Abstract
Congenital anomalies of the kidneys and urinary tract (CAKUT) constitute the most common cause of early-onset chronic kidney disease. In a previous study, we identified a heterozygous truncating variant in nuclear receptor-interacting protein 1 (NRIP1) as CAKUT causing via dysregulation of retinoic acid signaling. This large family remains the only family with NRIP1 variant reported so far. Here, we describe one additional CAKUT family with a truncating variant in NRIP1. By whole-exome sequencing, we identified one heterozygous frameshift variant (p.Asn676Lysfs*27) in an isolated CAKUT patient with bilateral hydroureteronephrosis and right grade V vesicoureteral reflux (VUR) and in the affected father with left renal hypoplasia. The variant is present twice in a heterozygous state in the gnomAD database of 125,000 control individuals. We report the second CAKUT family with a truncating variant in NRIP1, confirming that loss-of-function mutations in NRIP1 are a novel monogenic cause of human autosomal dominant CAKUT.
Keywords: CAKUT, NRIP1, whole-exome sequencing
1 ∣. INTRODUCTION
Congenital anomalies of the kidneys and urinary tract (CAKUT) comprise a large spectrum of congenital malformations that range from severe manifestations, such as renal agenesis, to milder conditions, for example, vesicoureteral reflux (Caruana & Bertram, 2015). CAKUT occurs in approximately 3–6 per 1000 live births. It constitutes the most common cause of chronic kidney disease in the first three decades of life (Vivante & Hildebrandt, 2016). Several lines of evidence in humans and mouse models indicate that CAKUT is often caused by recessive or dominant mutations in single (monogenic) genes (van der Ven et al., 2018). To date, genetic variants in more than 50 genes have been associated with CAKUT in humans, explaining 13%–20% of CAKUT cases (Bekheirnia et al., 2017; Kohl et al., 2021; van der Ven et al., 2018). Nuclear receptor-interacting protein 1 (NRIP1, also known as RIP140) is a nuclear receptor transcriptional co-factor, which acts as a transcriptional repressor by interacting with retinoic acid receptors (RARs) and retinoid receptors (RXRs) (Augereau et al., 2006). In a previous study, we identified a heterozygous truncating variant in NRIP1 as causing urinary tract malformations via dysregulation of retinoic acid signaling. In this large family with seven affected individuals, segregation of the NRIP1 variant followed an autosomal dominant inheritance pattern. This remains the only family with an NRIP1 mutation reported so far (Vivante et al., 2017). To test whether there are additional families harboring NRIP1 variants, we examined whole-exome sequencing (WES) data from 551 families with CAKUT (see Supplementary methods for detailed information of study participants and methods). Here, we describe one additional CAKUT family with a truncating NRIP1 variant. Moreover, a control-based analysis shows that rare NRIP1 missense variants in CAKUT patients are not likely pathogenic or might need more functional evidence.
