Summary
It is generally believed that environmental or cutaneous bacteria are the main origin of surgical infections. Therefore, measures to prevent postoperative infections focus on optimizing hygiene and improving asepsis and antisepsis. In a large cohort of patients with infections following major surgery, we identified that the causative bacteria are mainly of intestinal origin. Postoperative infections of intestinal origin were also found in mice undergoing partial hepatectomy. CCR6+ group 3 innate lymphoid cells (ILC3s) limited systemic bacterial spread. Such bulwark function against host invasion required the production of interleukin-22 (IL-22), which controlled the expression of antimicrobial peptides in hepatocytes, thereby limiting bacterial spread. Using genetic loss-of-function experiments and punctual depletion of ILCs, we demonstrate that the failure to restrict intestinal commensals by ILC3s results in impaired liver regeneration. Our data emphasize the importance of endogenous intestinal bacteria as a source for postoperative infection and indicate ILC3s as potential new targets.
Keywords: innate lymphoid cells, ILC3s, IL-22, antibacterial defense, antimicrobial peptides, surgical infection, sepsis, liver regeneration
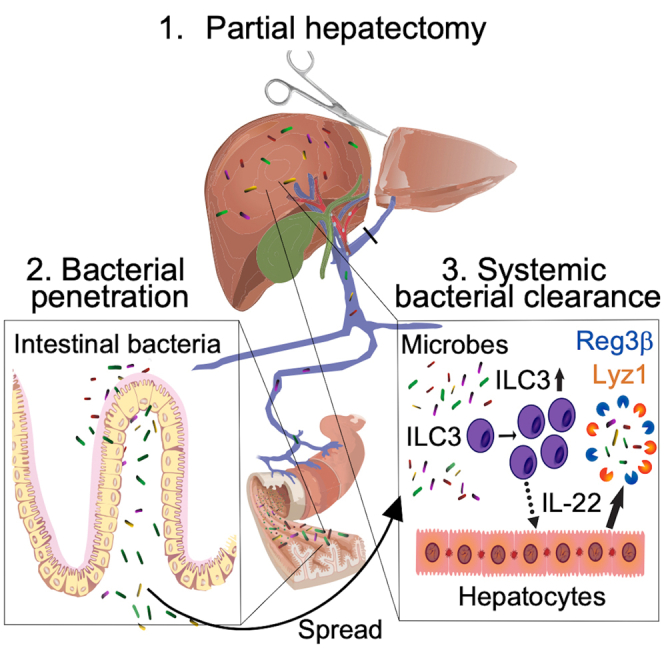
Graphical abstract
Highlights
-
•
Translocating intestinal bacteria cause surgery-related infections
-
•
Systemic dissemination of intestinal bacteria is restricted by ILC3s via IL-22
-
•
ILC3s promote antimicrobial peptide production in hepatocytes
-
•
The ILC3-controlled spread of intestinal bacteria limits liver regeneration
Jakob et al. report that surgical infections in humans have an intestinal microbial signature and, thus, an enteric origin. Modeling surgery in mice indicates that ILC3s control the systemic spread of intestinal microbes via IL-22 and induction of antimicrobial peptides in hepatocytes. Therefore, ILC3s may be targeted to limit surgery-associated infections.
Introduction
Infections following invasive interventions, such as in surgery, are the most frequent complication in humans1 and are independently associated with postoperative morbidity and mortality.2,3 The prevailing view has been that post-surgical infections are either caused by exogenous bacteria (e.g., colonizing the skin of the patient),4 by contamination of the operative field (e.g., by the surgeon), or by contaminated instruments and/or material.5 Accordingly, the vast majority of efforts to minimize surgical infections focused on reducing contamination and on depleting skin bacteria. However, even though many measures are taken, 10% of patients after surgery still develop infections suggesting alternative etiologies and mechanisms. Indirect evidence suggests that intestinal bacteria translocate across the intestinal epithelial barrier to systemic organs via the bloodstream following major surgery.6 Therefore, penetration of epithelial barriers and consequential bacterial spread may indicate failed anatomical containment of microbes.7
The mammalian intestine harbors the highest number of microorganisms of all barrier organs. Although required for many physiological functions, intestinal microorganisms represent a continuous threat to the host because of the potential to invade the organism.8 Due to its position, the liver has a pivotal role in microbial clearance because it constantly receives blood via the portal vein and is thus the primary draining hub of the intestine.9 This role becomes evident in hepatic diseases such as steatohepatitis, liver cirrhosis, and portal hypertension, in which the impaired hepatic function correlates with an increased translocation and systemic spread of intestinal bacteria.10 Thus, the overarching hypothesis in the current study is that infections after surgery are derived from the spread of bacteria from the intestine to the site of surgery. However, cellular mechanisms limiting systemic microbial dissemination after surgery remain unexplored.
Innate lymphoid cells (ILCs) emerge as first-line defenders at mucosal surfaces, fortifying the epithelial barrier against infections and mediating the containment of intestinal bacteria. ILCs enclose a heterogeneous group of innate immune cells that are analogous to T cells derived from the common lymphoid precursors. However, unlike T cells, ILCs lack rearranged antigen-specific receptors. ILCs can be developmentally and functionally classified in cytotoxic (natural killer [NK] cells) and helper-like ILCs, which can be further divided into three subpopulations: ILC1s, ILC2s, and ILC3s.11 Each subpopulation of ILCs is characterized by the expression of a lineage-specifying transcription factor, which determines the cell fate and the effector response.12 ILC3s are dependent on the transcription factor RORγt and can be divided into two subpopulations, CCR6+ and CCR6− ILC3s, with different characteristics.13,14 CCR6+ ILC3s are mainly localized in cryptopatches in the intestine and are major producers of interleukin (IL)-17 and IL-22 to protect against bacterial, viral, or fungal infections.15,16,17,18,19 In contrast, CCR6− ILC3s undergo a process that includes the loss of the lineage-specifying transcription factor RORγt, the downregulation of IL-22, and induction of T-bet instructing type 1 programs in these cells, including the production of interferon gamma (IFN-γ).14,20,21,22,23,24,25,26
At barrier surfaces, ILC3s are predominantly activated by myeloid cytokines, such as IL-1, IL-23, and glial cell line-derived neurotrophic factor (GDNF) to produce IL-22.17,18,27,28 The importance of IL-22 in mediating barrier function in steady state and during infection was mainly investigated in the gut, where intestinal epithelial cells expressing the IL-22 receptor are the main target of IL-22.29 IL-22 regulates antimicrobial peptide production of the Reg3 family members in Paneth cells, glycosylation of the epithelium, containment of intestinal bacteria, and proliferation of stem cells in the context of tissue repair and cancer. Moreover, IL-22 has a major role in protecting the barrier against infections with Citrobacter rodentium, Rotavirus, Salmonella, and Listeria monocytogens.17,18,27,29,30,31,32
ILCs are also present in systemic organs, such as the liver, spleen, and heart, and could potentially contribute to the fight against microbes by fine-tuning the local environment. The liver is armed with ILC3s in homeostatic conditions,33,34 in particular, NKp44− ILC3, which are the dominant hepatic ILC population in humans.35,36 Similar to what has been observed in the intestine and in the lungs, the effector cytokines of ILC3s in the liver are relevant for tissue homeostasis37,38 and liver regeneration post surgery.39 However, the contribution of liver ILC3s in the context of systemic antibacterial defense and outcomes after liver surgery, such as liver regeneration, remains unexplored.40
In this study, we reveal that microbes causing surgical infections in visceral surgeries, but in particular in liver surgery, have a common and mostly enteric profile suggesting their intestinal origin. Thus, we modeled major liver surgery in mice that revealed ILC-dependent passage of bacteria from the intestinal lumen to otherwise sterile systemic organs. By using multidimensional flow cytometry, we found a specific activation and proliferation of hepatic CCR6+ ILC3s upon liver surgery. Using state-of-the-art genetically modified mouse models, we show that ILC3s prevented the dissemination of intestinal bacteria to systemic organs. Mechanistically, ILC3s induced the production of antimicrobial peptides in the liver and thereby eliminated microbes from the circulation. As a consequence of intestinal bacterial dissemination, liver regeneration, as a surrogate of the local healing response, was impaired. Such findings indicate a pivotal role of systemic ILC3s for host protection and imply that ILC3s may be a target for future treatments to overcome intestinally derived systemic bacterial infections to ensure patients’ recovery.
Results
Surgical infections are caused by a defined set of predominantly intestinal microbes
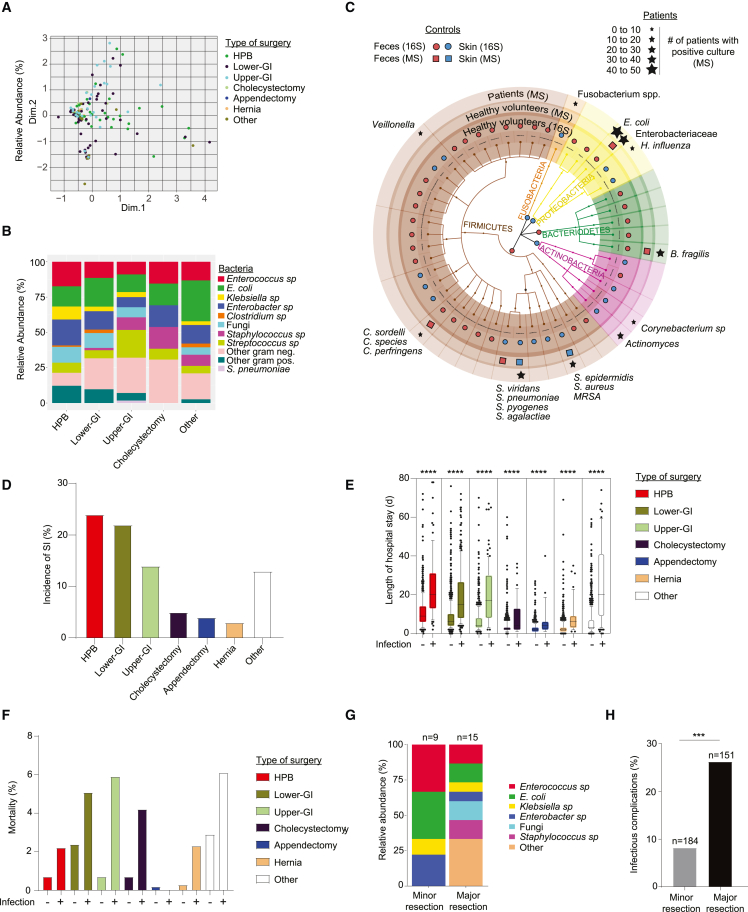
To characterize the microbes causing surgery-related infections (SIs), we determined the microorganisms in a multicenter cohort of patients who underwent general surgery. In a subset of patients with suspected SI, samples from the site of infection were cultured under aerobic and anaerobic conditions, and positive cultures were plated and analyzed using MALDI-TOF mass spectrometry within the routine clinical pipeline. The type and site of surgery did not predict microbial strains isolated from SI (Figure 1A). Regardless of the type of surgery, the main species identified were Enterococcus sp. and Enterobacteriaceae, such as Escherichia coli, and Clostridium sp. (Figure 1B). To associate these findings with potential patient-derived sources of bacteria (cutaneous versus intestinal), we analyzed feces and skin samples from healthy controls using MALDI-TOF and 16S rRNA sequencing techniques. Both approaches indicated that the microbes found in SI were mainly representative of the intestinal and, to a minor degree, the cutaneous microbiome of healthy controls (Figure 1C), suggesting host penetration of intestinally derived microbes. The highest incidence of SI was found in surgeries affecting the liver, the pancreas, and the biliary tracts (hepato-pancreatic-biliary surgery) as well as in surgeries of the small and large intestine (lower gastrointestinal surgery) (Figure 1D). Because infections and surgical outcome are strongly interlinked in most types of surgery,41 the same cohort was used to identify associated clinical consequences, such as length of hospital stay and mortality. Infections in these surgeries led to prolonged length of hospital stay (Figure 1E) and increased mortality (Figure 1F). Given the high incidence of infections and the clinical consequences in patients following liver surgery, the findings of this first cohort were validated in a second cohort of 335 patients undergoing major or minor liver surgery. Similar to the first cohort (Figure 1B), the catalog of microbes causing sepsis revealed a predominantly enteric microbial pattern (Figure 1G). Furthermore, the frequency of infectious complications (SI, sepsis) was remarkably higher in patients who underwent major liver resection compared with minor liver resections (Figure 1H). Together, these results suggest that, in abdominal operations, microbes with a potential intestinal origin cause most infections. Such findings argue for the penetration of microbiota from the intestine to systemic sites.
Figure 1.
A multicenter cohort revealed a limited but not surgery-specific composition of intestinal microbes causing surgical infections
A multicenter cohort of 3,515 patients from three Swiss surgical departments was analyzed.
(A and B) Microbial profile of surgical infections (SIs) found in patients with infectious complications. (A) Multiple correspondence analysis (MCA) plot of bacteria found in different surgical procedures. One dot represents one patient. (B) Distribution of bacterial species in SI cultures that underwent different surgical procedures as indicated.
(C) Overall representation of taxa significantly associated with either 96 fecal or 88 skin samples from the healthy volunteers analyzed by 16S rRNA amplicon sequencing are plotted and overlapped with bacterial strains that were identified with MALDI-TOF after culturing (mass spectrometry [MS]) of the fecal or skin samples from 10 healthy volunteers. The red closed circles (16S) or squares (MS) show taxa with increased abundances in fecal samples, and the blue circles/squares show taxa with increased abundances in the skin samples. Stars on top of the graph represent the identified culturable bacteria in SI in patients.
(D) Incidence of SI in different surgical procedures.
(E) Length of hospital stay and type of surgery for patients with or without SI. Each dot represents one patient. Errors bars indicate the median ± SD.
(F) Percentage of mortality related to each type of surgery based on the incidence of infection.
