Abstract
Objective
Automatically identifying patients at risk of immune checkpoint inhibitor (ICI)-induced colitis allows physicians to improve patientcare. However, predictive models require training data curated from electronic health records (EHR). Our objective is to automatically identify notes documenting ICI-colitis cases to accelerate data curation.
Materials and Methods
We present a data pipeline to automatically identify ICI-colitis from EHR notes, accelerating chart review. The pipeline relies on BERT, a state-of-the-art natural language processing (NLP) model. The first stage of the pipeline segments long notes using keywords identified through a logistic classifier and applies BERT to identify ICI-colitis notes. The next stage uses a second BERT model tuned to identify false positive notes and remove notes that were likely positive for mentioning colitis as a side-effect. The final stage further accelerates curation by highlighting the colitis-relevant portions of notes. Specifically, we use BERT’s attention scores to find high-density regions describing colitis.
Results
The overall pipeline identified colitis notes with 84% precision and reduced the curator note review load by 75%. The segment BERT classifier had a high recall of 0.98, which is crucial to identify the low incidence (<10%) of colitis.
Discussion
Curation from EHR notes is a burdensome task, especially when the curation topic is complicated. Methods described in this work are not only useful for ICI colitis but can also be adapted for other domains.
Conclusion
Our extraction pipeline reduces manual note review load and makes EHR data more accessible for research.
Keywords: deep learning, curation, information extraction, EHR
BACKGROUND AND SIGNIFICANCE
While immune checkpoint inhibitors (ICI) have improved cancer care, one of their main adverse events is ICI-induced colitis.1 Predicting patients that will likely experience ICI-induced colitis allows physicians to adapt care management. However, before supervised models to predict ICI colitis can be developed, training data from historical ICI colitis cases need to be curated from electronic health record (EHR) notes. Curation from notes is necessary because ICI colitis does not have clear diagnosis codes, and can be documented in a variety of ways (proctocolitis, ICI-associated diarrhea, diarrhea toxicity, etc.). Curating positive ICI colitis cases is an onerous task—keyword search identifies over 200 000 notes which need to be manually reviewed for accuracy before being imported into a database for more extensive expert curation of colitis episodes. A system which accurately identifies potential ICI colitis-related notes, and highlights relevant text, can facilitate and accelerate curators’ manual task.
Objective
The goal of the colitis curation task is to automatically identify EHR notes which refer to ICI colitis to create a dataset for predictive modeling. There are a couple of challenges associated with the automatic identification of ICI colitis from notes. First, ICI colitis can be described in multiple ways including descriptions of its varied symptoms such as diarrhea or bloody stool. Second, the dataset is highly imbalanced—ICI colitis notes have low incidence with only 1994 positive out of 23 313 notes in our training data. A less than 10% incidence rate of ICI colitis means the data pipeline must be tuned to not miss positive events, that is, it must have high recall. Third, many language models are pretrained to only use 512 tokens and most EHR notes exceed that length. Our pipeline must therefore account for splitting notes during classification. Finally, reading long notes during validation is taxing for curators and we need methods to highlight relevant regions in the text.
In this work, we present a deep-learning pipeline that accelerates manual curation by automatically identifying likely colitis-positive notes and reducing the note review burden. Additionally, we present a highlighting algorithm which uses BERT’s attention mechanism to identify relevant text within notes that are important for colitis identification. These highlighted regions can further accelerate curation by guiding curators during validation.
