Summary
Background
Dementia's diagnostic protocols are mostly based on standardised neuroimaging data collected in the Global North from homogeneous samples. In other non-stereotypical samples (participants with diverse admixture, genetics, demographics, MRI signals, or cultural origins), classifications of disease are difficult due to demographic and region-specific sample heterogeneities, lower quality scanners, and non-harmonised pipelines.
Methods
We implemented a fully automatic computer-vision classifier using deep learning neural networks. A DenseNet was applied on raw (unpreprocessed) data from 3000 participants (behavioural variant frontotemporal dementia-bvFTD, Alzheimer's disease-AD, and healthy controls; both male and female as self-reported by participants). We tested our results in demographically matched and unmatched samples to discard possible biases and performed multiple out-of-sample validations.
Findings
Robust classification results across all groups were achieved from standardised 3T neuroimaging data from the Global North, which also generalised to standardised 3T neuroimaging data from Latin America. Moreover, DenseNet also generalised to non-standardised, routine 1.5T clinical images from Latin America. These generalisations were robust in samples with heterogenous MRI recordings and were not confounded by demographics (i.e., were robust in both matched and unmatched samples, and when incorporating demographic variables in a multifeatured model). Model interpretability analysis using occlusion sensitivity evidenced core pathophysiological regions for each disease (mainly the hippocampus in AD, and the insula in bvFTD) demonstrating biological specificity and plausibility.
Interpretation
The generalisable approach outlined here could be used in the future to aid clinician decision-making in diverse samples.
Funding
The specific funding of this article is provided in the acknowledgements section.
Keywords: Computer vision, Reproducibility, Deep learning, Unprocessed MRI, Dementia
Research in context.
Evidence before this study
The authors reviewed the literature using traditional (e.g., PubMed) sources. While neurodegenerative disease status can be predicted from standardised neuroimaging data (MRI), the literature shows that lower quality scanners, non-harmonised processing pipelines, demographic, and region-specific sample heterogeneities induce biased results tempering generalisation.
Added value of this study
A raw-data based, automatic computer vision classifier using deep learning neural networks yielded reproducible and generalisable results across highly heterogeneous MRI testing databases of Alzheimer's disease, behavioural variant frontotemporal dementia, and healthy controls. This development led to a robust and interpretable classification pipeline of dementia subtypes. The replicability analysis was performed by testing the algorithm on diverse samples including 3T neuroimaging from the Global North, 3T neuroimaging data from Latin America, and routine 1.5T clinical images.
Implications of all the available evidence
This study may prove critical for future scalability and comparability across multiple global datasets, ultimately leading to the development of new clinical decision-making tools.
Introduction
In the global South, dementia prevalence is expected to increase rapidly in the next decades.1 Although patterns of regional brain atrophy can characterise neurodegenerative conditions such as frontotemporal dementia (FTD)2,3 and Alzheimer's disease (AD),4,5 multiple challenges hinder the implementation of massive, scalable, generalisable and automatic assessments using neuroimaging data outside the global North (and even within high income countries with larger health disparities). These challenges are larger in multicentre studies that include non-stereotypic samples (in terms of admixture, genetic, demographic, magnetic resonance imaging (MRI) signals, or cultural diversity), where computational models systematically fail to provide generalisations.6 Latin American Countries (LACs) constitute non-stereotypic samples7, 8, 9 and are also challenged by additional factors. In such regions with increased prevalence forecasts, there are relatively few qualified personnel (technicians and clinicians) due to hiring costs and training unavailability.7,8 In addition, current MRI guidelines and protocols are based on research-quality 3 tesla (T) acquisitions, such as that used in the Alzheimer's Disease Neuroimaging Initiative (ADNI)10 and the Neuroimaging in Frontotemporal Dementia (NIFD/LONI).11 However, clinical neuroimaging access in LACs is usually restricted to 1.5T, low-field MRI scanners, and heterogeneous recording parameters,7,8 generating high sources of variability and challenging current harmonisation procedures.12,13 In non-research-oriented sites assessing non-stereotypic populations, recruitment approaches tend to inadequately match for sociodemographic factors, increasing the challenges in relation to usability.7,8 Protocols to assess neuroimaging as a diagnostic tool for AD and FTD outside of the global North must therefore account for the aforementioned factors. To date, no previous approach has addressed these challenges (see Table 1 for the multiple gaps in the literature) and generalised findings to South vs North comparisons.
Table 1.
Literature review.
| Author | Samples | Methods | Results | Gaps addressed in this report |
|---|---|---|---|---|
| Odusami et al. (2022)14 | Healthy controls (CN) = 25 Subjective memory complaint (SCM) = 25 Early mild cognitive impairment (EMCI) = 25 Late MCI = 25 MCI = 13 Alzheimer's Disease (AD) = 25 |
Concatenated extracted features from MRI acquisitions using ResNet18 and DenseNet121 | 98.86% accuracy, 98.94% precision, and 98.89% recall in multiclass classification | No assessment of non-stereotypical samples Trained and tested with data from one source only (ADNI) No data augmentation No dementia subtype characterisation (all patients had the AD spectrum) No use of demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
| Razzak et al. (2022)15 | AD = 95 MCI = 138 CN = 146 |
PartialNet | Models averaged 99% accuracy | No assessment of non-stereotypical samples Image preprocessing not fully automatic Trained and tested with data from one source only (ADNI) No dementia subtype characterisation (all patients had the AD spectrum) No model interpretability analysis No use of demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
| Di Benedetto et al.(2022)16 | Database 1: Behavioural-variant frontotemporal dementia (bvFTD) = 50 CN = 110 Database 2: bvFTD = 29 CN = 24 |
Compared 3D CNN, vision transformers, and logistic regression | Up to 90% AUC | No assessment of non-stereotypical samples Preprocessed MRI using CAT12 toolbox No data augmentation No model interpretability analysis No dementia subtype characterisation (all patients were bvFTD) No use of demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
| Hosseini-Asl et al. (2018)17 | MCI = 70 AD = 70 CN = 70 |
3D CNN autoencoder | Up to 100% accuracy | Trained and tested with data from one source only (ADNI) Image preprocessing No dementia subtype characterisation (all patients had the AD spectrum) No model interpretability analysis No use of demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
| Qiu et al. (2020)18 | Database 1: CN = 229 AD = 118 Database 2: CN = 320 AD = 62 Database 3: CN = 73 AD = 62 Database 4: CN = 356 AD = 209 |
3D CNN | AUC up to 0.996 | No assessment of non-stereotypical samples No data augmentation Image preprocessing No dementia subtype characterisation All samples from high income country databases, no 1.5 Tesla testing samples Two databases were not demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
| Amini et al. (2021)19 | Low MCI = 690 Mild MCI = 236 Moderate MCI = 67 Severe MCI = 7 |
Quantum Matched-Filter Technique (QMFT) and CNN deep learning | Up to 96.7% accuracy | No assessment of non-stereotypical samples Trained and tested with data from one source only (ADNI) Image preprocessing No dementia subtype characterisation (all patients had the AD spectrum) No use of demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
| Zhang et al. (2021)20 | AD = 280 cMCI = 162 (MCI converters) ncMCI = 251 (MCI non-converters) CN = 275 |
Attentional CNN | Up to 97.35% accuracy | No assessment of non-stereotypical samples Trained and tested with data from one source only (ADNI) Image preprocessing No dementia subtype characterisation (all patients had the AD spectrum) No model interpretability analysis No use of demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
| Feng et al. (2020)21 | AD = 153 MCI = 157 CN = 159 |
3D-CNN | Up to 99.1% accuracy | No assessment of non-stereotypical samples Trained and tested with data from one source only (ADNI) No dementia subtype characterisation (all patients had the AD spectrum) No model interpretability analysis No use of demographically matched sample in age, sex, and years of education to discard possible biases across groups inflating classification accuracy |
Recent developments in computer vision stemming from the field of artificial intelligence provide an innovative solution to circumvent limitations in data acquisition and data heterogeneity. Deep learning-based visual artificial neural networks are characterised by flexibility in evaluating images without a priori expectations on conventional orientation, metric, or shape.22 Possible biases in the preprocessing pipeline selection steps such as image filtering, segmentation, rotation, and smoothing are eliminated because the input consists of unprocessed (raw) data, from which the most relevant features for automated classification of clinical status23 are extracted. Computer-assisted diagnostic approaches have already proven successful in several dementia domains,24,25 but there is a notable lack of results on the utility of using unprocessed neuroimaging data across diverse geographical regions and with non-standardised recording parameters (Table 1). These factors potentially bias the classification output, impacting reproducibility and generalisation that is crucially needed in global and clinical settings.
