Abstract
The growth of endophytic bacteria is influenced by the host plants and their secondary metabolites and activities. In this study, P. megaterium P-NA14 and P. megaterium D-HT207 were isolated from potato tuber and dendrobium stem respectively. They were both identified as Priestia megaterium. The antimicrobial activities and metabolites of both strains were explored. For antimicrobial activities, results showed that P. megaterium P-NA14 exhibited a stronger inhibition effect on the pathogen of dendrobium, while P. megaterium D-HT207 exhibited a stronger inhibition effect on the pathogen of potato. The supernatant of P. megaterium P-NA14 showed an inhibition effect only on Staphylococcus aureus, while the sediment of P. megaterium D-HT207 showed an inhibition effect only on Escherichia coli. For metabolomic analysis, the content of L-phenylalanine in P. megaterium P-NA14 was higher than that of P. megaterium D-HT207, and several key downstream metabolites of L-phenylalanine were associated with inhibition of S. aureus including tyrosine, capsaicin, etc. Therefore, we speculated that the different antimicrobial activities between P. megaterium P-NA14 and P. megaterium D-HT207 were possibly related to the content of L-phenylalanine and its metabolites. This study preliminarily explored why the same strains isolated from different hosts exhibit different activities from the perspective of metabolomics.
Subject terms: Microbiology, Bacteria, Environmental microbiology
Introduction
Endophytes are microorganisms that exist inside plants and are not harmful to their hosts. In recent years, endophytes have attracted extensive attention, since they play an important role in enhancing plant growth and maintaining plant health1,2. Bacterial endophytes can provide various benefits for host plants, especially in promoting growth and resisting pathogens3 or inducing the innate immune system of plants4. Several research findings have revealed that the endophytes can promote plant growth by fixing nitrogen, making nutrient acquisition from soil, producing antimicrobial metabolites and modulating phytohormones status of plants3,5. In addition, endophytes can protect plants against biotic and abiotic stresses6–8. The development of new products such as endophyte-based microbial formulations and inducers shows great potential to improve plant resistance and provide abundant sources of important secondary metabolites.
In recent years, endophytic bacterial diversity of various plant species has been studied. The most frequently isolated bacterial genera include Bacillus, Stenotrophomonas, Burkholderia and Pseudomonas, while Bacillus is one of the main genera9–11. The taxonomy of Bacillus has recently been revisited through comprehensive phylogenomic and comparative genomic approaches12. Bacillus megaterium was renamed as Priestia megaterium, which is a new separate genus from Bacillus. According to the reports, P. megaterium is considered as a potential biological control agent, which has antimicrobial activities and a variety of control effects on plant diseases13–15. P. megaterium can be isolated from various plants, such as alfalfa, black pepper, carrot, clover, cotton, cucumber, potato, wheat, ginseng, dendrobium, polygonatum sibiricum5,16,17. Three different mechanisms to promote the plant growth by P. megaterium have been described in previous studies. Firstly, P. megaterium can secrete organic acids that providing the foundation of phosphate solubilization18–20. Secondly, P. megaterium can be responsible for concentration changes of phytohormones and other regulators of plant growth21–23. Thirdly, P. megaterium can also act as a biopesticide or biocontrol agent24,25.
For instance, P. megaterium strain B388 was isolated from rhizosphere soil of pine at Jageshwar, District Almora, Uttarakhand in India. P. megaterium B388 inhibited the growth of phytopathogens such as Alternaria alternata and Fusarium oxysporum by producing the spreadable and volatile compounds26. Kong et.al demonstrated that the P. megaterium extracted from marine bacterium could be used as a biocontrol agent in 201027. The result of this study demonstrated that the P. megaterium from the Yellow Sea of East China exhibited significant activity in reducing infections caused by Aspergillus flavus on peanut seeds for the first time. At the same time, Xie et al. found that β-sitosterol, behenic acid, and phenylacetic acid extracted from P. megaterium L2 exhibited ideal antimicrobial activity against three gram-negative plant pathogen indicator bacteria as followed: Agrobacterium tumefaciens T-37, Erwinia carotovora EC-1 and Ralstonia solanacearum RS-228.
In this research, two P. megaterium strains from different hosts (potato and dendrobium) were used as the studied strains. Both strains were evaluated by 16S rDNA sequences analysis as well as genomic analysis. Meanwhile, the two strains were evaluated for antimicrobial activity against five phytopathogens and human pathogens, including Pectobacterium atroseptica, Athelia rolfsii, Staphylococcus aureus and Escherichia coli. To further explore the different metabolites and metabolic pathways of these two strains, the extracts were submitted to liquid chromatography-mass (LC–MS) for nontargeted metabolomics. The reasons of why the same species in different hosts showed the different antimicrobial activities were preliminarily explored from perspective of metabolomic.
Materials and methods
Identification of endophytic bacteria
16S rDNA molecular identification and phylogenetic tree analysis
Two endophytic strains of P-NA14 and D-HT207 were isolated from potato and dendrobium samples according to the method of our previous studies, respectively29,30. Extraction of endophytic bacteria DNA and sequencing was according to the method described by Wang et al. and Zhou et al.29,31. Related sequences were aligned and the phylogenetic tree was pictured using the maximum likelihood method with MEGA 7.0 software.
All plant materials used in this study comply with national guidelines. In addition, experimental research on endophytic bacteria was strictly comply with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Biochemical and physiological characterization
Based on Bergey’s Manual of Systematic Bacteriology, the identification of P. megaterium P-NA14 and P. megaterium D-HT207 was performed on morphological and biochemical characterization32. P. megaterium P-NA14 and P. megaterium D-HT207 were characterized based on the following biochemical tests: Hydrogen sulphide production test, Gelatin Liquefaction test, Methyl red test, Starch hydrolysis test, Urease test, Catalase test, Indole production test, Oxidase test, Voges-Proskauer test, Nitrate reduction test33,34.
High-throughput genome sequencing and genomic similarity analysis
DNA extracted from the P. megaterium P-NA14 and P. megaterium D-HT207 were subjected to whole genome sequencing by HiSeq × 10 platform (Illumina) following the supplier’s protocol (Illumina, UK), respectively.
The evaluation of similarity between the Coding sequence (CDS) of P. megaterium P-NA14 and P. megaterium D-HT207 was performed by BLAST. The protein sequences obtained were selected and aligned by GTDB-Tk v0.3.235. Values of average nucleotide identity (ANI) and average amino acid identity (AAI) were calculated using the program JSpecies, version 1.2.136. The sequences and annotations of other strains analyzed in this study were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov).
