Abstract
Cell reprogramming is a revolutionized biotechnology that offers a powerful tool to engineer cell fate and function for regenerative medicine, disease modeling, drug discovery, and beyond. Leveraging advances in biomaterials and micro/nanotechnologies can enhance the reprogramming performance in vitro and in vivo through the development of delivery strategies and the control of biophysical and biochemical cues. In this review, we present an overview of the state-of-the-art technologies for cell reprogramming and highlight the recent breakthroughs in engineering biomaterials with micro/nanotechnologies to improve reprogramming efficiency and quality. Finally, we discuss future directions and challenges for reprogramming technologies and clinical translation.
Keywords: cell reprogramming, biomaterials, nanotechnology, biophysical cues, biochemical cues, three-dimensional niches, tissue engineering, drug delivery
Graphical Abstrac
During mammalian development, cells progressively differentiate from a totipotent zygote into a terminally differentiated state with functionally and phenotypically distinct fates to form organized tissues and organs. Such cell fate specifications are precisely controlled and mediated by definitive transcriptional regulatory networks.1,2 Terminally differentiated somatic cells normally maintain their phenotype throughout the lifespan. Adult cells, however, may lose the capability to heal certain damaged tissues, such as the central nervous system,3,4 heart,5,6 and cartilage.7 Cellular reprogramming technology provides a revolutionizing approach that bypasses the rules of cell and developmental biology in cell fate determination, enabling the conversion of mature somatic cells to pluripotent cells or to other different cell lineages.8 Since the landmark work of reprogramming somatic cells into induced pluripotent stem cells (iPSCs) with a subset of specific transcription factors by Yamanaka in 2006,9 tremendous advances have been achieved not only in iPSC reprogramming and differentiation but also in other cell-reprogramming approaches such as direct or indirect cell reprogramming in vitro and in vivo.10-14 Such significant developments of reprogramming technologies have offered multiple powerful tools and strategies for regenerative medicine, disease modeling, and drug discovery.15-17
Biomaterials play imperative roles in treating disease and improving the quality of life for patients. Many natural and synthetic biomaterials have been engineered for diverse biomedical applications, including drug delivery,18,19 cell transplantation,20,21 tissue engineering,22,23 cancer therapy,24-26 device-based therapies, and medical diagnostics and imaging.27,28 The development of various biomaterials-based platforms have offered robust approaches to modulating cell reprogramming in vitro and in vivo.29-34 In particular, the physicochemical properties of biomaterials can be tailored by sophisticated chemistry and micro/nanoprocessing, not only for the delivery of reprogramming factors but also as tunable physicochemical cues on two-dimensional (2D) substrates or in three-dimensional (3D) microenvironments.32,35-38
Here, we review the development and current state of cell reprogramming, with a focus on engineered biomaterials-based drug-delivery strategies and biophysical and biochemical cues for modulating cell reprogramming, followed by a discussion on the future directions of this fast-growing field.
CELL REPROGRAMMING: DEVELOPMENT AND CURRENT STATE
Although Spemann initially proposed the concept which would later be coined as somatic cell nuclear transfer (SCNT) in 1938, it was not until 1952 that Briggs and King successfully developed the technique and demonstrated that introducing the nuclei of a blastula cell into an enucleated egg could give rise to a cleaved blastula.8 Ten years later, Gurdon transferred the nuclei from differentiated Xenopus intestinal epithelial cells into enucleated eggs, and this resulted in the formation of fully functional tadpoles.39 These findings revealed that the reversal of cell differentiation, now known as reprogramming, was feasible, which initiated mammalian cloning experiment using SCNT in 1996 and the derivation of SCNT-derived human embryonic stem cell lines in 2013.40,41
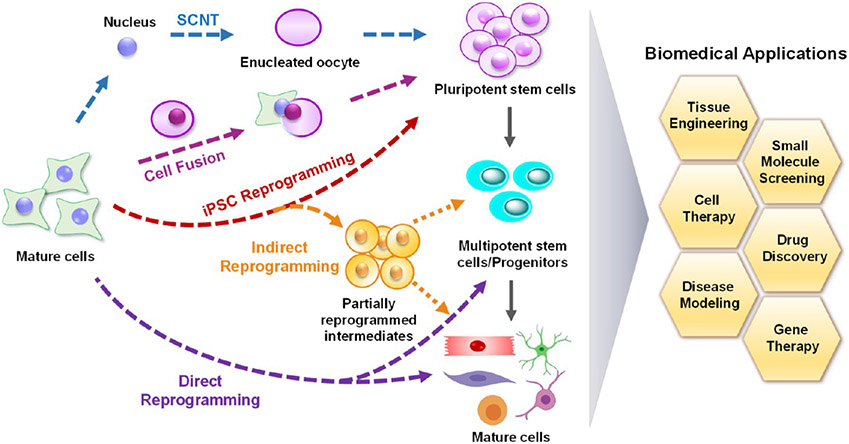
In comparison to SCNT and cell fusion, cell reprogramming offers a more robust approach through the ectopic expression of defined transcription factors (Figure 1). The concept of reprogramming regained wide interest in 2006 when Takahashi and Yamanaka demonstrated that they were able to generate iPSCs from mouse somatic cells via the combined expression of four transcriptions factors, Oct4, Sox2, Klf-4, and c-Myc (OSKM).9 It has been demonstrated that these iPSCs closely resemble embryonic stem cells (ESCs) in terms of karyotype, stemness phenotype, and differentiation capacity. This significant breakthrough highlighted the possibility of generating any desired cell type from a somatic cell through a combination of iPSC reprogramming and directed differentiation, and numerous pioneering studies have unraveled the mechanisms for cell reprogramming.42-44 Although human ESC-derived cells have been primarily used in clinical trials, iPSCs are considered superior to ESCs without the ethical concerns involving the destruction of embryos. It is encouraging that some preliminary clinical trials have recently launched using human iPSC cells (hiPSCs)-derived cells to treat diverse diseases, including macular degeneration, aplastic anemia, heart disease, endometriosis, Parkinson’s disease, and spinal cord injury.45-49 However, iPSCs can be associated with genetic and epigenetic abnormalities that may arise during the reprogramming process.50 As a result, there is an increased risk of tumorigenicity and immunogenicity when using iPSC-derived cells. Moreover, some additional hurdles, including unstable differentiation, low conversion efficiency, and lengthy differentiation still exist, thereby limiting the clinical translation of iPSC-based cell therapies.
Figure 1.
Cell-reprogramming strategies for biomedical applications. Schematic illustrating the process of SCNT, cell fusion, iPSC reprogramming, indirect reprogramming, and direct reprogramming. Cell reprogramming holds great promise for various biomedical applications, as indicated in the figure. SCNT, somatic cell nuclear transfer; iPSC, induced pluripotent stem cell.
To circumvent these issues, scientists have explored other methods of manipulating cell fate (Figure 1). Direct reprogramming is an alternative strategy that directly converts fully differentiated somatic cells into distinct cell types using transcription factors, microRNAs, or chemical compounds.10 In comparison to iPSC reprogramming, this method is considered faster and safer as cells do not proceed through an intermediate pluripotent state. The direct reprogramming was reported in the late 1980s that the forced expression of MyoD, a transcription factor in the skeletal muscle lineage, could induce mouse embryonic fibroblasts (MEFs) into stable myoblasts.51 Since then, numerous studies in the past two decades have shown that this method can be applied to obtain a wide variety of target cell types, including mouse or human hepatocytes,52,53 neurons,54-56 pancreatic β cells,57,58 renal tubular epithelial cells,59 immune T cells,60 embryonic Sertoli-like cells,61 endothelial cells,62 and cardiomyocytes.63-66 Direct reprogramming is a promising approach for personalized medicine as it has been shown that patient-derived cells recapitulate the disease phenotype, which offers great potential for disease modeling, drug screening, and personalized treatments. For example, direct neuron cell reprogramming has been used to study human disease progression in vitro, such as Alzheimer’s disease.67 Although direct reprogramming can be utilized to generate cells with a mature phenotype, this may not be ideal for clinical translation. Thus, more recently, direct reprogramming has been applied to generate various types of expandable stem cells or progenitors, such as neural stem cells,68 oligodendrocyte precursor cells,69 hepatic stem cells,70 hematopoietic stem cells,71,72 nephron progenitor-like cells,73 intestinal progenitor cells,74 skeletal muscle progenitor cells,75,76 and cardiac progenitor cells.77,78 By providing extensive proliferative capacities and restricted differentiation potentials, these directly reprogrammed progenitors have been shown to be more engraftable and desirable for transplantation in regenerative therapies.71 In addition, it is worth noting that directly reprogrammed cell populations usually have cellular heterogeneity. Thus, cell purification is commonly required to isolate the target cells from the rest of the population using fluorescence-activated cell sorting (FACS),71 immunopanning,69 or other sorting techniques.
Indirect reprogramming has been demonstrated as another reprogramming approach. This process entails converting somatic cells through a transient induction of a partially reprogrammed cell using the Yamanaka factors, followed by differentiation into the target cell type.79 Similar to direct reprogramming, this indirect approach does not give rise to pluripotent cells and thus bypasses the teratoma-forming potential that is typically associated with iPSCs. Using this indirect reprogramming method, several types of lineage-specific cells and multipotent progenitors have been generated from mouse and human cells.80-83
To date, most reprogramming studies have been conducted in vitro. Alternatively, several studies have shown that resident cells can be directly reprogrammed into another desired cell type in situ, such as mouse pancreatic cells,84-86 neurons,87-93 oligodendrocytes,94 cardiomyocytes,95-97 hepatocytes,98,99 retinal cells,100 epithelial cells,101 and lymphocytes,102 through the overexpression of lineage-specific transcription factors, microRNAs, small molecules, and other reprogramming factors. Such studies have shed light on an emerging field in exploiting in vivo lineage reprogramming to mend the broken tissues by harnessing and converting internal cells into targeted functional cells without cell transplantation, risks of contamination, and tumorigenesis.14 Notably, the in vivo microenvironment offers an optimal niche for enhancing direct cell reprogramming. Similarly, iPSC reprogramming has also been investigated in vivo.103-107 It has been reported that iPSCs derived in vivo demonstrate totipotency and a closer resemblance to ESCs in comparison to iPSCs generated in vitro.105 In contrast to the finding that DNA damaged and aged cells display lower reprogramming efficiencies in vitro, tissue injury and senescence promotes in vivo OSKM-mediated reprogramming.106,107
Although significant advances have been made for in vitro cell reprogramming during the past decade, in vivo reprogramming is still in the nascent stages. Moreover, numerous obstacles related to reprogramming efficiency, quality, safety, and controllability must be overcome before there is robust translation of reprogramming therapies. Promisingly, engineering proper delivery systems and microenvironmental cues via micro/nanotechnologies is expeditiously promoting vital breakthroughs in the field of cell reprogramming.
DELIVERY SYSTEMS FOR CELL REPROGRAMMING
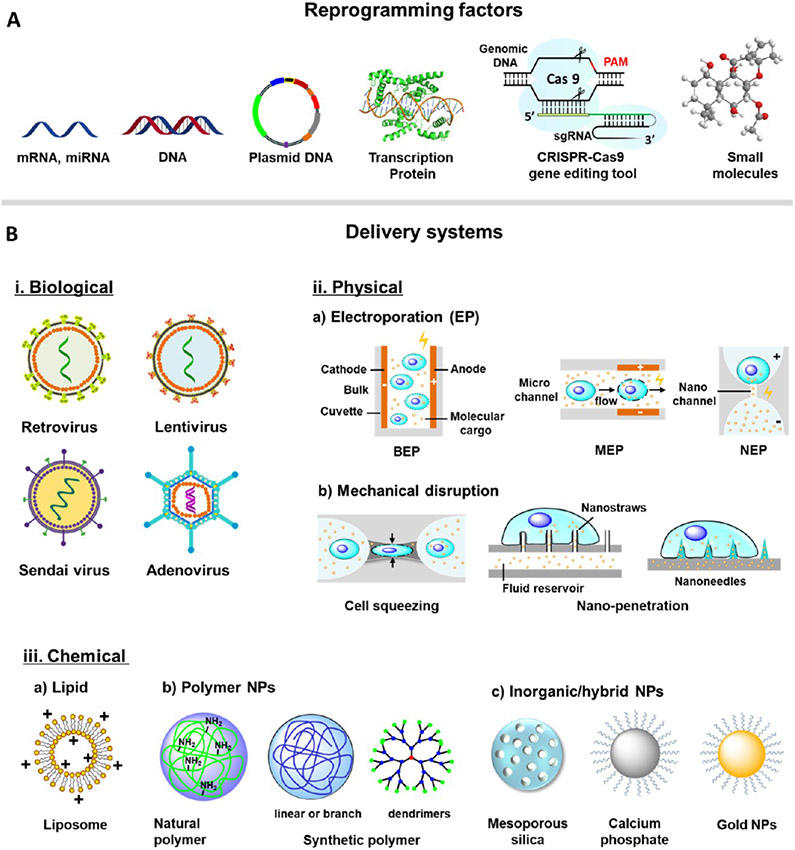
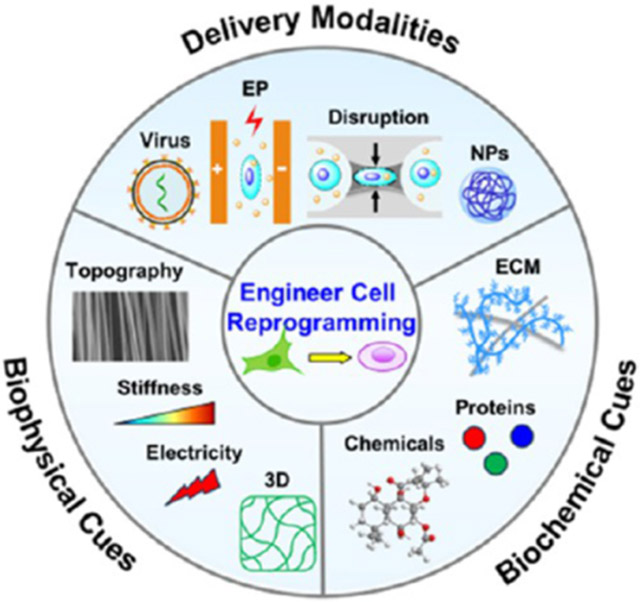
Various genetic and epigenetic regulators or reprogramming factors have been utilized for cell reprogramming (Figure 2A), including transcription factors in the form of proteins,11,108 episomal plasmids,109,110 messenger RNA (mRNA),111 micro-RNA (miRNA),112,113 small molecules,67,114,115 mini-circle DNA,116,117 synthetic transcription factors,118 and CRISPR-Cas9 gene-editing tools.119 It is important to note that although these reprogramming factors can yield differences in the reprogramming efficiency and reproducibility, this can potentially be improved upon using an optimal delivery system.120,121 Thus, the choice of a safe and effective delivery system is paramount in achieving successful reprogramming with specific reprogramming factors.122,123 Current delivery modalities for cell reprogramming can be classified into three groups: biological, physical, and chemical delivery systems (Figure 2B), and their comparisons are summarized in Table 1. Herein, we will focus on the recent advances of physical and chemical delivery systems, considered as biomaterials-based approaches in a broad sense, for cell reprogramming.
Figure 2.
Biomaterial- and micro/nanotechnology-based delivery approaches for cell reprogramming. (A) Cell reprogramming has been achieved using various reprogramming factors. (B) Three major delivery modalities that have been applied for cell reprogramming include biological, physical, and chemical methods. (i) Biological delivery relies on genetically engineered viruses to transfer DNA constructs into cells. (ii) Physical delivery methods include membrane-disruption approaches to deliver reprogramming factors into the cytoplasm or nucleus. (iii) Chemical delivery methods employ lipids, natural or synthetic polymers, and inorganic or hybrid carriers. sgRNA, single guide RNA; AAV, Adeno-associated virus; BEP, bulk electroporation; MEP, microchannel electroporation; NEP, nanochannel electroporation; nanopenetration, nanostructure-mediated membrane penetration; NPs, nanoparticles.
Table 1.
Comparison of Delivery Methods of Reprogramming Factors for Cell Reprogramming
| methods | biological | physical | chemical |
|---|---|---|---|
| advantages |
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|||
| limitations |
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
Biological Delivery Systems.
Viral delivery of reprogramming factors relies on genetically engineered viruses to transduce transcription-factor-encoding plasmids into cells, including integrative vectors based on retrovirus and lentivirus and nonintegrative vectors based on adenovirus, adenovirus-associated virus (AAV), and Sendai virus.64,124 Recombinant viral vectors are often preferred for cell reprogramming because of their high transduction efficiency. However, viral gene delivery usually has limited DNA packaging capacity, unpredictable gene dysfunction, cytotoxicity, immunogenicity, and tumorigenicity. Detailed information on virus-based delivery for iPSC and direct cell reprogramming can be found in several excellent review papers.11,14,120,125,126
Physical Delivery Systems.
Physical delivery strategies involve directly delivering reprogramming factors into the cytoplasm or the nucleus via membrane-disruption methods, including electroporation and mechanical disruption (e.g., cell squeezing and nanostructure penetration).
Electroporation.
