This work provides a compendium of early transcriptional responses to “common-gamma-chain” cytokines across all major immunocyte lineages of the mouse.
Abstract
“γc” cytokines are a family whose receptors share a “common-gamma-chain” signaling moiety, and play central roles in differentiation, homeostasis, and communications of all immunocyte lineages. As a resource to better understand their range and specificity of action, we profiled by RNAseq the immediate-early responses to the main γc cytokines across all immunocyte lineages. The results reveal an unprecedented landscape: broader, with extensive overlap between cytokines (one cytokine doing in one cell what another does elsewhere) and essentially no effects unique to any one cytokine. Responses include a major downregulation component and a broad Myc-controlled resetting of biosynthetic and metabolic pathways. Various mechanisms appear involved: fast transcriptional activation, chromatin remodeling, and mRNA destabilization. Other surprises were uncovered: IL2 effects in mast cells, shifts between follicular and marginal zone B cells, paradoxical and cell-specific cross-talk between interferon and γc signatures, or an NKT-like program induced by IL21 in CD8+ T cells.
Introduction
The immunologists and computational biologists of the Immunological Genome Project aim to generate an exhaustive definition of gene expression and regulatory networks of the mouse immune system. Here, we charted the primary transcriptional changes that result, in the major cell types of the immune system, from in vivo exposure to cytokines of the “common-gamma-chain” (γc) family.
γc cytokines play a fundamental role in the life cycle of immunocytes, ranging from early differentiation of bone marrow or thymic precursors to homeostatic control of peripheral pools, and to effector differentiation (Leonard et al., 2019; Barata et al., 2019; Spangler et al., 2015a; Ross and Cantrell, 2018; Goswami and Kaplan, 2011; Malek, 2008; Ma et al., 2006; Lin and Leonard, 2018). As a consequence, there is great interest in their therapeutic potential, with dozens of trials underway (Wolfarth et al., 2022). γc cytokines are usually considered, in part because of the circumstances of their discovery (Cosman et al., 1990; Leonard et al., 2019), to each have particular roles and “personalities”: IL4 was originally described as a B cell help factor produced by activated T helper 2 cells, which also induces Type2 differentiation during allergic states; IL2 was the first T cell–derived growth factor, later found to orchestrate the feedback loop involving T regulatory (Treg) cells; IL7 is a growth and differentiation factor for early lymphoid progenitors in the thymus and bone marrow; IL15 is another T and natural killer (NK) cell growth factor like IL2, but preferentially acting on cytotoxic cells. But beyond these overly simplistic core identities, each cytokine was later found to have more pleiotropic roles and range of action across wider varieties of cells, even outside the immune system (Leonard et al., 2019). In addition, there are paradoxical observations that do not fit the simple scenarios, for instance, NK cells respond to IL4 by making IFNγ (Morris et al., 2006) and ILCs make more IL2 transcript than CD4+ T cells.
These pleiotropic functions are channeled through a surprisingly common path (Lin and Leonard, 2019; Ross and Cantrell, 2018). Signaling by exogenous γc cytokines is routed through complexes composed of specific (“private”) receptor subunits, harnessing the γc protein subunit for signaling (Fig. 1 A). The latter is the primary signaling component of the complex, and upon cytokine binding activates receptor-associated JAK kinases. These then phosphorylate docking sites for signal transducers of the STAT family, which in turn translocate to the nucleus where they activate the transcription of target genes. There is some specificity in the relationships between γc cytokines and the JAK and STAT family members that transduce their signals, but it is not monomorphic: Several STATs can be activated by any one of the γc cytokines and there is overlap between them (Fig. 1 A). In parallel to STAT activation, γc cytokines trigger a number of other signaling pathways (MAPK, mTOR), inducing widespread changes in the phosphoproteome (Ross et al., 2016), in part because the cytoplasmic tails of the private receptor chains have some specific signaling capabilities (Ross and Cantrell, 2018; Wills-Karp and Finkelman, 2008).
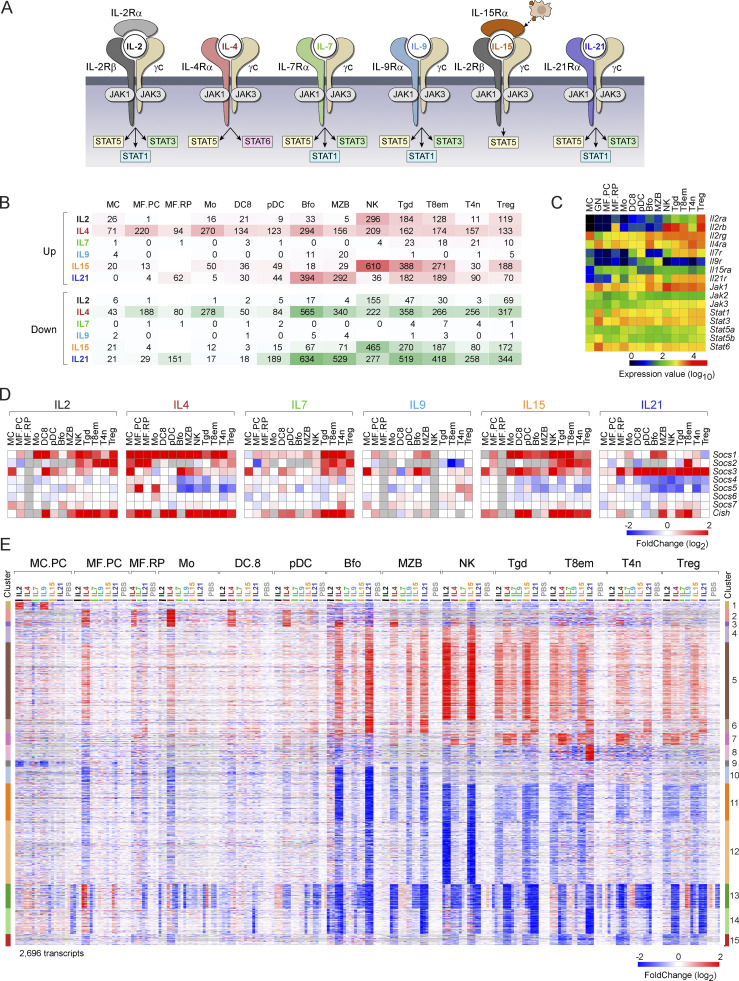
Figure 1.
Overview of early responses to γc cytokines. (A) A summary of the receptor and primary signal transducers for the γc cytokines used here (from Leonard et al., 2019). (B) Tally of the number of genes up- or downregulated (at arbitrary FC threshold of 2), 2 h after administration of indicated cytokines. See Table S2 for acronyms. (C) Expression of key receptors and signal transducers in the profiled cell-types (DEseq2 normalized values). (D) Induction of cytokine signal downregulators of the SOCS family (as FC relative to the mean of PBS controls in the matched experiments); gray cells: insufficient data. (E) Complete overview of 2,696 genes that significantly change in response to one or more γc cytokines, across all cell types. Activated and repressed clusters at right.
Although some studies have examined the transcriptional consequences of signals received from individual cytokines (Fontenot et al., 2005; Moro et al., 2022), there is no integrated perspective on the effects of γc cytokines on target cell transcriptomes, how cytokines compare, and how widely these effects are distributed across immunocytes. We have thus undertaken a systematic analysis of these signatures for six main cytokines of the γc family and across 14 different cell types in vivo. The results paint a different family portrait: great breadth to the signatures, as much repression as induction, more overlap between cytokines with essentially no cytokine-specific effects, responses observed with unanticipated cell/cytokine pairs, and even a novel cell state.
Results
Landscape of responses to γc cytokines: Overview
Several considerations determined the study design for this complete landscape of γc cytokine signatures. We elected to examine: (i) responses in a normal setting in vivo, unperturbed by tissue culture, and where cells are subject to all the intercellular signals they normally receive; (ii) early responses, when primary responses dominate and indirect effects would still be limited (accepting that this choice misses secondary amplifications and later stages of the responses); and (iii) a timepoint when differences in stability of the inducing cytokines or Ab-complexes would be minimized. We injected mice systemically with either IL2, IL4, IL7, IL9, IL15, or IL21, alone or conjugated within cytokine/anti-cytokine complexes for stabilization (Table S1; the dosages were chosen to match those used in functional experiments in the field, and should be considered high but not overwhelming). Peritoneal cells and splenocytes were harvested precisely 2 h later, and cell populations spanning the whole range of immunocyte lineages (Table S2) were purified by flow cytometry for low-input (ULI) population RNA sequencing (RNAseq), per ImmGen protocols. In all, 352 RNAseq datasets passing quality control thresholds were assembled, providing biological triplicates for virtually all cell/cytokine combinations. The results can be queried on the ImmGen website (https://www.immgen.org/Databrowser19/Cytokines.html).
A global perspective of the early impact of γc cytokine administrations is brought by Fig. 1 B, which presents the number of transcripts induced or repressed in each cell/cytokine combination (at arbitrary thresholds of FoldChange [FC] > 2 and t test P value <0.001). Several points are worth noting: (i) There is wide divergence in the extent of effects, very narrow for IL7 and IL9, and more extensive for the others. An independent repeat experiment confirmed that the low response observed with IL7 was not due to a faulty batch of cytokine. This result is consistent with the notion that IL7 is only involved in homeostatic control of cell survival for mature immunocytes, rather than inducing cell growth and differentiation. (ii) JAK/STAT signaling and transduction are expected to mainly activate transcription, but transcript downregulation was unexpectedly extensive, on par with upregulation, and showing roughly the same cell/cytokine distribution. (iii) Lymphoid cells were generally more responsive to γc cytokines than myeloid cells, although the latter did show clear responses, in particular to IL4, IL15, and IL21. (iv) IL4 performed as the most “universal” cytokine, affecting every type of immunocyte. These patterns were to a large extent a reflection of expression levels of the corresponding receptors (Fig. 1 C), indicating that differences in receptor quantity are a primary driver of cell specificity.
In virtually all biological inductive events, among the first responders are molecules that negatively feedback on the initiating signal. This is true of γc-induced responses, where negative regulators of the SOCS family are activated (Yoshimura et al., 2018). Here, different patterns were observed across cell types and cytokines (Fig. 1 D). Socs1 dominated after IL4, IL15, and to a lesser extent IL2 in lymphoid cells after IL7, but was mostly refractory to IL21. In contrast, IL21 was a prime inducer of Socs3. Cish showed yet another pattern. Since these Socs family members dampen signals from many cytokines and are non-redundant in their functional specificity (Linossi and Nicholson, 2015), the viable induction patterns observed here imply a nuanced diversification of regulatory feedback on broad cytokine signaling.
