Abstract
Inflammation is observed in many tumors, which affects metastasis, infiltration, and immune escape and causes poor differentiation of the cancer cells. However, the molecular basis underlying the relationship between inflammation and poor differentiation in tumors has not been identified. In this study, we demonstrate that angiopoietin‐like protein‐8 (ANGPTL8), which is induced by stress stimuli such as inflammation, is involved in the maintenance of the undifferentiated state of clear cell renal cell carcinoma (ccRCC) cells. ANGPTL8 is also involved in the production of chemokines that attract immune suppressor cells to the tumor microenvironment. ANGPTL8 sustains the continuous production of chemokines by activating the NF‐κB signaling pathway and maintains the undifferentiated state of ccRCC cells. Finally, ANGPTL8 is induced by STAT3 signaling, which is activated by immune cells in the tumor microenvironment. These results support a role for ANGPTL8 in determining the properties of ccRCC by hampering tumor cell differentiation and establishing the tumor microenvironment.
Keywords: angiopoietin‐like protein‐8, inflammation, renal cell carcinoma, tumor microenvironment, undifferentiation
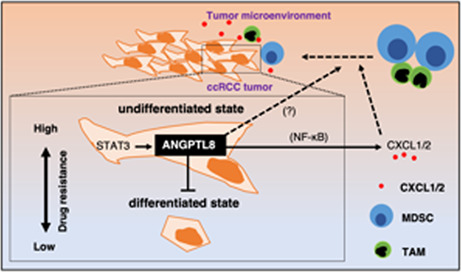
ANGPTL8 maintains the undifferentiated state in renal carcinoma and upregulates CXCL1 and CXCL2 to attract immune cells, which activates STAT3 signaling for ANGPTL8 induction in the tumor microenvironment.
Abbreviations
- ANGPTL8
angiopoietin‐like protein‐8
- ccRCC
clear cell renal cell carcinoma
- FBS
fetal bovine serum
- GO
gene ontology
- LTL
lotus tetragonolobus lectin
- MDSCs
myeloid‐derived suppressor cells
- RCC
renal cell carcinoma
- TAMs
tumor‐associated macrophages
- TCGA
The Cancer Genome Atlas
1. INTRODUCTION
Renal cell carcinoma (RCC) is the most common type of cancer, and it was estimated that there were more than 431,288 new cases of kidney cancer and 179,368 deaths worldwide in 2020. 1 Clear cell RCC (ccRCC) is a common histology accounting for approximately 70% of RCC cases and is associated with chronic tissue inflammation. 2 , 3 , 4 , 5 Systemic inflammation is frequently observed in the advanced stage of RCC. 6 , 7 , 8 Recent clinical studies have demonstrated that the expression of inflammation‐related genes and neutrophilic infiltration in tumor tissue are prognostic markers for metastatic RCC. 9 , 10 In addition, 10%–15% of RCCs are considered undifferentiated, more aggressive forms that exhibit increased inflammatory signals. 11 , 12 Inflammation is believed to prevent epithelial cell differentiation 13 , 14 , 15 ; however, the factors that regulate inflammation and RCC differentiation have not been identified.
Members of the angiopoietin‐like protein (ANGPTL) family contribute to angiogenesis and the maintenance of hematopoietic stem cells. 16 , 17 , 18 Previous studies have shown that ANGPTL 2, 3, 4, 6, and 7 are pro‐inflammatory factors that regulate tumor progression. 16 , 17 For example, ANGPTL2 is a factor that controls regeneration, differentiation, and the stem cell niche formation in various organs. 19 , 20 , 21 , 22 ANGPTL8 is a relatively small protein with a molecular weight of 22 kDa. ANGPTL8 interacts with ANGPTL3 and increases plasma levels of triglycerides. 23 Xu et al. (2021) reported that the expression of ANGPTL8 was associated with a poor prognosis in patients with ccRCC based on the analysis of data from The Cancer Genome Atlas (TCGA). 24 However, the role of ANGPTL8 in ccRCC has not been determined.
Inflammation induced by tumor cells establishes a tumor microenvironment consisting of tumor cells, extracellular matrix, mesenchymal fibroblasts, vascular endothelial cells, and immune cells. These factors contribute to tumor growth, infiltration, and metastasis. 25 The cells that make up the tumor microenvironment are induced by chemokines produced primarily by the tumor. Of these, chemokine (C‐X‐C motif) ligands CXCL1 and CXCL2 play an important role in the formation of the tumor microenvironment and are positively correlated with prognosis. 26 Immune cells, such as macrophages, contribute to the maintenance of the inflammatory tumor microenvironment by producing cytokines such as interleukin (IL)‐6 and tumor necrosis factor (TNF)‐α. Tumor‐promoting immune cells, such as myeloid‐derived suppressor cells (MDSCs) and tumor‐associated macrophages (TAMs), are recruited by CXCL1 and CXCL2. 27 , 28
In this study, we discovered that ANGPTL8 was highly correlated with inflammation in ccRCC. We demonstrated that ANGPTL8 maintained cancer cells in an undifferentiated state by inducing the expression of the CXCL1 and CXCL2 chemokines by activating the NF‐κB signaling pathway. We also found that ANGPTL8 was induced by the STAT3 signaling pathway. These results indicate that ANGPTL8 has an important role in ccRCC tumor progression by establishing the tumor microenvironment.
2. MATERIALS AND METHODS
2.1. Cell lines and cell cultures
Caki‐1 and KMRC‐1, human ccRCC cell lines, were purchased from the Japanese Collection of Research Bioresources Cell Bank. Caki‐1 cells were cultured in RPMI‐1640 medium (Wako) supplemented with 10% FBS and 1% penicillin/streptomycin, and KMRC‐1 cells were cultured in high‐glucose DMEM medium (Wako) supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were cultured at 37°C in a humidified incubator containing 5% CO2. For the tumor sphere assay, wild‐type or ANGPTL8 knockout (KO) Caki‐1 and KMRC‐1 cells (1 × 103) were seeded in 96‐well, Nunclon Sphere‐Treated, U‐Shaped‐Bottom Microplates (Thermo Fisher Scientific) and cultured for 5 days. For experiments evaluating STAT3, 2 × 105 Caki‐1 cells were seeded in six‐well dishes and treated with 20‐ng/mL IL‐6 or 10‐μM Stattic for 24 h.
2.2. Generation of ANGPTL8 ‐overexpressing cells
Caki‐1 and KMRC‐1 cells were transduced with the pLenti‐CMV‐MCS‐GFP puro vector containing FLAG‐tagged ANGPTL8 or the empty vector using FuGENE (Promega). The coding region of ANGPTL8 (RefSeq NM_018687.7) containing FLAG at the C‐terminus was replaced with GFP. Transfected cells were incubated with 2‐μg/m< puromycin to select ANGPTL8‐expressing cells. The expression levels of ANGPTL8 were confirmed using immunoblotting (Figure S2B). The cells infected with the empty vector (mock) were used as controls in experiments evaluating ANGPTL8‐overexpressing cells.
2.3. ANGPTL8 knockout cells
The LentiCRISPR v2 neo, psPAX2, and pCMV‐VSV‐G vectors were obtained from Addgene. The sequences of guide RNAs were cloned into the BsmBI sites of the LentiCRISPR v2 neo vector. The lentiviruses containing the guide RNAs were produced in 293 T cells cotransfected with the lentiCRISPR v2 neo, psPAX2, and pCMV‐VSV‐G. Caki‐1 and KMRC‐1 cells were infected with the lentivirus and cloned under G418 selection. Sequence analysis confirmed the genetic deletion in exon 1 of human ANGPTL8 (Figure S2A,B). The levels of ANGPTL8 were confirmed using immunoblotting (Figure S2C). Wild‐type cells were used as controls in experiments evaluating ANGPTL8 KO cells.
2.4. Real‐time quantitative RT‐PCR
Real‐time quantitative RT‐PCR was carried out as described. 29 Oligonucleotide primers are listed in Tables S1.