MATERIALS AND METHODS
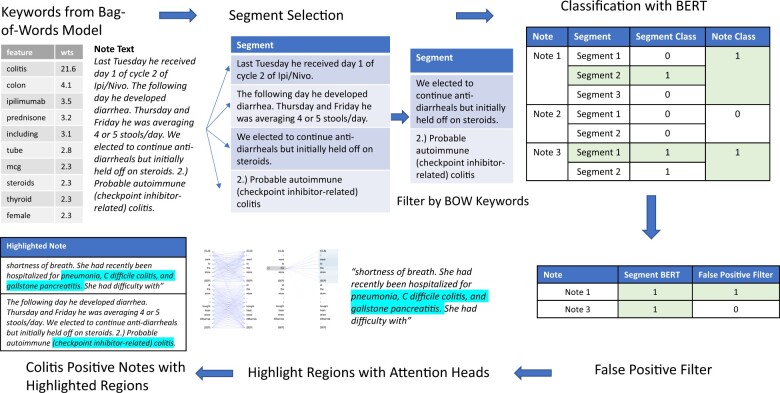
To reduce manual curation effort, we developed a data pipeline to accurately identify ICI colitis cases. Importantly, the pipeline should have a high recall, so curators do not miss cases, and moderate precision so curation time is not wasted on negative cases. Further, the system should highlight segments in the text which were used by the backend model in classifying cases. In this section, we first describe the dataset used to train and evaluate the model, followed by the classification and highlighting algorithms. The overview of the pipeline is given in Figure 1.
Figure 1.
Overview of the Data Pipeline.
Data
The ICI colitis cohort consisted of all Vanderbilt University Medical Center patients who received ICI treatment either at Vanderbilt or an external institution prior to December 31, 2020. The cohort consists of 3422 patients, of which, 703 patients were used to create a gold standard dataset. To create the gold standard, first the notes of the 703 patients were filtered using curator-provided keywords associated with colitis and its symptoms. Keyword filtering generated 23 313 possibly relevant notes for 703 patients. Curators then manually reviewed all 23 313 notes to indicate if they were true positives for ICI colitis or for one of its symptoms, that is, diarrhea or bloody stool. Curators consisted of expert reviewers with doctorates in biology as well as non-expert reviewers who were medical students.
Based on manual review of 23 313 notes, 1994 notes were positive for colitis within the diagnostic differential, 3906 were positive for the presence of diarrhea, and 548 were positive for the presence of bloody stool. Curators also highlighted the part of the note that was relevant for their determination of colitis or colitis-associated symptoms. We used this dataset for model selection, training, and validation. We used the standard 80:20 ratio to split our data into train (train + validation) and test sets. The training set had 14 920 notes, the validation set had 3730 notes, and the test set had 4663 notes. The test set is not exposed to the model during training and hence not used for fitting model weights.
After model development, the pipeline was applied to the remaining notes (of about 2700 patients) containing keywords which had not yet been manually reviewed and were “unseen” by the model—these notes were not used in model fitting. The unseen cohort had 147 853 notes, 128 314 of these contained colitis mention keywords, and 119 542 notes contained symptom keywords and provide our evaluation cohort.
Colitis Classification
In this section, we describe different classification methods for colitis prediction from EHR notes. We compared different approaches with increasing complexity, starting with bag-of-words (BOW)2 logistic regression, followed by distant supervision with Snorkel,3 and finally transformer-based models.4
Colitis Classification with BOW Model
The simplest colitis classification algorithm applied keyword search to identify positive notes. But, as seen in the labeled data above, this method had very low precision (0.08). Our next approach used a logistic regression classifier with a BOW feature encoding.2 In a BOW model, the words of the entire corpus are treated as binary features, and each input text is vectorized to denote the words it contains. We used the term frequency-inverse document frequency (TF-IDF)2 metric to rank the top 1000 words in the corpus. These words were used as features to train a logistic regression which classified a note as positive or negative for colitis. This approach had modest results.
To better understand the logistic regression model, we extracted the top 10 words from the BOW model that were predictive for colitis to compare against the curator-specified keywords. Unsurprisingly, the most predictive word was colitis. However, the remaining 9 words were not in the curators’ earlier-provided list. Some of them were semantically relevant such as Ipilimumab, colon, and steroids. But, others were broad terms such as including, MCG, and female. The latter terms might be a result of writing patterns. For example, colitis was often mentioned with potential side effects, and written as “potential side-effects including colitis,” which might have induced the model to give high feature importance to “including.”
