Abstract
Comprehensive cancer genome profiling (CGP) has been nationally reimbursed in Japan since June 2019. Less than 10% of the patients have been reported to undergo recommended treatment. Todai OncoPanel (TOP) is a dual DNA–RNA panel as well as a paired tumor–normal matched test. Two hundred patients underwent TOP as part of Advanced Medical Care B with approval from the Ministry of Health, Labour and Welfare between September 2018 and December 2019. Tests were carried out in patients with cancers without standard treatment or when patients had already undergone standard treatment. Data from DNA and RNA panels were analyzed in 198 and 191 patients, respectively. The percentage of patients who were given therapeutic or diagnostic recommendations was 61% (120/198). One hundred and four samples (53%) harbored gene alterations that were detected with the DNA panel and had potential treatment implications, and 14 samples (7%) had a high tumor mutational burden. Twenty‐two samples (11.1%) harbored 30 fusion transcripts or MET exon 14 skipping that were detected by the RNA panel. Of those 30 transcripts, 6 had treatment implications and 4 had diagnostic implications. Thirteen patients (7%) were found to have pathogenic or likely pathogenic germline variants and genetic counseling was recommended. Overall, 12 patients (6%) received recommended treatment. In summary, patients benefited from both TOP DNA and RNA panels while following the same indication as the approved CGP tests. (UMIN000033647).
Keywords: comprehensive genomic profiling, gene fusion, genomic medicine, germline finding, precision medicine
Todai OncoPanel (TOP) is a dual DNA‐RNA panel as well as a paired tumor‐normal matched test. Pharmaceuticals and Medical Devices Agency (PMDA) indications for comprehensive cancer genomic profiling were followed in this study. The percentage of patients who were given therapeutic or diagnostic recommendations was 61%, including patients with fusion or MET exon 14 skipping transcripts that led to recommendations.
Abbreviations
- CGP
comprehensive genomic profiling
- CI
confidence interval
- F1CDx
FoundationOne CDx
- F1LCDx
FoundationOne Liquid CDx
- FFPE
formalin‐fixed, paraffin‐embedded
- NOP
OncoGuide NCC Oncopanel
- PMDA
Pharmaceuticals and Medical Devices Agency
- TOP
Todai OncoPanel
- TRK
tropomyosin receptor kinase
1. INTRODUCTION
Cancer is the leading cause of death in Japan. While advancement in diagnosis and treatment have improved 5‐year survival of cancer by 10% in the last 10 years, the latest 5‐year survival rate is 64%. 1 With hopes of further improving survival, CGP tests have been applied in the clinic worldwide. In Japan, two CGP tests, NOP and F1CDx, have been nationally reimbursed in Japan since June 2019, and F1LCDx was later approved. OncoGuide NCC Oncopanel tests for 124 single nucleotide variants, indels, and amplifications and 13 gene fusions; it is a paired tumor–normal matched test and can distinguish somatic from germline variants. Both F1CDx and F1LCDx test for 309 single nucleotide variants, indels, copy number variants, and 36 gene rearrangements; F1CDx is a tumor‐only panel and cannot distinguish somatic from germline variants.
In Japan, the PMDA approves these CGP tests only for patients who have progressed with standard therapy, patients who are expecting to end standard therapy, or patients with cancers without established standard therapy. When patients underwent NOP or F1CDx as part of a clinical study and regardless of whether they received standard treatment, the percentage of patients who received recommended treatment was 13% and 14%, respectively. 2 , 3 However, less than 10% of the patients undergoing these CGP tests were given recommended treatments when indications were followed after approval. 4 , 5 , 6
The TOP is a dual DNA–RNA panel as well as a paired tumor–normal matched test. It tests for 464 gene alterations with its DNA panel and 365 fusion transcripts with its RNA panel. 7 Transcriptomic profiling has been shown to result in 12 of 38 patients benefiting from receiving recommended treatments. 8 We have previously reported that clinically relevant somatic alterations were identified in 32.2% (59/183) of patients by prospective TOP testing. However, the percentage of patients who received recommended treatment is unknown. In addition, testing was done as a clinical trial without following the PMDA indication.
