Abstract
Background
The use of markers has stimulated the development of more appropriate targeted therapies for chronic lymphocytic leukemia (CLL) and acute myeloid leukemia (AML). We assessed the use and prevalence of biological and genetic markers of CLL and AML in the homogeneous Hispanic population of Puerto Rico.
Methods
We used the Puerto Rico CLL/AML Population‐Based Registry, which combines information from linked databases. Logistic regression models were used to examine factors associated with biological and genetic testing.
Results
A total of 926 patients 18 years or older diagnosed with CLL (n = 518) and AML (n = 408) during 2011–2015 were included in this analysis. Cytogenetic testing (FISH) was reported for 441 (85.1%) of the CLL patients; of those, 24.0% had the presence of trisomy 12, 9.5% carried deletion 11q, 50.3% carried deletion 13q, and 6.3% carried deletion 17p. Regarding AML, patients with cytogenetics and molecular tests were considered to determine the risk category (254 patients), of which 39.8% showed poor or adverse risk. Older age and having more comorbidities among patients with CLL were associated with a lower likelihood of receiving a FISH test.
Conclusions
Although prognostic genetic testing is required for treatment decisions, the amount of testing in this Hispanic cohort is far from ideal. Furthermore, some tests were not homogeneously distributed in the population, which requires further exploration and monitoring. This study contributes to the field by informing the medical community about the use and prevalence of biological and genetic markers of CLL and AML. Similarly, it has the potential to improve the management of CLL and AML through benchmarking.
Keywords: acute myeloid leukemia (AML); genetic markers, chronic lymphocytic leukemia (CLL), Hispanics, Puerto Rico
We assessed the use and prevalence of biological and genetic markers of chronic lymphocytic leukemia (CLL) and acute myeloid leukemia (AML) in the homogeneous Hispanic population of Puerto Rico. In Puerto Rico, these tests are not performed consistently among patients with CLL and AML; however, the frequency of genetic testing was higher than that reported in other studies.

1. INTRODUCTION
Worldwide, leukemia remains one of the leading causes of cancer morbidity and mortality. Leukemia is subdivided into myeloid or lymphoid cells, depending on the starting location. Chronic lymphocytic leukemia (CLL) and acute myeloid leukemia (AML) are the most frequent types of leukemia among the elderly population. 1 Given the heterogeneity of CLL, some patients may live many years after diagnosis without the need for therapy, while others die within the first year from disease‐related complications. 1 AML is characterized by a group of phenotypic and genetically heterogeneous hematologic diseases, categorized by the clonal expansion of myeloid precursor with decreased differentiation capacity. 2
In Puerto Rico, leukemia is the ninth most common cancer, with an incidence rate of 10.2 per 100,000 population, and the eighth leading cause of cancer‐related death, with a mortality rate of 4.1 per 100,000 population. 3 When stratified by subtype, AML and CLL are the most frequently diagnosed types of leukemia, with an age‐adjusted incidence rate of 3.1 and 2.6, respectively. The Commonwealth of Puerto Rico is the largest US territory, with over 3.2 million population. 4 Puerto Ricans represent the second largest Hispanic population in the country, with more than 4 million living in the continental United States. Nearly 99% of the population living in Puerto Rico identify themselves as Hispanics. 4 Puerto Rico's population is older than the continental United States, with about 21.3% of the Puerto Rican population 65 years or older. 4 Puerto Rico faces a significant demographic shift due to migration to the continental United States and low fertility rates. 5 In Puerto Rico, more than 92% of the population is covered by health insurance, and most receive Medicaid or Medicare (60%). 4 Nearly 31.5% of Puerto Rico's population has private health insurance, including employer‐sponsored plans and plans purchased directly from insurers. Insurance companies cover cancer diagnostic procedures, including genetic testing, although patients could be responsible for out‐of‐pocket expenses depending on insurance coverage.
During the past decades, novel biomarkers have changed the way physicians treat patients with leukemia and assign targeted therapies. The use of markers in patients with CLL has provided important information on the prognosis of the disease and has stimulated the development of more appropriate targeted therapies. 6 Some of the most reliable molecular prognostic markers offered in routine diagnostic tests are the mutational status of the immunoglobulin heavy chain variable (IGHV) gene and those detected by the fluorescence in situ hybridization technique (FISH). 7 For AML, there are cytogenetic alterations producing fusion genes that encode aberrant proteins with altered functional characteristics. Polymerase chain reaction (PCR) test is recommended to detect leukemic cells during and after treatment because it has the highest analytic sensitivity. 8 Depending on the results of the chromosome tests, patients with AML are stratified into three categories that help to determine their prognosis and response to treatment. 9
The cytogenetic analysis of AML and CLL has become essential for the diagnosis, classification, prognostic stratification, and treatment guidance of the disease. 9 , 10 , 11 , 12 However, like most population‐based registries, the Puerto Rico Central Cancer Registry (PRCCR) does not collect extensive clinical information or cancer‐related biological and genetic markers, limiting the use of these registries to address critical research questions. Nevertheless, data from population‐based registries can be linked to different databases to expand the number of variables collected and increase their potential to address these critical research questions. To our best knowledge, no study has evaluated the use of these prognostic factors for CLL or AML in Puerto Rico, an aging Hispanic population. Therefore, in partnership with an external entity, we created the Puerto Rico CLL/AML Population‐Based Registry, which leverages the PRCCR capabilities to assess the pattern of use and prevalence of biological and genetic markers for CLL and AML and examine the factors associated with the administration of biological and genetic tests.
2. METHODS
2.1. Data sources
The PRCCR, one of the oldest population‐based cancer registries in the world, is responsible for collecting, analyzing, and publishing information on all cancer cases diagnosed and/or treated among residents of Puerto Rico. Since 1997, the PRCCR has been part of the Centers for Disease Control and Prevention's National Program of Cancer Registries and uses the Surveillance, Epidemiology, and End Results (SEER) program and the North American Association of Central Cancer Registries (NAACCR) standards for coding data. Furthermore, the PRCCR requests information from hospitals, outpatient clinics, pathology laboratories, and radiotherapy/chemotherapy sites throughout Puerto Rico. Over the years, the PRCCR improved data collection on cancer cases through electronic reporting, achieving complete information on more than 95% of cases since 2010. Additionally, the PRCCR–Health Insurance Linkage Database (PRCCR‐HILD) links the PRCCR database to Medicaid, Medicare, and private insurance data for residents of Puerto Rico and provides information about treatment, medical procedures, comorbidities, costs, and providers. This linkage has allowed us to conduct cancer care delivery research to better understand the patterns of cancer care on the island.