Colitis Classification with Distant Supervision
To improve model performance, we next explored if the keywords extracted from BOW model could be used in combination with curator keywords. There were around thirty curator keywords, which we grouped into 4 subsets based on the symptoms they described: diarrhea, bloody stool, abdominal pain, and dehydration. Each keyword group had different accuracies. To aggregate them, we leveraged the Snorkel library.3 Snorkel classifies with distant supervision by aggregating multiple inaccurate labeling functions (eg, keyword search) to produce high-quality labels. It trains a meta-learner which estimates the quality of each labeling function and weights them accordingly. It requires a minimum of 3 labeling functions. An example of a labeling function would be using keyword search to mark notes that contain “colitis” as positive and others as negative. Labeling functions can also use groups of words, for example, a note can be marked positive if it contains any of the words from the group [“colitis”, “colon”, “steroids”].
We compared 4 classification methods with Snorkel. The first method used the curator keywords grouped by the symptoms they described. So, one labeling function used all the diarrhea keywords to classify notes as positive, another used the bloody stool keywords, another used “colitis” for classification, and so on. The second method used the BOW keywords, with each labeling function corresponding to one of the ten BOW words. The third method, BOW+curator, used all the labelers from the first two methods. While the BOW did slightly better than the other two, the performance of all three methods were poor (Table 1). We hypothesized that the poor performance was due to multiple labeling functions having similar accuracies, making it difficult for Snorkel to aggregate. To ameliorate this issue in the fourth method, BOW+curator grouped, we reduced the number of labeling functions to 3: the first function did keyword search with “colitis” since it is the most influential word, the second function used all the curator keywords together, and the third function used all the BOW keywords. This grouping minimized the number of labelers, therefore reducing noise.
Table 1.
Snorkel performance with different keyword sets
| Precision |
Recall |
AUC |
||||
|---|---|---|---|---|---|---|
| Test set | Unseen | Test set | Unseen | Test set | Unseen | |
| Curator keywords | 0.38 | 0.11 | 0.07 | 0.02 | 0.53 | 0.51 |
| BOW keywords | 0.72 | 0.40 | 0.13 | 0.03 | 0.56 | 0.51 |
| BOW + Curator | 0.87 | 0.30 | 0.04 | 0.01 | 0.52 | 0.50 |
| BOW + Curator grouped | 0.79 | 0.64 | 0.99 | 1.00 | 0.98 | 0.98 |
Colitis Classification with Transformers
The issue with keyword search is that semantic context is lost. To address this problem, we used Bidirectional Encoder Representations from Transformer (BERT),4 a state-of-the-art natural language processing (NLP) model. Text inputs to BERT are first tokenized, where each token roughly corresponds to a word. The base BERT architecture is trained by randomly masking a subset of these tokens and then predicting the masked token. During this prediction task, BERT creates numerical representations called embeddings for each token, which can be used for downstream tasks. For classification, an additional layer is trained on top of the base architecture to classify each input text.
Longer Notes in BERT
A limitation of BERT is that it can only accept input sequences with a maximum length of 512 tokens since it was pretrained with that configuration. Adapting to longer token lengths will require training from scratch and has heavy computational load. Tokenized EHR notes are usually longer than 512 tokens and must be split into multiple segments. The classes from each segment are then aggregated to get the class for the EHR note.
Colitis and its symptoms may only be mentioned in certain parts of the notes, thus using the wrong subset of a note in the prediction can result in inaccuracies. Therefore, we considered multiple methods to analyze long clinical notes using BERT. First, we considered using the maximum value from a segment in the prediction as opposed to the majority vote (ie, if any segment was positive, the entire note was positive). Second, we considered if the model could be improved if only the segment containing colitis description were fed into the model. To test these variations, we compared 3 methods: (1) using all segments from a note, (2) randomly selecting segments from a note, and (3) using the highlighted sentences marked as relevant by the curator in the training dataset. The third method using the curator’s sentences outperformed the other two and motivated the need for automatic segment selection as an initial step in the pipeline.
