Abstract
Natural deep eutectic solvents (NADES) are a class of liquids with promising properties as components in pharmaceutical formulations, such as a low toxicity profile, biodegradability and versatility. Recently, their potential use as anti-biofilm agents has been proposed, due to their ability to solubilize and stabilize biological macromolecules. In the current work, the ability to break down biofilm matrix and the biofilm killing activity of three NADES of neutral pH were investigated against Staphylococcus aureus ATCC 6538 and Pseudomonas aeruginosa ATCC 9027 biofilms. The tested NADES were choline chloride:xylitol (ChX), choline chloride:glycerol (ChG) and betaine:sucrose (BS). Two of the NADES (ChX and ChG) significantly reduced the number of remaining viable cells of both bacterial species in pre-formed biofilm by 4–6 orders of magnitude, while the average biofilm biomass removal for all NADES was 27–67% (S. aureus) and 34–49% (P. aeruginosa). The tested NADES also inhibited biofilm formation of both bacterial species at concentrations at or below 0.5 x the minimal inhibitory concentration (MIC), possibly in part due to observed restrictions imposed by NADES on planktonic growth. These results demonstrate the potential value of neutral NADES as anti-biofilm agents in future antimicrobial preparations.
Keywords: Natural deep eutectic solvents (NADES), Biofilm, Green solvents, Anti-Biofilm activity
Abbreviations
- BS
betaine:sucrose (2:1)
- CFU
colony forming units
- ChG
choline chloride:glycerol (1:1)
- ChX
choline chloride:xylitol (5:2)
- EPS
extracellular polymeric substance
- MBEC
minimal biofilm eradication concentration
- MHB + gluc.
Mueller Hinton broth with 0.5% added glucose
- MIC
minimum inhibitory concentration
- NADES
natural deep eutectic solvents
- PBS
phosphate buffered saline;
- SEM
Scanning electron microscopy
- TSB
Tryptic soy broth
1. Introduction
Natural deep eutectic solvents (NADES) have rapidly become a focus in the search for new “green” media as alternatives to conventional organic solvents. Their applications as extraction and reaction media in fields such as the chemical industry have already been widely studied [[1], [2], [3], [4], [5]]. Recently, attention has grown for the potential use of NADES as solvents or excipients for pharmaceutical purposes due to their special properties [6,7]. NADES consist of supramolecular networks of two or more natural components, such as organic acids and bases, sugars or amino acids [8,9]. The deep eutectic mixture is formed when the components are mixed in a specific molar ratio, resulting in a liquid with significantly lower melting point than its individual constituents [10]. Some of the advantages of NADES include the low production cost, their adjustable physicochemical properties through changing the composition or water content, as well as their solubilizing properties [[11], [12], [13], [14]]. As the components are naturally abundant in living biological systems, the solvents have promising properties in pharmaceutical formulation, such as being biodegradable, environmentally friendly and presumably non-toxic [15]. Several studies have reported how NADES are able to extract, solubilize and stabilize biopolymers and other macromolecules, exemplified by bovine serum albumin, various enzymes and phenolic metabolites [[16], [17], [18], [19]]. In addition, a recent study found that certain NADES had biofilm matrix destruction properties [20].
Biofilms are three-dimensional matrices made up of microbial cells immersed in a self-generated extracellular polymeric substance (EPS). Biofilm formation poses a substantial challenge for efficient treatment of a range of skin and soft tissue infections in the human body, and is a well-known contributor to the development of chronic wounds [21]. The EPS is commonly composed of polysaccharides, proteins, and/or extracellular DNA, and provides protection against toxic substances, immune cells during infection, and inflammatory responses, as well as unexpected changes in the growth environment [[22], [23], [24]]. Therefore, the biofilm matrix can enhance the ability of an organism to survive exposure to antibiotics, potentially resulting in tolerance development and treatment failure [25]. Treatment strategies targeting the EPS, either by dissolving or disrupting the matrix, would be of potential great value in healing skin and soft tissue infections. Thus, a possible biofilm dissolving effect of NADES could lead to promising new antimicrobial formulations involving these solvents.
A previous study of biofilm matrix destruction properties of NADES did not include investigations in a biological system [20]. Therefore, the effects of NADES on bacterial biofilm should be expanded to in vitro biofilm models, in order to increase knowledge about novel potential applications of this group of solvents. The application of NADES in antimicrobial preparations is further actualized by studies reporting that these solvents could function synergistically, improving the antimicrobial activity of other compounds, e.g. catechins or porphyrins [26,27]. There are also indications that some NADES, in particular those with acidic properties, exhibit intrinsic antibacterial effects [28,29]. NADES with neutral pH are considered less toxic than their acidic counterparts, but are still able to dissolve biofilm components [20]. It would, therefore, be of interest to study the anti-biofilm effects of neutral NADES in particular.
In the present study, three NADES with neutral pH were investigated for their ability to break biofilm matrix structure and as biofilm formation inhibitors in different in vitro biofilm assays. Furthermore, their antibacterial effect in pre-formed biofilm was investigated. Neutral NADES were selected to avoid potential antimicrobial effects due to acid/base properties [30]. Pseudomonas aeruginosa and Staphylococcus aureus were chosen as model organisms, representative of Gram-negative and Gram-positive species, as they are both common biofilm-forming pathogens linked to topical infections. The selected model strains were sensitive to most common antibiotics.
2. Materials and methods
2.1. Materials
All materials were of analytical grade and were purchased from commercial sources.
2.2. Bacterial strains and growth medium
S. aureus ATCC 6538 and P. aeruginosa ATCC 9027 were cultivated at 37 °C in Tryptic Soy Broth (TSB) or on tryptic soy agar (TSA) plates unless otherwise stated. Original stocks of each strain were stored at −80 °C, and used to streak out bacterial cultures on master plates, which were stored at 4 °C for a maximum of two weeks before being discarded.
2.3. Neutral natural deep eutectic solvents
The NADES applied in this study were prepared as explained in a previous study [31]. In short, components were mixed and dissolved in warm water (50 °C), followed by evaporation of the water for 20 min in a rotary evaporator, resulting in a clear, viscous liquid. Table 1 gives an overview of the selected NADES, their constituents, and their measured water content. Unless otherwise stated, samples of NADES were diluted in the relevant growth media for the test organism. Concentrations are presented as % (v/v) due to the liquid nature of NADES.
Table 1.
Components of the natural deep eutectic solvents (NADES) included in the study, their molar ratios and measured water content when undiluted (from Ref. [31]).
| NADES | Molar ratio | Component 1 | Component 2 | Water content (% v/v) |
|---|---|---|---|---|
| ChG | 1:1 | Choline chloride | Glycerol | 15 |
| ChX | 5:2 | Choline chloride | Xylitol | 12 |
| BS | 2:1 | Betaine | Sucrose | 23 |
2.4. Minimum inhibitory concentration measurements and bacterial growth curves during exposure to NADES
The minimum inhibitory concentration (MIC) of each selected NADES at concentrations ranging from 5 to 35% was determined for both bacterial strains as a basis for further experiments. Over-night cultures of the strains in LB or TSB were prepared and incubated at 37 °C with shaking at 150 rpm prior to sample preparation. Inoculum (100 μl) was added to all samples in a 96-well round-bottom polystyrene microtiter plate, to a final volume of 200 μl and a starting OD600 of 0.01, in each well (Costar, Corning Inc., Kennebunk, USA). This was repeated for different starting inocula to investigate a possible inoculum effect of NADES. Plates were incubated at 37 °C for 18 h without shaking, followed by visual inspection to determine the MIC. Negative control (blank) wells (no bacteria added) and positive growth control wells were included in each plate, and the bacterial inoculum was controlled by plating of serial dilutions and colony counts.
