Tracking phagocytic tumor-associated myeloid cells in a novel autochthonous lung tumor model reveals key responses to neoplastic cell engulfment, including elevated oxidative phosphorylation and immune suppression. The phagocytic signature is also enriched in myeloid cells from human tumors.
Abstract
Phagocytosis is a key macrophage function, but how phagocytosis shapes tumor-associated macrophage (TAM) phenotypes and heterogeneity in solid tumors remains unclear. Here, we utilized both syngeneic and novel autochthonous lung tumor models in which neoplastic cells express the fluorophore tdTomato (tdTom) to identify TAMs that have phagocytosed neoplastic cells in vivo. Phagocytic tdTompos TAMs upregulated antigen presentation and anti-inflammatory proteins, but downregulated classic proinflammatory effectors compared to tdTomneg TAMs. Single-cell transcriptomic profiling identified TAM subset-specific and common gene expression changes associated with phagocytosis. We uncover a phagocytic signature that is predominated by oxidative phosphorylation (OXPHOS), ribosomal, and metabolic genes, and this signature correlates with worse clinical outcome in human lung cancer. Expression of OXPHOS proteins, mitochondrial content, and functional utilization of OXPHOS were increased in tdTompos TAMs. tdTompos tumor dendritic cells also display similar metabolic changes. Our identification of phagocytic TAMs as a distinct myeloid cell state links phagocytosis of neoplastic cells in vivo with OXPHOS and tumor-promoting phenotypes.
Introduction
Macrophages are innate immune cells that play crucial homeostatic, defensive, and reparative roles in all tissues of the body (Gessain et al., 2020). These cells respond to tissue environmental cues and possess a spectrum of activation states that contribute to their considerable heterogeneity (Mosser and Edwards, 2008). A key function of macrophages is to phagocytose extracellular material including pathogens, dead/dying cells, and neoplastic cells (Metchnikoff, 1883; Watanabe et al., 2019). The phagocytosis of apoptotic cells, termed efferocytosis, is critical for homeostasis, but can also suppress immune surveillance and promote tumor growth (A-Gonzalez et al., 2009; Hanayama et al., 2004; Mukundan et al., 2009; Roszer et al., 2011; Scott et al., 2001; Uribe-Querol and Rosales, 2020). Phagocytic macrophages at steady state have been associated with increased cytoskeletal rearrangements, phago-lysosomal activity, antigen presentation, and lipid metabolism (A-Gonzalez et al., 2017). Additionally, pathway analyses relating to phagocytosis have helped identify putative phagocytic macrophages across healthy and diseased tissue states (Jaitin et al., 2019; Lantz et al., 2020; Sanin et al., 2022). Despite these efforts, the functional identification and validation of phagocytic immune cells in the tumor microenvironment remains elusive, and thus it is unclear whether the engulfment of neoplastic cells by tumor-associated macrophages (TAMs) contributes to immune-permissive or immune-suppressive macrophage cell states.
TAMs are one of the most abundant immune cell types in tumors, and their numbers often inversely correlate with clinical outcome (Cassetta and Pollard, 2018). In lung cancer, TAMs can be derived from both tissue resident macrophages, such as alveolar macrophages (AMs) of primarily embryonic origins, and monocyte-derived macrophages, such as interstitial macrophages (IMs; Casanova-Acebes et al., 2021; Zhu et al., 2017). Understanding TAM heterogeneity has been an area of intense interest; however, how macrophage functions such as phagocytosis alter their phenotypes, the tumor microenvironment, and ultimately clinical outcomes remain poorly understood.
Macrophage metabolism greatly influences their phenotype and function (Koo and Garg, 2019; Wculek et al., 2022; Yan and Horng, 2020). In vitro treatment of macrophages with defined factors can either induce a proinflammatory/anti-tumor (M1) phenotype, which is dependent on glycolytic metabolism, or an anti-inflammatory/pro-tumor (M2) phenotype that is characterized by a more flexible metabolism, utilizing both glycolysis and mitochondrial oxidative phosphorylation (OXPHOS; Viola et al., 2019). The OXPHOS pathway is responsible for synthesizing and/or catabolizing metabolites or macromolecules like amino acids or lipids. Lipid accumulation in the cell, derived for example from phagocytosed material, needs to be either stored, effluxed, or catabolized (Yurdagul, 2021). During wound healing, fatty acids derived from phagocytosed cells can lead to increased OXPHOS to fuel reparative macrophage phenotypes (Zhang et al., 2019). Additionally, efferocytosis can induce macrophages to secrete suppressive cytokines (Yin and Heit, 2021). Therefore, efferocytosis and the consequent metabolic reprogramming of macrophages may play an important role in driving the phenotypes of TAMs in vivo.
TAMs are often described as having a wound-healing, anti-inflammatory phenotype, and the identification of functional and metabolic cell states of macrophages has improved our understanding of TAMs beyond the M1/M2 paradigm (Katzenelenbogen et al., 2020; Loke and Lin, 2022; Mulder et al., 2021; Sanin et al., 2022). Connecting the phagocytic and metabolic states of TAMs in the context of a growing tumor may uncover novel biological mechanisms in TAMs that could inform therapeutic design. Historically, studies of macrophage metabolism have employed short-term, in vitro assays, often using bone marrow–derived macrophages (BMDMs), and it is unclear whether these systems are relevant to TAMs in vivo (A-Gonzalez et al., 2017; Schulz et al., 2019; Zhang et al., 2019). Studies of macrophage identity and heterogeneity have shown that microenvironmental cues imprint phenotypic changes after chronic tissue residency (Guilliams et al., 2020; Gundra et al., 2017; Yona et al., 2013). Autochthonous mouse models, which rely upon engineered genetic mutations to drive tumor development in tissues, reproduce many features of human cancer (Singh et al., 2012). Importantly, these models provide unique systems to study how signals, like phagocytosis of neoplastic cells, influence TAM heterogeneity.
Here, we employed syngeneic and autochthonous mouse models of lung tumorigenesis to study the impact of phagocytosis of neoplastic cells on TAMs. We identify phagocytic TAMs as a distinct subset of macrophages in vivo, show that phagocytosis of neoplastic cells reprograms macrophage metabolism and sustains immune-suppressive TAM phenotypes, and uncover a unique phagocytic tumor (PHAT) signature enriched in OXPHOS genes. This signature is shared among monocytes, macrophages, and dendritic cells in murine lung tumors, and is enriched within phagocytic macrophages in previously described models of inflammation and cancer. The PHAT signature is enriched in lung tumor myeloid cells and correlates with poorer clinical outcome in several human cancer types. These expression signatures were validated by protein-level phenotyping and functional metabolic assays. Interestingly, non-phagocytic TAMs were phenotypically similar to macrophages in normal tissues, suggesting that engulfment of neoplastic cells is a major driver of TAM phenotypes. This work connects a key cellular function of TAMs—the phagocytosis of neoplastic cells—with changes in intracellular metabolism and TAM heterogeneity in lung tumors.
Results
Phagocytic TAMs have distinct phenotypes from non-phagocytic TAMs in a syngeneic transplant lung tumor model
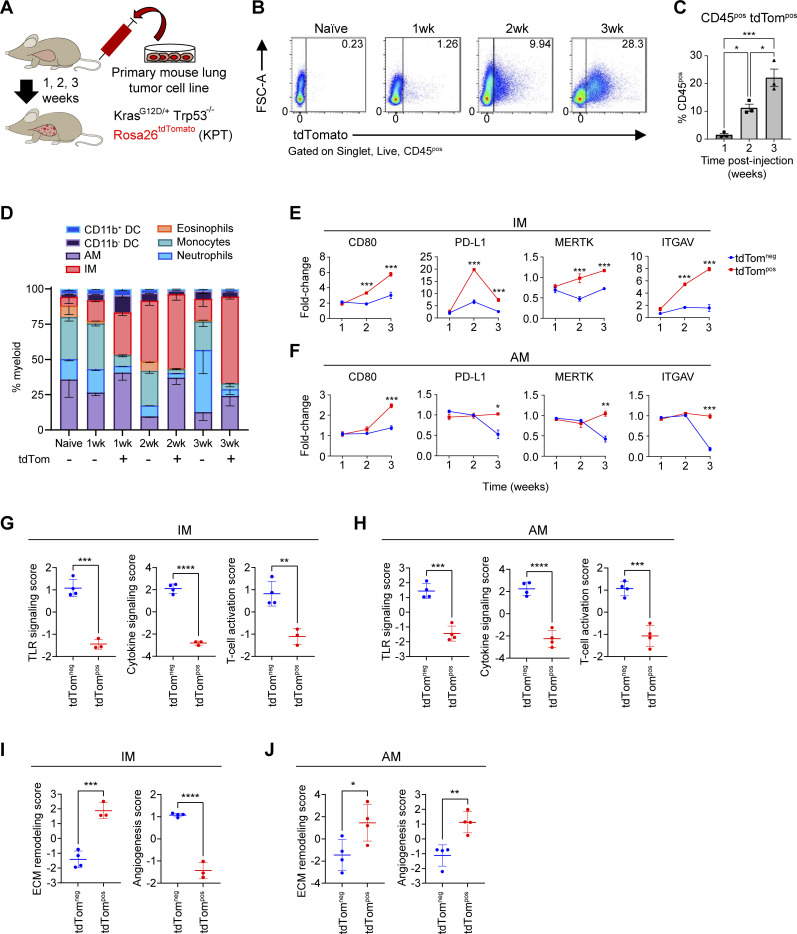
To initially investigate the phagocytosis of lung cancer cells by macrophages in vivo, we intravenously injected an oncogenic KrasG12D Trp53-deficient murine lung cancer cell line that expresses the red fluorescent protein tdTomato (tdTom) to seed orthotopic tumors in the lung (KPT cell line; Fig. 1 A). We identified CD45pos tdTompos immune cells as those that had recently phagocytosed cancer cells (Fig. 1 B). The frequency of CD45pos tdTompos cells significantly increased over time as did tdTom mean fluorescence intensity (MFI) in tumor-infiltrating CD45pos cells (Fig. 1, B and C). By 3 wk after transfer, alveolar macrophages (AMs; defined as CD64pos CD24neg CD11bneg SIGLEC-Fpos cells) and CD11bpos interstitial macrophages (IMs; defined as CD64pos CD24neg CD11bpos SIGLEC-Fneg cells) were the most abundant phagocytic cell type. IMs were the most numerous among tdTompos CD45pos myeloid cells, which is consistent with previously described models (Maier et al., 2020; Fig. 1 D). CD11bneg dendritic cells (DCs) and monocytes were also tdTompos, albeit at low frequencies. As expected, non-phagocytic B and T lymphocytes were tdTomneg within tumors (Fig. S1 A). Thus, in this syngeneic lung tumor model, IMs are the most prevalent phagocytes of neoplastic cells.
Figure 1.
TAMs have distinct phenotypes after phagocytosis in acute lung tumor mouse models. (A) Schematic of intravenous transplantation of a primary lung tumor cell line (KPT; SP110P) into recipient F1 C57BL/6J-129S1/Svlmj mice over a 3-wk time course. (B) Flow cytometry gating of CD45pos tdTompos and tdTomneg lung macrophages gated on cells at 1, 2, and 3 wk after i.v injection of KPT primary lung tumor cell line. (C) Ordinary one-way ANOVA (*, P ≤ 0.05; ***, P ≤ 0.001). Bar plots showing the percentage of CD45pos tdTompos cells in the acute lung tumor model after i.v. injection. (D) Myeloid cell composition within the tdTompos or tdTomneg immune cell fraction in naive and tumor-bearing mice. (E and F) Fold change MFI of immune-modulatory and phagocytosis cell surface markers on IMs (E) or AMs (F) relative to naive IMs or AMs, respectively. Unpaired t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). (G–J) Pathway enrichment score of immune-activation gene modules (G and H) and tissue-reorganization gene modules (I and J) for tdTompos or tdTomneg IMs and AMs based on analysis of focused panels of Nanostring mRNA probes. Unpaired t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). Data are representative of at least three independent experiments.
Figure S1.
