Abstract
Cell–cell interactions in the central nervous system play important roles in neurologic diseases. However, little is known about the specific molecular pathways involved, and methods for their systematic identification are limited. Here, we developed a forward genetic screening platform that combines CRISPR-Cas9 perturbations, cell coculture in picoliter droplets, and microfluidic-based fluorescence-activated droplet sorting to identify mechanisms of cell–cell communication. We used SPEAC-seq (systematic perturbation of encapsulated associated cells followed by sequencing), in combination with in vivo genetic perturbations, to identify microglia-produced amphiregulin as a suppressor of disease-promoting astrocyte responses in multiple sclerosis preclinical models and clinical samples. Thus, SPEAC-seq enables the high-throughput systematic identification of cell–cell communication mechanisms.
Multiple sclerosis (MS) is a chronic inflammatory disorder of the central nervous system (CNS) (1). Interactions among CNS-resident glial cells contribute to the pathogenesis of several neurologic diseases, including MS and its preclinical model experimental autoimmune encephalomyelitis (EAE) (2-13). Characterizing astrocyte–microglia interactions has the potential to identify candidate therapeutic targets for neurologic disorders. However, current methods do not causally link cellular cross-talk with molecular states (12, 14-16) and show a limited ability to detect transient cell–cell interactions mediated by surface or secreted factors. Approaches to study communication systematically between two or more cells of interest and identify the specific mechanisms involved are needed.
Forward genetic screening platforms, such as those based on the CRISPR-Cas9 system, are powerful tools to identify genes that control biologic processes of interest (17-24). However, limitations that are linked to the high-throughput coculture and screening of perturbed single cells hamper their use in studying cell–cell interactions. A forward genetic screening platform to study cell communication requires the establishment of cell–cell interactions mediated by surface or secreted factors in a controlled microenvironment. It further requires the detection of phenotypes that result from those interactions and the association of detected phenotypes with specific CRISPR-Cas9–induced genetic perturbations. Here, we report the development of systematic perturbation of encapsulated associated cells followed by sequencing (SPEAC-seq), a high-throughput platform that enables forward genetic screens of cell–cell interaction mechanisms.
Development of a droplet-based forward genetic cell–cell interaction screening platform
To establish a droplet microfluidic platform for the study of cell–cell interactions (Fig. 1A), we optimized the cell culture media to ensure droplet stability over time (fig. S1A) and confirmed that soluble factors did not transfer between droplets in 24 hours (fig. S1, B and C). Microfluidic co-flow of two aqueous suspensions (one per cell type) and oil (fig. S2, A and B) was used to generate picoliter water-in-oil droplets containing cell pairs, which were detected and sorted by using custom three-color optics and a dielectrophoretic microfluidic sorter (25) (Fig. 1, B and C; fig. S2, C and D; and movie S1).
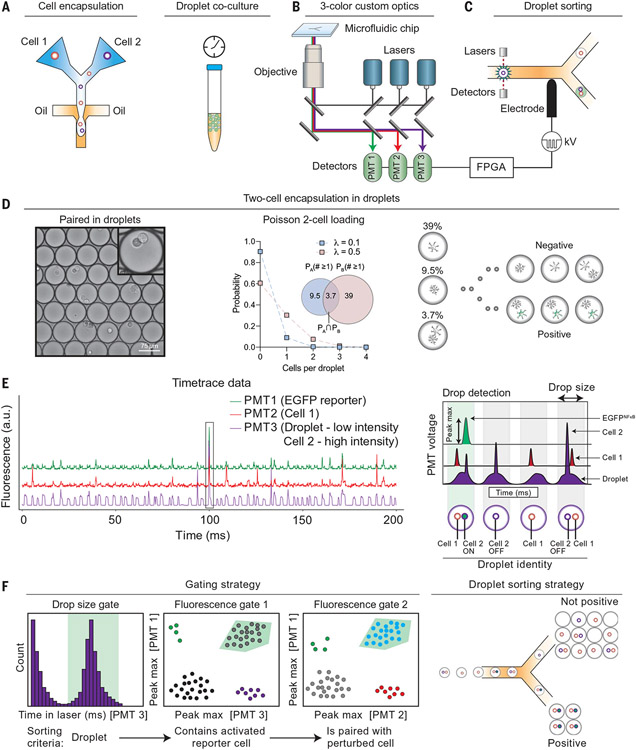
Fig. 1. Detection of cell–cell interactions in picoliter droplet vessels.
(A) Cells are coencapsulated by using microfluidics inside picoliter water-in-oil droplets. (B and C) (B) Cocultured cell pairs are monitored on the basis of their fluorescence by using a three-color custom droplet cytometric system and (C) sorted with dielectrophoresis to isolate cell–cell pairs. FPGA, field programmable gate array; PMT, photomultiplier tube. (D) Cells cocultured within droplets remain isolated from neighboring cell pairs and interact through direct contact and/or secreted soluble factors. Cell loading determines the probability that a drop contains each cell type; cell loading was set to favor a single cell containing a CRISPR-Cas9 perturbation. (E) Droplet cytometric time trace data showing presence of droplet (PMT3, low sustained intensity), cell 2 (PMT 3, sharp intensity peak), EGFP reporter (PMT1), and cell 1 (PMT2). An inert CY5 tracer dye was added to detect and gate drops of the correct size. The schematic (right) shows possible combinations of cell–cell pairings and their corresponding droplet fluorescence traces. a.u., arbitrary unit. (F) Gating strategy showing how cell–cell pairs were identified by sequentially gating drops that (i) were the correct size, (ii) contained an activated reporter cell (astrocyte), and (iii) were paired with the desired cell–cell pair (astrocyte–microglia) and sorted such that only drops containing two-cell combinations were studied.