Antimicrobial activity
Endophytic bacteria fermentation and extraction
The Fermentation conditions of P. megaterium P-NA14 and P. megaterium D-HT207 were exactly the same. Single colonies of those two strains were injected into 50 mL liquid YIM38 medium29 and cultured at 200 rpm at 28 °C until bacteria reached 107 mL−1 CFU (colony-forming unit), respectively. Then 20μL seed liquid was injected into 150 mL liquid YIM38 medium and cultured at the same conditions for 48 h, respectively. The culture liquid was centrifuged at 9500 rpm at 4 °C for 20 min. Then the supernatant and deposit were collected respectively.
The supernatant was mixed with an equal volume of ethyl acetate and collected by a separating funnel, then dried by rotary evaporator (RE-52AA; YARONG, China) at 38 °C. The sediment mixed with 15 mL of acetone for 12 h was filtered through a 0.22 μm nylon membrane filter and dried by a rotary evaporator. The extracts of supernatant and sediment were dissolved in 1 mL methanol and stored at − 20 °C prepared for experiments, respectively.
Antimicrobial activity screening
The antimicrobial activities of P. megaterium P-NA14 and P. megaterium D-HT207 were evaluated by five indicator strains as listed in Supplementary Table S1. Pectobacterium atroseptica was obtained from the Agricultural Culture Collection of China (ACCC). Athelia rolfsii was obtained from the Sanming Academy of Agricultural Sciences, Fujian Province, China. Staphylococcus aureus and Escherichia coli were deposited in Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College. Antimicrobial activities were tested by Kirby-Bauer test37. Twenty microliters of each sample were loaded on sterilized filter paper with a diameter of 6 mm. An equal amount of methanol was used as the negative control. Samples were cultured at 28 °C for 20 h. Antimicrobial activities were evaluated by measuring the diameter of the inhibition zones using an electronic digital caliper (0–150 mm).
Metabolomics comparative analysis
Metabolite extraction
The supernatant was dried and redissolved in 100µL resolution (acetonitrile: water = 1:1, v/v), then it was transferred to sample bottles for LC–MS/MS analysis after centrifugation at 13,000 rpm at 4 °C for 15 min. The samples of P. megaterium P-NA14 were named A14 group, and samples of P. megaterium D-HT207 were named A207 group during metabolomics analysis. Both samples were set as 6 parallel samples.
UPLC–TOF/MS analysis
Chromatographic separation of the metabolites was performed on ExionLC AD system (AB Sciex, USA) equipped with an ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm i.d., 1.7 µm; Waters, Milford, USA). The UPLC system was coupled to a quadrupole-time-of-flight mass spectrometer (Triple TOFTM5600+, AB Sciex, USA). The optimal conditions were set as listed (Supplementary Tables S2, S3). Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in acetonitrile: isopropanol (1:1, v/v). During the period of analysis, the rest samples were stored at 4 °C.
Data analysis
After UPLC-TOF/MS analysis, the raw data were imported into the Progenesis QI 2.3 (Waters Corporation, Milford, USA). The extracted data included the retention time (RT), mass-to-charge ratio (m/z) values, and peak intensity. Mass spectra of these metabolic features were identified by using the accurate mass fragments spectra and isotope ratio difference searching from reliable biochemical databases such as Metlin database (https://metlin.scripps.edu/). The internal standard was used for data QC (reproducibility), deleting variables with Relative Standard Deviation (RSD) > 30% in the QC samples.
Multivariate statistical and differential metabolites analysis
All multivariate statistical data analysis was performed using ropls (Version 1.6.2) from Bioconductor on Majorbio Cloud Platform (https://cloud.majorbio.com). Unsupervised principal component analysis (PCA), partial least squares discriminant analyses (PLS-DA) and orthogonal partial least squares discriminant analyses (OPLS-DA) were performed to distinguish the overall differences in metabolic profiles between groups and to find differential metabolites between groups. Variable importance in the projection (VIP) and fold change (FC) were calculated in the OPLS-DA model. P values were estimated with paired Student’s test on Single dimensional statistical analysis. Statistically significant among groups were selected with VIP > 1, FC > 2(or < 0.5), and p values < 0.05.
Differential metabolites among two groups were summarized and mapped into their biochemical pathways through metabolic enrichment and pathway analysis based on the KEGG database (http://www.genome.jp/kegg/). These metabolites can be classified according to the pathways they were involved in or the functions they performed. Scipy.stats (Python packages) (https://docs.scipy.org/doc/scipy/) was employed to identify statistically significantly enriched pathways using Fisher’s exact test.
Results
Identification of endophytic bacteria
16S rDNA molecular identification and phylogenetic tree analysis
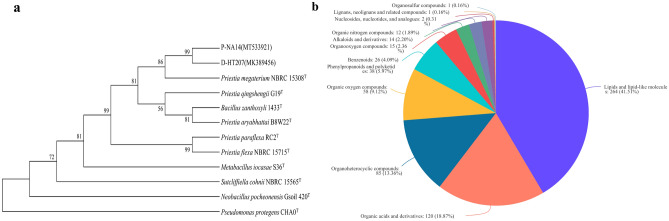
The 16S rDNA gene sequences of both strains were submitted to the NCBI GenBank to get the Accession Numbers. P. megaterium P-NA14 displayed 99.31% similarity with P. megaterium (Accession No. MT533921). P. megaterium D-HT207 displayed 99.12% similarity with P. megaterium (Accession No. MK389456). A phylogenetic tree was established for P. megaterium P-NA14 and P. megaterium D-HT207 to demonstrate their evolutionary relationship. The identification showed that P. megaterium P-NA14 and P. megaterium D-HT207 belonged to P. megaterium (Fig. 1a).
Figure 1.
(a) The phylogenetic tree of P. megaterium P-NA14 and P. megaterium D-HT207 was identified in this study. The identification showed that P. megaterium P-NA14 and P. megaterium D-HT207 belonged to the same species. (b) Classification of the identified metabolites annotated using the Metlin database.
Biochemical and physiological characterization
The results of the physiological and biochemical reactions of P. megaterium P-NA14 and P. megaterium D-HT207 turned up to be exactly the same although they were isolated from different hosts (Supplementary Table S4).
