Abstract
Several types of modifications have been proven to participate in the metabolism and processing of different RNA types, including non-coding RNAs (ncRNAs). N-6-methyladenosine (m6A) is a dynamic and reversible RNA modification that is closely involved in the ncRNA homeostasis, and serves as a crucial regulator for multiple cancer-associated signaling pathways. The ncRNAs usually regulate the epigenetic modification, mRNA transcription and other biological processes, displaying enormous roles in human cancers. In this review, we summarized the significant implications of m6A-ncRNA interaction in various types of cancers. In particular, the interplay between m6A and ncRNAs in cancer pathogenesis and therapeutic resistance are being widely recognized. We also discussed the relevance of m6A-ncRNA interaction in immune regulation, followed by the interference on cancer immunotherapeutic procedures. In addition, we briefly highlighted the computation tools that could identify the accurate features of m6A methylome among ncRNAs. In summary, this review would pave the way for a better understanding of the biological functions of m6A-ncRNA crosstalk in cancer research and treatment.
Keywords: Cancer, Immune regulation, N-6-Methyladenosine, Non-coding RNAs, Therapeutic response
Introduction
Nowadays, more than 100 different types of chemical modifications have been identified in the ribose nucleic acid (RNA), including messenger RNAs (mRNAs) and non-coding RNAs (ncRNAs).1,2 Amongst RNA modifications, N-6-methyladenosine (m6A) is the most prevalently and abundantly modified form present in RNA molecules. More importantly, m6A RNA modification have been proven to function as a promising regulatory layer that coordinate diverse aspects of RNA dynamics, such as RNA processing, translational efficiency and degradation.3 In recent years, a growing number of explorations have demonstrated the important biological significance of m6A epitranscriptome in cancer pathogenesis and highlighted the potential values of m6A regulators in therapeutic response and prognosis.
The ncRNAs, such as micro-RNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), have been revealed to regulate cell growth and development in vitro and in vivo, further contributing to many hallmarks of disease phenotypes, including cancers.4, 5, 6 Mounting evidence has demonstrated that the interactions between m6A modification and ncRNAs are extremely widespread.7 As the most prominent RNA modification, m6A is closely involved in ncRNA homeostasis, and serves as a crucial regulator participating in many physiological and pathological processes. Identification of m6A regulators have reported that m6A modification is critical throughout the whole ncRNA life cycle, such as miRNA biogenesis, lncRNA processing, and circRNA functions.8,9 More importantly, m6A modification affects the ncRNA-RNA and ncRNA-protein interactions that regulate and control their specific molecular functions, which is involved in cancer pathogenesis and therapeutic response.10
This review mainly provides an overview of m6A-ncRNA association in malignant progression, and discuss the recent reports that have provided novel insights into the underlying regulatory mechanisms of the m6A-ncRNA axis for therapy response. In addition, we also pointed some promising areas for future explorations, which might influence the potential clinical implications, specifically like the impact of m6A-ncRNA interaction on immune regulation. Evaluating the detailed molecular mechanisms and associated signaling pathways will help strengthen our understanding of m6A-ncRNA axis in cancer pathogenesis, offering promising therapeutic targets to improve the efficacy of cancer treatment.
Modulation of m6A methylation and demethylation
M6A modification, which was first discovered in the 1970s, has been confirmed to play extremely important functions in many biological processes through modulating the ncRNA metabolism and processing.11,12 Moreover, emerging findings have implicated the essential regulators underlying the biological importance of m6A methylome in cancers13, 14, 15 (Fig. 1). The canonical m6A motif is preferentially identified in a consensus sequence, consisting of RRACH (where R represents A or G, and H represents A, U or C).16 Several bioinformatic tools, such as m6Aboost17 and DeepPromise,18 have been developed to search for and discover the RRACH motif surrounding m6A methylation sites.
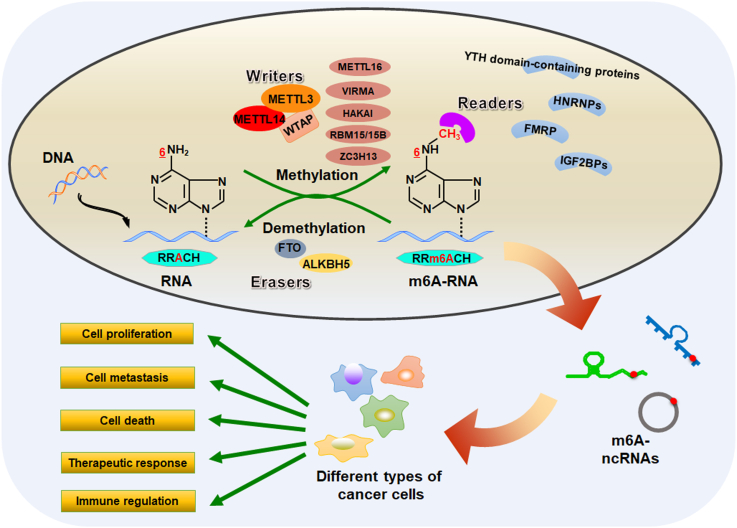
Figure 1.
Reversible m6A modification on ncRNAs. Schematic representation of known regulatory components of m6A modification in ncRNAs. The m6A modification is mainly facilitated by the functional association between RNA methyltransferases and demethylases. Meanwhile, the m6A modified sites can also be recognized by a group of specific RNA-binding proteins. These m6A regulatory proteins play a predominant role in executing m6A functions on ncRNA metabolism in cancer biology.
M6A modification is likely to be facilitated by the functional interaction between RNA methyltransferases and demethylases. As a dynamic reversible biological process, m6A modification is mainly catalyzed by a methyltransferase-like complex (MTC) consisting of at least three core subunits, methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14) and Wilms' tumor 1-associating protein (WTAP).19 In MTC complex, METTL3 is the catalytic subunit, while METTL14 functions as an RNA-binding platform.20,21 Serving as the primarily m6A writers, METTL3-METTL14-WTAP complex catalyzes the addition of a methyl group to the N6-position of adenine in the substrate RNA molecules, therefore producing RNA N6-adenosine methylation and effectively adding the m6A modification to mRNAs and ncRNAs.22 In addition, other MTC subunits, such as METTL16,23 VIRMA,24 HAKAI,13 RNA binding motif protein 15/15B (RBM15/15B)25 and zinc finger CCCH-type containing 13 (ZC3H13),26 have been recently identified as novel m6A writers that regulate multiple signaling pathways in m6A dependent manners, contributing to cancer progression.
The identification of m6A demethylases, AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO), have confirmed that the m6A modification can be reversibly demethylated.27 As m6A erasers, ALKBH5 and FTO could effectively reverse the m6A marks in substrates28 and may play important roles. For example, RNA demethylation by ALKBH5 or FTO alters the stability and trans-activity of UDP-glucuronosyltransferases B7 (UGT2B7) mRNA.29 Two potent small-molecule FTO inhibitors, CS1 and CS2, have been demonstrated to exhibit strong anti-tumor effects in multiple types of cancers.30 The meclofenamic acid (MA), another highly selective FTO inhibitor, could dramatically increase the cellular level of m6A in cervical cancer HeLa cells and suppress the cell growth.31 Recently, Huang and colleagues used the structure-based rational design to identify the compounds FB23 and FB23-2 as two promising FTO inhibitors.32 Mechanistically, FB23 and FB23-2 could inhibit the m6A demethylase activity of FTO by directly binding to FTO, leading to the suppression of proliferation and induction of apoptosis in human acute myeloid leukemia cells.32 However, the potential tumor-suppressing effect of FTO inhibitors have not been confirmed in the clinical trials.33 Therefore, it is of great importance to explore the clinical significance of these inhibitors for cancer therapy in the future. In addition, given that 9 homologs of AlkB protein, including ALKBH1-8 and FTO, have been discovered to date,34 additional explorations are warranted to understand the underlying roles of all AlkB family members in m6A demethylation.
The m6A modified sites can also be recognized by a group of specific RNA-binding proteins, including YT521-B homology (YTH) domain-containing proteins, eukaryotic initiation factor (eIF), heterogeneous nuclear ribonucleoproteins (HNRNPs), fragile X mental retardation protein (FMRP) and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs).35,36 These m6A reader proteins play a predominant role in executing m6A functions on RNA metabolism. It has been documented that the dysregulation of m6A reader proteins also participate in a variety of biological processes.37,38 Therefore, determine the underlying mechanisms of m6A regulators would be of great significance to elucidate their specific functions in RNA metabolism.
Crosstalk between M6A and noncoding RNAs in cancer pathogenesis
M6A methylation has multiple implications in regulating the ncRNA homeostasis. Aberrant regulatory machinery of m6A–ncRNA interaction have been associated with the pathophysiology of numerous diseases, especially cancers (Fig. 2). For example, METTL3 increased m6A level of lncRNA PCAT6, contributing to tumor growth and bone metastasis in prostate cancer.39 YTHDF2-mediated m6A-dependent lncAY stability facilitated HCC progression.40 FTO expression inversely correlated with miR-1266 during colorectal cancer progression.41 However, the biological significance of m6A-ncRNA interactions and the corresponding regulatory mechanisms in cancer pathogenesis and treatment remain incompletely understood.
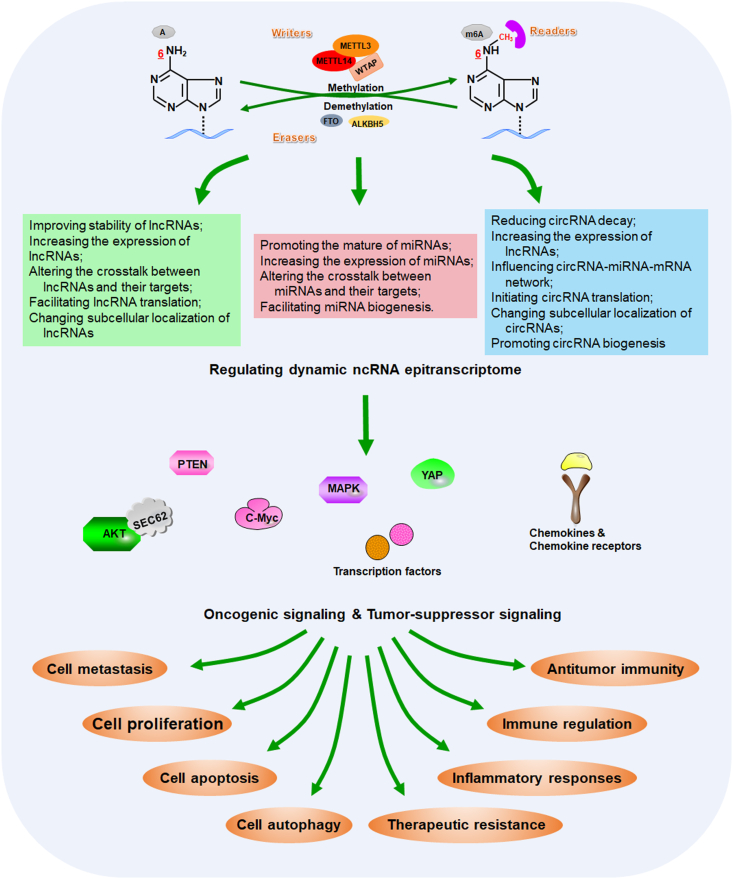
Figure 2.