2 ∣. A TRUNCATING NRIP1 VARIANT IN A FAMILY WITH CAKUT
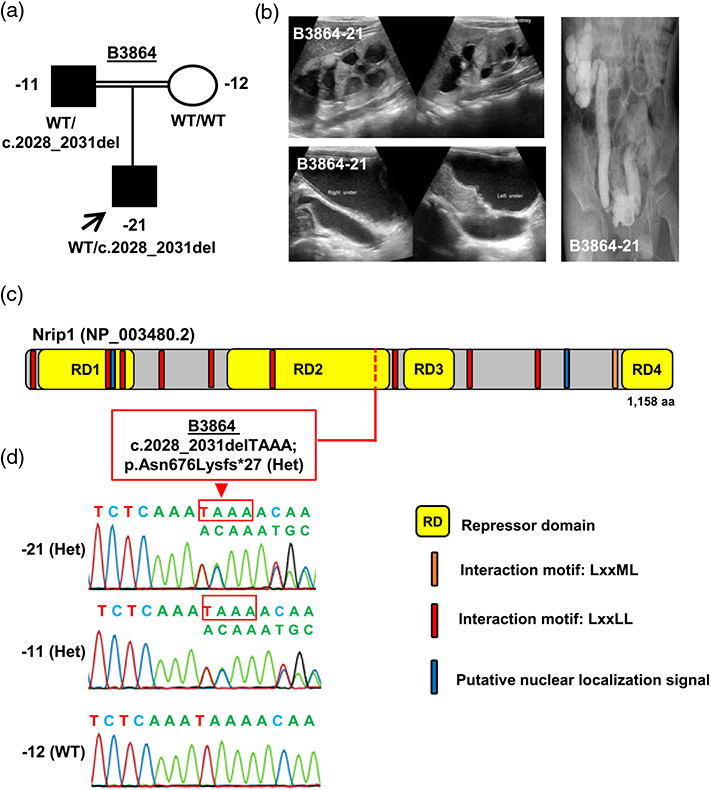
We identified one heterozygous frameshift NRIP1 variant (c.2028_2031delTAAA; p.Asn676Lysfs*27) in family B3864 (Figure 1a). The variant is present twice in a heterozygous state in the gnomAD database of 125,000 control individuals and was absent in an ethnically matched control with 324 middle eastern/Arabic individuals. Sanger confirmation revealed that the variant was inherited from the father (B3864-11) who presented with left renal hypoplasia (Figure 1a-d). Individual B3864-21 was an 11-year-old Arabic boy with neurogenic bladder, bilateral hydroureteronephrosis, and grade V VUR on the right side (Figure 1b). The renal ultrasonogram showed bilateral hyperechogenic renal parenchyma, and was suggestive of bilateral duplex systems with common renal pelvises (Figure 1b). His kidney function test indicated chronic kidney disease stage 3 (glomerular filtration rate (GFR) 58.8 ml/min/1.73 m2). No extra-renal syndromic manifestations were observed in the patient nor his father.
FIGURE 1.
Whole-exome sequencing identifies a heterozygous NRIP1 frameshift variant in a family with congenital anomalies of the kidneys and urinary tract (CAKUT). (a) Pedigree and genotype information for members of family B3864. Squares indicate males, circles females, filled symbols are affected individuals, and open symbols indicate healthy individuals. Proband is denoted by a black arrow. The proband (−21) and the affected father (−11) both have CAKUT and carried a heterozygous frameshift variant in NRIP1 (c.2028_2031delTAAA; p.Asn676Lysfs*27). (b) Diagnostic images of individual B3864-21. Ultrasound of the kidneys (upper panel) showing bilaterally hyperechoic renal parenchyma with common renal pelvises suggestive of duplex systems. Ultrasound of ureters (lower panel) showing bilateral hydroureteronephrosis. Micturating cystourethrogram (right panel) showing right VUR grade V. (c) Depicts the protein domain structure of human Nrip1 protein relative to the position of the index heterozygous NRIP1 variant c.2028_2031delTAAA, which leads to a frameshift and premature stop codon resulting in p. Asn676Lysfs*27. aa, amino acids. (d) Sequencing chromatograms of the heterozygous variant c.2028_2031delTAAA; p.Asn676Lysfs*27 detected in the affected proband and the affected father (−11), and WT sequence of NRIP1 in the mother (−12)
3 ∣. ADDITIONAL CAKUT FAMILIES WITH MISSENSE VARIANTS IN NRIP1
We identified three additional heterozygous NRIP1 missense variants with uncertain significance in three unrelated individuals with CAKUT. In particular, individual B633-21 with multicystic dysplastic left kidney carried a heterozygous missense variant (c.456A>C; p.Gln152His) (Table S1 and Figure S1). Individual A3460-21 with left renal agenesis harbored a heterozygous missense variant (c.970C>T; p.His324Tyr) (Table S1 and Figure S1). In individual A782-21 with right renal agenesis, we identified a heterozygous missense variant (c.1343G>A; p. Arg448Gln) (Table S1 and Figure S1). The three variants occurred 6, 0, and 2 times, respectively, as heterozygous variants in the gnomAD database of 125,000 control individuals. All three missense variants are in the repressor domains of the Nrip1 protein and show high evolutionary conservation (Table S1).