2.2. Creation of the Puerto Rico CLL/AML population‐based registry
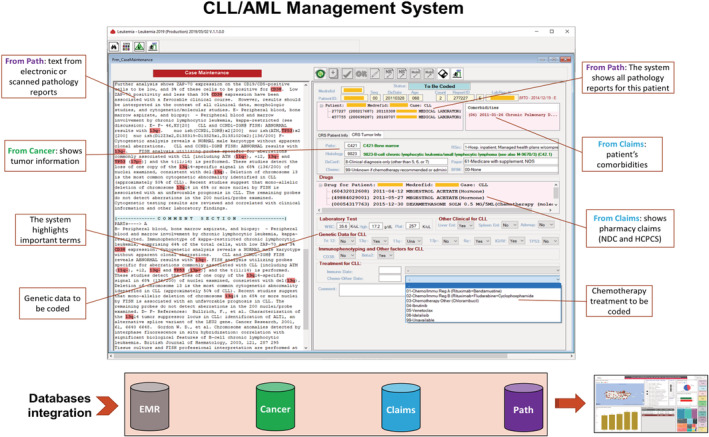
The PRCCR integrated a multidisciplinary team of oncologists, tumor registrars, epidemiologists, biostatisticians, and informatics to develop the Puerto Rico CLL/AML Population‐Based Registry software and database. The CLL/AML registry uses the PRCCR data and expands the number of clinical, biological, and genetic variables that are not collected regularly. After several meetings with experts, we determined the genetic markers, prognostic factors, laboratory tests, and treatment modalities needed for the project. We took advantage of pathology laboratories that report electronically using PathPlus, a PRCCR in‐house software with comprehensive case‐finding protocols to identify incident cases. An infrastructure with extensive algorithms was developed to search for specific CLL and AML‐related biomarkers in pathology reports. A solution was created in Visual Studio to manage variables related to CLL/AML, integrating data from PRCCR's cancer database, Pathology Reports database, electronic medical records (EMR), and PRCCR‐HILD. Furthermore, the Puerto Rico CLL/AML Population‐Based Registry database collected treatment, healthcare utilization, health insurance type, and a modified Charlson's comorbidity index described by Klabunde et al. 13 , 14 (see the CLL/AML Management System in Appendix 1). The oncologist trained a qualified tumor registrar to retrieve the variables of interest. Furthermore, the tumor registrar performed a quality control process to ensure the completeness of the diagnosis, tumor markers, and treatment information for the cases of CLL and AML.
2.3. Selection criteria
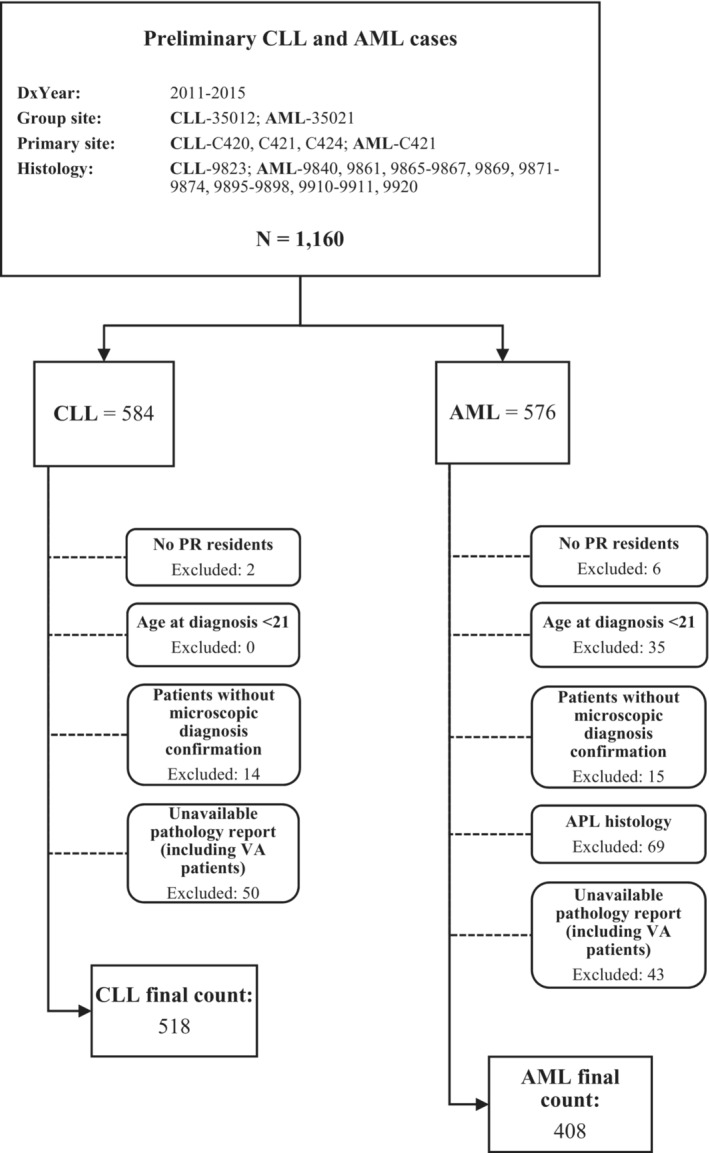
The study population consisted of cases reported to the PRCCR between January 1, 2011, and December 31, 2015, with a diagnosis of CLL (9823) and AML (9840, 9861, 9865–9867, 9869, 9871–9874, 9895–9897, 9898, 9910–9911, 9920), as defined by the International Classification of Diseases for Oncology, third edition (ICD‐O‐3). We also excluded (1) patients who were not residents of Puerto Rico at the time of diagnosis, (2) cases from the Veterans Health Administration (VHA) due to institutional restrictions of the VHA, (3) cases without diagnostic confirmation, and (4) cases incorrectly assigned as CLL or AML in the PRCCR database. The study cohort included 926 patients; 518 were patients with CLL, and 408 were patients with AML (see the cohort selection algorithm in Appendix 2).