Multiple functions of m6A modifications on ncRNAs in the control of cancer pathogenesis and treatment. As the most prevalently and abundantly modified form, m6A RNA modification have been proven to function as a promising regulatory layer that coordinate multiple steps of ncRNA homeostasis, such as ncRNA stability, ncRNA–target interaction and subcellular localization. Aberrant m6A–ncRNA machinery could result in dysregulation of cancer-associated signaling pathways, which is involved in a variety of biological behaviors in cancers, especially like cancer development, immune regulation and therapeutic response.
Impact of m6A on miRNA biogenesis and function
The miRNAs are the evolutionarily conserved small ncRNAs with a length of ∼23 nucleotides and play functional roles in diverse biological behaviors in cancers, including cell proliferation and stress response.42, 43, 44
The first step of miRNA biogenesis is the processing of longer precursors of miRNAs (pre-miRNAs) in the nucleus by the DROSHA microprocessor complex, composed of the RNase III endonuclease DROSHA and a cofactor DiGeorge syndrome critical region 8 (DGCR8).45,46 Increasingly, studies have established the biological function of m6A in promoting miRNA biogenesis. The m6A motif RRACH was over-represented in pre-miRNAs, and a single m6A site sufficiently drove pre-miRNA processing.47 Consistent with the viewpoint that site-specific m6A is associated with pre-miRNA methylation, two independent research groups demonstrated that m6A “reader” METTL3 enhanced the binding of DGCR8 to pre-miR-221/222 and pre-miR-92b, and positively regulated their mature forms, which concomitantly resulted in PTEN downregulation and pro-tumorigenic effects in bladder cancers48 and gallbladder cancers.49 In hepatocellular carcinoma (HCC), METTL3 overexpression triggered the malignant phenotypes of cancer cells by interacting with DGCR8 and positively modulating mature miR-873-5p. METTL3 was reported to promote the expression of miR-873-5p by increasing m6A abundance on pre-miR-873-5p in cells.50 In addition, METTL14 suppressed the metastasis capability of HCC cells through interaction with DGCR8.51 Mechanistically, decreased METTL14 negatively modulated the DGCR8 binding to pre-miR-126 in a m6A-dependent mechanism, and led to pre-miRNA accumulation and miR-126 downregulation. Consequently, the m6A status alters the cross talk between miRNAs and their target mRNAs. METTL3 was found to methylate pri-miR-1246, further promoting its maturation and downregulating its downstream target SPRED2 in the development of colorectal cancer (CRC).52 Likewise, upon treatment with cigarette smoke condensate, hypomethylated METTL3 increased the progression rate of pre-miR-25 to its mature form, miR-25-3p, via m6A-dependent pre-miRNA processing, which provoked the activation of oncogenic AKT signaling and thereby induced malignant phenotypes of pancreatic cancer cells.53 Additionally, the m6A induced cleavage of precursor miRNAs is impaired and subsequent miRNA biogenesis was reduced in cells when treated with the short interfering RNAs (siRNAs) specifically targeting RNA methyltransferase, ultimately weakening the metastatic propensities of lung cancer cells.54,55 All these findings reveal that acting as an important post-transcriptional modification, the METTL3/14-catalytic m6A modification is required for efficient initiating of miRNA biogenesis in cancer pathogenesis.
The members of DEAD box RNA helicases, for example DDX17, can serve as the co-factor of miRNA microprocessor, enhancing miRNA biogenesis through m6A modifications.56,57 Thus, it will be imperative to investigate the possible interaction between DEAD box RNA helicases and other regulatory proteins participating in m6A modifications of miRNAs. DDX3 interacted with m6A “eraser” ALKBH5 to demethylate certain miRNAs and ubiquitously regulated cell growth and proliferation,58 indicating that DDX3 functions as a partner of ALKBH5 to regulate dynamic miRNA epitranscriptome.
Another study supports the role of nuclear ‘‘reader’’ HNRNPA2B1 as a possible mediator of m6A-dependent miRNA processing events.59 In this study, HNRNPA2B1 was implicated as an adaptor to recruit the DROSHA microprocessor complex to pre-miRNAs, subsequently triggering their processing into mature miRNAs. Moreover, increased miRNA processing induced by HNRNPA2/B1 overexpression contributed to acquired resistance to tamoxifen and fulvestrant in breast cancer cells.60 Hence, HNRNPA2B1-depend m6A modification likely influences miRNA processing.61 However, this hypothesis needs to be proven using more systematic and large-scale studies.
Taken together, m6A-dependent control of miRNA expression and biosynthesis are linked to the development and progression of human cancers, and is associated with overall survival in cancer patients.10 Nevertheless, given that the underlying knowledge of RNA methylation in miRNA regulation is still in its infancy, additional valuable evidence for m6A regulatory patterns on the biogenesis and functions of miRNAs are worth further investigations in future trials.
miRNAs regulate m6A on mRNAs
Interestingly, miRNAs also regulate m6A modification through a sequence pairing mechanism. Notably, several miRNAs have been reported to exert its function by targeting the mRNA of m6A regulators. The predominant catalytic enzyme of m6A methyltransferase systems, METTL3, is responsible for increased m6A methylations of some carcinogenic mRNAs related to malignant transformation and therapeutic resistance.62,63 As described by Cai and colleague,64 miRNA let-7g reduced the expression of METTL3 mRNA by targeting its 3′-UTR. Hepatitis B X-interacting protein (HBXIP) upregulated METTL3 by interrupting the let-7g–METTL3 interaction, ultimately driving the breast cancer aggressiveness. Meanwhile, miR-4429 exerted a suppressive role in gastric cancer (GC) cells through targeting METTL3 and disturbing m6A-associated stabilization of oncogenic SEC62 mRNA.65 Similarly, miR-33a attenuated cell proliferation by directly targeting the 3′-UTR of METTL3 mRNA, and seemed like a promising therapeutic target in non-small-cell lung carcinoma.66 miR-186 was another effective therapeutic and prognostic biomarker in hepatoblastoma that targeted and downregulated METTL3 expression, which finally led to significant inhibition of aggressive cellular phenotype both in vitro and in vivo.67 Thus, regulating miRNAs–METTL3 signaling axis may be a potential targeting strategy, which in turn impairs the oncogenesis and improves the therapeutic effect.
The cross talk between miRNAs and RNA m6A “reader” proteins has been recently validated, to affect the biological behavior of cancer cells. YTHDF2 served as a m6A “reader” and selectively bound to m6A sites to mediate mRNA decay.68 Similarly, Yang et al found the negative correlation between miR-145 and YTHDF2 in HCC tissues and cells, where miR-145 targeted the 3′-UTR of YTHDF2 mRNA and strongly stabilized m6A-containing mRNA, ultimately decreasing the malignancy of HCC.69 In a recent study, proteomic analysis revealed m6A “reader” HNRNPF as putative hsa-miR-139-5p targets in thyroid cancer. Exogenous expression of hsa-miR-139-5p drastically reduced HNRNPF mRNA abundance, and affected the splicing events related to MAPK and AKT signaling cascades.70 In ovarian cancers, miR-744-5p directly downregulated mRNA and protein expressions of HNRNPC and activated the intrinsic apoptotic pathways. In addition, HNRNPC overexpression caused diminished miR-21 expression and AKT phosphorylation, further leading to the pro-apoptotic effects. Strikingly, these apoptosis-inducing effects can be obviously enhanced when cells received combination treatment of carboplatin and miR-744-5p.71 Moreover, exosomal miRNAs may reduce the magnitude of FTO methylation, thereby epigenetically affecting FTO expression and FTO-dependent mRNA splicing.72
In brief, all these data highlight that miRNAs display interesting cellular activities in m6A modification through targeting m6A regulatory factors, and lay the foundation for further investigations of m6A methylation patterns in cancer pathobiology.
Impact of m6A on lncRNA regulation and function
The discovery of lncRNAs in cancer cells has provided several key insights into the underlying molecular mechanisms of malignant phenotypes.73 Dysfunctional lncRNAs have been shown to play essential roles in cancer development and therapeutic response.74,75
It is well known that m6A modification is highly enriched within lncRNA sequences, and is strictly required for their biological functions.76 Several lncRNAs with m6A modifications display METTL3-dependent high stability and expression.77 As expected, the m6A modification was enriched in lncRNA FAM225A and improved its transcript stability.78 Short hairpin RNA (shRNA)-mediated silencing of METTL3 led to decreased m6A methylation of lncRNA FAM225A and further suppressed its oncogenic function in nasopharyngeal carcinoma. METTL3, along with METTL14, promoted m6A methylation of LNCAROD and improved its stability in head and neck squamous cell carcinoma (HNSCC). This subsequently stabilized LNCAROD role as a scaffold to promote YBX1-HSPA1A interaction, ultimately protecting oncoprotein YBX1 from proteasomal degradation.79 In prostate cancer cells, overexpression of VIRMA, the regulatory component of methyltransferase complex, significantly enhanced the stability and abundance of lncRNA CCAT1/2 methylated transcripts.80 Moreover, the unique profiles of m6A-related lncRNAs have been implicated as intriguing prognostic biomarkers for glioblastoma patients.81 Thus, clarifying the underlying mechanisms of m6A on lncRNA modulation is essential for future diagnostic and therapeutic inventions for malignant diseases.