Here, we report on the clinical utility of TOP as part of Advanced Medical Care B, a program approved by the Ministry of Health, Labour and Welfare that allows patients to participate in a clinical trial while being reimbursed for all other health‐care under the national health insurance coverage. The purpose of this program was to facilitate the transition from clinical trial to PMDA approval; therefore, the PMDA indication for CGP testing was followed.
2. MATERIALS AND METHODS
2.1. Patients
Between September 2018 and December 2019, 200 patients were recruited from The University of Tokyo Hospital, Yokohoma City University Hospital, Saitama Cancer Center, NTT Medical Center Tokyo, Dokkyo Medical University Hospital, Cancer Institute Hospital of Japanese Foundation for Cancer Research, Toranomon Hospital, Tokyo Medical and Dental University Hospital, Center Hospital of the National Center for Global Health and Medicine, Teikyo University Hospital, Yamanashi Prefectural Central Hospital, and Jichi Medical University Hospital. After approval by the institutional ethics review board (protocol #P2017017) at each participating institution, written informed consent was obtained from all patients.
2.2. Nucleic acid extraction and next‐generation sequencing
Genomic DNA target sequencing and RNA sequencing using cDNA capture were previously described. 7 Briefly, genomic DNA was isolated from FFPE samples using GeneRead DNA FFPE Kits (Qiagen). DNA quality was determined by the FFPE DNA QC Assay version 2, TaqMan Copy Number Reference Assay (Thermo Fisher Scientific). Total RNA was extracted from FFPE samples using RNeasy FFPE Kit (Qiagen). RNA quality was evaluated on a 2200 TapeStation (Agilent Technologies) to calculate DV200. DNA target sequencing library was prepared using the SureSelectXT custom kit (Agilent Technologies). Custom‐made probes were designed to hybridize and capture the genomic DNA of the target genes listed in Table S1 and intronic DNA of 4327 single nucleotide polymorphisms within the targeted gene regions. The cDNA library preparation was carried out using the TruSight RNA Pan‐Cancer Panel (Illumina). cDNA junction capture was undertaken using the SureSelect RNA Capture kit (Agilent Technologies) with custom‐made probes designed to hybridize and capture the junctional sequences of the target genes listed in Table S1. Next‐generation sequencing was carried out using the Next‐seq platform (Illumina) as previously described. 7
2.3. Quality assessment
Quality of TOP was determined by five parameters (number of unique reads, mean depth, percentage of reads on target, target exon coverage, and total reads) with the DNA panel and three parameters (total reads, coverage of housekeeping genes, and unique reads for housekeeping genes) for the RNA panel. Details of these parameters were described previously. 7
2.4. Data analysis for CGP
Todai OncoPanel evaluates somatic gene variants, copy number alterations, and germline findings with the DNA panel, and gene fusions and MET exon skipping by the RNA panel. Details of analysis pipelines were described previously. 7 Mutations were reported if they were somatic mutations within exons with an allele frequency of 5% or higher at a depth of 100× or higher. Tumor mutational burden was measured as the number of nonsynonymous and synonymous somatic mutations per megabase of exons surveyed. Gene fusions were considered pathogenic if the fusions were reported on COSMIC or OncoKB databases and if the exon–exon fusion points matched. Clinical actionability of TOP test findings were classified into different tiers as follows: 7 biomarkers recognized by PMDA as predictive of response to approved drugs as Tier 1, biomarkers recognized by the United States FDA as predictive of response to approved drugs or biomarkers used in clinical trials carried out in Japan as Tier 2, and biomarkers that were shown to predict drug resistance in clinical trials as Tier R (Table S2). In this study, reporting of germline findings was restricted to the following 23 genes: APC, BRCA1, BRCA2, CDH1, EPCAM, MEN1, MLH1, MSH2, MSH6, MUTYH, NF1, PALB2, PMS2, PTEN, RB1, RET, SDHB, STK11, TP53, TSC1, TSC2, WT1, and VHL. Gene variants reported in the 1000 Genomes Project or the Tohoku Medical Megabank Project to have allele frequency of over 1% in the general population were considered nonpathogenic and were not reported. 9 , 10 Data from this study has been deposited into MGeND (https://mgend.med.kyoto‐u.ac.jp/).