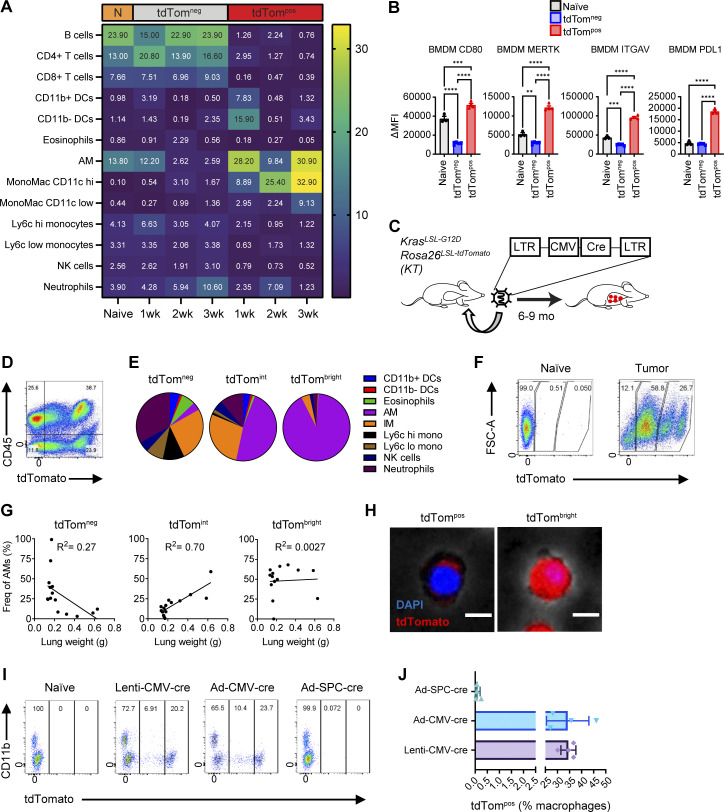
Identification of phagocytic versus transduced TAMs in syngeneic and autochthonous lung tumors in vivo. (A) Heatmap of immune cell frequencies within naive or syngeneic tumor-bearing mice. (B) MFIs of cell surface markers on tdTompos or tdTomneg BMDMs co-cultured with KPT SP110 cancer cells. (C) Schematic of lentiviral lung tumor initiation in KrasstopflG12D Rosa26stopfl-tdTomato mice. Ordinary one-way ANOVA (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). (D) Flow cytometry plot of total live cells isolated from KT tumor-bearing mice. (E) Pie charts of the frequencies of myeloid cell subsets gated on tdTom-negative (tdTomneg), tdTom-intermediate (tdTomint), or tdTom-bright (tdTombright) CD45pos CD11b/cpos cells. (F) Flow cytometry plots of tdTombright, tdTomint, and tdTomneg lung macrophages gated on CD64pos CD24neg cells at in autochthonous lung tumors. (G) Scatter plots of the frequencies of tdTomneg, tdTomint, or tdTombright AMs and lung weights with linear regression calculations. (H) Fluorescent cell imaging of AMs from autochthonous lung tumors sorted from tdTomint or tdTombright. Scale bar, 10 μm. (I) Flow cytometry plots of total macrophages gated on CD64pos CD11b/cpos CD45pos cells isolated from lungs of KTai65;SpcCreER mice transduced with indicated viral vectors. (J) Frequencies of tdTompos macrophages from mice in I. Data are representative of at least three independent experiments.
The expression of several key immune modulatory proteins (PD-L1 and CD80) and phagocytic receptors (MERTK and ITGAV) was quantified on TAMs by flow cytometry (Fig. 1, E and F). CD80 expression on tdTompos macrophages significantly increased over time on both AMs and IMs. tdTompos IMs had significantly higher expression of the immune checkpoint ligand PD-L1 than tdTomneg IMs at 2 and 3 wk after cell line transplantation. In contrast, PD-L1 was downregulated in tdTomneg AMs 3 wk after transfer, relative to naive AMs. Interestingly, MERTK and ITGAV expression on AMs followed a similar trend to PD-L1, in which tdTomneg AMs down-regulated cell surface expression. In contrast, the expression of MERTK and ITGAV on IMs steadily increased over time, thus highlighting key differences between tissue resident AMs and monocyte-derived IMs. Monocyte-derived macrophages that have phagocytosed KPT cells in vitro also upregulate CD80, MERTK, ITGAV, and PD-L1, consistent with the in vivo IM phenotypes (Fig. S1 B).
We next profiled tdTompos and tdTomneg AMs and IMs using a panel of mRNA probes for genes related to innate myeloid immunity. Phagocytic tdTompos IMs and AMs downregulated genes related to TLR signaling, cytokine signaling and T cell activation (Fig. 1, G and H). Relative to tdTomneg IMs, tdTompos IMs had higher expression of genes related to extracellular matrix (ECM) remodeling and lower expression of those related to angiogenesis. Conversely, tdTompos AMs had positive enrichment of both angiogenesis and ECM remodeling (Fig. 1, I and J). Together, these results suggest that phagocytosis of neoplastic cells alters TAM cell surface and transcriptional phenotypes in vivo in a manner distinct from those not actively engaged in phagocytosis, and that these phenotypes differ between IMs and AMs.
Tissue-resident AMs are the most prevalent phagocyte in autochthonous lung tumors
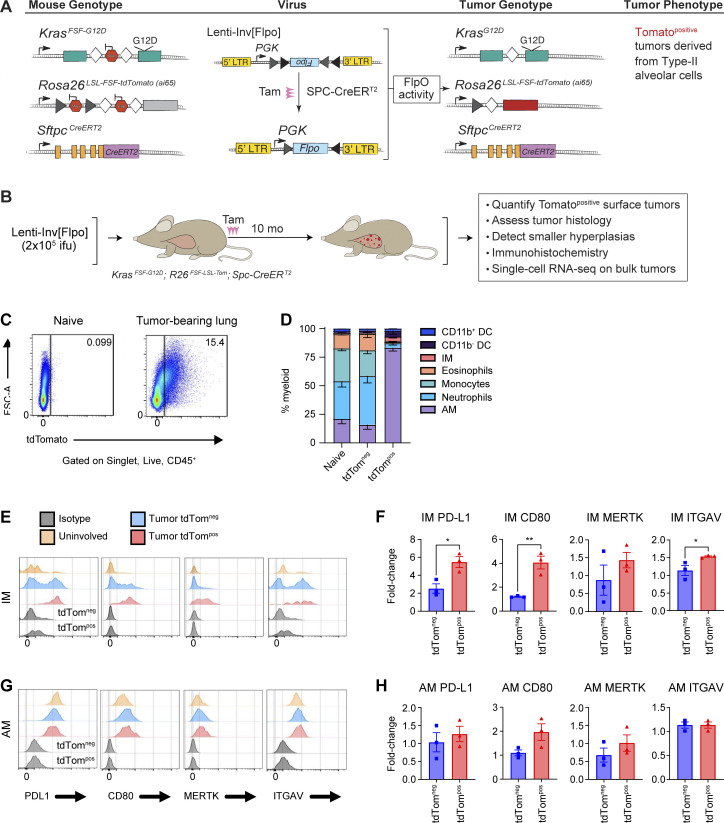
Genetically engineered mouse models recapitulate the genetics and histology of human lung tumors. Importantly, the slower growth of these tumors allows the microenvironment to develop over time, which more closely resembles the long-term tumor residency of macrophages in human tumors. Previous viral-Cre–induced lung cancer models have documented transduction of lung-resident AMs and consequently modification of conditional allele by Cre recombinase in AMs (Tippimanchai et al., 2018). Thus, we developed a novel model that specifically induces tdTomato protein expression and oncogenic alterations in neoplastic cells to enable tracking of phagocytic activity (Fig. 2, A and B). This autochthonous lung tumor model specifically induces genetic alterations in type II alveolar cells which are a cell type of origin of lung adenocarcinoma (Sutherland et al., 2014). Transduction of KrasFSF-G12D;Rosa26FSF-LSL-tdTomato(ai65);SftpcCreER (KTai65;SpcCreER) mice with a lentiviral vector containing an inverted FLPo flanked by pairs of heterotypic loxP sites enables tamoxifen-induced expression of oncogenic KRAS specifically in type II alveolar cells (Fig. 2 A; unpublished data). Nine months after tumor initiation, KTai65;SpcCreER mice developed numerous tdTomato-positive lung tumors. Phagocytic tdTompos myeloid cells could be visualized by flow cytometry similar to the cell transfer model (Fig. 2 B). Importantly, unlike in the Lenti-Cre initiated autochthonous lung tumor models (Fig. S1, C–H), tdTombright myeloid cells were not observed, consistent with recombination events only occurring in epithelial cells (Fig. 2 C).
Figure 2.
Alveolar macrophages are the primary phagocytic myeloid cell in autochthonous lung tumors. (A) Schematic of genetic recombination events in KTai65;SpcCreER mice to achieve specific oncogenic transformation and fluorescent labeling of type II alveolar cells. (B) Schematic detailing the formation and characterization of the epithelial-specific autochthonous mouse model. (C) Flow cytometry plot of macrophages isolated from naive lung or KTai65;SpcCreER tumors. (D) Stacked bar graph of myeloid cell frequencies from naive lung or KTai65;SpcCreER tumors. (E and G) Histogram plots of MFIs of surface markers in IMs (E) or AMs (G). (F and H) Fold-change expression of cell surface markers from IMs (F) or AMs (H) in KTai65;SpcCreER tumors compared to IMs or AMs from naive lung, respectively. Unpaired t test (*, P ≤ 0.05; **, P ≤ 0.01). Data are representative of three independent experiments.
Within the KTai65;SpcCreER model, the majority (∼70%) of tdTompos myeloid cells were AMs, whereas only 3% were IMs (Fig. 2 D). This suggests contrasting properties in myeloid surveillance in syngeneic transplant models, in which IMs accounted for 25–50% of the phagocytic myeloid population across the 3 wk of lung tumor growth (Fig. 1 D). Compared to the syngeneic model, similar trends were seen in the cell surface expression of PD-L1, CD80, MERTK, and ITGAV on tdTompos AMs and IMs; however, the fold-change differences were higher in IMs from syngeneic tumor models compared to the KTai65;SpcCreER model (Fig. 2, E–H; and Fig. 1, E and F). Moreover, tdTompos AMs from KTai65;SpcCreER tumors most closely resembled the tdTompos AMs from syngeneic tumors observed at 2 wk after transfer.
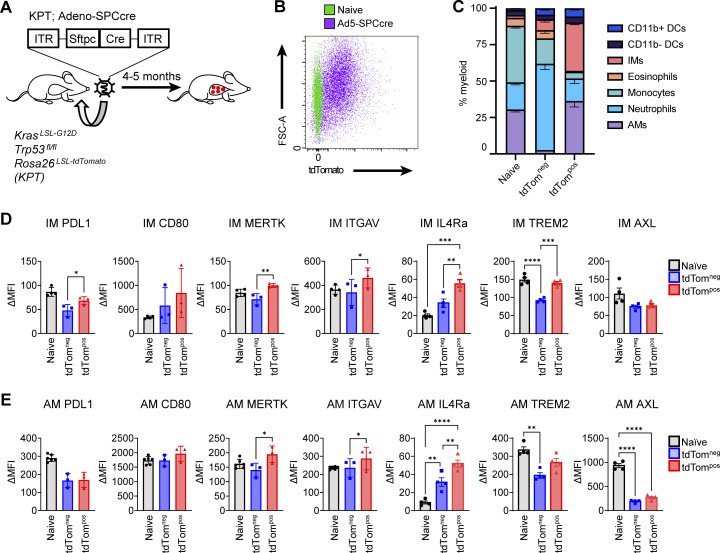
When lung tumors were initiated with Adeno-Spc-Cre in KrasLSL-G12D;Trp53fl/fl;Rosa26LSL-tdTomato (KPT) mice, there were also very few tdTombright AMs compared to when mice are transduced with non-specific viral vectors (Fig. S1, I and J). In these tumor-bearing KPT mice, there was only a slightly higher percentage of phagocytic AMs compared to IMs (Fig. S2, A–C). Thus, it appears that IMs play a larger phagocytic role in these tumors compared to tumors in the KTai65;SpcCreER model. However, the expression of key cell surface proteins on AMs and IMs closely resembled the phenotypes in the KTai65;SpcCreER model (Fig. S2, D and E). Collectively, the data from these models highlight the heterogeneity of phagocytic myeloid cells in the lung tumor microenvironment and provide insight into the phenotypes that correlate with the phagocytosis of neoplastic cells in vivo.
Figure S2.
Phagocytic TAMs have distinct cell surface phenotype in adenoviral autochthonous lung tumors in vivo. (A) Schematic of adenoviral lung tumor initiation in KrasLSL-G12D Trp53fl/fl Rosa26LSL-tdTomato mice. (B) Flow cytometry plots of tdTompos lung macrophages gated on CD64pos CD24neg cells isolated from autochthonous lung tumors. (C) Stacked bar graph of the frequencies of myeloid cell subsets gated on tdTompos or tdTomneg CD45pos CD11b/cpos cells in Adeno-SPCcre tumors. (D and E) MFIs of cell surface markers on tdTompos or tdTomneg IMs (D) or AMs (E) from autochthonous Ad-SPCcre lung tumors. Ordinary one-way ANOVA (*, P ≤ 0.05; **, P ≤ 0.01). Data are representative of at least three independent experiments.
