Abstract
Background
Knowledge of global clarithromycin (CLA)-resistant rates of Helicobacter pylori (H. pylori) is crucial for decision of the most appropriate eradication therapies with good clinical outcomes. Therefore, this review and meta-analysis aimed to evaluate the global prevalence of the CLA resistance in H. pylori to provide some guidance for selecting the first-line antibiotics.
Method
A comprehensive search was performed for relevant literature until April 2021 in PubMed, Embase, and Web of Science databases. Freeman-Tukey double arcsine transformation was performed to estimate the weighted pooled prevalence of resistance.
Results
The meta-analysis included 248 articles. The prevalence of CLA-resistant H. pylori was 27.53% (95% CI [25.41–29.69]). The heterogeneity between reports was significant (I2 = 97.80%, P < 0.01). The resistance rate increased from 24.28% in 2010–2017 to 32.14% in 2018–2021 (P < 0.01). Iran, with 38 articles, has the most report. Nevertheless, Switzerland, Portugal, and Israel had the highest resistance rates (67.16%, 48.11%, and 46.12%, respectively). The heterogeneity between the continents and the antimicrobial susceptibility methods also interpreted standard guidelines and breakpoints was insignificant (P > 0.05).
Conclusion
Overall CLA resistance rate was 27.53%, worldwide. The difference in CLA resistance rate among the included studies can be due to several reasons such as differences in antibiotic prescription rates in various geographic areas, use of different breakpoints or inaccurate criteria in performed studies, and the emergence of multidrug-resistant (MDR) strains.
Keywords: Clarithromycin, Meta-analysis, Antibiotic resistance, Helicobacter pylori
Introduction
Helicobacter pylori is one of the most successful human pathogens that affects approximately 50% of the population worldwide. In developing countries 70% to 90% of the population are infected by this bacterium (Arenas et al., 2019; Kocsmár et al., 2021). H. pylori infection is related to many gastric diseases, such as peptic ulcers, chronic gastritis, uninvestigated and functional dyspepsia and mucosa-associated lymphoid tissue lymphoma, and even increases the risk of gastric cancer (Savoldi et al., 2018). As for the high prevalence of the bacterium and its related diseases, proper treatment is very important. Today, standard treatment is a three-stage drug that consists of an acid neutralizer and two antibiotics, clarithromycin (CLA), and amoxicillin or metronidazole for 14 days (Hosseini et al., 2021).
However, treatment is difficult because the bacterium quickly develops resistance to the few antibiotics known to be effective (Park et al., 2016). The World Health Organization (WHO) has classified it among the 12 most resistant bacteria in the world (Essaidi et al., 2022). The increasing failure rate of eradication treatment due to the appearance of resistant H. pylori strains contributes to the worldwide prevalence of this infection and subsequent inflammatory and neoplastic disorders. Unfortunately, nowadays, the success of this treatment is less than 80% worldwide (Kocsmár et al., 2021; Hussein, Al-Ouqaili & Majeed, 2022).
CLA has been emerged as the basis for H. pylori treatment in combined therapy because of small effect on gastric acidity, its low minimal inhibitory concentration, and relatively good mucosal diffusion (Marques et al., 2020; Nishizawa & Suzuki, 2014). Due to extensive usage of CLA in some geographical regions, global prevalence rate of CLA resistance is increasing (Zou et al., 2020). In developing countries, CLA resistance and frequency of re-infection are factors that contribute to high worldwide prevalence of H. pylori infection and subsequent inflammatory and neoplastic disorders (Alarcón-Millán et al., 2016). In most European countries, as well as the rest of the world, the prevalence of CLA resistance has reached 20%. With rare exceptions, it is no longer recommended to include CLA in empirical treatment in regions where primary resistance to this antibiotic is 20% (Alarcón-Millán et al., 2016; Morilla et al., 2019).
Knowledge of global CLA-resistant rates of H. pylori is crucial for decision of the most appropriate eradication therapies with good clinical outcomes. Therefore, the aim of current review and meta-analysis is to evaluation of the global prevalence of the CLA resistance in H. pylori.
Method
Search strategy
A comprehensive search was conducted by two researchers in the online databases PubMed, Embase, and Web of Science until April 2021, using relevant keywords such as clarithromycin, antibiotic resistance, and H. pylori, as well as related MeSH terms (see Supplemental File 1 for the search syntax). The search syntax is available in Table 1.
Table 1. A systematic search including PubMed, Embase, and Web of Science with relevant keywords such as clarithromycin, antibiotic resistance, and Helicobacter pylori.
| First author (Reference) | Country | Enrollment time | Published year | Type of study | N. patients | Mean age | N. HP | N. Clarithromycin-resistant | AST method | Breakpoint |
|---|---|---|---|---|---|---|---|---|---|---|
| Horie et al. (2020) | Japan | 2005–2018 | 2020 | RET | 5,249 | 58.3 | 1300 | 426 | MIC | 1 |
| Haddadi et al. (2020) | Iran | 2020 | CS | 280 | 46 | 128 | 3 | DD | CLSI 2015 21 |
|
| Eisig et al. (2011) | Brazil | 2011 | PCS | 54 | 46.6 | 39 | 3 | MIC | 1 | |
| Aftab et al. (2016) | Bangladesh | 2014–2014 | 2015 | CS | 133 | 35.2 | 56 | 22 | MIC | 0.25 |
| Ortiz et al. (2019) | Honduras | 2013–2013 | 2019 | CS | 189 | 54 | 116 | 13 | MIC | 0.5 |
| Silva et al. (2018) | Portugal | 2013–2017 | 2018 | PCS | 74 | 14 | 58 | 7 | MIC | 1 |
| Almeida et al. (2014) | Portugal | 2009–2013 | 2014 | PCS | 180 | 43.4 | 180 | 90 | MIC | 1 |
| Ilie et al. (2011) | Romania | 2011 | CS | 100 | Range: 19–80 | 70 | 22 | DD | >20 CLSI 2010 |
|
| Vécsei et al. (2011) | Austria | 2007–2009 | 2011 | RET | 96 | 10.8 | 96 | 16 | MIC | 1 |
| Ranjbar & Chehelgerdi (2018) | Iran | 2016–2017 | 2018 | CS | 700 | Range: 3–72 | 526 | 335 | DD | 21 |
| Hamza et al. (2018) | Egypt | 2018 | CS | 150 | 20 | 12 | DD | 21 | ||
| Gong et al. (2020) | South Korea | 2017–2018 | 2020 | RET | 13 | 46 | 38 | MIC | 0.5 | |
| Wang et al. (2020) | China | 2018–2019 | 2020 | CS | 124 | 124 | 44 | MIC | 0.5 | |
| Su et al. (2022) | Taiwan | 2009–2019 | 2021 | RET | 87 | 13.5 | 65 | 15 | MIC | 1 |
| Sugimoto et al. (2017) | Japan | 2011–2015 | 2016 | RET | 111 | 55.2 | 111 | 90 | MIC | 1 |
| Abadi et al. (2011) | Iran | 2009–2009 | 2011 | CS | 210 | 40.7 | 197 | 89 | DD | 30 |
| Teh et al. (2014) | Malaysia | 2014 | CS | 110 | 102 | 7 | MIC | 1 | ||
| Peng et al. (2017) | China | 2013–2014 | 2017 | CS | 178 | 41.6 | 78 | 38 | MIC | 1 |
| Hashemi et al. (2019) | Iran | 2015–2016 | 2019 | CS | 150 | 157 | 38 | MIC | 1 | |
| Lauener et al. (2019) | Switzerland | 2013–2017 | 2019 | CS | 140 | 140 | 96 | MIC | 1 | |
| Domanovich-Asor et al. (2020) | Israel | 2015–2019 | 2020 | CS | 48 | 48 | 26 | MIC | 1 | |
| Wu et al. (2015) | Taiwan | 2010–2014 | 2015 | RET | 137 | 137 | 95 | MIC | 1 | |
| Vala et al. (2016) | Iran | 2011–2012 | 2016 | CS | 80 | 20 | 4 | MIC | 0.5 | |
| Omar et al. (2014) | Australia | 2014 | CS | 11 | 46.8 | 11 | 8 | MIC | 1 | |
| Vilaichone et al. (2016) | Thailand | 2013–2013 | 2016 | CS | 291 | 46.6 | 124 | 7 | MIC | 0.5 |
| Lee et al. (2014) | South Korea | 2003–2013 | 2014 | PCS | 2,202 | 52.9 | 475 | 147 | MIC | 1 |
| Lee et al. (2019) | South Korea | 2014–2018 | 2018 | PCS | 85 | 55.2 | 74 | 24 | MIC | 1 |
| Goudarzi et al. (2016) | Iran | 2014–2014 | 2016 | CS | 65 | 42 | 65 | 28 | MIC | 1 |
| Karpinski et al. (2015) | Poland | 1998–1999 2013–2014 |
2015 | CS | 108 | 108 | 9 | MIC | 1 | |
| Miyata et al. (2021) | Japan | 2007–2018 | 2020 | CS | 119 | 12 | 45 | 26 | MIC | 1 |
| Palmitessa et al. (2020) | Italy | 2017–2018 | 2020 | CS | 224 | 48.6 | 92 | 49 | MIC | 0.5 |
| Hung et al. (2021) | Taiwan | 2016–2019 | 2021 | RET | 197 | 54.8 | 62 | 9 | MIC | 1 |
| Miftahussurur et al. (2016) | Japan | 2012–2012 | 2016 | CS | 146 | 42.2 | 42 | 9 | MIC | 0.25 |
| Siddiqui et al. (2016) | Pakistan | 2008–2013 | 2016 | CS | 889 | 35.6 | 92 | 5 | MIC | 0.5 |
| Sugimoto et al. (2014) | Japan | 2009–2013 | 2014 | CS | 153 | 153 | 64 | MIC | 1 | |
| Jolaiya et al. (2020) | Nigeria | 2020 | CS | 492 | 104 | 41 | MIC | 0.5 | ||
| Pandya et al. (2014) | India | 2008–2011 | 2014 | CS | 125 | 80 | 47 | DD | 30 | |
| Lehours, Siffré & Mégraud (2011) | France | 2009–2009 | 2011 | CS | 127 | 43 | 26 | MIC | 0.5 | |
| Sun et al. (2018) | China | 2018 | CS | 49 | Range: 27–76 | 43 | 9 | MIC | 0.75 | |
| Dekhnich et al. (2018) | Russia | 2009–2017 | 2018 | CS | 783 | 51.8 | 276 | 16 | MIC | 0.5 |
| Sugimoto et al. (2020) | Japan | 2015–2019 | 2020 | RET | 307 | 62.3 | 307 | 102 | MIC | 1 |
| Siavoshi, Saniee & Malekzadeh (2018) | Iran | 2018 | CS | 450 | 44.1 | 104 | 37 | MIC | 2 | |
| Szadkowski, Zemlak & Muszynski (2018) | Poland | 2005–2015 | 2018 | CS | 154 | 55 | 15 | DD | 21 | |
| Costa, Soares & Goncalves (2017) | Portugal | 2012–2016 | 2017 | RET | 42 | 48.9 | 42 | 36 | DD | 17 |
| Aguilera-Correa et al. (2017) | Spain | 2016 | CS | 136 | 84 | 48 | MIC | 0.5 | ||
| Akar et al. (2021) | Turkey | 2018–2019 | 2021 | CS | 422 | 50 | 133 | 25 | MIC | 0.5 |
| Yula et al. (2013) | Turkey | 2010–2011 | 2012 | CS | 110 | 41.4 | 79 | 7 | MIC | 1 |
| Zhang et al. (2019) | China | 2015–2016 | 2018 | CS | 150 | 149 | 104 | MIC | 1 | |
| Macin et al. (2015) | Turkey | 2006–2012 | 2015 | CS | 311 | Range: 5–19 | 93 | 28 | MIC | 1 |
| Auttajaroon et al. (2019) | Thailand | 2017–2017 | 2019 | CS | 93 | 54.5 | 70 | 9 | MIC | 0.5 |
| Eghbali et al. (2016) | Iran | 2012–2013 | 2016 | CS | 89 | 53.6 | 89 | 5 | MIC | 1 |
| Wu et al. (2014) | Taiwan | 2014 | CS | 231 | 43 | 5 | MIC | 1 | ||
| Kocazeybek et al. (2019) | Turkey | 2014–2017 | 2019 | CS | 63 | 47.08 | 63 | 24 | MIC | 1 |
| Egli et al. (2020) | Switzerland | 2013–2017 | 2020 | CS | 76 | 76 | 49 | MIC | 1 | |
| Khani, Talebi Bezmin Abadi & Mohabati Mobarez (2019) | Iran | 2017–2018 | 2019 | CS | 81 | 56.8 | 61 | 13 | MIC | 0.5 |
| Morimoto et al. (2015) | Japan | 2014 | RET | 135 | 62.3 | 135 | 35 | MIC | 1 | |
| Alarcón-Millán et al. (2016) | Mexico | 2016 | CS | 144 | 48.3 | 45 | 8 | DD | 18 | |
| Tamayo et al. (2017) | Spain | 2013–2015 | 2017 | CS | 6,228 | 1986 | 349 | MIC | 1 | |
| Yoon et al. (2014) | South Korea | 2005–2010 | 2014 | RET | 204 | 52.5 | 212 | 18 | MIC | 1 |
| Miftahussurur et al. (2017) | Dominican | 2017 | CS | 158 | 47.1 | 64 | 2 | MIC | 8 | |
| Mohammad et al. (2011) | Iran | 2007–2007 | 2011 | CS | 263 | 84 | 19 | MIC | 1 | |
| Ha et al. (2019) | Vietnam | 2012–2017 | 2018 | CS | 185 | 42.3 | 104 | 56 | MIC | 1 |
| Tanih, Ndip & Ndip (2011) | South Africa | 2011 | CS | 254 | 44.5 | 200 | 40 | MIC | 1 | |
| Yeganeh et al. (2019) | Israel | 2016–2016 | 2019 | PCS | 218 | 42 | 218 | 96 | MIC | 1 |
| Liu et al. (2019) | China | 2010–2017 | 2019 | RET | 1,463 | 1463 | 296 | MIC | 0.5 | |
| Zhu et al. (2013) | China | 2002–2006 | 2012 | CS | 365 | 365 | 42 | MIC | 1 | |
| Farzi et al. (2019) | Iran | 2014–2015 | 2018 | CS | 97 | Ranging 10–70 | 40 | 14 | MIC | 0.25 |
| Abdollahi et al. (2019) | Iran | 2017–2018 | 2019 | CS | 191 | 38.2 | 63 | 20 | DD | 21 |
| Lee et al. (2019) | South Korea | 2015–2018 | 2018 | CS | 1,422 | 140 | 43 | MIC | 0.5 | |
| De Francesco et al. (2014) | Italy | 2011–2012 | 2014 | CS | 82 | 82 | 42 | MIC | 0.5 | |
| Seo et al. (2013) | South Korea | 1990–1994 2005–2009 |
2013 | CS | 91 | 11.8 | 91 | 10 | MIC | 1 |
| Kouitcheu Mabeku et al. (2019) | Cameroon | 2013–2015 | 2019 | CS | 140 | 140 | 19 | DD | 21 | |
| Yin et al. (2020) | China | 2016–2016 | 2016 | CS | 267 | 9.4 | 169 | 57 | MIC | 1 |
| Chen et al. (2018) | China | 2018 | CS | 12 | 12 | 6 | MIC | 1 | ||
| Kakiuchi et al. (2020) | Japan | 2018–2018 | 2020 | CS | 71 | 14.7 years | 23 | 7 | MIC | 0.5 |
| Cuadrado-Lavín et al. (2012) | Spain | 2010–2010 | 2011 | CS | 76 | 68 | 10 | MIC | 2 | |
| Gehlot et al. (2016) | India | 2011–2014 | 2015 | CS | 68 | Range: 18–86 | 68 | 8 | MIC | 0.5 |
| Ogata, Gales & Kawakami (2014) | Brazil | 2008–2009 | 2014 | CS | 77 | 11.1 | 77 | 16 | MIC | 1 |
| Eng et al. (2015) | Canada | 2012–2013 | 2015 | CS | 301 | 20 | 8 | MIC | 0.5 | |
| Alarcón et al. (2017) | Spain | 2007–2014 | 2017 | CS | 824 | 26 | 824 | 422 | MIC | 0.5 |
| Akhtereeva et al. (2018) | Russia | 2011–2013 | 2018 | CS | 76 | 13.6 | 30 | 9 | DD | 30 |
| Selgrad et al. (2013) | Germany | 2005–2012 | 2013 | RET | 436 | 51.7 | 159 | 12 | MIC | 1 |
| Gunnarsdottir et al. (2017) | Iceland | 2012–2013 | 2017 | PRO | 613 | 57 | 105 | 9 | MIC | 1 |
| Mahmoudi et al. (2017) | Iran | 2014–2015 | 2017 | CS | 90 | 9.4 | 32 | 7 | MIC | 1 |
| Shokrzadeh et al. (2011) | Iran | 2007–2008 | 2010 | CS | 92 | 45 ± 18 M 38 ± 14 F | 42 | 6 | MIC | 1 |
| Savari et al. (2010) | Iran | 2009–2009 | 2010 | CS | 191 | Range: 14–84 | 63 | 19 | DD | 21 |
| Shu et al. (2018) | China | 2012–2014 | 2018 | CS | 1,390 | 9.5 | 545 | 112 | MIC | 8 |
| Mosites et al. (2018) | USA | 2000–2016 | 2018 | CS | 763 | 52 | 800 | 238 | MIC | 1 |
| Parra-Sepúlveda et al. (2019) | Chile | 2005–2007 2015–2017 |
2019 | CS | 1,655 | 48.8 | 405 | 96 | DD | 21 |
| Fiorini et al. (2018) | Italy | 2010–2016 | 2018 | CS | 1,730 | 51.1 | 1424 | 114 | MIC | 0.5 |
| Shao et al. (2018) | China | 2013–2016 | 2017 | CS | 2,283 | 2283 | 519 | MIC | 1 | |
| Li et al. (2020) | China | 2019–2019 | 2021 | CS | 157 | 10.9 | 87 | 48 | MIC | 0.5 |
| Su et al. (2013) | China | 2010–2012 | 2013 | CS | 51,891 | 17731 | 3810 | MIC | 1 | |
| Hojsak et al. (2012) | Croatia | 2001–2010 | 2012 | RET | 2,313 | 12.9 | 168 | 20 | MIC | 1 |
| Hamidi et al. (2020) | Iran | 2017–2018 | 2020 | CS | 80 | 50.2 | 50 | 11 | MIC | 0.5 |
| An et al. (2013) | Korea | 2009–2012 | 2013 | RET | 165 | 165 | 20 | MIC | 1 | |
| Shiota et al. (2015) | USA | 2009–2013 | 2015 | CS | 656 | 128 | 6 | MIC | 1 | |
| Li et al. (2017) | China | 2009–2015 | 2017 | RET | 5,610 | 14 | 1746 | 286 | MIC | 1 |
| Bolor-Erdene et al. (2017) | Mongolia | 2011–2014 | 2017 | CS | 320 | 43.7 | 152 | 54 | MIC | 1 |
| Boehnke et al. (2017) | Peru | 2011–2013 | 2017 | CS | 109 | 76 | 27 | MIC | 0.5 | |
