Abstract
Penetrating flexible electrode arrays can simultaneously record thousands of individual neurons in the brains of live animals. However, it has been challenging to spatially map and longitudinally monitor the dynamics of large three-dimensional neural networks. Here we show that optimized ultraflexible electrode arrays distributed across multiple cortical regions in head-fixed mice and in freely moving rats allow for months-long stable electrophysiological recording of several thousand neurons at densities of about 1,000 neural units per cubic millimetre. The chronic recordings enhanced decoding accuracy during optogenetic stimulation and enabled the detection of strongly coupled neuron pairs at the million-pair and millisecond scales, and thus the inference of patterns of directional information flow. Longitudinal and volumetric measurements of neural couplings may facilitate the study of large-scale neural circuits.
The brain is a highly interconnected and dynamically evolving network composed of a large number of densely packed neurons. These physiologically and behaviourally relevant circuits span diverse temporal and spatial scales. Spikes, the action potentials from individual neurons, conduct basic information processing on the timescale of milliseconds, but changes in activity patterns that support adaptation, learning and related functions occur on timescales ranging from seconds to years. The complex neural circuits consist of both nearby groups of neurons and neurons distributed across multiple brain areas1. To fully observe, study and possibly augment these neural circuits, one needs to be able to measure large ensembles of neurons simultaneously at millisecond resolutions and over chronic timescales; resolve microscopic neural correlations to understand local information processing among neighbouring neurons; and record neurons at a high density across regions in a functional circuit to understand how information flows within and among regions as the brain processes sensory information, learns, remembers, makes decisions and takes action2,3.
The rapidly developing and growing body of available neurotechnology can meet some but not all of these stringent requirements. The development of fluorescent indicators and optical imaging techniques has enabled mapping of a large neuronal population within seconds and detection of neural dynamics at the microscale4, but detecting the millisecond dynamics in a sizable brain volume at the single-neuron resolution remains fundamentally challenging3,5. Recent state-of-the-art penetrating neural electrodes, both flexible6 and rigid7,8, have enabled simultaneous recording of hundreds to thousands of neurons, but three-dimensional (3D) volumetric mapping of a local circuit at high spatial resolutions remains challenging. This is in large part owing to the form factor of the electrodes2 and the resulting invasiveness to the brain, which impede their high-density implantation and undermine their recording stability and efficacy in both the short and long terms9.
Here we present ultraflexible electrode arrays for the volumetric mapping of neural networks in the brains of behaving rodents, microscopically and macroscopically, at the thousands-of-neurons scale and over chronic periods (several months). We take advantage of the biocompatibility of ultraflexible nanoelectronic thread electrodes (NETs)10, which afford orders-of-magnitude improvement in mechanical compliance, a seamless integration with neural tissue and stable longitudinal recordings of individual neurons. We demonstrate a scalable and flexible neural interface allowing for volumetric recordings at high-density of up to 1,000 units per mm3 of brain volume, as well as distributed recordings of several thousands of units. By analysing the timing of all spikes from the recorded neurons, we evaluated their pairwise correlation structures at the million-pair scale and at millisecond resolution, from which we inferred patterns of information flow in the neuronal population11. We also show that the large-scale, long-lasting electrophysiological mapping substantially improved decoding accuracy, elucidated populational plasticity and revealed chronic network stability. These results extend the spatiotemporal scales for measuring neuronal activity and underscore the importance of their use to interrogate complex microcircuitry and macrocircuitry in the brain.
Results
We constructed large-scale 3D electrode arrays from modularized devices in flexible arrangements. For this purpose, we designed and fabricated three types of 128-channel NET modules (Fig. 1a,b and Supplementary Video 1): type-I for distributed uniform coverage spanning an approximately 1 mm × 1 mm vertical plane; type-II for recordings at an oversampling spatial resolution that enhances single-unit isolation; and type-III for localized recordings in inhomogeneous anatomical structures such as the hippocampus. All these designs had an overall thickness of 1 μm, which provides unique structural ultraflexibility (Fig. 1c) and minimizes the chronic invasiveness to host tissues10,12 (Extended Data Fig. 1). The shank width tapered from 60 μm to 30 μm for type-I, 120 μm to 60 μm for type-II, and 100 μm to 35 μm for type-III. Importantly, these dimensions resulted in cross-sectional areas of the shanks of 30–100 μm2, which is comparable to the typical area of a soma, and smaller than most conventional neural electrodes by 1–2 orders of magnitude2. These markedly miniaturized footprints made it possible to implant NET modules at very high densities without occupying a large fraction of the extracellular space or substantially displacing and damaging the surrounding tissues10 (Extended Data Fig. 1). In our initial testing, we found that the type-II and type-III designs allow for the recording of individual neurons by multiple channels due to their high electrode density, and therefore perform better in single-unit isolation (Extended Data Fig. 2), while the type-I electrode design excels in maximizing the volumetric coverage of neural recording. We therefore deemed that the type-I design better serves the overall goal of this study and used only this design in our subsequent experiments. To accommodate large-scale electrophysiology in behaving rodents, we developed and implemented light-weight miniature connections for NETs (Fig. 1a and Extended Data Fig. 3) and a compact, robust head-mounted strategy based on a scalable neural data acquisition system6 (Methods). Combining the development of ultraflexible NETs, implantation and data acquisition techniques, we realized longitudinal intracortical recordings of up to 18 128-channel NET modules comprising 2,304 simultaneously monitored channels in behaving mice and rats (Methods).
Fig. 1 |. Overview of the large-scale modular NET technology.
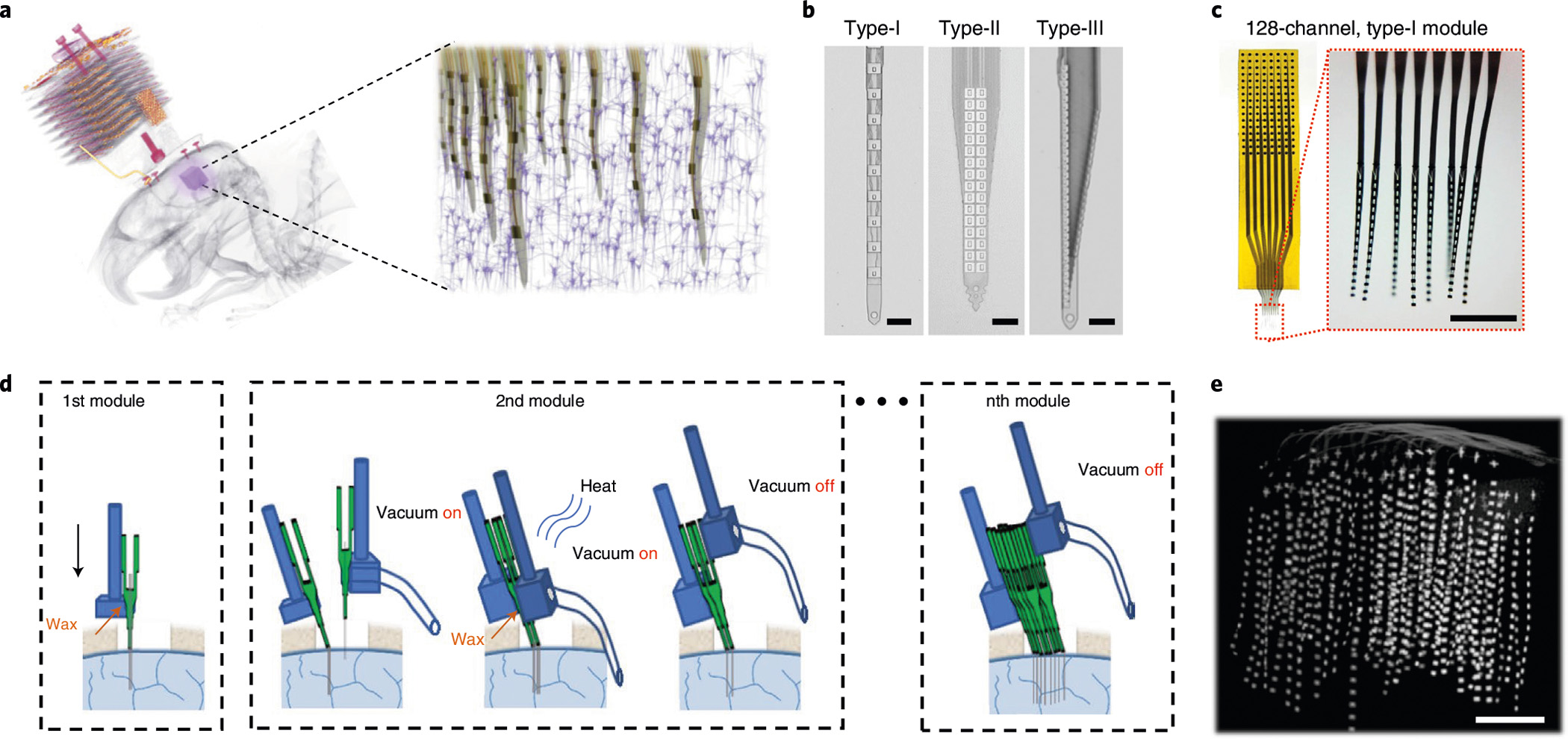
a, Left: a micro-CT scan of an implanted NET array in a rat brain consisting of 8 128-channel modules (1,024 channels in total) at high 3D density. The purple cube highlights the NET array. Right: schematics of the 3D NET array embedded in cortical tissues. b, Images of the three types of NET arrays used in the study. c, Photos of a 128-channel type-I NET module. Red dotted box highlights the released front end. Inset: 8-shank, 128-channel flexible section immersed in water. d, Schematics showing assembled NET modules (each containing 8 × 16 intracortical recording sites) implanted sequentially into the brain using a pair of stereotaxic micromanipulators, one module at a time. e, A micro-CT scan showing the volumetric distribution of an 8 × 8 × 16 (1,024-channel) NET array in a mouse visual cortex. The targeted module-to-module spacing is 150 μm. Scale bars, 50 μm (b), 500 μm (c) and 500 μm (e).
Building upon our previously published surgical procedures13,14, we further optimized a diverse set of parameters including the insertion speed and the spacing of insertion shuttles to enable high-throughput implantation of large numbers of NET modules. In typical surgeries, we sequentially inserted 8–18 128-channel NET modules, with module-to-module spacings as small as 150 μm, while minimizing damage to surface vasculature (Fig. 1d, Supplementary Methods and Fig. 1). In-situ micro-computed tomography (μCT) imaging of a representative implantation in mouse brain confirmed a relatively uniform volumetric deployment of 1,024 electrode contacts throughout the designated region of approximately 1 mm × 1 mm × 1 mm (Fig. 1e and Supplementary Video 2). With a contact density of about 1,000 mm−3, this implant represents the highest volumetric recording density we have achieved in this work.