Overlap between γc transcriptional signatures
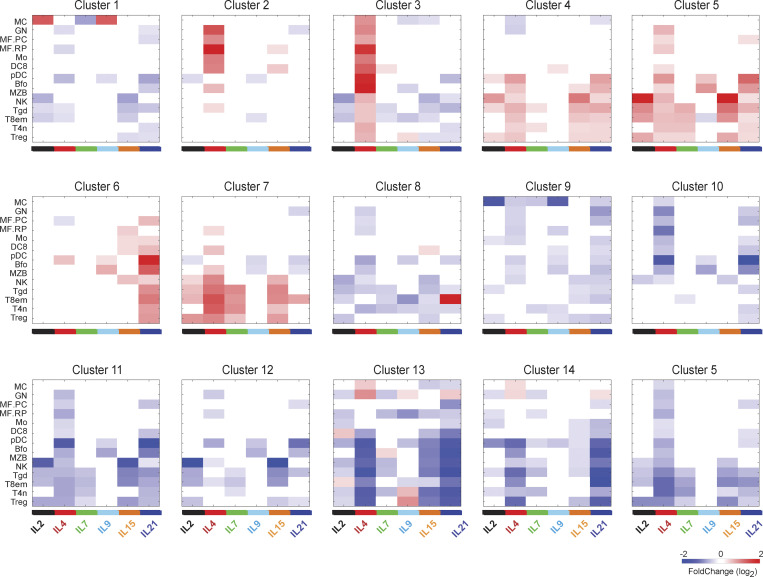
γc cytokines share signaling mechanisms (Fig. 1 A) but they have distinct biological effects, and the degree of overlap between their transcriptional signatures is poorly understood. To address this question, we identified a set of 2,696 genes whose expression was altered by at least one cytokine in at least one cell type (Table S3 and Fig. 1 E). This set represented a sizable proportion of the 15,511 genes expressed in those cell types (5–10% in any one cell), indicating that γc cytokines reshuffle, within hours, an important fraction of immunocyte transcriptomes. Several important observations stem from integrated overview of Fig. 1 E (see Fig. S1 for a more detailed view of each cluster). First, there was no constant signature for one cytokine that carried across most cell types and the effects were highly cell dependent. Second, there was a high degree of commonality between the responses, contrasting with the notion of γc cytokines as largely independent actors with individualized effects. Most response clusters were shared by several cytokines and/or several cell types (with the exception of Clusters 1 and 9, largely unique to mast cells [MCs], and 8 in effector/memory T8 [T8em]). In many cases, clusters were induced by one cytokine in one cell, by another cytokine elsewhere (e.g., Cluster 5). Widespread responsiveness was most striking for transcripts of Cluster 5, induced by every one of the γc cytokines in at least one cell type, but with preferential induction by different cytokines in different cells. The exception to the overlap rule was in Clusters 2 and 3, only induced by IL4. Only one instance of a selective response (to one cytokine in one cell type) was observed: Cluster 8 induced by IL21 in CD8+ Tem cells.
Figure S1.
Response of cluster genes to individual cytokines. Average of changes in gene expression in cell/cytokine pairs for the responsive clusters defined in Fig. 1 E. See Table S2 for acronyms.
Several of these clusters are worth dissecting further, as they inform on the type of cellular adaptations elicited by γc cytokines.
-
-
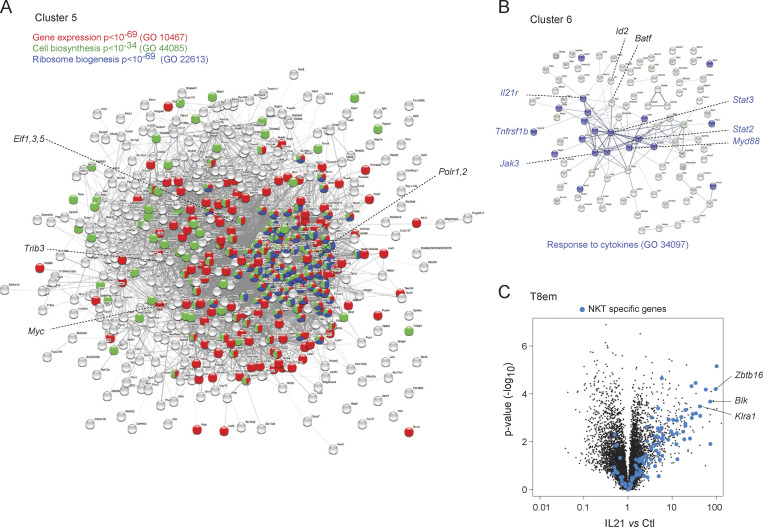
The largest cluster (Cluster 5, 601 transcripts, with the similar Cluster 4) was induced in every cell type tested, including MCs, representing a core response to γc cytokines. However, the most effective inducing cytokine(s) varied according to cell type (e.g., IL4, IL9, and IL21 in marginal zone B cells [MZB], IL2 and IL15 in NK cells, and IL4 and IL7 in naive CD4+ T cells [T4n]). IL4 induced Cluster 5 genes ubiquitously in all cells, albeit to different extents. GeneOntology analysis showed that Cluster 5 is a tightly clustered geneset that encompasses several cellular biosynthetic pathways (Fig. 2 A): ribosome biogenesis (115 genes, false discovery rate [FDR] < 10−71), cellular biosynthesis (168 genes, FDR < 10−35), and primary metabolic processes (331 genes, FDR < 10−44). Although regulators of gene expression were included, few sequence-specific transcription factors were found in Cluster 5 with the notable exception of Myc. Indeed, enrichment analysis showed that Myc targets were very highly represented in Cluster 5 (297 genes, P < 10−138), indicating that Myc is the major driver of this response. Thus, consistent with early work (Rapp et al., 1985; Miyazaki et al., 1995; Klemsz et al., 1989; Moro et al., 2022; Marchingo et al., 2020), γc cytokines rapidly reprogram cells for major metabolic and biosynthetic changes via Myc activation.
-
-
Cluster 2 (97 genes) was almost exclusively induced in myeloid cells and mostly by IL4. Very different from Cluster 5, it included mostly molecules involved in signaling (32 genes, FDR < 0.04), and a group of transcription factors (Irf4, Irf7, Mafb, Egr2) associated with early activation and cell differentiation.
-
-
Cluster 6 (109 genes) was almost exclusively stimulated in lymphoid cells and preferentially by IL21 (Fig. 1 D). It included 18 genes that are targets of STAT3, which is plausible because STAT3 is the major transducer of IL21 signals, and interestingly, Stat3 itself and a host of signaling molecules (Myd88, Jak3, Akt2, Irak4, Il6st, Ptpn1, Il12rb1, Tnfrsf1b—encodes TNFR2) and downstream transcriptional regulators (STAT2, Batf), with “response to stimulus” as a main ontology term (Fig. 2 B, 51 genes, FDR = 0.006). These may constitute a positive feed-forward loop, with IL21 heightening the potential to respond to γc and inflammatory cytokines (TNF or IL6) or TLR ligands. Together with the variegated effects on the SOCS family, these results indicate that an important effect of γc cytokines is to tune responses to cytokines (γc and other families) in a highly diversified manner.
-
-
Cluster 8 (122 genes) was the standout group of genes induced by IL21 quasi-exclusively in T8em cells. Several of these genes were actually repressed by other cytokines in other cells, e.g., by IL4 in B cells or monocytes (Fig. 1 E). We noted that Cluster 8 included Zbtb16 (a.k.a. PLZF), the determining transcription factor of invariant NKT cells. Indeed, a good fraction of signature genes that distinguish invariant NKT from other CD4+ T cells proved to be almost completely induced by IL21 in T8em cells (Fig. 2 C and Table S4): beyond Zbtb16 itself, several NKT-characteristic receptors of the Klr family, chemokines (Xcl1), and signaling molecules not commonly observed in CD8+ T cells (Lyn, Fcer1g, Blk). Thus, IL21 appears to induce a previously unreported differentiation path in T8em cells.
-
-
Cluster 14 contains a number of downregulated IFN signature genes (ISGs), indicating that the interplay between γc cytokines and the IFN system (further detailed below) is not exclusively synergistic.
Figure 2.
Composition of major response clusters. (A) Interaction map (String) of the main metabolic/biosynthetic Cluster 5 (from Fig. 1 E), annotated for major functional categories. (B) Cytokine signaling transcripts of Cluster 6. (C) Responses in CD8 T cells to IL21 (corresponding to Cluster 8), highlighted for transcripts that distinguish NKT from other CD4+ T cells. GO, GeneOntology.
Correlations between cytokines
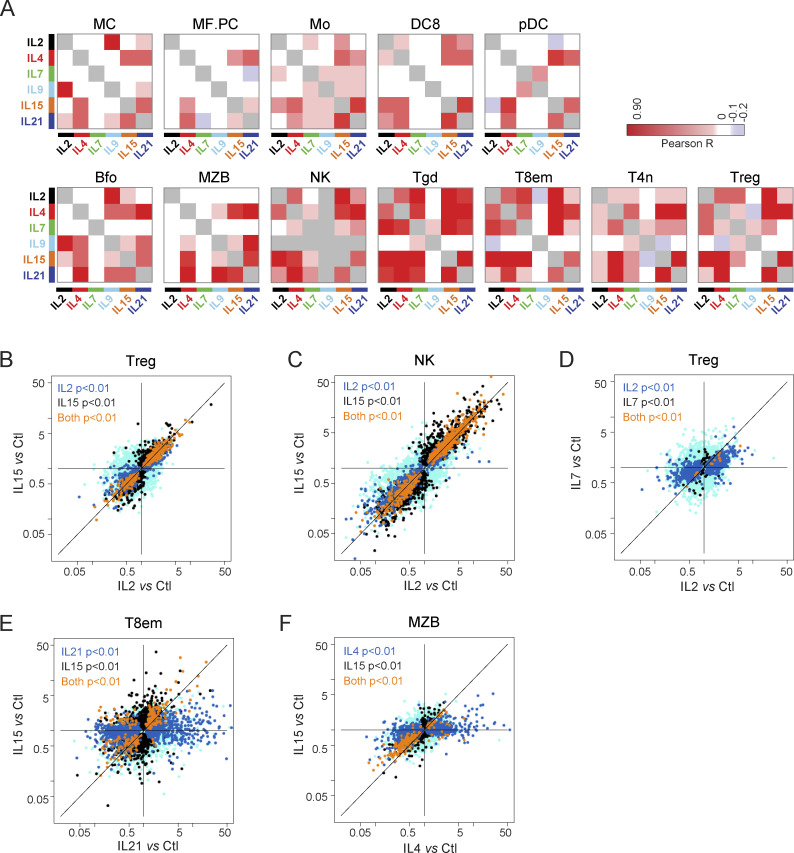
Most of the response clusters were affected by several γc cytokines (Fig. 1 E). For a more systematic appreciation of this overlap, we correlated the FC values for the different cytokines in each cell (Fig. 3 A). The correlations were almost all positive, with essentially no opposite effects. This near universality of positive correlations is logical when considering that γc cytokines exploit the same signaling pathways but are not compatible with models in which γc cytokines elicit antagonistic functional outcomes. FC/FC plots revealed further nuances to the overlap. In some cases, the signatures were quasi-identical, with only minor quantitative fluctuations (e.g., IL2 and IL15 in NK or Treg cells, Fig. 3, B and C), mirroring prior results in CD8+ T cells (Ring et al., 2012). In others, the genes affected were largely the same, but with a strong quantitative shift (for instance, IL7 affected mostly the same transcripts as IL2 in Tregs, but to a much lesser extent, Fig. 3 D). In yet others, the relationships were more complex (IL21 and IL15 in T8em, and IL4 and IL15 in MZB, Fig. 3, E and F).