| Ahmad, Zakaria & Mohamed (2011) | Malaysia | 2004–2007 | 2011 | CS | 777 | 187 | 4 | MIC | 1 | |
| Rasheed et al. (2014) | USA | 2011–2012 | 2014 | CS | 93 | 47.4 | 46 | 22 | MIC | 1 |
| Guo et al. (2019) | China | 2016–2017 | 2018 | CS | 346 | Range: 1–15 | 22 | 8 | MIC | 1 |
| Jiang et al. (2021) | China | 2017–2019 | 2021 | CS | 1,533 | 1533 | 721 | MIC | 0.5 | |
| Butenko et al. (2017) | Slovenia | 2011–2014 | 2017 | RET | 107 | 12 | 104 | 25 | MIC | 8 |
| Tveit et al. (2011) | Alaska | 2000–2008 | 2011 | CS | 1,181 | 51 | 531 | 159 | MIC | 1 |
| Tuan et al. (2019) | Vietnam | 2019 | CS | 206 | 45.3 | 55 | 14 | MIC | 8 | |
| Maev et al. (2020) | Russia | 2015–2018 | 2020 | CS | 27 | 27 | 3 | MIC | 0.5 | |
| Figueroa et al. (2012) | Colombia | 2012 | CS | 203 | 40 | 146 | 29 | MIC | 1 | |
| Kim et al. (2011) | Korea | 2008–2008 | 2011 | CS | 99 | 54.6 | 99 | 26 | MIC | 1 |
| Adeniyi et al. (2012) | Nigeria | 2012 | CS | 52 | Range: 10–90 | 43 | 3 | DD | 30 | |
| Yao et al. (2019) | Taiwan | 2013–2014 | 2019 | RET | 719 | 61.2 | 41 | 14 | MIC | 1 |
| Honma et al. (2019) | Japan | 2012–2015 | 2018 | CS | 1,298 | 14 | 13 | 5 | MIC | 1 |
| Bayati et al. (2019) | Iran | 2014–2015 | 2019 | CS | 170 | Range: 30–75 | 55 | 27 | MIC | 0.5 |
| Pichon et al. (2020) | France | 2012–2014 | 2020 | CS | 3 | 33.3 | 189 | 1 | MIC | 0.5 |
| Tanabe et al. (2018) | Japan | 2013–2016 | 2018 | RET | 1,355 | 212 | 50 | MIC | 1 | |
| Karabiber et al. (2014) | Turkey. | 2014 | CS | 159 | 98 | 23 | DD | 30 | ||
| Saracino et al. (2020) | Italy | 2009–2019 | 2020 | NA | 3,178 | 52.3 | 1646 | 553 | MIC | 0.5 |
| Liang et al. (2020) | Taiwan | 2013–2019 | 2020 | RET | 1,369 | 54.0 ± 11.9 | 1369 | 226 | MIC | 1 |
| Khademi et al. (2014) | Iran | 2011–2012 | 2014 | CS | 130 | 30 | 4 | MIC | 1 | |
| Milani et al. (2012) | Iran | 2010–2011 | 2012 | CS | 395 | 35 ± 19 | 112 | 16 | MIC | 1 |
| Famouri et al. (2018) | Iran | 2015–2018 | 2018 | CS | 102 | 8.65 ± 3.88 | 48 | 17 | MIC | 2 |
| Bruce et al. (2019) | Alaska | 1998–2006 | 2019 | PRO | 362 | 260 | 74 | MIC | 1 | |
| Park et al. (2020) | Korea | 2017–2019 | 2020 | PRO | 174 | 70 | 20 | MIC | 0.5 | |
| Binh et al. (2013) | Vietnam | 2008–2008 | 2013 | CS | 103 | 44.8 | 103 | 34 | MIC | 1 |
| Keshavarz Azizi Raftar et al. (2015) | Iran | 2013 | CS | 246 | 45.78 ± 16.23 | 95 | 32 | MIC | 1 | |
| Ang et al. (2016) | Singapore | 2000–2014 | 2016 | RET | 708 | 708 | 97 | MIC | 1 | |
| Gościniak et al. (2014) | Poland | 2008–2011 | 2014 | CS | 165 | 165 | 50 | MIC | 1 | |
| Wang et al. (2019) | China | 1998–2017 | 2019 | CS | 454 | 50.74 ± 10.942 | 100 | 31 | MIC | 1 |
| Bai et al. (2015) | China | 2013–2013 | 2015 | CS | 181 | 44.9 | 181 | 56 | MIC | 0.5 |
| Mégraud et al. (2021) | France | 2014–2018 | 2020 | CS | 951 | 52.4 ± 15.7 | 741 | 157 | MIC | 0.5 |
| Sadeghifard et al. (2013) | Iran | 2009–2010 | 2013 | CS | 50 | 50 | 16 | DD | 20 | |
| Bedoya-Gómez et al. (2020) | Colombia | 2019 | PRO | 115 | 41.8 | 61 | 5 | MIC | 0.5 | |
| Miftahussurur et al. (2016) | Japan | 2012–2015 | 2016 | PRO | 849 | 49.25 | 77 | 7 | MIC | 0.25 |
| Erkut et al. (2020) | Turkey | 2010–2011 | 2020 | PRO | 344 | 39.3 | 104 | 29 | MIC | 1 |
| Zhang et al. (2018) | China | 2013 | 2018 | CS | 394 | 136 | 10 | MIC | 1 | |
| Tsay et al. (2012) | Taiwan | 2005–2009 | 2011 | RET | 233 | 55.7 | 32 | 2 | MIC | 1 |
| Mascellino et al. (2018) | Italy | 2017 | 2020 | RET | 80 | 59 | 80 | 28 | MIC | 0.5 |
| Khoury et al. (2017) | Israel | 2012–2015 | 2017 | RET | 107 | 64 | 26 | MIC | 0.5 | |
| Saracino et al. (2020) | Italy | 2016–2019 | 2020 | RET | 270 | 51.4 | 221 | 202 | MIC | 0.5 |
| Lin et al. (2020) | Taiwan | 2008–2017 | 2019 | RET | 490 | 54.5 | 228 | 33 | MIC | 1 |
| Alfizah et al. (2014) | Malaysia | 2004–2007 | 2014 | CS | 99 | 161 | 2 | MIC | 1 | |
| Fasciana et al. (2015) | Italy | 2015 | CS | 100 | 100 | 25 | MIC | 0.5 | ||
| Ayala et al. (2011) | Mexico | 2002–2004 | 2011 | CS/PRO | 460 | 90 | 9 | MIC | 2 | |
| Picoli et al. (2014) | Brazil | 2011–2012 | 2014 | CS | 342 | 54 | 6 | MIC | 1 | |
| Larsen et al. (2013) | Norway | 2008–2009 | 2012 | CS | NA | 102 | 6 | MIC | 0.5 | |
| Kumar et al. (2020) | USA | 2009–2019 | 2019 | RET | 109 | 65 | 39 | MIC | 0.5 | |
| Khademi et al. (2013) | Iran | 2011–2012 | 2013 | CS | 260 | 45.8 ± 17.8 | 78 | 12 | MIC | 1 |
| Peretz et al. (2014) | Israel | 2011– 2012 | 2014 | CS | 176 | 85 | 20 | MIC | 1 | |
| Chung et al. (2012) | Korea | 2004–2007 | 2011 | CS | 185 | 50.7 ± 14.4 | 185 | 20 | MIC | 1 |
| Ghotaslou et al. (2013) | Iran | 2013 | CS | 123 | 35 ± 18 | 123 | 21 | DD | 30 | |
| Kostamo et al. (2011) | Finland | 2000–2008 | 2010 | RET | 3,045 | 62 | 1037 | 83 | MIC | 1 |
| Demiray-Gürbüz et al. (2017) | Turkey | 2006–2011 | 2016 | CS | 234 | 43.8 ± 14.0 | 114 | 32 | MIC | 1 |
| Agudo et al. (2011) | USA | 2008 | 2011 | CS | 118 | 118 | 42 | MIC | 1 | |
| Matta, Zambrano & Pazos (2018) | Colombia | 2018 | CS | 409 | 74 | 34 | MIC | 1 | ||
| Song et al. (2014) | China | 2008–2012 | 2014 | PRO/CS | 600 | 42.5 ± 13.2 | 600 | 225 | MIC | 0.5 |
| Wüppenhorst et al. (2014) | Germany | 2001–2012 | 2014 | PRO | 1,651 | 1523 | 475 | MIC | 1 | |
| Shi, Jiang & Zhao (2016) | China | 2016 | CS | 328 | 328 | 78 | MIC | 1 | ||
| Talebi Bezmin Abadi et al. (2012) | Iran | 2009–2010 | 2011 | CS | 170 | 38.6 | 150 | 51 | MIC | 1 |
| Boyanova et al. (2017) | Bulgaria | 2011–2016 | 2017 | CS | 233 | 59.1 | 233 | 60 | MIC | 0.5 |
| Manfredi et al. (2015) | Italy | 2011–2012 | 2015 | CS | 66 | 9.8 | 46 | 12 | MIC | 4 |
| Morilla et al. (2019) | Spain | 2004–2016 | 2019 | RET | 3,426 | 55.7 ± 16.9 | 1439 | 278 | MIC | 0.5 |
| Vekens et al. (2013) | Belgium | 2009–2010 | 2013 | PRO | 507 | 48.8 | 180 | 24 | MIC | 1 |
| Maleknejad et al. (2015) | Iran | 2012–2014 | 2015 | CS | 169 | 7.30 ± 3.12 | 21 | 1 | DD | 30 |
| Oleastro et al. (2011) | Portugal | 2000–2009 | 2011 | PRO | 1,115 | 10.17 ± 4.03 | 1115 | 387 | MIC | 1 |
| Zhang et al. (2015) | China | 2009–2010 2013–2014 |
2015 | PRO/CS | 1,555 | 42.4 | 1321 | 648 | MIC | 0.5 |
| Dargiene et al. (2018) | Lithuania | 2013–2015 | 2017 | CS | 297 | 32.85 | 79 | 2 | MIC | 0.5 |
| Liu et al. (2011) | China | 2009–2010 | 2011 | CS | 120 | 10.0 ± 5.8 | 73 | 62 | MIC | 1 |
| Liu et al. (2018) | China | 2010–2016 | 2017 | PRO | 1,117 | 960 | 247 | MIC | 1 | |
| Tang et al. (2020) | China | 2017–2019 | 2020 | CS | 400 | 44.7 | 117 | 52 | MIC | 0.5 |
| Bachir et al. (2018) | Algeria | 2012–2015 | 2017 | CS | 200 | 151 | 38 | MIC | 0.5 | |
| Seck et al. (2013) | Senegal | 2007–2009 | 2013 | CS | 108 | 45.3 | 108 | 1 | MIC | 1 |
| Karczewska et al. (2011) | Poland | 2006–2008 | 2011 | CS | 115 | 115 | 39 | MIC | 1 | |
| Lee et al. (2019) | South Korea | 2003–2018 | 2019 | PRO | 740 | 56.3 | 740 | 280 | MIC | 1 |
| Raaf et al. (2017) | Algeria | 2015–2016 | 2017 | PRO | 147 | 43 | 16 | DD | 17 | |
| Hansomburana et al. (2012) | Thailand | 2006–2008 | 2012 | PRO | 200 | 52.8 | 82 | 11 | MIC | 1 |
| Mirzaei et al. (2013) | Iran | 2011–2011 | 2013 | CS | 110 | 34 | 48 | 7 | MIC | 1 |
| Lee et al. (2013) | Korea | 2003–2012 | 2013 | PRO | 433 | 55.53 | 433 | 127 | MIC | 1 |
| Shokrzadeh et al. (2015) | Iran | 2010–2011 | 2014 | CS | 197 | 46 | 111 | 29 | MIC | 1 |
| Oporto et al. (2019) | Chile | 2018 | 2019 | CS | 229 | 50.68 | 44 | 18 | MIC | 0.5 |
| Aumpan et al. (2020) | Thailand | 2019 | 2020 | CS | 58 | 43.8 | 14 | 4 | MIC | 0.5 |
| Vilaichone et al. (2020) | Thailand | 2010–2015 | 2020 | CS | 1,178 | 41.5 | 357 | 7 | MIC | 0.5 |
| Cerqueira et al. (2011) | Portugal | 2011 | CS | NA | 33 | 21 | MIC | 1 | ||
| Binyamin et al. (2017) | Israel | 2015–2016 | 2017 | CS | 85 | 54 | 34 | MIC | 1 | |
| Camorlinga-Ponce et al. (2021) | Chile | 1997–2017 | 2021 | CS | 167 | 50.72 | 167 | 15 | MIC | 0.5 |
| Biernat et al. (2020) | Poland | 2016–2019 | 2020 | RET | 108 | 12.5 | 91 | 28 | MIC | 0.5 |
| Trespalacios et al. (2013) | Colombia | 2009–2011 | 2013 | CS | 256 | 276 | 42 | MIC | 1 | |
| Lok et al. (2020) | China | 2018–2019 | 2020 | CS | 176 | 48.4. | 65 | 34 | MIC | 0.5 |
| Bahmaninejad et al. (2021) | Iran | 2020–2020 | 2021 | CS | 100 | 50 | 33 | MIC | 1 | |
| Draeger et al. (2015) | Germany | 2004–2013 | 2015 | RET | 481 | 481 | 409 | MIC | 1 | |
| Zerbetto De Palma et al. (2017) | Argentina | 2011–2013 | 2015 | CS | 52 | 52 | 14 | MIC | 0.5 | |
| Boyanova et al. (2012) | Bulgaria | 2004–2010 | 2012 | CS | 519 | 52.16 | 519 | 93 | MIC | 1 |
| Tshibangu-Kabamba et al. (2020) | Congo | 2017–2018 | 2020 | CS | 220 | 45.3 ± 15.3 | 102 | 24 | MIC | 0.5 |
| Okuda et al. (2017) | Japan | 1997–2013 | 2016 | RET | 332 | 11.6 ± 3.4 | 76 | 33 | MIC | 1 |
| Vilaichone et al. (2013) | Thailand | 2004–2012 | 2013 | CS | 3,964 | 53.3 | 400 | 15 | MIC | 0.5 |
| Zhang et al. (2020) | China | 2017–2019 | 2020 | CS | 238 | 238 | 84 | MIC | 0.5 | |
| Zhang et al. (2020) | China | 2012–2014 | 2020 | CS | 79 | 9.7 ± 2.8 | 79 | 29 | MIC | 1 |
| Mansour et al. (2016) | France | 2009–2009 | 2015 | PRO | 149 | 53.65 | 42 | 12 | MIC | 1 |
| Kuo et al. (2021) | Taiwan | 2017–2020 | 2021 | CS | 64 | 53.8 | 41 | 38 | MIC | 0.5 |
| Miendje Deyi et al. (2011) | Belgium | 1990–2009 | 2011 | CS | 9,430 | 29.3 | 9430 | 524 | MIC | 1 |
| Han et al. (2016) | China | 2015–2015 | 2016 | CS | 325 | 47.2 | 325 | 65 | MIC | 1 |
| Bińkowska et al. (2018) | Italy | 2008–2016 | 2018 | CS | 170 | 170 | 29 | MIC | 1 | |
| Bachir et al. (2018) | Algeria | 2014–2016 | 2018 | PRO | 270 | 212 | 53 | MIC | 0.5 | |
| Hanafiah et al. (2019) | Malaysia | 2014–2015 | 2019 | CS | 288 | 52.41 ± 16.44 | 59 | 21 | MIC | 1 |
| Vazirzadeh et al. (2020) | Iran | 2018–2018 | 2020 | CS | 165 | 50:3 ± 15:5 | 83 | 21 | MIC | 0.5 |
| Rezaei, Abadi & Mobarez (2020) | Iran | 2015–2018 | 2019 | CS | 200 | 54 | 73 | 17 | MIC | 0.5 |
| Yakoob et al. (2013) | Pakistan | 2008– 2010 | 2013 | CS | 120 | 41 ± 13 | 47 | 17 | MIC | 1 |
| Gehlot et al. (2016) | India | 2011–2013 | 2016 | CS | 483 | 43 | 68 | 8 | MIC | 0.5 |
| Boyanova et al. (2013) | Bulgaria | 2007– 2012 | 2013 | RET | 588 | 588 | 118 | MIC | 1 | |
| Boyanova et al. (2015) | Bulgaria | 2012–2014 | 2015 | CS | 53 | 50.7 | 53 | 9 | MIC | 0.5 |
| Otth et al. (2011) | Chile | 2010 | CS | 240 | 54.5 ± 15.7 | 88 | 8 | MIC | 2 | |
| McNulty et al. (2012) | Uk | 2009–2010 | 2012 | CS | 2,063 | 241 | 86 | MIC | 1 | |
| Wang et al. (2018) | China | 2013–2014 | 2018 | CS | NA | 100 | 13 | MIC | 0.5 | |
| Alavifard et al. (2021) | Iran | 2017–2019 | 2020 | CS | 82 | 49.7 ± 3.33 | 82 | 36 | MIC | 0.5 |
| Regnath et al. (2017) | Germany | 2002–2015 | 2016 | RET | 582 | 12 years | 608 | 75 | MIC | 0.5 |
| Lu et al. (2019) | Taiwan | 1998–2018 | 2019 | RET | 70 | 13.2 ± 3.2 | 70 | 16 | MIC | 1 |
| Di Giulio et al. (2016) | Italy | 2010–2014 | 2015 | CS | 115 | 181 | 131 | MIC | 0.5 | |
| Enany & Abdalla (2015) | Egypt | 2015 | CS | 150 | 107 | 6 | DD | 40 | ||
| Trespalacios et al. (2015) | Colombia | 2014 | CS | 127 | 107 | 42 | MIC | 1 | ||
| Gatta et al. (2018) | Italy | 2010–2015 | 2018 | RET | 1,682 | 1325 | 478 | MIC | 0.5 | |
| Goudarzi et al. (2016) | Iran | 2015–2015 | 2016 | CS | 154 | 110 | 28 | MIC | 1 | |
| Bayati et al. (2020) | Iran | 2019 | CS | 170 | 30 ± 75. | 55 | 27 | MIC | 0.5 | |
| Dang et al. (2020) | Vietnam | 2014–2016 | 2020 | CS | 153 | 38.3 ± 10.7 | 153 | 111 | MIC | 1 |
| Phan et al. (2015) | Vietnam | 2012–2014 | 2014 | CS | 92 | 44.1 ± 13.4 | 92 | 39 | MIC | 1 |
| Khashei et al. (2016) | Iran | 2014–2014 | 2016 | CS | 318 | 41.5 | 100 | 20 | MIC | 1 |
| Shetty et al. (2019) | Australia | 2014–2017 | 2019 | CS | 180 | 46.2 ± 14 | 113 | 23 | MIC | 0.5 |
| Macías-García et al. (2017) | Spain | 2014–2016 | 2017 | PROCS | 217 | 64 | 76 | 17 | MIC | 1 |
| Farzi et al. (2019) | Iran | 2016–2017 | 2019 | CS | 160 | 46.5 ± 8.3 | 68 | 23 | MIC | 1 |
| Lyu et al. (2020) | China | 2016–2018 | 2020 | PRO | 1,113 | 43 | 791 | 271 | MIC | 0.5 |
| Shmuely et al. (2020) | Israel | 2013–2017 | 2020 | RET/CS | 128 | 45 | 128 | 70 | MIC | 256 |
| Ogata et al. (2013) | Brazil | 2008–2009 | 2013 | CS | 77 | 11.1 ± 3.9 | 77 | 15 | MIC | 2 |
| Abadi et al. (2011) | Iran | 2008–2010 | 2011 | CS | 147 | 34.5 | 147 | 32 | MIC | 1 |
| korn Vilaichone et al. (2017) | Thailand | 2016–2016 | 2017 | CS | 148 | 56.3 ± 13.3 | 50 | 1 | MIC | 0.5 |
| Ferenc et al. (2017) | Poland | 2011 and 2013 | 2016 | CS | 185 | 49 ± 16.8 | 67 | 37 | MIC | 1 |
| Azzaya et al. (2020) | Mongolia | 2014–2016 | 2020 | CS | 361 | 44.3 ± 13.4 | 361 | 108 | MIC | 0.5 |
| Mi et al. (2021) | China | 2018–2018 | 2021 | CS | 48 | 65 | 21 | MIC | 0.5 | |
| Boyanova et al. (2014) | Bulgaria | 2012–2013 | 2014 | CS | 50 | 50.5 | 50 | 11 | MIC | 0.5 |
| Boyanova et al. (2016) | Bulgaria | 2010–2015 | 2015 | CS | 299 | 47.3 | 299 | 84 | MIC | 0.5 |
| Megraud et al. (2021) | France | 2018–2019 | 2021 | PRO | 1,211 | 51.2 | 1211 | 259 | MIC | 0.5 |
| Megraud et al. (2013) | France | 2008–2009 | 2013 | PRO | 2,204 | 2204 | 431 | MIC | 1 | |
| Ducournau et al. (2016) | France | 2014–2015 | 2016 | CS | 984 | 51.5 ± 15.9 | 266 | 59 | MIC | 1 |
| Bouihat et al. (2017) | France | 2015–2016 | 2016 | PRO | 255 | 47.5 | 177 | 45 | MIC | 0.5 |
| Fernández-Reyes et al. (2019) | Spain | 2014–2017 | 2019 | PRO | 112 | 99 | 12 | MIC | 0.5 | |
| Saniee et al. (2018) | Iran | 2010–2017 | 2018 | CS | 985 | 218 | 75 | DD | 2 | |
| Mokhtar et al. (2019) | Malaysia | 2015–2016 | 2019 | CS | 352 | 52 | 13 | 4 | MIC | 0.5 |
| Montes et al. (2015) | Spain | 2008–2012 | 2014 | RET | 143 | 74 | 25 | MIC | 1 | |
| Deyi et al. (2019) | Belgium | 2015–2016 | 2019 | CS | 846 | 846 | 141 | MIC | 0.5 | |
| Tang et al. (2020) | China | 2016–2019 | 2020 | CS | NA | 301 | 201 | MIC | 0.5 | |
Study selection
All records obtained from online databases were imported into EndNote (Version 20), and duplicates were eliminated. M-H and S-K independently assessed the titles and abstracts; V-H-K resolved discrepancies. Studies were considered to be appropriate for the analysis if they presented data concerning the prevalence of H. pylori resistant to CLA. An English language restriction was imposed, while abstracts, conferences, case reports, case series, reviews, studies with unclear results, and duplicate articles were excluded from the analysis.
