Abstract
Emerging evidence has suggested that circular RNAs (circRNAs) have vital functions during the initiation and progression of various diseases. However, circRNA potential mechanisms in colorectal cancer (CRC) are largely unknown. Here, we sought to investigate the role and underlying regulatory mechanism of circ0104103 in CRC. circ0104103 was validated by quantitative RT‐PCR (qRT‐PCR) and Sanger sequencing. Gain‐ and loss‐of‐function assays in cell lines and mouse xenograft models were utilized to investigate the effects of circ0104103 in CRC. RNA pull‐down assays, RNA immunoprecipitation assays, bioinformatics analyses, RNA FISH, and luciferase reporter assays were used to elucidate the potential mechanism of circ0104103 in CRC. We identified circ0104103, which is strongly downregulated in CRC tissues and cell lines. Functional studies revealed that circ0104103 inhibited CRC cell growth, migration, and invasion both in vitro and in vivo. Mechanistically, circ0104103 binds to HuR, a functional RNA‐binding protein commonly expressed in CRC. HuR binds to the 3′UTR of LACTB mRNA to facilitate stabilization and increase its expression. Moreover, circ0104103 was verified as a competing endogenous RNA (ceRNA) via negative regulation of miR‐373‐5p to increase LACTB expression, resulting in inhibiting the occurrence and progression of CRC. Taken together, our study revealed that circ0104103 acts as a tumor suppressor and may be a novel biomarker and therapeutic target in CRC.
Keywords: CircRNA, colorectal cancer, HuR, LACTB, microRNA
Circ_0104103 interacts with HuR to promote the stability of LACTB mRNA, thereby enhancing its expression. Moreover, Circ_0104103 functions as a sponge of miR‐373‐5p to regulate LACTB expression. Due to its dual targeting effects on LACTB, targeting Circ_0104103 in combination with other therapies may achieve better therapeutic efficiency.
1. INTRODUCTION
Colorectal cancer (CRC) is an aggressive primary intestinal malignancy. Its incidence and mortality rank third among various cancers worldwide. 1 Because there are not enough sensitive biomarkers and effective screening methods, many patients with CRC are diagnosed with advanced disease and consequently suffer from poor 5‐year survival rates. 2 Although improvements in diagnosis and therapy have prolonged the survival of patients with CRC to a certain extent, the prognosis remains unfavorable owing to recurrence, metastasis, and drug resistance. 3 Hence, to provide better treatment strategies, new potential biomarkers and deeper elucidation of the mechanisms of CRC should be explored.
Circular RNAs (circRNAs) are highly conserved noncoding (nc)RNAs with closed‐loop structures that lack 5′ caps or 3′ poly(A) tails. 4 , 5 Emerging research has demonstrated that numerous dysregulated circRNAs in a variety of diseases, including tumors, can be used as biomarkers for monitoring tumor occurrence and development, such as colorectal cancer (CiRS‐7 and circHIPK3), 6 , 7 gastric cancer (circPVT1 and has_circ_0000096), 8 , 9 hepatocellular carcinoma (circMTO1 and circSMARCA5), 10 , 11 and bladder cancer (circMYLK and circITCH), 12 , 13 indicating that circRNAs have great potential as new tumor markers. Although some progress has been made in the study of circRNAs, the functions of circRNAs in CRC remain largely unclear and need to be further investigated.
Beta‐lactamase‐like (LACTB) is evolutionarily related to bacterial penicillin‐binding B‐lactamase proteins, ubiquitously expressed in the mitochondria of skeletal muscle, liver, and heart. 14 , 15 Xue et al. suggested that low LACTB expression promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. 16 Subsequently, the tumor‐suppressive functions of LACTB were also identified in breast cancer. 17 Moreover, our previous findings have demonstrated that LACTB is a novel epigenetically silenced tumor suppressor in CRC. 18
In this study, we characterized one novel circRNA derived from exons 3 to 5 of the LACTB gene (hsa_circ0104103). Furthermore, we identified that circ0104103 was an antioncogene that was downregulated in CRC tissues and cell lines. Mechanistically, circ0104103 interacts with human antigen R (HuR) to improve the mRNA stability of LACTB. Moreover, circ0104103 could sponge endogenous miR‐373‐5p to increase the expression of LACTB. Our findings demonstrated that circ0104103 could be a potential biomarker and therapeutic target for CRC.
2. MATERIALS AND METHODS
2.1. Cell culture
The human normal colorectal epithelial cell line NCM460 and the human CRC cell lines (HCT116, HT29, HCT8, SW480, SW620, DLD1 and CACO2) were obtained from the ATCC. NCM460 and DLD1 cells were cultured in RPMI 1640 medium (Gibco) containing 10% FBS (Gibco) and 1% penicillin–streptomycin (Gibco). SW620 and SW480 cells were grown in Leibovitz's L15 Medium (L15) (Gibco) with 10% FBS and 1% penicillin–streptomycin, and all other cells (HCT116, HT29, HCT8, and Caco2) were cultured in DMEM (Gibco) supplemented with 1% penicillin–streptomycin and 10% FBS. All cells were maintained in an incubator at 37°C with 5% CO2.
2.2. Patient samples
All matched samples (CRC tissue and adjacent nontumor tissue) were obtained from patients who received primary surgery at Nanjing First Hospital, Nanjing Medical University. Patients who received any radiotherapy or chemotherapy before surgery were excluded. The tissues were confirmed by experienced pathologists and immediately snap frozen after surgical removal. Informed consent was obtained from all participants, and the research was approved by the Ethics Committee on Human Research of the Nanjing First Hospital, Nanjing Medical University.
2.3. Vector construction and cell transfection
The small hairpin RNA (shRNA) of circ0104103 and circ0104103‐expressing vectors were designed by Genechem. Cells were grown to 30%–50% confluence in complete medium before being transiently infected with lentiviral vectors following the guidelines. To select the stable cell lines, cells were cultured in medium with 2 μg/mL puromycin. Two weeks later, the cells were harvested. Small interfering RNAs (si‐LACTB/si‐HuR and si‐NC) and microRNA mimics were designed by RiboBio. The plasmids were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer's guidelines. All sequences are listed in Table S1.