(G and H) A second cohort of 335 patients from the University Hospital of Vienna (Austria) undergoing minor or major liver surgery for colorectal liver metastasis was analyzed. (G) Identification of bacteria cultured from blood of patients with sepsis. (H) Frequency of SI in patients that underwent minor or major liver surgery. HPB, hepatopancreatobiliary surgery; GI, gastrointestinal. Normalized values were analyzed by Student’s t test to compare two experimental groups or by ANOVA to compare more than two groups in parallel. The p values are indicated as follows: ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
The innate immune system controls the systemic dissemination of intestinal bacteria independently of the intestinal microbial composition
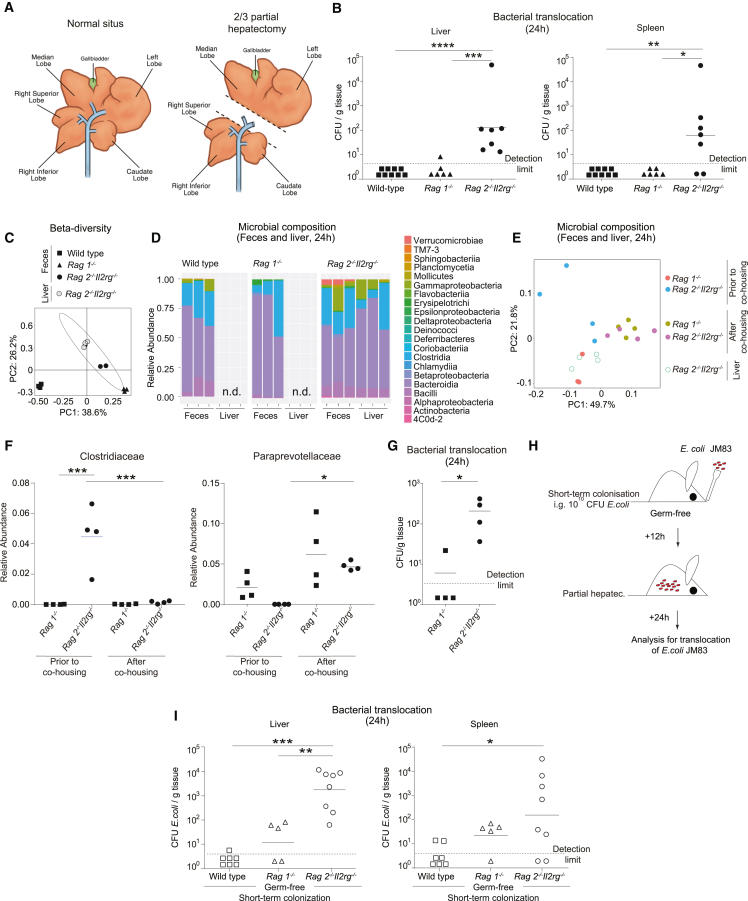
In the light of our human data that suggested that SIs are caused by intestinal bacteria (Figure 1), we first analyzed whether the gut permeability was altered following surgery by using an established murine model of two-third partial hepatectomy (PH) (Figure 2A). After intravenous (i.v.) injection of FITC-dextran and applying in vivo endomicroscopy into the small intestine, we found a specific increase in intestinal leakage induced by PH (Figure S1A), suggesting a surgery-dependent increase in gut permeability. To evaluate a potential spread of intestinal bacteria to systemic organs and to study the role of the immune system, surgery was not only performed in specific pathogen-free (SPF)-colonized wild-type C57BL/6 mice but also in Rag1−/− mice lacking adaptive lymphocytes (T cells and B cells) and alymphoid Rag2−/−Il2rg−/− mice lacking ILCs in addition to T and B cells. Using these genetically modified mouse models, systemic dissemination of bacteria was analyzed after 24 and 48 h post PH by culturing aerobic and anaerobic bacteria from mesenteric lymph nodes (MLNs), liver, and spleen. At both time points, we detected microbes in the liver and spleen of Rag2−/−Il2rg−/− mice (Figures 2B and S1B–S1D), whereas no bacteria could be cultured from the liver and spleen of wild-type and Rag1−/− mice, suggesting that components of the innate lymphoid compartment actively limit bacterial spread. Sanger sequencing of aerobic and anaerobic colonies identified E. coli and Enterobacter sp. as the main species in the liver and the spleen of Rag2−/−Il2rg−/− mice (Figure S1E), which usually populate the intestine and are among the most relevant bacterial species causing surgical infections in humans (Figure 1B). By whole-tissue 16S rRNA gene sequencing, the bacterial species of the classes Bacteroidia, Gammaproteobacteria, and Clostridia were identified in the liver of Rag2−/−Il2rg−/− mice, similar to the profile found in the intestine, suggesting microbial penetration from the intestine (Figures 2C and 2D). However, we also observed differences in intestinal microbial composition between different knockout mouse models, which may cause systemic bacterial spread. Therefore, Rag1−/− and Rag2−/−Il2rg−/− mice were co-housed for 3 weeks after weaning. Co-housing showed that the microbiome aligned independently of the genotype prior to PH, which was especially evident in the microbial families Clostridiaceae and Paraprevotellaceae (Figures 2E and 2F). However, even though the microbial profile was similar in both genotypes, bacterial systemic dissemination occurred only in Rag2−/−Il2rg−/− mice, excluding an impact of intestinal dysbiosis in the observed phenotype (Figure 2G). Next, germ-free Rag2−/−Il2rg−/−, Rag1−/−, and wild-type mice were mono-colonized with E. coli, the main translocating bacterium in previous experiments (Figure S1E). We gavaged 1010 CFU (colony-forming unit) E. coli JM83 (streptomycin-resistant strain was used for efficient tracking) before surgery (Figure 2H), and culturable streptomycin-resistant E. coli in tissues was measured. The bacterial titers in livers and spleens of Rag2−/−Il2rg−/− mice were significantly higher than in Rag1−/− mice, while no bacteria were found after mono-colonization of wild-type mice (Figure 2I). These results suggest that the lack of ILCs in Rag2−/−Il2rg−/− mice promotes the systemic dissemination of intestinal bacteria upon liver surgery.
Figure 2.
The ILC-compartment controls intestinal bacterial dissemination to systemic organs
(A–D) Specific pathogen-free (SPF) wild-type, Rag1−/−, or Rag2−/−Il2rg−/− mice underwent major surgery modeled by two-third partial hepatectomy (PH). (A) Schematic representation of two-third PH in mice. (B) Bacterial titers (colony-forming units [CFUs]) in the liver and the spleen in SPF wild-type, Rag1−/−, and Rag2−/−Il2rg−/− mice 24 h after PH. (C and D) Microbial composition within the feces and liver were determined by 16S rRNA amplicon analysis. (C) Principal-component analysis was done using Bray-Curtis distances on all operational taxonomic units (OTUs). (D) Microbial composition at class level.
(E–G) SPF Rag1−/− and Rag2−/−Il2rg−/− mice were co-housed at weaning for 3 weeks. Microbial composition determined by 16S rRNA amplicon sequencing in feces before or after co-housing. The liver tissue was analyzed 24 h after PH. (E) Principal-component analysis was done using Bray-Curtis distances on all OTUs. (F) Relative abundance of Clostridiaceae and Paraprevotellaceae in feces. (G) Bacterial titers in the liver were assessed 24 h post PH.
(H and I) Germ-free wild-type, Rag1−/−, and Rag2−/−Il2rg−/− mice were short-term colonized with 1010 CFU of E. coli JM83 12 h prior to PH. (H) Scheme of experimental setup. (I) E. coli titers (CFU) in the liver and spleen 24 h after PH. Geometric means for log scales and arithmetic means for linear scales are shown. Normalized values were analyzed by Student’s t test to compare two experimental groups or by ANOVA to compare more than two groups in parallel. Data in (C)–(G) are representative of at least n = 3 mice per group in two independent experiments, and data in (B) and (I) are pooled based on n = 5–9 mice from two or more independent experiments. The p values are indicated as follows: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. See also Figure S1.
PH stimulates hepatic CCR6+ ILC3s
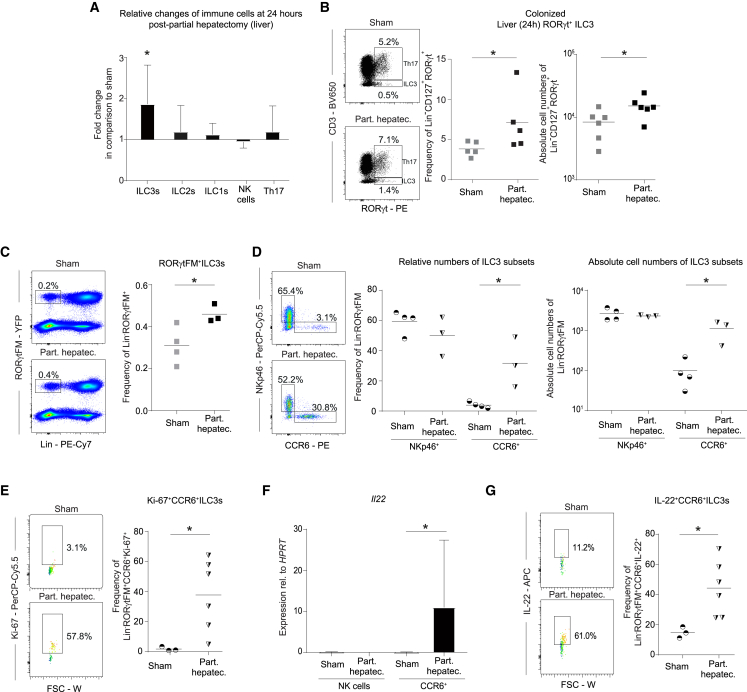
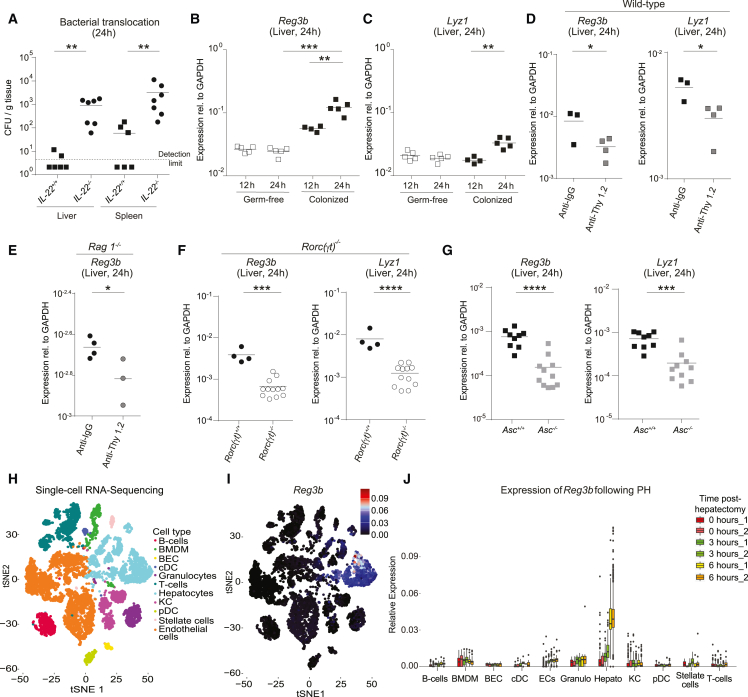
To dissect the immune response, which may limit systemic microbial dissemination in the context of liver surgery, we analyzed the innate lymphoid compartment in the small intestine, MLN, liver, and spleen by multidimensional flow cytometry (gating strategy liver [including antibody-validation for RORγt] Figures S2A and S2B, and small intestine Figure S3A). At 24 h post PH, we found RORγt+ ILC3s (Lin−CD127+RORγt+ cells) to be significantly decreased in the small intestine (Figures S3B and S3C) and increased in the liver (Figure 3A) compared with sham-operated mice, whereas no change was detected in the MLN and the spleen (Table S1). Other ILC subsets remained unchanged in all organs studied (liver, Figures 3A and S2C; small intestine, Figures S3B and S3C; MLN and spleen, Table S1). While the role of small-intestinal ILC3s for host-microbial compartmentalization has been previously described,29 the role of hepatic ILC3s remains elusive. In particular, the specific increase of liver ILC3s suggested targeted activation of these cells. Hepatic RORγt+ ILC3s expanded at 24 h post PH (Figure 3B) in relative and absolute cell numbers compared with sham-operated wild-type SPF but not germ-free mice (Figure S2D), suggesting that the exposure of microbes induced the increase in RORγt+ ILC3s. To gain a more precise view on the role of RORγt+ ILC3s, we used Rorc(γt)Cre-TgRosa26REYFP/+ mice, which label all RORγt-expressing cells. In steady state, we observed that the majority of these cells were NKp46+ CCR6− or were double-negative for NKp46 and CCR6. CCR6+ ILC3s were found in relatively low abundance (Figure S2E). The vast majority of hepatic ILC3s expressed the markers NK1.1 and CD90 (Thy1). Other markers known to be expressed on intestinal ILC3s, such as MHCII, CD4, Nrp-1, and NKG2D, were only expressed by a minor proportion (Figure S2E). As expected, hepatic RORγt ILC3s are developmentally dependent on the transcription factor RORγt (Figure S2F). By applying the PH-model in Rorc(γt)Cre-TgRosa26REYFP/+ mice, we observed a similar increase of labeled cells following PH (Figure 3C) as in colonized wild-type mice (Figure 3B). Specifically, we observed a profound increase in relative and absolute cell numbers of CCR6+ ILC3s at 24 h post PH, whereas NKp46+ ILC3s remained unaffected compared with sham-operated mice (Figure 3D). To elaborate whether this increase was due to recruitment or local expansion of CCR6+ ILC3s, we measured Ki-67 expression as marker for proliferative cells by flow cytometry. CCR6+ ILC3s showed a significant increase in proliferation post PH compared with sham-operated mice (Figure 3E). Additionally, we observed blasting of CCR6+ ILC3s, which is a consequence of cell activation. Consistent with this finding, we detected increased IL-22 mRNA (Figure 3F) and protein (Figure 3G) in CCR6+ ILC3s. To identify the spatial distribution of ILC3s in the liver, we performed confocal microscopy of Rorc(γt)Cre-Tg Rosa26REYFP/+ mice. By co-staining RORγtYFP cells with CD3 to exclude RORγt+ T cells, we observed a broad and random distribution of RORγtYFP CD3− ILCs within the liver and did not detect clusters of RORγtYFP CD3− ILCs in steady state as well as post PH (Figure S2G). Thus, PH induces proliferation and activation of hepatic RORγt+ ILC3s, a process dependent on the presence of microbiota.
