Abstract
A well-established role of cyclic GMP-AMP synthase (cGAS) is the recognition of cytosolic DNA, which is linked to the activation of host defense programs against pathogens via stimulator of interferon genes (STING)-dependent innate immune response. Recent advance has also revealed that cGAS may be involved in several noninfectious contexts by localizing to subcellular compartments other than the cytosol. However, the subcellular localization and function of cGAS in different biological conditions is unclear; in particular, its role in cancer progression remains poorly understood. Here we show that cGAS is localized to mitochondria and protects hepatocellular carcinoma cells from ferroptosis in vitro and in vivo. cGAS anchors to the outer mitochondrial membrane where it associates with dynamin-related protein 1 (DRP1) to facilitate its oligomerization. In the absence of cGAS or DRP1 oligomerization, mitochondrial ROS accumulation and ferroptosis increase, inhibiting tumor growth. Collectively, this previously unrecognized role for cGAS in orchestrating mitochondrial function and cancer progression suggests that cGAS interactions in mitochondria can serve as potential targets for new cancer interventions.
Subject terms: Cancer, Cell death
Introduction
Cyclic GMP-AMP synthase (cGAS) was originally discovered as a double-stranded DNA sensor that is a crucial component of innate immune defense against pathogens.1 Upon recognizing and binding to cytosolic microbial DNA, cGAS produces 2′3′-cyclic GMP-AMP (cGAMP), a messenger molecule that activates the protein adaptor stimulator of interferon genes (STING), consequently triggering innate immune response.2–5 Recent studies have extended the scope of known cGAS functions to including roles in cellular senescence, Huntington disease, and cancer autoimmune and inflammatory diseases through sensing native DNA.6–11 In addition, emerging evidence indicates that cGAS serves as a crucial factor in cancer progression,12,13 but its exact functions and mechanisms in the progression of different cancers remain unclear.
Cancer cells exhibit genomic instability and chromosomal abnormalities that commonly result in formation of cytosolic chromatin fragments and micronuclei. As a cytosolic DNA sensor, cGAS initiates antitumor immunity in both cell-autonomous and -nonautonomous manners by detecting tumor-derived DNA and activating the STING pathway.12 For example, cGAS association with micronuclei that arise from genome instability has been found to serve as a cell-intrinsic immune surveillance mechanism that can detect a range of neoplasia-inducing processes.14,15 Under stress states such as hypoxia, oxidative stress, and nutrient deprivation in cancer cells, chromatin fragments may be packaged into exosomes and subsequently activate cGAS-STING signaling in host antigen-presenting cells, which is important for the recruitment and expansion of tumor-infiltrating T cells.16 Therefore, some cancer cells can silence cGAS or STING to escape immune surveillance.
Recent work has revealed non-canonical functions of cGAS in subcellular compartments other than the cytosol.17–20 For instance, nuclear cGAS anchors to chromatin and suppresses homologous recombination-mediated DNA repair in response to genotoxic stress-induced DNA damage.19,21 These discoveries of the diverse functions of cGAS in cancer progression, dependent or independent of STING-mediated immune surveillance, establish cGAS as a major factor affecting cancer development. It is known that functional mitochondria support cancer cells by supplying energy and building blocks, and controlling redox homeostasis, oncogenic signaling and programmed cell death.22–26 cGAS has been shown to detect mitochondrial DNA (mtDNA) that leak from damaged mitochondria in the cytosol; however, the interaction between cGAS and mitochondria in cancer cells is still unclear.
Based on our previous exploration of cancer metabolism, mitochondrial function and cell death,22,27,28 we uncover here an unappreciated role of cGAS in mitochondria of hepatocellular carcinoma (HCC) cells and show that cGAS is localized to the outer mitochondrial membrane (OMM) to promote cancer progression. Using proteomics analysis and a series of knockdown and overexpression lines of HCC cells and xenograft mouse models, we find that cGAS is translocated into mitochondria via TOM70 where it interacts with dynamin-related protein 1 (DRP1) to facilitate its oligomerization. This interaction results in suppression of mitochondrial ROS accumulation and ferroptosis in cancer cells both in vitro and in vivo. These findings thus expand our understanding of cGAS function in cancer progression and provide a framework to guide future development of cancer treatments targeting its interactions in mitochondria.
Results
cGAS is localized in mitochondria of cancer cells
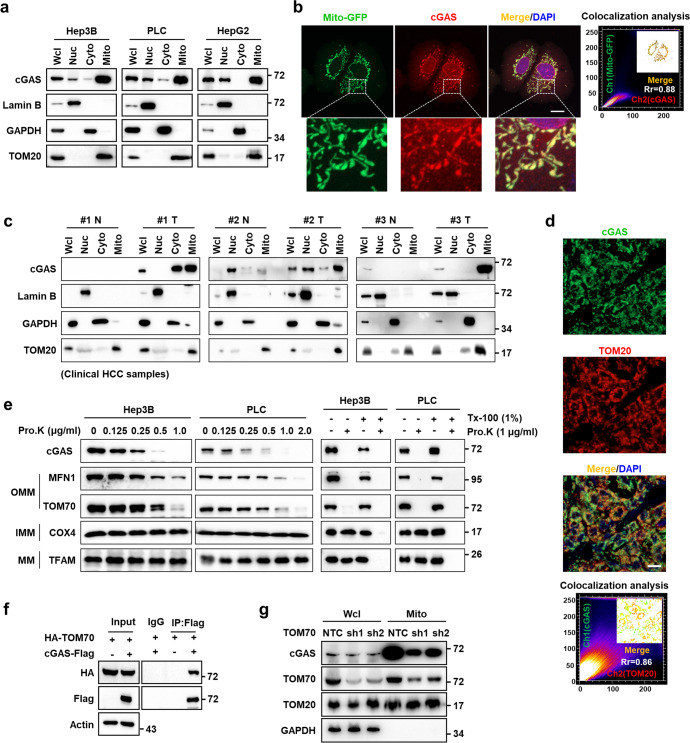
Due to the pivotal role of mitochondria in cancer development, we sought to identify mitochondrially translocated proteins with previously unrecognized contributions to HCC, using LC-MS/MS-based proteomics analysis. We found that cGAS, a known cytosolic and nuclear protein, was present in the mitochondrial fraction of Hep3B cells (Supplementary information, Fig. S1a). Western blotting analysis of the nuclear, cytosolic, and mitochondrial subcellular fractions further confirmed that cGAS was enriched in the mitochondria of Hep3B, PLC, and HepG2 cells (Fig. 1a). Confocal fluorescence microscopy with antibody against cGAS or mCherry-tagged cGAS also showed that cGAS was localized to mitochondria in Hep3B and PLC cells (Fig. 1b; Supplementary information, Fig. S1b–d), further supporting our LC-MS/MS data. We also observed the mitochondrial localization of cGAS in some other cancer cell lines, including A549, HT1080, PANC-1, MDA-MB-231 and U87 cells (Supplementary information, Fig. S1e). We then investigated the intracellular distribution of cGAS in clinical HCC tissues using both western blot and confocal microscopy and found that endogenous cGAS was distributed in mitochondria in clinical HCC samples (Fig. 1c, d). Similar results were observed in tumor tissues of a YAP-5SA-induced HCC mouse model (Supplementary information, Fig. S1f). Compared to adjacent healthy tissues obtained from clinical HCC patients, the expression of cGAS was significantly increased in tumor tissues (Supplementary information, Fig. S2a, b). Similar results were observed in YAP-5SA-induced mouse HCC samples (Supplementary information, Fig. S1f). Immunohistochemistry (IHC) analysis also showed a markedly increased expression of cGAS in HCC tissues compared with adjacent noncancerous tissues (Supplementary information, Fig. S2c). Importantly, high expression of cGAS predicted poor survival of HCC patients (Supplementary information, Fig. S2d), suggesting that mitochondrial cGAS likely contributes to cancer progression.