Here, we present a fully automatic pipeline for dementia subtype characterisation (AD and FTD) for stereotypic and non-stereotypic samples, assessing heterogeneous demography and acquisition parameters from 3 Tesla MRI raw data. This pipeline was tested on out-of-sample raw data from non-stereotypical populations with diverse MRI acquisition parameters, including clinical 1.5 Tesla MRI acquisitions. As most of the challenges in analysing neuroimaging data from non-stereotypic samples have not been addressed simultaneously in previous works, we summarise in Table 1 the previous gaps being tackled in this report. We collected raw online MRI datasets without specification to acquisition parameter, with and without demographic matching, to train DenseNet,26,27 a state-of-the-art deep learning neural network.28 Since a considerable number of samples is needed to obtain robust results,29 we augmented the input MRI images (3000 unmatched and 300 matched samples) ten times to obtain 30,000 unmatched and 3000 matched samples by employing several geometric transformations.30 After testing the data from standardised 3T data from large consortia, and with standardised 3T data from LACs, we then tested the classification's generalisation and reproducibility among routine clinical 1.5T MRI images from non-stereotypic populations in the scientific literature. As deep learning does not readily identify the main features involved in the classification, anatomical signatures were unveiled via occlusion sensitivity.31 Our approach systematically identified different regions across heterogeneous testing samples, enhancing model interpretability, and in doing so provided global North vs South generalisations and biological insights on shared brain anatomic differences across diverse populations.
We advanced a mixed hypothesis- and data-driven approach using traditional statistical approaches and deep learning, respectively. First, we hypothesised that the DenseNet binary classifications for bvFTD vs healthy controls (HC), AD vs HC, and bvFTD vs AD would yield high performance in the ADNI and NIFD/LONI databases. Second, we predicted that the generalisation and reproducibility across diverse LAC samples including (a) 3T and (b) 1.5T clinical image samples would be satisfactory. Third, we anticipated that the occlusion sensitivity analysis would confirm specific patterns of atrophy that characterise AD and FTD. By testing these hypotheses, we aimed to assess the robustness of our computer vision framework for characterising neurodegenerative diseases in non-harmonised data that would be scalable and generalisable to non-stereotypic and routine clinical settings.
Methods
Participants
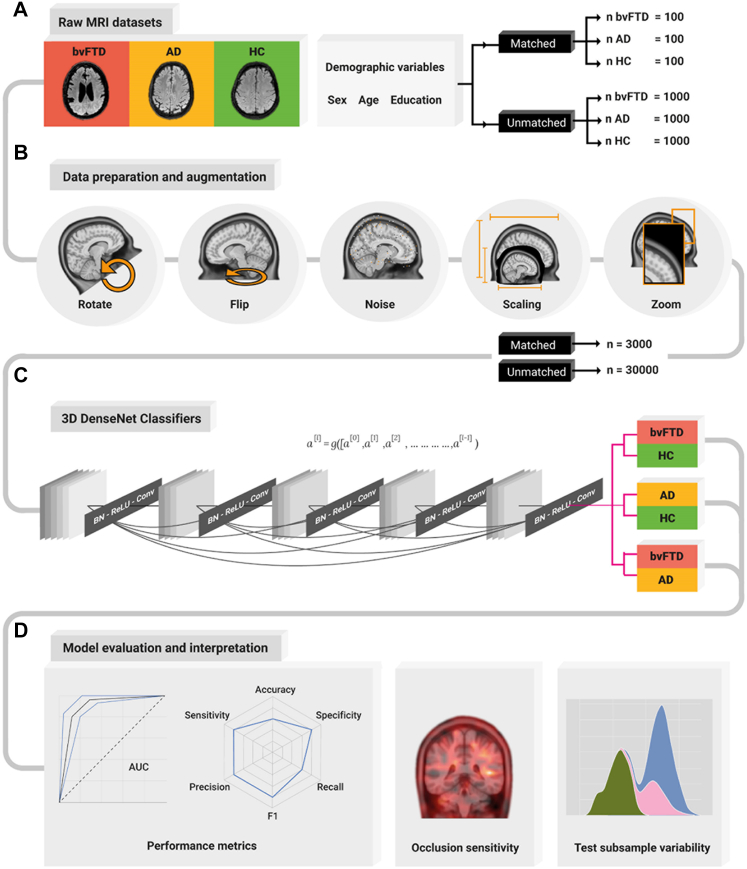
The sample included 3000 MRI scans (n = 1000 individuals with bvFTD, n = 1000 individuals with AD, and n = 1000 HC), without matching for demographic variables (i.e., age, sex, and education), referred onwards as the unmatched sample (Table 2). We also included a demographically matched sample of 100 individuals with bvFTD, 100 with AD, and 100 HC, referred hereafter as the matched sample (Table 2). The rationale behind creating separate samples was to account for the effect of possible demographic differences (i.e., sex, age, and education) that exist on average on patients with AD and with bvFTD. We used random sampling in the unmatched sample (n = 3000, before data augmentation) and stratified random sampling to create the matched sample (n = 300, before data augmentation, to control for age, sex, and education). Unprocessed (raw) 3D structural T1-weighted 3T MRIs were obtained from three online databases (hereafter ONLINE), and a Latin America and the Caribbean database comprising of 3T and 1.5T images (hereafter 3T LAC and 1.5T LAC, respectively). The online database consisted of data from the Alzheimer's Disease Neuroimaging Initiative (ADNI)32 (AD = 273 and HC = 301), the Neuroimaging in Frontotemporal Dementia (NIFD/LONI)11 (bvFTD = 362 and HC = 124), and the UNITED Consortium33 (bvFTD = 538, AD = 627, and HC = 475). The LAC sample included populations that are underrepresented in the scientific literature and was obtained from the Multi-Partner Consortium to Expand Dementia Research in Latin America (ReDLat).34, 35, 36, 37, 38 The LAC dataset comprised of 3T images (bvFTD = 50, AD = 50, and HC = 50) and 1.5 Tesla images from routine clinical assessment (bvFTD = 50, AD = 50, and HC = 50). See Supplementary Data 1 for image acquisition parameter details.
Table 2.
Demographic statistical results for the unmatched and matched samples.
| Variable | HCs US n = 1000 MS n = 100 |
bvFTD US n = 1000 MS n = 100 |
AD US n = 1000 MS n = 100 |
Statistics (all groups) | Post-hoc comparisons |
||
|---|---|---|---|---|---|---|---|
| Groups | p-value | ||||||
| Sex (F:M) | US | 477:523 | 464:536 | 561:439 | χ2 = 22.17, p < 0.05a | bvFTD-AD | 0.002b |
| HCs-bvFTD | n.sb | ||||||
| HCs-AD | 0.001b | ||||||
| MS | 48:52 | 50:50 | 53:47 | χ2 = 0.51, p = 0.77a | bvFTD-AD | n.sb | |
| HCs-bvFTD | n.sb | ||||||
| HCs-AD | n.sb | ||||||
| Age (years) | US | 72.14 (8.24) | 67.85 (6.35) | 78.12 (7.35) | F = 3.14, p = 0.03a, ηp2 = 0.09 | bvFTD-AD | 0.02c |
| HCs-bvFTD | n.sc | ||||||
| HCs-AD | 0.009c | ||||||
| MS | 73.25 (9.32) | 68.96 (11.63) | 75.51 (8.25) | F = 2.61, p = 0.07a, ηp2 = 0.06 | bvFTD-AD | n.sc | |
| HCs-bvFTD | n.sc | ||||||
| HCs-AD | n.sc | ||||||
| Years of education | US | 16.13 (4.75) | 14.96 (4.29) | 13.54 (3.89) | F = 2.44, p = 0.07a, ηp2 = 0.05 | bvFTD-AD | n.sc |
| HCs-bvFTD | n.sc | ||||||
| HCs-AD | n.sc | ||||||
| MS | 15.31 (5.35) | 14.75 (3.12) | 13.09 (4.51) | F = 3.24, p = 0.08a, ηp2 = 0.07 | bvFTD-AD | n.sc | |
| HCs-bvFTD | n.sc | ||||||
| HCs-AD | n.sc | ||||||
Results are presented as mean (SD). Demographic data was assessed through ANOVAs –except for sex, which was analyzed via Pearson's chi-squared (χ2) test. Effects sizes were calculated through partial eta squared (ηp2).
AD: Alzheimer's disease, bvFTD: behavioural variant of fronto-temporal dementia, HCs: healthy controls, MS: Matched sample, US: Unmatched sample.
p-values calculated via independent measures ANOVA.
p-values calculated via chi-squared test (χ2).
p-values calculated via Tukey's range test.
Across samples, clinical diagnoses were established by experts in dementia through an extensive neurological and neuropsychiatric examination comprising semi-structured interviews and standardised assessments, with current criteria for probable bvFTD,39 and NINCDS-ADRDA clinical criteria for AD40 (See Supplementary Data 2 for details). Patients did not present any vascular, psychiatric, or other neurological disorders. The inclusion of healthy subjects required confirmation of normal cognitive function, the absence of any disease, and a brain MRI free of lesions or significant white matter/atrophy changes. The respective IRB of each institution that contributed images to this study approved the acquisitions, and all the participants signed a consent form following the declaration of Helsinki. The methods were performed in accordance with relevant guidelines and regulations and approved by the committee of the ReDLat Multi-Partner Consortium members.34,35
Deep learning methods
Dataset split
The Deep Learning pipeline input were raw (i.e., with no preprocessing steps), 3D structural T1-weighted images of the unmatched sample and the matched sample from individuals with bvFTD, those with AD, and HC (Fig. 1A). Following best practices in deep learning,41 we split the dataset into 80% of the data for training and validation (3T images from the ONLINE database, bvFTD = 800, AD = 800, and HC = 800) and the 20% of the data to create three independent testing datasets: 3T ONLINE (bvFTD = 100, AD = 100, and HC = 100), 3T LAC (bvFTD = 50, AD = 50, and HC = 50), and 1.5T LAC (bvFTD = 50, AD = 50, and HC = 50). During training and validation in 80% of the data, we used k-fold (k = 5) cross validation (see Supplementary Data 3 for model training details). We intentionally created the full training set with the ONLINE dataset to evaluate the extent to which this method could generalise to LAC 3T and LAC 1.5T samples.