Data extraction
Our study included studies based on pre-defined criteria and evaluated as full-text articles. Two reviewers conducted the data extraction process independently (M-H, S-K). Any discrepancies were discussed and resolved by consensus of the two reviewers. The primary outcome of focus was the prevalence of clarithromycin-resistant Helicobacter pylori. Information extracted from each study included the first author’s name, year of publication, geographical location, antimicrobial susceptibility testing method, breakpoints for interpretation of the test results, sample size, and the number of clarithromycin-resistant H. pylori. All extracted data are available in an accompanying Supplemental File.
Quality assessment
Two reviewers (S-K and M-H) evaluated the quality of the studies using the Newcastle Ottawa Scale (NOS). In cases of disagreement, a third author (M-SH) was consulted to determine a consensus. The assessment of the studies was based on three criteria: selection, comparability, and exposure/outcome assessments.
Statistical analysis
For the present study, the sample size of isolates for antimicrobial susceptibility testing (AST) and the number of resistances to each antibiotic were used to calculate a weighted pooled resistance and their 95% confidence intervals. In order to prevent the exclusion of studies from the meta-analysis due to 0 or 100 resistance prevalence, the Inverse of Freeman-Tukey double arcsine transformation was conducted using Metaprop command in STATA software (version 17.1). A random-effects model was implemented to estimate pool proportions (Egger et al., 1997; Harbord et al., 2010). The I2 with a P ≤ 0.05 was used to identify significant heterogeneity. The presence of a small-study effect or publication bias was assessed using Egger’s linear regression test and Begg’s test (Harbord, Harris & Sterne, 2009). Subgroup analyses were conducted to determine the impact of the country, continent, publication year (2010–2017, 2018–2021), (AST) (Disc diffusion, Gradient methods), and breakpoints for interpretation of AST results on the variation.
Results
Descriptive statistics
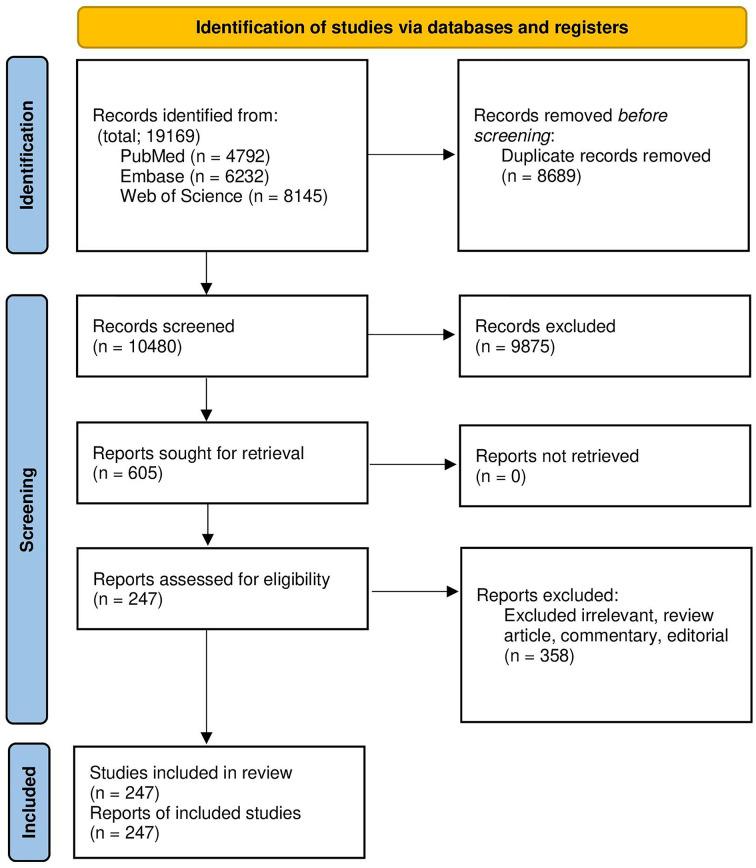
In this research, 19,169 records were acquired in EndNote version 20, a reference manager software. A total of 8,689 duplicated articles were then removed, leaving a total of 247 eligible studies that were included in the systematic review and meta-analysis. The screening and selection presage were summarized in the PRISMA flow chart (Fig. 1). Overly 20,936 H. pylori isolates have been investigated in included articles. More than half of the isolates were investigated in Asia (55.10% Isolated). Although most pieces were from Iran (38 articles), the highest number of isolates among the countries was that investigated from China (32,130 Isolates, 36.52% of total isolates). Description data are summarized in Table 2.
Figure 1. The study PRISMA flow diagram.
Table 2. Clarithromycin-resistant Helicobacter pylori prevalence. 95% Confidence Intervals (CI) were used. P ≤ 0.05 was considered statistically significant.
| No of article | Clar-resistant, Total isolates | Proportion (LCI, HCI) | Weight | I2 (P) | |
|---|---|---|---|---|---|
| Overall | 248 | 8736, 87991 | 27.53 (25.41, 29.69) | 100.00 | 97.80% (P = 0.00) |
| 2010–2017 | 143 | 12891, 60452 | 24.28 (21.7, 26.96) | 57.68 | 97.91% (P = 0.00) |
| 2018–2021 | 105 | 8045, 27476 | 32.14 (28.69, 35.69) | 42.32 | 97.24% (P = 0.00) |
| Iran | 38 | 1193, 3628 | 27.24 (21.68, 33.18) | 14.91 | 93.14% (P = 0.00) |
| Finland | 1 | 83, 1037 | 8.00 (6.43, 9.83) | 0.43 | NA |
| Chile | 4 | 137, 704 | 18.56 (8.47, 31.34) | 1.62 | 91.76% (P = 0.00) |
| Brazil | 4 | 40, 247 | 15.29 (9.79, 21.7) | 1.55 | 38.94% (P = 0.18) |
| Romania | 1 | 22, 70 | 31.43 (20.85, 43.63) | 0.40 | NA |
| Austria | 1 | 16, 96 | 16.67 (9.84, 25.65) | 0.41 | NA |
| France | 8 | 990, 4873 | 21.13 (15.26, 27.66) | 3.31 | 95.23% (P = 0.00) |
| Eastern Cape | 1 | 40, 200 | 20 (14.69, 26.22) | 0.42 | NA |
| Spain | 8 | 1161, 4650 | 27.41 (17.03, 39.18) | 3.30 | 98.22% (P = 0.00) |
| Malaysia | 5 | 38, 522 | 10.2 (1.59, 23.94) | 1.91 | 93.33% (P = 0.00) |
| Alaska | 2 | 233, 791 | 29.45 (26.31, 32.68) | 0.86 | NA |
| Korea | 5 | 213, 952 | 20.59 (12.26, 30.37) | 2.07 | 90.69% (P = 0.00) |
| Taiwan | 10 | 453, 2088 | 29.16 (15.9, 44.45) | 3.92 | 96.85% (P = 0.00) |
| Mexico | 2 | 17, 135 | 12.3 (7.14, 18.53) | 0.78 | NA |
| USA | 5 | 347, 1157 | 32.98 (17.21, 50.95) | 2.03 | 95.84% (P = 0.00) |
| Portugal | 5 | 541, 1428 | 48.11 (30.07, 66.41) | 1.97 | 95.52% (P = 0.00) |
| China | 32 | 8227, 32130 | 34.05 (29.33, 38.92) | 13.14 | 98.16% (P = 0.00) |
| Poland | 6 | 178, 601 | 29.77 (18.41, 42.52) | 2.42 | 90.49% (P = 0.00) |
| Belgium | 3 | 689, 10456 | 11.28 (3.95, 21.67) | 1.29 | NA |
| Turkey | 7 | 170, 684 | 25.78 (19.44, 32.67) | 3.22 | 76.74% (P = 0.00) |
| Croatia | 1 | 20, 168 | 11.9 (7.43, 17.79) | 0.42 | #VALUE! |
| Colombia | 5 | 152, 664 | 24.26 (12.96, 37.68) | 2.04 | 92.33% (P = 0.00) |
| Nigeria | 2 | 44, 147 | 28.22 (21.13, 35.86) | 0.78 | NA |
| Norway | 1 | 6, 102 | 5.88 (2.19, 12.36) | 0.41 | NA |
| Thailand | 7 | 54, 1097 | 6.24 (2.73, 10.86) | 2.73 | 81.45% (P = 0.00) |
| Bulgaria | 6 | 375, 1742 | 21.89 (18.2, 25.81) | 2.48 | 66.49% (P = 0.01) |
| UK | 1 | 86, 241 | 35.68 (29.64, 42.09) | 0.42 | NA |
| South Korea | 7 | 560, 1778 | 31.4 (19.68, 44.43) | 2.88 | 96.35% (P = 0.00) |
| Germany | 4 | 971, 2771 | 32.08 (6.55, 65.66) | 1.71 | 99.64% (P = 0.00) |
| Vietnam | 5 | 254, 507 | 45.72 (28.85, 63.11) | 2.02 | 93.56% (P = 0.00) |
| Senegal | 1 | 1, 108 | 0.93 (0.02, 5.05) | 0.41 | NA |
| Pakistan | 2 | 22, 139 | 13.33 (8.04, 19.63) | 0.78 | NA |
| Australia | 2 | 31, 124 | 23.47 (16.01, 31.75) | 0.67 | NA |
| Japan | 12 | 854, 2494 | 35.89 (27.02, 45.26) | 4.68 | 93.72% (P = 0.00) |
| India | 3 | 63, 216 | 25.25 (2.81, 59.01) | 1.19 | NA |
| Italy | 11 | 1663, 5367 | 40.38 (25.65, 56.04) | 4.55 | 99.12% (P = 0.00) |
| Israel | 6 | 272, 597 | 46.12 (35.66, 56.75) | 2.39 | 84.00% (P = 0.00) |
| Bangladesh | 1 | 22, 56 | 39.29 (26.5, 53.25) | 0.39 | NA |
| Canada | 1 | 8, 20 | 40.00 (19.12, 63.95) | 0.32 | NA |
| Argentina | 1 | 14, 52 | 26.92 (15.57, 41.02) | 0.38 | NA |
| Egypt | 2 | 18, 127 | 10.61 (5.53, 16.89) | 0.73 | NA |
| Singapore | 1 | 97, 708 | 13.70 (11.25, 16.46) | 0.43 | NA |
| Dominican | 1 | 2, 64 | 3.13 (0.38, 10.84) | 0.39 | NA |
| Iceland | 1 | 9, 105 | 8.57 (3.99, 15.65) | 0.41 | NA |
| Mongolia | 2 | 162, 513 | 31.54 (27.57, 35.64) | 0.84 | NA |
| Peru | 1 | 27, 76 | 35.53 (24.88, 47.34) | 0.40 | NA |
| Slovenia | 1 | 25, 104 | 24.04 (16.2, 33.41) | 0.41 | NA |
| Lithuania | 1 | 2, 79 | 2.53 (0.31, 8.85) | 0.40 | NA |
| Algeria | 3 | 107, 406 | 26.62 (21.42, 32.15) | 1.21 | NA |
| Russia | 3 | 28, 333 | 13.34 (2.11, 30.9) | 1.12 | NA |
| Honduras | 1 | 13, 116 | 11.21 (6.1, 18.4) | 0.41 | NA |
| Switzerland | 2 | 145, 216 | 67.16 (60.71, 73.31) | 0.81 | NA |
| Cameroon | 1 | 19, 140 | 13.57 (8.37, 20.38) | 0.41 | NA |
| Congo | 1 | 24, 102 | 23.53 (15.69, 32.96) | 0.41 | NA |
Note:
High confidence interval, HCI; low confidence interval, LCI; I-squared, I2; Degrees of freedom, DF.
Publication bias
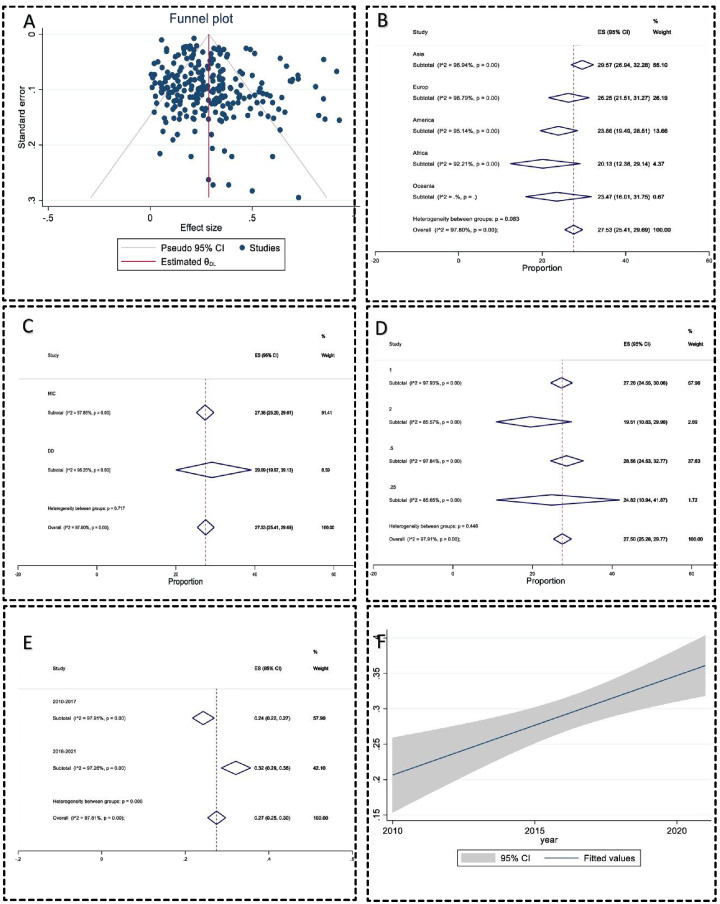
The publication bias was significant by the regression-based Egger test for small-study effects (P = 0.04), but Begg’s test for small-study effects was insignificant (P = 0.09). The Nonparametric trim-and-fill analysis of publication bias also did not change the effect size. The funnel plot also did not have significant evidence of publication bias (Fig. 2A). The sensitivity analysis or one leave-out method also had no significant bias.
Figure 2. Meta-analysis charts.
(A) The funnel plot of clarithromycin-resistant Helicobacter pylori prevalence did not have significant evidence of publication bias; (B) the subgroup analysis forest plot of clarithromycin-resistant Helicobacter pylori prevalence in different continents; (C) the subgroup analysis forest plot of clarithromycin-resistant Helicobacter pylori prevalence using different AST methods; (D) subgroup analysis forest plot of clarithromycin-resistant Helicobacter pylori prevalence in different breakpoints to interpret antimicrobial susceptibility test data; (E) subgroup analysis forest plot of clarithromycin-resistant Helicobacter pylori prevalence in years; (F) the regression analysis of clarithromycin-resistant Helicobacter pylori prevalence over years with 95% Confidence interval had a significant correlation 0.013 (95% CI [0.01–0.02]) (P < 0.001).
Meta-analysis
In 248 included studies, 20,936 isolates have been investigated, and 8,736 isolates have been reported as resistant. The pooled prevalence of CLA-resistance H. pylori was 27.53 (95% CI [25.41–29.6]). Heterogeneity between reports was significant (I2 = 97.80, P < 0.01). The heterogeneity between countries was substantial (P < 0.001). Switzerland, Portugal, and Israel had the highest resistance rates (67.16%, 48.11%, and 46.12%, respectively), and Senegal, Lithuania, and the Dominican Republic had the lowest resistance prevalence, 0.93%, 2.53%, and 3.13%, respectively) (Table 2). The heterogeneity between the continent subgroups was insignificant (P > 0.05) (Fig. 2B). The heterogeneity between the AST methods subgroup was insignificant (Fig. 2C). The breakpoints for the interpretation AST subgroup were insignificant (P > 0.05) (Fig. 2D). The CLA-resistant H. pylori prevalence increased from 24.28% in 2010–2017 to 32.14% in the 2018–2021 years period (P < 0.01) (Fig. 2E). All statistics are summarized in Table 2. The regression meta-analysis for resistance rate over the publication year had a significant correlation of 0.013 (95% CI [0.01–0.02]) (P < 0.001) (Fig. 2F).
Discussion
Over the past years, the treatment of H. pylori infections has been performed using the standard triple therapy regimen, including CLA, a proton pump inhibitor, with either metronidazole or amoxicillin (Gong et al., 2020). However, in recent years, it is revealed that some H. pylori isolates have developed resistance to CLA (Sanches et al., 2016). Therefore, the efficacy of the standard triple therapy regimen is in decline. In 2017, WHO listed the CLA-resistant H. pylori among antibiotic-resistant priority pathogens that need research and development of new antibiotics (Khani, Abadi & Mobarez, 2019). Globally, surveillance and being aware of the frequency of resistance to antibiotics among pathogens is critical, and obtained results can be helpful in different sections such as the design of screening or follow-up programs, and the development of antimicrobial stewardship programs (Azimi et al., 2019; Pormohammad, Nasiri & Azimi, 2019).
In the present systematic review and meta-analysis study, we surveyed and analyzed the worldwide prevalence of CLA resistance among H. pylori isolates from 2010 to 2021. The awareness of CLA resistance among different countries of the world and effective treatment of H. pylori infections are the main goal of the current study. The present systematic review and meta-analysis study included 247 eligible studies from 54 different countries. Our analyses revealed that the overall prevalence of clarithromycin-resistance H. pylori was 27.53%, worldwide.
Resistance to CLA among H. pylori is occur in two different levels including (1) a high level of resistance (MIC more than 64 mg l−1) and (2) a low level of resistance (0.5 ≤ MIC ≤ 1 mg l−1) (He et al., 2021). Point mutations, multidrug efflux pump systems, and synergistic effect of mutations in genes rpl22 (ribosomal protein L22) and infB (translation initiation factor IF-2) with 23S rRNA point mutations are the main CLA resistance mechanisms among H. pylori isolates (Marques et al., 2020; Li et al., 2021). Moreover, it is presumed that some outer-membrane proteins have a role in CLA resistance in H. pylori isolates (Marques et al., 2020). In the Western world and among developed countries, more than 90% of CLA resistance is related to point mutations in the peptidyl transferase region of the V domain of 23S rRNA gene (Mégraud, 2004). The main point mutations related to CLA resistance are A2142G, A2143G (adenine-to-guanine transition at either position 2142 or 2143), A2142C (adenine-to-cytosine transversion at position 2142), A2115G, A2144T, G2141A, G2144T, T2289C, T2717C, and C2694A (Gong et al., 2020; Marques et al., 2020; Li et al., 2021). Moreover, hp1181 and hp1184 mutations are associated with CLA resistance (Li et al., 2021). Mutation in the 2142 and 2143 positions leads to restricted resistance and different levels of resistance, respectively (Kim et al., 2020).
In the present research, more than half of the included studies were performed in Asia. These results demonstrated that CLA resistance is a main public health issue in most Asian countries. Among studies surveyed CLA resistance rates in 54 different countries, Switzerland (67.16%) and Senegal (0.93%) had the highest and lowest resistance rates, respectively. The high level of CLA resistance can be due to the following reasons: (1) inappropriate prescription and unregulated or widespread use of CLA, and (2) the use of CLA in other infections such as respiratory tract infections or intestinal parasites infections (Chen et al., 2017). Time trend analyses revealed that the CLA-resistant rates among H. pylori isolate increased from 24.28% in 2010–2017 to 32.14% in the 2018–2021 years’ period. An increase in CLA resistance rates is an alarming finding. In areas where CLA-resistance is more than 15%, it is recommended to perform susceptibility testing before prescribing the standard triple therapy regimen (Sanches et al., 2016; Abadi, 2017). Combination therapy with other drugs such as tinidazole can be helpful in the treatment of H. pylori infections. It is revealed that CLA combined with tinidazole can reduce the CLA resistance rate, decrease inflammatory reactions, and can effectively eliminate H. pylori infections (He et al., 2021). One of the limitations of this study was that we evaluated the CLA resistance rate only and the other antibiotics were not considered.
Conclusion
Our analysis revealed that CLA resistance rates varied among studies performed in different 54 countries. Altogether, results showed that the overall CLA resistance rate is 27.53%, worldwide. The difference in CLA resistance rate among the included studies can be due to several reasons such as differences in antibiotic prescription rates in various geographic areas, use of different MIC breakpoints or inaccurate criteria in performed studies, and the emergence of multidrug-resistant (MDR) strains. We performed a time trend analysis and the results revealed that the clarithromycin-resistance rates in increasing in recent years. Based on our findings, systematic surveillance, and proper monitoring of CLA resistance rates, as well as monitoring the use of CLA in patients, and performing the CLA susceptibility test before prescription may be critical actions for the inhibition and control of H. pylori infections.
Supplemental Information
Funding Statement
The authors received no funding for this work.
Contributor Information
Mohsen Heidary, Email: mohsenheidary40@gmail.com.