Figure 3.
Activated hepatic CCR6+ ILC3s proliferate in response to partial hepatectomy
(A) Cellular changes compared with sham-operated mice of different ILC subsets analyzed by flow cytometry in livers of SPF wild-type mice 24 h after PH (aritmetic mean ± SD). ILC1s, Lin−CD127+RORγt−Tbet+; ILC2s, Lin−CD127+RORγt−GATA+; ILC3s, Lin−CD127+RORγt+; NK cells, Lin−CD127−NKp46+; Th17 cells, CD19−CD3+RORγt+.
(B) Flow cytometric quantification and statistical analysis of liver RORγt+ILC3s 24 h after PH or sham surgery.
(C–G) Rorc(γt)Cre−TgRosa26REYFP/+ (RORγtFM) mice underwent PH and were analyzed by flow cytometry. (C and D) ILC3s and their subsets at 24 h post PH. (E) Proliferation of CCR6+ ILC3s was assessed by Ki-67 24 h after PH. (F) Sort purification of RORγt+ ILC3s and measurement of IL-22 by qPCR (aritmetic mean ± SD). (G) Flow cytometry analysis of IL-22 production by CCR6+ ILC3s. Geometric means for log scales and arithmetic means for linear scales are shown. Normalized values were analyzed by Student’s t test to compare two experimental groups or by ANOVA to compare more than two groups in parallel. Data in (A)–(G) are representative of n = 3–4 mice per group in two or more independent experiments. The p values are indicated as follows: ∗p ≤ 0.05. See also Figures S2 and S3 and Table S1.
ILC3s control the systemic dissemination of intestinal bacteria
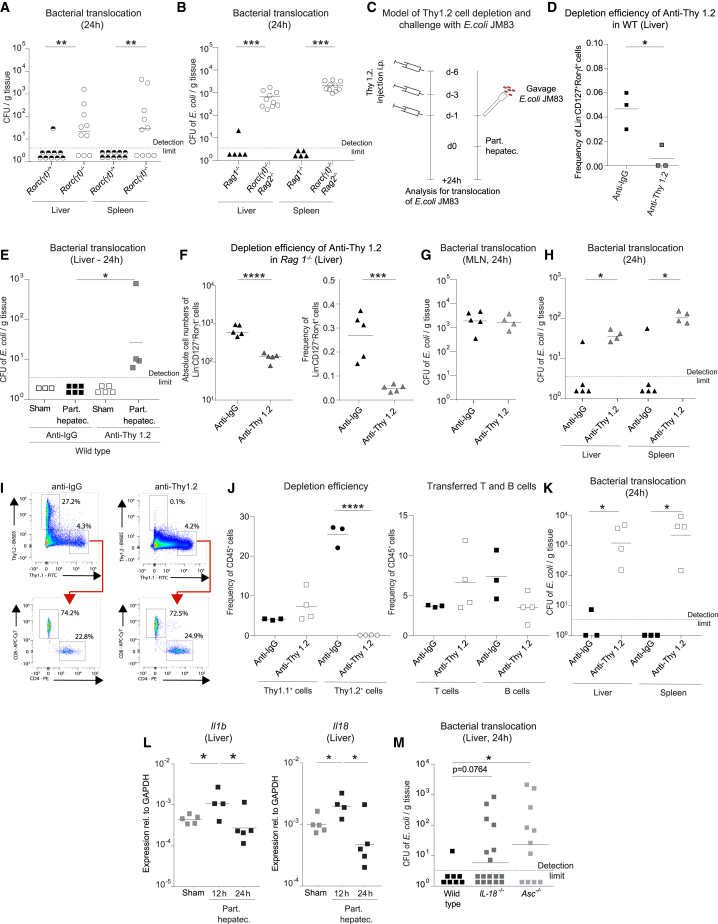
The finding that hepatic ILC3s were activated following PH led us to hypothesize that ILC3s may have a pivotal function in mediating immunity against bacterial spread. Because there is currently no in vivo mouse model for selective depletion of ILC3s in lymphoreplete mice, the contribution of ILC3s was assessed in Rorc(γt)−/− mice, which lack ILC3s and T helper (Th) 17 cells (but have all other T cells and ILCs subsets). Intestinal bacteria disseminated to the liver and spleen of Rorc(γt)−/− mice, whereas the liver and spleen of co-housed, littermate Rorc(γt)−/+ remained clean of microbes (Figure 4A). To further exclude a specific role of T cells in systemic antimicrobial defense, we performed the same experiments in Rorc(γt)−/− mice on a Rag2−/− background and compared them with Rag1−/− mice (Figure 4B). This approach allows us to link any functional defect specifically to ILC3s as Rorc(γt)−/−Rag2−/−mice specifically lack ILC3s compared with T cell-deficient Rag1−/− mice. We found a significant spread of intestinally delivered E. coli JM83 into systemic organs, such as the liver and the spleen, specifically in Rorc(γt)−/−Rag2−/− and not in Rag1−/− mice. To further exclude the role of Th17 cells in our model, we depleted CD4+-expressing cells in wild-type mice via two injections of anti-CD4 prior to PH (Figure S4A). ILC3s in the liver do not express CD4 (Figure S2E), and, hence, they are not affected by this depletion strategy. Following anti-CD4 injection and PH, we did not observe systemic microbial spread (Figure S4B). These data pinpoint the necessity of ILC3s in host defense mechanisms.
Figure 4.
ILC3s are required to prevent intestinal bacterial spread to liver and spleen
(A and B) (A) SPF Rorc(γt)−/+ and Rorc(γt)−/− littermate mice and (B) SPF Rag1−/− or Rorc(γt)−/−Rag2−/− mice underwent PH. Antibiotic-resistant E. coli JM83 was intragastrically (i.g.) delivered 12 h before PH for efficient trackability. E. coli titers (CFU) were assessed in the liver and spleen 24 h after PH.
(C) Experimental setup used in experiments for Thy 1.2 cell depletion using anti-Thy 1.2 or anti-immunoglobulin G (IgG) antibody administered 6 days, 3 days and 1 day prior to PH in SPF (D and E) wild-type or (F–H) Rag1−/− mice. Twelve hours before the PH or sham surgery, 1010 CFU of E. coli JM83 was i.g. delivered to the mice.
(D) Depletion efficiency of ILC3s in wild-type mice at day 0 (d0).
(E) E. coli titers (CFU) in the liver following Thy1.2 depletion in wild-type mice 24 h after PH.
(F) Depletion efficiency of ILC3s in Rag1−/− at d0.
(G and H) E. coli titers (CFU) in (G) MLN and (H) liver and spleen following Thy1.2 depletion in Rag1−/− mice 24 h after PH.
(I–K) Sort-purified CD19+ B cells and CD3+CD5+ T cells from spleen and MLN of Thy1.1 mice were transferred i.v. to Rag1−/− Thy1.2 recipient mice, and i.p. injection of anti-Thy1.2 was performed three times prior to PH. (I and J) Depletion efficiency of Thy 1.2+ cells and presence of T and B cells at d0. (K) E. coli titers (CFU) in the liver and spleen 24 h after PH.
(L) Expression of IL1b and IL18 in sham-operated or PH wild-type mice determined by RT-qPCR.
(M) Colonized wild-type, IL-18−/−, and Asc−/− underwent PH. Antibiotic-resistant E. coli JM83 was i.g. delivered 12 h before PH. E. coli titers (CFU) were assessed in the liver and spleen 24 h after PH. Geometric means for log scales and arithmetic means for linear scales are shown. Normalized values were analyzed by Student’s t test to compare two experimental groups or by ANOVA to compare more than two groups in parallel. Data in (D)–(K) are representative of n = 3–5 mice per group in two independent experiments, and data in (A), (B), and (M) are pooled based on n = 5–17 mice from two or more independent experiments. The p values are indicated as follows: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. See also Figure S4.
Given that the MLN are an integral part of the mucosal defense system, we tested whether the deficiency of MLN architecture could contribute to the bacterial dissemination observed in Rag2−/−Il2rg−/− and Ror(γt)−/−, which are equipped with primitive and undifferentiated secondary lymphoid structures.42 To study the role of ILCs without affecting secondary lymphoid structures, we injected anti-Thy1.2 antibody in wild-type mice to deplete ILC3s but also other cells expressing Thy1.2, including T cells (Figure 4C). The depletion efficiency of ILC3s was high as assessed by flow cytometry at the day of surgery (Figure 4D). Mice were then gavaged with E. coli JM83 and followed by PH 24 h later (Figure 4C). E. coli was found in the liver of operated but not sham-operated wild-type mice (Figure 4E), indicating the contribution of Thy1.2+-producing cells, including ILC3s, in the clearance of circulating bacteria. To delineate the contribution of ILCs, anti-Thy1.2 antibody injections were performed in Rag1−/− mice, where ILCs are the unique Th1.2+ lymphoid population.29 Depletion efficiency of hepatic and intestinal ILC3s in relative and absolute cell numbers was high, whereas other non-lymphoid tested cell types remained unaffected (Figures 4F and S4C). Bacterial CFU detected in the MLN post-surgery was equivalent in Rag1−/− mice following anti-Thy1.2 antibody administration compared with isotype control mice (Figures 4G and S1A). However, conversely to wild-type mice, bacteria were detected in the liver and spleen of ILC-depleted Rag1−/− mice (Figure 4H), indicating that the presence of bacteria in systemic organs is not the consequence of an increased bacterial load or their retention-capacity in the MLN.
It has been reported that the absence of T cells, such as in Rag−/− mice, leads to an expansion of ILC3s in the intestine, which may alter tissue physiology.43,44 To study ILC3s in the absence of T cells also in the liver, we phenotyped Rag1−/− mice in steady state and compared them with wild-type mice. Similar to what has been described in the intestine, we observed an increase in ILC3s and Ki67+ ILC3s in the liver of Rag1−/− mice,44 which may indicate that ILC3s have a compensatory role in T cell deficiency also in the liver (Figures S4D and S4E). To control for the absence of T cells and to generate lymphoreplete mice, we generated CD90-disparate chimeras as previously described.29 The lymphoid compartment of Rag1−/− (Thy1.2, CD90.2) mice was reconstituted by sort-purified B and T cell (Th1.1, CD90.1) transfer (Figure S4F). We confirmed that the T cell compartment was reconstituted and remained unaltered upon anti-Thy1.2 depletion (Figures 4I and 4J and S4G). Thereafter, the ILC compartment was depleted using anti-Thy1.2 (CD90.2) antibody prior to PH in a now lymphoreplete setting (Figures 4I and 4J). After ILC depletion, we found significant systemic bacterial spread to the liver and the spleen, suggesting that ILCs have a non-redundant role in clearing disseminated intestinal bacteria (Figure 4K). Collectively, our data strongly indicate that the loss of control of systemic bacterial dissemination clearly segregates with the presence of ILC3s (Figure S4H).
ILC3s are mainly activated by myeloid-derived cytokines, such as IL-1β and IL-18.34 By analyzing RNA-sequencing data of livers at steady state and after PH (Figure S5), we observed an increased expression of IL-1β and its receptor IL1r2 (Figure 4L, Figure S5), which are known functional regulators of ILCs in other tissues.45 To assess the potential functional relevance of these cytokines, we analyzed systemic bacterial dissemination following PH in IL-18−/− and Asc−/− mice (which lack active IL-1β and IL-18). The absence of active forms of both IL-1β and IL-18 in Asc−/− mice allows intestinal bacteria to disseminate to systemic organs (Figure 4M).
ILC3s control production of antimicrobial peptides in the liver
It has been shown that ILC3s induce antimicrobial peptide production at mucosal barriers, such as the intestine, via secretion of IL-22 and activation of epithelial cells expressing the IL-22 receptor.18,29,46 However, the mechanism by which ILC3s regulate systemic bacterial clearance is unclear. We first investigated in IL-22-deficient mice whether the cytokine IL-22 is required for systemic spread of bacteria following PH. Consistent with previous studies assigning an important function to IL-22 in promoting barrier immunity, we detected increased bacterial dissemination after PH (Figure 5A). To determine downstream responses in hepatocytes that express high levels of IL-22 receptor, we analyzed the transcriptional signatures of host-microbial defense genes in the livers of SPF wild-type mice. At 24 h post PH, expression of intestinal C-type regenerating islet derived-3 lectins (Reg3b) against gram-positive bacteria and lysozyme C (Lyz1) against gram-positive and gram-negative bacteria18,47 was elevated in the liver (Figures 5B and 5C) but not in the intestine (Figure S6). To investigate whether intestinal circulating microbes contribute to the upregulation of antimicrobial peptides, we performed the same experiments in germ-free mice. In contrast to colonized mice, no induction of the expression of the hepatic antimicrobial peptides Reg3b and Lyz1 was observed in germ-free mice (Figures 5B and 5C), indicating that exposure to bacteria is necessary for the upregulation of antimicrobial peptides. To determine the relevance of ILCs on hepatic antimicrobial peptide production after surgery, we administered anti-Thy 1.2 antibodies in wild-type mice or Rag1−/−. The expression of Reg3b and Lyz1 in the liver was decreased in both wild-type and Rag1−/− mice (Figures 5D and 5E). To exclude the contribution of ILC1s and ILC2s in the induction of antimicrobial peptide-encoding genes, expression of Reg3b and Lyz1 was assessed in Rorc(γt)−/− mice. Both Reg3b and Lyz1 were decreased in Rorc(γt)−/− mice (Figure 5F). Furthermore, the expression of antimicrobial peptides encoding genes was assessed in mice lacking active forms of Il-1β and IL-18 known to regulate ILC3s (Asc−/− mice; Figure 4J).48,49 We observed a decreased expression of Reg3b and Lyz1 in Asc−/− mice (Figure 5G), suggesting that Il-1β- and IL-18-mediated activation of ILC3s may be involved in regulating antimicrobial peptides in the liver. To delineate cell types responsible for antimicrobial peptide production, analysis of single-cell RNA-sequencing data of liver cells in steady state and in mice at different time points after PH was performed (Figure 5H). The expression and PH-induced upregulation of the representative antimicrobial peptide-encoding gene, Reg3b, was only observed in the hepatocyte cluster (Figure 5I) but not other major cell types in the liver (Figure 5J), indicating that ILC3s elicited an immune response characterized by specific expression of Reg3b in hepatocytes.