2.5. Immunoblot analysis and antibodies
Cells were homogenized in lysis buffer containing 1% NP‐40 and the complete protease inhibitor cocktail (Roche Diagnostics). Extracts prepared from the supernatants were separated using SDS‐PAGE, and the proteins were transferred to PVDF membranes. Immunodetection was performed using ECL Prime with the ImageQuant LAS 4000 mini system (GE Healthcare) according to the manufacturer's protocol. The antibodies are listed in the Supporting Information file. The ANGPTL8 antibody was kindly provided by Immuno‐Biological Laboratories.
2.6. Quantification of CXCL1 and CXCL2 and ANGPTL8 levels using ELISA
The cell culture medium was collected to quantify proteins using ELISA according to the manufacturer's instructions. Detailed information on the materials used for ELISA is provided in the Supporting Information file.
2.7. Reporter assay for NF‐κB activity
Cells (4 × 104) were seeded in 24‐well plates and cotransfected with the pGL4.32[luc2P/NF‐κB‐RE/Hygro]vector (Promega) and pRL‐TK vector (Promega) for normalization (total plasmid amount: 0.5 μg) using FuGENE (Promega). The cells were lysed with 100 μL of passive lysis buffer 24 h later. Luciferase activity was measured using the dual‐luciferase reporter assay system (Promega). Details on the materials used for the dual‐luciferase assay are shown in the Supporting Information file.
2.8. Chemotaxis assays
THP‐1 cells treated with PMA were resuspended in RPMI‐1640 medium containing 0.1% FBS at a final concentration of 2 × 105 cells/mL. The cells in suspension (300 μL) were seeded in the upper compartment of a transwell insert with a pore size of 8.0 μm (Corning). Caki‐1 cells (4 × 104 cells in 700 μL RPMI‐1640 medium containing 0.5% FBS) were seeded in the lower compartment. After 6 h of incubation, the cells migrating from the upper to the lower compartment were evaluated.
2.9. Cell proliferation and cell viability assays
Wild‐type or ANGPTL8 KO Caki‐1 and KMRC‐1 cells (3 × 103) were seeded in 96‐well plates, and the Cell Counting Kit‐8 (Dojindo) was used according to the manufacturer's instructions. Cell proliferation and viability after cisplatin treatment were evaluated. The experiment was performed in triplicate.
2.10. Scratch assay
Wild‐type or ANGPTL8 KO Caki‐1 cells (2 × 105) were seeded in a six‐well plate. The next day, a scratch was created at the bottom of the wells using a 840‐μm pipette tip (Labcon). The filling of the gap by the tumor cells was imaged at 0, 6, and 24 h using an EVOS FL fluorescence microscope (Thermo Fisher Scientific).
2.11. Promoter assay
Cells (4 × 104) were seeded in a 24‐well plate and cotransfected with the indicated promoter constructs and pRL‐TK vector for normalization (total plasmid amount: 0.5 μg) using FuGENE. Each promoter construct was created by inserting the promoter region approximately 1,500 bases upstream from the transcription start site for the chemokine (approximately 450 bases upstream from the transcription start site for the ANGPTL8 promoter) into the pGL4.10[luc2] vector (Promega). The binding sites in all promoter regions were identified using JASPAR (https://jaspar.genereg.net), and site‐directed mutagenesis was performed to create promoter constructs in which the transcription factor binding site was deleted. The primers used to create these constructs are listed in Table S1. The cells were lysed with 100 μL of passive lysis buffer 24 h later. Luciferase activity was measured using the dual‐luciferase reporter assay system. All samples were tested in triplicate from different wells, and the averages of the results were reported.
2.12. Flow cytometry analysis
Cells (5 × 105) were incubated with the indicated antibodies for 30 min at 4°C. After treatment, all cells were washed with PBS and analyzed using the FACS Calibur system and the CellQuest Pro software (BD Biosciences, Franklin Lakes). The antibodies are listed in the Supporting Information file.
2.13. RNA sequencing
The RNeasy Mini Kit was used to extract total RNA from Caki‐1 cells mock‐infected with the empty vector (n = 3), Caki‐1 cells overexpressing ANGPTL8 (n = 3), and Caki‐1 cells with ANGPTL8 KO (n = 3) in accordance with the manufacturer's instructions. The libraries were prepared using the TruSeq standard mRNA library kit (cat. no, 20020594; Illumina) and sequenced using NovaSeq 6000 (Illumina). The sequence reads were mapped to the human reference genome HG19 using TopHat2 version 2.1.0. The transcripts were assembled using Cufflinks version 2.2.1. Differentially expressed genes were determined using Cuffdiff. Gene ontology (GO) analysis using the CDS expression data from the RNA sequencing was conducted using Metascape. 30 The TRRUST was used to search for transcription factors that regulate enriched genes. 31
2.14. TCGA database analysis
The human RCC data were derived from TCGA Research Network, which can be found at http://cancergenome.nih.gov/. The dataset from this resource that supports the findings of the present study is available on cBioPortal (https://www.cbioportalorg/). Data were retrieved from the “Kidney Renal Clear Cell Carcinoma, TCGA, PanCancer Atlas” dataset in the cBioPortal dataset. Coexpression analysis of each mRNA gene and the ANGPTL8 mRNA was performed using the coexpression data obtained from cBioPortal. 32 , 33
2.15. Statistical analysis
All statistical analyses were performed using the Prism software version 8 (GraphPad). A two‐tailed Student's t‐test was used to compare two samples. The log‐rank test was used for the analysis of the Kaplan–Meier plots. All results were confirmed using at least three independent in vitro experiments. Data were shown as averages ± the standard error of the mean. Results with p‐values of <0.05 were considered statistically significant.
3. RESULTS
3.1. ANGPTL8 levels in tumor cells correlate with both systemic inflammation and poor prognosis in patients with clear cell renal cell carcinoma
Serum CRP, a marker of systemic inflammation, has been used as a prognostic marker in RCC. 34 Therefore, we initially investigated factors that correlate with CRP expression in ccRCC using TCGA database. The analysis of a ccRCC cohort from TCGA database revealed that ANGPTL8 mRNA levels were positively correlated with CRP mRNA levels in ccRCC tissues (Figure 1A). Therefore, we established ANGPTL8‐overexpressing Caki‐1 and KMRC‐1 cells, which are representative human ccRCC cell lines (Figure S1A). There was no difference in cell proliferation compared to the mock cells in both cell lines (Figure 1B and Figure S1B). We performed an RNA‐sequence analysis using Caki‐1 cells and found that chemotaxis‐related genes (CXCL1 and CXCL2) were significantly upregulated by GO analysis (Figure 1C,D). From these results, we hypothesized that ANGPTL8 expression in ccRCC may promote tumor cell progression by inducing CXCL chemokines.
FIGURE 1.
