Abstract
Campylobacter jejuni (C. jejuni) is the most common food-borne pathogen that causes human gastroenteritis in the United States. Consumption of contaminated poultry products is considered as the major source of human Campylobacter infection. An effective vaccine would be a promising alternative to antibiotic supplements to curb C. jejuni colonization in poultry gastrointestinal (GI) tract. However, the genetic diversity among the C. jejuni isolates makes vaccine production more challenging. Despite many attempts, an effective Campylobacter vaccine is not yet available. This study aimed to identify suitable candidates to develop a subunit vaccine against C. jejuni, which could reduce colonization in the GI tract of the poultry. In the current study, 4 C. jejuni strains were isolated from retail chicken meat and poultry litter samples and their genomes were sequenced utilizing next-generation sequencing technology. The genomic sequences of C. jejuni strains were screened to identify potential antigens utilizing the reverse vaccinology approach. In silico genome analysis predicted 3 conserved potential vaccine candidates (phospholipase A [PldA], TonB dependent vitamin B12 transporter [BtuB], and cytolethal distending toxin subunit B [CdtB]) suitable for the development of a vaccine. Furthermore, the expression of predicted genes during host-pathogen interaction was analyzed by an infection study using an avian macrophage-like immortalized cell line (HD11). The HD11 was infected with C. jejuni strains, and the RT-qPCR assay was performed to determine the expression of the predicted genes. The expression difference was analyzed using ΔΔCt methods. The results indicate that all 3 predicted genes, PldA, BtuB, and CdtB, were upregulated in 4 tested C. jejuni strains irrespective of their sources of isolation. In conclusion, in silico prediction and gene expression analysis during host-pathogen interactions identified 3 potential vaccine candidates for C. jejuni.
Key words: reverse vaccinology, Campylobacter jejuni; host-pathogen interaction; RT-qPCR; poultry
INTRODUCTION
Food-borne illness caused by Campylobacter is one of the major causes of human gastroenteritis (Kaakoush et al., 2015). Epidemiologic studies suggest that the majority of human infections are related to Campylobacter jejuni (C. jejuni) (Suzuki and Yamamoto, 2009; Skarp et al., 2016). In the United States, 1.3 million people are infected with Campylobacter annually, and the economic impact due to disease outbreaks is predicted to be around $6.9 billion annually (Tack et al., 2019; Scharff, 2020). Approximately half of food-borne Campylobacter outbreaks in human are associated with the consumption of contaminated poultry products (Friedman et al., 2004; Wilson et al., 2008; EFSA, 2011; Tack et al., 2019). Campylobacter colonizes up to 6 to 10 log10 CFU/g in GI tract of broilers (Sahin et al., 2001; Newell and Fearnley, 2003; Singh Dhillon et al., 2006) and their colonization does not cause any harm to birds (Beery et al., 1988; Gormley et al., 2014). This makes poultry a major reservoir host and the primary source of Campylobacter infection in humans (Fravalo et al., 2021). The quantitative risk assessment model by Romero-Barrios et al. (2013) had estimated that a 3 log10 CFU/g C. jejuni reduction in the cecal content of broiler could reduce the risk of human infection from consumption of chicken meat by 100%. The conventional approach of controlling pathogens utilizing antimicrobial products has become less effective due to widespread antibiotic resistance. Therefore, there is a critical need for an alternative intervention strategy to curb C. jejuni colonization in poultry to minimize human infection. The development of an effective vaccine might be a promising alternative for reducing C. jejuni colonization in poultry.
Previously, multiple strategies have been employed to develop a vaccine against C. jejuni. However, there was variation in the protection between experiments and limited cross-protection against heterogeneous challenge conditions were observed in the previously tested vaccine candidates (Ziprin et al., 2002; Wyszyńska et al., 2004; Neal-McKinney et al., 2014; Chintoan-Uta et al., 2015; Hodgins et al., 2015; Chintoan-Uta et al., 2016; Nothaft et al., 2016). Despite many attempts, an effective Campylobacter vaccine which can reduce colonization in ceca of poultry during heterogeneous challenge condition remains elusive. In order to tackle this problem, we utilized reverse vaccinology (RV) technology, a novel and emerging vaccine development strategy (Rappuoli, 2000), along with a host-pathogen interaction model via cell infection to identify conserved antigens among C. jejuni and Campylobacter coli, which can be potentially developed into vaccines. RV uses genomic sequences to identify potential antigens for vaccine production in silico (Sette and Rappuoli, 2010), reducing both the time and cost of vaccine development. This technique has been previously used to predict potential antigen candidates effectively for several pathogens (Staphylococcus aureus, Pseudomonas aeruginosa, Helicobacter pylori, Brucella melitensis) (Oprea and Antohe, 2013; Talukdar et al., 2014; Naz et al., 2015; Rashid et al., 2017; Vishnu et al., 2017) and developed a vaccine against Neisseria meningitidis serogroup B (Rappuoli, 2000).
Currently, 3 studies have utilized RV to predict potential vaccine candidates against C. jejuni (Meunier et al., 2016; Jain et al., 2019; Gupta and Kumar, 2020). Two of them utilized C. jejuni strain NCTC 11168 (human isolate) to predict the epitope-based vaccine intended to prevent human infection (Jain et al., 2019; Gupta and Kumar, 2020); however, selected candidates were not subjected to the development of the vaccine. Additionally, Meunier et al. (2016) intended to develop an avian C. jejuni vaccine using C. jejuni strain 81 to 176 (human isolate) to predict antigenic proteins and was tested in the broiler; but, the tested candidates were unable to provide the desired protection. Utilizing human isolates to predict the vaccine candidates for poultry might be less effective due to the genomic variation and low homology in the proteins among the clinical human isolates and chicken isolates (Epping et al., 2021; Truccollo et al., 2021; Audu et al., 2022). Thus, the aim of this study was to develop an effective vaccine that can reduce colonization of C. jejuni in ceca of poultry to reduce human food-borne illness. The objective was to predict potential C. jejuni vaccine candidates by utilizing the information of genome sequences of multiple C. jejuni strains isolated from the different sources of broiler chicken via in silico analysis and further analyze the expression of candidate genes during host-pathogen interactions to determine the pathogenic role of the predicted antigens.
MATERIALS AND METHODS
C. jejuni Genome Sequence and Protein Prediction
In this study, we utilized 4 C. jejuni genome sequences, which we had previously isolated from various poultry sources (Poudel et al., 2022a) and sequenced by Illumina and Oxford Nanopore techniques (Poudel et al., 2022b). Among them, 3 C. jejuni strains (MS2005, MS2058, and MS2074) were isolated from retail meat samples in Mississippi, and strain MS2167 was isolated from broiler feces. The information of genome sequences of 4 C. jejuni used in this study was available in NCBI GenBank (CP084080, CP084082, CP084084, and CP084085 (Poudel et al., 2022b).
Antigenic Protein Prediction
Vaxign (http://www.violinet.org/vaxign/; He et al., 2010) was used to find the subcellular localization, transmembrane helices, and adhesion probability of all predicted proteins for each strain. Vaxign utilizes pSORTb 2.0 to predict subcellular localization (Gardy et al., 2005), optimized HMMTOP based on a general hidden Markov model to predict transmembrane helix topology (Kall et al., 2007), and SPAAN to calculate the adhesion probability of protein (Sachdeva et al., 2005).