2.5. Advanced Medical Care B
This study was undertaken as Advanced Medical Care B after approval by the Ministry of Health, Labour and Welfare. Key inclusion criteria were: (1) pathologically diagnosed as a malignant solid tumor, (2) considered to be incurable, (3) already given or expected to finish standard therapy, (4) genomic testing is considered useful to guide therapy, (5) performance status 0 or 1, (6) tumor sample is sufficiently available for the testing, and (7) written informed consent is obtained. There were no exclusion criteria. The primary end‐point was the percentage of patients with gene alterations (mutations, copy number variations, gene fusions, and exon skipping) with therapeutic or diagnostic implications, identified by either the DNA or the RNA panel. The secondary end‐point was the percentage of patients who were given recommended treatment.
2.6. Statistical analysis
Analysis population was defined as all patients who were enrolled in this study. Patients whose sample was not analyzed by TOP DNA/RNA panel were excluded from evaluation of the clinical utility of DNA/RNA panel, respectively. Each end‐point was summarized by count and proportion. The exact 95% CI for proportion was calculated by the Clopper–Pearson method.
3. RESULTS
3.1. Patient characteristics
Two hundred patients consented to the study, and 120 were women. Mean age was 56.4 (95% CI, 54.3–58.4) and median age was 59 years. The ECOG performance status was 0 in 136 patients and 1 in 64 patients. Sixty‐six patients were current or former smokers, 118 were nonsmokers, and 16 were unknown. The five most frequent cancer types were ovarian cancer (21 samples), colorectal cancer (20 samples), endometrial cancer (19 samples), lung cancer, and cervical cancer (17 samples each) (Figure 1). More than one sample was analyzed for the following cancers: breast (15), stomach (10), pancreas (10), brain (9), soft tissue/bone (7), bile duct (6), kidney (6), prostate (5), esophagus (4), head and neck (4), retroperitoneum (4), unknown (4), duodenum (3), vulva (3), thymus (2), skin (2), and bladder (2).
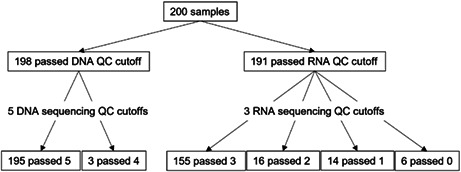
FIGURE 1.
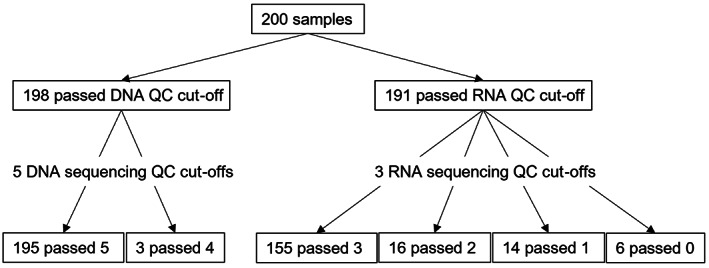
Number of samples that passed Todai OncoPanel DNA/RNA and sequencing quality control (QC) cut‐offs
3.2. Quality assessment of this study
For the DNA panel, two samples were excluded due to low quality of DNA (Figure 2). Of the 198 samples, 196 passed all five sequencing quality control parameters and two samples passed all five except for number of unique reads (<18,000,000 reads). For the RNA panel, nine samples were dropped due to low RNA yield (<50 ng) or quality (DV200 < 40%). Out of 191 samples, 155 samples passed all three sequencing qualification parameters (housekeeping gene coverage >70%, number of unique housekeeping gene reads >150,000 reads, number of total reads >20,000,000), 16 passed two, 14 passed one, and 6 did not pass any of the three parameters. The mean number of days between blood draw and DNA/RNA extraction was 7.6 days (95% CI, 6.8–8.5 days), between DNA/RNA extraction and sequencing was 9.1 days (95% CI, 8.6–9.6 days), and between sequencing to molecular tumor board (Expert Panel) was 18.8 days (95% CI, 18.1–19.4 days), resulting in total turnaround time of 35.5 days (95% CI, 34.3–36.6 days). Expert panels were held every 2 weeks.