Phagocytosis of neoplastic cells imprints distinct transcriptional patterns in monocytes and macrophages
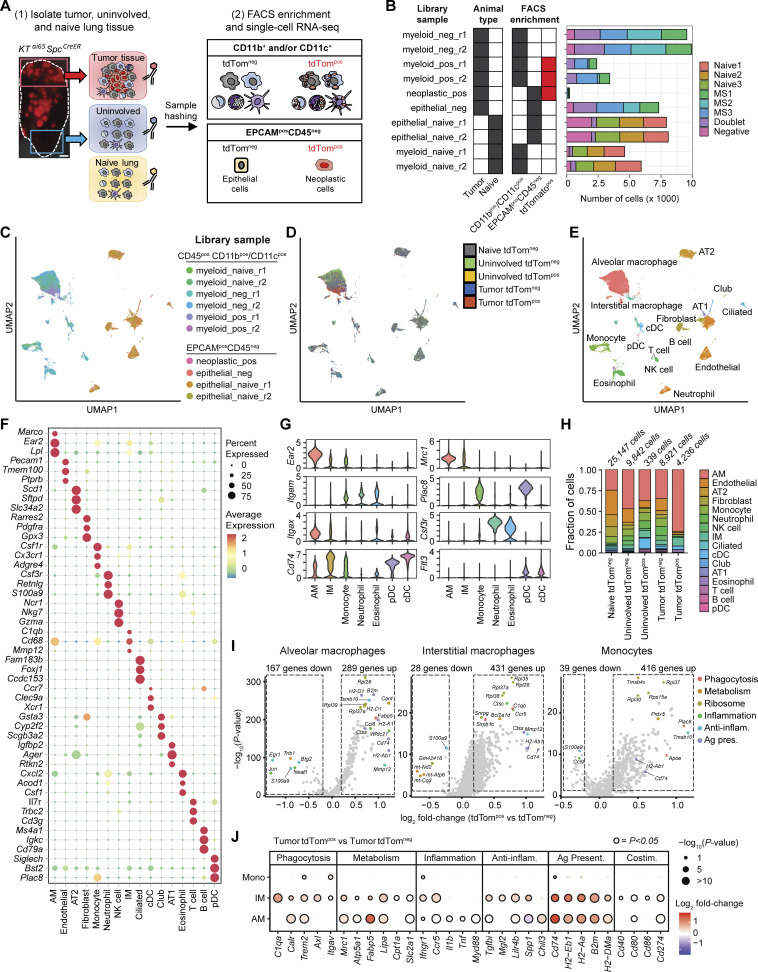
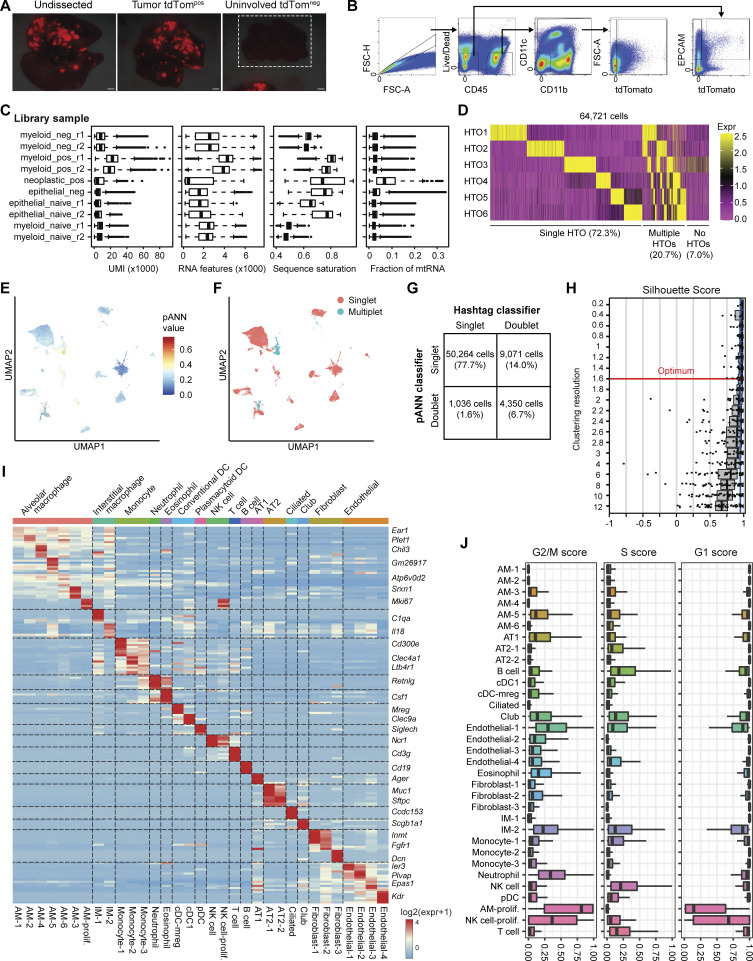
To investigate cell state changes that occur in phagocytic myeloid cells in lung tumors, we microdissected tumor and uninvolved tissue from KTai65;SpcCreER tumor-bearing mice, as well as lung tissue from tumor-naive mice, and performed single-cell RNA-sequencing (scRNA-seq) on sorted tdTompos and tdTomneg myeloid cells (enriched for myeloid cells; CD45pos CD11bpos and/or CD11cpos) and CD45neg EPCAMpos non-immune cells (Fig. 3 A and Fig. S3, A and B). Cell hashtagging using antibodies with conjugated oligonucleotide barcodes was used to multiplex cells from different animals and tissue regions, filter singlets, and enable direct comparisons between tdTompos and tdTomneg cells from tumor and uninvolved tissues (Fig. 3 B). After implementing a multi-step procedure for filtering transcriptome quality and removing multiplets (Fig. S3, C–G), we recovered 44,016 high-quality single-cell transcriptomes that could be assigned tissue and animal identities based on hashtag labeling (Fig. 3, C–E).
Figure 3.
Single-cell transcriptomic profiling identifies distinct and shared enrichment of metabolic and immune pathways in phagocytic tumor monocytes and macrophage subsets. (A) Schematic of scRNA-seq experiment detailing oligonucleotide-based sample hashing of microdissected tumors and adjacent normal tissue from KTai65;SpcCreER mice as well as FACS strategy for cell enrichment. Scale bar, 10 mm. (B) Stacked bar plot showing the number of single cells recovered from each library preparation and antibody hashtag classification. Hashtag labels are represented by animal identifiers. (C–E) UMAP projections of all cell types profiled by scRNA-seq. Cells are colored either by scRNA-seq library sample (C), by tissue of origin and tdTomato status (D), or by classified cell type (E). (F) Bubble plot showing expression of top three marker genes for each major cell type identified. Marker genes were determined by Wilcoxon test. (G) Violin plots of marker genes used to differentiate myeloid cell types. Marker genes were determined by Wilcoxon test. (H) Frequency of cell types for each grouping corresponding to tissue origin and phagocytic status. (I) Volcano plots of significantly enriched genes determined by MAST in tumor tdTompos AMs, IMs, or monocytes identified after differential expression analysis. Top differentially expressed genes are colored by gene function. Anti-inflam. = anti-inflammatory; Ag. present. = antigen presentation. (J) Differential expression of selected genes organized by function in tumor tdTompos versus tumor tdTomneg monocytes, IMs, and AMs. Anti-inflam. = anti-inflammatory; Ag. present. = antigen presentation; Costim. = costimulatory; P value = FDR-adjusted P value.
Figure S3.
Dissection, FACS, and scRNA-seq quality filtering of autochthonous KTai65;SpcCreER lung tumors. (A) Fluorescent images of whole lung and dissected lung isolated from SPCcreER mice. White box around the uninvolved tdTomneg image depicts uninvolved lung tissue, and a tdTompos tumor is shown in the same image to show light intensity settings of the fluorescence camera. Scale bar, 10 mm. (B) Gating strategy for cell sorting for scRNA-seq of tdTompos or tdTomneg myeloid and neoplastic cells from SPCcreER tumor-bearing mice or naive controls. (C) Sequencing quality metrics for each FACS-enriched 10X Chromium library preparation sample after removal of outlier and low-quality cell barcodes (see Materials and methods). Box plot lines correspond to the median and the interquartile range. (D) Heatmap showing log-normalized expression values of each of the six antibody hashtags used for each cell barcode. (E and F) UMAP projection of filtered cell barcodes colored by pArtificial Nearest Neighbors (pANN) score (E) and the resulting pANN classification of singlets vs. multiplets (F). (G) Confusion matrix showing the detection of cell barcode singlets and doublets by antibody-based sample hashing and by the pANN classifier. (H) Silhouette distribution plots for subsampled cells at different clustering resolution parameters of the Louvain algorithm, calculated using chooseR. Red line depicts optimal resolution parameter used for cell state classification. (I) Heatmap of all cell clusters identified and the averaged log-normalized expression of distinguishing marker genes. (J) Cell-cycle scores for G1, S, and G2/M phases of each cell state. Box plot lines correspond to the median and the interquartile range.
Graph-based clustering after optimizing cluster resolution identified 33 distinct cell states with unique expression profiles, including two actively proliferating cell states (Fig. S3, H–J). These grouped into 16 major cell types from myeloid, lymphoid, endothelial, or epithelial origins based on known cell markers (Fig. 3 F). We identified seven cell types of myeloid origin (Fig. 3 G). Although the identified myeloid cell types were present in all tissue types, AM, IM, monocyte, and DC cell types were enriched in sorted tdTompos myeloid cells from tumors (Fig. 3 H). This corroborated our flow cytometry results and underscores the role of these cell types in neoplastic cell engulfment.
To elucidate how cells that have engulfed neoplastic cells are transcriptionally distinct from other non-phagocytic myeloid cell types, we performed differential gene expression analyses comparing tumor-associated tdTompos monocytes, IMs, and AMs to their tumor-associated tdTomneg counterparts (Fig. 3 I). Genes involved in phagocytosis, metabolism, and co-stimulation were generally upregulated in tdTompos cells of all three types, although some genes associated with these functions were only significantly upregulated in IMs (e.g., Axl, Cpt1a, and Cd40; Fig. 3 J). Antigen presentation genes were upregulated in tdTompos AMs and IMs but not in tdTompos monocytes. Finally, certain mediators of inflammation (e.g., Il1b, Tnf) and tissue repair (e.g., Mgl2, Chil3) had contrasting expression patterns across cell types. These data suggest that phagocytosis of neoplastic cells induces conserved and distinct transcriptional programs in IMs, AMs, and monocytes in autochthonous lung tumors.
PHAT gene signature is associated with OXPHOS metabolism and translation
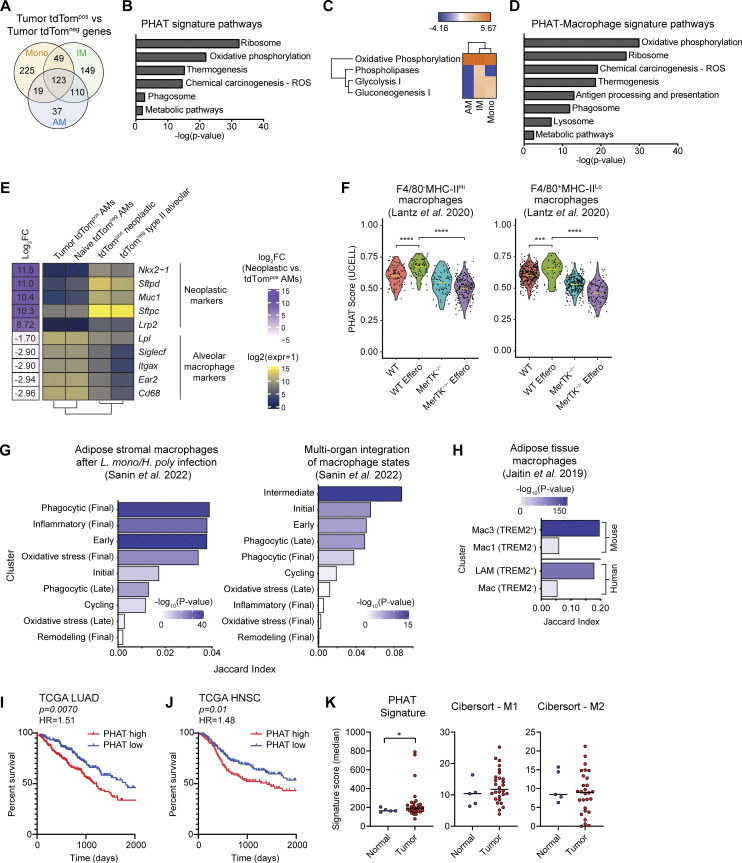
Next, we sought to identify a gene signature representing common transcriptional changes associated with phagocytosis of neoplastic cells. Because macrophages are the most prevalent tdTompos myeloid cell in KTai65;SpcCreER tumors and monocytes are the precursor to IMs, we analyzed the transcriptional changes occurring in AMs, IMs, and monocytes. tdTompos AMs, IMs, and monocytes shared a common gene signature of 123 upregulated genes, which we term the PHAT signature (Fig. 4 A and Tables S1, S2, S3, S4, and S5). A comparison of pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database with the PHAT signature identified significant enrichment for pathways relating to ribosomal translation, OXPHOS, phagosome, and metabolic pathways (Fig. 4 B). Additionally, Ingenuity Pathway Analysis revealed shared activation of the OXPHOS, phospholipases, glycolysis, and gluconeogenesis metabolic pathways in tdTompos macrophages and monocytes (Fig. 4 C). We also defined a 233-gene signature to uncover pathways shared by both macrophage populations, independent of monocytes, termed the PHAT-Macrophage signature. This analysis detected antigen processing/presentation and lysosomal pathways in addition to pathways identified from the overall PHAT signature (Fig. 4 D). Macrophage metabolism is known to be a key driver of the phenotypes and functions of TAMs and a source of TAM heterogeneity within the tumor microenvironment (Li et al., 2022; Wang et al., 2021; Xia et al., 2020). Our data identify a common metabolic gene signature of phagocytic monocytes and macrophages that have recently phagocytosed neoplastic cells in vivo.