RNA m6A modifications can induce the structural rearrangements of lncRNAs that sequester or expose the miRNA-binding sites.82 In HCC, m6A modification mediated by METTL3 resulted in LINC00958 upregulation via stabilization of its RNA transcript, consequently aggravating HCC lipogenesis and progression. Further biochemical analysis showed that LINC00958 contains several modification sites for m6A, which were indispensable for the recognition of miR-3619-5p.83 Likewise, METTL3 promoted lncRNA RHPN1-AS1 expression in a m6A-dependent manner, facilitating tumorigenicity and metastasis of ovarian cancer cells.84 Mechanistically, upregulated RHPN1-AS1 acts as a competing endogenous RNA (ceRNA) to sponge miR-596, finally increasing LETM1 expression and activating AKT signaling pathway. Another example involves lncRNA pseudogene, Olfr29-ps1, which could affect the myeloid-derived suppressor cells (MDSC)-targeted immunotherapy.85 It was found that m6A-modified Olfr29-ps1 enhanced its interaction with miR-214-3p and upregulated its target gene MyD88, thereby positively regulating the immunosuppressive function and differentiation of myeloid cells.
M6A modifications are responsible for the regulation of lncRNAs by providing the binding site for m6A “readers”. IGF2BP2 served as a critical “reader” for m6A-containing lncRNA DANCR and synergistically sustained pancreatic cancer pathogenesis and resistance.86 Meanwhile, LINC00278 was highly methylated by METTL3, which was required for YTHDF1 recognition and facilitated LINC00278 translation.87,88 The m6A mapping study has revealed that at least 78 m6A residues were distributed in lncRNA XIST sequences.89 METTL14-dependent m6A of XIST could be specifically recognized by YTHDF2, which led to XIST degradation and tumor inhibitory effects in CRC.90 Conversely, methylated lncRNA PVT1 could be demethylated by ALKBH5, thereby blocking the binding of m6A “reader” YTHDF2 to PVT1.91 In another study, YTHDF3 selectively bound to m6A-modifed lncRNA GAS5 and modulated GAS5 decay in a methylation-dependent manner. Surprisingly, GAS5 downregulation could further improve YAP-mediated transcription activation of YTHDF3.92 These results indicate a negative feedback loop between lncRNAs and m6A “readers” in tumor progression.
M6A methylation might also be involved in lncRNA regulation by regulating its subcellular localization.93,94 In line with this, Wu et al discovered that m6A-induced lncRNA RP11 triggered its nuclear accumulation in CRC cells. Furthermore, intranuclear lncRNA RP11 could post-translationally stimulate the expression of epithelial–mesenchymal transition (EMT)-related transcription factor Zeb1 by accelerating the mRNA degradation of two E3 ligases, Siah1 and Fbxo45.95
Therefore, these findings imply that the interaction between m6A epitranscriptome and lncRNA affects tumorigenesis and progression, which hence offers a potential interventional target for human cancers. Nonetheless, due to limited understanding of their functions and regulatory mechanisms, the relevance of m6A changes in lncRNAs requires additional validation, as well as functional follow-up studies.
LncRNAs regulate m6A on RNAs
Of note, RNA m6A modification can also be inversely regulated by lncRNAs, which is of great significance in modulation of cancer-associated signaling pathways and alteration of cellular fates. Particularly, several dysregulated lncRNAs exert their functions via regulation of methyltransferase complex.9 As elucidated by Sun et al, METTL14 could be recognized and recruited by LNC942 with the specific binding domain (+176 to +265).96 In this process, LNC942 revealed its pro-proliferation functions in breast cancer through promotion of METTL14-mediated m6A methylation and by elevating the stability of downstream targets CXCR4 and CYP1B1. In GC, lncRNA ARHGAP5-AS1 effectively recruited METTL3 to stimulate m6A modification of ARHGAP5. M6A-dependent stabilization of ARHGAP5 by ARHGAP5-AS1–METTL3 was responsible for impaired autophagy and acquired cisplatin (DDP) resistance.97 Meanwhile, LINC00470 promoted the degradation of PTEN mRNA on the METTL3-dependent pathway to facilitate GC cell proliferation and metastasis.98 In addition, lncRNA GATA3-AS served as a pivotal cis-regulatory element for the interaction between KIAA1429 and GATA3 nascent transcripts. With the guidance of GATA3-AS, KIAA1429 catalyzed m6A modification on the 3′-UTR of GATA3 pre-mRNA and in turn led to GATA3 downregulation in HCC cells.99
Both m6A “erasers” and “readers” are also decorated by lncRNAs. A number of studies has uncovered that RNA m6A demethylases rely on the lncRNA-associated regulatory mechanism to execute their functions.100 Elevation of long non-coding RNA just proximal to the X-inactive specific transcript (JPX) could promote the aerobic glycolysis and temozolomide chemoresistance in glioma cells. Mechanistically, lncRNA JPX has been found to interact with FTO demethylase and further enhance the FTO-dependent mRNA demethylation of phosphoinositide dependent kinase-1.101 The findings from Zhang's group demonstrated that lncRNA FOXM1-AS enhanced the interaction of ALKBH5 with FOXM1 pre-mRNA, resulting in the demethylated FOXM1 and oncogenic effects in glioblastoma.102 ALKBH5 could also interact with the lncRNA SOX2OT to confer temozolomide resistance in glioblastoma cells. In the SOX2OT-overexpressed condition, ALKBH5 promoted SOX2 expression by demethylating the SOX2 transcripts.103 Additionally, the oncopeptide RBRP encoded by LINC00266-1 was found to mainly bind to m6A “reader” IGF2BP1. RBRP recruited IGF2BP1 to recognize m6A-modified c-Myc transcripts and increased c-Myc oncogene expression.104 Moreover, lncRNA CCAT2 could significantly promote the tumorigenesis of esophageal squamous cell carcinoma by downregulating miR-200b to elevate the expression of miR-200b target IGF2BP2.105
At present, these reports have emphasized the importance of lncRNAs on the m6A epitranscriptome in the aggressive phenotype of human cancers. These findings have provided the evidence for lncRNAs' influence on m6A regulators. Whether lncRNAs directly mediate their activity and expression needs further experimental verification. Even though the specific roles of lncRNA LINRIS on the regulation of IGF2BP2 protein stabilization have been preliminarily reported,106 the underlying mechanisms are still under investigation.
Impact of m6A on circRNA signaling and function
Emerging studies have discovered circRNA as a novel endogenous ncRNAs. Unlike linear RNAs, the 3′- and 5′-terminals normally present in RNAs have been joined together, forming covalently closed continuous loop. This unique structure effectively prevents the degradation of circRNAs by ribonucleases or exonucleases, thus making them more stable.107 CircRNAs can function as miRNA sponges or critical regulators of their parental genes. Beyond that, several circRNAs can also serve as protein-coding RNA transcripts that encode functional proteins. Although the detailed mechanisms of circRNA biology remain to be elucidated, these well-known functions have identified them as potential biomarkers and therapeutic targets in human diseases, including cancers.108,109
CircRNAs, especially exon-derived circRNAs, are found to be m6A-modified in human transcriptome, which is enhanced by methyltransferase complex METTL3/14 and inhibited by demethylase FTO.110 Coincidentally, the consensus m6A recognition motif RRACH has been discovered in circRNA sequences by several independent research groups.111,112 The genome-wide mapping and analysis of m6A circRNAs displayed that m6A modification was frequently present in circRNAs,113 where Zhou et al identified more than one thousand m6A circRNAs in mammalian cells. The presence of m6A circRNAs was mainly dependent on the interaction between circRNAs and m6A “readers” YTHDF1/2, and could be decreased upon METTL3 depletion. Conversely, depleting m6A demethylases FTO and ALKBH5 induced a generalized increase in the abundance of m6A-containing circRNAs.114 In addition, the m6A circRNAs were differentially expressed in diverse cell types, thus confirming the cell-type-specific patterns of circRNA methylation.113 Meanwhile, Su et al used an m6A-seq approach to analyze the transcriptome-wide map of m6A circRNAs upon hypoxia stress, and found that hypoxia could significantly influence the m6A level of circRNAs and circRNAs abundance. Furthermore, m6A modification influenced the circRNA–miRNA-mRNA regulatory network involved in the hypoxia-driven signals in mammalian cells.115 In CRC cells, nuclear m6A “reader” YTHDC1 bound to m6A-modified circNSUN2 and facilitated its cytoplasmic export. Consequently, the increased circNSUN2 in the cytoplasm specifically interacted with IGF2BP2, thus stabilizing its downstream target HMGA2 and promoting CRC cell aggressiveness.116
Altered m6A regulators may contribute to the aberrant expression profiles of circRNAs seen in disease pathogenesis and progression.117,118 The majority of circRNAs are synthesized by a co-transcriptional back-splicing pattern.119,120 Moreover, increased m6A levels correlated with correct splicing,121 which may promote circRNA biogenesis. To prove this hypothesis, Tang et al demonstrated that the back-splicing occurred mostly at m6A sites, which were preferentially enriched near the start and stop codons in linear mRNAs.122 Additionally, ribonuclease P/MRP, an endoribonuclease complex, has been demonstrated to facilitate the circRNAs degradation. A subset of circRNAs carrying the m6A modification could be subjected to rapid endoribonucleolytic cleavage through m6A–YTHDF2–ribonuclease P/MRP dependent mechanism.123 A recent work conducted by Park and colleague showed that the endoribonucleolytic cleavage mediated by YTHDF2–HRSP12–ribonuclease P/MRP axis was applicable to m6A-containing circRNA decay. Knockdown of adaptor protein HRSP12 or ribonuclease P/MRP significantly abrogated the degradation of YTHDF2-bound circRNAs.124 YTHDF2 also recognized m6A-containing ncRNAs, and accelerated their destabilization via CCR4–NOT complex-mediated deadenylation pathway.125 Despite the important implications, the comprehensive molecular mechanisms of m6A RNA methylation responsible for the circRNA biogenesis and decay remain poorly understood, and thus warrant further research.