Fig. 1. cGAS is localized in mitochondria of cancer cells.
a Western blotting analysis of the protein levels of cGAS in subcellular fractions including nucleus, cytosol and mitochondria in HCC cell lines. Fractionation fidelity was verified by detection of lamin B in the nuclear fraction, GAPDH in the cytosolic fraction and TOM20 in the mitochondrial fraction. b Representative images of immunofluorescence staining for cGAS in Hep3B cells with Mito-GFP expression. The nucleus was stained with DAPI. Colocalization analysis of immunofluorescence images using the colocalization plugin, which calculates Pearson’s correlation coefficient. Scale bar, 20 μm. c Western blotting analysis of the protein levels of cGAS in subcellular fractions including nucleus, cytosol and mitochondria in paired adjacent noncancerous tissues and clinical HCC tissues. Fractionation fidelity was verified by detection of lamin B in the nuclear fraction, GAPDH in the cytosolic fraction and TOM20 in the mitochondrial fraction. d Representative images of immunofluorescence staining for cGAS and TOM20 in clinical HCC tissues. The nucleus was stained with DAPI. Colocalization analysis of immunofluorescence images using the colocalization plugin, which calculates Pearson’s correlation coefficient. Scale bar, 100 μm. e Proteinase K protection assay of cGAS in the mitochondria of Hep3B or PLC cells. Left: western blotting analysis of cGAS after incubation of purified mitochondria with the indicated concentration of proteinase K. Right: proteinase K protection assays were performed in the presence of the permeabilizing agent Triton X-100. Extent of digestion was determined by blotting for key intra-mitochondrial proteins (MFN1, TOM70, COX4, TFAM). OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane; MM, mitochondrial matrix. f Co-IP assay showing the protein interaction between cGAS and TOM70. Hep3B cells were infected with lentivirus carrying Flag-EV or cGAS-Flag and HA-TOM70 plasmids. Cell lysates were immunoprecipitated with an anti-Flag antibody, followed by western blotting analysis with antibodies against Flag and HA tags. g Western blotting analysis of the protein levels of cGAS in mitochondria of Hep3B cells with TOM70 knockdown. TOM20 and GAPDH served as the loading control.
Protein sequence analysis showed that cGAS carries two putative mitochondrial targeting sequence (MTS) from residues 161 to 200 and from residues 331 to 354, respectively, which are enriched with positively charged residues followed by hydrophobic residues residing near amphipathic helix (Supplementary information, Fig. S3a). To test their functionality, these two amino acid peptides (161–200 and 331–354 in full-length cGAS) were fused with GFP, respectively, and the results showed that only the fusion of cGAS 161–200 targeted GFP to the mitochondria in Hep3B cells (Supplementary information, Fig. S3a). In agreement with our data, residues 161–190 in cGAS functioning as MTS was also verified in THP-1 cells.29 Mitochondrial distribution of a cGAS variant that lacked the predicted MTS (161–190 in cGAS, named as ΔMTS-cGAS) was suppressed in Hep3B cells, suggesting that the predicted MTS was required for cGAS localization to mitochondria (Supplementary information, Fig. S3b–d).
To determine the sub-mitochondrial localization of cGAS, we isolated mitochondria from Hep3B and PLC cells for proteinase K protection assays. We observed that mitochondrial cGAS was sensitive to proteinase K digestion, indicating that cGAS was localized to the OMM (Fig. 1e). Immunoprecipitation (IP) with antibodies targeting Flag-tagged cGAS in Hep3B cells followed by MS analysis showed that TOM70 was associated with cGAS (Supplementary information, Table S1), which was confirmed by co-immunoprecipitation (co-IP) assays in Hep3B cells overexpressing both HA-tagged TOM70 and Flag-tagged cGAS (Fig. 1f). TOM70 is a major preprotein receptor of translocase on the OMM, and suppression of TOM70 by small hairpin RNAs (shRNAs) in Hep3B cells resulted in decreased enrichment of cGAS in mitochondria (Fig. 1g), indicating that TOM70 participated in cGAS translocation into mitochondria. However, our data showed that TOM20, another preprotein receptor that specifically recognizes the hydrophobic surface of the amphipathic helix, was not associated with cGAS (Supplementary information, Fig. S3e). Taken together, these results showed that cGAS was translocated to the OMM in cancer cells.
Mitochondrial cGAS inhibits ferroptosis
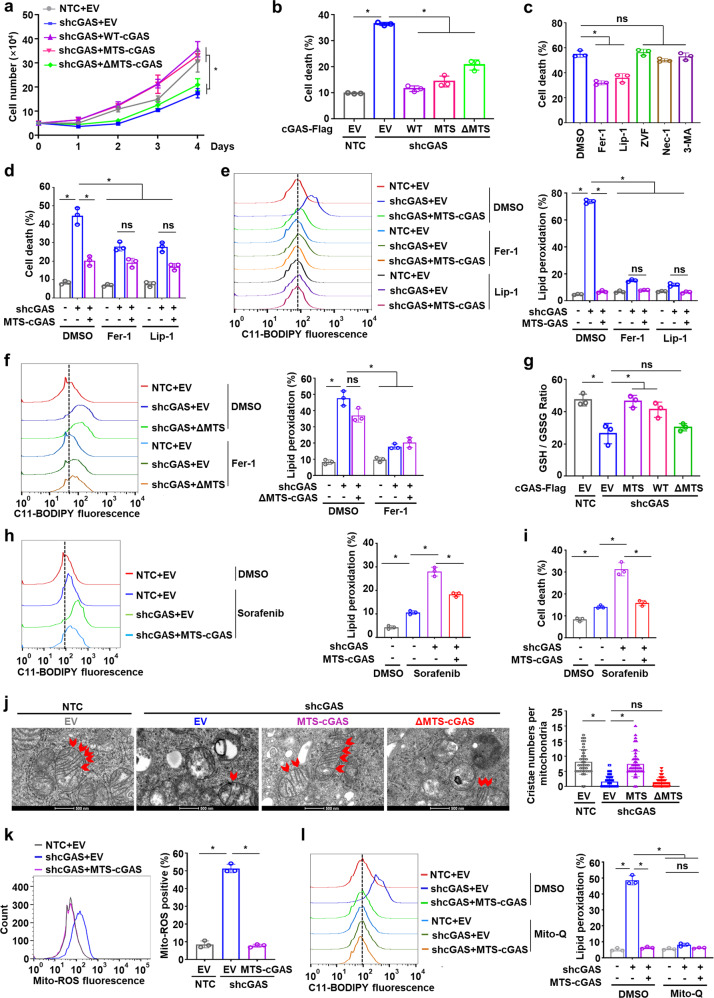
To better understand the effects of mitochondrial cGAS on HCC cell survival and growth, we knocked down cGAS (shcGAS) or knocked out cGAS (cGASKO) in Hep3B and PLC cells. We found that the cell growth was dramatically suppressed and the cell death was significantly increased by depletion of cGAS compared to the non-targeting controls (NTCs) or wild-type cells (Supplementary information, Fig. S4a–d). To explore the possible roles of mitochondria-localized cGAS in cancer cells, we generated MTS-fused cGAS (MTS-cGAS) which targeted cGAS specifically to the OMM (Supplementary information, Fig. S4e–g). Notably, we found that expression of MTS-cGAS and wild-type cGAS (WT-cGAS), but not ΔMTS-cGAS, markedly recovered the retarded cell growth and attenuated cell death in cells with cGAS knockdown (shcGAS targeting the 3′UTR of cGAS transcripts) (Fig. 2a, b). Mitochondria are the central initiators of the intrinsic apoptotic pathway and also participate in regulating other forms of cell death.24 To determine which mode of cell death is affected by mitochondrial cGAS, we examined the effects of inhibitors targeting different cell death pathways and found that the ferroptosis inhibitors ferrostatin-1 (Fer-1) and liproxstatin-1 (Lip-1) largely attenuated the cell death in Hep3B cells with endogenous cGAS knockdown and ΔMTS-cGAS expression (Fig. 2c). Moreover, MTS-cGAS expression remarkably reduced the cell death in cGAS-knockdown cells to a similar degree to Fer-1 and Lip-1 treatment (Fig. 2d). Consistent with these findings, expression of MTS-cGAS and WT-cGAS, but not ΔMTS-cGAS, attenuated the increase in cellular lipid peroxidation induced by shcGAS (Fig. 2e, f; Supplementary information, Fig. S4h, i). Glutathione (GSH) acts as a necessary cofactor for the normal function of the antioxidant enzyme GPX4 that reduces lipid hydroperoxides to lipid alcohols, through which GSH is oxidized to GSSG. We observed that expression of MTS-cGAS and WT-cGAS, but not ΔMTS-cGAS, reversed the decrease of GSH/GSSG ratio induced by shcGAS without affecting the protein levels of master regulators in the ferroptosis defense system including GSH and ubiquinone (CoQ) systems (Fig. 2g; Supplementary information, Fig. S4j, k). Similarly, expression of MTS-cGAS abolished the high levels of lipid peroxidation and cell death induced by cGAS knockdown in the cells treated with the ferroptosis inducer Sorafenib (Fig. 2h, i). Collectively, these results indicated that loss of mitochondrial cGAS could induce cell death in HCC cells via ferroptosis.
