Abstract
Cervical squamous cell carcinoma (CSCC) is one of the leading causes of cancer death in women worldwide. Patients with advanced cervical carcinoma always have a poor prognosis once resistant to cisplatin due to the lack of effective treatment. It is urgent to investigate the molecular mechanisms of cisplatin resistance. Circular RNAs (circRNAs) are known to exert their regulatory functions in a series of malignancies. However, their effects on CSCC remain to be elucidated. Here, we found that cytoplasmic circARHGAP5, derived from second and third exons of the ARHGAP5 gene, was downregulated in cisplatin‐resistant tissues compared with normal cervix tissues and untreated cervical cancer tissues. In addition, experiments from overexpression/knockdown cell lines revealed that circARHGAP5 could inhibit cisplatin‐mediated cell apoptosis in CSCC cells both in vitro and in vivo. Mechanistically, circARHGAP5 interacted with AU‐rich element RNA‐binding protein (AUF1) directly. Overexpression of AUF1 could also inhibit cell apoptosis mediated by cisplatin. Furthermore, we detected the potential targets of AUF1 related to the apoptotic pathway and found that bcl‐2‐like protein 11 (BIM) was not only negatively regulated by AUF1 but positively regulated by circARHGAP5, which indicated that BIM mRNA might be degraded by AUF1 and thereby inhibited tumor cell apoptosis. Collectively, our data indicated that circARHGAP5 directly bound to AUF1 and prevented AUF1 from interacting with BIM mRNA, thereby playing a pivotal role in cisplatin resistance in CSCC. Our study provides insights into overcoming cancer resistance to cisplatin treatment.
Keywords: AUF1, BIM, cervical cancer, circARHGAP5, CSCC
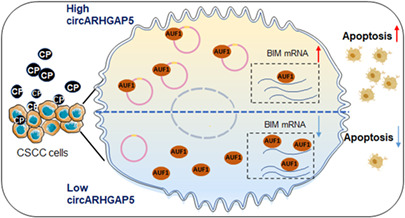
Circular RNA ARHGAP5 (circARHGAP5) inhibits cervical squamous cell carcinoma drug resistance. circARHGAP5 interacts with AU‐rich element RNA‐binding protein (AUF1) and attenuates AUF1 binding to bcl‐2‐like protein 11 (BIM) mRNA.
Abbreviation
- AGO2
argonaute RISC catalytic component 2
- ARE
AU‐rich element
- ARHGAP5
rho GTPase‐activating protein 5
- AUF1 (HNRNPD)
heterogeneous nuclear ribonucleoprotein D
- BIM (BCL2L11)
bcl‐2‐like protein 11
- circRNA
circular RNA
- CP
cisplatin
- CSCC
cervical squamous cell carcinoma
- HPV
human papillomavirus
- miRNA
microRNA
- p63
tumor protein P63
- qRT‐PCR
quantitative RT‐PCR
- RBP
RNA‐binding protein
- RNA‐seq
RNA sequencing
- RRM
RNA recognition motif
1. INTRODUCTION
Cervical cancer is the fourth most commonly malignant tumor and the fourth leading cause of cancer death in women worldwide. 1 Approximately 75% of cervical cancers are CSCC. 2 With the clinical advances in the past decades, cervical cancer is becoming a preventable disease because of the HPV vaccine and cytological screening. 3 , 4 , 5 However, it is still not eradicated because the coverage of HPV vaccination is still very low and unbalanced around the world. 6 , 7 , 8 Patients with advanced, recurrent, or metastatic cervical cancer have no choice but to accept CP‐based chemoradiotherapy. 9 , 10 But as for tumor heterogeneity and individual differences of patients in CP response, resistance to CP can be associated with poor prognosis. 9 Therefore, it is necessary to explore the molecular mechanisms of tumorigenesis and CP resistance for CSCC.
Tumor protein P63 functions as a key regulatory element involved in survival. 11 , 12 ∆Np63α is the predominant isoform expressed in cervical tissues, and is essential for regulation of epidermal morphogenesis and epithelial tissue homeostasis. 13 , 14 We have previously unveiled that ΔNp63α exerted antitumor functions including CP resistance in CSCC through regulating a series of its downstream direct protein and long noncoding RNA targets, but the functions and functional mechanisms of its direct circRNA in CSCC remain to be investigated. 14 , 15
Circular RNAs comprise a class of noncoding RNAs with a circular conformation produced from pre‐mRNAs through back‐splicing. 16 , 17 , 18 , 19 In contrast to linear RNAs, circRNAs are more stable and are resistant to RNase R digestion. 20 , 21 Some of the circRNAs are more abundant than their linear transcripts. 17 Furthermore, circRNAs are present in various animal genomes with tissue, cell type, and developmental specificity. 22 , 23 Recent studies have reported that circRNAs play increasingly vital pathological roles in multiple diseases including cancers, and dysregulation of circRNAs is found to be closely associated with biogenesis and progression of cancers. 19 , 22 For example, circE7 generated from HPV can lead to the suppression of cervical cancer progression. 24 Despite numerous functions of circRNAs in human cancers, much remains to be known about the functions and mechanisms of circRNAs involved in CSCC, especially their roles in drug resistance. The biological roles of circRNAs in CSCC require further investigation.
In this study, we identified that circARHGAP5 served as a regulatory target of ΔNp63α by RNA‐seq. Circular RNA ARHGAP5 was significantly downregulated in CSCC and positively correlated with ΔNp63α. Functionally, circARHGAP5 inhibited CSCC cell CP resistance by inducing cell apoptosis and preventing cell proliferation. Mechanistically, we provided evidence that circARHGAP5 interacted with AUF1 and showed the potential mechanism that circARHGAP5 could prevent AUF1 from degrading BIM mRNA, which thereby led to increased cell apoptosis and sensitivity to CP. Our data showed that circARHGAP5 might exert a potential tumor suppressor function in CSCC CP resistance.
2. MATERIALS AND METHODS
2.1. Tissues
Tissues were obtained from the First Affiliated Hospital of USTC, including 14 normal cervical tissues, 14 cervical cancer tissues, and 8 recurrent cervical cancer tissues resistant to CP. The normal cervical tissues were taken from patients with hysteromyoma undergoing hysterectomy. Cervical cancer tissues were acquired from cervical cancer patients with IB2‐IB3 (FIGO 2021) who accepted primary surgery. Tissues resistant to CP were acquired from cervix biopsy in patients who did not respond to CP therapy. This study was reviewed and approved by the Ethics Review Board of The First Affiliated Hospital of USTC. Written informed consent was obtained from each patient for this study (2022‐ky094).
2.2. Cell culture
Human cervical cancer SiHa, HeLa, and ME‐180 cells (ATCC) were cultured in DMEM (Hyclone) or McCoy's 5A medium (Gibco) with 10% (v/v) FBS (Gibco, Thermo Fisher Scientific), containing 100 units/ml penicillin and 100 mg/ml streptomycin (Hyclone).
2.3. RNA sequencing analysis
The total RNA samples (2 μg) were iron‐fragmented at 95°C and then subjected to end repair and 5′‐adaptor ligation. Differentially expressed circRNAs were identified using a t‐test (p < 0.05) combined with fold change. RNA sequencing was carried out as previously described. 15
2.4. RNA isolation and qRT‐PCR
Total RNA was isolated from cells or tissues by TRIzol reagent (Invitrogen). Reverse transcription was carried out using a PrimeScript RT Reagent Kit (Invitrogen) following the protocol, then qRT‐PCR was carried out (Roche) using a real‐time PCR instrument (Thermo Fisher Scientific). All the primers for detection are shown in Table S1.
2.5. RNase R resistance assay
Total RNA (5 μg) was treated with 8 U RNase R (Epicenter) in 50 μl total volume for 40 min, then analyzed by qRT‐PCR.
