Abstract
Market drugs, such as Food and Drug Administration (FDA) or European Medicines Agency (EMA)-approved drugs for specific indications provide opportunities for repurposing for newer therapeutics. This potentially saves resources invested in clinical trials that verify drug safety and tolerance in humans prior to alternative indication approval. Protein arginine methyltransferase 5 (PRMT5) overexpression has been linked to promoting the tumor phenotype in several cancers, including pancreatic ductal adenocarcinoma (PDAC), colorectal cancer (CRC), and breast cancer (BC), making PRMT5 an important target for cancer therapy. Previously, we showed that PRMT5-mediated methylation of the nuclear factor (NF)-κB, partially contributes to its constitutive activation observed in cancers. In this study, we utilized an AlphaLISA-based high-throughput screening method adapted in our lab, and identified one FDA-approved drug, Candesartan cilexetil (Can, used in hypertension treatment) and one EMA-approved drug, Cloperastine hydrochloride (Clo, used in cough treatment) that had significant PRMT5-inhibitory activity, and their anti-tumor properties were validated using cancer phenotypic assays in vitro. Furthermore, PRMT5 selective inhibition of methyltransferase activity was confirmed by reduction of both NF-κB methylation and its subsequent activation upon drug treatment. Using in silico prediction, we identified critical residues on PRMT5 targeted by these drugs that may interfere with its enzymatic activity. Finally, Clo and Can treatment have exhibited marked reduction in tumor growth in vivo. Overall, we provide basis for pursuing repurposing Clo and Can as anti-PRMT5 cancer therapies. Our study offers potential safe and fast repurposing of previously unknown PRMT5 inhibitors into clinical practice.
Keywords: Cancer Research, NF-kB, Pharmacology, PRMT5, PRMT5 inhibitors
Introduction
Repurposing of Food and Drug Administration (FDA)-approved drugs and market drugs from other countries than USA for indications apart from their primary intended therapeutic purpose has been a popular method in drug discovery and development. The gain in popularity for adopting this method is two-fold. Firstly, the safety, efficacy, formulation and toxicity profiles of a clinically approved drug are well established for a particular indication. This allows for valuable resources such as time and money to be saved by streamlining studies for testing a new indication based on existing data, such as preventing the need to conduct phase I clinical studies to understand its safety in humans. Thus, approval of repurposed drugs is ∼3–12 years faster and at ∼50% lower cost in comparison to their de novo counterparts. Secondly, compounds that were shelved in earlier clinical trials due to unfavorable results could still be potentially developed for other indications, thus preventing wastage of resources involved in unsuccessful clinical trials.1,2
Several successful examples of drugs that were in development or approved for treatment which were then repositioned for novel indications have been demonstrated over the years. Perhaps, the most famous example is Sildenafil (Viagra®), a phosphodiesterase 5 inhibitor first developed by Pfizer that is currently used to treat erectile dysfunction in adult males. Viagra® was originally being developed to treat hypertension in the mid-1980s, with the aim of vasodilating arteries and lessening blood pressure. Interestingly, in clinical trials, researchers observed that this drug was significantly more effective in treating erectile dysfunction than heart disease, thereby repositioning it for a new indication. Viagra® is currently prescribed to millions of people and has generated billions of dollars in revenue since it was approved by FDA in 1998. Interestingly, a further repurposing of Viagra® might be on the horizon since new trials have shown its potential as an anti-cancer drug.3 If shown to be effective, it will severely shorten the approval process for the new indication, while at the same time providing a cost-effective treatment option in cancers. Viagra® is just one of the many instances that have shown a successful repurposing of already approved drugs for cancers among other diseases; thus, providing a strong rationale for the study described here. We focus on adapting FDA-approved drugs and other market drugs, such as EMA-approved drugs, for potentially treating some of the deadliest cancers today namely, pancreatic ductal adenocarcinoma (PDAC), colorectal cancer (CRC), and breast cancer (BC).
CRC and PDAC are the third and fourth leading causes of deaths in men and women combined in the United States respectively. Similarly, BC is the second leading cause of death for women in the US, amounting to 14% of the total cancer-related deaths in females.4 The triple-negative (TNBC) subtype accounts for ∼15% of the cases and is marked by shorter survival and higher chance of recurrence than the other subtypes.5 For these three cancer types, chemotherapy and certain targeted therapies are currently available clinically. At the same time, several dozen novel small molecules/biologics are being developed, with some reaching clinical trials after several years in the discovery phase of drug development. Despite this, mortality due to these cancers continues to rise rapidly. Furthermore, the budget required to develop and bring these drugs to market as well as the treatment cost to suffering patients continues to be unimaginably high, creating a considerable challenge for developing cost-effective therapies at a rapid pace. It is imperative to design studies and identify critical factors that can be a subject of effective therapeutic strategies. Even more importantly, it would be highly beneficial to shorten the FDA approval process so that these treatments speedily reach the clinic while at the same time are effective, safe, and economical. Keeping these factors in mind, we have focused on identifying FDA-approved drugs that can target protein arginine methyltransferase 5 (PRMT5). A plethora of studies have highlighted the tumor-promoting role of PRMT5 in PDAC, CRC and BCs. PRMT5 is overexpressed in PDAC, CRC and BC6 and has been associated with increased cell proliferation, migration, and colony formation ability.6, 7, 8, 9 Furthermore, PRMT5 methylate critical substrate proteins such as nuclear factor kappa B (NF-κB), an important transcription factor that is constitutively activated in PDAC, CRC, and BC. We recently showed that PRMT5-mediated NF-κB activation is at least partially responsible for this constitutive activation and correlates with its activity as a tumor promoting agent.6,7 Furthermore, inhibition of PRMT5 using small molecule inhibitors slowed down tumor growth. Thus, PRMT5 has immense potential to be exploited as a therapeutic target in these cancers.
In this regard, we developed an AlphaLISA-based high-throughput screening (HTS) described previously10 to screen for FDA-approved drugs with anti-PRMT5 activity. Our goal is to screen a library of FDA or EMA approved compounds for their ability to inhibit PRMT5 and move this viable therapeutic target into clinic more rapidly, if found promising. In this process, we identified an FDA-approved drug, Candesartan cilexetil (Can), an antihypertensive drug, and Cloperastine hydrochloride (Clo), a cough suppressant commonly used in Europe and Japan, as promising candidates with anti-PRMT5 activity in cancers. We discovered that both Clo and Can inhibited PRMT5-mediated NF-κB methylation, activation, and its downstream target gene expression. Moreover, we demonstrated that Clo and Can reduced cancer cell proliferation and 3D spheroid growth in PDAC, CRC and BC cells. Interestingly, we observed that Clo or Can exhibited strong synthetic lethality with PDAC treatment drug, gemcitabine. Importantly, we confirmed that both Clo and Can could remarkably inhibit tumor growth in xenograft models in vivo, while without significantly affecting animals’ body weight. Interestingly, in silico model predicted the potential binding mechanism of Clo or Can with PRMT5, highlighting an important role for glutamine (E) 444 and phenylalanine (F) 327 in the binding process. Collectively, our studies suggest the promising nature of repurposing FDA-approved drugs to identify novel targets (for instance, PRMT5) and employ them for use in newer indications either as a sole or combinatory therapy, such as the cancer subtypes described in this study.
Materials and methods
Cell lines and reagents
PDAC (HPNE, PANC1, MiaPaCa2 and AsPC1) and BC (MDA-MB-231, MCF7, and BT20) cell lines were kind gifts from Pancreatic Cancer Signature Center and Dr. Britney Shea-Herbert respectively at Indiana University School of Medicine (IUSM). The CRC cell lines (HT29, HCT116, and DLD1) cell lines were obtained from American Type Culture Collection (Manassas, VA). Cell lines were authenticated using 9 Marker STR Profile (IDEXX Bioresearch, Columbia, MO) and were determined to be free of microbial contamination using SouthernBiotech Mycoplasma Detection Kit (Cat. No. 13100-01, Birmingham, AL, USA). PDAC (except AsPC1) and BC cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) (GE Healthcare, Chicago, IL), supplemented with 1% of penicillin/streptomycin and 5% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA). CRC cells and AsPC1 were grown in Roswell Park Memorial Institute Medium (RPMI) 1640 (GE Healthcare), with 1% penicillin/streptomycin and 5% FBS. A PANC1 shPRMT5 knockdown stable cell line was generated previously and used here.10 All cell lines were used within 6 passages. Cell lines were maintained in a sterile incubator at 37°C under 5% CO2.