2.4. Outcome variables
To assess the pattern of use of biological and genetic markers for CLL and AML in Puerto Rico, we identified the most relevant genetic and prognostic factors at the time of diagnosis. For CLL cases, we had the FISH test, which is used to identify trisomy 12, del(11q), del(13q), and del(17p). The immunoglobulin heavy chain variable region (IGHV) mutation test was used to identify IGHV mutation status. The status of the TP53 mutation was identified by PCR or FISH. For AML cases, we had information on the karyotype, PCR tests to identify TP53, CEBPA, FLT3, NPM1, c‐Kit, IdH 1&2, and flow cytometry to identify CD33. Furthermore, patients with AML were stratified into risk categories (favorable, intermediate, poor/adverse, not evaluated, and unknown) to determine prognosis and response to treatment, depending on cytogenetic markers 9 (see the description of selected markers for CLL and AML in Appendix 3).
2.5. Independent variables
The factors evaluated in the association of receiving the FISH or PCR test were sociodemographic characteristics at the time of diagnosis, including sex, age group (<50, 50–64, 65–79, ≥80 years), history of previous cancer, type of health insurance (private insurance, Medicaid, Medicare, or dually eligible for Medicare and Medicaid), and the modified Charlson's comorbidity index, classified as 0, 1, ≥2, and unknown comorbidities. 13 , 14
2.6. Statistical analysis
Descriptive statistics and frequency analyses were used to describe the variables of interest. We used logistic regression models to examine factors associated with the use of the FISH test among patients with CLL and the use of PCR (at least one of c‐Kit, TP53, IDH 1&2, NPM1, FLT3, or CEBPA) among patients with AML. The results of these models are presented in terms of adjusted odds ratios (aOR) and their 95% confidence intervals (CI). Statistical significance was based on two‐sided tests. All analyses were performed using Stata/SE version 15.1 statistical software (Stata Corp.). This study was approved by the Institutional Review Board (IRB) of the University of Puerto Rico Comprehensive Cancer Center (# 2018‐10‐04).
3. RESULTS
3.1. Characteristics of the cohort by leukemia subtype (CLL and AML)
A total of 926 patients were included in the analysis; of them, 518 had CLL, and 408 had AML. Both leukemia subtypes (CLL and AML) were more common among men and almost half of the patients were between 65 and 79 years old. More patients with AML (22.3%) had previous malignancy than patients with CLL (13.3%), and slightly more than half of patients with CLL and AML had a comorbidity index greater than zero. At the time of diagnosis, 29.3% of patients with CLL were enrolled in Medicare, and 24.3% of patients with AML were enrolled in Medicare‐Medicaid dual insurance (Table 1).
TABLE 1.
Description of the CLL/AML cohort by leukemia subtype: Puerto Rico, 2011–2015
| Characteristics | CLL (N = 518) | AML (N = 408) | ||
|---|---|---|---|---|
| Count | % | Count | % | |
| Sex | ||||
| Male | 306 | 59.1 | 208 | 51.0 |
| Female | 212 | 40.9 | 200 | 49.0 |
| Age group (years) | ||||
| <50 | 30 | 5.8 | 89 | 21.8 |
| 50–64 | 135 | 26.1 | 92 | 22.6 |
| 65–79 | 257 | 49.6 | 169 | 41.4 |
| 80+ | 96 | 18.5 | 58 | 14.2 |
| Previous cancer history | ||||
| No | 449 | 86.7 | 317 | 77.7 |
| Yes | 69 | 13.3 | 91 | 22.3 |
| Charlson comorbidity index | ||||
| 0 | 233 | 45.0 | 190 | 46.6 |
| 1 | 83 | 16.0 | 53 | 13.0 |
| ≥2 | 91 | 17.6 | 67 | 16.4 |
| Unknown | 111 | 21.4 | 98 | 24.0 |
| Insurance at diagnosis | ||||
| Private | 119 | 23.0 | 93 | 22.8 |
| Medicaid | 77 | 14.9 | 96 | 23.5 |
| Medicare/Medicaid | 117 | 22.6 | 99 | 24.3 |
| Medicare | 152 | 29.3 | 84 | 20.6 |
| Unknown/Other | 53 | 10.2 | 36 | 8.8 |
3.2. Prevalence of prognostic markers in CLL and AML
In general, the FISH test was reported in 85.1% of patients with CLL; among these, more than half carried deletion 13q and almost a quarter (24.0%) had the presence of trisomy 12. PCR testing was reported in 83.4% of patients with CLL, of those, 3.2% had the TP53 mutation. Meanwhile, the IGHV test was reported in 60.2% of patients with CLL, of which 57.1% of patients had mutated IGHV (Table 2).
TABLE 2.
Pattern of use of biological and genetic tests and prevalence of prognostic markers in chronic lymphocytic leukemia
| Biological and genetic tests | Total patients with test reported N (%) | Prognostic markers | Prevalence N (%) |
|---|---|---|---|
| FISH a | 441 (85.1%) | del(13q) | 222 (50.3%) |
| Tri12 b | 106 (24.0%) | ||
| del(11q) | 42 (9.5%) | ||
| del(17p) | 28 (6.4%) | ||
| PCR | 432 (83.4%) | TP53 c | 14 (3.2%) |
| IGHV mutation testing | 312 (60.2%) | IGHV | 178 (57.2%) |
At least one of trisomy 12, deletion 11q, deletion 13q, deletion 17p.
Trisomy 12 was reported only in 433 cases.
TP53 is detected using FISH or PCR.
We assigned AML risk categories only among the 254 patients who had reported cytogenetics and molecular tests (Table 3); of these, 18.5% had favorable risk, 30.7% had intermediate risk, 39.8% had poor or adverse risk, and 11.0% had unknown risk (Figure 1). Karyotype was reported in 265 AML patients. Among patients who had undergone karyotype testing, 64.2% had an abnormal karyotype and among those with an abnormal karyotype, 42.4% had a complex karyotype. The c‐Kit test was the highest PCR test reported, and among the patients who had the test, 86.7% had a c‐Kit mutation. Meanwhile, flow cytometry, used to identify CD33 was reported in 297 patients with AML, of whom almost all (92.3%) showed expression of CD33.
TABLE 3.
