Abstract
Group 3 innate lymphoid cells (ILC3s) play important roles in maintaining intestinal homeostasis by protecting the host from pathogen infections and tissue inflammation. The transcription factor PLZF (promyelocytic leukemia zinc finger), encoded by zinc finger BTB domain containing 16 (Zbtb16), is highly and transiently expressed in ILC precursors (ILCPs). However, the role of PLZF in regulating ILC3 development and function remains unknown. Here, we show that PLZF was specifically expressed in mature intestinal ILC3s compared with other ILC subsets. PLZF was dispensable for ILC3 development. However, PLZF deficiency in ILC3s resulted in increased innate interleukin-22 (IL-22) secretion and protection against gut infection and inflammation. Mechanistically, PLZF negatively regulated IL-22 expression by ILC3s in a cell-intrinsic manner by binding to the IL-22 promoter region for transcriptional repression. Together, our data suggest that PLZF restricts intestinal ILC3 function to regulate gut immune homeostasis.
Keywords: Biological sciences, Immunology, Innate immune cells, Innate lymphoid cells
Subject terms: Innate immune cells
Introduction
Group 3 innate lymphoid cells (ILC3s), characterized by the expression of the transcription factor RORγt (RAR-related orphan receptor gamma t), are derived from ILC precursors (ILCPs) marked by transient expression of PLZF [1, 2]. Based on their surface expression molecules, ILC3s are divided into three subsets: NKp46+ ILC3s, NKp46– ILC3s and Lti cells (lymphoid tissue inducer cells) [3]. ILC3s are enriched in the intestinal mucosal tissue and play important roles in maintaining intestinal homeostasis by protecting the host from pathogen infections and tissue inflammation [1, 4–7]. The protective effects of ILC3s in gut immune defense are considered to occur through the production of effector cytokines, including IL-22 and IL-17. IL-22 is critical in the control of pathogenic bacterial infection by inducing antimicrobial peptide production by intestinal epithelial cells [8]. In addition, IL-22 and IL-17 promote tissue repair by inducing epithelial cell proliferation and survival, therefore limiting tissue damage during gut inflammation, such as colitis [9, 10]. Dysregulation of ILC3s has been associated with inflammatory bowel disease and intestinal infection.
PLZF is specifically expressed in multiple innate-like lymphocyte lineages, including invariant NKT (iNKT) cells, γδT cells, and human mucosal-associated invariant T cells (MAIT) [11–14]. PLZF has been described as a key regulator of innate-like cell development, given that it is necessary for the acquisition of effector programs in these cells [2, 15]. In contrast, PLZF is not expressed in conventional αβT cell lineages, whether resting or activated, indicating tight lineage-specific control of expression [16]. Recent studies have demonstrated that PLZF is transiently expressed by ILCPs but not by mature ILCs using a transgenic mouse system expressing a GFP-Cre fusion protein under control of the endogenous PLZF gene [2]. Subsequent studies have shown that the deletion of PLZF markedly impaired the development and function of ILC2s [15]. However, whether PLZF plays a similar role in the development and function of ILC3s is unclear. Here, we show that PLZF is expressed specifically in mature ILC3s and acts as a negative regulator of IL-22 production by ILC3s. Our data suggest that PLZF restricts intestinal ILC3 function to regulate gut immune defense.
Results
The Zbtb16 gene is expressed in mature ILC3s
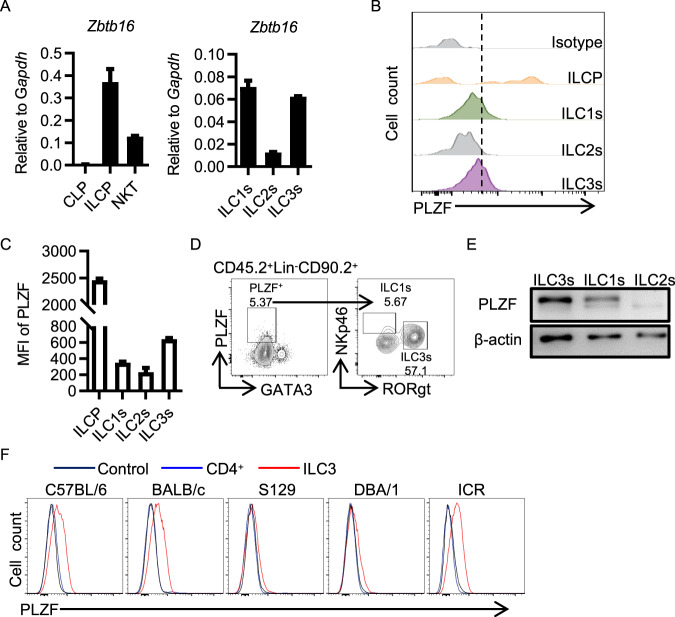
Previous studies have shown that PLZF is transiently expressed in ILCP and that its expression is downregulated with the development into mature ILCs [2, 15]. To obtain the dynamic expression pattern of PLZF during ILC3 development, we analyzed the expression of PLZF in ILC precursors in bone marrow (CLP, ILCP) and mature ILC subsets in the gut (ILC1s, ILC2s, ILC3s) (Supplementary Fig. 1A–C). Among these cells, ILCP appeared to express the highest level of PLZF at both the mRNA level and protein level, as measured by quantitative RT‒PCR and flow cytometry intracellular staining (Fig. 1A, B). With differentiation into distinct ILC subsets from ILCP, the expression level of PLZF was decreased dramatically, while ILC3s maintained a relatively high expression level of PLZF compared to ILC2s and ILC1s, as indicated by the highest MFI level in ILC3s (Fig. 1B, C). In addition, ILC3s accounted for approximately 57.1% of PLZF+ ILCs, indicating that ILC3s are the predominant subset expressing PLZF in mature ILCs (Fig. 1D). Approximately half of PLZF+ ILC3s were CCR6+ Lti cells (Supplementary Fig. 1D). To ensure PLZF expression in ILC3s, we took advantage of PLZF-deficient mice and determined PLZF expression. PLZF-deficient mice were generated by knock-in of an EGFP gene into exon 1 of the PLZF gene, the expression of which was disrupted in homozygous mice (Supplementary Fig. 2A). ILC3s were isolated from the small intestine of both WT mice and PLZF−/− mice by flow sorting and subjected to immunoblotting for PLZF detection. As expected, PLZF was highly expressed in NKT cells. Interestingly, PLZF was also expressed in WT ILC3s but not in PLZF-deficient ILC3s or ILC2s (Fig. 1E, Supplementary Fig. 2B). To rule out the possibility of genetic influence on PLZF expression, we analyzed PLZF expression in ILC3s from mice with different genetic backgrounds. As shown in Fig. 1F, PLZF was ubiquitously expressed in ILC3s from all mice with various genetic backgrounds. Collectively, these data indicate that PLZF is expressed in mature ILC3s. This finding prompted us to hypothesize that PLZF might have an important regulatory role in ILC3 lineage specification and function.