Fig. 1.
Deep Learning pipeline. (A) Number of raw MRI datasets employed in the analysis before data augmentation, with and without matching demographic variables (sex, age, and education) for the people with bvFTD, AD and the HC. (B) Data preparation and augmentation pipeline consisting of random volume rotations, random flipping, Gaussian noise addition, volume scaling and enhancing by a zoom transformation. This set of augmentations increased the sample size by a factor of 10. (C) 3D DenseNet network architecture consisting of a sequence of dense blocks and transition layers consisting of a Batch Normalization (BN), a rectified linear unit (ReLU), and a convolution transformation, ending in a prediction layer to produce the output. (D) Model evaluation interpretation, with the performance metrics consisting of the ROC curve and an AUC report, a radar plot showing the accuracy, sensitivity, specificity, precision, recall, and F1 metrics. An occlusion sensitivity analysis to obtain the most relevant parts of the images for the classification, and a test subsample variability analysis to assess sample heterogeneity. AD: Alzheimer's disease; BvFTD: behavioral-variant frontotemporal dementia; HC: healthy controls.
Data preparation and augmentation
Having the raw data as input, we augmented the sample size to increase model performance by training the models with more samples. One of the strengths of Deep Learning is the automatic feature extraction from raw data, increasing the method's flexibility and robustness that allows it to outperform traditional machine learning methods in biology and medicine applications.42 However, this advantage comes at a cost of requiring larger datasets of diverse examples to develop robust and flexible networks. Moreover, standard brain disorders association studies need the order of thousands of participants to obtain reproducible results.29 To bridge this gap, we employed data augmentation30 to increase the number of samples of each dataset 10 times by employing 4 different transformations. For this purpose, we applied random brain volume rotations (minimum degree = 0°, maximum = 180°) and brain volume flipping to increase input variability, Gaussian noise (mean = 0.0, SD = 0.1) incorporation to simulate low-quality acquisitions, and image zoom (1× to 1.5×) to focus on different brain areas each time. Employing this data augmentation process, our sample size increased to a total of 30,000 brain images for the unmatched sample and 3000 brain images for the matched sample. Finally, to normalise the neural network inputs we resized the images (Fig. 1B).
Deep neural network training
We trained three separate deep learning neural networks with only unprocessed MRI data as the input, one for the bvFTD vs HC classification, another for the AD vs HC classification, and a final for the bvFTD vs AD classification. The deep learning neural network employed was a DenseNet26 (121 architecture) adapted for 3D inputs. The DenseNet is a state-of-the-art deep learning convolutional neural network (CNN) employed in computer vision tasks aided by artificial intelligence. In this network architecture, each layer is connected to every other layer in a feed-forward fashion, producing networks that are substantially deeper, more accurate, and more efficient to train (Fig. 1C for the DenseNet diagram). See Supplementary Data 3 for neural network architecture details and Table S2 for model training parameters.
Multifeatured deep neural network training and sociodemographic control
To evaluate the importance of the demographic variables (i.e., sex, age, and education) in the classifications corresponding to the unmatched sample, we retrained the three neural networks (one for each binary classifications) and incorporated these demographic variables as new features. See Supplementary Data 4 for multifeatured neural network architecture details.
Ethics
The respective IRB of each institution that contributed images to this study approved the acquisitions, and all the participants signed a consent form following the declaration of Helsinki. The methods were performed in accordance with relevant guidelines and regulations and approved by the committee of the ReDLat Multi-Partner Consortium members.34,35 The ADNI and LONI data acquisition was conducted according to Good Clinical Practice guidelines, US 21CFR Part 50– Protection of Human Subjects, and Part 56 – Institutional Review Boards (IRBs)/Research Ethics Boards (REBs), and pursuant to state and federal HIPAA regulations. The UNITED consortium data had the approval of each IRB for the acquisitions and followed the declaration of Helsinki for participant consent. The informed consent of all participants was obtained.
Statistics
Sample size determination on dataset split in machine learning followed best practices.41 To obtain unbiased classification performance metrics irrespective of dataset imbalance we employed several estimators, namely the area under the curve (AUC) of the ROC curve, accuracy, sensitivity, specificity, precision, recall and F1. We obtained the confidence intervals by bootstrapping, using random subsamples with replacement on 5000 iterations. To assess statistical validation of our classification results, we assessed the performance by employing nonparametric permutation tests with sample label randomization.43 This allowed to assess the significance of results (FDR corrected) against a null distribution, and compare the ROC curves of different models being evaluated in the same testing dataset each time.44 The equality of the curve is tested at all operating points, and a reference distribution is generated by permuting the pooled ranks of the test scores for each classification. This allowed to statistically assess performance of our models against classification by chance, when training with a matched or an unmatched sample, when testing in different datasets, and when considering or not demographic variables in our unimodal vs multifeatured models. To interpret our model outputs, we ran occlusion sensitivity analyses.31 The occlusion sensitivity analysis is a technique in convolutional neural networks employed to understand what parts of an image are more relevant for deciding a classification output (Fig. 1D). See Supplementary Data 5 for model interpretation details. All neural network pipelines, including data augmentation and occlusion sensitivity analysis, were developed using the PyTorch-based API called Medical Open Network for Artificial Intelligence (MONAI).45 Data can be made available upon request on a case-by-case basis as allowed by the legislation and ethical approvals for a specific project.
Role of funders
The Funders did not have any role in study design, data collection, data analyses, interpretation, or writing of the report. No author has been paid to write this article by a pharmaceutical company or other agency. All authors were not precluded from accessing data in the study, and they accept responsibility to submit for publication.
Results
High classification rate and generalisation in the unmatched sample dataset
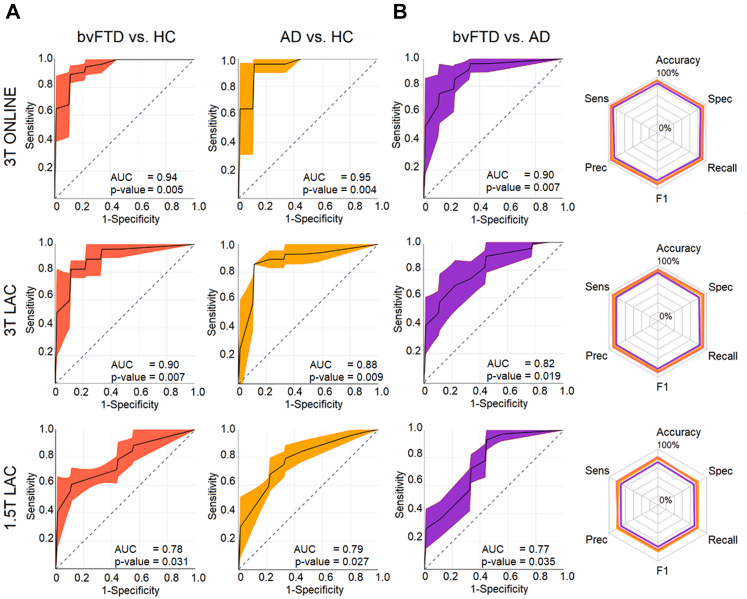
The results using the unmatched sample yielded a robust classification of bvFTD, AD and controls in both 3T dataset subsamples. The scores were highest in the 3T ONLINE testing dataset for the bvFTD vs HC classification (AUC: 0.94 ± 0.04 (p-value = 0.005 [permutation test, FDR corrected]); accuracy: 95% ± 1%; sensitivity: 94 ± 2%; specificity: 95% ± 1%; precision: 94% ± 3%; recall: 95% ± 2%; and F1: 94 ± 2%), for the AD vs HC classification (AUC: 0.95 ± 0.02 (p-value = 0.004 [permutation test, FDR corrected]); accuracy: 94% ± 2%, sensitivity: 96% ± 1%; specificity: 92% ± 2%; precision: 95% ± 2%; recall: 94% ± 1%; and F1: 94% ± 2%), and for the bvFTD vs AD classification (AUC: 0.90 ± 0.01 (p-value = 0.007 [permutation test, FDR corrected]); accuracy: 92% ± 1%; sensitivity: 91% ± 2%; specificity: 93% ± 2%; precision: 89% ± 1%; recall: 90% ± 1%; and F1: 89% ± 1%) (Fig. 2, first row).
Fig. 2.