Electroporation has become one of the most popular nonviral gene-delivery techniques since initially reported in 1982.127 Bulk electroporation (BEP) is commonly conducted with millimeter- to centimeter-sized chambers, where the cell suspension and molecules to be delivered are mixed in a conducting buffer solution between two electrodes. Electrical pulses are applied to induce transient permeability of cell membrane and diffusion/endocytosis for cytosolic cargo delivery.128 Electroporation-based methods can deliver a diverse range of cargos, including nucleic acids or genes (small interfering RNAs, miRNAs, mRNA, DNA plasmid), proteins or protein complexes, small molecules, and nanoparticles. This multifaceted capability provides the potential for a multitude of in vitro and less amenable to in vivo applications, such as gene editing,129 adoptive immunotherapy,130-132 regeneration medicine,133-135 cell reprogramming,136,137 and others.128,137,138
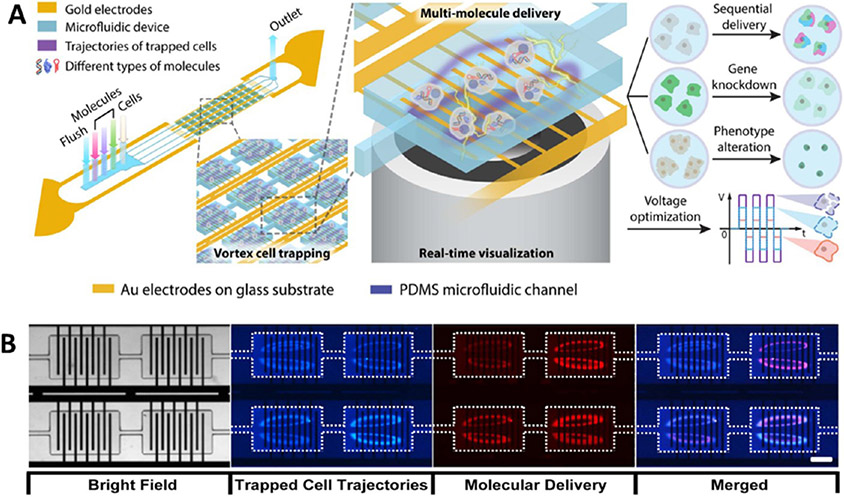
BEP-based delivery has been used to deliver episomal vectors to generate integration-free hiPSCs in vitro109,139 and to induce direct reprogramming into neurons140 and oligodendrocytes in vivo.94 Nevertheless, despite its simplicity, BEP-mediated transfection has drawbacks, including excessive cell damage caused by high electric fields, highly stochastic transfection, and low efficiency of intracellular delivery (diffusion dominated).141,142 To overcome these challenges, microfluidic-based electroporation technologies, such as microchannel electroporation (MEP), have been developed to operate at lower voltages, enabling the enhancement in efficiency and viability,143 single-cell143 or high-throughput molecular delivery,144 and precise dose control.138 Recently, another vortex-assisted microfluidic electroporation system, named microscale symmetrical electroporator array (μSEA), has been developed with integrated micropatterned planar electrodes in the vortex cell-trapping chamber and equipped with real-time visualization functionality (Figure 3). Such a system not only allows for sequential intracellular delivery of various molecules but also presents several other benefits, including dosage control, real-time visualization, multimolecular delivery, high transfection efficiency, and improved cell viability.123 However, the cargo size of the MEP approach is still limited because its cytosolic cargo delivery relies on diffusion and endocytosis.
Figure 3.
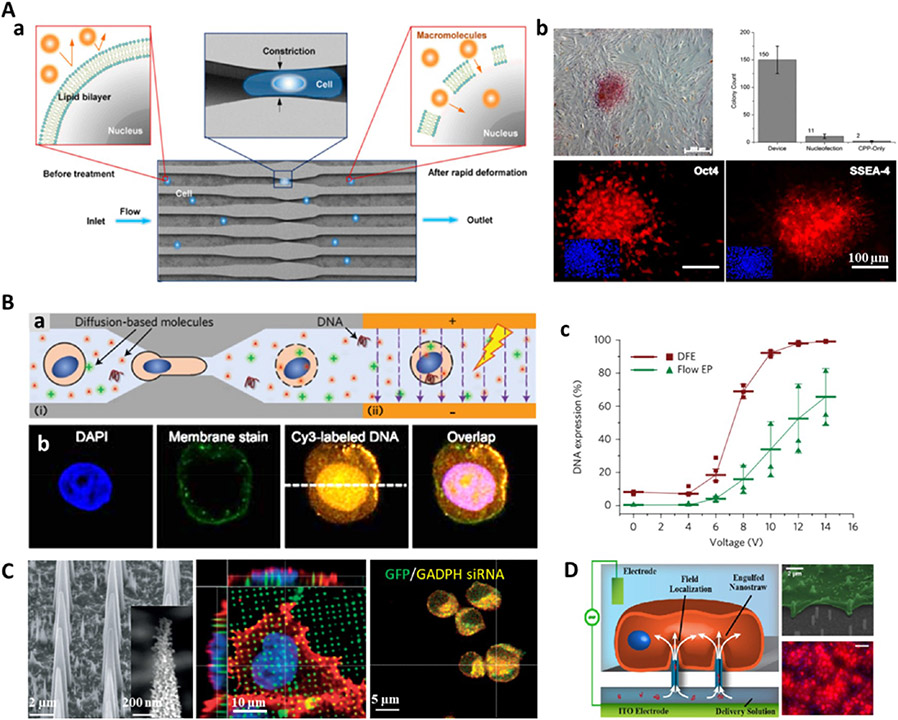
Microchannel or microfluidic electroporation for cargo delivery. (A) Vortex-assisted microfluidic-based electroporation system integrated with micropatterned planar electrodes in the vortex cell-trapping chamber and equipped with real-time visualization functionality. The patterned electrodes were merged within a single cell-trapping chamber with the following parameters: chamber length = 720 μm; chamber width = 480 μm; electrode length = 450 μm; and electrode width = 20 μm. (B) Membrane-impermeable fluorescent molecule delivered into trapped cells via electroporation, HEK 293 cells (blue), propidium iodide (red). Reprinted with permission from ref 123. Copyright 2017 Springer Nature.
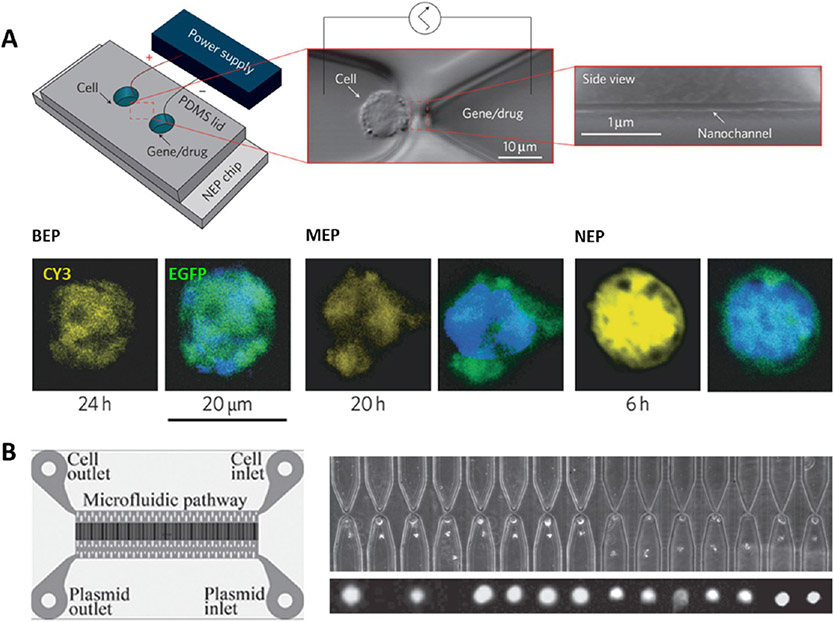
With advances in nanofabrication and engineering techniques, recently, nanoelectroporation (NEP) systems have been developed for efficient cell transfection.145 For example, a nanochannel electroporation device was designed with a microchannel–nanochannel–microchannel structure, and single cells were precisely positioned at the tip of the nanochannel via optical tweezers (Figure 4A).
Figure 4.
NEP-mediated delivery. (A) NEP device comprises a microchannel–nanochannel–microchannel structure and electrodes in the reservoirs. Single cells are precisely positioned using optical tweezers. NEP (90 nm diameter, 3 μm length for nanochannel) transfection of mouse embryonic fibroblasts with Cy3-labeled GFP plasmid shows higher transfection and faster gene expression than using BEP and MEP (5 μm) delivery. Reprinted with permission from ref 141. Copyright 2011 Springer Nature. (B) High-throughput NEP transfection of mouse embryonic fibroblasts with green fluorescent liposomes containing the reprogramming factor (OSKM plasmid). A large number of cells are loaded through centrifugal spinning method. Reprinted with permission from ref 146. Copyright 2014 Wiley.
In contrast to other electroporation methods, NEP can transfect cells faster and more efficiently at the single-cell level with a precise dosage of transfection agents (e.g., oligodeoxynucleotide, siRNA, quantum dots nanoparticles, GFP plasmid), while not affecting cell viability.141 However, it is hard to trap adherent cells by optical tweezers, and it is even less likely suitable for trapping large numbers of fast, adherent cells (e.g., MEFs) for cell reprogramming. Therefore, a more advanced high-throughput NEP biochip has been further developed by the same lab whereby cells were loaded using a centrifugal spinning method (Figure 4B). This technique greatly increased the cell loading efficiency from 10 to 80% compared to optical tweezers and allowed for a large number (i.e., 10 000 cells for a device) of MEF cells to be transfected with OSKM plasmid for mouse iPSC reprogramming.146 After NEP transfection, more than 90% of cells were alive and the transfection efficiency was significantly higher than that with BEP. However, further systematic characterization of specific gene and protein expressions of transfected cells should be performed to determine the final iPSC reprogramming efficiency and elucidate additional advantages of using nanoelectroporation for in vitro cell reprogramming.
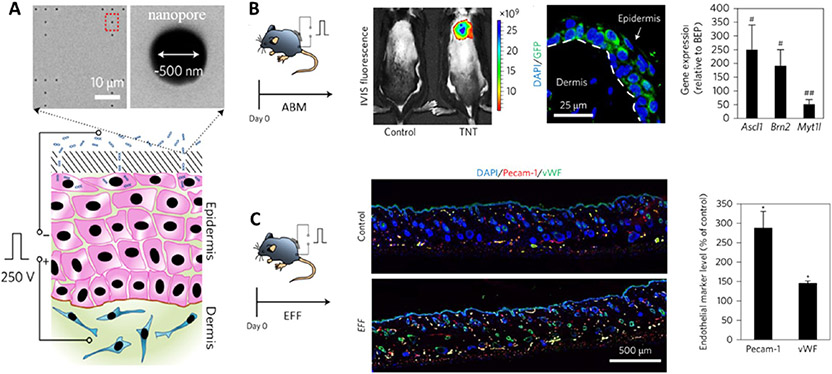
In addition, the NEP techniques also enable tissue nanotransfection (TNT) of nonviral cargos for rapid, highly effective, and deterministic in vivo reprogramming (Figure 5). The TNT platform could efficiently promote the reprogramming of mouse skin cells into neurons with the delivery of Ascl1/Brn2/Myt1l (ABM) plasmids. This approach elicited 50- to 250-fold greater gene expression in vivo compared with conventional bulk electroporation. In particular, via the delivery of Etv2/Foxc2/Fli1 (EFF) plasmids, skin cells could be reprogrammed into endothelial cells to increase vascularization and rescue skin tissue and whole limbs under necrotizing ischemia. Moreover, the TNT platform could also mediate RNA-based transfection of microRNA302/367 cluster to induce pluripotency of skin.136 Another nanotransfection approach is nanostraw electroporation, which can achieve high-throughput cell transfection with dosage control and high uniformity.147 Although the nanoscale electroporation platforms have displayed more efficient and enhanced transfection capabilities, some challenges remain to be addressed, including precisely targeted cell delivery in vivo and complex fabrication protocols that require special expertise for the manufacturing of miniaturized electroporation systems.
Figure 5.
Nanochannel electroporation mediates in vivo reprogramming. (A) Skin tissue nanotransfection through a nanochanneled device and a pulsed electric field to deliver reprogramming factors into skin cells. (B) Reprogramming skin cells into induced neurons with nanotransfection of Ascl1/Brn2/Myt1l (ABM) plasmids. Representative in vivo imaging system (IVIS) fluorescence of mouse skin treated with fluorescein-amidite-labeled DNA. GFP is a reporter of Ascl1 gene. (C) Reprogramming skin cells into induced endothelial cells via nanotransfection of Etv2/Foxc2/Fli1 (EFF) plasmids. Fluorescent staining of Pecam-1 and vWF show increased vascularization, and high-resolution laser speckle imaging shows enhanced perfusion in the EFF-treated area. Reprinted with permission from ref 136. Copyright 2017 Springer Nature.
Mechanical Disruption.
Mechanical disruption is an alternative method to disrupt the cell membrane for instantaneous delivery.122,148 Unlike electroporation methods, mechanically triggered cell disruption does not depend on electric fields, external materials, endocytosis, or cargo properties.122 This approach is particularly applicable to difficult to transfect cell types. Recently, the advances of microfluidics and nanotechnology have enabled mechanical-disruption approaches, such as cell squeezing and nanostructure-based penetration for cell reprogramming.
Cell squeezing permits the rapid deformation of cells flowing through a narrow microfluidic channel that is half to one-third of the cell’s diameter in size, thus leading to mechanical disruption of the plasma membrane that allows for the diffusion of various materials of interest into the cytosol.149-152 Particularly, microfluidic-mediated cell squeezing has been used to deliver OSKM recombinant proteins into human fibroblasts for iPSC reprogramming and showed a 10-fold improvement in colony formation compared to electroporation or cell-penetrating peptides (Figure 6A).152 To disrupt both the plasma membrane and nuclear envelope, electric-field-driven transport was combined with cell squeezing in a microfluidic device for the enhanced delivery of plasmid DNA (Figure 6B).144 A major advantage of the cell squeezing method is the use of simple devices to transfect diverse cargos into large numbers of cells within a short time period. However, current cell-squeezing methods have a critical limitation regarding inconsistent transfection efficiency for asynchronous cell populations with different sizes. Thus, more advanced squeezing devices need to be designed with a certain range of geometric constrictions to disrupt cells of different sizes.122
Figure 6.
Mechanical disruption mediates cell reprogramming. (A) (a) Cell squeezing device for intracellular delivery of macromolecules. (b) Human iPSC reprogramming in a cell squeezing device through the delivery of recombinant OSKM transcription factors into fibroblasts. Alkaline phosphatase staining showed the colony formation in a sample plate, and significantly higher numbers of colonies were present when compared to controls treated with nucleofection or cell-penetrating peptides (CPPs). Reprinted with permission from ref 152. Copyright 2013 National Academy of Sciences. (B) (a) Disruption and field-enhanced (DFE) delivery approach combines (i) cell squeezing and (ii) electroporation processes to disrupt both the plasma membrane and nuclear envelope for enhanced DNA delivery. (b) Immunofluorescent images show a CY3 fluorescence-labeled DNA plasmid was transferred into HeLa cells using DFE, and the fluorescent signal was present in both the cell nucleus and the cytoplasm. (c) DNA transfection efficiency at applied electric voltages. Reprinted with permission from ref 144. Copyright 2017 Springer Nature. (C) Porous silicon nanoneedles for intracellular co-delivery of GFP plasmid (green) and GAPDH-siRNA (yellow). Reprinted with permission from ref 157. Copyright 2015 Springer Nature. (D) Nanostraw electroporation mediated highly efficient intracellular delivery and transfection. Reprinted from ref 147. Copyright 2013 American Chemical Society.
In addition, several nanostructure-based penetration methods have demonstrated the potential for effective high-throughput delivery of an assortment of biomolecules into living cells, including nanowires,153 nanoneedles,154 nanospears,155 and nanostraws.147,156 The nanoscale devices can penetrate the plasma membrane, thus enabling the straightforward delivery of different biomolecules, loaded onto the surface or within the bulk structure, into the cytosol and nucleus.137 It has been demonstrated that porous silicon nanoneedles can efficiently deliver nucleic acids into the cytosol (greater than 90% efficiency), and in particular, in vivo neovascularization can also be achieved using the nanoneedle-mediated minimally invasive and localized delivery of angiogenic VEGF-165 gene (Figure 6C).157 Nanostraw is another alternative cell penetration approach to mediate cytosolic delivery of different biomolecules. Unlike other nanostructure-mediated delivery methods, microfluidic systems can be integrated with nanostraw platforms to allow permanent fluidic access and spatial and temporal control in delivery.158 Additionally, electroporation systems can be further combined to generate nanostraw electroporation devices (Figure 6D).147 These devices have been shown to greatly enhance plasmid transfection (~81%) for Chinese hamster ovary cells, who display relatively low (5–10%) transfection rates using nanostraw-mediated plasmid delivery. As such, this nanostraw electroporation system offers a powerful, high-throughput transfection platform with excellent dose, spatial, and temporal control, as well as high efficiency and cell viability. Although nanostructure-mediated delivery techniques remain in the early stage of rapid development, they have provided some exciting delivery platforms for a wide range of delivery applications and show great promise for the delivery of different reprogramming factors for in vitro and in vivo cell reprogramming. However, all the nanostructure-mediated penetrations usually lack a cell-specific targeting mechanism and cannot easily navigate 3D tissues, thus nanodevices are optimally suited for transfection into easily accessible 2D tissues such as the epidermis and retina.101
Chemical Delivery Systems.
Chemical delivery approaches, often involving nanoparticles or nanocarriers, are attractive in the drug- and gene-delivery field due to their suitable size for cell penetration, large surface area for loading molecular cargo, and protection of loaded substances.159 As a carrier for nonviral gene delivery, nanoparticles are usually regarded a advantageous to viral transduction in terms of larger transgene delivery, low immunogenicity, ease of transfection, low genetic integration, and minimal insertional mutagenesis risks,160 although the efficiency might be lower than that with virus-based methods. Nowadays, chemical-based nanocarriers, including lipids, polymers, and hybrid systems, have been widely applied to deliver plasmid DNA, miRNA, mRNA, recombinant proteins, and small molecules for diverse disease treatments161-164 and have been utilized for the delivery of reprogramming factors for cell reprogramming.
Lipid Nanoparticles.