Figure 4.
PHAT gene signature defined using an autochthonous model correlates with other phagocytosis-associated macrophage populations and with decreased survival in human LUAD. (A) Venn diagram of genes upregulated (fold change > 1.2; FDR < 0.05) in PHAT AMs, IMs, and monocytes. (B) DAVID KEGG Pathway analysis of 123 PHAT signature genes shared in tdTompos AM, IMs, and monocytes. (C) Ingenuity Pathway Analysis of metabolic pathway z-score enrichment above twofold threshold in tdTompos AMs, IMs, and monocytes relative to tdTomneg AMs, IMs, and monocytes. (D) DAVID KEGG Pathway analysis of 233 PHAT-Macrophage signature genes shared in tdTompos AM and IM. (E) Heatmap showing log-normalized RNA-seq expression values of neoplastic cell and alveolar macrophage markers for sorted tumor tdTompos AMs, naive tdTomneg AMs, tdTompos neoplastic cells, and tdTomneg type II alveolar cells. Log fold-change values between phagocytic tdTompos AMs and neoplastic cells were determined using DESeq2 and are listed to the left of the heatmap. (F) Violin plots depicting PHAT signature score for WT and MerTK−/− (kinase-deficient) efferocytic murine macrophage subsets co-cultured with apoptotic cells (Lantz et al. 2020). Statistical significance was determined by unpaired Wilcoxon test. (G) Left: Jaccard index values measuring marker gene overlap between the PHAT-IM signature and adipose stromal murine macrophage subsets after classical (Listeria monocytogenes) and type II (Heligmosomoides polygyrus) immune challenges (Sanin et al. 2022). Right: Jaccard index values measuring marker gene overlap between the PHAT signature and murine macrophage subsets integrated across nine tissues and multiple inflammatory conditions (Sanin et al. 2022). (H) Jaccard index values measuring marker gene overlap between the PHAT-IM signature and murine and human adipose tissue macrophage subsets (Jaitin et al. 2019). LAM = lipid-associated macrophage. P values for G and H were determined using Fisher’s exact test. (I and J) Kaplan–Meier survival curve of 396 LUAD patients (I) or 390 head and neck squamous cell carcinoma (HNSC) patients (J), stratified by high (>60th percentile) or low (<40th percentile) PHAT signature enrichment (TCGA). P values were calculated by Cox regression. HR = hazard ratio. (K) Enrichment of PHAT, M1 CIBERSORT, or M2 CIBERSORT signatures in myeloid cells isolated from tumor or normal tissue (UCSF Immunoprofiler dataset). One-tailed t test with two-sample unequal variance, tumor vs. normal.
To discriminate whether these genes originate from the phagocytes themselves or internalized neoplastic cells, we performed bulk RNA-seq on tdTompos AMs and tdTompos neoplastic cells from KTai65;SpcCreER tumors. Known marker genes for alveolar macrophages and type II alveolar cells/neoplastic cells showed expected cell-specific expression patterns. Importantly, tdTompos AMs had extremely low (>400-fold lower) relative expression of type II/neoplastic cell markers, suggesting there is minimal contribution of mRNA from engulfed neoplastic cells (Fig. 4 E). Therefore, the PHAT signature is comprised mainly of genes expressed by phagocytes.
Comparison of PHAT signatures to other in vivo macrophage datasets shows both conserved and context-dependent phagocytic macrophage states
Since our model enables characterization of phenotypic changes of immune cells following the in vivo engulfment of tdTompos neoplastic cells, we compared our functionally defined PHAT signatures to several studies of macrophage heterogeneity related to phagocytosis. One study directly follows efferocytosis-driven gene expression changes in peritoneal macrophages after isolation and co-culture with apoptotic cells; phagocytosis-dependent changes were defined by using a mouse model that expresses a kinase-deficient form of Mertk, termed MerTK−/−, thus preventing apoptotic cell uptake (Lantz et al., 2020). In this dataset, our PHAT signature score was higher in both the monocyte-derived F4/80neg MHCIIhi and tissue-resident F4/80pos MHClo macrophages co-cultured with apoptotic cells. However, MerTK−/− macrophages had decreased the PHAT scores, demonstrating that our signature is significantly upregulated in efferocytic macrophages (Fig. 4 F). Another study characterized monocyte-derived macrophage heterogeneity across a broad set of inflammatory states and defined certain cell clusters as “phagocytic” based on in vitro–defined key pathway genes (Sanin et al., 2022). We focused our signature to include only genes upregulated in the monocyte-derived IMs comparing tdTompos IMs relative to tdTomneg IMs, termed the PHAT-IM signature. In two different analyses, these putative phagocytosis-related clusters show enrichment for our PHAT-IM signature, but other differently labeled clusters also show enrichment (Fig. 4 G). This result highlights the importance of defining phenotypes functionally and suggests that although some pathways are conserved, the tumor context may drive phagocytic macrophages to resemble activation states in other inflammatory settings. Interestingly, a study of adipose tissue macrophages defined populations of lipid-rich Trem2pos macrophages in mouse (Mac3) and human (LAM; Jaitin et al., 2019). The PHAT-IM signature was highly enriched in LAMs compared to macrophages with low lipid content (Fig. 4 H). Additionally, flow cytometry revealed increased TREM2 expression on tdTompos AMs and IMs from autochthonous lung tumors (Fig. S2, D and E). Together, these comparisons show that gene expression changes in TAMs are shared in other non-tumor contexts, although certain changes are uniquely imprinted by the mechanism of macrophage phagocytosis, tissue milieu, and inflammatory status.
PHAT gene signature translates to human TAMs and clinical outcome in lung cancer
To investigate the clinical relevance of these phagocytic myeloid cells, we queried The Cancer Genome Atlas (TCGA) bulk gene expression datasets. Given the immune suppressive phenotypes of phagocytic TAMs, we anticipated that our PHAT signature could negatively correlate with clinical outcomes. We found that our PHAT signature could stratify patients with lung adenocarcinoma, with patients whose tumors were enriched for the PHAT signature having significantly decreased probability of survival (Fig. 4 I). Breast cancer, hepatocellular carcinoma, and colon adenocarcinoma patients with high PHAT signature had only slightly lower survival; however, the PHAT signature significantly stratified head and neck squamous cell carcinoma patients (Fig. 4 J and data not shown). These data suggest that certain molecular features are shared, although idiosyncrasies exist in different tumor settings.
To query our PHAT signature specifically within TAMs from human cancer, we investigated the enrichment of our PHAT signature in bulk RNA-seq dataset on sorted myeloid cells from tumors and normal lung tissue from non-small cell lung cancer patients (Combes et al., 2022). Importantly, the PHAT signature was significantly enriched in tumor-associated myeloid cells compared to myeloid cells from normal lung (Fig. 4 K). This contrasted with classical M1 and M2 myeloid gene signatures, which were not significantly enriched in tumor-associated myeloid cells compared to those from normal lung. Together, these findings demonstrate that our murine-derived PHAT signature associates with the gene expression of human tumor myeloid populations and connects the transcriptional changes that occur in phagocytic monocytes and macrophages in autochthonous mouse tumors with clinical outcome.
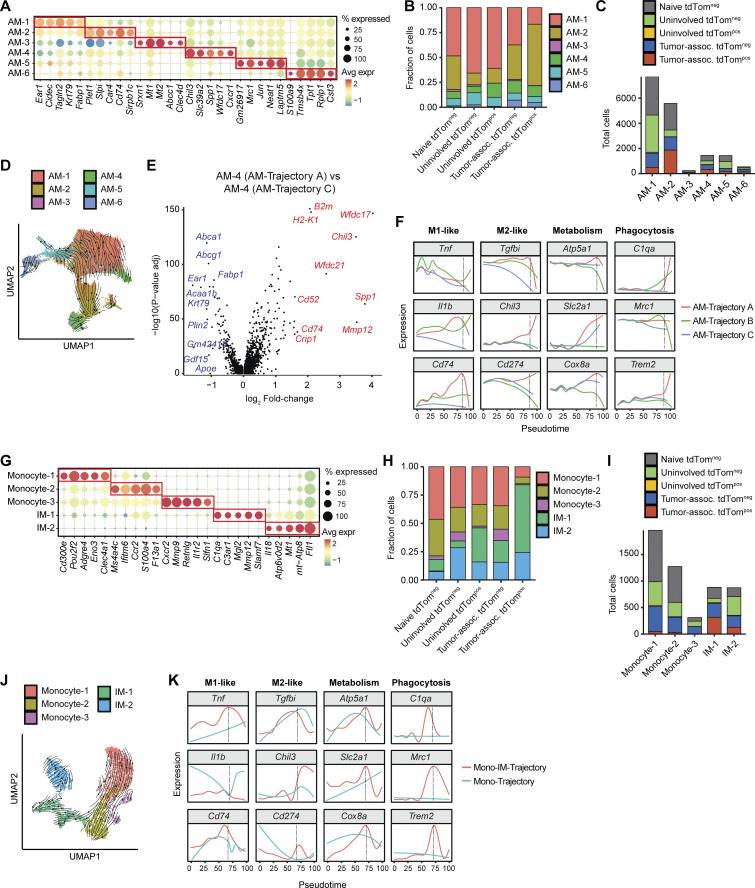
Phagocytic AMs mature through a distinct lineage associated with tissue remodeling
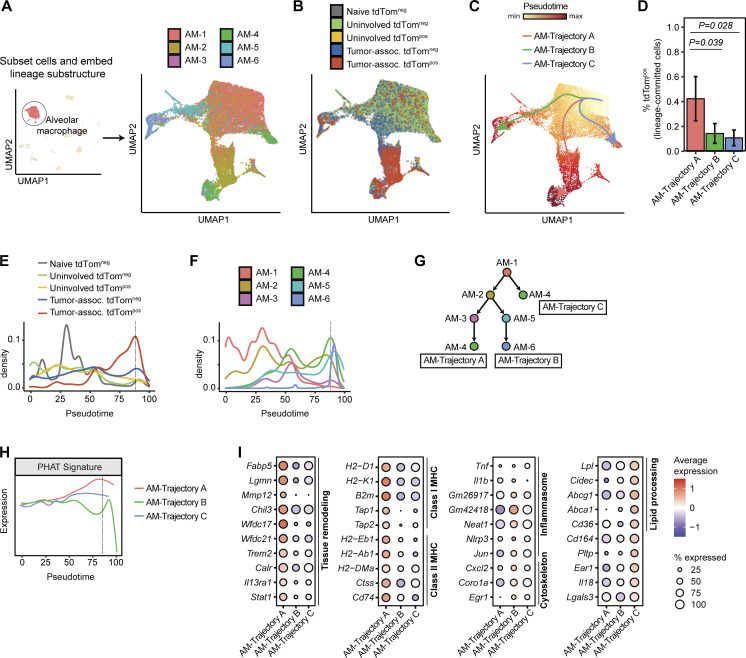
We next evaluated how phagocytosis influences AM maturation at single-cell resolution using trajectory inference and cell state composition analyses across tissue and phagocytic (tdTompos) status. Cell embedding using diffusion maps contextualized local maturation relationships between the six identified AM cell states within the lung microenvironment (Fig. 5 A and Fig. S4 A; Haghverdi et al., 2015). This embedding revealed that naive and tdTomneg AMs from uninvolved tissue and tumors grouped separately from tdTompos tumor-associated AMs, underscoring the unique transcriptional cell states driven by phagocytosis of neoplastic cells (Fig. 5 B). Furthermore, naive, uninvolved, and tumor-associated tdTomneg had a similar distribution of AM cell states, while the tumor-associated tdTompos AMs had a high proportion of AM-2 cells (Fig. S4 B). Trajectory inference using Slingshot predicted three trajectories of AMs (termed AM-Trajectory A, AM-Trajectory B, and AM-Trajectory C; Fig. 5 C; Street et al., 2018), and dynamic RNA velocity modeling confirmed the relative progression across AM cell states (Fig. S4 D). The majority of tdTompos tumor-associated AMs were positioned relatively late along the maturation continuum and were found in AM-Trajectory A (Fig. 5, D and E). Interestingly, the presence of tumor-associated tdTomneg AMs (but not AMs from normal lung) at later pseudotime values suggests that these cells may be phagocytic AMs that have degraded tdTomato.