Figure 3.
Cell-type-specific relationship between responses to γc cytokines. (A) Correlation matrices between responses to γc cytokines in different cell types. (B–F) FC/FC plots relating changes induced by two cytokines in the same cell. Transcripts whose changes meet a t test P value <0.01 (uncorrected) in one or both treatment conditions are highlighted in different colors. See Table S2 for acronyms.
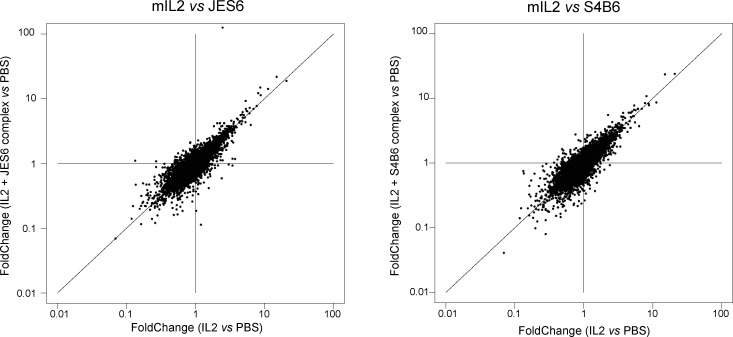
Those similarities raise the question of the known biological specificity. IL15 has the same signature in Tregs as IL2, a finding consistent with recent observations based on artificial cytokine surrogates (Yen et al., 2022), where surrogates for IL2, IL7, and IL15 had largely the same signatures. Note that free IL2 yielded the same changes as when injected as a complex with two different antibodies, JES6 and S4B6 (Fig. S2). Why, then, is IL2 uniquely required for Treg homeostasis? In physiological contexts, Tregs likely encounter these cytokines in different microenvironments, quantities, or interacting partners, which may be the answer. On the other hand, preliminary experiments showed that administration of IL2 and IL15, in the same systemic mode as used here, had different effects on Treg expansion 3 d later, suggesting that late-acting effects of cytokines matter, in addition to primary signatures. How this is achieved is unclear, but one might speculate that the functional divergence between IL2 and IL15 for Treg may relate to transpresentation of IL15 via the IL15Rα chain expressed by myeloid cells.
Figure S2.
Short-term responses to IL2/anti-IL2 complexes. Comparison of changes induced by pure recombinant mIL2 injected alone or complexed with two anti-IL2 antibodies (JES6 and S4B6) that allow IL2 signaling with preference for the CD25 or CD122 forms. Complexes were formed under the conditions of Spangler et al. (2015b), but we cannot be sure that no free IL2 dissociates after injection, possibly explaining the surprisingly similar profiles.
Relations between γc cytokine and IFN signatures
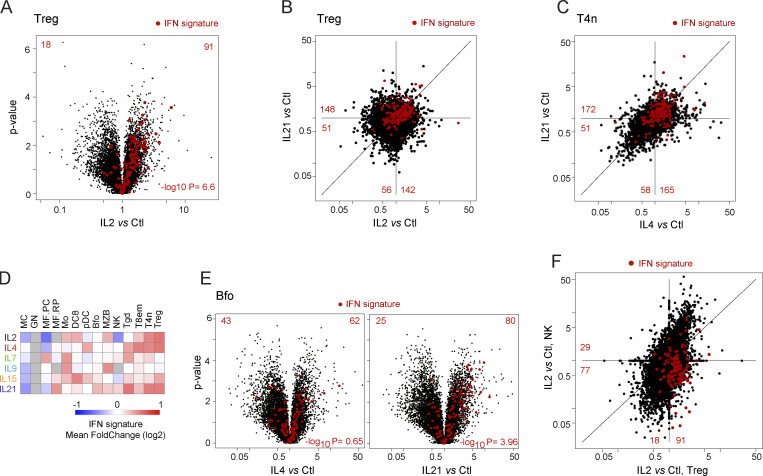
There is a recognized relationship between signaling pathways activated by γc cytokines and by Type-1 IFN, which enables lateral crosstalk between antimicrobial defenses tied to IFNs and the diverse immunologic functions molded by γc cytokines. Signaling by several γc cytokines involves STAT1, IFN’s principal transducer, and parallel signaling like MAP kinases or Akt can be activated by IFN and γc cytokines (Bezbradica and Medzhitov, 2009). It was thus interesting to assess the intersection between ISGs and γc signatures with ISGs. We leveraged our prior analysis of responses to Type-I IFNs in immunocytes (Mostafavi et al., 2016) that defined ISGs activated in the cell types analyzed here. ISG induction by γc cytokines proved pervasive. In Tregs, for instance, ISG expression was strongly biased after IL2 exposure (Fig. 4 A; P < 10−6). The same ISGs were often induced by several cytokines, e.g., by IL2 and IL21 in Tregs vs. IL4 and IL21 in T4n (Fig. 4, B and C), the latter unexpected since IL4 is not thought to induce Type-1 responses. In the integrated view of ISGs in each cytokine/cell pair (Fig. 4 D), ISG induction was clearest in T cells but also marked in some myeloid cells (Mo, DC8). Two paradoxical situations were uncovered. First, in follicular B cells (Bfo), IL21 induced ISGs, but IL4 was largely inert in this respect (Fig. 4 E), even though IL4 and IL21 had comparable effects in T4 cells (Fig. 4 C). If ISG induction is considered a proxy for STAT1 activation, the results imply that STAT1 can be unexpectedly engaged by the IL4 receptor but variably between cell types. More generally, it should be acknowledged that the reference schematic of STAT association drawn in Fig. 1 A was derived from a limited number of cell types and largely in vitro. Cell-specific STAT connections may contribute to the diversity of γc effects reflected in Fig. 1 E.
Figure 4.
Overlap between responses to Type-I IFN and γc cytokines. (A) Volcano plot of the response to IL2 in Tregs, with IFN signature genes (from Mostafavi et al., 2016) highlighted. (B) FC/FC plots comparing Treg responses to IL2 and IL21, with IFN signature genes highlighted. (C) As B, IL4, and IL21 in naive CD4+ T conventional cells. (D) Mean FC of IFN signature genes in all cytokine/cell pairs. See Table S2 for acronyms. (E) Volcano plots of responses to IL4 and IL21 in B cells, IFN signature genes highlighted. (F) As B, comparing responses to IL2 in Treg and NK cells.
An even more unexpected observation was made in NK cells: IL2 induced generally similar responses in Treg and NK cells, except for ISGs (Fig. 4 F). IL2 induced ISGs in Tregs but instead repressed them in NK cells (Fig. 4 F). This was not due to ineffective IFN signaling pathways in NK cells, which are strong IFN responders (Mostafavi et al., 2016). Rather, ISG downregulation by IL2 in NK cells suggests different signaling modalities and that STAT1 activated in the context of γc signaling may actually be repressive in NK cells, for instance by recruiting negative regulators in a cell-specific manner. These results further the notion of widespread cell-type specificity in how γc cytokines tune responses to other cytokine families, and more generally in the involvement of STAT molecules in γc signaling.
Up and down mechanisms
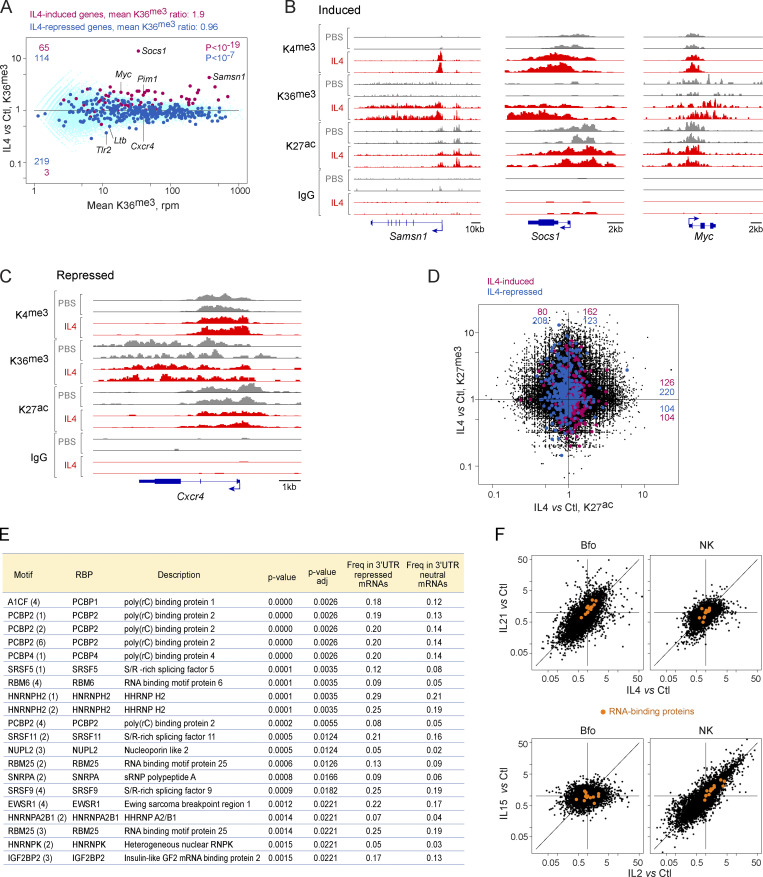
To delineate the types of changes induced by γc cytokines at the chromatin and transcriptional levels, we analyzed histone posttranscriptional modifications around cytokine-responsive genes, using CUT & RUN (Skene and Henikoff, 2017). Analyzing all cell type/cytokine combinations in this manner would be unrealistic, so we focused on IL4 effects in B cells, also after 2 h, probing histone methylation and acetylation, known to denote enhancers or promoter activity. Indeed, even at this early time point, a number of changes were observed. H3K36me3 deposition across gene bodies is a good proxy of actual transcriptional activity across a gene (Wagner and Carpenter, 2012) and was found generally increased (1.9-fold, averaging all genes with mRNA induced >4-fold, P < 10−12, Fig. 5 A, individual examples in B). In contrast, IL4-repressed genes showed a very modest reduction in H3K36me3 overall (mean = 0.95-fold, still significant at P < 10−6, Fig. 5 A, example in C). Thus, transcript upregulation seems to derive from rapidly induced transcription, but cessation of transcription is not the major root cause of γc cytokine-induced downregulation.
Figure 5.