Pattern of use of biological and genetic tests and prevalence of prognostic markers in acute myeloid leukemia
| Biological and genetic tests | Prognostic markers | Total patients with test reported N (%) | Prevalence of prognostic markers N (%) |
|---|---|---|---|
| PCR | c‐Kit | 308 (75.5%) | 267 (86.7%) |
| TP53 | 15 (3.7%) | 10 (66.7%) | |
| IDH 1&2 | 19 (4.7%) | 11 (57.9%) | |
| NPM1 | 144 (35.3%) | 38 (26.4%) | |
| FLT3 | 159 (39.0%) | 35 (22.0%) | |
| CEBPA | 144 (35.3%) | 21 (14.6%) | |
| Flow cytometry | CD33 | 297 (72.8%) | 274 (92.3%) |
| Cytogenetic test | Normal karyotype | 265 (64.6%) | 95 (35.9%) |
| Abnormal karyotype a | 170 (64.2%) | ||
| Complex karyotype | 72 (27.2%) |
Includes patients with complex karyotype.
FIGURE 1.
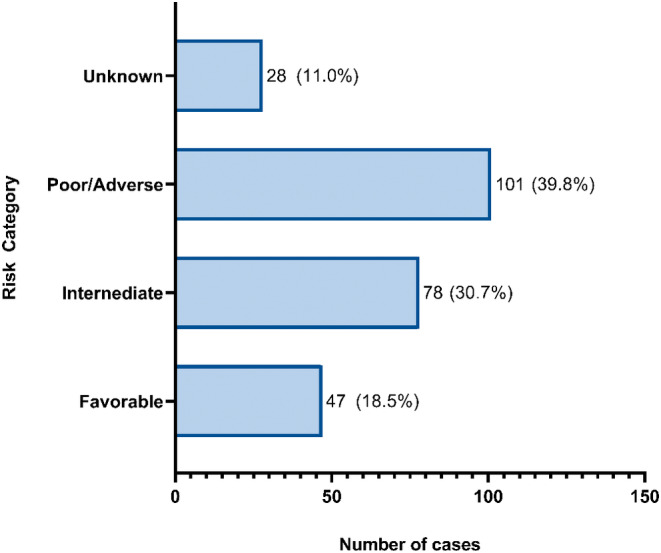
Distribution of risk category for AML cases.
3.3. Association between the use of prognostic tests/markers and patients' characteristics
Patients older than 74 years were 61% (aOR = 0.39; 95% CI: 0.23–0.67) less likely to have had a FISH test compared to those younger than 75 years. In addition, patients with a comorbidity index equal to one were 66% (aOR = 0.34; 95% CI: 0.17–0.69) less likely to be given FISH testing compared with those with a comorbidity index equal to zero. Meanwhile, Medicaid patients were less likely to be tested for TP53 compared to other types of insurance (p < 0.05) (Table 4). In terms of factors associated with undergoing IGHV testing, the analysis does not show any statistical associations between the different selected predictors and the performance of IGHV testing (p > 0.05) (data not shown).
TABLE 4.
Factors associated with performing FISH test among patients with chronic lymphocytic leukemia and PCR in patients with acute myeloid leukemia
| Characteristics | FISH test among patients with CLL | PCR test among patients with AML | ||||
|---|---|---|---|---|---|---|
| aOR | 95% CI | p‐value | aOR | 95% CI | p‐value | |
| Sex | ||||||
| Male | 1.00 | 1.00 | ||||
| Female | 1.30 | (0.77–2.20) | 0.322 | 1.05 | (0.65–1.71) | 0.838 |
| Age at dx, years | ||||||
| <75 | 1.00 | 1.00 | ||||
| ≥75 | 0.39 | (0.23–0.67) | 0.001 | 1.81 | (0.97–3.38) | 0.062 |
| Insurance in diagnosis | ||||||
| Private | 1.00 | 1.00 | ||||
| Medicaid | 0.54 | (0.22–1.32) | 0.176 | 0.84 | (0.42–1.69) | 0.619 |
| Medicare | 1.38 | (0.57–3.32) | 0.472 | 0.96 | (0.43–2.16) | 0.929 |
| Medicare/Medicaid | 0.74 | (0.32–1.72) | 0.485 | 1.36 | (0.61–3.03) | 0.450 |
| Others/Unknown | 0.38 | (0.11–1.33) | 0.131 | 0.51 | (0.19–1.39) | 0.189 |
| Charlson comorbidity index | ||||||
| 0 | 1.00 | 1.00 | ||||
| 1 | 0.34 | (0.17–0.69) | 0.003 | 0.82 | (0.38–1.78) | 0.621 |
| ≥2 | 0.48 | (0.23–1.01) | 0.052 | 1.25 | (0.54–2.90) | 0.610 |
| Unknown | 1.24 | (0.47–3.27) | 0.660 | 0.71 | (0.37–1.39) | 0.322 |
Abbreviation: aOR, Adjusted Odd Ratio.
When we analyzed factors associated with undergoing PCR among patients with AML, we did not find any significant statistical associations with any of the predictors included in the analysis (p > 0.05) (Table 4). Similarly, regarding factors associated with undergoing CD33 testing, the analysis does not show any associations between the different selected predictors and the performance of CD33 testing (p > 0.05) (data not shown). Furthermore, among patients older than 74 years, the likelihood of being tested for the karyotype is twice than that of younger patients (aOR = 2.04; 95% CI: 1.20–3.45). Similarly, patients with private insurance were twice as likely to have the karyotype tested compared to Medicaid patients with Medicaid (aOR = 2.06; 95% CI: 1.10–3.85) (data not shown).
4. DISCUSSION
In recent decades, the medical management of patients with CLL and AML has improved in diagnosis, prognosis, and monitoring, particularly in understanding genetic markers. 15 , 16 Genetic testing is a key tool to evaluate and guide treatment decisions among patients with CLL and AML. To our knowledge, this is the first study to assess the pattern of use and prevalence of biological and genetic markers of CLL and AML among a homogenous Hispanic population. In Puerto Rico, these tests are not performed consistently among patients with CLL and AML; however, the frequency of genetic testing was higher than that reported in other studies. 17 , 18 This difference might be attributable to the high coverage of health insurance and the adherence of healthcare professionals in Puerto Rico to recommended evidence‐based treatment guidelines. 19 However, the amount of testing in this cohort is still far from ideal, particularly today when these tests are required for treatment decisions. Therefore, this study is the first step to continue to monitor the management of CLL and AML among Hispanic populations.