The proteins were filtered by the following criteria: 1) surface exposed protein either present in the outer membrane or extracellular matrix, 2) adhesion probability score ≥ 0.5, and 3) no more than one transmembrane helixes (Monterrubio-López et al., 2015; Meunier et al., 2016). The surface-exposed proteins were selected because the host immune system targets these proteins to recognize and expel the pathogen. Adhesion probability of protein was considered an important factor for the vaccine candidate selection. During the bacterial invasion, the adhesion structure encounters the host immune system, and immunity against the adhesion structure helps to prevent further colonization and infection. Protein sequences having multiple transmembrane helixes were removed because they are difficult to express and purify (Monterrubio-López et al., 2015).
VaxiJen v2.0 (http://www.ddgpharmfac.net/VaxiJen/VaxiJen/VaxiJen.html; Doytchinova and Flower, 2007) was used to predict the antigenicity of the filtered proteins. VaxiJen software, based on RV, utilized a machine learning approach to predict antigenic protein sequence. This software utilizes the physicochemical properties to predict protein's antigenicity from the submitted amino acid sequences (Dalsass et al., 2019). A protein with an antigen score of ≥ 0.5 was selected for further analysis (Doytchinova and Flower, 2007). Proteins with an antigen score ≥ 0.5 are more likely recognized by the host immune system, making them potential vaccine candidates (Doytchinova and Flower, 2007; Monterrubio-Lopez et al., 2015), and were selected for further analysis.
Protein Conservation Prediction
To select vaccine candidates with the broadest spectrum, the predicted antigen proteins were aligned to genomes of 206 C. jejuni strains (including 18 C. jejuni from poultry strains; Supplemental Table S1), 18 C. jejuni strains isolated only from poultry (Supplemental Table S2), and 34 C. coli strains (Supplemental Table S3) available from NCBI using tblastn from BLAST+ v 2.2.31 (Camacho et al., 2009) via Geneious prime v2020.2.2 (Integrated Bioinformatic Solution, Auckland, North Island, New Zealand). Proteins with a pairwise identity ≥50% and minimum query coverage ≥ 80% were considered highly conserved and kept for further analysis. In addition, the candidate proteins were blast against Gallus gallus (taxid:9031) protein database from NCBI (https://blast.ncbi.nlm.nih.gov/) using blastp to identify any candidate proteins too similar (pairwise identity ≥80%) to a host protein.
Prediction of B-Epitope
BCPreds (http://ailab.ist.psu.edu/bcpred/) was used to predict B-cell epitopes for the selected candidate proteins. B-cell epitopes were selected utilizing 2 different algorithms, the amino acid pair method and string kernels. BCPreds predict antigenic linear nonoverlapping epitopes from the sequence of preselected amino acid sequences. All the preselected protein sequences from VaxiJen were analyzed, and epitopes with a score of > 0.8 (specificity > 80%) were considered B-epitope. In order to test the individual antigenicity of selected epitomes, they were further accessed utilizing VaxiJen v2.0 software. Epitope sequences having antigenic scores >0.5 were selected, and B-epitope density was calculated utilizing antigenic predicted epitopes (bcpreds score > 0.8 and VaxiJen >0.5) for the preselected protein sequence.
Selection of Vaccine Candidates
In order to remove the predicted homologous protein among 4 different C. jejuni strains (MS2005, MS2058, MS2074, and MS2167) and identify the previously tested vaccine candidate multiple alignment was done. A multiple alignment tool MAFFT (Multiple Alignment using Fast Fourier Transform) was utilized to align the predicted protein sequence obtained from the C. jejuni strains using Geneious prime v2020.2.2 (Integrated Bioinformatic Solution, Auckland, North Island, New Zealand). Protein sequences with >80% pairwise identity and >80% minimum query coverage were categorized as homologous protein sequence. Blastp analysis was performed to identify the name and function of the homologous proteins with a reference protein database of NCBI database (https://blast.ncbi.nlm.nih.gov/) with selected organism bacteria so that previously characterized and tested vaccine candidates can be removed from the final selection.
Gene Expression Analysis During Host-Pathogen Interaction
The expression analysis of the predicted gene candidates during host-pathogen interaction provides the crucial information about the role of candidate genes during infection. The expression levels of those candidate genes might be associated with the efficacy of developed vaccine. Therefore, the expression analysis of predicted potential antigenic vaccine candidates from in silico was performed during the host-pathogen interaction assay, utilizing following methods.
Bacterial Strain Selection and Culture Preparation
For the host-pathogen interaction assay, we selected 4 different C. jejuni strains, including MS2191, MS2074, ATCC29428, and ATCC33560, which were previously isolated from 4 different sources (broiler cloacal swab, retail chicken meat, human clinical, and bovine feces isolate, respectively; Poudel et al., 2022a) to test how the predicted candidate genes express in different strains of C. jejuni isolated from different sources during host-pathogen interaction. For the bacterial growth, the stock of the C. jejuni strains was cultured in double-strength blood-free Bolton broth (2 × BF-BB) (Oxoid, ThermoFisher Scientific, Waltham, MA) at 42°C for 48 h under microaerobic conditions (85% nitrogen, 10% carbon dioxide, and 5% oxygen) using Mart anaerobic jar with an Anoxomat II System (Mart Microbiology B. V., Lichtenvoorde, Oost Gelre, Netherlands). For the infection of the cells, the bacterial cell pellet was resuspended in Advanced Dulbecco's Modified Eagle Medium (Gibco Advanced DMEM, ThermoFisher Scientific, Grand Island, NY), supplemented with 1% fetal bovine serum (FBS) and OD600 was adjusted to 0.2 (∼8 log10 CFU/mL).
Cell Infection Model
The avian macrophage-like immortalized cell line (HD11) was used for the host-pathogen infection study of the C. jejuni strains. The HD11 cell line was suspended in an Advanced Dulbecco's Modified Eagle Medium supplemented with varying levels of 2.5 to 10% FBS (Gibco Fetal Bovine Serum, ThermoFisher Scientific, Grand Island, NY), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco Pen strep, ThermoFisher Scientific, Grand Island, NY), and 2 mM GlutaMax (Gibco GlutaMax-I, ThermoFisher Scientific, Grand Island, NY). The cell line was grown at 37°C with 5% CO2 and an 80 to 90% confluency (6.0 log10 cells/well) in 6-well tissue culture plates. Each of the C. jejuni strains, approximately at 8.3 log10 CFU/mL, was suspended in Advanced DMEM with 1% FBS and without antibiotics. Each C. jejuni suspension was used to infect a monolayer of the HD11 cells, to make the multiplicity of infection of 100 (Larson et al., 2008; Peng et al., 2018; Pogacar et al., 2020). Culture plates consisting of both C. jejuni and HD11 cells were incubated at 37°C in a 5% CO2 air atmosphere incubator (Forma Series 3 Water Jacketed CO2 Incubator, ThermoFisher, Marietta, OH) for 3 h. After infection and incubation, the nonattached bacteria and media were aspirated, and the wells still containing the attached bacteria and cells were washed 3 times using 1 mL of PBS. After washing, approximately 6.5 log10 CFU C. jejuni strains were attached (C. jejuni MS2191 [6.70 log10 CFU/well]; C. jejuni MS2074 [6.31 log10 CFU/well]; C. jejuni ATCC29428 [6.82 log10 CFU/well]; C. jejuni ATCC33560 [6.30 log10 CFU/well]). The cell pellet of the infected HD11 cells from each well was collected utilizing the cell scraper and stored at -80°C for further analysis.