ROC curves and radar plots for the unmatched sample. (A) ROC curves AUC for the bvFTD vs HC, AD vs HC, and bvFTD vs AD classification for the 3T ONLINE, 3T LAC, and 1.5T LAC datasets. First row: In the 3T ONLINE testing dataset, we obtained an AUC of 0.94 (p-value = 0.005 [permutation test, FDR corrected]), for the bvFTD vs HC classification, an AUC of 0.95 (p-value = 0.004 [permutation test, FDR corrected]) for the AD vs HC classification, and 0.90 (p-value = 0.007 [permutation test, FDR corrected]) for the bvFTD vs AD classification. Second row: In the 3T LAC testing dataset, we obtained an AUC of 0.90 (p-value = 0.007 [permutation test, FDR corrected]) for the bvFTD vs HC classification, an AUC of 0.88 (p-value = 0.009 [permutation test, FDR corrected]) for the AD vs HC classification, and 0.82 (p-value = 0.019 [permutation test, FDR corrected]) for the bvFTD vs AD classification. Third row: Lastly, in the 1.5T LAC testing dataset, we obtained an AUC of 0.78 (p-value = 0.031 [permutation test, FDR corrected]) for the bvFTD vs HC classification, an AUC of 0.79 (p-value = 0.027 [permutation test, FDR corrected]) for the AD vs HC classification, and 0.77 (p-value = 0.035 [permutation test, FDR corrected]) for the bvFTD vs AD classification. (B) Radar plots for the accuracy, sensitivity, specificity, precision, and recall performance metrics including each binary classification results. First row 3T ONLINE, second row 3T LAC, and third row 1.5T LAC. AD: Alzheimer's disease, AUC: Area Under the Curve, bvFTD: behavioral variant of fronto-temporal dementia, HCs: healthy controls, Sens: Sensitivity, Spec: Specificity. Prec: Precision. Sample size: (AD, n = 1000), (bvFTD, n = 1000), and (HC, n = 1000).
The 3T LAC testing dataset followed the ranking of accuracy, yielding good scores for the bvFTD vs HC classification (AUC: 0.90 ± 0.02 (p-value = 0.007 [permutation test, FDR corrected]); accuracy: 91% ± 2%, sensitivity: 91% ± 1%; specificity: 92% ± 2%; precision: 91% ± 1%; recall: 89% ± 2%; and F1: 90% ± 1%), for the AD vs HC classification, AUC: 0.88 ± 0.02 (p-value = 0.009 [permutation test, FDR corrected]); accuracy: 90% ± 2%, sensitivity: 89% ± 2%; specificity: 92% ± 2%; precision: 82% ± 3%; recall: 88% ± 1%; and F1: 85% ± 2%), and for the bvFTD vs AD classification (AUC: 0.82 ± 0.02 (p-value = 0.019 [permutation test, FDR corrected]); accuracy: 93% ± 2%, sensitivity: 93% ± 1%; specificity: 94% ± 1%; precision: 94% ± 1%; recall: 95% ± 1%; and F1: 94% ± 1%) (Fig. 2, second row).
Lastly, the 1.5T LAC testing dataset presented reduced but still relevant classification for the bvFTD vs HC classification (AUC: 0.78 ± 0.03 (p-value = 0.031 [permutation test, FDR corrected]); accuracy: 80% ± 1%, sensitivity: 79% ± 2%; specificity: 81% ± 2%; precision: 78% ± 1%; recall: 79% ± 2%; and F1: 78% ± 2%), for the AD vs HC classification (AUC: 0.79 ± 0.02 (p-value = 0.027 [permutation test, FDR corrected]); accuracy: 77% ± 2%, sensitivity: 77% ± 1%; specificity: 78% ± 2%; precision: 81% ± 1%; recall: 80% ± 2%; and F1: 81% ± 2%), and for the bvFTD vs AD classification (AUC: 0.77 ± 0.04 (p-value = 0.035 [permutation test, FDR corrected]); accuracy: 75% ± 2%, sensitivity: 73% ± 1%; specificity: 77% ± 2%; precision: 79% ± 1%; recall: 82% ± 2%; and F1: 81% ± 1%) (Fig. 2, third row).
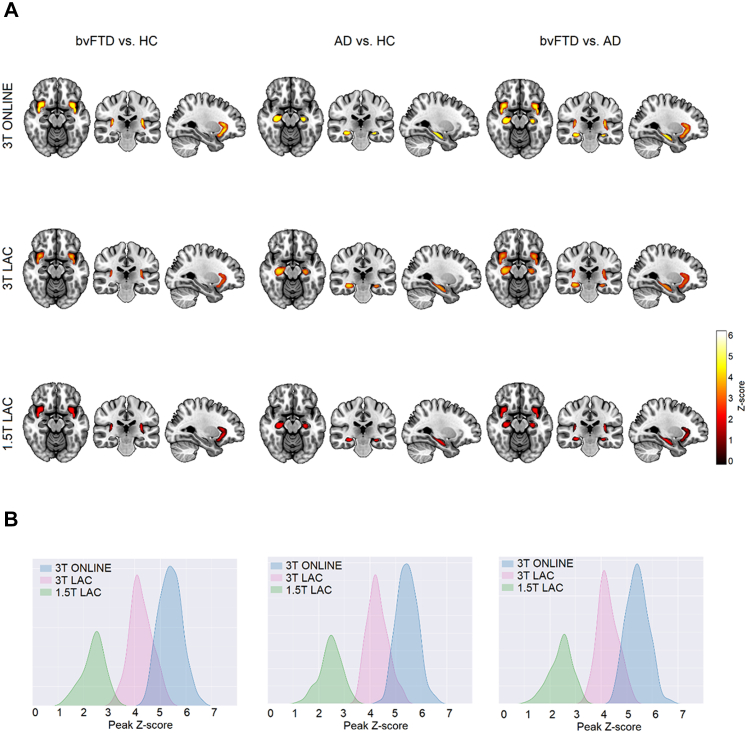
Occlusion sensitivity identifies critical brain regions in AD and bvFTD
The occlusion sensitivity analysis identified the most relevant brain areas for classifying each subject in the DenseNet. We statistically compared occlusion sensitivity maps among subject groups for each classification pair. For the bvFTD vs HC comparison, there were two significant clusters (α = 0.05, extent threshold 50, FDR corrected) with peaks located on the bilateral ventral anterior insula areas at MNI coordinates (−46, 4, −9) for the left hemisphere and (45, 4, −13) for the right hemisphere (Fig. 3A, first column). With respect to the AD vs HC comparison, we obtained significant clusters on bilateral hippocampus with peaks located at (−33, −19, −14) for the left hemisphere and (31, −21, −18) for the right hemisphere (Fig. 3A, second column). Finally, for the bvFTD vs AD comparison, the DenseNet identified both bilateral ventral anterior insula and both bilateral hippocampal areas (Fig. 3A, third column). The MNI coordinates of the peaks for the ventral anterior insula were (−44, 4, −9) for the left hemisphere and (46, 4, −14) for the right hemisphere, and for the hippocampus, (−33, −19, −15) for the left hemisphere and (31, −20, −19) for the right hemisphere.
Fig. 3.
Model interpretation for the unmatched sample. (A) Occlusion sensitivity analysis by normalizing each subject into MNI space and then averaging the results for bvFTD vs HC, AD vs HC, and bvFTD vs AD in the three testing samples (3T ONLINE, 3T LAC, and 1.5 LAC). For bvFTD vs HC, the DenseNet based its decision focusing on bilateral ventral anterior insula areas (first column). While for the AD vs HC comparison, the most relevant areas for the DenseNet bilateral hippocampal areas (second column). Finally, for the bvFTD vs AD comparison, the DenseNet looked on both ventral anterior insula and hippocampal areas (third column). The results were consistent across testing samples. (B) Test subsample variability analysis for the three databases and for the individual occlusion sensitivity maps of each subject, showing peak value dispersion to look for sample heterogeneity. AD: Alzheimer's Disease, bvFTD: behavioral-variant Frontotemporal Dementia, HC: Healthy controls. Sample size: (AD, n = 1000), (bvFTD, n = 1000), and (HC, n = 1000).
To evaluate the test subsample heterogeneity on the three databases we plotted the histograms showing the z-score distribution of the maximum peaks obtained from the individual occlusion sensitivity maps of each sample contained within the significant group comparison clusters. We obtained less variability with higher mean peak value for the 3T ONLINE sample for the three subject group comparisons, followed by a distribution with a lower mean peak value for the 3T LAC sample for the three subject group comparisons, and finally, a wider distribution with lower mean peak value for the 1.5 LAC sample for the three subject group comparisons. However, the differences in distribution variance were not statistically significant (See Supplementary Data 6 and Fig. 3B for details).
Replication approach in the matched sample dataset
When the DenseNet was trained with the matched sample dataset, a robust classification was also obtained in both 3T dataset test subsamples, and with reduced performance (but similar to the unmatched sample) metrics for the 1.5T subsample. Moreover, the occlusion sensitivity analysis replicated the same results on the matched sample. See Supplementary Data 7 for details. Furthermore, we checked the classification performance on an additional testing subsample having negligible age differences between groups. The change in performance was also not significant (p > 0.05 [AUC ROC difference test]), confirming that the model focused on image features unrelated with age differences (Supplementary Data 8).