Morteza Saki, Email: mortezasaki1981@gmail.com.
Additional Information and Declarations
Competing Interests
Morteza Saki is a PeerJ Academic Editor. The authors declare that they have no competing interests.
Author Contributions
Mohammad Sholeh conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Saeed Khoshnood conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Taher Azimi conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Jasem Mohamadi performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Vahab Hassan Kaviar conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Marzieh Hashemian conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Somayeh Karamollahi conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Nourkhoda Sadeghifard conceived and designed the experiments, performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Hedayat Heidarizadeh conceived and designed the experiments, performed the experiments, prepared figures and/or tables, and approved the final draft.
Mohsen Heidary conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Morteza Saki conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.
References
- Abadi (2017).Abadi ATB. Resistance to clarithromycin and gastroenterologist’s persistence roles in nomination for Helicobacter pylori as high priority pathogen by World Health Organization. World Journal of Gastroenterology. 2017;23(35):6379–6384. doi: 10.3748/wjg.v23.i35.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abadi et al. (2011).Abadi AT, Taghvaei T, Ghasemzadeh A, Mobarez AM. High frequency of A2143G mutation in clarithromycin-resistant Helicobacter pylori isolates recovered from dyspeptic patients in Iran. Saudi Journal of Gastroenterology: Official Journal of the Saudi Gastroenterology Association. 2011;17(6):396. doi: 10.4103/1319-3767.87181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abadi et al. (2011).Abadi AT, Taghvaei T, Mobarez AM, Carpenter BM, Merrell DS. Frequency of antibiotic resistance in Helicobacter pylori strains isolated from the northern population of Iran. The Journal of Microbiology. 2011;49(6):987–993. doi: 10.1007/s12275-011-1170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahi et al. (2019).Abdollahi H, Hashemzadeh M, Khoshnood S, Savari M. Characterization of Helicobacter pylori genotypes from Iranian patients with gastric clinical diseases: predominance of vacA s1a and cagA EPIYA-ABC genotypes. Gene Reports. 2019;16(1):100458. doi: 10.1016/j.genrep.2019.100458. [DOI] [Google Scholar]
- Adeniyi et al. (2012).Adeniyi BA, Lawal TO, Otegbayo JA, Oluwasola OA, Odaibo GN, Ola SO, Okolo CA, Akere A, Kehinde AO. Cultural characteristics and antibiotic susceptibility pattern of Helicobacter pylori isolated from dyspepsia patients. Gastroenterology Insights. 2012;4(2):e21. doi: 10.4081/gi.2012.e21. [DOI] [Google Scholar]
- Aftab et al. (2016).Aftab H, Miftahussurur M, Subsomwong P, Ahmed F, Khan AK, Yamaoka Y. Helicobacter pylori antibiotic susceptibility patterns in Bangladesh: emerging levofloxacin resistance. The Journal of Infection in Developing Countries. 2016;10(3):245–253. doi: 10.3855/jidc.7713. [DOI] [PubMed] [Google Scholar]
- Agudo et al. (2011).Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. Rapid detection of clarithromycin resistant Helicobacter pylori strains in Spanish patients by polymerase chain reaction-restriction fragment length polymorphism. Revista espanola de quimioterapia: publicacion oficial de la Sociedad Espanola de Quimioterapia. 2011;24(1):32. [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Correa et al. (2017).Aguilera-Correa JJ, Urruzuno P, Barrio J, Martinez MJ, Agudo S, Somodevilla A, Llorca L, Alarcón T. Detection of Helicobacter pylori and the genotypes of resistance to clarithromycin and the heterogeneous genotype to this antibiotic in biopsies obtained from symptomatic children. Diagnostic Microbiology and Infectious Disease. 2017;87(2):150–153. doi: 10.1016/j.diagmicrobio.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Ahmad, Zakaria & Mohamed (2011).Ahmad N, Zakaria WR, Mohamed R. Analysis of antibiotic susceptibility patterns of Helicobacter pylori isolates from Malaysia. Helicobacter. 2011;16(1):47–51. doi: 10.1111/j.1523-5378.2010.00816.x. [DOI] [PubMed] [Google Scholar]
- Akar et al. (2021).Akar M, Aydin F, Kayman T, Abay S, Karakaya E. Detection of Helicobacter pylori by invasive tests in adult dyspeptic patients and antibacterial resistance to six antibiotics, including rifampicin in Turkey. Is clarithromycin resistance rate decreasing? Turkish Journal of Medical Sciences. 2021;51(3):1455–1464. doi: 10.3906/sag-2101-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtereeva et al. (2018).Akhtereeva A, Morozova L, Faizullina R, Ivanovskaya K, Pozdeev O, Valeeva IK, Abdulkhakov S. Antibiotic susceptibility assessment of Helicobacter pylori isolates by disk-diffusion method. BioNanoScience. 2018;8(3):930–934. doi: 10.1007/s12668-018-0527-2. [DOI] [Google Scholar]
- Alarcón et al. (2017).Alarcón T, Urruzuno P, Martínez MJ, Domingo D, Llorca L, Correa A, López-Brea M. Antimicrobial susceptibility of 6 antimicrobial agents in Helicobacter pylori clinical isolates by using EUCAST breakpoints compared with previously used breakpoints. Enfermedades infecciosas y microbiologia clinica. 2017;35(5):278–282. doi: 10.1016/j.eimc.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Alarcón-Millán et al. (2016).Alarcón-Millán J, Fernández-Tilapa G, Cortés-Malagón EM, Castañón-Sánchez CA, De Sampedro-Reyes J, Cruz-del Carmen I, Betancourt-Linares R, Román-Román A. Clarithromycin resistance and prevalence of Helicobacter pylori virulent genotypes in patients from Southern México with chronic gastritis. Infection, Genetics and Evolution. 2016;44:190–198. doi: 10.1016/j.meegid.2016.06.044. [DOI] [PubMed] [Google Scholar]
- Alavifard et al. (2021).Alavifard H, Mirzaei N, Yadegar A, Baghaei K, Smith SM, Sadeghi A, Zali MR. Investigation of clarithromycin resistance-associated mutations and virulence genotypes of Helicobacter pylori isolated from Iranian population: a cross-sectional study. Current Microbiology. 2021;78(1):244–254. doi: 10.1007/s00284-020-02295-7. [DOI] [PubMed] [Google Scholar]
- Alfizah et al. (2014).Alfizah H, Norazah A, Hamizah R, Ramelah M. Resistotype of Helicobacter pylori isolates: the impact on eradication outcome. Journal of Medical Microbiology. 2014;63(5):703–709. doi: 10.1099/jmm.0.069781-0. [DOI] [PubMed] [Google Scholar]
- Almeida et al. (2014).Almeida N, Romaozinho JM, Donato MM, Luxo C, Cardoso O, Cipriano MA, Marinho C, Fernandes A, Calhau C, Sofia C. Helicobacter pylori antimicrobial resistance rates in the central region of Portugal. Clinical Microbiology and Infection. 2014;20(11):1127–1133. doi: 10.1111/1469-0691.12701. [DOI] [PubMed] [Google Scholar]
- An et al. (2013).An B, Moon BS, Kim H, Lim HC, Lee YC, Lee G, Kim SH, Park M, Kim JB. Antibiotic resistance in Helicobacter pylori strains and its effect on H. pylori eradication rates in a single center in Korea. Annals of Laboratory Medicine. 2013;33(6):415–419. doi: 10.3343/alm.2013.33.6.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang et al. (2016).Ang TL, Fock KM, Ang D, Kwek ABE, Teo EK, Dhamodaran S. The changing profile of Helicobacter pylori antibiotic resistance in Singapore: a 15-year study. Helicobacter. 2016;21(4):261–265. doi: 10.1111/hel.12291. [DOI] [PubMed] [Google Scholar]
- Arenas et al. (2019).Arenas A, Serrano C, Quiñones L, Harris P, Sandoval M, Lavanderos M, Sepúlveda R, Maquilón S, Echeverría A, Ríos CJSR. High prevalence of clarithromycin resistance and effect on Helicobacter pylori eradication in a population from Santiago, Chile: cohort study and meta-analysis. Scientific Reports. 2019;9(1):20070. doi: 10.1038/s41598-019-56399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumpan et al. (2020).Aumpan N, Vilaichone R-K, Gumnarai P, Sanglutong L, Ratanachu-Ek T, Mahachai V, Yamaoka Y. Prevalence and antibiotic resistance patterns of Helicobacter pylori infection in Koh Kong, Combodia. Asian Pacific Journal of Cancer Prevention: APJCP. 2020;21(5):1409–1413. doi: 10.31557/APJCP.2020.21.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auttajaroon et al. (2019).Auttajaroon J, Chotivitayatarakorn P, Yamaoka Y, Vilaichone RK. CYP2C19 genotype, CagA genotype and antibiotic resistant strain of Helicobacter pylori infection. Asian Pacific Journal of Cancer Prevention. 2019;20(4):1243–1247. doi: 10.31557/APJCP.2019.20.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala et al. (2011).Ayala G, Galván-Portillo M, Chihu L, Fierros G, Sánchez A, Carrillo B, Román A, López-Carrillo L, Silva-Sánchez J, Study Group Resistance to antibiotics and characterization of Helicobacter pylori strains isolated from antrum and body from adults in Mexico. Microbial Drug Resistance. 2011;17(2):149–155. doi: 10.1089/mdr.2010.0154. [DOI] [PubMed] [Google Scholar]
- Azimi et al. (2019).Azimi T, Maham S, Fallah F, Azimi L, Gholinejad Z. Evaluating the antimicrobial resistance patterns among major bacterial pathogens isolated from clinical specimens taken from patients in Mofid Children’s Hospital, Tehran, Iran: 2013–2018. Infection and Drug Resistance. 2019;12:2089–2102. doi: 10.2147/IDR.S215329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzaya et al. (2020).Azzaya D, Gantuya B, Oyuntsetseg K, Davaadorj D, Matsumoto T, Akada J, Yamaoka Y. High antibiotic resistance of Helicobacter pylori and its associated novel gene mutations among the Mongolian population. Microorganisms. 2020;8(7):1062. doi: 10.3390/microorganisms8071062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachir et al. (2018).Bachir M, Allem R, Benejat L, Tifrit A, Medjekane M, Drici AE-M, Megraud F, Douidi KT. Molecular detection of mutations involved in Helicobacter pylori antibiotic resistance in Algeria. Journal of Antimicrobial Chemotherapy. 2018;73(8):2034–2038. doi: 10.1093/jac/dky167. [DOI] [PubMed] [Google Scholar]
- Bachir et al. (2018).Bachir M, Allem R, Tifrit A, Medjekane M, Drici AE-M, Diaf M, Douidi KT. Primary antibiotic resistance and its relationship with cagA and vacA genes in Helicobacter pylori isolates from Algerian patients. Brazilian Journal of Microbiology. 2018;49(3):544–551. doi: 10.1016/j.bjm.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmaninejad et al. (2021).Bahmaninejad P, Ghafourian S, Mahmoudi M, Maleki A, Sadeghifard N, Badakhsh B. Persister cells as a possible cause of antibiotic therapy failure in Helicobacter pylori. JGH Open. 2021;5(4):493–497. doi: 10.1002/jgh3.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai et al. (2015).Bai P, Zhou LY, Xiao XM, Luo Y, Ding Y. Susceptibility of Helicobacter pylori to antibiotics in Chinese patients. Journal of Digestive Diseases. 2015;16(8):464–470. doi: 10.1111/1751-2980.12271. [DOI] [PubMed] [Google Scholar]
- Bayati et al. (2020).Bayati S, Alebouyeh M, Amirmozafari N, Ebrahimi Daryani N, Talebi M, Zali MR. Histological changes in refractory Helicobacter pylori infection and its relationship with increased levels of resistance to antibiotics and therapeutic regimens: one-year follow-up. APMIS. 2020;128(1):25–34. doi: 10.1111/apm.13001. [DOI] [PubMed] [Google Scholar]
- Bayati et al. (2019).Bayati S, Amirmozafari N, Alebouyeh M, Farzi N, Daryani NE, Zali MR. Antibiotic resistance among Helicobacter pylori strains isolated from patients with histopathological changes of the gastric tissue towards metronidazole, clarithromycin, and ciprofloxacin. Archives of Clinical Infectious Diseases. 2019;14(1):e55015. doi: 10.5812/archcid.55015. [DOI] [Google Scholar]
- Bedoya-Gómez et al. (2020).Bedoya-Gómez IJ, Alvarez-Aldana A, Moncayo-Ortiz JI, Guaca-González YM, Santacruz-Ibarra JJ, Arturo-Arias BL, Castaneda-Chávez LJ, Rodriguez DAL, Beltrán-Angarita L. Surveillance of the antimicrobial resistance rates of Helicobacter pylori ten years later in the Western Central Region, Colombia. Digestive Diseases. 2020;38(3):196–203. doi: 10.1159/000503381. [DOI] [PubMed] [Google Scholar]
- Biernat et al. (2020).Biernat MM, Bińkowska A, Łaczmański Ł, Biernat P, Krzyżek P, Gościniak G. Phenotypic and genotypic analysis of resistant Helicobacter pylori strains isolated from children with gastrointestinal diseases. Diagnostics. 2020;10(10):759. doi: 10.3390/diagnostics10100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binh et al. (2013).Binh TT, Shiota S, Nguyen LT, Ho DD, Hoang HH, Ta L, Trinh DT, Fujioka T, Yamaoka Y. The incidence of primary antibiotic resistance of Helicobacter pylori in Vietnam. Journal of Clinical Gastroenterology. 2013;47(3):233–238. doi: 10.1097/MCG.0b013e3182676e2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bińkowska et al. (2018).Bińkowska A, Biernat MM, Łaczmański Ł, Gościniak G. Molecular patterns of resistance among Helicobacter pylori strains in south-western Poland. Frontiers in Microbiology. 2018;9:3154. doi: 10.3389/fmicb.2018.03154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binyamin et al. (2017).Binyamin D, Pastukh N, On A, Paritsky M, Peretz A. Phenotypic and genotypic correlation as expressed in Helicobacter pylori resistance to clarithromycin and fluoroquinolones. Gut Pathogens. 2017;9(1):1–8. doi: 10.1186/s13099-017-0198-5. [DOI] [Google Scholar]
- Boehnke et al. (2017).Boehnke KF, Valdivieso M, Bussalleu A, Sexton R, Thompson KC, Osorio S, Reyes IN, Crowley JJ, Baker LH, Xi C. Antibiotic resistance among Helicobacter pylori clinical isolates in Lima, Peru. Infection and Drug Resistance. 2017;10:85. doi: 10.2147/IDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolor-Erdene et al. (2017).Bolor-Erdene M, Namdag B, Yamaoka Y, Jav S. Antibiotic resistance of Helicobacter pylori in Mongolia. The Journal of Infection in Developing Countries. 2017;11(11):887–894. doi: 10.3855/jidc.8619. [DOI] [PubMed] [Google Scholar]
- Bouihat et al. (2017).Bouihat N, Burucoa C, Benkirane A, Seddik H, Sentissi S, Al Bouzidi A, Elouennas M, Benouda A. Helicobacter pylori primary antibiotic resistance in 2015 in Morocco: a phenotypic and genotypic prospective and multicenter study. Microbial Drug Resistance. 2017;23(6):727–732. doi: 10.1089/mdr.2016.0264. [DOI] [PubMed] [Google Scholar]
- Boyanova et al. (2014).Boyanova L, Davidkov L, Gergova G, Kandilarov N, Evstatiev I, Panteleeva E, Mitov I. Helicobacter pylori susceptibility to fosfomycin, rifampin, and 5 usual antibiotics for H. pylori eradication. Diagnostic Microbiology and Infectious Disease. 2014;79(3):358–361. doi: 10.1016/j.diagmicrobio.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Boyanova et al. (2015).Boyanova L, Evstatiev I, Gergova G, Yaneva P, Mitov I. Linezolid susceptibility in Helicobacter pylori, including strains with multidrug resistance. International Journal of Antimicrobial Agents. 2015;46(6):703–706. doi: 10.1016/j.ijantimicag.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Boyanova et al. (2016).Boyanova L, Gergova G, Evstatiev I, Spassova Z, Kandilarov N, Yaneva P, Markovska R, Mitov I. Helicobacter pylori resistance to six antibiotics by two breakpoint systems and resistance evolution in Bulgaria. Infectious Diseases. 2016;48(1):56–62. doi: 10.3109/23744235.2015.1082035. [DOI] [PubMed] [Google Scholar]
- Boyanova et al. (2017).Boyanova L, Gergova G, Markovska R, Kandilarov N, Davidkov L, Spassova Z, Mitov I. Primary Helicobacter pylori resistance in elderly patients over 20 years: a Bulgarian study. Diagnostic Microbiology and Infectious Disease. 2017;88(3):264–267. doi: 10.1016/j.diagmicrobio.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Boyanova et al. (2012).Boyanova L, Ilieva J, Gergova G, Davidkov L, Spassova Z, Kamburov V, Katsarov N, Mitov I. Numerous risk factors for Helicobacter pylori antibiotic resistance revealed by extended anamnesis: a Bulgarian study. Journal of Medical Microbiology. 2012;61(1):85–93. doi: 10.1099/jmm.0.035568-0. [DOI] [PubMed] [Google Scholar]
- Boyanova et al. (2013).Boyanova L, Ilieva J, Gergova G, Evstatiev I, Nikolov R, Mitov I. Living in Sofia is associated with a risk for antibiotic resistance in Helicobacter pylori: a Bulgarian study. Folia Microbiologica. 2013;58(6):587–591. doi: 10.1007/s12223-013-0251-9. [DOI] [PubMed] [Google Scholar]
- Bruce et al. (2019).Bruce MG, Bruden D, Newbrough D, Hurlburt DA, Hennessy TW, Morris JM, Reasonover AL, Sacco F, McMahon BJ. The relationship between previous antimicrobial use, antimicrobial resistance and treatment outcome among Alaskans treated for Helicobacter pylori infection. GastroHep. 2019;1(4):172–179. doi: 10.1002/ygh2.352. [DOI] [Google Scholar]
- Butenko et al. (2017).Butenko T, Jeverica S, Orel R, Homan M. Antibacterial resistance and the success of tailored triple therapy in Helicobacter pylori strains isolated from Slovenian children. Helicobacter. 2017;22(5):e12400. doi: 10.1111/hel.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camorlinga-Ponce et al. (2021).Camorlinga-Ponce M, Gómez-Delgado A, Aguilar-Zamora E, Torres RC, Giono-Cerezo S, Escobar-Ogaz A, Torres J. Phenotypic and genotypic antibiotic resistance patterns in Helicobacter pylori strains from ethnically diverse population in Mexico. Frontiers in Cellular and Infection Microbiology. 2021;10:539115. doi: 10.3389/fcimb.2020.539115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira et al. (2011).Cerqueira L, Fernandes RM, Ferreira RM, Carneiro F, Dinis-Ribeiro M, Figueiredo C, Keevil CW, Azevedo NF, Vieira MJ. PNA-FISH as a new diagnostic method for the determination of clarithromycin resistance of Helicobacter pylori. BMC Microbiology. 2011;11(1):1–7. doi: 10.1186/1471-2180-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2017).Chen D, Cunningham SA, Cole NC, Kohner PC, Mandrekar JN, Patel R. Phenotypic and molecular antimicrobial susceptibility of Helicobacter pylori. Antimicrobial Agents and Chemotherapy. 2017;61(4):e02530–16. doi: 10.1128/AAC.02530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2018).Chen J, Ye L, Jin L, Xu X, Xu P, Wang X, Li H. Application of next-generation sequencing to characterize novel mutations in clarithromycin-susceptible Helicobacter pylori strains with A2143G of 23S rRNA gene. Annals of Clinical Microbiology and Antimicrobials. 2018;17(1):10. doi: 10.1186/s12941-018-0259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung et al. (2012).Chung JW, Lee GH, Jeong JY, Lee SM, Jung JH, Choi KD, Song HJ, Jung HY, Kim JH. Resistance of Helicobacter pylori strains to antibiotics in Korea with a focus on fluoroquinolone resistance. Journal of Gastroenterology and Hepatology. 2012;27(3):493–497. doi: 10.1111/j.1440-1746.2011.06874.x. [DOI] [PubMed] [Google Scholar]