Figure 5.
ILC3s regulate the hepatocyte-derived antimicrobial peptide production
(A) SPF Il22+/+ and Il22−/− mice underwent PH. Antibiotic-resistant E. coli JM83 was i.g. delivered 12h before PH. E. coli titers (CFU) were assessed in the liver and spleen 24 h after PH.
(B–G) Whole-tissue RNA was isolated from the liver and the expression of antimicrobial peptide-encoding genes was analyzed by RT-qPCR at the indicated time points. (B and C) SPF and germ-free wild-type mice. (D and E) Anti-Thy1.2 antibody or anti-IgG in (D) SPF wild-type and (E) SPF Rag 1−/− mice. (F) SPF Rorc(γt)+/+ and Rorc(γt)−/− mice. (G) Colonized Asc+/+ mice and Asc−/− mice.
(H) t-SNE dimensionality reduction visualization of single-cell RNA sequencing of purified liver cells showing the main cell types.
(I and J) Dropout corrected gene expression of Reg3b (aritmetic mean ± SD) at different time points after hepatectomy. Geometric means for log scales and arithmetic means for linear scales are shown. Normalized values were analyzed by Student’s t test to compare two experimental groups or by ANOVA to compare more than two groups in parallel. Data in (B)–(G) are representative of n = 3–12 mice per group in two or more independent experiments, and data in (A) are pooled based on n = 6–7 mice from two independent experiments. The p values are indicated as follows: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. See also Figures S5 and S6.
Failure in clearance of circulating microbes impairs liver regeneration
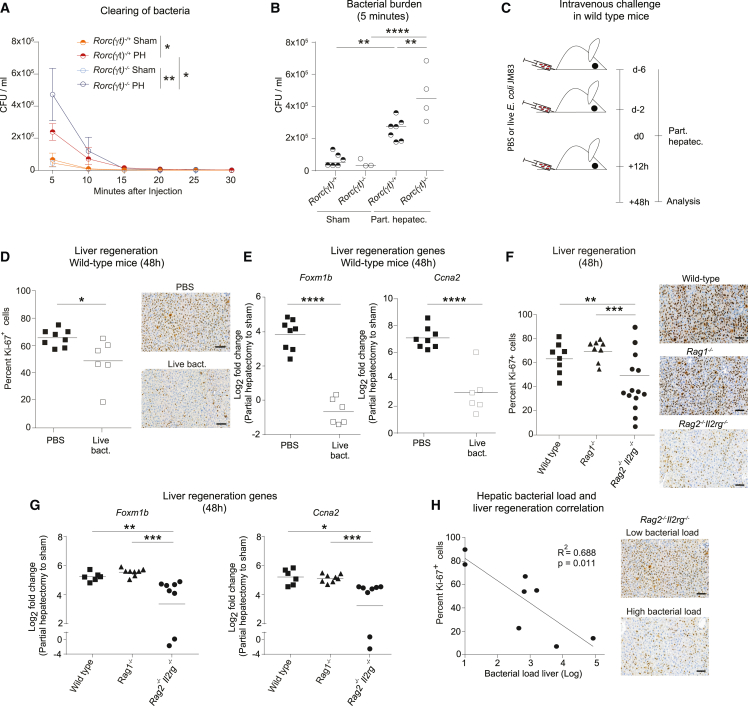
Based on the finding that absence of ILC3s led to systemic bacterial spread (Figures 2 and 4), we hypothesized that ILC3s participate in bacterial clearing mechanisms. In order to exclusively assess clearance of circulating bacteria and to avoid analyzing the role of the intestinal barrier, we measured bacterial titers in the blood of Rorc(γt)−/− and Rorc(γt)−/+ co-housed, littermate-control mice following i.v. delivery of E. coli (Figures 6A and 6B). This experimental setting showed that the systemic load of bacteria after PH was significantly increased and bacterial clearance was further delayed in Rorc(γt)−/− mice compared with their littermate heterozygous controls. These experiments reveal the relevance of bacterial clearance after PH and further indicate the functional relevance of ILC3s in this process.
Figure 6.
Colonization of the liver by circulating intestinal microbes impairs liver regeneration
(A) SPF Rorc(γt)−/+ and Rorc(γt)−/− littermate mice were subjected to sham or PH. Immediately after the operation, 107 CFU of E. coli JM83 were injected i.v. and E. coli titers (CFU; geometric mean ± SD) were assessed in the blood.
(B) E. coli titers (CFU) were assessed in the blood 5 min after PH.
(C) Experimental scheme: SPF wild-type mice were i.v. injected with 107 CFU E. coli JM83 6 and 2 days prior to and 12 h after PH (in D and E).
(D–H) Hepatic regeneration 48 h after surgery was assessed by analyzing the percentage of Ki-67+ hepatocytes or the expression of cell cycle regulatory genes (Foxm1b and Ccna2). Representative images are presented on the right and quantification of proliferation is presented on the left. (F–H) SPF wild-type, Rag1−/−, and Rag2−/−Il2rg−/− mice underwent PH. (H) Correlation of hepatic bacterial load and liver regeneration in SPF Rag2−/−Il2rg−/− mice. Scale bars in the pictures indicate 100 μm. Geometric means for log scales and arithmetic means for linear scales are shown. Data in (A) and (B) are representative of n = 3–7 mice per group in two independent experiments, and data in (D)–(H) are pooled based on n = 6–14 mice from two or more independent experiments. The p values are indicated as follows: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
Sepsis and SIs are the most important determinants of clinical outcome in human surgery (lancetglobalsurgery.org). Therefore, we evaluated whether circulating bacteria impair liver regeneration, as a surrogate of hepatic recovery after tissue loss, by measuring liver regeneration 48 h after PH. We i.v. injected E. coli before and after PH (Figure 6C) and assessed the indices of hepatocellular proliferation using the expression of Ki-67 and target genes regulating the cell cycle (Foxm1b and Ccna2). Hepatocellular proliferation (Figure 6D) and the hepatic expression of Foxm1b and Ccna2 (Figure 6E) were decreased in wild-type mice after systemic administration of bacteria compared with PBS controls. Next, we assessed whether the presence of disseminated bacteria observed in Rag2−/−Il2rg−/− mice (see Figure 2B) could similarly impair liver regeneration. Rag2−/−Il2rg−/− showed a decrease in liver regeneration compared with wild-type and Rag1−/− mice (Figures 6F and 6G). Furthermore, systemic bacteria in the liver in Rag2−/−Il2rg−/− mice were associated with failure in liver regeneration as illustrated by the negative correlation of hepatocellular proliferation with hepatic bacterial load (Figure 6H). Altogether, these data indicate that failure of the ILC3-controlled systemic dissemination of intestinal bacteria leads to impaired liver regeneration. Similar findings have been observed in patients undergoing surgery with infection-associated increase in mortality and length of hospital stay (Figures 1E and 1F).
Discussion
Clinical practice currently aims to prevent healthcare-associated infections by keeping the surgical environment as aseptic as possible. Data from a large patient cohort now demonstrate that a large proportion of surgical infections as well as post-surgery septicemia or sepsis have a predominant intestinal microbial signature and are, thus, likely to be endogenous by origin.
ILC3s have been implicated in instructing antibacterial immunity at mucosal sites. We now found that ILC3s play a crucial role in systemic antibacterial immunity to prevent SIs and that they are important regulators of the hepatic antimicrobial defense via the induction of antimicrobial peptides in hepatocytes. In this way, ILC3s promote the clearance of systemic translocated intestinal bacteria to ensure optimal liver regeneration. The finding that surgical infections are of intestinal origin and are prevented by ILC3s identifies new therapeutic targets that could be harnessed for therapy by modulating ILC3s’ function.
At barrier surfaces, ILC3 epithelial crosstalk has broad functional implications in responses to genotoxic stress,50 tissue regeneration,51 microbial compartmentalization,29 and nutrient uptake.52 The effects of ILC3s on epithelial cells are mainly mediated by the bystander production of the epitheliotropic cytokine IL-22. Interestingly, we found selective expansion of CCR6+ ILC3s but not CCR6− ILC3s after PH. This is consistent with published data showing that CCR6+ ILC3s are high producers of IL-22, whereas the cytokine is downregulated in CCR6− ILC3s.14 Hepatocytes that sense circulating bacteria from the portal blood have been reported to express high levels of IL-22 receptor, and recombinant IL-22 elicits protective effects against inflammation via strong activation of Stat3 signaling molecules.33 Such findings argue for similar downstream mechanisms of IL-22 beyond mucosal tissues. Similar to our results, ILC3s have been linked to systemic dissemination of intestinal bacteria to the liver and spleen in a sepsis model, in which IL-22 upregulates antimicrobial peptides in the intestine.53 Thus, ILC3 failure seems to be a broadly relevant patho-mechanism for systemic infections.
Similar to other ILCs, ILC3 activation is mediated by soluble factors, such as cytokines, metabolites, or inflammatory mediators.12 We detected upregulation of IL-1β and IL-18 as potential activating signals for liver ILC3s. Upon IL-1β activation, ILC3s induce the production of antimicrobial peptides, such as Reg3 and Lyz families, by intestinal epithelial cells to promote gut mucosal protection.29,54,55,56 Our results show that the expression of antimicrobial peptides in the liver requires both the presence of bacteria as well as ILC3s. Kupffer cells have been described to play an important role by clearing commensal microbes in the liver at steady state.7,9 Together with their phagocytic skills, Kupffer cells produce cytokines such as IL-1β in an inflammasome-dependent manner57 and could contribute to the activation of ILC3s. Future studies should address the exact activation signals of ILC3s.
Our study identifies that Enterobacteriaceae, which are facultative pathogens predominantly found in the intestine,58 are the main bacterial family causing SI. These pathobionts are known to cause disease in cases of disturbed intestinal homeostasis, especially in immunocompromised organisms.59 Enterobacteriaceae were the most abundant bacterial family found in wounds and the blood of surgical patients as well as in systemic organs of operated mice. 16S rRNA metagenomic sequencing of healthy controls showed traces of Proteobacteria in the blood,60,61 suggesting that members of particularly this phylum have the capability to enter the circulation. E. coli is a well-known Proteobacterium involved in secondary infections associated with liver disease and liver failure.10 Similar to our findings, E. coli reaches systemic organs, including the liver in case of liver failure.62 Another intestinal pathobiont, Enterococcus faecalis caused 10% of SIs in our patient cohort and also systemically translocated in our mouse model. E. faecalis is not displayed in the phylogenetic tree plot given its low frequency in the feces and high variability between volunteers. Despite low abundance in the intestinal biomass, bacteria such as intestinal E. faecalis have the ability to cause SI given it is highly resistant to antibiotics, forms biofilms, and produces highly efficient proteases to resist antimicrobial peptides.63
Healing responses, such as liver regeneration, are among the most relevant outcome parameters in surgery. We show that liver regeneration is dependent on the capability of the organism to clear systemic bacteria. The impact of the microbiota on liver regeneration has been shown before by the use of germ-free mice64 and antibiotics.65 Punctual or long-term depletion of microbes decreases liver regeneration and supports the assumption of an indirect effect of commensal bacteria through their metabolites66 for hepatocyte regeneration.