To validate this system, we first performed a time course analysis of cell survival by loading calcein-labeled cells into droplets and culturing them for 3, 24, 48, or 72 hours at 37°C (fig. S3A). We estimated cell survival rates of 95% and 80% by fluorescence-activated cell sorting and live/dead cell staining at 3 and 24 hours postencapsulation in droplets, respectively, which significantly decreased at 48 and 72 hours postencapsulation (fig. S3A). We then tested whether cells cultured in droplets responded to stimulation using transgenic astrocytes that express enhanced green fluorescent protein (EGFP) after nuclear factor kappa B (NF-κB) activation (26). Droplet-encapsulated NF-κB reporter astrocytes displayed dose-dependent EGFP expression in response to coencapsulation with increasing concentrations of the NF-κB–activating cytokines interleukin-1 beta (IL-1β) and tumor necrosis factor alpha (TNFα) (fig. S3B). We confirmed that astrocytes prestimulated with a subthreshold dose of IL-1β and TNFα (0.1 pg/ml) were susceptible to subsequent activation with proinflammatory cytokines including IL-6 and granulocyte-macrophage colony-stimulating factor (GM-CSF) (fig. S3C), as expected (27). Indeed, we detected NF-κB activation in transgenic NF-κB reporter astrocytes cultured in droplets that were loaded with conditioned media from lipopolysaccharide (LPS)–activated microglia, which contains IL-1β and TNFα (fig. S3, D and E). Time-course analyses detected EGFP expression 4 hours postencapsulation of NF-κB reporter astrocytes with microglia-conditioned media, with higher reporter activation detected 24 hours postencapsulation (fig. S3E).
Subsequently, we extended our studies to cell pairs to determine whether cues produced by one cell were sufficient to alter the cellular state of a cell cocultured in the same droplet. We developed and validated a system for the loading (Fig. 1D) and detection (Fig. 1E) of cell pairs in droplets using multiplexed labeling with cell permeant fluorescent dyes. Next, we optimized droplet-sorting parameters for the isolation of cell pairs that displayed reporter activation (Fig. 1F and fig. S4A). Preliminary experiments detected the up-regulation of EGFP expression in NF-κB reporter astrocytes that were coencapsulated in droplets with activated, but not resting, macrophages (fig. S4B).
SPEAC-seq identifies microglial suppressors of NF-κB signaling in astrocytes
We combined the droplet-based coculture system with CRISPR-Cas9 perturbations to establish SPEAC-seq, a platform for forward genetic screens of regulatory cell–cell interactions. First, we established that droplet-based culture of microglia in the presence of LPS recapitulates the transcriptional effects of microglial activation with LPS in culture plates (fig. S4C and data S1 and S2). In addition, we confirmed that lentiviral transduction or puromycin treatment did not significantly alter the microglial transcriptional responses that are associated with phagocytosis or other signaling pathways (fig. S4, C to H). Next, we confirmed that astrocyte treatment with a subthreshold dose of IL-1β and TNFα followed by incubation in droplets resulted in minimal background activation of NF-κB when astrocytes were cocultured for 24 hours, either with control media or with LPS-activated macrophages or microglia stably transduced with a nontargeting lentiviral CRISPR-Cas9 vector (fig. S5, A to G).
We used SPEAC-seq to identify microglial factors that are involved in the suppression of NF-κB activation in astrocytes (fig. S2, A to C) because NF-κB is an important driver of disease-promoting astrocyte responses (2, 3, 5, 28). We transduced primary mouse microglia with a genome-wide CRISPR-Cas9 library (29) and selected stably transduced cells with puromycin (Fig. 2A). We coencapsulated CRISPR-Cas9–transduced microglia for 24 hours with NF-κB reporter astrocytes suboptimally activated with IL-1β/TNFα (fig. S3B) and then sorted droplets that contained live cell pairs consisting of a microglia cell and an EGFP+ astrocyte (Fig. 2D). We calibrated droplet-sorting gates to capture EGFP reporter activation in astrocytes paired with CRISPR-Cas9–transduced microglia compared with nontargeting controls (Fig. 2D and fig. S5, D to G). We also developed a workflow to isolate single guide RNA (sgRNA) sequences that are stably incorporated into microglial genomic DNA from small numbers of sorted droplets by genomic DNA polymerase chain reaction (PCR) amplification and deep sequencing (Fig. 2E), which enabled the analysis of sgRNAs targeting microglial negative regulators of NF-κB activation in astrocytes. We filtered these SPEAC-seq hits against genes expressed in LPS-activated mouse microglia as detected by RNA sequencing (RNA-seq), which resulted in a list of 1061 candidate molecules (Fig. 2F and data S2 and S3). An analyis of the positive droplet fraction revealed known negative regulators of NF-κB signaling, including xenobiotic metabolism (11, 30), nuclear receptor activation (31, 32), and NRF2 signaling (27), as well as high concordance between sorted drops and the number of guide RNA sequences detected (Fig. 2F; fig. S5, G to K, and S6, A to C; and data S3 and S4). Conversely, the negative droplet fraction contained multiple nontargeting sgRNAs, which highlights the specificity of the droplet-sorting procedure (fig. S5K).