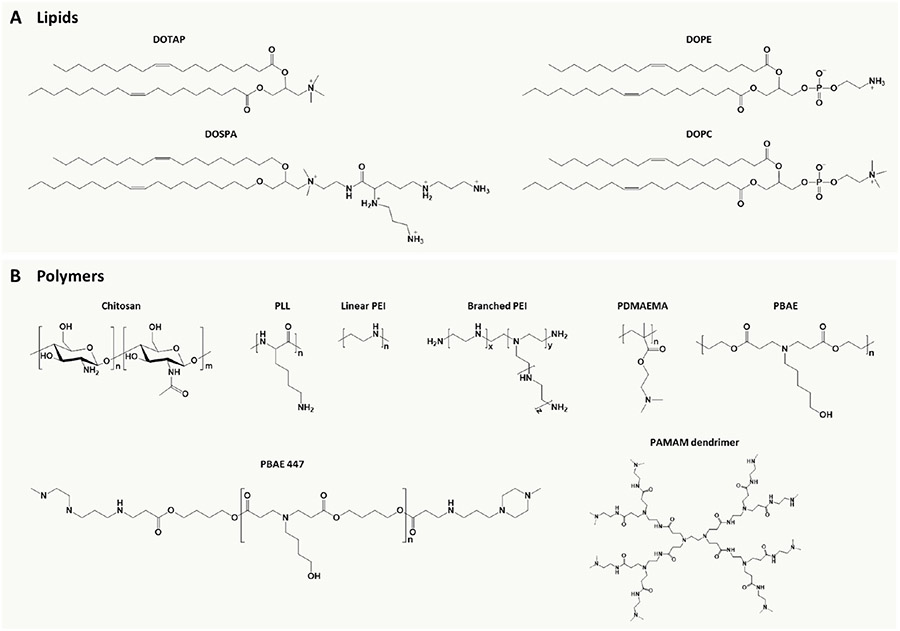
Lipids are amphiphilic molecules, consisting of hydrophobic tails and hydrophilic heads. As one of the most widely used nonviral gene carriers, lipid-based vectors/nanoparticles can deliver diverse cargos, including drugs, DNA, RNA, and genome-editing proteins into cells in an endocytosis-dependent manner.165-168 Cationic lipids (e.g., DOTAP and DOSPA) contain three structural components: a cationic headgroup, one or two hydrophobic tails, and a linking group between these domains, which are the main components that bind with negatively charged nucleic acids or molecules to form the lipid-based lipoplexes. In addition, neutral lipids (e.g., DOPE and DOPC) within the formulations can function as “helper lipids” to enhance nanoparticle stability and transfection activity.161
Currently, several lipid-based vectors have been developed and applied to achieve cell reprogramming (Figure 7A).111,113,169,170 Lipid-mediated repeated transfection was used to deliver expression plasmids containing the Yamanaka factors to reprogram MEFs into iPSCs, without evidence of plasmid integration, although the conversion efficiency was low (i.e., less than 0.0029%).171 iPSCs also can be reprogrammed from human fibroblasts with enhanced efficiency through the delivery of a modified RNA cocktail using cationic lipid carriers.111 In addition, lipid-based vectors can also be used to achieve in vivo cell reprogramming. It has been reported that the delivery of a single miRNA (miR-21) using DOPC/M-DOPE/Tween-80/DiO lipid nanoparticles can convert wound site macrophages to fibroblast-like cells in diabetic mice.172 Similarly, mice cardiac fibroblasts can be directly reprogrammed into cardiomyocytes in vitro and in vivo via lipid-based delivery of modified miRNAs.97 The miRNA-mediated reprogramming efficiency was significantly increased (i.e., by more than 10-fold) when combined with JAK inhibitor I. Therefore, further optimization of delivery methods that combine miRNA with small molecules for in vivo reprogramming may amplify their therapeutic implications.
Figure 7.
Chemical structures of representative nonviral vectors for cell reprogramming. (A) Cationic and neutral lipids. (B) Natural and synthetic cationic polymers. Abbreviations: DOTAP, 1,2-dioleoyl-3-trimethylammonium propane; DOSPA, 2,3-dioleyloxy-N-(2-(sperminecarboxamido)ethyl)-N,N-dimethyl-1-propanaminium; DOPE, dioleoylphosphatidylethanolamine; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; PLL, polylysine; PEI, polyethylenimine; PDMAEMA, poly((2-dimethylamino)ethyl methacrylate); PBAE, poly(β-amino ester); PAMAM, polyamidoamine.
Polymer Nanoparticles.
Polymer-based drug- and gene-delivery systems have been widely used to treat broad pathological conditions via less invasive or noninvasive routes.166,173 The most commonly used polymeric carriers are nanoparticles because they offer advantages such as chemical flexibility, stable formulation, excellent biocompatibility, facile preparation, and controlled release capability. Recently, delivery strategies based on natural and synthetic polymers have been adopted for cell reprogramming (Figure 7B).
Natural biopolymers like polysaccharides have been generally used as polymeric backbones for the formation of nanoparticles or as valuable drug- and gene-delivery carriers.174,175 For example, a positively charged polysaccharide was employed to deliver plasmid mixtures (encoding Oct4, Sox2, miR302/367) to generate hiPSCs from human umbilical cord mesenchymal stem cells.176 Similarly, the direct reprogramming of mouse fibroblasts into neural cells can be achieved by co-delivery of a group of plasmids (Ascl1, Brn4, Tcf3) with ethanediamine-modified Porphyra yezoensis polysaccharide (Ed-PYP) nanoparticles.177
Besides natural polymers, cationic synthetic polymers have immense chemical diversity and unlimited potential for functionalization.161 Several synthetic polymer nanoparticles have been used for cell reprogramming, such as those based on polyethylenimine,178 poly(β-amino ester),179 poly- (amidoamine) dendrimer,180 and cationic bolaamphiphile.181 Nontoxic cationic polyurethane–short-branch polyethylenimine (PU–PEI) nanoparticles have been developed to deliver Oct4/SirT1 plasmids into aged-related retinal pigment epithelium cells or light-injured rat retinas.178 For example, the PU–PEI-mediated Oct4/SirT1 gene transfer converted the retinal pigment epithelium cells into progenitor-like cells, which rescued retinal cell loss and improved electroretino-graphic responses in light-injured rat retinas. On the other hand, biodegradable polyurethanes with functional groups can provide a more robust platform for subsequent diverse modifications and thus can be potentially utilized for drug and gene delivery for cell reprogramming.182 Poly(β-amino ester) is another polymer that has been widely applied as a gene-delivery system. For example, it has been employed to deliver genes encoding disease-specific chimeric antigen receptors (CARs) in order to reprogram T cells for therapeutic purposes.102 The poly(β-amino ester) carriers also exhibited higher transfection efficiency of delivering a single polycistronic plasmid (pCAG-OSKMG) for human iPSC reprogramming, when compared with commercial transfection reagents.179 In addition, dendrimers such as poly(amidoamine) are promising nanocarriers due to their high loading capability, excellent biocompatibility, and functionalization capability. Positive charges on the terminal amine groups of poly- (amidoamine) dendrimers can promote gene loading, cell penetration, and endosomal escape during transfection, resulting in functional gene transfection.180 Furthermore, mouse iPSCs can be generated from MEFs using a single plasmid (pOSKM) delivered through arginine-terminated polyamidoamine dendrimer (G4Arg) nanoparticles, which exhibited a transfection efficiency (~15%) higher than that of commercially available carriers, such as Lipofectamine 2000 (~5%) or FuGENE HD (~8%).183
Taken together, synthetic materials not only enable the delivery of small molecules, proteins, and a variety of other compounds but also offer the potential for controlled nucleic acid transfection in vitro and in vivo, providing flexibility and possibility to manipulate cell fate. Next-generation drug- and gene-delivery systems with optimized kinetics will further improve targeted delivery and the efficiency of cell reprogramming.
Inorganic/Hybrid Nanoparticles.
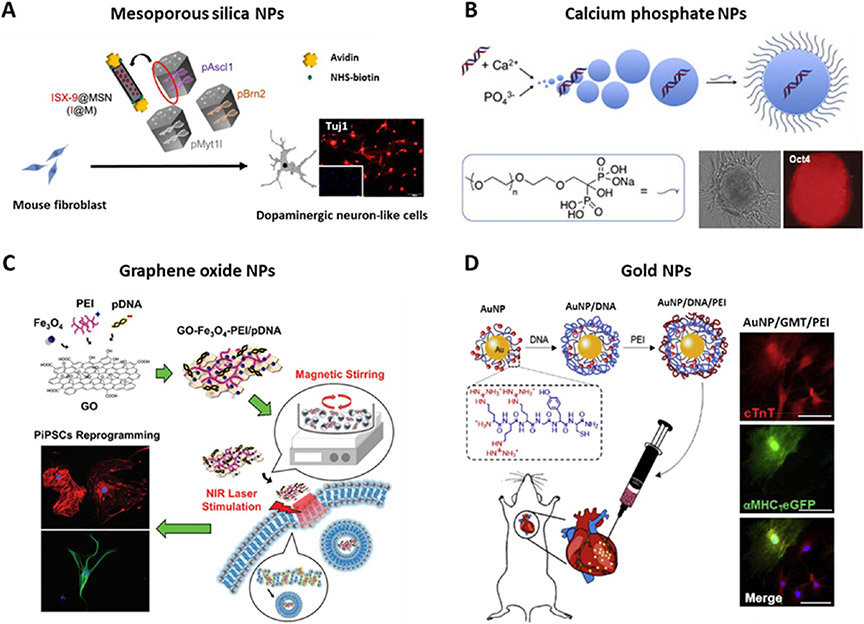
Inorganic and hybrid materials can serve as nonviral drug and gene carriers that generally possess versatile properties suitable for intracellular delivery, including diversity, multiple functionality, excellent biocompatibility, targeted delivery, and controlled release.184,185 Recently, several reports have used inorganic and hybrid particles for cell reprogramming (Figure 8), including mesoporous silica,186 calcium phosphate,187,188 graphene oxide,189,191 and gold-based nanoparticles.190 Mesoporous silica nanoparticles (MSNs) have been widely used as nanocarriers for gene and drug delivery, mainly due to their tailored mesoporous structure, high surface area ratio, biocompatibility, and functionalization capability.192 One study reported that avidin-capped MSNs adsorbed with three transcription factors (Ascl1/Brn2/Myt1l) could directly convert mouse fibroblasts into functional dopaminergic neuronlike cells. Interestingly, the co-delivery of a neurogenesis inducer, ISX-9, in the MSNs further enhanced the reprogramming efficiency.186 Therefore, this strategy of co-delivering small molecules and plasmids can also be applied to other delivery carriers to achieve enhanced reprogramming performance. In addition, calcium phosphate nanoparticles exhibited the efficient delivery of four plasmids encoding OSKM to generate iPSCs from human umbilical cord mesenchymal stem cells.187 The delivered nucleic acids formed ionic complexes with cationic calcium ions through their electrostatic interactions, which were easily transported into cells via ion-channel-mediated endocytosis. Interestingly, the hiPSC reprogramming can be accelerated upon incorporating calcium-phosphate-based hybrid nanoparticles into 3D collagen scaffolds, benefiting from the controlled release of OSKM genes and the biophysical regulation of the 3D niche.193 Additionally, gold nanoparticles have emerged as another promising platform for gene and drug delivery due to their high surface area, inertness, and easy fabrication.194 Cationic gold nanoparticles (AuNPs) are efficient carriers because they can interact with nucleic acids as well as the negatively charged plasma membrane.190,195 For example, it has been reported that in vitro and in vivo cardiac reprogramming can be achieved using hybrid AuNP-based nanoparticles. The AuNPs modified with arginine-rich peptide (RRRGYC) were mixed with GMT (Gata4, Mef2c, and Tbx5) plasmids and polyethylenimine (PEI) to make the AuNP/GMT/PEI hybrid nanocomplexes, which promoted the cardiac reprogramming efficiency of human and mouse fibroblasts. More importantly, upon injection into infarcted mouse models, the nanocomplexes enhanced the in vivo cardiac reprogramming outcome and cardiac function.190 Furthermore, hybrid materials such as graphene oxide (GO)–PEI complexes have been engineered to deliver mRNAs encoding OSKM for hiPSC generation.191 Intriguingly, GO–Fe3O4–PEI complexes were designed to achieve stimuli-sensitive gene delivery for the reprogramming of peripheral blood mononuclear cells into hiPSCs. Notably, the cell transfection and reprogramming efficiency were significantly improved by the combined treatment of magnetic stirring and near-infrared photothermal stimulation.189
Figure 8.
Inorganic/hybrid nanoparticle-mediated nonviral delivery for cell reprogramming. (A) Mesoporous silica nanoparticle mediated neuron reprogramming from mouse fibroblasts through the delivery of Ascl1, Brn2, and Myt1l plasmids. Reprinted with permission from ref 186. Copyright 2018 Springer Nature. (B) Calcium phosphate nanoparticle mediated iPSC reprogramming from human umbilical cord mesenchymal stem cells (HUMSCs), via co-delivering Oct4, Sox2, Klf4, and c-Myc plasmids. Reprinted with permission from refs 187 and 188. Copyright 2013 Wiley and 2011 Elsevier. (C) Graphene oxide nanoparticle mediated iPSC reprogramming from human peripheral blood mononuclear cells through the delivery of episomal plasmids (pCXLEhOCT3/4-shp53, pCXLE-hSK, and pCXLE-hUL). Iron oxide nanoparticle (Fe3O4)-decorated with graphene oxide (GO) were mixed with DNA and PEI to make GO–Fe3O4–PEI complexes. Reprinted with permission from ref 189. Copyright 2017 Wiley. (D) Gold nanoparticles delivering Gata4, Mef2c, and Tbx5 (GMT) plasmids mediated in vivo cardiac reprogramming in an infarcted heart. Modified AuNPs (arginine-rich AuNPs, RRRGYC-AuNPs) were mixed with DNA and then PEI to make a AuNP/GMT/PEI nanocomplex. Reprinted with permission from ref 190. Copyright 2019 Elsevier.
Despite the fact that inorganic or hybrid nanoparticles are promising as reprogramming delivery carriers, there are still some critical issues that need to be addressed. First, the loading capability and stability of genes or drugs are limited to a certain extent by surface adsorption. The second shortcoming is the possible aggregation of the inorganic nanoparticles loaded with nucleic acids, which may reduce the transfection efficiency. Another problem is the excretion of inorganic nanoparticles, and their accumulation in cells may be harmful; thus the in vivo safety of inorganic particles should be rigorously demonstrated before clinical translation.
Altogether, in addition to the reprogramming factors, the selection of an appropriate carrier among various biological, physical, or chemical delivery systems is critical to enable and enhance cell reprogramming. Undoubtedly, diverse drug- and gene-delivery strategies that have been developed for genome editing, tissue engineering, and disease therapies can also be tested and applied for cell reprogramming.19,122,123,196-199 From the perspective of clinical application, using non-gene-integrating platforms to generate reprogrammed cells is preferable to those that integrate into the genome. Moreover, considering the advantages and disadvantages summarized in Table 1 and translation potential, chemical carrier-mediated delivery systems, especially multifunctional, stimuli-sensitive drug-delivery systems,200,201 may be more promising for in vivo reprogramming therapies via the controlled local release of reprogramming factors and targeted delivery.
BIOPHYSICAL AND BIOCHEMICAL REGULATION OF CELL REPROGRAMMING
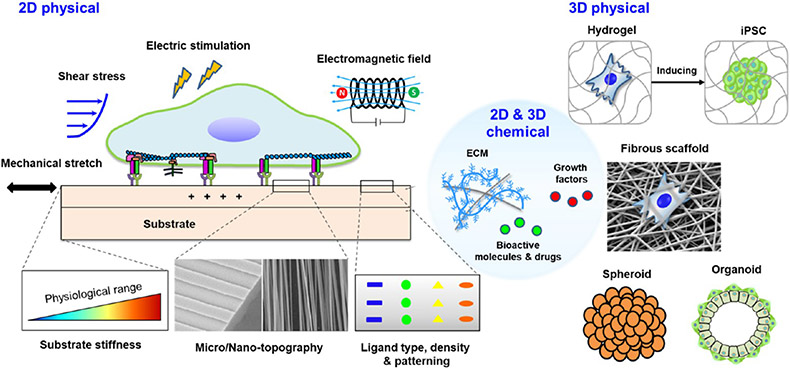
In addition to biochemical signals, biophysical factors are broadly involved in orchestrating morphogenesis, development, homeostasis, and tissue remodeling by modulating cell proliferation, migration, differentiation, lineage commitment, or apoptosis.202-204 Cells can convert extracellular biophysical cues into intracellular biochemical signals via mechanosensing and mechanotransduction, which in turn can regulate chromatin organization, gene expression, protein translation, and cell function.202,205,206 In the past decade, biomaterials and devices have been used to manipulate external biophysical and biochemical cues for cell reprogramming.33,34 By using advanced polymerization chemistries and micro- or nano-manufacturing techniques, biomaterials with desirable physical and chemical properties can be designed and fabricated.207 For elucidative purposes, we focus on the effects of microenvironmental factors, including micro/nanotopography, stiffness, extracellular force stimulation, 3D microenvironments, and biochemical modifications, on cell reprogramming (Figure 9).
Figure 9.
Various biophysical and biochemical cues for cell reprogramming. Biophysical factors consist of topographical cues, substrate stiffness, extracellular forces (including mechanical stretching, shear stress, electrical and electromagnetic forces), ligand density/patterning, and 3D microenvironments. Biochemical cues, such as extracellular matrix, growth factors, and small molecules, can be coated or immobilized onto 2D substrates and 3D scaffolds to modulate cell reprogramming.
Micro/Nanotopography.
Manipulation of micro/nano-scale surface topography has a profound influence on cell functions, such as cell morphology, adhesion, proliferation, migration, differentiation, and stemness maintenance.208-210 Although extensive research has demonstrated that topography can direct cell differentiation toward specific cell lineages,211,212 more recent studies have provided some insight into the role of topography in cell reprogramming.
Biomaterial topography, in the form of parallel microgrooves and aligned nanofibers, significantly improved iPSC generation through alterations in cell shape that resulted in an increase in histone acetylation and methylation and mesenchymal-to-epithelial transition (MET) promotion.213 Consistent with this observation, another study further confirmed that the MET process mediated topography-induced epigenetic changes during iPSC reprogramming from mouse somatic fibroblasts using graphene-coated substrates.214 These substrates, which have two-dimensional honeycomb structures, yielded almost 4 times the number of positive mouse iPSC colonies compared to glass surfaces. Apart from iPSC reprogramming, micro-topography can also influence the direct reprogramming process. For example, microgratings promoted the direct reprogramming of fibroblast into induced neurons by modulating neurite branching, neurite outgrowth, and gene expression.215,216 Similarly, the efficiency of direct cardiac reprogramming can be enhanced with topography modulation.217,218 Parallel microgrooves were found to be capable of not only modifying the epigenetic state (e.g., increased histone acetylation) but also regulating myocardin transcription factor A signaling to promote direct reprogramming into cardiomyocytes. Notably, microgrooved topography generated more mature cardiomyocyte-like cells, which may enhance functional performance after cell transplantation into an infarcted heart.217
In contrast to microtopography, nanotopography has demonstrated a more influential role in inducing differentiation of hiPSCs to a neuronal lineage,219 possibly due to the fact that these nanoscale features may more closely resemble the in vivo microenvironment. Upon examining how micro- and nanotopographical cues could influence direct reprogramming into induced dopaminergic neurons, it was determined that nanogrooved substrates further enhanced neuronal gene expression, maturation, and reprogramming efficiency through modulation of histone mark expression and MET.220 However, whether nanotopography can regulate direct reprogramming into other cell lineages remains to be further explored. The underlying mechanisms of topographical influence on cell reprogramming are not fully understood. However, the mechanisms that have been elucidated for topography-mediated cell differentiation may also be applicable in reprogramming. For example, the aforementioned studies suggest that both micro- and nanotopographical cues can influence cell and nuclear shape to alter the epigenetic state, gene expression, and thus cell fate.208-210 In addition, topography is strongly correlated to focal adhesion dynamics, leading to intracellular signaling transduction pathways triggered by topographic parameters of substrates. Moreover, topography-induced cytoskeletal changes can induce nuclear deformation that leads to altered nuclear mechanics and chromatin state, both of which can affect cell phenotype and function.221
Stiffness.