Planktonic growth curves in growth medium with added NADES were obtained at 37 °C with shaking at 150 rpm over a 24 h period, with absorbance measurements (OD600) every 10 min using a plate reader (Clariostar, BMG Labtech, Ortenberg, Germany), and were compared to untreated controls. In this assay, each NADES was added in two different sub-MIC concentrations (diluted in TSB growth medium) to sterile non-treated flat bottom polystyrene 96-well microplates (Tissue culture plates, VWR, USA) containing over-night cultures diluted in fresh growth medium (TSB) to a final starting optical density of OD600 = 0.01. Final concentrations of NADES were: ChX: 5% and 10%; ChG: 2.5% and 5%, BS: 10% and 20% (respectively). The plates were covered with sealing foil (Breathe-Easy membrane, Merck KGaA, Darmstadt, Germany) prior to measurements. Four biological replicates were performed for the growth curve experiments, each with four technical replicates for each growth condition for each bacterium.
2.5. Effects of NADES on preformed biofilm
2.5.1. Biofilm formation
P. aeruginosa and S. aureus biofilms were formed in a peg lid system (Calgary device) (Innovotech, Inc., Edmonton, Canada). Over-night cultures were prepared and incubated as described in section 2.4, and were then diluted hundred-fold in fresh growth medium and incubated for 3 h at 37 °C with shaking at 150 rpm. The precultures were then adjusted to a starting OD600 = 0.01, and 160 μl was added to each well of a 96-well microtiter plate (Innovotech, Inc., Edmonton, Canada). P. aeruginosa biofilm was grown in TSB over 24 h at 25 °C and shaking at 110 rpm. The S. aureus biofilm was formed in Mueller-Hinton broth with 0.5% added glucose (MHB + gluc.) over 72 h at 37 °C and 110 rpm shaking. In both cases, the incubation was performed in a humid environment by placing the plates in sealed-off containers with moist tissue paper.
2.5.2. Minimal biofilm eradication concentration (MBEC) and biofilm killing determination
Established biofilm (on pegs) was challenged with concentrations of NADES ranging from 10 to 30% in 96-well round bottom polystyrene microplates (Costar, Corning Inc., Kennebunk, USA), followed by incubation for 24 h under similar conditions as for biofilm formation (2.5.1). Untreated controls were included. In order to measure total biomass the remaining biofilm was stained with 0.3% (w/v) crystal violet for 20 min, then washed in phosphate buffered saline (PBS) and dissolved in 180 μl acetone:ethanol (1:3) over 10 min in an ultrasound bath (SONOREX RK 100, Bandelin electronic GmbH & Co KG, Berlin, Germany). Absorbance measurements at 570 nm were obtained in 96-well flat bottom polystyrene microplates (Costar, Corning Inc., Kennebunk, USA) using an endpoint measuring method in a plate reader (Clariostar, BMG Labtech, Ortenberg, Germany) (the canonical wavelength for crystal violet is 595 nm). Samples from P. aeruginosa biofilm were diluted ten-fold prior to measurements, whereas S. aureus biofilm samples were measured undiluted. The experiments were performed in three to five biological replicates, each with 16 technical replicates.
To investigate the number of remaining viable bacteria in biofilm following NADES treatment, biofilm was formed and challenged with NADES as described above. Instead of crystal violet staining, the biofilm was gently dislodged from the pegs into wells containing 200 μl 0.9% NaCl by placing the microplates on a metal surface in an ultrasound bath (SONOREX RK 100, Bandelin electronic GmbH & Co KG, Berlin, Germany) for 30 min. Each sample was then serial diluted (101–107 fold) in 0.9% NaCl and plated on TSA plates, followed by incubation at 37 °C for 18 h and counting of colony forming units (CFU). The viability was studied using three to four biological replicates, each with eight technical replicates.
2.6. Biofilm formation assay
The effects of NADES on biofilm formation were studied in liquid broth cultures in 96-well round bottom polystyrene microplates (Costar, Corning Inc., Kennebunk, USA). NADES were added at sub-MIC concentrations, corresponding to a final high sub-MIC (0.25 x - 0.5 x MIC) and low sub-MIC (0.1 x - 0.25 x MIC) concentration respectively, in the microtiter plate wells (Supplementary Table S1), after addition of an equal volume of preculture, grown as described in 2.5.1 and prediluted to OD600 = 0.02 (final starting OD600 = 0.01). The microplates were incubated for the same duration and temperature as previously described (2.5.1). Staining of the biofilm formed in the plate wells and the following absorbance measurements were performed as described in section 2.5.2, with the modification that the biofilm was dissolved in 150 μl of acetone:ethanol (1:3) and then diluted two-fold (S. aureus) or four-fold (P. aeruginosa) in acetone:ethanol (1:3) prior to measurements. The experiment was performed in four biological replicates of each strain, with 16 technical replicates.
2.7. Scanning electron microscopy imaging
Biofilm samples for scanning electron microscopy (SEM) imaging were fixed through a double fixation protocol. Biofilm was formed and challenged with NADES as described in section 2.5, then fixed twice with a solution containing 2% (v/v) glutaraldehyde and 4% (v/v) paraformaldehyde in 0.1 M HEPES buffer. The first fixation was performed as a single strength fix over 15 min at a temperature of 25 °C or 37 °C for the P. aeruginosa and S. aureus samples, respectively. The second fixation was done at room temperature, followed by refrigeration of the samples until imaging.
The fixed biofilm was dehydrated by 10 min equilibration in a sequence of ethanol concentrations (70, 80, 90 and 96%), then equilibrated four times for 15 min in 100% ethanol. The samples were critical point dried (BAL-TEC CPD 030, Critical Point Dryer, BAL-TEC AG, Liechtenstein), mounted on aluminum SEM stubs with silver epoxy, then sputter coated with 10 nm platinum (Cressington 308UHR, Ted Pella Incorporated, Redding, CA, USA). The samples were examined with a Hitachi scanning electron microscope (Hitachi S-4800 FEG, Tokyo, Japan).
2.8. Statistical analysis
Microsoft Excel (Microsoft Office 360) software was used for normalization of data prior to statistical analyses. The experimental data were obtained from at least three independent experiments with technical replicates, and were presented as mean ± standard deviation (SD). Statistical analyses were performed by using one-way or two-way ANOVA followed by the Tukey's multiple comparison test using GraphPad Prism version 9.1.0 for Windows (GraphPad Software, La Jolla, CA, USA; www.graphpad.com).
3. Results
3.1. Minimal inhibitory concentration (MIC) determination of NADES towards P. aeruginosa and S. aureus
The MICs of the three selected NADES were determined against planktonic cultures of P. aeruginosa ATCC 9027 and S. aureus ATCC 6538 (Table 2), both at a starting OD600 equal to 0.01 (equivalent to approximately 4*106 CFU/ml for P. aeruginosa and 1*107 CFU/ml for S. aureus, respectively). Note that the values are reported as % (v/v) due to the liquid nature of NADES. The lowest MIC was found for choline chloride:glycerol (ChG), with 20% against P. aeruginosa ATCC 9027 and 20–25% against S. aureus ATCC 6538, while betaine:sucrose (BS) was the apparently least effective, with an MIC of 30% against P. aeruginosa and no apparent inhibition (>35%) of S. aureus growth in the tested concentration range (5–35%). Interestingly, an inoculum effect was observed for all three NADES tested (Supplementary Table S2; no data available for BS with S. aureus, as no growth inhibition was observed in the tested concentration range, 5–35%).
Table 2.