Fig. 2. Mitochondria-localized cGAS inhibits ferroptosis.
a, b Cell growth analysis (a) and cell death analysis (b) of Hep3B cells with knockdown of endogenous cGAS and overexpression of cGAS fused with MTS (MTS-cGAS), wild-type cGAS (WT-cGAS), or cGAS mutant lacking the potential MTS (ΔMTS-cGAS). c Cell death analysis of Hep3B cells with endogenous cGAS knockdown and exogenous ΔMTS-cGAS overexpression treated with ferroptosis inhibitor Fer-1 (2 μM), Lip-1 (5 μM), apoptosis inhibitor Z-VAD-FMK (ZVF, 5 μM), necroptosis inhibitor Necrostatin-1 (Nec-1, 2 μM) or autophagy inhibitor 3-Methyladenine (3-MA, 2 mM), respectively. d Cell death analysis of Hep3B cells with the indicated genotypes treated with Fer-1 or Lip-1, respectively. e Lipid peroxidation analysis of Hep3B cells with the indicated genotypes treated with Fer-1 or Lip-1, respectively. Representative histogram data and statistical results are shown. f Lipid peroxidation analysis of Hep3B cells with the indicated genotypes treated with or without Fer-1. Representative histogram data and statistical results are shown. g Measurement of GSH/GSSG ratio in Hep3B cells with the indicated genotypes. h, i Lipid peroxidation analysis (h) and cell death analysis (i) of Hep3B cells with endogenous cGAS knockdown and exogenous MTS-cGAS overexpression treated with or without Sorafenib (10 μM). Representative histogram data and statistical results are shown. j Representative images of the mitochondria in Hep3B cells with the indicated genotypes under the electron microscope. Bar graphs depict the cristae numbers per mitochondria in the cells. Scale bars, 500 nm. The red arrows indicate the cristae in the mitochondria. k Measurement of mitochondrial ROS levels in Hep3B cells with the indicated genotypes. Representative histogram data and statistical results are shown. l Lipid peroxidation analysis of Hep3B cells with the indicated genotypes treated with or without Mito-Q (1 μM). Representative histogram data and statistical results are shown. For a–l, data are presented as mean ± SD of three independent experiments. P value was calculated by two-tailed unpaired Student’s t-test (b–l) or one-way analysis of variance (ANOVA) (a). *P ≤ 0.05; ns, not significant.
To further explore the mechanism by which the presence of mitochondrial cGAS prevented ferroptosis, we examined the mitochondrial morphology using the electron microscope and quantified the cristae number in mitochondria of the Hep3B cells with knockdown of endogenous cGAS and expression of MTS-cGAS or ΔMTS-cGAS. Compared to the NTC cells, the Hep3B cells with cGAS knockdown displayed damaged mitochondria with a disarranged structure of cristae, while expression of MTS-cGAS, but not ΔMTS-cGAS, significantly restored the damaged cristae structure in cGAS-knockdown cells (Fig. 2j), indicating that mitochondrial cGAS is at least partially responsible for the maintenance of mitochondrial quality. We then evaluated mitochondrial ROS levels, which are considered a leading cause of ferroptosis. Knockdown of endogenous cGAS led to abnormally high accumulation of mitochondrial ROS in Hep3B and PLC cancer cells, which was attenuated by MTS-cGAS expression (Fig. 2k; Supplementary information, Fig. S5a). Moreover, we observed that treatment with the mitochondria-targeted antioxidant Mito-Q abolished the high levels of lipid peroxidation and cell death in shcGAS cells similarly to overexpression of MTS-cGAS (Fig. 2l; Supplementary information, Fig. S5b, c). These results thus indicated that mitochondrial cGAS could suppress lipid peroxidation and ferroptosis by inhibiting the accumulation of excessive mitochondrial ROS.
Suppression of ferroptosis by mitochondrial cGAS is independent of STING activation
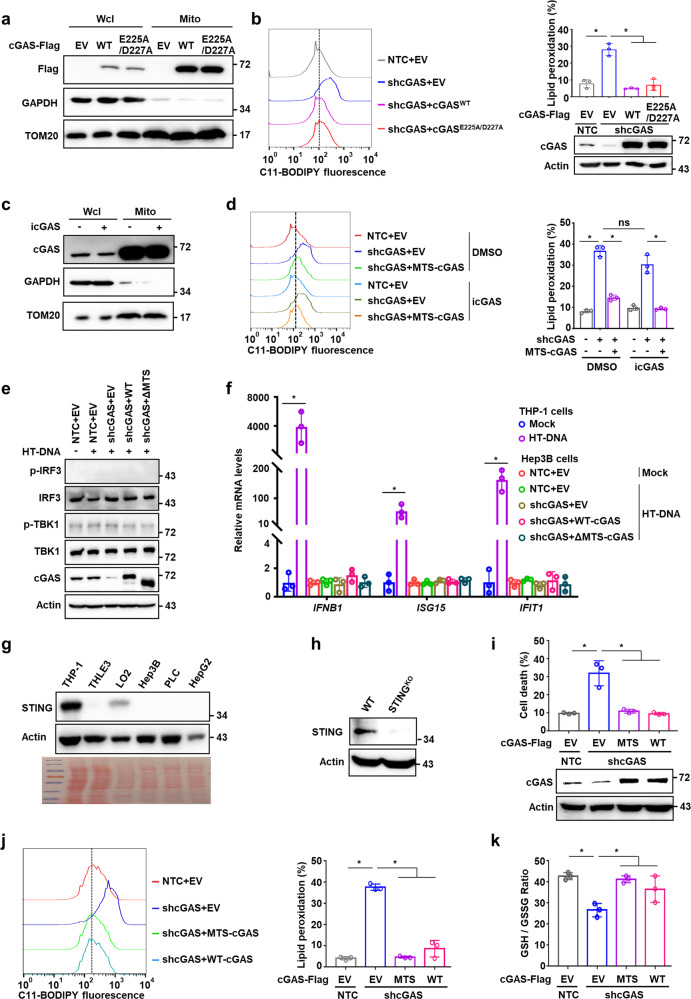
cGAS triggers innate immune response dependent on the activation of the adaptor STING.5 We next investigated whether cGAS-STING signaling is involved in mitochondrial cGAS-regulated ferroptosis. Intriguingly, expression of cGASE225A/D227A, a mutant incapable of binding magnesium necessary for synthesizing cGAMP and thus activating STING-dependent pathway, retained the mitochondrial localization and attenuated the high levels of cellular lipid peroxidation in Hep3B cells induced by shcGAS (Fig. 3a, b). Similarly, we observed that treatment with a cGAS catalytic inhibitor (PF-06928215) did not affect the mitochondrial localization of cGAS (Fig. 3c). Further lipid peroxidation analysis revealed that inhibition of cGAS catalytic activity did not influence cellular lipid peroxidation levels; however, MTS-cGAS expression attenuated the increase in cellular lipid peroxidation induced by shcGAS in the presence or absence of cGAS inhibitor (Fig. 3d). These data suggest that the cGAS localization and function in mitochondria were independent of its catalytic activity. Furthermore, we tested the phosphorylation of TBK1 and the mRNA levels of downstream target genes in cGAS-STING pathway in Hep3B cells under DNA stimulation condition. As a result, both the phosphorylation of TBK1 and IRF3 and the mRNA levels of the downstream target genes IFNB, ISG15 and IFIT1 were marginally detectable (Fig. 3e, f), suggesting distinct programs between mitochondrial function and DNA sensor function of cGAS. Comparison with monocytic cell line THP-1 showed that STING was expressed at substantially lower levels in both HCC cell lines and normal liver cells (Fig. 3g). Furthermore, depletion of STING in HCC cells did not affect the mitochondrial cGAS function in suppressing ferroptosis, as evidenced by the observation that MTS-cGAS expression could still abolish the increase in lipid peroxidation and cell death, and restore the decrease in GSH/GSSG ratio induced by shcGAS in STING-knockout Hep3B cells (Fig. 3h–k). These results thus indicated that mitochondrial cGAS could suppress lipid peroxidation and ferroptosis independent of the canonical cGAS-STING pathway.
