1 Introduction
I read with great interest the valuable article titled “Association between the miR-146a Rs2910164 Polymorphism and Childhood Acute Lymphoblastic Leukemia Susceptibility in an Asian Population” published in the October 2020 edition of the journal (Zou et al., 2020). The authors included six studies based on their inclusion criteria. Their main finding indicated that the CC genotype significantly increased the risk of childhood acute lymphoblastic leukemia (ALL) in the additive model (CC vs. GG: OR = 1.598; 95% CI: 1.003–2.545; p = 0.049). Also, the dominant model, recessive model, and allele model indicated a trend of increasing risk for childhood ALL. However, there are some issues in the data extraction and meta-analysis that affect the results and must be noticed. Here, I aim to comment on the issues and provide accurate results through conducting a new meta-analysis. First, in Devanandan’s study, the genotyping method has been incorrectly recorded (Jemimah Devanandan et al., 2019). Second, in Xue’s study and Pei’s study, the frequency of the GG genotype has been defined as the frequency of the CC genotype and the frequency of the G allele has been defined as the frequency of the C allele, and vice versa (Xue et al., 2019; Pei et al., 2020). This has caused mistakes in statistical analysis and result interpretation. Third, in Pei’s study, all the participants were Taiwanese, not Taiwanese and Chinese (Pei et al., 2020). Therefore, in order to correct the findings of the meta-analysis by Zou et al. (2020), I used STATA 17.0 and CMA 3.0 software applications to conduct a meta-analysis based on the information reported in the original studies.
2 Results of my current meta-analysis
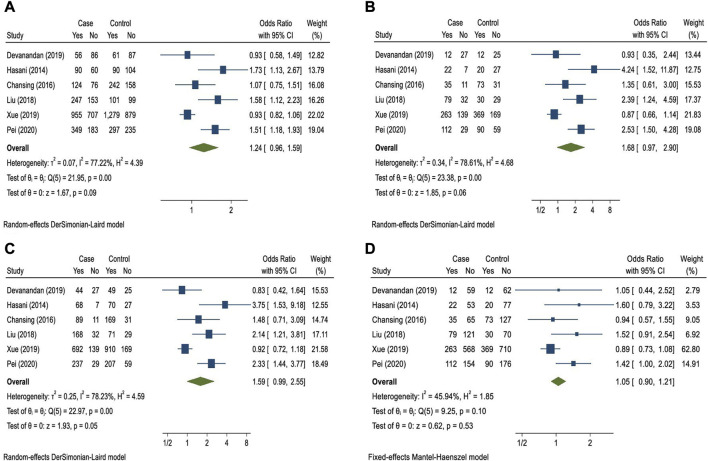
I present the correct characteristics of the included studies in Table 1 (Hasani et al., 2014; Chansing et al., 2016; Liu et al., 2018; Jemimah Devanandan et al., 2019; Xue et al., 2019; Pei et al., 2020). Based on the heterogeneity results of the meta-analysis of the association between rs2910164 and childhood ALL, except for the recessive model, the random effects model was used for meta-analysis. I obtained a pooled OR of 1.24 (95% CI: 0.96–1.59; p = 0.09) for the C allele in the allele model, 1.59 (95% CI: 0.99–2.55; p = 0.05) for the CC + CG genotype in the dominant model, 1.05 (95% CI: 0.90–1.21; p = 0.53) for the CC genotype in the recessive model, and 1.68 (95% CI: 0.97–2.90; p = 0.06) for the CC genotype in the additive model (Figure 1 and Supplementary Table S1). In all models, there was no significant association between the rs2910164 polymorphism and childhood ALL risk. Based on sensitivity analysis, removing the studies one by one from the included list showed that when Xue’s study is removed, the overall effect size of the different models changes significantly. Also, when Devanandan’s study is removed, the overall effect size of the dominant model changes significantly (Supplementary Figure S1 and Supplementary Table S2). According to the results of the funnel plot, Begg’s test, and Egger’s test, no publication bias was observed (Supplementary Figure S2 and Supplementary Table S3).
TABLE 1.