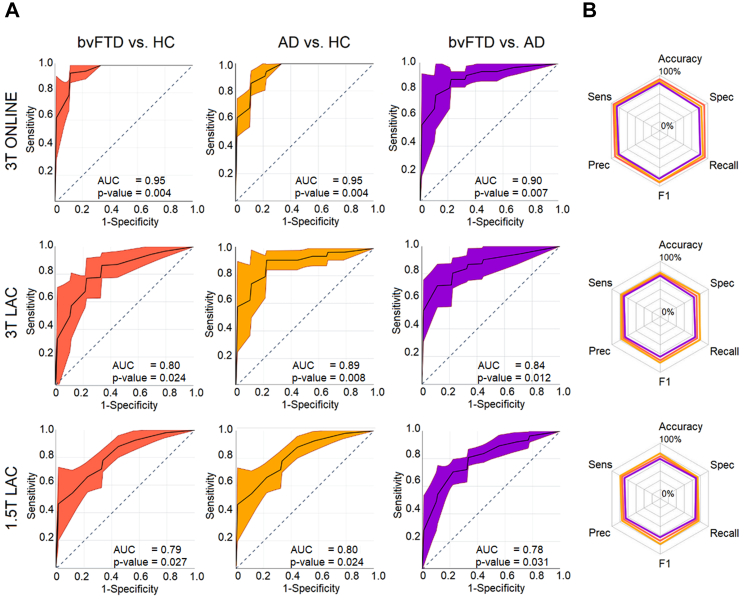
Multi-feature results incorporating sociodemographic data
Lastly, we trained a multi-feature model incorporating sociodemographic data on the unmatched sample. This model yielded high classification results overall on each test subsample dataset (Table 3 and Fig. 4 for details). Differences in classification performance between the multifeatured model and the unimodal unmatched counterpart were not significant (Supplementary Data 9), suggesting that sociodemographic information, although useful, is only a minor contributor for the classification accuracy.
Table 3.
Multifeatured performance metrics.
| Sample | Classification | AUC | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | Recall (%) | F1 (%) |
|---|---|---|---|---|---|---|---|---|
| 3T ONLINE | bvFTD vs HC | 0.95 ± 0.03 | 95 ± 2 | 96 ± 2 | 95 ± 3; | 94 ± 2 | 93 ± 2 | 93 ± 2 |
| AD vs HC | 0.95 ± 0.02 | 94 ± 1 | 93 ± 3 | 95 ± 2 | 93 ± 2 | 94 ± 3 | 94 ± 1 | |
| bvFTD vs AD | 0.90 ± 0.02 | 91 ± 3 | 92 ± 2 | 90 ± 3 | 91 ± 2 | 92 ± 3 | 91 ± 2 | |
| 3T LAC | bvFTD vs HC | 0.90 ± 0.04 | 90 ± 2 | 91 ± 2 | 90 ± 2 | 91 ± 2 | 92 ± 3 | 91 ± 2 |
| AD vs HC | 0.89 ± 0.03 | 90 ± 1 | 90 ± 2 | 89 ± 2 | 88 ± 2 | 90 ± 1 | 89 ± 2 | |
| bvFTD vs AD | 0.84 ± 0.03 | 85 ± 2 | 85 ± 1 | 86 ± 3 | 88 ± 2 | 86 ± 2 | 87 ± 2 | |
| 1.5 LAC | bvFTD vs HC | 0.80 ± 0.02 | 81 ± 2 | 81 ± 2 | 82 ± 2 | 79 ± 1 | 82 ± 2 | 81 ± 2 |
| AD vs HC | 0.80 ± 0.03 | 80 ± 2 | 82 ± 1 | 79 ± 2 | 78 ± 1 | 83 ± 1 | 81 ± 2 | |
| bvFTD vs AD | 0.79 ± 0.05 | 81 ± 2 | 79 ± 2 | 82 ± 2 | 81 ± 1 | 79 ± 3 | 80 ± 2 |
AD: Alzheimer's disease, bvFTD: behavioural variant of fronto-temporal dementia, HC: Healthy control.
Fig. 4.
ROC curves and radar plots for the unmatched sample multifeatured model.(A) AUC results the bvFTD vs HC, AD vs HC, and bvFTD vs AD classification for the 3T ONLINE, 3T LAC, and 1.5T LAC datasets. First row: In the 3T ONLINE testing dataset, we obtained an AUC of 0.95 (p-value = 0.004 [permutation test, FDR corrected]) for the bvFTD vs HC classification, an AUC of 0.95 (p-value = 0.004 [permutation test, FDR corrected]) for the AD vs HC classification, and 0.90 (p-value = 0.007 [permutation test, FDR corrected]) for the bvFTD vs AD classification. Second row: In the 3T LAC testing dataset, we obtained an AUC of 0.80 (p-value = 0.024 [permutation test, FDR corrected]) for the bvFTD vs HC classification, an AUC of 0.89 (p-value = 0.008 [permutation test, FDR corrected]) for the AD vs HC classification, and 0.84 (p-value = 0.012 [permutation test, FDR corrected]) for the bvFTD vs AD classification. Third row: Lastly, in the 1.5T LAC testing dataset, we obtained an AUC of 0.78 (p-value = 0.027 [permutation test, FDR corrected]) for the bvFTD vs HC classification, an AUC of 0.80 (p-value = 0.024 [permutation test, FDR corrected]) for the AD vs HC classification, and 0.78 (p-value = 0.031 [permutation test, FDR corrected]) for the bvFTD vs AD classification. (B) Radar plots for the accuracy, sensitivity, specificity, precision, and recall performance metrics including each binary classification results. First row 3T ONLINE, second row 3T LAC, and third row 1.5T LAC. AD: Alzheimer's disease, AUC: Area Under the Curve, bvFTD: behavioral variant of fronto-temporal dementia, HCs: healthy controls, Prec: Precision, Sens: Sensitivity, Spec: Specificity. Sample size: (AD, n = 1000), (bvFTD, n = 1000), and (HC, n = 1000).
Discussion
We developed a successful computer-vision-based pipeline for dementia subtype characterisation based on unprocessed MRI imaging across diverse samples and scanners. In contrast with previous works (Table 1), we used raw MRI data, assessed of non-stereotypical samples, trained the deep learning with different databases (including low-field scanners), used data augmentation to increase our sample size, classified dementia subtypes instead of just comparing dementia against healthy controls, evaluated the effects of demographic variables that may inflate classification performance, and used occlusion sensitivity for providing model interpretability. The results, combining hypothesis and data-driven approaches quantified the effectiveness of the DenseNet neural network in classifying bvFTD vs HC, AD vs HC, and bvFTD vs AD across heterogeneous testing datasets. Importantly, the model generalised to classify images from geographically diverse settings and even to routine clinical imaging with low-field MRI scanner acquisitions. Our approach proved robust to potential confounding of demographic variables, by constructing separate datasets (i.e., matched and unmatched samples) and also by incorporating the demographic variables in the multifeatured model. Finally, the occlusion sensitivity analysis confirmed (a) the involvement of critical anatomic regions for each disease, and (b) the identification of same regions across models. Both results are biologically plausible and demonstrate generalisation of the classification results. Overall, these set of findings highlight the potential future application of this pipeline in massive, scalable, automatic, and non-harmonised settings for the clinical decision-making process – particularly in the global South where dementia rates will increase rapidly.
A unique aspect of present result is the power to generalise the classification to underrepresented populations and to lower-quality clinical images. The neuropathology of neurodegenerative disorders in high income countries may manifest differently than in other regions due to varied social, cultural, and regional contexts.46,47 The genetic,48 cognitive,49 and brain structural and functional network variability,50,51 together with socioeconomic disparities can induce heterogeneous presentations47 of AD and bvFTD that challenging global approaches to classification. Moreover, the neuroimaging tools employed in LAC are currently limited due to poor MR protocol harmonisation and employment of low-field scanners for routine clinical assessments.52 Against this background, the generalisation and reproducibility across diverse LAC samples including 3T (research-oriented) and 1.5T (clinical-oriented) scans as testing datasets produced adequate results differentiating subject groups. While the performance of the DenseNet was lower in the LAC samples compared to the HIC initiatives, such as ADNI and NIFD/LONI, it was still adequate in terms of predictive values and considering the high heterogeneity in the sample evidenced by our test subsample variability analysis. Furthermore, it allowed to weigh the importance of image quality in the quest for global diagnostic protocols. The test subsample variability analysis showed higher results heterogeneity for the 1.5T LAC database, followed by the 3T LAC, and lastly, the 3T ONLINE database. Even if the classification rate was lower for clinical images, the accuracy level was robust enough to provide complementary probabilistic information to assist with the clinical decision-making process. This work features a deep neural network model bringing reproducible and generalisable results across highly heterogeneous MRI testing databases. This study may prove critical for future scalability and comparability across multiple global datasets.