2.6. Vector construction, cell transfection, and plasmids
The circARHGAP5 target shRNAs were cloned into pLKO.1 puro vector construct stable cell lines. Overexpression plasmid of circARHGAP5 and linear mutations of circARHGAP5 were cloned into pcDNA3.0 vector. The transfection of siRNAs and plasmids was undertaken using the Lipofectamine 2000 kit (Invitrogen). The shRNA and siRNA sequences are listed in Table S2.
2.7. Cell viability assay
Cells (1 × 103) were seeded in 100 μl complete culture media in 96‐well plates. After 24 h, the CP gradient concentration diluted with complete medium was treated and then detected by CCK‐8 assay (Dojindo Laboratories).
2.8. Apoptosis assay
A total of 3 × 105 cells were plated each well of 6‐well plates. The cells were grown up to 70% confluency and treated with IC50 drug concentrations of CP for 48 h; the IC50 values for SiHa cells is 8 μg/ml and 9 μg/ml for ME‐180 cells, diluted with complete medium. Total cells were collected, washed twice in binding buffer, and stained with allophycocyanin labeled with annexin V (BioLegend) and propidium iodide as per the manufacturer's guidelines.
2.9. Colony formation assay
The number of all cells per well was counted and cells were kept in uniform distribution. For SiHa and ME‐180 cells, 1000 cells were split into 6‐well plates separately and in triplicates, and then treated with IC50 drug concentrations of CP. Cells were allowed to grow for 10 days in 5% CO2 incubators before being stained with 0.5% Crystal Violet Staining Solution (Solarbio).
2.10. Xenograft and orthotopic models of cancer in mice
Animal experiments were carried out as described previously. 14 , 15 Five‐week‐old female nude mice (Experiment Animal Center of Shanghai) (each group, n = 5) were subcutaneously injected with 6 × 106 ME‐180/NC, ME‐180/sh1‐circARHGAP5, or ME‐180/sh2‐circARHGAP5 cells in 0.1 ml PBS containing 20% Matrigel. When the tumor volumes reached 50–100 mm3, tumor‐bearing mice then received a tail vein injection of CP (3 mg/kg body weight, every 3 days). Fifteen days after treatment, the mice were killed to determine tumor volumes and were photographed.
2.11. RNA pull‐down assay
Cells (1 × 107) were incubated with 5′‐biotinylated antisense oligo probes against circARHGAP5 backsplice junction region or scramble probes at 4°C overnight. A total of 100 μl washed Dynabeads M‐280 Streptavidin (11206D; Thermo Fisher Scientific) were added to each binding reaction. Finally, the retrieved proteins were used for mass spectrometry or western blot analysis. Probe sequences are shown in Table S3.
2.12. RNA immunoprecipitation
Cells (1 × 107) were incubated with 2 μg FLAG (F1804; Sigma), AGO2 (SAB4200085; Sigma), anti‐AUF1 (P45‐99469; Thermo), or IgG primary Abs for 4 h at room temperature. Protein G was then added to each sample, then analyzed by qRT‐PCR and western blot.
2.13. RNA FISH‐immunofluorescence microscopy
To detect colocalization of circARHGAP5 with indicated proteins, cells were incubated with primary Ab (1:200 dilution, P45‐99469; Thermo Fisher Scientific) for 4 h at room temperature, which was followed by a reaction with Alexa Fluor 488‐conjugated secondary Abs (1:100 dilution; Life Technologies). Subsequently, hybridization using Cy‐3‐conjugated circARHGAP5 probes was carried out at 37°C in the dark overnight and the nuclei were stained with DAPI (Dojindo Laboratories). Visualization of the staining was carried out with a Zeiss LSM 880 confocal laser scanning microscope. Probe sequence is shown in Table S3.
2.14. Western blot analysis
For western blots, samples were separated on SDS‐PAGE gels. Membranes were incubated with Ab anti‐FLAG (1:1000 dilution, 20,543‐1‐AP; Proteintech), AUF1 (1:2000 dilution, P45‐99469; Thermo Fisher Scientific), AGO2 (1:1000, SAB4200085; Sigma), BIM (1:600 dilution, Proteintech, 22,037‐1‐AP), or anti‐β‐actin (1:1000 dilution, HC201; TransGen).
2.15. Statistical analysis
All of the experiments were repeated three times. Prism software (GraphPad Software 8) was used for all statistical analyses. Quantitative data of all the experiments were expressed as mean ± SD from three biological replicates. Student's two‐tailed t‐test or ANOVA was used to assess the statistical significance of the difference. *p < 0.05, **p < 0.01, ***p < 0.001.
2.16. Data
The accession number for the RNA‐seq data reported in this paper is GEO: GSE135257.
3. RESULTS
3.1. Identification of circARHGAP5 in CSCC cells
To identify and characterize direct circRNA targets of ΔNp63α in CSCC, we performed RNA‐seq in two CSCC cell lines, ME‐180/shΔNp63α and SiHa/ΔNp63α, with respective controls. 14 A total of 193 differentially expressed circRNAs were identified in two stable cell line groups (cut‐off of two‐fold changes and p < 0.05) (Figure 1A). Integrative analysis showed that three circRNAs were differentially expressed in two cell groups (Figure 1B). Then we undertook qRT‐PCR to further validate these three circRNAs in SiHa/ΔNp63α and ME‐180/shΔNp63α cells, and found that only two circRNA candidates, circARHGAP5 and circHERC4, were consistent with the result of circRNA deep sequencing (Figures 1C and S1A). We finally focused on circARHGAP5, also annotated as hsa_circ_0031584 in circBase, as it had a higher abundance of reads and expression levels than circHERC4 and was positively associated with ΔNp63α. In addition, qRT‐PCR showed that underexpression or overexpression of ΔNp63α did not affect the mRNA level of the ARHGAP5 gene, which showed that regulation of ΔNp63α on circARHGAP5 expression was independent on the host ARHGAP5 gene (Figure S1B). Our results indicated that circARHGAP5 was an abundant circRNA in CSCC and was positively correlated with ΔNp63α in CSCC.
FIGURE 1.
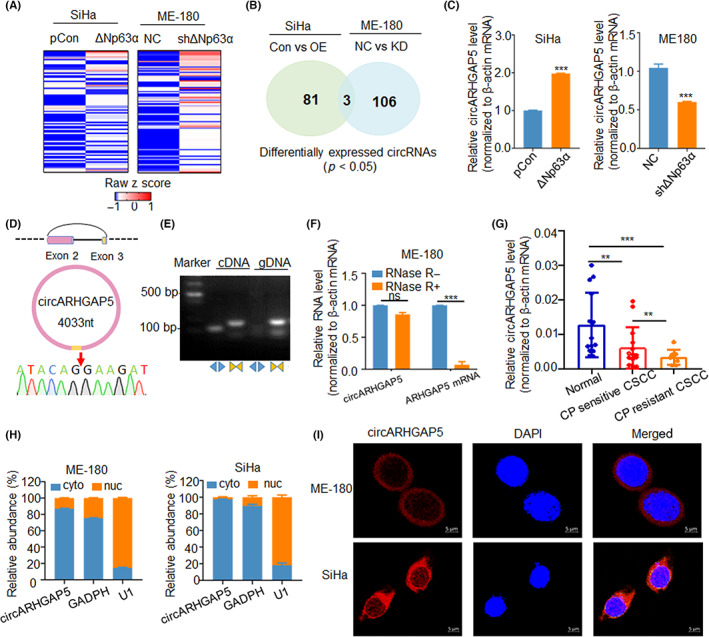
Identification and characterization of circular RNA ARHGAP5 (circARHGAP5) in cervical squamous cell carcinoma (CSCC) cells and tissues. (A) Heatmap of circRNA expression profile in SiHa/Con with SiHa/ΔNp63α and ME‐180/NC with ME‐180/shΔNp63α stable cell lines. (B) Overlap of 193 differentially expressed circRNAs in these two groups of cells. (C) Verification of circARHGAP5 expression levels in the above two groups of cells. (D) Sanger sequencing of backsplice junction site for circARHGAP5. (E) PCR analysis for circARHGAP5 in cDNA and genomic DNA (gDNA) with divergent and convergent primers. (F) Relative RNA levels of circARHGAP5 and ARHGAP5 with or without RNase R digestion. (G) Expression level of circARHGAP5 in normal cervical tissues and untreated and cisplatin (CP)‐resistant CSCC tissues. (H) Nuclear (nuc) and cytoplasmic (cyto) levels of circARHGAP5 in ME‐180 and SiHa cells. GADPH served as cytoplasmic control, while U1 served as nuclear control. (I) FISH assay of circARHGAP5 (red) in ME‐180 and SiHa cells. The location of circARHGAP5 (red) in SiHa and ME‐180 cells was determined by FISH assay. DAPI‐stained nuclei are blue. Scale bar = 5 μm. **p < 0.01, ***p < 0.001. Con, control; NC, negative control; ns, not significant; OE, overexpression.