FIGURE 2.
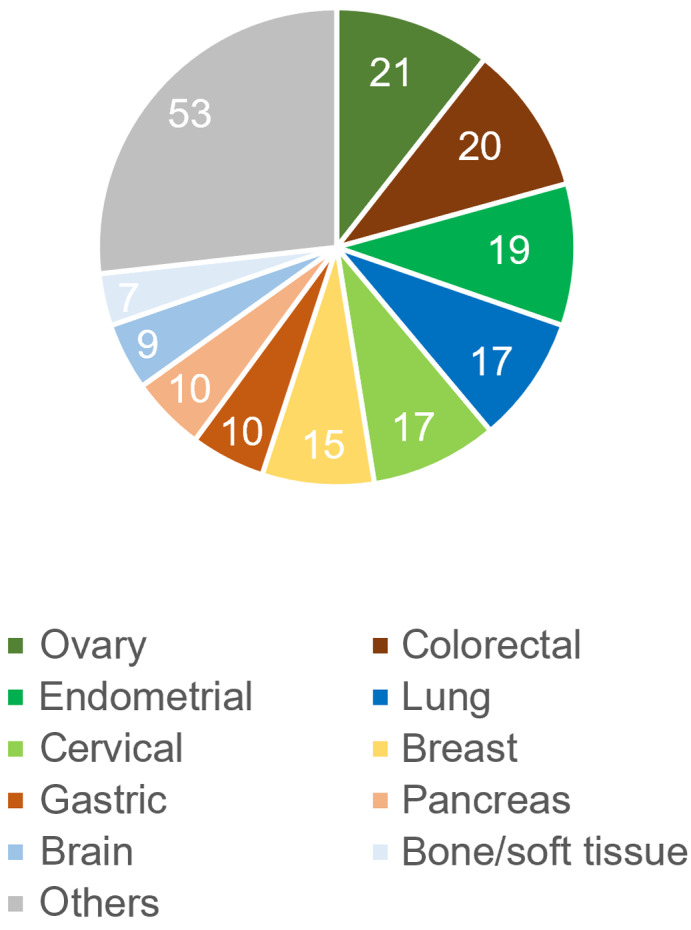
Number of samples that underwent Todai OncoPanel genomic testing, by cancer type
3.3. Significance of TOP DNA panel
Gene alterations were detected in all 198 samples. The five most frequent genes with pathogenic or likely pathogenic alterations were TP53 (101, 51.0%), KRAS (36, 18.2%), FGFR3 (20, 10.1%), PIK3CA (20, 10.1%), and APC (17, 8.6%) (Figure 3). Therapeutic or diagnostic recommendations were made in 117 of 198 patients (59.1%; 95% CI 51.9%–66.0%). Tiers 1, 2, or R (Table S2) were detected in 104 (52.5%), tumor mutational burden of 10/Mb or higher was seen in 14 (7.1%), and diagnostic significance was seen in 15 (7.6%, Table 1). The genomic data for all patients are shown in Table S3.
FIGURE 3.