2.4. Western blot analysis
Cell proteins were harvested using RIPA lysis buffer (Beyotime Biotechnology) supplemented with PMSF, protein inhibitors, and phosphatase inhibitors (KeyGEN). Then, the concentration of protein was quantified, separated by SDS‐PAGE, and transferred PVDF membranes (Millipore). Then, the membranes were blocked with 5% skim milk and probed with specific antibodies at 4°C overnight. Protein expression was visualized using the bioimaging system ECL Plus (Millipore) after incubation with secondary antibodies. The detailed information of antibodies used in the research were as follows: GAPDH (Proteintech, 60,004‐1‐Ig, 1:2,000), MMP9 (Proteintech, 10,375‐2‐AP, 1:1,000), PCNA (Proteintech, 10,205‐2‐AP, 1:2,000), BCL2 (Proteintech, 12,789‐1‐AP, 1:2000), P53 (Proteintech, 10,442‐1‐AP, 1:2000), LACTB (Abcam, ab131171,1:2,000), HuR (Abcam, ab200342, 1:2000), goat antimouse IgG, peroxidase conjugated, H + L (Biosharp, 1:5,000), and goat antirabbit IgG, peroxidase conjugated, H + L (Biosharp, 1:5,000).
2.5. In vivo experiments
All animal experiments were performed in compliance with a protocol approved by the Animal Care Committee of Nanjing Medical College. In the subcutaneous tumor xenograft experiment, five‐week‐old male BALB/c nude mice were randomly divided into two groups with six mice in each group. Stably infected HCT116 cells or negative control (5 × 106 cells/0.2 mL PBS) were inoculated into the left and right flanks of nude mice, respectively. Xenografts were determined every 5 days with caliper measurements, and tumor volumes were calculated with the following formula: (L × W2)/2. Four weeks later, the mice were killed. The volumes and weights of tumors were measured.
For the metastasis assay, five‐week‐old male BALB/c nude mice were randomly divided into three groups with four mice in each group. Next, 1 × 106 HCT116 cells stably infected with sh‐circ0104103#2, circ0104103 or empty vectors suspended in 100 μL PBS were injected into nude mice via the tail vein. After 5 weeks of injection, all mice were killed, and their lungs were surgically removed and stained by H&E staining.
2.6. Immunohistochemistry
All samples were collected, embedded in paraffin, and cut into 4‐μM slices. For antigen retrieval, the sections were treated with fractionated ethanol and distilled water and 3% H2O2 for 30 min, followed by incubation with 10% goat serum for 30 min to prevent binding of nonspecific antibodies. After washing, sections were incubated with specific antibodies. According to the manufacturer's protocol, after staining with diaminobenzidine, all sections were dehydrated and sealed. All images were visualized and recorded under a microscope (Olympus). The details of the antibodies used in the research are as follows: MMP9 (Proteintech, 10,375‐2‐AP, 1:100), PCNA (Proteintech, 10,205‐2‐AP, 1:200), BCL2 (Proteintech, 12,789‐1‐AP, 1:100), P53 (Proteintech, 10,442‐1‐AP, 1:100), and LACTB (Abcam, ab244455,1:200).
2.7. RNA pull‐down assay
RNA pull‐down assays were performed using a Magnetic RNA Protein Pull‐Down Kit (Thermo Fisher Scientific) following the manufacturer's instructions. Biotin‐labeled circ0104103 RNA probes were designed by RiboBio as follows: CCTAAACCTTGCTCTATTGT. Briefly, the biotin‐labeled circ0140103 RNA probes were incubated with prewashed streptavidin‐agarose beads and incubated with cell lysates at 4°C overnight. Eluted proteins were analyzed by western blot.
2.8. RNA immunoprecipitation assay
The EZ Magna RNA immunoprecipitation Kit (Millipore) was used according to the manual. In brief, magnetic beads were preincubated with anti‐HuR antibody (ab200342). HCT‐116 cells were lysed in RNA immunoprecipitation (RIP) lysis buffer and incubated with beads at 4°C overnight. Next, RNA was purified, and the relative expression of circ0104103 and LACTB was measured by qRT‐PCR and normalized to the relative expression of circ‐0104103 and LACTB in input samples.
2.9. Dual‐luciferase reporter assay
A dual‐luciferase reporter assay was used to evaluate the direct binding between miR‐373‐5p and circ0104103 and LACTB. Briefly, the circ0104103 and LACTB 3′UTR fragments containing a wild‐type (Wt) or mutant (Mut) binding site of miR‐373‐5p were amplified by qRT‐PCR and inserted into the PmirGLO vector (GeneCreat). Then, 293 T cells were cotransfected with sh‐miR‐373‐5p/sh‐NC with Wt or Mut circ0104103 and LACTB firefly luciferase reporter vectors. After transfection for 48 h, the results were measured using a Dual Luciferase Assay System (Promega).
2.10. Statistical analysis
All statistical analyses were carried out using SPSS 19.0 software (IBM), and images were acquired with GraphPad Prism 7 software. Differences between the groups were assessed by applying Student's paired or unpaired t‐test or one‐way analysis of variance (ANOVA), respectively. p < 0.05 indicated that the difference was statistically significant.
A complete description of the methods, including RNA extraction and quantitative real‐time PCR, colony formation assay, EdU assay, wound healing assay, Transwell invasion assays, in situ hybridization, RNA FISH, and subcellular fractionation, are available in the Appendix S1.