These large-scale electrode arrays enabled dense volumetric samplings of electrophysiological activity in the implanted brain regions. In Fig. 2, we show that a representative example of 1,024-channel recording in an approximately 1 mm3 volume in rat visual cortex provided high-fidelity 3D reconstruction of local field potentials (LFPs, Fig. 2a and Supplementary Video 3) and enabled the isolation of 987 units (sorted using Mountainsort15, Fig. 2b; see Methods for details). We note that a sorted unit could be a single unit putatively representing the activity of one neuron, or a multi unit representing the activity of a small group of neighbouring neurons (see Discussion and Methods). We further evaluated the million pairwise correlations among all sorted units, identified about 40,000 significantly correlated pairs, and determined the temporal bias in the correlation at the millisecond resolution, which indicated preferred firing sequence (see Methods). These measurements detected the activity and resolved the temporal correlation in the activity of approximately 5–10% of the total active neurons in the recorded volume, which is on par with the high-density recording reported by Neuropixels probes reported recently16. Furthermore, our surgical strategy allows us to flexibly implant NET modules at multiple sites across brain regions. In n = 5 animals, we had on average 1,058 simultaneously recording contacts per animal, yielding 1.25 units per contact and a mean signal to noise ratio (SNR) of 5.51. The representative recording performance and individual variations in unit yield, amplitude and SNR are summarized in Fig. 2c. Out of the total of 6,598 sorted units, 2,854 are single units, representing a 43.3% single-unit yield. In the following sections, we demonstrate the unique capabilities of this versatile neural interface in a variety of brain regions, implantation densities and channel counts (Supplementary Table 1).
Fig. 2 |. Overview of the recording performance.
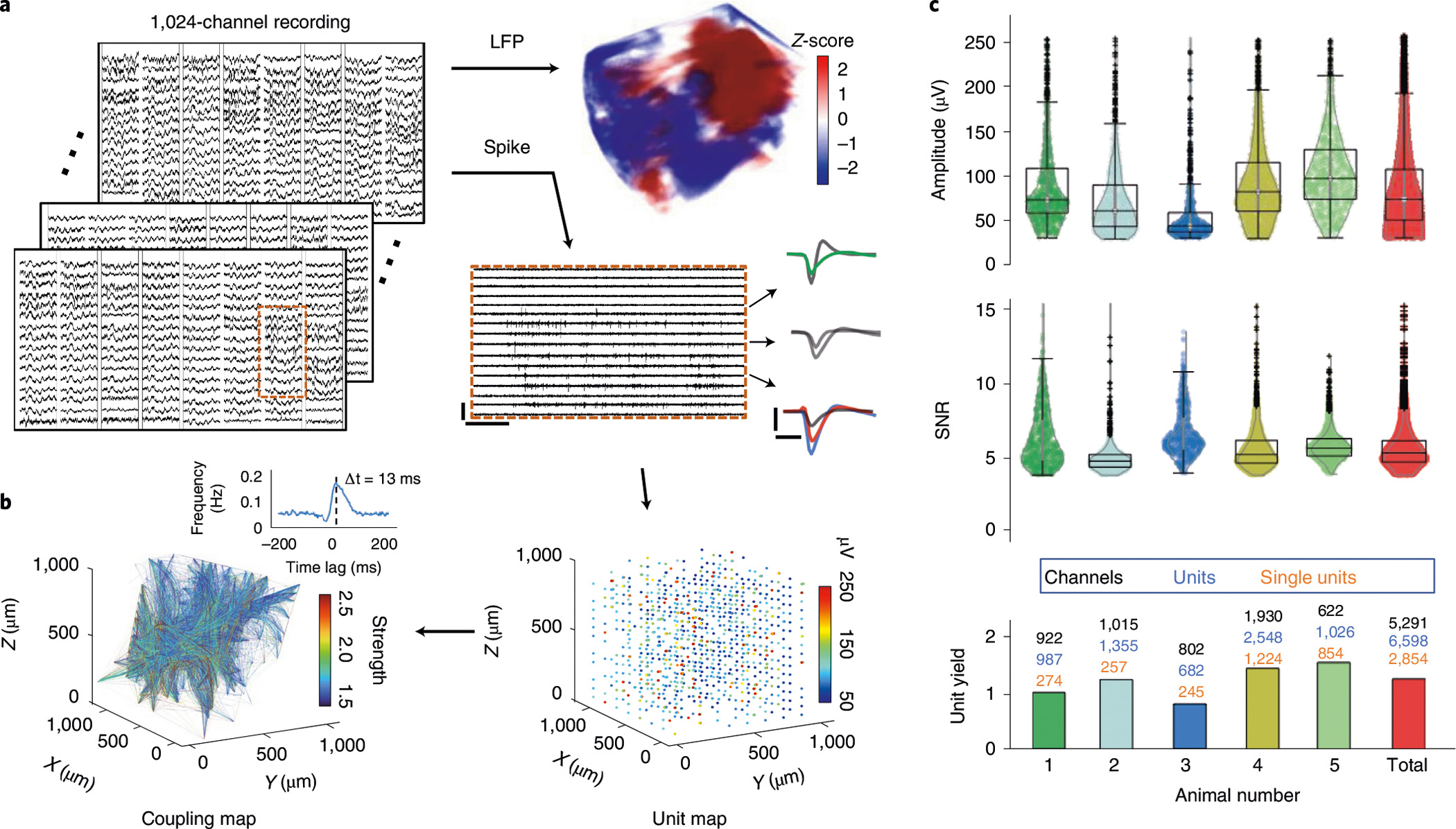
a, Representative recordings from 1,024 channels in an animal (#1 in c) showing both LFP (bandpass filtered at (8 12) Hz) and spikes (high-pass filtered at 300 Hz). Scale bars, 250 μV (vertical) and 0.3 s (horizontal). Typical waveforms are depicted. Scale bars, 100 μV (vertical) and 1 ms (horizontal). b, Volumetric recording enables reconstruction of unit location (right) and mapping of pairwise correlation (left). The amplitude of units and the coupling strength are colour coded. Inset: a representative cross-correlogram showing the detection of temporal bias at the millisecond resolution. c, Distribution of unit amplitude, SNR and unit yields of n = 5 implanted animals. The average of all units is shown in the last column. All units are shown in the violin plots.
High-density volumetric electrophysiological mapping of the visual cortex
To demonstrate the ability to provide high-resolution measurements across all three dimensions, we implanted NET arrays at high volumetric densities for 3D electrophysiological mapping in specific brain regions. Figure 3 shows one example of 10 type-I NET modules implanted in a mouse visual cortex (n = 1) at a module-to-module spacing of 200 μm, which gave a relatively uniform 3D distribution of 1,280 contact sites (1,036 remained connected after surgery) in an approximately 1 mm × 1.8 mm × 1 mm brain volume (Fig. 3a,b). To showcase the recording capacity in resolving local neural dynamics, to the awake animal we presented visual stimuli of drifting gratings at various angles, detected LFPs and spiking activity, evaluated the response of each sorted unit to visual stimuli and quantified their respective tuning curves to the grating angles (Fig. 3c). In a representative session at 6 weeks post implantation, we isolated 1,355 units simultaneously, at a yield of 1.31 units per recording site (Fig. 3d). We observed that about 40% of the units were significantly modulated by the visual stimuli and showed orientation selectivity index (OSI) > 0.5 (Fig. 3d)17. The spatial organization of the preferred directions in this cortical region revealed a classic ‘salt-and-pepper’ micro-architecture (Fig. 3e) consistent with previous studies using optical imaging18.
Fig. 3 |. Volumetric high-density mapping of the visual cortex in an awake head-fixed mouse.
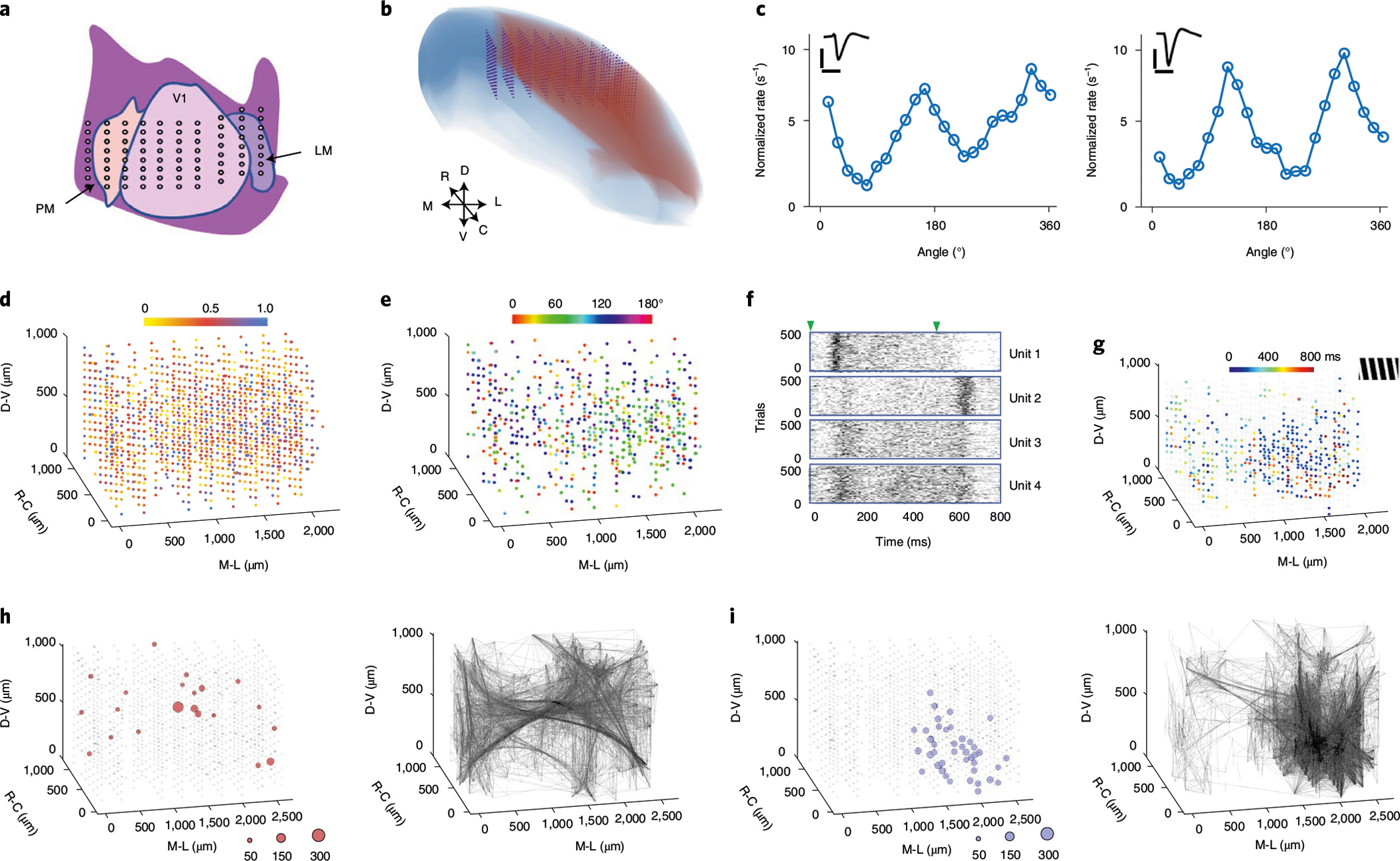
a, Schematic representation of the targeted implantation location in the visual cortex. LM, lateromedial area; V1, primary visual cortex; PM, posteromedial area. Each dot represents a 16-channel shank. b, Reconstruction of expected locations of all 8 × 10 recording shanks (1,280 channels). Blue, neocortex of right hemisphere; red, visual area19; purple, NET arrays. D, V, R, C, M and L denote the orientations of dorsal, ventral, anterior, posterior, medial and lateral, respectively. c, Two sorted units recorded by the same contact in V1 showing distinct orientation tuning curves. Inset: unit waveforms. Scale bars, 1 ms (horizontal) and 100 μV (vertical). d, Colour-coded OSI for all recorded units (1,355 in total). All units are individually plotted as a sphere superimposed on the closest contact site. e, Preferred angle of grating for units with OSI > 0.5. f, Spike raster of 500 trials from representative units highlighting different temporal responses to grating stimuli. Triangles mark the start and end of the gratings. g, The peak firing time from the most active 30% of units under a stimulus of 165° drifting grating (shown in the upper right inset). h,i, Spatial distribution of the super nodes (left) and the strong couplings (right) without (h) and with (i) visual stimulation. The diameter of the circles is scaled to the number of couplings made at each node.