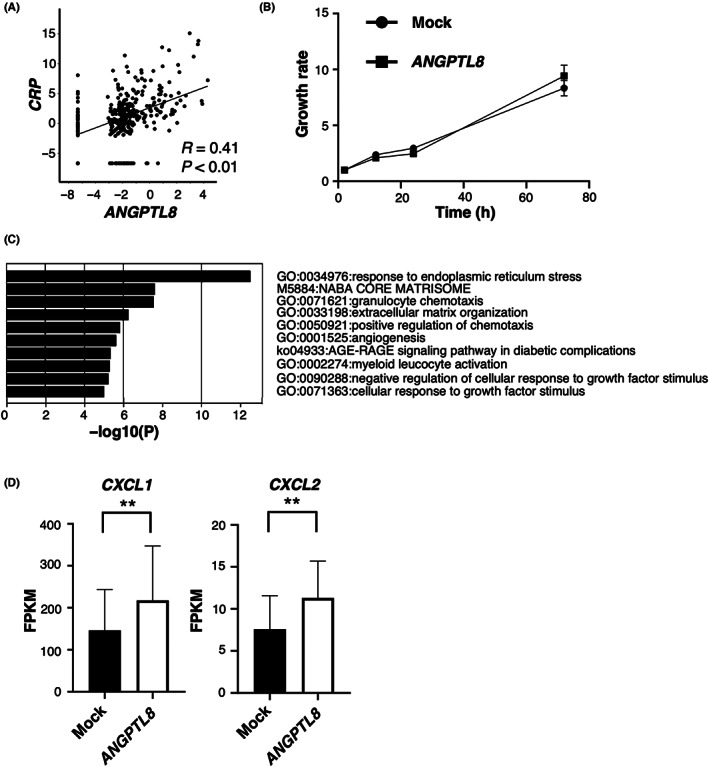
ANGPTL8 in clear cell renal cell carcinoma (ccRCC) cells correlates with poor RCC prognosis and inflammation (A) Correlation of ANGPTL8 and CRP expression in ccRCC based on the analysis of The Cancer Genome Atlas database (n = 354). Each value is presented on a log scale. Pearson's product–moment correlation analysis. (B) Cell proliferation of ANGPTL8‐overexpressing Caki‐1 cells and control Caki‐1 cells with normal ANGPTL8 expression cells (mock). (C) Ranking of the top 10 upregulated gene ontology (GO) terms in Caki‐1 cells following ANGPTL8 expression based on the GO analysis of the CDS expression data of RNA sequencing. (D) CXCL1 and CXCL2 expression in ANGPTL8‐overexpressing Caki‐1 cells by RNA‐sequence analysis. Each gene (n = 3) is presented as fragments per kilobase of exon million mapped reads (FPKM). **p < 0.01; unpaired Student's t test.
3.2. ANGPTL8 knockout in clear cell renal cell carcinoma cells not only decreases chemotaxis‐related gene expression but increases urogenital development‐related gene expression
Next, we established ANGPTL8 KO Caki‐1 and KMRC‐1 cells using the CRISPR/Cas9 system to confirm whether ANGPTL8 KO in ccRCC cells reduces the expression of CXCLs (Figure S2A–C). There was no difference in cell proliferation between the wild‐type and ANGPTL8 KO cells (Figure 2A and Figure S2D). We performed an RNA‐sequence analysis of ANGPTL8 KO Caki‐1 cells and found that chemotaxis‐related genes (CXCL1 and CXCL2) were markedly decreased in ANGPTL8 KO cells based on GO analysis (Figure 2B,C, and Figure S2E). Next, we examined the genes that were upregulated in ANGPTL8 KO cells to determine the effect of ANGPTL8 deletion in ccRCC cells. Surprisingly, we found that several genes associated with the development of the urogenital system were enriched among a total of 342 genes upregulated in ANGPTL8 KO Caki‐1 cells (Figure 2D and Figure S2F). Therefore, we next examined the relationship between ANGPTL8 and the development of the urogenital system in ccRCC.
FIGURE 2.
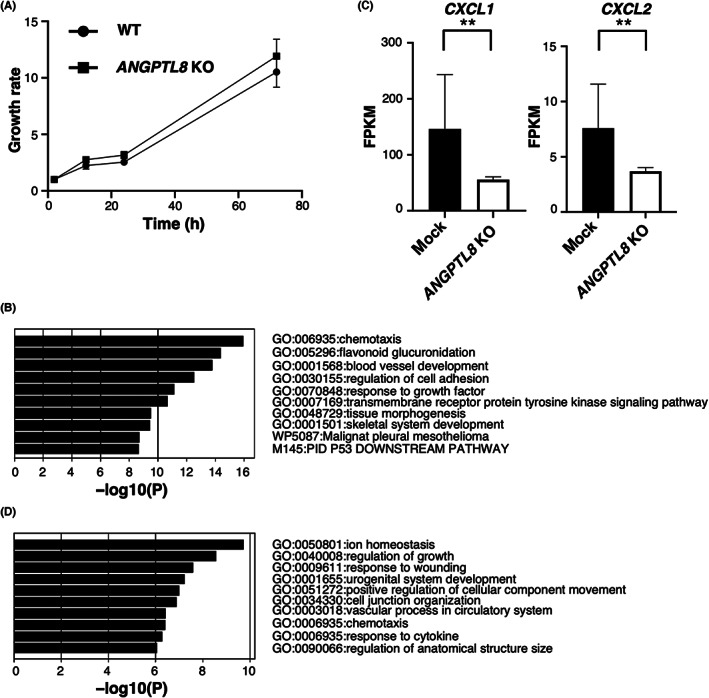
Knockout of ANGPTL8 in clear cell renal cell carcinoma (ccRCC) cells downregulates CXCLs and upregulates urogenital development‐related genes. (A) Cell proliferation assay of wild‐type and ANGPTL8 knockout (KO) Caki‐1 cells. (B) Ranking of the top 10 upregulated gene ontology (GO) terms in Caki‐1 cells following ANGPTL8 KO based on GO analysis of CDS expression data of RNA sequencing data. (C) CXCL1 and CXCL2 levels in ANGPTL8 KO Caki‐1 cells by RNA‐sequence analysis. Each gene (n = 3) is presented as fragments per kilobase of exon million mapped reads (FPKM). (D) Ranking of the top 10 downregulated GO terms in Caki‐1 cells following ANGPTL8 KO based on the GO analysis of the CDS expression data of RNA sequencing data. **p < 0.01; unpaired Student's t‐test.
3.3. Knockout of ANGPTL8 in clear cell renal cell carcinoma cells promotes differentiation to proximal tubule‐like cells
Based on RNA‐sequence analysis, we observed increased mRNA levels associated with urogenital development, such as FGFR2, GATA2, and HOXD13, in ANGPTL8 KO Caki‐1 cells compared with the mock cells (Figure 3A). In contrast, the mRNA levels of CD44 and MET, which are considered undifferentiated cell markers in RCC, 35 were decreased in ANGPTL8 KO Caki‐1 cells (Figure 3B). These results were also confirmed at the protein level, except for FGFR2, which was not detected in KMRC‐1 cells (Figure 3C,D). However, except for MYC, the expression levels of several cancer stem markers in RCC, 35 , 36 , 37 , 38 including NANOG, SOX2, and POU5F1, were not significantly decreased (Figure S3A). These results suggest that ANGPTL8 is not involved in the acquisition of the cancer stem cell phenotype. The staining of Lotus tetragonolobus lectin (LTL), a marker of proximal tubules, was significantly increased in both ANGPTL8 KO Caki‐1 and KMRC‐1 cells compared with the wild‐type cells (Figure 3E and Figure S3B). In contrast, CD44 and MET expression were decreased in both ANGPTL8 KO Caki‐1 and KMRC‐1 cells compared to the wild‐type cells (Figure 3D and Figure S3C). The expression levels of E‐cadherin, a distal tubule marker, and PODXL, a glomerular epithelial cell marker, were slightly upregulated in ANGPTL8 KO Caki‐1 cells compared with the wild‐type cells (Figure S3D,E). It is generally known that RCC cells exhibit acquired cisplatin resistance. As CD44 is associated with resistance to cisplatin in other cancer cells, 39 , 40 , 41 we examined whether ANGPTL8 KO in ccRCC cells reduces cisplatin resistance. ANGPTL8 KO Caki‐1 cells showed decreased viability to cisplatin compared to the wild‐type cells. (Figure S3F,G). Next, we examined the effect of ANGPTL8 KO on sphere formation in both cell lines. ANGPTL8 KO Caki‐1 and KMRC‐1 cells exhibited attenuated sphere formation compared with the wild‐type cells (Figure 3F,G and Figure S3I,J). In addition, ANGPTL8 KO Caki‐1 cells showed markedly reduced migratory ability compared to the wild‐type cells (Figure 3H,I and Figure S3H). These findings suggest that ANGPTL8 suppresses differentiation and increases drug resistance in ccRCC cells.
FIGURE 3.