Fig. 1.
The Zbtb16 gene is highly expressed in ILC3s. A Zbtb16 mRNA expression was analyzed by real-time RT‒PCR (n = 3). CLP and ILCP cells were sorted from bone marrow (BM); NKT cells were sorted from the liver; and ILC subsets were sorted from the large intestine (LI) lamina propria lymphocytes (LPLs). B Representative flow cytometry histograms for PLZF expression on ILCP from BM and ILC subsets from LI LPLs (n = 3). C Mean fluorescence intensity (MFI) of PLZF on ILCP from BM and ILC subsets from LI LPLs (n = 3). D PLZF expression in wild-type (WT, C57BL/6) ILCs from LI LPLs. Numbers adjacent to boxed areas indicate the relative percentage of gated populations. E Western blotting was performed on ILC subsets from the LPLs to assess PLZF expression. F Representative flow cytometry histograms for PLZF expression on CD4+ T cells and ILC3s from mice with different genetic backgrounds (as indicated). A–F Data are representative of at least two independent experiments. Data are represented as the mean ± SEM. Error bars show SEM. Cell subtypes were identified as BM CLP (Lin−CD127+α4β7−Flt3+), BM ILCP (Lin−CD127+α4β7+PD1+), liver NKT (CD45+CD3+NK1.1+), NK (CD3−NK1.1+), LPL ILC1s (Lin−CD45.2hiCD90.2midNK1.1+), LPL ILC2s (Lin−CD45.2hiCD90.2midKLRG1+), and LPL ILC3s (Lin−CD45.2midCD90.2hi). Lineage markers include B220, CD19, CD3, CD5, CD11b, Gr-1, and CD11c
PLZF is dispensable for ILC3 development
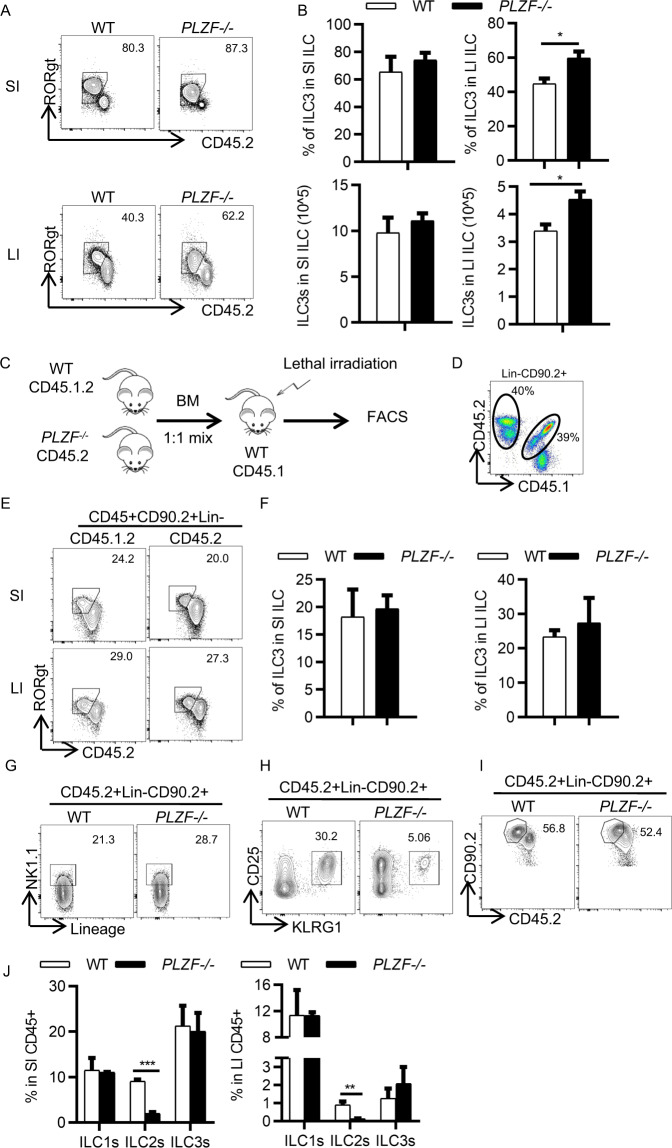
To address whether PLZF plays a role in regulating ILC3 development, we examined the number of ILC3 cells in both WT mice and PLZF-deficient (PLZF−/−) mice. Compared with the littermate WT controls, the PLZF−/− mice did not show any differences in the frequency and absolute cell number of total ILCs in either the small intestine or large intestine (Supplementary Fig. 3A, B). However, we observed that the frequency and number of ILC3s were increased in the large intestine but not the small intestine (Fig. 2A, B). Of note, there was no difference in ILC3 subsets between the WT and PLZF−/− mice (Supplementary Fig. 3C). To determine the possible cause for the increased ILC3s in the large intestine, we examined cell proliferation by Ki-67 staining and apoptosis by Caspase-3. Interestingly, we found that ILC3s from the PLZF−/− mice exhibited higher proliferation than those from the WT mice (Supplementary Fig. 3D). However, there was no difference in apoptosis between WT and PLZF−/− ILC3s (Supplementary Fig. 3E). Given that PLZF is also expressed in other cell types, including NKT cells, γδT cells, MAIT cells, and myeloid cells, we wanted to determine whether the increased ILC3s in the large intestine in the PLZF−/− mice are due to cell-intrinsic effects. To determine this, we generated competitive bone marrow chimeric mice by transferring PLZF−/− (CD45.2+) or WT (CD45.2+) bone marrow cells mixed with an equal ratio (1:1) of WT (CD45.1.2+) bone marrow cells into lethally irradiated CD45.1 recipient mice (Fig. 2C). Donor chimerism of ILC3s in both the small intestine and large intestine was analyzed by measuring the ratio of the CD45.2 and CD45.1.2 markers (Fig. 2D). The percentage of ILC3s derived from CD45.2+ PLZF−/− donor cells was comparable to that from the competitor CD45.1.2+ WT donor cells in both the small intestine and large intestine (Fig. 2E, F). These results suggest that PLZF is not required for ILC3 development. To further confirm this, we isolated ILC progenitors (ILCPs) from the bone marrow of both WT and PLZF−/− mice by flow sorting and transferred them into Rag2−/−Il2rg−/− recipient mice, which lack mature T cells, B cells, and ILCs. ILC subsets were analyzed after 8 weeks of reconstitution. Consistent with the bone marrow chimera data, the percentage of ILC3s derived from PLZF−/− ILCPs was similar to that from WT ILCPs (Fig. 2I, J). ILC1s also showed no difference between the WT and PLZF−/− groups (Fig. 2G, J). In contrast, ILC2s derived from PLZF−/− ILCPs were significantly reduced compared to those from WT ILCPs, which is in accordance with a previous report [2] (Fig. 2H, J). Taken together, these results suggest that PLZF is dispensable for ILC3 development.
Fig. 2.