Minimum inhibitory concentration (% v/v) of the NADES ChX, ChG and BS against Pseudomonas aeruginosa ATCC 9027 and Staphylococcus aureus ATCC 6538, both at starting OD600 = 0.01. ChX: choline chloride:xylitol; ChG: choline chloride:glycerol; BS: betaine:sucrose.
| ChX | ChG | BS | |
|---|---|---|---|
| P. aeruginosa ATCC 9027 | 20% | 20% | 30% |
| S. aureus ATCC 6538 | 25% | 20–25% | >35% |
3.2. Effect on biofilm biomass and biofilm killing effect of NADES
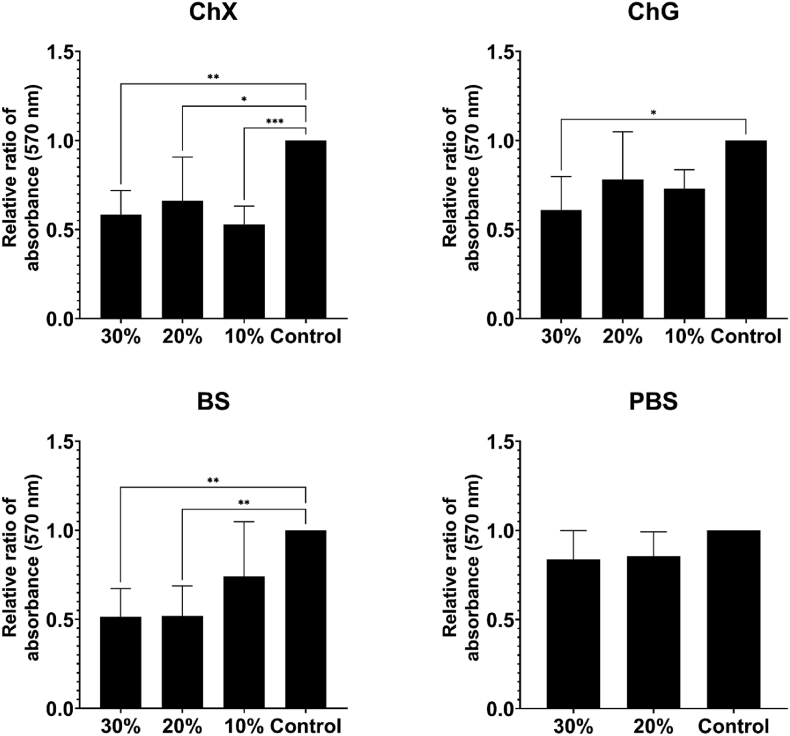
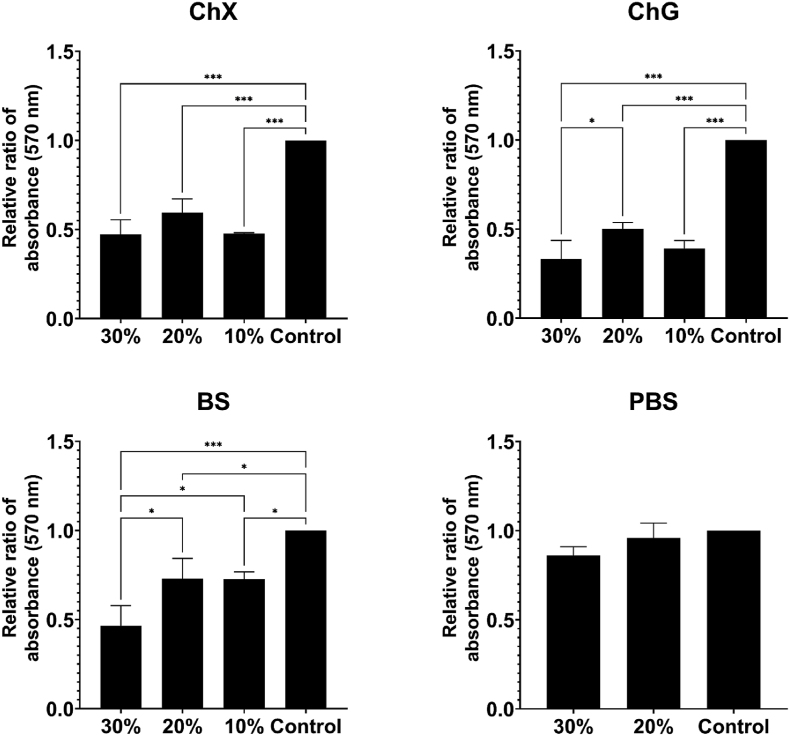
Pre-established biofilms were challenged with the selected NADES, and Fig. 1, Fig. 2 portray the effects on biofilm biomass of the tested NADES against P. aeruginosa ATCC 9027 and S. aureus ATCC 6538, respectively. The biofilm was stained with crystal violet prior to absorbance measurements, and a low absorbance value would correspond to a higher degree of biofilm biomass removal (lower residual biofilm). Statistically significant (p < 0.05–0.001) disruption of the biofilm relative to untreated control was found for choline chloride:xylitol (ChX) (all concentrations), betaine:sucrose (BS) (20% and 30%), and choline chloride:glycerol (ChG) (30%) against both bacterial strains (Fig. 1, Fig. 2), as well as for ChG (10% and 20%) and BS (10%) against S. aureus (Fig. 2). However, none of the NADES were able to eradicate all biofilm biomass at the tested concentrations. To control for the possible effects of a reduced volume of growth medium in the samples containing 20–30% NADES, samples added identical concentrations (v/v) of PBS were included. PBS samples did not affect the preformed biofilm significantly (Fig. 1, Fig. 2). For S. aureus the various NADES treatments lead to an on average 27–67% removal of biofilm biomass compared to control (Fig. 2), while for P. aeruginosa NADES treatment resulted in an average biofilm disruption of 34–49% (Fig. 1).
Fig. 1.
Biofilm biomass of Pseudomonas aeruginosa ATCC 9027 after 24 h challenge with NADES ChX, ChG and BS on pre-established biofilm, evaluated through staining the biofilm with crystal violet and measuring absorbance intensity (570 nm) (n = 3). Control samples challenged with 30% and 20% PBS (v/v, bottom right) were included to exclude effects on viability from a reduced volume of growth media in the samples containing 20–30% NADES. The indicated control was sample not added neither NADES nor PBS. Samples were diluted ten-fold in acetone:ethanol (1:3) prior to measurement. Results were normalized to control prior to statistical analyses, which were performed using one-way ANOVA. Error bars represent the standard deviation (SD) of normalized data from five independent experiments. Asterisks (*) denote significant differences, *p < 0.05, **p < 0.01 and ***p < 0.001.
Fig. 2.
Biofilm biomass of Staphylococcus aureus ATCC 6538 after 24 h challenge with NADES ChX, ChG and BS on pre-established biofilm, evaluated through staining the biofilm with crystal violet and measuring absorbance intensity (570 nm) (n = 5). PBS samples at 30% and 20% (v/v) were included to exclude effects on viability from a reduced volume of growth media in the samples containing 20–30% NADES. The indicated control sample was not added neither NADES nor PBS. Results were normalized to control prior to statistical analyses, which were performed using one-way ANOVA. Error bars represent the standard deviation (SD) of normalized data from five independent experiments. Asterisks (*) denote significant differences, *p < 0.05, **p < 0.01 and ***p < 0.001.