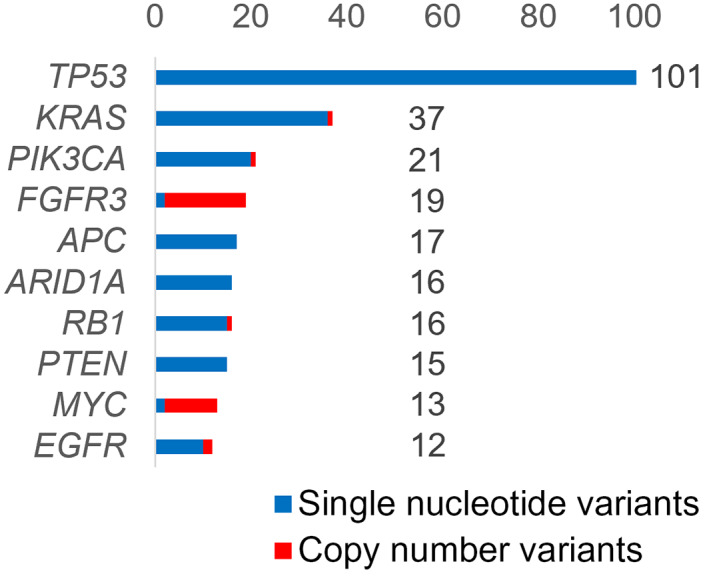
Percentage of samples that underwent Todai OncoPanel genomic testing, with each gene alteration
TABLE 1.
Diagnostic significance seen in 15 samples from Todai OncoPanel DNA panel
| Cancer type | Pathology | Diagnostic significance |
|---|---|---|
| Ovary | Endometrioid | Mutation profile more compatible with endometrioid than high grade serous |
| Soft tissue | Spindle cell/sclerosing rhabdomyosarcoma | Presence of MYOD1 mutation confirmed the diagnosis |
| Thyroid | Undifferentiated | PTEN mutation and haploid type more compatible with oncocytic follicular thyroid carcinoma |
| Kidney | Undetermined | Presence of FH mutation suggests FH‐deficient papillary renal cell cancer |
| Soft tissue | Undetermined | NF2 mutation suggests mesothelioma |
| Prostate | Adenocarcinoma | AR amplification suggests resistance to androgen therapy |
| Brain | Anaplastic astrocytoma | Presence of TERT mutation and absence of TP53 or IDH mutation or 1p/19q codeletion is more compatible with glioblastoma |
| Kidney | Undetermined | Presence of FH mutation suggests FH‐deficient papillary renal cell cancer |
| Breast | Ductal carcinoma | Presence of ESR1 mutation suggests resistance to hormone therapy |
| Soft tissue | Undetermined | Presence of PIK3CA mutation suggests mucinous sarcoma |
| Unknown primary | Neuroendocrine | Presence of ERBB2 amplification suggests gastric cancer and possible anti‐HER2 therapy |
| Brain | Glioblastoma | Presence of PTEN and TERT mutations confirms the pathological diagnosis |
| Duodenum | Adenocarcinoma | Presence of BAP1 mutation suggests bile duct carcinoma |
| Cervix | Adenocarcinoma | Presence of PGR and PTEN mutations suggests endometrial cancer |
| Unknown primary | Undetermined | Presence of BCOR mutation suggests renal cell carcinoma |
3.4. Significance of TOP RNA panel
Thirty fusion transcripts or MET exon skipping were detected in 22 of 191 samples (11.5%) (Table 2). Nine fusion transcripts were recurrent and 18 were novel. Of the 18 novel fusion transcripts, 6 were determined to be likely pathogenic and 12 were variants of unknown significance. Pathogenic or likely pathogenic fusion transcripts were detected in 13 of 191 samples (6.8%). Treatment recommendations were made for four fusions (FGFR1‐SET1, BRCA1‐EFCAB5, ZC3HAV1‐BRAF, SNRNP70‐NTRK3) and two tumors with MET exon 14 skipping. In addition, four gene fusions had diagnostic significance (CIC‐DUX4 in soft tissue sarcoma of the brain, NAB2‐STAT6 in solitary fibrous tumor of the lung, MYB‐NFIB in adenoid cystic carcinoma, and AIP‐SETD2 suggestive of BAP1 loss in malignant pleural mesothelioma). Overall, therapeutic or diagnostic recommendations were made in 117 patients using the DNA panel and 8 patients using the RNA panel. There was overlap in 5 patients; therefore, 120 of 198 patients (61%) were given therapeutic or diagnostic recommendations.