PLZF is not required for ILC3 development. A, B Small intestine (SI) and large intestine (LI) LPLs were isolated from WT or PLZF−/− mice (n = 4–6). A Representative flow cytometry plots for ILC3s (CD45.2midRORγt+) gated on CD45+Lin−CD90.2+. B Frequencies of ILC3s in CD45+Lin−CD90.2+ cells and total numbers of ILC3s in WT and PLZF−/− mice. C–F Competitive bone marrow chimeric mice were generated, and ILC3s were analyzed 8 weeks later (n = 3). C Experimental scheme for bone marrow transfer. BM from the CD45.2 PLZF−/− or CD45.1.2 WT mice were mixed 1:1 and transplanted into lethally irradiated recipients (CD45.1+). Eight weeks after engraftment, the donor BM-derived ILC3s (CD45.2+ PLZF−/− and CD45.1.2+ WT) were analyzed by flow cytometry. D Representative flow cytometry plots for LI LPLs from bone marrow chimeric mice. E Representative flow cytometry plots for ILC3 -gated on CD45+Lin−CD90.2+ from LPLs of bone marrow chimeric mice. F Frequencies of ILC3s in SI and LI LPLs from bone marrow chimeric mice. G ILCP cells were sorted from the bone marrow of WT or PLZF−/− mice and transplanted into lethally irradiated Rag2−/−Il2rg−/− recipient mice. The development of ILCs was analyzed by flow cytometry 8 weeks later (n = 4). Representative flow cytometry plots for ILC1s (G), ILC2s (H), and ILC3s (I) in SI LPLs. J Frequencies of ILC1s, ILC2s and ILC3s in SI and LI LPLs of recipient mice. Data are represented as the mean ± SEM. Error bars show SEM. *P < 0.05; **P < 0.01
PLZF regulates gut immunity
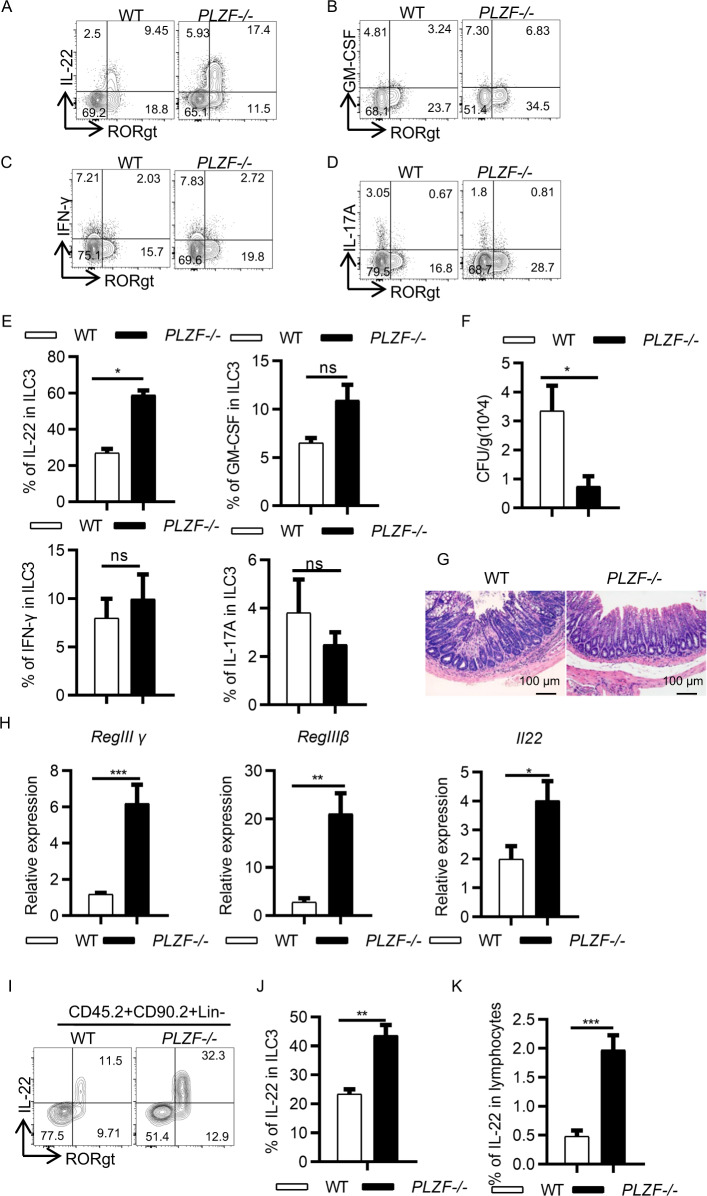
Next, we sought to determine whether PLZF regulates ILC3 activation and function. First, we examined cytokine production by ILC3s in the WT and PLZF−/− mice. We observed that ILC3s from the PLZF−/− mice produced significantly higher amounts of IL-22, but not GM-CSF, IFN-g, and IL-17A, than those from the WT control mice (Fig. 3A–E). We also examined the activation markers and homing receptors on ILC3s, including CD69, MHC-II, CCR9, and CCR6. Interestingly, none of them were different between the WT and PLZF−/− ILC3s (Supplementary Fig. 4). IL-22 plays an important role in host defense during pathogenic bacterial infection and inflammation. Given the increased IL-22 production in ILC3 cells from the PLZF−/− mice, we hypothesized that loss of PLZF would protect mice from Citrobacter rodentium, a mouse pathogen that mimics human pathogenic Escherichia coli infection. To determine this, we infected both WT and PLZF−/− mice with C. rodentium. First, we found that the expression level of PLZF was decreased in ILC3s when mice were infected with C. rodentium (Supplementary Fig. 5A). Compared to the PLZF−/− mice, the WT mice had approximately 5-fold more bacterial colony-forming units (CFUs) of C. rodentium in their feces (Fig. 3F). Consistently, the PLZF−/− mice exhibited improved pathohistological changes, as indicated by fewer infiltrated cells in HE staining (Fig. 3G). Since IL-22 can act on epithelial cells to induce the production of antimicrobial peptides, we also determined the expression of RegIIIβ and RegIIIγ. Notably, the expression levels of genes encoding antimicrobial peptides were all increased in the PLZF−/− mice, in line with the elevated expression level of IL-22 (Fig. 3H). The protein level of IL-22 expression by ILC3s was also examined in both WT and PLZF−/− mice. Consistently, the frequency and cell number of IL-22+ ILC3s were markedly increased in the PLZF−/− mice compared to the WT mice (Fig. 3I–K).
Fig. 3.