Figure 5.
Phagocytic AMs have distinct expression of genes involved in tissue remodeling in vivo. (A and B) UMAP projection of AMs following spectral embedding using diffusion maps. Cells are colored by AM cell state (A) or tissue and phagocytic status (B). (C) Trajectory inference of AMs using Slingshot. (D). Frequency of tdTompos cells for each maturation trajectory identified by Slingshot (n = 3 animals per group, Wilcoxon rank sum test). (E and F) Density plots across pseudotime for AMs grouped by tissue and phagocytic status (E) or by AM cell state (F). Dotted black line depicts the pseudotime value with highest frequency of tdTompos tumor cells. (G) Schematic depicting the relative maturation relationship between AM cell states as inferred by Slingshot and RNA velocity. (H) Density plot of expression of PHAT signature across pseudotime for each AM trajectory. Dotted black line depicts the pseudotime value with highest frequency of tdTompos tumor cells. (I) Bubble plots showing differentially expressed genes (Log2FC > 1.2 and P < 0.05) for each AM trajectory. Statistical significance between different trajectories was determined using MAST.
Figure S4.
Alveolar macrophage and interstitial macrophage transcriptional heterogeneity differs across lung tissue type and phagocytosis status. (A) Top five marker genes defining each AM cell state from KTai65;SpcCreER tumors and naive tissue. Marker genes were determined by Wilcoxon test. (B) Frequency of each AM cell state delineated by tissue and tdTom cell sorting status. (C) Absolute numbers of each monocyte and IM cell state, delineated by tissue and tdTom (phagocytic) status. (D) UMAP projection showing trajectory inference of AMs using dynamic RNA velocity modeling. Arrows represent the direction of AM maturation. (E) Volcano plot showing differential expression (MAST) between AM-4 cell state between AM-Trajectory A and AM-Trajectory C. Top 10 genes upregulated in AM-Trajectory A and AM-Trajectory C are shown in red and blue, respectively. (F) Density plot of selected genes across pseudotime for each AM trajectory. (G) Top five marker genes defining each monocyte and IM cell state from SPCcreER tumors and naive tissue. Marker genes were determined by Wilcoxon test. (H) Frequency of each monocyte and IM cell state delineated by tissue and tdTom (phagocytic) status. (I) Absolute numbers of each AM cell state, delineated by tissue and tdTom (phagocytic) status. (J) UMAP projecting showing trajectory inference of Mono-IM trajectories using dynamic RNA velocity modeling. Arrows represent the direction of Mono-IM maturation. (K) Density plot of selected genes across pseudotime for each IM-Mono trajectory.
To define the transcriptional phenotypes of tdTompos AMs between trajectories, we analyzed the position of AM cell states across pseudotime (Fig. 5 F). All three AM-Trajectories initiate from the AM-1 state (Fig. 5 G). While AM-Trajectory A and AM-Trajectory B then differentiate toward the AM-2 state (Plet1high and MHC class IIhigh) before diverging toward AM-3/AM-4 (tissue remodeling) or AM-5/AM-6 (inflammasome activation/cytoskeletal organization) states, AM-Trajectory C, which has the highest fraction of AMs from naive and uninvolved lung, directly transitions from AM-1 to AM-4. AM-4 appears in AM-Trajectory A and C, and differential expression testing identified higher immunosuppressive genes in AM-4 cells from Trajectory A, suggesting that convergence of AM maturation may arise from different environmental stimuli (Fig. S4 E).
Cells from AM-Trajectory A had the highest expression of PHAT signature genes, revealing shared molecular programs between different types of phagocytic macrophages (Fig. 5 H). Also, cells from AM-Trajectory A expressed higher levels of tissue remodeling regulators (e.g., Fabp5, Lgmn, Mmp12, Trem2, Calr, Il13ra1) and M2-like genes (e.g., Tgfbi, Chil3, Cd274) compared to AM-Trajectory B and AM-Trajectory C (Fig. 5 I and Fig. S4 F). This downregulation of inflammasome genes (e.g., Tnf, Neat1, Nlrp3, Jun, Il18) and upregulation of antigen tissue remodeling genes is indicative of an M2-like skewing of TAMs correlating with phagocytosis. However, TAMs within AM-Trajectory A also upregulate antigen presentation genes commonly associated with M1 macrophage phenotypes. This is in line with mixed M1/M2 TAM phenotypes commonly observed in human TAMs (Larionova et al., 2020). Metabolic genes, such as Atp5a1, a subunit of Complex V of the electron transport chain, and Slc2a1, which encodes the glucose transporter GLUT1, have high, sustained expression throughout AM maturation in Trajectories A and C but not in Trajectory B, suggesting utilization of both OXPHOS and glycolytic metabolism by cells from AM-Trajectory A and AM-Trajectory C (Fig. S4 F). In summary, trajectory analysis of AMs, combined with fluorescent tracking of neoplastic engulfment, highlights the diversity of AM responses and suggests that neoplastic engulfment promotes AM cell states with increased metabolic gene expression, tissue repair, and anti-inflammation properties as well as an inability to sustain proinflammatory gene expression.
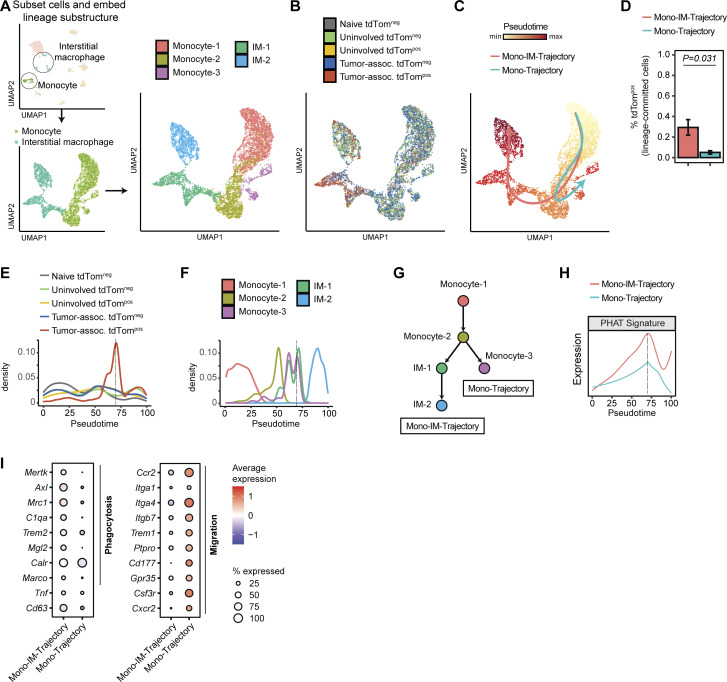
Phagocytic IMs diverge from monocytes and exhibit plasticity after engulfment
To understand how phagocytosis influences monocyte and IM functions, we embedded these populations in a lower-dimensional space using diffusion maps and then performed trajectory inference (Fig. 6, A and C). tdTompos monocytes/IMs accumulated separately from cells in naive and uninvolved tissues along the differentiation continuum, suggesting that differential transcriptional regulation occurs in cells that have phagocytosed neoplastic cells. Tumor-associated IMs, which could be further classified into two cell states based on the expression of complement system genes (e.g., C1qa and C3ar1) or oxidative stress genes (e.g., Ftl1 and Mt1; Fig. S4 G), accounted for a higher proportion (∼80%) of tumor tdTompos monocytes/IMs compared to monocytes (∼20%; Fig. S4, H and I). Trajectory inference using Slingshot predicted two trajectories (Mono-IM-Trajectory and Mono-Trajectory) and ordered cells by pseudotime. The directionality of the trajectory was confirmed using RNA velocity (Fig. S4 J). The Mono-IM-Trajectory contained a significantly higher percentage of tdTompos cells compared to the Mono-Trajectory (Fig. 6 D). Like tumor-associated tdTompos AMs, the majority of tdTompos monocytes/IMs were associated with later pseudotime values (Fig. 6, E and F).
Figure 6.
Phagocytic interstitial macrophages acquire differentiated metabolic and immune states from monocytes. (A and B) UMAP projections of monocytes and interstitial macrophages following spectral embedding using diffusion maps, highlighting cell state (A) and tissue and phagocytic status (B). (C) Trajectory inference of monocytes and IMs using Slingshot. (D) Frequency of tdTompos cells for each maturation trajectory identified by Slingshot (n = 3 animals per group, Wilcoxon rank sum test). (E and F) Density plots across pseudotime for monocytes and IMs grouped by tissue and phagocytic status (E) and cell state (F). Dotted black line depicts the pseudotime value with highest frequency of tdTompos tumor cells. (G) Schematic depicting the relative maturation relationship between Mono-IM cell states as inferred by Slingshot and RNA velocity. (H) Density plot of expression of PHAT signature across pseudotime for each IM trajectory. Dotted black line depicts the pseudotime value with highest frequency of tdTompos tumor cells. (I) Bubble plots showing differentially expressed genes (log2FC > 1.2 and P < 0.05) for each monocyte/IM trajectory. Statistical significance between different trajectories was determined using MAST.
Both trajectories initiate from the Cd300ehigh monocyte state (Monocyte-1) and an intermediate Monocyte-2 cell state, followed by a bifurcation into a terminal Monocyte-3 state (Mono-Trajectory) or into two IM states (Mono-IM-Trajectory; Fig. 6, F and G). Trajectory inference predicted that IM-1, which accounts for the majority (∼65%) of tumor-associated phagocytic monocytes/IMs, precedes IM-2 in maturation and is less widely distributed across non-tumor tissues (Fig. 6 G and Fig. S4 I). The PHAT signature gene expression was highest in cells along the actively phagocytic Mono-IM trajectory (Fig. 6 H). Genes associated with phagocytosis were more highly expressed in the Mono-IM-Trajectory compared to the Mono-Trajectory, while genes associated with migration were more highly expressed in the Mono-Trajectory (Fig. 6 I). Markers of phagocytosis (e.g., C1qa, Mrc1, Trem2) and glucose metabolism (e.g., Atp5a1, Slc2a1, Cox8a) also correlated with the highest enrichment of tumor-associated tdTompos monocytes/IMs (Fig. S4 K), but expression of these genes was downregulated at later pseudotime values. In contrast, phagocytic AMs have persistent upregulation of the same genes. We observed a similar transient upregulation of pro-inflammatory (M1-like) markers (e.g., Tnf and Cd74) across pseudotime, though mixed expression patterns were observed in anti-inflammatory (M2-like) genes, as Chil3 was upregulated in both cell types, but Tgfbi and Cd274 were upregulated and eventually downregulated in more mature states (Fig. S4 K). This reveals potential differences in the response of AMs and IMs to neoplastic cell engulfment. Together, the transcriptional patterns associated with phagocytosis in IMs suggest that tumor-associated IMs exhibit greater transcriptional plasticity throughout maturation than AMs and do not sustain expression of immune, phagocytic, and metabolic markers that peak during phagocytosis of neoplastic cells.
Phagocytic dendritic cells acquire an mregDC-like phenotype
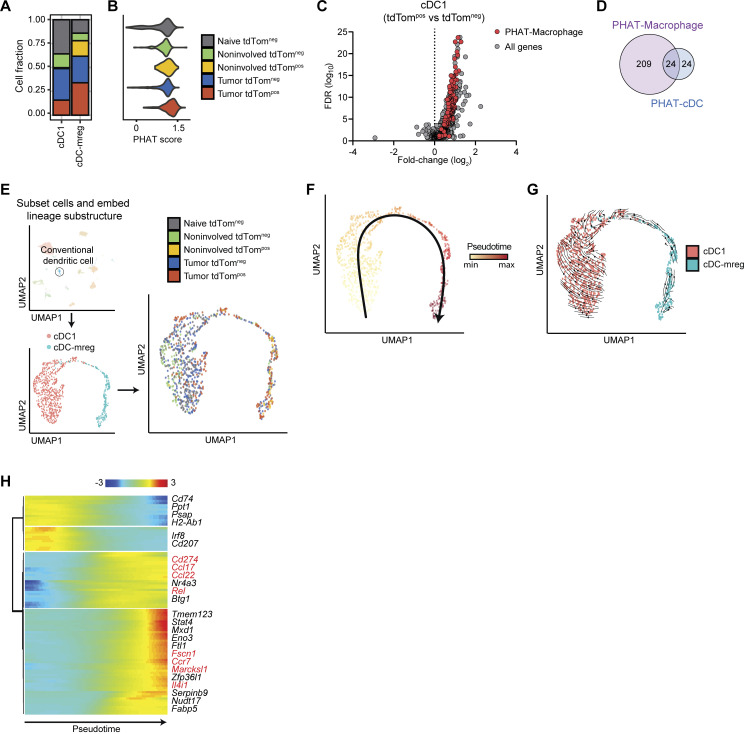
Although conventional dendritic cells (cDCs) represented a minor proportion of tdTompos cells, understanding phagocytic phenotypes in cDCs is relevant because they are potent antigen presenting cells that play a central role in innate immunity and priming adaptive immune responses (Murphy et al., 2016). A population of tumor-associated cDCs classified as “mregDCs” are transcriptionally distinct from previously described cDC1 and cDC2s and have been characterized by their expression of maturation and immunoregulatory molecules (Maier et al., 2020). From our scRNA-seq dataset, we identified Xcr1-cDC and Ccl22-cDC cell states that correspond to putative cDC1 and mregDCs, respectively (Fig. S3 I). Although we identified cDC2 cells in the lung by flow cytometry (CD11bpos DCs in Fig. 2 D), we did not observe cDC2 cells in our scRNA-seq data likely due to their relatively low frequency. mregDCs had almost double the percentage of tumor associated tdTompos cells compared to cDC1s (Fig. 7 A). mregDCs also contained the highest percentage of tdTompos cells from uninvolved tissue of any myeloid cell type, suggesting a potential migratory capacity of these cells from tumor tissue once they engulf neoplastic cells. Both findings are consistent with previous studies that report increased antigen uptake and high migratory potential in tumor-associated mregDCs and reveal that mregDCs increase these functions after phagocytosing neoplastic cells (Kryczanowsky et al., 2016; Maier et al., 2020; Ness et al., 2021). The PHAT gene signature, defined in monocytes and macrophages, was most highly expressed in tdTompos cDCs, particularly cDC1s, compared to tdTomneg cDCs demonstrating that this signature could be applied beyond monocytes and macrophages (Fig. 7, B and C). Overall, half of the cDC DEGs overlapped with the PHAT-Macrophage signature (Fig. 7 D).