Several latest reports revealed that methylated circRNAs display protein-encoding potential in a cap-independent pattern, and their translated products can offer a novel perspective for a variety of pathophysiological contexts.126,127 Interestingly, a single m6A site sufficiently initiated circRNA translation in eukaryotic cells by directly recruiting the eukaryotic translation initiation factors (eIFs) and m6A regulators.128 Especially, the direct binding of 5′UTR m6A to eIF3 subsequently promoted translation in the absence of cap-binding factor eIF4E.129 Upon specific environmental stress, the m6A in 5′UTR of RNA transcripts was reported to regulate the ribosomal scanning and start codon selection, thereby driving the non-classical cap-independent translation mechanism.130 This concept has been strongly enhanced using m6A-seq method, and demonstrates that circRNAs can be efficiently translated using short sequences carrying the consensus m6A residues in human cells.131,132 For example, circ-ZNF609 was previously demonstrated to contain an open reading frame and can undergo translation.133 The translation ability of circ-ZNF609 could be effectively controlled through recognition by m6A “reader” YTHDF3 and eIF4G2.134 The circE7 generated from oncogenic human papillomaviruses (HPVs) was preferentially m6A-modified and translated to produce E7 oncoprotein, which then accelerated the phenotype transformation of cervical carcinoma CaSki cells.135 Therefore, all these findings suggest that m6A-mediated translation initiation can serve as a potential driving mechanism for circRNA translation, enlightening the need to characterize the complexity of translated circRNAs in the pathological changes of cancer cells.
Although m6A epitranscriptional modification has been confirmed for its enrichment and biological importance in modulating circRNAs in cancer pathogenesis,136 the underlying regulatory networks of m6A-circRNAs have not been fully elucidated so far. More detailed evaluations are needed to clarify the putative relationships between m6A-epitranscriptomes and circRNA-dependent signaling events. Particularly, it is of interest to note that m6A methylated circRNAs have permanently extended our attention towards cancer pathological behaviors, especially in tumorigenesis and treatment.
CircRNAs regulate m6A on RNAs
Interestingly, abnormal level of circRNAs might also regulate m6A abundance in cancer cells and further influence cancer phenotypes. CircRNAs can function as efficient miRNA sponges by competitively binding to miRNA response elements (MREs) to suppress their activity, resulting in increased levels of miRNA targets.137,138 Given the potential regulatory roles of miRNAs on m6A modification,139 we hypothesize that to some extent, circRNAs might modulate m6A modification in miRNA-dependent manner. A recent study established the circRNA–miRNA–METTL14 interaction network in kidney renal clear cell carcinoma (KIRC) supporting this hypothesis. The findings from this research showed that circRNAs regulated m6A methyltransferase METTL14 by sponging multiple miRNAs involved in the occurrence and progression of KIRC.140 However, another study reported the direct interaction between circRNAs and m6A regulators. In dysfunction astrocytes, overexpression of circSTAG1 abolished the translocation of ALKBH5 into the nucleus by capturing ALKBH5, and further promoted the m6A-mediated degradation of FAAH transcripts.141 These studies preliminarily provided proof-of-concept evidence for the functional roles of circRNAs on m6A modulation, and shed novel insights into the development of preventive strategy for cancers. Nevertheless, the potential regulatory mechanisms need further clarification with research on the functional link between circRNAs and m6A epitranscriptional modification.
Interplay of m6A-ncRNA related to immune system
The cells in innate and adaptive immune system have been reported to exert regulatory effects on the development and progression of cancer. In addition, m6A modification has been implied as a vital regulator of the immune system and to be involved in various aspects of immune-related disorders, including cancer.142,143 M6A methylation of N6-adenosine frequently occurs in immunological disorders-associated ncRNAs, including miRNAs, and consequently changing their mechanisms of action in the immune response144 (Table 1). As an important regulator for m6A modification, miR-142 overexpression had been proven to improve the HCC patients' prognosis through interference in the abundance of tumor-infiltrating immune cells.145 Thus, the relationship between m6A-modifiers and ncRNAs can potentially be used to reprogram the tumor microenvironment (TME) by modulating the immune-associated signatures, hence, interfering with the cancer immunotherapeutic procedures.146,147
Table 1.
M6A-ncRNA interaction in the regulation of cancer immune response.
| M6A-ncRNAs | Immune regulation | Biological behaviors | Refs |
|---|---|---|---|
| MiR-142 | Affecting immune cell infiltration | Functioning as m6A regulator | 145 |
| 11 m6A-related lncRNAs | Modulating immune checkpoint inhibitors | Involving in immunotherapy | 149 |
| Demethylated Lnc-Dpf3 | Suppression of dendritic cell migration | Prevention of aberrant inflammatory responses | 150 |
| 244 m6A-modified lncRNAs | Regulating cellular immune system | Regulators for pro-inflammatory and anti-inflammatory response | 151 |
| M6A-related lncRNA signature | Affecting immune cell infiltration | A promising immune prognostic model | 142 |
| M6A modified circRNAs | Blunting the immunogenicity of circRNAs | Inhibiting innate immunity | 156 |
| CircZbtb20 | Promoting m6A-demethylation of Nr4a1 mRNA that is required for function of innate lymphoid cells | Maintaining innate lymphoid cell homeostasis | 157 |
| M6A-modified circNDUFB2 | Recruiting immune cells into tumor microenvironment | Facilitating anti-tumor immunity | 158 |
Several recent reports discussed the promising roles of lncRNA–m6A interaction networks in host immune response, and suggested probable clinical applications.148,149 Upregulated C–C motif chemokine receptor 7 (CCR7) inhibited the degradation of lnc-Dpf3 by promoting its demethylation, resulting in suppression of dendritic cell migration and prevention of aberrant inflammatory responses.150 Li et al evaluated the m6A modification pattern of lncRNAs in pro-inflammatory (M1-L) and anti-inflammatory (M2-L) cells. 244 lncRNAs were differentially methylated with m6A between M1-L and M2-L cells. Moreover, these differentially m6A-modified lncRNAs mainly participated in the regulation of cellular immune system and signal transduction.151 Through comprehensive analyses of the profiles of m6A-related lncRNAs in the immune regulation of HCC, Yu et al identified a m6A-related lncRNA signature as a promising immune prognostic model.152 Moreover, higher levels of several immune cells, such as activated memory CD4+ T cells, CD8+ T cells and follicular T-helper cells, were shown to infiltrate the TME of high-risk patients, revealing that higher density of tumor-infiltrating immune cells may affect the response to immunotherapy. Therefore, clarifying the biological roles of m6A–lncRNA interactions in immune regulation might elucidate important scientific knowledge in the understanding of novel therapeutic and prognostic targets for cancer patients.
It was presented that the frequently expressed circRNAs in eukaryotic cells makes them potential therapeutic biomarkers for the antitumor immunity.153 M6A modification has been revealed to significantly blunt the immunogenicity of circRNAs in human cells, and serves as potent contributor in regulating circRNA-associated immune signaling.154 In line with the observations, recent studies have demonstrated that antitumor immunity can be effectively controlled by m6A methylation-dependent processes. Loss of YTHDF1 in dendritic cells increased the production of interferon-γ (IFN-γ), followed by the activation of cytotoxic T cells and tumor inhibition.155 Meanwhile, YTHDF2 interacted with m6A sites in circRNAs to suppress the antigen-specific T cell activation and antitumor immunity in vivo.156 Liu et al demonstrated the essential roles of circZbtb20 for maintenance of innate lymphoid cell homeostasis. Mechanistically, circZbtb20 enhanced ALKBH5-mediated demethylation of Nr4a1 mRNA, improving Nr4a1 stability and its downstream Notch2 signaling, which were required for the functions of innate lymphoid cells.157 During the progression of non-small-cell lung cancer (NSCLC), m6A-modified circNDUFB2 facilitated the anti-tumor immunity by eliciting the polyubiquitination-mediated degradation of IGF2BPs.158 Collectively, these data show that interfering with m6A-modified circRNAs may be efficient in improving the immunotherapy outcomes in cancer patients.
M6A-ncRNA interaction in the regulation of cancer therapeutic resistance
Therapeutic resistance has been identified as a major obstacle to the treatment of patients with malignancies. Cancer therapeutic resistance is a complex phenomenon that can be influenced by multiple mechanisms, such as tumor heterogeneity, epigenetic modification, genetic alteration, etc. Emerging studies have revealed that m6A–ncRNAs interaction serves as an important biological process involved in the treatment of multiple human diseases, including cancers.159,160 In recent years, the functions of m6A–ncRNA interaction in cancer therapeutic resistance regulation have been evaluated, and the specific molecular mechanisms have been partially discovered.161 Regulation of m6A–ncRNA-dependent signaling events can successfully limit cell growth and overcome therapeutic resistance of cancer cells,162,163 suggesting that understanding of m6A–ncRNAs association might be useful for proposing promising strategies for cancer treatment (Table 2). However, because of the unexplored mechanisms underlying the interaction between m6A and ncRNAs, the application of RNA modification regulatory factors in cancer therapy is limited in clinical practice.
Table 2.