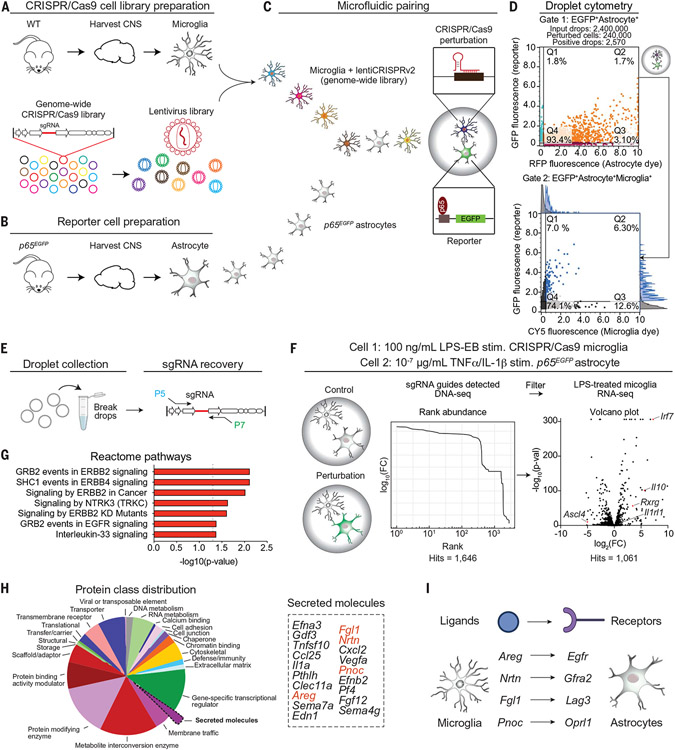
Fig. 2. SPEAC-seq identifies microglial factors that limit astrocyte proinflammatory responses.
(A) Microglia were isolated from wild-type (WT) B6 mice and transduced with a pooled genome-wide lentiviral CRISPR-Cas9 library (78,637 sgRNA sequences) by low-MOI spinfection to generate a single mutation in each cell. (B) Astrocytes were isolated from p65EGFP reporter mice and paired in droplets with a single CRISPR-Cas9–perturbed microglial cell for 24 hours. (C) CRISPR-Cas9–based perturbations in microglia that resulted in NF-κB activation in astrocytes after 24 hours were screened by using a high-throughput microfluidic fluorescence-activated cell sorting platform. (D) Identification of activated cell pairs after 24 hours by using a three-color, dual-gating strategy. Representative gating strategy: The upper gate identifies EGFP+ astrocytes (activated NF-κB), and the bottom gate identifies EGFP+ primary astrocytes paired with a single perturbed microglial cell. Fluorescence histograms in the bottom droplet cytometry panel show the distribution within each channel. RFP, red fluorescent protein. (E) Droplet sorting of cell pairs, genomic DNA extraction, and sgRNA recovery through PCR was used to generate a library for Illumina sequencing. (F) Experimental schematic (left). Analysis of guides detected in the genomic DNA of microglia from sorted droplets containing an EGFP+ astrocyte (middle). SPEAC-seq hits were filtered against an RNA-seq database of LPS-activated primary mouse microglia (right). Volcano plot represents expression of LPS treatment relative to vehicle treatment, n = 3 per group. EB, Escherichia coli 0111:B4; FC, fold change. (G) Pathways detected by SPEAC-seq that limit astrocyte NF-κB activation discovered through bioinformatic analysis. (H and I) Analysis of secreted signals perturbed in microglia enriched in SPEAC-seq data (H) and the cognate astrocyte receptors that transduce the signals of several candidate genes (I).
We then analyzed the SPEAC-seq dataset to identify microglial factors that suppress proinflammatory astrocyte responses. SPEAC-seq detected physiologically relevant candidate molecules expressed by microglia in four independent published bulk or single-cell RNA sequencing (scRNA)–seq microglial datasets (27, 33-35). All but one (1060 out of 1061, 99.9%) of SPEAC-seq hits had been previously detected in microglia (fig. S7, A to C, and data S5). Next, we performed a gene ontology analysis of genes that, upon their CRISPR-Cas9–driven perturbation in microglia, led to NF-κB activation in astrocytes; these analyses identified transcriptional signatures linked to growth factor signaling (Fig. 2G). To focus on candidate proteins involved in regulatory pathways that control cell–cell communication, we analyzed secreted molecules (Fig. 2H) and identified four candidate growth factors expressed by microglia (Areg, Nrtn, Fgl1, and Pnoc) (fig. S7, B to D) that signal through four independent receptors (epidermal growth factor receptor [Egfr], Lag3, Gfra2, Oprl1) expressed by astrocytes (Fig. 2I).