3.2. Characteristics of circARHGAP5 in CSCC cells and tissues
Circular RNA ARHGAP5 is generated from the second and third exons of the human ARHGAP5 gene, consisting of 4033 nt, and the backsplice junction site of circARHGAP5 was validated by Sanger sequencing (Figure 1D). Additionally, PCR results showed that circARHGAP5 was amplified in cDNA by divergent primers and no product could be detected from genomic DNA as template (Figure 1E). To further evaluate the stability of circARHGAP5, RNase R digestion assay was used to confirm that circARHGAP5 harbored a closed stable loop structure and was RNase R resistant (Figure 1F). We suspected that circARHGAP5 played a role in CSCC CP resistance and then detected the expression of circARHGAP5 in normal cervix tissues and untreated and CP‐resistant cervical cancer tissues. The qRT‐PCR results showed that circARHGAP5 was downregulated in CP‐resistant cervical cancer tissues compared with others, suggesting circARHGAP5 might serve as a tumor suppressor and could be associated with CP resistance very closely (Figure 1G). In addition, circARHGAP5 was predominately localized in the cytoplasm in ME‐180 and SiHa cells by nuclear‐cytoplasmic fractionation and FISH (Figure 1H,I). These results indicated that circARHGAP5 was a bona fide circRNA localized in cytoplasm and it was downregulated in CP‐resistance CSCC tissues.
3.3. Circular RNA ARHGAP5 inhibited CSCC CP resistance in vivo and in vitro
One of the hotspots in CSCC treatment is CP resistance. We have previously reported that ΔNp63α exerted antitumor functions, including CP resistance, by modulating its protein and noncoding RNA targets in CSCC. 14 , 25 To investigate whether circARHGAP5 as a ΔNp63α circRNA target also played functional roles in CP resistance in CSCC, we constructed SiHa and ME‐180 stable cells with circARHGAP5 knockdown by shRNAs targeting the circRNA junction sites. The qRT‐PCR results showed that the level of circARHGAP5 was significantly decreased in stable shcircARHGAP5 CSCC cells (Figure 2A), while neither of the shRNAs had an effect on the linear ARHGAP5 mRNA (Figure 2B). We then used the CCK‐8 assay with gradient concentration of CP and colony formation with the IC50 concentration of the drug in two CSCC cell lines to detect cell viability following circARHGAP5 knockdown. Results showed that knockdown of circARHGAP5 led to an increase in cell viability of CSCC cells in the context of CP (Figure 2C,D). As apoptosis is one of the indicators of CP sensitivity, we also assessed the effect of circARHGAP5 knockdown on apoptosis of CSCC cells with IC50 concentration of CP. Flow cytometry analysis showed that the apoptotic rates of CSCC cells in sh‐circARHGAP5 groups were significantly lower than the control group cells in both ME‐180 and SiHa cells (Figure 2E). These phenomena are also consistent in HeLa cells (Figure S2A–D). Subsequently, in vitro studies showed that inhibition of circARHGAP5 significantly promoted proliferation and suppressed apoptosis in CSCC cells, which indicated that knockdown of circARHGAP5 increased CP resistance.
FIGURE 2.
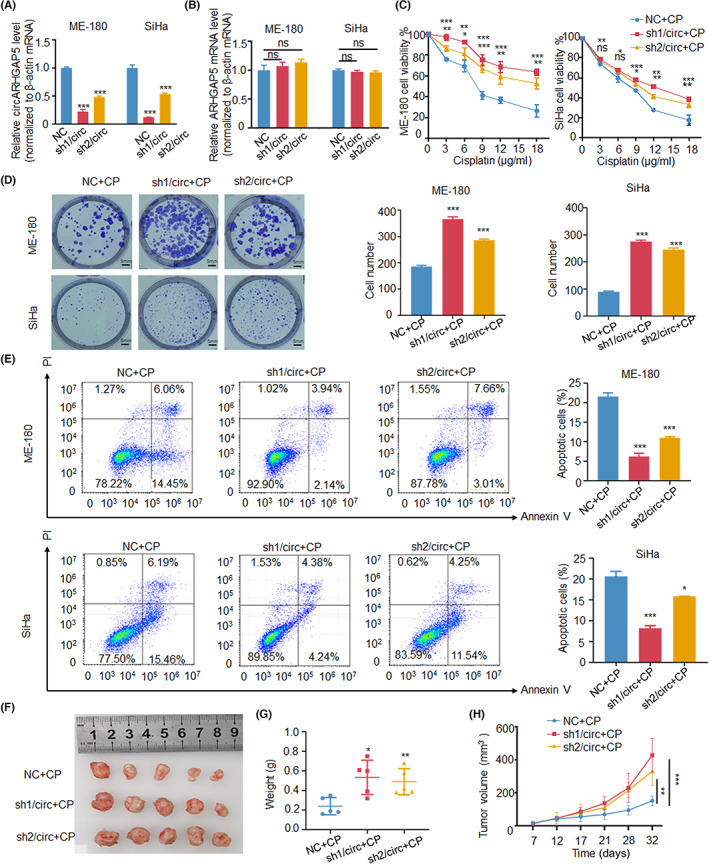
Knockdown of circular RNA ARHGAP5 (circARHGAP5) promoted cervical squamous cell carcinoma drug resistance in vivo and in vitro. (A) Knockdown efficiency of circARHGAP5 shRNAs in SiHa/NC with SiHa/shcircARHGAP5s and ME‐180/NC with ME‐180/shcircARHGAP5s stable cells. (B) Relative change levels of ARHGAP5 in the above two groups of cells. (C) CCK‐8 assays with gradient concentration of cisplatin (CP) in the above two groups of cells. (D, E) Colony formation and apoptosis assays with IC50 concentration of drug cisplatin in the two groups of cells. IC50 for SiHa is 8 μg/ml, and 9 μg/ml for ME‐180. (F) Representative pictures of tumors. (G, H) Weight and volume of the xenograft tumors in the ME‐180/Con and ME180/shcircARHGAP5s cells after 15 days of injection of cisplatin. *p < 0.05, **p < 0.01, ***p < 0.001. Con, control; NC, negative control; ns, not significant; PI, propidium iodide.
Moreover, in vivo experiments showed that decreased levels of circARHGAP5 could markedly promote CP resistance of CSCC. Cisplatin was injected into subcutaneous tumor tissues of nude mice with stable ME‐180/shNC and ME‐180/shcircARHGAP5 cells. The tumor tissues in the knockdown group had heavier weight and larger volume than the control group (Figure 2F–H). These data suggested that knockdown of circARHGAP5 increased the CP resistance with the progression of CSCC.