High-throughput genome sequencing and genomic similarity analysis
The complete genome of P. megaterium P-NA14 has a total size of 4 929 750 bp assembled in one scaffold, with a 37.84% G + C content. The complete genome of P. megaterium D-HT207 has a total size of 5 108 847 bp assembled in one scaffold, with a 38.15% G + C content. The genomes from P. megaterium P-NA14 and P. megaterium D-HT207 were deposited in the NCBI GenBank database, under the accession number CP109761-CP109767 and CP109859-CP109862, respectively. Supplementary Table S5 demonstrated the comparison between the genome sequence of P. megaterium P-NA14 and P. megaterium D-HT207.
The phylogenomic analysis based on the concatenated alignment of 120 orthologous proteins showed that the P. megaterium P-NA14 and P. megaterium D-HT207 were identified as P. megaterium (Supplementary Table S6). ANI and AAI represent the average nucleotide and the amino acid identity, respectively, of all orthologous genes shared between any two genomes and offer robust resolution between strains of the same or closely related species38,39. The values of ANI and AAI between P. megaterium P-NA14 and P. megaterium D-HT207 genomes were 97.34% and 96.7% above the 96% threshold, respectively (Table 1). Furthermore, the ANI and AAI values indicated that P. megaterium P-NA14 and P. megaterium D-HT207 had the closest evolutionary relationship with P. megaterium NBRC 15308. This result indicated that genomes of P. megaterium P-NA14 and P. megaterium D-HT207 belonged to the same species.
Table 1.
ANI and AAI values (%) between two strains and type strains of phylogenetically related species.
| Genome A | Genome B | Mean ANI (%) | Mean AAI (%) |
|---|---|---|---|
| P-NA14 | D-HT207 | 96.7 | 97.34 |
| P-NA14 | P. megaterium NBRC 15308 | 96.8 | 97.24 |
| P-NA14 | B. aryabhattai B8W22 | 95.4 | 96.58 |
| P-NA14 | B. flexus NBRC 15715 | 75.2 | 73.86 |
| D-HT207 | P-NA14 | 96.7 | 97.34 |
| D-HT207 | P. megaterium NBRC 15308 | 96.5 | 97.18 |
| D-HT207 | B. aryabhattai B8W22 | 95.6 | 96.66 |
| D-HT207 | B. flexus NBRC 15715 | 75.4 | 74.47 |
The highest ANI and AAI values between two strains and type strains of phylogenetically related species are in bold text.
Antimicrobial activity
The antimicrobial activities of P. megaterium P-NA14 and P. megaterium D-HT207 against five different pathogens were evaluated. The results showed inhibition diameters of P. megaterium P-NA14 and P. megaterium D-HT207 (Table 2). P. megaterium P-NA14 and P. megaterium D-HT207 showed antimicrobial activity on plant pathogens but showed different activity on S. aureus and E. coli. For P. atroseptica, P. megaterium D-HT207 showed higher activity than P. megaterium P-NA14, and the supernatant of P. megaterium D-HT207 possessed the strongest antimicrobial activity. For A. rolfsii, P. megaterium P-NA14 showed higher activity than P. megaterium D-HT207, and the supernatant of P. megaterium P-NA14 possessed the strongest antimicrobial activity. The supernatant of P. megaterium P-NA14 showed inhibition against S. aureus ATCC 29213, while the sediment of P. megaterium D-HT207 showed inhibition against E. coli. P. megaterium P-NA14 and P. megaterium D-HT207 showed no inhibition against S.aureus ATCC 33591.
Table 2.
Inhibition effect of fermentation broth of strain P-NA14 and strain D-HT207 on the different pathogens.
| Strain | Active fraction | P. atroseptica (mm) | A. rolfsii (mm) | S. Aureus ATCC 29213 (mm) | S. Aureus ATCC 33591 (mm) | E. coli (mm) |
|---|---|---|---|---|---|---|
| P-NA14 | Supernatant | 10.43 ± 0.19 | 12.88 ± 0.26 | 11.23 ± 0.38 | – | – |
| Sediment | 13.53 ± 1.11 | 8.06 ± 0.39 | – | – | – | |
| D-HT207 | Supernatant | 14.19 ± 1.62 | 10.23 ± 0.30 | – | – | – |
| Sediment | 12.14 ± 0.52 | 8.95 ± 0.34 | – | – | 10.78 ± 0.29 |
Metabolomics comparative analysis
To evaluate the metabolite changes of P. megaterium P-NA14 and P. megaterium D-HT207, the metabolic compounds of each sample were analyzed using an untargeted metabolomics approach. The results indicated that 4363 and 4881 metabolites were detected by the POS (positive) and NEG (negative) models, respectively. According to annotation in Metlin database, 437 compounds in POS and 389 compounds in NEG were defined as common compounds for A14 group and A207 group. After data preprocessing, 636 metabolites obtained from level-two identification were annotated to the Metlin database, which were matched and classified into 12 superclasses, including 264 lipids and lipid-like molecules; 120 organic acids; 85 organoheterocyclic compounds; 58 organic oxygen compounds; 38 phenylpropanoids and polyketides; 26 benzenoids; 15 organooxygen compounds; 14 alkaloids and derivatives; 12 organic nitrogen compounds; 2 nucleosides, nucleotides and analogues; 1 lignans, neolignans and related compounds and 1 organosulfur compounds (Fig. 1b).
Multivariate statistical analysis
Principal component analysis (PCA)
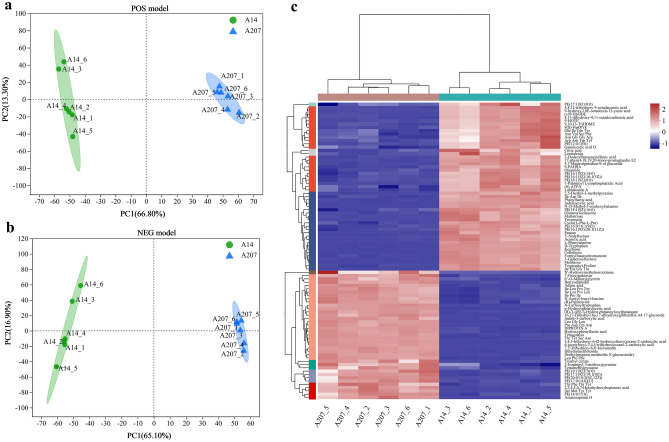
To get a preliminary understanding of the metabolites difference between A14 group and A207 group, the metabolic data matrix for A14 group and A207 group were analyzed using PCA plot. The PCA scores plot showed that the QC clustered tightly together (Supplementary Fig. S1), which indicated the QC repeatability was good and the stability of the analysis system was high.