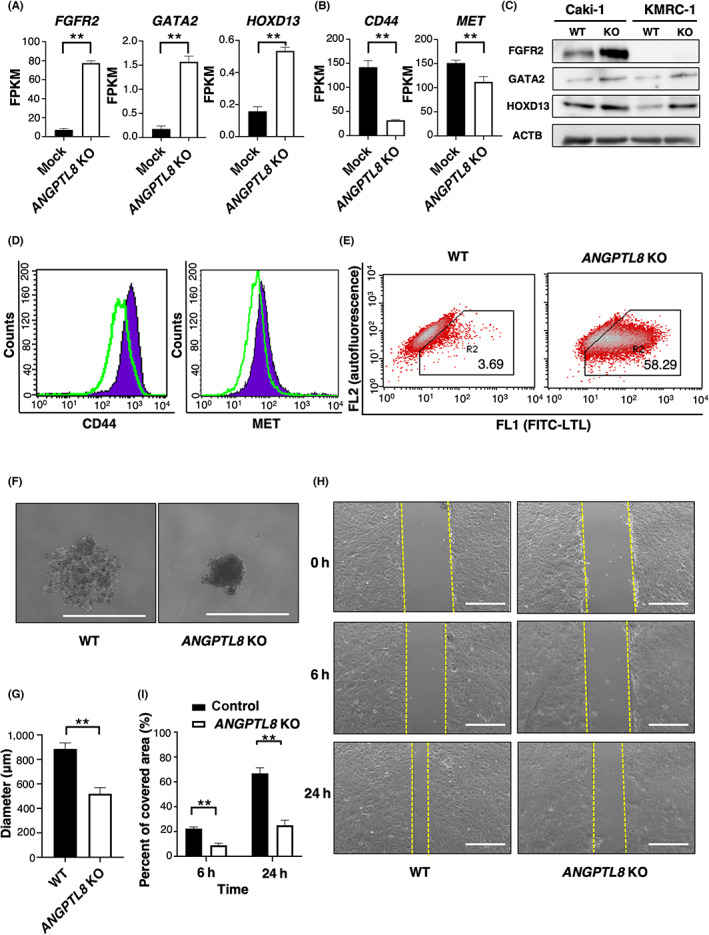
Knockout of ANGPTL8 in clear cell renal cell carcinoma (ccRCC) cells promotes a differential state in proximal tubule‐like cells. (A) FGFR2, GATA2, and HOXD13 levels in ANGPTL8 knockout (KO) Caki‐1 cells by RNA‐sequence analysis. Each gene (n = 3) is presented as fragments per kilobase of exon million mapped reads (FPKM). (B) CD44 and MET levels in ANGPTL8 KO Caki‐1 cells by RNA‐sequence analysis. Each gene (n = 3) is presented as fragments per kilobase of exon million mapped reads (FPKM). (C) FGFR2, GATA2, HOXD13, and ACTB protein levels in ANGPTL8 KO Caki‐1 and KMRC‐1 cells using immunoblotting. (D) FACS analysis of cell surface CD44 and MET in wild‐type Caki‐1 cells and ANGPTL8 KO Caki‐1 cells. Green line, ANGPTL8 KO Caki‐1 cells. Purple area, wild‐type Caki‐1 cells. (E) FACS analysis of LTL staining in wild‐type Caki‐1 and ANGPTL8 KO Caki‐1 cell cultures. (F) Representative images of the colony formation assay in wild‐type and ANGPTL8 KO Caki‐1 cells. Scale bar, 1,000 μm. (G) Quantitative analysis of the colony formation assay shown in Figure 3F. (H) Representative image showing cell motility of the wild‐type and ANGPTL8 KO Caki‐1 cells at 0, 6, and 24 h. Left, wild‐type Caki‐1 cells. Right, ANGPTL8 KO Caki‐1 cells. Scale bar, 500 μm. (I) Quantitative analysis of cell motility at indicated times using the cell migration assay shown in Figure 3H (n = 3). Displayed values are presented as the percentage of the range at 0 h, which is set as 100%. **p < 0.01; unpaired two‐tailed Student's t‐test.
3.4. The cancer‐promoting chemokines CXCL1 and CXCL2 are regulated by ANGPTL8
Inflammation in the cancer microenvironment is recognized as a factor in cancer cell undifferentiation, as described in the Introduction. 14 , 42 RNA‐sequence analysis showed that ANGPTL8 KO in Caki‐1 cells reduced the expression level of chemotaxis‐related genes (Figure 2B). The CXCL1 and CXCL2 proteins secreted by cancer cells are important factors for the malignant phenotype by attracting immunosuppressive cells, such as MDSCs and TAMs. 27 , 28 Nishida et al. reported the prognostic value of CXCL1 and CXCL2 in ccRCC. 5 Therefore, we measured CXCL1 and CXCL2 mRNA levels in ANGPTL8 KO Caki‐1 cells using quantitative RT‐PCR. The expression of CXCL1 and CXCL2 was significantly downregulated in both ANGPTL8 KO Caki‐1 and ANGPTL8 KO KMRC‐1 cells compared with the wild‐type cells (Figure S4A,B). The CXCL1 and CXCL2 protein levels in the cell supernatant were also markedly decreased in ANGPTL8 KO Caki‐1 compared to the wild‐type cells (Figure 4A). In addition, CXCL1 and CXCL2 expression was partially reversed in ANGPTL8 KO Caki‐1 cells transfected with the ANGPTL8 overexpression vector (Figure 4A). We hypothesized that by deleting ANGPTL8, differentiated cells might no longer having difficulty in going to an undifferentiated state despite ANGPTL8 rescuing. Therefore, we speculated that chemokine expression might not be fully restored. We also observed that CXCL1 and CXCL2 mRNA levels in ccRCC were positively correlated with ANGPTL8 expression in the analysis of TCGA KIRC dataset (Figure 4B). In contrast, we confirmed that there was no relationship between these chemokines and ANGPTL8 mRNA in other cancers, such as melanoma, colorectal cancer, pancreatic cancer, glioblastoma, and stomach cancer, by analyzing other TCGA datasets.
FIGURE 4.
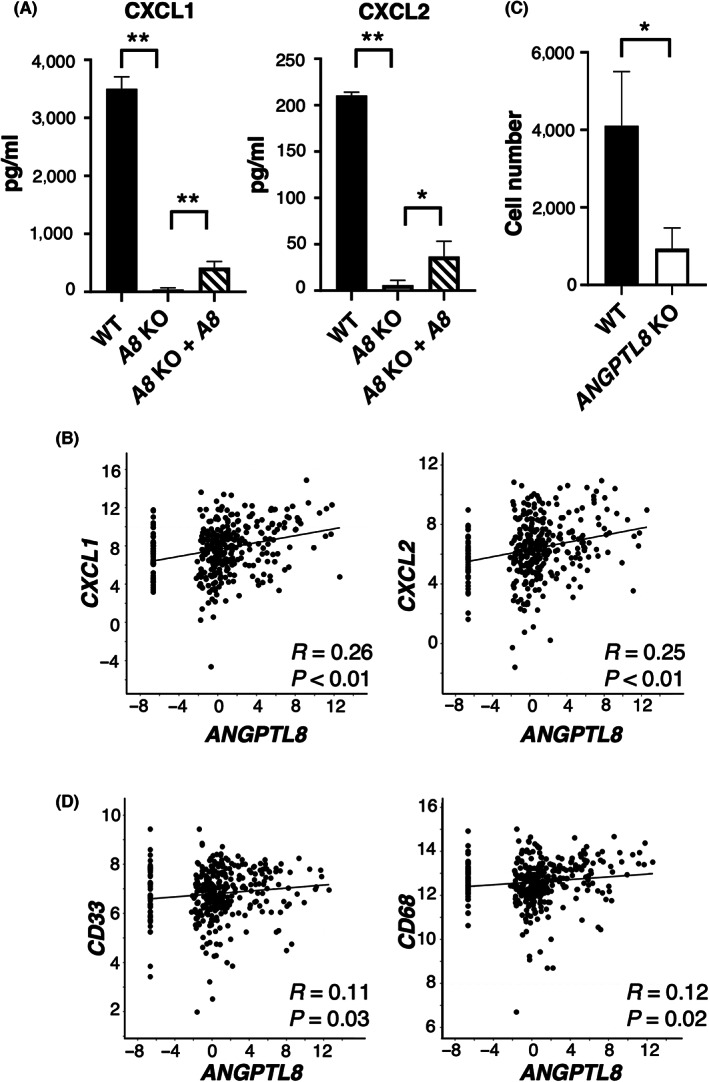
ANGPTL8 regulates CXCL1 and CXCL2 and attracts immune cells, such as MDSC and TAM. (A) Comparison of CXCL1 and CXCL2 levels in the culture medium of wild‐type, ANGPTL8 knockout (KO), and ANGPTL8 KO rescued by the ANGPTL8‐overexpressing vector in Caki‐1 cells. Data represents means ± SD from three experiments. (B) Correlation of the ANGPTL8 and CXCL1, and CXCL2 expression in clear cell renal cell carcinoma (ccRCC) by analysis of The Cancer Genome Atlas (TCGA) database (n = 354). Each value is presented in log scale. Pearson's product–moment correlation analysis. (C) Migration of THP‐1 cells co‐cultured with wild‐type or ANGPTL8 KO Caki‐1 cells (n = 3). (D) Correlation of ANGPTL8 and CD33 or CD68 expression in ccRCC by analysis of TCGA database (n = 354). Each value is presented on the log scale. Pearson's product–moment correlation analysis. *p < 0.05, **p < 0.01; unpaired two‐tailed Student's t‐test.