Substrate stiffness is one of the most widely applied physical cues to modulate stem cell differentiation.222-224 For example, soft substrates promote the differentiation of human MSCs into neurons or adipocytes, whereas osteogenesis is favored on stiffer substrates.225 Matrix stiffness can also modulate the cell-reprogramming process. Recently, it has been demonstrated that soft polyacrylamide hydrogels with a stiffness of 100 Pa enhanced mouse iPSC reprogramming from MEFs on the surface of the hydrogel,226 whereas another study reported that the optimal stiffness for mouse and human iPSC generation was between 300 and 600 Pa in 3D polyethylene glycol (PEG) hydrogels.227
Mechanistic studies have revealed that substrate stiffness can alter cell adhesion and cytoskeletal reorganization, via mechanotransductive mechanisms, to activate signaling pathways that modulate gene expression and, thus, cell behavior.222,228 Although several intracellular mechanosensitive signaling cascades are regulated by matrix stiffness, such as AKT/YAP/RUNX2,229 p190RhoGAP,230 and YAP/TAZ mechanotransduction,29,231 the underlying molecular mechanisms of how stiffness regulates cell reprogramming remain unknown and require further investigation.
Extracellular Force Stimulation.
Extracellular mechanical forces can influence organ development and function in health and disease by triggering a series of cytoskeletal-mediated mechanotransduction and intracellular signaling cascades.202,232,233 Recently, extracellular forces including shear stress, mechanical stretching, and electromagnetic forces have been used as effective biophysical cues to modulate cell reprogramming.
Extracellular fluids, including blood plasma, interstitial fluid, and lymph, are crucial in the exchange of substances and the maintenance of the microenvironment. Fluid shear stress is an essential mechanical factor in flowing liquid that can induce cytoskeletal organization remodeling, trigger mechanosensitive gene expression, and alter cell behavior, demonstrating an important role in vascular homeostasis, vascular remodeling, cardiac development, atherogenesis,234,235 and cancer metastasis.145,236 In addition, fluid movement enhances convective mixing and transport. Compared to conventional adherent culture, faster and more efficient mouse iPSC reprogramming can be achieved in a stirred suspension bioreactor with retroviral transduction of transcription factors (i.e., Oct4, Sox2, and Klf4, with or without c-Myc), displaying more than a 10-fold increase in efficiency compared to that with the adherent culture.237 Fluid shear stress present within a dynamic culture suppressed differentiation and provided the additional benefit of selective aggregate formation, enabling reprogrammed cells to flourish. As a result, dynamic suspension-induced reprogramming is a promising and scalable approach for basic research and clinical uses.238 Interestingly, 2D culture with dynamic mixing at the mid to late phase of reprogramming also demonstrates significant improvement of mouse iPSC reprogramming, which was found to be attributed to the enhancement of cell proliferation.239
Mechanical stress and strain in vivo play fundamental roles in guiding cell behavior and tissue homeostasis.240 At the cellular level, the mechanical stretch has demonstrated crucial regulatory effects on cell spreading, growth, lineage commitment, and stem cell differentiation on 2D substrates.241,242 In addition, externally applied stretch (uniaxial, biaxial, and equiaxial) has been utilized to engineer functional and mature tissues in 3D scaffolds, such as in heart tissue,243,244 skeletal muscles,245 and blood vessels.246,247 Mechanical stretch can also be used for in vivo tissue regeneration, for example, by activating hair stem cells and eliciting macrophage recruitment and M2 phenotype polarization to facilitate hair regeneration in a strain/duration-dependent manner.248 Recent studies have shown that mechanical stretch has a positive effect on reprogramming. When dermal fibroblasts transduced with OSKM were seeded onto flexible membranes that were mechanically stretched, 3 and 5 times more human iPSC colonies were obtained upon equiaxial stretching for 4 days at 3 and 8% strain, respectively, than with the unstretched groups.249 However, cyclic mechanical stretch did not improve the yield of direct cardiac reprogramming.218 Therefore, the effects of mechanical stretch may be dependent on specific signaling pathways involved in the reprogramming process. In addition, various parameters of stretching such as mode, magnitude, rate, frequency, duration, continuity, and relaxing period may have profound effects on cell signaling and reprogramming, which is worth further studies.
Moreover, the mechanical forces driven by electromagnetic or electric fields can improve cellular reprogramming. For example, extremely low-frequency electromagnetic fields (EL-EMF) significantly enhanced the reprogramming efficiency from mouse fibroblasts to iPSCs. EL-EMF induced dynamic regulation of epigenetic changes through the activation of a histone lysine methyltransferase.250 In addition, electro-magnetized AuNPs provided an excellent interface to deliver EMF for cell reprogramming. Application of EMF to cells cultured on AuNPs directly reprogrammed fibroblasts into induced dopaminergic neurons both in vitro and in vivo. EMF stimulation specifically activated the histone acetyltransferase Brd2, leading to histone H3K27 acetylation and neuron-specific gene activation.195 Furthermore, a triboelectrical stimulation system can be combined with nonviral gene delivery to directly convert fibroblasts into functional neuronal cells with higher efficiency.251 The triboelectrical stimulation promoted dynamic changes of intracellular calcium levels in reprogrammed cells, including increased calcium influx and ERK1/2 pathway activation. These electromagnetic or electric stimulation systems can potentially serve as electroceuticals to provide less invasive and non-chemical-based methods to facilitate in vivo reprogramming or in situ direct cell conversion.
3D Microenvironment.
3D microenvironments can provide biomimetic systems to direct cell fate, including stemness maintenance, organogenesis, and tissue development.252-254 Of particular interest are biomaterials-mediated and self-assembled cellular 3D microenvironments that can serve as powerful platforms to modulate cell reprogramming and highlight the clinical potential of reprogramming technologies.
3D biomaterial-based microenvironments enable robust spatial and temporal control of biophysical and biochemical cues to modulate cell fate both in vitro and in vivo. It has been demonstrated that mouse and human iPSC reprogramming can be enhanced in 3D PEG-based hydrogels by optimizing parameters such as stiffness, degradability, and biochemical composition.227 It was determined that cell conversion could be vastly improved using highly degradable gels with a stiffness of 600 Pa. This 3D approach resulted in a more than 3-fold increase in the reprogramming efficiency compared to 2D environments, as a result of accelerating the MET process and increasing epigenetic remodeling. In addition, electrospun fibrous scaffolds, which have been widely applied as biomimetic microenvironments for biomedical applications, can also promote in situ stem cell neuronal reprogramming, resulting in neural network formation and brain engraftment.216
Furthermore, several studies have shown that reprogramming cells in 3D microenvironments can enhance cell phenotypic functions. Chitosan substrate-mediated gene delivery of a naked Foxd3 plasmid can reprogram human fibroblasts into neural crest stem-like cells that self-assemble into 3D cellular spheroids. Notably, the Foxd3-transfected spheroids demonstrated significantly greater functional rescue of the CNS in impaired zebrafish models compared to that with dispersed cells.255 In addition, under Matrigel-embedded 3D culture conditions, the combined expression of Gata6, Cdx2, Hnf4a, and Foxa3 directly converted mouse fibroblasts into fetal intestine-derived progenitor cells (FIPCs) and allowed for the formation of spherical organoids, which further developed into budding organoids.74 Particularly, the derived intestinal organoids regenerated colonic tissue upon transplantation, demonstrating great promise for the treatment of intestinal diseases. Therefore, these organoids, which consist of 3D structures with a heterogeneous cell composition, may serve as biomimetic systems that can recapitulate specific gene and protein expression, tissue morphology, and physiological functions. In the same study, it was determined that the defined transcription factors also induced human FIPCs from human dermal fibroblasts and human umbilical vein endothelial cells.74 However, in contrast to mouse iFIPCs, human iFIPCs did not generate budding organoids, suggesting additional factors or functional 3D niches may be needed.
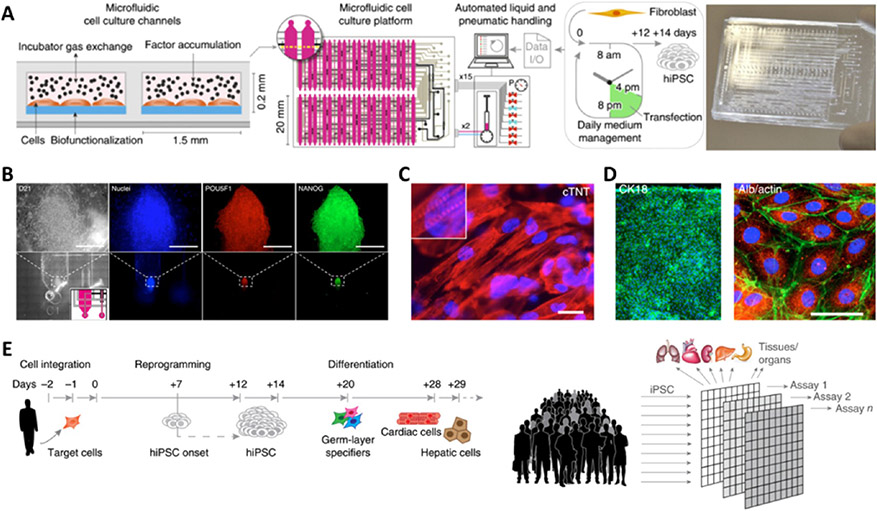
Organ-on-a-chip is a burgeoning technique and powerful tool for disease modeling, drug discovery, toxicology research, and regenerative medicine.256 Organ-on-a-chip systems are manufactured with advanced micro/nanofabrication techniques based on microfluidics, materials, and tissue engineering, enabling the design of customized cellular and tissue microenvironments with precise perfusion, mechanical and structural control, electrical stimulation, less probability of contamination, and high-throughput analysis.257 The engineered architectures can recreate partial or all functions of native tissues, such as biological tissue barriers, parenchymal tissue functions, and multiorgan interactions. In particular, diverse 3D organoids, including liver,258 pancreatic islet,259 lung,260 and brain,261 that have been developed within organon-a-chip systems through in situ iPSC differentiation have demonstrated complex tissue and organ-specific functions. Most recently, organ-on-a-chip has shown great potential and superiority in cell reprogramming through the confinement of the microenvironment. Through the manipulation of the concentration and the sequential delivery of reprogramming factors (mixed mRNAs encoding transcription factors) and small molecules, an automated microfluidic platform dramatically improved human iPSC reprogramming with a 50-fold increase in efficiency (Figure 10).262,263 In the same platform, the generated hiPSCs could be precisely differentiated into functional cardiomyocytes and hepatocytes. The ability to perform both cell reprogramming and differentiation of patient-specific cells within these devices hold great promise for personalized drug screening, bringing us closer to more personalized medicine.
Figure 10.
Cellular reprogramming and differentiation on microfluidic chips. (A) Automated microfluidic platform for highly efficient hiPSC reprogramming by sequential delivery of mRNAs of POU5F1, SOX2, KLF4, c-MYC, NANOG, LIN28. (B) hiPSC colony formation within a microfluidic channel and the expression of pluripotency markers. (C) Differentiation of primary μ-hiPSCs into cardiomyocytes. (D) Differentiation of primary μ-hiPSCs into hepatic cells. (E) Vision of high-throughput biological assays via integrated cell reprogramming and differentiation in a microfluidic chip platform. Reprinted with permission from ref 262. Copyright 2016 Springer Nature.
Biochemical Modifications.
Biochemical cues such as extracellular matrix (ECM) components, small molecules, growth factors, and other signaling molecules regulate not only stem cell fate but also cell reprogramming.254 ECM and its derivatives are commonly used as hydrogels or bioactive molecules immobilized onto 2D substrates and 3D scaffolds. One study demonstrated that fibrin and a fibrin–collagen composite were superior to Matrigel and collagen I in supporting indirect cardiac reprogramming.264 It was determined that matrix identity and tractional forces played major roles in cell reprogramming, whereas matrix mechanics, matrix microstructure, and cell proliferation influenced the process to a lesser extent.264 To interrogate the effect of ligand identity, ligand density, and substrate modulus on cell reprogramming, ECM-coated polyacrylamide substrates can be developed to decouple physicochemical features.265 In addition, PEG hydrogels can be modified with specific bioactive molecules to modulate specific cellular responses while preventing unwanted adsorption of proteins. PEG hydrogels modified with high concentrations of laminin and RGD peptide nearly doubled the efficiency of induced cardiomyocyte-like cells from fibroblasts.265 Therefore, cellular reprogramming can be enhanced using customized engineered materials with immobilized biosignals.
Small molecules and growth factors have been widely explored to enhance or induce either iPSC reprogramming114,266 or direct reprogramming into various cell types, including neurons,67 cardiomyocytes,66 pancreatic β-like cells,267 and hepatocytes.268 Indeed, these small molecules and proteins can be mixed or conjugated, as ECM molecules, onto 2D cell culture substrates and 3D scaffolds to influence cell fate. Of note, such biochemical stimuli can be endowed and controlled by the defined delivery systems and can also be integrated with biophysical cues to maximize cell reprogramming, especially for in situ reprogramming.
FUTURE PERSPECTIVES
Cell reprogramming is a highly valuable method for engineering cell fate. This comprehensive review highlights the recent progress in engineering biomaterials with advanced micro/nanofabrication techniques to provide delivery platforms and biophysical and biochemical cues for enhancing in vitro and in vivo cell reprogramming. However, many challenges remain to be addressed before the clinical application of reprogramming technologies, including efficiency, quality, safety issues, in situ targeting, and the underlying signaling mechanisms. To address these challenges, multidisciplinary collaborative research from the fields of materials science, bioengineering, biology, and medicine would be required.
Optimized delivery systems need to be developed or selected from a diverse group of drug- and gene-delivery strategies to achieve the desired reprogramming outcome. First, reproducibility and higher efficiency will be the critical criteria in developing the optimal delivery methods for cell reprogramming. For in vivo reprogramming, if specific parts of the body, organs or tissues, or a subpopulation of cells are targeted for cell type conversion, a drug-delivery system incorporated with a specific ligand, antibodies, peptides, or aptamers can be developed. Second, as small molecules offer powerful tools to manipulate cell fate by inducing or enhancing cell reprogramming, the co-delivery of nucleic acids with small molecules or the multidelivery of chemical cocktails can be developed to promote cell reprogramming in vitro and in vivo. Third, smart and stimuli-responsive delivery systems can be custom-designed to achieve more efficient in situ targeted delivery of reprogramming factors, which is an emerging area of biomaterials research. Fourth, biophysical and biochemical cues can also be incorporated into the drug-delivery system to enhance cell reprogramming, yet the optimal conditions for reprogramming remain to be clarified.
Biophysical manipulation provides a promising approach to regulate cell fate and improve the reprogramming process. First, the majority of biophysical cues (including topography, stiffness, and extracellular force) and biochemical cues (including ECM, soluble factors, and small molecules) have been employed for cell reprogramming individually, which demonstrates the feasibility of these approaches and helps elucidate the underlying mechanisms. Combining such cues may synergistically enhance reprogramming efficiency, but it is a challenging task to figure out the optimal conditions with so many possible combinations, crosstalk, and potential side effects. In addition, temporal regulation of the reprogramming process at the early or late phase may require different microenvironmental factors. If sufficient experimental data can be collected and specific experiments can be designed to address the combinatory effects, a machine learning approach can be used to optimize the reprogramming conditions and protocol. Second, to make it relevant to in vivo conditions, reprogramming in 3D scaffolds with desired and integrated cues will be a key step to generate and apply reprogramming-mediated engineered tissues in vitro or in situ. Third, whether and how biophysical factors may influence in vivo reprogramming is not well understood and deserves further investigation in the future.
Advances in other cutting-edge technologies will continue to expand the frontiers of cell reprogramming. First, microfluidic platforms enable the delivery of reprogramming factors in a more controlled and efficient way from the perspectives of composition, dosage, and spatiotemporal features, which can also offer well-defined nanotopographical and mechanical cues, including fluid shear stress, cyclic strain, and compression. It is expected that the orchestrated interplay of the controlled delivery and combined physical and chemical cues can further enhance cell reprogramming. Meanwhile, the microfluidic environment allows directed generation of functional cells in the same platform, which creates a model of “tissue/organ reprogramming-on-a-chip”. Second, 3D printing holds great promise for engineering tissues and organs for disease modeling and regenerative medicine.269 3D printing enables the manipulation of the microenvironment with spatially defined mechanical, structural, and chemical properties, thus, making it suitable to generate 3D complex niches for cell reprogramming. Third, single-cell sequencing analysis will provide insight into the heterogeneous cellular responses and fate determination during the reprogramming process270,271 and help elucidate the signal pathways, molecular identity, and underlying mechanisms by which biophysical and biochemical cues regulate cell reprogramming. Fourth, CRISPR/Cas9 genome-editing platforms provide powerful tools not only for genetic correcting of hereditary diseases272-274 but also for somatic cell reprogramming, as demonstrated in recent studies for the derivation of iPSCs275 and neurons.276 Viral-free CRISPR/Cas9 delivery systems have shown promising results, which may enable more in vivo applications. Hence, simultaneous cell reprogramming and CRISPR/Cas9 gene editing, through the co-delivery of both gene-editing systems and reprogramming factors, may guide the design of next-generation cell-reprogramming technologies.
The advancement and integration of cell reprogramming and biotechnologies present exciting opportunities for research and technology translation in these interdisciplinary frontiers. We expect that the rational design and development of delivery systems and biomaterials that control microenvironmental cues will accelerate the translation of reprogramming technologies for cell engineering and therapies.
ACKNOWLEDGMENTS
The authors are supported in part by a grant from the National Institutes of Health (HL121450, to S.L.), the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH under the Ruth L. Kirschstein National Research Service Award (T32AR059033, to J.S.), UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Innovation Award (to S.L.), and UCLA startup funding.