Demographic characteristics had little effect on overall model performance, thus highlighting the neural network's capability to drive its outputs by focusing on atrophy features alone across heterogenous datasets. This was the case for both the unmatched sample analysis and the replication testing approach with a demographically matched sample. The classification performance in the smaller matched sample was slightly lower but not statistically different from the larger unmatched sample. Also, we assessed the performance of the models on imbalanced testing datasets, to reflect real world scenarios in which the prevalence of AD is higher than the prevalence of bvFTD. Results were not affected by sample imbalance (Supplementary Data 10). The multi-feature model incorporating sociodemographic data in the unmatched sample resulted in slightly higher performance metrics, but again, not reaching statistical significance in its differences with the unimodal counterpart. This is relevant for diagnostic purposes, as real-world clinical data is often demographically heterogeneous.
The present framework provides advantages to overcome traditional limitations when training deep neural networks.23,41 First, data augmentation enabled us to increase the sample size by a factor of ten while adding data variability that is much needed to train deep learning models.29 By employing random geometric transformations, we produced dataset inputs reproducing diverse acquisition parameters. By incorporating Gaussian noise in some training samples, we mimicked noise in low-field acquisitions. Thus, training with this augmented dataset, a more flexible model was able to handle heterogeneous testing datasets from LAC samples as well as 1.5T recordings. Second, unlike traditional deep learning black-box type models,42 occlusion sensitivity provided insights on why the model was producing its outputs, successfully identifying critical brain hubs for bvFTD and AD pathophysiology in the classification process. Finally, unlike a recent study in dementia subtype characterisation based on raw MRI,53 we trained a multi-feature model by concatenating image embeddings with demographic features. The performance improvement in this more comprehensive model was not statistically significant, showing that the DenseNet was classifying the subject groups primarily based on imaging features which are not driven by demographics. An additional feature importance analysis (Fig. S3) evidenced that in the AD vs bvFTD comparison, the demographic factors have a more salient effect, although lower than the brain features. This is expected since AD and bvFTD have important differences in age,54,55 sex54,55 and years of education.54,56 All in all, these pipelines and processes allowed to increase the performance and interpretability.
The occlusion sensitivity analysis identified specific patterns of atrophy that characterise AD and bvFTD. These disease-specific regional peaks were observed across different testing databases comprising heterogeneous populations, acquisition parameters, sample size, and demographic matching. For the detection of people with AD, the DenseNet mainly identified the left and right hippocampus, consistent with several reported source of evidence.4,57 When comparing AD variants, the hippocampal atrophy is more systematic than neocortical affectation which is dependent on AD subtype.4 The hippocampus is a hub early compromised in AD due to tau deposition, facilitating subsequent amyloid pathology spread along the default mode network (DMN)58, 59, 60 (note that 60 is a preprint). Global intensity of tau-PET in fact predicts the rate of subsequent atrophy.5 Moreover, direct associations exists between memory-specific impairments occurring early in the disease and hippocampal atrophy.61 An ADNI-based MRI longitudinal study evidenced hippocampal atrophy as the hallmark signature of progression from mild cognitive impairment (MCI) to AD.57 However, other studies have questioned this finding,62,63 in which sample or preprocessing heterogeneity may have affected the specificity of the results. In our work, results were tested in different samples without using image preprocessing and found reproducible atrophy patterns, although with an expected lower accuracy in non-stereotypical samples from Latin America. In contrast, for bvFTD the DenseNet highlighted the left and right ventral anterior insula. This is a key structure affected in the early-stages of frontotemporal degeneration,13,64, 65, 66, 67, 68, 69, 70, 71, 72 leading to impairments in action self-control.73 In particular, a study showed that the ventral anterior insula was specifically affected in bvFTD, in contrast to the dorsal anterior insula that is mostly affected in non-fluent/agrammatic variant of primary progressive aphasia.74 The insula is a critical functional75 and structural hub76 across compromised networks in bvFTD.2,75,77 Graph theory analysis in insular networks has shown a reduction in global network degree and efficiency that predicts behavioural deficits.78,79 Specific bvFTD deficits in social-emotional abilities has been linked to the insular hub in the salience80 and the allostatic interoceptive networks.2,75,77 The present results, pointing to hippocampus (in AD) and insula (in bvFTD), were completely data-driven, without any a-priori region selection, highlighting the method's robustness to identify core disease-specific pathophysiological regions, and open a new agenda to test this approach in other neurodegenerative and neurological conditions. Future assessments may help to identify contributions at the individual level, given that the approach can help to provide personalised probabilistic diagnosis by inputting new MRI scans into the model.
Limitations and further assessments
Our work features some limitations and opens a new agenda for further research. First, while the sample size was larger than most of previous studies without considering data augmentation,53,81, 82, 83, 84, 85 deep learning models may need even larger amounts of data to further test its generalizability at a more global scale. However, the performance metrics obtained while training with the matched sample (n = 100, without augmentation) and unmatched samples (n = 1000, without augmentation) showed negligible differences. Moreover, results were replicated in different datasets with very heterogenous settings, suggesting that the sample size was robust enough to reproduce relevant patterns across studies. Second, the subject-level classification was made on two neurodegenerative conditions. Further exploration of earlier stages (MCI in AD; prodromal in FTD), cognitive decline and severity, as well other neurodegenerative conditions will provide additional challenging scenarios to test the framework. Third, the multi-feature approach suggests that sociodemographic differences (age, gender, and education), although relevant, are not strong drivers of the classification, making the approach disease-specific and generalisable to non-matched databases. However, future studies should include other related aspects as socioeconomic status, social determinants of health, and ethnicity. Fourth, we employed a standard DenseNet-121 architecture. Future studies may explore architecture modifications to test possible performance improvements. Fifth, although tested samples here replicated similar results across different databases, even more heterogeneous populations may yield classificatory differences in which additional model training would be necessary. Finally, the future assessment of additional relevant imaging signatures (fMRI, DTI, PET) combined with non-imaging biomarkers (plasma, cognition, retina, EEG, others) may be incorporated to create a multi-modal deep learning approach.
Conclusions
A fully automatic deep learning framework was developed based on raw (unpreprocessed) MRI data and tested on non-stereotypical, heterogeneous datasets to test result generalisability. We employed datasets acquired under different MRI fields and parameters, and demographically matched (and unmatched) data to test for possible biases that may affect model performance in out-of-sample validation. Our method produced a result that generalised across North vs South heterogeneous samples. The findings across different testing databases highlight the robustness of the DenseNet and underscores its potential as an architecture to be employed in underrepresented, diverse, and clinical settings, where costly biomarkers are unavailable. Moreover, by employing model interpretability analysis via occlusion sensitivity we confirmed the specific anatomical structures differentiating the affectation in AD and bvFTD. In the future, this approach may be tested as a clinical decision support framework with the aim of providing affordable and personalised dementia assessments.
Contributors
Sebastian Moguilner: conceptualisation; data curation, underlying data review, formal analysis, writing – original draft, writing – review & editing, decision to submit the manuscript. Robert Whelan: supervision, review & editing. Hieab Adams: review & editing. Victor Valcour: review & editing. Enzo Tagliazucchi: underlying data review & editing. Agustín Ibáñez: conceptualisation; supervision; review & editing, decision to submit the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
Data can be made available upon request on a case-by-case basis as allowed by the legislation and ethical approvals for a specific project. All source code and libraries used to process the data and obtain the results, including neural networks pipelines, data augmentation and occlusion sensitivity analysis, are uploaded in the following open science foundation (OSF) link: https://osf.io/nsy7x/
Declaration of interests
Dr Victor Valcour has a grant from the US National Institutes of Health which is not related to this paper. The other authors have no conflicts of interest to declare.
Acknowledgments
S.M. is an Atlantic Fellow for Equity in Brain Health at the Global Brain Health Institute (GBHI) and is supported with funding from GBHI, Alzheimer's Association, and Alzheimer's Society (GBHI ALZ UK-21-721776). A.I. is partially supported by grants of Takeda CW2680521; CONICET; FONCYT-PICT (2017-1818, 2017-1820); ANID/FONDECYT Regular (1210195, 1210176, 1220995); ANID/FONDAP (15150012); ANID/FONDEF (ID20I10152 and ID22I10029), ANID/PIA/ANILLOS ACT210096; and the Multi-Partner Consortium to Expand Dementia Research in Latin America (ReDLat), funded by the National Institutes of Aging of the National Institutes of Health under award number R01AG057234, an Alzheimer's Association grant (SG-20-725707-ReDLat), the Rainwater Foundation, and the GBHI.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104540.