Microglia–astrocyte AREG-EGFR signaling limits astrocyte pathogenic activities in EAE
To evaluate the regulatory role of each candidate pathway uncovered by SPEAC-seq in the context of inflammation, we applied a cell type–specific in vivo Perturb-seq approach (36). We designed lentiviral vectors to coexpress sgRNAs that target receptors of interest with RNA-encoded barcodes in the Cas9 open reading frame (Fig. 3A); the expression of Cas9 and the RNA barcode was driven by the GFAP promoter that is active in astrocytes and detectable by scRNA-seq (12, 13, 27, 37). We designed an sgRNA against each receptor predicted to be activated by the microglial ligands that were identified by SPEAC-seq (Egfr, Gfra2, Lag3, Oprl1), as well as a nontargeting control, and used an equimolar cocktail of each virus to transduce the CNS of mice. After induction of EAE by immunization with MOG35–55, we sorted EGFP+ astrocytes by flow cytometry, performed scRNA-seq (fig. S8, A and B), and analyzed both the sgRNA and transcriptional profile of perturbed cells (Fig. 3B and fig. S8C). When compared to the sgScrmbl control-transduced cells, astrocytes harboring sgRNAs that target Egfr, Gfra2, Lag3, or Oprl1 showed increased NF-κB transcriptional activation (Fig. 3C). However, Egfr targeting led to the strongest activation of IL-1β/TNFα signaling, which promotes NF-κB–driven transcriptional astrocyte responses that are associated with EAE and MS (Fig. 3D; fig. S8, D to F; and data S6) (27). Moreover, Egfr also showed higher expression in astrocytes in previously published scRNA-seq datasets than Gfra2, Lag3, and Oprl1 (27) (fig. S9, A and B). The EGFR ligand identified by SPEAC-seq was Areg, which encodes amphiregulin. Notably, Areg showed higher expression than Nrtn, Fgl1, and Pnoc in stimulated mouse microglia (fig. S7, B and C). Thus, we investigated the effects of AREG-EGFR signaling on microglia–astrocyte interactions.
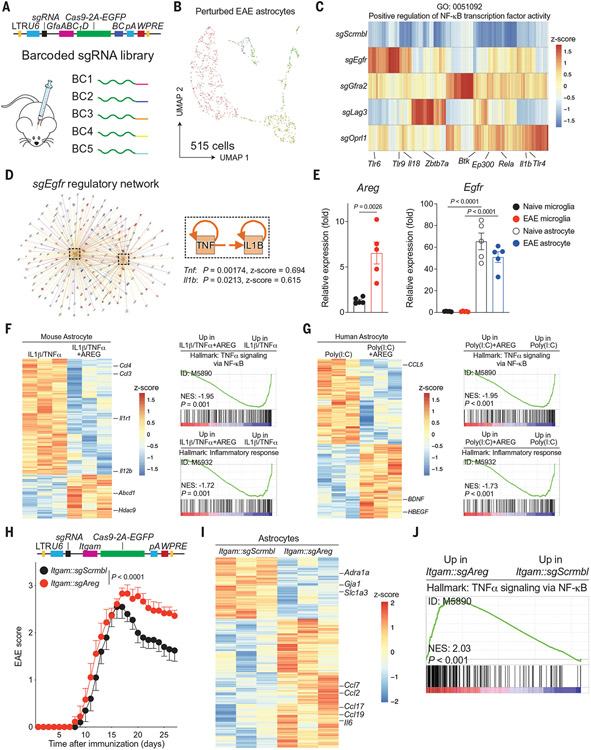
Fig. 3. Microglial AREG limits astrocyte proinflammatory responses.
(A) Construction of a barcoded lentiviral library for in vivo Perturb-seq analysis of candidate astrocyte receptors. (B) UMAP plot of astrocytes captured by Perturb-seq from n = 4 EAE mice. (C) Analysis of NF-κB signaling activation as a function of Perturb-seq–based knockdown of candidate astrocyte receptors. (D) Qiagen IPA network analysis showing that EGFR signaling limits TNFα and IL-1β-driven NF-κB signals. Right-tailed Fisher’s exact test. (E) Egfr and Areg expression determined by qPCR in primary astrocytes and microglia from naïve or EAE mice. n = 5 per group. Unpaired two-tailed t test. (F and G) Analysis of the transcriptional effects of AREG in primary mouse (F) or human (G) astrocytes pretreated with proinflammatory cytokines and recombinant AREG. n = 3 per group. (H) EAE disease course in mice transduced with Itgam::Cas9 lentiviruses coexpressing sgAreg or sgScrmbl. n = 14 sgScrmbl, n = 12 sgAreg mice. Experiment repeated three times. Two-way repeated measures analysis of variance (ANOVA). LTR, long terminal repeat. (I) Volcano plot of differential gene expression analyzed by RNA-seq of astrocytes isolated from EAE mice transduced with Itgam::sgAreg versus Itgam::sgScrmbl. n = 3 mice per group. (J) GSEA preranked analysis of RNA-seq data comparing NF-κB signaling in astrocytes isolated from Itgam::sgAreg versus Itgam::sgScrmbl microglia. NES, normalized enrichment score.