Automatic Segment Selection to Reduce Note Length
Selecting the segment of the note that contains colitis description for classification improved model performance. However, for new datasets, we would not have access to curator-highlighted sentences and must identify the relevant segments automatically. A simple segment identification method is to filter by keyword search. For positive colitis notes, all segments that contained any word from a specified keyword list were selected as inputs to the model. For negative notes, a random sample of segments were selected, and the number of segments per note was the same on average for positive and negative notes.
To determine the ideal segment selection approach, we compared segment filtering with curator-specified keywords and BOW keywords. During empirical evaluation, the curator-specified keyword pipeline had higher precision while the BOW keywords had higher recall. Recall is very important because of the low incidence rates of colitis (1994 positive in 23 313 notes). While the curator-specified keywords are more semantically related to colitis, the model needed a broader training set of positive note segments to learn to discriminate colitis-positive notes. However, during inference, filtering the test set with curator-specified keywords improves performance, making the precision comparable to the curator keyword model, without sacrificing recall. Hence, our segment selection hybrid model is trained on BOW keyword-filtered segments and then applied to a curator keyword-filtered dataset. We compared the performance of this hybrid model against the baseline model that uses all segments, the curator keyword model, and the BOW keyword model (Table 2).
Table 2.
BERT performance with different segment selection strategies
| All segments | Curator keywords | BOW keywords | BOW + Curator | |
|---|---|---|---|---|
| Precision | 0.76 | 0.71 | 0.57 | 0.72 |
| Recall | 0.96 | 0.97 | 0.98 | 0.98 |
| Training time (h) | 2.5 | 1 | 1.5 | 1.5 |
We applied the segment selection model to the unseen cohort of 128 314 notes containing colitis keywords. After reviewing the first 1080 notes, the curators found an 18% false positive rate, that is, 194 notes were not colitis cases and most of these mentioned colitis as a potential side-effect at therapy initiation. A reason for the higher false positive rate of this model could be imperfect training data—non-expert reviewers sometimes marked the potential side-effect colitis mention as positive.
Secondary Prediction to Filter False Positives
To further reduce the curator review burden, we trained a second model using the 1080 reviewed notes (mentioned above) to identify and filter false positive notes from the notes identified in the segment-selected model. These notes were reviewed by expert curators to ensure we had a high-quality dataset. After reviewing another small sample of 20 notes output from the false positive filter, curators found that notes that did not contain the word “colitis” were not relevant. Our final workflow (Figure 1) then was to filter data by curator keywords, apply the segment selection model to get positive notes, apply the false positive filtering model, and then send the positive notes containing the word “colitis” for curator review.
We built two data pipelines, one for identifying colitis mention notes, and another for colitis symptom notes. The pipeline for the symptom notes was similar, except the BOW keywords were extracted from a model trained to identify diarrhea and bloody stool, a separate manually reviewed set of 1266 notes output from the symptom segment classifier were used to train the false positive filter, and we did not search for the word “colitis” in the last step. We evaluate the pipeline by manually verifying the number of true positives in the classified data set, that is, precision.5 Curators also looked at a small sample of negative notes and found them to be true negatives and hence did not review the remaining notes.