For AlphaLISA HTS: the substrate, histone H4 peptide unmethylated at arginine (R) 3 (unmeH4R3) was purchased from AnaSpec (Fremont, CA). The 23-amino acid H4R3 peptide was biotinylated at its N’ terminus: SGRGKGGKGLGKGGAKRHRKVLRGG-K(biotin)-NH2. S-adenosyl methionine (SAM) was used as a methyl group donor (New England Biolabs, Ipswich, MA). Compound libraries comprising of FDA-approved drugs, synthetic and semi-synthetic natural products as well as bioactives were collectively purchased from Analyticon Discovery (Rockville, MD), MilliporeSigma (Burlington, MA) and Microsource Discovery Systems Inc (Gaylordsville, CT). Acceptor beads specific for dimethylated H4R3, Donor beads with a streptavidin tag, assay buffer, OptiPlate™ 384-well white bottom plates, TopSeal™-A film to seal the plates and EnVision® Multilabel Reader were sourced from PerkinElmer (Waltham, MA). Clo and Can were sourced from Sigma–Aldrich (St. Louis, MO), both in powder form and dissolved in appropriate dimethyl sulfoxide (DMSO) stocks for experimental use.
Primary antibodies for symmetric dimethylation at arginine (R) 30 subunit of p65 (detected by generating a customized polyclonal primary antibody in collaboration with GenScript Inc, Piscataway, NJ, at 1:750) and β-actin (Sigma–Aldrich, Cat # A5316, at 1:5000) with their corresponding secondary antibodies were used.
AlphaLISA-based HTS
A detailed description of optimization of the AlphaLISA-based HTS assay protocol has already been published previously.10 Briefly, substrate/SAM mixture (60 nM unmethylated H4R3 and 200 μM SAM) was added to the respective wells, following which stock of library compounds was pipetted into the wells. PRMT5 enzyme preparation purified using co-immunoprecipitation experiments in 293 cells overexpressing Flag-tagged PRMT5 was then added to the wells, as described earlier.10 No compound was added to wells with positive controls that denoted “maximal signal” in the reaction. For negative control wells, no enzyme or compound was added which denoted the “background signal”. The plates were incubated at room temperature (R.T.) for 1 h followed by addition of Acceptor beads specific for dimethylated H4R3 (1:50 dilution of the 5 mg/ml stock) into all wells of the assay plates and incubated at R.T. for 1 h. Finally, Donor beads (1:50 dilution of the 5 mg/mL stock) were added to the plates and incubated at R.T. before Alpha signal was then quantified using the EnVision® reader.
Enzyme-linked immunoassay (ELISA) & conditioned media assay
The PANC1, HT29, or MDA-MB-231 cells were seeded into 12-well plates, cultivated to 90% confluency, treated with either Clo or Can at their IC50s for 24 h. The media were collected as described before.11 Floating cells were pelleted at 3000 g at 4°C, for 10 min, and the supernatant was aliquoted into sterile tubes and either used immediately or stored at −80°C. The media were then analyzed with ELISA assays for interleukin 8 (IL8) and tumor necrosis alpha (TNFα). Assays were performed as manufacturer suggested using kits including standard curve comparisons (TNFα, Cat# 76175-416; IL8, Cat # 76271-190) (Peprotech East Windsor, NJ).
MTT [(3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide)] assay
Cells were grown to 60% density in clear flat-bottom 96-well plates (Corning Inc, Corning, NY) for 24 h and treated with a range of concentrations of either Clo or Can respectively, followed by incubation for 4 days. 10 μL of MTT dye (Sigma–Aldrich) was added to each well and incubated for 2 h at 37°C. The medium: MTT mixture was then removed and 100 μL DMSO was added per well. Absorbance was measured using Synergy H4 Multi-Mode Reader (BioTek Instruments, Winooski, VT).
Three-dimensional (3D) colony formation assay
PANC1 and MDA-MB-231 (1000 cells) as well as HT29 (250 cells) were seeded per well within ultra-low attachment plates (Corning Inc) in media comprising 3% growth factor reduced Matrigel (BD Biosciences, Franklin Lakes, NJ).12 Clo and Can were added in a concentration range from 5–100 μM to 5–120 μM respectively after seeding for 48 h. After 72 h of drug treatment, the cells were stained using 10% Alamar blue dye (Fisher) and fluorescence was quantified using a Synergy H4 Reader.
Western blotting
PDAC, CRC, and BC cells were pelleted post treatment with either Clo or Can in phosphate buffered saline (PBS). Pellets were lysed with lysis buffer [(10 mM Tris-Cl pH 8.0, 1 mM EDTA, 1% Triton X, 0.1% sodium deoxycholate, 0.1% SDS (sodium dodecyl sulfate), 14 mM NaCl, 1 mM phenylmethylsulfonyl fluoride]. Protein concentration for each sample was determined using Protein Assay Reagent (Bio-Rad, Hercules, CA). Equal protein concentrations were run on a 10% SDS-PAGE (polyacrylamide gel electrophoresis) gel and transferred using polyvinylidene fluoride membrane (Thermo Fisher Scientific). Membranes were exposed to respective primary and secondary antibodies at dilutions and times based on manufacturer's instructions. Anti-Caspase-3 (Cell Signaling Technology, Danvers, MA, Cat# 14220, at 1:2000), anti-tubulin (Cell Signaling Technology, Danvers, MA, Cat# 10376, at 1:2500), anti-Lamin (Rosemont, IL, Cat# 12987-1-AP, at 1:1500), anti–NF–κB p65 (Santa Cruz Biotechnology, Inc. Dallas, TX, Cat# sc-109, at 1:4500). Protein signal was detected using enhanced chemiluminescence (ECL) reagent (PerkinElmer).
Nuclear fractionation
Fractionation experiments were conducted as previously described13 and according to the manufacturer's instructions for the nuclear extract kit (Active Motif, Carlsbad, CA, Cat# 40010). Briefly, cells were grown to about 80% confluency, then treated with either Clo or Can at its respective IC50 for 24 h. Cells were then washed with ice-cold 1 × PBS, collected and pelleted for 5 min at 500 rpm. Cell pellets were gently resuspended in 1 × hypotonic buffer and incubated for 15 min on ice. After centrifuging for 30 s at 14,000×g, the cell pellet was further lysed in Complete Lysis Buffer, incubated on ice for 30 min and centrifuged for 10 min at 14,000×g to collect the nuclear fraction. The samples were then subjected to Western blot analyses.
NF-κB luciferase assay
The NF-κB luciferase lentiviral construct pLA-NFκBmCMV-luc-H4-puro (containing five tandem copies of the NF-κB site from the IP10 gene)14 was introduced in the respective cell lines using Lipofectamine™ LTX Reagent and PLUS Reagents (Thermo Fisher Scientific). Luciferase activity was measured after 48 h (with or without drug treatment) using Reporter Lysis Buffer kit (Promega, Madison, WI) as per manufacturer's instructions and the Synergy H4 Reader.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Following treatment with either Clo or Can for 48 h, total RNA was isolated from the respective cell lines using TRIzol® as described earlier.6 First strand complementary DNA was generated using SuperScript III First-Strand Synthesis Kit (Invitrogen, Carlsbad, CA). qPCR was subsequently executed using FastStart Universal SYBR Green kit (Roche, Indianapolis, IN). Primers were designed using the Primer Express 3.0 software (Thermo Fisher Scientific). IL8-F TCCTGATTTCTGCAGCTCTGT; IL8-R AAATTTGGGGTGGAAAGGTT; TNF-F TGGCCCAGGCAGTCAGA; TNF-R GGTTTGCTACAACATGGGCTACA NFKBIA-F GCTGAAGAAGGAGCGGCTACT; NFKBIA-R TCGTACTCCTCGTCTTTCATGGA; TRAF1-F CCCCAGAGTGAAACCAACGT; TRAF1-R CTTGGGTGACTGCAGTTTGCT.
Docking analysis
Chain A of PRMT5 protein in 4X61.pdb from the Protein Databank was used for docking. All docking experiments were performed using the Glide program (version 7.6) of the Schrodinger suite in standard precision mode.15, 16, 17 The two compounds, Clo and Can, were docked into the PRMT5 active site under two conditions: (i) Docking the compound into PRMT5 active site in presence of SAM; (ii) Docking the compound into completely empty PRMT5 active site. Figures were prepared with Maestro version 11.6 18.