TABLE 2.
Gene fusions detected from Todai OncoPanel RNA panel
| Cancer type | Fusion | Significance | Recurrent/novel | Pathogenicity |
|---|---|---|---|---|
| Ovary | FGFR1‐SET | Therapeutic | Novel | Likely |
| Vulva |
BRCA1‐EFCAB5 PAX3‐FOXO1 |
Therapeutic |
Novel Recurrent |
Likely Likely |
| Prostate |
ZC3HAV1‐BRAF BRD4‐CALR3 |
Therapeutic |
Recurrent Novel |
Pathogenic Unknown |
| Soft tissue |
SNRNP70‐NTRK3 ERCC1‐URI1 |
Therapeutic |
Novel Novel |
Likely Unknown |
| Brain | CIC‐DUX4 | Diagnostic | Recurrent | Pathogenic |
| Meninges | NAB2‐STAT6 | Diagnostic | Recurrent | Pathogenic |
| Pleura | AIP‐SETD2 | Diagnostic | Novel | Likely |
| Salivary gland |
MYB‐NFIB NFIB‐MYB |
Diagnostic | Recurrent | Pathogenic |
| Duodenum | PTPRK‐RSPO3 | Recurrent | Pathogenic | |
| Ovary | CCDC6‐ANK3 | Recurrent | Likely | |
| Soft tissue | SS18‐SSX2 | Recurrent | Likely | |
| Colorectal | TANC2‐CA4 | Recurrent | Likely | |
| Ovary | RB1‐FBXO28 | Novel | Likely | |
| Retroperitoneum | HMGA2‐C12orf29 | Novel | Unknown | |
| Head and neck |
HMGA2‐DPYD NFIA‐METAP1D NF1A‐SLC38A11 RAVER2‐MDM2 |
Novel Novel Novel Novel |
Unknown Unknown Unknown Unknown |
|
| Ovary | IMPDH1‐KMT2C | Novel | Unknown | |
| Cervix | EIF3E‐EMC2 | Novel | Unknown | |
| Bile duct | PTPRK‐BRSK2 | Novel | Unknown | |
| Skin | IPP‐MAST2 | Novel | Unknown | |
| Duodenum |
B2M‐RABGAP1L PTPRK‐SNX3 |
Novel Novel |
Unknown Unknown |
3.5. Significance of the germline findings
Todai OncoPanel is a matched tumor–normal panel, which allows discrimination of somatic from germline variants. Pathogenic or likely pathogenic germline variants were detected in 13 of 198 samples (6.6%), and recommendations were made for genetic counseling (Table 3). BRCA1, BRCA2, and TP53 were the genes in which pathogenic or likely pathogenic variants were detected in more than one patient, that is, four, three, and three patients, respectively.
TABLE 3.
Germline variants detected from peripheral blood white blood cells
| Gene | Number of patients and cancer type |
|---|---|
| BRCA1 | 4 (All were ovary) |
| BRCA2 | 3 (Breast, pancreas, cecum) |
| TP53 | 3 (Glioblastoma, lung adenocarcinoma, osteosarcoma) |
| PALB2 | 1 (Breast) |
| RET | 1 (Liposarcoma) |
| VHL | 1 (Clear cell renal cell carcinoma) |
3.6. Number of patients who received recommended treatment
Overall, 12 of 198 patients (6.1%; 95% CI, 3.2%–10.3%) received recommended treatments. Specifically, from the DNA panel, EGFR mutation was detected in four patients with lung cancer, KIT mutation was detected in two patients (gastric cancer and retroperitoneal cancer), ESR1 and ERBB2 mutations were detected in breast cancer patients, and BRCA1 and MSH6 mutations were detected in ovarian cancer and cancer of unknown primary patients, respectively, and patients received treatments accordingly. MET exon 14 skipping and NTRK fusion were detected in lung cancer and soft tissue sarcoma patients, respectively, using the RNA panel.