Appendix A. Supplementary data
References
- 1.Mukadam N., Sommerlad A., Huntley J., Livingston G. Population attributable fractions for risk factors for dementia in low-income and middle-income countries: an analysis using cross-sectional survey data. Lancet Glob Health. 2019;7(5):e596–e603. doi: 10.1016/S2214-109X(19)30074-9. [DOI] [PubMed] [Google Scholar]
- 2.Migeot J.A., Duran-Aniotz C.A., Signorelli C.M., Piguet O., Ibanez A. A predictive coding framework of allostatic-interoceptive overload in frontotemporal dementia. Trends Neurosci. 2022;45(11):838–853. doi: 10.1016/j.tins.2022.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olney N.T., Spina S., Miller B.L. Frontotemporal dementia. Neurol Clin. 2017;35(2):339–374. doi: 10.1016/j.ncl.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Migliaccio R., Agosta F., Possin K.L., et al. Mapping the progression of atrophy in early- and late-onset Alzheimer's disease. J Alzheimers Dis. 2015;46(2):351–364. doi: 10.3233/JAD-142292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Joie R., Visani A.V., Baker S.L., et al. Prospective longitudinal atrophy in Alzheimer's disease correlates with the intensity and topography of baseline tau-PET. Sci Transl Med. 2020;12(524):5732. doi: 10.1126/scitranslmed.aau5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene A.S., Shen X., Noble S., et al. Brain-phenotype models fail for individuals who defy sample stereotypes. Nature. 2022;609(7925):109–118. doi: 10.1038/s41586-022-05118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parra M.A., Baez S., Sedeño L., et al. Dementia in Latin America: paving the way toward a regional action plan. Alzheimers Dement. 2021;17(2):295–313. doi: 10.1002/alz.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parra M.A., Baez S., Allegri R., et al. Dementia in Latin America: assessing the present and envisioning the future. Neurology. 2018;90(5):222–231. doi: 10.1212/WNL.0000000000004897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santamaria-Garcia H., Moguilner S., Rodriguez-Villagra O.A., et al. The impacts of social determinants of health and cardiometabolic factors on cognitive and functional aging in Colombian underserved populations. Geroscience. 2023 doi: 10.1007/s11357-023-00755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen R.C., Aisen P.S., Beckett L.A., et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NIFD is the nickname for the frontotemporal lobar degeneration neuroimaging initiative (FTLDNI A, which was funded by the NIA and NINDS to characterize longitudinal clinical and imaging changes in FTLD.
- 12.Jovicich J., Barkhof F., Babiloni C., Herholz K., Mulert C. Harmonization of neuroimaging biomarkers for neurodegenerative diseases: a survey in the imaging community of perceived barriers and suggested actions. Alzheimers Dement. 2019;11(1):69–73. doi: 10.1016/j.dadm.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedeno L., Piguet O., Abrevaya S., et al. Tackling variability: a multicenter study to provide a gold-standard network approach for frontotemporal dementia. Hum Brain Mapp. 2017;38(8):3804–3822. doi: 10.1002/hbm.23627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odusami M., Maskeliūnas R., Damaševičius R. An intelligent system for early recognition of Alzheimer's disease using neuroimaging. Sensors. 2022;22(3):740. doi: 10.3390/s22030740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razzak I., Naz S., Ashraf A., Khalifa F., Bouadjenek M.R., Mumtaz S. Mutliresolutional ensemble PartialNet for Alzheimer detection using magnetic resonance imaging data. Int J Intell Syst. 2022;2022(37):6613–6630. doi: 10.1002/int.22856. [DOI] [Google Scholar]
- 16.Di Benedetto M., Carrara F., Tafuri B., et al. Deep networks for behavioral variant frontotemporal dementia identification from multiple acquisition sources. Comput Biol Med. 2022;148:105937. doi: 10.1016/j.compbiomed.2022.105937. [DOI] [PubMed] [Google Scholar]
- 17.Hosseini-Asl E., Ghazal M., Mahmoud A., et al. Alzheimer's disease diagnostics by a 3D deeply supervised adaptable convolutional network. Front Biosci (Landmark Ed) 2018;23(3):584–596. doi: 10.2741/4606. [DOI] [PubMed] [Google Scholar]
- 18.Qiu S., Joshi P.S., Miller M.I., et al. Development and validation of an interpretable deep learning framework for Alzheimer's disease classification. Brain. 2020;143(6):1920–1933. doi: 10.1093/brain/awaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amini M., Pedram M., Moradi A., Ouchani M. Diagnosis of Alzheimer's disease severity with fMRI images using robust multitask feature extraction method and convolutional neural network (CNN) Comput Math Methods Med. 2021;2021:5514839. doi: 10.1155/2021/5514839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Zeng B., Gao A., Feng X., Liang D., Long X. A 3D densely connected convolution neural network with connection-wise attention mechanism for Alzheimer's disease classification. Magn Reson Imaging. 2021;78:119–126. doi: 10.1016/j.mri.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Feng W., Halm-Lutterodt N.V., Tang H., et al. Automated MRI-based deep learning model for detection of Alzheimer's disease process. Int J Neural Syst. 2020;30:2050032. doi: 10.1142/S012906572050032X. [DOI] [PubMed] [Google Scholar]
- 22.Shen D., Wu G., Suk H. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19(1):221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen M., Verspoor K., Jenkinson M., Law M., Abbott D.F., Jackson G.D. Artificial intelligence for clinical decision support in neurology. Brain Commun. 2020;2(2):96. doi: 10.1093/braincomms/fcaa096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huys Q.J., Maia T.V., Frank M.J. Computational psychiatry as a bridge from neuroscience to clinical applications. Nat Neurosci. 2016;19(3):404. doi: 10.1038/nn.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed M.R., Zhang Y., Feng Z., Lo B., Inan O.T., Liao H. Neuroimaging and machine learning for dementia diagnosis: recent advancements and future prospects. IEEE Rev Biomed Eng. 2019;12(1):19–33. doi: 10.1109/RBME.2018.2886237. [DOI] [PubMed] [Google Scholar]
- 26.Huang G., Liu Z., Van der Maaten L., Weinberger K. IEEE Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks. CVPR 2017. [Google Scholar]
- 27.Moguilner S., Ibanez A. 2023. Deep learning classification based on raw MRI images. Chapter preprint. [DOI] [Google Scholar]
- 28.Remedios L.W., Lingam S., Remedios S.W., et al. Comparison of convolutional neural networks for detecting large vessel occlusion on computed tomography angiography. Med Phys. 2021;48(10):6060–6068. doi: 10.1002/mp.15122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marek S., Tervo-Clemmens B., Calabro F.J., et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603(7902):654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chlap P., Min H., Vandenberg N., Dowling J., Holloway L., Haworth A. A review of medical image data augmentation techniques for deep learning applications. J Med Imaging Radiat Oncol. 2021;65:545–563. doi: 10.1111/1754-9485.13261. [DOI] [PubMed] [Google Scholar]
- 31.Dyrba M., Hanzig M., Altenstein S., et al. Improving 3D convolutional neural network comprehensibility via interactive visualization of relevance maps: evaluation in Alzheimer's disease. Alzheimers Res Ther. 2021;13(1):191. doi: 10.1186/s13195-021-00924-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber C.J., Carrillo M.C., Jagust W., et al. The worldwide Alzheimer's Disease Neuroimaging Initiative: ADNI-3 updates and global perspectives. Alzheimers Dement. 2021;7(1):122. doi: 10.1002/trc2.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams H., Evans T.E., Terzikhan N. The uncovering neurodegenerative insights through ethnic diversity consortium. Lancet Neurol. 2019;18(10):915. doi: 10.1016/S1474-4422(19)30324-2. [DOI] [PubMed] [Google Scholar]
- 34.Ibanez A., Yokoyama J.S., Possin K.L., et al. The multi-partner consortium to expand dementia research in Latin America (ReDLat): driving multicentric research and implementation science. Front Neurol. 2021;12:631722. doi: 10.3389/fneur.2021.631722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibanez A., Parra M.A., Butlerfor C., Latin America and the Caribbean Consortium on Dementia (LAC-CD) The Latin America and the Caribbean Consortium on Dementia (LAC-CD): from networking to research to implementation science. J Alzheimers Dis. 2021;82(s1):S379–S394. doi: 10.3233/JAD-201384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maito M.A., Santamaría-García H., Moguilner S., et al. Classification of Alzheimer's disease and frontotemporal dementia using routine clinical and cognitive measures across multicentric underrepresented samples: a cross sectional observational study. Lancet Reg Health Am. 2023;17:100387. doi: 10.1016/j.lana.2022.100387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herzog R., Rosas F.E., Whelan R., et al. Genuine high-order interactions in brain networks and neurodegeneration. Neurobiol Dis. 2022;175:105918. doi: 10.1016/j.nbd.2022.105918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prado P., Moguilner S., Mejía J.A., et al. Source space connectomics of neurodegeneration: one-metric approach does not fit all. Neurobiol Dis. 2023;179:106047. doi: 10.1016/j.nbd.2023.106047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rascovsky K., Salmon D.P., Lipton A.M., et al. Rate of progression differs in frontotemporal dementia and Alzheimer disease. Neurology. 2005;65(3):397–403. doi: 10.1212/01.wnl.0000171343.43314.6e. [DOI] [PubMed] [Google Scholar]