Taking advantage of the high spatial and temporal resolution of the 3D electrophysiological mapping, we further demonstrate the ability to map the spatiotemporal structure of the network responses to visual stimuli. Temporally, we resolve the diverse and distinct temporal patterns of units responding to the stimuli: the time course of firing rate variation change from neuron to neuron (Fig. 3f, Supplementary Fig. 2 and Video 4). Spatially, we map the 3D location of neurons and their response time: units near the lateromedial area (LM) are most likely to have the highest firing rate at either the beginning or the end of each trial, while units that respond in the middle of the trials are distributed throughout the recording regions (Fig. 3g). This spatiotemporal pattern of neural dynamics was similar across all tested stimuli (Supplementary Fig. 3).
The high temporal resolution of electrophysiological recordings at the timescale of milliseconds, together with large-scale parallel recordings at the scale of 1,000s of channels, permitted a massive mapping of correlation structures at a high temporal resolution. We computed the pairwise cross-correlations between the spiking times of each pair of units and analysed the change in pairwise correlation across the two brain states of visual stimuli on and off from the same recording session (Supplementary Fig. 4). In the presence of visual stimuli, the mean correlation strength was significantly enhanced (unpaired two-sided t-test, P < 10−15), while the mean latency of the cross-correlation peak decreased (Wilcoxon rank sum test, P < 10−15). In particular, the number of short-range coupling (<1 mm) increased, resulting in a decrease of the mean distance of short-range coupling when visual stimuli were presented (unpaired two-sided t-test, P < 10−15). As a comparison, we computed the same pairwise cross-correlations using only single units and found that their distributions share similar features (Supplementary Fig. 4; see Discussion).
To further demonstrate our ability to map and evaluate the changes in the neural correlation structures, we compared the spatial patterns of coupling with and without visual stimulation. To capture and visualize the major information flow in this population, we then extracted the strongly correlated unit pairs (selected by meeting two criteria: correlation strength >2σ above baseline and peak latency <20 ms; see Methods). We refer to these as ‘strongly coupled pairs’ hereafter. The presence of visual stimuli changed the spatial distribution of strongly coupled pairs from being distributed through the recording volume to being spatially concentrated near the LM regions with stimulation (Fig. 3h,i). Consistently, the ‘super nodes’, defined as units with numbers of strong couplings that exceed 2σ from the mean value, altered their distribution in a similar manner. These results highlight the ability to resolve local network communications by high-density volumetric recording.
Visual decoding using high-density volumetric recordings
The large number of units being recorded also allowed us to decode stimulus orientation with high accuracy. We modified and applied to our data a recently published linear decoder designed for an optically recorded neural population20 (see Methods). To optimize the decoding accuracy, we tested a number of parameters, including the number of units, the time period used for averaging spiking activities in each trial and the number of the trials used for training (Supplementary Figs. 5 and 9). We found that the optimal setting yielded a mean error of 2.3° when we used all 40 trials of all the 1,355 units and a time period of 200–600 ms from the onset of stimuli for averaging (Fig. 4a). The number of neurons we simultaneously recorded and the resulting decoding precision exceed what has been previously demonstrated by electrophysiological recordings and approach the state-of-the-art large-scale two-photon imaging that resolved and used over 1,000 neurons20. Consistent with previous studies20, the ability to record a large population of neurons is evidently the key to precise decoding. As shown in Fig. 4b, increasing the number of included units reduced the decoding error.
Fig. 4 |. Visual decoding using large-scale neural recordings.
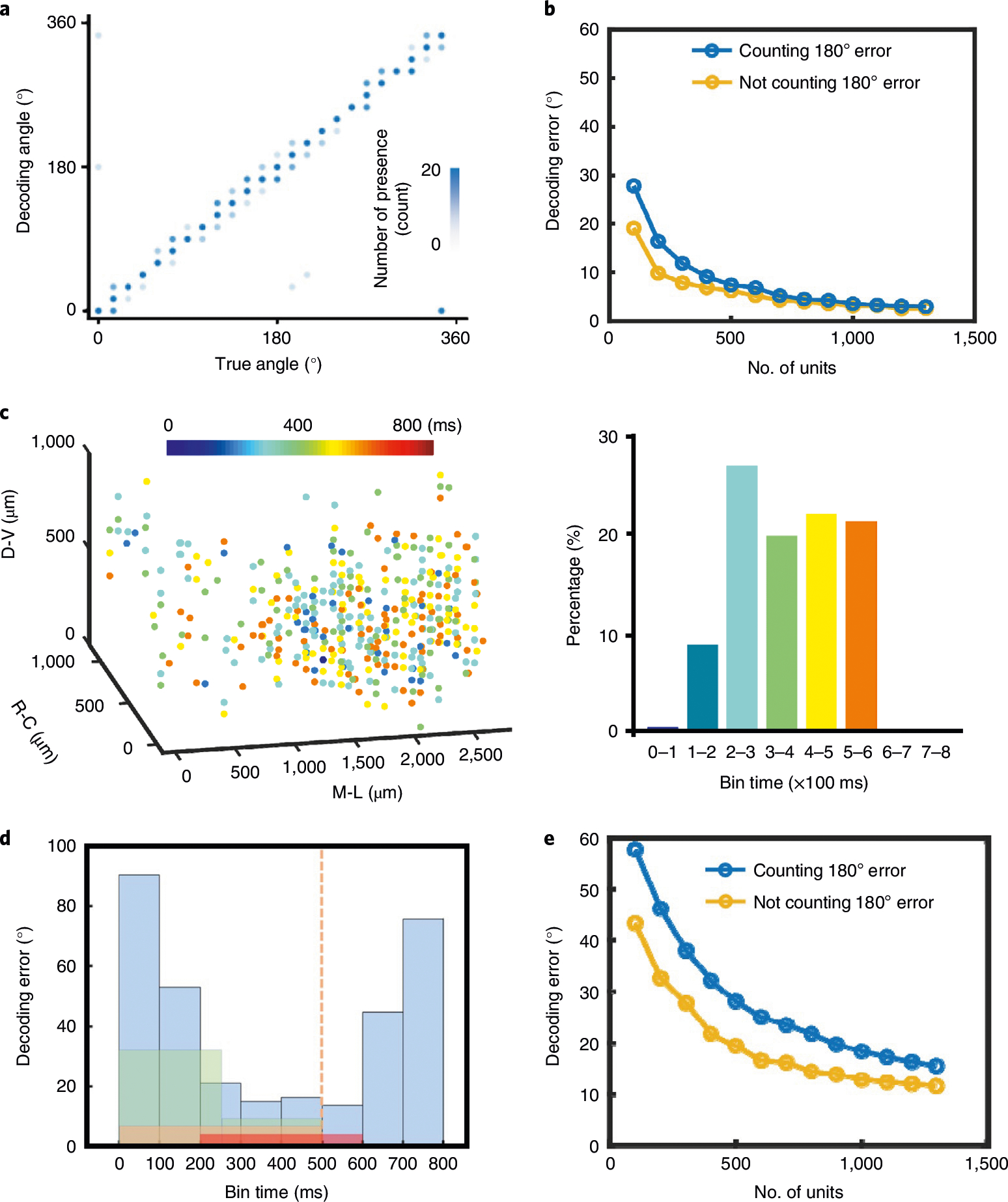
a, True versus decoded orientation of the drifting gratings. Contrast is scaled to the number of presence. b, Scaling of the mean decoding error with the number of recorded units using the time segment of 200–600 ms and 40 training trials. Stimuli were presented in the interval 0–500 ms. Decoding errors were computed in two ways: counting or not counting the 180° difference in the angles of drifting gratings. c, Spatiotemporal distribution of the units with the top 30% prediction power. Colours code the time at their maximum prediction power. Bars (right) present their population percentage. d, Mean decoding error as a function of selected time segment used for training. Colours mark different binning methods. Blue, every 100 ms; green, 0–250 ms and 250–500 ms; orange, 1–500 ms; red, 200–600 ms. The orange dashed line denotes the finish of visual stimulation. e, Scaling of the decoding error with the number of recorded units using the period of 300–400 ms and 40 training trials.
We then investigated how decoding accuracy changes with recording duration and time segment. We first determined the time segment after stimulation at which each unit had the highest predictive power (Fig. 4c and Supplementary Fig. 6). Interestingly, very few units were most predictive in the first 100 ms segment, few were in the 100–200 ms segment, while substantial numbers of units became predictive in the segments after 200 ms. This is consistent with the lag of ~100 ms from the onset of visual stimuli to changes in firing rates as shown in Fig. 3f. Subsequently, all 100 ms segments within 200–600 ms resulted in similar decoding performance (Fig. 4d). Although the very short 100 ms time window used here for decoding led to considerably larger error, the decoding error decreased sharply with the unit number and became relatively low once the unit count reached ~1,000 (Fig. 4e). This result suggests that large-scale neural recordings could enable a viable avenue to improve decoding temporal resolutions at affordable cost of errors.
Simultaneous optogenetic stimulation and volumetric electrophysiological mapping
NET arrays are compatible with optical methods10,21 and therefore allow for electrophysiological mapping under controlled modulation by optogenetic tools in the same brain region. Here we demonstrate the potential inherent in combining large-scale recording and optogenetic manipulations to assess the diversity and temporal profile of neural response. We implanted 8 type-I NET modules, 1,024 channels in total in V1 of a Thy1-mhChR2-EYFP mouse (n = 1) and an optic fibre near LM in V1, 400 μm away from the array and at a depth of 500 μm (Fig. 5a, Methods). Recording while applying optical stimulation, we determined the individual responses of a population of 656 units in the 1 mm3 recording volume recorded on all 8 128-channel modules (Fig. 5b).
Fig. 5 |. Simultaneous volumetric recording and optogenetic stimulation.