RNA Extraction and cDNA Synthesis
The total RNA was extracted from the infected cell pellets containing both prokaryotic and eukaryotic cells using Zymo Quick-Miniprep Plus (Zymo Research, Irvine, CA) with bead beating using ZR Bashing Beads (Zymo Bashing Beads). On column DNase I treatment was done following the manufacturer's protocol to eliminate the genomic DNA contamination. RNA quality, quantity, and purity were assessed via 1% gel electrophoresis and NanoDrop One spectrophotometer (Thermo Scientific, Wilmington, MA). A total of 1000 ng of the extracted RNA from the infected cells was reversely transcribed to first-stand cDNA using SuperScript VILO MasterMix (ThermoFisher Scientific, Carlsbad, CA). Similarly, prokaryotic RNA was extracted from bacterial cultures of all 4 C. jejuni strains suspended in cell culture medium without HD11 cell line (control; untreated group), and a total of 20 ng of prokaryotic RNA was used for the first-strand cDNA synthesis.
RT-qPCR
RT-qPCR assay was conducted using QuantStudio 3 (Applied Biosystems, Waltham, MA) and the following program: an initial denaturation step at 95°C for 2 min, followed by 40 cycles at 95°C for 5 sec and 60°C for 20 sec. The mixture of each assay contained 1 µL of diluted cDNA (ranged 5–15 times), 5 µL of 2 × PowerTrack SYBR Green Master Mix (ThermoFisher Scientific, Carlsbad, CA), 0.25 µL of each forward and reverse primer with 10 µM concentration, and 3.5 µL of nuclease-free water to make a final volume of 10 µL.
Normalization of Prokaryotic RNA Amount in Total RNA Sample
Due to the limited and varied bacterial RNA content among the infected host cells, the first-strand cDNA product of pure C. jejuni extracted RNA (described in “RNA Extraction and cDNA Synthesis”) was used to generate a standard curve of 16S rRNA gene for estimating the prokaryotic RNA content in each extracted total RNA in the treated samples (RNA obtained from infected cells). The standard curve of 16S rRNA gene was conducted with 10-fold serial dilutions of first-strand cDNA product utilizing prokaryotic RNA obtained from C. jejuni strains and Campylobacter-specific 16S rRNA primer pair via the RT-qPCR assay. In order to estimate the bacterial RNA content in treated samples, cycle threshold value (Ct-value) of the 16S rRNA gene was obtained from the first-strand cDNA product from each treated sample. The actual bacterial RNA amounts for each treated sample were estimated using the equation obtained from the standard curve via RT-qPCR. The estimated bacterial RNA amount present in the treated samples ranged between 0.03 and 0.06 ng/µL. In order to make a similar input amount of RNA from both treated and untreated samples, untreated samples were further diluted based on the predicted amount of RNA present in the treated samples. The normalized samples were used for analyzing the expression level of the vaccine candidate genes.
Candidate Gene Expression
The gene-specific primers for each gene of interest were designed using Geneious prime v2020.2.2 (Integrated Bioinformatic Solution, Auckland, North Island, New Zealand). The information about primer and primer sequences are listed in Table 1. In order to verify the amplicon size and primer specificity, the standard RT-PCR was conducted using Eppendorf Master cycler ep gradient system (Eppendorf, Enfield, CT). The condition used for the RT-PCR was an initial heat step 95°C for 2 min, followed by 35 cycling steps: 95°C for 30 sec and 60°C for 30 sec, 72°C for 45 sec, followed by a final extension step at 72°C for 5 min. The correctly amplified PCR product was cleaned up using GeneJET PCR purification Kit (ThermoFisher Scientific, Waltham, MA) and cloned into pGEM-T Easy vector (Promega, Madison, WI) following the manufactures protocol. Five independent recombinant plasmid DNAs for each gene of interest were isolated and sent to Eurofins Genomics LLC (Louisville, KY) for Sanger sequencing, and sequencing results were aligned with the original target gene sequence using Geneious prime v2020.2.2 (Integrated Bioinformatic Solution, Auckland, North Island, New Zealand) to confirm the specificity of each designed primer pair.
Table 1.
List of primers used in RT-qPCR analysis.
| Transcript | Primer |
||||
|---|---|---|---|---|---|
| Target transcript | Orientation | Sequence (5′–3′) | Length (nt) | Tm (°C) | Amplicon size (bp) |
| CdtB | Forward | TCCTGTAATTGCATAATCAAGAGTCC | 26 | 61.4 | 206 |
| Reverse | CAGATGTAGGAGCAATTATCACAGC | 25 | 62.9 | ||
| PldA | Forward | AAGCAATGGCAAGGGAGATGAG | 22 | 62.7 | 201 |
| Reverse | TCATCGCCCAAATACGCTAAATTC | 24 | 61.6 | ||
| BtuB | Forward | TACGCAATGTAATCAGCATAGAAGG | 25 | 61.3 | 217 |
| Reverse | GTTACACCATGGGAATTATCAAGAAC | 26 | 61.4 | ||
| Campylobacter 16S1 | Forward | GATGAAGCTTTTAGCTTGCTAGAAGTGG | 28 | 65.0 | 165 |
| Reverse | GTCTCATCCTACACCGAAAAACTTTCC | 27 | 65.0 | ||
Accession number: LS483362.1.
The expression changes of predicted vaccine candidate genes in the bacteria-cell infection study were analyzed using RT-qPCR, with the assay mixture and condition as described previously. For each tested C. jejuni strain, 3 biological replicates were analyzed, and 3 technical replicates were conducted in RT-qPCR assay for each sample. The expression change of the interested transcript was determined based on the ΔΔCt method (Livak and Schmittgen, 2001). Campylobacter-specific 16S rRNA gene was used as a reference gene for normalization. The paired Student t test was used to compare the control (RNA obtained from C. jejuni not involved in cell-infection) and treatment (RNA obtained from C. jejuni, which was involved in cell-infection) using GraphPad prism 9 (GraphPad Software, San Diego, CA).
RESULTS
In Silico Prediction of Vaccine Candidate
Of the 7,132 proteins (1,820, 1,933, 1,629, and 1,759 from MS2005, MS2058, MS2074, and MS2167, respectively), 30 proteins passed the selection criteria for Vaxigen (Supplemental Table S4). C. jejuni strains MS2005 and MS2058 possess 1,820 and 1,933 protein-coding sequences (CD), respectively and from each strain, 7 protein sequences meet the applied selection criteria of localization, adhesion probability, and transmembrane helixes. C. jejuni strain MS2074 consists of 1,629 CDs and 10 proteins, which meet the selection criteria. C. jejuni strain MS2167 consists of 1,750 CDs and possesses 6 protein sequences that meet the selection criteria (Supplemental Table S4). The antigenicity of the preselected proteins was tested using VaxiJen v2.0. The antigenic score for C. jejuni strains MS2005, MS2058, MS2074, and MS2167 ranged from 0.36 to 0.72 (Supplemental Table S4). Of the 30 preselected protein sequences, 27 were classified as antigenic (VaxiJen antigenic probability > 0.5).