Highlighting regions
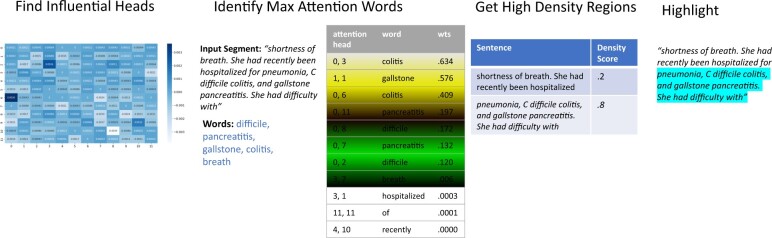
To further accelerate the curation task, we wanted the system to highlight the sections within notes which were used to classify colitis. To this end, we used BERT’s attention heads to identify significant regions. Briefly, BERT has 12 layers, each of which has 12 attention heads, for a total of 144 attention heads. Heads focus on different patterns and regions. For example, prior work has found that some heads pay attention to (i) matching tokens (same words) in other sentences, (ii) to tokens surrounding the current token, or (iii) to sentence boundaries.6,7 Each head computes a self-attention matrix for each input segment, where an attention value is calculated for every pair of tokens in the segment. The attention value of token B from token A denotes how important B is for providing context to A. While tokens mostly correspond to words, if a word is outside BERT’s vocabulary, it is split into multiple tokens. Since we are interested in word-level attention, the attention from split words must be aggregated. To aggregate attention, attention values from a split word’s tokens are averaged and attentions to split tokens are summed. This preserves the property of attentions from a word summing to one.7 So, for each word in a segment with 512 words, each attention head has 512 attention values. That is 512x512 attention values for each of the 144 attention heads to evaluate.
As a simple baseline for selecting words for highlighting, we used the TF-IDF metric and selected the top scoring 5 words. While TF-IDF had decent performance, it does not make use of the classification model’s mechanisms. To this end, we used the method described by Clark et al.8 which averaged a word’s attention score from all 144 heads and then selected the top scoring 5 words for highlighting.8 Additionally, prior work has also shown that all heads are not equally helpful in prediction.7,9 To address this, we employed a third algorithm which first identified influential attention heads and then focused on words relevant to our task to find other significant words. To identify influential heads, we employed the algorithm described by Michel et al.,9 where the importance of a head was quantified as the difference in loss on the test set when the head is masked during inference. We used this leave-one-out strategy to measure the difference in accuracy on the test set and only considered the heads which led to a decrease in accuracy. This reduced the number of heads from 144 to 47 for the colitis mention pipeline and to 57 for the symptom mention pipeline. Next for each input segment, we looked at important keywords such as colitis, diarrhea, and stool, and for each of the influential heads, selected the words that the keywords paid the most attention to and had an attention score of at least 0.001. Lower attention scores usually denoted prepositions. Boundary tokens were excluded during this process since they are not relevant for highlighting.
Once influential words were identified using one of the above 3 algorithms, we needed to identify regions in the input sentences that contained these words. We divided sentences into regions of 10 words and then scored each region by the number of segment-specific influential words it contained, normalized by region length (ie, 10). Regions with a score of 0.5 or greater, that is, high-density regions, were highlighted (Figure 2). Curators validated our highlighting algorithm by manually reviewing a random set of 10 patients with 79 colitis-positive notes. For evaluating symptoms, an additional 37 notes were reviewed for a total of 116 notes. Out of these 116 notes, 79 were positive for diarrhea and 29 were positive for bloody stool.
Figure 2.
Highlighting Algorithm.
RESULTS
In this section, we present results in the order it was described in the methods. First, we present the results of classification, going from keyword search to the BERT classification model. We then present the results of the highlighting algorithms.
Keyword search with Snorkel for Colitis Classification
The simplest colitis classification model was using the BOW logistic regression, which had an AUC5 of 0.78 on the test set. To further improve performance, we extracted the most positive predictive words and used them for colitis classification with Snorkel. Table 1 shows the results of using Snorkel to label data using different sets of keywords on the test set of 4663 notes as well as on the unseen cohort of 128 314 notes. The BOW keywords had a slightly better AUC than that of the curator keywords and BOW+curator on the test set, but all 3 performed poorly on the larger unseen set. A possibility for the low performance of the curator keywords could be that the individual labeler functions had balanced performance on the training set and hence Snorkel’s learner could not easily aggregate the data. On the other hand, when we used only 3 labelers, grouping keywords into BOW, curator, or colitis keyword, Snorkel had improved performance, because the colitis function had significantly better performance than the other two. However, the precision for this method on the unseen set was still only 0.64, which would lead to significant curator burden for filtering out false positives.