3. RESULTS
3.1. Expression and characterization of circ0104103 in colorectal cancer
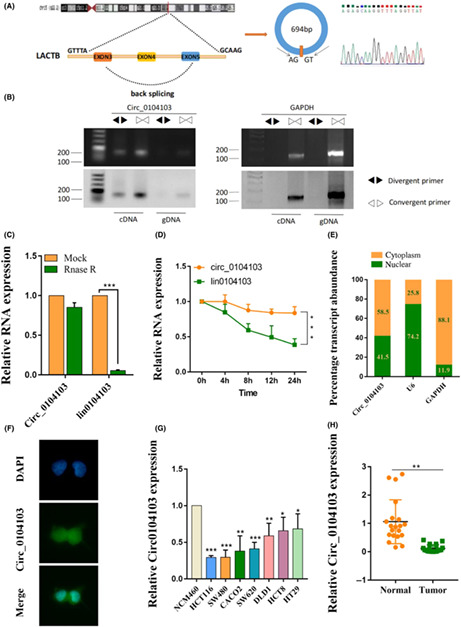
circ0104103 (hsa_circ0104103) is derived from the beta‐lactamase‐like (LACTB) gene exons 3 to 5, whose spliced mature sequence length is 694 bp (Figure 1A). To further confirm the characterization of circ0104103, we examined the head‐to‐tail splicing of circ0104103 by qRT‐PCR with convergent primers for linear 0104103 and divergent primers for circ0104103. The results showed that circ0104103 was only amplified in cDNA but not genomic DNA (gDNA) (Figure 1B). Next, we evaluated the stability and localization of circ0104103. RNase R and actinomycin D treatment assays revealed that circ0104103 was more stable than linear 0104103 (Figure 1C,D). Moreover, subcellular fractionation assays and FISH assays showed that circ0104103 was localized in both the nucleus and cytoplasm (Figure 1E,F). Next, we examined the expression level of circ0104103 in CRC cell lines, and the results showed that circ0104103 was significantly downregulated in CRC cell lines (HCT116, SW480, CACO2, SW620, DLD1, HCT8, and HT29) compared with normal colon mucosal epithelial cells (NCM460) (Figure 1G). Similar results were also observed in CRC compared with matched normal tissues (Figure 1H). ISH assays also revealed that circ0104103 expression levels in CRC tissues were lower than the circ0104103 expression levels in adjacent normal tissues (Figure S1). Finally, we analyzed the relationship between circ0104103 expression and the clinicopathologic features of CRC. We divided the enrolled patients into two groups based on circ0104103 expression. A χ2‐test was used to analyze the distribution differences of different clinical features between the two groups. As shown in Table S3, circ0104103 expression was correlated with tumor invasion depth and TNM stage.
FIGURE 1.
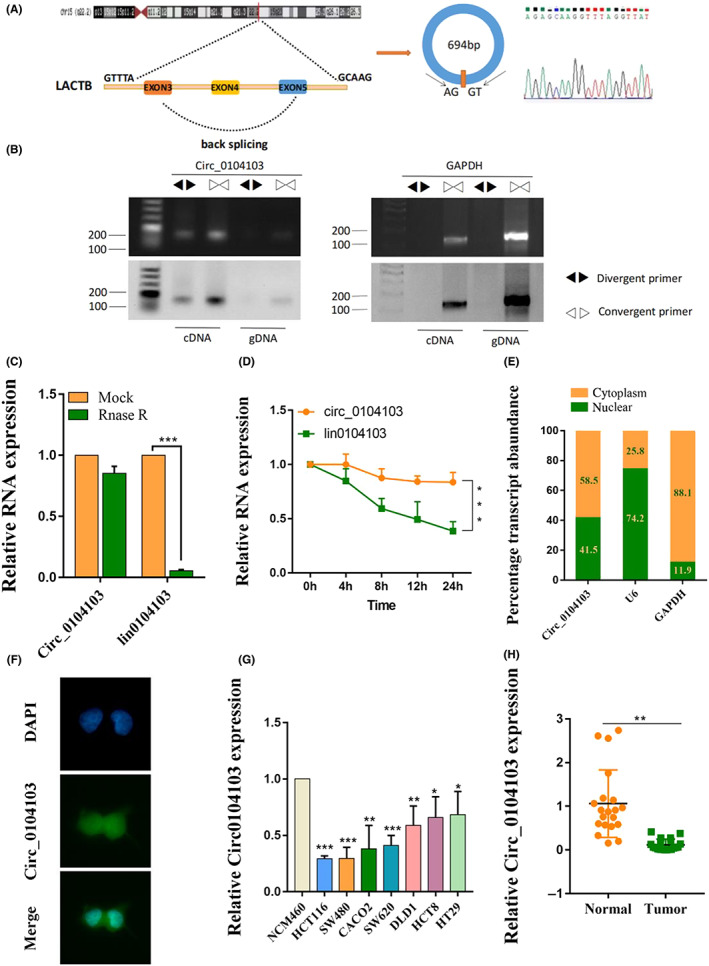
Expression and characterization of Circ0104103 in colorectal cancer (CRC). (A) Schematic illustration demonstrating the structure of Circ_0104013 and Alu elements in the flanking sequence. (B) RT‐PCR identified the presence of Circ0104103 in the HCT116 cells. Divergent primers amplified Circ0104103 in cDNA but not in genomic DNA. GAPDH was used as the negative control. (C, D) Quantitative RT‐PCR (qRT‐PCR) detected the expression of Circ0104103 and lin0104103 in CRC cells with or without RNase R or actinomycin D treatment. (E) Relative Circ0104103 expression levels in nuclear and cytosolic fractions of HCT116 cells. Nuclear controls: U6, cytosolic controls: GAPDH. (F) FISH assay identifying the subcellular location of Circ0104103 in the HCT116 cells. (G, H) qRT‐PCR for the expression of Circ0104103 in CRC cell lines and tissues. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.2. circ0104103 inhibits colorectal cancer cell growth in vitro
To explore the functional roles of circ0104103 in CRC, we established lentiviral‐mediated stable circ0104103 expression in HCT116 and HCT8 cells. In addition, the circ0104103‐silencing vector was also constructed to knock down circ0104103 in cell lines. qRT‐PCR assays were carried out to verify the lentiviral transduction efficiency (Figure S2). By performing colony formation and EdU assays, we found that interference with circ0104103 promoted HCT116 and HCT8 cell growth, whereas overexpression of circ0104103 inhibited CRC cell growth (Figure 2A,B). Furthermore, wound healing and Transwell assays revealed that silencing circ0104103 significantly increased the migration and invasion abilities of CRC cells. In contrast, circ0104103 overexpression inhibited migration and invasion abilities (Figure 2C,D). Western blot assay results further demonstrated that cell proliferation‐related protein PCNA, metastasis‐related protein MMP9, and apoptosis‐related protein BCL2 were all decreased in circ0104103 overexpressed cells, but on the contrary in sh‐circ0104103#2 cells. Moreover, the overexpression of circ0104103 led to increased expression of the cycle‐related protein P53, whereas silencing circ0104103 decreased its expression (Figure 2E). Altogether, the above results demonstrated that circ0104103 inhibits CRC cell growth in vitro.
FIGURE 2.