Tumor efficacy study
NSG mice were obtained from the In Vivo Therapeutics Core at Indiana University School of Medicine. After acclimation for 7 days, NSG mice (5–6 weeks old) were xenografted with Mycoplasma-free PANC1 or HT29 cells subcutaneously (1 × 107 PANC1, 3 × 106 HT29 cells used per mouse in 0.2 mL of a 1:1 mix of serum free DMEM media, or serum free RPMI media and Matrigel). Five mice were randomized in each group when tumor volumes reached ∼50 mm3. Mice were treated with either vehicle control (10% DMSO, 20% PEG400, 5% Tween 80 and 65% sterile water), or 100 mg/kg Can or Clo through oral gavage daily. Tumor volumes were measured every other day per week and body weights were measured twice a week. The study was performed in accordance with the guidelines and standards of the Institutional Animal Care and Use Committee (IACUC) and under the approved animal protocol # 19026 MD/R by Indiana University.
Kaplan–Meier plot analysis
The normalized gene expression profile and clinical data for PDAC and CRC datasets were downloaded from cBioPortal using the R package ‘cgdsr’. Patients were divided into two groups of high expression and low expression based on the median expression level. Kaplan–Meier method was used to estimate the overall survival (OS) curves of those two groups of patients, and the statistical difference between the survival curves was compared using the log-rank test. A Cox proportional-hazards model was fitted to estimate the hazard ratio between two groups. P value less than 0.05 was considered as statistical significance.
For BC data, PAM50 classification data were acquired from KM Plot source. Analysis excluded biased arrays, removed redundant samples, and checked for proportional hazard samples. Patients were divided into two groups of high expression and low expression based on the median expression level. Kaplan–Meier method was used to estimate the OS curves of those two groups of patients, and the statistical difference between the survival curves was compared using the log-rank test. A Cox proportional-hazards model was fitted to estimate the hazard ratio between two groups. P value less than 0.05 was considered as statistical significance.
Statistical analysis
Prism 6 software (GraphPad, La Jolla, CA) was used to perform the statistical analysis. Statistics for the Kaplan–Meier survival curves were calculated using log-rank test. A two-tailed Student's t-test was used to compare two means and check for significant differences. All statistics were done using data from triplicate experiments and P value < 0.05 was considered to be statistically significant. Results are presented as mean ± standard deviation (SD) or mean ± standard error of mean (SEM), as specified. Percent inhibition for compounds derived from the AlphaLISA HTS was calculated using the following formula: [({Avg. maximum reading – Inhibitor reading}/Avg. maximum reading) × 100]. Compusyn was used to conduct Chou-Talalay analysis including Bliss and HSA analysis (ComboSyn Inc, Paramus, NJ).
Results
PRMT5 is a negative prognostic factor in PDAC, CRC and BC patients
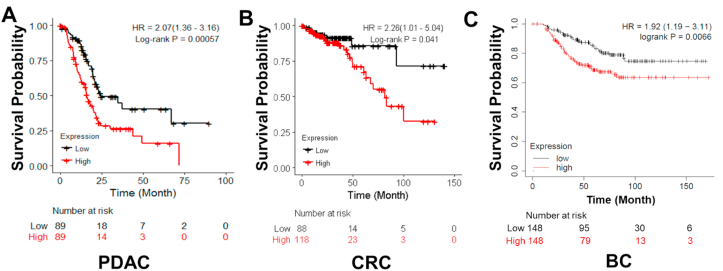
PRMT5 has been widely implicated as a tumor promoter in several cancers, including PDAC, CRC, and BC.6, 7, 8, 9 We recently found that PRMT5 can function as a critical promoter of the tumor phenotype, at least partly due to its increased expression, resulting methyltransferase activity, leading to activation of NF-κB pathway, and contributing to its constitutive signaling in PDAC and CRC 6. In this study, we conducted a retrospective analysis to check for the correlation between PRMT5 protein expression and patient survival in these cancers. Using TCGA open access databases, we constructed Kaplan–Meier plots for each of these cancers and stratified the patient datasets based on low and high PRMT5 expression in the tumor tissue. The resource of data and analysis method are fully described in the Method section. As shown in Figure 1, we clearly observe that high PRMT5 expression is associated with poor survival in PDAC, CRC, and BC patients, as compared to patients in which PRTM5 was lowly expressed. It is worth noting that in BC patients, PRMT5 overexpression is particularly strongly associated with poor OS in basal subtype (TNBC subtype). These observations, in combination with the previously documented role of PRMT5 as a critical tumor promoter, reiterate the potential therapeutic significance of inhibiting PRMT5 in cancers. This led towards our efforts to probe for FDA-approved drugs and market drugs in Europe and Japan that could selectively inhibit PRMT5 and have been described in the following sections, thereby having the capacity to move faster through the clinical trial process with potential candidate compounds.
Figure 1.
Kaplan–Meier plots in (A) PDAC, (B) CRC and (C) BC patient cohorts respectively, denoting that patient survival negatively correlated with PRMT5 expression in these groups of patients. Overall, PRMT5-high group had significant less overall survival (OS) than PRMT5-low group in all three cancer types. Time indicates the months after chemotherapy. Low vs. high: P = 0.00057 (PDAC); P = 0.041 (CRC); P = 0.0066. P-value less than 0.05 is considered to be statistically significant. The Kaplan–Meier plot analysis is fully described in the Method section.
Identification of top two marketed drugs with PRMT5-inhibitory activity in vitro using AlphaLISA-based HTS
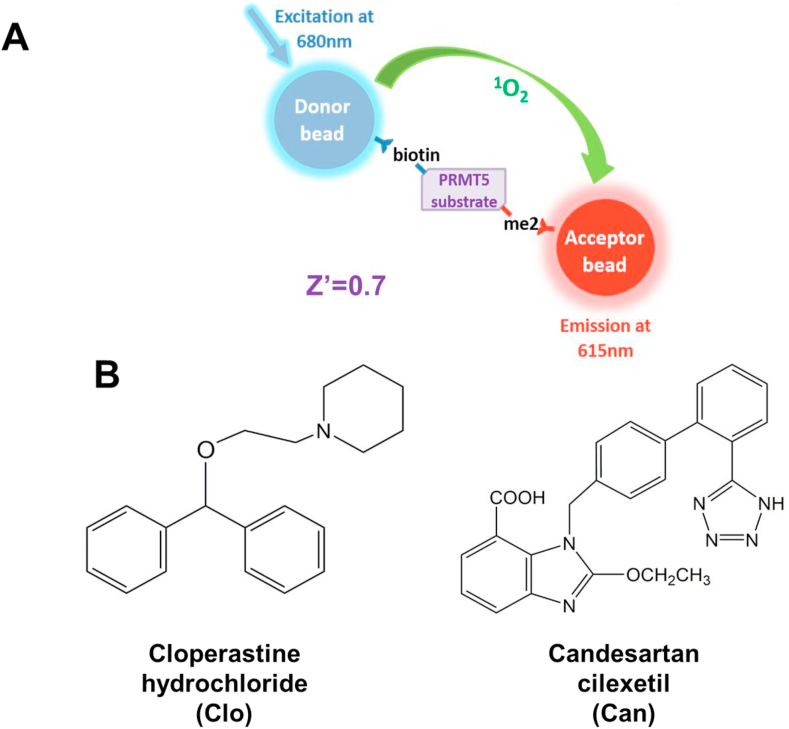
In order to design a drug screen to identify compounds that had the potential to inhibit PRMT5 methyltransferase activity, we utilized the AlphaLISA-based HTS assay. Optimization of the AlphaLISA protocol to be used has been published by our lab in great detail previously.10 The principle of this assay is illustrated in Figure 2A and utilizes a well-known PRMT5 substrate H4R3 that is biotinylated, the methyl group donor SAM, PRMT5 and Acceptor/Donor beads. Addition of PRMT5 and SAM enables symmetric dimethylation of biotin-H4R3 to form biotin-H4R3-me2. Acceptor beads with a specific antibody tag for dimethylated H4R3 recognize the symmetric dimethylated mark and bind to it. Donor beads have a streptavidin tag that allows for binding to biotinylated H4R3. Excitation at 680 nm emits a singlet oxygen from the Donor beads that is accepted by the Acceptor beads bound to the same peptide on the dimethyl mark, in turn causing an emission of light at 615 nm. This emitted AlphaLISA signal is directly proportional to the methyltransferase activity of PRMT5, thus allowing us to quantify its enzymatic activity.
Figure 2.