Amphiregulin is reported to control inflammation in the periphery (38-42) and in the CNS during stroke (43), which suggests that it is induced in response to trauma and/or inflammation. Indeed, we detected increased Areg expression in microglia at peak EAE, 17 days after disease induction (Fig. 3E). Consistent with our SPEAC-seq data, Egfr was expressed at higher levels in astrocytes than in microglia (Fig. 3E). We validated these findings by immunostaining, which detected the up-regulation of microglial AREG levels during EAE (fig. S10A). The microglial expression of the microbiome-controlled EGFR ligand TGFα was reduced during EAE (fig. S10, B and C) (11), which suggests a role for Areg+ microglia in limiting immunopathology during CNS inflammation. Indeed, the comparison of Areg+ and Tgfa+ microglia by scRNA-seq and RABID-seq (12) revealed largely nonoverlapping microglial populations with distinctive transcriptional signatures and upstream regulators, which participated in different cell–cell interaction networks (fig. S10, C and D).
To evaluate the functional impact of AREG signaling in astrocytes, we first evaluated the effect of AREG treatment on primary mouse or human astrocytes activated with proinflammatory cytokines in vitro. In both serum-free and serum-containing mouse or human astrocytes, AREG decreased the activation of proinflammatory pathways that are associated with EAE and MS pathogenesis detected by quantitative PCR (qPCR) and bulk RNA-seq (Fig. 3, F and G; fig. S10, E to G; and data S7). We detected a similar anti-inflammatory effect of AREG in astrocytes grown in adherent cultures or in droplets or when astrocytes were isolated after AREG intracranial injection (fig. S10, H to K, and data S8 to 11).
To investigate the function of Areg+ microglia in vivo, we used a CRISPR-Cas9–expressing lentivirus under the control of the Itgam promoter administered through intracerebroventricular injection 1 week before EAE induction (11, 12). Consistent with an anti-inflammatory role for microglial AREG, sgAreg-targeted mice displayed a significant worsening of EAE when compared to controls (Fig. 3H and fig. S11A). No effects were detected in the number of CNS-resident cells, recruited proinflammatory monocytes, or CNS-recruited or splenic T cell subsets (fig. S11, B to D), despite the increased activation of microglia after Areg inactivation (fig. S11, E and F, and data S12), which suggests that AREG+ microglia limit local proinflammatory signals within the CNS. Moreover, the genetic inactivation of Fgl1, Nrtn, or Pnoc in microglia did not alter EAE development (fig. S11G). Similarly, EAE was not modified in Cd4::Cre;Areg(f/f) mice, in line with previous reports of Areg knockdown in regulatory T cells (44), whereas Areg−/− complete knockout mice displayed a worsening of EAE similar to the one detected after microglia-specific Areg knockdown (fig. S12, A to F). Consistent with our SPEAC-seq data, astrocytes and microglia isolated from Itgam::sgAreg mice or Areg−/− mice and analyzed by RNA-seq or qPCR displayed increased NF-κB signaling relative to controls, concomitant with the activation of pathways associated with astrocyte pathogenic activities in EAE (Fig. 3, I and J; fig. S12, G and H; and data S13). Thus, microglial AREG signaling by means of EGFR suppresses astrocyte pathogenic activities during EAE and potentially MS.
Astrocyte-derived IL-33 induces AREG expression in ST2+ microglia
We hypothesized that a cue associated with CNS pathology induced microglial Areg expression during EAE. Previous studies identified IL-33 as a suppressor of EAE (45) and an inducer of Areg expression (38, 43). IL-33 is an alarmin (46), a class of molecules released from cells in the context of tissue damage (47). To determine whether IL-33 regulates microglia–astrocyte interactions mediated by amphiregulin, we first reanalyzed a RABID-seq dataset (12) and identified microglia–astrocyte interactions associated with Areg+ and Areg−microglia during peak EAE. These data identified astrocyte-triggered IL-33 signaling as a putative upstream regulator of Areg+ microglia during EAE (Fig. 4A). To validate this RABID-seq prediction, we stimulated primary microglia in vitro with recombinant IL-33, which detected increased microglial Areg/AREG expression (Figs. 4, B and C) and a transcriptional response comparable to the one induced by IL-33 in microglia during development (fig. S13A) (10, 48). These findings suggest a regulatory feedback loop in which astrocyte-produced IL-33 induces the microglial expression of Areg, which then acts on astrocytes to suppress disease-promoting responses.
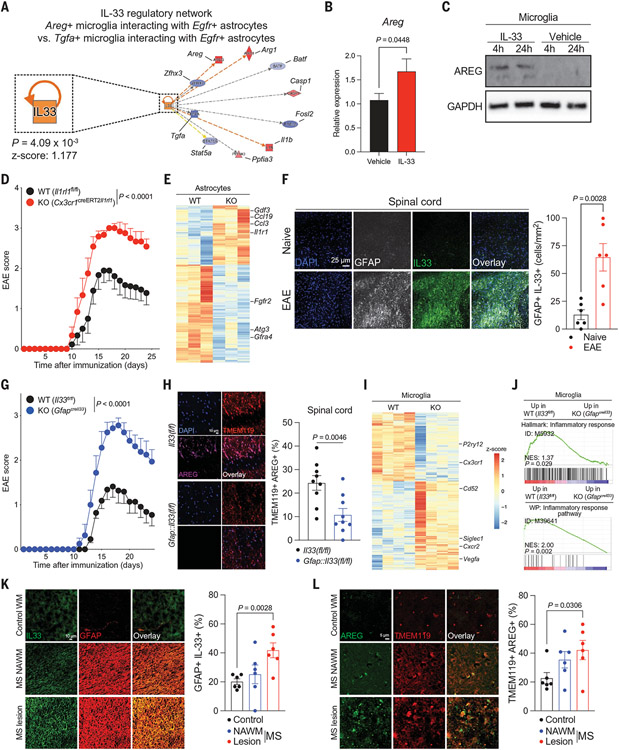
Fig. 4. IL-33-ST2 signaling controls an astrocyte–microglia regulatory circuit.