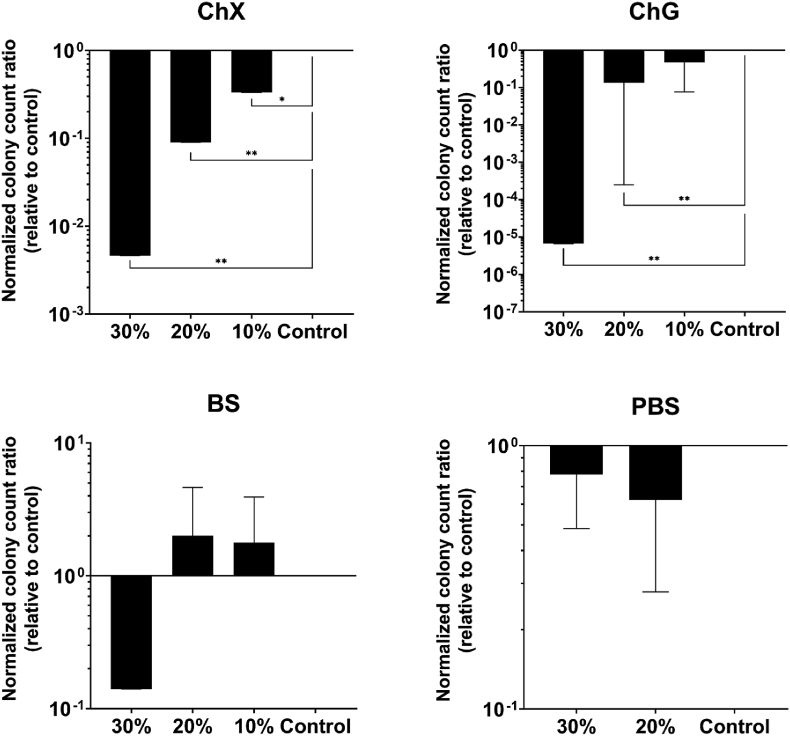
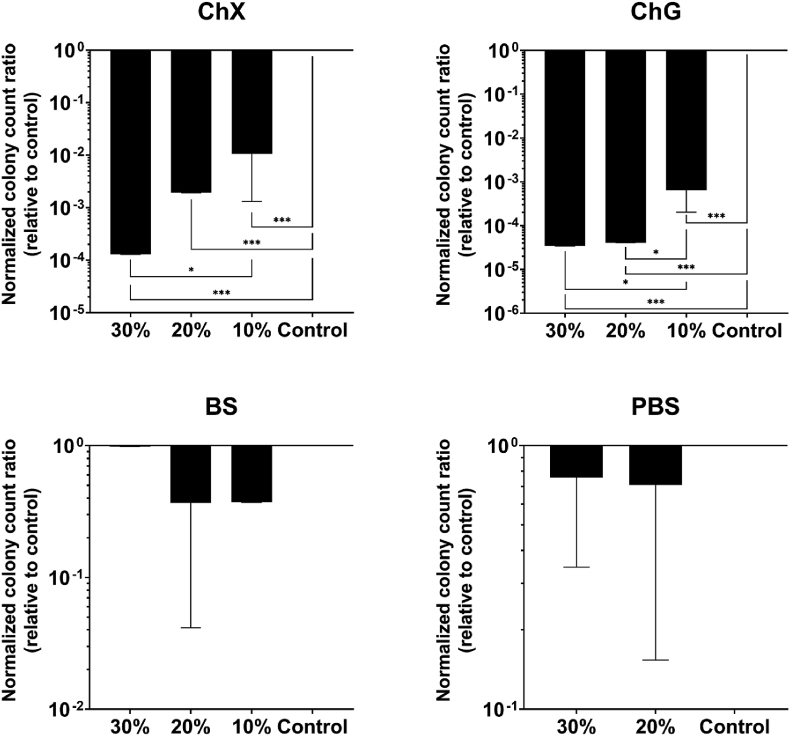
Although crystal violet staining provides insight into the amount of biomass eradicated by NADES treatment, it does not shed light on the effects of NADES on bacterial viability. Therefore, cell viability within the biofilm was investigated post treatment with NADES. The number of viable (CFU/ml) P. aeruginosa ATCC 9027 and S. aureus ATCC 6538 cells after 24 h challenge of the biofilm with the various NADES is presented in Fig. 3, Fig. 4, respectively. Except for 10% ChG with P. aeruginosa, all other tested concentrations of ChX and ChG significantly affected the number of viable cells of both species (p < 0.05–0.001) compared to control, with an average log10 reduction of 5–6 observed in the case of ChG (30%) against P. aeruginosa (Fig. 3), and average log10 reduction of 4–5 for ChG (20% and 30%) against S. aureus (Fig. 4). None of the organisms were susceptible to BS (Fig. 3, Fig. 4), with no significant change (p < 0.05) in viability following exposure (Fig. 3). Again, no significant change in number of viable cells was observed in the samples added PBS to 20% or 30% (Fig. 3, Fig. 4). Note the log-scale change in value for the vertical axis between the experiments with different NADES.
Fig. 3.
Viable Pseudomonas aeruginosa bacteria (CFU/ml) in biofilm after challenge with NADES ChX, ChG and BS for 24 h (n = 3), relative to control. PBS samples at 30% and 20% (v/v) were included to exclude effects on viability from a reduced volume of growth media in the samples containing 20–30% NADES. In the indicated control sample, neither NADES nor PBS was added. Results were normalized to the control prior to statistical analyses, which were performed using one-way ANOVA. Error bars represent the standard deviation (SD) of normalized data from four independent experiments. Asterisks (*) denote significant differences, *p < 0.05, **p < 0.01 and ***p < 0.001.
Fig. 4.
Viable Staphylococcus aureus bacteria (CFU/ml) in biofilm after challenge with NADES ChX, ChG and BS for 24 h (n = 4), relative to control. PBS samples at 30% and 20% (v/v) were included to exclude effects on viability from a reduced volume of growth media in the samples containing 20–30% NADES. In the indicated control sample, neither NADES nor PBS was added. Results were normalized to the control prior to statistical analyses, which were performed using one-way ANOVA. Error bars represent the standard deviation (SD) of normalized data from four independent experiments. Asterisks (*) denote significant differences, *p < 0.05, **p < 0.01 and ***p < 0.001.
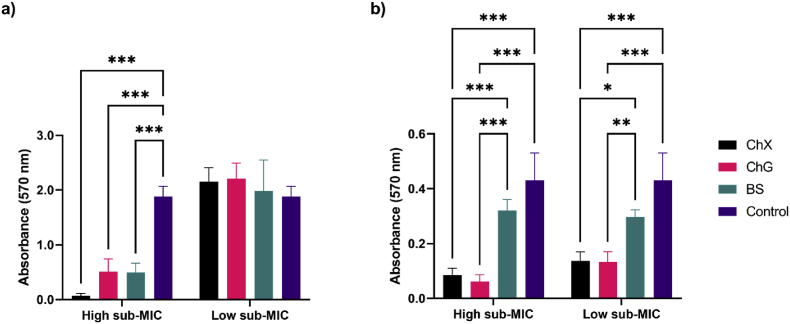
3.3. Effects of NADES on biofilm formation
The effects of NADES ChX, ChG and BS on P. aeruginosa and S. aureus biofilm formation are shown in Fig. 5, with applied concentrations of NADES at or below 0.5 x the determined MIC values at starting OD600 = 0.01 (Table 2). Two concentrations were used for each NADES, a high sub-MIC concentration (0.25 x – 0.5 x MIC) and a low sub-MIC concentration (0.1–0.25 x MIC) (Supplementary Table S1; Supplementary Table S2). The high sub-MIC concentrations of ChX and ChG significantly reduced biofilm formation for both strains relative to control (Fig. 5a and b), whereas the BS high sub-MIC sample only affected P. aeruginosa biofilm formation significantly (p < 0.001) (Fig. 5a). None of the low sub-MIC samples (0.1 x - 0.25 x MIC) were able to inhibit biofilm formation of P. aeruginosa (Fig. 5a), whereas ChX and ChG at low sub-MIC concentrations significantly (p < 0.05–0.001) affected S. aureus biofilm formation relative to control (Fig. 5b). For all three NADES, increasing the concentration from low sub-MIC to high sub-MIC levels had a significant inhibitory effect on P. aeruginosa biofilm formation (Supplementary Table S3). This effect was not observed for S. aureus.
Fig. 5.