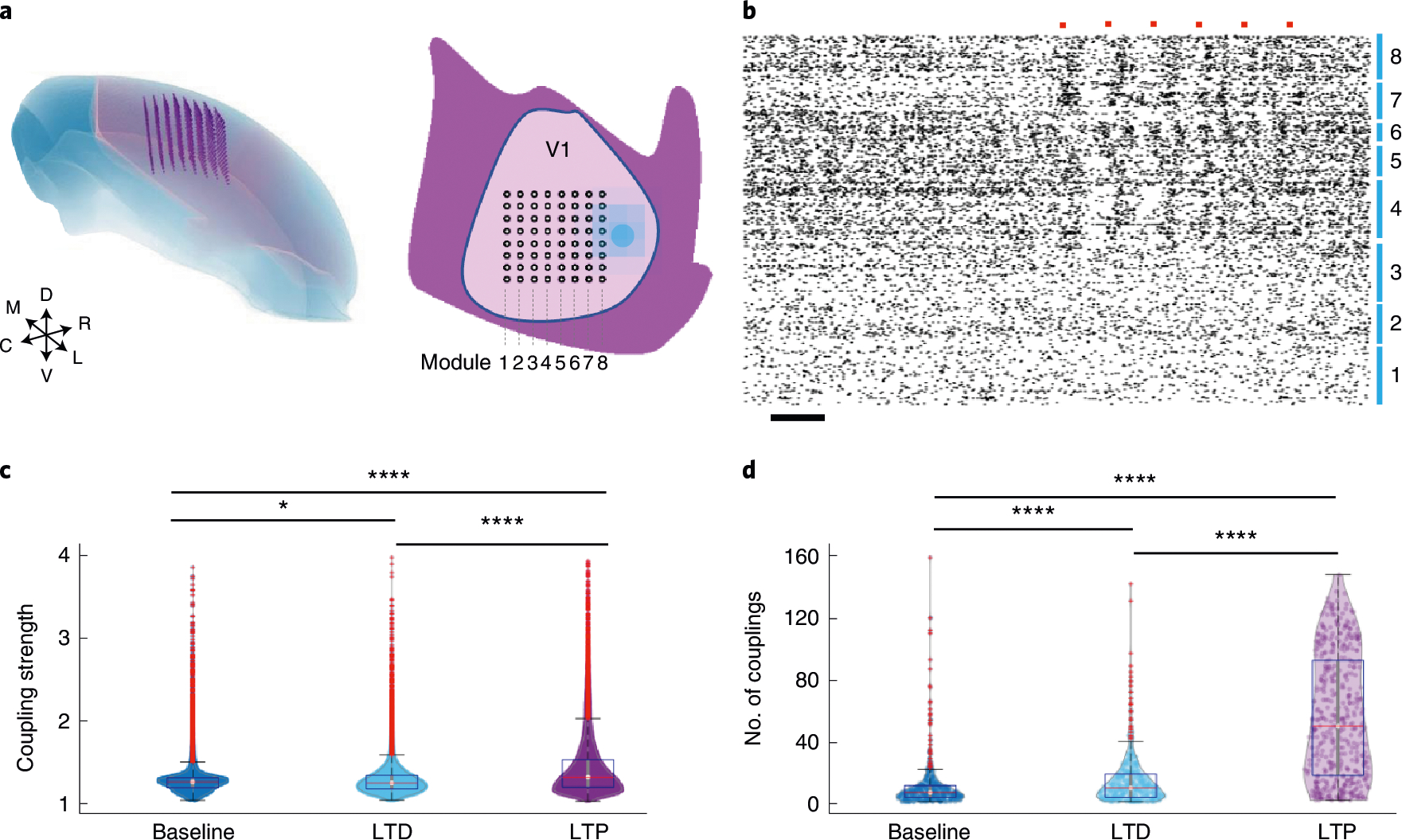
a, Schematic showing the targeted locations of the 8 × 8 × 16 recording sites in 3D (left), and the surface implantation locations for NET shanks (8 × 8) and the optical fibre (filled dot) in V1 (right). b, Spike raster of all 686 units recorded. Numbers mark the recording modules as in a. Orange dots mark the presence of optical stimulation. Scale bar, 0.1 s. c,d, Distribution of the couplings and nodes at baseline (blue), after optical LTD (cyan) and LTP (purple) protocols. Boxplots show the median, upper and lower 25%, and outliers. All data are shown in the violin plots. *P < 0.05, **P < 0.01, ****P < 0.0001, unpaired two-sided t-test, Bonferroni corrected.
Taking advantage of the ability to spatiotemporally resolve neural coupling structures, we characterize the responses to stimulation protocols designed to induce synaptic plasticity. We employed the previously published optogenetic protocols for the induction of long-term depression (LTD) and long-term potentiation (LTP)22. We mapped and quantified the location and strength distributions of strongly coupled pairs, the spatial and degree distributions of super nodes using the same aforementioned criteria and then determined their changes induced by applying the two protocols. We found that both LTP and LTD protocols resulted in enhancement of coupling strength and the number of couplings (node degree) at each node, while the LTP protocol led to stronger increases in both measures (Fig. 5c,d and Supplementary Fig. 7). Furthermore, while the LTD protocol had a relatively small effect on the spatial and degree distribution of the super nodes, the LTP protocol resulted in a remodelling of the locations of the super nodes (Supplementary Fig. 7). These results showcase the compatibility of volumetric electrophysiological mapping with optogenetic stimulation and its capacity in resolving the stimulation-induced plasticity in a neighbouring population of neurons.
Mapping distributed cortical activity in behaving mice
Our technology also makes it possible to resolve network activity spanning multiple brain regions. We implanted a mouse (n = 1) with 18 type-I NET modules distributed across both hemispheres of the neocortex (Methods) and assessed the neural responses to sensory stimulations. Figure 6 shows representative recordings from 18 NET modules in the motor, sensory and visual regions (Fig. 6a and Supplementary Videos 5 and 6). The module-to-module spacing was approximately 150–600 μm within each region, with exceptions listed in Supplementary Table 1. In a representative recording session, we resolved a total of 2,548 units, yielding 1.32 units per recording site and mild variations in unit yield across regions (Fig. 6b). We presented visual stimulation to the animal, video-recorded the animal’s spontaneous whisker and limb motion and investigated how these behavioural states changed and correlated with neural activity in different cortical regions (Fig. 6c). Visual stimulation modulated the spiking rate of neurons in the visual cortex. Whisker motion was associated with an increase in spike rate in all cortical areas and as expected, and neurons in the barrel cortex had the strongest response. Spontaneous locomotion was similarly associated with increased rates in the motor cortex and other cortical regions. We then quantified the correlations and latency between the neural activity in the three regions with these three behavioural markers (Fig. 6d,e). As one may intuitively expect, neural activity in the sensory cortex showed a stronger correlation with both whisker and limb motion, while activities in the visual cortex had the strongest correlation during grating stimuli. As a general observation, the correlations between the three cortical regions and the three behaviour markers are statistically different (unpaired two-sided t-test, P < 0.001, Bonferroni corrected), except that neural recordings in the motor and visual cortices had comparable correlations with limb activity (Fig. 6d). Neural activity led whisker and limb movement by around 200 ms and lagged the visual stimulus by about 100 ms (Fig. 6e).
Fig. 6 |. Large-scale distributed recording in awake head-fixed mice and behaviour decoding.
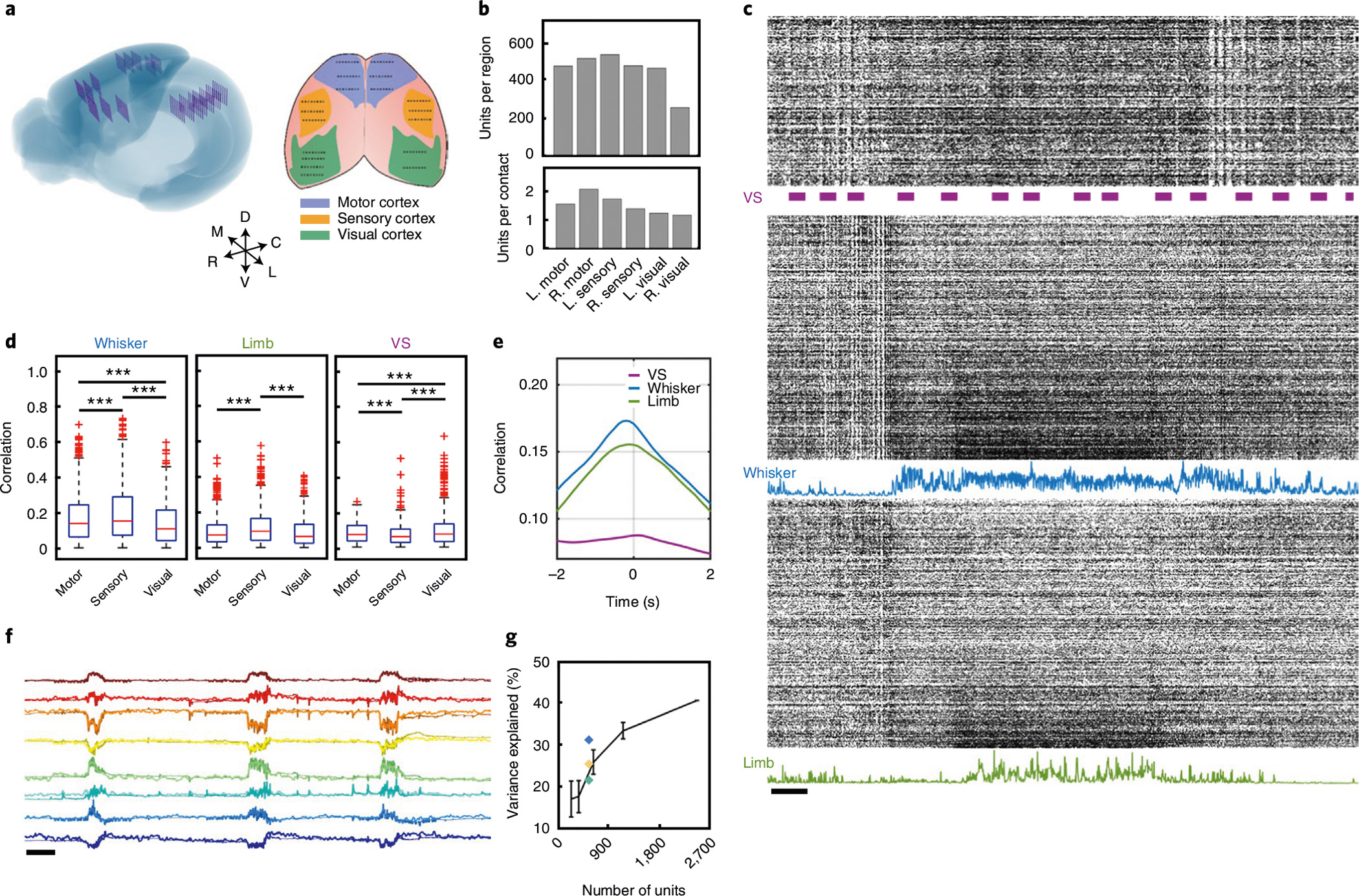
a, Left: schematic showing reconstructed locations of all 144 recording shanks across the neocortex of mouse brain. Right: schematic representation of the surface implantation location with target brain region colour coded, each dot representing a 16-channel shank. b, Quantification of unit yield across six brain regions in a typical recording session. c, Representative spike raster of the 2,548 units recorded from the NET array. Three segments depict the units in the visual (top), sensory (middle) and motor (bottom) cortices. Bars mark the presence of visual stimuli. Blue and green curves plot the magnitude of the whisker deflection and limb motion, respectively. Scale bar, 1 s. d, Boxplots (median, upper and lower 25%, and outliers) showing the correlation between neural activity and three behavioural markers: whisker motion (left), limb motion (middle) and the presence of visual stimuli (right). Unpaired two-sided t-test, ***P < 0.001, Bonferroni corrected. e, Mean correlation of spike activity of all units and three biomarkers as a function of time latency. f, Prediction of eight latent behavioural states extracted from the face video with neural activity. Darker colours mark the real traces, and lighter colours mark the predicted traces. Scale bar, 10 s. g, Prediction performance (mean ± s.d., n = 5) as a function of brain regions used for decoding. Blue, motor area; yellow, sensory area; green, visual area; black, scaling of the prediction performance with the number of recorded units randomly selected from all regions.