VOCABULARY
- embryonic stem cells
pluripotent stem cells derived from the undifferentiated inner cell mass of a blastocyst, an early stage embryo
- somatic cell nuclear transfer
a technique involving the transfer of somatic cell nucleus into the cytoplasm of an enucleated egg
- cell differentiation
a process whereby a cell changes from an immature to more specialized phenotype
- iPSC reprogramming
induced conversion of somatic cells into pluripotent stem cells
- direct reprogramming
induced conversion of a mature cell into another cell type without proceeding through an pluripotent state
- indirect reprogramming
transient induction of a partially reprogrammed cell using the Yamanaka factors, followed by differentiation into the target cell type
- transcription factor
a protein that can regulate gene transcription by binding to a specific DNA sequence
Footnotes
The authors declare no competing financial interest.
Contributor Information
Jun Fang, Department of Bioengineering and Department of Medicine, University of California, Los Angeles, Los Angeles, California 90095, United States.
Yuan-Yu Hsueh, Department of Bioengineering, University of California, Los Angeles, Los Angeles, California 90095, United States; Division of Plastic Surgery, Department of Surgery, College of Medicine, National Cheng Kung University Hospital, Tainan 70456, Taiwan.
Jennifer Soto, Department of Bioengineering and Department of Medicine, University of California, Los Angeles, Los Angeles, California 90095, United States.
Wujin Sun, Department of Bioengineering and Center for Minimally Invasive Therapeutics (C-MIT), California NanoSystems Institute, University of California, Los Angeles, Los Angeles, California 90095, United States.
Jinqiang Wang, Department of Bioengineering and Center for Minimally Invasive Therapeutics (C-MIT), California NanoSystems Institute, University of California, Los Angeles, Los Angeles, California 90095, United States.
Zhen Gu, Department of Bioengineering, Center for Minimally Invasive Therapeutics (C-MIT), California NanoSystems Institute, and Jonsson Comprehensive Cancer Center, University of California, Los Angeles, Los Angeles, California 90095, United States.
Ali Khademhosseini, Department of Bioengineering, Center for Minimally Invasive Therapeutics (C-MIT), California NanoSystems Institute, Department of Chemical and Biomolecular Engineering, and Department of Radiology, University of California, Los Angeles, Los Angeles, California 90095, United States.
Song Li, Department of Bioengineering, Department of Medicine, and Center for Minimally Invasive Therapeutics (C-MIT), California NanoSystems Institute, University of California, Los Angeles, Los Angeles, California 90095, United States.
REFERENCES
- (1).Boyer LA; Lee TI; Cole MF; Johnstone SE; Levine SS; Zucker JR; Guenther MG; Kumar RM; Murray HL; Jenner RG; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Smith ZD; Sindhu C; Meissner A Molecular Features of Cellular Reprogramming and Development. Nat. Rev. Mol. Cell Biol 2016, 17, 139–154. [DOI] [PubMed] [Google Scholar]
- (3).Horner PJ; Gage FH Regenerating the Damaged Central Nervous System. Nature 2000, 407, 963–970. [DOI] [PubMed] [Google Scholar]
- (4).Anderson MA; Burda JE; Ren YL; Ao Y; O’Shea TM; Kawaguchi R; Coppola G; Khakh BS; Deming TJ; Sofroniew MV Astrocyte Scar Formation Aids Central Nervous System Axon Regeneration. Nature 2016, 532, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Laflamme MA; Murry CE Heart Regeneration. Nature 2011, 473, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tzahor E; Poss KD Cardiac Regeneration Strategies: Staying Young at Heart. Science 2017, 356, 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Makris EA; Gomoll AH; Malizos KN; Hu JC; Athanasiou KA Repair and Tissue Engineering Techniques for Articular Cartilage. Nat. Rev. Rheumatol 2015, 11, 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Briggs R; King TJ Transplantation of Living Nuclei from Blastula Cells into Enucleated Frogs Eggs. Proc. Natl. Acad. Sci. U. S. A 1952, 38, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Takahashi K; Yamanaka S Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [DOI] [PubMed] [Google Scholar]
- (10).Xu J; Du Y; Deng H Direct Lineage Reprogramming: Strategies, Mechanisms, and Applications. Cell Stem Cell 2015, 16, 119–134. [DOI] [PubMed] [Google Scholar]
- (11).Takahashi K; Yamanaka S A Decade of Transcription Factor-Mediated Reprogramming to Pluripotency. Nat. Rev. Mol. Cell Biol 2016, 17, 183–193. [DOI] [PubMed] [Google Scholar]
- (12).Rowe RG; Daley GQ Induced Pluripotent Stem Cells in Disease Modelling and Drug Discovery. Nat. Rev. Genet 2019, 20, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Soldner F; Jaenisch R Stem Cells, Genome Editing, and the Path to Translational Medicine. Cell 2018, 175, 615–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Srivastava D; DeWitt N In Vivo Cellular Reprogramming: The Next Generation. Cell 2016, 166, 1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Mertens J; Marchetto MC; Bardy C; Gage FH Evaluating Cell Reprogramming, Differentiation and Conversion Technologies in Neuroscience. Nat. Rev. Neurosci 2016, 17, 424–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cherry ABC; Daley GQ Reprogramming Cellular Identity for Regenerative Medicine. Cell 2012, 148, 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Grskovic M; Javaherian A; Strulovici B; Daley GQ Induced Pluripotent Stem Cells-Opportunities for Disease Modelling and Drug Discovery. Nat. Rev. Drug Discov 2011, 10, 915–929. [DOI] [PubMed] [Google Scholar]
- (18).Caldorera-Moore M; Peppas NA Micro- and Nano-technologies for Intelligent and Responsive Biomaterial-Based Medical Systems. Adv. Drug Deliv. Rev 2009, 61, 1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cucchiarini M; Madry H Biomaterial-Guided Delivery of Gene Vectors for Targeted Articular Cartilage Repair. Nat. Rev. Rheumatol 2019, 15, 18–29. [DOI] [PubMed] [Google Scholar]
- (20).Mitrousis N; Fokina A; Shoichet MS Biomaterials for Cell Transplantation. Nat. Rev. Mater 2018, 3, 441–456. [Google Scholar]
- (21).Higuchi A; Suresh Kumar S; Benelli G; Ling Q-D; Li H-F; Alarfaj AA; Munusamy MA; Sung T-C; Chang Y; Murugan K Biomaterials Used in Stem Cell Therapy for Spinal Cord Injury. Prog. Mater. Sci 2019, 103, 374–424. [Google Scholar]
- (22).Lutolf MP; Hubbell JA Synthetic Biomaterials as Instructive Extracellular Microenvironments for Morphogenesis in Tissue Engineering. Nat. Biotechnol 2005, 23, 47–55. [DOI] [PubMed] [Google Scholar]
- (23).Sadtler K; Singh A; Wolf MT; Wang XK; Pardoll DM; Elisseeff JH Design, Clinical Translation and Immunological Response of Biomaterials in Regenerative Medicine. Nat. Rev. Mater 2016, 1, 16040. [Google Scholar]
- (24).Riley RS; June CH; Langer R; Mitchell MJ Delivery Technologies for Cancer Immunotherapy. Nat. Rev. Drug Discov 2019, 18, 175–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gu L; Mooney DJ Biomaterials and Emerging Anticancer Therapeutics: Engineering the Microenvironment. Nat. Rev. Cancer 2016, 16, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wang H; Mooney DJ Biomaterial-Assisted Targeted Modulation of Immune Cells in Cancer Treatment. Nat. Mater 2018, 17, 761–772. [DOI] [PubMed] [Google Scholar]
- (27).Langer R; Tirrell DA Designing Materials for Biology and Medicine. Nature 2004, 428, 487–492. [DOI] [PubMed] [Google Scholar]
- (28).Peppas NA; Langer R New Challenges in Biomaterials. Science 1994, 263, 1715–1720. [DOI] [PubMed] [Google Scholar]
- (29).Brusatin G; Panciera T; Gandin A; Citron A; Piccolo S Biomaterials and Engineered Microenvironments to Control Yap/Taz-Dependent Cell Behaviour. Nat. Mater 2018, 17, 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Leijten J; Khademhosseini A From Nano to Macro: Multiscale Materials for Improved Stem Cell Culturing and Analysis. Cell Stem Cell 2016, 18, 20–24. [DOI] [PubMed] [Google Scholar]
- (31).Lv LW; Tang YM; Zhang P; Liu YS; Bai XS; Zhou YS Biomaterial Cues Regulate Epigenetic State and Cell Functions-A Systematic Review. Tissue Eng., Part B 2018, 24, 112–132. [DOI] [PubMed] [Google Scholar]
- (32).Crowder SW; Leonardo V; Whittaker T; Papathanasiou P; Stevens MM Material Cues as Potent Regulators of Epigenetics and Stem Cell Function. Cell Stem Cell 2016, 18, 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang YX; Bi Y; Gao SR Epigenetic Regulation of Somatic Cell Reprogramming. Curr. Opin. Genet. Dev 2017, 46, 156–163. [DOI] [PubMed] [Google Scholar]
- (34).Wong SY; Soto J; Li S Biophysical Regulation of Cell Reprogramming. Curr. Opin. Chem. Eng 2017, 15, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Dvir T; Timko BP; Kohane DS; Langer R Nanotechnological Strategies for Engineering Complex Tissues. Nat. Nanotechnol 2011, 6, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Peer D; Karp JM; Hong S; FarokHzad OC; Margalit R; Langer R Nanocarriers as an Emerging Platform for Cancer Therapy. Nat. Nanotechnol 2007, 2, 751–760. [DOI] [PubMed] [Google Scholar]
- (37).Abdeen AA; Saha K Manufacturing Cell Therapies Using Engineered Biomaterials. Trends Biotechnol. 2017, 35, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Mitragotri S; Lahann J Physical Approaches to Biomaterial Design. Nat. Mater 2009, 8, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Gurdon JB The Developmental Capacity of Nuclei Taken from Intestinal Epithelium Cells of Feeding Tadpoles. J. Embryol. Exp. Morphol 1962, 10, 622–640. [PubMed] [Google Scholar]
- (40).Campbell KH; McWhir J; Ritchie WA; Wilmut I Sheep Cloned by Nuclear Transfer from a Cultured Cell Line. Nature 1996, 380, 64–66. [DOI] [PubMed] [Google Scholar]
- (41).Tachibana M; Amato P; Sparman M; Gutierrez NM; Tippner-Hedges R; Ma H; Kang E; Fulati A; Lee HS; Sritanaudomchai H; et al. Human Embryonic Stem Cells Derived by Somatic Cell Nuclear Transfer. Cell 2013, 153, 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Buganim Y; Faddah DA; Jaenisch R Mechanisms and Models of Somatic Cell Reprogramming. Nat. Rev. Genet 2013, 14, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ma H; Morey R; O’Neil RC; He YP; Daughtry B; Schultz MD; Hariharan M; Nery JR; Castanon R; Sabatini K; et al. Abnormalities in Human Pluripotent Cells Due to Reprogramming Mechanisms. Nature 2014, 511, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kim K; Doi A; Wen B; Ng K; Zhao R; Cahan P; Kim J; Aryee MJ; Ji H; Ehrlich LIR; et al. Epigenetic Memory in Induced Pluripotent Stem Cells. Nature 2010, 467, 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Haake K; Ackermann M; Lachmann N Concise Review: Towards the Clinical Translation of Induced Pluripotent Stem Cell-Derived Blood Cells-Ready for Take-Off. Stem Cells Transl. Med 2019, 8, 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Akabayashi A; Nakazawa E; Jecker NS The World’s First Clinical Trial for an Aplastic Anemia Patient with Thrombocytopenia Administering Platelets Generated from Autologous IPS Cells. Int. J. Hematol 2019, 109, 239–240. [DOI] [PubMed] [Google Scholar]
- (47).Rosati J; Ferrari D; Altieri F; Tardivo S; Ricciolini C; Fusilli C; Zalfa C; Profico DC; Pinos F; Bernardini L; et al. Establishment of Stable iPS-Derived Human Neural Stem Cell Lines Suitable for Cell Therapies. Cell Death Dis. 2018, 9, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Higuchi A; Kumar SS; Benelli G; Alarfaj AA; Munusamy MA; Umezawa A; Murugan K Stem Cell Therapies for Reversing Vision Loss. Trends Biotechnol. 2017, 35, 1102–1117. [DOI] [PubMed] [Google Scholar]
- (49).Higuchi A; Ku NJ; Tseng YC; Pan CH; Li HF; Kumar SS; Ling QD; Chang Y; Alarfaj AA; Munusamy MA; et al. Stem Cell Therapies for Myocardial Infarction in Clinical Trials: Bioengineering and Biomaterial Aspects. Lab. Invest 2017, 97, 1167–1179. [DOI] [PubMed] [Google Scholar]
- (50).Liang G; Zhang Y Genetic and Epigenetic Variations in iPSCs: Potential Causes and Implications for Application. Cell Stem Cell 2013, 13, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Davis RL; Weintraub H; Lassar AB Expression of a Single Transfected cDNA Converts Fibroblasts to Myoblasts. Cell 1987, 51, 987–1000. [DOI] [PubMed] [Google Scholar]
- (52).Sekiya S; Suzuki A Direct Conversion of Mouse Fibroblasts to Hepatocyte-like Cells by Defined Factors. Nature 2011, 475, 390–393. [DOI] [PubMed] [Google Scholar]
- (53).Huang PY; Zhang LD; Gao YM; He ZY; Yao D; Wu ZT; Cen J; Chen XT; Liu CC; Hu YP; et al. Direct Reprogramming of Human Fibroblasts to Functional and Expandable Hepatocytes. Cell Stem Cell 2014, 14, 370–384. [DOI] [PubMed] [Google Scholar]
- (54).Vierbuchen T; Ostermeier A; Pang ZP; Kokubu Y; Sudhof TC; Wernig M Direct Conversion of Fibroblasts to Functional Neurons by Defined Factors. Nature 2010, 463, 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Caiazzo M; Dell’Anno MT; Dvoretskova E; Lazarevic D; Taverna S; Leo D; Sotnikova TD; Menegon A; Roncaglia P; Colciago G; et al. Direct Generation of Functional Dopaminergic Neurons from Mouse and Human Fibroblasts. Nature 2011, 476, 224–227. [DOI] [PubMed] [Google Scholar]
- (56).Mertens J; Paquola ACM; Ku MC; Hatch E; Bohnke L; Ladjevardi S; McGrath S; Campbell B; Lee H; Herdy JR; et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 2015, 17, 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Thorel F; Nepote V; Avril I; Kohno K; Desgraz R; Chera S; Herrera PL Conversion of Adult Pancreatic α-cells to β-cells after Extreme β-cell Loss. Nature 2010, 464, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Chakravarthy H; Gu X; Enge M; Dai X; Wang Y; Damond N; Downie C; Liu K; Wang J; Xing Y; et al. Converting Adult Pancreatic Islet Alpha Cells into Beta Cells by Targeting Both Dnmt1 and Arx. Cell Metab. 2017, 25, 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kaminski MM; Tosic J; Kresbach C; Engel H; Klockenbusch J; Muller AL; Pichler R; Grahammer F; Kretz O; Huber TB; et al. Direct Reprogramming of Fibroblasts into Renal Tubular Epithelial Cells by Defined Transcription Factors. Nat. Cell Biol 2016, 18, 1269–1280. [DOI] [PubMed] [Google Scholar]
- (60).Hakelien AM; Landsverk HB; Robl JM; Skalhegg BS; Collas P Reprogramming Fibroblasts to Express T-Cell Functions Using Cell Extracts. Nat. Biotechnol 2002, 20, 460–466. [DOI] [PubMed] [Google Scholar]
- (61).Buganim Y; Itskovich E; Hu YC; Cheng AW; Ganz K; Sarkar S; Fu DD; Welstead GG; Page DC; Jaenisch R Direct Reprogramming of Fibroblasts into Embryonic Sertoli-like Cells by Defined Factors. Cell Stem Cell 2012, 11, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Lee S; Park C; Han JW; Kim JY; Cho K; Kim EJ; Kim S; Lee SJ; Oh SY; Tanaka Y; et al. Direct Reprogramming of Human Dermal Fibroblasts Into Endothelial Cells Using ER71/ETV2. Circ. Res 2017, 120, 848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Ieda M; Fu JD; Delgado-Olguin P; Vedantham V; Hayashi Y; Bruneau BG; Srivastava D Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Miyamoto K; Akiyama M; Tamura F; Isomi M; Yamakawa H; Sadahiro T; Muraoka N; Kojima H; Haginiwa S; Kurotsu S; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103. [DOI] [PubMed] [Google Scholar]
- (65).Fu JD; Stone NR; Liu L; Spencer CI; Qian L; Hayashi Y; Delgado-Olguin P; Ding S; Bruneau BG; Srivastava D Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. Stem Cell Rep. 2013, 1, 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Cao N; Huang Y; Zheng JS; Spencer CI; Zhang Y; Fu JD; Nie BM; Xie M; Zhang ML; Wang HX; et al. Conversion of Human Fibroblasts into Functional Cardiomyocytes by Small Molecules. Science 2016, 352, 1216–1220. [DOI] [PubMed] [Google Scholar]
- (67).Hu WX; Qiu BL; Guan WQ; Wang QY; Wang M; Li W; Gao LF; Shen L; Huang Y; Xie GC; et al. Direct Conversion of Normal and Alzheimer’s Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell 2015, 17, 204–212. [DOI] [PubMed] [Google Scholar]