Effect of NADES ChX, ChG and BS in sub-MIC concentrations corresponding to high sub-MIC (0.25 x - 0.5 x) and low sub-MIC (0.1 x - 0.25 x) concentrations, respectively, on P. aeruginosa (a) and S. aureus (b) biofilm formation (n = 4). Absorbance (570 nm) was measured after staining the biofilm with crystal violet. S. aureus samples were diluted two-fold, whereas P. aeruginosa samples were diluted four-fold in acetone:ethanol (1:3) prior to measurements. Statistics were performed using two-way ANOVA. Error bars represent the standard deviation (SD) of normalized data from four independent experiments. Asterisks (*) denote significant reduction in formed biofilm, *p < 0.05, **p < 0.01 and ***p < 0.001. Statistics for comparisons between samples across the two different NADES concentrations are shown in Supplementary Table S3. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
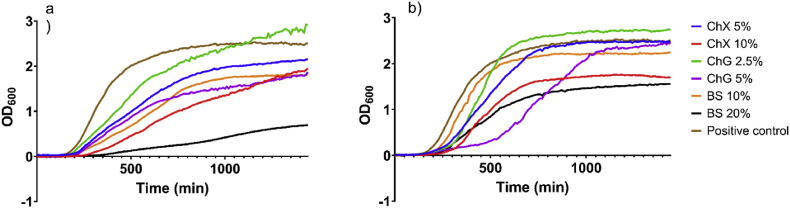
In order to determine if NADES ChX, ChG and BS had an impact on planktonic bacterial growth rates, which may potentially also affect initial phases of biofilm formation, growth curve experiments were performed at two different sub-MIC concentrations of each compound (Fig. 6). The slope value for each growth curve was calculated during the exponential growth phase (as identified by log-transformation of the growth curve) for each individual culture condition, and was used as a measure for the bacterial growth rate (Supplementary Table S4). Also, delay of growth for the various NADES treatments relative to control was quantified, by determining the time to reach OD600 = 0.20 (corresponding to mid-exponential growth for the untreated control samples), and the time to reach OD600 = 0.50, respectively (Supplementary Table S4). All NADES samples inhibited the growth of the bacteria to a significant degree at the highest sub-MIC concentration tested (relative to control) both for P. aeruginosa and S. aureus, and for P. aeruginosa also at the lower sub-MIC concentrations, leading to significant changes in exponential phase growth rate (Supplementary Tables S4 and S5). In addition, experiments investigating the time to reach exponential growth phase (time to reach OD600 = 0.20 and 0.50, respectively) for each culture condition showed that NADES ChX and BS at the highest tested sub-MIC concentrations conferred a significant growth delay for P. aeruginosa (both at OD600 = 0.20 and OD600 = 0.50), while NADES ChX and ChG at the highest tested sub-MIC concentrations conferred a significant growth delay for S. aureus at OD600 = 0.50 (Supplementary Tables S4 and S6).
Fig. 6.
Growth curves (OD600) of P. aeruginosa ATCC 9027 (a) and S. aureus ATCC 6538 (b), in the presence of NADES ChX, ChG and BS at different concentrations. OD600 of each culture was measured every 10 min over 24 h (n = 4). Values for slope and delay of growth relative to control are shown in Supplementary Table S4, and statistical analysis using two-way ANOVA is provided in Supplementary Tables S5 and S6.
3.4. Scanning electron microscopy (SEM) imaging of biofilms challenged with NADES
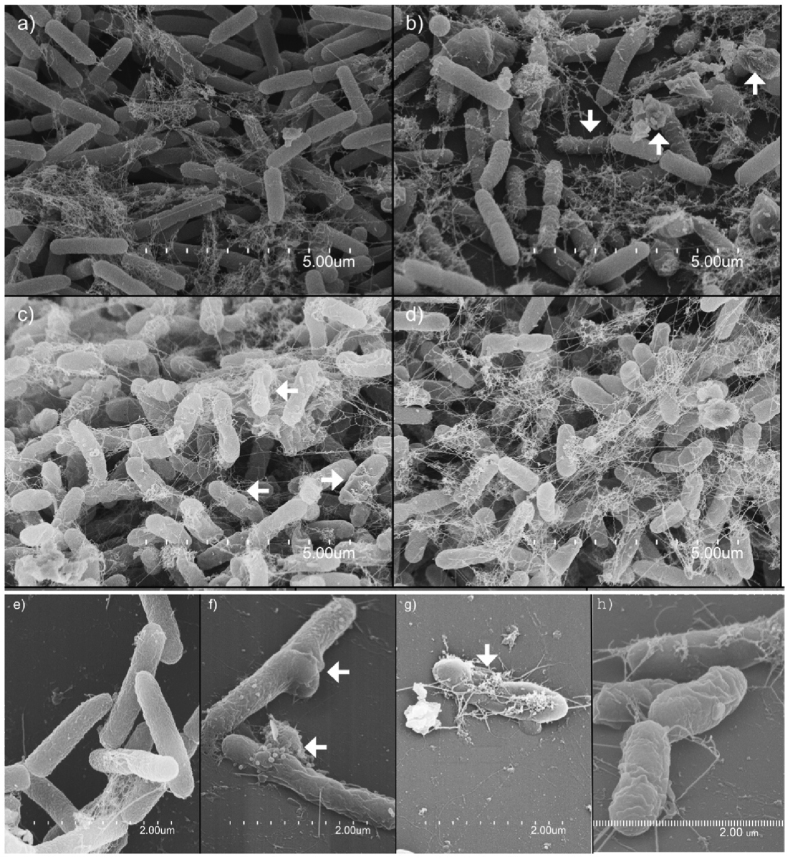
P. aeruginosa ATCC 9027 and S. aureus ATCC 6538 biofilms challenged with NADES were analysed by scanning electron microscopy (SEM), and representative images are shown in Fig. 7, Fig. 8, respectively. The experiment was performed twice and at least nineteen images were analysed for each treatment for each strain. Challenge with NADES was performed identically to the procedure used for the MBEC assay, employing the Calgary device model (3.2), and imaging was performed directly on the plastic pegs where biofilm was formed, at the section corresponding to the liquid-air interface. Images of the samples with 30% concentration of NADES were chosen, to clearly observe potential differences between samples and control (Fig. 7a and e and Fig. 8a and e shows untreated P. aeruginosa and S. aureus biofilm, respectively).
Fig. 7.
Scanning electron microscopy (SEM) images of P. aeruginosa biofilm: a) and e) without treatment, b) and f) after treatment with BS 30%, c) and g) after treatment with ChG 30%, d) and h) after treatment with ChX 30%. Examples of damaged bacteria after treatment with BS 30% and with ChG 30% are indicated by white arrows. White dotted lines are space bars indicating size.