The samples from the same group clustered, respectively. The A14 group was clustered together while A207 group was located on the opposite branch. Both the tested groups were well separated in the PCA plot, showing a remarkable variation in metabolism between two groups (Fig. 2a,b). The clustering heatmap also revealed significant variation in metabolites between A14 and A207 groups (Fig. 2c).
Figure 2.
(a,b) PCA scores plot of A14 and A207 group in positive model and negative model. (c) Top 90 metabolites clustering heatmap of all samples using HCA, the horizontal direction were the samples, and the longitudinal direction were the identified metabolites.
Partial least squares-discriminant analysis (PLS-DA)
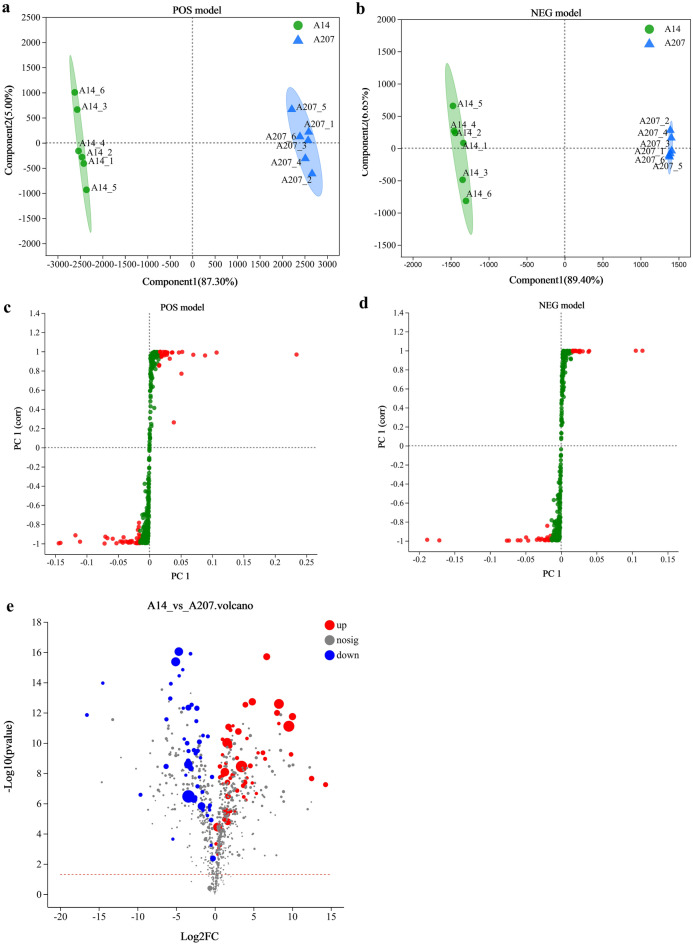
Similar to the PCA analysis, PLS-DA was used to maximize the slight difference between the samples, which was useful to detect differential metabolites. The six parallel samples of A14 group were distributed within the A14 group cluster, and six parallel samples of A207 group were within the A207 group cluster. In both positive and negative models, two clusters were clearly distinguished from the score plot, and the distribution trend was identical to the PCA results, indicating that there was a distinct difference between the A14 group and A207 group (Fig. 3a,b). A permutation test with 200 iterations was performed to evaluate the possible overfitting of the PLS-DA models. The R2 and Q2 were extremely close to 1, indicating the high accuracy in model fitting. The results indicated that the PLS-DA models had a great predictive capacity and might be performed for further metabolites variance analysis.
Figure 3.
(a,b) PLS-DA scores plot derived from A14 and A207 each group in positive ion mode and negative ion mode. (c,d) S-plot based on OPLS-DA model (positive ion mode and negative ion mode). (e) Volcano map of differential metabolites between A14 group and A207 group. Each point in the figure represents a metabolite.
Differential metabolites analysis
OPLS-DA is a multivariate statistical analysis method with supervised pattern recognition, and can solve the problem that PCA is not sensitive to the variables with little correlation. The variables with VIP values larger than 1 were highlighted with red dots. These red dots were relevant variables to separate the two groups (Fig. 3c,d).
The statistics of differential metabolites between two endophytes were shown in Fig. 3e. There were 90 differential metabolites, which contained 51 up-regulated metabolites and 39 down-regulated metabolites (Supplementary Table S7), and 13 of the 90 differential metabolites were annotated to KEGG metabolic pathways, including cellobiose, melibiose, L-Phenylalanine, aconitic acid, maltotriose, 9(S)-HpOTrE, citric acid, phenyllactic acid, 9,10,13-TriHOME, adipic acid, 6''-O-Malonylglycitin, hydroxyphenyllactic acid, p-Hydroxyphenylacetic acid (Table 3).
Table 3.
Significant differential metabolites between the comparison groups of A14 group and A207 group.
| No. | Metabolite | Formula | FC (A14/A207) | VIP (OPLS-DA) | P value | RT | Mode | M/Z |
|---|---|---|---|---|---|---|---|---|
| 1 | Cellobiose | C12H22O11 | 1043.85 | 5.13 | 1.80E−12 | 0.72 | Neg | 387.11 |
| 2 | Melibiose | C12H22O11 | 756.16 | 11.97 | 7.87E−12 | 0.65 | Neg | 377.09 |
| 3 | l-Phenylalanine | C9H11NO2 | 310.95 | 9.56 | 2.62E−13 | 1.59 | Pos | 166.09 |
| 4 | Aconitic acid | C6H6O6 | 90.02 | 1.58 | 1.12E−09 | 1.05 | Neg | 173.01 |
| 5 | Maltotriose | C18H32O16 | 14.72 | 2.05 | 3.69E−08 | 0.65 | Pos | 527.16 |
| 6 | 9(S)-HpOTrE | C18H30O4 | 13.30 | 1.36 | 3.68E−07 | 5.87 | Neg | 309.21 |
| 7 | Citric acid | C6H8O7 | 5.95 | 1.04 | 3.24E−06 | 0.68 | Neg | 191.02 |
| 8 | Phenyllactic acid | C9H10O3 | 3.38 | 3.99 | 8.67E−12 | 3.17 | Neg | 165.06 |
| 9 | 9,10,13-TriHOME | C18H34O5 | 3.23 | 3.44 | 3.36E−07 | 4.98 | Neg | 329.23 |
| 10 | Adipic acid | C6H10O4 | 0.35 | 1.37 | 3.24E−11 | 1.17 | Neg | 145.05 |
| 11 | 6''-O-Malonylglycitin | C25H24O13 | 0.25 | 1.13 | 1.79E−08 | 0.98 | Pos | 497.11 |
| 12 | Hydroxyphenyllactic acid | C9H10O4 | 0.20 | 2.76 | 5.09E−13 | 2.39 | Neg | 181.05 |
| 13 | P-Hydroxyphenylacetic acid | C8H8O3 | 0.11 | 1.32 | 1.27E−16 | 2.90 | Neg | 151.04 |
FC was the mean ratio between the A14 group and A207 group. Positive numbers represented that increasing in the A14 group compared with the A207 group, and negative numbers mean a decrease.