Figure 7.
Tumor-associated tdTompos dendritic cells acquire an mregDC phenotype. (A) Cell frequency plots for each cDC cell state, delineated by tissue and phagocytic status. (B) Violin plot showing PHAT signature scores for cDC grouped by tissue and phagocytic status. (C) Volcano plot of differentially expressed genes between tdTompos and tdTomneg DC1s (cDC.Xcr1). PHAT-Macrophage signature genes are highlighted in red. Statistical significance was determined using MAST. (D) Venn diagram showing overlapping genes between PHAT-cDC signature and PHAT-Macrophage signature. (E–G) UMAP projections of cDCs, highlighting cDC subsets (E, left), tissue/phagocytic status (E, right), pseudotime-based trajectory inference using Slingshot (F), and dynamic RNA velocity trajectory inference (G). (H) Heatmap of genes that are differentially expressed along cDC trajectory. Gene groupings were determined by hierarchical clustering of genes by expression pattern. Genes highlighted in red depict mreg-DC marker genes reported by Maier et al., (2020). Statistical significance of genes across the single cDC trajectory was determined by Wald test after fitting the data using a generalized additive model.
Trajectory inference of cDCs predicted maturation from cDC1s toward mregDCs (Fig. 7, E–G). Analysis of differentially expressed genes across pseudotime not only revealed a concomitant expression shift from DC1 markers (e.g., Ppt1 and Irf8) toward mregDC markers (e.g., Ccl22, Fscn1, and Ccr7), but also an increase in proinflammatory signaling mediators (e.g., Stat4 and Rel) despite higher Cd274 and lower class II MHC (Fig. 7 H). Therefore, tumor-associated tdTompos cDCs in lung tumors from KTai65;SpcCreER mice appear to acquire an mregDC-like phenotype upon engulfment of neoplastic cells in vivo. By tracking neoplastic engulfment in cDCs, our findings reveal an association between phagocytosis and the coordination of migratory, inflammatory, and immunoregulatory functions in cDCs.
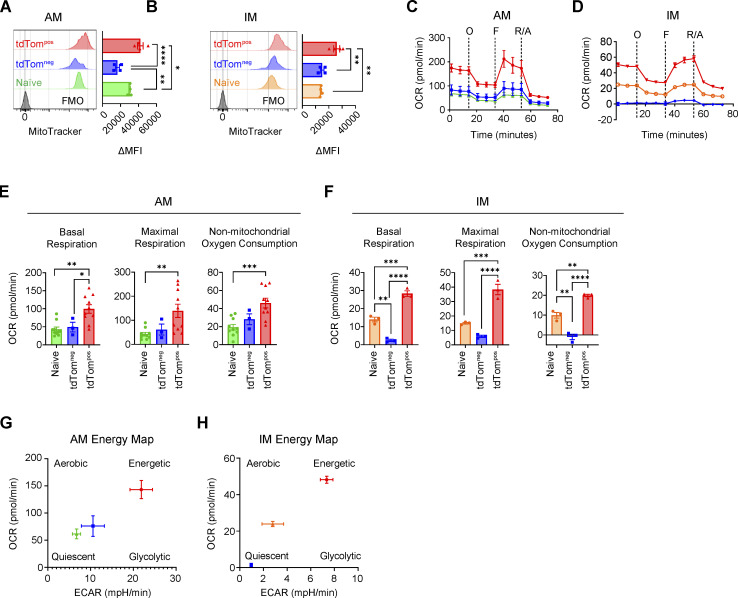
Phagocytic TAMs have increased mitochondrial content, oxygen consumption, and expression of OXPHOS-related genes
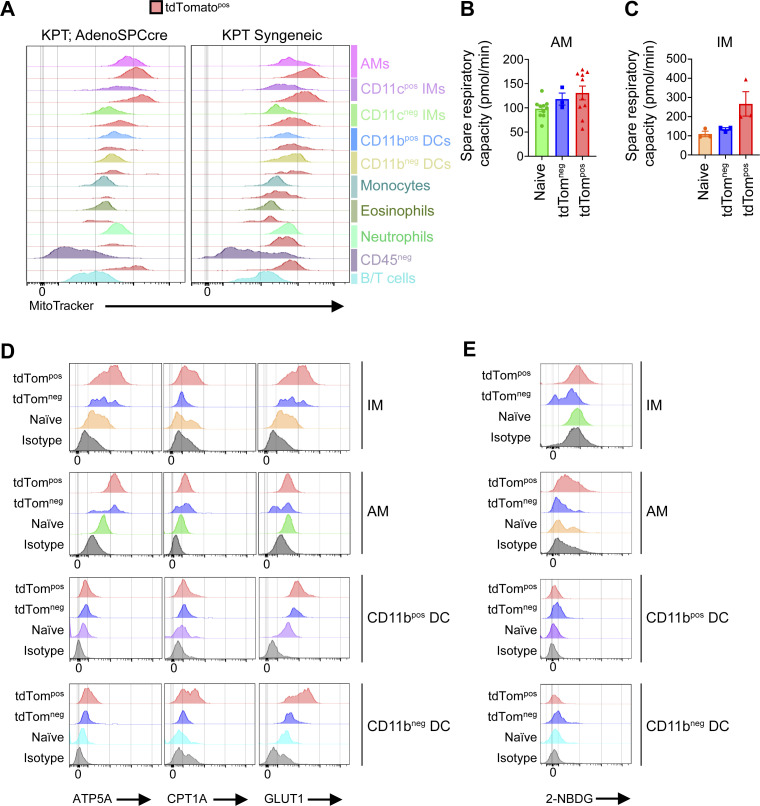
To understand the functional consequences of the upregulated metabolic gene expression signatures in tumor-associated myeloid cells, we investigated the metabolic features of phagocytic tumor-associated myeloid cells from both the syngeneic and autochthonous KPT:Adeno-SpcCre lung tumor models. Mitochondria are the location of OXPHOS metabolism and their abundance has been shown to affect macrophage phenotypes and functions (Tur et al., 2017). Intracellular staining of mitochondria in myeloid cells (CD11bpos and/or CD11cpos) revealed significantly increased mitochondrial content in tdTompos AM, IM, and DCs compared to tdTomneg cells in both tumor models, suggesting a potential functional role of mitochondrial metabolism (Fig. S5 A and Fig. 8, A and B). Therefore, we next performed extracellular flux analysis of sorted macrophages from autochthonous tumors to measure metabolic activity (Fig. 8, C and D). tdTompos AMs and IMs had significantly increased oxygen consumption rate (OCR) at baseline and at maximal respiration (induced by FCCP treatment), indicative of ATP generation via the electron transport chain in mitochondria (Fig. 8, E and F). However, no significant differences in the levels of spare respiratory capacity were detected despite increased OCR (Fig. S5, B and C). Non-mitochondrial oxygen consumption was significantly higher in tdTompos AMs and IMs compared to tdTomneg AMs and IMs (Fig. 8, E and F). Naive AMs and IMs had modest OCR and tdTomneg AM respiration levels resembled that of the naive AMs. Interestingly, tdTomneg IMs have nearly undetectable levels of cellular respiration. This suggests that IMs that are not activated by phagocytosis of neoplastic cells are in a metabolically quiescent state, unlike AMs which have relatively higher baseline levels of oxygen consumption.
Figure S5.
Phagocytic tumor monocytes and macrophage subsets have increased mitochondrial content and cellular respiration. (A) MFI of MitoTracker staining within tdTompos (red) or tdTomneg immune cell populations isolated from autochthonous KPT;AdenoSPCcre or KPT syngeneic lung tumors. (B and C) Spare respiratory capacity calculated from basal OCR and OCR after oligomycin treatment in AMs (B) or IMs (C). (D) Histogram plots of intracellular flow cytometry staining of metabolic enzymes (ATP5A, CPT1A, and GLUT1) by MET-FLOW in IMs, AMs, CD11bpos DCs, or CD11bneg DCs sorted from KT;Adeno-SPCcre lung tumors or naive lungs. (E) Histogram plots showing intracellular accumulation of 2-NBDG quantified by flow cytometry in IMs, AMs, CD11bpos DCs, or CD11bneg DCs sorted from KT;Adeno-SPCcre tumors or naive lung. Data are representative of three independent experiments.
Figure 8.
Phagocytic TAMs have increased oxygen consumption ex vivo. (A and B) MitoTracker staining of tdTompos or tdTomneg AMs (A) or IMs (B) sorted by FACS from Adeno-SPCcre lung tumors. (C and D) Extracellular flux analysis of the OCR of sorted AMs (C) or IMs (D) with inhibitors or agonists of the OXPHOS pathway. (E and F) Quantification of the basal, maximal, or non-mitochondrial respiration in sorted tdTompos or tdTomneg AMs (E) or IMs (F). (G and H) Extracellular flux quantification of basal OCR and ECAR for sorted AMs (D) or IMs (E) isolated from AdenoSPCcre lung tumors ex vivo. Ordinary one-way ANOVA (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). Data are representative of three independent experiments.
The extracellular acidification rate (ECAR) is indicative of the secretion of lactic acid, a by-product of the glycolytic pathway. ECAR was dramatically elevated in tdTompos AMs and IMs compared to their respective naive or tdTomneg counterparts. A concomitant increase in both OCR and ECAR at baseline suggests that phagocytic tdTompos macrophages have increased glycolysis, in addition to increased OXPHOS (Fig. 8, G and H). This combinatorial increase in OCR and ECAR is indicative of an elevated energetic state in phagocytic tdTompos macrophages. Interestingly, naive IMs have a skewing toward aerobic respiration and lack of glycolytic respiration indicating an energetically quiescent state.