Fig. 3. Mitochondrial cGAS suppresses ferroptosis independent of STING activation.
a Western blotting analysis of the protein levels of wild-type cGAS (cGASWT) or cGAS mutant (cGASE225A/D227A) with cGAMP synthase deficiency in mitochondria of Hep3B cells. Fractionation fidelity was verified by detection of GAPDH in the cytosolic fraction and TOM20 in the mitochondrial fraction. b Lipid peroxidation analysis of Hep3B cells with knockdown of endogenous cGAS and overexpression of the indicated cGAS (cGASWT or cGASE225A/D227A). Representative histogram data and statistical results are shown. Western blotting showed the protein levels of cGAS in Hep3B cells with the indicated genotypes. β-Actin served as the loading control. c Western blotting analysis of the protein levels of cGAS in mitochondrial fraction of Hep3B cells treated with or without cGAS catalytic inhibitor PF-06928215 (icGAS, 5 μM). Fractionation fidelity was verified by detection of GAPDH in the cytosolic fraction and TOM20 in the mitochondrial fraction. d Lipid peroxidation analysis of Hep3B cells with endogenous cGAS knockdown and exogenous MTS-cGAS overexpression treated with icGAS. Representative histogram data and statistical results are shown. e Western blot detection of p-IRF3, IRF3, p-TBK1, and TBK1 in Hep3B cells with knockdown of endogenous cGAS and overexpression of wild-type cGAS (WT-cGAS) or cGAS mutant (ΔMTS-cGAS) transfected with HT-DNA (2 mg/mL) for 6 h. f qRT-PCR analysis of the mRNA levels of IFNB1, ISG15 and IFIT1 genes in THP-1 cells or Hep3B cells with the indicated genotypes. g Western blotting analysis of the protein levels of STING in the indicated cell lines. β-Actin served as the loading control and Ponceau S staining is shown at the bottom. h Western blotting analysis of the protein levels of STING in the wild-type or STING-knockout Hep3B cells. i–k Cell death analysis (i), lipid peroxidation analysis (j) or GSH/GSSG ratio analysis (k) in STING-knockout Hep3B cells with the indicated cGAS genotypes. Western blotting analysis of the protein levels of cGAS in Hep3B cells with the indicated genotypes. β-Actin served as the loading control (i). For b, d, f and i–k, data are presented as mean ± SD of three independent experiments. P value was calculated by two-tailed unpaired Student’s t-test (b, d, f, i–k). *P ≤ 0.05; ns, not significant.
Mitochondrial cGAS associates with DRP1 and facilitates its oligomerization
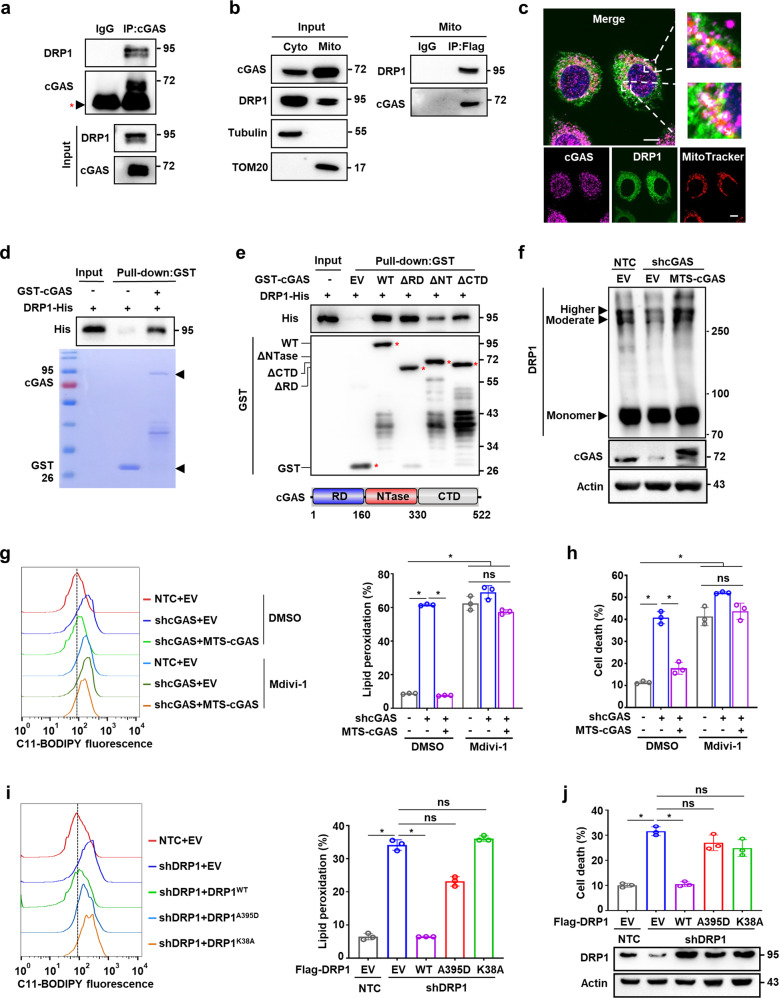
To explore the possible mechanisms by which mitochondrial cGAS suppresses ferroptosis and excessive ROS accumulation, we first screened our MS data for proteins that interacted with cGAS in mitochondria (Supplementary information, Table S1). Among the candidates, we focused on long-chain-fatty-acid-CoA ligase 4 (ACSL4), dynamin-like 120 kDa protein (OPA1), and dynamin-1-like protein (DRP1) which are reported to be involved in regulating ferroptosis or mitochondrial redox. Co-IP using antibodies targeting cGAS or Flag-tagged cGAS showed that it strongly interacted with DRP1, but not ACSL4 or OPA1 (Fig. 4a; Supplementary information, Fig. S6a, b). Co-IP using isolated mitochondrial protein fractions (Fig. 4b) in conjunction with immunofluorescence microscopy (Fig. 4c) verified the association between cGAS and DRP1 in mitochondria. Pull-down assays using glutathione S-transferase (GST)-fused cGAS and His-fused DRP1 expressed and purified from E. coli suggested that cGAS could directly interact with DRP1 (Fig. 4d). Notably, co-IP and pull-down assays showed that cGASE225A/D227A mutant had no effect on the association of cGAS with DRP1 (Supplementary information, Fig. S6c, d). Further pull-down assays using a series of cGAS truncation variants revealed that the NTase domain of cGAS was required for the interaction between cGAS and DRP1 (Fig. 4e). Taken together, these data demonstrate the direct association between cGAS and DRP1 in the mitochondria of HCC cells.