Next, we examined whether ANGPTL8 expression by ccRCCs attracted immune cells. The number of migrated THP‐1 cells was significantly decreased when cultured with ANGPTL8 KO Caki‐1 compared with the wild‐type cells (Figure 4C). We also confirmed from TCGA database that the mRNA levels of CD33, a marker for MDSC, and CD68, a marker for TAM, were positively correlated with that of ANGPTL8 in ccRCCs (Figure 4D). Based on these results, we hypothesized that the expression of ANGPTL8 in ccRCCs may attract immune cells, such as MDSCs or TAMs, by upregulating CXCL1 and CXCL2 and contribute to the establishment of the tumor microenvironment to maintain undifferentiated ccRCC cells.
3.5. ANGPTL8 induces CXCL1 and CXCL2 through NF‐κB
We next examined the mechanism through which ANGPTL8 induces chemokine expression. Based on the results of our RNA‐seq experiments, we examined the transcription factors that may be involved in chemokine expression. From the results of the analysis using TRRUST, we focused on NF‐κB, which was significantly upregulated in ANGPTL8‐overexpressing cells (Figure 5A,B). The activity of NF‐κB in ANGPTL8 KO Caki‐1 and KMRC‐1 cells was significantly reduced compared with the wild‐type cells (Figure 5C and Figure S5A). We also confirmed that phosphorylation of p65, a representative marker for NF‐κB activation, was decreased in ANGPTL8 KO Caki‐1 cells (Figure 5D). Deletion of the NF‐κB site in the promoter region of CXCL1 and CXCL2 resulted in reduced transcriptional activity of these chemokines (Figure S5B). These results indicate that ANGPTL8 activates NF‐κB to induce CXCL1 and CXCL2.
FIGURE 5.
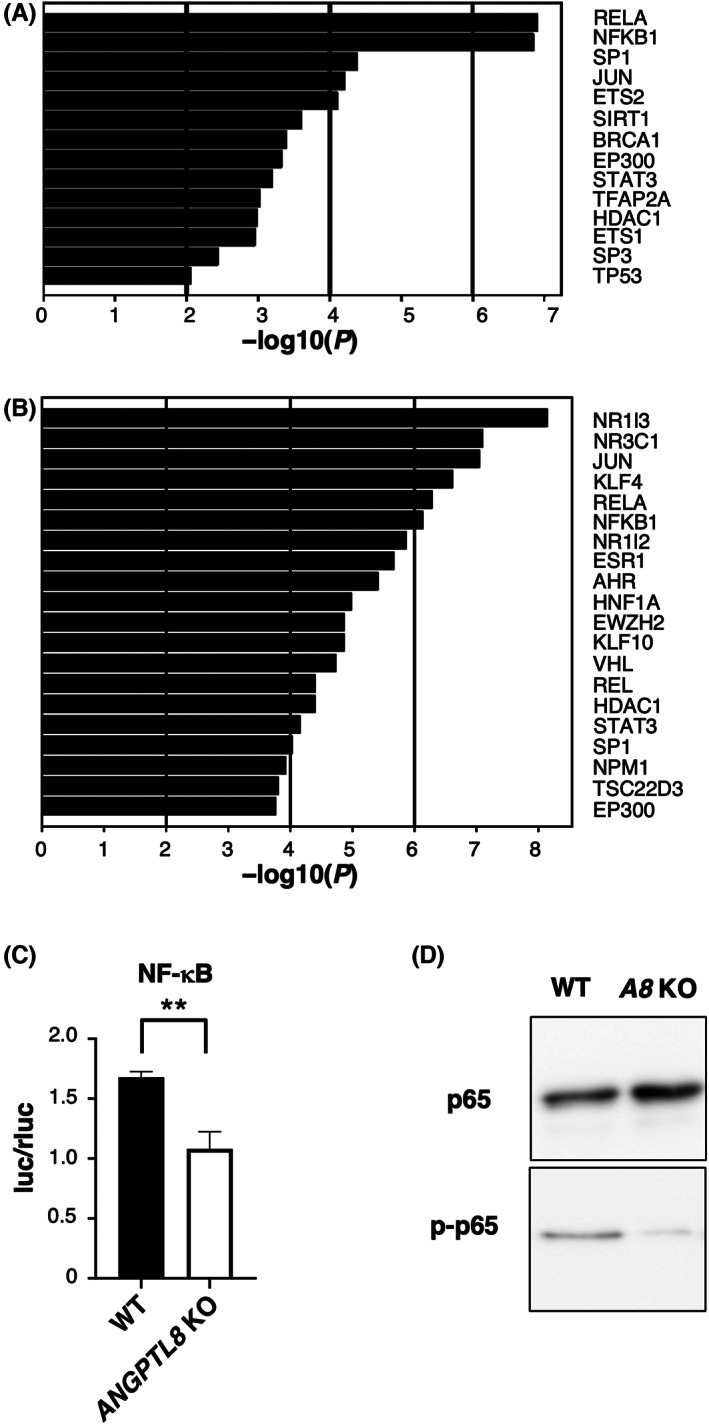
ANGPTL8 activates NF‐κB signaling pathway (A) Ranking of the top 14 upregulated transcription factor targets in ANGPTL8 overexpressing Caki‐1 cells by the enrichment analysis of the CDS expression data of the RNA sequencing. (B) Ranking of the top 20 downregulated transcription factor targets in ANGPTL8 knockout (KO) Caki‐1 cells by the enrichment analysis of the CDS expression data of the RNA sequencing. (C) Relative comparison of luciferase activity between wild‐type and ANGPTL8 KO Caki‐1 cells transfected with an NF‐κB expression plasmid. Data represent means ± SD from three experiments. (D) Representative images of the immunoblotting analysis of p65 and p‐p65 protein levels in wild‐type and ANGPTL8 KO Caki‐1 cells. **p < 0.01; unpaired two‐tailed Student's t‐test.