- Costa, Soares & Goncalves (2017).Costa S, Soares JB, Goncalves R. Efficacy and tolerability of culture-guided treatment for Helicobacter pylori infection. European Journal of Gastroenterology & Hepatology. 2017;29(11):1258–1263. doi: 10.1097/MEG.0000000000000960. [DOI] [PubMed] [Google Scholar]
- Cuadrado-Lavín et al. (2012).Cuadrado-Lavín A, Salcines-Caviedes JR, Carrascosa MF, Mellado P, Monteagudo I, Llorca J, Cobo M, Campos MR, Ayestarán B, Fernández-Pousa A. Antimicrobial susceptibility of Helicobacter pylori to six antibiotics currently used in Spain. Journal of Antimicrobial Chemotherapy. 2012;67(1):170–173. doi: 10.1093/jac/dkr410. [DOI] [PubMed] [Google Scholar]
- Dang et al. (2020).Dang NQH, Ha TMT, Nguyen S-T, Le NDK, Nguyen TMT, Nguyen TH, Pham TTH. High rates of clarithromycin and levofloxacin resistance of Helicobacter pylori in patients with chronic gastritis in the south east area of Vietnam. Journal of Global Antimicrobial Resistance. 2020;22:620–624. doi: 10.1016/j.jgar.2020.06.007. [DOI] [PubMed] [Google Scholar]
- Dargiene et al. (2018).Dargiene G, Kupcinskas J, Jonaitis L, Vezbavicius M, Kadusevicius E, Kupcinskiene E, Frandsen TH, Kucinskiene R, Kupcinskas L, Andersen LP. Primary antibiotic resistance of Helicobacter pylori strains among adults and children in a tertiary referral centre in Lithuania. APMIS. 2018;126(1):21–28. doi: 10.1111/apm.12752. [DOI] [PubMed] [Google Scholar]
- De Francesco et al. (2014).De Francesco V, Zullo A, Giorgio F, Saracino I, Zaccaro C, Hassan C, Ierardi E, Di Leo A, Fiorini G, Castelli V, Lo Re G, Vaira D. Change of point mutations in Helicobacter pylori rRNA associated with clarithromycin resistance in Italy. Journal of Medical Microbiology. 2014;63(3):453–457. doi: 10.1099/jmm.0.067942-0. [DOI] [PubMed] [Google Scholar]
- Dekhnich et al. (2018).Dekhnich N, Ivanchik N, Kozlov R, Alimov A, Steshits A, Kirsov P, Pandav K. Dynamics of antimicrobial resistance of Helicobacter pylori isolates in the Smolensk region of Russian Federation. Helicobacter. 2018;23(6):e12545. doi: 10.1111/hel.12545. [DOI] [PubMed] [Google Scholar]
- Demiray-Gürbüz et al. (2017).Demiray-Gürbüz E, Yılmaz Ö, Olivares AZ, Gönen C, Sarıoğlu S, Soytürk M, Tümer S, Altungöz O, Şimşek İ, Perez Perez GI. Rapid identification of Helicobacter pylori and assessment of clarithromycin susceptibility from clinical specimens using FISH. The Journal of Pathology: Clinical Research. 2017;3(1):29–37. doi: 10.1002/cjp2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyi et al. (2019).Deyi VYM, Burette A, Ntounda R, Elkilic O, Cadranel S, Bontems P, Hallin M. Update of primary Helicobacter pylori resistance to antimicrobials in Brussels, Belgium. Diagnostic Microbiology and Infectious Disease. 2019;95(4):114875. doi: 10.1016/j.diagmicrobio.2019.114875. [DOI] [PubMed] [Google Scholar]
- Di Giulio et al. (2016).Di Giulio M, Di Campli E, Di Bartolomeo S, Cataldi V, Marzio L, Grossi L, Ciccaglione AF, Nostro A, Cellini L. In vitro antimicrobial susceptibility of Helicobacter pylori to nine antibiotics currently used in Central Italy. Scandinavian Journal of Gastroenterology. 2016;51(3):263–269. doi: 10.3109/00365521.2015.1092577. [DOI] [PubMed] [Google Scholar]
- Domanovich-Asor et al. (2020).Domanovich-Asor T, Motro Y, Khalfin B, Craddock HA, Peretz A, Moran-Gilad J. Genomic analysis of antimicrobial resistance genotype-to-phenotype agreement in Helicobacter pylori. Microorganisms. 2020;9(1):2. doi: 10.3390/microorganisms9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draeger et al. (2015).Draeger S, Wüppenhorst N, Kist M, Glocker E-O. Outcome of second-and third-line Helicobacter pylori eradication therapies based on antimicrobial susceptibility testing. Journal of Antimicrobial Chemotherapy. 2015;70(11):3141–3145. doi: 10.1093/jac/dkv223. [DOI] [PubMed] [Google Scholar]
- Ducournau et al. (2016).Ducournau A, Bénéjat L, Sifré E, Besséde E, Lehours P, Mégraud F. Helicobacter pylori resistance to antibiotics in 2014 in France detected by phenotypic and genotypic methods. Clinical Microbiology and Infection. 2016;22(8):715–718. doi: 10.1016/j.cmi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Egger et al. (1997).Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghbali et al. (2016).Eghbali Z, Mojtahedi A, Moien Ansar M, Fakhrieh Asl S, Aminian K. Detection of 23SrRNA mutations strongly related to clarithromycin resistance in Helicobacter pylori strains isolated from patients in the North of Iran. Jundishapur Journal of Microbiology. 2016;9(2):e29694. doi: 10.5812/jjm.29694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli et al. (2020).Egli K, Wagner K, Keller PM, Risch L, Risch M, Bodmer T. Comparison of the diagnostic performance of qPCR, sanger sequencing, and whole-genome sequencing in determining clarithromycin and levofloxacin resistance in Helicobacter pylori. Frontiers in Cellular and Infection Microbiology. 2020;10:596371. doi: 10.3389/fcimb.2020.596371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisig et al. (2011).Eisig JN, Silva FM, Barbuti RC, Navarro-Rodriguez T, Moraes-Filho JPP, Pedrazzoli J., Jr Helicobacter pylori antibiotic resistance in Brazil: clarithromycin is still a good option. Arquivos de Gastroenterologia. 2011;48(4):261–264. doi: 10.1590/S0004-28032011000400008. [DOI] [PubMed] [Google Scholar]
- Enany & Abdalla (2015).Enany S, Abdalla S. In vitro antagonistic activity of Lactobacillus casei against Helicobacter pylori. Brazilian Journal of Microbiology. 2015;46(4):1201–1206. doi: 10.1590/S1517-838246420140675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng et al. (2015).Eng NF, Ybazeta G, Chapman K, Fraleigh NL, Letto R, Altman E, Diaz-Mitoma F. Antimicrobial susceptibility of Canadian isolates of Helicobacter pylori in Northeastern Ontario. Canadian Journal of Infectious Diseases and Medical Microbiology. 2015;26(3):137–144. doi: 10.1155/2015/853287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkut et al. (2020).Erkut M, Uzun DY, Kaklıkkaya N, Fidan S, Yoğun Y, Coşar AM, Akyıldız E, Topbaş M, Özgür O, Arslan M. Sociodemographic characteristics and clinical risk factors of Helicobacter pylori infection and antibiotic resistance in the Eastern Black Sea region of Turkey. The Turkish Journal of Gastroenterology. 2020;31(3):221. doi: 10.5152/tjg.2020.18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaidi et al. (2022).Essaidi I, Bounder G, Jouimyi RM, Boura H, Elyounsi I, Kheir F-Z, Benomar H, Badre W, Zerouali K, Maachi F. Comparative study of Helicobacter pylori resistance to clarithromycin and metronidazole and its association with epidemiological factors in a moroccan population. Asian Pacific Journal of Cancer Prevention. 2022;23(8):2755–2761. doi: 10.31557/APJCP.2022.23.8.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famouri et al. (2018).Famouri F, Emadoleslami MS, Riahi R, Saneian H, Nasri P. The sensitivity of H. pylori in gastric tissue samples of children and adolescents to various antibiotics in center of Iran. International Journal of Pediatrics. 2018;6(12):8685–8696. doi: 10.22038/ijp.2018.33647.2974. [DOI] [Google Scholar]
- Farzi et al. (2019).Farzi N, Behzad C, Hasani Z, Alebouyeh M, Zojaji H, Zali MR. Characterization of clarithromycin heteroresistance among Helicobacter pylori strains isolated from the antrum and corpus of the stomach. Folia Microbiologica. 2019;64(2):143–151. doi: 10.1007/s12223-018-0637-9. [DOI] [PubMed] [Google Scholar]
- Farzi et al. (2019).Farzi N, Yadegar A, Sadeghi A, Asadzadeh Aghdaei H, Marian Smith S, Raymond J, Suzuki H, Zali MR. High prevalence of antibiotic resistance in Iranian Helicobacter pylori isolates: importance of functional and mutational analysis of resistance genes and virulence genotyping. Journal of Clinical Medicine. 2019;8(11):2004. doi: 10.3390/jcm8112004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasciana et al. (2015).Fasciana T, Calà C, Bonura C, Di Carlo E, Matranga D, Scarpulla G, Manganaro M, Camilleri S, Giammanco A. Resistance to clarithromycin and genotypes in Helicobacter pylori strains isolated in Sicily. Journal of Medical Microbiology. 2015;64(11):1408–1414. doi: 10.1099/jmm.0.000163. [DOI] [PubMed] [Google Scholar]
- Ferenc et al. (2017).Ferenc S, Gnus J, Kościelna M, Kinda M, Yarka A, Stewart L, Witkiewicz W. High antibiotic resistance of Helicobacter pylori and its effect on tailored and empiric eradication of the organism in Lower Silesia, Poland. Helicobacter. 2017;22(2):e12365. doi: 10.1111/hel.12365. [DOI] [PubMed] [Google Scholar]
- Fernández-Reyes et al. (2019).Fernández-Reyes M, Tamayo E, Rojas-Rengifo D, Fischer W, Carrasco-García E, Alonso M, Lizasoain J, Bujanda L, Cosme Á, Montes M. Helicobacter pylori pathogenicity and primary antimicrobial resistance in Northern Spain. European Journal of Clinical Investigation. 2019;49(8):e13150. doi: 10.1111/eci.13150. [DOI] [PubMed] [Google Scholar]
- Figueroa et al. (2012).Figueroa M, Cortés A, Pazos Á, Bravo LE. Sensibilidad in vitro a amoxicilina y claritromicina de Helicobacter pylori obtenido de biopsias gástricas de pacientes en zona de bajo riesgo para cáncer gástrico. Biomédica. 2012;32(1):32–42. doi: 10.7705/biomedica.v32i1.454. [DOI] [PubMed] [Google Scholar]
- Fiorini et al. (2018).Fiorini G, Zullo A, Saracino IM, Pavoni M, Vaira D. Antibiotic resistance pattern of Helicobacter pylori strains isolated in Italy during 2010–2016. Scandinavian Journal of Gastroenterology. 2018;53(6):661–664. doi: 10.1080/00365521.2018.1464596. [DOI] [PubMed] [Google Scholar]
- Gatta et al. (2018).Gatta L, Scarpignato C, Fiorini G, Belsey J, Saracino I, Ricci C, Vaira D. Impact of primary antibiotic resistance on the effectiveness of sequential therapy for Helicobacter pylori infection: lessons from a 5-year study on a large number of strains. Alimentary Pharmacology & Therapeutics. 2018;47(9):1261–1269. doi: 10.1111/apt.14597. [DOI] [PubMed] [Google Scholar]
- Gehlot et al. (2016).Gehlot V, Mahant S, Mukhopadhyay AK, Das K, Alam J, Ghosh P, Das R. Low prevalence of clarithromycin-resistant Helicobacter pylori isolates with A2143G point mutation in the 23S rRNA gene in North India. Journal of Global Antimicrobial Resistance. 2016;6:39–43. doi: 10.1016/j.jgar.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Gehlot et al. (2016).Gehlot V, Mahant S, Mukhopadhyay AK, Das K, De R, Kar P, Das R. Antimicrobial susceptibility profiles of Helicobacter pylori isolated from patients in North India. Journal of Global Antimicrobial Resistance. 2016;5(Suppl. 1):51–56. doi: 10.1016/j.jgar.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Ghotaslou et al. (2013).Ghotaslou R, Milani M, Akhi MT, Hejazi MS, Nahaei MR, Hasani A, Sharifi Y. Relationship between drug resistance and cagA Gene in Helicobacter pylori. Jundishapur Journal of Microbiology. 2013;6(10):321. doi: 10.5812/jjm.8480. [DOI] [Google Scholar]
- Gong et al. (2020).Gong EJ, Ahn JY, Kim JM, Lee SM, Na HK, Lee JH, Jung KW, Choi KD, Kim DH, Song HJ, Lee GH, Kim SW, Jung H-Y. Genotypic and phenotypic resistance to clarithromycin in Helicobacter pylori strains. Journal of Clinical Medicine. 2020;9(6):1930. doi: 10.3390/jcm9061930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong et al. (2020).Gong EJ, Ahn JY, Kim JM, Lee SM, Na HK, Lee JH, Jung KW, Choi KD, Kim DH, Song HJ, Lee GH, Kim SW, Jung HY. Genotypic and phenotypic resistance to clarithromycin in Helicobacter pylori strains. Journal of Clinical Medicine. 2020;9(6):1930. doi: 10.3390/jcm9061930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gościniak et al. (2014).Gościniak G, Biernat M, Grabińska J, Bińkowska A, Poniewierka E, Iwańczak B. The antimicrobial susceptibility of Helicobacter pylori strains isolated from children and adults with primary infection in the Lower Silesia Region, Poland. Polish Journal of Microbiology. 2014;63(1):57. doi: 10.5114/aoms.2013.36917. [DOI] [PubMed] [Google Scholar]
- Goudarzi et al. (2016).Goudarzi M, Heidary M, Azad M, Fazeli M, Goudarzi H. Evaluation of antimicrobial susceptibility and integron carriage in Helicobacter pylori isolates from patients. Gastroenterology and Hepatology from Bed to Bench. 2016;9(Suppl1):S47–S52. [PMC free article] [PubMed] [Google Scholar]
- Goudarzi et al. (2016).Goudarzi M, Seyedjavadi SS, Fazeli M, Roshani M, Azad M, Heidary M, Navidinia M, Goudarzi H. Identification of a novel cassette array in integronbearing Helicobacter pylori strains isolated from Iranian patients. Asian Pacific Journal of Cancer Prevention. 2016;17(7):3309–3315. doi: 10.14456/apjcp.2016.93. [DOI] [PubMed] [Google Scholar]
- Gunnarsdottir et al. (2017).Gunnarsdottir AI, Gudjonsson H, Hardardottir H, Jonsdottir KD, Bjornsson ES. Antibiotic susceptibility of Helicobacter pylori in Iceland. Infectious Diseases. 2017;49(9):647–654. doi: 10.1080/23744235.2017.1317359. [DOI] [PubMed] [Google Scholar]
- Guo et al. (2019).Guo C, Liu F, Zhu L, Wu F, Cui G, Xiong Y, Wang Q, Yin L, Wang C, Wang H, Wu X, Zhang Z, Chen Z. Analysis of culturable microbiota present in the stomach of children with gastric symptoms. Brazilian Journal of Microbiology. 2019;50(1):107–115. doi: 10.1007/s42770-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha et al. (2019).Ha TMT, Le PTQ, Phan TN, Tran TNH. Characterisation of point mutations in domain V of the 23S rRNA gene of clinical Helicobacter pylori strains and clarithromycin-resistant phenotype in central Vietnam. Journal of Global Antimicrobial Resistance. 2019;16:87–91. doi: 10.1016/j.jgar.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Haddadi et al. (2020).Haddadi MH, Negahdari B, Asadolahi R, Bazargani A. Helicobacter pylori antibiotic resistance and correlation with cagA motifs and homB gene. Postgraduate Medicine. 2020;132(6):512–520. doi: 10.1080/00325481.2020.1753406. [DOI] [PubMed] [Google Scholar]
- Hamidi et al. (2020).Hamidi S, Badmasti F, Sadeghpour Heravi F, Safapoor MH, Mohammad Ali Tabrizi A, Ghorbani M, Azizi O. Antibiotic resistance and clonal relatedness of Helicobacter pylori strains isolated from stomach biopsy specimens in northeast of Iran. Helicobacter. 2020;25(2):e12684. doi: 10.1111/hel.12684. [DOI] [PubMed] [Google Scholar]
- Hamza et al. (2018).Hamza D, Elhelw R, Elhariri M, Ragab E. Genotyping and antimicrobial resistance patterns of Helicobacter pylori in human and dogs associated with A2142G and A2143G point mutations in clarithromycin resistance. Microbial Pathogenesis. 2018;123(7):330–338. doi: 10.1016/j.micpath.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Han et al. (2016).Han R, Lu H, Jiang M-W, Tan K-W, Peng Z, Hu J-L, Fang D-C, Lan C-H, Wu X-L. Multicenter study of antibiotic resistance profile of H. pylori and distribution of CYP2C19 gene polymorphism in rural population of Chongqing, China. Gastroenterology Research and Practice. 2016;2016:8547686. doi: 10.1155/2016/8547686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafiah et al. (2019).Hanafiah A, Binmaeil H, Ali RAR, Rose IM, Lopes BS. Molecular characterization and prevalence of antibiotic resistance in Helicobacter pylori isolates in Kuala Lumpur, Malaysia. Infection and Drug Resistance. 2019;12:3051. doi: 10.2147/IDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansomburana et al. (2012).Hansomburana P, Anantapanpong S, Sirinthornpunya S, Chuengyong K, Rojborwonwittaya J. Prevalence of single nucleotide mutation in clarithromycin resistant gene of Helicobacter pylori: a 32-months prospective study by using hybridization real time polymerase chain reaction. Journal of the Medical Association of Thailand. 2012;95(3):S28–S35. [PubMed] [Google Scholar]
- Harbord, Harris & Sterne (2009).Harbord RM, Harris RJ, Sterne JAC. Updated tests for small-study effects in meta-analyses. The Stata Journal. 2009;9(2):197–210. doi: 10.1177/1536867X0900900202. [DOI] [Google Scholar]
- Harbord et al. (2010).Harbord R, Harris RJ, Sterne JA, Steichen T. METABIAS: stata module to test for small-study effects in meta-analysis. 2010. https://econpapers.repec.org/software/bocbocode/s404901.htm https://econpapers.repec.org/software/bocbocode/s404901.htm
- Hashemi et al. (2019).Hashemi SJ, Sheikh AF, Goodarzi H, Yadyad MJ, Seyedian SS, Aslani S, Assarzadegan MA. Genetic basis for metronidazole and clarithromycin resistance in Helicobacter pylori strains isolated from patients with gastroduodenal disorders. Infection and Drug Resistance. 2019;12:535–543. doi: 10.2147/IDR.S192942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2021).He C, Kong F, Zhu X, Kong F, Zhao W, Liu Y, Wang K, Yang J. Clinical effect of clarithromycin combined with tinidazole on Helicobacter pylori-related gastritis and its influence on COX-2 expression. BioMed Research International. 2021;2021:1–9. doi: 10.1155/2021/4171019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hojsak et al. (2012).Hojsak I, Kos T, Dumancic J, Misak Z, Jadresin O, Jaklin Kekez A, Lukic Grlic A, Kolacek S. Antibiotic resistance of Helicobacter pylori in pediatric patients—10 years’ experience. European Journal of Pediatrics. 2012;171(9):1325–1330. doi: 10.1007/s00431-012-1722-8. [DOI] [PubMed] [Google Scholar]
- Honma et al. (2019).Honma H, Nakayama Y, Kato S, Hidaka N, Kusakari M, Sado T, Suda A, Lin Y. Clinical features of Helicobacter pylori antibody-positive junior high school students in Nagano Prefecture, Japan. Helicobacter. 2019;24(2):e12559. doi: 10.1111/hel.12559. [DOI] [PubMed] [Google Scholar]
- Horie et al. (2020).Horie R, Handa O, Ando T, Ose T, Murakami T, Suzuki N, Sendo R, Imamoto E, Itoh Y. Helicobacter pylori eradication therapy outcome according to clarithromycin susceptibility testing in Japan. Helicobacter. 2020;25(4):e12698. doi: 10.1111/hel.12698. [DOI] [PubMed] [Google Scholar]
- Hosseini et al. (2021).Hosseini RS, Rahimian G, Shafigh MH, Validi M, Khaledi M, Gholipour AJAE. Correlation between clarithromycin resistance, virulence factors and clinical characteristics of the disease in Helicobacter pylori infected patients in Shahrekord, Southwest Iran. AMB Express. 2021;11(1):1–9. doi: 10.1186/s13568-021-01310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung et al. (2021).Hung KT, Yang SC, Wu CK, Wang HM, Yao CC, Liang CM, Tai WC, Wu KL, Kuo YH, Lee CH, Chuah SK. Eradication rates for esomeprazole and lansoprazole-based 7-day non-bismuth concomitant quadruple therapy for first-line anti-Helicobacter pylori treatment in real world clinical practice. Infection and Drug Resistance. 2021;14:1239–1246. doi: 10.2147/IDR.S304711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein, Al-Ouqaili & Majeed (2022).Hussein RA, Al-Ouqaili MT, Majeed YH. Detection of clarithromycin resistance and 23SrRNA point mutations in clinical isolates of Helicobacter pylori isolates: phenotypic and molecular methods. Saudi Journal of Biological Sciences. 2022;29(1):513–520. doi: 10.1016/j.sjbs.2021.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilie et al. (2011).Ilie M, Popa M, Chifiriuc MC, Baltac A, Constantinescu G, Tãnãsescu C. Helicobacter pylori cultivation from gastric biopsies and susceptibility to antibiotics used in empirical therapy. Roumanian Archives of Microbiology and Immunology. 2011;70(2):60–64. [PubMed] [Google Scholar]