The interaction between m6A and ncRNA in cancer therapeutic resistance.
| M6A regulators | NcRNAs | Functions | Therapeutic responses | Cancers | Refs |
|---|---|---|---|---|---|
| METTL3 | MiR-221-3p | Stimulating pri-miR-221-3p maturation | Sustaining doxorubicin resistance | Breast cancer | 164 |
| METTL3 | MiR-4443 | Interfering METTL3-mediated FSP1 m6A methylation | Conferring cisplatin resistance | Lung cancer | 165 |
| METTL3 | LncRNA MALAT1 | Enhancing MALAT1 stability | Conferring cisplatin resistance | Lung cancer | 63 |
| METTL3 | LOC554202 | Promoting FOXO3 methylation and its downstream LOC554202 | Inhibiting sorafenib resistance | Hepatocellular carcinoma | 166 |
| METTL3 | LINC00922 | Enhancing TFAP2C stability and promoting LINC00922 transcription | Promoting doxorubicin resistance | Osteosarcoma | 168,169 |
| METTL3 | LncRNA ARHGAP5-AS1 | Increasing M3TTL3-mediated ARHGAP5 stabilization | Conferring cisplatin resistance | Gastric cancer | 97 |
| METTL3 | CircCUX1 | Enhancing circCUX1 stability | Reversing radiotherapy resistance | Hypopharyngeal squamous cell carcinoma | 170 |
| METTL3 | CircRNA-SORE | Increasing m6A level of circRNA-SORE | Promoting sorafenib resistance | Hepatocellular carcinoma | 171 |
| ALKBH5 | Circ_0072083 | Increasing ALKBH5 expression | Promoting temozolomide resistance | Glioma | 173 |
| FTO | LncRNA HLA-F-AS1 | Activation STAT3 and inducing HLA-F-AS1 expression | Facilitating doxorubicin resistance | Breast cancer | 174,175 |
| HNRNPA2/B1 | Results in a significant change in multiple miRNAs | Promoting tamoxifen resistance | Breast cancer | 60 |
The RNA “writer” METTL3 has been proven to regulate miRNA metabolism, contributing to therapeutic resistance in cancer cells. In MCF-7 breast cancer cells, METTL3 overexpression stimulated pre-miR-221-3p maturation in a m6A-dependent manner, reduced cell apoptosis and sustained doxorubicin resistance.164 Surprisingly, miR-4443 bound METTL3 and downregulated its activities, suggesting that METTL3 was a direct target of miR-4443. High level of exosome-derived miR-4443 conferred DDP resistance in NSCLC cell line A549-R via interference of METTL3-mediated m6A methylation of FSP1.165 Accumulating evidence confirmed the important roles of lncRNAs in cancer therapeutic resistance through sponging of miRNAs and regulating their downstream targets. METTL3-mediated m6A modification of lncRNA MALAT1 markedly enhanced its stability, functioned as miR-1914-3p sponge and subsequently enhanced the expression of downstream target YAP, thereby promoting lung cancer metastasis and DDP resistance.63 In addition, forkhead box O3 (FOXO3)-induced lncRNA LOC554202 bound to miR-485-5p and impaired its tumor-promoting function in HCC.166 Since FOXO3 is a downstream target of METTL3, Lin et al found that METTL3 knockdown enhanced sorafenib resistance of HCC by abolishing the FOXO3 methylation and its downstream signaling.167 Moreover, using lncRNA microarray, Gu and colleague analyzed lncRNA expression profiles and identified the highly-expressed LINC00922 in doxorubicin-resistant osteosarcoma cells MG63/DXR.168 Mechanistic assays indicated that METTL3-mediated m6A methylation enhanced the stability of transcription factor activating protein 2 gamma (TFAP2C) in resistant sarcoma cells.169 Concurrently, overexpressed TFAP2C promoted the transcription of LINC00922, contributing to chemotherapy resistance.168 In GC, lncRNA ARHGAP5-AS1 recruited METTL3 to accelerate m6A modification of ARHGAP5 mRNA and promote its stabilization in the cytoplasm. Blocking lncRNA ARHGAP5-AS1/ARHGAP5 signaling axis significantly decreased DDP resistance and improved therapeutic outcome, which might provide a promising strategy to overcome chemoresistance.97 In addition, m6A-mediated circCUX1 stabilization relies on the m6A reader METTL3. Knockdown of METTL3 by siRNA significantly reversed the expression of oncogenic circCUX1, thereby promoting cell apoptosis and reversing radiotherapy resistance in hypopharyngeal squamous cell carcinoma (HPSCC).170 Similarly, METTL3 depletion resulted in decreased m6A methylation of circRNA-SORE, thereby deactivating the Wnt/β-catenin pathway and increasing sorafenib sensitivity in HCC.171 However, the detailed regulation and function of circRNAs on METTL3 in treatment resistance are largely unknown and remain to be explored.
Serving as a representative RNA “eraser”, the demethylase ALKBH5 can lead to the demethylation of cancer-associated ncRNAs, affecting the tumorigenicity and treatment efficacy.172 Exosomal circ_0072083 competitively sponges miR-1252-5p to enhance its ALKBH5 targeting in temozolomide-resistant glioma cells U251/TR and U87/TR. The circ_0072083 silence evidently decreased the ALKBH5 expression and reduced ALKBH5-mediated demethylation of nanog homeobox (NANOG), facilitating apoptosis and suppressing growth of temozolomide-resistant cells in vivo and in vitro.173 The m6A “eraser” FTO facilitated doxorubicin resistance of breast cancer cells by controlling the activation of signal transducer and activator of transcription 3 (STAT3) signaling.174 Moreover, STAT3-induced lncRNA HLA-F-AS1 enhanced cell viability and induced poor therapeutic effect.175 In addition, the m6A “reader” HNRNPA2/B1 was shown to be remarkably upregulated in tamoxifen-resistant breast cancer LCC9 cells. Whole genome miRNA profiling revealed that overexpression of HNRNPA2/B1 resulted in a significant change in multiple miRNAs, affecting several signal pathways associated with endocrine therapy resistance, such as transforming growth factor β (TGFβ) signaling.60 Taken together, these findings support the fundamental roles of m6A–ncRNAs interaction in the processes that contribute to therapy resistance in human cancers.
Databases for m6A-ncRNA analysis in cancer research
Nowadays, identifying the accurate features of the m6A methylome among mammalian systems is still a major challenge. Especially, there are significant requirements for new computational tools to analyze the data from methylated RNA immunoprecipitation combined with RNA sequencing (MeRIP-seq) to gain further insight into the functional links between RNA methylation and ncRNAs.13 In recent years, some available algorithms have been developed to annotate and identify the interaction networks between m6A modifications and ncRNAs (Table 3). These user-friendly databases enable more convenient acquisition of m6A–ncRNA interaction by using different computational methods, which can be used in additional research. The MethylTranscriptome DataBase (MeT-DB) is the first comprehensive resource for interrogating m6A methyltranscriptome from all published MeRIP-seq datasets. More importantly, this database also offers a genome browser to query and visualize the context-specific m6A with miRNAs and their target genes.176,177 The website RMBase offers a variety of interfaces and graphic visualizations to facilitate analyses of the relationships between m6A modification and multiple types of genes, including miRNAs and lncRNAs. This web-tool can also be an applicable strategy for mapping RNA modifications in normal tissues and cancer cells.178,179 Recently, M6A2Target was constructed to identify potential lncRNA targets and regulators of m6A RNA methylation.180 Furthermore, the hierarchical model MeTCluster, developed by Cui et al, can systematically uncover and cluster the m6A methylation peaks in lncRNAs with high accuracy and good sensitivity.181 Similarly, MeTDiff is another novel computational tool for predicting the differential m6A methylation sites on human lncRNAs with higher sensitivity and specificity.182 Utilizing these two algorithms, researchers confirmed that the methylation density with less peak enrichment are more likely to be distributed at the 5′-end of lncRNAs, while those with higher peak enrichment tend to cluster at the 3′-end of lncRNAs. These results indicate that m6A's functions in lncRNA regulation could be location specific. In addition, a newly constructed tool, LncVar, can be used to systematically integrate the effects of single nucleotide polymorphisms (SNPs) in m6A regions on lncRNAs from six species (Homo sapiens, Mus musculus, Danio rerio, Caenorhabditis elegans, Drosophila melanogaster and Arabidopsis thaliana).183 CVm6A software proposes a quantitative detection method to visualize the cell-dependent m6A patterns in mRNAs and lncRNAs. In particular, CVm6A can also be used to distinguish the unique m6A profiles in cancer and non-cancer cells.184 The study by Liu and colleague presents another computational framework, LITHOPHONE, for scanning the entire human lncRNAome for all possible lncRNA m6A sites. Using the Ensemble predictor algorithm, they can achieve the best performance for identifying lncRNA methylation sites in a non-biased way.185 Circbank, designed by Liu's group, is a comprehensive database for circRNA with the standard nomenclature, which can alleviate the terminological confusion for both bioinformatic and experimental circRNA research. Of note, Circbank can also visualize the difference of m6A modification between circRNAs and their host genes.186 Although other m6A-associated algorithms, such as m6AVar,187 REPIC,188 SRAMP189 and WHISTLE,190 have not been utilized to detect the m6A peaks in ncRNAs currently, these databases would provide more strategies for comparison, eventually leading to a more accurate landscape of the m6A methylation in transcriptome. Moreover, some challenges exist across these algorithms, where there is no clear-cut golden standard and non-overlapping predicted behaviors,107 should be addressed with further high-quality studies. Consequently, addressing these challenges is important for using bioinformatics approaches to appropriately validate the prediction findings.
Table 3.
The databases and analysis tools for m6A-ncRNA interaction in cancer research and treatment.
| Databases | Characteristics | URL | Refs |
|---|---|---|---|
| MeT-DB | Interrogating the context-specific m6A with the binding sites of miRNAs | http://compgenomics.utsa.edu/MeTDB/ | 177 |
| RMBase | Analyzing the relationships between m6A modification and miRNAs/lncRNAs | http://rna.sysu.edu.cn/rmbase/ | 179 |
| M6A2Target | Confirming lncRNAs as potential targets of m6A regulators | http://m6a2target.canceromics.org | 180 |
| MeTCluster | Clustering the specific m6A methylation peaks in lncRNAs | http://compgenomics.utsa.edu/metcluster | 181 |
| MeTDiff | Predicting the m6A methylation sites on human lncRNAs | https://github.com/compgenomics/MeTDiff | 182 |
| LncVar | Systematically integrating the statistics of SNPs in m6A regions on lncRNAs | http://bioinfo.ibp.ac.cn/LncVar | 183 |
| CVm6A | Exploring the cell-dependent m6A patterns in lncRNAs | http://gb.whu.edu.cn:8080/CVm6A | 184 |
| LITHOPHONE | Scanning the entire human lncRNAome for all possible lncRNA m6A sites | http://180.208.58.19/lith/ | 185 |
| Circbank | Visualizing the difference of m6A modification between circRNAs and its host gene | http://www.circbank.cn/ | 186 |
Conclusion and future remarks
RNA m6A modification draws much attention, and has diverse regulatory roles on ncRNAs, such as their biogenesis, degradation, expression, etc. Growing evidence shows that the m6A–ncRNA regulatory networks are prevalent features of cancer cells and likely to have important biological functions in disease development and treatment. More detailed studies on the association between m6A status and ncRNAs would provide the novel insights on how their interactions affect tumorigenesis, immune regulations and therapeutic responses. Considering that various molecular pathways during cancer pathogenesis can be triggered following m6A–ncRNA signaling, a better understanding of the underlying regulatory mechanisms in different cell-types and stimuli remains to be achieved. Meanwhile, the clarification of m6A–ncRNA regulatory networks may also have clinical implications and prognostic significance, which is an under-studied field demanding further investigations. More importantly, it will be worthwhile determining how cancer cells can accurately respond to different therapeutic strategies mediated by the m6A–ncRNA signaling pathways. Eventually, comprehensive and detailed knowledge of these issues will open new therapeutic avenues to effectively intervene in the m6A–ncRNA signaling for cancer cell elimination.