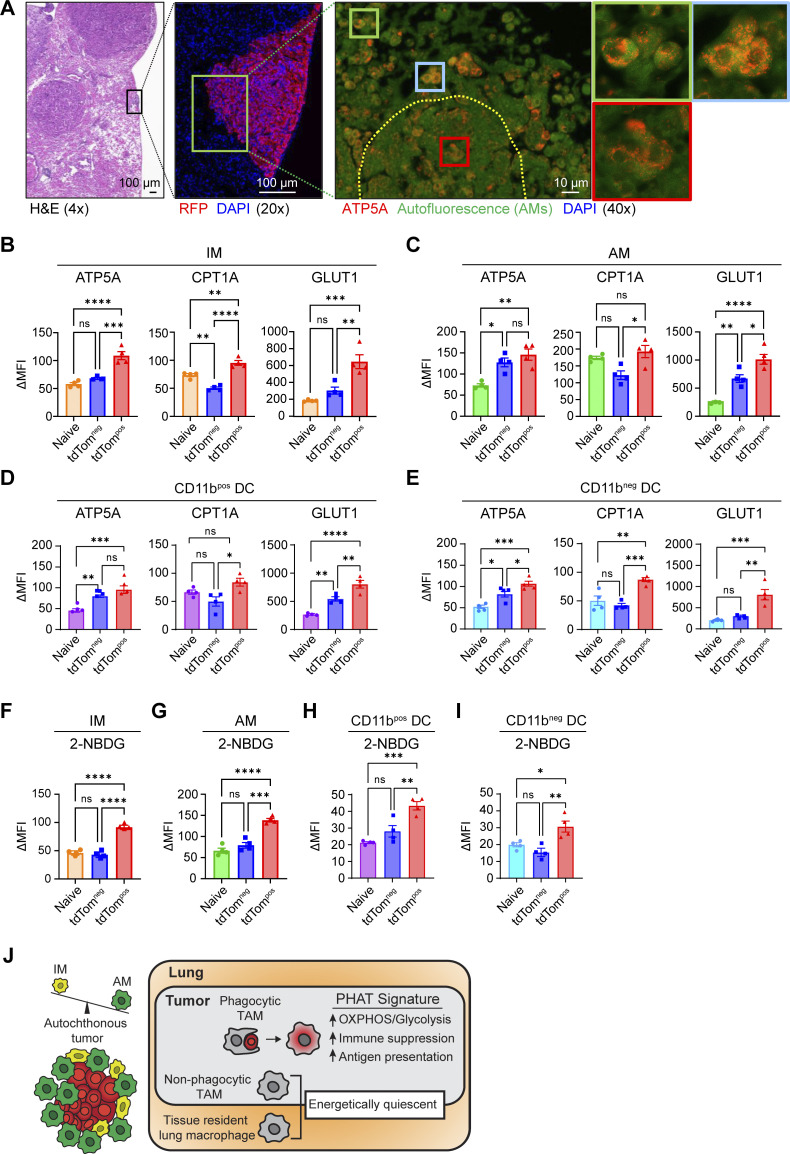
The final step of cellular respiration involves the production of ATP by the mitochondrial ATP synthase, which consists of several subunits, including the protein, ATP5A (Jonckheere et al., 2012). We used ATP5A as a marker of mitochondria and cellular respiration in histological samples from autochthonous lung tumors. To evaluate the spatial localization of macrophages with increased metabolic activity, we performed immunofluorescence staining for ATP5A in autochthonous lung tumors. AMs localized near and within the tumors had increased expression of ATP5A compared to AMs in normal adjacent tissue (Fig. 9 A). To further validate these metabolic features at the single-cell protein level, we quantified the levels of key metabolic enzymes relating to OXPHOS and glycolysis utilizing metabolic flow cytometry (METFLOW) on sorted tdTompos and tdTomneg myeloid cells from autochthonous lung tumors (Ahl et al., 2020). tdTompos IMs AMs and cDCs had significantly increased expression of proteins associated with key metabolic pathways, including GLUT1 (glycolysis) ATP5A (OXPHOS) and CPT1A (fatty acid oxidation [FAO]; Fig. 9, B–E; and Fig. S5 D). Higher expression of CPT1A and ATP5A suggest that mitochondrial lipid transport and respiration is active in phagocytic TAMs and cDCs. Higher expression of GLUT1 is indicative of the sensing and transport of glucose into the cell for glycolysis.
Figure 9.
Phagocytic tumor macrophages upregulate both OXPHOS and glycolytic metabolism in vivo. (A) H&E and immunofluorescence imaging of autochthonous lung tumors stained for RFP, ATP5A, and DAPI. (B–E) Quantification of ΔMFI from tdTomneg and tdTompos IM (B), AM (C), CD11b-pos DC (D), and CD11b-neg DC (E) after intracellular MET-FLOW staining for metabolic enzymes. (F–I) Quantification of ΔMFI from tdTomneg and tdTompos IM (F), AM (G), CD11b-pos DC (H), and CD11b-neg DC (I) after intracellular quantification of 2-NBDG using flow cytometry. (J) Schematic detailing the predominance of AM populations found in autochthonous lung tumors and the metabolic and immune phenotypes of the PHAT signature found in phagocytic TAMs. Ordinary one-way ANOVA (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., P > 0.05). Data are representative of three independent experiments.
In parallel to METFLOW, we measured the glycolytic activity of phagocytic tumor-associated myeloid cells by treating these cells ex vivo with a fluorescently labeled, non-catalyzable glucose analog (2-NBDG). Glycolytic activity was significantly higher in tdTompos AMs, IMs, and cDCs compared to tdTomneg and naive lung macrophages (Fig. 9, F–I; and Fig. S5 E). These data demonstrate that the transcriptional signatures relating to OXPHOS and glycolytic metabolism produce altered metabolic states in phagocytic TAMs and DCs. Taken together, these data identify tdTompos macrophages as a distinct TAM state in lung tumors and highlight OXPHOS metabolism as a key feature that correlates with immune suppressive phenotypes and functions in lung tumors (Fig. 9 J).
Discussion
In this study, we used a novel autochthonous genetic lung tumor model that minimizes undesirable, direct labeling of immune cells to identify, quantify, and profile phagocytic myeloid cells at the transcript, protein, and functional levels. Prior studies characterizing TAMs in vivo have not specifically isolated phagocytic cells and most have used either cell transfer models, in which the dominant phagocytic cell is monocyte derived, or autochthonous models that include macrophages that are directly transformed (Casanova-Acebes et al., 2021; Gordon et al., 2017). Our models provide a unique path to define how phagocytosis alters tumor-associated myeloid cells. We find that IMs engulf more neoplastic cells in syngeneic transplant models, while AMs are the dominant phagocytic cell type in autochthonous lung tumors. This could be due to localization of AMs in the alveolar space versus IM localization in the lung parenchyma and the differential source of tumors, either seeding from the blood or initiated from transformed alveolar type II cells.
Within autochthonous lung tumors, we also found key differences between AMs and IMs in the type of response to the engulfment of neoplastic cells in vivo. The inducible response observed in IMs supports prior findings of increased plasticity of monocytes and monocyte-derived macrophages compared to tissue resident macrophages such as AMs, which harbor poised enhancer landscapes shaped by early lung development and/or local microenvironment cues (Lavin et al., 2014). Our scRNA-seq analysis of phagocytic IMs partially correlated with previous studies identifying phagocytic monocyte-derived macrophages in inflamed and cancer tissues (Sanin et al., 2022). The discrepancy of overlap between their signature and ours could be a result of our defining actively phagocytosing cells versus enrichment of genes associated with phagocytosis. Furthermore, we were able to corroborate gene expression of phagocytic TAMs at the transcriptional level with protein detection within AMs and IMs in autochthonous tumors. For example, PD-L1 surface expression was upregulated in tdTompos IMs but was also detected at high levels in AMs irrespective of phagocytic status, consistent with a described tolerogenic role for most tissue resident macrophages (Fig. 2, E and F; and Fig. S2, D and E; Allard et al., 2018). One possible explanation for this is that AMs actively phagocytose lipid-rich lung surfactant at steady-state and in doing so, have high expression of genes involved in tolerogenic pathways at baseline (de Aguiar Vallim et al., 2017).
While our model identifies tdTompos macrophages that have recently internalized neoplastic cells, it cannot explicitly distinguish between macrophages that have degraded the tdTomato fluorophore or have never phagocytosed a neoplastic cell. Trajectory inference of AMs and IMs, allowed us to model the diverse activation paths of macrophages during tissue homeostasis and in the presence of tumors. Notably, the upregulation of Cd274 (which encodes PD-L1) in AM-Trajectory A revealed an association between trajectory-specific neoplastic cell engulfment and immunosuppression (Fig. S4 E). Of the three AM trajectories, the highest percentage of tdTompos AMs were in AM-Trajectory A, suggesting that neoplastic engulfment drives AM maturation toward the cell states enriched in tissue remodeling genes, culminating in a Chil3high AM state (AM-4; Fig. 5 I and Fig. S4 A). Our results suggest that the AM-4 cell state, despite having a lower fraction of tdTompos cells than the AM-2 state, derives from actively phagocytic AMs and represents an imprinted consequence in AMs following phagocytosis of neoplastic cells. AM-4 cells were also detected in AM-Trajectory C, although AM-4 cells in AM-Trajectory A had higher immune-suppressive gene expression. The high fraction of AMs in naive and uninvolved tissues in AM-Trajectory C suggests that certain homeostatic processes may polarize AM maturation toward phenotypes similar to those that arise after neoplastic cell engulfment.
In addition to defining cell type–specific phagocytosis-driven TAM phenotypes, we identified a common 123-gene phagocytic signature. Importantly, this PHAT signature was enriched in human lung cancer tumor-associated myeloid cells and correlates with a lower probability of survival in patients with lung adenocarcinoma (Fig. 4 I). In human tumors, direct identification of myeloid cells that have recently phagocytosed neoplastic cells is not feasible. However, one study used morphological features to characterize putative phagocytic large TAMs (L-TAM) and compared them to non-phagocytic small TAMs (s-TAM; Donadon et al., 2020). The enrichment of lipid metabolic pathways in L-TAMs is consistent with our findings in tdTompos TAMs. Interestingly, our PHAT signature better represents human TAM gene expression than traditional M1 or M2 phenotypes suggesting that gene signatures associated with macrophage functions, such as the phagocytosis of neoplastic cells, more closely identifies TAMs in clinical samples.
Phagocytic tdTompos cDCs were identified both in tumors and adjacent tissues (Fig. 7), and Ccl22 mregDCs were enriched in phagocytic cells and represented a more differentiated state compared to Xcr1 cDC1s, consistent with other studies (Maier et al., 2020). These cells represented the majority of tdTompos cells from non-tumor tissue, potentially due to their enhanced migratory capacity. Additionally, tdTompos cDCs have significant enrichment of the PHAT Signature, in which OXPHOS metabolism is a key feature. Functional assays are consistent with phagocytic DCs being more metabolically active, with increased OXPHOS. These DCs have higher mitochondrial content and ATP5A and CPT1A expression, as well as increased glycolysis, with higher glucose uptake and GLUT1 expression. A link has been proposed between lipid-fueled FAO and cancer-associated DCs with tolerogenic functions in tumors (Herber et al., 2010; Zhao et al., 2018). Thus, we provide further evidence that engulfment of neoplastic cell may promote a tolerogenic mregDC program which is sustained by metabolic reprogramming.
OXPHOS metabolism is associated with immune suppressive phenotypes of macrophages in vitro (Jin et al., 2014; Viola et al., 2019). Macrophages can acquire lipids from the engulfment of dying cells (Tabas and Bornfeldt, 2016; A-Gonzalez et al., 2017); these lipids are transferred to the mitochondria to replenish the TCA cycle with acetyl-CoA for the generation of NADH and FADH2 to support the electron transport chain (ETC; Mehta et al., 2017). Our functional metabolic assays demonstrated that phagocytic tumor-associated AMs and IMs undergo metabolic reprogramming toward increased OXPHOS metabolism. Earlier reports demonstrated that type-2 cytokines in vitro or in vivo can maintain an intact TCA cycle coupled with mitochondrial OXPHOS (Jha et al., 2015). Recent analyses of macrophage metabolism describe how lipid accumulation occurs within macrophages in chronic inflammatory environments due to defective ETC when tissue oxygen levels are low, ultimately leading to an inefficient ETC and subsequent inhibition of OXPHOS metabolism (Eming et al., 2021). However, we find that the OXPHOS metabolism is intact in phagocytic TAMs (Fig. 8, C–H). Thus, phagocytosed material, including lipids derived from neoplastic cells, can be catabolized via FAO and the TCA cycle in TAMs from autochthonous tumors. Although we connect OXPHOS metabolism with phagocytosis in TAMs, more work will be needed to elucidate the requirement of this energetic shift to sustain TAM phenotypes and functions in vivo and, most importantly, how these phenotypes change with therapeutic intervention.
The connection of TAM and DC phagocytosis with metabolic changes toward increased OXPHOS and an immune suppressive/tumor-promoting phenotype links previous studies defining metabolic phenotypes of phagocytic macrophages in vitro with characterization of TAM metabolism in vivo (Lantz et al., 2020; Li et al., 2022). Similarly, disease associated microglia (DAM) and lipid-associated macrophages (LAM) have been shown to utilize lipid metabolic pathways and OXPHOS and share significant gene expression overlap with each other (Jaitin et al., 2019; Keren-Shaul et al., 2017). Interestingly, we found significant transcriptional similarity between PHAT macrophages and lipid-rich Trem2pos LAMs (Fig. 4 I). This finding underscores a recurrent theme among macrophages in inflammatory microenvironments, in which the magnitude of lipid metabolism and oxidation may direct macrophage fate.
Lipid metabolism has been reported to be essential for macrophage alternative activation of macrophages and efficient efferocytosis (Huang et al., 2014). In line with the downstream catabolism of lipids potentially derived from recently phagocytosed cells, key components of the ETC are significantly upregulated in tdTompos TAMs including ATP5A, a subunit of the complex V of the ETC, and CPT1A, a critical transporter of fatty acids from the cytosol into the mitochondria (Fig. 9, B–D). Using immunofluorescence, we show that ATP5A is detected at higher levels in TAMs that are in close proximity to neoplastic cells in tumor tissue, showing in situ the increased metabolic state of macrophages positioned to phagocytose cancer cells. These data suggest that myeloid cell phagocytosis of neoplastic cells results in dynamic metabolic changes that could inform the design of novel therapies targeting TAM functions and induce macrophage repolarization.