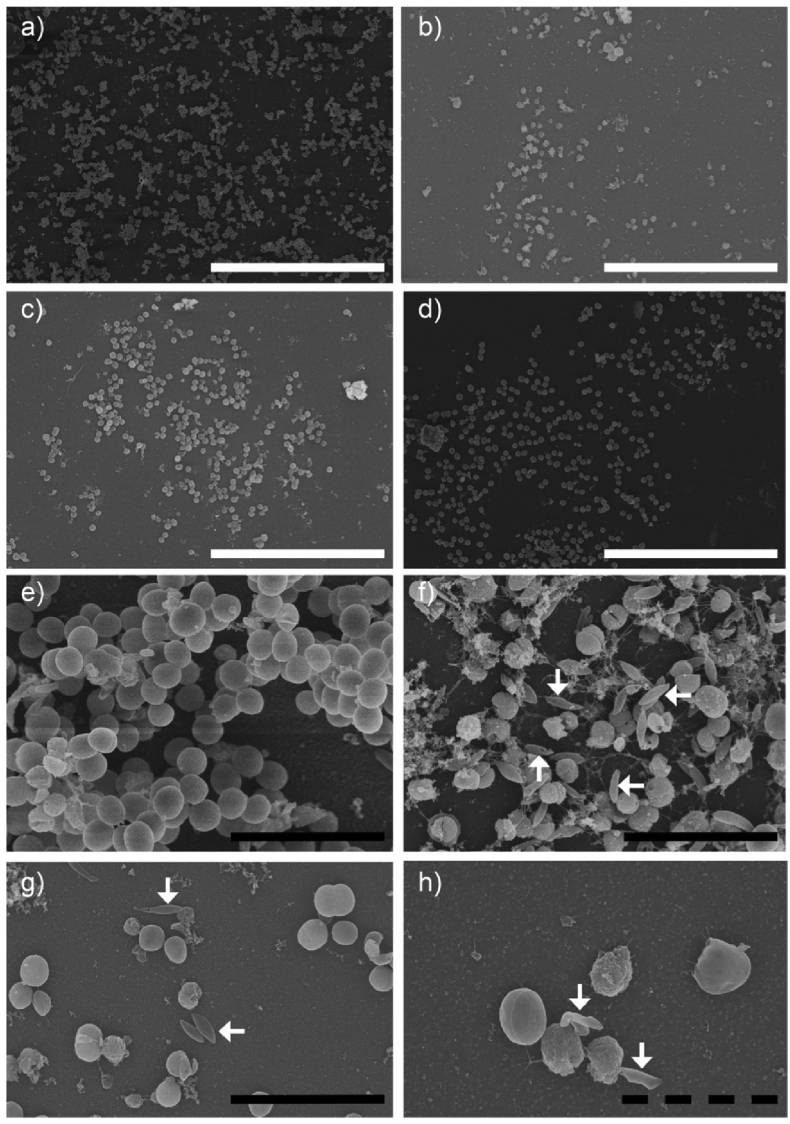
Fig. 8.
Scanning electron microscopy (SEM) images of S. aureus biofilm: a) and e) without treatment, b) and f) after treatment with NADES BS 30%, c) and g) after treatment with ChG 30% and d) and h) after treatment with ChX 30%. White arrows indicate cell morphological changes after NADES treatment. Space bars: white = 20 μM, black = 5 μM and dotted line = 2 μM.
The images of P. aeruginosa biofilm exposed to 30% BS (Fig. 7b and f) and 30% ChG (Fig. 7c and g) indicated damages to the surface of the cells compared to untreated control (Fig. 7a and e). Changes in the morphology (i.e. irregularities on the surface) of the treated cells could be observed for both 30% BS and 30% ChG treatments (Fig. 7f and g). Cell damage was not observed in the ChX samples. Changes in the morphology (i.e. irregularities on the surface) of the treated cells could also be observed (Fig. 7f and g). However, the biofilms were clearly not fully eradicated following NADES treatment (Fig. 7b, c and d), with the SEM images showing intact residual biofilm matrix after challenge with all tested NADES at 30%, in line with the MBEC results measuring biofilm biomass using crystal violet staining (Fig. 1, Fig. 2).
Images of S. aureus biofilm treated with NADES (Fig. 8a–d) are in line with results from the MBEC experiments (Fig. 1, Fig. 2), which indicate some biofilm removal compared to controls. The morphology of S. aureus ATCC 6538 cells was affected by all three different NADES, e.g. as shown for 30% BS (Figs. 8f), 30% ChG (Figs. 8g) and 30% ChX (Fig. 8h) compared to control (Fig. 8a and e). The morphological effect was thereby more pronounced for S. aureus ATCC 6538 in the BS samples than for P. aeruginosa ATCC 9027.
4. Discussion
Biofilms are notoriously more difficult to treat with antimicrobials than planktonic cells, and new treatment strategies for biofilm destruction are needed. NADES have emerged as promising solvents for pharmaceutical formulations and may have a place in novel antimicrobial preparations targeting biofilm. This work investigated the potential value of neutral NADES as anti-biofilm agents in future antimicrobial formulations, employing S. aureus and P. aeruginosa as representative indicator organisms.
MIC values of the selected NADES toward the indicator bacteria were determined prior to the anti-biofilm assays (Table 2). Both ChX and ChG were found to show anti-bacterial activity, inhibiting growth of the test organisms at concentrations 20–25% v/v in growth medium. The antimicrobial activities of ChX and ChG have also been investigated in previous studies. Radosevic et al. (2018) reported no inhibition for ChX against S. aureus and P. aeruginosa in similar concentrations as those used here [29]. However, that study utilized a disk diffusion method, meaning that the antimicrobial activity is highly dependent on diffusion of the NADES into the surrounding solid medium. The high viscosity of most NADES could hamper diffusion, thus possibly resulting in a lower antimicrobial effect in a disk diffusion method compared to MIC determination employing broth microdilution methods, which also better reflect the use of NADES as antimicrobial and anti-biofilm agents. In a short communication by Ref. [32]; ChG did not exhibit antimicrobial properties [32]. That study did however not specify the tested concentrations of NADES, nor describe the method for MIC determination, and the results are therefore not necessarily comparable. Our experiments therefore to our knowledge constitute the first demonstration of antibacterial activity of neutral NADES.
Growth of both indicator organisms was considerably affected in the presence of the highest sub-MIC concentrations of each NADES used for the growth curve experiments (corresponding to 0.25 x – 0.67 x MIC for the different NADES) relative to control, both with respect to growth rate (all tested NADES, both bacteria) and delay of exponential phase growth (ChX and BS for P. aeruginosa, ChX and ChG for S. aureus) (Fig. 6; Supplementary Tables S4–S6). The lower sub-MIC NADES concentrations (corresponding to 0.125 x – 0.33 x MIC) had a significant effect on growth rate only for P. aeruginosa (all tested NADES), and did not significantly delay entry into exponential growth for any of the two bacteria tested. Although it should be noted that the concentrations used to assay planktonic growth corresponded to slightly different fold MICs for each of the respective NADES, the results clearly showed that the tested neutral NADES, even at sub-MIC concentrations, could inhibit the growth of the bacteria, which is supported by a previous study on other types of NADES [33].
To our knowledge, the effect of NADES on viability of bacterial cells within a biofilm has never been subject to previous study. Here it was observed that although complete eradication of biofilm biomass was not observed at tested concentrations for any of the three NADES investigated, the number of remaining viable biofilm cells of both test organisms was significantly affected after challenge of established biofilm with ChG and ChX (except for the lowest concentration of ChG with P. aeruginosa; Fig. 3, Fig. 4). A 4-6-range log10 reduction in viable cells was seen for the 30% ChG sample against both species, corresponding to a 99.99%–99.9999% reduction in viable bacteria, showing that specific NADES can exhibit high antibacterial efficacy, also against bacteria within a protective biofilm.
An alternative interpretation of this data could be ChG and ChX inducing biofilm dispersal, with biofilm cells becoming planktonic during challenge, and the exact cellular and molecular mechanism(s) responsible for the reduction in viable bacteria within the biofilm remain to be mapped. However, clearly these NADES are able to penetrate into the biofilm, and all tested NADES appeared to have effects on cell morphology (Fig. 7, Fig. 8). Penetration into the biofilm may potentially lead to direct interactions of NADES with bacterial cell surface structures, and/or possibly induce osmotic shock of the cells due to the hyperosmolar NADES environment. The compounds applied in this study would have an expected osmolality from 350 to >3000 mOsm/kg (based on previous measurements of 10–15 fold diluted NADES), depending on the type of NADES and concentration [31]. In line with this, the SEM images (Fig. 7, Fig. 8) showed clear damage to the bacterial membrane and changes in size and morphology of the cells. Also, a previous study in our group identified similar shrinkage to artificial cell membranes in the presence of NADES [31].