The sequences of vaccine candidate proteins were aligned against the Gallus gallus proteins to find candidates with high similarity to the host; none of the selected proteins were found homologous to the host (Supplemental Table S4). The candidate sequences were also aligned to genomes of C. jejuni and C. coli strains available from NCBI. Poorly conserved candidates (<80% present among the C. jejuni and C. coli strains) were removed from future analysis. Strains MS2005, MS2058, and MS2167 consist of 5 highly conserved protein sequences (TonB dependent vitamin B12 transporter [BtuB], major outer membrane porin [MoMP] or PorA, Phospholipase A (PldA), flagellar capping protein [FliD], and Cytolethal distending toxin subunit B [CdtB]). Strain MS2074 consists of 6 highly conserved proteins (BtuB, MoMP, or PorA, PldA, FliD, flagellar hook protein [FlgE], and CdtB; Table 2 and Supplemental Table S4). Although extracellular CdtB protein was poorly conserved (66.5% present) among all C. jejuni strains, it was considered a potential candidate because it was highly conserved (88.89%) among the poultry specific C. jejuni strains and (91.18%) among C. coli strains, so protein CdtB was added back to conserved list. A total of 21 proteins were obtained while analyzing the 4 C. jejuni strains MS2005, MS2058, MS2074, and MS2167 (Table 2).
Table 2.
List of potential vaccine candidates selected from the analysis of C. jejuni using different bioinformatic software.
| Annotated protein ID | Protein size (aa) | Localization | Adhesin probability | Antigen probability | Sharing (%) among C. jejuni | Sharing (%) among C. jejuni strains from poultry origin | Sharing (%) among C. coli | MAFFT alignment | Protein name |
|---|---|---|---|---|---|---|---|---|---|
| MS2005_GHPAHEKB_01319 | 329 | OMP | 0.659 | 0.566 | 97.09 | 94.44 | 100 | 98.80% | Phospholipase A |
| MS2058_ICDGEIKM_01045 | 329 | OMP | 0.646 | 0.553 | 97.57 | 94.44 | 100 | ||
| MS2074_CJDKBBOE_01490 | 329 | OMP | 0.670 | 0.580 | 97.09 | 94.44 | 100 | ||
| MS2167_CDNINIFE_01244 | 329 | OMP | 0.659 | 0.566 | 97.09 | 94.44 | 100 | ||
| MS2074_CJDKBBOE_01412 | 418 | OMP | 0.744 | 0.622 | 99.03 | 100 | 100 | 81.4% | MoMP or PorA |
| MS2058_ICDGEIKM_00883 | 424 | OMP | 0.664 | 0.631 | 99.03 | 100 | 100 | ||
| MS2167_CDNINIFE_01337 | 425 | OMP | 0.722 | 0.642 | 99.03 | 100 | 100 | ||
| MS2005_GHPAHEKB_01469 | 431 | OMP | 0.863 | 0.642 | 99.03 | 100 | 100 | ||
| MS2074_CJDKBBOE_00243 | 545 | Extracellular | 0.852 | 0.653 | 100 | 100 | 100 | FlgE | |
| MS2167_CDNINIFE_00321 | 642 | Extracellular | 0.801 | 0.724 | 99.51 | 100 | 100 | 97% | FliD |
| MS2005_GHPAHEKB_00382 | 643 | Extracellular | 0.799 | 0.690 | 99.51 | 100 | 100 | ||
| MS2058_ICDGEIKM_00093 | 643 | Extracellular | 0.799 | 0.690 | 99.51 | 100 | 100 | ||
| MS2074_CJDKBBOE_00715 | 643 | Extracellular | 0.774 | 0.690 | 99.15 | 100 | 100 | ||
| MS2074_CJDKBBOE_00067 | 706 | OMP | 0.652 | 0.614 | 96.60 | 100 | 100 | 94.8% | BtuB |
| MS2058_ICDGEIKM_01297 | 715 | OMP | 0.665 | 0.610 | 96.60 | 88.89 | 100 | ||
| MS2005_GHPAHEKB_01062 | 718 | OMP | 0.689 | 0.608 | 96.60 | 88.89 | 100 | ||
| MS2167_CDNINIFE_00988 | 718 | OMP | 0.686 | 0.607 | 96.60 | 88.89 | 100 | ||
| MS2005_GHPAHEKB_00842 | 265 | Extracellular | 0.654 | 0.5867 | 66.50 | 88.89 | 91.18 | 100% | CdtB |
| MS2058_ICDGEIKM_01516 | 265 | Extracellular | 0.654 | 0.5867 | 66.50 | 88.89 | 91.18 | ||
| MS2074_CJDKBBOE_00273 | 265 | Extracellular | 0.654 | 0.5867 | 66.50 | 88.89 | 91.18 | ||
| MS2167_CDNINIFE_00770 | 265 | Extracellular | 0.654 | 0.5867 | 66.50 | 88.89 | 91.18 |
Abbreviation: OMP, outer membrane protein.
Antigenic protein sequences obtained were analyzed for the number and density B-epitopes. The conserved protein sequences from C. jejuni strain MS2005, MS2058, MS2074, and MS2167 contain BCPred B-epitopes ranging from 2 to 13 and density of 0.008 to 0.020. For C. jejuni strains, MS2005, MS2058, MS2074, and MS2167 consist of amino acid pair antigenic B-epitope numbers between 3 and 11 and density between 0.009 and 0.015. The epitope number and density are summarized in Supplemental Table S4.