Development of AlphaLISA HTS screen to identify specific PRMT5 inhibitors. (A) Principle of AlphaLISA technique adapted to HTS. Biotinylated histone H4 (a well-known PRMT5 substrate) was mixed with methyl donor, S-adenosyl-l-methionine (SAM) and PRMT5 enzyme. Symmetric demethylation of third arginine (R3) of H4R3 by PRMT5 on biotin-H4 is recognized by Acceptor beads with an antibody tag specific for this methylation site. Furthermore, Donor beads have a streptavidin tag that bind to biotin tag on H4. Excitation at 680 nm induces Donor beads to emit a singlet oxygen, which is accepted by Acceptor beads bound to the dimethylated site to emit a chemiluminescent signal, thus allowing for quantifying PRMT5 methyltransferase activity. (B) Cloperastine hydrochloride (Clo) and Candesartan cilexetil (Can) were identified as two top FDA-approved hits from AlphaLISA HTS.
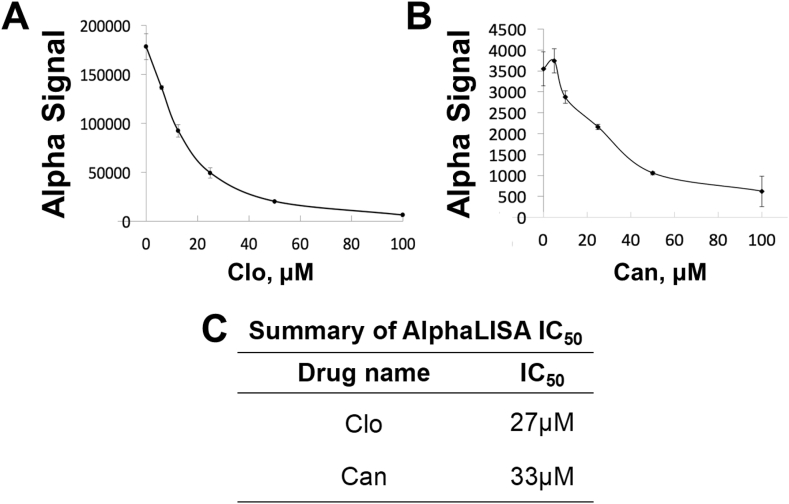
Using this PRMT5-specific AlphaLISA assay, we screened for chemical libraries containing FDA or EMA-approved compounds as described in the Methods section. Upon calculating percent inhibition from initial screening studies, we identified Clo and Can as the two top PRMT inhibitors (Fig. 2B). We further validated their ability to reduce the AlphaLISA signal by using concentration-dependent curves for these drugs. As expected, we observed a decrease in AlphaLISA signal with increasing concentrations of Clo (Fig. 3A) and Can (Fig. 3B) respectively. The respective IC50s are summarized in Figure 3C.
Figure 3.
Determination of IC50 using AlphaLISA assay, showing a concentration-dependent decrease in Alpha signal for (A) Clo and (B) Can. (C) Table summarizing AlphaLISA IC50 values for Clo and Can.
Clo is used for the treatment of cough.19 On the other hand, Can is an angiotensin II receptor blocker used in the treatment of hypertension.20 These hits were exciting to explore for alternative indications as these drugs are already approved by the FDA or EMA. This in turn would expedite the timeline for clinical trials if found to have promising therapeutic implications.
Effect of Clo and Can treatment on NF-κB activation and methylation
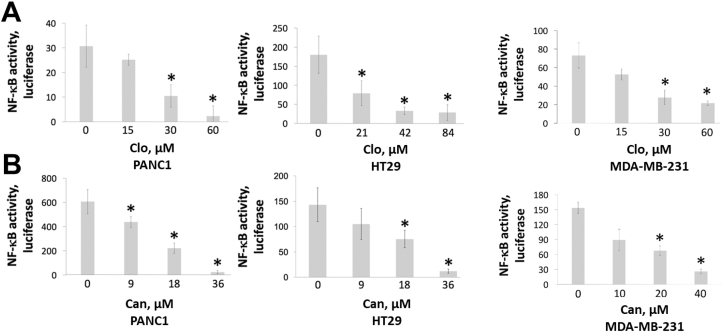
Previously, we presented that PRMT5 activates NF-κB through arginine methylation.6,7 We would like to further determine whether Clo and Can block NF-κB activity following inhibition of PRMT5. To achieve goal, we conducted NF-κB luciferase assays in three representative cell lines, PANC1, HT29, and MDA-MB-231. Overall, we noted that there was a concurrent decrease in the NF-κB activity, with increasing doses of both Clo (Fig. 4A) and Can (Fig. 4B).
Figure 4.
NF-κB luciferase assay, depicting reduction in NF-κB activation with increasing concentrations of (A) Clo and (B) Can, respectively in PDAC, CRC and BC cells. Data represent means ± S.D. for three independent experiments. ∗P < 0.05 vs. control group (0 μM).
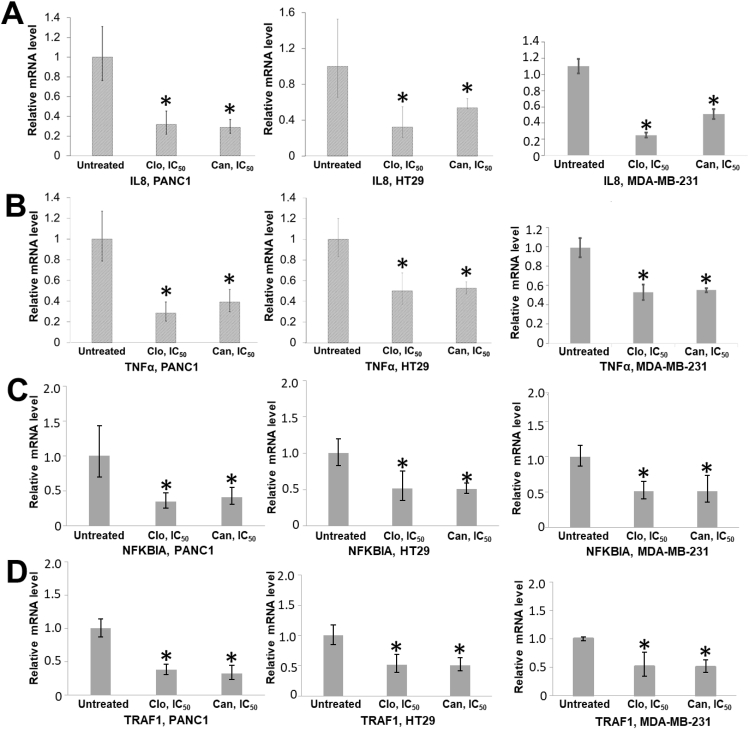
Furthermore, NF-κB activation is a critical step in its function as a transcription factor that leads to activation of genes implicated in promoting many hallmarks of cancer.21,22 Thus, we were curious to determine if decrease in NF-κB activation due to potential PRMT5 inhibition by Clo and Can (Fig. 5) affected its function as a transcription factor and correlated with decreased downstream target gene expression. Therefore, we treated PANC1, HT29 and MDA-MB-231 cells with Clo and Can respectively, followed by qRT-PCR to check for expression of well-known NF-κB target genes implicated in proinflammation and cancers, such as IL8, TNFα, NFKBIA, and TRAF1.7 Data demonstrated a significant decrease in mRNA expression of all genes in the drug-treated cancer cell lines, as compared to untreated control (Fig. 5).
Figure 5.
RT-qPCR assay, showing reduction in activation of NF-kB dependent genes, IL8 (A), TNFα (B), NFKBIA (C), and TRAF1 (D) in PANC1, HT29, and MDA-MB-231 cells, upon treatment with Clo and Can respectively. Data represent means ± S.D. for three independent experiments. ∗P < 0.05 vs. Untreated vehicle control group.