We thereafter sought to leverage the distributed electrophysiological mapping to decode the animal’s spontaneous behavioural states. We took videos of the animal’s facial motions as an ensemble of behaviours and compressed and deconstructed the videos into eight latent variables using a convolutional autoencoder23 (Supplementary Video 7). We then used the recorded neural activity in the three cortical regions to predict these variables (Fig. 6f) by employing a single-layer long short-term memory (LSTM) regression model24. We found that the neural activity of the 2,574 units from all recorded cortical regions could be used to predict a large fraction of the variance in facial-motion latent states (R2 = 0.41). This prediction was, not surprisingly, most accurate when there were substantial facial motions (Supplementary Video 7). While neural signals from all cortical regions encoded the variance of these latent states, activities in the motor cortex explained the largest fraction of the variance (R2 = 0.31, Fig. 6g). Furthermore, when we selected a random subset of units in all regions for decoding, the variance that could be explained had a steep scaling with the number of units used in the decoder (R2 = 0.16 for decoding using 288 random units; Fig. 6g). These results illustrate that the abilities to record a large population of neurons and to record across multiple brain regions enable accurate decoding of complex behaviours.
Chronic stability in recording and network mapping
The ability to chronically map a neuronal population and their network coupling depends on the implanted electrode array being functional and minimally perturbative to the host tissues. We evaluated the longitudinal recording performance in two mice (n = 2) implanted with 16 and 5 128-channel NET modules and for 145 days and 290 days, respectively. The average value of the impedance per contact site increased after implantation, stabilized after about a month and remained stable for the duration of the experiment (Fig. 7a). Importantly, the average signal amplitude and the SNR also remained stable after the initial variability following implantation (Fig. 7b,c). The averaged unit yield (Fig. 7d) was 1.3 units per contact after semi-automatic sorting and curation15, about 60% of which were putative single units (see Methods). Furthermore, we did not observe notable degradation in recording quality up to 290 d.
Fig. 7 |. Chronic stability of large-scale recordings by NETs, and evaluation of network stability over time.
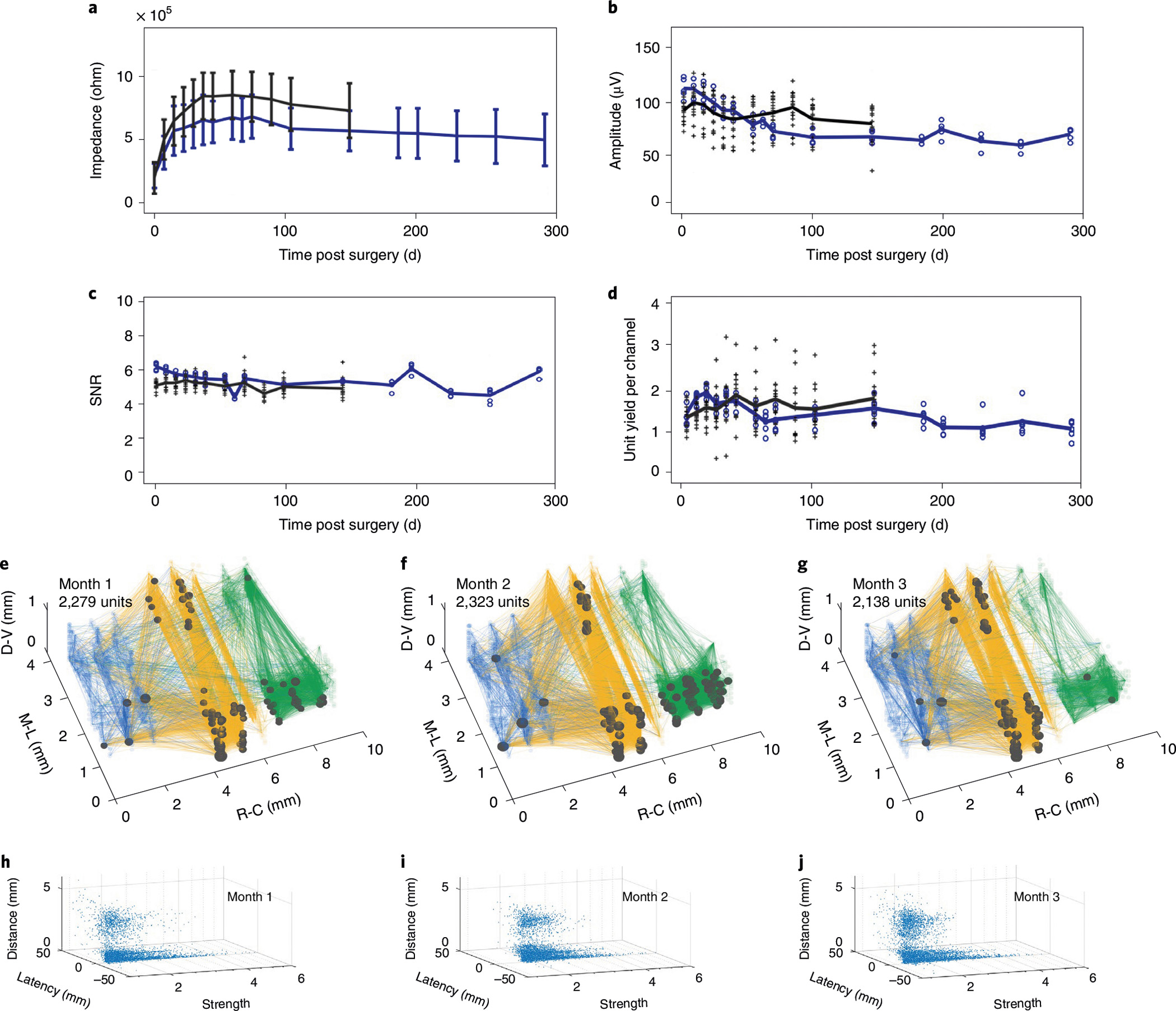
a–d, Performance and stability of unit recordings from 21 128-channel modules over time. The averaged values of impedance (a), peak-to-valley amplitude (b), SNR (c) and single- and multi-unit yield (d) remained stable for the experimental duration, except for the initial changes within 60 days after implantation. Each cross and dot presents the module-averaged values from 16 modules that were tracked for 145 days and from the other 5 modules that were tracked for 290 days, respectively. Black and blue lines depict the average values of all 16 and 5 modules, respectively. Error bars in a denote s.d. e–g, Cortical network stability over 3 months demonstrated by the spatial distribution of the strong couplings (lines) and super nodes (dots) under visual stimuli. The size of the dots indicates the number of couplings from the node. Measurement time and total unit number are indicated. h–j, Distribution of the coupling strength, latency and separation between the pairs under visual stimulation at 1 (h), 2 (i) and 3 (j) months post implantation, respectively.
In addition to the classic evaluations of recoding performance as described above, we employed the coupling patterns of neural activity recorded in 18 × 128 channels at a specific behavioural state to assess the longitudinal recording stability. Figure 7e–g show the network coupling map over 3 months spanning visual, sensory and motor cortices in both hemispheres when the animal was under the same visual stimulus (n = 1). The total number of units resolved, the spatial distribution of the super nodes and strong couplings showed similar patterns. Consistently, the distribution of coupling strength, latency and separation between the pairs (Fig. 7h–j) had minimal changes over 3 months. These results underscore the capability of ultraflexible NET arrays to stably record individual neurons and map functional couplings in a large-scale network at high fidelity and longitudinally.
Discussion
Neural electrodes are widely used in neuroscience studies but often have substantially larger sizes compared with the sizes of cells and vasculature in the brain, which make implanting many of these probes exceedingly invasive. For example, typical microwire or silicon neural probes have cross-sectional areas on the order of thousands of square micrometres2, whereas those of neuronal soma and capillaries are on the order of hundreds of square micrometres2. Recent studies have shown that reduction of the implant size has a marked potential for the high-density implantation of neural probes that have cellular-scale feature sizes10,12,25. The ultraflexible NET probes used in this work have cross-sectional areas on the order of 10–100 μm2, resulting in considerably less tissue displacement during chronic implantation, and therefore possibly allow for higher density of implantation2 than what has been possible so far. Furthermore, the substantially reduced mechanical mismatch between ultraflexible NET probes and brain tissue also enhances their long-term biocompatibility and facilitates large-scale implantation by eliminating any mechanical coupling between modules10. This is important for multiple-probe implantations.
Conventional silicon neural probes can record over a large planar region at hundreds of channels26, and more recent developments7,8 take advantage of integrated circuits built underneath the recording sites and have enabled high-density recording along the depth of the probe. However, these devices are challenging to scale at a high implantation density along lateral dimensions owing to their large cross-sectional areas. Previously demonstrated thicker polymer electrode arrays6,27 allowed for large-scale recordings at multiple distributed brain regions and showed long-term recording capabilities, but their size limits the implantation density, and clear tissue responses were found around the probe6.
The system we show exploits multiple new features involving electrode designs, the recording system and surgical approaches. Electrode designs are tailored to fit heterogeneous brain structures in a scalable manner. The modular device-headstage construction allows for the selective implantation of electrode arrays at designated locations according to specific needs. The biocompatibility of ultraflexible NETs makes possible long-term stable recording. These technical advances allow for the scaling-up of unit recordings to the thousands-of-neuron scale in chronic implantations in behaving animal brains. Our densest implantations reached approximately 1,000 recording sites per mm3 of cortical volume. Assuming an average neuronal density of 100,000 mm−3 (refs. 2,3), this corresponds to one recording site out of 100 neurons. Considering the estimate that less than about 10–20% of all neurons are electrophysiologically active18, it is possible that the volumetric recordings shown here can capture up to 5–10% of all neuronal activities in the volume. We show that a high-density neural recording clearly improved the decoding accuracy in a visual stimulation experiment and examined the dynamic responses of the neural processes.
It should be noted that the resolution of lithography limits the total number of contacts on each shank and involves a trade-off between the density and coverage of the NET electrodes. Designs with dense recording contacts can better resolve single units but provide a limited recording coverage (Extended Data Fig. 2). We chose a type-I design to maximize volumetric recording coverage at the cost of a moderate single-unit yield (43.3%; see Results). Technically, although ‘single units’ provide the most straightforward evidence to interpret the activity of individual neurons, multi units also carry meaningful information and should not be simply excluded, especially when recording a large population. Fundamentally, a ‘multi unit’ is a cluster of spikes fired by more than one neuron, which is a critical caveat when one focuses on the functions of one or a few neurons. It is, however, less of an issue when the analysis involves a large population. Taking the correlation analysis in this work as an example, the significant correlation between units (single or multi) can determine the presence of functionally connected neuron pairs and, importantly, the structure of correlation between neuronal populations. In the decoding analysis (Fig. 4), we also found that both single units and multi units contributed to the prediction power. The importance of including all units is supported by a number of recent works20,28,29 in which populational spike data were directly used without classification.
Lastly but importantly, being able to record the precise timing of spikes of local or global neural populations allowed us to assess the pairwise functional couplings that can help understand the underlying microcircuits. When the activity of two units is temporally correlated, it suggests a strong likelihood of direct or indirect anatomical connections. Measuring these correlations in the form of a spike-train cross-correlogram requires detecting the pairwise co-activation of neurons at fine spatial and temporal resolutions. Standard electrophysiological recording techniques afford the necessary temporal resolution to infer the coupling, but it is conventionally challenging to measure volumetric neuronal populations. The capability to densely evaluate large-scale couplings provides a direct and quantitative means to investigate critical phenomena at multiple timescales in the nervous system, such as circuit plasticity and decision making.
Methods
NET array design, fabrication and assembly
Three NET array designs, all consisting of 128 individually addressed recording sites at an overall device thickness of 1 μm, were used in the study. Type-I module has 8 shanks, 16 contacts per shank at the contact size of 25 μm × 25 μm and pitch of 60 μm. The shank spacing is 150 μm centre to centre. Type-II module has 4 shanks at the shank space of 300 μm centre to centre. Each shank contains two rows of 16 20 μm × 20 μm contacts at the centre-to-centre spacing of 35 μm both horizontally and vertically. Type-III module has 4 shanks at the shank space of 300 μm centre to centre. Each shank contains one row of 32 15 μm × 15 μm contacts at the centre-to-centre spacing of 20 μm vertically.