(A) IL-33 regulates Areg+ microglial interactions with Egfr+ astrocytes determined by RABID-seq during peak EAE. (B and C) IL-33 induces the expression of Areg/AREG in primary microglia. n = 15 to 18 per condition (qPCR). Unpaired two-tailed t test. (D) EAE curve of Cx3cr1::CreERT2Il1rl1 mice (ST2 knockout [KO]) and controls. Two-way repeated measures ANOVA. n = 9 control, n = 6 KO. Experiment repeated three times. (E) Analysis of astrocytes isolated from Cx3cr1::CreERT2Il1rl1 mice by RNA-seq. n = 3 per group. (F) Quantification of IL-33 in GFAP+ astrocytes by immunostaining. n = 6 images from n = 3 mice per group. Unpaired two-tailed t test. (G) EAE curve of GfapIl33 mice and controls. n = 11 control, n = 8 KO. Experiment repeated twice. Two-way repeated measures ANOVA. DAPI, 4',6-diamidino-2-phenylindole; GFAP, glial fibrillary acidic protein. (H) Immunostaining analysis of microglial AREG expression in GfapI133 mice. n = 3 mice per group, n = 9 images. Unpaired two-tailed t test. TMEM, transmembrane protein. (I and J) RNA-seq analyses of microglia isolated from GfapIl33 mice. n = 3 per group. (K) Analysis of IL33+ astrocytes by immunostaining in MS patient CNS samples. n = 3 patients per condition, n = 6 images. Unpaired two-tailed t test. NAWM, normal-appearing white matter; WM, white matter. (L) Analysis of AREG+ microglia by immunostaining in MS patient CNS samples. n = 3 patients per condition, n = 6 images. Unpaired two-tailed t test.
To investigate the role of IL-33 in the control of microglia in vivo, we bred mice harboring a floxed allele of the IL-33 receptor ST2 (encoded by Il1rl1) with mice expressing tamoxifen-inducible Cre recombinase in microglia, which generated Cx3cr1::CreERT2;Il1rl1(f/f) mice. We treated Cx3cr1::CreERT2;Il1rl1(f/f) mice with tamoxifen and one month later induced EAE through active immunization with MOG35–55; Il1rl1(f/f) mice treated with tamoxifen were used as controls. Consistent with our in vitro data, Il1rl1 deletion in microglia resulted in EAE worsening (Fig. 4D), concomitant with decreased microglial Areg expression and increased proinflammatory responses in astrocytes and microglia; no significant changes were detected in the number of CNS-resident or recruited cells or in splenic T cell subsets (Fig. 4E; fig. S13, B to F; and data S14 and S15).
IL-33 expression was increased in astrocytes during EAE; Il33 expression in astrocytes was higher than in microglia (Fig. 4F and fig. S14A). Thus, to investigate the role of astrocyte-produced IL-33 in the control of microglial responses during EAE, we bred mice that harbored a floxed allele of Il33 with mice that expressed Cre recombinase in astrocytes, which generated Gfap::Cre;Il33(f/f) mice. IL-33 inactivation in astrocytes in Gfap::Cre;Il33(f/f) mice resulted in EAE worsening (Fig. 4G and fig. S14B) concomitant with decreased microglial AREG expression (Fig. 4H and fig. S14C to F) and increased microglial NF-κB signaling (Fig. 4, I and J, and data S16). No significant changes were detected in the numbers of peripheral or CNS-recruited T cells or in the total numbers of CNS-resident cells (fig. S14, G to I). Moreover, we detected no change in the number of CNS-recruited AREG+ regulatory T cells (Tregs) in Gfap::Cre;Il33(f/f) mice compared with controls (fig. S14, J and K). Collectively, these findings identify a regulatory feedback loop that is mediated by amphiregulin and IL33-ST2 signaling, which limits CNS pathology driven by microglia–astrocyte interactions in EAE.
Additionally, we investigated IL-33-ST2–driven AREG expression in microglia and astrocytes in MS samples. We detected increased IL33+GFAP+ cells in MS patient lesions compared with control samples (Fig. 4K). Similarly, the reanalysis of scRNA-seq datasets (27, 33) detected increased IL33 expression in astrocytes (fig. S15, A and B), in agreement with our in vitro data (fig. S15C) and increased microglial ST2-driven mitogen-activated protein kinase signaling, which is reported to induce AREG expression (49), in MS patient samples when compared with controls (fig. S15, D and E, and data S17) (33). Indeed, we detected more AREG+ microglia in MS lesions when compared with control samples (Fig. 4L). Moreover, astrocytes exhibiting an increased EGFR activation signature depicted decreased NF-κB activation (fig. S15F), whereas MS patient astrocytes showed decreased EGFR expression (fig. S15G), in support of an anti-inflammatory role of amphiregulin signaling in astrocytes. Thus, increased IL-33 production by astrocytes in MS triggers microglial AREG production by means of ST2 signaling to limit disease-promoting astrocyte activities (fig. S16).