- Jiang et al. (2021).Jiang ZD, He BS, Zhang ZY, Wang SK, Ran D, Wang ZB. Analysis of the primary and post-treatment antibiotic resistance of Helicobacter pylori in the Nanjing area. Current Pharmaceutical Biotechnology. 2021;22(5):682–685. doi: 10.2174/1389201021666200722162613. [DOI] [PubMed] [Google Scholar]
- Jolaiya et al. (2020).Jolaiya TF, Fowora MA, Onyekwere C, Ugiagbe R, Agbo II, Lesi O, Ndububa DA, Adekanle O, Njom HA, Idowu A, Adeleye IA, Bamidele M, Ngoka FN, Palamides PF, Smith SI. Efflux pump mediated antibiotic resistance in clinical isolates of Helicobacter pylori from South West Nigeria. Journal of Gastroenterology and Hepatology Research. 2020;9(4):3283–3289. doi: 10.17554/j.issn.2224-3992.2020.09.944. [DOI] [Google Scholar]
- Kakiuchi et al. (2020).Kakiuchi T, Hashiguchi K, Imamura I, Nakayama A, Takamori A, Okuda M, Matsuo M. Assessment of a novel method to detect clarithromycin-resistant Helicobacter pylori using a stool antigen test reagent. BMC Gastroenterology. 2020;20(1):1–6. doi: 10.1186/s12876-020-01549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabiber et al. (2014).Karabiber H, Selimoglu MA, Otlu B, Yildirim O, Ozer A. Virulence factors and antibiotic resistance in children with Helicobacter pylori gastritis. Journal of Pediatric Gastroenterology and Nutrition. 2014;58(5):608–612. doi: 10.1097/MPG.0000000000000273. [DOI] [PubMed] [Google Scholar]
- Karczewska et al. (2011).Karczewska E, Wojtas-Bonior I, Sito E, Zwolińska-Wcisło M, Budak A. Primary and secondary clarithromycin, metronidazole, amoxicillin and levofloxacin resistance to Helicobacter pylori in southern Poland. Pharmacological Reports. 2011;63(3):799–807. doi: 10.1016/S1734-1140(11)70592-8. [DOI] [PubMed] [Google Scholar]
- Karpinski et al. (2015).Karpinski TM, Andrzejewska E, Eder P, Linke K, Szkaradkiewicz A. Evaluation of antimicrobial resistance of Helicobacter pylori in the last 15 years in West Poland. Acta Microbiologica et Immunologica Hungarica. 2015;62(3):287–293. doi: 10.1556/030.62.2015.3.6. [DOI] [PubMed] [Google Scholar]
- Keshavarz Azizi Raftar et al. (2015).Keshavarz Azizi Raftar S, Moniri R, Saffari M, Razavi Zadeh M, Arj A, Mousavi SGA, Mirzaei Ghazi Kalayeh H, Dastehgoli K. The Helicobacter pylori resistance rate to clarithromycin in Iran. Microbial Drug Resistance. 2015;21(1):69–73. doi: 10.1089/mdr.2014.0104. [DOI] [PubMed] [Google Scholar]
- Khademi et al. (2014).Khademi F, Faghri J, Moghim S, Esfahani BN, Fazeli H, Poursina F, Adibi P, Madhi M, Safaei HG. The study of mutation in 23S rRNA resistance gene of Helicobacter pylori to clarithromycin in patients with gastrointestinal disorders in Isfahan—Iran. Advanced Biomedical Research. 2014;3:98. doi: 10.4103/2277-9175.129368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi et al. (2013).Khademi F, Faghri J, Poursina F, Esfahani BN, Moghim S, Fazeli H, Adibi P, Mirzaei N, Akbari M, Safaei HG. Resistance pattern of Helicobacter pylori strains to clarithromycin, metronidazole, and amoxicillin in Isfahan, Iran. Journal of Research in Medical Sciences: The Official Journal of Isfahan University of Medical Sciences. 2013;18(12):1056. [PMC free article] [PubMed] [Google Scholar]
- Khani, Abadi & Mobarez (2019).Khani S, Abadi ATB, Mobarez AM. Clarithromycin-susceptible but virulent Helicobacter pylori strains infecting Iranian patients’ stomachs. Infection and Drug Resistance. 2019;12:3415. doi: 10.2147/IDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khani, Talebi Bezmin Abadi & Mohabati Mobarez (2019).Khani S, Talebi Bezmin Abadi A, Mohabati Mobarez A. Clarithromycin-susceptible but virulent Helicobacter pylori strains infecting Iranian patients’ stomachs. Infection and Drug Resistance. 2019;12:3415–3420. doi: 10.2147/IDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khashei et al. (2016).Khashei R, Dara M, Bazargani A, Bagheri Lankarani K, Taghavi A, Moeini M, Dehghani B, Sohrabi M. High rate of A2142G point mutation associated with clarithromycin resistance among Iranian Helicobacter pylori clinical isolates. APMIS. 2016;124(9):787–793. doi: 10.1111/apm.12567. [DOI] [PubMed] [Google Scholar]
- Khoury et al. (2017).Khoury J, Geffen Y, Shaul R, Sholy H, Chowers Y, Saadi T. Secondary antibiotic resistance of Helicobacter pylori isolates in Israeli children and adults. Journal of Global Antimicrobial Resistance. 2017;10(1):182–185. doi: 10.1016/j.jgar.2017.05.017. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2011).Kim JY, Kim NY, Kim SJ, Baik GH, Kim GH, Kim JM, Nam RH, Kim HB, Lee DH, Jung HC, Song IS. Regional difference of antibiotic resistance of Helicobacter pylori strains in Korea. The Korean Journal of Gastroenterology. 2011;57(4):221–229. doi: 10.4166/kjg.2011.57.4.221. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2020).Kim YM, Lee KH, Kim J-H, Park SY, Song YG, Jeon SY, Park H. Is only clarithromycin susceptibility important for the successful eradication of Helicobacter pylori? Antibiotics. 2020;9(9):589. doi: 10.3390/antibiotics9090589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocazeybek et al. (2019).Kocazeybek B, Sakli MK, Yuksel P, Demirci M, Caliskan R, Sarp TZ, Saribas S, Demiryas S, Kalayci F, Cakan H. Comparison of new and classical point mutations associated with clarithromycin resistance in Helicobacter pylori strains isolated from dyspeptic patients and their effects on phenotypic clarithromycin resistance. Journal of Medical Microbiology. 2019;68(4):566–573. doi: 10.1099/jmm.0.000944. [DOI] [PubMed] [Google Scholar]
- Kocsmár et al. (2021).Kocsmár É, Buzás GM, Szirtes I, Kocsmár I, Kramer Z, Szijártó A, Fadgyas-Freyler P, Szénás K, Rugge M, Fassan M, Kiss A, Schaff Z, Röst G, Lotz G. Primary and secondary clarithromycin resistance in Helicobacter pylori and mathematical modeling of the role of macrolides. Nature Communications. 2021;12(1):1–12. doi: 10.1038/s41467-021-22557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- korn Vilaichone et al. (2017).korn Vilaichone R, Gamnarai P, Subsomwong P, Uchida T, Yamaoka Y, Mahachai V. High fluoroquinolone resistant strains of Helicobacter pylori in the Golden triangle. Asian Pacific Journal of Cancer Prevention: APJCP. 2017;18(2):455–458. doi: 10.22034/APJCP.2017.18.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostamo et al. (2011).Kostamo P, Veijola L, Oksanen A, Sarna S, Rautelin H. Recent trends in primary antimicrobial resistance of Helicobacter pylori in Finland. International Journal of Antimicrobial Agents. 2011;37(1):22–25. doi: 10.1016/j.ijantimicag.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Kouitcheu Mabeku et al. (2019).Kouitcheu Mabeku LB, Eyoum Bille B, Tepap Zemnou C, Tali Nguefack LD, Leundji H. Broad spectrum resistance in Helicobacter pylori isolated from gastric biopsies of patients with dyspepsia in Cameroon and efflux-mediated multiresistance detection in MDR isolates. BMC Infectious Diseases. 2019;19(1):880. doi: 10.1186/s12879-019-4536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2020).Kumar S, Sangitha R, Nachamkin I, Metz DC. Resistance patterns of refractory Helicobacter pylori infection in a referral centre in the Delaware Valley. GastroHep. 2020;2(1):6–12. doi: 10.1002/ygh2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo et al. (2021).Kuo C-J, Lee C-H, Chang M-L, Lin C-Y, Lin W-R, Su M-Y, Chiu C-H, Tseng C-N, Wu Y-S, Chiu C-T. Multidrug resistance: the clinical dilemma of refractory Helicobacter pylori infection. Journal of Microbiology, Immunology and Infection. 2021;54(6):1184–1187. doi: 10.1016/j.jmii.2021.03.006. [DOI] [PubMed] [Google Scholar]
- Larsen et al. (2013).Larsen AL, Ragnhildstveit E, Moayeri B, Eliassen L, Melby KK. Resistance rates of metronidazole and other antibacterials in Helicobacter pylori from previously untreated patients in Norway. APMIS. 2013;121(4):353–358. doi: 10.1111/apm.12009. [DOI] [PubMed] [Google Scholar]
- Lauener et al. (2019).Lauener FN, Imkamp F, Lehours P, Buissonniere A, Benejat L, Zbinden R, Keller PM, Wagner K. Genetic determinants and prediction of antibiotic resistance phenotypes in Helicobacter pylori. Journal of Clinical Medicine. 2019;8(1):53. doi: 10.3390/jcm8010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2014).Lee JY, Kim N, Kim MS, Choi YJ, Lee JW, Yoon H, Shin CM, Park YS, Lee DH, Jung HC. Factors affecting first-line triple therapy of Helicobacter pylori including CYP2C19 genotype and antibiotic resistance. Digestive Diseases and Sciences. 2014;59(6):1235–1243. doi: 10.1007/s10620-014-3093-7. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2013).Lee JW, Kim N, Kim JM, Nam RH, Chang H, Kim JY, Shin CM, Park YS, Lee DH, Jung HC. Prevalence of primary and secondary antimicrobial resistance of Helicobacter pylori in Korea from 2003 through 2012. Helicobacter. 2013;18(3):206–214. doi: 10.1111/hel.12031. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2019).Lee JY, Kim N, Nam RH, In Choi S, Lee JW, Lee DH. Primary and secondary antibiotic resistance of Helicobacter pylori in Korea from 2003 to 2018. Helicobacter. 2019;24(6):e12660. doi: 10.1111/hel.12660. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2019).Lee JW, Kim N, Nam RH, Lee SM, Kwon YH, Sohn SD, Kim JM, Lee DH, Jung HC. Favorable outcomes of culture-based Helicobacter pylori eradication therapy in a region with high antimicrobial resistance. Helicobacter. 2019;24(2):e12561. doi: 10.1111/hel.12561. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2019).Lee KH, Park SY, Jeong SJ, Kim J-H, Jeong SH, Kang I-M, Song YG. Can aminoglycosides be used as a new treatment for Helicobacter pylori? In vitro activity of recently isolated Helicobacter pylori. Infection & Chemotherapy. 2019;51(1):10–20. doi: 10.3947/ic.2019.51.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehours, Siffré & Mégraud (2011).Lehours P, Siffré E, Mégraud F. DPO multiplex PCR as an alternative to culture and susceptibility testing to detect Helicobacter pylori and its resistance to clarithromycin. BMC Gastroenterology. 2011;11(1):1–5. doi: 10.1186/1471-230X-11-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2020).Li J, Deng J, Wang Z, Li H, Wan C. Antibiotic resistance of Helicobacter pylori strains isolated from pediatric patients in Southwest China. Frontiers in Microbiology. 2020;11:621791. doi: 10.3389/fmicb.2020.621791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2021).Li X-H, Huang Y-Y, Lu L-M, Zhao L-J, Luo X-K, Li R-J, Dai Y-Y, Qin C, Huang Y-Q, Chen H. Early genetic diagnosis of clarithromycin resistance in Helicobacter pylori. World Journal of Gastroenterology. 2021;27(24):3595–3608. doi: 10.3748/wjg.v27.i24.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2017).Li L, Ke Y, Yu C, Li G, Yang N, Zhang J, Li Y. Antibiotic resistance of Helicobacter pylori in Chinese children: a multicenter retrospective study over 7 years. Helicobacter. 2017;22(3):e12373. doi: 10.1111/hel.12373. [DOI] [PubMed] [Google Scholar]
- Liang et al. (2020).Liang C-M, Tai W-C, Hsu P-I, Wu D-C, Kuo C-H, Tsay F-W, Lee C-L, Chen K-Y, Chuah S-K. Trend of changes in antibiotic resistance in Helicobacter pylori from 2013 to 2019: a multicentre report from Taiwan. Therapeutic Advances in Gastroenterology. 2020;13:1756284820976990. doi: 10.1177/1756284820976990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin et al. (2020).Lin T-F, Wu D-C, Tsay F-W, Tsai K-W, Tsai T-J, Peng N-J, Kao S-S, Chen W-C, Chen Y-H, Hsu P-I. Reverse hybrid therapy achieves a similar eradication rate as standard hybrid therapy for Helicobacter pylori infection. Journal of the Chinese Medical Association. 2020;83(3):233–237. doi: 10.1097/JCMA.0000000000000256. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2018).Liu D-S, Wang Y-H, Zeng Z-R, Zhang Z-Y, Lu H, Xu J-M, Du Y-Q, Li Y, Wang J-B, Xu S-P. Primary antibiotic resistance of Helicobacter pylori in Chinese patients: a multiregion prospective 7-year study. Clinical Microbiology and Infection. 2018;24(7) doi: 10.1016/j.cmi.2017.11.010. 780.e5–780.e8. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2019).Liu DS, Wang YH, Zhu ZH, Zhang SH, Zhu X, Wan JH, Lu NH, Xie Y. Characteristics of Helicobacter pylori antibiotic resistance: data from four different populations. Antimicrobial Resistance & Infection Control. 2019;8(1):192. doi: 10.1186/s13756-019-0632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2011).Liu G, Xu X, He L, Ding Z, Gu Y, Zhang J, Zhou L. Primary antibiotic resistance of Helicobacter pylori isolated from Beijing children. Helicobacter. 2011;16(5):356–362. doi: 10.1111/j.1523-5378.2011.00856.x. [DOI] [PubMed] [Google Scholar]
- Lok et al. (2020).Lok C-H, Zhu D, Wang J, Ren Y-T, Jiang X, Li S-J, Zhao X-Y. Phenotype and molecular detection of clarithromycin and levofloxacin resistance in Helicobacter pylori clinical isolates in Beijing. Infection and Drug Resistance. 2020;13:2145–2153. doi: 10.2147/IDR.S249370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2019).Lu HH, Lai FP, Lo HY, Sheu BS, Yang YJ. Increasing antimicrobial resistance to clarithromycin and metronidazole in pediatric Helicobacter pylori infection in southern Taiwan: a comparison between two decades. Helicobacter. 2019;24(5):e12633. doi: 10.1111/hel.12633. [DOI] [PubMed] [Google Scholar]
- Lyu et al. (2020).Lyu T, Cheung KS, Ni L, Guo J, Mu P, Li Y, Yang Q, Yu X, Lyu Z, Wu J. High prevalence and risk factors of multiple antibiotic resistance in patients who fail first-line Helicobacter pylori therapy in southern China: a municipality-wide, multicentre, prospective cohort study. Journal of Antimicrobial Chemotherapy. 2020;75(11):3391–3394. doi: 10.1093/jac/dkaa315. [DOI] [PubMed] [Google Scholar]
- Macías-García et al. (2017).Macías-García F, Llovo-Taboada J, Díaz-López M, Bastón-Rey I, Domínguez-Muñoz JE. High primary antibiotic resistance of Helicobacter pylori strains isolated from dyspeptic patients: a prevalence cross-sectional study in Spain. Helicobacter. 2017;22(6):e12440. doi: 10.1111/hel.12440. [DOI] [PubMed] [Google Scholar]
- Macin et al. (2015).Macin S, Demir H, Ozen H, Yuce A, Akyon Y. Determination of Helicobacter pylori antibiotic resistance patterns in pediatric gastroenterology patients: the Hacettepe experience. The Turkish Journal of Pediatrics. 2015;57(3):254–257. [PubMed] [Google Scholar]
- Maev et al. (2020).Maev IV, Andreev DN, Govorun VM, Ilina EN, Kucheryavyy YA, Oganesian TS, Melnikova EV, Zayratyants OV, Parfenova TV, Dzhedzheia LV, Kirillova NV, Maevskaya EA, Fomenko AK, Lobanova EG, Zaborovskii AV, Kriukov KA. Antibiotic resistance of Helicobacter pylori in the European part of the Russian Federation: first results. Terapevticheskiy Arkhiv. 2020;92(8):24–28. doi: 10.26442/00403660.2020.08.000761. [DOI] [PubMed] [Google Scholar]
- Mahmoudi et al. (2017).Mahmoudi S, Mamishi S, Banar M, Keshavarz Valian S, Bahador A, Najafi M, Farahmand F, Pourakbari B. Antibiotic susceptibility of Helicobacter pylori strains isolated from Iranian children: high frequency of A2143G point mutation associated with clarithromycin resistance. Journal of Global Antimicrobial Resistance. 2017;10(1):131–135. doi: 10.1016/j.jgar.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Maleknejad et al. (2015).Maleknejad S, Mojtahedi A, Safaei-Asl A, Taghavi Z, Kazemnejad E. Primary antibiotic resistance to Helicobacter pylori strains isolated from children in Northern Iran: a single center study. Iranian Journal of Pediatrics. 2015;25(6):308. doi: 10.5812/ijp.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi et al. (2015).Manfredi M, Gismondi P, Maffini V, Bizzarri B, Fornaroli F, Madia C, Salerno A, Cangelosi AM, de’Angelis GL. Primary antimicrobial susceptibility changes in children with Helicobacter pylori infection over 13 years in Northern Italy. Gastroenterology Research and Practice. 2015;2015:717349. doi: 10.1155/2015/717349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour et al. (2016).Mansour KB, Fendri C, Battikh H, Garnier M, Zribi M, Jlizi A, Burucoa C. Multiple and mixed Helicobacter pylori infections: comparison of two epidemiological situations in Tunisia and France. Infection, Genetics and Evolution. 2016;37(e43370):43–48. doi: 10.1016/j.meegid.2015.10.028. [DOI] [PubMed] [Google Scholar]
- Marques et al. (2020).Marques AT, Vítor JM, Santos A, Oleastro M, Vale FF. Trends in Helicobacter pylori resistance to clarithromycin: from phenotypic to genomic approaches. Microbial Genomics. 2020;6(3):e000344. doi: 10.1099/mgen.0.000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques et al. (2020).Marques AT, Vítor JMB, Santos A, Oleastro M, Vale FF. Trends in Helicobacter pylori resistance to clarithromycin: from phenotypic to genomic approaches. Microbial Genomics. 2020;6(3):614735. doi: 10.1099/mgen.0.000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascellino et al. (2018).Mascellino MT, Oliva A, De Angelis M, Pontone S, Porowska B. Helicobacter pylori infection: antibiotic resistance and eradication rate in patients with gastritis showing previous treatment failures. New Microbiologica. 2018;41(4):306–309. [PubMed] [Google Scholar]
- Matta, Zambrano & Pazos (2018).Matta AJ, Zambrano DC, Pazos AJ. Punctual mutations in 23S rRNA gene of clarithromycin-resistant Helicobacter pylori in Colombian populations. World Journal of Gastroenterology. 2018;24(14):1531–1539. doi: 10.3748/wjg.v24.i14.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty et al. (2012).McNulty C, Lasseter G, Shaw I, Nichols T, D’Arcy S, Lawson A, Glocker E. Is Helicobacter pylori antibiotic resistance surveillance needed and how can it be delivered? Alimentary Pharmacology & Therapeutics. 2012;35(10):1221–1230. doi: 10.1111/j.1365-2036.2012.05083.x. [DOI] [PubMed] [Google Scholar]
- Mégraud (2004).Mégraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53(9):1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mégraud et al. (2021).Mégraud F, Alix C, Charron P, Bénéjat L, Ducournau A, Besséde E, Lehours P. Survey of the antimicrobial resistance of Helicobacter pylori in France in 2018 and evolution during the previous 5 years. Helicobacter. 2021;26(1):e12767. doi: 10.1111/hel.12767. [DOI] [PubMed] [Google Scholar]