4. DISCUSSION
We report here the performance of TOP given as part of Advanced Medical Care B, where patients were selected by the same indication as the CGP panels approved by PMDA. The study was carried out between September 2018 and December 2019, a period immediately before and after PMDA approved NOP and F1CDx in June 2019.
Compared with the data from the national registry, more patients had gynecological cancers in this study, possibly due to the expertise of the principal investigator (K.O.). 6 However, the frequency of pathogenic or likely pathogenic gene alterations was similar to that seen in the same database (TP53, KRAS, APC, CDKN2A, and KMT2D were the top five genes in the national database). Therefore, our cohort might have had an increased number of patients with germline findings compared with the general population, but likely had a similar percentage of patients who would potentially receive therapeutic or diagnostic recommendations.
A major issue with CGP testing is that, although actionable gene alterations are often found, it does not translate to patients receiving novel treatment. The same trend was seen in our study, as 59% of the patients received therapeutic or diagnostic recommendations and 6% received recommended treatment. However, this does not imply that only the 12 patients benefited from undergoing TOP, as 15 patients also saw a diagnostic benefit. In addition, because TOP is a tumor–normal matched panel, 7% of the patients were found to have (likely) pathogenic germline findings. In addition, our previous study, 7 undertaken in 2017–2018, showed that Tier 1 or 2 was detected in only 32%. The increase to 52% in this study could be mainly attributable to differences in cancer type but also the time of study, as evidence continuously accumulates.
One way to increase the percentage of patients who receive treatment recommendations is to use an RNA panel, with which gene fusions and rearrangements can be detected. Using the TOP RNA panel, six patients received treatment recommendations and four fusions had diagnostic significance. More studies are needed to improve fusion gene databases so that gene fusions can be better annotated. Furthermore, exon skipping other than MET and expression analysis can be incorporated in the RNA pipeline but was beyond the scope of this study. The benefits of transcriptomic profiling have been previously shown and could be incorporated into TOP in the future. 8
Adding an RNA panel to a conventional DNA panel will increase assay cost, but the increment may be small compared with the overall cost of the oncogene panel test. Accurate and sensitive detection of fusion genes by the TOP RNA panel, combined with better prediction of fusion pathogenicity obtained by RNA expression profiling, should lead to treatment recommendations for more patients in the future.
There are limitations to this study. This study was undertaken during a period immediately before and after PMDA approved NOP and F1CDx. Since then, evidence level classification (Table S2) has been published as a guidance 11 and implemented nationwide, and the number of clinical trials has increased. Furthermore, because evidence levels and clinical trial information continuously change, the precise, up‐to‐date performance of CGP panels are difficult to determine. One notable trial missing during this period is the BELIEVE trial, a national umbrella/basket off‐label trial that began registration in October 2019 and has accrued 317 patients as of April 2022. Furthermore, tropomyosin receptor kinase (TRK) inhibitors were not approved for NTRK fusion positive tumors and pembrolizumab was not yet approved for tumor mutational burden high tumors. In addition, genetic counseling was recommended for patients with pathogenic or likely pathogenic variants in one of 23 genes. The number of genes that are candidates for disclosure has increased according to a national guideline. 12 However, because the number of patients with gynecological cancers was high in our study, this might have offset the limited number of genes to yield a reasonable secondary finding rate between 5%–10%.
In summary, TOP resulted in 61% of the patients receiving therapeutic or diagnostic recommendations and 6% of the patients receiving recommended treatment. The TOP RNA panel led to detection of 30 fusion transcripts or MET exon skipping in 12% of the patients and therapeutic or diagnostic recommendations in 10 of 191 patients. This will likely increase with better annotation of fusion genes and an increase in basket trials.