Limitations of the study
Because of the observational nature and the size of the clinical cohorts, it was not possible to correct for specific parameters. However, the data show that even though virtually all patients received antibiotics, enteric bacteria represent the largest fraction of culturable microbes causing infections. Sampling of liver tissue in the postoperative period would be ideal to investigate clearing mechanisms during liver regeneration in humans, which is impossible from an ethical aspect. While we found a significant increase of CCR6+ ILC3s in the liver, we also observed a decrease in small-intestinal ILC3s, which implies regulation of these cells in different anatomic compartments. Currently, there is no animal model available to selectively deplete ILC3s in different anatomic sites to causally prove the liver-specific role of liver-resident ILC3s.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse: CCR6-PE | Biolegend | Cat# 129804; RRID: AB_1279137 |
| Anti-mouse CD3-BV650 | Biolegend | Cat# 100229; RRID: AB_11204249 |
| Anti-mouse CD3-BV785 | Biolegend | Cat# 100231; RRID: AB_11218805 |
| Anti-mouse CD3-PE-Cy7 | Biolegend | Cat# 152314; RRID: AB_2629847 |
| Anti-mouse CD3-eFluor660 | Invitrogen | Cat# 50-0032-82; RRID:AB_10598657 |
| Anti-mouse CD31-BV480 | BD Bioscience | Cat# 746260; RRID: AB_2743592 |
| Anti-mouse CD4-APC-eFluor780 | eBioscience | Cat# 47-0041-82; RRID: AB_11218896 |
| Anti-mouse CD4-BV785 | Biolegend | Cat# 100551; RRID: AB_11218992 |
| Anti-mouse CD4-PE | eBioscience | Cat# 12-0042-82; RRID: AB_465510 |
| Anti-mouse CD5-PE-Cy7 | Biolegend | Cat# 100622; RRID: AB_2562773 |
| Anti-mouse CD8- APC Cy7 | BioLegend | Cat# 100714; RRID: AB_312753 |
| Anti-mouse CD11b-APC-Cy7 | Biolegend | Cat# 101226; RRID: AB_830642 |
| Anti-mouse CD11c PE-CF594 | BD Bioscience | Cat# 562454; RRID: AB_2737617 |
| Anti-mouse CD19-BV421 | Biolegend | Cat# 115537; RRID: AB_10895761 |
| Anti-mouse CD19- BV785 | Biolegend | Cat# 115543; RRID: AB_11218994 |
| Anti-mouse CD19- PE-Cy7 | Biolegend | Cat# 115520; RRID: AB_313655 |
| Anti-mouse CD44- APC Cy7 | Biolegend | Cat# 103028; RRID: AB_830785 |
| Anti-mouse CD103 Biotinylate | BD Bioscience | Cat# 557493; RRID: AB_396730 |
| Anti-mouse CD62L BV510 | Biolegend | Cat# 104441; RRID: AB_2561537 |
| Anti-mouse CD45-BUV395 | BD Bioscience | Cat# 564279; RRID: AB_2651134 |
| Anti-mouse CD90 – BV605 | Biolegend | Cat# 140317; RRID: AB_11203724 |
| Anti-mouse CD127-FITC | eBioscience | Cat# 11-1271-82; RRID: AB_465195 |
| Anti-mouse CX3CR1-FITC | Biolegend | Cat# 149020; RRID: AB_2565703 |
| Fixable Viability dye – efluor450 | eBioscience | Cat# 65-0863-14 |
| Fixable Viability dye – efluor506 | eBioscience | Cat# 65-0866-14 |
| Anti-mouse FceRIα-PE-Cy7 | Biolegend | Cat# 134318; RRID: AB_10640122 |
| Anti-mouse Foxp3-AF700 | eBioscience | Cat# 56-5773-82; RRID: AB_1210557 |
| Anti-mouse F4/80-APC | Biolegend | Cat# 123116; RRID: AB_893481 |
| Anti-mouse Gata3-AF647 | BD Bioscience | Cat# 560068; RRID: AB_1645316 |
| Anti-GFP AF488 | Thermo Fisher Scientific | Cat# A-213111; RRID: AB_221477 |
| Anti-mouse I-A/I-E-AF700 | Biolegend | Cat# 107622; RRID: AB_493727 |
| Anti-mouse I-A/I-E-BV650 | Biolegend | Cat# 107641; RRID: AB_2565975 |
| Anti-mouse Il-22-APC | eBioscience | Cat# 17-7222-82; RRID: AB_10597583 |
| Anti-mouse KI-67 Percp-Cy55 | BD Bioscience | Cat# 561284; RRID: AB_10611574 |
| Anti-mouse Ly6C-BV421 | Biolegend | Cat# 128032; RRID: AB_2562178 |
| Anti-mouse Ly6G-PE-Cy7 | Biolegend | Cat# 127618; RRID: AB_1877261 |
| Anti-mouse NK1.1-APC | Biolegend | Cat# 108720; RRID: AB_2132713 |
| Anti-mouse Neuropilin1-PE | Biolegend | Cat# 145204; RRID: AB_2561928 |
| Anti-mouse Nkp46-PercP-Cy5.5 | Biolegend | Cat# 137610; RRID: AB_10641137 |
| Anti-mouse RORγt-PE | eBioscience | Cat# 12-6981-82; RRID: AB_10807092 |
| Anti-mouse Siglec F-PE | Biolegend | Cat# 155506; RRID: AB_2750235 |
| Streptavidin-PE-Cy7 | Bioscience | Cat# 557598; RRID: AB_10049577 |
| Anti-mouse Tbet-PECy7 | eBioscience | Cat# 25-5825-82; RRID: AB_11042699 |
| Anti-mouse Thy 1.1 | eBioscience | Cat# 11-0902-82; RRID: AB_465154 |
| Anti-mouse Thy 1.2 | Biolegend | Cat# 140318; RRID: AB_2650924 |
| Rat IgG1 isotype control PE | Thermo Fisher Scientific | Cat# 12-4301-81; RRID:AB_470046 |
| Monoclonal anti-mouse Ki-67 (Tec3 Ab) | Dako, Glostrup Denkmark | Cat# M7249 |
| InVivoMAb rat anti-mouse IgG2b | BioXcell | Cat# BE0090; RRID: AB_1107780 |
| InVivoMAb anti-mouse CD4 | BioXcell | Cat# BE0003-1; RRID:AB_1107636 |
| InVivoMAb anti-mouse CD90.2 (30H12) | BioXcell | Cat# BE0066; RRID: AB_1107682 |
| Chemicals, peptides, and recombinant proteins | ||
| Formaldehydlösung 4% neutral | SAV Liquid Production GmbH | Cat# FN-5000-4-1 |
| Percoll | Sigma-Aldrich | Cat# P1644-1L |
| Dulbeccos PBS (w/o calcium and magnesium) (DPBS) | Gibco | Cat# 4190-094 |
| Dulbeccos PBS (w calcium and magnesium) (PBS) | Gibco | Cat# D8662 |
| High Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat# 4368813 |
| DNAse | Sigma-Aldrich | Cat# DN25-5g |
| Collagenase D | Sigma-Aldrich | Cat# 11088882001 |
| Dispase | Corning | Cat# 354235 |
| DNAse I | Roche | Cat# 05952077103 |
| Collagenase IV | Worthington | Cat# LS0004186 |
| Collagenase IA | Sigma | Cat# C9891 |
| Collagenase VIII | Sigma | Cat# C2139 |
| Trizol | Thermo Fisher Scientific | Cat# 15596026 |
| HEPES | Gibco | Cat# 15630-056 |
| BSA | Sigma-Aldrich | Cat# A7030-100G |
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | Cat# 41965062 |
| Iscove’s Modified Dulbecco’s Medium (IMDM) | Gibco | Cat# 21980032 |
| HEPES (1 M) | Gibco | Cat# 15630080 |
| Fetal bovine serum | Pan-Biotech | Cat# P30-3602 |
| Chloroform | Sigma Aldrich | Cat# 288306-100ML |
| 70 kD Fluorescein isothiovyanate (FITC)-dextran | Sigma | Cat# 46945-100MG |
| MacConkey II agar | BD BBL | Cat# 212306 |
| Schaedler Agar | BD BBL | Cat# 212189 |
| Sheep blood defibrinated | Oxoid | Cat# SR0051E |
| Kanamycin sulfate | Sigma-Aldrich | Cat# 60615 |
| Vitamin-K3 (Menadion) | Sigma-Aldrich | Cat# M5625 |
| Columbia blood agar base | Oxoid | Cat# CM331 |
| GC agar base | BD Difco | Cat# 228950 |
| Hemoglobin | Oxoid | Cat# LP0053 |
| Difco supplement VX | BD Difco | Cat# 233541 |
| Brain heart infusion | Oxoid | Cat# CM1135 |
| Hemin | Sigma-Aldrich | Cat# 51280-1G |
| Vitamin-K1 | Sigma-Aldrich | Cat# 95271-1G |
| Bacto Tryptone | BD | Cat# 211699 |
| Bacto Yeast Extract | Gibco | Cat# 212750 |
| Sodium chloride | Sigma-Aldrich | Cat# S9625-5KG |
| Agar | Sigma-Aldrich | Cat# 05039-500G |
| Sakura Finetek™ Tissue-Tek™ O.C.T. Compound | Sakura | Cat# 12351753 |
| SuperFrost® Plus | VWR | Cat# 631-9483 |
| Triton X-100 | Sigma-Aldrich | Cat# X100-100ML |
| Saccharose | Sigma-Aldrich | Cat# S9378-500G |
| Fluoromount-G | Invitrogen | Cat# 00-4958-02 |
| Mouse Serum | Life Science | Cat# S-012L |
| EDTA | Thermo Fisher Scientific | Cat# AM9912 |
| Lysozyme | Sigma | Cat# 62970-5G-F |
| Streptomycin | Sigma-Aldrich | Cat# S6501 |
| Gelatin from cold water fish skin (GCWFS) | Sigma | Cat# G7041 |
| DAPI | Carl Roth | Cat# 6843.1 |
| Normal Mouse Serum | Invitrogen | Cat# 10410 |
| Critical commercial assays | ||
| Blocking Kit VE Avidin/Biotin | Vector laboratories | Cat# SP-2001 |
| FoxP3 staining kit | Thermo Fisher Scientific | Cat# 00-5521-00 |
| Cytofix/cytoperm Buffer | BD Bioscience | Cat# 554722 |
| miRNeasy Micro Kit | Qiagen | Cat# 217084 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368813 |
| SYBR™ Green PCR Master Mix | Applied Biosystems | Cat# 4334973 |
| Taqman™ Universal Master Mix | Thermo Fisher | Cat# 4440039 |
| QIAamp DNA stool kit | Qiagen | Cat# 51604 |
| Qubit dsDNS BR Assay-Kit | Thermo Fisher Scientific | Cat# Q32853 |
| Qubit dsDNS HS Assay-Kit | Thermo Fisher Scientific | Cat# Q32854 |
| Deposited data | ||
| Bulk RNA-seq (of liver pre- and post-partial hepatectomy) | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158174 | code: cdunemgsdnixjyb |
| Experimental models: Organisms/strains | ||
| C57BL/6J (wild-type) | Envigo | N/A |
| B6.PL-Thy1a/CyJ | RRID:IMSR_JAX:000406 | |
| B6.FVB-Tg(Rorc-cre)tm1Litt/J | Eberl et al.16 | N/A |
| B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | Srinivas et al.67 | RRID:IMSR_JAX:006148 |
| B6.129P2-Rorctm1Litt/J | Sun et al.68 | RRID:IMSR_JAX:007571 |
| B6.129S7-Rag1tm1Mom/J | Mombaerts et al.69 | RRID:IMSR_JAX:002216 |
| Rag2−/− | Hao et al.70 | N/A |
| IGIF−/−(IL-18−/−) | N/A | |
| B6.Il22−/− | Kreymborg et al.71 | N/A |
| Pycard−/−(Asc−/−) | Drexler et al.72 | N/A |
| Oligonucleotides | ||
| See Table S2 for Oligonucleotides and RNA sequences | N/A | |
| Software and algorithms | ||
| FlowJo10.4 | Tree Star |
https://www.flowjo.com RRID: SCR_008520 |
| R studio | R foundation |
https://www.r-project.org RRID: SCR_000432 |
| Prism 8 | GraphPad software, Inc. |
www.graphpad.com RRID: SCR_002798 |
| ImageJ 1.8.0 | Image J Software | https://imagej.nih.gov/ij/ |
| Leica Application Suite X | Leica Microsystems |
https://www.leica-microsystems.com RRID: SCR_013679 |
| Qiime | QIIME development team | https://qiime2.org |
| Imaris | Oxford Instruments | https://imaris.oxinst.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Guido Beldi (guido.beldi@insel.ch).
Materials availability
This study did not generate new unique reagents. Mouse lines generated for this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Patient data
Two cohorts of patients were included for the analysis. One dataset includes the patients from a prospective interventional trial, which aimed to test the impact of the introduction of structured communication on surgical infections as the main outcome parameter (STOP? Trial, NCT02428179). The institutional review board of the Canton of Bern, and Canton Zürich, Switzerland have approved this study (EK ZH 2015-0232, KEK 161/14). The study included all consecutive patients except if patients refused to participate in clinical studies. The outcome parameters SSI was prospectively recorded in the context of a Swiss Surveillance Program (SWISSNOSO) that includes regular monitoring to ensure quality of inclusion of patients and assessment of the endpoints. Median age of included patients was 58 years and 1985 patients (56,5%) were male. Data on cultured bacteria were available from 2 participating centers from which the data were included.
All consecutive patients undergoing liver resections for colorectal liver metastasis between 2001 and 2016 at the University hospital of Vienna, Austria, were retrospectively recorded (n = 335). The institutional review board of the Medical University of Vienna, Austria has approved this study (424/2010; 2032/2013, NCT01700231, NCT02118545). Infectious complications were summarized with a possible event of an infection during the postoperative course. Major liver resection was defined as resection of three or more liver segments according to the Brisbane 2000 nomenclature.73 All bacterial cultures of septic patient were manually reviewed and recorded. Bacterial species were identified by MALDI-TOF mass spectrometry. Other bacteria in Figure 1B include: n = 2 Citrobacter spp, n = 1 Fusobacterium spp, n = 1 Clostridium sordelli, n = 1 Streptococcus spp.
Ethical approval
The analysis of patient’ data has been approved by the ethics committee of the ethical committee Bern and University of Vienna. The clinical studies were conducted in compliance with the protocol, the current version of the Declaration of Helsinki, the ICH-GCP, the human research act as well as other locally relevant legal and regulatory requirements. Experiments were done with Institutional Animal Care and Use Committee approval and in strict accord with good animal practice as defined by the Office of Laboratory Animal Welfare.
Mice
Specific-pathogen-free (SPF) wild-type C57BL/6J RccHsd (Envigo), Rag1−/−,69 Rag2−/−,70 Rag2−/−Il2rg−/−,69,74 Rorc(γt)−/−,16 IL22−/−71 animals were bred and housed in individual ventilated cages in the Animal Facilities of the University of Bern (Switzerland), Charite-Universitätsmedizin Berlin (Germany).
Rorc(γt)−/−Rag2−/− and Thy1.1 mice were kindly provided by Prof. Chiara Romagniani (German Rheumatism Research Centre-a Leibniz Institute, Berlin, Germany). The IGIF−/− (IL-18−/−) were kindly provided by Kevin Maloy (University of Oxford, UK) and Pycard−/− (Asc−/−) were kindly provided by Prof. N. Fasel (University of Lausanne, Department of Biochemistry, Institute for Arthritis Research, Switzerland)72 and both, housed under colonized conditions at the Clean Mouse Facility of the University of Bern, Switzerland.
Germ-free wild-type C57BL/6J, Rag1−/−69 and Rag2−/−Il2rg−/−69,74 mice were rederived by axenic two-cell embryo transfer as previously described75 and bred and maintained in flexible film isolators at the Clean Mouse Facility of the University of Bern, Switzerland. Germ-free status was routinely confirmed by extensive aerobic and anaerobic culture as well as DNA stain using SYTOX green (Invitrogen, Gaithersburg, MD) and Gram staining (Harleco, EMD Millipore Corporation, Billerica, MA) of caecal contents to detect unculturable contamination.
All experiments were performed in the morning on 8- to 12-week-old adult female mice, supplied with a 12 hour light/dark cycle at 22°C, and fed ad libitum with chow and water. All animal procedures were carried out in accordance to the guidelines for the care and use of laboratory animals, and the experimental protocol received approval by the Animal Care Committee of the Canton of Bern, Switzerland, the local animal care committees (LAGeSo) Berlin, Germany.