All these modules were fabricated using a planar microfabrication method and featured a multi-layer architecture and a sacrificial layer under the flexible section of the device. The fabrication procedures were detailed in our previous publication10. Several refinements were implemented to enable large-scale recording. To facilitate shuttle device assembly, microtrenches were fabricated using SU8–100 on top of the device and were later used to align shuttle wires with NET shanks before surgery. The microtrenches were also used to confine the movement of the shuttle wires when they were retracted after implantation13. To minimize the dimension and weight of the carrier chip, silicon wafers of 200 μm thickness (University Wafer) were used as the device substrate. A 500-μm-pitch ball grid array was patterned on the device and used as the bonding pads to connect to 50-μm-thick customized flexible printed circuits (FPCs) by soldering. The FPC interfaces with modular stacking headstages of 128 channels per module (SpikeGadgets, technical specification can be found at https://spikegadgets.com/products/64-1024/). The total weight of eight stackable headstages (128 channels per module, 1,024 channels in total) is 16 g, which can be carried by free-moving rats (Extended Data Fig. 3). Mice implanted with multiple modules only carry FPCs and NETs. The weight per module is about 0.12 g (silicon carrier chip per NET module is about 0.05 g; FPC and epoxy between each module is about 0.07 g per module), providing a total weight of about 0.96 g for eight modules. Mice electrophysiological recordings were performed during head fixation, which provided weight supports for the modular stacking headstages. Each FPC piece was connected to a stackable headstage module for data acquisition and was disconnected after measurements.
After bonding to the FPC, the NET module was released from the fabrication substrate. The gold contacts were electrochemically deposited with poly(3,4-ethylenedioxythiophene)-poly(styrenesulfonate) (PEDOT:PSS) in an aqueous solution at a concentration of 0.14% w/v PEDOT and 0.05% w/v PSS to lower the impedance6. Constant current was sequentially applied to each individual site for PEDOT:PSS deposition using an electroplating board (Intan RHD Electroplating Board) at a current of 10 μA and 1 s cycle duration until the impedance at 1 kHz dropped below 100 kΩ. Tungsten microwires (Ø25 μm), which were used as shuttle wires, were electrochemically sharpened and manually placed into the pre-fabricated microtrenches on the NET module13. The released NETs shanks were then temporarily attached to the tungsten microwires with polyethylene glycol (PEG m.w. 35,000) through a manual process as previously described13. The assembly device was ready for implantation after drying in ambient conditions for 1 min.
Animal and surgical implantation
Six wild-type adult male mice (three C57B6 and three CD1, 12 weeks old), one transgenetic male mouse (Thy1-mhChR2-EYFP) and two male rats (Sprague Dawley) were used in the experiments (Supplementary Table 1). The animals were housed at the Animal Resources Center at the University of Texas at Austin at a 12-hour light/dark cycle at 22 °C and with food and water ad libitum.
During surgical implantation, the animals was anaesthetized using isoflurane (3% for induction and 1–2% for maintenance) in medical-grade oxygen. The skull was exposed and prepared by scalping the crown and removing the fascia, and then scoring with the tip of a scalpel blade. The targets were then marked. Craniotomy was performed with a surgical drill over the target area. For distributed implantation, craniotomy at multiple different regions was performed together before the insertion. The skull was then cleaned with phosphate-buffered saline (PBS) and allowed to dry before implantation. The exposed brain was covered by PBS-soaked gelfoam to keep the tissue moisturized.
All assembled NET modules were implanted sequentially into the brain using a pair of stereotaxic micromanipulators, one module at a time in a similar manner as previously described (Supplementary Fig. 1)13. The first assembled NET module was attached to a stereotaxic micromanipulator through a customized vacuum holder and inserted into the targeted brain region. A digital encoder (Digital lab standard, Stoelting) was used to measure the implantation coordinates in three dimensions. Once the target depth was reached, the artificial cerebrospinal fluid (ACSF) was dripped onto the brain surface and the microtrenches to dissolve the PEG (30 s). The shuttle wires were then manually retracted one at a time, while the flexible NET shanks were left embedded in tissue. The carrier chip was then moved away from the implantation site without perturbing the implanted section and held still by the stereotaxic micromanipulator at the planned location for skull fixation. The second NET module was then attached to another stereotaxic micromanipulator and implanted using the same procedures as described above. After the shuttle wires were retracted, the carrier chip was moved to the stacking position next to the previous module. A small amount of adhesive was placed into the gap between the two modules to attach them. After the adhesive was set, the second module was detached from the manipulator, allowing the identical implantation procedure to be repeated for the rest of the modules. The implantation procedure took about 10–15 min per module (128 channels). In some cases, a small number of individual wires were selectively withdrawn before insertion on the basis of the vessel distribution in the targeted brain region to reduce disruption of surface vasculature.
After all the NET modules were implanted and affixed, a thin layer of silicone gel (Dow-Corning 3–4680) was applied to seal the craniotomies, and Kwik-Sil adhesive (World Precision Instruments) was used to fill the gaps between the NET carrier chips and provide support for them. After the skull was cleaned and dried, a layer of cyanoacrylate was applied over the skull. A thick layer of C&B-Metabond (Parkell) was applied over the cyanoacrylate and the Kwik-Sil adhesive to cement carrier chips to the skull. For mice, a custom-made head plate was attached to the skull using extra cement. For rats, addition dental acrylic was applied around the carrier chips to reinforce mounting of the array. The modular recording headstages were connected to the FPCs and enclosed by a 3D-printed cover cage.
All procedures complied with the National Institutes of Health guidelines for the care and use of laboratory animals and were approved by the University of Texas Institutional Animal Care and Use Committee (IACUC) under protocol AUP 201700211 and by the Rice University IACUC under protocol 1486751-1.
Large-scale electrophysiological recording and spike sorting
The in vivo impedance of all implanted modules was measured before each recording session using an Intan 128-channel RHD2164 evaluation system at 1 kHz, with a silver wire in the contralateral hemisphere of the brain as the grounding reference. The mice were head fixed, and the rats were anaesthetized during the measurement.
Electrophysiological data were then recorded using SpikeGadgets modular headstages at a sampling rate of 20 kHz, with the bare Ag in the contralateral hemisphere of the brain as the grounding reference. Each headstage module supports 128 channels, and the data acquisition system simultaneously records up to 10 modules. For recordings from more than ten modules, two systems were used simultaneously and synchronized by a shared digital input. The data from each system were streamed via a micro-HDMI cable to a computer for visualization and storage.
The recordings were performed on awake head-fixed mice on a custom-made running wheel, or on freely moving rats in a customized recording arena. Each animal was recorded once a week for the first 2 months after implantation, twice a month for the following 2 months, and once a month for the rest of the monitoring period. Each recording session lasted about 1 hour.
Spike sorting was performed in Mountainsort15. Common median referencing30 was applied to the raw data from each shank to reduce common mode noise, such as motion artefacts. The spike detection threshold was set to four times the standard deviation, and the adjacency radius was set to 100 μm. A further curation was then performed to reject noise clusters. The remaining unit clusters need to meet the following criteria simultaneously: firing rate > 0.1 Hz, full width at half maximum (FWHM) of the mean waveform at 0.15 ms < FWHM < 0.75 ms, valley to peak time (VPT) of the mean waveform at 0.15 ms < VPT < 0.85 ms and waveform distribution (peak amplitude sum of the top three waveforms contributes more than 60% of the peak amplitude sum of waveforms across the entire module) of sorted units. Putative single units were identified using the criterion that the fraction of spikes with inter-spike intervals under 2 ms was less than 1% of the total firing events31.
To process local field potentials (Fig. 2a and Supplementary Video 3), signal from each channel was first filtered into 5 frequency bands: delta (1–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), beta (12–30 Hz) and gamma (30–110 Hz). Z-score of the filtered data at each band was calculated and clamped to be within −2.5 to +2.5. These values from each channel were then interpolated at every 4 ms in a 3D volume enclosed by the entire array. Volume rendering was then performed across time in Vispy32.
Visual stimulation
Visual stimulation was conducted on awake head-fixed mice. A monitor (45 cm × 35 cm) was placed in front of the animal’s contralateral eye to the implanted NET array at a distance of 15 cm, subtending a 30 × 20° visual angle. Full-field drifting gratings (Psychtoolbox) were presented on the monitor at 1 of 24 directions evenly spaced at 15° at a spatial frequency of 0.05 cycles per degree and a temporal frequency of 2 Hz. Each direction was presented for a duration of 0.5 s. A set of 24 directions were presented in random order with a grey-screen interstimulus interval randomly distributed between 0.3 and 1.1 s. An additional 30 s grey-screen resting period was added upon the completion of each set of 24 directions, and the procedure was repeated 78 times. Each recording session lasted about 72 min. Transistor-transistor logic (TTL) pulses were used to synchronize the stimuli and the neural recording. The animals were always returned to the same field of view to the screens at the beginning of every session.
Optogenetic stimulation
Optical stimulation was conducted on awake head-fixed mice during simultaneous electrical recording using an implanted optical fibre (Thorlabs, Ø150 μm) and a 473 nm laser (LRS-0473 DPSS, Laserglow) for excitation. TTL pulses were used to synchronize the stimulation and neural recording. No photoelectric artefacts were detected during the stimulation.
After the animals were acclimated to head fixation, baseline recordings were performed and one of the following three stimulation protocols were applied.
10 Hz.
Optical stimulation at 5 power levels (0.1, 0.5, 1, 6, 10 mW mm−2) was induced subsequently for 15 min each. Stimulation at each power contains 180 pulse trains at 4 s intertrain intervals, and each pulse train was at 10 Hz, 5% duty cycle and 1 s duration. There was a 10 min resting period between power switches.
LTD induction.
Optical LTD was induced for 15 min at 10 mW mm−2, 1 Hz and 2% duty cycle. After the induction, the animal was recorded for another 15 min.
LTP induction.
Optical LTP was induced for 15 min using 5 pulse trains at 3 min intertrain intervals, and each pulse train was at 100 Hz, 20% duty cycle and 1 s duration. After the induction, the animal was recorded for another 15 min.
Motion videography
Two high-speed cameras were used to capture videos from an awake head-fixed mouse running on a treadmill at 100 frames per second (fps). One camera targeted the front limb from the front view of the mouse, while the other camera zoomed in on the facial area to target the pupil and whisker from the side view. Infrared LEDs were used for infrared video acquisition in darkness. The electrical recording was performed simultaneously and synchronized by TTL signals from the cameras.
Visual response processing
Visual stimulation under drifting gratings.
The response of each recorded unit to a certain orientation of the drifting gratings was calculated by averaging the total spike counts among all trials when the grating was presented. The preferred orientation, θpref, was determined as the orientation that elicited the highest number of spikes. The orientation tuning curve was fitted as the sum of two Gaussians centred on θpref and θpref + π, of different amplitudes A1 and A2, an equal width σ and a constant baseline B. The orientation selectivity index (OSI) was calculated as the depth of modulation from the preferred orientation to its orthogonal orientation θortho = θpref + π/2, as OSI = (θortho − θpref)/(θortho + θpref). The neural responses at θpref and θortho used in the above calculation were after baseline subtraction.
Decoding.
