gr.193995.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35(4):467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An J.J., Gharami K., Liao G.Y., et al. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134(1):175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berkovits B.D., Mayr C. Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522(7556):363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mbita Z., Hull R., Mbele M., et al. Expression Analysis of RbBP6 in human cancers: a Prospective biomarker. Anti Cancer Drugs. 2019;30(8):767–773. doi: 10.1097/CAD.0000000000000809. [DOI] [PubMed] [Google Scholar]
- 58.Vorlová S., Rocco G., Lefave C.V., et al. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell. 2011;43(6):927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Giammartino D.C., Li W., Ogami K., et al. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3' UTRs. Genes Dev. 2014;28(20):2248–2260. doi: 10.1101/gad.245787.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masamha C.P., Xia Z., Yang J., et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510(7505):412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye C., Zhou Q., Hong Y., et al. Role of alternative polyadenylation dynamics in acute myeloid leukaemia at single-cell resolution. RNA Biol. 2019;16(6):785–797. doi: 10.1080/15476286.2019.1586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Y., Li Z., Ozsolak F., et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40(17):8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiang Y., Ye Y., Lou Y., et al. Comprehensive characterization of alternative polyadenylation in human cancer. J Natl Cancer Inst. 2018;110(4):379–389. doi: 10.1093/jnci/djx223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu Y., Chen L., Chen C., et al. Crosstalk between alternative polyadenylation and miRNAs in the regulation of protein translational efficiency. Genome Res. 2018;28(11):1656–1663. doi: 10.1101/gr.231506.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liaw H.H., Lin C.C., Juan H.F., et al. Differential microRNA regulation correlates with alternative polyadenylation pattern between breast cancer and normal cells. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garaulet D.L., Zhang B., Wei L., et al. miRNAs and neural alternative polyadenylation specify the virgin behavioral state. Dev Cell. 2020;54(3):410–423. doi: 10.1016/j.devcel.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brumbaugh J., Di Stefano B., Wang X., et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell. 2018;172(1–2):106–120. doi: 10.1016/j.cell.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brooks G.A. The science and translation of lactate shuttle theory. Cell Metabol. 2018;27(4):757–785. doi: 10.1016/j.cmet.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 69.El Hassouni B., Granchi C., Vallés-Martí A., et al. The dichotomous role of the glycolytic metabolism pathway in cancer metastasis: interplay with the complex tumor microenvironment and novel therapeutic strategies. Semin Cancer Biol. 2020;60:238–248. doi: 10.1016/j.semcancer.2019.08.025. [DOI] [PubMed] [Google Scholar]
- 70.Fu Y., Sun Y., Li Y., et al. Differential genome-wide profiling of tandem 3' UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21(5):741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fragata I., Blanckaert A., Dias Louro M.A., et al. Evolution in the light of fitness landscape theory. Trends Ecol Evol. 2019;34(1):69–82. doi: 10.1016/j.tree.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 72.Cho Y., Kang H.G., Kim S.J., et al. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ. 2018;25(10):1781–1795. doi: 10.1038/s41418-018-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen H., Yao J., Bao R., et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer. 2021;20(1) doi: 10.1186/s12943-021-01322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curinha A., Oliveira Braz S., Pereira-Castro I., et al. Implications of polyadenylation in health and disease. Nucleus. 2014;5(6):508–519. doi: 10.4161/nucl.36360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosenberg L.H., Lafitte M., Quereda V., et al. Therapeutic targeting of casein kinase 1 delta in breast cancer. Sci Transl Med. 2015;7(318) doi: 10.1126/scitranslmed.aac8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Busse T.M., Roth J.J., Wilmoth D., et al. Copy number alterations determined by single nucleotide polymorphism array testing in the clinical laboratory are indicative of gene fusions in pediatric cancer patients. Genes Chromosomes Cancer. 2017;56(10):730–749. doi: 10.1002/gcc.22477. [DOI] [PubMed] [Google Scholar]