- (68).Han DW; Tapia N; Hermann A; Hemmer K; Hoing S; Arauzo-Bravo MJ; Zaehres H; Wu GM; Frank S; Moritz S; et al. Direct Reprogramming of Fibroblasts into Neural Stem Cells by Defined Factors. Cell Stem Cell 2012, 10, 465–472. [DOI] [PubMed] [Google Scholar]
- (69).Yang N; Zuchero JB; Ahlenius H; Marro S; Ng YH; Vierbuchen T; Hawkins JS; Geissler R; Barres BA; Wernig M Generation of Oligodendroglial Cells by Direct Lineage Conversion. Nat. Biotechnol 2013, 31, 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Yu B; He ZY; You P; Han QW; Xiang D; Chen F; Wang MJ; Liu CC; Lin XW; Borjigin U; et al. Reprogramming Fibroblasts into Bipotential Hepatic Stem Cells by Defined Factors. Cell Stem Cell 2013, 13, 328–340. [DOI] [PubMed] [Google Scholar]
- (71).Cheng H; Ang HYK; EL Farran CA; Li P; Fang HT; Liu TM; Kong SL; Chin ML; Ling WY; Lim EKH; et al. Reprogramming Mouse Fibroblasts into Engraftable Myeloerythroid and Lymphoid Progenitors. Nat. Commun 2016, 7, 13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Batta K; Florkowska M; Kouskoff V; Lacaud G Direct Reprogramming of Murine Fibroblasts to Hematopoietic Progenitor Cells. Cell Rep. 2014, 9, 1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Vanslambrouck JM; Woodard LE; Suhaimi N; Williams FM; Howden SE; Wilson SB; Lonsdale A; Er PX; Li J; Maksimovic J; et al. Direct Reprogramming to Human Nephron Progenitor-like Cells Using Inducible PiggyBac Transposon Expression of SNAI2-EYA1-SIX1. Kidney Int. 2019, 95, 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Miura S; Suzuki A Generation of Mouse and Human Organoid-Forming Intestinal Progenitor Cells by Direct Lineage Reprogramming. Cell Stem Cell 2017, 21, 456–471. [DOI] [PubMed] [Google Scholar]
- (75).Bar-Nur O; Gerli MFM; Di Stefano B; Almada AE; Galvin A; Coffey A; Huebner AJ; Feige P; Verheul C; Cheung P; et al. Direct Reprogramming of Mouse Fibroblasts into Functional Skeletal Muscle Progenitors. Stem Cell Rep. 2018, 10, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Ito N; Kii I; Shimizu N; Tanaka H; Takeda S Direct Reprogramming of Fibroblasts into Skeletal Muscle Progenitor Cells by Transcription Factors Enriched in Undifferentiated Subpopulation of Satellite Cells. Sci. Rep 2017, 7, 8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Zhang Y; Cao N; Huang Y; Spencer CI; Fu JD; Yu C; Liu K; Nie B; Xu T; Li K; et al. Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts. Cell Stem Cell 2016, 18, 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Lalit PA; Salick MR; Nelson DO; Squirrell JM; Shafer CM; Patel NG; Saeed I; Schmuck EG; Markandeya YS; Wong R; et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell Stem Cell 2016, 18, 354–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Lujan E; Wernig M An indirect Approach to Generating Specific Human Cell Types. Nat. Methods 2013, 10, 44–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Wang LH; Wang LL; Huang WH; Su HX; Xue YT; Su ZH; Liao BJ; Wang HT; Bao XC; Qin DJ; et al. Generation of Integration-Free Neural Progenitor Cells from Cells in Human Urine. Nat. Methods 2013, 10, 84–89. [DOI] [PubMed] [Google Scholar]
- (81).Szabo E; Rampalli S; Risueno RM; Schnerch A; Mitchell R; Fiebig-Comyn A; Levadoux-Martin M; Bhatia M Direct Conversion of Human Fibroblasts to Multilineage Blood Progenitors. Nature 2010, 468, 521–526. [DOI] [PubMed] [Google Scholar]
- (82).Kurian L; Sancho-Martinez I; Nivet E; Aguirre A; Moon K; Pendaries C; Volle-Challier C; Bono F; Herbert JM; Pulecio J; et al. Conversion of Human Fibroblasts to Angioblast-like Progenitor Cells. Nat. Methods 2013, 10, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Margariti A; Winkler B; Karamariti E; Zampetaki A; Tsai TN; Baban D; Ragoussis J; Huang Y; Han JDJ; Zeng LF; et al. Direct Reprogramming of Fibroblasts into Endothelial Cells Capable of Angiogenesis and Reendothelialization in Tissue-Engineered Vessels. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 13793–13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Xiao X; Guo P; Shiota C; Zhang T; Coudriet GM; Fischbach S; Prasadan K; Fusco J; Ramachandran S; Witkowski P; et al. Endogenous Reprogramming of Alpha Cells into Beta Cells, Induced by Viral Gene Therapy, Reverses Autoimmune Diabetes. Cell Stem Cell 2018, 22, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Zhou Q; Brown J; Kanarek A; Rajagopal J; Melton DA In Vivo Reprogramming of Adult Pancreatic Exocrine Cells to Beta-Cells. Nature 2008, 455, 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Xiao XW; Guo P; Shiota C; Zhang T; Coudriet GM; Fischbach S; Prasadan K; Fusco J; Ramachandran S; Witkowski P; et al. Endogenous Reprogramming of Alpha Cells into Beta Cells, Induced by Viral Gene Therapy, Reverses Autoimmune Diabetes. Cell Stem Cell 2018, 22, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Niu WZ; Zang T; Zou YH; Fang SH; Smith DK; Bachoo R; Zhang CL In Vivo Reprogramming of Astrocytes to Neuroblasts in the Adult Brain. Nat. Cell Biol 2013, 15, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).De la Rossa A; Bellone C; Golding B; Vitali I; Moss J; Toni N; Luscher C; Jabaudon D In Vivo Reprogramming of Circuit Connectivity in Postmitotic Neocortical Neurons. Nat. Neurosci 2013, 16, 193–200. [DOI] [PubMed] [Google Scholar]
- (89).Torper O; Pfisterer U; Wolf DA; Pereira M; Lau S; Jakobsson J; Bjorklund A; Grealish S; Parmar M Generation of Induced Neurons via Direct Conversion In Vivo. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 7038–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Grande A; Sumiyoshi K; Lopez-Juarez A; Howard J; Sakthivel B; Aronow B; Campbell K; Nakafuku M Environmental Impact on Direct Neuronal Reprogramming In Vivo in the Adult Brain. Nat. Commun 2013, 4, 2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Guo ZY; Zhang L; Wu Z; Chen YC; Wang F; Chen G In Vivo Direct Reprogramming of Reactive Glial Cells into Functional Neurons after Brain Injury and in an Alzheimer’s Disease Model. Cell Stem Cell 2014, 14, 188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Su ZD; Niu WZ; Liu ML; Zou YH; Zhang CL In Vivo Conversion of Astrocytes to Neurons in the Injured Adult Spinal Cord. Nat. Commun 2014, 5, 3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Torper O; Ottosson DR; Pereira M; Lau S; Cardoso T; Grealish S; Parmar M In Vivo Reprogramming of Striatal NG2 Glia into Functional Neurons that Integrate into Local Host Circuitry. Cell Rep. 2015, 12, 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).El Waly B; Cayre M; Durbec P Promoting Myelin Repair through In Vivo Neuroblast Reprogramming. Stem Cell Rep. 2018, 10, 1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Qian L; Huang Y; Spencer CI; Foley A; Vedantham V; Liu L; Conway SJ; Fu JD; Srivastava D In Vivo Reprogramming of Murine Cardiac Fibroblasts into Induced Cardiomyocytes. Nature 2012, 485, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Song K; Nam YJ; Luo X; Qi X; Tan W; Huang GN; Acharya A; Smith CL; Tallquist MD; Neilson EG; et al. Heart Repair by Reprogramming Non-Myocytes with Cardiac Transcription Factors. Nature 2012, 485, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Jayawardena TM; Egemnazarov B; Finch EA; Zhang LN; Payne JA; Pandya K; Zhang ZP; Rosenberg P; Mirotsou M; Dzau VJ MicroRNA-Mediated In Vitro and In Vivo Direct Reprogramming of Cardiac Fibroblasts to Cardiomyocytes. Circ. Res 2012, 110, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Rezvani M; Espanol-Suner R; Malato Y; Dumont L; Grimm AA; Kienle E; Bindman JG; Wiedtke E; Hsu BY; Naqvi SJ; et al. In Vivo Hepatic Reprogramming of Myofibroblasts with AAV Vectors as a Therapeutic Strategy for Liver Fibrosis. Cell Stem Cell 2016, 18, 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Song GQ; Pacher M; Balakrishnan A; Yuan QG; Tsay HC; Yang DK; Reetz J; Brandes S; Dai Z; Putzer BM; et al. Direct Reprogramming of Hepatic Myofibroblasts into Hepatocytes In Vivo Attenuates Liver Fibrosis. Cell Stem Cell 2016, 18, 797–808. [DOI] [PubMed] [Google Scholar]
- (100).Ueki Y; Wilken MS; Cox KE; Chipman L; Jorstad N; Sternhagen K; Simic M; Ullom K; Nakafuku M; Reh TA Transgenic Expression of the Proneural Transcription Factor Ascl1 in Muller Glia Stimulates Retinal Regeneration in Young Mice. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Kurita M; Araoka T; Hishida T; O’Keefe DD; Takahashi Y; Sakamoto A; Sakurai M; Suzuki K; Wu J; Yamamoto M; et al. In Vivo Reprogramming of Wound-Resident Cells Generates Skin Epithelial Tissue. Nature 2018, 561, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Smith TT; Stephan SB; Moffett HF; McKnight LE; Ji WH; Reiman D; Bonagofski E; Wohlfahrt ME; Pillai SPS; Stephan MT In Situ Programming of Leukaemia-Specific T Cells Using Synthetic DNA Nanocarriers. Nat. Nanotechnol 2017, 12, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Gao X; Wang XT; Xiong WH; Chen JH In Vivo Reprogramming Reactive Glia into iPSCs to Produce New Neurons in the Cortex Following Traumatic Brain Injury. Sci. Rep 2016, 6, 22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Doeser MC; Scholer HR; Wu GM Reduction of Fibrosis and Scar Formation by Partial Reprogramming In Vivo. Stem Cells 2018, 36, 1216–1225. [DOI] [PubMed] [Google Scholar]
- (105).Abad M; Mosteiro L; Pantoja C; Canamero M; Rayon T; Ors I; Grana O; Megias D; Dominguez O; Martinez D; et al. Reprogramming In Vivo Produces Teratomas and iPS Cells with Totipotency Features. Nature 2013, 502, 340–345. [DOI] [PubMed] [Google Scholar]
- (106).Mosteiro L; Pantoja C; Alcazar N; Marion RM; Chondronasiou D; Rovira M; Fernandez-Marcos PJ; Munoz-Martin M; Blanco-Aparicio C; Pastor J; et al. Tissue Damage and Senescence Provide Critical Signals for Cellular Reprogramming In Vivo. Science 2016, 354, aaf4445. [DOI] [PubMed] [Google Scholar]
- (107).Chiche A; Le Roux I; von Joest M; Sakai H; Aguin SB; Cazin C; Salam R; Fiette L; Alegria O; Flamant P; et al. Injury-Induced Senescence Enables In Vivo Reprogramming in Skeletal Muscle. Cell Stem Cell 2017, 20, 407–414. [DOI] [PubMed] [Google Scholar]
- (108).Zhou HY; Wu SL; Joo JY; Zhu SY; Han DW; Lin TX; Trauger S; Bien G; Yao S; Zhu Y; et al. Generation of Induced Pluripotent Stem Cells using Recombinant Proteins. Cell Stem Cell 2009, 4, 381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Yu JY; Hu KJ; Smuga-Otto K; Tian SL; Stewart R; Slukvin II; Thomson JA Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science 2009, 324, 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Wen W; Zhang JP; Xu J; Su RJ; Neises A; Ji GZ; Yuan WP; Cheng T; Zhang XB Enhanced Generation of Integration-Free iPSCs from Human Adult Peripheral Blood Mononuclear Cells with An Optimal Combination of Episomal Vectors. Stem Cell Rep. 2016, 6, 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Warren L; Manos PD; Ahfeldt T; Loh YH; Li H; Lau F; Ebina W; Mandal PK; Smith ZD; Meissner A; et al. Highly Efficient Reprogramming to Pluripotency And Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell 2010, 7, 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Anokye-Danso F; Trivedi CM; Juhr D; Gupta M; Cui Z; Tian Y; Zhang YZ; Yang WL; Gruber PJ; Epstein JA; et al. Highly Efficient Mirna-Mediated Reprogramming of Mouse and Human Somatic Cells to Pluripotency. Cell Stem Cell 2011, 8, 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Miyoshi N; Ishii H; Nagano H; Haraguchi N; Dewi DL; Kano Y; Nishikawa S; Tanemura M; Mimori K; Tanaka F; et al. Reprogramming of Mouse and Human Cells to Pluripotency Using Mature microRNAs. Cell Stem Cell 2011, 8, 633–638. [DOI] [PubMed] [Google Scholar]
- (114).Hou PP; Li YQ; Zhang X; Liu C; Guan JY; Li HG; Zhao T; Ye JQ; Yang WF; Liu K; et al. Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 2013, 341, 651–654. [DOI] [PubMed] [Google Scholar]
- (115).Li X; Zuo XH; Jing JZ; Ma YT; Wang JM; Liu DF; Zhu JL; Du XM; Xiong L; Du YY; et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 2015, 17, 195–203. [DOI] [PubMed] [Google Scholar]
- (116).Jia FJ; Wilson KD; Sun N; Gupta DM; Huang M; Li ZJ; Panetta NJ; Chen ZY; Robbins RC; Kay MA; et al. A Nonviral Minicircle Vector for Deriving Human iPS Cells. Nat. Methods 2010, 7, 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Narsinh KH; Jia FJ; Robbins RC; Kay MA; Longaker MT; Wu JC Generation of Adult Human Induced Pluripotent Stem Cells Using Nonviral Minicircle DNA Vectors. Nat. Protoc 2011, 6, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Black JB; Gersbach CA Synthetic Transcription Factors for Cell Fate Reprogramming. Curr. Opin. Genet. Dev 2018, 52, 13–21. [DOI] [PubMed] [Google Scholar]
- (119).Weltner J; Balboa D; Katayama S; Bespalov M; Krjutskov K; Jouhilahti EM; Trokovic R; Kere J; Otonkoski T Human Pluripotent Reprogramming with CRISPR Activators. Nat. Commun 2018, 9, 2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Higuchi A; Ling QD; Kumar SS; Munusamy MA; Alarfaj AA; Chang Y; Kao SH; Lin KC; Wang HC; Umezawa A Generation of Pluripotent Stem Cells without the Use of Genetic Mmaterial. Lab. Invest 2015, 95, 26–42. [DOI] [PubMed] [Google Scholar]
- (121).Engel JL; Ardehali R Direct Cardiac Reprogramming: Progress and Promise. Stem Cells Int. 2018, 2018, 1435746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Stewart MP; Sharei A; Ding X; Sahay G; Langer R; Jensen KF In Vitro and Ex Vivo Strategies for Intracellular Delivery. Nature 2016, 538, 183–192. [DOI] [PubMed] [Google Scholar]
- (123).Ouyang MX; Hill W; Lee JH; Hur SC Microscale Symmetrical Electroporator Array as a Versatile Molecular Delivery System. Sci. Rep 2017, 7, 44757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Gonzalez F; Boue S; Belmonte JCI Methods for Making Induced Pluripotent Stem Cells: Reprogramming a La Carte. Nat. Rev. Genet 2011, 12, 231–242. [DOI] [PubMed] [Google Scholar]
- (125).Xu J; Du YY; Deng HK Direct Lineage Reprogramming: Strategies, Mechanisms, and Applications. Cell Stem Cell 2015, 16, 119–134. [DOI] [PubMed] [Google Scholar]
- (126).Shi Y; Inoue H; Wu JC; Yamanaka S Induced Pluripotent Stem Cell Technology: A Decade of Progress. Nat. Rev. Drug Discov 2017, 16, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Neumann E; Schaeferridder M; Wang Y; Hofschneider PH Gene-Transfer into Mouse Lyoma Cells by Electroporation in High Electric-Fields. EMBO J. 1982, 1, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Chang L; Li L; Shi J; Sheng Y; Lu W; Gallego-Perez D; Lee LJ Micro-/Nanoscale Electroporation. Lab Chip 2016, 16, 4047–4062. [DOI] [PubMed] [Google Scholar]
- (129).Chu VT; Weber T; Wefers B; Wurst W; Sander S; Rajewsky K; Kuhn R Increasing the Efficiency of Homology-Directed Repair for CRISPR-Cas9-Induced Precise Gene Editing in Mammalian Cells. Nat. Biotechnol 2015, 33, 543–548. [DOI] [PubMed] [Google Scholar]
- (130).Zhao Y; Moon E; Carpenito C; Paulos CM; Liu X; Brennan AL; Chew A; Carroll RG; Scholler J; Levine BL; et al. Multiple Injections of Electroporated Autologous T Cells Expressing a Chimeric Antigen Receptor Mediate Regression of Human Disseminated Tumor. Cancer Res. 2010, 70, 9053–9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Kalos M; June CH Adoptive T Cell Transfer for Cancer Immunotherapy in the Era of Synthetic Biology. Immunity 2013, 39, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Chang L; Gallego-Perez D; Zhao X; Bertani P; Yang Z; Chiang CL; Malkoc V; Shi J; Sen CK; Odonnell L; et al. Dielectrophoresis-Assisted 3D Nanoelectroporation for Non-Viral Cell Transfection in Adoptive Immunotherapy. Lab Chip 2015, 15, 3147–3153. [DOI] [PubMed] [Google Scholar]