Fig. 4. Mitochondrial cGAS associates with DRP1 and facilitates its oligomerization.
a Co-IP assay showing the protein interaction between cGAS and DRP1. Cell lysate of Hep3B cells was immunoprecipitated with an anti-cGAS antibody, followed by western blotting analysis with antibodies against cGAS and DRP1. Red *, IgG heavy chain. b Co-IP assay showing the protein interaction between cGAS and DRP1 in the mitochondria of Hep3B cells. Hep3B cells were transfected with cGAS-Flag plasmid. The mitochondria were isolated, and lysates were immunoprecipitated with an anti-Flag antibody, followed by western blotting analysis with antibodies against cGAS and DRP1. c Representative images of immunofluorescence staining for cGAS, DRP1 and MitoTracker in Hep3B cells. The nucleus was stained with DAPI. Scale bars, 10 μm. d Pull-down assay showing the protein interaction between GST-cGAS and DRP1-His. GST or 6× His coding sequence was cloned into the N-terminus of cGAS or the C-terminus of DRP1, respectively. GST-tagged cGAS and 6× His-tagged DRP1 proteins were purified from E. coli and incubated in vitro, followed by western blot analysis with antibody against GST or His tag. e Pull-down assay showing the protein interaction between GST-cGAS with different domain truncations and DRP1-His. GST-tagged wild-type cGAS or cGAS truncations (GST-cGASΔRD, GST-cGASΔNTase, and GST-cGASΔCTD) and 6× His-tagged DRP1 proteins were purified from E. coli and incubated in vitro, followed by western blot analysis with antibody against GST or His tag. Red *, GST or GST fusion proteins. Bottom: diagram of cGAS protein (NCBI accession number NP_612450.2). Amino (N)-terminal regulatory domain (RD), central nucleotidyl transferase (NTase) domain, and carboxyl (C)-terminal domain (CTD) are indicated. f PAGE distinguished oligomerization states of DRP1 protein in Hep3B cells with endogenous cGAS knockdown and exogenous MTS-cGAS overexpression, including higher- and moderate-order oligomers and monomer. g Lipid peroxidation analysis of Hep3B cells with the indicated genotypes treated with or without DRP1 inhibitor Mdivi-1 (50 μM). Representative histogram data and statistical results are shown. h Cell death analysis of Hep3B cells with the indicated genotypes treated with or without Mdivi-1 (50 μM). i Lipid peroxidation analysis of MTS-cGAS-overexpressing Hep3B cells with the indicated DRP1 genotypes. Representative histogram data and statistical results are shown. j Cell death analysis of MTS-cGAS-overexpressing Hep3B cells with the indicated DRP1 genotypes. Western blotting showed the protein levels of DRP1 in Hep3B cells with the indicated genotypes. β-Actin served as the loading control. For g–j, data are presented as mean ± SD of three independent experiments. P value was calculated by two-tailed unpaired Student’s t-test. *P ≤ 0.05; ns, not significant.
In mammalian cells, the cytosolic DRP1 oligomerizes on the OMM in response to specific cellular cues and further constricts the membrane, leading to mitochondrial fission.30 Detection of DRP1 oligomerization by crosslinking experiments revealed that knockdown of cGAS suppressed the DRP1 oligomerization, resulting in reduced levels of intermediately and highly oligomerized DRP1 in Hep3B and PLC cells. However, DRP1 oligomer levels were markedly restored by MTS-cGAS overexpression (Fig. 4f; Supplementary information, Fig. S6e, f), suggesting that mitochondrial cGAS is required for DRP1 oligomerization.
DRP1 activity is necessary for mitochondrial respiratory regulation and mitochondrial quality surveillance.31 Inhibition of DRP1 by a specific inhibitor, Mdivi-1, abolished the effects of MTS-cGAS on alleviating shcGAS-induced mitochondrial ROS accumulation (Supplementary information, Fig. S6g). Consistent with this result, in the presence of Mdivi-1, MTS-cGAS expression could not reduce the elevated lipid peroxidation and cell death levels induced by shcGAS (Fig. 4g, h), suggesting that DRP1 functions with mitochondrial cGAS in the suppression of ferroptosis and mitochondrial ROS accumulation. In Hep3B cells overexpressing MTS-cGAS, silencing of DRP1 (shDRP1) led to the accumulation of mitochondrial ROS, as well as increased lipid peroxidation and cell death, which was abolished by the expression of wild-type DRP1, but not the DRP1A395D or DRP1K38A mutant that is defective in mitochondrial targeting or GTPase activity, respectively32–34 (Fig. 4i, j; Supplementary information, Fig. S6h). Taken together, these data showed that the interaction with DRP1 to facilitate its oligomerization on the OMM is necessary for mitochondrial cGAS to suppress lipid peroxidation and ferroptosis.
Mitochondrial cGAS promotes tumor progression via suppressing ferroptosis in vivo
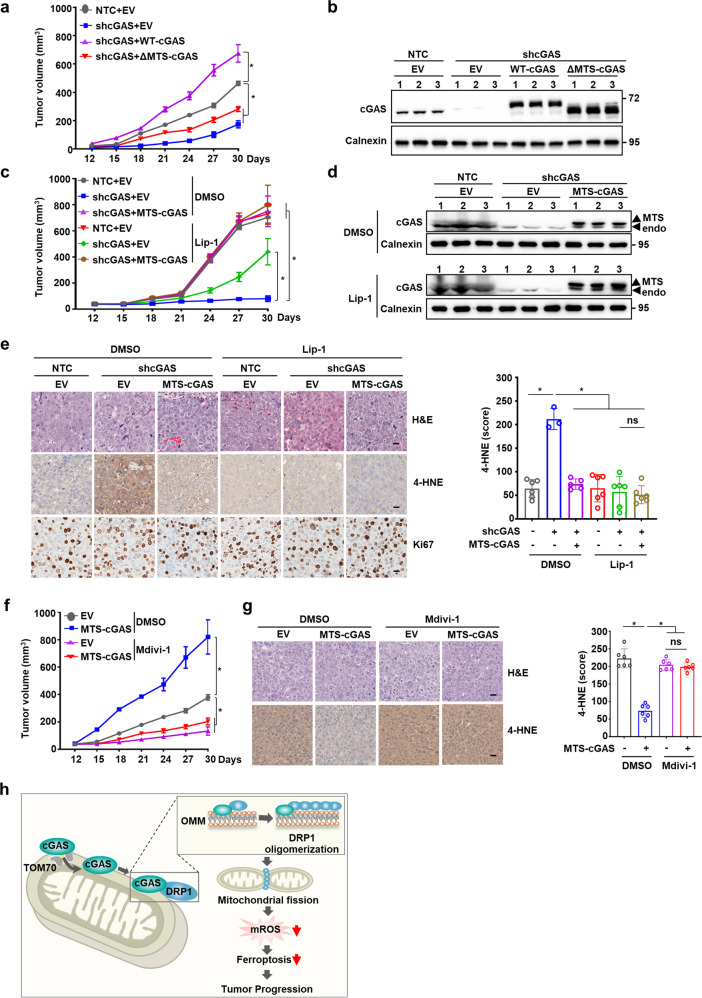
We next investigated the effects of mitochondrial cGAS inhibition as an intervention for HCC progression in vivo. Consistent with our observations in cell cultures, cGAS knockdown significantly suppressed tumor growth of Hep3B cells in mouse xenograft model, which was completely rescued by the restored expression of WT-cGAS, but not ΔMTS-cGAS, suggesting that the mitochondria-localized cGAS is important for tumor growth in HCC (Fig. 5a, b; Supplementary information, Fig. S7a). Further, MTS-cGAS overexpression markedly restored the tumor growth suppressed by shcGAS, suggesting that mitochondria-localized cGAS could play a major role in HCC tumorigenesis (Fig. 5c, d; Supplementary information, Fig. S7b). More importantly, treatment with ferroptosis inhibitor, Lip-1, resulted in significant rescue of tumor growth suppressed by shcGAS in Hep3B xenograft mice, suggesting that ferroptosis was indeed involved in cGAS-mediated tumor growth (Fig. 5c, d; Supplementary information, Fig. S7b). Our findings that cGAS was largely localized in the mitochondria of HCC cells (Fig. 1a) and that MTS-cGAS overexpression could remarkably restore the tumor growth suppressed by shcGAS, provided solid evidence that cGAS localization in mitochondria enhanced tumor growth by suppressing ferroptosis in vivo. Furthermore, IHC staining using antibody against 4-HNE, a lipid peroxidation marker, revealed that both MTS-cGAS overexpression and Lip-1 treatment significantly decreased lipid peroxidation in Hep3B xenograft tumor tissues, without affecting cell proliferation indicated by staining with Ki67 marker (Fig. 5e; Supplementary information, Fig. S7c). Collectively, these results demonstrated that mitochondrial cGAS serves as a major factor in promoting HCC tumor growth by suppressing ferroptosis in vivo.