KEGG pathway analysis for differential metabolites
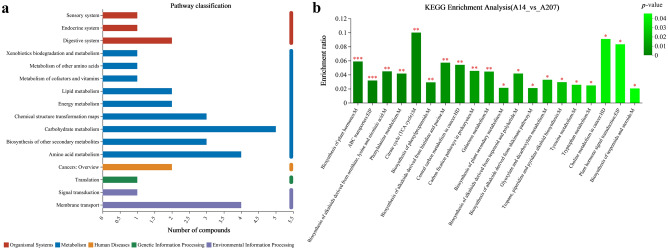
KEGG enrichment analysis showed that 90 differential metabolites were annotated into 46 pathways and divided into 16 categories. The differential metabolites were mainly enriched in metabolism, organismal systems, environmental information processing, cellular processes (Fig. 4a). There were 20 metabolic pathways (p < 0.05) that showed higher enrichment ratio (Fig. 4b), including carbohydrate metabolism, energy metabolism, amino acid metabolism, biosynthesis of other secondary metabolites, chemical structure transformation maps, membrane transport, signal transduction, cancer: overview.
Figure 4.
(a) Classification of differential metabolites between the A14 group and A207 group by KEGG. (b) The abscissa represents the pathway name. The ordinate represents the enrichment ratio, indicating the ratio of the metabolite number to the background number annotated to the pathway. The column color gradient indicates the significance of enrichment. *** indicates p < 0.001, ** indicates p < 0.01, * indicates p < 0.05.
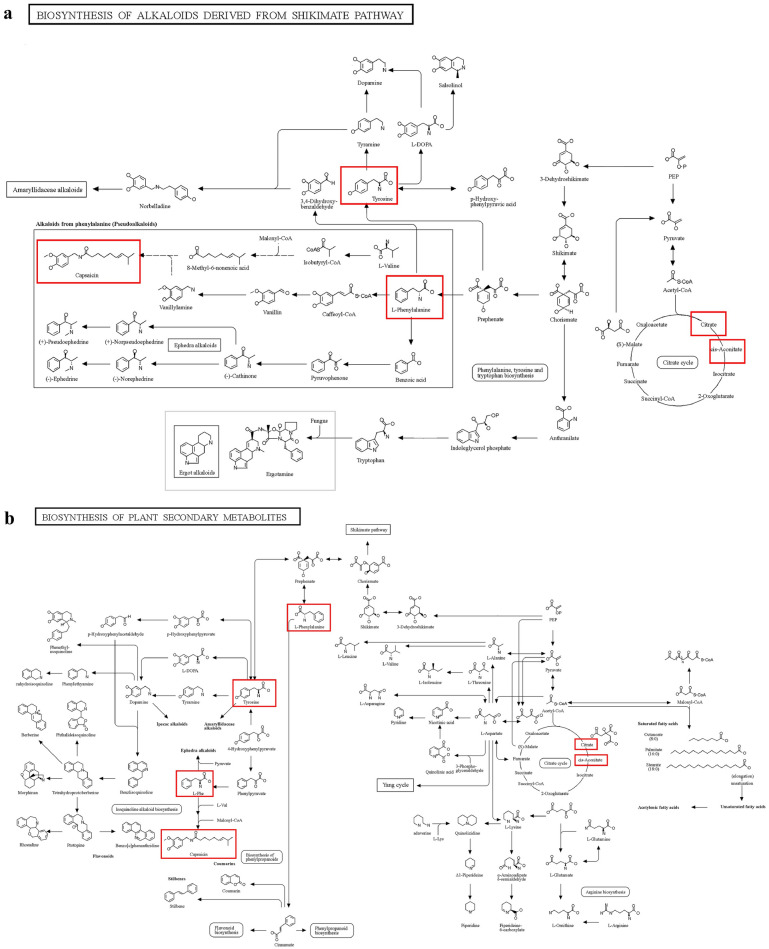
Two pathways showed the most significant differences (p < 0.001): Biosynthesis of plant hormones (map01070) and ABC transporters (map02010). In addition, the pathways that might explain the different antimicrobial activities of the two endophytes are as follows: biosynthesis of plant secondary metabolites (map01060); biosynthesis of alkaloids derived from shikimate pathway (map01063); biosynthesis of alkaloids derived from ornithine, lysine and nicotinic acid (map01064); biosynthesis of alkaloids derived from terpenoid and polyketide (map01066); biosynthesis of amino acids (map01230). The involved differential metabolites were aconitic acid, citric acid and L-Phenylalanine in this study. In those pathways, aconitic acid and citric acid were mainly involved in the tricarboxylic acid cycle. L-Phenylalanine might affect several downstream metabolites which could affect the antimicrobial activities of two endophytes: capsaicin, tyrosine, piperine, tomatine and geraniol. L-Phenylalanine was closer to related downstream metabolites in the biosynthesis of plant secondary metabolites pathway and biosynthesis of alkaloids derived from shikimate pathway (Fig. 5).
Figure 5.
(a) Biosynthesis of plant secondary metabolites pathway. (b) Biosynthesis of alkaloids derived from shikimate pathway.
Discussion
In recent years, endophytes have attracted extensive attention, since they play an important role in enhancing plant growth and maintaining plant health. Endophytic bacterial diversity of various plant species has been studied. P. megaterium, as one of the common endophytes, shows antimicrobial activities and a variety of control effects on plant diseases. In this research, antimicrobial activities against five pathogens of two P. megaterium strains have been explored. The possible reason for the different antimicrobial activities of the two P. megaterium strains was explored by analyzing metabolites with the metabolomic analysis and KEGG metabolic pathway annotation40,41. The present study provided a reference for the biological control of dendrobium phytopathogens and potato phytopathogens. In addition, the related metabolic pathways will provide a theoretical basis for the subsequent exploration of metabolites in P. megaterium that are isolated from others hosts.