PLZF deficiency protects mice from C. rodentium infection. A–E Six- to eight-week-old WT or PLZF−/− mice were sacrificed, and LI LPLs were isolated for analysis (n = 3). A–D Representative flow cytometry plots for IL-22 (A), GM-CSF (B), IFN-γ (C), and IL-17A (D) produced by ILC3s gated on CD45.2+Lin−CD90.2+. E Frequencies of each cytokine in ILC3s in WT and PLZF−/− mice. F–I Six- to eight-week-old WT or PLZF−/− mice were infected with C. rodentium (n = 3–6). Samples were collected and analyzed at the indicated time points. F Bacterial load after C. rodentium infection on Day 6. G Representative image of H&E staining of colon sections from C. rodentium–infected mice at Day 12 p.i. Scale bar is 100 μm. H Relative mRNA expression of Reg3β, Reg3γ and IL-22 in colonic tissue homogenates of infected mice was analyzed by real-time RT‒PCR. I Representative flow cytometry plots for IL-22-expressing ILC3s in large intestine LPLs after C. rodentium infection on Day 6. J, K Frequencies of IL-22 in ILC3s and lymphocytes in the large intestine LPLs. A‒K Data are representative of at least two independent experiments. Data are represented as the mean ± SEM. Error bars show SEM. *P < 0.05; **P < 0.01; ***P < 0.01
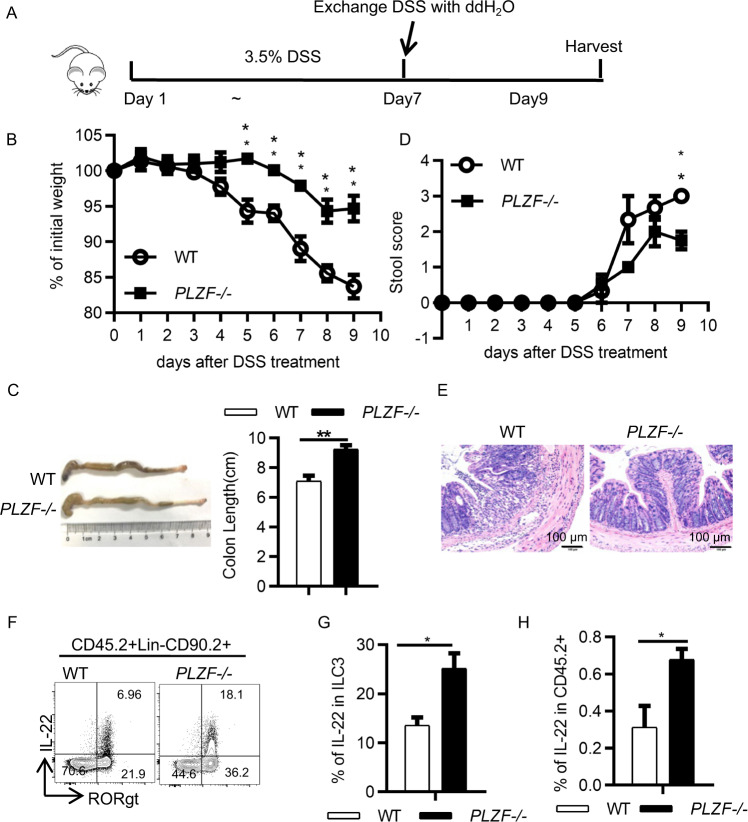
To further determine the role of PLZF in ILC3s during gut inflammation caused by tissue damage, we used a DSS-induced innate colitis model (Fig. 4A). Similar to the findings in the C. rodentium-infected mice, the expression level of PLZF in ILC3s was also decreased in the mice with colitis (Supplementary Fig. 5B). These data suggest that pathogenic signals from infection or inflammation can regulate PLZF expression levels. Given that IL-23 is an important disease signal that activates ILC3s, we examined the effect of IL-23 on PLZF expression. Interestingly, IL-23 stimulation resulted in the downregulation of PLZF in ILC3s (Supplementary Fig. 5C). The PLZF−/− mice had less weight loss, longer colon length, and lower clinical score (Fig. 4B–D). In accordance with these findings, the PLZF−/− mice exhibited decreased inflammation, as indicated by fewer infiltrated cells in HE staining (Fig. 4E). Accordingly, an increased frequency of IL-22+ ILC3s was observed in the PLZF−/− mice (Fig. 4F–H). Together, these data indicate that PLZF plays a critical role in regulating ILC3 function during gut infection and inflammation.
Fig. 4.
Loss of PLZF protects mice from DSS-induced colitis. A Experimental scheme for the DSS-induced acute colitis model. Acute colitis was induced in 6- to 8-week-old WT or PLZF−/− mice with 3.5% DSS in the drinking water for 7 days and with sterile water for 2 days, and then, the mice were sacrificed at Day 9. B–D Colitis severity is shown by weight loss (B), colon length (C), and stool score (D). E Representative image of H&E staining of colon sections from the DSS-treated mice at Day 9. Scale bar is 100 μm. F Representative flow cytometry plots for IL-22-expressing ILC3s. G, H Frequencies of IL-22+ in ILC3s and CD45.2+ lymphocytes in lamina propria lymphocytes. A–H Data are representative of at least two independent experiments. Data are represented as the mean ± SEM. Error bars show SEM. *P < 0.05; **P < 0.01; ***P < 0.001
PLZF intrinsically regulates ILC3 function
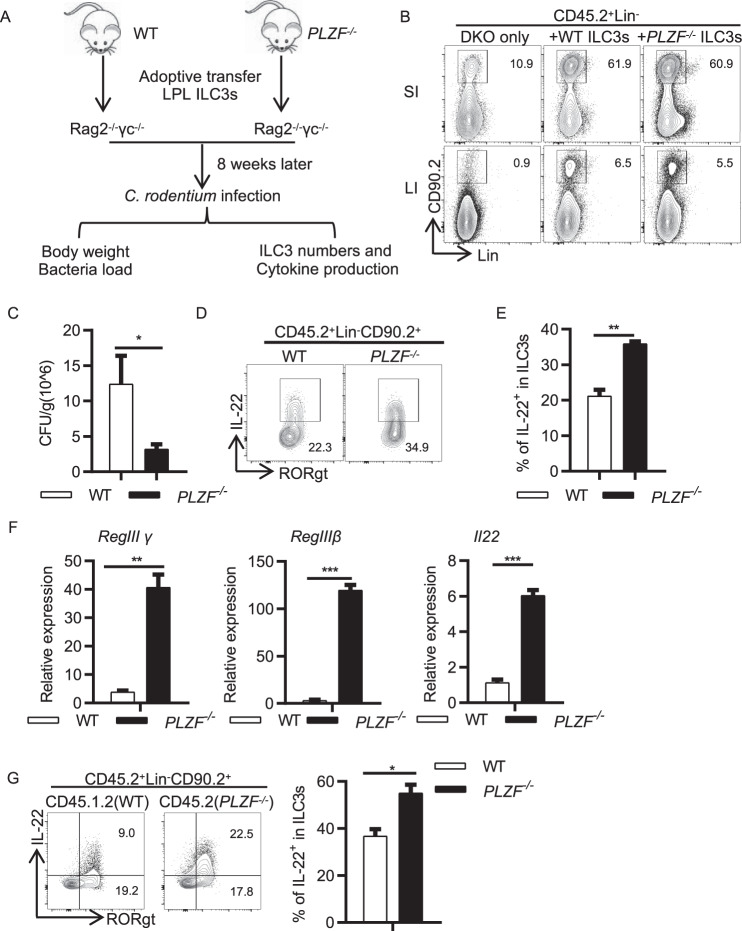
To further determine whether PLZF regulates ILC3 function in a cell-intrinsic manner, we adoptively transferred gut ILC3s purified from either WT mice or PLZF−/− mice into Rag2−/−Il2rg−/− recipient mice, which lack T, B, NK, and ILC cells. ILC3s in the intestine were examined 8 weeks after transfer (Fig. 5A). Although a small amount of residual ILC3s remained in the small intestine of Rag2−/−Il2rg−/− recipient mice, ILC3s were successfully reconstituted in both the small intestine and large intestine (Fig. 5B). The number of ILC3s was not different in the mice receiving PLZF−/− ILC3s compared to those receiving WT ILC3s (Fig. 5B). To further determine whether PLZF regulates gut immunity, we infected these mice with C. rodentium. After 4 days of infection, the mice receiving PLZF−/− ILC3s had a significantly decreased bacterial load in their feces (Fig. 5C). IL-22 production was dramatically increased in PLZF−/− ILC3s compared to WT ILC3s (Fig. 5D, E). Consistently, the expression of antimicrobial peptides was also elevated in the PLZF−/− mice (Fig. 5F). Moreover, the cell-intrinsic effect of PLZF on IL-22 production was confirmed in mixed bone marrow chimeric mice by transferring PLZF−/− (CD45.2+) and WT (CD45.1.2+) bone marrow cells (1:1 ratio) into lethally irradiated CD45.1 recipient mice (Fig. 5G). Collectively, these data suggest that PLZF regulates gut defense against C. rodentium infection in an ILC3-specific manner.