It has previously been proposed that Gram-positive bacteria may be more resistant to NADES than Gram-negative species due to the differences in cell wall structure [34]. A hypothesised method of action for NADES is hydrogen bond- or electrostatic interactions of the NADES components with polysaccharides and peptides in the cell wall [9,35]. As the Gram-positive cell wall is composed of a thicker layer of these macromolecules than the Gram-negative wall, resistance towards NADES could potentially be increased in Gram-positive species. The MIC assay showed that all three NADES tested inhibited growth by the Gram-negative bacterium P. aeruginosa at somewhat lower concentrations than for S. aureus (Gram-positive) (Table 2). This could support the proposed theory, although other factors could also be involved.
Crystal violet staining is a well-known output parameter for total biofilm biomass, staining both bacterial cells and the surrounding EPS. Results in this study indicated that the three tested NADES were able to remove some of the established biofilm, although only in the range 27–67% (Fig. 1, Fig. 2). Nava-Ocampo et al. [20] reported that two out of seven tested NADES were able to solubilize EPS and other biomolecules present in biofilms, thus exhibiting biofilm matrix destruction properties in an in vitro setting [20]. The study did not investigate the same NADES as the present work, although choline chloride and betaine were components in some of the tested solvents. Among these, only the NADES composed of betaine and urea was reported to have biofilm matrix destruction properties. In the present study however, both the NADES containing betaine (BS) and NADES containing choline chloride (ChG and ChX) showed some degree of biofilm matrix destruction property compared to control. NADES ChG and ChX both affected biofilm cell viability (Fig. 3, Fig. 4) and planktonic bacterial growth rate (Fig. 6; Supplementary Tables S4–S5). BS, however, did not significantly affect the viability of preformed biofilm, thus appearing less effective as an anti-biofilm agent compared to ChX or ChG, as primarily shown by the MBEC cell viability results (Fig. 3, Fig. 4). As mentioned previously, many NADES are of a highly viscous nature, and among the NADES tested in the present work, BS has a considerably higher viscosity than the other solvents (reported to be 874 mPa S undiluted in previous measurements by our group [31]). A high viscosity environment has previously been found to increase the tolerance of P. aeruginosa and S. aureus to antibiotics [36,37], and it can at present not be excluded that the poor anti-biofilm properties of BS could in part be due to its high viscosity.
In addition, planktonic growth was clearly affected by the presence of NADES, even at concentrations below the MIC, as shown by the growth curve assays (Fig. 6; Supplementary Tables S4–S6). In a biofilm culture, bacteria are in an equilibrium between being in planktonic form and residing in the biofilm [37]. Also, during the initial stages of a biofilm formation assay, the bacterial cells are primarily in planktonic growth. Therefore, the inhibition effect of NADES on planktonic growth could potentially over time affect the biofilm formation capacity, and, although the assays performed for biofilm formation and planktonic growth were different in setup (e.g. different growth media, shaking frequency, NADES concentrations) one can not exclude that the observed reduction in biofilm formation imposed by NADES at least partly could be due to growth restrictions of the planktonic bacteria, in addition to any potential direct inhibition of biofilm formation mechanisms per se.
5. Conclusions
To the best of our knowledge, this is the first study on anti-biofilm activity of NADES in a biological model system. In this paper, neutral NADES ChX and ChG were demonstrated to have anti-biofilm properties, by exhibiting bactericidal activity against P. aeruginosa and S. aureus cells in pre-established bacterial biofilm. Furthermore, biofilm formation was inhibited at sub-MIC concentrations of NADES, and the inhibitory effect observed by NADES on bacterial planktonic growth could potentially contribute to this anti-biofilm effect. There was no clear evidence of strong biofilm matrix destruction properties or biofilm eradication by neutral NADES within the present data, and the potential use of NADES in this context needs further investigation.
Credit author statement
Helene Liepelt Nystedt: Conceptualization, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Krister Gjestvang Grønlien: Investigation, Methodology, Visualization, Writing – review & editing. Rebekka Rekkedal Rolfsnes: Investigation, Methodology, Writing – review & editing. Hanne Cecilie Winther-Larsen: Supervision, Resources, Writing – review & editing. Ole Andreas Løchen Økstad: Supervision, Resources, Investigation, Methodology, Funding acquisition, Writing – review & editing. Hanne Hjorth Tønnesen: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
This work was supported by internal grants from the Department of Pharmacy and the Faculty of Mathematics and Natural Sciences, University of Oslo, to HHT and OAØ. This research did not receive any specific external grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful to Veronika Smith and Truls Corneliussen Rasmussen, Department of Pharmacy, University of Oslo and Antje Hofgaard, Department of Biosciences, University of Oslo, for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2023.100114.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Fernández M.d.l.Á., Boiteux J., Espino M., Gomez F.J.V., Silva M.F. Natural deep eutectic solvents-mediated extractions: the way forward for sustainable analytical developments. Anal Chim Acta. 2018;1038:1–10. doi: 10.1016/j.aca.2018.07.059. [DOI] [PubMed] [Google Scholar]
- 2.Kamarza M., Dezaldi A., Ida Z., Elsa K. Green extraction of palmitic acid from palm oil using betaine-based natural deep eutectic solvents. Int. J. Technol. 2018;9(2):335–344. doi: 10.14716/ijtech.v9i2.1008. [DOI] [Google Scholar]
- 3.Gómez A., Biswas A., Tadini C., Furtado R., Alves C., Cheng H. Use of natural deep eutectic solvents for polymerization and polymer reactions. J Braz Chem Soc. 2019;30(4) doi: 10.21577/0103-5053.20190001. [DOI] [Google Scholar]
- 4.Ma Y., Li Y., Ali S., Li P., Zhang W., Rauch M.C.R., Willot S.J.P., Ribitsch D., Choi Y.H., Alcalde M., Hollmann F., Wang Y. Natural deep eutectic solvents as performance additives for peroxygenase catalysis. ChemCatChem. 2020;12(4):989–994. doi: 10.1002/cctc.201901842. [DOI] [Google Scholar]
- 5.Hikmawanti N.P.E., Ramadon D., Jantan I., Mun’im A. Natural deep eutectic solvents (Nades): phytochemical extraction performance enhancer for pharmaceutical and nutraceutical product development. Plants (Basel) 2021;10(10):2091. doi: 10.3390/plants10102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedro S.N., Freire M.G., Freire C.S.R., Silvestre A.J.D. Deep eutectic solvents comprising active pharmaceutical ingredients in the development of drug delivery systems. Expet Opin Drug Deliv. 2019;16:497–506. doi: 10.1080/17425247.2019.1604680. [DOI] [PubMed] [Google Scholar]