Removal of Similar and Previously Tested Protein
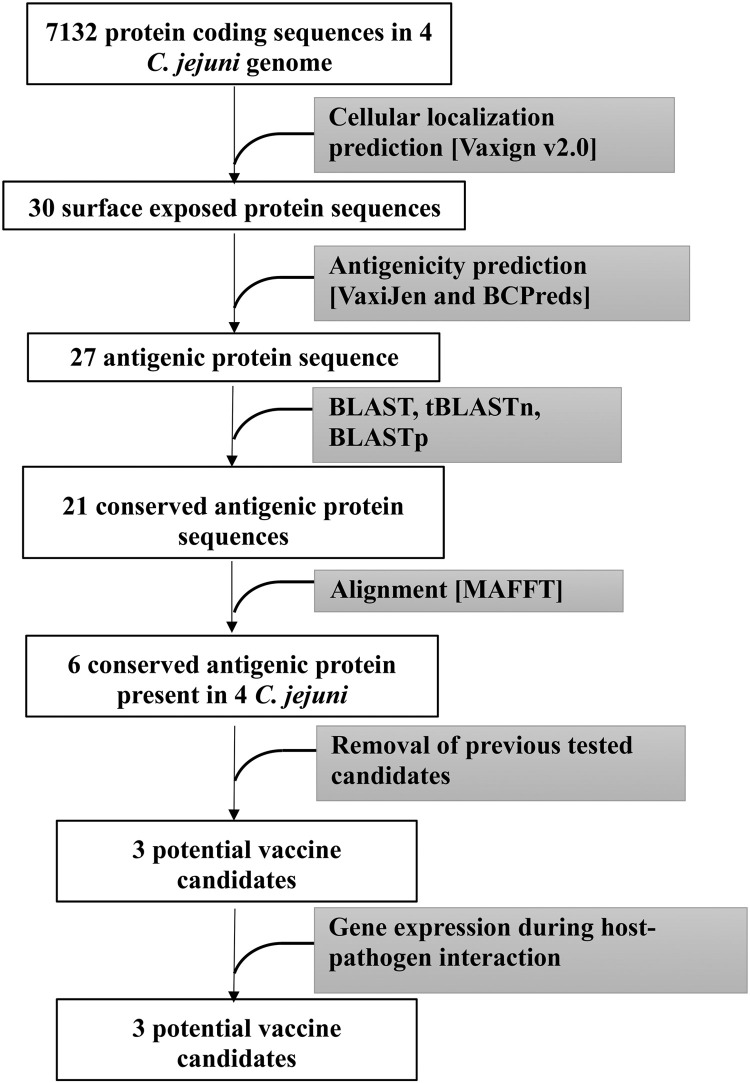
Antigenic protein sequences predicted from 4 different C. jejuni strains were aligned using MAFFT alignment. From 21 conserved antigenic proteins, 6 highly conserved candidates were selected: 3 extracellular proteins (FlgE, FliD, and CdtB), and 3 outer membrane proteins (PldA, MoMP, and BtuB) (Table 2). The proteins FlgE, FliD, and MoMP were removed from further analysis due to a lack of cross-protection and short-lived protection during previous in-vivo experiments (de Zoete et al., 2007; Isalm et al., 2010; Meunier et al., 2017). Finally, 3 potential vaccine candidates, PldA, BtuB, and CdtB were obtained (Table 2 and Figure 1).
Figure 1.
A flow chart summarizing the methodology utilized in this study to identify the potential antigenic vaccine candidates against Campylobacter jejuni.
Expression Analysis of Selected Vaccine Candidates
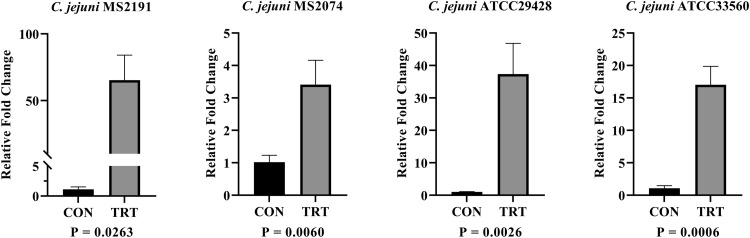
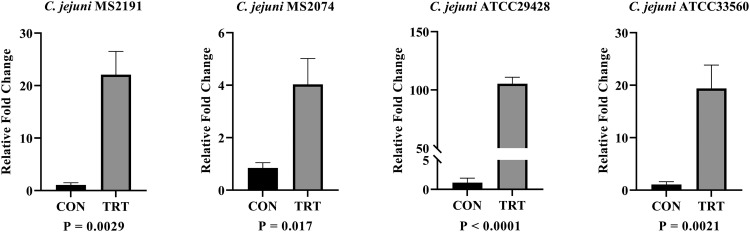
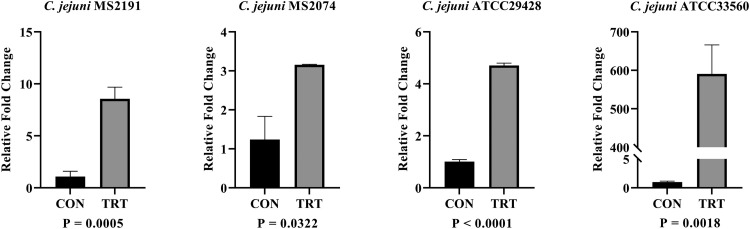
The expression analysis during host-pathogen interaction helps to understand the immunogenicity of the in silico predicted candidates. Therefore, the expression analysis of predicted potential antigenic vaccine candidates from in silico was performed through the host-pathogen interaction assay. The results of gene expression analysis during host-pathogen interaction were summarized in Figure 2, Figure 3, Figure 4. During the infection of HD11 cells by C. jejuni strains, there was significant upregulation (P ≤ 0.05) of all 3 vaccine candidate genes suggesting that these genes might have a significant impact on the bacterial pathogenesis. The PldA gene, which plays a major role in the hemolytic activity, was significantly upregulated by 3.41- to 65.41-fold in all 4 C. jejuni strains (MS2191, MS2074, ATCC33560, ATCC29428). The expression of PldA gene was highest in C. jejuni strain MS2191 isolated from cloacal swab of broiler followed by strain ATCC29428 (human clinical isolate), ATCC33560 (bovine feces isolate), and MS2074 (chicken meat isolate). Compared to other isolates, C. jejuni isolated from chicken meat showed the lowest expression of PldA gene during interaction with the host-cell (Figure 2). BtuB gene, an outer membrane vitamin B12 transporter, was upregulated by 4.03- to 105.4-fold in all 4 tested C. jejuni strains. The expression of BtuB gene was highest in C. jejuni strain ATCC29428 compared to strains isolated from the other sources. Similar to PldA, BtuB expression was also lower in C. jejuni strain MS2074 isolated from chicken meat samples compared to other strains (Figure 3). CdtB, an extracellular protein responsible for producing toxins, was found to have a higher variation in the expression among the isolates; its upregulation ranged from 3.15- to 590.04-fold among tested strains. The expression of CdtB gene was much higher in C. jejuni strain ATCC33560 compared to other strains tested, whereas the expression level was similar other 3 strains (MS2074, MS2191, and ATCC29428) ranging between 3.15 and 8.550 (Figure 4).
Figure 2.
Expression analysis of PldA gene in 4 C. jejuni strains (A) MS2191, (B) MS2074, (C) ATCC29428, and (D) ATCC33560, during host-pathogen infection study via RT-qPCR. Relative fold change in vaccine candidate gene PldA during the infection of HD11 cells. Each vertical bar represents mean relative fold change ± standard error of mean (n =3). The expression change of transcript was determined based on ΔΔCt methods. Abbreviations: CON, untreated group; TRT, treatment group.
Figure 3.
Expression analysis of BtuB gene in 4 C. jejuni strains (A) MS2191, (B) MS2074, (C) ATCC29428, and (D) ATCC33560, during host-pathogen infection study via RT-qPCR. Relative fold change in vaccine candidate gene BtuB during the infection of HD11 cells. Each vertical bar represents mean relative fold change ± standard error of mean (n =3). The expression change of transcript was determined based on ΔΔCt methods. Abbreviations: CON, untreated group; TRT, treatment group.
Figure 4.
Expression analysis of CdtB gene in 4 C. jejuni strains (A) MS2191, (B) MS2074, (C) ATCC29428, and (D) ATCC33560, during host-pathogen infection study via RT-qPCR. Relative fold change in vaccine candidate gene CdtB during the infection of HD11 cells. Each vertical bar represents mean relative fold change ± standard error of mean (n =3). The expression change of transcript was determined based on ΔΔCt methods. Abbreviations: CON, untreated group; TRT, treatment group.