Fig. 5. Mitochondrial cGAS promotes tumor progression by suppressing ferroptosis.
a Tumor growth analysis of Hep3B xenografts with knockdown of endogenous cGAS and overexpression of wild-type cGAS (WT-cGAS) or cGAS mutant (ΔMTS-cGAS). The indicated Hep3B cells were subcutaneously injected into male nude mice (n = 5 for each group). Tumor sizes were measured starting from 12 days after inoculation. b Western blotting analysis of the protein levels of cGAS in Hep3B xenograft tumors with the indicated genotypes. Calnexin served as the loading control. c Tumor growth analysis of Hep3B xenografts with the indicated genotypes treated with or without Lip-1. The indicated Hep3B cells were subcutaneously injected into male nude mice (n = 5 for each group). Lip-1 (10 mg/kg) was injected intraperitoneally every other day. Tumor sizes were measured starting from 12 days after inoculation. d Western blotting analysis of the protein levels of cGAS in Hep3B xenograft tumors with the indicated genotypes treated with or without Lip-1. Calnexin served as the loading control. e Representative images of IHC staining for 4-HNE or Ki67 in the Hep3B xenograft tumors with the indicated genotypes treated with or without Lip-1. Scale bars, 20 μm. 4-HNE staining was quantitated using the Histo-score (H-score) in IHC assays. f Tumor growth analysis of Hep3B xenografts with endogenous cGAS knockdown and exogenous MTS-cGAS overexpression treated with or without Mdivi-1. The indicated Hep3B cells were subcutaneously injected into male nude mice (n = 5 for each group). Mdivi-1 (25 mg/kg) was injected intraperitoneally every other day. Tumor sizes were measured starting from 12 days after inoculation. g Representative images of IHC staining for 4-HNE in the Hep3B xenograft tumors with endogenous cGAS knockdown and exogenous MTS-cGAS overexpression treated with or without Mdivi-1. Scale bars, 20 μm. 4-HNE staining was quantitated using the H-score in IHC assays. h Schematic cartoon showing that mitochondria-localized cGAS interacts with DRP1 to facilitate its oligomerization and function, thus preventing mitochondrial ROS accumulation and ferroptosis. For a, c and e–g, data are presented as mean ± SEM. P value was calculated by two-tailed unpaired Student’s t-test (e, g), or two-way ANOVA (a, c, f). *P ≤ 0.05; ns, not significant.
Further investigation of the role of DRP1 in mitochondrial cGAS-mediated tumor growth showed that MTS-cGAS overexpression facilitated the in vivo tumor growth of Hep3B xenografts with endogenous cGAS knockdown, which was abolished by treatment with the DRP1 inhibitor, Mdivi-1 (Fig. 5f; Supplementary information, Fig. S7d, e). IHC staining with anti-4-HNE antibody revealed that Mdivi-1 treatment also abolished the suppressive effects of MTS-cGAS expression on lipid peroxidation in xenograft tumor tissues (Fig. 5g). Thus, our data demonstrate that mitochondria-localized cGAS promotes tumorigenesis by suppressing ferroptosis in a DRP1-dependent manner (Fig. 5h).
Discussion
In previous studies, cGAS has been well established to serve as a sensor for foreign DNA linked to the orchestration of innate immune defense against pathogens. However, more recent studies revealed non-canonical functions of cGAS that depend on its subcellular localization to different targets, and its localization in the micronuclei, nucleus and cytoplasmic membrane has been observed under different contexts.14,19,20 During mitosis, cGAS enters the nucleus and associates with chromatin DNA to mediate cellular senescence induced by DNA damage.6 In HeLa cells cGAS localized in the nucleus to prevent autoreactivity.35 cGAS has also been shown to interact with nucleosomes and mitotic chromosomes in cells with mitotic aberrations, inducing apoptosis by mediating the accumulation of phosphorylated IRF3.36 In phagocytes, the presence of cGAS in the plasma membrane has been demonstrated as a critical factor for cellular response to genotoxic stress and viral infections.20 A study using Huntington disease striatal cells also found that cGAS was enriched in lysosomes and mediated autophagy.11 In this study, we detected a remarkable pool of cGAS in the mitochondria of HCC cells. Our results further indicate that cGAS suppresses excessive ROS accumulation and ferroptosis induction by interacting with DRP1 to facilitate its oligomerization on the OMM (Fig. 5h). Taken together, these diverse studies show that the subcellular localization of cGAS determines its function and can control cell fate in different cell types.
Ferroptosis is a form of non-apoptotic cell death driven by iron-dependent lipid peroxidation and targeted induction of ferroptosis has been proposed as potential therapeutic strategy for cancers.37–39 At least three systems have been identified for their function in protecting cells from ferroptosis: 1) GPX4 uses reduced GSH to detoxify cytosolic and mitochondrial lipid hydroperoxides40,41; 2) FSP1 (also called AIFM2) functions as an oxidoreductase to reduce CoQ to ubiquinol (CoQH2) mainly on the plasma membrane42,43; and 3) DHODH also greatly contributes to detoxifying lipid peroxides in mitochondria.23 Mitochondria are highly dynamic organelles that orchestrate numerous essential metabolic processes, redox balance, and calcium homeostasis in response to different intracellular and environmental cues.44 An increasing body of evidence supports a strong link between mitochondrial dysfunction and ferroptosis.24,45,46 The presumable link between ferroptosis and mitochondria is the excessive ROS accumulation derived from mitochondrial respiration that gets out of control. In this regard, we observed that depletion of cGAS in HCC cells results in the increased accumulation of ROS in the mitochondria, leading to extensive lipid peroxidation and ferroptosis. Importantly, the overexpression of mitochondria-tethered cGAS abolishes the heightened induction of cell ferroptosis and inhibition of tumor growth induced by depletion of endogenous cGAS, indicating that mitochondrial cGAS contributes to keeping the mitochondrial function in order and thus suppressing ferroptosis.
The quality and quantity of mitochondria are controlled by the dynamic changes of fusion and fission which are coordinated by the dynamin-related GTPases MFN1/2 and OPA1, and cytosolic DRP1 and its receptors on the OMM, respectively.47,48 The dysregulation of fusion and fission impaired the mitochondrial function including mitochondrial respiration, leading to abnormal cell fate decision.49–51 We found that loss of mitochondrial cGAS impairs DRP1 oligomerization and can further lead to excessive mitochondrial ROS levels and ferroptosis. We further demonstrate that cGAS facilitates tumor growth via inhibition of lipid peroxidation and ferroptosis in a DRP1-dependent manner in vivo, linking cGAS to mitochondrial quality control and cell fate decision.
Taken together, our results reveal that mitochondria-localized cGAS promotes cancer progression by facilitating DRP1 oligomerization to suppress excessive ROS production and ferroptosis, suggesting that targeting mitochondrial cGAS may be an effective potential strategy for the development of HCC interventions.
Materials and Methods
Cell culture
Human HCC cells (Hep3B, PLC, and HepG2), A549, HT1080, PANC-1, MDA-MB-231, U87 and HEK293T cell lines were purchased from the American Type Culture Collection. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin. All cell lines were tested for mycoplasma contamination. The cells were cultured in 10 cm cell culture dish and then transferred into a 6-well plate for mitochondrial ROS, lipid peroxidation and cell death measurements. The reagents used in this study are summarized in Supplementary information, Table S2.
Plasmid construction and establishment of stable cell lines
Expression plasmids encoding Flag-, HA-, mCherry-, or GFP-tagged wild-type, mutant or truncated human cGAS, TOM70, TOM20, DRP1, ACSL4 and OPA1 were constructed with the mammalian expression vector pSin-EF2-Puro. shRNAs against TOM70 in pLKO.1 vector were commercially purchased (Sigma-Aldrich). shRNAs against cGAS or DRP1 (3′-UTR or 5′-UTR) were constructed into the pLKO.1 vector. The target sequences of all shRNAs used in this study are summarized in Supplementary information, Table S3. The mitochondria-targeted cGAS variant (MTS-cGAS) was generated by fusing the sequence encoding the MTS of the cytochrome c oxidase subunit 4 isoform 1 (COX4) to the N-terminus of cGAS. The gRNA sequences targeting cGAS and STING (hcGAS, 5′-ATATTCTTGTAGCTCAAT-3′ and 5′-ATTGAGCTACAAGAATAT-3′; hSTING, 5′-ATCCATCCATCCCGTGTCCC-3′ and 5′-GGGACACGGGATGGATGGAT-3′) were cloned into lenti-guide vector. These constructs were co-transfected with lentiCas9-blast plasmid and packaging plasmids pMD2.G and psPAX2 into HEK293T cells to generate lentivirus. The plasmids were co-transfected with viral packaging plasmids into HEK293T cells using PEI (Polysciences). Culture medium containing virus particles was collected 48 h post transfection and added into culture medium of Hep3B or PLC cells in the presence of 8 μg/mL polybrene (Sigma-Aldrich). The infected cells were selected by 0.5 µg/mL puromycin to establish stable cells.