Method details
Partial hepatectomy
All surgical procedures were performed under laminar flow and under sterile conditions using general anesthesia with isoflurane (Nicholas Piramal (I) Limited, London, UK). The details of the partial hepatectomy model in mice were previously described.76 Briefly, anesthetized mice were immobilized in a supine position and the abdomen was entered through a midline incision. After exposure of liver lobes, partial hepatectomy was performed by central ligature (Vicryl 4-0, Ethicon, Johnson & Johnson, Spreitenbach, Switzerland) of the median and left lobe in order to achieve a standard two-third hepatectomy. The ligated liver lobes were surgically removed, weighted, and further processed. The laparotomy was then closed with a two-layer running suture (Prolene 6-0, Ethicon, Johnson & Johnson, Spreitenbach, Switzerland). During the procedure, the intestine was rinsed with saline to avoid drying-out. Resuscitation of the intraoperative fluid loss was achieved by putting saline into the abdominal cavity at the end of the operation. Analgesia with buprenorphine (Reckitt Benckiser AG, Switzerland) was administered at the beginning of the surgical intervention as well as ad libitum during the postoperative course by subcutaneous injection. At the time of sacrifice, mice were anesthetized by isoflurane inhalation, lethal blood samples were then taken from the inferior vena cava and livers were surgically removed for further analyses. In sham operated animals, a midline incision was performed, the liver was mobilized, the abdominal cavity was rinsed and the abdomen was closed with a two-layer running suture.
Microbial community analysis
The microbial community presented in the fecal and skin samples from 5 healthy volunteers were collected every second day for 2 months. Human fecal and skin swabs samples or murine fecal and liver samples were analyzed by 16S rRNA amplicon sequencing on the lonTorrent PGM™ platform, as previously described.77 Briefly, microbial DNA was extracted from fecal and skin samples using the QIAamp DNA stool kit (Qiagen) according to the manufacturer’s instructions78 with an additional step for efficient Gram positive bacteria lysis using 20 mg/mL lysozyme (Sigma); 20mM Tris⋅HCl, pH 8.0; 2mM EDTA; 1.2% Triton buffer. Concentrations of purified DNA were measured using Qubit (Thermo Fisher Scientific). DNA was pooled at a concentration of 26pM and was sequenced for the V5/V6 region of 16S rRNA genes using a multiplex approach with the barcoded forward fusion primer 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG BARCODE ATTAGATACCCYGGTAGTCC-3′ in combination with the reverse fusion primer 5′-CCTCTCTATGGGCAGTCGGTGATACG AGCTGACGACARCCATG-3′, in IonTorrent PGM system according to the manufacturer’s instructions (ThermoFisher).79,80 Data was further analyzed using QIIME pipeline after filtering out low quality (accuracy of base calling; q < 25) samples. Operational taxonomic units (OTUs) were picked using UCLUST with a 97% sequence identity threshold followed by taxonomy assignment using the latest GreenGenes database (http://greengenes.secondgenome.com/). Alpha diversity (that describes the number of different taxa within a sample) and beta diversity (that delineates differences between samples (i.e. “between habitat” diversity) were calculated using the phyloseq pipeline in R (v3.4).81,82 The non-parametric Mann-Whitney U-tests was used to compare alpha diversity between samples and Adonis from vegan package to assess the effects of groups for beta diversity via phyloseq in R.81,82 Taxonomic differences at phylum and genus levels between tested groups were identified using the “multivariate analysis by linear models” (MaAsLin) R package.83 Only taxa present in at least 50% of samples and OTUs comprising more than 0.001% of relative abundance were analyzed in MaAsLin. A p < 0.05 and a false discovery rate (FDR; Benjamini-Hochberg correction) of q < 0.05 were set as cut-off values for significance. All the relevant codes for running the MaAsLin in R platform are available in Dr. Huttenhower’s group webpage (https://huttenhower.sph.harvard.edu). Phylogenetic tree was plot using GraPhlAn.84
Identification of culturable bacteria from human skin and fecal samples
Skin swabs and fecal samples were freshly collected from 10 healthy volunteers. Samples were streaked on MacConkey agar, chocolate (CHOC) agar, Columbia blood agar under aerobic conditions and Schaedler-protein K-blood agar (with and without Kanamycin) under anaerobic conditions and cultured for 48 hours at 37°C. Bacterial colonies were morphologically differentiated and identified by matrix-assisted laser desorption time-of-flight (MALDI-TOF) mass spectrometry.
Assessment of systemic bacterial dissemination in mice
To determine the presence of bacteria in the liver, spleen, and mesenteric lymph nodes (MLN), the respective organs were harvested under sterile conditions and plated on LB, and BHI (brain-heart infusion)-hemin-protein K-blood agar plates as indicated. LB agar plates were cultured for 24 hours at 37°C. BHI-blood agar plates were cultured under anaerobic conditions for 48 hours at 37°C. Aerobic and anaerobic single isolated colonies were identified by full 16S rRNA sequencing and collectedly quantified (Sanger sequencing-Microsynth). LB-enriched with streptomycin plates was used to quantify the dissemination of E. coli JM83.
Bacterial culture
Escherichia coli (E. coli) K-12 strain JM83 (F-(lac-proAB) phi80, (lacZ)M15 ara rpsL thi lambda-; streptomycin-resistant)85 was cultured overnight in Luria Broth (LB) medium supplemented with Streptomycin (100μg/mL) at 37°C, shaking at 200 rpm. To prepare bacterial inocula for gavage or intravenous (i.v.) injection, bacterial cultures were centrifuged for 10 min at 4000 g and washed twice with sterile PBS. The required dose was resuspended in 500 μL of sterile PBS.
Bacterial delivery
Intravenous injection was performed 6 and 2 days prior to partial hepatectomy. 107 CFU live E. coli JM83 were i.v. injected in the tail vein. An additional injection of live bacteria was performed 12 hours after partial hepatectomy. For experiments to evaluate bacterial clearance in the liver (Figures 6A and 6B): 107 CFU live E. coli JM83 were i.v. injected into the tail vein immediate after PH. Blood was repetitively drawn and plated at indicated time points.
For efficient trackability of intestinal bacteria - in the indicated experiments - SPF or germ-free mice were intragstric (i.g.) gavaged with 1010 CFU of E. coli JM83 12 hours prior PH.
Cellular isolation
The livers were flushed by injection of cold DPBS into the portal vein. Thereafter, the livers were harvested, cut into small pieces with a scalpel and collected for 30 minutes with shaking at 37°C. The medium was then filtered through a 100μm gauge cell strainer into a 50 mL conical tube. Cells were resuspended with cold wash buffer [1X DPBS (GIBCO, Life Technologies), 2% Hepes (Sigma Aldrich) and 2% fetal calf serum (FCS)]. Samples were centrifuged at 1250g for 10 mins at room temperature. The supernatant was discarded and centrifuged until it became clear. Isolation of cells was achieved by gradient centrifugation using Percoll (GE Healthcare). The cell suspension was resuspended in 40% Percoll (Sigma) solution and layered on top of an 80% Percoll solution. Gradient centrifugation was carried out (2000 g, 20 min, 20°C, no brake). Lymphocytes were collected from the interphase, washed with IMDM (10% FCS) and centrifuged (600 g, 7 min). To isolate leukocytes from the lamina propria of small intestine, intestines were removed from the mouse and placed in ice-cold DPBS. Residual fat and Peyer’s patches were also removed. The intestine was opened longitudinally and sectioned into 2 cm segments. The tissue was washed once in ice-cold DPBS followed by four washes of 8 min in 15 mL of DPBS (5 mM EDTA, 10 mM HEPES) with shaking at 37°C to detach epithelial cells. Residual tissue was then washed for 8 min in 15 mL of IMDM containing 10% FCS at 37°C before being minced and digested in 15 mL of IMDM containing 1 mg/mL collagenase type VIII (Sigma) and 10 U/ml DNase I (Roche) with shaking at 37°C for 20–30 min (small intestine) or 30–40 min (colon). The resulting cell suspension was passed through a cell strainer (100 μm) and washed with 20 mL of IMDM 10% FCS. Cells were centrifuged (600 g, 7 min, 4°C) and resuspended in 300 μL of FACS buffer (DPBS 2% FCS 2mM EDTA 0.01% NaN3). Intestines were processed individually. Spleens and lymph nodes were cut into small pieces and digested in IMDM (2% FCS) containing collagenase type IA (1 mg/mL, Sigma) and DNase I (10 U/ml, Roche) at 37°C for 30 min. Cellular suspensions were also passed through a cell strainer (40 μm) and washed with IMDM (2% FCS, 2mM EDTA). Cells were finally resuspended in FACS buffer and counted using a Neubauer counting chamber in Trypan blue.
Flow cytometry
After isolation, cells were washed once with DPBS before being stained with fixable viability dye (eBioscience) diluted in DPBS for 30 min on ice. Single cell suspensions were sequentially incubated with primary/biotin- and fluorescence-coupled antibodies diluted in FACS buffer. Cells surface staining was performed by incubating them for 20 minutes at 4°C. Intracytoplasmic staining was performed in unstimulated cells using the Cytofix/Cytoperm staining kit (BD). Intranuclear staining was performed using the Foxp3 staining kit (eBioscience). The following mouse-specific conjugated antibodies were used: CD127-FITC (A7R34, eBioscience), CD19-BV421, -BV785, -PE-Cy7 (6D5, Biolegend), CD3-BV650, -BV785, -PE-Cy7 (17A2, Biolegend), FceRIα-PE-Cy7 (MAR1, Biolegend) CD4-BV785 (RM4-5, Biolegend), CD45-BUV395 (30-F11, BD Bioscience), CD90-BV605 (53-2.1, Biolegend), CD62L-BV510 (MEL-14), CD5-PE-Cy7 (53-7.3, Biolegend), Foxp3-AF700 (FJK-16s, eBioscience), Gata3-A647 (TWAJ, BD), Neuropilin1-PE (3E12, Biolegend), NK1.1-AF647 (PK136, Biolegend), NKp46-PerCP-Cy55 (19A1.4, Biolegend, 29A1.a, eBioscience), RORγt-PE (B2D, eBioscience), CCR6-PE (29-2L17, Biolegend), T-bet-PE-Cy7 (4B10, eBioscience), fixable viability dye -efluor 450 or -efluor 506 (ebioscience), IL-22 APC (IL22JOP, eBioscience), KI-67 PerCP-Cy55 (B56, ebioscience), CD44-APC-Cy7 (Im7, Biolegend), CX3CR1-FITC (SA011F11, Biolegend), Siglec F-PE (E50-2440, Biolegend), CD103 Biotinylate (M290, BD), CD11b-APC-Cy7 (M1/70, Biolegend), CD11c-PE-CF594 (HL3, BD), F4/80-APC (BM8, Biolegend), MHC-II(I-A/I-E)-AF700, -BV650 (M5/114.15.2, Biolegend), Ly6C-BV421 (HK1.4, Biolegend), Ly6G-PE-Cy7 (1A8, Biolegend), Streptavin PE-Cy7 (BD). LSR Fortessa (BD) was used for flow cytometry and data were analyzed with FlowJo software (Treestar Data Analysis Software).
Thy1.2 or CD4+ T cell depletion
Anti-CD90.2 mAb (Thy1.2, 30H12) and anti-CD4 mAb (GK1.5) was purchased from BioXCell (West Lebanon, NH). Depletion mAb or isotype mAb (rat anti-mouse IgG) treatments were administered by i.p. injection 6 days, 3 days and 1 day prior to the surgery at a dose of 250 μg/mouse.29
CD90-disparate chimeras
CD90-disparate chimeras were generated as previously described.29 Briefly, 40–60 106 FACS sort-purified CD19+ B cells and CD3+CD5+ T cells from C57BL/6 Thy1.1 mice of spleen and MLN were transferred i.v. injection to Rag1−/− Thy1.2 recipient mice. Reconstitution was permitted for 8–10 weeks post-transfer. I.p. injection of anti-Thy1.2 was performed three times prior to partial hepatectomy.
RNA isolation
Tissues were collected and snap-frozen using liquid nitrogen. Thawed tissues were immediately homogenized (Retsch bead-beater) in 500 μL of Nucleozol reagent. Chloroform (200 μL) was added, samples were mixed, and centrifuged (12,000 g, 15 min, 4°C). The upper phase was collected and RNA was precipitated with ice-cold isopropanol by centrifugation (12,000 g, 10 min, 4°C). The RNA pellet was washed with 75% ethanol, dried and resuspended in RNase-free water. RNA concentrations and purity were analyzed using a Nanodrop2000 (Thermo Scientific).
RT-qPCR
Genomic DNA in RNA samples was digested using the DNA-free kit (Ambion) and RNA was reverse-transcribed into cDNA using Superscript III reverse transcriptase according to Invitrogen protocols. RT-qPCR was performed using 384 well-plates with 50–100 ng of cDNA per well containing the SsoFast EvaGreen Supermix (Biorad) and gene-specific primers. All reactions were run in triplicate. Samples were normalized to the expression of TBP for TaqMan and GAPDH for SYBR Green by calculating 2ˆ(-deltaCt).
Primer sequences:
GAPDH (Gapdh) forward: 5′ - CAT CAA GAA GGT GGT GAA GC -3′.
GAPDH (Gapdh) reverse: 5′ - CCT GTT GCT GTA GCC GTA TT -3′.
REG3B (Reg3beta) forward: 5′ – GCA GAA CCC AAT GGA GGT GG -3′.
REG3B (Reg3beta) reverse: 5′ – CAC CCA GGG ATG TGA GAA GAG -3′.
REG3G (Reg3gamma) forward: 5′ – TTC CTG TCC TCC ATG ATC AAA -3′.
REG3G (Reg3gamma) reverse: 5′ – CAT CCA CCT CTG TTG GGT TC -3′.
LYZ1 (Lysozyme 1) forward: 5′ – CTT GTC ACT CCT CAC CCC TG -3′.
LYZ1 (Lysozyme 1) reverse: 5′ – AGC CGT TCC CCT TCC AAT G -3′.