DISCUSSION
Reverse Vaccinology Approach
Despite the utmost necessity to control C. jejuni colonization in poultry to reduce human infection, there are no effective antibiotic alternative strategies for reducing colonization in the gastrointestinal tract of poultry. Previous attempts to make an effective vaccine to reduce colonization of C. jejuni in poultry have been unsuccessful (Pumtang-on et al., 2021). The utilization of conventional strategies for developing the vaccine against C. jejuni in poultry has not led to a successful vaccine that can produce both immunogenicity and reduce colonization of bacteria in the ceca of poultry. Reverse vaccinology might be a suitable strategy for the development of a vaccine against C. jejuni. RV is a new vaccine developmental strategy that helps in the rapid identification and refinement of the antigenic vaccine candidates using the genome sequence of microorganisms (Moxon et al., 2019). Previously, the vaccine candidate predicted using RV technology was directly utilized for immunogenicity testing via vaccine development (Rappuoli, 2000; Meunier et al., 2016; Meunier et al., 2017). Even though RV technology helps us identify and narrow down the potential vaccine candidate, the possibility of obtaining a successful vaccine is low (Meunier et al., 2016; Meunier et al., 2017). Developing the vaccine without knowing how these candidate genes express and function during the interaction with the host might have potentially increased the risk of vaccine candidate failure, increasing the cost, and the time for developing a vaccine, ultimately leading to low efficacy for vaccine development. To circumvent this issue, we introduced an additional screening step, that is, gene expression analysis during host-pathogen interaction, to further validate the vaccine candidates after in silico prediction. The gene expression analysis during host-pathogen interactions helps to understand the role of the gene during the pathogenesis of bacteria, as well as to quantify the expression level of genes, helping to determine the best suitable candidate for the development of the vaccine. Therefore, in the current study, we combined the in silico and gene expression analysis during host-pathogen interaction assay to identify potential vaccine candidates.
In the current study, to select the universal vaccine candidate against C. jejuni, the genomes of multiple C. jejuni strains obtained from different poultry sources were analyzed to predict the conserved surface-exposed antigenic proteins. Furthermore, we utilized 4 C. jejuni strains obtained from 4 different sources (broiler cloacal swab, retail chicken meat, human clinical, and bovine feces isolate) for analyzing vaccine candidate expression during host-pathogen interaction to identify the candidate that significantly upregulated in all tested strains so that the selected candidate would provide protection against C. jejuni originated from a broad host range (wild animals, birds, food animals, and pet animals). Since poultry can get infected from different sources, C. jejuni can get colonized in the birds' gastrointestinal tract, ultimately leading to C. jejuni exposure to humans. Finally, combining the in silico approach and gene expression analysis, we identified 3 potential vaccine candidates: BtuB (an outer membrane protein), CdtB, and PldA (extracellular proteins).
Candidate Protein Description
BtuB is a barrel-shaped outer-membrane protein responsible for the transportation of vitamin B12, an essential nutrient and cofactor necessary for numerous microbial metabolic pathways (Fang et al., 2017). Outer membrane BtuB protein is directly connected to the TonB-dependent system in the inner membrane of the cell. Recently, the TonB-dependent transporters (TBDTs) have gained attention in developing the vaccine for gram-negative bacteria, and this transporter has all the essential characteristics for successful vaccine development (Wang et al., 2021) These finding collectively suggest that conserved antigen BtuB could be further used to develop a universal vaccine against genetically diverse C. jejuni strains.
Cytolethal-distending toxin consists of 3 subunits CdtA, CdtB, and CdtC, which help in the production of toxins and the pathogenesis of Campylobacter. CdtB was identified as the potential antigen candidate for the construction of subunit vaccines, and it was predicted to be located extracellularly. Along with Campylobacter, this toxin was produced by other enteric and nonenteric bacterial pathogens (Escherichia coli, Shigella spp., Actinobacillus actinomycetemcomitans, and Haemophilus ducreyi) (Svensson et al., 2002; Thelestam and Frisan, 2004; Boesze-Battaglia et al., 2017; Meza-Segura et al., 2017). Furthermore, the prevalence of toxin-producing gene CdtB was present in greater than 90% of poultry isolates (Talukder et al., 2008; Findik et al., 2011; Nouri Gharajalar et al., 2020).
The third antigenic protein identified in this study was Phospholipase A (PldA), which plays a major role in hemolytic activity and helps bacterial colonization (Ziprin et al., 1999; Ziprin et al., 2002). Although PldA had not been previously tested as a potential vaccine candidate via developing a vaccine, this antigen was also previously identified as the potential vaccine candidate by in silico analysis utilizing the C. jejuni 81 to 176 strain (Meunier et al., 2016). Finally, this gene is yet to be tested as a vaccine candidate.
These findings collectively suggest that conserved antigenic proteins BtuB, CdtB, and PldA could be further used to develop a universal vaccine against genetically diverse C. jejuni strains. Although these proteins selected here were predicted and identified as antigenic through in silico analysis and gene expression analysis during host-pathogen interaction, we still need to determine the efficacy of these vaccine candidates during in vivo experiments.
CONCLUSION
In conclusion, this research identifies 3 potential antigenic vaccine candidates which have the potential to develop effective vaccines against C. jejuni and C. coli in poultry. We expect that these selected candidates may help reduce C. jejuni and C. coli colonization in the poultry gastrointestinal tract and subsequently help reduce C. jejuni and C. coli infections in humans. Furthermore, this study added a valuable step for screening the vaccine candidates utilizing RV technology, which can be utilized for selecting the vaccine candidate for other pathogens.
ACKNOWLEDGMENTS
This publication is a contribution of the Mississippi Agricultural and Forestry Experiment Station, under US Department of Agriculture, Hatch project accession number MIS-322430/NE1942. This material is based on work that is supported by the US Poultry and Egg Association, Award No. #724. The authors thank Attila Karsi of Mississippi State University and Matt Scott of Texas A&M University for the editorial assistance on this manuscript. The authors thank Matthew K. Ross for providing the avian macrophage-like immortalized cell (HD11) for the research.
Availability of Data: The authors confirm that the data supporting the finding of this study are available within the article and its additional materials.
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2023.102592.