A previously published method was applied to decode the grating angles using a locally linear decoder20. Neural activity was regressed onto ‘super neurons’ using MATLAB’s ‘fitrlinear’ function. These super neurons were von Mises tuning curves (n = 24) with peaks equally spaced along 360° and with σ = 0.1, vk = exp((cos(θ − θk)−1)/σ) and θk = 360(k − 1)/n, where σ controls the tuning width of super neuron, k denotes super neuron number which spans from 1 to n, θk is the preferred angle of the super neuron, and vk specifies the response of the super neuron as a function of angle θ. The transformation from unit activities to super neurons was fit on training trials using lasso regularization with search of optimal penalization parameter, and the super neuron responses on test trials were then predicted. The decoding error for each trial was defined as the difference between the true angle and the decoded angle. For the case in which a 180° angle difference was not counted, the errors that were larger than 180° were corrected as 360° minus the value. The mean value of all pairs defined the overall decoding error. For each predictor (or each unit), the maximum value of the initial linear coefficient estimates among all 24 super neurons (or all orientations) was selected as the individual prediction power.
Feeding into the decoder.
Spike counts of 800 ms following the stimulus onset were binned into segments of different sizes ranging from 10 to 500 ms. For most of the analysis, a total of 1,872 trials were randomly split 50/50 into training and testing sets. In the study of number of trials vs decoding accuracy, a random selection of the trials was assigned to the test sets and the training sets remained as 50% of the trials. In the study of temporal distribution of individual unit prediction power (Fig. 4c and Supplementary Fig. 6), all the trials were chronologically split 50/50. Switching the two sections between training and testing provided a similar result.
Functional correlation and coupling
The calculation of functional coupling followed the same methods from ref. 31. Basically, the correlograms of all unit pairs were calculated in 0.5 ms bins at a lag range of −200 ms to 200 ms and smoothed with a 3 ms boxcar filter. Correlations were considered significant if two consecutive bins of the correlogram were above a threshold set at three standard deviations across the spike count of all bins of the shuffled correlogram computed for the same pair (one spike train was shuffled by adding ±400 ms random jitter to each spike). The minimum total spike count across all bins for the correlograms to be included was 5,000. The correlation strength of a pair was measured by the normalized amplitude of the correlogram, which was defined as the peak value averaged over three bins around the peak and normalized by the average value across all bins of the shuffled correlogram. The lag was defined as the bin location of the peak in the correlogram.
Automatic extraction of mouse behaviours
The behavioural variables were extracted from the videos through DeepLabCut33 and FaceMap34. Specifically, DeepLabCut was used to automatically extract the positions of the front limb as a point source and four points on the pupil’s circumference with equal distances. The centre position and size of the pupil were further derived from these four points. FaceMap was used to automatically extract the motion energy of the whisking. The whisker signal was defined as the total motion energy summed over all pixels in a manually defined region covering the whisker pad.
Motion correlation and decoding
The spike train of all units was binned into 100 ms segments and aligned with the behavioural variables. The cross correlation was then calculated between binned spike activity of each unit and different behavioral variables using MATLAB’s ‘xcorr’ functions at ±4 s time lag.
To decode the motion of the animal, eight latent variables were extracted from regions of interest in each video frame using the convolutional autoencoder described in ref. 23. The spike train of all units was binned into 30 ms segments and aligned with downsampled video frames as well as their corresponding latent variables. Sixteen bins of spiking activity before the current video frame, one bin aligned with the current frame and eight bins after the current video frame were used to decode the eight latent variables of the frame. Decoding was performed with single-layer LSTM regression24 that minimizes the mean squared error of true versus decoded latent variables. One hundred models of different hyperparameters (summarized in Supplementary Table 2) were searched during the model fitting, and the best models were selected using Bayesian optimization with Gaussian process prior. Eighty percent of the data was used for training, and 10% was used for model validation. The decoding quality was defined as variance explained (R2, coefficient of determination) for the remaining 10% test set. The model fitting was repeated each time when we selected a subset of units for decoding in Fig. 6f,g.
Micro-CT
The micro-CT image in Fig. 1a was acquired by scanning the upper body of the perfused rat after fixation in 4% paraformaldehyde solution with Xradia MicroCT scanner at ×10 objective and 90 kV. The reconstruction was done by Xradia Reconstructor at voxels of 47.6 μm. The image in Fig. 1e was acquired by scanning the dissected brain tissue with NSI scanner at 180 kV. The reconstruction was done by Xradia Reconstructor at voxels of 2.19 μm.
Histological analysis
The mice were perfused intracardially using oxygenated ACSF at 4 °C and then with a 4% paraformaldehyde solution in 0.02 M PBS. A solution of 30% sucrose/4% paraformaldehyde was used to soak the brains overnight. Brain tissue was sliced using a Leica CM1950 cryostat (Leica Microsystems) into 20 μm slices, which were then washed for three 5 min sessions before being incubated for 30 min in a sodium citrate solution (85 °C to 95 °C, 0.01 M in H2O) for antigen retrieval. The slices were washed again for three 5 min sessions, incubated in a blocking solution, permeabilized (0.5% Triton X-100 and 10% normal goat serum (Sigma-Aldrich) in PBS) for 3 hours at 25 °C, washed again for four 5 min sessions and then incubated with one-step fluorophore-conjugated antibodies at 4 °C for 24 hours. To immunochemically stain neurons, Alexa Fluor 568-conjugated anti-NeuN antibody (ab207282, abcam) was used.
Data and computational scale
In the study, we conducted a total of 102 1-hour sessions of neural recording at a sampling rate of 20 kHz in five animals, yielding 13 TB of raw data. Approximately 1,530 CPU (Intel Xeon 2680 v2) hours were spent on spike sorting. Approximately 2,100 CPU hours and 1,700 GPU (Nvidia GeForce GTX 1080 Ti) hours were spent on all the post analysis.
Extended Data
Extended Data Fig. 1 |. Immunohistochemistry analysis showing the tissue-NET interface at a high implantation density.
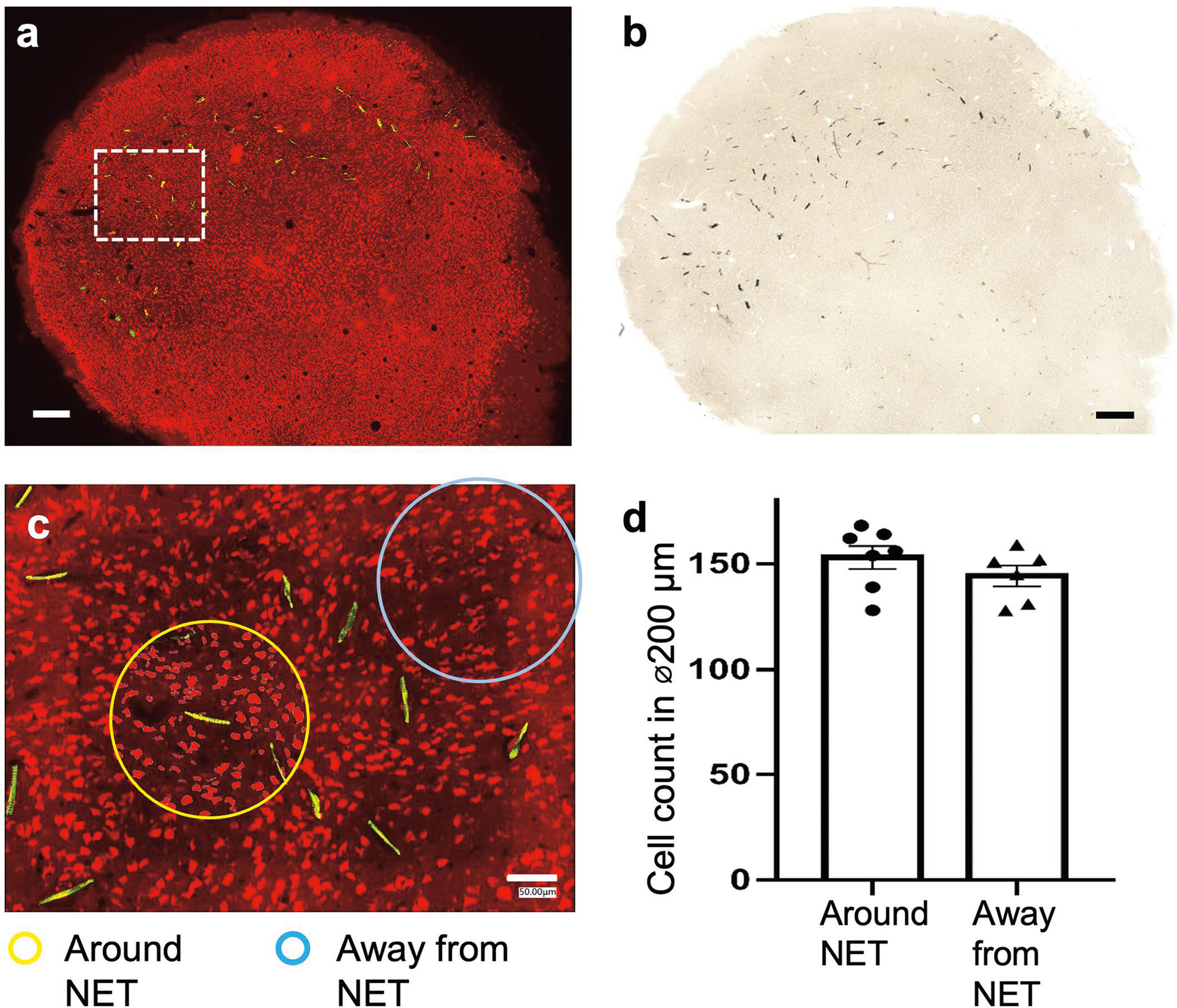
One mouse was implanted in visual cortex with 10 type-I NET modules, at inter-shank spacings of 150 μm and inter-module spacings of 250 μm. Fluorescent (a) and brightfield (b) microscopy show no observable scarring in the implanted regions. Neuron: red; NETs: yellow. (c), We counted numbers of neurons in seven randomly-chosen 200-μm-diameter regions centering around NETs (yellow circled regions as an example) and five 200-μm-diameter randomly-chosen regions away from NETs (cyan circled regions as an example). We found no significant difference between the two groups using unpaired T test, p = 0.275 (d), suggesting no significant neuronal loss induced by dense NET implants. These results are consistent with our prior studies using single or few NETs (10, 13). Scalebars: 250 μm (a and b) and 50 μm (c).
Extended Data Fig. 2 |. Example recording performances of type-II (A) and –III (B) NET electrode designs.