Discussion
Here, we report the development of SPEAC-seq, a platform based on microfluidics, cell coculture in droplets, CRISPR-Cas9 genetic perturbations, and droplet sorting, which enables forward genetic screens for the identification of cell communication mechanisms. In future applications, SPEAC-seq could be coupled with genome-wide analyses of the epigenome (50) or the transcriptome (51, 52) to study their regulation by cell–cell interactions, or with antibody- (53) or small molecule- (54, 55) barcoded libraries to identify therapeutic modulators of cell–cell communication. In addition, SPEAC-seq could incorporate other types of CRISPR-Cas9–driven perturbations, such as those targeting the epigenetic status (56), transcriptional activation or repression (57, 58), or RNA editing (59) of interacting cells. Future SPEAC-seq developments may also involve serum- or lentivirus-free methods to minimize potential perturbations to the cells under investigation, including the activation of microglia as a result of droplet encapsulation.
We applied SPEAC-seq to the study of negative regulators of microglia–astrocyte interactions. Recent studies established that microglial IL-1/TNF/C1q (9, 60), VEGF-B (11), and Sema4D or EphrinB3 (12) signaling induce astrocytes neurotoxic and proinflammatory responses. Although microbiome-driven microglial TGFα production was reported to limit astrocyte proinflammatory responses (11), little is known about cell–cell communication circuits that limit disease-promoting glial responses. Using SPEAC-seq, we identified a regulatory negative feedback loop that is driven by microglia–astrocyte interactions, which are mediated by amphiregulin and IL33-ST2 signaling, that limits astrocyte NF-κB-driven proinflammatory responses that promote CNS pathology in EAE and, potentially, MS. IL-33 has been linked to both pro- and anti-inflammatory functions, but its role in driving amphiregulin expression in peripheral immune cells has been extensively documented (43, 61-63). We did not detect notable changes in CNS-recruited AREG+ Tregs in response to astrocyte-derived IL-33, which suggests that IL-33 produced by astrocytes has limited effects on the modulation of adaptive immunity in the CNS and highlights the need to further investigate stimulus-induced astrocytes in neurologic and other CNS diseases (64).
IL-33-ST2 signaling participates in astrocyte-induced microglial synaptic engulfment during neurodevelopment, controlling phenotypes that are associated with startle reflex and seizures (10, 48). Similarly, axon guidance cues Sema4D and EphrinB3, which participate in the physiologic control of developmental patterning, also act as mediators of microglia–astrocyte pathologic cross-talk in the context of CNS inflammation (12). Moreover, complement, a mediator of synaptic pruning during CNS development (65), contributes to CNS pathology in the context of neurologic disorders (65-67). Together, these data point to the importance of reactivation of developmental programs in neurologic diseases. We developed a method to define molecular mechanisms of cell–cell communication and identified IL-33-ST2 signaling as a microglia–astrocyte circuit that limits CNS pathology through the amphiregulin-mediated control of astrocyte responses.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Plasencia and L. Li for technical assistance and all members of the Quintana laboratory for helpful advice and discussions. We thank R. Krishnan for technical assistance with flow cytometry. We thank R. Sankowski for providing published data. We thank the NeuroTechnology Studio at Brigham and Women’s Hospital for providing NextSeq550 and LSM710 instrument access.
Funding:
This work was supported by grants NS102807, ES02530, ES029136, and AI126880 from the NIH; RG4111A1 and JF2161-A-5 from the NMSS; RSG-14-198-01-LIB from the American Cancer Society; and PA-1604-08459 from the International Progressive MS Alliance to F.J.Q. M.A.W. was supported by NIH (R01MH130458, R00NS114111, F32NS101790), a training grant from the NIH and Dana-Farber Cancer Institute (T32CA207201), a traveling neuroscience fellowship from the Program in Interdisciplinary Neuroscience at Brigham and Women’s Hospital, the Women’s Brain Initiative at Brigham and Women’s Hospital, and U19AI133524 (NIH). I.C.C. was supported by K22AI152644 and DP2AI154435 from the NIH. H.-G.L. was supported by a Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1A6A3A14039088). G.P. is a trainee in the Medical Scientist Training Program funded by NIH T32GM007356. B.M.A. was supported by grant K12CA090354 from NIH. H.Y. was supported by the Crohn’s and Colitis Foundation Research Fellowship Award 937437. S.E.J.Z. held a postdoctoral studentship from the FRQS/MSSC Partnership Award. A.P. holds the T1 Canada Research Chair in MS and is funded by the Canada Institute of Health Research, the NMSS, and the Canadian Foundation for Innovation. C.D. holds a professorship grant from the Excellence Program for Outstanding Female Scientists from the Helmholtz Association and is supported by a Research Division (TDI) at Helmholtz Zentrum München, the German Center for Diabetes Research (DZD), through a membership in the CRC1054 of the Deutsche Forschungsgemeinschaft (B11), and through an award of the EFSD/JDRF/Lilly Programme on Type 1 Diabetes Research 2020. J.-H.L. was supported by the NRF of Korea and the Ministry of Education (2022R1A6A3A03071157). Y.-C.K. was supported by the NMSS FG-2007-36929. T.I. was supported by the EMBO postdoctoral fellowship (ALTF: 1009-2021).