Author contributions
Conceptualization: Yuanliang Yan and Zhijie Xu; Data curation: Yuanliang Yan and Zhijie Xu; Formal analysis: Qiuju Liang and Xinxin Ren; Funding acquisition: Yuanliang Yan and Zhijie Xu; Investigation: Yuan Cai and Bi Peng; Methodology: Yuan Cai and Bi Peng; Project administration: Zhijie Xu; Resources: Zhijie Xu; Software: Xi Chen, Xiang Wang and Qiaoli Yi; Supervision: Yuanliang Yan and Zhijie X; Validation: Yuanliang Yan and Zhijie X; Visualization: Yuanliang Yan and Zhijie X; Roles/Writing - original draft: Yuanliang Yan; Writing - review & editing: Jinwu Peng and Zhijie Xu.
Conflict of interests
The authors declare no conflict of interests.
Funding
This work was supported by The National Natural Science Foundation of China (No. 81803035); The China Postdoctoral Science Foundation (No. 2021T140754 and 2020M672521); The Natural Science Foundation of Hunan Province, China (No. 2020JJ5934); and The Postdoctoral Science Foundation of Central South University, China (No. 248485).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Uddin M.B., Wang Z., Yang C. The m(6)A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol Cancer. 2021;20(1) doi: 10.1186/s12943-021-01356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z., Huang J., Gao M., et al. Current perspectives on the clinical implications of oxidative RNA damage in aging research: challenges and opportunities. Geroscience. 2021;43(2):487–505. doi: 10.1007/s11357-020-00209-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Z., Peng B., Cai Y., et al. N6-methyladenosine RNA modification in cancer therapeutic resistance: current status and perspectives. Biochem Pharmacol. 2020;182 doi: 10.1016/j.bcp.2020.114258. [DOI] [PubMed] [Google Scholar]
- 4.Yi Y.C., Chen X.Y., Zhang J., et al. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-01233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Y., Chen X., Wang X., et al. The effects and the mechanisms of autophagy on the cancer-associated fibroblasts in cancer. J Exp Clin Cancer Res. 2019;38(1) doi: 10.1186/s13046-019-1172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju Q., Zhao Y.J., Ma S., et al. Genome-wide analysis of prognostic-related lncRNAs, miRNAs and mRNAs forming a competing endogenous RNA network in lung squamous cell carcinoma. J Cancer Res Clin Oncol. 2020;146(7):1711–1723. doi: 10.1007/s00432-020-03224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coker H., Wei G., Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):310–318. doi: 10.1016/j.bbagrm.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C., Fu J., Zhou Y. A review in research progress concerning m6A methylation and immunoregulation. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu G.M., Zeng H.D., Zhang C.Y., et al. Identification of METTL3 as an adverse prognostic biomarker in hepatocellular carcinoma. Dig Dis Sci. 2021;66(4):1110–1126. doi: 10.1007/s10620-020-06260-z. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., Lin Y., Shu Y., et al. Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-01207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S., Liu F., Wu Z., et al. Contribution of m6A subtype classification on heterogeneity of sepsis. Ann Transl Med. 2020;8(6) doi: 10.21037/atm.2020.03.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L., Liu W.K., Du X.W., et al. Large-scale transcriptome analysis identified RNA methylation regulators as novel prognostic signatures for lung adenocarcinoma. Ann Transl Med. 2020;8(12) doi: 10.21037/atm-20-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T., Kong S., Tao M., Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng Y., Guan R., Hong W., et al. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann Transl Med. 2020;8(6) doi: 10.21037/atm.2020.03.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H., Bai Y., Lu X., et al. N6-methyladenosine associated prognostic model in hepatocellular carcinoma. Ann Transl Med. 2020;8(10) doi: 10.21037/atm-20-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenjaroenpun P., Wongsurawat T., Wadley T.D., et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 2021;49(2) doi: 10.1093/nar/gkaa620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Körtel N., Rücklé C., Zhou Y., et al. Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning. Nucleic Acids Res. 2021;49(16) doi: 10.1093/nar/gkab485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z., Zhao P., Li F., et al. Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Briefings Bioinf. 2020;21(5):1676–1696. doi: 10.1093/bib/bbz112. [DOI] [PubMed] [Google Scholar]
- 19.He Y., Xing J., Wang S., et al. Increased m6A methylation level is associated with the progression of human abdominal aortic aneurysm. Ann Transl Med. 2019;7(24) doi: 10.21037/atm.2019.12.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Feng J., Xue Y., et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534(7608):575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 21.Liu X., Qin J., Gao T., et al. Analysis of METTL3 and METTL14 in hepatocellular carcinoma. Aging (Albany NY) 2020;12(21):21638–21659. doi: 10.18632/aging.103959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fazi F., Fatica A. Interplay between N (6)-methyladenosine (m(6)A) and non-coding RNAs in cell development and cancer. Front Cell Dev Biol. 2019;7 doi: 10.3389/fcell.2019.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satterwhite E.R., Mansfield K.D. RNA Methyltransferase METTL16: Targets and Function. Wiley Interdiscip Rev RNA. 2021 doi: 10.1002/wrna.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue Y., Liu J., Cui X., et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4 doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong P.J., Shao Y.C., Yang Y., et al. Analysis of N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as tumor suppressor genes in breast cancer. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.578963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J., Yang D., Ma X.X. Immune infiltration-related N6-methyladenosine RNA methylation regulators influence the malignancy and prognosis of endometrial cancer. Aging (Albany NY) 2021;13(12):16287–16315. doi: 10.18632/aging.203157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang L., Zhang M., Wu J., et al. Exploring diagnostic m6A regulators in endometriosis. Aging (Albany NY) 2020;12(24):25916–25938. doi: 10.18632/aging.202163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toh J.D.W., Crossley S.W.M., Bruemmer K.J., et al. Distinct RNA N-demethylation pathways catalyzed by nonheme iron ALKBH5 and FTO enzymes enable regulation of formaldehyde release rates. Proc Natl Acad Sci U S A. 2020;117(41):25284–25292. doi: 10.1073/pnas.2007349117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ondo K., Isono M., Nakano M., et al. The N(6)-methyladenosine modification posttranscriptionally regulates hepatic UGT2B7 expression. Biochem Pharmacol. 2021;189 doi: 10.1016/j.bcp.2020.114402. [DOI] [PubMed] [Google Scholar]
- 30.Su R., Dong L., Li Y., et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38(1):79–96. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., Yan J., Li Q., et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43(1):373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y., Su R., Sheng Y., et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35(4):677–691. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang Q., Shao B., Zhou Q., et al. RNA N (6)-methyladenosine in cancer metastasis: roles, mechanisms, and applications. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.681781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G., Yan Y., Cai Y., et al. ALKBH1-8 and FTO: potential therapeutic targets and prognostic biomarkers in lung adenocarcinoma pathogenesis. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.633927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 36.Sokpor G., Xie Y., Nguyen H.P., et al. Emerging role of m(6) A methylome in brain development: implications for neurological disorders and potential treatment. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.656849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y., Shi Y., Shen H., et al. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020;13(1) doi: 10.1186/s13045-020-00872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng B., Yan Y., Xu Z. The bioinformatics and experimental analysis of AlkB family for prognosis and immune cell infiltration in hepatocellular carcinoma. PeerJ. 2021;9 doi: 10.7717/peerj.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang C., Yin C., Lin K., et al. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med. 2021;11(6) doi: 10.1002/ctm2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M.H., Fu L.S., Zhang F., et al. LncAY controls BMI1 expression and activates BMI1/Wnt/β-catenin signaling axis in hepatocellular carcinoma. Life Sci. 2021;280 doi: 10.1016/j.lfs.2021.119748. [DOI] [PubMed] [Google Scholar]
- 41.Shen X.P., Ling X., Lu H., et al. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur Rev Med Pharmacol Sci. 2018;22(23):8220–8226. doi: 10.26355/eurrev_201812_16516. [DOI] [PubMed] [Google Scholar]
- 42.Nasr M.A., Salah R.A., Abd Elkodous M., et al. Dysregulated microRNA fingerprints and methylation patterns in hepatocellular carcinoma, cancer stem cells, and mesenchymal stem cells. Front Cell Dev Biol. 2019;7 doi: 10.3389/fcell.2019.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiss T., Giles C.B., Tarantini S., et al. Nicotinamide mononucleotide (NMN) supplementation promotes anti-aging miRNA expression profile in the aorta of aged mice, predicting epigenetic rejuvenation and anti-atherogenic effects. GeroScience. 2019;41(4):419–439. doi: 10.1007/s11357-019-00095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju Q., Zhao Y.J., Dong Y., et al. Identification of a miRNA-mRNA network associated with lymph node metastasis in colorectal cancer. Oncol Lett. 2019;18(2):1179–1188. doi: 10.3892/ol.2019.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dexheimer P.J., Cochella L. MicroRNAs: from mechanism to organism. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vishlaghi N., Lisse T.S. Dicer- and bulge stem cell-dependent microRNAs during induced anagen hair follicle development. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alarcón C.R., Lee H., Goodarzi H., et al. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han J., Wang J.Z., Yang X., et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin R., Zhan M., Yang L., et al. Deoxycholic acid modulates the progression of gallbladder cancer through N(6)-methyladenosine-dependent microRNA maturation. Oncogene. 2020;39(26):4983–5000. doi: 10.1038/s41388-020-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J., Ishfaq M., Xu L., et al. METTL3/m(6)A/miRNA-873-5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating Keap1/Nrf2 pathway. Front Pharmacol. 2019;10 doi: 10.3389/fphar.2019.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma J.Z., Yang F., Zhou C.C., et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 52.Peng W., Li J., Chen R., et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38(1) doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J., Bai R., Li M., et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-09712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H., Deng Q., Lv Z., et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1108-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Yang L., Ma Y., Han W., et al. Proteinase-activated receptor 2 promotes cancer cell migration through RNA methylation-mediated repression of miR-125b. J Biol Chem. 2015;290(44):26627–26637. doi: 10.1074/jbc.M115.667717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ngo T.D., Partin A.C., Nam Y. RNA specificity and autoregulation of DDX17, a modulator of microRNA biogenesis. Cell Rep. 2019;29(12):4024–4035. doi: 10.1016/j.celrep.2019.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu K.J. The role of miRNA biogenesis and DDX17 in tumorigenesis and cancer stemness. Biomed J. 2020;43(2):107–114. doi: 10.1016/j.bj.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah A., Rashid F., Awan H.M., et al. The DEAD-Box RNA helicase DDX3 interacts with m(6)A RNA demethylase ALKBH5. Stem Cell Int. 2017;2017 doi: 10.1155/2017/8596135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alarcón C.R., Goodarzi H., Lee H., et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klinge C.M., Piell K.M., Tooley C.S., et al. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-45636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu B., Su S., Patil D.P., et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9(1) doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choe J., Lin S., Zhang W., et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561(7724):556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin D., Guo J., Wu Y., et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12(1) doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Cai X., Wang X., Cao C., et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 65.He H., Wu W., Sun Z., et al. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem Biophys Res Commun. 2019;517(4):581–587. doi: 10.1016/j.bbrc.2019.07.058. [DOI] [PubMed] [Google Scholar]