Molecular mechanisms underlying responses to γc cytokines. Chromatin immunoprecipitation and sequencing (CUT&RUN) was used to analyze chromatin structure in Bfo, 2 h after in vivo administration of IL4 (or PBS control). (A) Integrated signal for H3.K36me3 across gene bodies, plotting mean signal vs. IL4/control FC at each locus. Red highlights: genes with IL4-induced mRNA >4-fold; blue: genes with IL4-repressed mRNA <0.25-fold. (B) Representative tracks across three genes whose mRNAs are induced by IL4 for H3.K4me3 (promoters), H3.K36me3, and H3.K27ac; precipitation with non-specific IgG shown as a control. (C) As B, for a representative repressed gene. (D) Relative changes in H3.K27ac (active enhancer) and H3.K27me3 (closed enhancer) after IL4 treatment. (E) Over-representation, computed with reference to oRNAment tables, in sequence motifs for RNA-binding proteins in the 3′UTRs of genes repressed by γc cytokines (1,055 Cluster 10–13 genes, Fig. 1 E), relative to 2,979 IL4-neutral taken as background reference. P value of the difference in frequencies. (F) Expression of transcripts encoding the RNA-binding proteins from E, shown as orange highlights, in Bfo and NK cells, treated with IL4 or IL21, and IL2 or IL15, respectively.
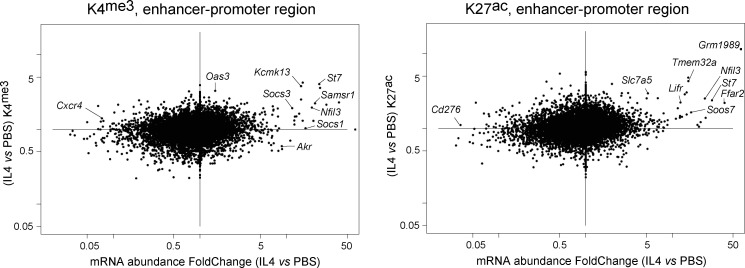
Changes in enhancer and promoter activity were reflected by H3K4me3 (active promoters), H3K4me1, and H3K27ac (active enhancers) deposition in the −2 kb > +200 region relative to the transcriptional start site, which encompasses the promoter and a sizeable fraction of enhancer elements. Some induced genes showed a sharp increase in promoter-region H3K4me3 deposition (e.g., Samsn1 in Fig. 5 B), others (Socs1) showed displaced H3K4me3 deposition. In general, increased H3K4me3 and H3K27ac marks were observed for induced genes, especially those with the strongest mRNA induction (Fig. S3). Reciprocal changes in the abundance of H3K27ac and H3K27me3 (denoting active and repressed enhancers, respectively) were observed at IL4-induced and -repressed loci (Fig. 5 D), indicating that enhancer activity is also adjusting. In conclusion, different mechanisms are brought into play for the early response to γc cytokines: for induced genes, transcriptional induction accompanied by activation of enhancer and promoter elements. For repressed genes, much more modest changes in transcriptional activity were observed, suggesting that the widespread decrease in mRNA abundance may initially result from destabilization, although coordinated dampening of enhancer/promoter activity also sets in, ensuring longer-lasting effects. Although we cannot formally extrapolate to all other cell/cytokine pairs, the similarity in signaling pathways and the overlapping signatures suggest that these conclusions reached for IL4 in B cells will likely apply to other cells and γc cytokines.
Figure S3.
IL4-induced changes in marks of promoter and enhancer activity. H3K4me3 and H3K27ac signals were integrated over the −2,000 to +200 bp region around genes’ transcription start site. Their IL4-induced changes (y-axis) were plotted against corresponding changes in mRNA abundance.
Several mechanisms can provoke selective mRNA decay: deadenylation of polyA tails and decapping followed by exonuclease digestion, and specific endonucleases including those targeting A-U rich elements (ARE) in 3′UTRs. No relevant changes were observed in mRNAs encoding the CRR4-NOT complex, polyA-specific ribonucleases (Pan, Parn), decapping enzymes (Dcp1/2), or the Zpf36, Rc3h1 (Roquin) or Zc3h12a (REGNASE-1) endonucleases. ARE motifs were not enriched in 3′UTRs of downregulated vs. upregulated transcripts (24.4 and 26.6%, respectively). On the other hand, we observed that 3′UTR regions of repressed mRNAs (Clusters 10–13) were enriched in binding motifs for a set of RNA-binding proteins (RBP), relative to expression-matched γc -neutral transcripts (23 proteins at FDR < 0.05; Fig. 5 E). Enrichment was most marked for the polyC-binding proteins and S/R-rich splicing factors families, both of which have pleiotropic roles in transcriptional regulation (Makeyev and Liebhaber, 2002; Twyffels et al., 2011). Substantiating the notion that these factors are involved in the rapid repression of transcripts after γc cytokine exposure, they were themselves induced by γc cytokines with a cell/cytokine specificity that correlated with the strength of repression: induced by IL4 and 21 in Bfo, but not by IL2 or IL15; induced by IL2 and IL15 in NKs, but not by IL4 or 21 (Fig. 5 F). Thus, both motif overrepresentation and correlation between RBP induction and the repression of Clusters10-13 suggest that these RNA-binding proteins are one of the mechanisms through which γc cytokines rapidly repress a substantial portion of the transcriptome.
Cell-specific aspects: Surprising responses in MCs
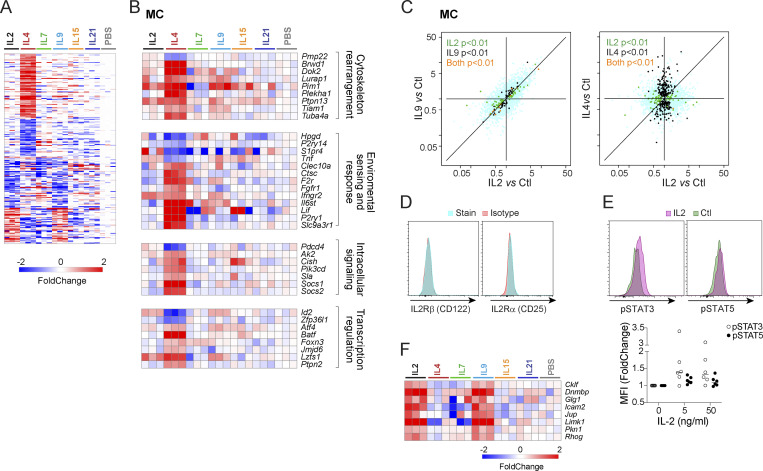
MCs exhibited two distinct responses (Fig. 6 A). The strongest followed IL4 treatment, in keeping with IL4’s documented pleiotropic role supporting MC survival, increasing adhesion, and modifying MC responses to activating stimuli (McLeod et al., 2015). IL4-regulated transcripts fell into several broad categories, indicating that IL4 is not solely a trophic factor for MCs but instead broadly modulates their phenotype (Fig. 6 B): some associated with cytoskeleton rearrangements and the coordination of cellular movement (Lurap1, Tuba4a, and Tiam1; Castellanos-Montiel et al., 2020; Cheng et al., 2017; Kobayashi et al., 2013), receptors and signaling molecules for cytokines and growth factors (Ifngr2, Il6st, Fgfr1, Pik3cd), or danger signals (F2r, Clec10a, P2ry1), the cytokine Lif, and Ctsc, which encodes cathepsin C, required for the maturation of the key MC granule-associated protease β-tryptase (Le et al., 2011). Conversely, Tnf was downregulated along with Hpgd, encodes 15-Hydroxyprostaglandin dehydrogenase, an enzyme associated with MC effector function.
Figure 6.
Unexpected responses to γc cytokines in MCs. (A) Heatmap of all significant changes (FC < 0.5 or > 2, with t test P < 0.01) induced by at least one γc cytokine in MCs, as log2 FC. (B) As A, focused on responses to IL4. (C) FC/FC plot comparing changes induced by IL2, IL9, and IL4 in MCs; t test P values color-coded as in Fig. 3 B. (D) Flow cytometric analysis of both chains of the conventional IL2 receptor on peritoneal MCs. (E) Flow cytometric detection of phosphorylated-STAT3 or STAT5, 30 min after in vitro treatment of peritoneal MCs with IL2 (5 ng/ml); control are parallel culture wells with no IL2; the results from several experiments are compiled at right (change in Mean Fluorescence Intensity [MFI] relative to untreated cells). (F) Focused heatmap of IL2 and IL9 responsive transcripts in MCs.
The other genesets, corresponding to the MC-specific Clusters 1 and 9 of Fig. 1 E, were similarly up- or downregulated by both IL2 and IL9 (Fig. 6, A and B). That MCs respond to IL9 is well known (Mukai et al., 2018), but the response to IL2 was unexpected, if only because no mRNA for Il2ra and Il2rb can be detected in MCs (see ImmGen data browsers). We verified the absence of either IL2Rα or IL2Rβ on peritoneal MCs (Fig. 6 D). To validate that MCs were truly responding to IL2, and not to another cytokine systemically induced by IL2, we challenged peritoneal cavity MCs with IL2 in vitro and evaluated the phosphorylation of STAT5, the major transducer for IL2 (Lin and Leonard, 2019; Ross and Cantrell, 2018). Indeed, STAT5 phosphorylation increased markedly within 30 min (Fig. 6 E). Transcripts within the shared IL2 and IL9 cluster included genes associated with cell adhesion and migration (Glg1, Icam2, Jup, Cklf, Dnmbp, and Pkn1), and the GTPases Rhog and Limk1, which promote actin remodeling (Nadella et al., 2009; Vigorito et al., 2003; Fig. 6 F), suggesting that the MC response to IL2 and IL9 could be related to cellular motility. Given the quasi-identical response, we speculate that IL2 signals via the IL9 receptor in MCs (although this property is not shared by MZB cells) respond most to IL9 but not to IL2 (Fig. 1 B). Overall, the changes described here show MCs sensing signals from other immunocytes and mounting a fine-tuned response to these signals. This degree of coordination departs from the simplistic view of MCs as simple “time-bombs” waiting to be ignited by IgE ligands or other surface sensors.