4 ∣. NEGATIVE CONTROL EVALUATION FOR RARE NRIP1 VARIANTS
To distinguish CAKUT-associated variants from benign rare variants, we screened for NRIP1 variants in a negative control cohort of 520 samples of individuals who had nephrotic syndrome rather than CAKUT. The same evaluation criteria were applied to NRIP1 variants in this negative control cohort as in the CAKUT case cohort (see Supplementary methods). Following evaluation, no truncating variants were observed in the control cohort. However, four rare heterozygous NRIP1 missense variants were detected (Table S2). The four variants were rare in the gnomAD database and were deemed probably damaging by three in-silico programs (PolyPhen-2, MutationTaster, and SIFT) (Table S2). The missense variant (c.456A>C; p.Gln152His) that was identified in the individual B633-21, was also found in the control group. The four individuals harboring the NRIP1 variants in the control cohort have been carefully phenotyped to exclude CAKUT. In silico analyses incorporating evolutionary conservation failed to distinguish inferred functional differences between CAKUT-associated and control variants (Table S2). Furthermore, no domain specificity was observed for the missense variants identified in CAKUT cases and controls (Figure S2).
Informed consent was obtained from the patients discussed in the report.
5 ∣. DISCUSSION
In family B3864, the proband inherited the loss-of-function variant (p. Asn676Lysfs*27) from the affected father. This family is the second CAKUT family with a truncating variant in NRIP1. In our previous study, we identified a heterozygous truncating variant (p.Trp93fs*) in NRIP1, which segregated with seven affected individuals in the family. Mice heterozygous for a null allele of Nrip1 showed dysplastic kidneys with cystic dilations, severe hydoureter with hydronephrosis, and ureterocele (Vivante et al., 2017), which are consistent with the spectrum of CAKUT phenotypes in families with NRIP1 variants.
When we screened for rare NRIP1 variants in a negative control cohort to distinguish CAKUT-associated variants from benign rare variants, we found that missense mutations were detected in three out of 551 families in the CAKUT group versus four out of 520 families in the negative control group. In silico analyses incorporating evolutionary conservation failed to distinguish potential functional differences between CAKUT-associated and control variants. We also tested the hypothesis that the control versus CAKUT cohort difference could be protein domain-specific, but no domain specificity was observed (as shown in Figure S2). Taken together, these findings suggest that it is unlikely that missense variants of NRIP1 cause CAKUT.
Nrip1 was first identified as a hormone-recruited nuclear receptor transcriptional cofactor (Cavailles et al., 1995). However, Nrip1 was found to be interacting with and regulating the function of additional nuclear receptors such as retinoic acid receptor alpha (RARA), retinoid X receptor alpha (RXRA), peroxisome proliferator activated receptor alpha (PPARA), and vitamin D receptor (VDR) (Nautiyal, 2017). The recruitment of Nrip1 to nuclear receptors is accomplished by nine LXXLL motifs (as indicated in Figure 1c) and a 10th LXXML motif (as indicated in Figure 1c). Christian et al. characterized four distinct autonomous repression domains (RD1–RD4) in Nrip1, which provide platforms for different corepressor complexes (Christian et al., 2004). We previously demonstrated that NRIP1 serves as a feedback inhibitor for retinoic acid signaling in the nephric duct and ureter development, and that loss of this feedback inhibition leads to congenital renal and urinary tract malformations (Vivante et al., 2017). All three missense variants identified in the CAKUT group were located in the repressor domains. To completely resolve the pathogenicity of these missense mutations observed in CAKUT families, further comprehensive in vitro or in vivo functional characterization is needed.