- 66.Du M., Zhang Y., Mao Y., et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482(4):582–589. doi: 10.1016/j.bbrc.2016.11.077. [DOI] [PubMed] [Google Scholar]
- 67.Cui X., Wang Z., Li J., et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53(3) doi: 10.1111/cpr.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou J., Zhang H., Liu J., et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Z., Li J., Feng G., et al. MicroRNA-145 modulates N(6)-methyladenosine levels by targeting the 3'-untranslated mRNA region of the N(6)-methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292(9):3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montero-Conde C., Graña-Castro O., Martín-Serrano G., et al. Hsa-miR-139-5p is a prognostic thyroid cancer marker involved in HNRNPF-mediated alternative splicing. Int J Cancer. 2020;146(2):521–530. doi: 10.1002/ijc.32622. [DOI] [PubMed] [Google Scholar]
- 71.Kleemann M., Schneider H., Unger K., et al. MiR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-27438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Melnik B.C. Milk: an epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. J Transl Med. 2015;13 doi: 10.1186/s12967-015-0746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Y., Zhang H., Ju Q., et al. Comprehensive analysis of survival-related lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in gastric cancer. Front Genet. 2021;12 doi: 10.3389/fgene.2021.610501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan Y., Xu Z., Chen X., et al. Novel function of lncRNA ADAMTS9-AS2 in promoting temozolomide resistance in glioblastoma via upregulating the FUS/MDM2 ubiquitination axis. Front Cell Dev Biol. 2019;7 doi: 10.3389/fcell.2019.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ou C., Sun Z., He X., et al. Targeting YAP1/LINC00152/FSCN1 signaling axis prevents the progression of colorectal cancer. Adv Sci. 2020;7(3) doi: 10.1002/advs.201901380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J., Guo S., Piao H.Y., et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75(3):379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Visvanathan A., Patil V., Abdulla S., et al. N(6)-methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes. 2019;10(2) doi: 10.3390/genes10020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng Z.Q., Li Z.X., Zhou G.Q., et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79(18):4612–4626. doi: 10.1158/0008-5472.CAN-19-0799. [DOI] [PubMed] [Google Scholar]
- 79.Ban Y., Tan P., Cai J., et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14(6):1282–1296. doi: 10.1002/1878-0261.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barros-Silva D., Lobo J., Guimarães-Teixeira C., et al. VIRMA-dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers. 2020;12(4) doi: 10.3390/cancers12040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang W., Li J., Lin F., et al. Identification of N(6)-methyladenosine-related lncRNAs for patients with primary glioblastoma. Neurosurg Rev. 2021;44(1):463–470. doi: 10.1007/s10143-020-01238-x. [DOI] [PubMed] [Google Scholar]
- 82.McCown P.J., Wang M.C., Jaeger L., et al. Secondary structural model of human MALAT1 reveals multiple structure-function relationships. Int J Mol Sci. 2019;20(22) doi: 10.3390/ijms20225610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuo X., Chen Z., Gao W., et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13(1) doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J., Ding W., Xu Y., et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) 2020;12(5):4558–4572. doi: 10.18632/aging.102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shang W., Gao Y., Tang Z., et al. The pseudogene Olfr29-ps1 promotes the suppressive function and differentiation of monocytic MDSCs. Cancer Immunol Res. 2019;7(5):813–827. doi: 10.1158/2326-6066.CIR-18-0443. [DOI] [PubMed] [Google Scholar]
- 86.Hu X., Peng W.X., Zhou H., et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27(6):1782–1794. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu S., Zhang L., Deng J., et al. A novel micropeptide encoded by Y-linked LINC00278 links cigarette smoking and AR signaling in male esophageal squamous cell carcinoma. Cancer Res. 2020;80(13):2790–2803. doi: 10.1158/0008-5472.CAN-19-3440. [DOI] [PubMed] [Google Scholar]
- 88.Banday A.R., Papenberg B.W., Prokunina-Olsson L. When the smoke clears m(6)A from a Y chromosome-linked lncRNA, men get an increased risk of cancer. Cancer Res. 2020;80(13):2718–2719. doi: 10.1158/0008-5472.CAN-20-0961. [DOI] [PubMed] [Google Scholar]
- 89.Patil D.P., Chen C.K., Pickering B.F., et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang X., Zhang S., He C., et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen S., Zhou L., Wang Y. ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20 doi: 10.1186/s12935-020-1105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ni W., Yao S., Zhou Y., et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jacob R., Zander S., Gutschner T. The dark side of the epitranscriptome: chemical modifications in long non-coding RNAs. Int J Mol Sci. 2017;18(11) doi: 10.3390/ijms18112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dinescu S., Ignat S., Lazar A.D., et al. Epitranscriptomic signatures in lncRNAs and their possible roles in cancer. Genes. 2019;10(1) doi: 10.3390/genes10010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu Y., Yang X., Chen Z., et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun T., Wu Z., Wang X., et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39(31):5358–5372. doi: 10.1038/s41388-020-1338-9. [DOI] [PubMed] [Google Scholar]
- 97.Zhu L., Zhu Y., Han S., et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10(6) doi: 10.1038/s41419-019-1585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yan J., Huang X., Zhang X., et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521(4):887–893. doi: 10.1016/j.bbrc.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 99.Lan T., Li H., Zhang D., et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang X., Zhang J., Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11(8):4909–4921. [PMC free article] [PubMed] [Google Scholar]
- 101.Li X.D., Wang M.J., Zheng J.L., et al. Long noncoding RNA just proximal to X-inactive specific transcript facilitates aerobic glycolysis and temozolomide chemoresistance by promoting stability of PDK1 mRNA in an m6A-dependent manner in glioblastoma multiforme cells. Cancer Sci. 2021;112(11):4543–4552. doi: 10.1111/cas.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang S., Zhao B.S., Zhou A., et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu B., Zhou J., Wang C., et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020;11(5) doi: 10.1038/s41419-020-2540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu S., Wang J.Z., Chen D., et al. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-15403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu X., Fan Y., Liu Y., et al. Long non-coding RNA CCAT2 promotes the development of esophageal squamous cell carcinoma by inhibiting miR-200b to upregulate the IGF2BP2/TK1 axis. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.680642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y., Lu J.H., Wu Q.N., et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu Z., Yan Y., Zeng S., et al. Circular RNAs: clinical relevance in cancer. Oncotarget. 2017;9(1):1444–1460. doi: 10.18632/oncotarget.22846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haque S., Ames R.M., Moore K., et al. circRNAs expressed in human peripheral blood are associated with human aging phenotypes, cellular senescence and mouse lifespan. GeroScience. 2020;42(1):183–199. doi: 10.1007/s11357-019-00120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu X., Xiao Y., Ma J., et al. Circular RNA: a novel potential biomarker for skin diseases. Pharmacol Res. 2020;158 doi: 10.1016/j.phrs.2020.104841. [DOI] [PubMed] [Google Scholar]
- 110.Meng S., Zhou H., Feng Z., et al. Epigenetics in neurodevelopment: emerging role of circular RNA. Front Cell Neurosci. 2019;13 doi: 10.3389/fncel.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma S., Chen C., Ji X., et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1) doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun H.D., Xu Z.P., Sun Z.Q., et al. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-27837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou C., Molinie B., Daneshvar K., et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20(9):2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Diallo L.H., Tatin F., David F., et al. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie. 2019;164:45–52. doi: 10.1016/j.biochi.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 115.Su H., Wang G., Wu L., et al. Transcriptome-wide map of m(6)A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genom. 2020;21(1) doi: 10.1186/s12864-020-6462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen R.X., Chen X., Xia L.P., et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang G., Li S., Yang N., et al. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 118.Ji P., Wu W., Chen S., et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26(12):3444–3460. doi: 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- 119.Xiao M.S., Ai Y., Wilusz J.E. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30(3):226–240. doi: 10.1016/j.tcb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tatomer D.C., Wilusz J.E. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol Cell. 2017;66(1):1–2. doi: 10.1016/j.molcel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 121.Tang C., Klukovich R., Peng H., et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2018;115(2):E325–E333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang C., Xie Y., Yu T., et al. m(6)A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020;30(3):211–228. doi: 10.1038/s41422-020-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee Y., Choe J., Park O.H., et al. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 2020;36(3):177–188. doi: 10.1016/j.tig.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 124.Park O.H., Ha H., Lee Y., et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74(3):494–507. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 125.Du H., Zhao Y., He J., et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7 doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu W., Ji P., Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020;21(1) doi: 10.1186/s13059-020-02018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kong S., Tao M., Shen X., et al. Translatable circRNAs and lncRNAs: driving mechanisms and functions of their translation products. Cancer Lett. 2020;483:59–65. doi: 10.1016/j.canlet.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 128.Das A., Gorospe M., Panda A.C. The coding potential of circRNAs. Aging (Albany NY) 2018;10(9):2228–2229. doi: 10.18632/aging.101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meyer K.D., Patil D.P., Zhou J., et al. 5' UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lei M., Zheng G., Ning Q., et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang Y., Fan X., Mao M., et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Q., Liu H., Liu Z., et al. Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genet. 2020;240:33–39. doi: 10.1016/j.cancergen.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 133.Legnini I., Di Timoteo G., Rossi F., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Di Timoteo G., Dattilo D., Centrón-Broco A., et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020;31(6) doi: 10.1016/j.celrep.2020.107641. [DOI] [PubMed] [Google Scholar]