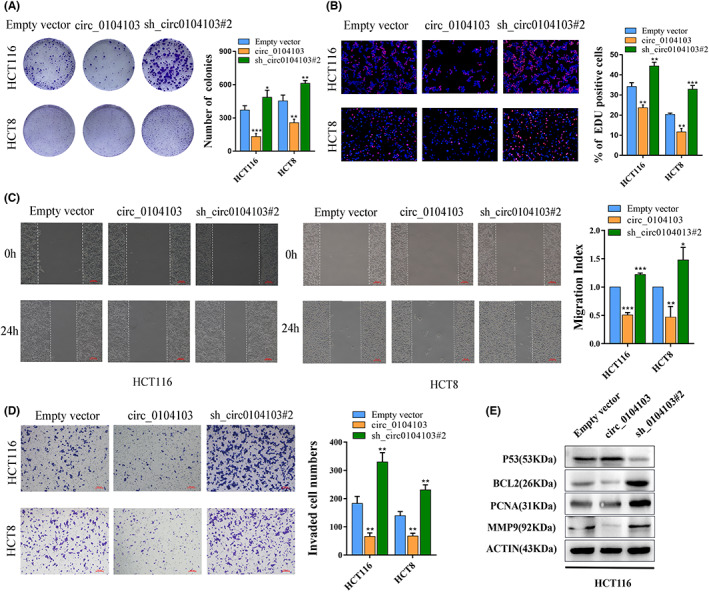
Circ0104103 inhibits colorectal cancer (CRC) cell growth and metastasis in vitro. (A) HCT116 and HCT8 cells infected with circ0104103 or sh_circ0104103#2 were seeded into six‐well plates. The number of colonies was counted on the 14th day after seeding. (B) EdU assays were used to determine the cell proliferation ability of infected cells. (C) Representative images of wound healing assays performed using HCT116 cells and HCT8 cells after Circ0104103 was silenced or overexpressed. (D)Transwell assays were used to determine the invasion abilities of infected cells. (E) Immunoblot analysis of MMP9, PCNA, BCL2, and P53 protein expression in circ0104103‐overexpressing HCT116 cells and circ0104103 silencing HCT116 cells. Scale bar = 100 μm. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.3. circ0104103 inhibits colorectal cancer cell growth and metastasis in vivo
To further confirm whether circ0104103 affects CRC growth and metastasis in vivo, we constructed tumor xenograft nude mouse models as described in the methods. Four weeks later, the mice were killed. As shown in Figure 3A, the sizes and weights of the tumors from the circ0104103 group were significantly smaller than the sizes and weights of the tumors of the empty vector group, whereas intratumoral injection of sh‐circ0104103#2 significantly increased the sizes and weights of the tumors. For lung metastasis, more and fewer metastatic nodules on the lung surfaces were observed in the circ0104103 knockdown and overexpression groups, respectively, than in the empty vector groups (Figure 3B). Moreover, we performed immunohistochemical staining, and the results revealed that the expression of MMP9, P53, PCNA, and BCL2 in subcutaneous tumors was consistent with previous western blot results (Figure 3C). In conclusion, the above results are consistent with our in vitro findings and indicate that circ0104103 inhibits tumor growth and metastasis in vivo.
FIGURE 3.
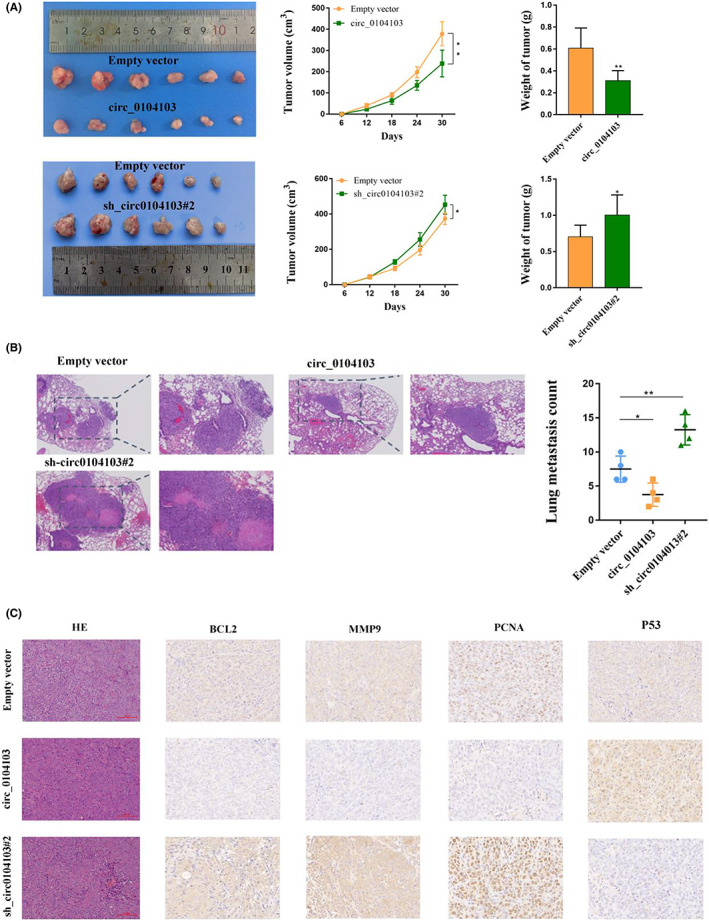
Circ0104103 inhibits colorectal cancer (CRC) cell growth in vivo. (A) Representative image of tumors formed in nude mice from empty vector, sh‐ circ0104103#2 vector, and circ0104103 overexpression vector groups and the tumor volume growth curves and weight of different groups. (B) Representative H&E staining and the number of lung metastatic nodules in nude mice in the three groups. (C) Representative images for H&E‐staining, BCL2, MMP9, P53, and PCNA immunostaining of tumor samples from the different groups. *p < 0.05, **p < 0.01, and ***p < 0.001.
In addition, we also observed that compared with the empty vector groups and circ0104103 groups, more blood vessels could be seen in the H&E staining images of subcutaneous tumors in the sh‐circ0104103#2 groups (Figure S3A). Then, we performed immunohistochemistry (IHC) assays to label blood vessels by CD34, and the results were also consistent with H&E staining (Figure S3B). The results suggest that circ0104103 may inhibit the angiogenesis of CRC cells.