In addition, we also verified the conclusion that circARHGAP5 could inhibit CP resistance with the progression of CSCC by constructing overexpression plasmid of circARHGAP5 among SiHa and ME‐180 cells. The qRT‐PCR results showed the overexpression efficiency of circARHGAP5 in overexpressed circARHGAP5 CSCC cells (Figure 3A), but no effect on the linear ARHGAP5 mRNA (Figure 3B). The CCK‐8 assay with gradient concentration of CP was also used in two CSCC cell lines to detect cell viability following circARHGAP5 overexpression. Results showed that overexpression of circARHGAP5 led to a decrease in cell viability of CSCC cells in the context of CP (Figure 3C). Flow cytometry analysis showed that the apoptotic rates of CSCC cells in overexpressed‐circARHGAP5 groups were significantly higher than the control group cells in both ME‐180 and SiHa cells with CP (Figure 3D). To sum up, results showed that overexpression of circARHGAP5 significantly suppressed proliferation and promoted apoptosis in CSCC cells, which indicated that overexpression of circARHGAP5 inhibited CP resistance. These phenomena were also consistent in HeLa cells (Figure S2E–H). In conclusion, we proved that circARHGAP5 could inhibit CP resistance in CSCC cells. We also detected that circARHGAP5 affected cell viability of CSCC without CP; results showed that circARHGAP5 itself might also function as a tumor suppressor of cervical cancer cell proliferation (Figure S3).
FIGURE 3.
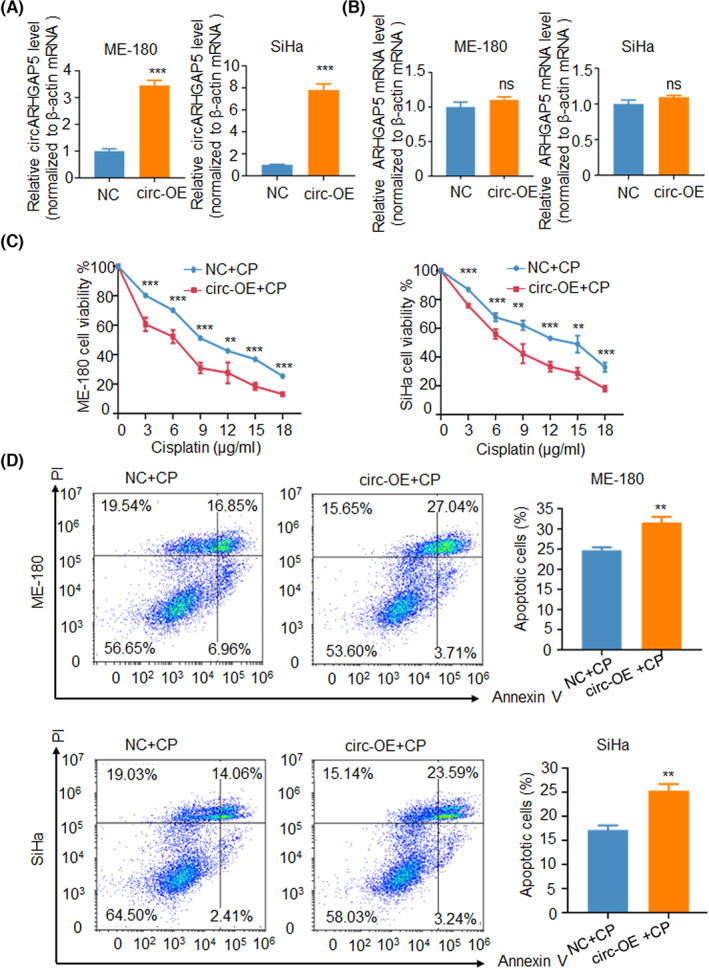
Overexpression (OE) of circular RNA ARHGAP5 (circARHGAP5) inhibited cervical squamous cell carcinoma drug resistance. (A) Overexpression efficiency of circARHGAP5 in ME‐180/NC and ME‐180/OE‐circARHGAP5 cells, and SiHa/NC and SiHa/OE‐circARHGAP5 cells. (B) Relative change in ARHGAP5 levels in NC/circ‐OE ME‐180 and SiHa cells. (C) CCK‐8 assay with gradient concentration of cisplatin (CP) in NC/circ‐OE ME‐180 cells and SiHa cells. (D) Apoptosis assays with IC50 concentration of drug cisplatin in NC/circ‐OE ME‐180 cells and SiHa cells. IC50 for SiHa is 8 μg/ml, and 9 μg/ml for ME‐180. *p < 0.05, **p < 0.01, ***p < 0.001. NC, negative control; ns, not significant; PI, propidium iodide.
3.4. Circular RNA ARHGAP5 interacted with AUF1 in CSCC
Circular RNAs exert their functions primarily through four mechanisms: miRNA sponges, protein decoys/scaffolds, transcriptional regulators, and translation templates. 19 To investigate the underlying functional mechanism of circARHGAP5, we first explored whether circARHGAP5 could act as a miRNA sponge at first due to its cytoplasmic localization. It has been demonstrated that circRNAs serving as miRNA sponges can form a circRNA‐AGO2‐miRNA complex, hence we undertook the AGO2 RIP, 26 , 27 , 28 which showed that circARHGAP5 was not significantly enriched by AGO2 compared with control, suggesting that circARHGAP5 might not serve as a miRNA sponge (Figure 4A). To explore whether circARHGAP5 exerted functions through interactions with RBP, we used the RNA pull‐down assay with circARHGAP5‐specific probes to identify circARHGAP5‐associated proteins (Figure 4B). The qRT‐PCR analysis confirmed that the antisense probe for circARHGAP5 could enrich circARHGAP5, and then we detected a specific band through silver staining of western blot gels, followed by mass spectrometry for the specific band (Figure 4C,D). Among the top predicted protein candidates, AUF1 was the only protein that was validated to interact with circARHGAP5 (Figure 4E; Table S4). We then confirmed this result using AUF1 RIP and further validated that AUF1 could interact with circARHGAP5 (Figure 4F). Notably, circARHGAP5 could interact with AUF1 in SiHa cells as well (Figure 4G,H). In addition, FISH‐immunofluorescence analysis in ME‐180 and SiHa cell lines showed that circARHGAP5 colocalized with AUF1 in the cytoplasm (Figure 4I). These data revealed that circARHGAP5 could physically interact with AUF1 in cytoplasm.
FIGURE 4.
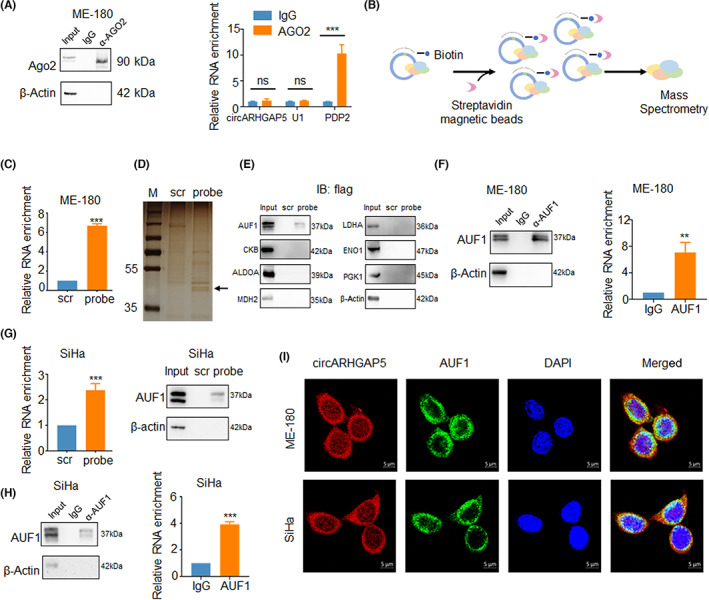
Circular RNA ARHGAP5 (circARHGAP5) interacted with AUF1 in cervical squamous cell carcinoma. (A) Ago2 RIP for detection of circARHGAP5 in ME‐180 cells. PDP2 acted as a positive control, while U1 was a negative control. (B) Model of RNA pull‐down assay. (C) Enrichment of circARHGAP5 of RNA pull‐down in ME‐180 cells. (D) Silver staining of proteins binding with circARHGAP5. (E) Western blot of RNA pull‐down for detecting the binding protein of top enrichment proteins of mass spectrometry. (F–H) RNA pull‐down and RIP efficiency of AUF1 protein and enrichment of circARHGAP5 in ME‐180 and SiHa cells. Protein efficiency was validated through western blot, RNA enrichment was validated by quantitative RT‐PCR. (I) FISH of circARHGAP5 (red) along with immunofluorescence staining (IF) of AUF1 (green) in ME‐180 and SiHa cells. IF/FISH assay showed that circARHGAP5 was colocated with AUF1 in ME‐180 and SiHa cells. Scale bar, 5 μm. **p < 0.01, ***p < 0.001. IB, immunoblot; M, marker; ns, not significant; scr, scramble.