Appendix. Supplementary materials
REFERENCES
- Audu B.J., Norval S., Bruno L., Meenakshi R., Marion M., Forbes K.J. Genomic diversity and antimicrobial resistance of Campylobacter spp. from humans and livestock in Nigeria. J. Biomed. Sci. 2022;29:1–15. doi: 10.1186/s12929-022-00786-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beery J.T., Hugdahl M.B., Doyle M.P. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 1988;54:2365–2370. doi: 10.1128/aem.54.10.2365-2370.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K., Walker L.P., Dhingra A., Kandror K., Tang H.Y., Shenker B.J. Internalization of the active subunit of the Aggregatibacter actinomycetemcomitans cytolethal distending toxin is dependent upon cellugyrin (synaptogyrin 2), a host cell non-neuronal paralog of the synaptic vesicle protein, synaptogyrin 1. Front. Cell. Infect. Microbiol. 2017;7:469. doi: 10.3389/fcimb.2017.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintoan-Uta C., Cassady-Cain R.L., Al-Haideri H., Watson E., Kelly D.J., Smith D.G.E., Sparks N.H.C., Kaiser P., Stevens M.P. Superoxide dismutase SodB is a protective antigen against Campylobacter jejuni colonization in chickens. Vaccine. 2015;33:6206–6211. doi: 10.1016/j.vaccine.2015.09.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintoan-Uta C., Cassady-Cain R.L., Stevens M.P. Evaluation of flagellum-related proteins FliD and FspA as subunit vaccines against Campylobacter jejuni colonization in chickens. Vaccine. 2016;34:1739–1743. doi: 10.1016/j.vaccine.2016.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsass M., Brozzi A., Medini D., Rappuoli R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 2019;10:113. doi: 10.3389/fimmu.2019.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoete M.R., van Putten J.P.M., Wagenaar J.A. Vaccination of chickens against Campylobacter. Vaccine. 2007;25:5548–5557. doi: 10.1016/j.vaccine.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Panel on Biological Hazards (BIOHAZ) Scientific opinion on Campylobacter in broiler meat production: control options and performance objectives and/or targets at different stages of the food chain. EFSA J. 2011;9:2105. [Google Scholar]
- Epping L., Walther B., Piro R.M., Knüver M.T., Huber C., Thürmer A., Flieger A., Fruth A., Janecko N., Wieler L.H., Stingl K., Semmler T. Genome-wide insights into population structure and host specificity of Campylobacter jejuni. Sci. Rep. 2021;11:1–15. doi: 10.1038/s41598-021-89683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H., Kang J., Zhang D. Microbial production of vitamin B12: a review and future perspectives. Microb. Cell Fact. 2017;16:1–14. doi: 10.1186/s12934-017-0631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findik A., Ica T., Onuk E.E., Percin D., Kevenk T.O., Ciftci A. Molecular typing and cdt genes prevalence of Campylobacter jejuni isolates from various sources. Trop. Anim. Health Prod. 2011;43:711–719. doi: 10.1007/s11250-010-9758-0. [DOI] [PubMed] [Google Scholar]
- Fravalo P., Kooh P., Mughini-Gras L., David J., Thébault A., Cadavez V., Gonzales-Barron U. Risk factors for sporadic Campylobacteriosis: a systematic review and meta-analysis. Microb. Risk Anal. 2021;17 [Google Scholar]
- Friedman C.R., Hoekstra R.M., Samuel M., Marcus R., Bender J., Shiferaw B., Reddy S., Ahuja S.D., Helfrick D.L., Hardnett F., Carter M., Anderson B., Tauxe R.V. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 2004;38:S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- Gardy J.L., Laird M.R., Chen F., Rey S., Walsh C.J., Ester M., Brinkman F.S.L. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Gormley F.J., Bailey R.A., Watson K.A., McAdam J., Avendaño S., Stanley W.A., Koerhuis A.N.M. Campylobacter colonization and proliferation in the broiler chicken upon natural field challenge is not affected by the bird growth rate or breed. Appl. Environ. Microbiol. 2014;80:6733–6738. doi: 10.1128/AEM.02162-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Kumar A. Designing an efficient multi-epitope vaccine againstCampylobacter jejuni using immunoinformatic and reverse vaccinology approach. Microb. Pathog. 2020;147 doi: 10.1016/j.micpath.2020.104398. [DOI] [PubMed] [Google Scholar]
- He Y., Xiang Z., Mobley H.L.T. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J. Biomed. Biotechnol. 2010:297505. doi: 10.1155/2010/297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins D.C., Barjesteh N., St. Paul M., Ma Z., Monteiro M.A., Sharif S. Evaluation of a polysaccharide conjugate vaccine to reduce colonization by Campylobacter jejuni in broiler chickens. BMC Res. Notes. 2015;8:204. doi: 10.1186/s13104-015-1203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A., Raghupathy R., Albert M.J. Recombinant PorA, the major outer membrane protein of Campylobacter jejuni, provides heterologous protection in an adult mouse intestinal colonization model. Clin. Vaccine Immunol. 2010;17:1666–1671. doi: 10.1128/CVI.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Singh S., Verma S.K., Jain A. Genome-wide prediction of potential vaccine candidates for Campylobacter jejuni using reverse vaccinology. Interdiscip. Sci. Comput. Life Sci. 2019;11:337–347. doi: 10.1007/s12539-017-0260-5. [DOI] [PubMed] [Google Scholar]
- Kaakoush N.O., Castaño-Rodríguez N., Mitchell H.M., Man S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015;28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E.L.L. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007;35:429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson C.L., Shah D.H., Dhillon A.S., Call D.R., Ahn S., Haldorson G.J., Davitt C., Konkel M.E. Campylobacter jejuni invade chicken LMH cells inefficiently and stimulate differential expression of the chicken CXCLi1 and CXCLi2 cytokines. Microbiology. 2008;154:3835–3847. doi: 10.1099/mic.0.2008/021279-0. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meunier M., Guyard-Nicodème M., Hirchaud E., Parra A., Chemaly M., Dory D. Identification of novel vaccine candidates against Campylobacter through reverse vaccinology. J. Immunol. Res. 2016;2016:5715790. doi: 10.1155/2016/5715790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier M., Guyard-Nicodème M., Vigouroux E., Poezevara T., Beven V., Quesne S., Bigault L., Amelot M., Dory D., Chemaly M. Promising new vaccine candidates against Campylobacter in broilers. PLoS One. 2017;12:1–14. doi: 10.1371/journal.pone.0188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza-Segura M., Zaidi M.B., Maldonado-Puga S., Huerta-Cantillo J., Chavez-Dueñas L., Navarro-Garcia F., Estrada-Garcia T. Cytolethal distending toxin-producing Escherichia coli strains causing severe diarrhea in young Mexican children. JMM Case Rep. 2017;4 doi: 10.1099/jmmcr.0.005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterrubio-López G.P., González-Y-Merchand J.A., Ribas-Aparicio R.M. Identification of novel potential vaccine candidates against Tuberculosis based on reverse vaccinology. Biomed Res. Int. 2015;2015:1–16. doi: 10.1155/2015/483150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon R., Reche P.A., Rappuoli R. Editorial: reverse vaccinology. Front. Immunol. 2019;10:2776. doi: 10.3389/fimmu.2019.02776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz A., Awan F.M., Obaid A., Muhammad S.A., Paracha R.Z., Ahmad J., Ali A. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: a reverse vaccinology-based approach. Infect. Genet. Evol. 2015;32:280–291. doi: 10.1016/j.meegid.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Neal-McKinney J.M., Samuelson D.R., Eucker T.P., Nissen M.S., Crespo R., Konkel M.E. Reducing Campylobacter jejuni colonization of poultry via vaccination. PLoS One. 2014;9:1–19. doi: 10.1371/journal.pone.0114254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell D.G., Fearnley C. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 2003;69:4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H., Davis B., Lock Y.Y., Perez-Munoz M.E., Vinogradov E., Walter J., Coros C., Szymanski C.M. Engineering the Campylobacter jejuni N-glycan to create an effective chicken vaccine. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri Gharajalar S., Hassanzadeh P., Hosseinali Nejad N. Molecular detection of Campylobacter species and cytolethal distending toxin isolated from chicken livers in Tabriz. Comp. Immunol. Microbiol. Infect. Dis. 2020;71 doi: 10.1016/j.cimid.2020.101474. [DOI] [PubMed] [Google Scholar]