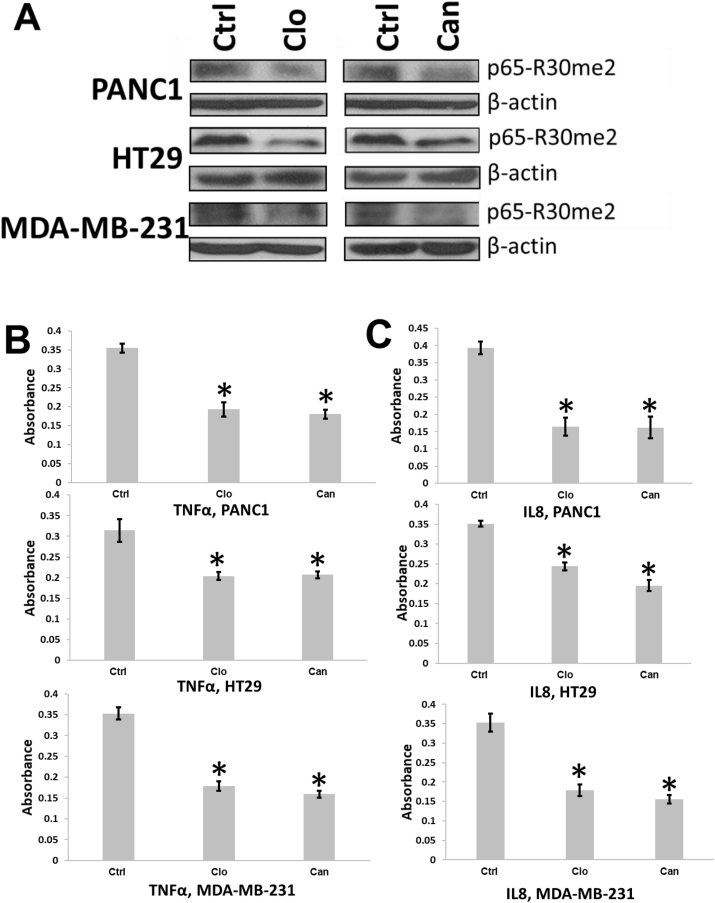
Furthermore, we expect that treatment with PRMT5 inhibitors such as Clo and Can would inhibit PRMT5-mediated methylation of NF-κB. We generated a specific antibody to detect the arginine (R) 30 dimethyl tag introduced by PRMT5 in collaboration with GenScript Inc. Using this antibody, we conducted Western blotting studies to detect effect of Clo and Can on R30 dimethylation in the panel of cancer cells. As shown in Figure 6, treatment with both drugs led to reduction in the dimethylation levels at R30 in PANC1, HT29 and MDA-MB-231 cells, as compared to untreated control. Also, both drugs reduced TNFα and IL8 expression as determined by ELISA analysis of the cell culture media (Fig. 6B, C). Furthermore, reduction in the nuclear fraction of p65-me2 was observed upon treatment with either Clo or Can (Fig. S1).
Figure 6.
Treatment of Clo or Can decreased p65me2 and reduced NF-κB target gene expression in cancer cells. (A) Western blots, showing that treatment of Clo and Can decreased symmetric demethylation of p65, a PRMT5 substrate, in PDAC, CRC and BC cells. (B, C) ELISA assays, showing that secretion of TNFα (B) or IL8 (C) was significantly reduced in the condition media from cancer cells treated with either Clo or Can.
Taken together, the above results confirm that Clo and Can inhibit PRMT5-mediated NF-κB methylation and activation, and thereby, reduce NF-κB target genes expression, such as IL8 and TNFα in multiple cancer cell lines.
Viability of cancer cell lines with Clo and Can treatment
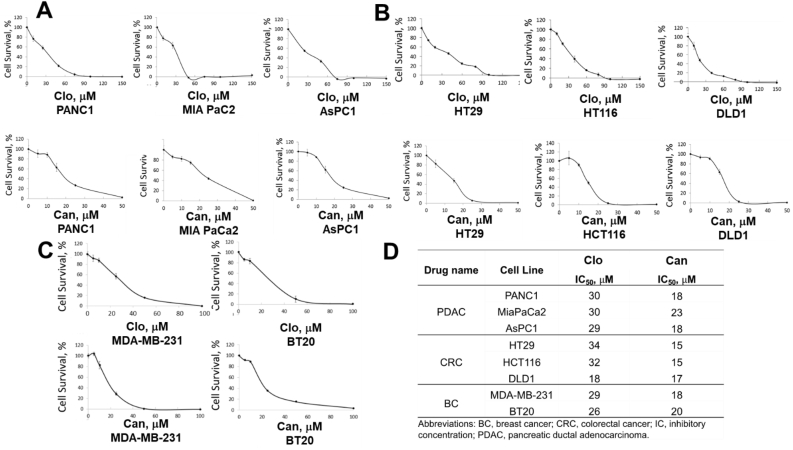
Since AlphaLISA protocol is a non-cell, plate-based assay, the effect of Clo and Can in cell-based in vitro assays was evaluated. Using the MTT assay, we determined the effect of these drugs on cell viability of additional well-known PDAC, CRC and TNBC cell lines. With increasing concentrations of Clo as well as Can, there was a concurrent decrease in cell viability of PDAC cells (PANC1, MiaPaCa2 and AsPC1) with an IC50 of ∼30 μM and ∼20 μM respectively for both these drugs (Fig. 7A, D). CRC cell lines (HT29, HCT116 and DLD1) respond similarly to Clo and Can with IC50s of 18–34 μM for Clo and 15–17 μM for Can (Fig. 7B, D). Similarly, IC50 values for TNBC cells (MDA-MB-231 and BT20) ranged in 26–29 μM and 18–20 μM for Clo and Can respectively (Fig. 7C, D). Additionally, to assess whether the cancer cell death upon Clo and Can treatment was due to apoptosis, we further determined the procaspase 3 and cleaved caspase 3 levels by Western Blotting. As shown in Figure S2, upon treatment with either Clo or Can, no significant procaspase 3 degradation nor production of cleaved caspase 3 was observed, suggesting that apoptosis is not the major cell death program that leads to cancer cell death by Clo or Can treatment. Overall, this study highlighted the efficacy of these drugs in inhibiting cancer cell viability of PDAC, CRC and BC cell lines that express high levels of PRMT5.
Figure 7.
MTT assay, showing that increasing concentrations of Clo and Can inhibited cell viability in (A) PDAC cells (PANC1, MiaPaCa2 and AsPC1), (B) CRC cells (HT29, HCT116, and DLD1) and (C) TNBC cells (MDA-MB-231 and BT20), respectively. (D) Summary table listing IC50 values from (A–C). Data represent means ± S.D. for three independent experiments.
To test whether knocking down PRMT5 with shRNA would desensitize the cancer cells to Clo or Can treatment, we used our previously established PANC1-shScramble and PANC1-shPRMT510 as the model. As shown in Figure S3A and B, Clo or Can had less cell killing effect on PANC1-shPRMT5 cells or normal PDAC control HPNE cells, suggesting that Clo and Can function through PRMT5 and cancer cells with high PRMT5 (such as PANC1-shScramble cells) are more sensitive to Clo and Can treatment as compared to low PRMT5 cancer cells (such as PANC1-shPRMT5 cells) or normal control cells. Interestingly, Wei et al reported that using targeted CRISPR screening they identified PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells.23 To test the possible combinatory effect of Clo or Can with gemcitabine on PDAC cells, we conducted Chou-Talalay experiments. As shown in Figure S3C, Clo or Can exhibited strong synthetical lethality with gemcitabine in three typical PDAC cell lines: PANC1, MIA PaCa2, and AsPC1. This finding further affirmed the potential application of Clo or Can in PDAC combinatory therapy in the future.
3D culture growth of cancer cell lines with Clo and Can treatment
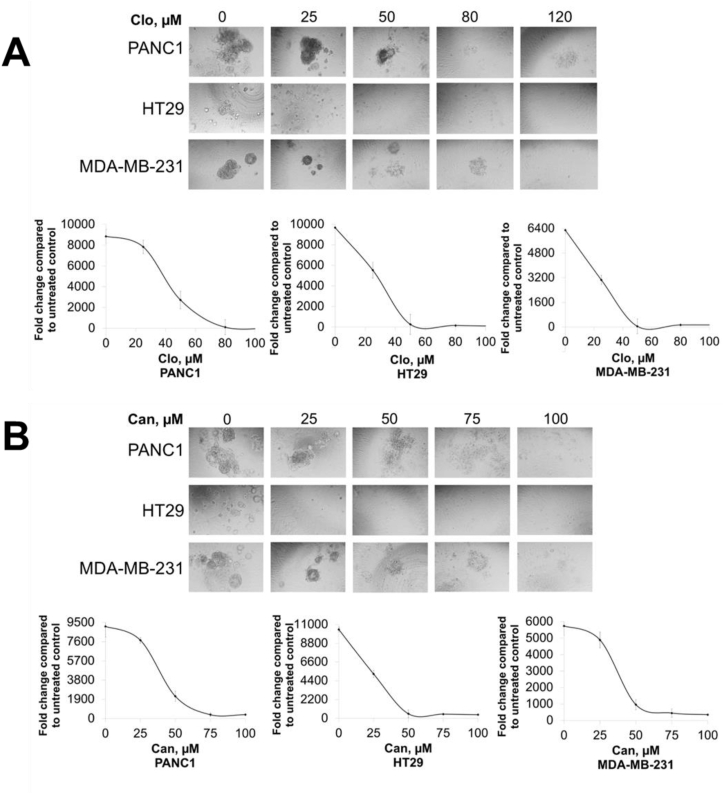
Recent validation studies with 3D spheroidal cultures demonstrate their ability to closely mimic critical characteristics of the in vivo tumor microenvironment.24,25 These properties contribute to the proliferation and metastatic potential of cancer cells. Hence, we tested if treatment with either Clo or Can had an inhibitory effect on the 3D colony growth in PDAC, CRC, and TNBC spheroids. We observed that increasing concentrations of both Clo (Fig. 8A) and Can (Fig. 8B) caused a marked decrease in the extent of 3D spheroid growth in representative PDAC, CRC and TNBC cell lines, PANC1, HT29 and MDA-MB231 respectively, highlighting the promising efficacy of both these drugs to limit the 3D culturing potential of these cells.