As an RBP, AUF1 comprises a family of proteins consisting of four related isoforms including p37, p40, p42, and p45, containing two tandem RRMs. 29 , 30 Additionally, AUF1 could recognize ARE sequences within different transcripts and control their stability. 31 To investigate which of the motifs is responsible for the circRNA–protein interaction, we designed different truncations of AUF1 (Figure 5A,B). RNA pull‐down results showed that the binding sites of circARHGAP5 to AUF1 were RRM1 and RRM2 of isoforms of p37 (Figure 5C). In summary, circARHGAP5 bound with the p37 isoform of AUF1 through the binding sites RRM1 and RRM2.
FIGURE 5.
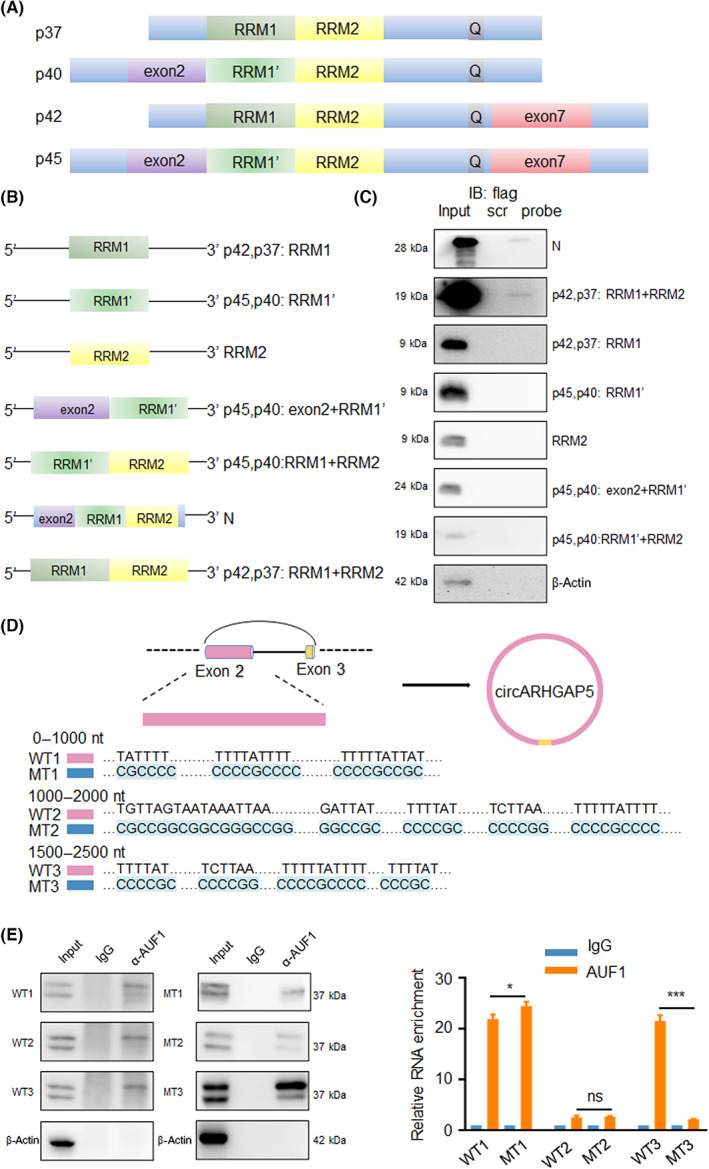
Circular RNA ARHGAP5 (circARHGAP5) bound to RNA recognition motif 1 (RRM1) and RRM2 of AUF1 in cervical squamous cell carcinoma. (A) Structure of AUF1 with four isoforms. (B) Model of truncations of AUF1. (C) Western blot of RNA pull‐down assay for detecting the truncations of AUF1 binding to circARHGAP5 in ME‐180 cells. (D) Model of circARHGAP5 truncations. (E) RIP efficiency of AUF1 protein and enrichment of circARHGAP5 (WTs and mutants [MTs]) in ME‐180 cells. Protein efficiency was validated through western blot, RNA enrichment was validated by quantitative RT‐PCR. *p < 0.05, ***p < 0.001. ns, not significant
To continue to ascertain AUF1 binding sites of circARHGAP5, we then predicted the binding sites of circARHGAP5 and AUF1 (rbpmap.technion.ac.il). We input convergent and divergent sequences of circARHGAP5 into the website and there were 15 potential binding sites of circARHGAP5 and AUF1 all located at exon 2 (z‐score > 2.5) (Table S5). As the sequence of circARHGAP5 is too long and most of the potential binding sites were focused on exon 2, we designed circARHGAP5 into three linear truncations as 1–1000 nt (wt1 and mt1), 1000–2000 nt (wt2 and mt2), and 1500–2500 nt (wt3 and mt3) of exon 2 (Figure 5D,E). The RNA‐immunoprecipitation showed that AUF1 binding sites of circARHGAP5 might be located in the 1500–2500 nt sequence of exon 2 (Figure 5E).
3.5. AUF1 inhibited CSCC drug resistance in vitro
To further investigate the functions of AUF1 in CSCC, we constructed overexpression vector of AUF1 in ME‐180 and SiHa cells. The qRT‐PCR and western blot results showed that the expression of AUF1 was significantly increased when transfected with overexpression vector (Figure 6A,B). The CCK‐8 assay with gradient concentration of CP revealed that overexpression of AUF1 significantly increased cell viability in ME‐180 and SiHa cells (Figure 6C,D). Flow cytometry analysis of apoptosis with IC50 concentration of CP also showed that overexpression of AUF1 led to decrease of apoptosis in CSCC cells (Figure 6E). Taken together, these results suggested that overexpression of AUF1 might promote proliferation and inhibit apoptosis of CSCC cells in the context of CP treatment.
FIGURE 6.
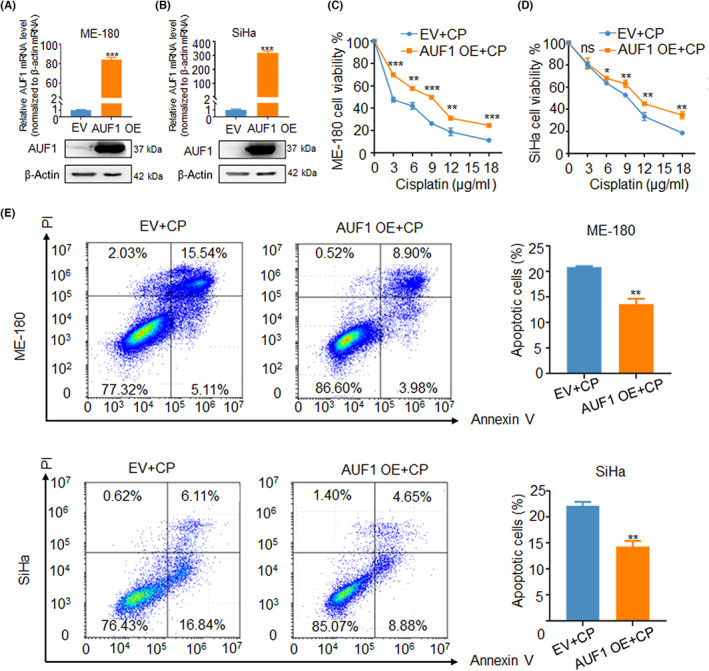
AUF1 inhibited cervical squamous cell carcinoma drug resistance in vitro. (A, B) Overexpression (OE) efficiency of AUF1 upon mRNA and protein levels in ME‐180 and SiHa cells. (C, D) CCK‐8 assay with gradient concentration of cisplatin (CP) in AUF1/EV and AUF1/OE upon ME‐180 and SiHa cells. (E) Apoptosis assay with IC50 concentration of drug cisplatin in the two groups of cells. IC50 for SiHa is 8 μg/ml, and 9 μg/ml for ME‐180. *p < 0.05, **p < 0.01, ***p < 0.001. EV, empty vector; ns, not significant; PI, propidium iodide.