- 7.Zainal-Abidin M.H., Hayyan M., Ngoh G.C., Wong W.F., Looi C.Y. Emerging frontiers of deep eutectic solvents in drug discovery and drug delivery systems. J Contr Release. 2019;316:168–195. doi: 10.1016/j.jconrel.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Choi Y.H., van Spronsen J., Dai Y., Verberne M., Hollmann F., Arends I.W.C.E., Witkamp G.-J., Verpoorte R. Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology? Plant Physiol. 2011;156(4):1701–1705. doi: 10.1104/pp.111.178426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai Y., van Spronsen J., Witkamp G.-J., Verpoorte R., Choi Y.H. Natural deep eutectic solvents as new potential media for green technology. Anal Chim Acta. 2013;766:61–68. doi: 10.1016/j.aca.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Friesen J.B., McAlpine J.B., Lankin D.C., Chen S.-N., Pauli G.F. Natural deep eutectic solvents: properties, applications, and perspectives. J Nat Prod. 2018;81(3):679–690. doi: 10.1021/acs.jnatprod.7b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai Y., Witkamp G.-J., Verpoorte R., Choi Y.H. Tailoring properties of natural deep eutectic solvents with water to facilitate their applications. Food Chem. 2015;187:14–19. doi: 10.1016/j.foodchem.2015.03.123. [DOI] [PubMed] [Google Scholar]
- 12.Aroso I.M., Paiva A., Reis R.L., Duarte A.R.C. Natural deep eutectic solvents from choline chloride and betaine – physicochemical properties. J Mol Liq. 2017;241:654–661. doi: 10.1016/j.molliq.2017.06.051. [DOI] [Google Scholar]
- 13.Savi L.K., Carpiné D., Waszczynskyj N., Ribani R.H., Haminiuk C.W.I. Influence of temperature, water content and type of organic acid on the formation, stability and properties of functional natural deep eutectic solvents. Fluid Phase Equil. 2019;488:40–47. doi: 10.1016/j.fluid.2019.01.025. [DOI] [Google Scholar]
- 14.Mustafa N.R., Mustafa N., Spelbos V., Witkamp G.-J., Verpoorte R., Choi Y. Solubility and stability of some pharmaceuticals in natural deep eutectic solvents-based formulations. Molecules. 2021;2645 doi: 10.3390/molecules26092645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paiva A., Craveiro R., Aroso I., Martins M., Reis R.L., Duarte A.R.C. Natural deep eutectic solvents – solvents for the 21st century. ACS Sustainable Chem Eng. 2014;2(5):1063–1071. doi: 10.1021/sc500096j. [DOI] [Google Scholar]
- 16.Morrison H.G., Sun C.C., Neervannan S. Characterization of thermal behavior of deep eutectic solvents and their potential as drug solubilization vehicles. Int J Pharm. 2009;378(1–2):136–139. doi: 10.1016/j.ijpharm.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 17.Dai Y., Witkamp G.-J., Verpoorte R., Choi Y.H. Natural deep eutectic solvents as a new extraction media for phenolic metabolites in Carthamus tinctorius L. Anal Chem. 2013;85(13):6272–6278. doi: 10.1021/ac400432p. [DOI] [PubMed] [Google Scholar]
- 18.Li N., Wang Y., Xu K., Huang Y., Wen Q., Ding X. Development of green betaine-based deep eutectic solvent aqueous two-phase system for the extraction of protein. Talanta. 2016;152:23–32. doi: 10.1016/j.talanta.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 19.Knudsen C., Bavishi K., Viborg K.M., Drew D.P., Simonsen H.T., Motawia M.S., Møller B.L., Laursen T. Stabilization of dhurrin biosynthetic enzymes from Sorghum bicolor using a natural deep eutectic solvent. Phytochemistry. 2020;170 doi: 10.1016/j.phytochem.2019.112214. 112214-112214. [DOI] [PubMed] [Google Scholar]
- 20.Nava-Ocampo M.F., Fuhaid L.A., Verpoorte R., Choi Y.H., van Loosdrecht M.C.M., Vrouwenvelder J.S., Witkamp G.J., Farinha A.S.F., Bucs S.S. Natural deep eutectic solvents as biofilm structural breakers. Water Res. 2021;201 doi: 10.1016/j.watres.2021.117323. 117323-117323. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y.-K., Cheng N.-C., Cheng C.-M. Biofilms in chronic wounds: pathogenesis and diagnosis. Trends Biotechnol. 2019;37(5):505–517. doi: 10.1016/j.tibtech.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Allison D.G. The biofilm matrix. Biofouling. 2003;19(2):139–150. doi: 10.1080/0892701031000072190. [DOI] [PubMed] [Google Scholar]
- 23.Ma L., Conover M., Lu H., Parsek M.R., Bayles K., Wozniak D.J. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5(3) doi: 10.1371/journal.ppat.1000354. e1000354-e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonawane J.M., Rai A.K., Sharma M., Tripathi M., Prasad R. Microbial biofilms: recent advances and progress in environmental bioremediation. Sci Total Environ. 2022;824 doi: 10.1016/j.scitotenv.2022.153843. 153843-153843. [DOI] [PubMed] [Google Scholar]
- 25.Thi M.T.T., Wibowo D., Rehm B.H.A. Pseudomonas aeruginosa biofilms. Int J Mol Sci. 2020;21(22):1–25. doi: 10.3390/ijms21228671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wikene K.O., Bruzell E., Tønnesen H.H. Improved antibacterial phototoxicity of a neutral porphyrin in natural deep eutectic solvents. J Photochem Photobiol B Biol. 2015;148:188–196. doi: 10.1016/j.jphotobiol.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 27.Zhou P., Tang D., Zou J., Wang X. An alternative strategy for enhancing stability and antimicrobial activity of catechins by natural deep eutectic solvents. LWT. 2022;112558 doi: 10.1016/j.lwt.2021.112558. [DOI] [Google Scholar]
- 28.Wikene K.O., Rukke H.V., Bruzell E., Tønnesen H.H. Investigation of the antimicrobial effect of natural deep eutectic solvents (NADES) as solvents in antimicrobial photodynamic therapy. J Photochem Photobiol B Biol. 2017;171:27–33. doi: 10.1016/j.jphotobiol.2017.04.030. [DOI] [PubMed] [Google Scholar]
- 29.Radošević K., Čanak I., Panić M., Markov K., Bubalo M.C., Frece J., Srček V.G., Redovniković I.R. Antimicrobial, cytotoxic and antioxidative evaluation of natural deep eutectic solvents. Environ Sci Pollut Res Int. 2018;25(14):14188–14196. doi: 10.1007/s11356-018-1669-z. [DOI] [PubMed] [Google Scholar]
- 30.Minor T.E., Marth E.H. Loss of viability by Staphylococcus aureus in acidified media. I. Inactivation by several acids, mixtures of acids, and salts of acids. J Milk Food Technol. 1972;35(4):191–196. [Google Scholar]
- 31.Nystedt H.L., Grønlien K.G., Tønnesen H.H. Interactions of natural deep eutectic solvents (NADES) with artificial and natural membranes. J Mol Liq. 2021;328 doi: 10.1016/j.molliq.2021.115452. [DOI] [Google Scholar]
- 32.Hayyan M., Hashim M.A., Hayyan A., Al-Saadi M.A., AlNashef I.M., Mirghani M.E.S., Saheed O.K. Are deep eutectic solvents benign or toxic? Chemosphere. 2013;90(7):2193–2195. doi: 10.1016/j.chemosphere.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Jurić T., Mićić N., Potkonjak A., Milanov D., Dodić J., Trivunović Z., Popović B.M. The evaluation of phenolic content, in vitro antioxidant and antibacterial activity of Mentha piperita extracts obtained by natural deep eutectic solvents. Food Chem. 2021;362 doi: 10.1016/j.foodchem.2021.130226. 130226-130226. [DOI] [PubMed] [Google Scholar]
- 34.Hou X.-D., Liu Q.-P., Smith T.J., Li N., Zong M.-H. Evaluation of toxicity and biodegradability of cholinium amino acids ionic liquids. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zdanowicz M., Wilpiszewska K., Spychaj T. Deep eutectic solvents for polysaccharides processing. A review. Carbohydr. Polym. 2018;200:361–380. doi: 10.1016/j.carbpol.2018.07.078. [DOI] [PubMed] [Google Scholar]
- 36.Kostenko V., Ceri H., Martinuzzi R.J. Increased tolerance of Staphylococcus aureus to vancomycin in viscous media. FEMS Immunol Med Microbiol. 2007;51(2):277–288. doi: 10.1111/j.1574-695X.2007.00300.x. [DOI] [PubMed] [Google Scholar]
- 37.Omar A., Wright J.B., Schultz G., Burrell R., Nadworny P. Microbial biofilms and chronic wounds. Microorganisms. 2017;5(1):9. doi: 10.3390/microorganisms5010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.