- (133).Saijilafu; Hur EM; Zhou FQ Genetic Dissection of Axon Regeneration via In Vivo Electroporation of Adult Mouse Sensory Neurons. Nat. Commun 2011, 2, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).dal Maschio M; Ghezzi D; Bony G; Alabastri A; Deidda G; Brondi M; Sato SS; Zaccaria RP; Di Fabrizio E; Ratto GM; et al. High-Performance and Site-Directed in Utero Electroporation by a Triple-Electrode Probe. Nat. Commun 2012, 3, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Pinyon JL; Tadros SF; Froud KE; Wong ACY; Tompson IT; Crawford EN; Ko M; Morris R; Klugmann M; Housley GD Close-Field Electroporation Gene Delivery Using the Cochlear Implant Electrode Array Enhances the Bionic Ear. Sci. Transl. Med 2014, 6, 233ra54. [DOI] [PubMed] [Google Scholar]
- (136).Gallego-Perez D; Pal D; Ghatak S; Malkoc V; Higuita-Castro N; Gnyawali S; Chang LQ; Liao WC; Shi JF; Sinha M; et al. Topical Tissue Nano-Transfection Mediates Non-Viral Stroma Reprogramming and Rescue. Nat. Nanotechnol 2017, 12, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Larouche J; Aguilar CA New Technologies to Enhance In Vivo Reprogramming for Regenerative Medicine. Trends Biotechnol. 2019, 37, 604. [DOI] [PubMed] [Google Scholar]
- (138).Shi JF; Ma YF; Zhu J; Chen YX; Sun YT; Yao YC; Yang ZG; Xie J A Review on Electroporation-Based Intracellular Delivery. Molecules 2018, 23, No. 3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Okita K; Matsumura Y; Sato Y; Okada A; Morizane A; Okamoto S; Hong H; Nakagawa M; Tanabe K; Tezuka K; et al. A More Efficient Method to Generate Integration-Free Human iPS Cells. Nat. Methods 2011, 8, 409–412. [DOI] [PubMed] [Google Scholar]
- (140).Rouaux C; Arlotta P Direct Lineage Reprogramming of Post-Mitotic Callosal Neurons into Corticofugal Neurons In Vivo. Nat. Cell Biol 2013, 15, 214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Boukany PE; Morss A; Liao WC; Henslee B; Jung H; Zhang X; Yu B; Wang X; Wu Y; Li L; et al. Nanochannel Electroporation Delivers Precise Amounts of Biomolecules into Living Cells. Nat. Nanotechnol 2011, 6, 747–754. [DOI] [PubMed] [Google Scholar]
- (142).Geng T; Lu C Microfluidic Electroporation for Cellular Analysis and Delivery. Lab Chip 2013, 13, 3803–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Valero A; Post JN; van Nieuwkasteele JW; ter Braak PM; Kruijer W; van den Berg A Gene Transfer and Protein Dynamics in Stem Cells Using Single Cell Electroporation in a Microfluidic Device. Lab Chip 2008, 8, 62–67. [DOI] [PubMed] [Google Scholar]
- (144).Ding XY; Stewart MP; Sharei A; Weaver JC; Langer RS; Jensen KF High-Throughput Nuclear Delivery and Rapid Expression of DNA via Mechanical and Electrical Cell-Membrane Disruption. Nat. Biomed. Eng 2017, 1, No. 0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Yang Y; Kulangara K; Sia J; Wang L; Leong KW Engineering of a Microfluidic Cell Culture Platform Embedded with Nanoscale Features. Lab Chip 2011, 11, 1638–1646. [DOI] [PubMed] [Google Scholar]
- (146).Gao KL; Li L; He LN; Hinkle K; Wu Y; Ma JY; Chang LQ; Zhao X; Perez DG; Eckardt S; et al. Design of a Microchannel-Nanochannel-Microchannel Array Based Nanoelectroporation System for Precise Gene Transfection. Small 2014, 10, 1015–1023. [DOI] [PubMed] [Google Scholar]
- (147).Xie X; Xu AM; Leal-Ortiz S; Cao Y; Garner CC; Melosh NA Nanostraw-Electroporation System for Highly Efficient Intracellular Delivery and Transfection. ACS Nano 2013, 7, 4351–4358. [DOI] [PubMed] [Google Scholar]
- (148).Stewart MP; Langer R; Jensen KF Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chem. Rev 2018, 118, 7409–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Lee J; Sharei A; Sim WY; Adamo A; Langer R; Jensen KF; Bawendi MG Nonendocytic Delivery of Functional Engineered Nanoparticles into the Cytoplasm of Live Cells Using a Novel, High-Throughput Microfluidic Device. Nano Lett. 2012, 12, 6322–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Kollmannsperger A; Sharei A; Raulf A; Heilemann M; Langer R; Jensen KF; Wieneke R; Tampe R Live-Cell Protein Labelling with Nanometre Precision by Cell Squeezing. Nat. Commun 2016, 7, 10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Szeto GL; Van Egeren D; Worku H; Sharei A; Alejandro B; Park C; Frew K; Brefo M; Mao S; Heimann M; et al. Microfluidic Squeezing for Intracellular Antigen Loading in Polyclonal B-Cells as Cellular Vaccines. Sci. Rep 2015, 5, 10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Sharei A; Zoldan J; Adamo A; Sim WY; Cho N; Jackson E; Mao S; Schneider S; Han MJ; Lytton-Jean A; et al. A Vector-Free Microfluidic Platform for Intracellular Delivery. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Shalek AK; Robinson JT; Karp ES; Lee JS; Ahn DR; Yoon MH; Sutton A; Jorgolli M; Gertner RS; Gujral TS; et al. Vertical Silicon Nanowires as a Universal Platform for Delivering Biomolecules into Living Cells. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Wang Y; Yang Y; Yan L; Kwok SY; Li W; Wang Z; Zhu X; Zhu G; Zhang W; Chen X; et al. Poking Cells for Efficient Vector-Free Intracellular Delivery. Nat. Commun 2014, 5, 4466. [DOI] [PubMed] [Google Scholar]
- (155).Xu XB; Hou S; Wattanatorn N; Wang F; Yang Q; Zhao CZ; Yu X; Tseng HR; Jonas SJ; Weiss PS Precision-Guided Nanospears for Targeted and High-Throughput Intracellular Gene Delivery. ACS Nano 2018, 12, 4503–4511. [DOI] [PubMed] [Google Scholar]
- (156).Xu XB; Yang C; Wattanatorn N; Zhao CZ; Chiang NH; Jonas SJ; Weiss PS Multiple-Patterning Nanosphere Lithography for Fabricating Periodic Three-Dimensional Hierarchical Nanostructures. ACS Nano 2017, 11, 10384–10391. [DOI] [PubMed] [Google Scholar]
- (157).Chiappini C; De Rosa E; Martinez JO; Liu X; Steele J; Stevens MM; Tasciotti E Biodegradable Silicon Nanoneedles Delivering Nucleic Acids Intracellularly Induce Localized In Vivo Neovascularization. Nat. Mater 2015, 14, 532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).VanDersarl JJ; Xu AM; Melosh NA Nanostraws for Direct Fluidic Intracellular Access. Nano Lett. 2012, 12, 3881–3886. [DOI] [PubMed] [Google Scholar]
- (159).Petros RA; DeSimone JM Strategies in the Design of Nanoparticles for Therapeutic Applications. Nat. Rev. Drug Discov 2010, 9, 615–627. [DOI] [PubMed] [Google Scholar]
- (160).Behzadi S; Serpooshan V; Tao W; Hamaly MA; Alkawareek MY; Dreaden EC; Brown D; Alkilany AM; Farokhzad OC; Mahmoudi M Cellular Uptake of Nanoparticles: Journey Inside the Cell. Chem. Soc. Rev 2017, 46, 4218–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet 2014, 15, 541–555. [DOI] [PubMed] [Google Scholar]
- (162).Foldvari M; Chen DW; Nafissi N; Calderon D; Narsineni L; Rafiee A Non-Viral Gene Therapy: Gains and Challenges of Non-Invasive Administration Methods. J. Controlled Release 2016, 240, 165–190. [DOI] [PubMed] [Google Scholar]
- (163).Hajj KA; Whitehead KA Tools for Translation: Non-Viral Materials for Therapeutic mRNA Delivery. Nat. Rev. Mater 2017, 2, 17056. [Google Scholar]
- (164).Zhou ZX; Liu XR; Zhu DC; Wang Y; Zhang Z; Zhou XF; Qiu NS; Chen XS; Shen YQ Nonviral Cancer Gene Therapy: Delivery Cascade and Vector Nanoproperty Integration. Adv. Drug Deliv. Rev 2017, 115, 115–154. [DOI] [PubMed] [Google Scholar]
- (165).Savla R; Browne J; Plassat V; Wasan KM; Wasan EK Review and Analysis of FDA Approved Drugs Using Lipid-Based Formulations. Drug Dev. Ind. Pharm 2017, 43, 1743–1758. [DOI] [PubMed] [Google Scholar]
- (166).Kakkar A; Traverso G; Farokhzad OC; Weissleder R; Langer R Evolution of Macromolecular Complexity in Drug Delivery Systems. Nat. Rev. Chem 2017, 1, No. 0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Tseng YC; Mozumdar S; Huang L Lipid-Based Systemic Delivery of siRNA. Adv. Drug Deliv. Rev 2009, 61, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Zuris JA; Thompson DB; Shu Y; Guilinger JP; Bessen JL; Hu JH; Maeder ML; Joung JK; Chen ZY; Liu DR Cationic Lipid-Mediated Delivery of Proteins Enables Efficient Protein-based Genome Editing In Vitro And In Vivo. Nat. Biotechnol 2015, 33, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Subramanyam D; Lamouille S; Judson RL; Liu JY; Bucay N; Derynck R; Blelloch R Multiple Targets of miR-302 and miR-372 Promote Reprogramming of Human Fibroblasts to Induced Pluripotent Stem Cells. Nat. Biotechnol 2011, 29, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Judson RL; Babiarz JE; Venere M; Blelloch R Embryonic Stem Cell-Specific MicroRNAs Promote Induced Pluripotency. Nat. Biotechnol 2009, 27, 459–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Okita K; Nakagawa M; Hyenjong H; Ichisaka T; Yamanaka S Generation of Mouse Induced Pluripotent Stem Cells without Viral Vectors. Science 2008, 322, 949–953. [DOI] [PubMed] [Google Scholar]
- (172).Sinha M; Sen CK; Singh K; Das A; Ghatak S; Rhea B; Blackstone B; Powell HM; Khanna S; Roy S Direct Conversion of Injury-Site Myeloid Cells to Fibroblast-like Cells of Granulation Tissue. Nat. Commun 2018, 9, 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Pack DW; Hoffman AS; Pun S; Stayton PS Design and Development of Polymers for Gene Delivery. Nat. Rev. Drug Discov 2005, 4, 581–593. [DOI] [PubMed] [Google Scholar]
- (174).Raemdonck K; Martens TF; Braeckmans K; Demeester J; De Smedt SC Polysaccharide-Based Nucleic Acid Nano-formulations. Adv. Drug Deliv. Rev 2013, 65, 1123–1147. [DOI] [PubMed] [Google Scholar]
- (175).Khan W; Hosseinkhani H; Ickowicz D; Hong PD; Yu DS; Domb AJ Polysaccharide Gene Transfection Agents. Acta Biomater. 2012, 8, 4224–4232. [DOI] [PubMed] [Google Scholar]
- (176).Deng WW; Cao X; Chen JJ; Zhang ZJ; Yu QT; Wang Y; Shao GB; Zhou J; Gao XD; Yu JN; et al. Microrna Replacing Oncogenic Klf4 and C-Myc for Generating iPS Cells via Cationized Pleurotus eryngii Polysaccharide-Based Nanotransfection. ACS Appl. Mater. Interfaces 2015, 7, 18957–18966. [DOI] [PubMed] [Google Scholar]
- (177).Yu QT; Chen JJ; Deng WW; Cao X; Wang Y; Zhou J; Xu WQ; Du P; Wang Q; Yu JN; et al. Direct Reprogramming of Mouse Fibroblasts into Neural Cells via Porphyra Yezoensis Polysaccharide Based High Efficient Gene Co-Delivery. J. Nanobiotechnol 2017, 15, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Peng CH; Cherng JY; Chiou GY; Chen YC; Chien CH; Kao CL; Chang YL; Chien Y; Chen LK; Liu JH; et al. Delivery of Oct4 and SirT1 with Cationic Polyurethanes-Short Branch PEI to Aged Retinal Pigment Epithelium. Biomaterials 2011, 32, 9077–9088. [DOI] [PubMed] [Google Scholar]
- (179).Montserrat N; Garreta E; Gonzalez F; Gutierrez J; Eguizabal C; Ramos V; Borros S; Belmonte JCI Simple Generation of Human Induced Pluripotent Stem Cells Using Poly-beta-Amino Esters as the Non-Viral Gene Delivery System. J. Biol. Chem 2011, 286, 12417–12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Astruc D; Boisselier E; Ornelas C Dendrimers Designed for Functions: from Physical, Photophysical, and Supramolecular Properties to Applications in Sensing, Catalysis, Molecular Electronics, Photonics, and Nanomedicine. Chem. Rev 2010, 110, 1857–1959. [DOI] [PubMed] [Google Scholar]
- (181).Khan M; Narayanan K; Lu HF; Choo Y; Du C; Wiradharma N; Yang YY; Wan ACA Delivery of Reprogramming Factors into Fibroblasts for Generation of Non-Genetic Induced Pluripotent Stem Cells Using a Cationic Bolaamphiphile as a Non-Viral Vector. Biomaterials 2013, 34, 5336–5343. [DOI] [PubMed] [Google Scholar]
- (182).Fang J; Ye SH; Shankarraman V; Huang YX; Mo XM; Wagner WR Biodegradable Poly(ester urethane)urea Elastomers with Variable Amino Content for Subsequent Functionalization with Phosphorylcholine. Acta Biomater. 2014, 10, 4639–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Wang C; Zhu K; Li J; Lai H; Wang C; Lai H Reprogramming Fibroblasts to Pluripotency using Arginine-Terminated Polyamidoamine Nanoparticles Based Non-Viral Gene Delivery System. Int. J. Nanomed 2014, 9, 5837–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Xu ZP; Zeng QH; Lu GQ; Yu AB Inorganic Nanoparticles as Carriers for Efficient Cellular Delivery. Chem. Eng. Sci 2006, 61, 1027–1040. [Google Scholar]
- (185).He CB; Lu JQ; Lin WB Hybrid Nanoparticles for Combination Therapy of Cancer. J. Controlled Release 2015, 219, 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Chang JH; Tsai PH; Wang KY; Wei YT; Chiou SH; Mou CY Generation of Functional Dopaminergic Neurons from Reprogramming Fibroblasts by Nonviral-Based Mesoporous Silica Nanoparticles. Sci. Rep 2018, 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Cao X; Deng WW; Qu R; Yu QT; Li J; Yang Y; Cao Y; Gao XD; Xu XM; Yu JN Non-Viral Co-Delivery of the Four Yamanaka Factors for Generation of Human Induced Pluripotent Stem Cells via Calcium Phosphate Nanocomposite Particles. Adv. Funct. Mater 2013, 23, 5403–5411. [Google Scholar]
- (188).Giger EV; Puigmarti-Luis J; Schlatter R; Castagner B; Dittrich PS; Leroux JC Gene Delivery with Bisphosphonate-Stabilized Calcium Phosphate Nanoparticles. J. Controlled Release 2011, 150, 87–93. [DOI] [PubMed] [Google Scholar]
- (189).Chiang MY; Lin YZ; Chang SJ; Shyu WC; Lu HE; Chen SY Direct Reprogramming of Human Suspension Cells into Mesodermal Cell Lineages via Combined Magnetic Targeting and Photothermal Stimulation by Magnetic Graphene Oxide Complexes. Small 2017, 13, 1700703. [DOI] [PubMed] [Google Scholar]
- (190).Chang Y; Lee E; Kim J; Kwon YW; Kwon Y; Kim J Efficient In Vivo Direct Conversion of Fibroblasts into Cardiomyocytes Using a Nanoparticle-Based Gene Carrier. Biomaterials 2019, 192, 500–509. [DOI] [PubMed] [Google Scholar]
- (191).Choi HY; Lee TJ; Yang GM; Oh J; Won J; Han J; Jeong GJ; Kim J; Kim JH; Kim BS; et al. Efficient mRNA Delivery with Graphene Oxide-Polyethylenimine for Generation of Footprint-Free Human Induced Pluripotent Stem Cells. J. Controlled Release 2016, 235, 222–235. [DOI] [PubMed] [Google Scholar]
- (192).Zhou YX; Quan GL; Wu QL; Zhang XX; Niu BY; Wu BY; Huang Y; Pan X; Wu CB Mesoporous Silica Nanoparticles for Drug and Gene Delivery. Acta Pharm. Sin. B 2018, 8, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Deng WW; Cao X; Wang QT; Wang Y; Chen JJ; Yu QT; Zhang ZJ; Zhou J; Xu WQ; Du P; et al. Prolonged Three-Dimensional Co-Delivery of Yamanaka Factors for Cell Reprogramming. ACS Appl. Mater. Interfaces 2016, 8, 19916–19927. [DOI] [PubMed] [Google Scholar]
- (194).Ghosh P; Han G; De M; Kim CK; Rotello VM Gold Nanoparticles in Delivery Applications. Adv. Drug Deliv. Rev 2008, 60, 1307–1315. [DOI] [PubMed] [Google Scholar]
- (195).Yoo J; Lee E; Kim HY; Youn DH; Jung J; Kim H; Chang YJ; Lee W; Shin J; Baek S; et al. Electromagnetized Gold Nanoparticles Mediate Direct Lineage Reprogramming into Induced Dopamine Neurons In Vivo for Parkinson’s Disease Therapy. Nat. Nanotechnol 2017, 12, 1006–1014. [DOI] [PubMed] [Google Scholar]
- (196).Yin H; Kauffman KJ; Anderson DG Delivery Technologies for Genome Editing. Nat. Rev. Drug Discov 2017, 16, 387–399. [DOI] [PubMed] [Google Scholar]
- (197).Shi DQ; Xu XQ; Ye YQ; Song K; Cheng YX; Di J; Hu QY; Li JX; Ju HX; Jiang Q; et al. Photo-Cross-Linked Scaffold with Kartogenin-Encapsulated Nanoparticles for Cartilage Regeneration. ACS Nano 2016, 10, 1292–1299. [DOI] [PubMed] [Google Scholar]
- (198).Biswas A; Liu Y; Liu TF; Fan GP; Tang Y Polyethylene Glycol-Based Protein Nanocapsules for Functional Delivery of A Differentiation Transcription Factor. Biomaterials 2012, 33, 5459–5467. [DOI] [PubMed] [Google Scholar]
- (199).Biswas A; Joo KI; Liu J; Zhao MX; Fan GP; Wang P; Gu Z; Tang Y Endoprotease-Mediated Intracellular Protein Delivery Using Nanocapsules. ACS Nano 2011, 5, 1385–1394. [DOI] [PubMed] [Google Scholar]
- (200).Zhang YQ; Yu JC; Bomba HN; Zhu Y; Gu Z Mechanical Force-Triggered Drug Delivery. Chem. Rev 2016, 116, 12536–12563. [DOI] [PubMed] [Google Scholar]
- (201).Torchilin VP Multifunctional, Stimuli-Sensitive Nano-particulate Systems for Drug Delivery. Nat. Rev. Drug Discov 2014, 13, 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Kumar A; Placone JK; Engler AJ Understanding the Extracellular Forces that Determine Cell Fate and Maintenance. Development 2017, 144, 4261–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Higuchi A; Ling QD; Chang Y; Hsu ST; Umezawa A Physical Cues of Biomaterials Guide Stem Cell Differentiation Fate. Chem. Rev 2013, 113, 3297–3328. [DOI] [PubMed] [Google Scholar]