Antimicrobial activities of P. megaterium P-NA14 and P. megaterium D-HT207
In this research, the results of 16S rDNA gene sequencing and genomic analysis showed that P. megaterium P-NA14 and P. megaterium D-HT207 belonged to P. megaterium, and they had the closest evolutionary relationship with P. megaterium NBRC 15308. Previous studies have demonstrated that the genus Bacillus is well known for the natural production of secondary metabolites with antimicrobial activities and shows a very strong biocontrol potential, which works against phytopathogens such as Burkholderia solanacearum and Fusarium oxysporum42,43. Previous studies showed that the antimicrobial activity of P. megaterium D-HT207 against A. rolfsii was the highest among the endophytes isolated from dendrobium29, the antimicrobial activity of P. megaterium P-NA14 against P. atroseptica ACCC 19901 was the highest among the endophytes isolated from potato30. According to the results of our antibacterial test, P. megaterium P-NA14 showed a stronger inhibition effect on A. rolfsii than P. megaterium D-HT207. P. megaterium D-HT207 showed a stronger inhibition effect on P. atroseptica ACCC 19901 than P. megaterium P-NA14. Meanwhile, the antimicrobial activities of the two strains against S. aureus and E. coli were opposite. Interestingly, P. megaterium showed antimicrobial activity against both S. aureus and E. coli isolated from dairy waste, mango pulp waste and oral microflora44,45. At present, there is no research on the mechanism of different activities of strains isolated from different hosts46,47. It was speculated that in the long-term coevolution process of endophytic bacteria and host, the stimulation to their original host pathogens will gradually weaken, but the inhibitive activity to exogenous pathogens will be stronger.
The differential metabolites
According to recent reports, the secondary metabolites of plant endophytes would be affected by different hosts. Dobrzanski et al. compared the adaptive responses of bacteria belonging to the same species that came from two different ecosystems (water and soil). The final balance of ROS produced was different between the two strains, but the related metabolic pathways was still not described yet48. A slight difference in the plant growth-promoting genes among the four Bacillus cereus strains isolated from different hosts (forest, haw, wheat, and alfalfa) was observed. The results of specific genes analysis showed that the differential genes were related to amino acid, carbohydrate and coenzyme transport and metabolism49,50. The differential metabolites of P. megaterium P-NA14 and P. megaterium D-HT207 were analyzed by UPLC-QTOF-MS, L-phenylalanine in P. megaterium P-NA14 is up-regulated compared to P. megaterium D-HT207. In plants, L-phenylalanine was found to be a precursor to a multitude of plant metabolites that are important in plant growth, nutrition, reproduction, resistance, and environmental response51–53. The increase of L-phenylalanine content may be the key reason for the differential changes of downstream metabolites.
Related downstream metabolites of L-phenylalanine in the KEGG pathways
With the analysis of KEGG database54,55. L-phenylalanine might affect the synthesis of capsaicin, tyrosine, piperine, tomatine, and geraniol. Capsaicin and tyrosine were the closest downstream metabolites to L-phenylalanine in pathways (Fig. 5). Recent research showed that capsaicin had stronger antimicrobial activity on S. aureus than E. coli56. Capsaicin inhibited the NorA efflux pump of S. aureus and reduced the invasiveness of S. aureus57. Tyrosine affects the production of downstream glycopeptide antibiotics through the biosynthesis of vancomycin group antibiotics (map01055), which had an inhibitory effect on S. aureus. Piperine showed antimicrobial activity on S. aureus instead of E. coli. The tomatine and geraniol exhibited higher antimicrobial activity and MIC values of S. aureus than E. coli3,58. The antimicrobial activity of these five downstream metabolites against S. aureus is the same as that of P. megaterium P-NA14, indicating for the first time that the antimicrobial activity of P. megaterium against S. aureus may be related to the secondary metabolites. But the reason for the different inhibitory activity against E. coli still needs further investigation.
Conclusion
The results confirmed in this study that the difference in antimicrobial activity between P. megaterium P-NA14 and P. megaterium D-HT207 was related to downstream metabolites affected by L-phenylalanine. Previous reports showed downstream metabolites affected by higher content of L-phenylalanine exhibited higher antimicrobial activity against S. aureus than E. coli. In summary, the antimicrobial activities of the same species isolated from different hosts turn out different. This study has proved that the secondary metabolites of plant endophytes were affected by the host, and it might be affected at the level of hormones, enzymes and genes. To clarify how the host affects endophytic bacterial products, it should be analyzed not only in metabolomics but also in target compounds and genomics levels.
Supplementary Information
Acknowledgements
The study was supported by the Knowledge innovation program funding of institute of food science and technology, Chinese Academy of Agricultural Sciences (125161015000150013), Agricultural product quality and safety risk assessment project (GJFP20210502).
Author contributions
B.F., F.Z.W., J.M.L. and Y.T.L. contributed conception and design of the study. S.S.W., N.J., C.L. and Y.F.S. participated in plant collection and endophytic bacteria isolation. S.Y.L., J.M.L., Y.T.L. and J.S. performed the experiments. Y.T.L. and J.M.L. performed the statistical analysis and were major contributors to writing the manuscript. All authors read and approved the final manuscript.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jia-Meng Liu and Yan-Tian Liang.
Contributor Information
Bei Fan, Email: caasBFan@163.com.
Feng-Zhong Wang, Email: caasFZWang@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-32337-6.