- Megraud et al. (2021).Megraud F, Bruyndonckx R, Coenen S, Wittkop L, Huang T-D, Hoebeke M, Bénéjat L, Lehours P, Goossens H, Glupczynski Y. Helicobacter pylori resistance to antibiotics in Europe in 2018 and its relationship to antibiotic consumption in the community. Gut. 2021;70(10):1815–1822. doi: 10.1136/gutjnl-2021-324032. [DOI] [PubMed] [Google Scholar]
- Megraud et al. (2013).Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62(1):34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- Mi et al. (2021).Mi M, Wu F, Zhu J, Liu F, Cui G, Wen X, Hu Y, Deng Z, Wu X, Zhang Z. Heterogeneity of Helicobacter pylori strains isolated from patients with gastric disorders in Guiyang, China. Infection and Drug Resistance. 2021;14:535–545. doi: 10.2147/IDR.S287631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miendje Deyi et al. (2011).Miendje Deyi VY, Bontems P, Vanderpas J, De Koster E, Ntounda R, Van den Borre C, Cadranel S, Burette A. Multicenter survey of routine determinations of resistance of Helicobacter pylori to antimicrobials over the last 20 years (1990 to 2009) in Belgium. Journal of Clinical Microbiology. 2011;49(6):2200–2209. doi: 10.1128/JCM.02642-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahussurur et al. (2017).Miftahussurur M, Cruz M, Subsomwong P, Abreu JAJ, Hosking C, Nagashima H, Akada J, Yamaoka Y. Clarithromycin-based triple therapy is still useful as an initial treatment for Helicobacter pylori infection in the Dominican Republic. The American Journal of Tropical Medicine and Hygiene. 2017;96(5):1050. doi: 10.4269/ajtmh.16-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahussurur et al. (2016).Miftahussurur M, Shrestha PK, Subsomwong P, Sharma RP, Yamaoka Y. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiology. 2016;16(1):256. doi: 10.1186/s12866-016-0873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahussurur et al. (2016).Miftahussurur M, Syam AF, Nusi IA, Makmun D, Waskito LA, Zein LH, Akil F, Uwan WB, Simanjuntak D, Wibawa IDN. Surveillance of Helicobacter pylori antibiotic susceptibility in Indonesia: different resistance types among regions and with novel genetic mutations. PLOS ONE. 2016;11(12):e0166199. doi: 10.1371/journal.pone.0166199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani et al. (2012).Milani M, Ghotaslou R, Somi MH, Rafeey M, Akhi MT, Nahaei MR, Hasani A, Sharifi Y. The status of antimicrobial resistance of Helicobacter pylori in Eastern Azerbaijan, Iran: comparative study according to demographics. Journal of Infection and Chemotherapy. 2012;18(6):848–852. doi: 10.1007/s10156-012-0425-4. [DOI] [PubMed] [Google Scholar]
- Mirzaei et al. (2013).Mirzaei N, Poursina F, Faghri J, Talebi M, Khataminezhad MR, Hasanzadeh A, Safaei HG. Prevalence of resistance of Helicobacter pylori strains to selected antibiotics in Isfahan, Iran. Jundishapur Journal of Microbiology. 2013;6(5):1. doi: 10.5812/jjm.6342. [DOI] [Google Scholar]
- Miyata et al. (2021).Miyata E, Kudo T, Ikuse T, Tokita K, Arai N, Oka I, Kyodo R, Sato M, Hosoi K, Jimbo K, Aoyagi Y, Ohtsuka Y, Shimizu T. Eradication therapy for Helicobacter pylori infection based on the antimicrobial susceptibility test in children: a single-center study over 12 years. Helicobacter. 2021;26(1):e12764. doi: 10.1111/hel.12764. [DOI] [PubMed] [Google Scholar]
- Mohammad et al. (2011).Mohammad K, Maryam B, Abbas D, Sadegh G-D. Clarithromycin resistance and 23S rRNA mutations in Helicobacter pylori isolates in Iran. African Journal of Microbiology Research. 2011;5(8):853–856. doi: 10.5897/AJMR10.120. [DOI] [Google Scholar]
- Mokhtar et al. (2019).Mokhtar NA, Muttaqillah NAS, Hussin S, Soh TST. Helicobacter pylori infection: prevalence, demographic characteristics, clarithromycin resistance and evaluation of the in-house rapid urease test in Sungai Buloh Hospital, Malaysia. Sains Malaysiana. 2019;48(12):2675–2682. doi: 10.17576/jsm-2019-4812-08. [DOI] [Google Scholar]
- Montes et al. (2015).Montes M, Villalon FN, Eizaguirre FJ, Delgado M, Muñoz-Seca IM, Fernández-Reyes M, Pérez-Trallero E. Helicobacter pylori infection in children. Antimicrobial resistance and treatment response. Helicobacter. 2015;20(3):169–175. doi: 10.1111/hel.12187. [DOI] [PubMed] [Google Scholar]
- Morilla et al. (2019).Morilla AM, Álvarez-Argüelles ME, Duque JM, Armesto E, Villar H, Melón S. Primary antimicrobial resistance rates and prevalence of Helicobacter pylori infection in the north of Spain. A 13-year retrospective study. Gastroenterology & Hepatology. 2019;42(8):476–485. doi: 10.1016/j.gastrohep.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Morilla et al. (2019).Morilla AM, Álvarez-Argüelles ME, Duque JM, Armesto E, Villar H, Melón S. Primary antimicrobial resistance rates and prevalence of Helicobacter pylori infection in the north of Spain. A 13-year retrospective study. Gastroenterología y Hepatología. 2019;42(8):476–485. doi: 10.1016/j.gastrohep.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Morimoto et al. (2015).Morimoto N, Takeuchi H, Nishida Y, Morisawa M, Yoshikawa T, Morita T, Morimoto M, Sugimoto C, Matsumura Y, Sugiura T. Clinical application of the diversilab microbial typing system using repetitive sequence-based PCR for characterization of Helicobacter pylori in Japan. Journal of Clinical Laboratory Analysis. 2015;29(3):250–253. doi: 10.1002/jcla.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosites et al. (2018).Mosites E, Bruden D, Morris J, Reasonover A, Rudolph K, Hurlburt D, Hennessy T, McMahon B, Bruce M. Antimicrobial resistance among Helicobacter pylori isolates in Alaska, 2000–2016. Journal of Global Antimicrobial Resistance. 2018;15(Suppl 1):148–153. doi: 10.1016/j.jgar.2018.06.016. [DOI] [PubMed] [Google Scholar]
- Nishizawa & Suzuki (2014).Nishizawa T, Suzuki H. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Frontiers in Molecular Biosciences. 2014;1:19. doi: 10.3389/fmolb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, Gales & Kawakami (2014).Ogata SK, Gales AC, Kawakami E. Antimicrobial susceptibility testing for Helicobacter pylori isolates from Brazilian children and adolescents: comparing agar dilution, E-test, and disk diffusion. Brazilian Journal of Microbiology. 2014;45(4):1439–1448. doi: 10.1590/S1517-83822014000400039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata et al. (2013).Ogata SK, Godoy APO, da Silva Patricio FR, Kawakami E. High Helicobacter pylori resistance to metronidazole and clarithromycin in Brazilian children and adolescents. Journal of Pediatric Gastroenterology and Nutrition. 2013;56(6):645–648. doi: 10.1097/MPG.0b013e31828b3669. [DOI] [PubMed] [Google Scholar]
- Okuda et al. (2017).Okuda M, Kikuchi S, Mabe K, Osaki T, Kamiya S, Fukuda Y, Kato M. Nationwide survey of Helicobacter pylori treatment for children and adolescents in Japan. Pediatrics International. 2017;59(1):57–61. doi: 10.1111/ped.13038. [DOI] [PubMed] [Google Scholar]
- Oleastro et al. (2011).Oleastro M, Cabral J, Ramalho PM, Lemos PS, Paixão E, Benoliel J, Santos A, Lopes AI. Primary antibiotic resistance of Helicobacter pylori strains isolated from Portuguese children: a prospective multicentre study over a 10 year period. Journal of Antimicrobial Chemotherapy. 2011;66(10):2308–2311. doi: 10.1093/jac/dkr293. [DOI] [PubMed] [Google Scholar]
- Omar et al. (2014).Omar M, Crowe A, Tay CY, Hughes J. Expressions of P-glycoprotein in treatment-resistant Helicobacter pylori patients. Journal of Applied Biomedicine. 2014;12(4):263–269. doi: 10.1016/j.jab.2014.02.001. [DOI] [Google Scholar]
- Oporto et al. (2019).Oporto M, Pavez M, Troncoso C, Cerda A, Hofmann E, Sierralta A, Rios E, Coppelli L, Barrientos L. Prevalence of infection and antibiotic susceptibility of Helicobacter pylori: an evaluation in public and private health systems of Southern Chile. Pathogens. 2019;8(4):226. doi: 10.3390/pathogens8040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz et al. (2019).Ortiz V, Estevez-Ordonez D, Montalvan-Sanchez E, Urrutia-Argueta S, Israel D, Krishna US, Romero-Gallo J, Wilson KT, Peek RM, Dominguez R, Morgan DR. Helicobacter pylori antimicrobial resistance and antibiotic consumption in the low-resource Central America setting. Helicobacter. 2019;24(4):e12595. doi: 10.1111/hel.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otth et al. (2011).Otth L, Wilson M, Fernández H, Otth C, Toledo C, Cárcamo V, Rivera P, Ruiz L. Isolation of Helicobacter pylori in gastric mucosa and susceptibility to five antimicrobial drugs in Southern Chile. Brazilian Journal of Microbiology. 2011;42(2):442–447. doi: 10.1590/S1517-83822011000200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmitessa et al. (2020).Palmitessa V, Monno R, Panarese A, Cuppone R, Burattini O, Marangi S, Curlo M, Fumarola L, Petrosillo A, Parisi A, Capozzi L, Bianco A, Lippolis A. Evaluation of antibiotic resistance of Helicobacter pylori strains isolated in Bari, Southern Italy, in 2017-2018 by phenotypic and genotyping methods. Microbial Drug Resistance. 2020;26(8):909–917. doi: 10.1089/mdr.2019.0262. [DOI] [PubMed] [Google Scholar]
- Pandya et al. (2014).Pandya HB, Agravat HH, Patel JS, Sodagar NRK. Emerging antimicrobial resistance pattern of Helicobacter pylori in central Gujarat. Indian Journal of Medical Microbiology. 2014;32(4):408–413. doi: 10.4103/0255-0857.142256. [DOI] [PubMed] [Google Scholar]
- Park et al. (2016).Park JY, Dunbar KB, Mitui M, Arnold CA, Lam-Himlin DM, Valasek MA, Thung I, Okwara C, Coss E, Cryer B, Doern CD. Helicobacter pylori clarithromycin resistance and treatment failure are common in the USA. Digestive Diseases and Sciences. 2016;61(8):2373–2380. doi: 10.1007/s10620-016-4091-8. [DOI] [PubMed] [Google Scholar]
- Park et al. (2020).Park JY, Shin T-S, Kim JH, Yoon HJ, Kim BJ, Kim JG. The prevalence of multidrug resistance of Helicobacter pylori and its impact on eradication in Korea from 2017 to 2019: a single-center study. Antibiotics. 2020;9(10):646. doi: 10.3390/antibiotics9100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Sepúlveda et al. (2019).Parra-Sepúlveda C, Merino JS, Sáez-Carrillo K, González C, García-Cancino A. Antibiotic resistance surveillance of Helicobacter pylori at the Biobío region (Chile) in a decade. Arquivos de Gastroenterologia. 2019;56(4):361–366. doi: 10.1590/s0004-2803.201900000-72. [DOI] [PubMed] [Google Scholar]
- Peng et al. (2017).Peng X, Song Z, He L, Lin S, Gong Y, Sun L, Zhao F, Gu Y, You Y, Zhou L, Zhang J. Gastric juice-based real-time PCR for tailored Helicobacter pylori treatment: a practical approach. International Journal of Medical Sciences. 2017;14(6):595–601. doi: 10.7150/ijms.18996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz et al. (2014).Peretz A, Paritsky M, Nasser O, Brodsky D, Glyatman T, Segal S, On A. Resistance of Helicobacter pylori to tetracycline, amoxicillin, clarithromycin and metronidazole in Israeli children and adults. The Journal of Antibiotics. 2014;67(8):555–557. doi: 10.1038/ja.2014.38. [DOI] [PubMed] [Google Scholar]
- Phan et al. (2015).Phan TN, Santona A, Tran TNH, Cappuccinelli P, Rubino S, Paglietti B. High rate of levofloxacin resistance in a background of clarithromycin-and metronidazole-resistant Helicobacter pylori in Vietnam. International Journal of Antimicrobial Agents. 2015;45(3):244–248. doi: 10.1016/j.ijantimicag.2014.10.019. [DOI] [PubMed] [Google Scholar]
- Pichon et al. (2020).Pichon M, Tran CT, Motillon G, Debiais C, Gautier S, Aballea M, Cremniter J, Vasseur P, Tougeron D, Garcia M. Where to biopsy to detect Helicobacter pylori and how many biopsies are needed to detect antibiotic resistance in a human stomach. Journal of Clinical Medicine. 2020;9(9):2812. doi: 10.3390/jcm9092812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picoli et al. (2014).Picoli SU, Mazzoleni LE, Fernández H, De Bona LR, Neuhauss E, Longo L, Prolla JC. Resistance to amoxicillin, clarithromycin and ciprofloxacin of Helicobacter pylori isolated from Southern Brazil patients. Revista do Instituto de Medicina Tropical de São Paulo. 2014;56(3):197–200. doi: 10.1590/S0036-46652014000300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pormohammad, Nasiri & Azimi (2019).Pormohammad A, Nasiri MJ, Azimi T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: a systematic review and meta-analysis. Infection and Drug Resistance. 2019;12:1181. doi: 10.2147/IDR.S215329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaf et al. (2017).Raaf N, Amhis W, Saoula H, Abid A, Nakmouche M, Balamane A, Ali Arous N, Ouar-Korichi M, Vale FF, Bénéjat L. Prevalence, antibiotic resistance, and MLST typing of Helicobacter pylori in Algiers. Algeria Helicobacter. 2017;22(6):e12446. doi: 10.1111/hel.12446. [DOI] [PubMed] [Google Scholar]
- Ranjbar & Chehelgerdi (2018).Ranjbar R, Chehelgerdi M. Genotyping and antibiotic resistance properties of Helicobacter pylori strains isolated from human and animal gastric biopsies. Infection and Drug Resistance. 2018;11:2545–2554. doi: 10.2147/IDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed et al. (2014).Rasheed F, Campbell BJ, Alfizah H, Varro A, Zahra R, Yamaoka Y, Pritchard DM. Analysis of clinical isolates of Helicobacter pylori in Pakistan reveals high degrees of pathogenicity and high frequencies of antibiotic resistance. Helicobacter. 2014;19(5):387–399. doi: 10.1111/hel.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnath et al. (2017).Regnath T, Raecke O, Enninger A, Ignatius R. Increasing metronidazole and rifampicin resistance of Helicobacter pylori isolates obtained from children and adolescents between 2002 and 2015 in southwest Germany. Helicobacter. 2017;22(1):e12327. doi: 10.1111/hel.12327. [DOI] [PubMed] [Google Scholar]
- Rezaei, Abadi & Mobarez (2020).Rezaei S, Abadi ATB, Mobarez AM. Metronidazole-resistant Helicobacter pylori isolates without rdxA mutations obtained from Iranian dyspeptic patients. New Microbes and New Infections. 2020;34(7):100636. doi: 10.1016/j.nmni.2019.100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghifard et al. (2013).Sadeghifard N, Seidnazari T, Ghafourian S, Soleimani M, Maleki A, Qomi MA, Sekawi Z. Survey in Iran of clarithromycin resistance in Helicobacter pylori isolates by PCR-RFLP. Southeast Asian Journal of Tropical Medicine & Public Health. 2013;44(1):89–95. [PubMed] [Google Scholar]
- Sanches et al. (2016).Sanches BS, Martins GM, Lima K, Cota B, Moretzsohn LD, Ribeiro LT, Breyer HP, Maguilnik I, Maia AB, Rezende-Filho J, Meira AC, Pinto H, Alves E, Mascarenhas R, Passos R, de Souza JD, Trindade OR, Coelho LG. Detection of Helicobacter pylori resistance to clarithromycin and fluoroquinolones in Brazil: a national survey. World Journal of Gastroenterology. 2016;22(33):7587. doi: 10.3748/wjg.v22.i33.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saniee et al. (2018).Saniee P, HosseinI F, Kadkhodaei S, Siavoshi F, Khalili-Saman S. Helicobacter pylori multidrug resistance due to misuse of antibiotics in Iran. Archives of Iranian Medicine. 2018;21(7):283–288. [PubMed] [Google Scholar]
- Saracino et al. (2020).Saracino IM, Fiorini G, Zullo A, Pavoni M, Saccomanno L, Vaira D. Trends in primary antibiotic resistance in H. pylori strains isolated in Italy between 2009 and 2019. Antibiotics. 2020;9(1):26. doi: 10.3390/antibiotics9010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracino et al. (2020).Saracino IM, Pavoni M, Zullo A, Fiorini G, Saccomanno L, Lazzarotto T, Antonelli G, Cavallo R, Borghi C, Vaira D. Rifabutin-based triple therapy or bismuth-based quadruple regimen as rescue therapies for Helicobacter pylori infection. European Journal of Internal Medicine. 2020;81(11):50–53. doi: 10.1016/j.ejim.2020.06.029. [DOI] [PubMed] [Google Scholar]
- Savari et al. (2010).Savari M, Abdolahi H, Zahedi M, Darvishmoghadam S, Hayat Bakhshe Abasi M. Antibiotic-resistance patterns of Helicobacter pylori isolates obtained from patients in Kerman-2009. Journal of Kerman University of Medical Sciences. 2010;17(1):73–82. [Google Scholar]
- Savoldi et al. (2018).Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli EJG. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology. 2018;155(5):1372–1382.e17. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seck et al. (2013).Seck A, Burucoa C, Dia D, Mbengue M, Onambele M, Raymond J, Breurec S. Primary antibiotic resistance and associated mechanisms in Helicobacter pylori isolates from Senegalese patients. Annals of Clinical Microbiology and Antimicrobials. 2013;12(1):1–5. doi: 10.1186/1476-0711-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selgrad et al. (2013).Selgrad M, Meissle J, Bornschein J, Kandulski A, Langner C, Varbanova M, Wex T, Tammer I, Schluter D, Malfertheiner P. Antibiotic susceptibility of Helicobacter pylori in central Germany and its relationship with the number of eradication therapies. European Journal of Gastroenterology & Hepatology. 2013;25(11):1257–1260. doi: 10.1097/MEG.0b013e3283643491. [DOI] [PubMed] [Google Scholar]
- Seo et al. (2013).Seo JH, Jun JS, Yeom JS, Park JS, Youn HS, Ko GH, Baik SC, Lee WK, Cho MJ, Rhee KH. Changing pattern of antibiotic resistance of Helicobacter pylori in children during 20 years in Jinju, South Korea. Pediatrics International. 2013;55(3):332–336. doi: 10.1111/ped.12048. [DOI] [PubMed] [Google Scholar]
- Shao et al. (2018).Shao Y, Lu R, Yang Y, Xu Q, Wang B, Ye G. Antibiotic resistance of Helicobacter pylori to 16 antibiotics in clinical patients. Journal of Clinical Laboratory Analysis. 2018;32(4):e22339. doi: 10.1002/jcla.22339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty et al. (2019).Shetty V, Lamichhane B, Tay CY, Pai GC, Lingadakai R, Balaraju G, Shetty S, Ballal M, Chua EG. High primary resistance to metronidazole and levofloxacin, and a moderate resistance to clarithromycin in Helicobacter pylori isolated from Karnataka patients. Gut Pathogens. 2019;11(1):1–8. doi: 10.1186/s13099-019-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Jiang & Zhao (2016).Shi J, Jiang Y, Zhao Y. Promising in vitro and in vivo inhibition of multidrug-resistant Helicobacter pylori by linezolid and novel oxazolidinone analogues. Journal of Global Antimicrobial Resistance. 2016;7:106–109. doi: 10.1016/j.jgar.2016.07.016. [DOI] [PubMed] [Google Scholar]
- Shiota et al. (2015).Shiota S, Reddy R, Alsarraj A, El-Serag HB, Graham DY. Antibiotic resistance of Helicobacter pylori among male United States veterans. Clinical Gastroenterology and Hepatology. 2015;13(9):1616–1624. doi: 10.1016/j.cgh.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuely et al. (2020).Shmuely H, Topaz S, Berdinstein R, Yahav J, Melzer E. High metronidazole and clarithromycin resistance of Helicobacter pylori isolated from previously treated and naïve patients. The Israel Medical Association Journal: IMAJ. 2020;22(10):562–566. [PubMed] [Google Scholar]