Figure 8.
3D spheroid assay signifying a substantial decrease in spheroid growth in PDAC, CRC, BC cells with increasing concentrations of (A) Clo and (B) Can, respectively. Representative pictures were captured under 10 x magnification. Data represent means ± S.D. for three independent experiments.
Predicted binding of Clo and Can to PRMT5
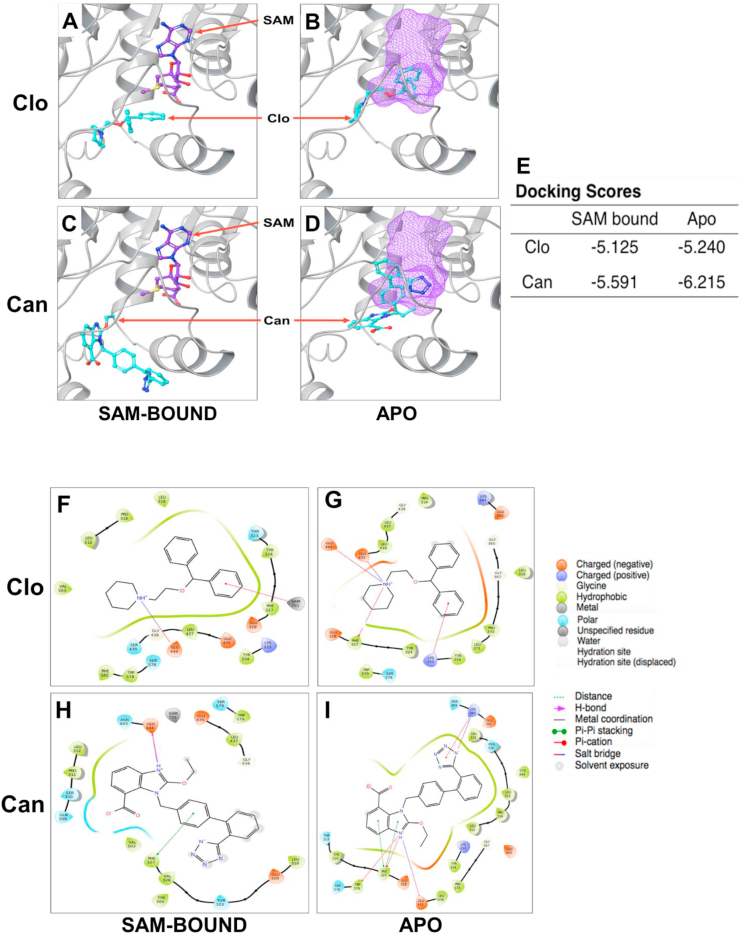
To predict the potential binding mechanism of Clo or Can with PRMT5, we employed in silico docking studies to identify residues on PRMT5 structure that are critical in binding interactions of Clo and Can with PRMT5. The best energetically favorable interactions for Clo in presence and absence of the methyl donor SAM have been depicted in Figure 9A and B. It is interesting to note here that Clo partially overlaps with the SAM binding site in the absence of SAM. This could be a possible mechanism by which Clo could interfere with SAM binding to PRMT5, thereby inhibiting its methyltransferase activity. If we consider binding scores for both these conditions, they are very close to each other and thus it is hard to predict which condition is more favorable (Fig. 9E). Similarly, for Can, we observed that the binding pose did not show much overlap in the area where SAM was bound to PRMT5. However, in the absence of SAM, Can showed some overlap, and could interfere through a similar mechanism as Clo proposed earlier (Fig. 9C, D). Binding affinity for the Apo form was higher than the SAM-bound condition, suggesting that the former might be a more energetically favorable and thus likely occurrence compared to the latter (Fig. 9E).
Figure 9.
Docking studies of Clo or Can with PRMT5, showing that (A) in SAM-bound condition, Clo bound to a site distinct from SAM on PRMT5, (B) in apo condition (in the absence of SAM), several binding residues for Clo with PRMT5 overlapped with SAM, suggesting that Clo could block PRMT5 activity by interfering with binding several similar residues as SAM. (C, D) Similar observations were observed for the Can-PRMT5 interaction in the presence and absence of SAM, respectively. (E) Table, listing the binding affinities of PRMT5 interactions in SAM bound and Apo-PRMT5 conditions with Clo and Can. Clo showed favorable binding energies under both conditions, Can showed more binding affinity in the Apo-PRMT5 condition (−6.215 kcal/mol). Ligand affinity map highlighting PRMT5 residues involved in the SAM-bound condition and apo condition for Clo (F, G) and Can (H, I) respectively. Key PRMT5 residues such as E444 (part of catalytic cleft) and F327 (critical for substrate binding) were shown to be important in both docking conditions for Clo and Can. Symbol of bonds and charges are described at the side of the figure. The location of SAM is depicted as a molecular surface in purple wireframe in (B, D).
We also plotted ligand affinity maps to better understand which PRMT5 residues potentially interact with the Clo (Fig. 9F, G) and Can (Fig. 9H, I) respectively, in presence and absence of SAM. Notably, two PRMT5 residues that stood out from these docking studies were E444 and F327. The E444 residue belongs to the catalytic cleft of PRMT5 and plays a key role in the methyltransferase activity of the enzyme. On the other hand, the F327 residue plays a crucial role for PRMT5 product specificity.26 Overall, these ligand affinity maps hint towards residues that play a prime role in the enzyme–drug interactions and serve as a basis for designing future studies to validate these findings in vitro.
Effect of Clo and Can treatment on tumor growth
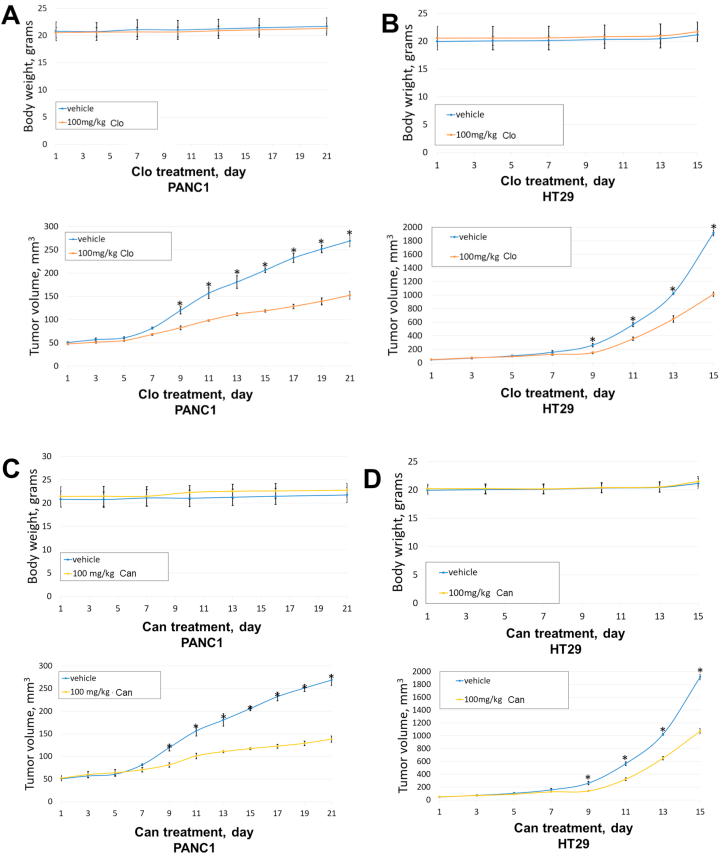
Finally, due to the anti-cancer activity Clo and Can in monolayer and in 3D in vitro, the efficacy of Clo and Can was evaluated in vivo for tumor growth inhibition. Either PANC1 or HT29 cells were subcutaneously implanted into the flanks of NSG mice and then treated with 100 mg/kg Can or Clo, or vehicle daily for the duration of the experiment. Both body weight and tumor size were monitored during this process. We show that treatment with Clo (Fig. 10A, B) and Can (Fig. 10C, D) did not significantly affect the body weight of the mice indicating that the treatment was well-tolerated. With both FDA or EMA approved agents at least 50%–60% reduction in tumor volume was observed (Fig. 10), suggesting strong anti-tumor efficacy as a single agent in vivo.