Cellular fractionation isolation and mitochondria purification
Mitochondria, nucleus, and cytosol were isolated from cells mainly using a published protocol with minor changes.52 Briefly, cells were resuspended and homogenized in ice-cold mitochondria isolation buffer (1.5 mM MgCl2, 10 mM KCl, 5 μM trichostatin A, 10 mM Tris-HCl, pH 7.4, 50 mM nicotinamide and protease inhibitors). Nuclei were pelleted by centrifugation at 1200× g for 10 min at 4 °C and were lysed in nuclear buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 500 mM NaCl, 0.2% NP-40, 10 mM β-mercaptoethanol, 10% glycerol) supplemented with protease inhibitor cocktail by sonication. The supernatant containing mitochondrial fraction was centrifuged again at 4000× g for 20 min at 4 °C to separate mitochondrial fraction from other cytosolic fractions. The supernatant was transferred to a fresh tube as cytosolic fractions. The pellet, mainly containing mitochondria, was washed twice with isolation buffer and then centrifuged again at 10,000× g for 15 min at 4 °C to pellet mitochondria. The mitochondrial fraction was lysed in mitochondria lysis buffer and centrifuged at 13,000× g for 10 min. The supernatants containing the nuclear proteins, cytosolic proteins and mitochondrial proteins were then subjected to immunoblot analysis.
To predict MTS, sequences of cGAS from several species were downloaded from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/protein) and UniProt database (https://www.uniprot.org/), and multiple sequence alignment was performed using the online tool Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Proteinase K protection assay
Purified mitochondria were resuspended in ice-cold mitochondria isolation buffer and incubated with different concentrations of proteinase K for 30 min on ice, in the presence or absence of 1% Triton X-100. Samples were then subjected to immunoblot analysis. TOM70 and MFN1 were used as markers of the OMM, COX4 and TFAM were used to as markers of the inner mitochondrial membrane (IMM) and the mitochondrial matrix (MM), respectively.
Western blotting analysis
Cells were lysed using RIPA buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 0.1% SDS, and 1% NP-40) supplemented with protease inhibitor cocktail. Equal amounts of cell lysates were boiled and fractionated by SDS-PAGE. Primary antibodies used for immunoblotting included anti-cGAS (Abcam), and antibodies to DRP1, TOM20, TOM70, MFN1, COX4, TFAM, GAPDH, lamin B1, β-actin, tubulin and calnexin (Proteintech). HRP-conjugated anti-mouse or anti-rabbit secondary antibodies (Bio-Rad) were used, and signals were determined by the Western ECL Substrate (Tanon). The antibodies used in this study are summarized in Supplementary information, Table S2.
Co-IP
For co-IP, cells were lysed with low-salt IP buffer (0.5% NP-40, 50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 2 mmol/L EDTA, 1.5 mmol/L MgCl2) supplemented with protease inhibitor cocktail for 1–2 h on ice, and then centrifuged at 12,000× g for 10 min at 4 °C. The supernatants were incubated with the indicated antibody overnight at 4 °C followed by incubation with protein A/G-conjugated beads for 4 h. The beads were then washed with IP buffer and boiled in SDS-PAGE loading buffer. Protein samples were analyzed by western blotting.
qRT-PCR
Total RNA was extracted from cells using TRIzol reagent (Life Technologies) following the manufacturer’s guidelines. Reverse transcripts were generated using an HiScript® II 1st Strand cDNA Synthesis Kit (Vazyme), while real-time reverse-transcription PCR (RT-PCR) was performed using SYBR Green master mix (Vazyme) on a LightCycler®96 (Roche). Primer sequences are shown in Supplementary information, Table S4. All samples were normalized to 18S RNA.
Pull-down assay
The cDNA encoding DRP1 was cloned into the pET-22b (+) vector (Novagen). The cDNA encoding cGAS or truncated cGAS was cloned into the pGEX-4T-1 vector (GE Healthcare) by the 5× Ligation-Free Cloning Master Mix (ABM). The target proteins were induced with 0.5 mM IPTG for 18 h at 16 °C. Purified His-tagged proteins and GST-fused proteins were incubated in pull-down assay buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.02 mM EDTA, 0.1% Triton X-100). After incubation, the beads were pelleted and washed with pull-down assay buffer, followed by elution of proteins. The proteins were detected by western blotting.
LC-MS/MS and data analysis
The proteins of the isolated mitochondria from the Hep3B cells were digested into the peptides. The peptides were prepared using a preparation kit (Thermo Fisher, 90057), and analyzed on a QE-plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with an EASY-nLC 1200 nano liquid chromatography (HPLC) system (Thermo Fisher Scientific, Waltham, MA, USA). All data were further analyzed using proteomics discovery software (Thermo Scientific Proteome Discoverer 2.2.) against the human UniProt database. The original data for LC-MS/MS-based proteomics are available via ProteomeXchange with identifier PXD036920.
Crosslinking assay
DSS crosslinking was performed to detect the DRP1 oligomerization. In brief, equal numbers of cells were washed with ice-cold PBS twice, followed by suspension in conjugation buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 8.0) with a final concentration of 1 mM DSS (Sangon Biotech). After incubation at 37 °C for 30 min, the samples were boiled and analyzed by western blotting.
Immunofluorescence staining
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde for 20 min and then washed in PBS. Cells were permeabilized with 0.1% Triton X-100 for 20 min, followed by blocking with 5% BSA for 30 min. Primary antibodies, including anti-cGAS (1:50), anti-TOM20 (1:100) and anti-DRP1 (1:100) antibodies were diluted in blocking solution and incubated with cells in a humidified chamber overnight at 4 °C. Anti-rabbit secondary antibodies conjugated to Texas Red (1:200) and anti-mouse secondary antibody conjugated to FITC (1:200) were used. The cells were stained with DAPI (Sigma-Aldrich) for 5 min for the detection of nuclei. Representative microphotographs were acquired using a Zeiss800 laser scanning confocal microscope (Zeiss) and analyzed by ZEN microscope imaging software (Zeiss). For MitoTracker staining, cells were incubated with 200 nM MitoTracker Red FM at 37 °C for 30 min, then washed 3 times with DMEM, and finally processed for immunofluorescence staining.
Cell death assay and lipid peroxidation measurement
Cell death was measured by flow cytometry analysis with propidium iodide (Sigma-Aldrich) staining according to the manufacturer’s instructions. Briefly, cells were seeded at a density of 50% 24 h before being treated with different reagents. Then, cells were harvested (including floating dead cells) and stained with 5 μg/mL propidium iodide. After incubation for 10 min, the propidium iodide-positive dead cell population was analyzed with a flow cytometer. To analyze lipid peroxidation, cells were incubated with 5 μM C11-BODIPY 581/591 (Invitrogen) for 30 min in a humidified incubator (at 37 °C, 5% CO2). Then, cells were harvested, and lipid peroxidation levels were analyzed with a flow cytometer. Data were analyzed using FlowJo Software.
Mitochondrial ROS measurement
Mitochondrial ROS levels were measured using MitoSOX Red fluorescence assay (Life Technologies) following the manufacturers’ instructions. In brief, cells were stained with 5 µM MitoSOX at 37 °C for 30 min and examined by a flow cytometer. Data were analyzed using FlowJo Software.