Distinct cytokine responsiveness of Bfo and MZB cells
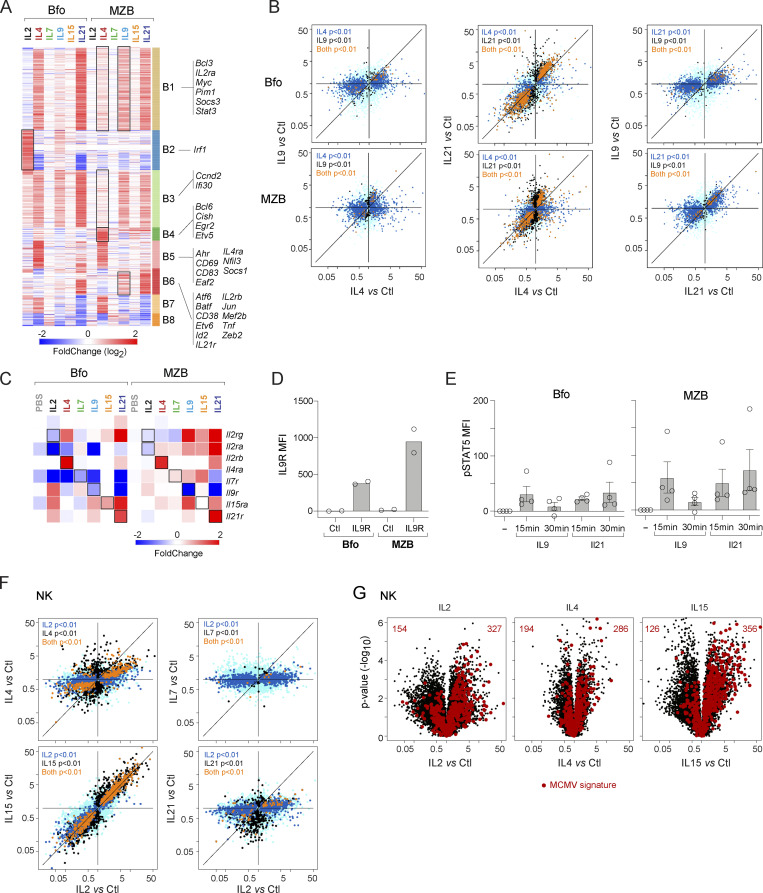
B cells respond to IL4 and IL21, resulting in activation, proliferation, immunoglobulin class switching recombination, and plasma cell differentiation (Lin and Leonard, 2018; Tangye and Ma, 2020). The most striking observation from the present dataset was the difference in the transcriptional response between MZB and Bfo (Fig. 1 E and Fig. 7 A). The response to IL2 in Bfo was essentially absent in MZB. Bfo showed a strong and highly correlated response to IL4 and IL21, while the induction of many IL4-driven genes was much lower in MZB (Fig. 7, A and B). Instead, MZB responded to IL9, a little-studied cytokine in B cells. These changes are likely linked to the physiological roles of these subsets, as Bfo can undergo selection and affinity-based maturation and selection in germinal centers, a process regulated by IL4 and IL21 (Ozaki et al., 2002), while MZB is located outside the lymphoid follicles and is specialized to rapidly differentiate to plasma cells in the absence of direct T cell help.
Figure 7.
Unexpected responses to γc cytokines in B and NK cells. (A) Reclustered heatmap of all significant upregulations (FC > 2, with t test P < 0.01) induced by at least one γc cytokine in Bfo or MZB. Black squares indicate clusters differentially induced in Bfo and MZB. (B) FC/FC plots relating changes induced by two cytokines in either Bfo or MZB cells; color-coding of statistical significance as in Fig. 3 B. (C) Profound changes induced in transcripts encoding cytokine receptors in Bfo or MZB. (D) Flow cytometric detection (shown as MFI) of IL9R on Bfo or MZB cells—each dot is an independent experiment. (E) Flow cytometric detection of phosphorylated STAT5 after in vitro treatment of splenic B cells with IL9 or IL21. Control (no cytokine) MFI was subtracted from the test sample value. Each dot is an independent experiment. (F) FC/FC plot comparing changes induced by IL2 and other γc cytokines in NK cells; t test P values color-coded as in Fig. 3 B. (G) Changes elicited by γc cytokines in NK cells, with highlights corresponding to the signature of MCMV infection in NK (from Bezman et al., 2012).
Analysis of genes upregulated in any Bfo or MZB sample revealed eight distinct clusters (B1–8; Fig. 7 A). Cluster B1 was elicited by IL4 and IL21 in Bfo and IL9 and IL21 in MZB and is characterized by Myc and Stat3 upregulation, as well as the known target of IL9 signaling in T cells, Bcl3 (Richard et al., 1999). The loss of IL4 responsiveness in MZB was selective, as Clusters B4 (transcription factors Bcl6, Etv5, and Egr2) and B5 included IL4-induced genes associated with activation (Cd69, Cd83), and the transcription factors Ahr and Nfil3, a known target of IL4 in B cells (Kashiwada et al., 2010). Cluster B6 genes were responsive to IL21 in Bfo, but to IL9 and IL21 MZB, and include distinct transcriptional regulators (Jun, Atf6, Mef2b, Batf, and Zeb2). These data indicate considerable diversity of transcriptional programming of mature B cell subsets after short γc exposure.
A distinctive feature was the modulation of the γc receptor family by the cytokines themselves (Fig. 7 C), as previously noted in vitro (Leonard et al., 2019). Il4ra and Il21r were potently induced by their ligands in Bfo and MZB, suggesting a feed-forward response. In contrast, Il9r expression was repressed by IL9 and IL21. IL21 also induced the Il2ra (Bfo, MZB) and Il2rb (MZB only) components of IL2R suggesting a rationale for synergistic activation of mouse B cells with IL21 and IL2, as has been reported for human B cells (Berglund et al., 2013).
IL9R expression has been documented on memory B cells (Takatsuka et al., 2018), B1 cells (Vink et al., 1999), and MZB (Kleiman et al., 2015), although its function on MZBs is unknown. Analysis of the cell surface expression of IL9R confirmed higher abundance on MZB compared with Bfo (Fig. 7 D). The IL9/IL9R complex has been proposed to signal via STAT1, 3, or 5 (Kleiman et al., 2015). We compared the levels of STAT5 phosphorylation after exposure of Bfo or MZB to IL9 or IL21. In keeping with the higher IL9R abundance, MZB showed higher pSTAT5 than Bfo (Fig. 7 E). In contrast to the IL21 response that was stable or increased from 15 to 30 min, signaling through the IL9R was transient, returning to a basal state after 30 min (Fig. 7 E). The mechanism for this abrupt cessation of signaling is not clear as IL9 elicits less expression of SOCS family members than IL4 or IL21 (Fig. 1 D).
The very similar responses elicited by IL4 and IL21 suggest that their STAT intermediates may be broader than previously thought (STAT6 for IL4, STAT3 and STAT5 for IL21) and that many of the distinct functions of each cytokine observed in vivo may be more a matter of geographically preferred availability, rather than of intrinsically different properties. Intriguingly, IL9 can substitute for some of the functions of IL4 and IL21 in MZB, further supporting the specialized function of this B cell subset. CD4 T cells and ILCs are proposed to be physiological sources of IL9 (Angkasekwinai and Dong, 2021), although whether they are the source of IL9 for MZB activation, as well as the longer-term cell biological consequences of IL9 signaling, remains to be investigated.
NK cells are the most reactive to a broad swath of γc cytokines
The IL2Rβ-γc dimer transduces signals from both IL2 and IL15 (Giri et al., 1994). Accordingly, there was a high concordance between IL2- and IL15-induced signals (Fig. 7 F). Many genes induced in NK cells after IL2 and IL15 injection were elevated in early mouse cytomegalovirus (MCMV) infection (Bezman et al., 2012; Fig. 7 G). This overlap suggests that an important component of the antiviral response in NK cells is actually in response to the rapid production of IL15 by DCs and granulocytes.
IL4 also induced the MCMV signature, albeit less intensely than IL2 or IL15 (Fig. 7 G), and IL4 generally had the strongest effect after IL2 and IL15 on NK cells. It induced much of the IL2-responsive contingent, again less strongly (Fig. 7 F). This observation contrasted with previous conceptions about IL4 in NK cells, for instance, that it antagonizes proliferation induced by IL15 in vitro (Brady et al., 2010). On the other hand, in vivo administration of IL4 induced IFN (Morris et al., 2006). Thus, IL4 may have a dual action in NK cells, serving as an IL2/IL15 analog, but also specific (inhibitory) effects via the transcripts that it uniquely induces (e.g., Dusp10, Dlk2, Fig. 7 F).
Discussion
Aside from providing a rich reference of signatures and genesets, we feel that these results bring a new perspective on the realm of γc cytokines, the changes in chromatin structure, transcription, and mRNA stability that they rapidly elicit. A number of specific points have been discussed above and will not be repeated, but the overarching implications are worth highlighting.
Foremost are the notions of fluidity and overlap. There is no specific signature of any one cytokine that applies across all cells. Instead, there was much sharing of effects: What IL2 achieves can also be done by several other cytokines, and in several instances, two cytokines had exactly the same impact on a given cell. Genesets activated by a cytokine in one cell type are elicited by another cytokine in a different cell type. In addition, many cytokines influence their own and others’ receptors or signaling regulators (especially SOCS family downmodulators), allowing a great deal of nuance to be introduced into the directionality and scale of the response. It is not clear how to reconcile these observations with the more sharply delineated “functional personalities” that have been attributed to γc cytokines over the past decades. Immunocytes may normally be exposed to cytokines in a far more context-specific manner than the broad exposure achieved here, but our collective grasp of γc cytokine function may have focused more on the distinguishing aspects of responses, tending to overlook similarities.
The survey performed here investigated the early and most direct responses to avoid secondary and indirect signals. We cannot formally exclude such effects, which are likely minor at the 2 h timepoint used: the fastest-acting IFNs take 1–2 h to fully elevate mRNA levels of ISGs. Thus, cell–cell communication cascades would need to act extremely fast to secondarily affect other mRNAs in bystander cells. For realism, the study was performed at a single dose, chosen to match those commonly employed for in vivo administration in experiments that aim to modify cell homeostasis or differentiation. We cannot rule out that cell- or cytokine-specific responses would have appeared at higher doses. We also did not attempt to use equimolar doses, which would be difficult in the absence of biodiffusion or stability data.
This choice of design did also miss late responses, which develop further over time (Moro et al., 2022). It is thus possible that, after an early stage of commonality, responses to γc cytokines branch out further and become more individualized than observed here. Another scenario that might account for the overlap between cytokine signatures is of cell-autonomous indirect effects: the primary response to injected ILx in one cell modifies the signaling circuits (e.g., by SOCS induction) used by an ongoing response to cytokine ILy in the same cell. In doing so, ILx may appear to control a set of genes that are really ILy targets. More generally, the ultimate physiological effect of a cytokine in vivo does not solely result from its own signature, but from the highly combinatorial consequences that it has, via modulation of other receptors and regulators and interaction with other signals, on the entire cell–cell interaction network. One also expects that a cell’s response to a cytokine will change as it differentiates. It is also possible that alternative receptors are involved, beyond those portrayed in Fig. 1 A (i.e., the IL13ra1 subunit for IL4). The present results provide a starting point to unravel this complexity.
Another key aspect was the centrality and generality of Myc-controlled phenomena. The involvement of Myc downstream of γc cytokines, especially those with strong trophic effects, has been known for some time (e.g., Rapp et al., 1985; Miyazaki et al., 1995; Klemsz et al., 1989; Moro et al., 2022), but Myc’s dominant presence was perhaps surprising. All γc cytokines, in one cell or the other, initiated to some degree the major changes in cell metabolism and biosynthesis that Myc controls (Marchingo et al., 2020).
One final aspect to mention is the connection between γc cytokines and Type-1 IFN signaling. The two generally reside in different sections of textbooks. IFN belongs to the innate immune system, induced by viral infection, and more generally by activation of innate sensors, while γc cytokines mostly manage the crosstalk in adaptive responses. Here, we found that many γc cytokines induced essentially all IFN-responsive genes in some cells, although some antagonistic effects were observed as well (most strikingly, the inhibition of most ISG by IL2 in NK cells). Thus, the γc-responsive and IFN-responsive networks are highly connected, inscribing γc cytokines in the broadest range of immune responses.