Segment-Selected BERT for Colitis Classification
We next present the results of the BERT colitis classification model with different segment selection strategies to account for the maximum sequence length of 512 tokens. Table 2 shows the results on the test set of the different segment selection strategies. The fourth method, training on BOW keywords and filtering the test set with curator keywords, performed the best with the highest recall which was important for colitis classification due to the imbalanced dataset. It also decreased the training time to 1.5 hours from the 2.5 hours needed to train using all segments.
The curation pipeline for colitis mention notes with results is shown in Figure 3. The segment-selected BERT algorithm had a precision of 0.72 and recall of 0.98, while the false positive filter, trained on 864 notes had a precision of 0.92 and recall of 0.79 on the test set of 162 notes. The overall pipeline had a precision of 0.92. The colitis pipeline started with 128K unseen notes based on curator keyword search, which was reduced to 14K by segment-selected BERT, further reduced to 12K by false positive filter BERT, and finally, only 8K contained the word “colitis” and were reviewed by the curators. We thus decreased the note review load from 128K to 8K.
Figure 3.
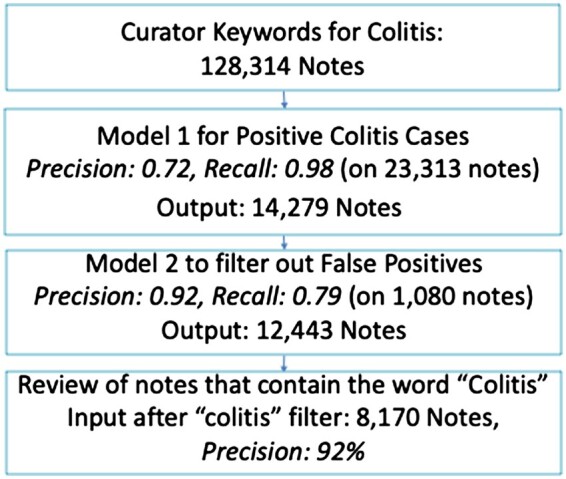
Colitis mention results.
The symptom pipeline (Figure 4), that is, classifying if a note mentioned diarrhea or bloody stool, had an overall precision of 0.84. The segment-selected BERT algorithm had a precision of 0.77 and recall of 0.96, while the false positive filter BERT trained on 945 notes had a precision of 0.97 and recall of 0.98 on the test set of 249 notes. The unseen cohort for this pipeline started with 119K based on curator keyword search, which was reduced to 40K by the segment-selected BERT, and further reduced to 30K by the false positive filter BERT, which were manually reviewed.
Figure 4.
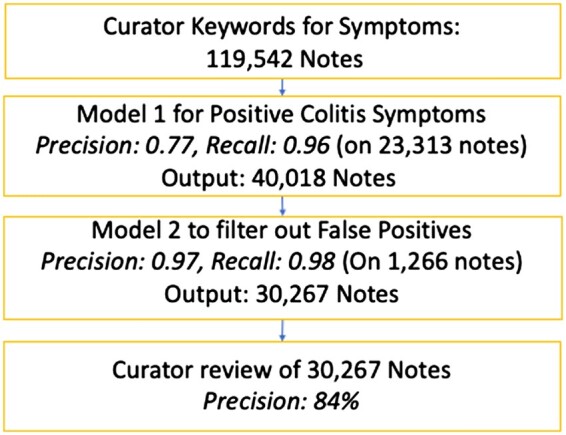
Symptom mention results.
Highlighting accuracy
We next present the results of different highlighting algorithms to guide curators in reviewing the correctness of colitis notes. For each manually evaluated note, the curators indicated if the pipeline had highlighted all relevant regions. Additionally, curators noted relevant regions that were missed. The curator-reviewed notes with annotations were used as the gold standard for the comparison of different highlighting algorithms such as TF-IDF, average attention, and focused attention (ie, looking at attention values of words relevant to the problem). Table 3 shows the results of 10 patients with 79 notes. While highlighting was applied to all the notes, curators were asked to provide feedback on a small subset so as not to add extra time to their task. The focused attention highlighting algorithm was able to capture all relevant regions for all 79 notes, while TF-IDF and average attention highlighted all regions in 73% and 67% of notes, respectively.