Fig. 5.
PLZF regulates ILC3 function and gut immunity in a cell-intrinsic manner. A Experimental scheme for ILC3 adoptive transfer. ILC3s (Lin−CD45midCD90hi) sorted from the large intestine LPLs of WT and PLZF−/− mice were transplanted into Rag2−/−Il2rg−/− recipients (1 × 10^5 ILC3s per recipient), and recipients were subjected to C. rodentium infection by gavage 8 weeks later (n = 3–4). B Representative flow cytometry plots showing ILC3 reconstitution in the small intestine and the large intestine. C Bacterial load after C. rodentium infection on Day 6. D Representative flow cytometry plots for IL-22-expressing ILC3s from the large intestine are shown from the recipients. E Frequencies of IL-22+ in ILC3s. F Relative mRNA expression of Reg3γ, Reg3β and IL-22 in colonic tissue homogenates of infected recipients on Day 6 p.i. was analyzed by real-time RT‒PCR. G Bone marrow from the CD45.2 PLZF−/− or CD45.1.2 WT mice were mixed 1:1 and transplanted into lethally irradiated recipients (CD45.1+). IL-22 production by ILC3s from donor BM-derived ILC3s (CD45.2+ PLZF−/− and CD45.1.2+ WT) was determined by flow cytometry after 8 weeks of transfer. A–G Data are representative of at least two independent experiments. Data are represented as the mean ± SEM. Error bars show SEM. *P < 0.05; **P < 0.01; ***P < 0.001
PLZF controls ILC3 function and gut immunity in an IL-22-dependent manner
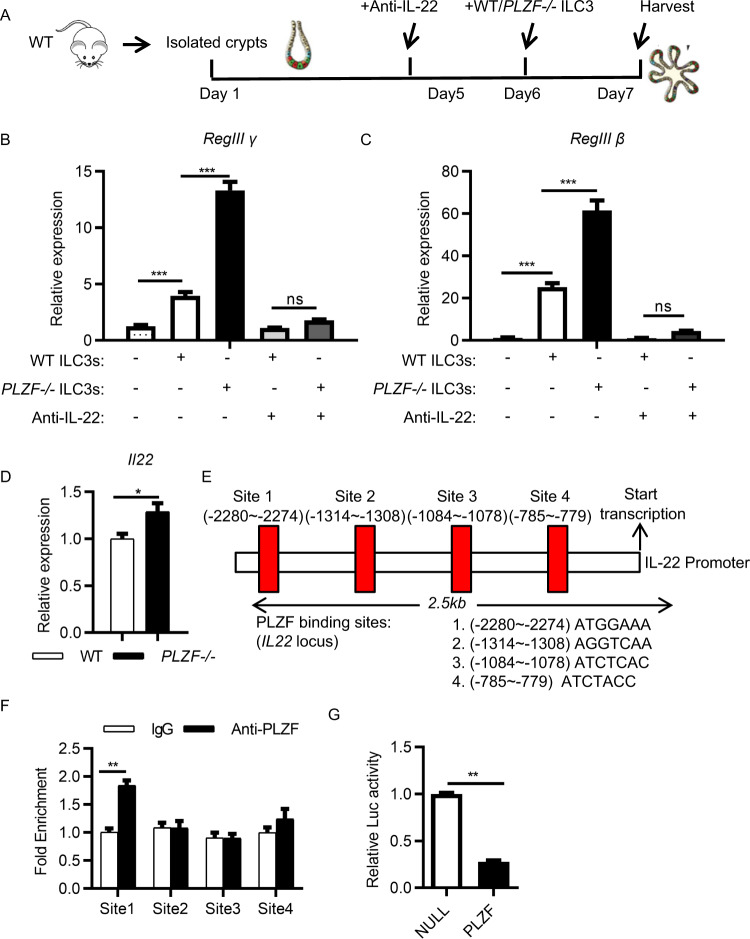
To determine whether IL-22 is the molecular link between PLZF-dependent ILC3 activation and intestinal epithelial integrity, we employed a multitissue organoid culture system. Epithelial organoids were cocultured with ILC3s purified from either WT mice or PLZF−/− mice in the presence of an anti-IL-22 blocking antibody (Fig. 6A). We observed that the addition of WT ILC3s significantly increased the expression of the antimicrobial peptide genes RegIIIγ and RegIIIβ in epithelial organoids. However, PLZF−/− ILC3s exhibited a stronger effect on the induction of epithelial reactivity gene expression than WT ILC3s (Fig. 6B, C). Notably, the addition of anti-IL-22 antibody blocked the induction of epithelial reactivity genes by both WT and PLZF−/− ILC3s and abolished the difference between WT and PLZF−/− ILC3s (Fig. 6B, C). These data suggest that PLZF regulates gut immunity by regulating IL-22 production by ILC3s.
Fig. 6.