3.4. circ0104103 enhances beta‐lactamase‐like expression in colorectal cancer
circ0104103 is generated from exons 3–5 of the LACTB gene. Our previous findings demonstrated that LACTB can function as a tumor suppressor in CRC, indicating that LACTB has potential as a therapeutic target for CRC. Subsequently, we performed qRT‐PCR and western blot assays, and the results showed that interference with circ01040103 significantly decreased the expression of LACTB in HCT116 and HCT8 cells, whereas overexpression of circ01040103 increased it (Figure 4A,B). IHC assays indicated that LACTB expression was strongly increased in xenograft tumor tissues formed by circ0104103‐overexpressing cells but not in sh‐circ0104103#2 cells. Correlation analysis revealed that LACTB expression is positively associated with circ0104103 in 30 colorectal cancer tissues (Figure S4A). IHC assays revealed that LACTB expression levels in CRC tissues were lower than the LACTB expression levels in adjacent normal tissues (Figure 4C).
FIGURE 4.
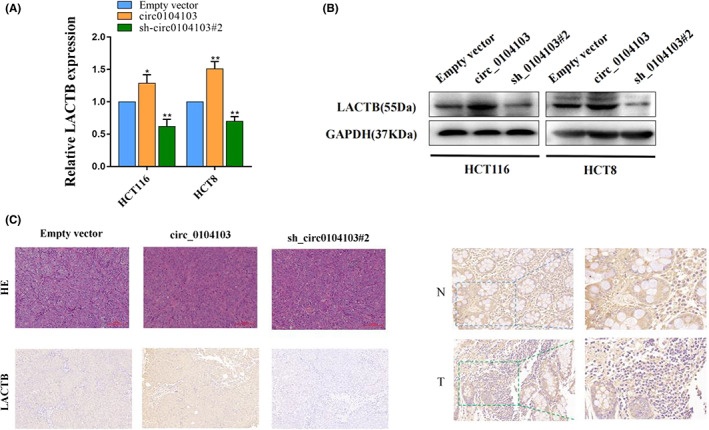
Circ0104103 enhances beta‐lactamase‐like (LACTB) expression in colorectal cancer (CRC). (A, B) Quantitative RT‐PCR (qRT‐PCR) and western blot assays detected the expression of LACTB in HCT116 and HCT8 cell lines with circ0104103 knockdown or overexpression. (C) LACTB expression was detected by immunohistochemistry (IHC) xenograft tumor tissues. LACTB protein levels in CRC tissues and corresponding normal tissues was detected by IHC. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.5. circ0104103 facilitates the stability of beta‐lactamase‐like mRNA by recruiting HuR
To investigate the potential mechanisms by which circ0104103 enhances LACTB expression in CRC, we then performed RNA pulldown assays using a biotin‐labeled circ0104103 probe, and the results verified that circ0104103 could interact with HuR from HCT116 cell extracts (Figure 5A). In addition, RIP assay results revealed that circ0104103 and LACTB were significantly enriched in complexes precipitated by the antiHuR antibody, suggesting that the HuR protein bound to circ0104103 and LACTB directly in CRC (Figure 5B). Next, we designed three small interfering RNAs (siRNAs) targeting HuR (Figure 5C). qRT‐PCR and western blot assay results showed that interference with HuR significantly decreased the expression of LACTB in HCT116 cells (Figure 5D,F). In addition, HuR knockdown reversed the upregulation of LACTB expression caused by circ0104103 overexpression (Figure 5E,G). Moreover, HuR knockdown significantly reduced the mRNA stability of LACTB, whereas overexpressed circ0104103 increased the remaining LACTB mRNA (Figure S4B). Together, we conclude that circ0104103 interacts with HuR to promote the stability of LACTB mRNA, thereby enhancing its expression.
FIGURE 5.
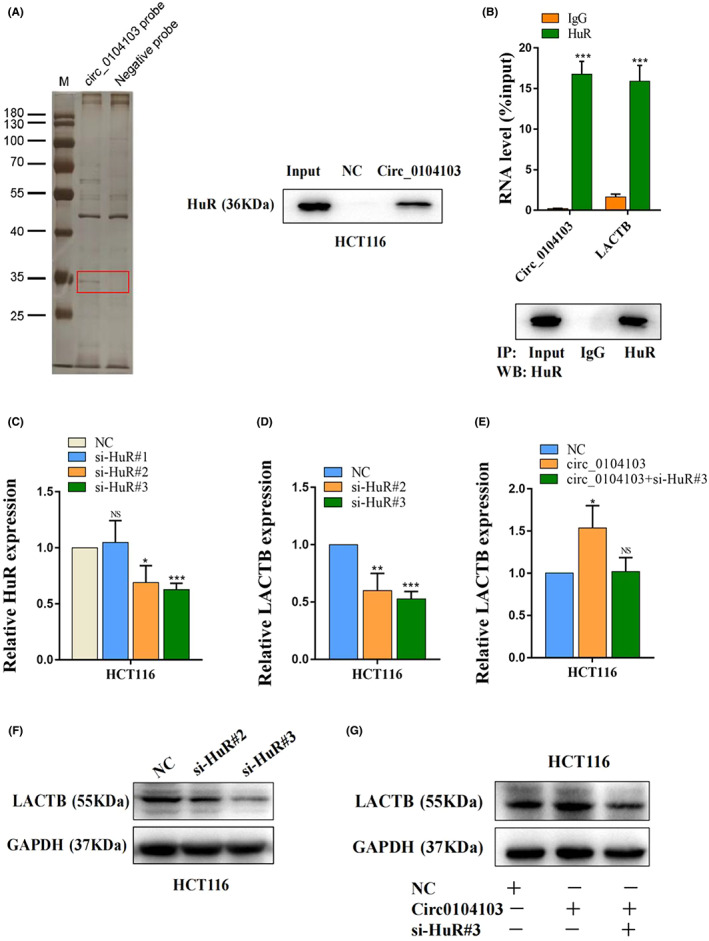
Circ0104103 facilitates stability of beta‐lactamase‐like (LACTB) mRNA by recruiting HuR. (A) RNA pull‐down assays were used to examine the interaction of Circ0104103 and HuR in HCT‐116 cells. (B) RNA immunoprecipitation with an anti‐HuR antibody was used to assess endogenous HuR binding to RNA in HCT‐116 cells; IgG was used as the control. Circ0104103 and LACTB levels were determined by quantitative RT‐PCR (qRT‐PCR) and presented as fold enrichment in HuR relative to input. RIP efficiency of HuR protein was detected by western blot. (C) HuR were quantified by qRT‐PCR after transfection of HuR siRNAs in HCT116 cells. (D–F) qRT‐PCR and western blotting assays detected the expression of LACTB in HCT116 cells with HuR knockdown. (E–G) LACTB expression was detected by qRT‐PCR and western blotting in HCT116 cells with indicated treatment. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.6. circ0104103 upregulates beta‐lactamase‐like expression by sponging miR‐373‐5p
Because circRNAs expressed in the cytoplasm act mainly as miRNA sponges, we further investigated whether circ0104103 can sponge miRNAs to regulate LACTB. Bioinformatics analysis predicted that miR‐373‐5p can interact with circ0104103 (Figure 6A). Moreover, miR‐373‐5p was increased with circ0104103 knockdown, while its level was decreased with circ0104103 overexpression in both HCT116 and HCT8 cells (Figure 6B). Then, we carried out a dual‐luciferase reporter assay to further explore whether circ0104103 and LACTB could interact with miR‐373‐5p directly. A dual‐luciferase reporter assay indicated that cotransfection of the wild‐type circ0104103 luciferase vector (WT‐Circ0104103) and the wild‐type LACTB luciferase vector (WT‐LACTB) with the miR‐373‐5p mimic, but not the mutant circ0104103 vector (MUT‐circ0104103) or the mutant LACTB vector (MUT‐LACTB), significantly decreased the luciferase activity (Figure 6C). Finally, correlation analysis revealed that miR‐373‐5p expression is negatively associated with circ0104103 and LACTB in 30 colorectal cancer tissues (Figure S5). The above results indicated that circ0104103 and LACTB directly bind to miR‐373‐5p.