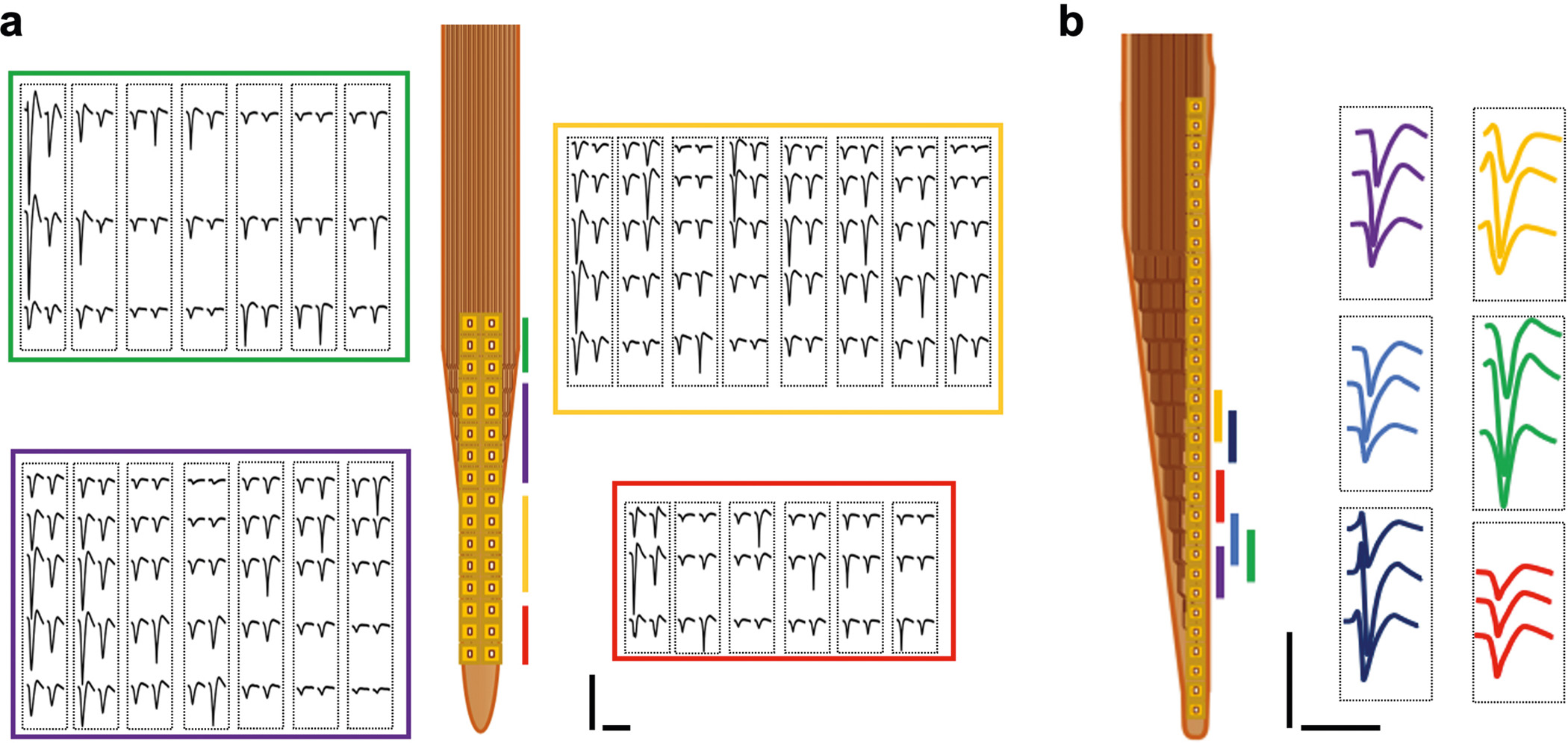
These two designs offer high electrode density that allows for the recording of individual neurons by multiple channels and therefore perform better in single unit isolation. Color code corresponds to the location of the recording sites. Each dashed box outlines the waveforms of a single unit recorded on the corresponding sites. Scale bars: 100 μV (vertical) and 2 ms (horizontal).
Extended Data Fig. 3 |. Modular recording system interfacing with NETs.
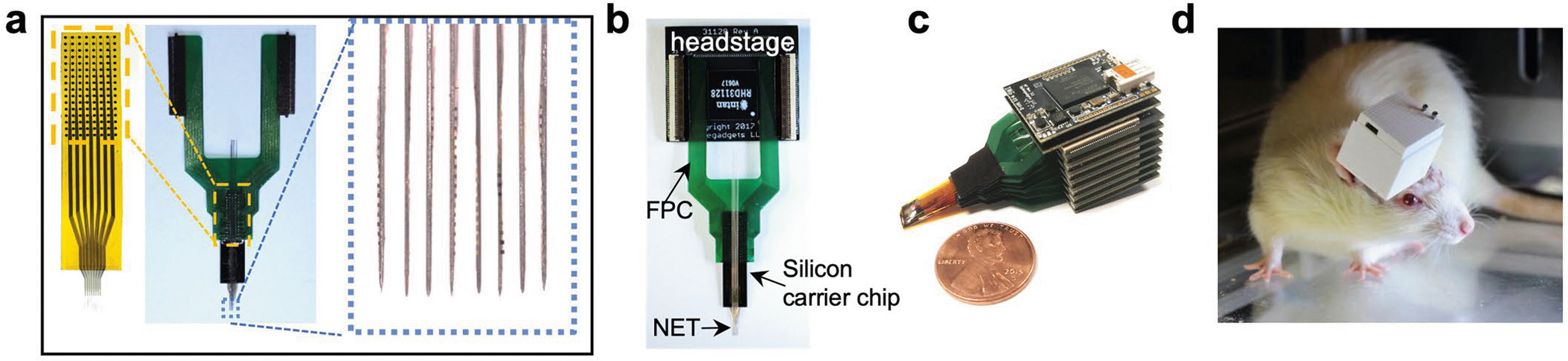
(a), A ready-to-implant 128-channel NET module connected to a flexible printed circuit (FPC). Left panel shows the NET module and ball grid array (BGA) through which the connection to FPC was made. Right panel shows that individual NET shanks was attached to tungsten microwires for implantation. (b), Photo of a 128-channal stackable headstage connected to NET through an FPC. (c), Photo of a 1024-channel system composed of eight stackable modules. (d), Photo of a free-moving rat carrying a 1024-channel NET array and stackable headstages as shown in c. 3D printed case enclosed all headstages.
Supplementary Material
Acknowledgements
We thank M. Karlsson and M. Borius for input about neural recording electronics, C. Kemere for input about motion decoding, V. Yip for assistance with devices fabrication and assembly and A. Li for assistance with tests of the decoding model. This work was funded by the National Institute of Neurological Disorders and Stroke grants R01NS102917 (to C.X.), U01NS115588 (to C.X. and L.F.), R01NS109361(to L.L.) and UF1 NS107667 (to L.F); the National Heart, Lung and Blood Institute grant K25HL140153 (to L.L.); the Welch Foundation Research grant #F-1941-20170325 (to C.X.); and the Howard Hughes Medical Institute (to L.F.)
Footnotes
Competing interests
C.X., L.L. and Z.Z. are co-inventors on a patent filed by The University of Texas (WO2019051163A1, 14 March 2019) on the ultraflexible neural electrode technology described in this study. L. F., L.L. and C.X. hold equity ownership in Neuralthread Inc., an entity that is licensing this technology. All other authors declare no competing interests.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
Mountainsort3, which we used for spike sorting, is not maintained anymore, but Mountainsort4 is available at https://github.com/magland/ml_ms4alg. Deeplabcut, for animal-posture extraction, is available at https://github.com/DeepLabCut/DeepLabCut. Facemap, for animal-posture extraction, is available at https://github.com/MouseLand/facemap. Visual stimulus decoder was adapted into Matlab code, but Python code is available from the original author at https://github.com/MouseLand/stringer-et-al-2019. BehaveNet for video latent extraction is available at https://github.com/themattinthehatt/behavenet. The LSTM neural decoder is available at https://github.com/KordingLab/Neural_Decoding. The remaining analyses were done with custom Matlab routines, which are available from the corresponding authors on reasonable request.
Extended data is available for this paper at https://doi.org/10.1038/s41551-022-00941-y.
Peer review information Nature Biomedical Engineering thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-022-00941-y.
Data availability
The main data supporting the results of this study are available within the paper and its Supplementary Information. The datasets generated during the study are too large to be publicly shared. Sample data are available from the corresponding authors on reasonable request.
References
- 1.Braitenberg V & Schüz A Anatomy of the Cortex: Statistics and Geometry (Springer, 1991). [Google Scholar]
- 2.Kleinfeld D et al. Can one concurrently record electrical spikes from every neuron in a mammalian brain? Neuron 103, 1005–1015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marblestone AH et al. Physical principles for scalable neural recording. Front. Comput. Neurosci. 7, 137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen TW et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adam Y et al. All-optical electrophysiology reveals brain-state dependent changes in hippocampal subthreshold dynamics and excitability. Nature 569, 413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung JE et al. High-density, long-lasting, and multi-region electrophysiological recordings using polymer electrode arrays. Neuron 101, 21–31.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun JJ et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinmetz NA et al. Neuropixels 2.0: a miniaturized high-density probe for stable, long-term brain recordings. Science 372, eabf4588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He F, Lycke R, Ganji M, Xie C & Luan L Ultraflexible neural electrodes for long-lasting intracortical recording. iScience 23, 101387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan L et al. Ultraflexible nanoelectronic probes form reliable, glial scar-free neural integration. Sci. Adv. 3, e1601966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown EN, Kass RE & Mitra PP Multiple neural spike train data analysis: state-of-the-art and future challenges. Nat. Neurosci. 7, 456–461 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Wei X et al. Nanofabricated ultraflexible electrode arrays for high-density intracortical recording. Adv. Sci. 5, 1700625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Z et al. Parallel, minimally-invasive implantation of ultra-flexible neural electrode arrays. J. Neural Eng. 16, 035001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung JE et al. Chronic implantation of multiple flexible polymer electrode arrays. J. Vis. Exp. 152, e59957 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung JE et al. A fully automated approach to spike sorting. Neuron 95, 1381–1394.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegle JH et al. Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niell CM & Stryker MP Highly selective receptive fields in mouse visual cortex. J. Neurosci. 28, 7520–7536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohki K, Chung S, Ch’ng YH, Kara P & Reid RC Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433, 597–603 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Stringer C, Michaelos M, Tsyboulski D, Lindo SE & Pachitariu M High-precision coding in visual cortex. Cell 184, 2767–2778.e15 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z et al. Nanoelectronic coating enabled versatile multifunctional neural probes. Nano Lett. 17, 4588–4595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nabavi S et al. Engineering a memory with LTD and LTP. Nature 511, 348–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batty E et al. BehaveNet: nonlinear embedding and Bayesian neural decoding of behavioral videos. Advances in Neural Information Processing Systems 15706–15717 (2019). [Google Scholar]
- 24.Glaser JI et al. Machine learning for neural decoding. eNeuro 7, ENEURO.0506–19.2020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozai TDY et al. Ultrasmall implantable composite microelectrodes with bioactive surfaces for chronic neural interfaces. Nat. Mater. 11, 1065–1073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berenyi A et al. Large-scale, high-density (up to 512 channels) recording of local circuits in behaving animals. J. Neurophysiol. 111, 1132–1149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musk E & Neuralink An integrated brain-machine interface platform with thousands of channels. J. Med. Internet Res. 21, e16194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kay K et al. Constant sub-second cycling between representations of possible futures in the hippocampus. Cell 180, 552–567.e25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trautmann EM et al. Accurate estimation of neural population dynamics without spike sorting. Neuron 103, 292–308 e294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolston JD, Gross RE & Potter SM Common median referencing for improved action potential detection with multielectrode arrays. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2009, 1604–1607 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Dhawale AK et al. Automated long-term recording and analysis of neural activity in behaving animals. eLife 6, e27702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Combrisson E et al. Visbrain: a multi-purpose GPU-accelerated open-source suite for multimodal brain data visualization. Front. Neuroinform. 13, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathis A et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Stringer C et al. Spontaneous behaviors drive multidimensional, brainwide activity. Science 364, 255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results of this study are available within the paper and its Supplementary Information. The datasets generated during the study are too large to be publicly shared. Sample data are available from the corresponding authors on reasonable request.