3.6. Circular RNA ARHGAP5 compromised AUF1 degradation of BIM mRNA to promote cell apoptosis
In order to further explore the regulatory functions between circARHGAP5 and AUF1, we first tested whether changing circARHGAP5 levels might alter AUF1 levels. Results showed that knockdown of circARHGAP5 or overexpression of AUF1 did not significantly affect mRNA levels of AUF1 or the level of circARHGAP5 in CSCC cells (Figure 7A,B). As AUF1 is predominantly known to promote mRNA decay of genes, 32 we suspected that interaction of circARHGAP5–AUF1 could influence AUF1‐mediated downstream mRNA decay. Due to the functions of both circARHGAP5 and AUF1 in cell apoptosis in CSCC stated above, we wondered whether the downstream gene of AUF1 was related to apoptotic. We selected nine of the most common apoptotic regulators. 33 , 34 We thus detected the level of several genes related to cell apoptosis upon AUF1 overexpression in ME‐180 and SiHa cells. The qRT‐PCR results showed that BIM was the only target that was negatively related with AUF1 in both ME‐180 and SiHa cells with the consistent trend, which suggested that BIM might be a downstream target of AUF1 (Figure 7C,D).
FIGURE 7.
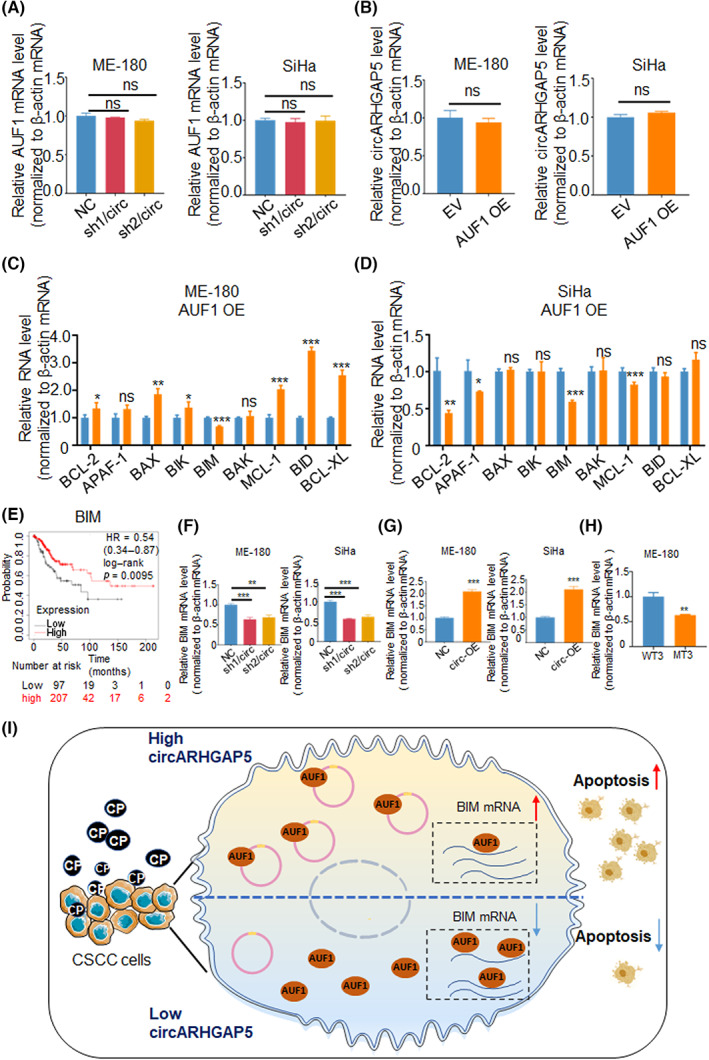
Circular RNA ARHGAP5 (circARHGAP5) compromised AUF1 degradation of BIM mRNA to promote cell apoptosis. (A) Relative AUF1 mRNA in NC and shcircARHGAP5 stable cells upon ME‐180 and SiHa cervical squamous cell carcinoma (CSCC) cells. (B) Relative circARHGAP5 levels in AUF1/EV and AUF1/overexpressed (OE) upon ME‐180 and SiHa cells.(C, D) Relative mRNA levels of several apoptosis‐related genes in AUF1/EV and AUF1/OE upon ME‐180 and SiHa cells. (E) Survival analysis of BIM in CSCC from Kaplan–Meier Plotter. (F) Relative BIM mRNA in the above NC and shcircARHGAP5 stable cells upon ME‐180 and SiHa cells. (G) Quantitative RT‐PCR showed relative BIM mRNA after circARHGAP5 overexpression in ME‐180 and SiHa cells. (H) Relative BIM mRNA in circARHGAP5‐mutated (MT) ME‐180 cells. (H) Proposed mechanism of circARHGAP5‐regulated cisplatin (CP) resistance. CircARHGAP5 derived from exons 2–3 of ARHGAP5, mainly located in cytoplasm, and then work as a sponge for AUF1 protein, which resulted in the decreased number of “free” AUF1 proteins that bind to BIM mRNAs to promote cell apoptosis in cisplatin resistance. *p < 0.05, **p < 0.01, ***p < 0.001. EV, empty vector; NC, negative control; ns, not significant.
As AUF1 can regulate mRNA decay of target gene, public cross‐linking and immunoprecipitation high throughput sequencing (CLIP‐seq)data that revealed the bindings of BIM mRNA to AUF1 proteins (Table S6). 35 We suspected that circARHGAP5 might affect AUF1 binding to BIM mRNA. First, we found that high expression levels of BIM mRNA predicted better prognosis in CSCC (Figure 7E). In addition, we examined the correlation between circARHGAP5 and BIM mRNA. Results showed that knockdown of circARHGAP5 suppressed the mRNA level of BIM in both ME‐180 and SiHa cells, whereas overexpression of circARHGAP5 promoted the BIM mRNA level (Figure 7F,G). Additionally, qRT‐PCR showed that the BIM mRNA level was lower when 1500–2500 nt of exon 2 (AUF1 binding sites) of circARHGAP5 was deleted, which might suggest that circARHGAP5 did not bind to AUF1 at that time and that “free” AUF1 increased, which may bind to BIM, finally led to decrease of BIM (Figure 7H). Taken together, these data suggested that BIM was positively related with circARHGAP5 and negatively with AUF1 in the progression of CSCC, which suggested that BIM might act as the downstream target for interaction of circARHGAP5–AUF1. To sum up the above results, we made a potential working model that showed that interaction between circARHGAP5 and AUF1 would reduce the degradation of BIM and release BIM to increase CP‐mediated cell apoptosis in CSCC cells (Figure 7I).
4. DISCUSSION
Patients with recurrent, progressive, and metastatic cervical cancer have an obviously poor overall prognosis once resistant to CP. 36 Research on circRNA in CP resistance has attracted increasing attention in recent years. 37 In this study, we found that circARHGAP5, a direct RNA target positively regulated by ΔNp63α, contributed to inducing platinum‐mediated apoptosis. We have also identified that circARHGAP5 could interact with AUF1, and overexpression of AUF1 could also inhibit the CP‐mediated cell apoptosis. Furthermore, BIM was one of the AUF1 potential targets, and BIM mRNA might serve as a downstream target of AUF1. Therefore, we first revealed that circARHGAP5 regulated by ΔNp63α could inhibit CP resistance in CSCC through AUF1‐dependent degradation of pro‐apoptotic BIM mRNA.