FIGURE 6.
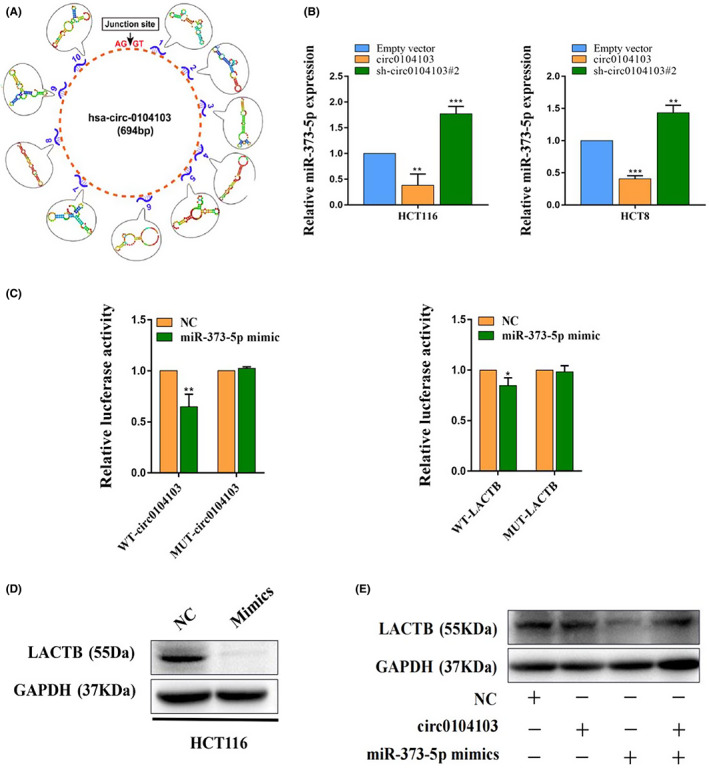
Circ0104103 upregulates beta‐lactamase‐like (LACTB) expression by sponging miR‐373‐5p. (A) Bioinformatics analysis predicted miR‐373‐5p can interact with Circ0104103. (B) MiR‐373‐5p expression was detected by quantitative RT‐PCR (qRT‐PCR) in HCT116 and HCT8 cells after circ0104103 silenced or overexpressed. (C) Dual‐luciferase reporter assays were conducted with wild‐type and mutant‐type (putative binding sites for miR‐373‐5p) luciferase report vectors. (D, E) Beta‐lactamase‐like (LACTB) expression was detected by western blotting in HCT116 cells with indicated treatment. *p < 0.05, **p < 0.01, and ***p < 0.001.
In addition, we found that miR‐373‐5p overexpression significantly reduced LACTB protein levels in CRC cells (Figure 6D). Western blot assays also showed that overexpression of circ0104103 significantly upregulated LACTB protein levels, whereas cotransfection of the circ0104103 overexpression vector and miR‐373‐5p mimic eliminated this effect (Figure 6E). Overall, the above results suggested that circ0104103 might function as a sponge of miR‐373‐5p to regulate LACTB expression.
To identify whether the antitumour effects of circ0104103 were dependent on sponging miR‐373‐5p, several rescue assays were carried out. The results indicated that circ0104103 overexpression could inhibit the invasion, migration, and proliferation of CRC cells. However, cotransfection of circ0104103 and the mimic of miR‐373‐5p may counteract this effect (Figure 7A–D).
FIGURE 7.
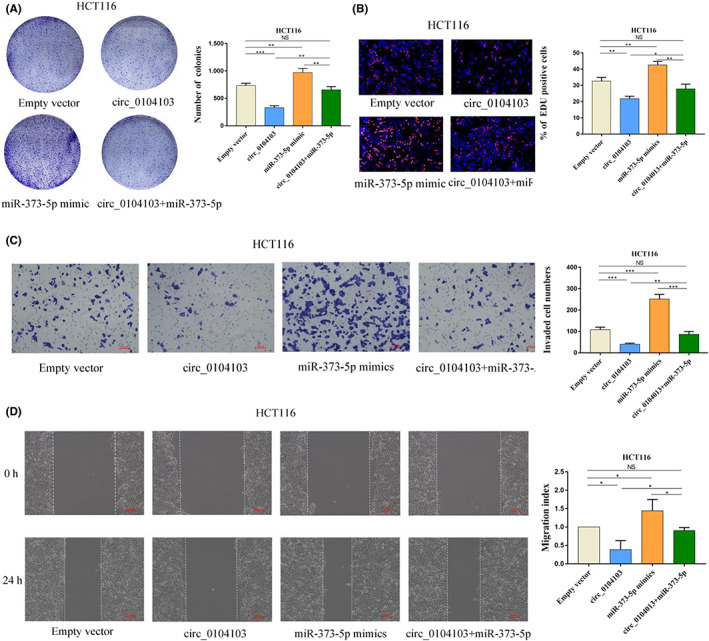
Circ0104103 upregulates beta‐lactamase‐like (LACTB) expression by sponging miR‐373‐5p. (A) Colony formation assays of Circ0104103 overexpressed colorectal cancer (CRC) cells with transfection of miR‐373‐5p mimics. (B) Representative images of EdU assays performed using HCT116 cells with indicated treatment. (C) Representative images of Transwell invasion assays performed using CRC cells with indicated treatment. (D) Representative images of wound healing assays performed using HCT116 cells with indicated treatment. Scale bar = 100 μm. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.7. Tumor‐suppressing functions of circ0104103 are dependent on beta‐lactamase‐like
To determine whether the tumor‐suppressing functions of circ0104103 are dependent on LACTB in CRC, we designed one small interfering RNA (siRNA) targeting LACTB to perform several rescue experiments in circ0104103‐overexpressing cells. As expected, the stimulative effect of circ0104103 overexpression on the cell proliferation, colony formation, migration, and invasion abilities of HCT116 cells was reversed in the presence of si‐LACTB (Figure 8A–D). In addition, western blotting results demonstrated that LACTB knockdown reversed the downregulation of the protein levels of MMP9, BCL2, and PCNA and the upregulation of the protein level of P53 caused by circ0104103 overexpression (Figure 8E). Overall, the data suggested that circ0104103 exerted its antitumour effects by regulating LACTB expression (Figure 8F).