Il22 (IL-22) forward: 5′ – ATG AGT TTT TCC CTT ATG GGG AC-3′
Il22 (IL-22) reverse: 5′ – GCT GGA AGT TGG ACA CCT CAA-3′.
DEFA21 (alpha defensin 21) forward: 5′ – AGG CTG TGT CTG TCT CCT TTG -3′.
DEFA21 (alpha defensin 21) reverse: 5′ – TGC AAG CAT CCA TCA CAC TGG -3′.
Il1B (IL-1β, forward: 5′ - ACCTGTCCTGTGTAATGAAAGACG -3′.
Il1B (IL-1β) reverse: 5′ - TGGGTATTGCTTGGGATCCA -3′.
Il18 (IL-18) forward: 5’ - ACA ACT TTG GCC GAC TTC AC -3′.
Il18 (IL-18) reverse: 5′ - TGG ATC CAT TTC CTC AAA GG -3′.
For the assessment of hepatocyte proliferation, the quantitative RT–qPCR with TaqMan gene expression assays were used. Reagents were used according to the standard protocols (Applied Biosystem). Mouse probes used include CCNA2 (cyclin A2) (Mm00438063_m1), FOXM1B (Mm00514925_m1), and TBP (Mm01277042_m1).
Single cell RNA-seq analysis
The single cell RNA-seq (scRNA-seq) data was downloaded from Sanchez-Taltavull et al.86 The acquired dataset consisted on dropout corrected gene expression of different time points post partial hepatectomy, together with the cell classification and t-SNE coordinates. Cells classified as doublets or unknown were removed.
RNA-seq analysis
The RNA concentration was determined using a Bioanalyzer 2100 (Agilent). Library preparation was performed using TruSeq RNA sample preparation v2 kit (TruSeq Stranded mRNA Sample Preparation, Illumina). Libraries were sequenced by Illumina HiSeq 2500 on the 100 bp single-end mode. Between 12.4 and 17.0 mio read pairs were obtained per sample and the quality of the reads was assessed using fastqc v.0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were mapped to the mouse reference genome (ensembl m38, build 75) using Tophat v. 2.0.11.87 We then used htseq-count v. 0.6.188 to count the number of reads per gene, and DESeq2 v. 1.4.589 to test for differential expression between groups of samples. The output of DESeq2 was used to perform gene set enrichment analysis (GSEA) using the SetRank method. The key principle of this algorithm is that it discards gene sets that have initially been flagged as significant, if their significance is only due to the overlap with another gene set. It calculates the p-value of a gene set using the ranking of its genes in the ordered list of p-values as calculated by DESeq2. Genes were scored according to an absolute fold-change ≥ 2 and adjusted p-value of < 0.01. The proteins corresponding to the obtained gene sets were searched against the version 10 of the STRING database90 to display functional protein-association networks. Interactions were considered with a STRING confidence score ≥ 0.4 (medium and high confidence). Multiple rounds of iteration of the k-means clustering method were performed.
Immunohistochemical analysis of liver
Immunohistochemistry for hepatocyte proliferation using Ki-67 stain was performed on paraffin-embedded liver sections (3 μm). Fresh liver tissue was fixed with 4% Formaldehyde. Paraffin-embedded tissue sections were then dried, deparaffinized, and rehydrated, followed by blocking of endogenous peroxidase with 3% H2O2 (Sigma H-1009; Sigma- Aldrich Chemie GmbH, Germany) in DPBS. Antigen was retrieved by heating the slides for 10 min. Diluted monoclonal mouse anti-Ki-67 antibody (TEC-3 antibody [Dako Code M7249]; Dako, Glostrup, Denmark) was then applied and slides incubated overnight at room temperature in a humidified chamber. Then, biotinylated rabbit anti-mouse IgG ([Code E0464]; Dako, Glostrup, Denmark) was added, followed by a brief incubation with 3,3′-diaminobenzidine substrate (DAB+; [Code K3467]; Dako, Glostrup, Denmark). The tissue sections were counterstained in hematoxylin. Total numbers of Ki-67-labeled hepatocytes were finally determined by counting positively stained hepatocyte nuclei of the whole stained slides in a blinded manner by an independent observer. Percentage of positive proliferating cells (Ki-67+ cells) was finally calculated for each animal.
Immunofluorescence analysis of liver
Livers were perfused via the left ventricle with DPBS (Sigma Aldrich) and 2% PFA. Afterwards, liver samples were fixed using 2% PFA for 12 hours at 4°C and dehydrated with 30% sucrose at 4°C overnight. Tissues were embedded in OCT compound (Sakura Finetek) and stored at −80°C. Serial 30 mm sections were cut on a CM3050S cryostat (Leica) and adhered to Superfrost Plus object slides (VWR). After rehydration with DPBS for 10 min at room temperature, sections were permeabilized and blocked with DPBS containing 1% BSA (Sigma Aldrich), 1% GCWFS (Sigma Aldrich), 0.3% Triton-X 100 (Sigma Aldrich) and 1% normal mouse serum (Life Technologies) for 1 hour at room temperature. Staining was performed in blocking buffer at 4°C overnight. Sections were washed with DPBS and mounted with Fluoromount-G (Invitrogen). Acquisition was performed on a Leica Stellaris 5 confocal microscope with LAS X software. The following antibodies were used for staining: anti-mouse CD31 BV480 (Clone: MEC 13.3; BD Biosciences), anti-GFP AF488 (Invitrogen), anti-mouse CD3 eFlour660 (Clone: 17A2; Invitrogen) and DAPI (Carl Roth).
Confocal endomicroscopy experiments
Partial hepatectomy was performed immediately prior to imaging. After the operation, 70 kD Fluorescein isothiovyanate (FITC)-dextran (10 μg/ml, Sigma) in PBS was intravenously injected into the tail vein to provide real-time contrast. Immediately after injection, the terminal ileum was incised and a confocal miniprope (Cellvizio, Mauna Kea Technologies, Paris, France) was introduced into the small intestine. The fluorescence intensity outside:inside was determined using ImageJ software (NIH) calculating the ratios of extravascular to intravascular fluorescence.
Quantification and statistical analysis
Statistical analysis
Dot plots with a logarithmic scale show the geometric mean of each experimental group in addition to the individual samples represented as single data points. Dot plots with a linear scale show the arithmetic mean. Normalized values were analyzed by Student’s t test or one-way ANOVA. p-values are indicated as follows: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
Acknowledgments
We thank members of the research laboratory of visceral surgery and medicine, Prof. S. Ganal-Vonarburg, and Prof. A.J. Macpherson for critical input and support. We thank I. Lendenmann and I. Büchi for the technical support. The clean mouse facility of the Department of Biomedical Research kindly housed and provided the GF mice. Prof. Francesca Ronchi facilitated the use of IGIF (IL-18)−/− and Pycard−/− (Asc−/−) mice. Prof. Chiara Romagniani and Dr. Christina Stehle provided us with Rorc(γt)−/−Rag2−/− and Thy1.1 mice. M.O.J. received funding by the ARS Bridge to academic career grant. M.O.J., G.B., and M.G.A. received funding by the Swiss National Science Foundation (grant ID: M.O.J. 184425; G.B. 166594, 156882; M.G.A. 168012). M.G.A. is supported by the German Research Foundation (DFG; project number GO3241/1-1 SPP 2330, 324392634-TRR 221) and the Max Planck Society (Max Planck Research Groups). C.S.N.K. is supported by the European Research Council Starting Grant (ERCEA; 803087) and the German Research Foundation (DFG; project ID 259373024; CRC/TRR 167, FOR2599 project 5, B05, project ID 375876048; CRC/TRR 241, KL 2963/5-2, SPP1937, KL 2963/2-1, and KL 2963/3-1 to C.S.N.K.). A.D. is supported by grants from the European Research Council (ERC AdG #101055309: ILCAdapt) and the German Research Foundation (SPP1937 [DI764/9-2]; FOR2599 [project A5, DI764/10-2], CRC 1444 [project #427826188; project 11]; CRC-TRR 241 [project #375876048; project A01], CRC-TRR 156 [project A02]; CRC-TRR 84 [projects A01 und A06]). A.D. is the Einstein Professor for Microbiology (Einstein Foundation Berlin).
Author contributions
M.O.J., D.S., L.S., and M.G.A. performed the animal experiments. M.O.J., M.G.A., and G.B. made the experimental plan of investigation. M.O.J., C.S.N.K., M.G.A., and G.B. analyzed the experimental data and wrote the manuscript. M.G.A., B.Y., and T.S. carried out the microbiota profiling analysis on fecal, liver, and skin human and murine samples. D.S.T., B.Y., R.D.L., and C.M. performed and interpreted the bioinformatic analysis. Y.O. and G.G. performed the liver ILC3 histology. I.M., D.C., D.S., A.D., and G.G. provided input during work discussions and provided guidance on the concept and design. G.B. and F.T. conceived, designed, and were responsible for the clinical study. P.S. and D.P. provided and interpreted the human data on liver surgeries. M.O.J. and M.G.A. created the figures and wrote the methods. All authors edited and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112269.
Contributor Information
Manuel O. Jakob, Email: manuel.jakob@ymail.com.
Guido Beldi, Email: guido.beldi@insel.ch.
Supplemental information
Data and code availability
-
•
Bulk-RNA Seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resource table. The patient’ data that support the findings of this study are available from the lead contact upon reasonable request. Microscopy data reported in this paper will be shared by the lead contact upon request until 02/2033.
-
•
Data were analysed using the standard DESeq2 and Qiime pipelines, or with the stated variations. User scripts will be shared upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Magill S.S., O'Leary E., Janelle S.J., Thompson D.L., Dumyati G., Nadle J., Wilson L.E., Kainer M.A., Lynfield R., Greissman S., et al. Changes in prevalence of health care-associated infections in U.S. Hospitals. N. Engl. J. Med. 2018;379:1732–1744. doi: 10.1056/NEJMoa1801550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akashi K., Yokoyama Y., Mizuno T., Abe T., Fukaya M., Asahara T., Nagino M. Association between preoperative muscle mass and intraoperative bacterial translocation in patients undergoing hepatectomy, pancreatoduodenectomy, and esophagectomy. Ann. Surg Oncol. 2019;26:4805–4813. doi: 10.1245/s10434-019-07707-y. [DOI] [PubMed] [Google Scholar]
- 3.Allegranzi B., Bischoff P., de Jonge S., Kubilay N.Z., Zayed B., Gomes S.M., Abbas M., Atema J.J., Gans S., van Rijen M., et al. New WHO recommendations on preoperative measures for surgical site infection prevention: an evidence-based global perspective. Lancet Infect. Dis. 2016;16:e276–e287. doi: 10.1016/S1473-3099(16)30398-X. [DOI] [PubMed] [Google Scholar]
- 4.Allegranzi B., Zayed B., Bischoff P., Kubilay N.Z., de Jonge S., de Vries F., Gomes S.M., Gans S., Wallert E.D., Wu X., et al. New WHO recommendations on intraoperative and postoperative measures for surgical site infection prevention: an evidence-based global perspective. Lancet Infect. Dis. 2016;16:e288–e303. doi: 10.1016/S1473-3099(16)30402-9. [DOI] [PubMed] [Google Scholar]
- 5.Dai T., Kharkwal G.B., Tanaka M., Huang Y.Y., Bil de Arce V.J., Hamblin M.R. Animal models of external traumatic wound infections. Virulence. 2011;2:296–315. doi: 10.4161/viru.2.4.16840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishigaki E., Abe T., Yokoyama Y., Fukaya M., Asahara T., Nomoto K., Nagino M. The detection of intraoperative bacterial translocation in the mesenteric lymph nodes is useful in predicting patients at high risk for postoperative infectious complications after esophagectomy. Ann. Surg. 2014;259:477–484. doi: 10.1097/SLA.0b013e31828e39e8. [DOI] [PubMed] [Google Scholar]
- 7.Balmer M.L., Slack E., de Gottardi A., Lawson M.A.E., Hapfelmeier S., Miele L., Grieco A., Van Vlierberghe H., Fahrner R., Patuto N., et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci. Transl. Med. 2014;6:237ra66. doi: 10.1126/scitranslmed.3008618. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee S., Hooper L.V. Antimicrobial defense of the intestine. Immunity. 2015;42:28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 9.McDonald B., Zucoloto A.Z., Yu I.L., Burkhard R., Brown K., Geuking M.B., McCoy K.D. Programing of an intravascular immune firewall by the gut microbiota protects against pathogen dissemination during infection. Cell Host Microbe. 2020;28:660–668.e4. doi: 10.1016/j.chom.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Albillos A., de Gottardi A., Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J. Hepatol. 2020;72:558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. Innate lymphoid cells: 10 years on. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Klose C.S.N., Artis D. Innate lymphoid cells control signaling circuits to regulate tissue-specific immunity. Cell Res. 2020;30:475–491. doi: 10.1038/s41422-020-0323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawa S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F., Di Santo J.P., Eberl G. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 14.Klose C.S.N., Kiss E.A., Schwierzeck V., Ebert K., Hoyler T., d'Hargues Y., Göppert N., Croxford A.L., Waisman A., Tanriver Y., Diefenbach A. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 15.Gladiator A., Wangler N., Trautwein-Weidner K., LeibundGut-Landmann S. Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defense against fungal infection. J. Immunol. 2013;190:521–525. doi: 10.4049/jimmunol.1202924. [DOI] [PubMed] [Google Scholar]