3.6. ANGPTL8 is induced by STAT3 in clear cell renal cell carcinoma cells
Although we have demonstrated that ANGPTL8 maintains undifferentiation and upregulates CXCL1 and CXCL2 expression, we have not determined how ANGPTL8 is induced in ccRCC. The transcription factors regulating genes correlated with ANGPTL8 in the TCGA KIRC dataset were searched using TRRUST. We found that STAT3 may be important for ANGPTL8 induction (Figure 6A). We identified several STAT3‐binding sites in the promoter region of human ANGPTL8 (up to 1300 bp upstream) (Figure S6A). IL‐6 is a cytokine that activates STAT3, and ANGPTL8 tends to be induced by IL‐6 stimulation in Caki‐1 and KMRC‐1 cells (Figure 6B and Figure S6B). In contrast, Stattic, a STAT3 inhibitor, significantly reduced ANGPTL8 expression in Caki‐1 and KMRC‐1 cells (Figure 6C and Figure S6C). Furthermore, the reporter assay confirmed that ANGPTL8 was directly regulated by STAT3 (Figure 6D and Figure S6D). These results indicate that the induction of ANGPTL8 in ccRCC is regulated by STAT3.
FIGURE 6.
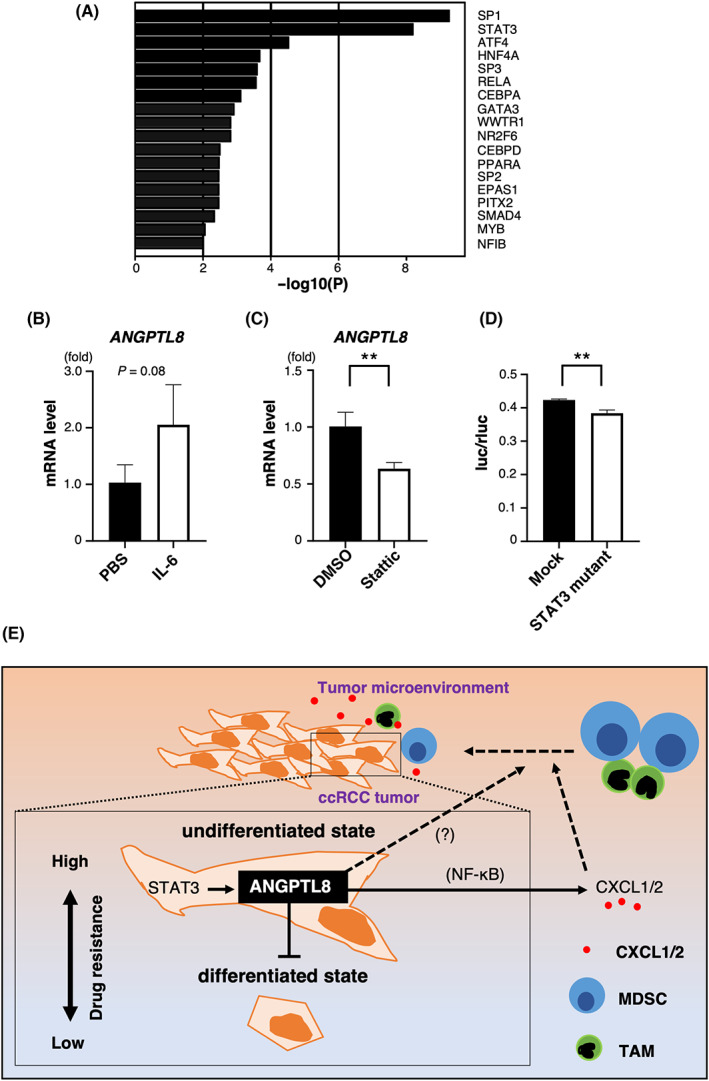
STAT3 induces ANGPTL8 expression (A) Ranking of the top 18 transcription factors that are highly correlated with ANGPTL8 by enrichment analysis of the top 3,000 genes in The Cancer Genome Atlas TCGA KIRC dataset. (B) Relative ANGPTL8 expression in Caki‐1 cells treated with PBS or human recombinant IL‐6. Data from the PBS‐treated cultures was set as 1. Data represent means ± SD from three experiments, p = 0.08; unpaired two‐tailed Student's t‐test. (C) Relative ANGPTL8 expression in Caki‐1 cells treated with DMSO or Stattic, which is a STAT3 inhibitor. Data from the DMSO‐treated cultures was set as 1. Data represent means ± SD from three experiments, **p < 0.01 unpaired two‐tailed Student's t‐test. (D) Reporter assay for the STAT3‐binding site in the promoter region of ANGPTL8. Comparison of STAT3‐binding sites with and without mutation. The lack of mutation is shown as mock. Data represent means ± SD from three experiments, **p < 0.01; unpaired two‐tailed Student's t‐test. (E) ANGPTL8 maintains the undifferentiated state in renal carcinoma and upregulates CXCL1 and CXCL2 to attract immune cells, which activates STAT3 signaling for ANGPTL8 induction in the tumor microenvironment.
4. DISCUSSION
In this study, we demonstrated that ANGPTL8 inhibits differentiation in ccRCC and upregulates CXCL1 and CXCL2 by activating the NF‐κB pathway. In addition, we found that ANGPTL8 was induced by STAT3. Our results reveal a novel role for ANGPTL8 in ccRCC cancer progression (Figure 6E). In addition, our results support an earlier study by Xu et al., who reported that ANGPTL8 was involved in the poor prognosis of patients with ccRCC based on TCGA analysis. 24
The protein structure of ANGPTL1–ANGPTL7 includes an N‐terminal coiled‐coil domain and a C‐terminal fibrinogen‐like domain; however, ANGPTL8 is the only member of the ANGPTL family that lacks a C‐terminal fibrinogen‐like domain and appears to exhibit functions different from those of other ANGPTL family members. In fact, the main role of ANGPTL8 is to interact with ANGPTL3 and increase the plasma levels of triglycerides and non‐esterified fatty acids. 23 A previous study reported that ANGPTL8 was expressed in hepatocellular carcinoma, 43 suggesting that ANGPTL8 may contribute to cancer progression.
Angiopoietin‐like protein‐8 family members are secretory proteins that activate signaling pathways in an autocrine or a paracrine manner. 17 Therefore, we measured ANGPTL8 levels in the culture medium of ccRCC cells using a Human ANGPTL8 Assay Kit (IBL). We confirmed that ANGPTL8 protein was significantly increased in the medium of ANGPTL8‐overexpressing Caki‐1 cells, whereas it was barely detectable in the culture medium of mock Caki‐1 cells (Figure S5C). In contrast, stimulation with the culture medium from ANGPTL8 overexpressing Caki‐1 cells did not activate NF‐κB in ANGPTL8 KO Caki‐1 cells (Figure S5D). Several studies showed that intracellular, rather than extracellular ANGPTL8, activated intracellular signaling pathways. 44 , 45 Therefore, we hypothesize that intracellular ANGPTL8 is important for ccRCC differentiation, although further study is needed.
ANGPTL8 is highly expressed in the liver and adipocytes, whereas it is not abundantly expressed in healthy kidneys or RCCs. 24 In contrast, analysis of TCGA database revealed that ANGPTL8 mRNA levels were associated with ccRCC progression. 24 These results suggest that the induction of ANGPTL8 in ccRCC may be important for ccRCC development and that ANGPTL8 levels in ccRCC may represent a prognostic marker.
We demonstrated that ANGPTL8 upregulates CXCL1 and CXCL2 expression through the NF‐κB pathway. Zhang et al. previously reported that ANGPTL8 suppresses NF‐κB activity in response to TNF‐α. 44 Therefore, we also examined the relationship between ANGPTL8 and TNF‐α in ccRCC. We confirmed that the activity of NF‐κB was reduced in ANGPTL8‐overexpressing Caki‐1 cells compared with mock Caki‐1 cells, which was consistent with their findings (Figure S5E). Therefore, we hypothesized that ANGPTL8 has a dual function, which includes repressing NF‐κB in TNF‐α signaling under inflammatory conditions and activating NF‐κB under normal conditions to maintain undifferentiation in ccRCC cells. The phosphorylation of p65 was weak in ANGTPL8 KO cells (Figure 5D), suggesting that ANGPTL8 might be involved in IKK activation, which is responsible for the S536 phosphorylation of p65. However, we did not determine the detailed mechanism by which ANGPTL8 activates NF‐κB signaling in ccRCC, which will be a subject of future studies.