PLZF controls ILC3 function in an IL-22-dependent manner. A–C Crypts were isolated from WT mice and cultured for 5 days. The crypts were cocultured with sorted WT or PLZF−/− ILC3s from the large intestine with or without an anti-IL22 blocking antibody. Samples were collected and analyzed at Day 7. A Experimental scheme for ILC3 and organoid coculture. B, C Reg3γ and Reg3β mRNA expression in organoids was analyzed by real-time RT‒PCR (n = 3). D Relative mRNA expression of IL-22 in sorted ILC3s from WT or PLZF−/− large intestine LPLs was analyzed by real-time RT‒PCR (n = 3). E PLZF directly binds sites upstream of the Il22 promoter. PLZF binding sites of the Il22 promoter were predicted. F ChIP‒qPCR analysis of PLZF binding is shown at the indicated sites, with a rabbit IgG control. ILC3s (Lin−CD45.2midCD90.2hi) were sorted from the large intestine LPLs of Rag2−/− mice. G HEK293T cells were cotransfected with IL22 promoter-driven luciferase reporter plasmid plus control or PLZF expression plasmid as indicated. Luciferase activity was assayed 24 h after transfection. A–G Data are representative of at least two independent experiments. Data are represented as the mean ± SEM. Error bars show SEM. *P < 0.05; **P < 0.01; ***P < 0.001
Next, we sought to investigate the molecular mechanisms underlying PLZF regulation of IL-22 production. We first examined whether IL-22 expression is regulated at the transcriptional level by PLZF. We found that Il22 mRNA was increased significantly in PLZF−/− ILC3s compared to that in WT ILC3s, indicating that IL-22 is regulated by PLZF at the transcriptional level (Fig. 6D). Previous studies have shown that RORγt, AhR, and STAT3 are critical transcription factors for IL-22 expression in ILC3s. We examined the expression levels of RORγt and found no difference between WT and PLZF−/− ILC3s (Supplementary Fig. 6A). To determine whether AhR or STAT3 is involved in regulating IL-22 expression, we used small molecule inhibitors for AhR (AhR antagonist, Millipore) and STAT3 (STA-21, Selleck) to treat ILC3s. Both the AhR antagonist and STA-21 significantly inhibited IL-22 expression by ILC3s. However, they did not abolish the increased IL-22 production by PLZF−/− ILC3s (Supplementary Fig. 6B, C). These results ruled out the possibility that PLZF regulated IL-22 expression by interacting with RORγt, AhR, and STAT3. Hence, we reason that PLZF might regulate IL-22 transcription directly in ILC3s. Previous studies have shown that PLZF binding sites contain consensus binding motifs [17, 18] (Supplementary Fig. 7). Through bioinformatic analysis of the Il22 promoter, we found four putative binding sites for PLZF upstream of the Il22 transcription start site (Fig. 6E). Thus, to investigate whether PLZF interacts with these potential binding sites in the Il22 locus, we performed chromatin immunoprecipitation (ChIP) assays. We observed a significant interaction of PLZF with the Il22 promoter at site 1 in ILC3 cells sorted from the small intestine (Fig. 6F). Next, we examined the functional effects of the interaction of PLZF with its bound DNA sequence using a reporter plasmid carrying firefly luciferase under the control of the Il22 promoter. Transfection with the PLZF expression vector resulted in a significant reduction in luciferase activity (Fig. 6G). Together, these results suggest that PLZF directly regulates IL-22 transcription.
Discussion
Here, we showed that PLZF is specifically expressed in mature intestinal ILC3s. PLZF is dispensable for ILC3 development but controls ILC3 function in a cell-intrinsic manner. PLZF inhibits innate IL-22 production in ILC3s and is an essential regulator of gut immune homeostasis.
Previous studies have shown that PLZF is highly expressed in ILCP but not in mature ILC3s [2]. In our study, we found that ILC3s did express PLZF but at lower levels than their progenitor-ILCP. We confirmed PLZF protein expression in ILC3s by both immunoblotting and intracellular staining. The discrepancy might be due to the weak expression efficiency of the GFP-reporter system under the control of the endogenous PLZF gene. PLZF plays an important role in ILC2 development and function [15]. Our data showed that PLZF played a dispensable role in regulating ILC3 development but suppressed IL-22 production by ILC3s. The mechanisms by which PLZF regulates ILC3 function are mediated through transcriptional regulation, which is different from that in ILC2s because ILC2s do not express PLZF. ILC3s are considered a major source of IL-22 in vivo, which is important in maintaining intestinal epithelial integrity and inhibiting pathogenic bacterial expansion. Therefore, regulation of ILC3-derived IL-22 production is critical for intestinal tissue homeostasis [19]. Downregulation of PLZF expression in mature ILC3s originating from ILCPs might reflect a physiological adaptation for ILC3s to increase their IL-22 production for host defense under steady state. Under C. rodentium infection and colitis conditions, PLZF expression was further reduced to enhance ILC3 function and facilitate bacterial clearance and tissue repair. Therefore, PLZF is an important regulator of ILC3s for gut defense during infection and inflammation.
Our data show that PLZF inhibited IL-22 production by ILC3s in the gut. The transcription factors RORγt, AhR, and STAT3 are the master transcriptional regulators of IL-22 expression in ILC3s [9, 20–22]. This finding prompted us to consider whether PLZF controls IL-22 production by interacting with these transcription factors. Interestingly, the expression level of RORγt was not different between WT and PLZF−/− ILC3s. In addition, small molecule inhibitors of AhR and STAT3 did not diminish the increased IL-22 level in PLZF−/− ILC3s. These data rule out the possibility of PLZF interacting with these transcription factors. PLZF has been reported to have multiple layers of transcriptional regulation in innate lymphocytes, including binding to genes encoding cytokine receptors, adhesion receptors, and transcription factors [23]. Accordingly, our data show that PLZF could bind to the IL22 locus directly and regulate IL22 transcription. Thus, PLZF inhibits IL-22 production in ILC3s through transcriptional control.
In summary, PLZF was identified as a negative regulator of ILC3 function by inhibiting IL-22 production. The downregulation of PLZF expression from ILCPs to mature ILC3s might be important to maintain high level of innate IL-22 to maintain gut homeostasis under steady-state conditions. During gut infection and inflammation, PLZF expression was further reduced in ILC3s, which resulted in enhanced IL-22 production in the gut and is thus beneficial to host gut defense against diseases.
Materials and methods
Mice
PLZF-deficient mice were a gift from Dr. Xingxu Huang at Shanghai Tech University, and the mice had been described previously [24]. Rag2−/−Il2rg−/− mice were a gift from Dr. Xiaohuan Guo at Tsinghua University. CD45.1, C57BL/6, BALB/c, S129, DBA, and ICR mice were purchased from Shanghai Laboratory Animal Center (SLAC). All mice were bred and maintained in individually ventilated cages on a strict 12-hour day-night cycle under specific pathogen-free conditions in accredited animal facilities. Eight- to ten-week-old gender-matched littermate control mice were used as controls for all experiments. All animal experiments were performed in compliance with the Guide for the Care and Use of Laboratory Animals, approved by the Shanghai Jiao Tong University School of Medicine Institutional Animal Care and Use Committees (IACUC).