- 16.Eberl G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., Littman D.R. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 17.Hernández P.P., Mahlakoiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F., Gronke K., Ryffel B., Hoelscher C., Dumoutier L., et al. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 19.Sonnenberg G.F., Monticelli L.A., Elloso M.M., Fouser L.A., Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vonarbourg C., Mortha A., Bui V.L., Hernandez P.P., Kiss E.A., Hoyler T., Flach M., Bengsch B., Thimme R., Hölscher C., et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rankin L.C., Groom J.R., Chopin M., Herold M.J., Walker J.A., Mielke L.A., McKenzie A.N.J., Carotta S., Nutt S.L., Belz G.T. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sciumé G., Hirahara K., Takahashi H., Laurence A., Villarino A.V., Singleton K.L., Spencer S.P., Wilhelm C., Poholek A.C., Vahedi G., et al. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 2012;209:2331–2338. doi: 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernink J.H., Peters C.P., Munneke M., te Velde A.A., Meijer S.L., Weijer K., Hreggvidsdottir H.S., Heinsbroek S.E., Legrand N., Buskens C.J., et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 24.Cella M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K.M., Doherty J.M., Mills J.C., Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abt M.C., Lewis B.B., Caballero S., Xiong H., Carter R.A., Sušac B., Ling L., Leiner I., Pamer E.G. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe. 2015;18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song C., Lee J.S., Gilfillan S., Robinette M.L., Newberry R.D., Stappenbeck T.S., Mack M., Cella M., Colonna M. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 2015;212:1869–1882. doi: 10.1084/jem.20151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynders A., Yessaad N., Vu Manh T.P., Dalod M., Fenis A., Aubry C., Nikitas G., Escalière B., Renauld J.C., Dussurget O., et al. Identity, regulation and in vivo function of gut NKp46+RORgammat+ and NKp46+RORgammat- lymphoid cells. EMBO J. 2011;30:2934–2947. doi: 10.1038/emboj.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibiza S., García-Cassani B., Ribeiro H., Carvalho T., Almeida L., Marques R., Misic A.M., Bartow-McKenney C., Larson D.M., Pavan W.J., et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature. 2016;535:440–443. doi: 10.1038/nature18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnenberg G.F., Monticelli L.A., Alenghat T., Fung T.C., Hutnick N.A., Kunisawa J., Shibata N., Grunberg S., Sinha R., Zahm A.M., et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goto Y., Obata T., Kunisawa J., Sato S., Ivanov I.I., Lamichhane A., Takeyama N., Kamioka M., Sakamoto M., Matsuki T., et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345 doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanash A.M., Dudakov J.A., Hua G., O'Connor M.H., Young L.F., Singer N.V., West M.L., Jenq R.R., Holland A.M., Kappel L.W., et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37:339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zenewicz L.A., Yancopoulos G.D., Valenzuela D.M., Murphy A.J., Karow M., Flavell R.A. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klose C.S.N., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 35.Macpherson A.J., Heikenwalder M., Ganal-Vonarburg S.C. The liver at the nexus of host-microbial interactions. Cell Host Microbe. 2016;20:561–571. doi: 10.1016/j.chom.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Forkel M., Berglin L., Kekäläinen E., Carlsson A., Svedin E., Michaëlsson J., Nagasawa M., Erjefält J.S., Mori M., Flodström-Tullberg M., et al. Composition and functionality of the intrahepatic innate lymphoid cell-compartment in human nonfibrotic and fibrotic livers. Eur. J. Immunol. 2017;47:1280–1294. doi: 10.1002/eji.201646890. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto A., Kanai T., Mikami Y., Chu P.S., Nakamoto N., Ebinuma H., Saito H., Sato T., Yagita H., Hibi T. IL-22-producing RORgammat-dependent innate lymphoid cells play a novel protective role in murine acute hepatitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q., Zhou J., Zhang B., Tian Z., Tang J., Zheng Y., Huang Z., Tian Y., Jia Z., Tang Y., et al. Hepatitis B virus induces IL-23 production in antigen presenting cells and causes liver damage via the IL-23/IL-17 axis. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kudira R., Malinka T., Kohler A., Dosch M., de Agüero M.G., Melin N., Haegele S., Starlinger P., Maharjan N., Saxena S., et al. P2X1-regulated IL-22 secretion by innate lymphoid cells is required for efficient liver regeneration. Hepatology. 2016;63:2004–2017. doi: 10.1002/hep.28492. [DOI] [PubMed] [Google Scholar]
- 40.Okita Y., Shiono T., Yahagi A., Hamada S., Umemura M., Matsuzaki G. Interleukin-22-Induced antimicrobial phospholipase A2 group IIA mediates protective innate immunity of nonhematopoietic cells against Listeria monocytogenes. Infect. Immun. 2016;84:573–579. doi: 10.1128/IAI.01000-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghaferi A.A., Birkmeyer J.D., Dimick J.B. Variation in hospital mortality associated with inpatient surgery. N. Engl. J. Med. 2009;361:1368–1375. doi: 10.1056/NEJMsa0903048. [DOI] [PubMed] [Google Scholar]
- 42.Macpherson A.J., Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 43.Sawa S., Lochner M., Satoh-Takayama N., Dulauroy S., Bérard M., Kleinschek M., Cua D., Di Santo J.P., Eberl G. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 44.Mao K., Baptista A.P., Tamoutounour S., Zhuang L., Bouladoux N., Martins A.J., Huang Y., Gerner M.Y., Belkaid Y., Germain R.N. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 2018;554:255–259. doi: 10.1038/nature25437. [DOI] [PubMed] [Google Scholar]
- 45.Melo-Gonzalez F., Hepworth M.R. Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology. 2017;150:265–275. doi: 10.1111/imm.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanos S.L., Bui V.L., Mortha A., Oberle K., Heners C., Johner C., Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bel S., Pendse M., Wang Y., Li Y., Ruhn K.A., Hassell B., Leal T., Winter S.E., Xavier R.J., Hooper L.V. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357:1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eberl G., Colonna M., Di Santo J.P., McKenzie A.N.J. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muñoz M., Eidenschenk C., Ota N., Wong K., Lohmann U., Kühl A.A., Wang X., Manzanillo P., Li Y., Rutz S., et al. Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity. 2015;42:321–331. doi: 10.1016/j.immuni.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Gronke K., Hernández P.P., Zimmermann J., Klose C.S.N., Kofoed-Branzk M., Guendel F., Witkowski M., Tizian C., Amann L., Schumacher F., et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature. 2019;566:249–253. doi: 10.1038/s41586-019-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindemans C.A., Calafiore M., Mertelsmann A.M., O'Connor M.H., Dudakov J.A., Jenq R.R., Velardi E., Young L.F., Smith O.M., Lawrence G., et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talbot J., Hahn P., Kroehling L., Nguyen H., Li D., Littman D.R. Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature. 2020;579:575–580. doi: 10.1038/s41586-020-2039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duffin R., O'Connor R.A., Crittenden S., Forster T., Yu C., Zheng X., Smyth D., Robb C.T., Rossi F., Skouras C., et al. Prostaglandin E(2) constrains systemic inflammation through an innate lymphoid cell-IL-22 axis. Science. 2016;351:1333–1338. doi: 10.1126/science.aad9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L., Fouts D.E., Stärkel P., Hartmann P., Chen P., Llorente C., DePew J., Moncera K., Ho S.B., Brenner D.A., et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19:227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvarado D.M., Chen B., Iticovici M., Thaker A.I., Dai N., VanDussen K.L., Shaikh N., Lim C.K., Guillemin G.J., Tarr P.I., Ciorba M.A. Epithelial indoleamine 2,3-dioxygenase 1 modulates aryl hydrocarbon receptor and notch signaling to increase differentiation of secretory cells and alter mucus-associated microbiota. Gastroenterology. 2019;157:1093–1108.e11. doi: 10.1053/j.gastro.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Domingues R.G., Hepworth M.R. Immunoregulatory sensory circuits in group 3 innate lymphoid cell (ILC3) function and tissue homeostasis. Front. Immunol. 2020;11:116. doi: 10.3389/fimmu.2020.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Negash A.A., Ramos H.J., Crochet N., Lau D.T.Y., Doehle B., Papic N., Delker D.A., Jo J., Bertoletti A., Hagedorn C.H., Gale M., Jr. IL-1beta production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamada N., Chen G.Y., Inohara N., Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chow J., Mazmanian S.K. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Païssé S., Valle C., Servant F., Courtney M., Burcelin R., Amar J., Lelouvier B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56:1138–1147. doi: 10.1111/trf.13477. [DOI] [PubMed] [Google Scholar]
- 61.Schierwagen R., Alvarez-Silva C., Madsen M.S.A., Kolbe C.C., Meyer C., Thomas D., Uschner F.E., Magdaleno F., Jansen C., Pohlmann A., et al. Circulating microbiome in blood of different circulatory compartments. Gut. 2019;68:578–580. doi: 10.1136/gutjnl-2018-316227. [DOI] [PubMed] [Google Scholar]
- 62.Sorribas M., Jakob M.O., Yilmaz B., Li H., Stutz D., Noser Y., de Gottardi A., Moghadamrad S., Hassan M., Albillos A., et al. FXR modulates the gut-vascular barrier by regulating the entry sites for bacterial translocation in experimental cirrhosis. J. Hepatol. 2019;71:1126–1140. doi: 10.1016/j.jhep.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 63.Nešuta O., Buděšínský M., Hadravová R., Monincová L., Humpoličková J., Čeřovský V. How proteases from Enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides. Pathog. Dis. 2017;75 doi: 10.1093/femspd/ftx091. [DOI] [PubMed] [Google Scholar]
- 64.Cornell R.P., Liljequist B.L., Bartizal K.F. Depressed liver regeneration after partial hepatectomy of germ-free, athymic and lipopolysaccharide-resistant mice. Hepatology. 1990;11:916–922. doi: 10.1002/hep.1840110603. [DOI] [PubMed] [Google Scholar]
- 65.Wu X., Sun R., Chen Y., Zheng X., Bai L., Lian Z., Wei H., Tian Z. Oral ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology. 2015;62:253–264. doi: 10.1002/hep.27791. [DOI] [PubMed] [Google Scholar]
- 66.Uchimura Y., Fuhrer T., Li H., Lawson M.A., Zimmermann M., Yilmaz B., Zindel J., Ronchi F., Sorribas M., Hapfelmeier S., et al. Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity. 2018;49:545–559.e5. doi: 10.1016/j.immuni.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213x-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Z., Unutmaz D., Zou Y.R., Sunshine M.J., Pierani A., Brenner-Morton S., Mebius R.E., Littman D.R. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 69.Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., Papaioannou V.E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 70.Hao Z., Rajewsky K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 2001;194:1151–1164. doi: 10.1084/jem.194.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kreymborg K., Etzensperger R., Dumoutier L., Haak S., Rebollo A., Buch T., Heppner F.L., Renauld J.C., Becher B. IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but not required for the development of autoimmune encephalomyelitis. J. Immunol. 2007;179:8098–8104. doi: 10.4049/jimmunol.179.12.8098. [DOI] [PubMed] [Google Scholar]
- 72.Drexler S.K., Bonsignore L., Masin M., Tardivel A., Jackstadt R., Hermeking H., Schneider P., Gross O., Tschopp J., Yazdi A.S. Tissue-specific opposing functions of the inflammasome adaptor ASC in the regulation of epithelial skin carcinogenesis. Proc. Natl. Acad. Sci. USA. 2012;109:18384–18389. doi: 10.1073/pnas.1209171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pang Y.Y. The Brisbane 2000 terminology of liver anatomy and resections. HPB 2000; 2:333–39. HPB. 2002;4:99–100. doi: 10.1080/136518202760378489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colucci F., Soudais C., Rosmaraki E., Vanes L., Tybulewicz V.L., Di Santo J.P. Dissecting NK cell development using a novel alymphoid mouse model: investigating the role of the c-abl proto-oncogene in murine NK cell differentiation. J. Immunol. 1999;162:2761–2765. [PubMed] [Google Scholar]
- 75.Slack E., Hapfelmeier S., Stecher B., Velykoredko Y., Stoel M., Lawson M.A.E., Geuking M.B., Beutler B., Tedder T.F., Hardt W.D., et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inderbitzin D., Studer P., Sidler D., Beldi G., Djonov V., Keogh A., Candinas D. Regenerative capacity of individual liver lobes in the microsurgical mouse model. Microsurgery. 2006;26:465–469. doi: 10.1002/micr.20271. [DOI] [PubMed] [Google Scholar]
- 77.Yilmaz B., Spalinger M.R., Biedermann L., Franc Y., Fournier N., Rossel J.B., Juillerat P., Rogler G., Macpherson A.J., Scharl M. The presence of genetic risk variants within PTPN2 and PTPN22 is associated with intestinal microbiota alterations in Swiss IBD cohort patients. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H., Limenitakis J.P., Fuhrer T., Geuking M.B., Lawson M.A., Wyss M., Brugiroux S., Keller I., Macpherson J.A., Rupp S., et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whiteley A.S., Jenkins S., Waite I., Kresoje N., Payne H., Mullan B., Allcock R., O'Donnell A. Microbial 16S rRNA ion tag and community metagenome sequencing using the ion torrent (PGM) platform. J. Microbiol. Methods. 2012;91:80–88. doi: 10.1016/j.mimet.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 80.Yilmaz B., Juillerat P., Øyås O., Ramon C., Bravo F.D., Franc Y., Fournier N., Michetti P., Mueller C., Geuking M., et al. Microbial network disturbances in relapsing refractory Crohn's disease. Nat. Med. 2019;25:323–336. doi: 10.1038/s41591-018-0308-z. [DOI] [PubMed] [Google Scholar]
- 81.McMurdie P.J., Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac. Symp. Biocomput. 2012:235–246. [PMC free article] [PubMed] [Google Scholar]
- 82.Callahan B.J., Sankaran K., Fukuyama J.A., McMurdie P.J., Holmes S.P. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Research. 2016;5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Asnicar F., Weingart G., Tickle T.L., Huttenhower C., Segata N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 86.Beldi G., Brodie T., Zindel J., Dommann N., Surewaard B.G.J., Keogh A., Melin N., Buechi I., Tombolini R., Candinas D., et al. Single cell chronoatlas of regenerating mouse livers reveals early Kupffer cell proliferation. bioRxiv. 2021 doi: 10.1101/2021.06.09.447699. Preprint at. [DOI] [Google Scholar]
- 87.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anders S., Pyl P.T., Huber W. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Bulk-RNA Seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resource table. The patient’ data that support the findings of this study are available from the lead contact upon reasonable request. Microscopy data reported in this paper will be shared by the lead contact upon request until 02/2033.
-
•
Data were analysed using the standard DESeq2 and Qiime pipelines, or with the stated variations. User scripts will be shared upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.