Material and methods
Mice and cytokine treatments
6–8-wk-old C57BI/6J mice were obtained from the Jackson Laboratory and housed in the specific pathogen–free facility at Harvard Medical School, following animal protocols approved by the Harvard Medical School Institutional Animal Use and Care Committee (protocol IS00001257). Cytokines or cytokine/anti-cytokine complexes were injected i.v. (normalized to the mouse’s weight), per Table S1, controls from the same lots of mice were injected with PBS, and the mice were euthanized exactly 2 h after injection.
Cell preparation and sorting
Overall, the data were generated in three main batches. Each batch included cells from cytokine-treated mice and PBS-treated controls. The ImmGen protocol (http://www.immgen.org) for the 14-cell set preparation was followed. Briefly, peritoneal cavity lavage was harvested after euthanasia by i.p. injection and aspiration of 10 ml FACS buffer (Phenol red-free DMEM, 2% FBS, 0.1% azide, and 10 mM Hepes, pH 7.9). Splenic tissue was homogenized thoroughly through a 100-μm filter, centrifuged, and erythrocytes lysed in ACK lysing buffer (Lonza, 1 ml per spleen) for 3 min at 4C, spun, washed, and resuspended in FACS buffer. Cells were stained and double-sorted per the standard ImmGen 14-cell set (Table S2), the second sort directly into a LoBind tube containing 5 μl TCL buffer (Qiagen) with 1% vol/vol β-mercaptoethanol. Immediately after the sort, cells were kept on ice for 5 min, spun down, and frozen on dry ice.
MC culture and activation
For MC flow cytometry analysis, peritoneal cell suspensions were obtained by lavage of the peritoneal cavity with 7 ml HBSS (Corning) containing 1 mM EDTA (Boston Bioproducts) followed by erythrocyte lysis using RBC lysis buffer (Thermo Fisher Scientific Scientific). The following antibodies/clones were used: FcεR1a (MAR-1; BioLegend); CD117 (2B8; BioLegend); CD11b (M1/70; BioLegend); CD25 (3C7; BioLegend); CD122 (TM-β1; BioLegend); and phospho-STAT5 (Y694; BioLegend). For analysis of STAT5 phosphorylation, unfractionated peritoneal suspensions were incubated at 37°C for 30 min with 50 ng/μl IL2 (PeproTech). Intracellular phospho-STAT5 staining was conducted using True-Phos Perm buffer (BioLegend), according to the manufacturer’s published protocol. Suspensions were assessed on a five-laser LSR II Fortessa using the BD FACSDiva software (BD Biosciences). Downstream data analysis of MCs (FcεR1a+ CD117+ CD11b−) within the suspensions was conducted using FlowJo.
B cell culture and activation
Single-cell suspensions were produced from 9-wk-old female C57BL/6 mice and either (i) stained with B220-APC (clone 200; eBioscience), CD19-BB700 (clone 1D3; Becton Dickinson), CD21-BV421 (clone 7E9; BioLegend), CD23-PECy7 (clone B3B4; eBioscience), Fixable viability dye eFluor 780 (Thermo Fisher Scientific), and with or without IL9R-PE (S18011D; BioLegend); or (ii) incubated at 37°C in RPMI with 10% FCS alone (15 min) or supplemented with 15 ng/ml IL9 (R&D Systems) or IL21 (a gift from ZymoGenetics Inc., Seattle, WA, USA) for 15 or 30 min. The cells were immediately washed with PBS, stained with Fixable viability dye eFluor 780, washed again, then fixed with BD Phosflow Lyse/Fix buffer, and permeabilized with BD Perm buffer III according to supplier instructions. Cells were stained with B220-APC, CD19-PE (in house, clone ID3), CD21-BV421, CD23-PECy7, phosphor-STAT5-PerCP eFluor710 (clone SRBCZX; eBioscience). Samples were run on a BD Fortessa X-20 flow cytometer and analyzed in FlowJo.
Gene expression profiling by population RNAseq
After the final sort of 1,000 cells directly into 5 μl TCL buffer, Smart-seq2 libraries were prepared as previously described (Picelli et al., 2013, 2014) with slight modifications. Briefly, total RNA was captured and purified on RNAClean XP beads (Beckman Coulter). Polyadenylated mRNA was then selected using an anchored oligo (dT) primer (5′-AAGCAGTGGTATCAACGCAGAGTACT30VN-3′) and converted to cDNA via reverse transcription. First-strand cDNA was subjected to limited PCR amplification followed by transposon-based fragmentation using the Nextera XT DNA Library Preparation Kit (Illumina). Samples were then PCR-amplified for 18 cycles using barcoded primers such that each sample carries a specific combination of eight base Illumina P5 and P7 barcodes and pooled together prior to sequencing. Paired-end sequencing was performed on an Illumina NextSeq500 using 2 × 25 bp reads.
Population RNAseq and computational analysis
Data preprocessing and quality control
Reads were aligned to the mouse genome (GENCODE GRCm38/mm10 primary assembly and gene annotations vM25; https://www.gencodegenes.org/mouse/release_M25.html) with STAR 2.7.3a (Dobin et al., 2013). The ribosomal RNA gene annotations were removed from GTF (General Transfer Format) file. The gene-level quantification was calculated by featureCounts. Raw reads count tables were normalized by median of ratios method with DESeq2 package from Bioconductor (Love et al., 2014) and then converted to GCT and CLS format.
Samples with less than 1 million uniquely mapped reads, or having less than 8,000 genes with >10 reads, or with Transcript Integrity score <45 were removed from the data set prior to downstream analysis and excluded from normalization to mitigate the effect of poor-quality samples on normalized counts. All samples were also screened for contamination by using known cell-type-specific transcripts (per ImmGen ULI RNAseq and microarray data). In practice, the acceptable threshold was set at 1 of typical gene expression of contaminant cell types. Retaining such samples can create structure in the data, and/or generate false distances between samples. In addition, biological replicates were analyzed for Pearson correlation to identify poor-quality samples and remove them from the data set. Pearson correlation was calculated on transcripts with an average of >5 reads or below the 99th percentile for number of reads in the dataset to avoid outlier effects. Any replicates that did not exhibit a correlation of 0.9 or greater were removed from the data set prior to downstream analysis. Finally, the RNA integrity for all samples was measured by median Transcript Integrity across mouse housekeeping genes with RSeQC 2.6.4 (Wang et al., 2016).
Selection of expressed genes
This exploration was performed over a large number of different cell types with different transcriptomes and needed to encompass all the transcripts expressed at levels compatible with quantitatively robust assessment, yet not be confounded by noise stemming from low expression levels in other cells. In practice, genes were retained for consideration in one cell type if they had an expression >20 in at least one treatment condition in that cell. Some genes yield intrinsically high variance even in untreated datasets. These “noisy” genes were removed from consideration (on a per cell-type basis) if their coefficient of variation in that cell was >1.
Basic change metrics, correlation analysis, and signature sets
FC and t test P values in cytokine-exposed cells relative to control were calculated vs. PBS control cells generated in the same batch, only on gene.cell pairs that passed the expression criteria above. For gene–cell pairs that did not pass the expression level or noise criteria, FCs were set to 1 and –log10 (P values) to zero.
Selection of individual cytokine signatures (Table S3, website)
For robustness, while leveraging the power of confirmatory responses in different cells or different cytokines, signature genes were selected as (i) [FC >1.8 and t test –log10(P value) > 3 (nominal)] or (ii) [FC >1.5 and t test –log10(P value) > 2] if the same gene showed [FC >3 and –log10(P value) > 4] in another cytokine/cell combination. Note that these signature tables include all genes responsive in at least one of the cell types, but these are not necessarily responsive in all cell types.
Cluster analysis
A geneset used to support the global cross-comparisons of cytokine effects was selected in several steps. Given the number of cytokine and cell-type combinations, identifying differentially expressed genes was not as straightforward as in simpler study designs. We exploited the power of the combined datasets to identify weaker effects intrinsically validated because occurring repeatedly in several cell types or cytokines, while also capturing effects that might occur only in one cytokine/cell-type combination, but not polluting differentially expressed gene identification with genes flagged because of noise in one of the combinations. In practice, we opted for a combination of selection criteria: (i) transcripts with nominal t test P values <10−4 (cytokine vs. PBS treated) in at least one cytokine/cell combination; (ii) transcripts showing a combined (FC > 2 [or < 0.5] and P value <0.01) in a single cytokine/cell-type (3,506 genes). From this initial set, additional genes were added for consideration if they correlated strongly to clusters of the original genes, and other genes were dropped from consideration if they had high inter-replicate variance (these likely corresponded to experimental variance as they were readily detected by clustering across the FC matrix). Altogether, the process condensed to 2,696 reliably variable genes, grouped into 15 clusters (Fig. 1 E)
During the analysis, a distinct cluster of genes showed exclusive induction by IL15 in Red Pulp Macrophages (MFrp). It proved to be wholly composed of transcripts that are exclusively expressed in T cells in normal conditions—indeed, all transcripts overexpressed in T cells proved to be overexpressed in all three of the MFrp/IL15 samples (genes like Lck, the whole Cd3 cluster, Trac, Trbc1, Cd5, Bcl11b, CD8b, Lef1, Tcf7, and Zap70). Inspection of the flow cytometry sorting plots did not reveal any perturbation in these sorts, nor any indication of a contamination. Genes belonging to this cluster were removed from consideration in macrophages. We surmised that IL15 treatment has increased the phagocytosis of T cells by macrophages in the red pulp, and this cluster was removed for consideration in MFs.
IFN signatures
ISGs were obtained from the pan-immunocyte IFN responses established in Mostafavi et al. (2016). When comparing responses in different cells, a “Common IFN signature” was used (selected as FC >2 in all immunocytes). When determining cell-specific responses (i.e., in B, NK, or Treg cells), we also included genes responsive at FC > 2 in that cell type.
NKT signature
The NKT signature used in Fig. 2 C (182 genes) was obtained from the ImmGen Population Comparison data browser, selecting genes with a difference in mean expression between NKT cells on one side, and naive CD4+ T, activated CD4+ T and Tregs on the other (mean FC between these groups ranging from 1.57 to 1068, median = 12) in ImmGen ULI RNAseq data.
RNA-binding protein motif analysis
We selected a set of 1,055 IL4-repressed genes (members of Fig. 1 Clusters 10, 11, 12, or 13) with a strict IL4 FC <2 in both Bfo and NK cells, and an IL4-neutral reference geneset (2,979 genes with 0.87 < FC < 1.14). RBP target motifs in mouse transcriptomes were downloaded from the oRNAment database (Benoit Bouvrette et al., 2020). The tables were filtered to only consider 3′UTR regions, with motif similarity score >0.8 and unpaired probability <0.2. The enrichment analysis was performed by Fisher’s exact test (FDR-adjusted P value <0.05), comparing the repressed and neutral genesets.