- Shokrzadeh et al. (2015).Shokrzadeh L, Alebouyeh M, Mirzaei T, Farzi N, Zali MR. Prevalence of multiple drug-resistant Helicobacter pylori strains among patients with different gastric disorders in Iran. Microbial Drug Resistance. 2015;21(1):105–110. doi: 10.1089/mdr.2014.0081. [DOI] [PubMed] [Google Scholar]
- Shokrzadeh et al. (2011).Shokrzadeh L, Jafari F, Dabiri H, Baghaei K, Zojaji H, Alizadeh AH, Aslani MM, Zali MR. Antibiotic susceptibility profile of Helicobacter pylori isolated from the dyspepsia patients in Tehran. Saudi Journal of Gastroenterology. 2011;17(4):261–264. doi: 10.4103/1319-3767.82581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu et al. (2018).Shu X, Yin G, Liu M, Peng K, Zhao H, Jiang M. Antibiotics resistance of Helicobacter pylori in children with upper gastrointestinal symptoms in Hangzhou, China. Helicobacter. 2018;23(3):e12481. doi: 10.1111/hel.12481. [DOI] [PubMed] [Google Scholar]
- Siavoshi, Saniee & Malekzadeh (2018).Siavoshi F, Saniee P, Malekzadeh R. Effective antimicrobial activity of rifabutin against multidrug-resistant Helicobacter pylori. Helicobacter. 2018;23(6):e12531. doi: 10.1111/hel.12531. [DOI] [PubMed] [Google Scholar]
- Siddiqui et al. (2016).Siddiqui TR, Ahmed W, Arif A, Bibi S, Khan A. Emerging trends of antimicrobial resistance in Helicobacter pylori isolates obtained from Pakistani patients: the need for consideration of amoxicillin and clarithromycin. Journal of Pakistan Medical Association. 2016;66(6):710–716. [PubMed] [Google Scholar]
- Silva et al. (2018).Silva GM, Silva HM, Nascimento J, Goncalves JP, Pereira F, Lima R. Helicobacter pylori antimicrobial resistance in a pediatric population. Helicobacter. 2018;23(5):e12528. doi: 10.1111/hel.12528. [DOI] [PubMed] [Google Scholar]
- Song et al. (2014).Song Z, Zhang J, He L, Chen M, Hou X, Li Z, Zhou L. Prospective multi-region study on primary antibiotic resistance of Helicobacter pylori strains isolated from Chinese patients. Digestive and Liver Disease. 2014;46(12):1077–1081. doi: 10.1016/j.dld.2014.08.038. [DOI] [PubMed] [Google Scholar]
- Su et al. (2022).Su D-J, Chang M-H, Yang J-C, Ni Y-H, Hsu H-Y, Wu J-F. Fourteen-day sequential therapy is superior to 7-day triple therapy as first-line regimen for Helicobacter pylori infected children. Journal of the Formosan Medical Association. 2022;121(1):202–209. doi: 10.1016/j.jfma.2021.03.001. [DOI] [PubMed] [Google Scholar]
- Su et al. (2013).Su P, Li Y, Li H, Zhang J, Lin L, Wang Q, Guo F, Ji Z, Mao J, Tang W, Shi Z, Shao W, Mao J, Zhu X, Zhang X, Tong Y, Tu H, Jiang M, Wang Z, Jin F, Yang N, Zhang J. Antibiotic resistance of Helicobacter pylori isolated in the Southeast Coastal Region of China. Helicobacter. 2013;18(4):274–279. doi: 10.1111/hel.12046. [DOI] [PubMed] [Google Scholar]
- Sugimoto et al. (2020).Sugimoto M, Hira D, Murata M, Kawai T, Terada T. Effect of antibiotic susceptibility and CYP3A4/5 and CYP2C19 genotype on the outcome of vonoprazan-containing Helicobacter pylori eradication therapy. Antibiotics. 2020;9(10):645. doi: 10.3390/antibiotics9100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto et al. (2017).Sugimoto M, Sahara S, Ichikawa H, Kagami T, Ban H, Otsuka T, Andoh A, Furuta T. Four-times-daily dosing of rabeprazole with sitafloxacin, high-dose amoxicillin, or both for metronidazole-resistant infection with Helicobacter pylori in Japan. Helicobacter. 2017;22(1):e12319. doi: 10.1111/hel.12319. [DOI] [PubMed] [Google Scholar]
- Sugimoto et al. (2014).Sugimoto M, Uotani T, Sahara S, Ichikawa H, Yamade M, Sugimoto K, Furuta T. Efficacy of tailored Helicobacter pylori eradication treatment based on clarithromycin susceptibility and maintenance of acid secretion. Helicobacter. 2014;19(4):312–318. doi: 10.1111/hel.12128. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2018).Sun L, Talarico S, Yao L, He L, Self S, You Y, Zhang H, Zhang Y, Guo Y, Liu G, Salama NR, Zhang J. Droplet digital PCR-based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. Journal of Clinical Microbiology. 2018;56(9):e00019–18. doi: 10.1128/JCM.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szadkowski, Zemlak & Muszynski (2018).Szadkowski A, Zemlak M, Muszynski J. Effectiveness of Helicobacter pylori eradication established on the basis of examination of antibiotic resistance of the bacteria. Przeglad Gastroenterology Review. 2018;13(2):93–98. doi: 10.5114/pg.2018.75821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebi Bezmin Abadi et al. (2012).Talebi Bezmin Abadi A, Ghasemzadeh A, Taghvaei T, Mobarez AM. Primary resistance of Helicobacter pylori to levofloxacin and moxifloxacine in Iran. Internal and Emergency Medicine. 2012;7(5):447–452. doi: 10.1007/s11739-011-0563-1. [DOI] [PubMed] [Google Scholar]
- Tamayo et al. (2017).Tamayo E, Montes M, Fernandez-Reyes M, Lizasoain J, Ibarra B, Mendarte U, Zapata E, Mendiola J, Perez-Trallero E. Clarithromycin resistance in Helicobacter pylori and its molecular determinants in Northern Spain, 2013–2015. Journal of Global Antimicrobial Resistance. 2017;9:43–46. doi: 10.1016/j.jgar.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Tanabe et al. (2018).Tanabe H, Yoshino K, Ando K, Nomura Y, Ohta K, Satoh K, Ichiishi E, Ishizuka A, Otake T, Kohgo Y. Vonoprazan-based triple therapy is non-inferior to susceptibility-guided proton pump inhibitor-based triple therapy for Helicobacter pylori eradication. Annals of Clinical Microbiology and Antimicrobials. 2018;17(1):1–7. doi: 10.1186/s12941-018-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2020).Tang X, Chen X, Shen Y, Yang T, Hu R, Debowski AW, Stubbs KA, Benghezal M, Marshall BJ, Li H. Primary antibiotic resistance of Helicobacter pylori among a Chinese Tibetan population. Future Microbiology. 2020;15(14):1353–1361. doi: 10.2217/fmb-2020-0206. [DOI] [PubMed] [Google Scholar]
- Tang et al. (2020).Tang X, Shen Y, Hu R, Yang T, Benghezal M, Li H, Tang H. Re-assessment of the disk diffusion technique for routine antimicrobial susceptibility testing for Helicobacter pylori. Helicobacter. 2020;25(4):e12703. doi: 10.1111/hel.12703. [DOI] [PubMed] [Google Scholar]
- Tanih, Ndip & Ndip (2011).Tanih NF, Ndip LM, Ndip RN. Characterisation of the genes encoding resistance to metronidazole (rdxA and frxA) and clarithromycin (the 23S-rRNA genes) in South African isolates of Helicobacter pylori. Annals of Tropical Medicine & Parasitology. 2011;105(3):251–259. doi: 10.1179/136485911X12899838683485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh et al. (2014).Teh X, Khosravi Y, Lee WC, Leow AH, Loke MF, Vadivelu J, Goh KL. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLOS ONE. 2014;9(7):e101481. doi: 10.1371/journal.pone.0101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trespalacios et al. (2013).Trespalacios AA, Otero W, Caminos JE, Mercado MM, Ávila J, Rosero LE, Arévalo A, Poutou-Piñales RA, Graham DY. Phenotypic and genotypic analysis of clarithromycin-resistant Helicobacter pylori from Bogotá DC, Colombia. Journal of Microbiology. 2013;51(4):448–452. doi: 10.1007/s12275-013-2465-6. [DOI] [PubMed] [Google Scholar]
- Trespalacios et al. (2015).Trespalacios AA, Rimbara E, Otero W, Reddy R, Graham DY. Improved allele-specific PCR assays for detection of clarithromycin and fluoroquinolone resistant of Helicobacter pylori in gastric biopsies: identification of N87I mutation in GyrA. Diagnostic Microbiology and Infectious Disease. 2015;81(4):251–255. doi: 10.1016/j.diagmicrobio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay et al. (2012).Tsay FW, Tseng HH, Hsu PI, Wang KM, Lee CC, Chang SN, Wang HM, Yu HC, Chen WC, Peng NJ. Sequential therapy achieves a higher eradication rate than standard triple therapy in Taiwan. Journal of Gastroenterology and Hepatology. 2012;27(3):498–503. doi: 10.1111/j.1440-1746.2011.06885.x. [DOI] [PubMed] [Google Scholar]
- Tshibangu-Kabamba et al. (2020).Tshibangu-Kabamba E, Ngoma-Kisoko PDJ, Tuan VP, Matsumoto T, Akada J, Kido Y, Tshimpi-Wola A, Tshiamala-Kashala P, Ahuka-Mundeke S, Mumba Ngoy D. Next-generation sequencing of the whole bacterial genome for tracking molecular insight into the broad-spectrum antimicrobial resistance of Helicobacter pylori clinical isolates from the Democratic Republic of Congo. Microorganisms. 2020;8(6):887. doi: 10.3390/microorganisms8060887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan et al. (2019).Tuan VP, Narith D, Tshibangu-Kabamba E, Dung HDQ, Viet PT, Sokomoth S, Binh TT, Sokhem S, Tri TD, Ngov S, Tung PH, Thuan NPM, Truc TC, Phuc BH, Matsumoto T, Fauzia KA, Akada J, Trang TTH, Yamaoka Y. A next-generation sequencing-based approach to identify genetic determinants of antibiotic resistance in Cambodian Helicobacter pylori clinical isolates. Journal of Clinical Medicine. 2019;8(6):858. doi: 10.3390/jcm8060858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tveit et al. (2011).Tveit AH, Bruce MG, Bruden DL, Morris J, Reasonover A, Hurlburt DA, Hennessy TW, McMahon B. Alaska sentinel surveillance study of Helicobacter pylori isolates from Alaska Native persons from 2000 to 2008. Journal of Clinical Microbiology. 2011;49(10):3638–3643. doi: 10.1128/JCM.01067-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vala et al. (2016).Vala MH, Eyvazi S, Goudarzi H, Sarie HR, Gholami M. Evaluation of clarithromycin resistance among Iranian Helicobacter pylori isolates by E-test and real-time polymerase chain reaction methods. Jundishapur Journal of Microbiology. 2016;9(5):e29839. doi: 10.5812/jjm.29839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazirzadeh et al. (2020).Vazirzadeh J, Falahi J, Moghim S, Narimani T, Rafiei R, Karbasizadeh V. Molecular assessment of resistance to clarithromycin in Helicobacter pylori strains isolated from patients with dyspepsia by fluorescent in situ hybridization in the center of Iran. BioMed Research International. 2020;2020(1):1–7. doi: 10.1155/2020/2304173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vécsei et al. (2011).Vécsei A, Innerhofer A, Graf U, Binder C, Giczi H, Hammer K, Bruckdorfer A, Hirschl AM, Makristathis A. Helicobacter pylori eradication rates in children upon susceptibility testing based on noninvasive stool polymerase chain reaction versus gastric tissue culture. Journal of Pediatric Gastroenterology and Nutrition. 2011;53(1):65–70. doi: 10.1097/MPG.0b013e318210586d. [DOI] [PubMed] [Google Scholar]
- Vekens et al. (2013).Vekens K, Vandebosch S, De Bel A, Urbain D, Mana F. Primary antimicrobial resistance of Helicobacter pylori in Belgium. Acta Clinica Belgica. 2013;68(3):183–187. doi: 10.2143/ACB.3233. [DOI] [PubMed] [Google Scholar]
- Vilaichone et al. (2020).Vilaichone R-K, Aumpan N, Ratanachu-ek T, Uchida T, Tshering L, Mahachai V, Yamaoka Y. Population-based study of Helicobacter pylori infection and antibiotic resistance in Bhutan. International Journal of Infectious Diseases. 2020;97(38):102–107. doi: 10.1016/j.ijid.2020.05.077. [DOI] [PubMed] [Google Scholar]
- Vilaichone et al. (2013).Vilaichone R-K, Gumnarai P, Ratanachu-ek T, Mahachai V. Nationwide survey of Helicobacter pylori antibiotic resistance in Thailand. Diagnostic Microbiology and Infectious Disease. 2013;77(4):346–349. doi: 10.1016/j.diagmicrobio.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Vilaichone et al. (2016).Vilaichone RK, Ratanachu-Ek T, Gamnarai P, Chaithongrat S, Uchida T, Yamaoka Y, Mahachai V. Extremely high prevalence of metronidazole-resistant Helicobacter pylori strains in mountain people (Karen and Hmong) in Thailand. The American Journal of Tropical Medicine and Hygiene. 2016;94(4):717–720. doi: 10.4269/ajtmh.15-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2019).Wang D, Guo Q, Yuan Y, Gong Y. The antibiotic resistance of Helicobacter pylori to five antibiotics and influencing factors in an area of China with a high risk of gastric cancer. BMC Microbiology. 2019;19(1):1–10. doi: 10.1186/s12866-019-1517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2020).Wang YH, Wang FF, Gong XL, Yan LL, Zhao QY, Song YP, Zhao RL, He YJ, Zhou L, Liu DS, Xie Y. Genotype profiles of Helicobacter pylori from gastric biopsies and strains with antimicrobial-induced resistance. Therapeutic Advances in Gastroenterology. 2020;13:1756284820952596. doi: 10.1177/1756284820952596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2018).Wang B, Zhao Q, Yin W, Yuan Y, Wang X, Wang Y-H, Wang H, Ye W, Chen S, Guo H-l. In-vitro characterisation of a novel antimicrobial agent, TNP-2092, against Helicobacter pylori clinical isolates. Swiss Medical Weekly. 2018;148(2930):w14630. doi: 10.4414/smw.2018.14630. [DOI] [PubMed] [Google Scholar]
- Wu et al. (2015).Wu IT, Chuah SK, Lee CH, Liang CM, Lu LS, Kuo YH, Yen YH, Hu ML, Chou YP, Yang SC, Kuo CM, Kuo CH, Chien CC, Chiang YS, Chiou SS, Hu TH, Tai WC. Five-year sequential changes in secondary antibiotic resistance of Helicobacter pylori in Taiwan. World Journal of Gastroenterology. 2015;21(37):10669–10674. doi: 10.3748/wjg.v21.i37.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2014).Wu JY, Wang SS, Lee YC, Yamaoka Y, Graham DY, Jan CM, Wang WM, Wu DC. Detection of genotypic clarithromycin-resistant Helicobacter pylori by string tests. World Journal of Gastroenterology. 2014;20(12):3343–3349. doi: 10.3748/wjg.v20.i12.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüppenhorst et al. (2014).Wüppenhorst N, Draeger S, Stüger HP, Hobmaier B, Vorreiter J, Kist M, Glocker E-O, ResiNet Study Group Prospective multicentre study on antimicrobial resistance of Helicobacter pylori in Germany. Journal of Antimicrobial Chemotherapy. 2014;69(11):3127–3133. doi: 10.1093/jac/dku243. [DOI] [PubMed] [Google Scholar]
- Yakoob et al. (2013).Yakoob J, Abid S, Jafri W, Abbas Z, Mumtaz K, Hamid S, Ahmed R. Low rate of recurrence of Helicobacter pylori infection in spite of high clarithromycin resistance in Pakistan. BMC Gastroenterology. 2013;13(1):1–7. doi: 10.1186/1471-230X-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao et al. (2019).Yao C-C, Kuo C-M, Hsu C-N, Yang S-C, Wu C-K, Tai W-C, Liang C-M, Wu K-L, Huang C-F, Bi K-W. First-line Helicobacter pylori eradication rates are significantly lower in patients with than those without type 2 diabetes mellitus. Infection and Drug Resistance. 2019;12:1425. doi: 10.2147/IDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeganeh et al. (2019).Yeganeh M, Paritsky M, On A, Azrad M, Roshrosh H, Moalem R, Peretz A. Characteristics of antibiotic resistance of Helicobacter pylori among adult Arab and Jewish populations in Northern Israel. Microbial Drug Resistance. 2019;25(1):103–107. doi: 10.1089/mdr.2018.0148. [DOI] [PubMed] [Google Scholar]
- Yin et al. (2020).Yin G, Bie S, Gu H, Shu X, Zheng W, Peng K, Zhao H, Li F, Chen B, Botchway BOA, Fang M, Jiang M. Application of gene chip technology in the diagnostic and drug resistance detection of Helicobacter pylori in children. Journal of Gastroenterology and Hepatology. 2020;35(8):1331–1339. doi: 10.1111/jgh.14980. [DOI] [PubMed] [Google Scholar]
- Yoon et al. (2014).Yoon K-H, Park SW, Lee SW, Kim BJ, Kim JG. Clarithromycin-based standard triple therapy can still be effective for Helicobacter pylori eradication in some parts of the Korea. Journal of Korean Medical Science. 2014;29(9):1240–1246. doi: 10.3346/jkms.2014.29.9.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yula et al. (2013).Yula E, Nagiyev T, Kaya ÖA, İnci M, Çelik MM, Köksal F. Detection of primary clarithromycin resistance of Helicobacter pylori and association between cagA+ status and clinical outcome. Folia Microbiologica. 2013;58(2):141–146. doi: 10.1007/s12223-012-0192-8. [DOI] [PubMed] [Google Scholar]
- Zerbetto De Palma et al. (2017).Zerbetto De Palma G, Mendiondo N, Wonaga A, Viola L, Ibarra D, Campitelli E, Salim N, Corti R, Goldman C, Catalano M. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires City. Microbial Drug Resistance. 2017;23(3):351–358. doi: 10.1089/mdr.2015.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2019).Zhang XY, Shen WX, Chen CF, Sheng HH, Cheng H, Li J, Hu F, Lu DR, Gao HJ. Detection of the clarithromycin resistance of Helicobacter pylori in gastric mucosa by the amplification refractory mutation system combined with quantitative real-time PCR. Cancer Medicine. 2019;8(4):1633–1640. doi: 10.1002/cam4.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2020).Zhang S, Wang X, Wise MJ, He Y, Chen H, Liu A, Huang H, Young S, Tay CY, Marshall BJ. Mutations of Helicobacter pylori RdxA are mainly related to the phylogenetic origin of the strain and not to metronidazole resistance. Journal of Antimicrobial Chemotherapy. 2020;75(11):3152–3155. doi: 10.1093/jac/dkaa302. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2020).Zhang Y, Wen Y, Xiao Q, Zheng W, Long G, Chen B, Shu X, Jiang M. Mutations in the antibiotic target genes related to clarithromycin, metronidazole and levofloxacin resistance in Helicobacter pylori strains from children in China. Infection and Drug Resistance. 2020;13:311. doi: 10.2147/IDR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2018).Zhang J, Zhong J, Ding J, Shi J, Tang T, Liu Q, Huang H, Dai L, Yang N. Simultaneous detection of human CYP2C19 polymorphisms and antibiotic resistance of Helicobacter pylori using a personalised diagnosis kit. Journal of Global Antimicrobial Resistance. 2018;13:174–179. doi: 10.1016/j.jgar.2017.12.018. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang Y-X, Zhou L-Y, Song Z-Q, Zhang J-Z, He L-H, Ding Y. Primary antibiotic resistance of Helicobacter pylori strains isolated from patients with dyspeptic symptoms in Beijing: a prospective serial study. World Journal of Gastroenterology: WJG. 2015;21(9):2786. doi: 10.3748/wjg.v21.i9.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2013).Zhu Z, Huang D, Xie Y, Liu L, Lu N. Characterization of 23S rRNA gene mutation in primary and secondary clarithromycin-resistant Helicobacter pylori strains from East China. Turkish Journal of Gastroenterology. 2013;24(1):5–9. doi: 10.4318/tjg.2013.0525. [DOI] [PubMed] [Google Scholar]
- Zou et al. (2020).Zou Y, Qian X, Liu X, Song Y, Song C, Wu S, An Y, Yuan R, Wang Y, Xie YJH. The effect of antibiotic resistance on Helicobacter pylori eradication efficacy: a systematic review and meta-analysis. Helicobacter. 2020;25(4):e12714. doi: 10.1111/hel.12714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.