Electron microscopy
For mitochondrial ultrastructure analysis, Hep3B cells were fixed for 12 h at 4 °C in 2.5% glutaraldehyde. After being rinsed in PBS, the cells were post-fixed in 2% ferrocyanide-reduced osmium tetroxide. Cells were then stained with 1% uranyl acetate aqueous solution and dehydrated in ascending concentrations of ethanol, passed through acetone and embedded in Epon resin. Ultrathin sections were finally counterstained with 2% uranyl acetate and lead citrate. Samples were analyzed using a transmission electron microscope (120 kV; Tecnai G2 Spirit, FEI).
Cell line-derived xenograft model
All animal studies were conducted with approval from the Animal Research Ethics Committee of South China University of Technology. Five-week-old male nude mice (BALB/c) were randomly assigned to experimental groups. For xenograft experiments, equal numbers of Hep3B cells with indicated genotype mixed with Matrigel were injected subcutaneously into nude mice (SJA Laboratory Animal Company). For Lip-1 or Mdivi-1 treatment, when the tumor volume reached 50–100 mm3, the mice were randomly divided into different treatment groups (n = 5). Lip-1 or Mdivi-1 was dissolved in dimethyl sulfoxide (DMSO) and diluted in PBS. Lip-1 or DMSO was intraperitoneally injected daily at a dose of 10 mg/kg. Mdivi-1 or DMSO was intraperitoneally injected every other day at a dose of 25 mg/kg. The tumor volume was measured every 3 days after inoculation using digital calipers and calculated using the equation: volume = length × width2 × 0.52.
Clinical human HCC specimens
Snap-frozen HCC lesions and the corresponding adjacent noncancerous tissues, which were at least 2 cm distant from the edge of the tumors, were collected from 42 HCC patients in the First Affiliated Hospital of University of Science and Technology of China. Proteins were extracted from paired HCC and noncancerous tissues and detected by immunoblotting. Formalin-fixed, paraffin-embedded primary HCC specimens obtained from 54 patients were randomly selected from the archives of the First Affiliated Hospital of University of Science and Technology of China.
IHC
The IHC procedure was performed according to standard protocols. In brief, fresh tumor tissues were fixed in 10% neutral buffered formalin, and stored in 70% ethanol at 4 °C. Paraffin-embedded tissues were sliced into 4-μm thick sections for hematoxylin and eosin (H&E) or IHC staining. The sections were incubated with anti-4-HNE (Abcam, 1:400), anti-Ki67 (Abcam, 1:200) or anti-cGAS (Cell Signaling Technology, 1:200) antibodies overnight at 4 °C. After incubation with HRP-conjugated secondary antibodies (MXB Biotechnology), the sections were visualized with DAB. Images were randomly obtained at 200× magnification using Leica AperioCS2 and quantification was performed with HALO software.
Ethics statement
For use of the clinical materials for research purposes, prior written informed consent from the patients and study approval from the Institutional Research Ethics Committee of The First Affiliated Hospital of University of Science and Technology of China were obtained (2022-BE(H)-149).
Statistical analysis
All experimental data are presented as mean ± SD or mean ± SEM as stated from at least three independent experiments. Student’s t-test between two groups and two-way ANOVA across multiple groups were used to calculate P-values by GraphPad Prism 7. Statistical significance is displayed as *P < 0.05.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (91957203, 81930083, 82130087, 82072656, 82192893, 81821001), the National Key R&D Program of China (2018YFA0800303, 2022YFA1304504), the Chinese Academy of Sciences (XDB39000000), Hefei Comprehensive National Science Center Institute of Health and Medicine Project (DJK-LX-2022001).
Author contributions
P.G. conceived and supervised the study. X.Z., S.Q., P.G., and H.Z. designed the experiments. S.Q., X.Z., X.M., S.L., X.Q., H.L., J.C., Y.Z., M.W., and Z.Y. performed and analyzed the experiments. P.G., H.Z., X.Z., and S.Q. wrote the manuscript. All the authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Shiqiao Qiu, Xiuying Zhong, Xiang Meng.
Contributor Information
Xiuying Zhong, Email: zxywawj@ustc.edu.cn.
Huafeng Zhang, Email: hzhang22@ustc.edu.cn.
Ping Gao, Email: pgao2@ustc.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-023-00788-1.
References
- 1.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai J, et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 2019;176:1447–1460.e14. doi: 10.1016/j.cell.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 6.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gluck S, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017;19:1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 9.Paul BD, Snyder SH, Bohr VA. Signaling by cGAS-STING in neurodegeneration, neuroinflammation, and aging. Trends Neurosci. 2021;44:83–96. doi: 10.1016/j.tins.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ablasser A, Chen ZJ. cGAS in action: Expanding roles in immunity and inflammation. Science. 2019;363:eaat8657. doi: 10.1126/science.aat8657. [DOI] [PubMed] [Google Scholar]
- 11.Sharma M, Rajendrarao S, Shahani N, Ramirez-Jarquin UN, Subramaniam S. Cyclic GMP-AMP synthase promotes the inflammatory and autophagy responses in Huntington disease. Proc. Natl. Acad. Sci. USA. 2020;117:15989–15999. doi: 10.1073/pnas.2002144117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 2020;10:26–39. doi: 10.1158/2159-8290.CD-19-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018;215:1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding SM, et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu M, et al. ATM inhibition enhances cancer immunotherapy by promoting mtDNA leakage and cGAS/STING activation. J. Clin. Invest. 2021;131:e139333. doi: 10.1172/JCI139333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domizio JD, et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022;603:145–151. doi: 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakhoum SF, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 20.Barnett KC, et al. Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell. 2019;176:1432–1446.e11. doi: 10.1016/j.cell.2019.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H, et al. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 2019;38:e102718. doi: 10.15252/embj.2019102718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao C, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bock FJ, Tait SWG. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020;21:85–100. doi: 10.1038/s41580-019-0173-8. [DOI] [PubMed] [Google Scholar]
- 25.Kleih M, et al. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 2019;10:851. doi: 10.1038/s41419-019-2081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasan K, Werner M, Chandel NS. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 2020;32:341–352. doi: 10.1016/j.cmet.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong XY, et al. Mitochondrial dynamics is critical for the full pluripotency and embryonic developmental potential of pluripotent stem cells. Cell Metab. 2019;29:979–992.e4. doi: 10.1016/j.cmet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Zhang T, et al. ENO1 suppresses cancer cell ferroptosis by degrading the mRNA of iron regulatory protein 1. Nat. Cancer. 2022;3:75–89. doi: 10.1038/s43018-021-00299-1. [DOI] [PubMed] [Google Scholar]
- 29.Li T, et al. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science. 2021;371:eabc5386. doi: 10.1126/science.abc5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabara LC, Morris JL, Prudent J. The complex dance of organelles during mitochondrial division. Trends Cell Biol. 2021;31:241–253. doi: 10.1016/j.tcb.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt K, et al. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab. 2018;27:657–666.e5. doi: 10.1016/j.cmet.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Chang CR, et al. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J. Biol. Chem. 2010;285:32494–32503. doi: 10.1074/jbc.M110.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volkman HE, Cambier S, Gray EE, Stetson DB. Tight nuclear tethering of cGAS is essential for preventing autoreactivity. Elife. 2019;8:e47491. doi: 10.7554/eLife.47491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zierhut C, et al. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell. 2019;178:302–315.e23. doi: 10.1016/j.cell.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockwell BR, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedmann AJP, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bersuker K, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doll S, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 44.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao M, et al. Role of mitochondria in ferroptosis. Mol. Cell. 2019;73:354–363.e3. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng J, Conrad M. The metabolic underpinnings of ferroptosis. Cell Metab. 2020;32:920–937. doi: 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 47.Ni HM, Williams JA, Ding WX. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015;4:6–13. doi: 10.1016/j.redox.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song J, Herrmann JM, Becker T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 2021;22:54–70. doi: 10.1038/s41580-020-00300-2. [DOI] [PubMed] [Google Scholar]
- 49.Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 50.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herkenne S, et al. Developmental and tumor angiogenesis requires the mitochondria-shaping protein Opa1. Cell Metab. 2020;31:987–1003.e8. doi: 10.1016/j.cmet.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.