Footnotes
Competing interests: M.A.W., I.C.C., and F.J.Q. have filed a provisional patent application for the SPEAC-seq platform outlined in this work. D.A. has contributed to scientific advisory boards at Pfizer, Regeneron, Takeda, Nemagene, and the Kenneth Rainin Foundation; none of these activities is related to the work presented in this work. L.M.S. is also now affiliated with Seismic Therapeutic, which is not related to the work presented in this study.
Data and materials availability: Sequencing data were deposited into GEO under the SuperSeries accession no. GSE200457. All other materials will be made available upon request.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Reich DS, Lucchinetti CF, Calabresi PA, N. Engl. J. Med 378, 169–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han RT, Kim RD, Molofsky AV, Liddelow SA, Immunity 54, 211–224 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Linnerbauer M, Wheeler MA, Quintana FJ, Neuron 108, 608–622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prinz M, Masuda T, Wheeler MA, Quintana FJ, Annu. Rev. Immunol 39, 251–277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanmarco LM, Polonio CM, Wheeler MA, Quintana FJ, J. Exp. Med 218, e20202715 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schirmer L, Schafer DP, Bartels T, Rowitch DH, Calabresi PA, Trends Immunol. 42, 228–247 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khakh BS, Deneen B, Annu. Rev. Neurosci 42, 187–207 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Khakh BS, Cell 185, 220–220.e1 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Liddelow SA et al. , Nature 541, 481–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vainchtein ID et al. , Science 359, 1269–1273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothhammer V et al. , Nature 557, 724–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark IC et al. , Science 372, eabf1230 (2021).33888612 [Google Scholar]
- 13.Wheeler MA et al. , Cell 176, 581–596.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giladi A et al. , Nat. Biotechnol 38, 629–637 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Vento-Tormo R et al. , Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqual G et al. , Nature 553, 496–500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S et al. , Cell 160, 1246–1260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow RD et al. , Nat. Methods 16, 405–408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaFleur MW et al. , Nat. Commun 10, 1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manguso RT et al. , Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan D et al. , Science 359, 770–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wucherpfennig KW, Cartwright AN, Curr. Opin. Immunol 41, 55–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin H, Xue W, Anderson DG, Nat. Rev. Clin. Oncol 16, 281–295 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Dixit A et al. , Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark IC, Thakur R, Abate AR, Lab Chip 18, 710–713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everhart MB et al. , J. Immunol 176, 4995–5005 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Wheeler MA et al. , Nature 578, 593–599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sofroniew MV, Nat. Rev. Neurosci 16, 249–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doench JG et al. , Nat. Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothhammer V et al. , Nat. Med 22, 586–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glass CK, Saijo K, Nat. Rev. Immunol 10, 365–376 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Clark IC et al. , Nature 614, 326–333 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda T et al. , Nature 566, 388–392 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y et al. , Nat. Med 26, 131–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond TR et al. , Immunity 50, 253–271.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin X et al. , Science 370, eaaz6063 (2020).33243861 [Google Scholar]
- 37.Sanmarco LM et al. , Nature 590, 473–479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monticelli LA et al. , Proc. Natl. Acad. Sci. U.S.A 112, 10762–10767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaiss DMW, Gause WC, Osborne LC, Artis D, Immunity 42, 216–226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnan S et al. , Proc. Natl. Acad. Sci. U.S.A 115, 10738–10743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minutti CM et al. , Immunity 50, 645–654.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsou AM et al. , Nature 611, 787–793 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito M et al. , Nature 565, 246–250 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Pohar J et al. , Eur. J. Immunol 52, 1335–1349 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang HR et al. , Eur. J. Immunol 42, 1804–1814 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Yang D, Han Z, Oppenheim JJ, Immunol. Rev 280, 41–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gadani SP, Walsh JT, Smirnov I, Zheng J, Kipnis J, Neuron 85, 703–709 (2015). [DOI] [PubMed] [Google Scholar]
- 48.He D et al. , Immunity 55, 159–173.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Funakoshi-Tago M et al. , Cell. Signal 20, 1679–1686 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Demaree B et al. , Nat. Commun 12, 1583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein AM et al. , Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macosko EZ et al. , Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoeckius M et al. , Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodnow RA Jr., Dumelin CE, Keefe AD, Nat. Rev. Drug Discov 16, 131–147 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Cochrane WG et al. , ACS Comb. Sci 21, 425–435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwon DY, Zhao YT, Lamonica JM, Zhou Z, Nat. Commun 8, 15315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konermann S et al. , Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeo NC et al. , Nat. Methods 15, 611–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abudayyeh OO et al. , Nature 550, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guttenplan KA et al. , Cell Rep. 31, 107776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiering C et al. , Nature 513, 564–568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arpaia N et al. , Cell 162, 1078–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burzyn D et al. , Cell 155, 1282–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee HG, Wheeler MA, Quintana FJ, Nat. Rev. Drug Discov 21, 339–358 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stevens B et al. , Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Pandey MK et al. , Nature 543, 108–112 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Sekar A et al. , Nature 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.