- (204).Higuchi A; Ling QD; Kumar SS; Chang Y; Alarfaj AA; Munusamy MA; Murugan K; Hsu ST; Umezawa A Physical Cues of Cell Culture Materials Lead the Direction of Differentiation Lineages of Pluripotent Stem Cells. J. Mater. Chem. B 2015, 3, 8032–8058. [DOI] [PubMed] [Google Scholar]
- (205).Kirby TJ; Lammerding J Emerging Views of the Nucleus as a Cellular Mechanosensor. Nat. Cell Biol 2018, 20, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Geiger B; Spatz JP; Bershadsky AD Environmental Sensing through Focal Adhesions. Nat. Rev. Mol. Cell Biol 2009, 10, 21–33. [DOI] [PubMed] [Google Scholar]
- (207).Darnell M; Mooney DJ Leveraging Advances in Biology to Design Biomaterials. Nat. Mater 2017, 16, 1178–1185. [DOI] [PubMed] [Google Scholar]
- (208).Dalby MJ; Gadegaard N; Tare R; Andar A; Riehle MO; Herzyk P; Wilkinson CDW; Oreffo ROC The Control of Human Mesenchymal Cell Differentiation Using Nanoscale Symmetry and Disorder. Nat. Mater 2007, 6, 997–1003. [DOI] [PubMed] [Google Scholar]
- (209).McMurray RJ; Gadegaard N; Tsimbouri PM; Burgess KV; McNamara LE; Tare R; Murawski K; Kingham E; Oreffo ROC; Dalby MJ Nanoscale Surfaces for the Long-Term Maintenance of Mesenchymal Stem Cell Phenotype and Multipotency. Nat. Mater 2011, 10, 637–644. [DOI] [PubMed] [Google Scholar]
- (210).Tsimbouri PM; McMurray RJ; Burgess KV; Alakpa EV; Reynolds PM; Murawski K; Kingham E; Oreffo ROC; Gadegaard N; Dalby MJ Using Nanotopography and Metabolomics to Identify Biochemical Effectors of Multipotency. ACS Nano 2012, 6, 10239–10249. [DOI] [PubMed] [Google Scholar]
- (211).Ireland RG; Simmons CA Human Pluripotent Stem Cell Mechanobiology: Manipulating the Biophysical Microenvironment for Regenerative Medicine and Tissue Engineering Applications. Stem Cells 2015, 33, 3187–3196. [DOI] [PubMed] [Google Scholar]
- (212).Dalby MJ; Gadegaard N; Oreffo ROC Harnessing Nanotopography and Integrin-Matrix Interactions to Influence Stem Cell Fate. Nat. Mater 2014, 13, 558–569. [DOI] [PubMed] [Google Scholar]
- (213).Downing TL; Soto J; Morez C; Houssin T; Fritz A; Yuan FL; Chu JL; Patel S; Schaffer DV; Li S Biophysical Regulation of Epigenetic State and Cell Reprogramming. Nat. Mater 2013, 12, 1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Yoo J; Kim J; Baek S; Park Y; Im H; Kim J Cell Reprogramming into the Pluripotent State Using Graphene Based Substrates. Biomaterials 2014, 35, 8321–8329. [DOI] [PubMed] [Google Scholar]
- (215).Kulangara K; Adler AF; Wang H; Chellappan M; Hammett E; Yasuda R; Leong KW The Effect of Substrate Topography on Direct Reprogramming of Fibroblasts to Induced Neurons. Biomaterials 2014, 35, 5327–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Carlson AL; Bennett NK; Francis NL; Halikere A; Clarke S; Moore JC; Hart RP; Paradiso K; Wernig M; Kohn J; et al. Generation and Transplantation of Reprogrammed Human Neurons in the Brain Using 3D Microtopographic Scaffolds. Nat. Commun 2016, 7, 10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Morez C; Noseda M; Paiva MA; Belian E; Schneider MD; Stevens MM Enhanced Efficiency of Genetic Programming Toward Cardiomyocyte Creation through Topographical Cues. Biomaterials 2015, 70, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Sia JR; Yu PZ; Srivastava D; Li S Effect of Biophysical Cues on Reprogramming to Cardiomyocytes. Biomaterials 2016, 103, 1–11. [DOI] [PubMed] [Google Scholar]
- (219).Pan F; Zhang M; Wu G; Lai Y; Greber B; Scholer HR; Chi L Topographic Effect on Human Induced Pluripotent Stem Cells Differentiation towards Neuronal Lineage. Biomaterials 2013, 34, 8131–8139. [DOI] [PubMed] [Google Scholar]
- (220).Yoo J; Noh M; Kim H; Jeon NL; Kim BS; Kim J Nanogrooved Substrate Promotes Direct Lineage Reprogramming of Fibroblasts to Functional Induced Dopaminergic Neurons. Biomaterials 2015, 45, 36–45. [DOI] [PubMed] [Google Scholar]
- (221).Wang K; Bruce A; Mezan R; Kadiyala A; Wang LY; Dawson J; Rojanasakul Y; Yang Y Nanotopographical Modulation of Cell Function through Nuclear Deformation. ACS Appl. Mater. Interfaces 2016, 8, 5082–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Engler AJ; Sen S; Sweeney HL; Discher DE Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- (223).Wen JH; Vincent LG; Fuhrmann A; Choi YS; Hribar KC; Taylor-Weiner H; Chen SC; Engler AJ Interplay of Matrix Stiffness and Protein Tethering in Stem Cell Differentiation. Nat. Mater 2014, 13, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Das RK; Gocheva V; Hammink R; Zouani OF; Rowan AE Stress-Stiffening-Mediated Stem-Cell Commitment Switch in Soft Responsive Hydrogels. Nat. Mater 2016, 15, 318–325. [DOI] [PubMed] [Google Scholar]
- (225).Discher DE; Mooney DJ; Zandstra PW Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science 2009, 324, 1673–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Choi B; Park KS; Kim JH; Ko KW; Kim JS; Han DK; Lee SH Stiffness of Hydrogels Regulates Cellular Reprogramming Efficiency through Mesenchymal-to-Epithelial Transition and Stemness Markers. Macromol. Biosci 2016, 16, 199–206. [DOI] [PubMed] [Google Scholar]
- (227).Caiazzo M; Okawa Y; Ranga A; Piersigilli A; Tabata Y; Lutolf MP Defined Three-Dimensional Microenvironments Boost Induction of Pluripotency. Nat. Mater 2016, 15, 344–352. [DOI] [PubMed] [Google Scholar]
- (228).van Helvert S; Storm C; Friedl P Mechanoreciprocity in Cell Migration. Nat. Cell Biol 2018, 20, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Yuan HH; Zhou YX; Lee MS; Zhang YZ; Li WJ A Newly Identified Mechanism Involved in Regulation of Human Mesenchymal Stem Cells by Fibrous Substrate Stiffness. Acta Biomater. 2016, 42, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Kshitiz; Hubbi ME; Ahn EH; Downey J; Afzal J; Kim DH; Rey S; Chang C; Kundu A; Semenza GL; et al. Matrix Rigidity Controls Endothelial Differentiation and Morphogenesis of Cardiac Precursors. Sci. Signaling 2012, 5, ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Nardone G; Oliver-De La Cruz J; Vrbsky J; Martini C; Pribyl J; Skladal P; Pesl M; Caluori G; Pagliari S; Martino F; et al. Yap Regulates Cell Mechanics by Controlling Focal Adhesion Assembly. Nat. Commun 2017, 8, 15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Vining KH; Mooney DJ Mechanical Forces Direct Stem Cell Behaviour in Development and Regeneration. Nat. Rev. Mol. Cell Biol 2017, 18, 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Przybyla L; Muncie JM; Weaver VM Mechanical Control of Epithelial-to-Mesenchymal Transitions in Development and Cancer. Annu. Rev. Cell Dev. Biol 2016, 32, 527–554. [DOI] [PubMed] [Google Scholar]
- (234).Tzima E; Irani-Tehrani M; Kiosses WB; Dejana E; Schultz DA; Engelhardt B; Cao GY; DeLisser H; Schwartz MA A Mechanosensory Complex that Mediates the Endothelial Cell Response to Fluid Shear Stress. Nature 2005, 437, 426–431. [DOI] [PubMed] [Google Scholar]
- (235).Davies PF Hemodynamic Shear Stress and the Endothelium in Cardiovascular Pathophysiology. Nat. Clin. Pract. Cardiovasc. Med 2009, 6, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Wirtz D; Konstantopoulos K; Searson PC The Physics of Cancer: The Role of Physical Interactions and Mechanical Forces in Metastasis. Nat. Rev. Cancer 2011, 11, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Shafa M; Day B; Yamashita A; Meng G; Liu SY; Krawetz R; Rancourt DE Derivation of iPSCs in Stirred Suspension Bioreactors. Nat. Methods 2012, 9, 465–466. [DOI] [PubMed] [Google Scholar]
- (238).Amit M; Laevsky I; Miropolsky Y; Shariki K; Peri M; Itskovitz-Eldor J Dynamic Suspension Culture for Scalable Expansion of Undifferentiated Human Pluripotent Stem Cells. Nat. Protoc 2011, 6, 572–579. [DOI] [PubMed] [Google Scholar]
- (239).Sia J; Sun R; Chu J; Li S Dynamic culture improves cell reprogramming efficiency. Biomaterials 2016, 92, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Humphrey JD; Dufresne ER; Schwartz MA Mechanotransduction and Extracellular Matrix Homeostasis. Nat. Rev. Mol. Cell Biol 2014, 15, 802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Cui YD; Hameed FM; Yang B; Lee K; Pan CQ; Park S; Sheetz M Cyclic Stretching of Soft Substrates Induces Spreading and Growth. Nat. Commun 2015, 6, 6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (242).He L; Si G; Huang JH; Samuel ADT; Perrimon N Mechanical Regulation of Stem-Cell Differentiation by the Stretch-Activated Piezo Channel. Nature 2018, 555, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Mihic A; Li J; Miyagi Y; Gagliardi M; Li SH; Zu J; Weisel RD; Keller G; Li RK The Effect of Cyclic Stretch on Maturation and 3D Tissue Formation of Human Embryonic Stem Cell-Derived Cardiomyocytes. Biomaterials 2014, 35, 2798–2808. [DOI] [PubMed] [Google Scholar]
- (244).Ott HC; Matthiesen TS; Goh SK; Black LD; Kren SM; Netoff TI; Taylor DA Perfusion-Decellularized Matrix: Using Nature’s Platform to Engineer a Bioartificial Heart. Nat. Med 2008, 14, 213–221. [DOI] [PubMed] [Google Scholar]
- (245).Rao LJ; Qian Y; Khodabukus A; Ribar T; Bursac N Engineering Human Pluripotent Stem Cells into a Functional Skeletal Muscle Tissue. Nat. Commun 2018, 9, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Niklason LE; Gao J; Abbott WM; Hirschi KK; Houser S; Marini R; Langer R Functional Arteries Grown in Vitro. Science 1999, 284, 489–493. [DOI] [PubMed] [Google Scholar]
- (247).Riehl BD; Park JH; Kwon IK; Lim JY Mechanical Stretching for Tissue Engineering: Two-Dimensional and Three-Dimensional Constructs. Tissue Eng., Part B 2012, 18, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Chu SY; Chou CH; Huang HD; Yen MH; Hong HC; Chao PH; Wang YH; Chen PY; Nian SX; Chen YR; et al. Mechanical Stretch Induces Hair Regeneration through the Alternative Activation of Macrophages. Nat. Commun 2019, 10, 1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (249).Kim YM; Kang YG; Park SH; Han MK; Kim JH; Shin JW; Shin JW Effects of Mechanical Stimulation on the Reprogramming of Somatic Cells into Human-Induced Pluripotent Stem Cells. Stem Cell Res. Ther 2017, 8, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (250).Baek S; Quan X; Kim S; Lengner C; Park JK; Kim J Electromagnetic Fields Mediate Efficient Cell Reprogramming into a Pluripotent State. ACS Nano 2014, 8, 10125–10138. [DOI] [PubMed] [Google Scholar]
- (251).Jin Y; Seo J; Lee JS; Shin S; Park HJ; Min S; Cheong E; Lee T; Cho SW Triboelectric Nanogenerator Accelerates Highly Efficient Nonviral Direct Conversion and In Vivo Reprogramming of Fibroblasts to Functional Neuronal Cells. Adv. Mater 2016, 28, 7365–7374. [DOI] [PubMed] [Google Scholar]
- (252).Fatehullah A; Tan SH; Barker N Organoids as an In Vitro Model of Human Development and Disease. Nat. Cell Biol 2016, 18, 246–254. [DOI] [PubMed] [Google Scholar]
- (253).Rossi G; Manfrin A; Lutolf MP Progress and Potential in Organoid Research. Nat. Rev. Genet 2018, 19, 671–687. [DOI] [PubMed] [Google Scholar]
- (254).Lutolf MP; Gilbert PM; Blau HM Designing Materials to Direct Stem-Cell Fate. Nature 2009, 462, 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (255).Tseng TC; Hsieh FY; Dai NT; Hsu SH Substrate-Mediated Reprogramming of Human Fibroblasts into Neural Crest Stem-like Cells and Their Applications in Neural Repair. Biomaterials 2016, 102, 148–161. [DOI] [PubMed] [Google Scholar]
- (256).Zhang BY; Korolj A; Lai BFL; Radisic M Advances in Organ-on-a-Chip Engineering. Nat. Rev. Mater 2018, 3, 257–278. [Google Scholar]
- (257).Wnorowski A; Yang HX; Wu JC Progress, Obstacles, and Limitations in the Use of Stem Cells in Organ-on-a-Chip Models. Adv. Drug Deliv. Rev 2019, 140, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (258).Wang YQ; Wang H; Deng PW; Chen WW; Guo YQ; Tao TT; Qin JH In Situ Differentiation and Generation of Functional Liver Organoids from Human iPSCs in a 3D Perfusable Chip System. Lab Chip 2018, 18, 3606–3616. [DOI] [PubMed] [Google Scholar]
- (259).Tao T; Wang Y; Chen W; Li Z; Su W; Guo Y; Deng P; Qin J Engineering Human Islet Organoids from iPSCs Using an Organ-on-Chip Platform. Lab Chip 2019, 19, 948–958. [DOI] [PubMed] [Google Scholar]
- (260).Gkatzis K; Taghizadeh S; Huh D; Stainier DYR; Bellusci S Use of Three-Dimensional Organoids and Lung-on-a-Chip Methods to Study Lung Development, Regeneration and Disease. Eur. Respir. J 2018, 52, 1800876. [DOI] [PubMed] [Google Scholar]
- (261).Wang YQ; Wang L; Zhu YJ; Qin JH Human Brain Organoid-on-a-Chip to Model Prenatal Nicotine Exposure. Lab Chip 2018, 18, 851–860. [DOI] [PubMed] [Google Scholar]
- (262).Luni C; Giulitti S; Serena E; Ferrari L; Zambon A; Gagliano O; Giobbe GG; Michielin F; Knobel S; Bosio A; et al. High-Efficiency Cellular Reprogramming with Microfluidics. Nat. Methods 2016, 13, 446–452. [DOI] [PubMed] [Google Scholar]
- (263).Giulitti S; Pellegrini M; Zorzan I; Martini P; Gagliano O; Mutarelli M; Ziller MJ; Cacchiarelli D; Romualdi C; Elvassore N; et al. Direct Generation of Human Naive Induced Pluripotent Stem Cells from Somatic Cells in Microfluidics. Nat. Cell Biol 2019, 21, 275–286. [DOI] [PubMed] [Google Scholar]
- (264).Kong YP; Carrion B; Singh RK; Putnam AJ Matrix Identity and Tractional Forces Influence Indirect Cardiac Reprogramming. Sci. Rep 2013, 3, 3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (265).Kong YP; Rioja AY; Xue XF; Sun YB; Fu JP; Putnam AJ A Systems Mechanobiology Model to Predict Cardiac Reprogramming Outcomes on Different Biomaterials. Biomaterials 2018, 181, 280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (266).Huangfu DW; Maehr R; Guo WJ; Eijkelenboom A; Snitow M; Chen AE; Melton DA Induction of Pluripotent Stem Cells by Defined Factors is Greatly Improved by Small-Molecule Compounds. Nat. Biotechnol 2008, 26, 795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (267).Zhu SY; Russ HA; Wang XJ; Zhang ML; Ma TH; Xu T; Tang SB; Hebrok M; Ding S Human Pancreatic Beta-like Cells Converted from Fibroblasts. Nat. Commun 2016, 7, 10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Zhu SY; Rezvani M; Harbell J; Mattis AN; Wolfe AR; Benet LZ; Willenbring H; Ding S Mouse Liver Repopulation with Hepatocytes Generated from Human Fibroblasts. Nature 2014, 508, 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (269).Murphy SV; Atala A 3D Bioprinting of Tissues and Organs. Nat. Biotechnol 2014, 32, 773–785. [DOI] [PubMed] [Google Scholar]
- (270).Zhao T; Fu Y; Zhu JL; Liu YF; Zhang Q; Yi ZX; Chen S; Jiao ZG; Xu XC; Xu JQ; et al. Single-Cell RNA-Seq Reveals Dynamic Early Embryonic-like Programs during Chemical Reprogramming. Cell Stem Cell 2018, 23, 31–45. [DOI] [PubMed] [Google Scholar]
- (271).Biddy BA; Kong WJ; Kamimoto K; Guo C; Waye SE; Sun T; Morris SA Single-Cell Mapping of Lineage and Identity in Direct Reprogramming. Nature 2018, 564, 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Li L; Hu S; Chen XY Non-Viral Delivery Systems for CRISPR/Cas9-Based Genome Editing: Challenges and Opportunities. Biomaterials 2018, 171, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (273).Long CZ; McAnally JR; Shelton JM; Mireault AA; Bassel-Duby R; Olson EN Prevention of Muscular Dystrophy in Mice by CRISPR/Cas9-Mediated Editing of Germline DNA. Science 2014, 345, 1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (274).Amoasii L; Hildyard JCW; Li H; Sanchez-Ortiz E; Mireault A; Caballero D; Harron R; Stathopoulou TR; Massey C; Shelton JM; et al. Gene Editing Restores Dystrophin Expression in a Canine Model of Duchenne Muscular Dystrophy. Science 2018, 362, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (275).Liu P; Chen M; Liu Y; Qi LS; Ding S CRISPR-Based Chromatin Remodeling of the Endogenous Oct4 or Sox2 Locus Enables Reprogramming to Pluripotency. Cell Stem Cell 2018, 22, 252–261. [DOI] [PubMed] [Google Scholar]
- (276).Black JB; Adler AF; Wang HG; D’Ippolito AM; Hutchinson HA; Reddy TE; Pitt GS; Leong KW; Gersbach CA Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell 2016, 19, 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]