4.1. Prognostic markers in CLL
FISH testing to identify genetic abnormalities has proved to be relevant in assessing the prognosis of patients with CLL. 20 However, consistent with previous studies, 17 , 18 the age group (<75 years vs. ≥75 years) was an independent predictor of FISH testing. Our findings show that the older patients with CLL are less likely to undergo FISH testing, which is critical to determine treatment modalities. Furthermore, patients with CLL with a comorbidity index greater than zero were less likely to have FISH testing than those with a comorbidity index equal to zero. Although more research is needed to understand these disparities, this may indicate that physicians tend to assess prognostic factors more in the youngest patients due to better outcomes in this population. Nonetheless, according to National Comprehensive Cancer Network (NCCN) guidelines, the FISH panel is recommended for all CLL patients, regardless of age, comorbidities, or other patient characteristics. Whereas elderly patients are less likely to tolerate intensive regimens, more conservative therapies are considered. 21 For example, for older and/or comorbid patients, currently approved therapies or clinical trials remain options to improve their quality of life. 21 , 22 , 23 , 24 Future studies are needed to monitor these patterns, since the evaluation of these markers is essential to determine the long‐term prognosis and treatment of patients, regardless of age and comorbidities. Our results show that, excluding del(11q) and trisomy 12, most abnormalities detected by FISH among patients with CLL were similar to those reported by the scientific literature. 25 , 26 In Puerto Rico, del(11q) was found in 9.5% of patients with CLL, which is lower compared to other studies (18%–20%). 25 , 27 Del(11q) has been associated with shorter disease progression and survival. 25 Like previous studies, Trisomy12 was the second most common chromosomal abnormality identified. Trisomy 12 was found in 24.0% of patients with CLL in Puerto Rico, which is higher than in other populations with CLL (14%–16%). 25 , 28 , 29 , 30 However, a recent study using the Connect CLL Registry reported that Trisomy12 was present in 21% of CLL cases in the US. 31 Possible reasons for this difference could be attributed to the cohort of patients examined, the methods and probes used, and the number of neoplastic B‐cells in the sample analyzed. 32 Further research is warranted to understand this pattern since few studies compare genetic abnormalities among different populations.
Furthermore, the scientific literature has suggested that patients with unmutated IGHV have a worse prognosis than those with mutated IGHV; the status of IGHV and the TP53 mutations influence the choice of therapy for patients with CLL. 33 , 34 , 35 IGHV is one of the most important molecular prognostic markers for CLL; however, only 60% of CLL patients had this test in Puerto Rico. Additionally, 57.1% of patients with CLL had a mutated IGHV, which is slightly lower than that reported in other studies (60%–70%). 36 , 37 Meanwhile, in our study, among patients with CLL who underwent the PCR test, only 3.2% had a mutation in TP53, compared to other studies reporting between 5% and 12%. 38 , 39 , 40
4.2. Prognostic markers in AML
For patients with AML, no statistical association was found between selected predictors and those undergoing PCR testing (TP53, CEBPA, FLT3, NPM1, c‐Kit, IDH 1&2). It is important to consider that not all genetic tests were reported for patients with AML. TP53 and IDH 1&2 were reported in only 15 and 19 patients, respectively. Although these results must be interpreted with caution due to the limited sample size, we found a similar prevalence of CEBPA, FLT3, NPM1, and c‐Kit‐related mutations, as in previous studies. 41 , 42 , 43 , 44 , 45 One of the reasons for the reported low testing rate for IDH 1&2 could be that they were first used around 2009, close to the study period. 46 Presently, the performance of these tests is important because there are new drugs targeting patients with IDH1 and IDH2 mutations. 47 In our study, the FLT3 mutation was found in 21.9% of cases of AML. This is slightly lower than the 27% reported in a large study by the United Kingdom Medical Research Council. 42 Additionally, c‐Kit was found to be expressed in 86% of AML cases, which is slightly higher than the 60%–80% reported by Malaise et al. 44 Among patients with AML whose karyotype was reported, 36% have a normal karyotype. This finding is consistent with other studies that reported a normal karyotype in 40% to 48% of patients with AML. 48 , 49 , 50 , 51 However, a complex karyotype (≥3 clonal abnormalities) was detected in 27% of the patients who received the test, which is higher than that found in the previous studies. 50 , 52 , 53 A possible explanation for these results is that more than 55% of the patients in this study with AML were 65 years or older. Studies have shown that older patients have more complex karyotypes than younger patients.
4.3. Strengths and limitations
This study has some limitations that must be acknowledged. First, we could not evaluate relevant clinical information such as physical examination, blood test results, the Rai's/Binet's staging systems, and other tests such as B2 microglobulin. Second, genetic markers could be underreported for various reasons, including failure to be reported to the PRCCR (PRCCR does not collect this clinical data regularly) or not being documented in the pathology reports. 54 Third, only 20% of the study cohort has been linked to EMR. This limitation has been documented in other studies that use population‐based registry data and clinical trial data. 49 , 52 However, it does not affect the generalizability of our results since most of the information for the scope of this study was obtained from the pathology reports database, which was complete and EMR was only used to complement the dataset. Eventually, we expect more physicians to report to the PRCCR through EMR, improving the completeness of the data, particularly physical examination and blood tests. Despite these limitations, our findings highlight the importance of testing for prognostic genetic markers for all patients with CLL and AML and suggest the need for increased awareness and knowledge regarding the value of this genetic information at the time of diagnosis in Puerto Rico. The database developed for this project proved to be an invaluable resource to characterize and monitor the use of biological and genetic markers for CLL and AML in Puerto Rico and potentially could be modified for other cancer sites.
5. CONCLUSIONS
Our findings show the potential of the Puerto Rico CLL/AML Population‐Based Registry database to estimate and assess the pattern of use of these biological markers to guide treatment decisions, monitor outcomes, and improve the management among patients with CLL and AML in Puerto Rico. The recommended genetic tests performed among our cohort of Hispanics are inadequate, raising concerns about the treatment decision among this population. This study adds to the scientific literature on CLL and AML among Hispanic populations and could guide public policy and control efforts for these conditions and related morbidities in this population. More studies are needed to understand these patterns and assess the importance of the characteristics of the physician/health system in the performance of these tests.