Flow cytometry and cell sorting
Antibodies used for staining followed by flow cytometry analysis or cell sorting are listed in Supplementary Table 1. Dead cells were identified using the LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Thermo Fisher Scientific). Fc receptor blockade was performed using purified anti-mouse CD16/CD32 before surface staining. A Foxp3 staining kit (Thermo Fisher Scientific) was used for intracellular transcription factor staining. The eBioscienceTM Intracellular Fixation & Permeabilization kit was used for intracellular cytokine staining. Flow cytometry data were collected using an LSRFortessa X20 and FACSymphony A3 (BD Bioscience) and analyzed by FlowJo software (Tree Star, Inc.). For detection of cytokine production, intestinal LPLs were treated with 50 ng/ml PMA (Sigma) and 500 ng/ml ionomycin (Sigma) for 3 h. Then, 2 μg/ml brefeldin A (Sigma) was added to the culture for at least 3 h before the cells were harvested for analysis. ILC3s and ILCP were sorted using BD FACSAria III. ILCPs from bone marrow were sorted as lineage−CD127+α4β7+PD-1+, and ILC3s from intestine were sorted as lineage−CD45.2midCD90.2hiKLRG1−.
Isolation of intestinal lamina propria lymphocytes (LPLs)
The isolation of intestinal LPLs was performed as previously described [7]. Briefly, small and large intestines were dissected. Fat tissues were removed carefully from the intestines. Intestines were dissected longitudinally and subsequently cut into several pieces, followed by washing with cold phosphate-buffered saline (PBS). For removal of epithelial cells, the intestines were then incubated successively with 1 mM dithiothreitol (DTT)-PBS once for 10 min at room temperature (RT) and with 30 mM ethylenediaminetetraacetic acid (EDTA)-PBS twice for 10 min at 37 °C while shaking at 250 rpm. The tissues were then digested with DNase I (Sigma; 150 µg/ml) and collagenase VIII (Sigma; 300 U/ml) in RPMI 1640 medium (Thermo Fisher Scientific) at 37 °C in a 5% CO2 incubator for 1.5 h. The digested tissues were homogenized by vigorous shaking and filtered with a 100 µm cell strainer. Mononuclear cells were then harvested from the interphase of an 80% and 40% Percoll (GE Healthcare Life Sciences) gradient after spinning at 2500 rpm for 20 min at RT.
Bone marrow chimeras
Bone marrow cells from the femur and tibia of donor CD45.2 mice (WT or PLZF−/−) were collected, mixed with WT CD45.1.2 bone marrow cells at a 1:1 ratio, and resuspended in PBS at a concentration of 3 × 107 cells per mL. A total of 6 × 106 cells in 200 μl of PBS were injected intravenously into lethally irradiated recipient mice (CD45.1). After 6–8 weeks, bone marrow reconstitution was verified by staining peripheral blood cells with anti-mouse CD45.1 (clone A20) and anti-mouse CD45.2 (clone 104) antibodies (Biolegend). Reconstituted mice were subjected to analysis of the constitution of ILCs in the small and large intestine.
ILCP and ILC3 transfer model
Bone marrow cells from the femurs and tibias of donor mice (WT and PLZF−/−) were collected and resuspended in PBS. Lineage+ cells were removed using the EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (Stem Cell) before ILCPs were sorted by BD FACSAria III. We intravenously injected 1000 ILCP cells in 200 μl of PBS into Rag2−/−Il2rg−/−recipient mice. After 6–8 weeks, reconstituted mice were subjected to analysis of the constitution of ILCs in the small and large intestine. For the ILC3 transfer model, we intravenously injected 1 × 105 ILC3 cells in 200 μl of PBS into Rag2−/−Il2rg−/−recipient mice. After 6–8 weeks, reconstituted mice were infected with C. rodentium and analyzed further.
C. rodentium infection
C. rodentium was cultured overnight in Luria-Bertani broth. Mice were orally gavaged with 5 × 109 colony forming units (CFU) of C. rodentium in PBS. Feces and tissue samples were collected, weighed, homogenized, serially diluted and plated to determine the CFU. Some tissue samples were also collected and analyzed by real-time RT‒PCR and flow cytometry.
DSS-induced acute colitis
Sex-matched PLZF WT and PLZF−/− mice at the age of 6–8 wk were fed 3.5% DSS (MP Biomedical) in normal drinking water for 7 days, followed by normal drinking water for another 2 days to induce acute colitis. Mouse weights were monitored every day during the colitis model. The humane endpoint for the DSS study, in accordance with Institutional Animal Care and Use Committee recommendations from Shanghai Jiao Tong University, was the loss of 25–30% of initial body weight. Scoring for stool consistency was performed as previously described. In brief, stool scores were determined as follows: (0) well-formed pellets; (1) soft but still formed; (2) semiformed stools that adhered to the anus; and (3) liquid stools that adhered to the anus. Some tissue samples were also collected and analyzed by real-time RT‒PCR and flow cytometry.
Detection of mRNA by real-time RT‒PCR
RNA was isolated with TRIzol reagent (Invitrogen). cDNA was synthesized using a GoScript™ Reverse Transcription kit (Promega). Real-time PCR was performed using SYBR Green Master Mix reagents (Roche), and primer mixtures were used for real-time PCR (Supplementary Table 2). Reactions were run with chromatin immunoprecipitation (ChIP) coupled with quantitative PCR. The mRNA results are displayed as relative expression values normalized to Gapdh.
Histological analysis
For H&E staining, ∼3 mm of the large intestine was fixed in 4% paraformaldehyde and embedded in paraffin. Then, sections were stained with H&E for histological analysis. Sections were scored in a blinded manner on a scale from 0 to 4, based on previous reports [25]. In brief, 0, no inflammatory cell infiltration or occasional inflammatory cells in the lamina propria with no mucosal damage; 1, mild inflammatory cells in the lamina propria with mild mucosal damage; 2, moderate inflammatory cells in the lamina propria with moderate mucosal damage; 3, marked inflammatory cells in the lamina propria with marked mucosal damage; and 4, transmural extension of inflammatory cells in the lamina propria with extensive mucosal damage and extension into deeper structures of the bowel wall.
Western blot and immunoblot analysis
Sorted cells were lysed with radioimmunoprecipitation assay buffer with a protease inhibitor mixture (Roche) on ice for 30 min. Supernatants were collected after centrifugation at 13,000 rpm for 15 min at 4 °C. Western blot analysis was performed using the indicated antibodies, including anti-PLZF (1:1000) from Abcam, anti-β-actin (1:2000; Santa Cruz), and anti-rabbit IgG (1:2000; CST).
Reporter gene assay
Lipofectamine 2000 transfection reagent (Invitrogen) was used to transfect 293 T cells with IL22 promoter containing pGL3-basic reporter construct, expression vectors for control and PLZF, and Renilla luciferase was included as an internal control. Cells were lysed 24 h after transfection with 100 μl per well of 1 × passive lysis buffer. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). The firefly luciferase signal was normalized to cotransfected Renilla luciferase activity in the same sample.