Table 3.
Highlighting Algorithm Results
| Accuracy | |
|---|---|
| TF-IDF | 58 (73%) |
| Average attention | 53 (67%) |
| Focused attention | 79 (100%) |
The focused attention algorithm was applied to the symptom pipeline as well. The accuracy of highlighting for diarrhea was 97% and for bloody stool was 72%. For these cases, certain words such as loose stools and melena were not highlighted because they were not necessarily attended to by keywords such as diarrhea and stool.
DISCUSSION
We have presented a pipeline for identifying colitis-positive notes which considerably reduces curation burden. We have shown that for large EHR notes, narrowing down to relevant segments prior to inputting data into the model improves performance. While logistic regression had a modest performance for classification (AUC of 0.78), the predictive keywords were useful for segment selection. This result indicates that while keywords are not effective for prediction, they are useful for segment identification. The colitis mention pipeline had a precision of 92% and reduced the note review load by 93.4% (128K–8K). The symptoms pipeline had a precision of 84% and reduced the note review load by 75% (119K–30K). The slightly lower performance for the symptom model can be attributed to imperfect training data, since the initial set of 23 313 had false positive labels which were curated by non-expert reviewers. Even so, our models greatly accelerated data curation on an unseen cohort.
Additionally, we compared the usage of different keyword sets to classify the dataset with Snorkel. Based on our limited comparisons, Snorkel performs the best when there are fewer labeling functions and at least two functions label a sample. The curator keywords covered different samples and had high accuracies because of the high number of negative samples, and Snorkel was unable to effectively aggregate these, leading to the low recall. The BOW keyword functions had more overlap, leading to slight increase in recall. Finally, grouping all keywords into three functions, one corresponding to BOW keywords, one for curator keywords, and one for the word colitis, had the best recall. This improvement might be because the positive samples got at least two votes, allowing Snorkel to discriminate better. However, the precision was still lower than BERT with segment selection.
We used the standard train/test split of 80:20 for evaluating our algorithms during model selection. We further validated our results on a large, unseen cohort of 128 314 notes, which improves the robustness of our results. However, adapting and implementing this method at other institutions might still require finetuning the models with their datasets due to differences in language and documentation styles.
Finally, we presented an algorithm for highlighting relevant regions using BERT’s attention mechanism. Selecting important heads and then choosing words that were attended to by focused keywords performed the best. Both the baseline methods, TF-IDF and average attention picked up regions related to colitis, such as descriptions of gastrointestinal symptoms, but missed mentions of colitis. These methods can still be used in the absence of curator-provided keywords, albeit with slightly lower recall.
Our pipeline helps researchers quickly curate ICI-induced colitis cases. It is intended for clinical research use and not necessarily geared towards point-of-care. The results of our pipeline are manually reviewed by curators to ensure correctness. It is possible that our pipeline biases curators towards colitis in false positive cases. We try to mitigate this bias by showing the curator the highlighted regions used by the model for the prediction. The curator can thus check if the note actually mentions colitis or if it was marked positive for some other reason.
Researchers can use the data curated by our pipeline to build algorithms to predict if a patient will develop colitis. These predictive algorithms could be integrated into the EHR for point-of-care. The quality of the data output by our pipeline will then affect the downstream predictive model. Our pipeline has high recall, meaning there are very few false negative cases. For cases where we fail to predict colitis, the quality of care will remain as it is now. For cases where we accurately predict colitis, care can potentially improve to prevent colitis or provide prior mitigation measures. For false positives, changing the patient’s care could lead to a decreased efficiency. One way to indicate the possibility of false positives to providers is to show them confidence intervals of the probability of the patient developing colitis as well as show features that increase the patient’s probability of colitis.