Low cell input CUT&RUN
A low-input (10,000 primary immunocytes) of the CUT&RUN technique (Skene and Henikoff, 2017) was developed in close collaboration with Epicypher and applied here. Briefly, spleens of IL4- or PBS-treated mice were prepared and sorted as above, except that a LIVE/DEAD fixable dead cell stain (# L34971; Thermo Fisher Scientific) was used as a viability marker. After staining (Table S2, www.immgen.org/img/Protocols/ImmGen%2014%20Cell%20Set.pdf), cell suspensions were washed and lightly fixed in 200 μl of 0.1% formaldehyde in PBS (freshly diluted from 37% stock; # 252549; Sigma-Aldrich) for exactly 1 min at room temperature, before quenching with 10 μl 2.5 M glycine in 200 μl. Cells were then washed in FACS medium, and 8 × 104 Bfo cells were sorted, with the same gating strategies as above. Sort volume was measured and diluted with an equal volume of 2X Nuclei Extraction buffer [40 mM Hepes, 20 mM KCl, 0.2% Triton X-100, 40% Glycerol, 2 mM DTT, 1 mM Spermidine, 2X Roche Complete Protease Inhibitor (# 11873580001; Millipore Sigma)] and freshly supplemented to 2X KDAC inhibitor cocktail (2 μM trichostatin A 1 mM sodium butyrate, 1 mM nicotinamide in 70% DMSO). Samples were then frozen in an isopropanol-filled container at −80°C. CUT&RUN (Skene and Henikoff, 2017) was performed in batch mode at EpiCypher. Briefly, samples were thawed on ice and diluted to 105 cells/ml in 1X Nuclei Extraction buffer. 10 μl of activated Concanavalin A beads, 2 μl of 1:50 (vol/vol) SNAP-CUTANA K-MetStat Panel, and 0.5 µg of primary antibody [rabbit IgG (13-0042; Lot 20335004-04; EpiCypher), H3K4me1 (701763; Lot 2135869; Thermo Fisher Scientific), H3K4me3 (13-0041; Lot 210760004-01; EpiCypher), H3K27me3 (MA5-11198; Lot VL3152691; Thermo Fisher Scientific), H3K36me3 (Lot GR3386101-1; ab9050; Abcam), CTCF (13-2014; Lot 21195001-01; EpiCypher), H3K27ac (CST 8173S; Lot 8), and H3.3 (Active Motif 91191; Lot 25820004)] were added per reaction (104 cells) and incubated overnight. The next day, the beads were washed in 250 μl Digitonin Buffer [20 mM pH 7.5 Hepes, 150 mM NaCl, 0.5 mM Spermidine, 1X Roche Complete mini, 0.01% digitonin] twice prior to the addition of 5 μl CUTANA pAG-MNase (20X) in 50 μl of Digitonin Buffer per reaction. Beads were again washed twice in 250 μl Digitonin Buffer and suspended in 50 μl Digitonin Buffer. For chromatin digestion, CaCl2 was added to 2 mM in each reaction to activate the MNase. 33 μl of High Salt Stop Buffer [750 mM NaCl, 26.4 mM EDTA, 5.28 mM EGTA, 66 µg/ml RNase A, 66 µg/ml Glycogen] was added to each reaction to terminate the MNase activity after a 2-h incubation at 4°C. 20 pg CUTANA E.coli Spike-in DNA (EpiCypher 18-1401) was added per sample. Samples were incubated for 10 mins at 37°C to release the cleaved chromatin. CUT&RUN-enriched DNA was isolated from the Concanavalin A beads and cleaned up using 2:1 (Bead:DNA) ratio of Serapure beads. Libraries were prepared using a CUTANA CUT&RUN Library Prep Kit (#14-1001; EpiCypher) and sequenced on Illumina NextSeq 500/550 (paired end 2 × 75 bp read). All autoCUT&RUN steps were optimized and performed on Tecan Freedom EVO robotics platforms with gentle rocking for incubation steps and magnetic capture for medium exchange/washing steps.
Fastq data were adaptor trimmed using Trim Galore v0.6.6. They were aligned to mm 10 reference genome using bowtie2 v.2.3.4.3 with parameters -X 700 -I 10. Non-unique, duplicated, and ChrM reads were removed using SAMTools view and Picard MarkDuplicates. BEDTools with the option intersect -v was used to filter reads that did not overlap with ENCODE blacklist. Reads that overlapped with gene bodies were identified and quantified using bedtools intersect from BEDTools and GRCm38.06 gene body annotations from Ensembl release 67. Peaks were called using SEACR (Meers et al., 2019) with the option “norm” and “relaxed” for CTCF, H3K4me3, and H3K27ac while “norm” and “stringent” were used for H3.3, H3K27me3, H3K36me3, and H3K4me1. Genome coverage tracks were generated using deeptools2 with parameters bamCoverage—binSize 50—normalizeUsing CPM—ignoreDuplicates—extendReads and visualized using IGV v2.10.2.
Statistics
Statistical treatment of the gene expression data is as described above. All tests were two-sided. t test was used to compare cell activation, P < 0.05 was considered significant.
Online supplementary material
Fig. S1 shows the averaged response relative to control, for all cells and all cytokines, of genes that belong to the clusters defined in Fig. 1 E, and Fig. S2 shows the transcriptional responses in Treg cells after administration of IL2/anti-IL2 complexes. Fig. S3 shows changes in posttranslational modifications of histone proteins that denote activity at promoters (H3-K4me3) or enhancers (H3-K27Ac). Table S1 shows the form, origin, and dose of cytokines administered. Table S2 lists the cell types analyzed and how they were sorted. Table S3 gives the average FC, relative to the mean of cells from vehicle-injected mice, after injection of all cytokines and in all cell types. Table S4 shows the changes induced by IL21 in CD8+ T cells, for genes that belong to the canonical NKT signature.
Supplementary Material
shows the form, origin, and dose of cytokines administered.
lists the cell types analyzed and how they were sorted.
gives the average FC, relative to the mean of cells from vehicle-injected mice, after injection of all cytokines and in all cell types.
shows the changes induced by IL21 in CD8+ T cells, for genes that belong to the canonical NKT signature.
Acknowledgments
We thank Drs. C. Garcia, W. Leonard, Paul Anderson, and D. Cantrell for discussion and advice; K. Hattori, A. Ortiz-Lopez, C. Araneo, D. Ischiu, and A. Wood for help with mice and cell sorting; and the EpiCypher team (C. Lin, M. Marunde, D. Maryanski) for fruitful collaboration.
This study was funded by a resource grant from National Institutes of Health/National Institute of Allergy and Infectious Diseases to the ImmGen consortium (AI072073). S.L. Nutt was supported by a National Health and Medical Research Council Fellowship (1155342).
Author contributions: A. Baysoy, K. Seddu, T. Salloum, C.A. Dawson, J. Lee, and J. Tellier performed experiments and generated the data; A. Baysoy, K. Seddu, T. Salloum, C.A. Dawson, J. Lee, S. Gal-oz, H. Ner-Gaon, J. Tellier, A. Millan, A. Sasse, B. Brown, and C. Benoist analyzed the data; L. Lanier, T. Shay, S. Nutt, D.F. Dwyer, and C. Benoist provided oversight; C. Benoist, B. Brown, L. Lanier, T. Shay, and S. Nutt secured funding, and all authors participated in writing.
Data availability
Results are displayed on the ImmGen website (https://www.immgen.org/Databrowser19/Cytokines.html). Primary data have been deposited to the NCBI Gene Expression Omnibus (GSE180020). Custom code is available from the corresponding author upon reasonable request.
References
- Angkasekwinai, P., and Dong C.. 2021. IL-9-producing T cells: potential players in allergy and cancer. Nat. Rev. Immunol. 21:37–48. 10.1038/s41577-020-0396-0 [DOI] [PubMed] [Google Scholar]
- Barata, J.T., Durum S.K., and Seddon B.. 2019. Flip the coin: IL-7 and IL-7R in health and disease. Nat. Immunol. 20:1584–1593. 10.1038/s41590-019-0479-x [DOI] [PubMed] [Google Scholar]
- Benoit Bouvrette, L.P., Bovaird S., Blanchette M., and Lécuyer E.. 2020. oRNAment: A database of putative RNA binding protein target sites in the transcriptomes of model species. Nucleic Acids Res. 48:D166–D173. 10.1093/nar/gkz986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, L.J., Avery D.T., Ma C.S., Moens L., Deenick E.K., Bustamante J., Boisson-Dupuis S., Wong M., Adelstein S., Arkwright P.D., et al. 2013. IL-21 signalling via STAT3 primes human naive B cells to respond to IL-2 to enhance their differentiation into plasmablasts. Blood. 122:3940–3950. 10.1182/blood-2013-06-506865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezbradica, J.S., and Medzhitov R.. 2009. Integration of cytokine and heterologous receptor signaling pathways. Nat. Immunol. 10:333–339. 10.1038/ni.1713 [DOI] [PubMed] [Google Scholar]
- Bezman, N.A., Kim C.C., Sun J.C., Min-Oo G., Hendricks D.W., Kamimura Y., Best J.A., Goldrath A.W., Lanier L.L., and Immunological Genome Project Consortium . 2012. Molecular definition of the identity and activation of natural killer cells. Nat. Immunol. 13:1000–1009. 10.1038/ni.2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, J., Carotta S., Thong R.P., Chan C.J., Hayakawa Y., Smyth M.J., and Nutt S.L.. 2010. The interactions of multiple cytokines control NK cell maturation. J. Immunol. 185:6679–6688. 10.4049/jimmunol.0903354 [DOI] [PubMed] [Google Scholar]
- Castellanos-Montiel, M.J., Chaineau M., and Durcan T.M.. 2020. The neglected genes of ALS: Cytoskeletal dynamics impact synaptic degeneration in ALS. Front. Cell. Neurosci. 14:594975. 10.3389/fncel.2020.594975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X.N., Shao M., Li J.T., Wang Y.F., Qi J., Xu Z.G., and Shi D.L.. 2017. Leucine repeat adaptor protein 1 interacts with Dishevelled to regulate gastrulation cell movements in zebrafish. Nat. Commun. 8:1353. 10.1038/s41467-017-01552-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosman, D., Lyman S.D., Idzerda R.L., Beckmann M.P., Park L.S., Goodwin R.G., and March C.J.. 1990. A new cytokine receptor superfamily. Trends Biochem. Sci. 15:265–270. 10.1016/0968-0004(90)90051-C [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot, J.D., Rasmussen J.P., Gavin M.A., and Rudensky A.Y.. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6:1142–1151. 10.1038/ni1263 [DOI] [PubMed] [Google Scholar]
- Giri, J.G., Ahdieh M., Eisenman J., Shanebeck K., Grabstein K., Kumaki S., Namen A., Park L.S., Cosman D., and Anderson D.. 1994. Utilization of the beta and gamma chains of the IL-2 receptor by the novel cytokine IL-15. EMBO J. 13:2822–2830. 10.1002/j.1460-2075.1994.tb06576.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, R., and Kaplan M.H.. 2011. A brief history of IL-9. J. Immunol. 186:3283–3288. 10.4049/jimmunol.1003049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada, M., Levy D.M., McKeag L., Murray K., Schröder A.J., Canfield S.M., Traver G., and Rothman P.B.. 2010. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc. Natl. Acad. Sci. USA. 107:821–826. 10.1073/pnas.0909235107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman, E., Salyakina D., De Heusch M., Hoek K.L., Llanes J.M., Castro I., Wright J.A., Clark E.S., Dykxhoorn D.M., Capobianco E., et al. 2015. Distinct transcriptomic features are associated with transitional and mature B-cell populations in the mouse spleen. Front. Immunol. 6:30. 10.3389/fimmu.2015.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemsz, M.J., Justement L.B., Palmer E., and Cambier J.C.. 1989. Induction of c-fos and c-myc expression during B cell activation by IL-4 and immunoglobulin binding ligands. J. Immunol. 143:1032–1039. 10.4049/jimmunol.143.3.1032 [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Tsubosaka Y., Hori M., Narumiya S., Ozaki H., and Murata T.. 2013. Prostaglandin D2-DP signaling promotes endothelial barrier function via the cAMP/PKA/Tiam1/Rac1 pathway. Arterioscler. Thromb. Vasc. Biol. 33:565–571. 10.1161/ATVBAHA.112.300993 [DOI] [PubMed] [Google Scholar]