It is worth noting that multiple circRNAs can be generated from the same host gene as different isoforms, and in some cases, the biogenesis of each circRNA, even derived from the same locus, is independently regulated in different tissues with distinct functions. 38 Circular RNA ARHGAP5 was first reported to function in adipocyte metabolism. 38 There were two circular isoforms of circARHGAP5, named circARHGAP5‐1 and circARHGAP5‐2, arising from different exons, and the sequence of circARHGAP5‐1 in a previous study concurs with circARHGAP5 identified in this context. However, the molecular roles of circARHGAP5s regulating adipogenesis are still poorly understood. In our analysis, we explored one circular isoform of circARHGAP5 in CSCC and found it is involved in CSCC CP resistance by interacting with RNA‐binding proteins as AUF1. Therefore, we have expanded the functions of circARHGAP5 in disease and revealed the mechanism of circARHGAP5.
Recent evidence indicates that circRNAs were involved in tumorigenesis, and abnormal expression of circRNAs could led to progression of disease. 26 , 39 , 40 , 41 Platinum‐based chemoradiotherapy has been used in patients with cervical cancer as an addition to surgery to improve their outcome. 42 , 43 Cisplatin can induce cell apoptosis by multiple intertwined signaling pathways. 44 , 45 , 46 In chemoresistance of cervical cancer, circRNAs could serve as future therapeutic biomarkers. 47 There is growing evidence that circRNAs can regulate the chemosensitivity of chemotherapy with CP in cervical cancer. For example, studies have shown that circMTO1 could promote cervical cancer cell tumor progression and CP resistance through sponging miR‐6893, and hsa_circ_0023404 inhibits autophagy‐mediated cell death to confer chemoresistance of cervical cancer. 48 , 49 In this context, we first determined that circARHGAP5 is regulated by ΔNp63α, which could exert CP resistance in CSCC, and decreased circARHGAP5 in CSCC cells was antiapoptotic in CP resistance, which could provide cells with time for DNA repair and homeostasis re‐establishment.
AUF1 is among the first ARE RNA‐binding proteins to be purified, cloned, and studied for its complex regulation of mRNA targets. 50 , 51 AUF1 can bind to many ARE mRNAs like cyclin D1 and p21 and assembles some factors to recruit the mRNA degradation machinery. 32 AUF1 has also been shown to play a vital and complex role in cancer progression and chemoresistance. 52 , 53 , 54 For example, AUF1 can enhance the chemotherapeutic drug resistance of hepatoma cells and among breast cancer patients. 55 , 56 However, the functions and mechanisms of AUF1 in the development and progression of CSCC are still obscure. In this study, we proved that AUF1 could interact with circARHGAP5 and overexpression of AUF1 could suppress CP‐mediated apoptosis of CSCC.
In our study, results showed that circARHGAP5 and AUF1 did not have a direct effect on each other. Therefore, we hypothesized that circARHGAP5 might directly influence the binding of AUF1 and its mRNA targets. As the biological functions of circARHGAP5 in CSCC drug resistance led to cell apoptosis, we found that BIM was positively related with circARHGAP5, but negatively with AUF1. BIM is a protein that belongs to the Bcl‐2 family, which is a well‐known pro‐apoptotic molecule. 57 , 58 BIM can initiate the intrinsic apoptotic pathway and its overexpression inhibited tumor growth and drug resistance. 59 Previous studies have shown that BIM can sensitize some kinds of cancer to drugs, and also in CSCC, high BIM expression is a potential prognostic marker as well as a chemotherapeutic target. 60 , 61 In this study, we found that AUF1 might play a critical role in altering BIM mRNA stability in apoptosis. In addition, we suspected that circARHGAP5 directly bound AUF1 and prevented AUF1 from interacting with BIM mRNA, enabling decreases in BIM mRNA, and in turn inhibiting apoptosis in drug resistance.
However, there are some limitations in our study. We need more experiments to expand the knowledge of CP resistance and additional studies will be necessary to deepen our knowledge of the target gene of AUF1.
In conclusion, we identified and characterized circARHGAP5 as an essential circRNA target regulated by ΔNp63α that was downregulated in CP‐resistant tissues. Circular RNA ARHGAP5 might work as a sponge for AUF1 protein, which could result in decreased numbers of AUF1 proteins that bind to BIM mRNAs to promote cell apoptosis in CP resistance, while the number of AUF1 proteins would not decrease. Thus, our findings provide an insight into understanding the progression of CSCC drug resistance, and circARHGAP5 can be used as a diagnostic biomarker and potential therapeutic approach in CSCC CP resistance.
AUTHOR CONTRIBUTIONS
YZ and LC supported and supervised the study. SSD and LLQ wrote the manuscript. LWL collected the CSCC tissues. SSD, LLQ, and HYL performed all the experimental validation assays. ZHX, YJL, and YYW analyzed data. LLQ, SSD, and LWL performed the animal experiments. All authors discussed the results and commented on the manuscript.
ACKNOWLEDGEMENTS
We would like to thank Lab Ge Shan (University of Science and Technology of China) for technical assistance.
FUNDING INFORMATION
This work was supported by the National Natural Science Foundation of China (81902632, 32270590 and 82172773); the National Key R&D Program of China (2022YFC2403400 and 2019YFA0802600), Novel Medicine Funding of USTC (WK9110000104); Anhui Provincial Key Research and Development Projects (2022e07020013); and Beijing Science and Technology Innovation Medical Development Foundation (KC2021‐JX‐0186‐143).
CONFLICT OF INTEREST
The authors have no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an institutional review board: This study was conducted with the approval from the Institutional Review Board of the first Affiliated Hospital of USTC (approval no. 2022‐ky094).
Informed consent: The present study was conducted in accordance with the principles of the Declaration of Helsinki and written informed consent about the therapy and participation in the research work was obtained from all patients before surgery.
Registry and the registration no. of the study/trial: N/A.
Animal studies: The use of mice was approved by the Animal Care and Use Committee of the USTC University (USTCACUC1801017).
Supporting information
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.
Figure S1.
Figure S2.
Figure S3.
Deng S, Qian L, Liu L, et al. Circular RNA ARHGAP5 inhibits cisplatin resistance in cervical squamous cell carcinoma by interacting with AUF1 . Cancer Sci. 2023;114:1582‐1595. doi: 10.1111/cas.15723
Sisi Deng and Lili Qian contributed equally to this work and share first authorship.
Contributor Information
Liang Chen, Email: anqingcl@ustc.edu.cn.