FIGURE 8.
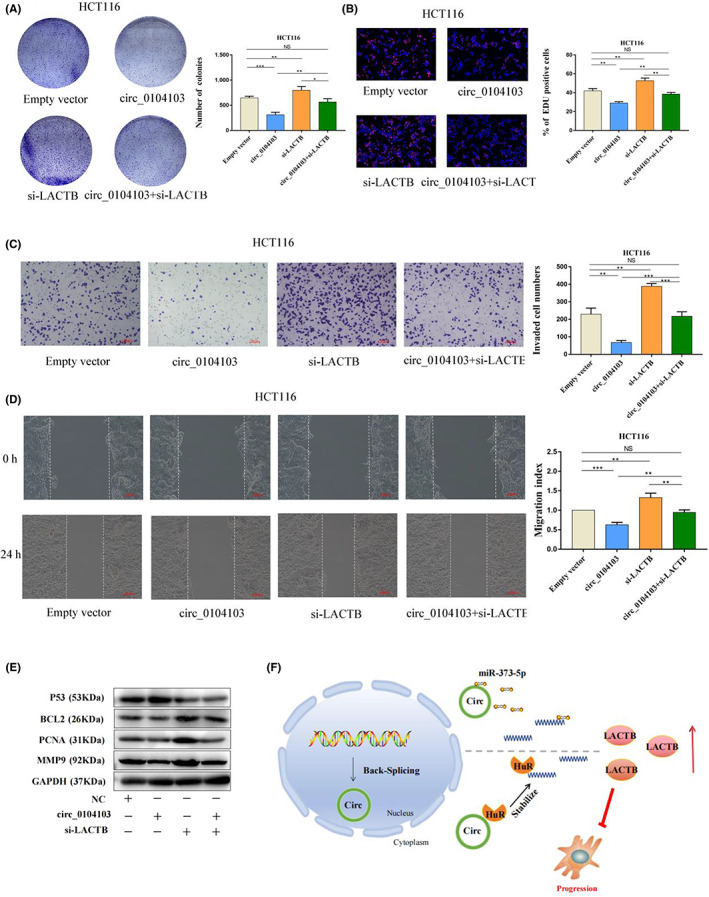
Tumor‐suppressing functions of Circ0104103 are dependent on beta‐lactamase‐like (LACTB). (A) Colony formation assays demonstrated that overexpression of circ0104103 inhibited cancer cell growth. LACTB knockdown could abolish growth inhibition caused by circ0104103. (B) EdU assays showed that LACTB knockdown abolished the decreased proliferation rates of colorectal cancer (CRC) cells caused by circ0104103. (C, D) Transwell assays and wound healing assays demonstrated that LACTB knockdown abolished the decreased abilities of migration and invasion caused by circ0104103. (E) Immunoblot analysis of MMP9, PCNA, BCL2, and P53 protein expression in HCT116 cells with indicated treatment. (F) Schematic of the proposed mechanism of circ0104103 in CRC. Scale bar = 100 μm. *p < 0.05, **p < 0.01, and ***p < 0.001.
4. DISCUSSION
Recently, the functions of circRNAs have attracted much attention, and an increasing number of studies have suggested that circRNAs have a crucial role in cancer development and progression. Due to their abundant expression and high stability, circRNAs are suitable as biomarkers for disease diagnosis and prognosis. 19 , 20 In this study, we identified circ0104103 as a stable circular transcript without a poly‐A tail. In addition, we found that circ0104103 was expressed at low levels in CRC tissues and cell lines. Functional experiments showed that circ0104103 significantly inhibited the growth and metastasis of CRC cells in vivo and in vitro. Mechanistically, we found that circ0104103 was localized in both the nucleus and cytoplasm. circ0104103 interacts with HuR to promote the stability of LACTB mRNA, thereby enhancing its expression. Moreover, circ0104103 functions as a sponge of miR‐373‐5p to regulate LACTB expression. Due to its dual targeting effects on LACTB, targeting circ0104103 in combination with other therapies may achieve better therapeutic efficiency.
As an evolutionarily conserved RNA‐binding protein, HuR is widely expressed in various tissues of the body. 21 Studies have revealed that HuR is closely related to the occurrence and development of inflammatory diseases and tumors, such as cell division, cell senescence, immune cell activation, and other life activities. 22 , 23 HuR regulates its stability and translation efficiency by specifically binding to target mRNA, thereby affecting the expression level of target genes in cells. 24 , 25 In the present study, we proved the interaction between circ0104103/LACTB and HuR by performing RNA pull‐down and RIP assays. Rescue experiments demonstrated that HuR facilitates its stability by binding to LACTB mRNA and thus regulating its protein level expression in CRC cells.