AUTHOR CONTRIBUTIONS
Karen J Ortiz‐Ortiz: Conceptualization (equal); data curation (supporting); formal analysis (supporting); funding acquisition (lead); investigation (equal); methodology (equal); supervision (equal); visualization (supporting); writing – original draft (equal). Carlos Ruben Torres‐Cintrón: Conceptualization (equal); formal analysis (equal); investigation (equal); methodology (equal); project administration (equal); visualization (equal); writing – original draft (equal). Tonatiuh Suárez Ramos: Data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); writing – original draft (equal). Maira Castañeda‐Avila: Formal analysis (equal); investigation (equal); methodology (equal); writing – original draft (equal). Luis Cotto Santana: Investigation (equal); validation (equal); writing – review and editing (equal). Guillermo Tortolero‐Luna: Conceptualization (equal); investigation (equal); project administration (equal); supervision (equal); writing – review and editing (equal).
FUNDING INFORMATION
Financial support for the study was provided by AbbVie Corp. AbbVie participated in the review and approval of the publication. This study was partially supported by the National Program of Cancer Registries (NPCR), Centers for Disease Control and Prevention (CDC; (award number: NU58DP007164) to the Puerto Rico Central Cancer Registry (PRCCR)).
CONFLICT OF INTEREST
Dr. Ortiz‐Ortiz and Dr. Tortolero‐Luna reported receiving grants from AbbVie Corp. related to the submitted work and grants from Merck & Co outside of the submitted work. No other disclosures were reported.
ETHICS STATEMENT
This article does not contain studies with human participants performed by any of the authors. This study was approved by the Institutional Review Board (IRB) of the University of Puerto Rico Comprehensive Cancer Center (# 2018‐10‐04).
APPENDIX 1. CLL/AML management system
APPENDIX 2. Cohort selection algorithm
APPENDIX 3. Description of selected markers for CLL and AML
| Marker | Description |
|---|---|
| Chronic lymphocytic leukemia | |
| del(13q) | 13q deletion is associated with a more favorable outcome. 55 |
| Trisomy 12 | This common aberration at the time of diagnosis is associated with intermediate prognostic risk. 28 |
| del(11q) | 11q contains the ATM gene, associated with a least favorable outcome, correlated with non‐mutated IGHV genes. 56 |
| del(17p) | The genetic abnormality involves the TP53 gene and del(11q), associated with a less favorable outcome, correlated with non‐mutated IGHV genes. 38 |
| TP53 | The tumor‐suppressor protein p53 is associated with a poor prognosis. Mutations of TP53 are also associated with poor prognosis independently of the presence of del(17p). It is the strongest prognostic and predictive marker guiding treatment decisions and is associated with markedly decreased survival and impaired response to chemoimmunotherapy. 38 |
| IGHV | The immunoglobulin heavy chain variable region (IGHV) is used to determine the risk group in newly diagnosed cases. Mutated IGHV is associated with a more indolent clinical course, while cases with unmutated IGHV genes often behave aggressively. 35 |
| Acute myeloid leukemia | |
| Cytogenetics (karyotype) | Important for the diagnosis and classification of AML and some are associated with distinctive clinicopathologic features, have prognostic significance, and/or influence the choice of therapy. 57 |
| c‐kit | Associated with a poor prognosis. 58 |
| TP53 | Aberrations of TP53 are associated with an exceedingly adverse prognosis. TP53 mutations and deletions that include the TP53 locus have a different prognostic impact, with only mutations but not deletions significantly influencing the survival of these patients. 59 |
| IdH 1 | Occur concurrently with NPM1 mutations, associated with CEBPA and absence of FLT3 abnormalities. It is associated with worse outcomes among intermediate risk diseases. 60 , 61 |
| IdH 2 | The prognostic effect is inconsistent. 61 |
| NPM1 | One of the most commonly mutated genes is associated with a better risk prognosis. 62 |
| FLT3 | The most frequent genetic alteration and a poor prognostic factor. 63 |
| CEBPA | Associated with lower relapse rate, improved survival, and a favorable risk category. 64 |
| CD33 | CD33 is an excellent myeloid marker and is commonly used for the diagnosis of AML. 65 |
Ortiz‐Ortiz KJ, Torres‐Cintrón CR, Suárez Ramos T, Castañeda‐Avila MA, Cotto Santana LA, Tortolero‐Luna G. Patterns of use of biological and genetic markers for chronic lymphocytic leukemia and acute myeloid leukemia in Puerto Rico. Cancer Med. 2023;12:6889‐6901. doi: 10.1002/cam4.5482
DATA AVAILABILITY STATEMENT
This study includes data from the Puerto Rico Central Cancer Registry (PRCCR). Therefore, data from this study will not be available because of the confidentiality agreement between PRCCR and the authors. Nevertheless, investigators could obtain the data through PRCCR following the confidentiality procedures upon reasonable request. PRCCR data can be requested through the following site: http://www.rcpr.org/Datos‐de‐Cancer/Acceso‐a‐Datos.
REFERENCES
- 1. Mertens D, Stilgenbauer S. Prognostic and predictive factors in patients with chronic lymphocytic leukemia: relevant in the era of novel treatment approaches? J Clin Oncol. 2014;32:869‐872. [DOI] [PubMed] [Google Scholar]
- 2. Medeiros BC, Pandya BJ, Hadfield A, et al. Treatment patterns in patients with acute myeloid leukemia in the United States: a cross‐sectional, real‐world survey. Curr Med Res Opin. 2019;35:927‐935. [DOI] [PubMed] [Google Scholar]
- 3. Torres‐Cintrón CR, Alvarado‐Ortiz M, Román‐Ruiz Y, Ortiz‐Ortiz KJ, Zavala‐Zegarra D, Tortolero‐Luna G. Cancer in Puerto Rico, 2012–2016. San Juan; 2020. [Google Scholar]
- 4. US Census Bureau . American Community Survey 5‐Year Estimates, 2015‐2019. US Census Bureau; 2020. [Google Scholar]
- 5. Hinojosa J, Meléndez E, Severino K Population Decline and School Closure in Puerto Rico. (2019).