We observed that ANGPTL8 KO in ccRCC cells promoted differentiation. In contrast, overexpressing ANGPTL8 in ccRCC cells did not significantly promote undifferentiation. These results indicate that excessive levels of ANGPTL8 are not needed to inhibit the differentiation of ccRCC cells, although some ANGPTL8 expression may be required for normal renal cancer cell function.
We demonstrated that ANGPTL8 was induced via the STAT3 signaling pathway. In recent reports, STAT3 has been shown to contribute to RCC progression and metastasis. 46 , 47 , 48 Our results suggest one mechanism by which STAT3 activity affects ccRCC progression through ANGPTL8 signaling. IL‐6 is a major STAT3 activator and is produced by many immune cells, such as MDSCs and TAMs. 49 , 50 Therefore, MDSCs and/or TAMs in the tumor microenvironment may promote ANGPTL8 induction in ccRCCs. In addition, it was reported that ANGPTL8 was induced by the endoplasmic reticulum (ER) stress pathway. We confirmed that ANGPTL8 was induced in Caki‐1 cells by tunicamycin, which is an ER stress inducer (Figure S6E), suggesting that ANGPTL8 is induced in the tumor microenvironment in ccRCC cells by hypoxia and malnutrition. We hypothesize that ANGPTL8 is also induced by ER stress in ccRCCs and induces CXCL1 and CXCL2 to attract MDSCs and TAMs. These factors combine to create an aggressive tumor microenvironment in ccRCC.
In summary, we demonstrate a role for ANGPTL8 in maintaining ccRCC cell properties. Based on these findings, we propose that downregulation of ANGPTL8 promotes the differentiation of ccRCC cells. These studies may lead to new therapeutic strategies to prevent ccRCC progression.
AUTHOR CONTRIBUTIONS
TM, TD, and ME designed the experiments. TM, TD, KO, and KS performed the experiments and/or provided advice and technical expertise. TM, TD, KO, NF, and ME wrote the manuscript. All authors amended the manuscript.
FUNDING INFORMATION
This work was supported by the Scientific Research Fund of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grants 20 K08530, 21 K07972, and 22 K08296).
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Reviewer Board: N/A.
Informed Consent: N/A.
Registry and the registration no. of the study/trial: N/A.
Animal studies: N/A.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Figure S5.
Figure S6.
Table S1:
Table S2:
ACKNOWLEDGMENTS
We thank Ms S. Miyoshi for her technical assistance.
Matsukawa T, Doi T, Obayashi K, Sumida K, Fujimoto N, Endo M. ANGPTL8 links inflammation and poor differentiation, which are characteristics of malignant renal cell carcinoma. Cancer Sci. 2023;114:1410‐1422. doi: 10.1111/cas.15700
DATA AVAILABILITY STATEMENT
The RNA sequencing data have been deposited in the NCBI Gene Expression Omnibus and can be accessed using the accession number GSE203429 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203429).
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Chung CJ, Bao BY, Lin YC, et al. Polymorphism of nucleotide binding domain‐like receptor protein 3 (NLRP3) increases susceptibility of total urinary arsenic to renal cell carcinoma. Sci Rep. 2020;10(1):6640. doi: 10.1038/s41598-020-63469-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gudowska‐Sawczuk M, Kudelski J, Mroczko B. The role of chemokine receptor CXCR3 and its ligands in renal cell carcinoma. Int J Mol Sci. 2020;21(22):8582. doi: 10.3390/ijms21228582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoefflin R, Harlander S, Schafer S, et al. HIF‐1alpha and HIF‐2alpha differently regulate tumour development and inflammation of clear cell renal cell carcinoma in mice. Nat Commun. 2020;11(1):4111. doi: 10.1038/s41467-020-17873-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nishida J, Momoi Y, Miyakuni K, et al. Epigenetic remodelling shapes inflammatory renal cancer and neutrophil‐dependent metastasis. Nat Cell Biol. 2020;22(4):465‐475. doi: 10.1038/s41556-020-0491-2 [DOI] [PubMed] [Google Scholar]
- 6. Hu Q, Gou Y, Sun C, et al. The prognostic value of C‐reactive protein in renal cell carcinoma: a systematic review and meta‐analysis. Urol Oncol. 2014;32(1):50.e51‐50.e58. doi: 10.1016/j.urolonc.2013.07.016 [DOI] [PubMed] [Google Scholar]
- 7. Hu H, Yao X, Xie X, et al. Prognostic value of preoperative NLR, dNLR, PLR and CRP in surgical renal cell carcinoma patients. World J Urol. 2017;35(2):261‐270. doi: 10.1007/s00345-016-1864-9 [DOI] [PubMed] [Google Scholar]
- 8. Chang Y, An H, Xu L, et al. Systemic inflammation score predicts postoperative prognosis of patients with clear‐cell renal cell carcinoma. Br J Cancer. 2015;113(4):626‐633. doi: 10.1038/bjc.2015.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Vivar Chevez AR, Finke J, Bukowski R. The role of inflammation in kidney cancer. Adv Exp Med Biol. 2014;816:197‐234. doi: 10.1007/978-3-0348-0837-8_9 [DOI] [PubMed] [Google Scholar]
- 10. Wang T, Lu R, Kapur P, et al. An empirical approach leveraging Tumorgrafts to dissect the tumor microenvironment in renal cell carcinoma identifies missing link to prognostic inflammatory factors. Cancer Discov. 2018;8(9):1142‐1155. doi: 10.1158/2159-8290.CD-17-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bakouny Z, Braun DA, Shukla SA, et al. Integrative molecular characterization of sarcomatoid and rhabdoid renal cell carcinoma. Nat Commun. 2021;12(1):808. doi: 10.1038/s41467-021-21068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones TD, Eble JN, Wang M, Maclennan GT, Jain S, Cheng L. Clonal divergence and genetic heterogeneity in clear cell renal cell carcinomas with sarcomatoid transformation. Cancer. 2005;104(6):1195‐1203. doi: 10.1002/cncr.21288 [DOI] [PubMed] [Google Scholar]
- 13. Chong LW, Chou RH, Liao CC, et al. Saturated fatty acid induces cancer stem cell‐like properties in human hepatoma cells. Cell Mol Biol (Noisy‐le‐Grand). 2015;61(6):85‐91. [PubMed] [Google Scholar]
- 14. Mengoni M, Braun AD, Gaffal E, Tuting T. The aryl hydrocarbon receptor promotes inflammation‐induced dedifferentiation and systemic metastatic spread of melanoma cells. Int J Cancer. 2020;147(10):2902‐2913. doi: 10.1002/ijc.33252 [DOI] [PubMed] [Google Scholar]
- 15. Schaffner F, Ray AM, Dontenwill M. Integrin alpha5beta1, the fibronectin receptor, as a pertinent therapeutic target in solid tumors. Cancers (Basel). 2013;5(1):27‐47. doi: 10.3390/cancers5010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carbone C, Piro G, Merz V, et al. Angiopoietin‐like proteins in angiogenesis, inflammation and cancer. Int J Mol Sci. 2018;19(2):431. doi: 10.3390/ijms19020431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Endo M. The roles of ANGPTL families in cancer progression. J UOEH. 2019;41(3):317‐325. doi: 10.7888/juoeh.41.317 [DOI] [PubMed] [Google Scholar]
- 18. Kadomatsu T, Endo M, Miyata K, Oike Y. Diverse roles of ANGPTL2 in physiology and pathophysiology. Trends Endocrinol Metab. 2014;25(5):245‐254. doi: 10.1016/j.tem.2014.03.012 [DOI] [PubMed] [Google Scholar]