In conclusion, by the discovery of a second CAKUT family with a truncating variant in NRIP1, we confirm heterozygous loss-of-function variants in NRIP1 as a monogenic cause of human autosomal dominant CAKUT. A control-based analysis shows that rare NRIP1 missense variants in CAKUT patients are not likely pathogenic.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the families and study individuals for their contribution. Friedhelm Hildebrandt is supported by the William E. Harmon Professor of Pediatrics. He is also supported by the Begg Family Foundation. We also thank the Yale Center for Mendelian Genomics for whole-exome sequencing analysis (U54HG006504). This research was supported by grants from the National Institutes of Health to Friedhelm Hildebrandt. (DK076683, DK088767, and DK068306).
Footnotes
CONFLICT OF INTEREST
Friedhelm Hildebrandt is a cofounder and Scientific Advisory Committee (S.A.C.) member and holds stock in Goldfinch-Bio. All other authors declare that they have no competing financial interests.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher's website.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Augereau P, Badia E, Carascossa S, Castet A, Fritsch S, Harmand PO, Jalaguier S, & Cavaillès V (2006). The nuclear receptor transcriptional coregulator RIP140. Nuclear Receptor Signaling, 4, e024. 10.1621/nrs.04024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekheirnia MR, Bekheirnia N, Bainbridge MN, Gu S, Coban Akdemir ZH, Gambin T, Janzen NK, Jhangiani SN, Muzny DM, Michael M, Brewer ED, Elenberg E, Kale AS, Riley AA, Swartz SJ, Scott DA, Yang Y, Srivaths PR, Wenderfer SE, … Lamb DJ (2017). Whole-exome sequencing in the molecular diagnosis of individuals with congenital anomalies of the kidney and urinary tract and identification of a new causative gene. Genetics in Medicine, 19(4), 412–420. 10.1038/gim.2016.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana G, & Bertram JF (2015). Congenital anomalies of the kidney and urinary tract genetics in mice and men. Nephrology (Carlton), 20(5), 309–311. 10.1111/nep.12402 [DOI] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner PJ, & Parker MG (1995). Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. The EMBO Journal, 14(15), 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Tullet JM, & Parker MG (2004). Characterization of four autonomous repression domains in the corepressor receptor interacting protein 140. The Journal of Biological Chemistry, 279(15), 15645–15651. 10.1074/jbc.M313906200 [DOI] [PubMed] [Google Scholar]
- Kohl S, Habbig S, Weber LT, & Liebau MC (2021). Molecular causes of congenital anomalies of the kidney and urinary tract (CAKUT). Molecular and Cellular Pediatrics, 8(1), 2. 10.1186/s40348-021-00112-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal J (2017). Transcriptional coregulator RIP140: An essential regulator of physiology. Journal of Molecular Endocrinology, 58(3), R147–R158. 10.1530/JME-16-0156 [DOI] [PubMed] [Google Scholar]
- van der Ven AT, Connaughton DM, Ityel H, Mann N, Nakayama M, Chen J, Vivante A, Hwang DY, Schulz J, Braun DA, Schmidt JM, Schapiro D, Schneider R, Warejko JK, Daga A, Majmundar AJ, Tan W, Jobst-Schwan T, Hermle T, … Hildebrandt F (2018). Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. The Journal of the American Society of Nephrology, 29(9), 2348–2361. 10.1681/ASN.2017121265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivante A, & Hildebrandt F (2016). Exploring the genetic basis of early-onset chronic kidney disease. Nature Reviews Nephrology, 12(3), 133–146. 10.1038/nrneph.2015.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivante A, Mann N, Yonath H, Weiss AC, Getwan M, Kaminski MM, Bohnenpoll T, Teyssier C, Chen J, Shril S, van der Ven AT, Ityel H, Schmidt JM, Widmeier E, Bauer SB, Sanna-Cherchi S, Gharavi AG, Lu W, Magen D, … Hildebrandt F (2017). A dominant mutation in nuclear receptor interacting protein 1 causes urinary tract malformations via dysregulation of retinoic acid signaling. The Journal of the American Society of Nephrology, 28(8), 2364–2376. 10.1681/ASN.2016060694 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.