CONCLUSION
Data curation is a bottleneck for many informatics research pipelines. Building automated curation tools can accelerate this process. While there are many NLP tools for prediction and sentence completion, customizing them for data extraction is nontrivial. Tuning a curation pipeline requires a tight working loop between data scientists and curators with domain expertise,10 as demonstrated by this work. We show that identifying relevant text to be input into the model has an impact on performance. Applying similar methods to other extraction domains can be greatly beneficial for democratizing research data.
Contributor Information
Protiva Rahman, Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Cheng Ye, Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Kathleen F Mittendorf, Vanderbilt Ingram Cancer Center, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Michele Lenoue-Newton, Vanderbilt Ingram Cancer Center, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Christine Micheel, Vanderbilt Ingram Cancer Center, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Jan Wolber, Pharmaceutical Diagnostics, GE Healthcare, Chalfont St Giles, UK.
Travis Osterman, Vanderbilt Ingram Cancer Center, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Daniel Fabbri, Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
FUNDING
The GE Healthcare and Vanderbilt University Medical Center partnership.
AUTHOR CONTRIBUTIONS
Conception and design: Protiva Rahman and Daniel Fabbri. Collection and assembly of data: Cheng Ye, Kathleen F. Mittendorf, and Michele LeNoue-Newton. Data analysis: Protiva Rahman and Daniel Fabbri. Interpretation of Results: All authors. Manuscript writing: All authors. Final approval of manuscript: All authors. Accountable for manuscript: All authors.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this article cannot be shared publicly due to the presence of Protected Health Information. The code will be shared on reasonable request to the corresponding author.
REFERENCES
- 1. Karamchandani DM, Chetty R.. Immune checkpoint inhibitor-induced gastrointestinal and hepatic injury: pathologists’ perspective. J Clin Pathol 2018; 71 (8): 665–71. [DOI] [PubMed] [Google Scholar]
- 2. Martineau J, Finin T. Delta TFIDF: an improved feature space for sentiment analysis. In: proceedings of the International AAAI Conference on Web and Social Media, Vol. 3, No. 1; March 20, 2009: 258–61; California, USA. [Google Scholar]
- 3. Ratner A, Bach SH, Ehrenberg H, Fries J, Wu S, Ré C. Snorkel: rapid training data creation with weak supervision. In: proceedings of the VLDB Endowment. International Conference on Very Large Data Bases, Vol. 11, No. 3; November, 2017; NIH Public Access: 269; Munich, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. In: proceedings of NAACL-HLT; 2019: 4171–86. [Google Scholar]
- 5. Davis J, Goadrich M. The relationship between precision-recall and ROC curves. In: proceedings of the 23rd International Conference on Machine Learning; June 25, 2006: 233–40; Pennsylvania, USA.
- 6. Michel P, Levy O, Neubig G. Are sixteen heads really better than one? In: Advances in Neural Information Processing Systems, Vol. 32;2019; Vancouver, Canada. [Google Scholar]
- 7. Li R, Xiao W, Wang L, Carenini G. Human interpretation and exploitation of self-attention patterns in transformers: a case study in extractive summarization. arXiv, arXiv:2112.05364, December 10, 2021, preprint: not peer reviewed.
- 8. Clark K, Khandelwal U, Levy O, Manning CD. What does Bert look at? An analysis of Bert's attention. In: proceedings of the 2019 ACL Workshop BlackboxNLP: Analyzing and Interpreting Neural Networks for NLP; August 2019: 276–86. [Google Scholar]
- 9. Bolotova V, Blinov V, Zheng Y, Croft WB, Scholer F, Sanderson M. Do people and neural nets pay attention to the same words: studying eye-tracking data for non-factoid QA evaluation. In: proceedings of the 29th ACM International Conference on Information & Knowledge Management; October 19, 2020: 85–94; Galway, Ireland.
- 10. Rahman P, Nandi A, Hebert C.. Amplifying domain expertise in clinical data pipelines. JMIR Med Inform 2020; 8 (11): e19612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article cannot be shared publicly due to the presence of Protected Health Information. The code will be shared on reasonable request to the corresponding author.