FUNDING INFORMATION
This study was supported in part by a grant from the Program for an Integrated Database of Clinical and Genomic Information (JP19kk0205016) from the Japan Agency for Medical Research and Development (AMED). Sequencing of the clinical specimens was carried out in a branch laboratory of Sysmex Corporation within The University of Tokyo.
CONFLICT OF INTEREST
Y.S. and Y.Y received a research fund from Sysmex Corporation. H.K., S.K., K.T, H.A., H.M., and K.O. received research funds and patent royalties from Konica Minolta, Inc. outside this work. K.M., S.T., S.I., and K.O. received research funds and lecture fees from Chugai Pharmaceutical Co., Ltd. S.I. received research funds from ACT Genomics. H.A., H.M., K.M., and K.O. are journal editors. The other authors have no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by the Institutional Ethics Review Board: #P2017017.
Informed consent: Written informed consent was obtained from all patients.
Registry and registration no. of the study/trial: UMIN000033647.
Animal studies: N/A.
Supporting information
Table S1.
Table S2.
Table S3.
ACKNOWLEDGMENTS
We thank the members of the Clinical Research Promotion Center at the University of Tokyo Hospital for their support in this trial. We thank Kunihiro Nishimura and Takashi Aoki of Xcoo, Inc. for annotation of TOP.
Kage H, Shinozaki‐Ushiku A, Ishigaki K, et al. Clinical utility of Todai OncoPanel in the setting of approved comprehensive cancer genomic profiling tests in Japan. Cancer Sci. 2023;114:1710‐1717. doi: 10.1111/cas.15717
REFERENCES
- 1. National Cancer Center Japan . Cancer Statistics in Japan 2021. 2022. https://ganjoho.jp/public/qa_links/report/statistics/2021_en.html
- 2. Sunami K, Ichikawa H, Kubo T, et al. Feasibility and utility of a panel testing for 114 cancer‐associated genes in a clinical setting: a hospital‐based study. Cancer Sci. 2019;110:1480‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takeda M, Takahama T, Sakai K, et al. Clinical application of the FoundationOne CDx assay to therapeutic decision‐making for patients with advanced solid tumors. Oncologist. 2021;26:e588‐e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sunami K, Naito Y, Aimono E, et al. The initial assessment of expert panel performance in core hospitals for cancer genomic medicine in Japan. Int J Clin Oncol. 2021;26:443‐449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Komine KSK, Naito Y, Amano T, et al. Chronological Improvement in Precision Oncology Implementation in Japan. ESMO Congress 2021; 2021:S583‐S620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Center for Cancer Genomics and Advanced Therapeutics . Cancer genomic medicine and comprehensive genomic profiling. 2022. https://for‐patients.c‐cat.ncc.go.jp/library/statistics/
- 7. Kohsaka S, Tatsuno K, Ueno T, et al. Comprehensive assay for the molecular profiling of cancer by target enrichment from formalin‐fixed paraffin‐embedded specimens. Cancer Sci. 2019;110:1464‐1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodon J, Soria JC, Berger R, et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med. 2019;25:751‐758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuriyama S, Yaegashi N, Nagami F, et al. The Tohoku Medical Megabank Project: design and Mission. J Epidemiol. 2016;26:493‐511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Genomes Project Consortium , Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naito Y, Aburatani H, Amano T, et al. Clinical practice guidance for next‐generation sequencing in cancer diagnosis and treatment (edition 2.1). Int J Clin Oncol. 2021;26:233‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kyoto University School of Public Health Genetic Councelor Course . Comprehensive tumor genomic profiling: Materials for review of secondary findings, Ver 1.0. 2021. http://sph.med.kyoto‐u.ac.jp/gccrc/pdf/k101E_kentousiryo_v1.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Table S2.
Table S3.