Figure 10.
Tumor inhibition effect of Clo or Can in xenograft mouse model in vivo. NSG mice were xenografted with mycoplasma-free cancer cells subcutaneously (1 × 107 PANC1, 3 × 106 HT29). When tumor volumes reached ∼50 mm3 mice were treated with either vehicle control (10% DMSO, 20% PEG400, 5% Tween 80 and 65% sterile water), or 100 mg/kg Clo or Can through oral gavage daily. Tumor volumes were measured daily following the first week and body weights were measured twice a week. ∗P < 0.05 vehicle vs. 100 mg/kg Clo, ∗P < 0.05 vehicle vs. 100 mg/kg Can.
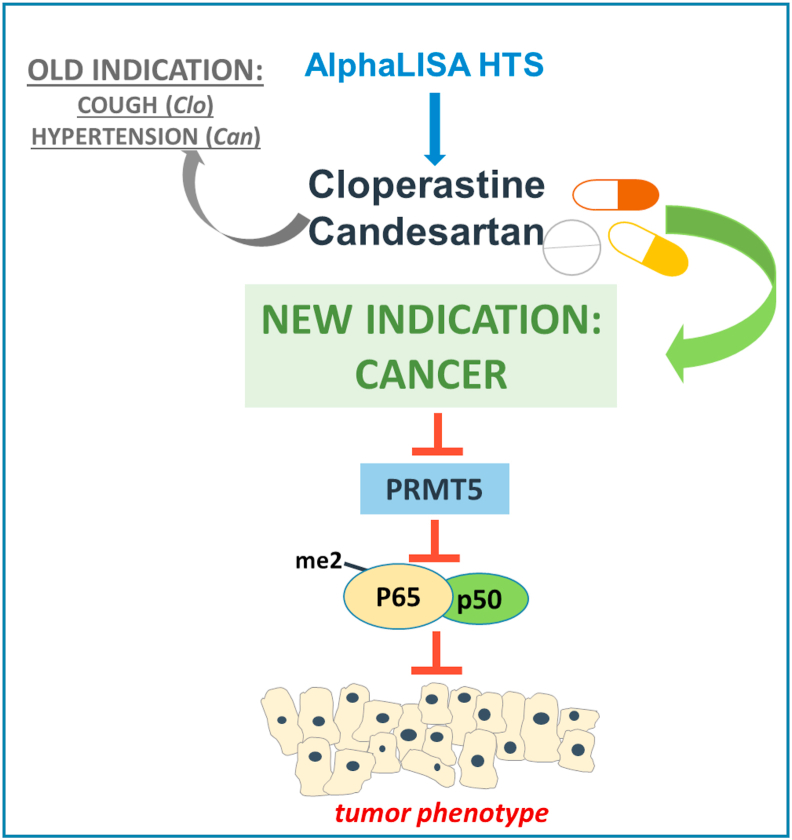
Hypothetical model
As illustrated in Figure 11, in this study, we successfully identified one FDA-approved drug and one EMA-approved drug, Can and Clo as potential inhibitors of PRMT5, a bona fide drug target in several kinds of cancer. Both drugs offer potential for use in new indications via their ability to inhibit PRMT5 in PDAC, CRC and BC cells and in turn, inhibit the activation of its critical substrate, NF-κB, a known tumor promoting factor. This effect may eventually lead to a decrease in the tumor phenotype, thereby highlighting the potential of Clo and Can, the cough suppressant and anti-hypertension drugs, respectively, to be repurposed for new indications in cancer patients.
Figure 11.
Hypothetical model. We illustrate the experimental approach of employing AlphaLISA-based HTS to identify two FDA-approved drugs, Clo and Can. Both drugs offer potential for use in new indications via their ability to inhibit PRMT5-mediated NF-κB activation, therefore, the expression of its downstream genes in PDAC, CRC and BC. This effect may eventually lead to an inhibition of tumor phenotype, thereby highlighting the potential of Clo and Can to be repurposed for new indications in cancer patients.
Discussion
In the past decades, the cost of drug development process has remained sky-high. From early discovery to the time a drug is approved for use in the clinic, it costs about a billion dollars and takes a total of 12–15 years. This is quite astounding as diseases such as cancer continue to rise at a rapid pace with each passing year. It is estimated that cancer will overtake heart disease to become the number one cause of disease-related deaths in US in the upcoming decade. The cost of treatment also continues to rise to counterbalance billions of dollars invested in drug development, thereby increasing the brunt on cancer patients. It is imperative to devise cost and time-effective strategies that help to find better treatment options for this deadly disease.
In this study, we have employed a strategy of identifying drugs already approved for certain indications by the FDA or have been used as common therapeutics elsewhere globally, such as Europe and Japan, and repurposing them to be used in three of the deadliest cancers that currently exist: PDAC, CRC and BC. This strategy of repositioning approved drugs has been successfully used in the past to accelerate identification of new indications. Specifically, in the cancer field, a successful example of drug repurposing is Everolimus, which was originally identified as an immunosuppressant and later repurposed to be used in a rare form of advanced pancreatic neuroendocrine tumors.27 This and other examples fueled our efforts to identify FDA-approved drugs or EMA-approved drugs to be repurposed for cancer by inhibiting PRMT5 activity.
PRMT5 has emerged as one of the most pursued targets in recent times. Indeed, a slew of latest drug discovery efforts by our lab and others have identified small molecule inhibitors against PRMT5.7,28 Indeed, our compound PR5-LL-CM01 has proven effective in reducing PRMT5 activity in cancers compared to normal cells shown via MTT.10 However, it will be at least many years before these small molecules come close to clinical use as they have to go through the drug approval process. Repurposing FDA-approved drugs instead seems like a lucrative option in comparison to this method. Furthermore, the wide interest in PRMT5 as a therapeutic target can be attributed to numerous studies identifying its contributions in promoting the cancer phenotype, including PDAC, CRC, BC, etc.
In this study, we highlight two market drugs, Clo and Can identified using AlphaLISA PRMT5-specific HTS technique that is developed in our lab. Clo (brand names Hustazol, Nitossil, Seki), is an antitussive and antihistamine that is marketed as a cough suppressant in Japan, Hong Kong, and in some European countries. It was first introduced in 1972 in Japan, and then in Italy in 1981. The precise mechanism of action of Clo functioning as a cough suppressant is not fully clear, but several different biological activities have been identified for the drug, For example, ligand of the σ1 receptor,29 GIRK channel blocker30 which may be involved in or responsible for the antitussive efficacy of Clo. In contrast, the FDA drug we identified, Can (Brand name: Atacand; Genetic name: Candesartan), is a quite different drug. Can is used to treat high blood pressure (hypertension). Lowering high blood pressure helps prevent strokes, heart attacks, and kidney problems. Can belongs to a class of drugs called angiotensin receptor blockers (ARBs). It works by relaxing blood vessels so blood can flow more easily. This medication is also used to treat heart failure.
In this study, we found both Clo and Can not only reduced cell proliferation, 3D colony growth, and tumor growth in PDAC, CRC and BC, but also correlated at least partly via the reduction of NF-κB activation, leading to decreased expression of typical NF-κB target genes, like TNFα and IL8. Overall, Can's IC50s are ∼20 μM for PDAC, ∼15 μM for CRC, and ∼18 μM for BC (Fig. 7) which is slightly more potent than Clo, whose IC50s are ∼30 μM in PDAC, ∼18–34 μM in CRC, and ∼26 μM in BC (Fig. 7). Furthermore, in our in silico study, we suggest that E444 and F327 are the two sites that are crucial to the binding of Clo or Can to PRMT5. E444 locates at the active site of PRMT5, and is known to play a key role in PRMT5's methyltransferase activity, while F327 is essential for PRMT5 product specificity.25 These data, together with our promising in vivo tumor inhibition result (Fig. 10), bring forth the promise of repositioning these FDA approved drugs as a treatment option in PDAC, CRC and BC, by inhibiting PRMT5 (Fig. 11). Additionally, to address the potential for Can or Clo inhibiting NF-κB activity through means other than PRMT5 inhibition we utilized PANC1 PRMT5 knockdown cells generated previously10 and performed MTTs with or without Can and Clo in comparison to HPNE control cells and PANC1 wild-type cells. As shown in Figure S3, IC50s of Can and Clo had increased cell survival in normal and shPRMT5 cells. We acknowledge that shPRMT5 stable cells showed slightly less robust insensitivity to Clo or Can than HPNE normal control cells (Fig. S2B). This could be due to partial (∼70–80%) instead of 100% PRMT5 knockdown in PANC1-shPRMT5 stable cells (Fig. S2A). In the future, a CRISPR-mediated PRMT5 KO cell line might generate more comparable Clo or Can insensitivity to that of normal HPNE control cells.