- 135.Zhao J., Lee E.E., Kim J., et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang L., Hou C., Chen C., et al. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hansen T.B., Jensen T.I., Clausen B.H., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 138.Ren S., Lin P., Wang J., et al. Circular RNAs: promising molecular biomarkers of human aging-related diseases via functioning as an miRNA sponge. Mol Ther Methods Clin Dev. 2020;18:215–229. doi: 10.1016/j.omtm.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.He L., Li H., Wu A., et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Q., Zhang H., Chen Q., et al. Identification of METTL14 in kidney renal clear cell carcinoma using bioinformatics analysis. Dis Markers. 2019;2019 doi: 10.1155/2019/5648783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang R., Zhang Y., Bai Y., et al. N(6)-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatr. 2020;88(5):392–404. doi: 10.1016/j.biopsych.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 142.Li L.J., Fan Y.G., Leng R.X., et al. Potential link between m(6)A modification and systemic lupus erythematosus. Mol Immunol. 2018;93:55–63. doi: 10.1016/j.molimm.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 143.Zhang M., Song J., Yuan W., et al. Roles of RNA methylation on tumor immunity and clinical implications. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.641507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sekar D., Lakshmanan G. Methylation of N6-adenosine (m6A) modification in miRNAs and its implications in immunity. Epigenomics. 2020;12(13):1083–1085. doi: 10.2217/epi-2020-0131. [DOI] [PubMed] [Google Scholar]
- 145.Lin Y., Yao Y., Wang Y., et al. PD-L1 and immune infiltration of m(6)A RNA methylation regulators and its miRNA regulators in hepatocellular carcinoma. BioMed Res Int. 2021;2021 doi: 10.1155/2021/5516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kumar S., Nagpal R., Kumar A., et al. Immunotherapeutic potential of m6A-modifiers and microRNAs in controlling acute myeloid leukaemia. Biomedicines. 2021;9(6) doi: 10.3390/biomedicines9060690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jin Y., Wang Z., He D., et al. Analysis of m6A-related signatures in the tumor immune microenvironment and identification of clinical prognostic regulators in adrenocortical carcinoma. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.637933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Y., Wang Y., Luo W., et al. Roles of long non-coding RNAs and emerging RNA-binding proteins in innate antiviral responses. Theranostics. 2020;10(20):9407–9424. doi: 10.7150/thno.48520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang H., Meng Q., Ma B. Characterization of the prognostic m6A-related lncRNA signature in gastric cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.630260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu J., Zhang X., Chen K., et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019;50(3):600–615. doi: 10.1016/j.immuni.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 151.Li Q., Wen S., Ye W., et al. The potential roles of m(6)A modification in regulating the inflammatory response in microglia. J Neuroinflammation. 2021;18(1) doi: 10.1186/s12974-021-02205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yu Z.L., Zhu Z.M. Comprehensive analysis of N6-methyladenosine -related long non-coding RNAs and immune cell infiltration in hepatocellular carcinoma. Bioengineered. 2021;12(1):1708–1724. doi: 10.1080/21655979.2021.1923381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yan L., Chen Y.G. Circular RNAs in immune response and viral infection. Trends Biochem Sci. 2020;45(12):1022–1034. doi: 10.1016/j.tibs.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Paramasivam A., Vijayashree Priyadharsini J. Novel insights into m6A modification in circular RNA and implications for immunity. Cell Mol Immunol. 2020;17(6):668–669. doi: 10.1038/s41423-020-0387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Han D., Liu J., Chen C., et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(7743):270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen Y.G., Chen R., Ahmad S., et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76(1):96–109. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu B., Liu N., Zhu X., et al. Circular RNA circZbtb20 maintains ILC3 homeostasis and function via Alkbh5-dependent m(6)A demethylation of Nr4a1 mRNA. Cell Mol Immunol. 2021;18(6):1412–1424. doi: 10.1038/s41423-021-00680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li B., Zhu L., Lu C., et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12(1) doi: 10.1038/s41467-020-20527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang X., Wei X., Yang J., et al. Epitranscriptomic regulation of insecticide resistance. Sci Adv. 2021;7(19) doi: 10.1126/sciadv.abe5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Q., He W., Wan G. Methyladenosine modification in RNAs: classification and roles in gastrointestinal cancers. Front Oncol. 2021;10 doi: 10.3389/fonc.2020.586789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Peng L., Yuan X., Jiang B., et al. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 2016;37(3):2779–2788. doi: 10.1007/s13277-015-4663-9. [DOI] [PubMed] [Google Scholar]
- 162.Huang H., Weng H., Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37(3):270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.He L., Li J., Wang X., et al. The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J Cell Mol Med. 2018;22(10):4630–4639. doi: 10.1111/jcmm.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pan X., Hong X., Li S., et al. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med. 2021;53(1):91–102. doi: 10.1038/s12276-020-00510-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Song Z., Jia G., Ma P., et al. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021;276 doi: 10.1016/j.lfs.2021.119399. [DOI] [PubMed] [Google Scholar]
- 166.Yang L., Deng W.L., Zhao B.G., et al. FOXO3-induced lncRNA LOC554202 contributes to hepatocellular carcinoma progression via the miR-485-5p/BSG axis. Cancer Gene Ther. 2022;29(3-4):326–340. doi: 10.1038/s41417-021-00312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lin Z., Niu Y., Wan A., et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39(12) doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gu Z., Zhou Y., Cao C., et al. TFAP2C-mediated LINC00922 signaling underpins doxorubicin-resistant osteosarcoma. Biomed Pharmacother. 2020;129 doi: 10.1016/j.biopha.2020.110363. [DOI] [PubMed] [Google Scholar]
- 169.Wei J., Yin Y., Zhou J., et al. METTL3 potentiates resistance to cisplatin through m(6) A modification of TFAP2C in seminoma. J Cell Mol Med. 2020;24(19):11366–11380. doi: 10.1111/jcmm.15738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu P., Fang X., Liu Y., et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12(4) doi: 10.1038/s41419-021-03558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xu J., Wan Z., Tang M., et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cai Y., Wu G., Peng B., et al. Expression and molecular profiles of the AlkB family in ovarian serous carcinoma. Aging (Albany NY) 2021;13(7):9679–9692. doi: 10.18632/aging.202716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ding C., Yi X., Chen X., et al. Warburg effect-promoted exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma. J Exp Clin Cancer Res. 2021;40(1) doi: 10.1186/s13046-021-01942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang Y., Cheng Z., Xu J., et al. Fat mass and obesity-associated protein (FTO) mediates signal transducer and activator of transcription 3 (STAT3)-drived resistance of breast cancer to doxorubicin. Bioengineered. 2021;12(1):1874–1889. doi: 10.1080/21655979.2021.1924544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu D., Jia H., Zhang Z., et al. STAT3-induced HLA-F-AS1 promotes cell proliferation and stemness characteristics in triple negative breast cancer cells by upregulating TRABD. Bioorg Chem. 2021;109 doi: 10.1016/j.bioorg.2021.104722. [DOI] [PubMed] [Google Scholar]
- 176.Liu H., Flores M.A., Meng J., et al. MeT-DB: a database of transcriptome methylation in mammalian cells. Nucleic Acids Res. 2015;43(Database issue):D197–D203. doi: 10.1093/nar/gku1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu H., Wang H., Wei Z., et al. MeT-DB V2.0: elucidating context-specific functions of N6-methyl-adenosine methyltranscriptome. Nucleic Acids Res. 2018;46(D1):D281–D287. doi: 10.1093/nar/gkx1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sun W.J., Li J.H., Liu S., et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016;44(D1):D259–D265. doi: 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xuan J.J., Sun W.J., Lin P.H., et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46(D1):D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Deng S., Zhang H., Zhu K., et al. M6A2Target: a comprehensive database for targets of m6A writers, erasers and readers. Briefings Bioinf. 2021;22(3) doi: 10.1093/bib/bbaa055. [DOI] [PubMed] [Google Scholar]
- 181.Cui X., Meng J., Zhang S., et al. A hierarchical model for clustering m(6)A methylation peaks in MeRIP-seq data. BMC Genom. 2016;17(suppl 7) doi: 10.1186/s12864-016-2913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cui X., Zhang L., Meng J., et al. MeTDiff: a novel differential RNA methylation analysis for MeRIP-Seq data. IEEE ACM Trans Comput Biol Bioinf. 2018;15(2):526–534. doi: 10.1109/TCBB.2015.2403355. [DOI] [PubMed] [Google Scholar]
- 183.Chen X., Hao Y., Cui Y., et al. LncVar: a database of genetic variation associated with long non-coding genes. Bioinformatics. 2017;33(1):112–118. doi: 10.1093/bioinformatics/btw581. [DOI] [PubMed] [Google Scholar]
- 184.Han Y., Feng J., Xia L., et al. CVm6A: a visualization and exploration database for m(6)As in cell lines. Cells. 2019;8(2) doi: 10.3390/cells8020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu L., Lei X., Fang Z., et al. LITHOPHONE: improving lncRNA methylation site prediction using an ensemble predictor. Front Genet. 2020;11 doi: 10.3389/fgene.2020.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu M., Wang Q., Shen J., et al. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16(7):899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zheng Y., Nie P., Peng D., et al. m6Avar: a database of functional variants involved in m6A modification. Nucleic Acids Res. 2018;46(D1):D139–D145. doi: 10.1093/nar/gkx895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu S., Zhu A., He C., et al. REPIC: a database for exploring the N(6)-methyladenosine methylome. Genome Biol. 2020;21(1) doi: 10.1186/s13059-020-02012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhou Y., Zeng P., Li Y.H., et al. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44(10) doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chen K., Wei Z., Zhang Q., et al. WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res. 2019;47(7) doi: 10.1093/nar/gkz074. [DOI] [PMC free article] [PubMed] [Google Scholar]