References
- 1.Nisa H, et al. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 2015;82:50–59. doi: 10.1016/j.micpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Yadav, G. & Meena, M. Bioprospecting of endophytes in medicinal plants of Thar Desert: An attractive resource for biopharmaceuticals. Biotechnol. Rep. (Amst).30, e00629 (2021). [DOI] [PMC free article] [PubMed]
- 3.Pandey SS, et al. Plant probiotics—Endophytes pivotal to plant health. Microbiol. Res. 2022;263:127148. doi: 10.1016/j.micres.2022.127148. [DOI] [PubMed] [Google Scholar]
- 4.Morales-Cedeño LR, et al. Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: fundamentals, methods of application and future perspectives. Microbiol. Res. 2021;242:126612. doi: 10.1016/j.micres.2020.126612. [DOI] [PubMed] [Google Scholar]
- 5.Afzal I, Shinwari ZK, Sikandar S, Shahzad S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019;221:36–49. doi: 10.1016/j.micres.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Issa, A. et al. Impacts of Paraburkholderia phytofirmans strain PsJN on tomato (Lycopersicon esculentum L.) under high temperature. Front. Plant Sci.9, 1397 (2018). [DOI] [PMC free article] [PubMed]
- 7.Molina-Montenegro MA, Acuña-Rodríguez IS, Torres-Díaz C, Gundel PE, Dreyer I. Antarctic root endophytes improve physiological performance and yield in crops under salt stress by enhanced energy production and Na+ sequestration. Sci. Rep. 2020;10(1):5819. doi: 10.1038/s41598-020-62544-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berlec A. Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci. Int. J. Exp. Plant Biol. 2012;193–194:96–102. doi: 10.1016/j.plantsci.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Collinge DB, Jensen B, Jørgensen HJ. Fungal endophytes in plants and their relationship to plant disease. Curr. Opin. Microbiol. 2022;69:102177. doi: 10.1016/j.mib.2022.102177. [DOI] [PubMed] [Google Scholar]
- 10.Santoyo G, Moreno-Hagelsieb G, Orozco-Mosqueda MC, Glick BR. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016;183:92–99. doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Biedendieck R, Knuuti T, Moore SJ, Jahn D. The, "beauty in the beast"-the multiple uses of Priestia megaterium in biotechnology. Appl. Microbiol. Biotechnol. 2021;105(14–15):5719–5737. doi: 10.1007/s00253-021-11424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta, R.S., Patel, S., Saini, N. & Chen S. Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol.70(11), 5753–5798 (2020). [DOI] [PubMed]
- 13.Alvarez F, et al. The plant-associated Bacillus amyloliquefaciens strains MEP2 18 and ARP2 3 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J. Appl. Microbiol. 2012;112(1):159–174. doi: 10.1111/j.1365-2672.2011.05182.x. [DOI] [PubMed] [Google Scholar]
- 14.Yang W. Components of rhizospheric bacterial communities of barley and their potential for plant growth promotion and biocontrol of Fusarium wilt of watermelon. Braz. J. Microbiol. 2019;50(3):749–757. doi: 10.1007/s42770-019-00089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayakumar A, Nair IC, Radhakrishnan EK. Environmental adaptations of an extremely plant beneficial Bacillus subtilis Dcl1 identified through the genomic and metabolomic analysis. Microb. Ecol. 2021;81(3):687–702. doi: 10.1007/s00248-020-01605-7. [DOI] [PubMed] [Google Scholar]
- 16.Kłosowski G, Mikulski D, Pielech-Przybylska K. Pyrazines biosynthesis by Bacillus strains isolated from natto fermented soybean. Biomolecules. 2021;11(11):1736. doi: 10.3390/biom11111736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajan L, Chakraborty K, Chakraborty RD. Pharmacological properties of some mangrove sediment-associated bacillus isolates. Arch. Microbiol. 2021;203(1):67–76. doi: 10.1007/s00203-020-01999-5. [DOI] [PubMed] [Google Scholar]
- 18.Kang SM, et al. Phosphate solubilizing Bacillus megaterium mj1212 regulates endogenous plant carbohydrates and amino acids contents to promote mustard plant growth. Indian J. Microbiol. 2014;54(4):427–433. doi: 10.1007/s12088-014-0476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X, et al. Development of a biologically based fertilizer, incorporating Bacillus megaterium A6, for improved phosphorus nutrition of oilseed rape. Can. J. Microbiol. 2013;59(4):231–236. doi: 10.1139/cjm-2012-0579. [DOI] [PubMed] [Google Scholar]
- 20.Singh RK, et al. Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: A comprehensive study of plant-microbe interactions for growth enhancement in Saccharum spp. BMC Plant Biol. 2022;20(1):220. doi: 10.1186/s12870-020-02400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty U, Chakraborty B, Basnet M. Plant growth promotion and induction of resistance in camellia sinensis by Bacillus megaterium. J. Basic Microbiol. 2006;46(3):186–195. doi: 10.1002/jobm.200510050. [DOI] [PubMed] [Google Scholar]
- 22.Feng, F. et al. Enhanced degradation of chlorpyrifos in rice (Oryza sativa L.) by five strains of endophytic bacteria and their plant growth promotional ability. Chemosphere.184, 505–513 (2017). [DOI] [PubMed]
- 23.Solanki MK, et al. Characterization of mycolytic enzymes of Bacillus strains and their bio-protection role against Rhizoctonia solani in tomato. Curr. Microbiol. 2012;65(3):330–336. doi: 10.1007/s00284-012-0160-1. [DOI] [PubMed] [Google Scholar]
- 24.Kim YC, Anderson AJ. Rhizosphere pseudomonads as probiotics improving plant health. Mol. Plant Pathol. 2018;19(10):2349–2359. doi: 10.1111/mpp.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mannaa, M. & Kim, K. D. Biocontrol activity of volatile-producing Bacillus megaterium and Pseudomonas protegens against aspergillus and penicillium spp. predominant in stored rice grains: study II. Mycobiology. 46(1), 52–63 (2018). [DOI] [PMC free article] [PubMed]
- 26.Trivedi P, Pandey A. Plant growth promotion abilities and formulation of Bacillus megaterium strain B 388 (MTCC6521) isolated from a temperate Himalayan location. Indian J. Microbiol. 2008;48(3):342–347. doi: 10.1007/s12088-008-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong Q, Shan S, Liu Q, Wang X, Yu F. Biocontrol of Aspergillus flavus on peanut kernels by use of a strain of marine Bacillus megaterium. Int. J. Food Microbiol. 2010;139(1–2):31–35. doi: 10.1016/j.ijfoodmicro.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 28.Xie Y, et al. Isolation and identification of antibacterial bioactive compounds from Bacillus megaterium L2. Front. Microbiol. 2021;12:645484. doi: 10.3389/fmicb.2021.645484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang SS, et al. Diversity of culture-independent bacteria and antimicrobial activity of culturable endophytic bacteria isolated from different Dendrobium stems. Sci. Rep. 2019;9(1):10389. doi: 10.1038/s41598-019-46863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu JM, et al. Antimicrobial activity against phytopathogens and inhibitory activity on solanine in potatoes of the endophytic bacteria isolated from potato tubers. Front. Microbiol. 2020;11:570926. doi: 10.3389/fmicb.2020.570926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou SQ, et al. A rapid method for extracting DNA from actinomycetes by chelex-100. Biotechnol. Bull. 2010;20:123–125. [Google Scholar]
- 32.Holt, J. G., Noel R. K., & Peter, HA. S. Bergey's manual of determinative bacterology. 31–35 (1994).