- Oprea M., Antohe F. Reverse-vaccinology strategy for designing T-cell epitope candidates for Staphylococcus aureus endocarditis vaccine. Biologicals. 2013;41:148–153. doi: 10.1016/j.biologicals.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Peng M., Tabashsum Z., Patel P., Bernhardt C., Biswas D. Linoleic acids overproducing Lactobacillus casei limits growth, survival, and virulence of Salmonella typhimurium and enterohaemorrhagic Escherichia coli. Front. Microbiol. 2018;9:1–14. doi: 10.3389/fmicb.2018.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacar M.S., Langerholc T., Mičetić-Turk D., Možina S.S., Klančnik A. Effect of Lactobacillus spp. On adhesion, invasion, and translocation of Campylobacter jejuni in chicken and pig small-intestinal epithelial cell lines. BMC Vet. Res. 2020;16:1–14. doi: 10.1186/s12917-020-2238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel S., Li T., Arick M.A., II, Hsu C.-Y, Thrash A., Sukumaran A.T., Adhikari P., Kiess A.S., Zhang L. Complete genome sequence of four Campylobacter jejuni isolated from retail chicken and broiler feces. Microbiol. Resour. Announc. 2022;11 doi: 10.1128/mra.00898-22. e0089822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel S., Li T., Chen S., Zhang X., Cheng W.-H., Sukumaran A.T., Kiess A.S., Zhang L. Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter isolated from broilers and broiler meat raised without antibiotics. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.00251-22. e0025122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumtang-On P., Mahony T.J., Hill R.A., Vanniasinkam T. A systematic review of Campylobacter jejuni vaccine candidates for chickens. Microorganisms. 2021;9:1–48. doi: 10.3390/microorganisms9020397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R. Reverse vaccinology. Curr. Opin. Microbiol. 2000;3:445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- Rashid M.I., Naz A., Ali A., Andleeb S. Prediction of vaccine candidates against Pseudomonas aeruginosa: an integrated genomics and proteomics approach. Genomics. 2017;109:274–283. doi: 10.1016/j.ygeno.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Romero-Barrios P., Hempen M., Messens W., Stella P., Hugas M. Quantitative microbiological risk assessment (QMRA) of food-borne zoonoses at the European level. Food Control. 2013;29:343–349. [Google Scholar]
- Sachdeva G., Kumar K., Jain P., Ramachandran S. SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics. 2005;21:483–491. doi: 10.1093/bioinformatics/bti028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin O., Zhang Q., Meitzler J.C., Harr B.S., Morishita T.Y., Mohan R. Prevalence, antigenic specificity, and bactericidal activity of poultry anti-Campylobacter maternal antibodies. Appl. Environ. Microbiol. 2001;67:3951–3957. doi: 10.1128/AEM.67.9.3951-3957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff R.L. Food attribution and economic cost estimates for meat- and poultry-related illnesses. J. Food Prot. 2020;83:959–967. doi: 10.4315/JFP-19-548. [DOI] [PubMed] [Google Scholar]
- Sette A., Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Dhillon A., Shivaprasad H.L., Schaberg D., Wier F., Weber S., Bandli D. Campylobacter jejuni infection in broiler chickens. Avian Dis. 2006;50:55–58. doi: 10.1637/7411-071405R.1. [DOI] [PubMed] [Google Scholar]
- Skarp C.P.A., Hänninen M.L., Rautelin H.I.K. Campylobacteriosis: the role of poultry meat. Clin. Microbiol. Infect. 2016;22:103–109. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Yamamoto S. Campylobacter contamination in retail poultry meats and by-products in Japan: a literature survey. Food Control. 2009;20:531–537. doi: 10.1292/jvms.71.255. [DOI] [PubMed] [Google Scholar]
- Svensson L.A., Henning P., Lagergård T. The cytolethal distending toxin of Haemophilus ducreyi inhibits endothelial cell proliferation. Infect. Immun. 2002;70:2665. doi: 10.1128/IAI.70.5.2665-2669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack D.M., Marder E.P., Griffin P.M., Cieslak P.R., Dunn J., Hurd S., Scallan E., Lathrop S., Muse A., Ryan P., Smith K., Tobin-D'Angelo M., Vugia D.J., Holt K.G., Wolpert B.J., Tauxe R., Geissler A.L. Preliminary incidence and trends of infections with pathogens transmitted commonly through food —food-borne diseases active surveillance network, 10 US sites, 2015–2018. Am. J. Transplant. 2019;19:1859–1863. doi: 10.15585/mmwr.mm6816a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S., Zutshi S., Prashanth K.S., Saikia K.K., Kumar P. Identification of potential vaccine candidates against Streptococcus pneumoniae by reverse vaccinology approach. Appl. Biochem. Biotechnol. 2014;172:3026–3041. doi: 10.1007/s12010-014-0749-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder K.A., Aslam M., Islam Z., Azmi I.J., Dutta D.K., Hossain S., Nur-E-Kamal A., Nair G.B., Cravioto A., Sack D.A., Endtz H.P. Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J. Clin. Microbiol. 2008;46:1485–1488. doi: 10.1128/JCM.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelestam M., Frisan T. Cytolethal distending toxins. Rev. Physiol. Biochem. Pharmacol. 2004;152:111–133. doi: 10.1007/s10254-004-0030-8. [DOI] [PubMed] [Google Scholar]
- Truccollo B., Whyte P., Burgess C., Bolton D. Genetic characterization of a subset of Campylobacter jejuni isolates from clinical and poultry sources in Ireland. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnu U.S., Sankarasubramanian J., Gunasekaran P., Rajendhran J. Identification of potential antigens from non-classically secreted proteins and designing novel multitope peptide vaccine candidate against Brucella melitensis through reverse vaccinology and immunoinformatic approach. Infect. Genet. Evol. 2017;55:151–158. doi: 10.1016/j.meegid.2017.09.015. [DOI] [PubMed] [Google Scholar]
- Wang J., Xiong K., Pan Q., He W., Cong Y. Application of TonB-dependent transporters in vaccine development of gram-negative bacteria. Front. Cell. Infect. Microbiol. 2021;10:589115. doi: 10.3389/fcimb.2020.589115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.J., Gabriel E., Leatherbarrow A.J.H., Cheesbrough J., Gee S., Bolton E., Fox A., Fearnhead P., Hart C.A., Diggle P.J. Tracing the source of Campylobacteriosis. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszyńska A., Raczko A., Lis M., Jagusztyn-Krynicka E.K. Oral immunization of chickens with avirulent Salmonella vaccine strain carrying C. jejuni 72Dz/92 cjaA gene elicits specific humoral immune response associated with protection against challenge with wild-type Campylobacter. Vaccine. 2004;22:1379–1389. doi: 10.1016/j.vaccine.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Ziprin R.L., Hume M.E., Young C.R., Harvey R.B. Inoculation of chicks with viable non-colonizing strains of Campylobacter jejuni: evaluation of protection against a colonizing strain. Curr. Microbiol. 2002;44:221–223. doi: 10.1007/s00284-001-0088-3. [DOI] [PubMed] [Google Scholar]
- Ziprin R.L., Young C.R., Stanker L.H., Hume M.E., Konkel M.E. The absence of cecal colonization of chicks by a mutant of Campylobacter jejuni not expressing bacterial fibronectin-binding protein. Avian Dis. 1999;43:586–589. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.