- 6. International CLL‐IPI working group . An international prognostic index for patients with chronic lymphocytic leukaemia (CLL‐IPI): a meta‐analysis of individual patient data. Lancet Oncol. 2016;17:779‐790. [DOI] [PubMed] [Google Scholar]
- 7. Rosenquist R, Cortese D, Bhoi S, Mansouri L, Gunnarsson R. Prognostic markers and their clinical applicability in chronic lymphocytic leukemia: where do we stand? Leuk Lymphoma. 2013;54:2351‐2364. [DOI] [PubMed] [Google Scholar]
- 8. Wertheim GBW. Molecular characterization and testing in acute myeloid leukemia. J Hematop. 2015;8:177‐189. [Google Scholar]
- 9. Lagunas‐Rangel FA, Chávez‐Valencia V, Gómez‐Guijosa MÁ, Cortes‐Penagos C. Acute myeloid leukemia—genetic alterations and their clinical prognosis. Int J Hematol Stem Cell Res. 2017;11:329‐339. [PMC free article] [PubMed] [Google Scholar]
- 10. Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgVH status and immunophenotyping. Leukemia. 2007;21:2442‐2451. [DOI] [PubMed] [Google Scholar]
- 11. Kröber A, Bloehdorn J, Hafner S, et al. Additional genetic high‐risk features such as 11q deletion, 17p deletion, and V3‐21 usage characterize discordance of ZAP‐70 and VH mutation status in chronic lymphocytic leukemia. J Clin Oncol. 2006;24:969‐975. [DOI] [PubMed] [Google Scholar]
- 12. Wang ML, Bailey NG. Acute myeloid leukemia genetics risk stratification and implications for therapy. Arch Pathol Lab Med. 2015;139:1215‐1223. [DOI] [PubMed] [Google Scholar]
- 13. D'Hoore W, Bouckaert A, Tilquin C. Practical considerations on the use of the charlson comorbidity index with administrative data bases. J Clin Epidemiol. 1996;49:1429‐1433. [DOI] [PubMed] [Google Scholar]
- 14. Klabunde CN, Potosky AL, Legler JM, Warren JL. Development of a comorbidity index using physician claims data. J Clin Epidemiol. 2000;53:1258‐1267. [DOI] [PubMed] [Google Scholar]
- 15. Yun X, Zhang Y, Wang X. Recent progress of prognostic biomarkers and risk scoring systems in chronic lymphocytic leukemia. BMC. 2020;8:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prada‐Arismendy J, Arroyave JC, Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63‐76. [DOI] [PubMed] [Google Scholar]
- 17. Seymour EK, Ruterbusch JJ, Beebe‐Dimmer JL, Schiffer CA. Real‐world testing and treatment patterns in chronic lymphocytic leukemia: a SEER patterns of care analysis. Cancer. 2019;125:135‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mato A, Nabhan C, Kay NE, et al. Prognostic testing patterns and outcomes of chronic lymphocytic leukemia patients stratified by fluorescence In situ hybridization/cytogenetics: a real‐world clinical experience in the connect CLL registry. Clin Lymphoma Myeloma Leuk. 2018;18:114.e2‐124.e2. [DOI] [PubMed] [Google Scholar]
- 19. Ortiz‐Ortiz KJ, Tortolero‐Luna G, Ríos‐Motta R, et al. Use of adjuvant chemotherapy in patients with stage III colon cancer in Puerto Rico: a population‐based study. PLoS One. 2018;13:e0194415. doi: 10.1371/journal.pone.0194415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cramer P, Hallek M. Prognostic factors in chronic lymphocytic leukemia—what do we need to know? Nat Rev Clin Oncol. 2011;8:38‐47. [DOI] [PubMed] [Google Scholar]
- 21. Pinilla‐Ibarz J, Emole J. Chronic lymphocytic leukemia in the elderly, which investigations are necessary: a map for the practicing oncologist. Cancer Control. 2015;22:7‐16. [DOI] [PubMed] [Google Scholar]
- 22. Stauder R, Eichhorst B, Hamaker ME, et al. Management of chronic lymphocytic leukemia (CLL) in the elderly: a position paper from an international Society of Geriatric Oncology (SIOG) task force. Ann Oncol off J Eur Soc Med Oncol. 2017;28:218‐227. [DOI] [PubMed] [Google Scholar]
- 23. Mozaheb Z. Treating the elderly patient with chronic lymphocytic leukemia: current and emerging options. Blood Lymphat Cancer Targets Ther. 2014;4:9‐14. [Google Scholar]
- 24. Barrientos JC. Management of chronic lymphocytic leukemia in the elderly. Cancer Control. 2015;22:17‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910‐1916. [DOI] [PubMed] [Google Scholar]
- 26. Wierda WG, Byrd JC, Abramson JS, et al. Chronic lymphocytic leukemia/small lymphocytic lymphoma, version 4.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2020;18:185‐217. [DOI] [PubMed] [Google Scholar]
- 27. Amaya‐Chanaga CI, Rassenti LZ. Biomarkers in chronic lymphocytic leukemia: clinical applications and prognostic markers. Best Pract Res Clin Haematol. 2016;29:79‐89. [DOI] [PubMed] [Google Scholar]
- 28. Abruzzo LV, Herling CD, Calin GA, et al. Trisomy 12 chronic lymphocytic leukemia expresses a unique set of activated and targetable pathways. Haematologica. 2018;103:2069‐2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stilgenbauer S, Bullinger L, Lichter P, Döhner H. Genetics of chronic lymphocytic leukemia: genomic aberrations and VH gene mutation status in pathogenesis and clinical course. Leukemia. 2002;16:993‐1007. [DOI] [PubMed] [Google Scholar]
- 30. Durak B, Akay OM, Aslan V, et al. Prognostic impact of chromosome alterations detected by FISH in Turkish patients with B‐cell chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2009;188:65‐69. [DOI] [PubMed] [Google Scholar]
- 31. Mato A, Nabhan C, Kay NE, et al. Real‐world clinical experience in the connect® chronic lymphocytic leukaemia registry: a prospective cohort study of 1494 patients across 199 US centres. Br J Haematol. 2016;175:892‐903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Autore F, Strati P, Laurenti L, Ferrajoli A. Morphological, immunophenotypic, and genetic features of chronic lymphocytic leukemia with trisomy 12: a comprehensive review. Haematologica. 2018;103:931‐938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Del Giudice I, Foà R. Another step forward in the 20‐year history of IGHV mutations in chronic lymphocytic leukemia. Haematologica. 2019;104:219‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Curovic Rotbain EC, Da Cunha‐Bang C, Hjalgrim H, et al. IGHV mutation status and outcome in CLL patients treated with chemoimmunotherapy: a Nationwide analysis. Blood. 2017;130:1742. [Google Scholar]