- 33.Derguine-Mecheri, L., Kebbouche-Gana, S. & Djenane, D. Biosurfactant production from newly isolated Rhodotorula sp.YBR and its great potential in enhanced removal of hydrocarbons from contaminated soils. World J. Microbiol. Biotechnol.37(1), 18 (2021). [DOI] [PubMed]
- 34.Nagkirti, P. D., Engineer, A. S. & Dhakephalkar, P. K. Xylanimonas oleitrophica sp. nov., a novel petroleum hydrocarbon degrading bacterium isolated from an Indian oil reservoir. Antonie Van Leeuwenhoek.114(2), 129–136 (2021). [DOI] [PubMed]
- 35.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36(6):1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U S A. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kronvall G, Giske CG, Kahlmeter G. Setting interpretive breakpoints for antimicrobial susceptibility testing using disk diffusion. Int. J. Antimicrob. Agents. 2011;38(4):281–290. doi: 10.1016/j.ijantimicag.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 2005;187(18):6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laux M, et al. New plastomes of eight Ipomoea species and four putative hybrids from Eastern Amazon. PLoS ONE. 2022;17(3):e0265449. doi: 10.1371/journal.pone.0265449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang B, et al. Genomic analysis of Bacillus megaterium NCT-2 reveals its genetic basis for the bioremediation of secondary salinization soil. Int. J. Genomics. 2020;2020:4109186. doi: 10.1155/2020/4109186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Shao L, Zhu J, Li H, Duan H. Comparative analysis of tuberous root metabolites between cultivated and wild varieties of Rehmannia glutinosa by widely targeted metabolomics. Sci. Rep. 2021;11(1):11460. doi: 10.1038/s41598-021-90961-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radhakrishnan R, Hashem A, Abd-Allah EF. Bacillus: a biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017;8:667. doi: 10.3389/fphys.2017.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus as potential probiotics: status, concerns, and future perspectives. Front. Microbiol. 2017;8:1490. doi: 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel AK, Deshattiwar MK, Chaudhari BL, Chincholkar SB. Production, purification and chemical characterization of the catecholate siderophore from potent probiotic strains of Bacillus spp. Bioresour. Technol. 2009;100(1):368–373. doi: 10.1016/j.biortech.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Al-Thubiani ASA, et al. Identification and characterization of a novel antimicrobial peptide compound produced by Bacillus megaterium strain isolated from oral microflora. Saudi Pharm. J. 2018;26(8):1089–1097. doi: 10.1016/j.jsps.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinski A, Betekhtin A, Hupert-Kocurek K, Mur LAJ, Hasterok R. Defining the genetic basis of plant endophytic bacteria interactions. Int. J. Mol. Sci. 2019;20(8):1947. doi: 10.3390/ijms20081947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji X, Xia Y, Zhang H, Cui JL. The microscopic mechanism between endophytic fungi and host plants: From recognition to building stable mutually beneficial relationships. Microbiol. Res. 2022;261:127056. doi: 10.1016/j.micres.2022.127056. [DOI] [PubMed] [Google Scholar]
- 48.Dobrzanski T, et al. Bacillus megaterium strains derived from water and soil exhibit differential responses to the herbicide mesotrione. PLoS ONE. 2018;13(4):e0196166. doi: 10.1371/journal.pone.0196166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng Q, et al. Comparative genomic and functional analyses of four sequenced Bacillus cereus genomes reveal conservation of genes relevant to plant-growth-promoting traits. Sci. Rep. 2018;8(1):17009. doi: 10.1038/s41598-018-35300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan, L., Pan, K., Li, Y., Yi, B. & Gao, B. Comparative transcriptome analysis of Alpinia oxyphylla Miq. reveals tissue-specific expression of flavonoid biosynthesis genes. BMC Genom. Data.22(1), 19 (2021). [DOI] [PMC free article] [PubMed]
- 51.Guo YW, et al. L-Phenylalanine alters the privileged secondary metabolite production in the marine-derived fungus Trichoderma erinaceum F1–1. J. Nat. Prod. 2020;83(1):79–87. doi: 10.1021/acs.jnatprod.9b00710. [DOI] [PubMed] [Google Scholar]
- 52.Maeda H, Yoo H, Dudareva N. Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat. Chem. Biol. 2011;7(1):19–21. doi: 10.1038/nchembio.485. [DOI] [PubMed] [Google Scholar]
- 53.Koca N, Karaman Ş. The effects of plant growth regulators and L-phenylalanine on phenolic compounds of sweet basil. Food Chem. 2015;166:515–521. doi: 10.1016/j.foodchem.2014.06.065. [DOI] [PubMed] [Google Scholar]
- 54.Xue X, et al. Metabolomics-based analysis of flavonoid metabolites in Chinese jujube and sour jujube fruits from different harvest periods. J. Food Sci. 2022;87(9):3752–3765. doi: 10.1111/1750-3841.16290. [DOI] [PubMed] [Google Scholar]
- 55.Pal T, Padhan JK, Kumar P, Sood H, Chauhan RS. Comparative transcriptomics uncovers differences in photoautotrophic versus photoheterotrophic modes of nutrition in relation to secondary metabolites biosynthesis in Swertia chirayita. Mol. Biol. Rep. 2018;45(2):77–98. doi: 10.1007/s11033-017-4135-y. [DOI] [PubMed] [Google Scholar]
- 56.Orlo E, Russo C, Nugnes R, Lavorgna M, Isidori M. Natural methoxyphenol compounds: Antimicrobial activity against foodborne pathogens and food spoilage bacteria, and role in antioxidant processes. Foods. 2021;10(8):1807. doi: 10.3390/foods10081807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalia NP, et al. Capsaicin, a novel inhibitor of the NorA efflux pump, reduces the intracellular invasion of Staphylococcus aureus. J. Antimicrob. Chemother. 2012;67(10):2401–2408. doi: 10.1093/jac/dks232. [DOI] [PubMed] [Google Scholar]
- 58.Aćimović M, et al. Chemical characterization and antibacterial activity of essential oil of medicinal plants from eastern serbia. Molecules. 2022;25(22):5482. doi: 10.3390/molecules25225482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.