Intestinal organoid culture
Mouse colons were opened longitudinally and cut into 1-cm-long pieces. The colon pieces were washed four times in a 50 mL tube containing 20 mL of ice-cold PBS with quick gentle shaking, followed by 30 min incubation in 5 mL of DPBS containing 2 mM EDTA at 4 °C with gentle shaking. Crypts were released by shaking the tube for 20 s in 5 mL of ice-cold dissociation buffer (55 mM d-sorbitol and 43 mM sucrose in DPBS), passed through a 100-μm cell strainer and collected by centrifugation at 150 g for 5 min at 4 °C. The isolated crypts were mixed with ice-cold unsolidified growth factor-reduced Matrigel (Corning) at 200 crypts per 50 μL of Matrigel and then placed at the center of each well in a 24-well plate. Following polymerization of the Matrigel for 15 min at 37 °C, each well was supplemented with 500 μL of advanced DMEM/F12 medium (Thermo Fisher Scientific) containing 200 ng/mL EGF (Thermo Fisher Scientific, PMG8041), 100 ng/mL Noggin (PeproTech, 250-38), 100 ng/mL WNT3A (R&D Systems, 1324-WN/CF) and 1 μg/mL R-spondin 1 (PeproTech, 120-38) and incubated at 37 °C for further analysis. The circumferences of the colonic organoids were recorded daily. For organoid coculture experiments, a total of 200 crypts were mixed with 5 × 103 flow cytometry-sorted ILC3s from WT and PLZF−/− mice cultured with or without anti-IL-22 antibody (10 µg/mL). Following polymerization of the Matrigel (15 min at 37 °C), crypts were cultured in advanced DMEM/F12 medium plus 200 ng/mL EGF, 100 ng/mL Noggin and 100 ng/mL WNT3A. The cocultured cells were collected and further analyzed by real-time RT‒PCR.
Chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) assays
FACS-sorted ILC3s were fixed with 1% formaldehyde (Sigma) and fragmented by sonication to 100–500 bp. Anti-PLZF antibody (Abcam) or rabbit IgG (Abcam) was then used for immunoprecipitation. After being washed and reverse crosslinked, the pulldown DNA, as well as the input DNA, was purified by ethanol precipitation and quantified using real-time PCR. PLZF binding motifs containing cis-regulatory elements were identified using the JASPAR database.
Statistical analysis
Unless otherwise noted, the Kolmogorov–Smirnov test was performed for normality tests. Statistical analysis was then performed using the unpaired Student’s t test as parametric tests or Mann–Whitney test as nonparametric tests on individual biological samples using the GraphPad Prism program. Data from these experiments are presented as the mean values ± SEMs. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary information
Acknowledgements
We thank Dr. Xingxu Huang from Shanghai Tech University for the PLZF-deficient mice and Dr. Xiaohuan Guo from Tsinghua University for the Rag2−/−Il2rg−/− mice. We thank all the members from the Shen laboratory for their help and suggestions. We thank the Flow Cytometry Core in Shanghai Institute of Immunology and Animal Facility at Shanghai Jiao Tong University School of Medicine for service and assistance.
Author contributions
YX, HZ, and SW performed the experiments and analyzed the data. JL, HL, DW, YZ, HN and XS performed flow cytometry and in vitro experiments. JS and LS wrote the manuscript and conceived, designed, and supervised the project.
Funding
This study was supported by grant 2020YFA0509200 (to LS) from the Ministry of Science and Technology of China, grant 81971487 (to LS) from the National Natural Science Foundation of China, grants 20ZR1430200 and 20142202300 (to LS) from the Science and Technology Commission of Shanghai Municipality, grant 20194Y0275 (to JS) from the Shanghai Municipal Health Commission, and the Shanghai Frontiers Science Center of Cellular Homeostasis and Human Diseases.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yaru Xu, Huasheng Zhang.
Contributor Information
Jiping Sun, Email: sunjiping@shsmu.edu.cn.
Lei Shen, Email: lshen@shsmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-023-00975-5.
References
- 1.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 2.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–5. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 4.Melo-Gonzalez F, Hepworth MR. Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology. 2017;150:265–75. doi: 10.1111/imm.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernández PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, et al. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–34. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu J, Heller JJ, Guo X, Chen ZE, Fish K, Fu YX, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–9. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 9.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–72. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JS, Tato CM, Joyce-Shaikh B, Gulen MF, Cayatte C, Chen Y, et al. Interleukin-23-Independent IL-17 production regulates intestinal epithelial permeability. Immunity. 2015;43:727–38. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishizuka IE, Constantinides MG, Gudjonson H, Bendelac A. The innate lymphoid cell precursor. Annu Rev Immunol. 2016;34:299–316. doi: 10.1146/annurev-immunol-041015-055549. [DOI] [PubMed] [Google Scholar]
- 12.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF (Zbtb16) directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. PNAS. 2009;106:12453–8. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. 2016;17:1300–11. doi: 10.1038/ni.3565. [DOI] [PubMed] [Google Scholar]
- 15.Verhoef PA, Constantinides MG, McDonald BD, Urban JF, Jr, Sperling AI, Bendelac A. Intrinsic functional defects of type 2 innate lymphoid cells impair innate allergic inflammation in promyelocytic leukemia zinc finger (PLZF)-deficient mice. J Allergy Clin Immunol. 2016;137:591–600. doi: 10.1016/j.jaci.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raberger J, Schebesta A, Sakaguchia S, Bouchero N, Blomberg KE, Berglof A, et al. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. PNAS. 2008;105:17919–24. [DOI] [PMC free article] [PubMed]
- 17.Li JY, English MA, Ball HJ, Yeyati PL, Waxman S, Licht JD. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger prote. JBC. 1997;272:22447–55. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 18.Sitterlin D, Tiollais P, Transy C. The RAR alpha-PLZF chimera associated with Acute Promyelocytic Leukemia has retained a sequence-specific DNA-binding domain. Oncogene. 1997;14:1067–74. [DOI] [PubMed]
- 19.Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol. 2016;17:179–86. doi: 10.1038/ni.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Bostick JW, Zhou L. Regulation of innate lymphoid cells by aryl hydrocarbon receptor. Front Immunol. 2017;8:1909. doi: 10.3389/fimmu.2017.01909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity. 2014;40:25–39. doi: 10.1016/j.immuni.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn YO, Blazar BR, Miller JS, Verneris MR. Lineage relationships of human interleukin-22-producing CD56+ RORgammat+ innate lymphoid cells and conventional natural killer cells. Blood. 2013;121:2234–43. doi: 10.1182/blood-2012-07-440099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, et al. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. PNAS. 2016;113:7602–7. [DOI] [PMC free article] [PubMed]
- 24.Yin S, Yu J, Hu B, Lu C, Liu X, Gao X, et al. Runx3 mediates resistance to intracellular bacterial infection by promoting IL12 signaling in group 1 ILC and NCR+ILC3. Front Immunol. 2018;9:2101. doi: 10.3389/fimmu.2018.02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–91. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.