Certainly, amidst these promising results, one of the next logic steps would be to verify the potential of these drugs as PRMT5 inhibitors toward the path of clinical trials in cancer patients. Furthermore, in addition to using Clo or Can as single agents, combination studies of current standard-of-care regiments can be used to determine the potential of enhancing the current standard of care. If the results are promising, we hope to eventually move these drugs to late phase clinical trials to understand their real-time potential with the ultimate goal of FDA approval to repurpose these drugs to treat cancer patients, and perhaps other PRMT5-related diseases as well. In regard to this avenue, the recent study by Wei et al indicated CRISPR/cas9 knockdown of PRMT5 sensitized PDAC cells to gemcitabine-based DNA damage.23 Our pilot test of Clo or Can in combination with gemcitabine in three PDAC cell lines by Chou-Talalay experiment and HSA analysis showed overall strong synergistic lethality (Fig. S3C), suggesting the promising potential in the future to pursue Can and Clo not just as single agents but combinatorial therapies.
Conflict of interests
The authors declare conflicting financial interest; Alternative use of Cloperastine and Candesartan in cancer has been filed for provisional patent on 12/31/2020 by TL, LP, and MM. TL is the founder of EQon Pharmaceuticals, LLC.
Funding
This work was supported by grants from Indiana Center for Technology and Science Innovation (CTSI), USA (No. 2286230 to TL) and Indiana Drug Discovery Alliance (IDDA), USA (No. 2286233 to TL), both are funded in part by National Institutes of Health, USA (No. UL1TR002529); National Institutes of Health, USA (No. 1R01GM120156-01A1 to TL; No. R03 CA223906-01 to TL). This work was also supported by National Institutes of Health, USA (No. P41-GM103426 and DP2OD007237 to REA), National Science Foundation, USA (No. CHE060073N to REA), and National Institutes of Health, USA (No. R01 CA069202 to ZYZ). MLF and MRK were supported by IUSCCC Cancer Center, USA (No. P30 CA082709), National Institutes of Health, USA (No. R01CA167291 and R01CA254110). MRK was also supported by National Institutes of Health, USA (No. R01CA205166, R01CA231267, and R01HL140961). MLF was also supported by National Institutes of Health, USA (No. R01CA211098, U01HL143403, and NF180045). MLF and MRK were additionally supported by the Riley Children's Foundation, USA.
Acknowledgements
We would like to thank Dr. Britney Shea-Herbert for her generous gift for the breast cancer cell lines.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.04.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Talevi A. Drug repositioning: current approaches and their implications in the precision medicine era. Expert Rev Precis Med Drug Dev. 2018;3(1):49–61. [Google Scholar]
- 2.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 3.Pantziarka P., Bouche G., Meheus L., et al. The repurposing drugs in oncology (ReDO) project. Ecancermedicalscience. 2014;8:442. doi: 10.3332/ecancer.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA - Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.Collignon J., Lousberg L., Schroeder H., et al. Triple-negative breast cancer: treatment challenges and solutions. Breast Cancer (Dove Med Press) 2016;8:93–107. doi: 10.2147/BCTT.S69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei H., Wang B., Miyagi M., et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc Natl Acad Sci U S A. 2013;110(33):13516–13521. doi: 10.1073/pnas.1311784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prabhu L., Chen L., Wei H., et al. Development of an AlphaLISA high throughput technique to screen for small molecule inhibitors targeting protein arginine methyltransferases. Mol Biosyst. 2017;13(12):2509–2520. doi: 10.1039/c7mb00391a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang K., Zielinska A.E., Shaaban A.M., et al. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21(12):3498–3513. doi: 10.1016/j.celrep.2017.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B., Dong S., Zhu R., et al. Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3. Oncotarget. 2015;6(26):22799–22811. doi: 10.18632/oncotarget.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prabhu L., Wei H., Chen L., et al. Adapting AlphaLISA high throughput screen to discover a novel small-molecule inhibitor targeting protein arginine methyltransferase 5 in pancreatic and colorectal cancers. Oncotarget. 2017;8(25):39963–39977. doi: 10.18632/oncotarget.18102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin M., Mundade R., Hartley A.V., et al. Using VBIM technique to discover ARMC4/ODAD2 as a novel negative regulator of NF-κB and a new tumor suppressor in colorectal cancer. Int J Mol Sci. 2022;23(5):2732. doi: 10.3390/ijms23052732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arpin C.C., Mac S., Jiang Y., et al. Applying small molecule signal transducer and activator of transcription-3 (STAT3) protein inhibitors as pancreatic cancer therapeutics. Mol Cancer Therapeut. 2016;15(5):794–805. doi: 10.1158/1535-7163.MCT-15-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley A.V., Wang B., Jiang G., et al. Regulation of a PRMT5/NF-κB axis by phosphorylation of PRMT5 at serine 15 in colorectal cancer. Int J Mol Sci. 2020;21(10):3684. doi: 10.3390/ijms21103684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dermawan J.K.T., Gurova K., Pink J., et al. Quinacrine overcomes resistance to erlotinib by inhibiting FACT, NF-κB, and cell-cycle progression in non-small cell lung cancer. Mol Cancer Therapeut. 2014;13(9):2203–2214. doi: 10.1158/1535-7163.MCT-14-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halgren T.A., Murphy R.B., Friesner R.A., et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 16.Friesner R.A., Banks J.L., Murphy R.B., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 17.Small-Molecule Drug Discovery Suite 2017-3: Glide. Schrödinger, LLC; New York, NY: 2017. version 7.6. [Google Scholar]
- 18.Schrödinger Release 2017-3: Maestro. Schrödinger, LLC; New York, NY: 2017. version 11.3. [Google Scholar]
- 19.Catania M.A., Cuzzocrea S. Pharmacological and clinical overview of cloperastine in treatment of cough. Therapeut Clin Risk Manag. 2011;7:83–92. doi: 10.2147/TCRM.S16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClellan K.J., Goa K.L. Candesartan cilexetil. Drugs. 1998;56(5):847–869. doi: 10.2165/00003495-199856050-00013. [DOI] [PubMed] [Google Scholar]
- 21.Baud V., Karin M. Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8(1):33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naugler W.E., Karin M. NF- kappaB and cancer—identifying targets and mechanisms. Curr Opin Genet Dev. 2008;18(1):19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei X., Yang J., Adair S.J., et al. Targeted CRISPR screening identifies PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells. Proc Natl Acad Sci U S A. 2020;117(45):28068–28079. doi: 10.1073/pnas.2009899117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fennema E., Rivron N., Rouwkema J., et al. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013;31(2):108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Heylman C., Sobrino A., Shirure V.S., et al. A strategy for integrating essential three-dimensional microphysiological systems of human organs for realistic anticancer drug screening. Exp Biol Med (Maywood) 2014;239(9):1240–1254. doi: 10.1177/1535370214525295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonysamy S., Bonday Z., Campbell R.M., et al. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci U S A. 2012;109(44):17960–17965. doi: 10.1073/pnas.1209814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu E., Marincola P., Oberg K. Everolimus in the treatment of patients with advanced pancreatic neuroendocrine tumors: latest findings and interpretations. Therap Adv Gastroenterol. 2013;6(5):412–419. doi: 10.1177/1756283X13496970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan-Penebre E., Kuplast K.G., Majer C.R., et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 29.Gregori-Puigjané E., Setola V., Hert J., et al. Identifying mechanism-of-action targets for drugs and probes. Proc Natl Acad Sci U S A. 2012;109(28):11178–11183. doi: 10.1073/pnas.1204524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung K.F., Widdicombe J. vol. 187. Springer Science & Business Media; 2008. (Pharmacology and Therapeutics of Cough). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.