Recently, an increasing number of studies have shown that circRNAs function as competitive inhibitors of microRNAs (miRNAs) and then regulate their expression to participate in various cancer biological processes. 26 miRNAs are a large class of short (~22 nt) noncoding RNAs that are closely associated with cancer progression. circRNAs can adjust miRNA activities by competing for miRNA‐binding sites. For instance, circRNA‐MYLK bound to miR‐29a to promote bladder carcinoma metastasis by inducing EMT. 12 circRNA AKT3 upregulates PIK3R1 by sponging miR‐198 to enhance the resistance of gastric cancer to cisplatin. 27 Our previous study also revealed that circHIPK3 promotes CRC occurrence and progression by sponging miR‐7. 6 In our study, we confirmed that circ0104103 is partly located in the cytoplasm, which provides theoretical support that circ0104103 sponges miR‐373‐5p in the cytoplasm. Meanwhile, we validated the targeting relationship between circ0104103/LACTB and miR‐373‐5p via a dual‐luciferase reporter assay, proving that circ0104103 could regulate LACTB expression by binding to miR‐373‐5p in CRC cells. In addition, functional assays verified that miR‐373‐5p could reverse the inhibitory effect of circ0104103 on CRC cell proliferation, migration, and invasion.
To determine whether the antitumour effect of circ0104103 depends on sponging LACTB, some rescue experiments were carried out. The results showed that cotransfection of circ0104103‐expressing vectors and si‐LACTB may counteract the inhibition of invasion, migration, and proliferation of CRC cells caused by circ0104103 overexpression. These results confirmed that circ0104103 exerted its antitumour effects by interacting with HuR and regulating the miR‐373‐5P/LACTB axis.
In summary, our study is the first to identify circ0104103 as a novel tumor suppressor in CRC. circ0104103 exerts its effects by upregulating LACTB expression through interacting with HuR and acting as a sponge for miR‐373‐5p. Our results suggest a strategy for targeting circ0104103 as a potential biomarker and a therapeutic target in CRC.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ETHICS STATEMENT
The research was approved by the Ethics Committee on Human Research of the Nanjing First Hospital, Nanjing Medical University.
Informed Consent. N/A.
Registry and the Registration No. of the study/trial. N/A.
All animal experiments were performed in compliance with a protocol approved by the Animal Care Committee of Nanjing Medical College.
Supporting information
Appendix S1.
Figures S1–S5.
Tables S1–S3.
ACKNOWLEDGMENTS
This project was supported by grants from The National Nature Science Foundation of China (No. 81972806), Jiangsu Provincial Key Research and Development Plan (BE2019614), Elderly Health Research Project of Jiangsu Province (Grant No. LR2021017), and the Specialized Cohort Research Project of Nanjing Medical University (NMUC2021013A).
Tan P, Sun H, Xu M, et al. Circular RNA circ0104103 inhibits colorectal cancer progression through interactions with HuR and miR‐373‐5p. Cancer Sci. 2023;114:1396‐1409. doi: 10.1111/cas.15695
Pei Tan, Huiling Sun and Mu Xu contributed equally to this work.
Contributor Information
Shukui Wang, Email: sk_wang@njmu.edu.cn.
Yuqin Pan, Email: panyuqin01@163.com.
DATA AVAILABILITY STATEMENT
Data are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 2. Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nishihara R, Wu K, Lochhead P, et al. Long‐term colorectal‐cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369(12):1095‐1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rybak‐Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870‐885. [DOI] [PubMed] [Google Scholar]
- 5. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR‐7. Cell Death Dis. 2018;9(4):417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Weng W, Wei Q, Toden S, et al. Circular RNA ciRS‐7‐a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918‐3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208‐219. [DOI] [PubMed] [Google Scholar]
- 9. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214‐1227. [DOI] [PubMed] [Google Scholar]
- 11. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA‐9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151‐1164. [DOI] [PubMed] [Google Scholar]
- 12. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305‐317. [DOI] [PubMed] [Google Scholar]
- 13. Yang C, Yuan W, Yang X, et al. Circular RNA circ‐ITCH inhibits bladder cancer progression by sponging miR‐17/miR‐224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peitsaro N, Polianskyte Z, Tuimala J, et al. Evolution of a family of metazoan active‐site‐serine enzymes from penicillin‐binding proteins: a novel facet of the bacterial legacy. BMC Evol Biol. 2008;8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith TS, Southan C, Ellington K, Campbell D, Tew DG, Debouck C. Identification, genomic organization, and mRNA expression of LACTB, encoding a serine beta‐lactamase‐like protein with an amino‐terminal transmembrane domain. Genomics. 2001;78(1–2):12‐14. [DOI] [PubMed] [Google Scholar]
- 16. Xue C, He Y, Zhu W, et al. Low expression of LACTB promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. Am J Transl Res. 2018;10(12):4152‐4162. [PMC free article] [PubMed] [Google Scholar]
- 17. Keckesova Z, Donaher JL, De Cock J, et al. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature. 2017;543(7647):681‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeng K, Chen X, Hu X, et al. LACTB, a novel epigenetic silenced tumor suppressor, inhibits colorectal cancer progression by attenuating MDM2‐mediated p53 ubiquitination and degradation. Oncogene. 2018;37(41):5534‐5551. [DOI] [PubMed] [Google Scholar]
- 19. López‐Jiménez E, Rojas AM, Andrés‐León E. RNA sequencing and prediction tools for circular RNAs analysis. Adv Exp Med Biol. 2018;1087:17‐33. [DOI] [PubMed] [Google Scholar]
- 20. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428‐442. [DOI] [PubMed] [Google Scholar]
- 21. Giammanco A, Blanc V, Montenegro G, et al. Intestinal epithelial HuR modulates distinct pathways of proliferation and apoptosis and attenuates small intestinal and colonic tumor development. Cancer Res. 2014;74(18):5322‐5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mubaid S, Ma JF, Omer A, et al. HuR counteracts miR‐330 to promote STAT3 translation during inflammation‐induced muscle wasting. Proc Natl Acad Sci USA. 2019;116(35):17261‐17270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ostareck DH, Ostareck‐Lederer A. RNA‐binding proteins in the control of LPS‐induced macrophage response. Front Genet. 2019;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H, Zhao X, Yun W, Chen LH, Li ST. Effect of inhibiting p38 on HuR involving in β‐AChR post‐transcriptional mechanisms in denervated skeletal muscle. Cell Mol Neurobiol. 2019;39(7):1029‐1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. García‐Mauriño SM, Rivero‐Rodríguez F, Velázquez‐Cruz A, et al. RNA binding protein regulation and cross‐talk in the control of AU‐rich mRNA fate. Front Mol Biosci. 2017;4:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42‐51. [DOI] [PubMed] [Google Scholar]
- 27. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR‐198 suppression. Mol Cancer. 2019;18(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1.
Figures S1–S5.
Tables S1–S3.
Data Availability Statement
Data are available from the corresponding author upon reasonable request.