Ying Zhou, Email: caddiezy@ustc.edu.cn.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Small W, Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer. 2017;123(13):2404‐2412. [DOI] [PubMed] [Google Scholar]
- 3. Das M. WHO launches strategy to accelerate elimination of cervical cancer. Lancet Oncol. 2021;22(1):20‐21. [DOI] [PubMed] [Google Scholar]
- 4. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169‐182. [DOI] [PubMed] [Google Scholar]
- 5. Lei J, Ploner A, Elfstrom KM, et al. HPV vaccination and the risk of invasive cervical cancer. N Engl J Med. 2020;383(14):1340‐1348. [DOI] [PubMed] [Google Scholar]
- 6. Lemp JM, De Neve JW, Bussmann H, et al. Lifetime prevalence of cervical cancer screening in 55 low‐ and middle‐income countries. JAMA. 2020;324(15):1532‐1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruni L, Diaz M, Barrionuevo‐Rosas L, et al. Global estimates of human papillomavirus vaccination coverage by region and income level: a pooled analysis. Lancet Glob Health. 2016;4(7):e453‐e463. [DOI] [PubMed] [Google Scholar]
- 8. Oberlin AM, Rahangdale L, Chinula L, Fuseini NM, Chibwesha CJ. Making HPV vaccination available to girls everywhere. Int J Gynaecol Obstet. 2018;143(3):267‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amable L. Cisplatin resistance and opportunities for precision medicine. Pharmacol Res. 2016;106:27‐36. [DOI] [PubMed] [Google Scholar]
- 10. Ghosh S. Cisplatin: the first metal based anticancer drug. Bioorg Chem. 2019;88:102925. [DOI] [PubMed] [Google Scholar]
- 11. Mangiulli M, Valletti A, Caratozzolo MF, et al. Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 2009;37(18):6092‐6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G, Knight R. p63 in epithelial development. Cell Mol Life Sci. 2008;65(20):3126‐3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moses MA, George AL, Sakakibara N, et al. Molecular mechanisms of p63‐mediated squamous cancer pathogenesis. Int J Mol Sci. 2019;20(14):3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Y, Liu H, Wang J, et al. DeltaNp63alpha exerts antitumor functions in cervical squamous cell carcinoma. Oncogene. 2020;39(4):905‐921. [DOI] [PubMed] [Google Scholar]
- 15. Liu H, Zhu C, Xu Z, et al. lncRNA PART1 and MIR17HG as DeltaNp63alpha direct targets regulate tumor progression of cervical squamous cell carcinoma. Cancer Sci. 2020;111(11):4129‐4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869‐881.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428‐442. [DOI] [PubMed] [Google Scholar]
- 18. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675‐691. [DOI] [PubMed] [Google Scholar]
- 19. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475‐490. [DOI] [PubMed] [Google Scholar]
- 20. Suzuki H, Tsukahara T. A view of pre‐mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331‐9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49‐57. [DOI] [PubMed] [Google Scholar]
- 23. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333‐338. [DOI] [PubMed] [Google Scholar]
- 24. Zheng SR, Zhang HR, Zhang ZF, et al. Human papillomavirus 16 E7 oncoprotein alters the expression profiles of circular RNAs in Caski cells. J Cancer. 2018;9(20):3755‐3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu H, Luo H, Zhang W, et al. Molecular mechanisms of cisplatin resistance in cervical cancer. Drug des Devel Ther. 2016;10:1885‐1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang G, Ling Y, Mehrpour M, et al. Autophagy‐associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non‐small cell lung cancer progression via destabilizing IGF2BPs and activating anti‐tumor immunity. Nat Commun. 2021;12(1):295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384‐388. [DOI] [PubMed] [Google Scholar]
- 29. Choi YJ, Yoon JH, Chang JH. Crystal structure of the N‐terminal RNA recognition motif of mRNA decay regulator AUF1. Biomed Res Int. 2016;2016:3286191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moore AE, Chenette DM, Larkin LC, Schneider RJ. Physiological networks and disease functions of RNA‐binding protein AUF1. Wiley Interdiscip Rev RNA. 2014;5(4):549‐564. [DOI] [PubMed] [Google Scholar]
- 31. White EJ, Matsangos AE, Wilson GM. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA. 2017;8(2):e1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1(3):457‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carneiro BA, EL‐Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17(7):395‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Galluzzi L, Vitale I, Abrams JM, et al. Molecular definitions of cell death subroutines: recommendations of the nomenclature committee on cell death 2012. Cell Death Differ. 2011;19(1):107‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoon JH, De S, Srikantan S, et al. PAR‐CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cohen AC, Roane BM, Leath CA 3rd. Novel therapeutics for recurrent cervical cancer: moving towards personalized therapy. Drugs. 2020;80(3):217‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mu Q, Lv Y, Luo C, et al. Research Progress on the functions and mechanism of circRNA in cisplatin resistance in tumors. Front Pharmacol. 2021;12:709324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arcinas C, Tan W, Fang W, et al. Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat Metab. 2019;1(7):688‐703. [DOI] [PubMed] [Google Scholar]
- 39. Liu J, Li D, Luo H, Zhu X. Circular RNAs: the star molecules in cancer. Mol Aspects Med. 2019;70:141‐152. [DOI] [PubMed] [Google Scholar]
- 40. Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng X, Huang M, Xing L, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple‐negative breast cancer. Mol Cancer. 2020;19(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liontos M, Kyriazoglou A, Dimitriadis I, Dimopoulos MA, Bamias A. Systemic therapy in cervical cancer: 30 years in review. Crit Rev Oncol Hematol. 2019;137:9‐17. [DOI] [PubMed] [Google Scholar]
- 43. Masadah R, Rauf S, Pratama MY, et al. The role of microRNAs in the cisplatin‐ and radio‐resistance of cervical cancer. Cancer. 2021;13(5):1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lorusso D, Petrelli F, Coinu A, Raspagliesi F, Barni S. A systematic review comparing cisplatin and carboplatin plus paclitaxel‐based chemotherapy for recurrent or metastatic cervical cancer. Gynecol Oncol. 2014;133(1):117‐123. [DOI] [PubMed] [Google Scholar]
- 45. Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31(15):1869‐1883. [DOI] [PubMed] [Google Scholar]
- 46. Skowron MA, Melnikova M, Van Roermund JG, et al. Multifaceted mechanisms of cisplatin resistance in Long‐term treated urothelial carcinoma cell lines. Int J Mol Sci. 2018;19(2):590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wen X, Liu S, Sheng J, Cui M. Recent advances in the contribution of noncoding RNAs to cisplatin resistance in cervical cancer. PeerJ. 2020;8:e9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen M, Ai G, Zhou J, Mao W, Li H, Guo J. circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR‐6893. Biomed Pharmacother. 2019;117:109064. [DOI] [PubMed] [Google Scholar]
- 49. Guo J, Chen M, Ai G, Mao W, Li H, Zhou J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR‐5047. Biomed Pharmacother. 2019;115:108957. [DOI] [PubMed] [Google Scholar]
- 50. Brewer G. An a + U‐rich element RNA‐binding factor regulates c‐myc mRNA stability in vitro. Mol Cell Biol. 1991;11(5):2460‐2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang W, Wagner BJ, Ehrenman K, et al. Purification, characterization, and cDNA cloning of an AU‐rich element RNA‐binding protein, AUF1. Mol Cell Biol. 1993;13(12):7652‐7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113‐126. [DOI] [PubMed] [Google Scholar]
- 53. Cheadle C, Fan J, Cho‐Chung YS, et al. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gao Y, Wang W, Cao J, et al. Upregulation of AUF1 is involved in the proliferation of esophageal squamous cell carcinoma through GCH1. Int J Oncol. 2016;49(5):2001‐2010. [DOI] [PubMed] [Google Scholar]
- 55. Zhang T, Guan G, Zhang J, et al. E2F1‐mediated AUF1 upregulation promotes HCC development and enhances drug resistance via stabilization of AKR1B10. Cancer Sci. 2022;113(4):1154‐1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Al‐Tweigeri T, AlRaouji NN, Tulbah A, et al. High AUF1 level in stromal fibroblasts promotes carcinogenesis and chemoresistance and predicts unfavorable prognosis among locally advanced breast cancer patients. Breast Cancer Res. 2022;24(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kelly PN, White MJ, Goschnick MW, et al. Individual and overlapping roles of BH3‐only proteins Bim and bad in apoptosis of lymphocytes and platelets and in suppression of thymic lymphoma development. Cell Death Differ. 2010;17(10):1655‐1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ouyang P, An W, Chen R, et al. IL‐37 promotes cell apoptosis in cervical cancer involving Bim upregulation. Onco Targets Ther. 2019;12:2703‐2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shukla S, Saxena S, Singh BK, Kakkar P. BH3‐only protein BIM: An emerging target in chemotherapy. Eur J Cell Biol. 2017;96(8):728‐738. [DOI] [PubMed] [Google Scholar]
- 60. Kim BW, Cho H, Ylaya K, et al. Bcl‐2‐like protein 11 (BIM) expression is associated with favorable prognosis for patients with cervical cancer. Anticancer Res. 2017;37(9):4873‐4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Florent R, Weiswald LB, Lambert B, et al. Bim, puma and Noxa upregulation by Naftopidil sensitizes ovarian cancer to the BH3‐mimetic ABT‐737 and the MEK inhibitor Trametinib. Cell Death Dis. 2020;11(5):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.
Figure S1.
Figure S2.
Figure S3.