- Le, Q.T., Gomez G., Zhao W., Hu J., Xia H.Z., Fukuoka Y., Katunuma N., and Schwartz L.B.. 2011. Processing of human protryptase in mast cells involves cathepsins L, B, and C. J. Immunol. 187:1912–1918. 10.4049/jimmunol.1001806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, W.J., Lin J.X., and O’Shea J.J.. 2019. The γc family of cytokines: Basic biology to therapeutic ramifications. Immunity. 50:832–850. 10.1016/j.immuni.2019.03.028 [DOI] [PubMed] [Google Scholar]
- Lin, J.X., and Leonard W.J.. 2018. The common cytokine receptor γ chain family of cytokines. Cold Spring Harb. Perspect. Biol. 10:a028449. 10.1101/cshperspect.a028449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.X., and Leonard W.J.. 2019. Fine-tuning cytokine signals. Annu. Rev. Immunol. 37:295–324. 10.1146/annurev-immunol-042718-041447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linossi, E.M., and Nicholson S.E.. 2015. Kinase inhibition, competitive binding and proteasomal degradation: Resolving the molecular function of the suppressor of cytokine signaling (SOCS) proteins. Immunol. Rev. 266:123–133. 10.1111/imr.12305 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, A., Koka R., and Burkett P.. 2006. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu. Rev. Immunol. 24:657–679. 10.1146/annurev.immunol.24.021605.090727 [DOI] [PubMed] [Google Scholar]
- Makeyev, A.V., and Liebhaber S.A.. 2002. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA. 8:265–278. 10.1017/S1355838202024627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek, T.R. 2008. The biology of interleukin-2. Annu. Rev. Immunol. 26:453–479. 10.1146/annurev.immunol.26.021607.090357 [DOI] [PubMed] [Google Scholar]
- Marchingo, J.M., Sinclair L.V., Howden A.J., and Cantrell D.A.. 2020. Quantitative analysis of how Myc controls T cell proteomes and metabolic pathways during T cell activation. Elife. 9:e53725. 10.7554/eLife.53725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, J.J., Baker B., and Ryan J.J.. 2015. Mast cell production and response to IL-4 and IL-13. Cytokine. 75:57–61. 10.1016/j.cyto.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers, M.P., Tenenbaum D., and Henikoff S.. 2019. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 12:42. 10.1186/s13072-019-0287-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, T., Liu Z.J., Kawahara A., Minami Y., Yamada K., Tsujimoto Y., Barsoumian E.L., Permutter R.M., and Taniguchi T.. 1995. Three distinct IL-2 signaling pathways mediated by bcl-2, c-myc, and lck cooperate in hematopoietic cell proliferation. Cell. 81:223–231. 10.1016/0092-8674(95)90332-1 [DOI] [PubMed] [Google Scholar]
- Moro, A., Gao Z., Wang L., Yu A., Hsiung S., Ban Y., Yan A., Sologon C.M., Chen X.S., and Malek T.R.. 2022. Dynamic transcriptional activity and chromatin remodeling of regulatory T cells after varied duration of interleukin-2 receptor signaling. Nat. Immunol. 23:802–813. 10.1038/s41590-022-01179-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, S.C., Orekhova T., Meadows M.J., Heidorn S.M., Yang J., and Finkelman F.D.. 2006. IL-4 induces in vivo production of IFN-gamma by NK and NKT cells. J. Immunol. 176:5299–5305. 10.4049/jimmunol.176.9.5299 [DOI] [PubMed] [Google Scholar]
- Mostafavi, S., Yoshida H., Moodley D., LeBoité H., Rothamel K., Raj T., Ye C.J., Chevrier N., Zhang S.Y., Feng T., et al. 2016. Parsing the interferon transcriptional network and its disease associations. Cell. 164:564–578. 10.1016/j.cell.2015.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai, K., Tsai M., Saito H., and Galli S.J.. 2018. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol. Rev. 282:121–150. 10.1111/imr.12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadella, K.S., Saji M., Jacob N.K., Pavel E., Ringel M.D., and Kirschner L.S.. 2009. Regulation of actin function by protein kinase A-mediated phosphorylation of Limk1. EMBO Rep. 10:599–605. 10.1038/embor.2009.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, K., Spolski R., Feng C.G., Qi C.F., Cheng J., Sher A., Morse H.C. III, Liu C., Schwartzberg P.L., and Leonard W.J.. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science. 298:1630–1634. 10.1126/science.1077002 [DOI] [PubMed] [Google Scholar]
- Picelli, S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., and Sandberg R.. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 10:1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- Picelli, S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., and Sandberg R.. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9:171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Rapp, U.R., Cleveland J.L., Brightman K., Scott A., and Ihle J.N.. 1985. Abrogation of IL-3 and IL-2 dependence by recombinant murine retroviruses expressing v-myc oncogenes. Nature. 317:434–438. 10.1038/317434a0 [DOI] [PubMed] [Google Scholar]
- Richard, M., Louahed J., Demoulin J.B., and Renauld J.C.. 1999. Interleukin-9 regulates NF-kappaB activity through BCL3 gene induction. Blood. 93:4318–4327. 10.1182/blood.V93.12.4318 [DOI] [PubMed] [Google Scholar]
- Ring, A.M., Lin J.X., Feng D., Mitra S., Rickert M., Bowman G.R., Pande V.S., Li P., Moraga I., Spolski R., et al. 2012. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat. Immunol. 13:1187–1195. 10.1038/ni.2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, S.H., and Cantrell D.A.. 2018. Signaling and function of interleukin-2 in T lymphocytes. Annu. Rev. Immunol. 36:411–433. 10.1146/annurev-immunol-042617-053352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, S.H., Rollings C., Anderson K.E., Hawkins P.T., Stephens L.R., and Cantrell D.A.. 2016. Phosphoproteomic analyses of interleukin 2 signaling reveal integrated JAK kinase-dependent and -independent networks in CD8(+) T cells. Immunity. 45:685–700. 10.1016/j.immuni.2016.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene, P.J., and Henikoff S.. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 6:e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler, J.B., Moraga I., Mendoza J.L., and Garcia K.C.. 2015a. Insights into cytokine-receptor interactions from cytokine engineering. Annu. Rev. Immunol. 33:139–167. 10.1146/annurev-immunol-032713-120211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler, J.B., Tomala J., Luca V.C., Jude K.M., Dong S., Ring A.M., Votavova P., Pepper M., Kovar M., and Garcia K.C.. 2015b. Antibodies to interleukin-2 elicit selective T cell subset potentiation through distinct conformational mechanisms. Immunity. 42:815–825. 10.1016/j.immuni.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka, S., Yamada H., Haniuda K., Saruwatari H., Ichihashi M., Renauld J.C., and Kitamura D.. 2018. IL-9 receptor signaling in memory B cells regulates humoral recall responses. Nat. Immunol. 19:1025–1034. 10.1038/s41590-018-0177-0 [DOI] [PubMed] [Google Scholar]
- Tangye, S.G., and Ma C.S.. 2020. Regulation of the germinal center and humoral immunity by interleukin-21. J. Exp. Med. 217:e20191638. 10.1084/jem.20191638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyffels, L., Gueydan C., and Kruys V.. 2011. Shuttling SR proteins: More than splicing factors. FEBS J. 278:3246–3255. 10.1111/j.1742-4658.2011.08274.x [DOI] [PubMed] [Google Scholar]
- Vigorito, E., Billadeu D.D., Savoy D., McAdam S., Doody G., Fort P., and Turner M.. 2003. RhoG regulates gene expression and the actin cytoskeleton in lymphocytes. Oncogene. 22:330–342. 10.1038/sj.onc.1206116 [DOI] [PubMed] [Google Scholar]
- Vink, A., Warnier G., Brombacher F., and Renauld J.C.. 1999. Interleukin 9-induced in vivo expansion of the B-1 lymphocyte population. J. Exp. Med. 189:1413–1423. 10.1084/jem.189.9.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J., and Carpenter P.B.. 2012. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13:115–126. 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Nie J., Sicotte H., Li Y., Eckel-Passow J.E., Dasari S., Vedell P.T., Barman P., Wang L., Weinshiboum R., et al. 2016. Measure transcript integrity using RNA-seq data. BMC Bioinformatics. 17:58. 10.1186/s12859-016-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills-Karp, M., and Finkelman F.D.. 2008. Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci. Signal. 1:pe55. 10.1126/scisignal.1.51.pe55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfarth, A.A., Dhar S., Goon J.B., Ezeanya U.I., Ferrando-Martínez S., and Lee B.H.. 2022. Advancements of common gamma-chain family cytokines in cancer immunotherapy. Immune Netw. 22:e5. 10.4110/in.2022.22.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, M., Ren J., Liu Q., Glassman C.R., Sheahan T.P., Picton L.K., Moreira F.R., Rustagi A., Jude K.M., Zhao X., et al. 2022. Facile discovery of surrogate cytokine agonists. Cell. 185:1414–1430.e19. 10.1016/j.cell.2022.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, A., Ito M., Chikuma S., Akanuma T., and Nakatsukasa H.. 2018. Negative regulation of cytokine signaling in immunity. Cold Spring Harb. Perspect. Biol. 10:a028571. 10.1101/cshperspect.a028571 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the form, origin, and dose of cytokines administered.
lists the cell types analyzed and how they were sorted.
gives the average FC, relative to the mean of cells from vehicle-injected mice, after injection of all cytokines and in all cell types.
shows the changes induced by IL21 in CD8+ T cells, for genes that belong to the canonical NKT signature.
Data Availability Statement
Results are displayed on the ImmGen website (https://www.immgen.org/Databrowser19/Cytokines.html). Primary data have been deposited to the NCBI Gene Expression Omnibus (GSE180020). Custom code is available from the corresponding author upon reasonable request.