- 35. Crombie J, Davids MS. IGHV mutational status testing in chronic lymphocytic leukemia. Am J Hematol. 2017;92:1393‐1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaidano G, Rossi D. The mutational landscape of chronic lymphocytic leukemia and its impact on prognosis and treatment. Hematology. 2017;2017:329‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Molica S, Shanafelt TD, Giannarelli D, et al. The chronic lymphocytic leukemia international prognostic index (CLL‐IPI) predicts time to first treatment in early CLL: independent validation in a prospective cohort of early stage patients. Am J Hematol. 2017;91:1090‐1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campo E, Cymbalista F, Ghia P, et al. TP53 aberrations in chronic lymphocytic leukemia: an overview of the clinical implications of improved diagnostics. Haematologica. 2018;103:1956‐1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zenz T, Eichhorst B, Busch R, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473‐4479. [DOI] [PubMed] [Google Scholar]
- 40. Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122‐2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079‐1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kottaridis PD, Gale RE, Frew ME, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the united king. Blood. 2001;98:1752‐1759. [DOI] [PubMed] [Google Scholar]
- 43. Chotirat S, Thongnoppakhun W, Promsuwicha O, Boonthimat C, Auewarakul CU. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol. 2012;5:5. doi: 10.1186/1756-8722-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malaise M, Steinbach D, Corbacioglu S. Clinical implications of c‐kit mutations in acute myelogenous leukemia. Curr Hematol Malig Rep. 2009;4:77‐82. [DOI] [PubMed] [Google Scholar]
- 45. Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636‐3643. [DOI] [PubMed] [Google Scholar]
- 46. Horbinski C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 2013;125:621‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu X, Gong Y. Isocitrate dehydrogenase inhibitors in acute myeloid leukemia. Biomark Res. 2019;7:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322‐2333. [PubMed] [Google Scholar]
- 49. Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from cancer and leukemia group B (CALGB 8461). Blood. 2002;100:4325‐4336. [DOI] [PubMed] [Google Scholar]
- 50. Schoch C, Kern W, Schnittger S, Büchner T, Hiddemann W, Haferlach T. The influence of age on prognosis of de novo acute myeloid leukemia differs according to cytogenetic subgroups. Haematologica. 2004;89(9):1082‐1090. [PubMed] [Google Scholar]
- 51. Jahns‐Streubel G, Braess J, Schoch C, et al. Cytogenetic subgroups in acute myeloid leukemia differ in proliferative activity and response to GM‐CSF. Leukemia. 2001;15:377‐384. [DOI] [PubMed] [Google Scholar]
- 52. Lazarevic V, Hörstedt A‐S, Johansson B, et al. Incidence and prognostic significance of karyotypic subgroups in older patients with acute myeloid leukemia: the Swedish population‐based experience. Blood Cancer J. 2014;4:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bendari M, Khoubila N, Cherkaoui S, et al. Acute myeloid leukemia (AML) in elderly: cytogenetic characteristics of patients treated at hematology and pediatric oncology center in Casablanca. Open Access Maced J Med Sci. 2018;6:2328‐2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rack KA, van den Berg E, Haferlach C, et al. European recommendations and quality assurance for cytogenomic analysis of haematological neoplasms: reponse to the comments from the francophone Group of Hematological Cytogenetics (GFCH). Leukemia. 2020;34:2262‐2264. doi: 10.1038/s41375-020-0736-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nabhan C, Raca G, Lynn Wang Y. Predicting prognosis in chronic lymphocytic leukemia in the contemporary era. JAMA Oncol. 2015;1:965‐974. [DOI] [PubMed] [Google Scholar]
- 56. Guarini A, Marinelli M, Tavolaro S, et al. Atm gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica. 2012;97:47‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moarii M, Papaemmanuil E. Classification and risk assessment in AML: integrating cytogenetics and molecular profiling. Hematology. 2017;2017:37‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ayatollahi H, Shajiei A, Sadeghian MH, et al. Prognostic importance of C‐KIT mutations in Core binding factor acute myeloid leukemia: a systematic review. Hematol Oncol Stem Cell Ther. 2017;10:1‐7. [DOI] [PubMed] [Google Scholar]
- 59. Prochazka KT, Pregartner G, Rücker FG, et al. Clinical implications of subclonal TP53 mutations in acute myeloid leukemia. Haematologica. 2019;104:516‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang M, Yang C, Zhang L, Schaar DG. Molecular mutations and their cooccurrences in cytogenetically normal acute myeloid leukemia. Stem Cells Int. 2017;2017:1‐11. doi: 10.1155/2017/6962379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patnaik MM, Hanson CA, Hodnefield JM, et al. Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: a Mayo Clinic study of 277 patients. Leukemia. 2012;26:101‐105. [DOI] [PubMed] [Google Scholar]
- 62. Rau R, Brown P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity. Hematol Oncol. 2009;27:171‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chillón MC, Fernández C, García‐Sanz R, et al. FLT3‐activating mutations are associated with poor prognostic features in AML at diagnosis but they are not an independent prognostic factor. Hematol J. 2004;5:239‐246. [DOI] [PubMed] [Google Scholar]
- 64. Pastore F, Kling D, Hoster E, et al. Long‐term follow‐up of cytogenetically normal CEBPA‐mutated AML. J Hematol Oncol. 2014;7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Walter RB, Appelbaum FR, Estey EH, Bernstein ID. Acute myeloid leukemia stem cells and CD33‐targeted immunotherapy. Blood. 2012;119:6198‐6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study includes data from the Puerto Rico Central Cancer Registry (PRCCR). Therefore, data from this study will not be available because of the confidentiality agreement between PRCCR and the authors. Nevertheless, investigators could obtain the data through PRCCR following the confidentiality procedures upon reasonable request. PRCCR data can be requested through the following site: http://www.rcpr.org/Datos‐de‐Cancer/Acceso‐a‐Datos.