Characteristics of six studies included in the present meta-analysis.
| SNP | First author-year | Country | Continent | Genotyping method | Sample size | Genotype and allele distribution | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||||||||||
| Case | Control | GG | CG | CC | G | C | GG | CG | CC | G | C | |||||
| rs2910164 | Devanandan-2019 | India | Asian | TaqMan | 71 | 74 | 27 | 32 | 12 | 86 | 56 | 25 | 37 | 12 | 87 | 61 |
| Hasani-2014 | Iran | Asian | T-ARMS-PCR | 75 | 97 | 7 | 46 | 22 | 60 | 90 | 27 | 50 | 20 | 104 | 90 | |
| Xue-2019 | China | Asian | SNaPshot | 831 | 1,079 | 139 | 429 | 263 | 707 | 955 | 169 | 541 | 369 | 879 | 1,279 | |
| Chansing-2016 | Thailand | Asian | PCR-RFLP | 100 | 200 | 11 | 54 | 35 | 76 | 124 | 31 | 96 | 73 | 158 | 242 | |
| Pei-2020 | Taiwan | Asian | PCR-RFLP | 266 | 266 | 29 | 125 | 112 | 183 | 349 | 59 | 117 | 90 | 235 | 297 | |
| Liu-2018 | China | Asian | PCR-RFLP | 200 | 100 | 32 | 89 | 79 | 153 | 247 | 29 | 41 | 30 | 99 | 101 | |
FIGURE 1.
Forest plots for the allele (A), additive (B), dominant (C), and recessive (D) models of the relationship between the miR-146a rs2910164 polymorphism and the risk of childhood acute lymphoblastic leukemia.
3 Conclusion
I conducted a meta-analysis based on information derived from six studies included in the meta-analysis by Zou et al. (2020) to assess the efficacy of the miR-146a rs2910164 polymorphism on childhood ALL risk. According to analysis, there was no significant association between the rs2910164 polymorphism and childhood ALL in all models. Based on sensitivity analysis after removing Xue’s study, childhood ALL risk was significantly increased in allele (C vs. G), additive (CC vs. GG), dominant (CC + CG vs. GG), and recessive (CC vs. CG + GG) models. Also, after removing Devanandan’s study, childhood ALL risk was significantly increased in the dominant model (CC + CG vs. GG). The results of the funnel plots, Egger’s test, and Begg’s test suggested that there is no obvious publication bias.
Author contributions
AN designed the research, performed the statistical analysis, and wrote the manuscript. AN took primary responsibility for the final content.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1134659/full#supplementary-material
References
- Chansing K., Pakakasama S., Hongeng S., Thongmee A., Pongstaporn W. (2016). Lack of Association between the mir146a polymorphism and susceptibility to Thai childhood acute lymphoblastic leukemia. Asian Pac. J. Cancer Prev. 17 (5), 2435–2438. 10.7314/APJCP.2016.17.5.2435 [DOI] [PubMed] [Google Scholar]
- Hasani S-S., Hashemi M., Eskandari-Nasab E., Naderi M., Omrani M., Sheybani-Nasab M. (2014). A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: A preliminary report. Tumor Biol. 35 (1), 219–225. 10.1007/s13277-013-1027-1 [DOI] [PubMed] [Google Scholar]
- Jemimah Devanandan H., Venkatesan V., Scott J. X., Magatha L. S., Durairaj Paul S. F., Koshy T. (2019). MicroRNA 146a polymorphisms and expression in Indian children with acute lymphoblastic leukemia. Lab. Med. 50 (3), 249–253. 10.1093/labmed/lmy074 [DOI] [PubMed] [Google Scholar]
- Liu X., Liu L., Cao Z., Guo B., Mingwei L. (2018). Association between miR146a (rs2910164) G> C polymorphism and susceptibility to acute lymphoblastic leuke-mia in children. Chin. J. Appl. Clin. Pediatr. 33 (3), 200–202. 10.3760/cma.j.issn.2095-428X.2018.03.010 [DOI] [Google Scholar]
- Pei J-S., Chang W-S., Hsu P-C., Chen C-C., Chin Y-T., Huang T-L., et al. (2020). Significant association between the MiR146a genotypes and susceptibility to childhood acute lymphoblastic leukemia in Taiwan. Cancer genomics and proteomics 17 (2), 175–180. 10.21873/cgp.20178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Yang X., Hu S., Kang M., Chen J., Fang Y. (2019). A genetic variant in miR-100 is a protective factor of childhood acute lymphoblastic leukemia. Cancer Med. 8 (5), 2553–2560. 10.1002/cam4.2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Yin J., Ye Z., Zeng Q., Tian C., Wang Y., et al. (2020). Association between the miR-146a Rs2910164 polymorphism and childhood acute lymphoblastic leukemia susceptibility in an asian population. Front. Genet. 11, 886. 10.3389/fgene.2020.00886 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.