- 40.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association Workgroups on Diagnostic Guidelines for Alzheimer's Disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poldrack R.A., Baker C.I., Durnez J., et al. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat Rev Neurosci. 2017;18(2):115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ching T., Himmelstein D.S., Beaulieu-Jones B.K., et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15(141):2017. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venkatraman E.S. A permutation test to compare receiver operating characteristic curves. Biometrics. 2000;56:1134–1138. doi: 10.1111/j.0006-341x.2000.01134.x. [DOI] [PubMed] [Google Scholar]
- 44.Mason S.J., Graham N.E. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: statistical significance and interpretation. Q J R Meteorol Soc. 2002;128:2145–2166. [Google Scholar]
- 45.Alyward Sea, MONAI Consortium MONAI: medical open network for AI, Zenodo, 2022. 2020. https://monai.medium.com/about
- 46.Alladi S., Hachinski V. World dementia: one approach does not fit all. Neurology. 2018;91(6):264–270. doi: 10.1212/WNL.0000000000005941. [DOI] [PubMed] [Google Scholar]
- 47.Ibanez A. The mind's golden cage and cognition in the wild. Trends Cogn Sci. 2022;26(12):1031–1034. doi: 10.1016/j.tics.2022.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrari R., Hernandez D.G., Nalls M.A., et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 2014;13(7):686–699. doi: 10.1016/S1474-4422(14)70065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye B.S., Choi S.H., Han S.H., et al. Clinical and neuropsychological comparisons of early-onset versus late-onset frontotemporal dementia: a CREDOS-FTD study. J Alzheimers Dis. 2015;45(2):599–608. doi: 10.3233/JAD-141044. [DOI] [PubMed] [Google Scholar]
- 50.Noh Y., Jeon S., Lee J.M., et al. Anatomical heterogeneity of Alzheimer disease: based on cortical thickness on MRIs. Neurology. 2014;83(21):1936–1944. doi: 10.1212/WNL.0000000000001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ossenkoppele R., Pijnenburg Y.A., Perry D.C., et al. The behavioural/dysexecutive variant of Alzheimer's disease: clinical, neuroimaging and pathological features. Brain. 2015;138(Pt 9):2732–2749. doi: 10.1093/brain/awv191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arbabshirani M.R., Plis S., Sui J., Calhoun V.D. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. Neuroimage. 2017;145:137–165. doi: 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J., Qing Z., Liu R., et al. Deep learning-based classification and voxel-based visualization of frontotemporal dementia and Alzheimer's disease. Front Neurosci. 2021;14(626):154. doi: 10.3389/fnins.2020.626154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bang J., Spina S., Miller B.L. Frontotemporal dementia. Lancet. 2015;386(10004):1672–1682. doi: 10.1016/S0140-6736(15)00461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rascovsky K., Hodges J.R., Knopman D., et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sattler C., Toro P., Schönknecht P., Schroder J. Cognitive activity, education and socioeconomic status as preventive factors for mild cognitive impairment and Alzheimer's disease. Psychiatry Res. 2012;196(1):90–95. doi: 10.1016/j.psychres.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Frankó E., Joly O. Evaluating Alzheimer's disease progression using rate of regional hippocampal atrophy. PLoS One. 2013;8(8):71354. doi: 10.1371/journal.pone.0071354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu M., Sporns O., Saykin A.J. The human connectome in Alzheimer disease - relationship to biomarkers and genetics. Nat Rev Neurol. 2021;17(9):545–563. doi: 10.1038/s41582-021-00529-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanz Perl Y., Zamora-Lopez G., Montbrió E., et al. The impact of regional heterogeneity in whole-brain dynamics in the presence of oscillations. Netw Neurosci. 2022:1–42. doi: 10.1162/netn_a_00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanz Perl Y., Fittipaldi S., Gonzalez Campo C., et al. Model-based whole-brain perturbational landscape of neurodegenerative diseases. bioRxiv. 2022 doi: 10.1101/2022.09.26.509612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritter A., Hawley N., Banks S.J., Miller J.B. The association between Montreal cognitive assessment memory scores and hippocampal volume in a neurodegenerative disease sample. J Alzheimers Dis. 2017;58(3):695–699. doi: 10.3233/JAD-161241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dubois B., Feldman H.H., Jacova C., et al. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol. 2014;13(6):614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 63.Ferreira D., Pereira J.B., Volpe G., Westman E. Subtypes of Alzheimer's disease display distinct network abnormalities extending beyond their pattern of brain atrophy. Front Neurol. 2019;10:524. doi: 10.3389/fneur.2019.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santamaría-García H., Ogonowsky N., Baez S., et al. Neurocognitive patterns across genetic levels in behavioral variant frontotemporal dementia: a multiple single cases study. BMC Neurol. 2022;22(1):454. doi: 10.1186/s12883-022-02954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gonzalez-Gomez R., Ibanez A., Moguilner S., Initiative F.L.D.N. Multiclass characterization of frontotemporal dementia variants via multimodal brain network computational inference. Netw Neurosci. 2023;7(1):322–350. doi: 10.1162/netn_a_00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ibáñez A. Brain oscillations, inhibition and social inappropriateness in frontotemporal degeneration. Brain. 2018;141(10):e73. doi: 10.1093/brain/awy233. [DOI] [PubMed] [Google Scholar]
- 67.Baez S., Kanske P., Matallana D., et al. Integration of intention and outcome for moral judgment in frontotemporal dementia: brain structural signatures. Neurodegener Dis. 2016;16(3-4):206–217. doi: 10.1159/000441918. [DOI] [PubMed] [Google Scholar]
- 68.Baez S., Manes F., Huepe D., et al. Primary empathy deficits in frontotemporal dementia. Front Aging Neurosci. 2014;6:262. doi: 10.3389/fnagi.2014.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ibañez A., Manes F. Contextual social cognition and the behavioral variant of frontotemporal dementia. Neurology. 2012;78(17):1354–1362. doi: 10.1212/WNL.0b013e3182518375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ibanez A., Billeke P., de la Fuente L., Salamone P., Garcia A.M., Melloni M. Reply: towards a neurocomputational account of social dysfunction in neurodegenerative disease. Brain. 2017;140(3):e15. doi: 10.1093/brain/aww316. [DOI] [PubMed] [Google Scholar]
- 71.Legaz A., Abrevaya S., Dottori M., et al. Multimodal mechanisms of human socially reinforced learning across neurodegenerative diseases. Brain. 2022;145(3):1052–1068. doi: 10.1093/brain/awab345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ibanez A., Schulte M. Situated minds: conceptual and emotional blending in neurodegeneration and beyond. Brain. 2020;143(12):3523–3525. doi: 10.1093/brain/awaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seeley W. Anterior insula degeneration in frontotemporal dementia. Brain Struct Funct. 2010;214(5):465–475. doi: 10.1007/s00429-010-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mandelli M.L., Vitali P., Santos M., et al. Two insular regions are differentially involved in behavioral variant FTD and nonfluent/agrammatic variant PPA. Cortex. 2016;74(1):149–157. doi: 10.1016/j.cortex.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Birba A., Santamaría-García H., Prado P., et al. Allostatic-interoceptive overload in frontotemporal dementia. Biol Psychiatry. 2022;92(1):54–67. doi: 10.1016/j.biopsych.2022.02.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mahoney C.J., Simpson I.J.A., Nicholas J.M., et al. Longitudinal diffusion tensor imaging in frontotemporal dementia. Ann Neurol. 2015;77(1):33–46. doi: 10.1002/ana.24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salamone P.C., Legaz A., Sedeño L., et al. Interoception primes emotional processing: multimodal evidence from neurodegeneration. J Neurosci. 2021;41(19):4276–4292. doi: 10.1523/JNEUROSCI.2578-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filippi M., Basaia S., Canu E., et al. Brain network connectivity differs in early-onset neurodegenerative dementia. Neurology. 2017;89(1):1764–1772. doi: 10.1212/WNL.0000000000004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sedeño L., Couto B., García-Cordero I., et al. Brain network organization and social executive performance in frontotemporal dementia. J Int Neuropsychol Soc. 2016;22(1):250–262. doi: 10.1017/S1355617715000703. [DOI] [PubMed] [Google Scholar]
- 80.Seeley W.W. The salience network: a neural system for perceiving and responding to homeostatic demands. J Neurosci. 2019;39:9878–9882. doi: 10.1523/JNEUROSCI.1138-17.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cohen D.S., Carpenter K.A., Jarrell J.T., Huang X., Alzheimer’s Disease Neuroimaging Initiative Deep learning-based classification of multi-categorical Alzheimer's disease data. Curr Neurobiol. 2019;10(3):141–147. [PMC free article] [PubMed] [Google Scholar]
- 82.Iizuka T., Fukasawa M., Kameyama M. Deep-learning-based imaging-classification identified cingulate island sign in dementia with Lewy bodies. Sci Rep. 2019;9(1):8944. doi: 10.1038/s41598-019-45415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jo T., Nho K., Saykin A.J. Deep learning in Alzheimer's disease: diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci. 2019;11:220. doi: 10.3389/fnagi.2019.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oh K., Chung Y.C., Kim K.W., Kim W.S., Oh I.S. Classification and visualization of Alzheimer's disease using volumetric convolutional neural network and transfer learning. Sci Rep. 2019;9(1):18150. doi: 10.1038/s41598-019-54548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vieira S., Pinaya W.H., Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: methods and applications. Neurosci Biobehav Rev. 2017;74(Pt A):58–75. doi: 10.1016/j.neubiorev.2017.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.