- 19. Horiguchi H, Endo M, Kawane K, et al. ANGPTL2 expression in the intestinal stem cell niche controls epithelial regeneration and homeostasis. EMBO J. 2017;36(4):409‐424. doi: 10.15252/embj.201695690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takano A, Fukuda T, Shinjo T, et al. Angiopoietin‐like protein 2 is a positive regulator of osteoblast differentiation. Metabolism. 2017;69:157‐170. doi: 10.1016/j.metabol.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 21. Tanoue H, Morinaga J, Yoshizawa T, et al. Angiopoietin‐like protein 2 promotes chondrogenic differentiation during bone growth as a cartilage matrix factor. Osteoarthr Cartil. 2018;26(1):108‐117. doi: 10.1016/j.joca.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 22. Yu Z, Yang W, He X, et al. Endothelial cell‐derived angiopoietin‐like protein 2 supports hematopoietic stem cell activities in bone marrow niches. Blood. 2022;139(10):1529‐1540. doi: 10.1182/blood.2021011644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abu‐Farha M, Sriraman D, Cherian P, et al. Circulating ANGPTL8/betatrophin is increased in obesity and reduced after exercise training. PLoS One. 2016;11(1):e0147367. doi: 10.1371/journal.pone.0147367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu F, Tian D, Shi X, Sun K, Chen Y. Analysis of the expression and prognostic potential of a novel metabolic regulator ANGPTL8/betatrophin in human cancers. Pathol Oncol Res. 2021;27:1609914. doi: 10.3389/pore.2021.1609914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marozzi M, Parnigoni A, Negri A, et al. Inflammation, extracellular matrix remodeling, and Proteostasis in tumor microenvironment. Int J Mol Sci. 2021;22(15):8102. doi: 10.3390/ijms22158102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang X, Hao J, Tan YQ, Zhu T, Pandey V, Lobie PE. CXC chemokine signaling in progression of epithelial ovarian cancer: Theranostic perspectives. Int J Mol Sci. 2022;23(5):2642. doi: 10.3390/ijms23052642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katoh H, Wang D, Daikoku T, Sun H, Dey SK, Dubois RN. CXCR2‐expressing myeloid‐derived suppressor cells are essential to promote colitis‐associated tumorigenesis. Cancer Cell. 2013;24(5):631‐644. doi: 10.1016/j.ccr.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taki M, Abiko K, Baba T, et al. Snail promotes ovarian cancer progression by recruiting myeloid‐derived suppressor cells via CXCR2 ligand upregulation. Nat Commun. 2018;9(1):1685. doi: 10.1038/s41467-018-03966-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doi T, Hojo H, Ohba S, et al. Involvement of activator protein‐1 family members in beta‐catenin and p300 association on the genome of PANC‐1 cells. Heliyon. 2022;8(2):e08890. doi: 10.1016/j.heliyon.2022.e08890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist‐oriented resource for the analysis of systems‐level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han H, Shim H, Shin D, et al. TRRUST: a reference database oh human transcriptional regulatory interactions. Sci Rep. 2015;5:11432. doi: 10.1038/srep11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401‐404. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steffens S, Kohler A, Rudolph R, et al. Validation of CRP as prognostic marker for renal cell carcinoma in a large series of patients. BMC Cancer. 2012;12(1):399. doi: 10.1186/1471-2407-12-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fendler A, Bauer D, Busch J, et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat Commun. 2020;11(1):929. doi: 10.1038/s41467-020-14700-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu W, Wang B, Wan F, et al. SOX2 and SOX12 are predictive of prognosis in patients with clear cell renal cell carcinoma. Oncol Lett. 2018;15(4):4564‐4570. doi: 10.3892/ol.2018.7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang CS, Tang SJ, Lee MH, Chang Wang CC, Sun GH, Sun KH. Galectin‐3 promotes CXCR2 to augment the stem‐like property of renal cell carcinoma. J Cell Mol Med. 2018;22(12):5909‐5918. doi: 10.1111/jcmm.13860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rasti A, Mehrazma M, Madjd Z, Abolhasani M, Saeednejad Zanjani L, Asgari M. Co‐expression of cancer stem cell markers OCT4 and NANOG predicts poor prognosis in renal cell carcinomas. Sci Rep. 2018;8(1):11739. doi: 10.1038/s41598-018-30168-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hueber PA, Waters P, Clark P, Eccles M, Goodyer P. PAX2 inactivation enhances cisplatin‐induced apoptosis in renal carcinoma cells. Kidney Int. 2006;69(7):1139‐1145. doi: 10.1038/sj.ki.5000136 [DOI] [PubMed] [Google Scholar]
- 40. Ishimoto T, Nagano O, Yae T, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(−) and thereby promotes tumor growth. Cancer Cell. 2011;19(3):387‐400. doi: 10.1016/j.ccr.2011.01.038 [DOI] [PubMed] [Google Scholar]
- 41. Trivedi R, Dihazi GH, Eltoweissy M, Mishra DP, Mueller GA, Dihazi H. The antioxidant protein PARK7 plays an important role in cell resistance to cisplatin‐induced apoptosis in case of clear cell renal cell carcinoma. Eur J Pharmacol. 2016;784:99‐110. doi: 10.1016/j.ejphar.2016.04.014 [DOI] [PubMed] [Google Scholar]
- 42. Schwitalla S, Fingerle AA, Cammareri P, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem‐cell‐like properties. Cell. 2013;152(1–2):25‐38. doi: 10.1016/j.cell.2012.12.012 [DOI] [PubMed] [Google Scholar]
- 43. Dong XY, Pang XW, Yu ST, et al. Identification of genes differentially expressed in human hepatocellular carcinoma by a modified suppression subtractive hybridization method. Int J Cancer. 2004;112(2):239‐248. doi: 10.1002/ijc.20363 [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y, Guo X, Yan W, et al. ANGPTL8 negatively regulates NF‐kappaB activation by facilitating selective autophagic degradation of IKKgamma. Nat Commun. 2017;8(1):2164. doi: 10.1038/s41467-017-02355-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Z, Wu H, Dai L, et al. ANGPTL8 enhances insulin sensitivity by directly activating insulin‐mediated AKT phosphorylation. Gene. 2020;749:144707. doi: 10.1016/j.gene.2020.144707 [DOI] [PubMed] [Google Scholar]
- 46. Huynh J, Chand A, Gough D, Ernst M. Therapeutically exploiting STAT3 activity in cancer ‐ using tissue repair as a road map. Nat Rev Cancer. 2019;19(2):82‐96. doi: 10.1038/s41568-018-0090-8 [DOI] [PubMed] [Google Scholar]
- 47. Zhang D, Yang XJ, Luo QD, et al. EZH2 enhances the invasive capability of renal cell carcinoma cells via activation of STAT3. Mol Med Rep. 2018;17(3):3621‐3626. doi: 10.3892/mmr.2017.8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng JM, Zhou HX, Yu HY, et al. By increasing the expression and activation of STAT3, sustained C5a stimulation increases the proliferation, migration, and invasion of RCC cells and promotes the growth of Transgrafted tumors. Cancer Manag Res. 2021;13:7607‐7621. doi: 10.2147/CMAR.S326352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro‐environment in tumor progression: the role of tumor‐associated macrophages. Crit Rev Oncol Hematol. 2008;66(1):1‐9. doi: 10.1016/j.critrevonc.2007.07.004 [DOI] [PubMed] [Google Scholar]
- 50. Radharani NNV, Yadav AS, Nimma R, et al. Tumor‐associated macrophage derived IL‐6 enriches cancer stem cell population and promotes breast tumor progression via Stat‐3 pathway. Cancer Cell Int. 2022;22(1):122. doi: 10.1186/s12935-022-02527-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Figure S5.
Figure S6.
Table S1:
Table S2:
Data Availability Statement
The RNA sequencing data have been deposited in the NCBI Gene Expression Omnibus and can be accessed using the accession number GSE203429 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203429).