Materials and methods
Mouse strains
Autochthonous mouse models were performed at Stanford animal facilities and used KrasLSL-G12D;Trp53flox/flox;Rosa26LSL-tdTomato (KPT) mice and KrasFSF-G12D;Rosa26FSF-LSL-tdTomato(ai65);SftpcCreER (KTai65;SpcCreER) mice. KrasLSL-G12D (Jackson et al., 2005; Jackson et al., 2001; Tuveson et al., 2004), Trp53flox/flox (Jax# 008462; Marino et al., 2000; Jonkers et al., 2001), Rosa26LSL-tdTomato (Jax# 007905; Madisen et al., 2010), KrasFSF-G12D (Jax# 023590; Young et al., 2011), Rosa26FSF-LSL-tdTomato(ai65) (Jax# 021875; Madisen et al., 2015), and SftpcCreER (Jax# 028054; Rock et al., 2011) have been previously characterized. Intravenous cell transfer experiments were conducted at Amgen animal facilities and used 8-12-wk-old F1 (first filial generation) hybrid C57BL/6J and 129S1/Svlmj mice (JAX Strain#: 101043; B6129SF1/J) using a primary lung tumor cell line derived from a KPT autochthonous lung tumor. All animal experimental protocols performed at Amgen were approved by the IACUC of Amgen and were conducted in accordance with the guidelines set by the Association for Assessment and Accreditation of Laboratory Animal Care. The Stanford Institute of Medicine Animal Care and Use Committee approved all animal studies and procedures performed at Stanford University.
Lenti-PGK-Cre and Ad5-SPCcre generation
Lentiviral plasmids were co-transfected with packaging vectors (delta8.2 and VSV-G) into 293T cells using PEI. The supernatant was collected at 36 and 48 h after transfection, ultracentrifuged at 25,000 rpm for 90 min, and resuspended in PBS. Tumors were initiated by intratracheal infection of mice with lentiviral or adenoviral vectors expressing either inverted-FLPo or Cre recombinase (as indicated for KTai65;SpcCreER or KPT-AdenoSPCcre tumor models). Ad5-SPCcre adenoviral vectors were purchased from University of Iowa Carver College of Medicine Viral Vector Core, deposited by Dr. Anton Berns from Netherlands Cancer Institute (VVC-Berns-1168). For Ad5-SPCcre transductions, mice were given an intratracheal dose of 1 × 109 TFU in 60 μL of PBS.
Lenti-InFlpo generation
Lentiviral backbone with inverted FLPo recombinase (Lenti-InFlpo), was generated through modification of pLL3.3 lentiviral vector by addition of PGK promoter and inverted FLPo recombinase surrounded by pairs of heterotypic lox sites (LoxP and Lox 2272). These additional fragments were generated by Integrated DNA Technologies and assembled through NEB HiFi DNA assembly system (E2621). In this design, there are two different lox sites (LoxP and Lox 2272) on each side of FLPo Recombinase. Identical lox sites (one on each side of FLPo recombinase) are facing each other and they invert the FLPo recombinase as a result of Cre mediated recombination. To prevent expression of FLPo expression in the absence of Cre recombination even when the Lentiviral vector integrates next to an endogenous promoter, we included an HSV TK poly A between the 3′ LoxP and Lox 2272 sites (unpublished data).
Tumor dissociation and cell sorting
Primary tumors were dissected and dissociated as previously published (Yu et al., 2016). Briefly, (n = 3 tumor-bearing mice and n = 3 tumor-naive mice) using an enzymatic digestion mixture of 1.5 mg/ml of Collagenase A (Sigma-Aldrich: COLLA-RO) and 200 units/ml DNaseI in DPBS (with Ca2+/Mg2+) plus 5% heat-inactivated fetal bovine serum (HIFBS) and 10 mM HEPES. Lungs were inflated with digestion mixture and incubated at 37°C for 30 min shaking at 120 rpm. The lungs were mashed and sequentially strained, washed with media (RPMI, no Glut containing 10% HIFBS), and centrifuged at 450 ×g through 100-, 70-, and 50-μm cell strainers. The resulting cell suspensions were treated with ACK RBC lysis solution (A1049201; Gibco) for 5 min at room temperature and washed/centrifuged with media. For cell sorting, antibody labeled cells were sorted based on detection of indicated antibody fluorescence into 1.7-ml Eppendorf tubes containing media and placed on ice for downstream applications.
Flow cytometry and staining
After cell counting, at least 1 × 106 cells per sample were stained with Near-IR Fixable LIVEDEAD dye (L10119; Life Technologies) according to the manufacturer’s instructions. Cells were then incubated in FACS buffer (554657; BD) containing 1% TruStain FcX anti-mouse CD16/32 antibody (101320; Biolegend) for 15 min on ice. Subsequently, cells were washed with FACS buffer, centrifuged, and stained with immunophenotyping antibodies (Anti-CD11b, clone: M1/70; Anti-CD45, clone: 30-F11; Anti-CD11c, clone: N418; Anti-Ly6G, clone: 1A8; Anti-I-A/I-E, clone: M5/114.15.2; Anti-CD64, clone: X54-5/7.1; Anti-CD24a, clone: M1/69; Anti-PD-L1, clone: 10F.9G2; Anti-CD80, clone: 16-10A1; Anti-MERTK, clone: 2B10C42; Anti-ITGAV, clone: RMV-7; Anti-IL-4Ra, clone: mIL4R-M1; Anti-TREM2, clone: FAB17291A; Anti-AXL, clone: MAXL8DS; Anti-EpCAM, clone: G8.8) for 25 min on ice. After staining, cells were washed with FACS buffer and either analyzed on the flow cytometer or cell sorter directly or fixed with DPBS containing 2% paraformaldehyde for 10 min at room temperature, protected from light. Cells were then washed with FACS buffer and stored at 4°C until analyzed on the flow cytometer. For Metabolic Flow cytometry (METFLOW), staining procedures were described elsewhere (Ahl et al., 2020). Briefly, cell suspensions were stained with cell surface antibodies as detailed above then cells were fixed and permeabilized using eBioscience Foxp3/Transcription Factor Staining Buffer Set (Cat# 00-5523-00; Invitrogen) according to manufacturer’s instructions. Cells were then washed with cold DPBS and centrifuged at 450 ×g, 5 min, three times. Subsequently, cells were stained with intracellular antibodies (ATP5A, clone: EPR13030(B); CPT1A, clone: 8F6AE9; GLUT1, clone: EPR3915) in permeabilization buffer for 1 h at room temperature. Then, cells were washed once with permeabilization buffer and then once with cold DPBS. Samples were acquired on a X-30 FACSymphony (BD) with FACS Diva Version (BD, Version 8.0.1) software. Analysis was performed using FlowJo (Treestar, version 10.5.2). TdTomato-positive and -negative population percentages only displayed cell populations above a 5% threshold of CD11bpos and/or CD11cpos cells.
Nanostring gene expression analyses
RNA was purified from ∼200,000 FACS sorted tdTompos or tdTomneg lung macrophages (AMs: CD45pos Ly6Gneg MHCIIpos SSC-Ahigh CD64pos CD24neg CD11blow; IMs: CD45pos Ly6Gneg MHCIIpos SSC-Ahigh CD64pos CD24neg CD11bhigh) using the Qiagen Rneasy kit, per the manufacturer's recommendations. RNA quantification of selected genes was performed using the nCounter myeloid innate immunity panel for mouse consisting of 770 target genes from 19 pathways with housekeeping and control genes (NanoString Technologies). Pathway scores and P values were calculated from quality-controlled, normalized raw expression data using the nSolver software (v4.0) with the nCounter Advanced Analysis add-on (v2.0.134).
Phagocytosis in vitro co-culture assay
BMDMs were derived per previous well-established protocol (Weischenfeldt and Porse, 2008). Briefly, whole bone marrow was flushed from hindlimb bones of C57BL/6 mice (female, between 6–8 wk of age) using PBS. Red blood cells were lysed (R7757-100ML; Sigma-Aldrich) and remaining bone marrow cells were cultured in BMDM media (DMEM plus 10% HIFBS and 1% penicillin/streptomycin media that contains 100 ng/ml recombinant murine M-CSF; Peprotech: 315-02) for 6 d with a media change on day 3. Fully differentiated BMDM were detached, counted, and plated into 12-well plates at 2 million cells per well (n = 5) and allowed to settle for 2 h. KPT cancer cells (cell clone: SP110P) were counted and resuspended in BMDM media to overlay onto BMDMs at 1:1 ratio. The co-culture was incubated at 37°C overnight (16–18 h), at which point cells were detached and subject to flow cytometry–based analyses to evaluate the status of their surface marker expression and intracellular metabolic activity (see Flow cytometry and staining; and Metabolic characterization using extracellular flux analysis and cellular dyes).
Metabolic characterization using extracellular flux analysis and cellular dyes
ECAR (mpH/min) and OCR (pmol/min) were measured using the XFe96 extracellular flux analyzer (Agilient Technologies). Glycolytic function correlated with ECAR and mitochondrial respiration correlated with OCR. 200,000 cells were plated per well in complete media (RPMI 10% HIFBS 1× pen/strep/glut) in a 96-well plate after cell sorting and PBS wash. The assay was performed using XF Assay Modified Media with L-glutamine (2 mM; 103579-100; Agilient Technologies), sodium pyruvate (1 mM; 103578-100; Agilient Technologies) with 11 mM glucose (103577-100; Agilient Technologies). Mitochondrial respiration was measured using the mitochondrial stress test kit (103015-100; Agilient Technologies), by sequentially adding oligomycin (2 µM), carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (1.5 µM), and rotenone and antimycin A (1 µM). For measurement of glycolysis using a dye as described elsewhere (Bala et al., 2021), 200,000 isolated tumor cells were resuspended in complete media containing 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose (2-NBDG) at 60 µM for 1 h at 37°C. Cells were then analyzed for via flow cytometry for fluorescence intensity on the FITC detector. For mitochondrial staining, MitoTracker Deep Red FM (M22426; Invitrogen) was used per the manufacturer’s recommendations and analyzed for MFI via flow cytometry.
Histology and immunofluorescence
Tumor tissue samples were fixed in 10% neutral buffered formalin for 12–24 h at room temperature. Fixed samples were embedded in paraffin blocks, and 5-μm sections were cut. H&E and staining procedures were performed by Pathology core at Stanford University. Fluorescent staining was performed with anti-RFP (600-401-379; 1:200, Rockland Immunochemicals) with secondary anti-rabbit Alexa Flour 647 Ab, ATP5A (ab196198, 1:200; Abcam) with secondary anti-rabbit Cy3 Ab. Pictures were taken on Keyence BZ-X710 using ×4 PlanFluor NA 0.13 PhL, ×20 PlanApo NA 0.75, and ×40 PlanFlour NA 0.60 Ph2 objective lens.
Sample preparation and FACS for scRNA-seq
Single cell suspensions of lung tumor cells were prepared as described above. Sample hashing was performed using TotalSeq-A (BioLegend) reagents. Specifically, single cell suspensions were stained with oligo-tagged antibodies specific for B2M and CD45 at a concentration of 1:100 for 25 min on ice and subsequently washed and stained for surface markers as described above. Prior to sorting on the FACS machine, adjacent normal and tumor samples of the same biological replicate were combined equally and myeloid cells (Singlet Live CD45+CD11b+CD11c+) and epithelial/neoplastic cells (Singlet Live CD45-EpCAM+) cells were sorted based on their tdTomato fluorescence (tdTompos or tdTomneg).
scRNA-seq library preparation and sequencing
Sorted single-cell suspensions were pooled into scRNA-seq sample groups according to the sample hashtag labeling strategy to identify cells by animal and tissue type of origin. Samples were centrifuged for 400 ×g × 7 min and resuspended in RPMI + 10% FBS at a concentration of 400–1,000 cells/μl for samples containing over 50,000 cells and at the resuspension concentration (200–300 cells/μl) for samples with <50,000 cells. Cell encapsulation and transcriptomic library preparation were performed using the 10X Genomics Chromium 3′ V3 chemistry according to manufacturer’s instructions (protocol CG000185 RevD). Targeted cell recovery was 10,000 cells for samples with over 50,000 cells sorted, and 46.6 μl of cell resuspension was loaded for samples with <50,000 cells sorted.
After encapsulation of cells and hydrogel beads into Gel Bead-In Emulsions and reverse transcription, 11–12 cDNA amplification cycles were performed with standard 10X Genomics gene expression cDNA primers with 2 nM HTO additive primer to amplify native transcripts and hashtag oligo (HTO) barcodes, respectively. SPRI clean-up of cDNA amplification products was performed to separate HTO products (in the supernatant) from gene expression (GEX) products (in the pellet). GEX products were then subject to fragmentation, end-repair, A-tailing, adapter ligation, and an additional round of 11-15 PCR cycles to attach library-specific 8-bp i7 indexes. After double-sided SPRI bead purification, this resulted in the generation of ∼500 base pair libraries for next-generation sequencing. Libraries were quantified using Agilent TapeStation 4200 D5000 high-sensitivity screen tapes.
HTO libraries were constructed using the BioLegend TotalSeq-A HTO protocol (https://www.biolegend.com/en-us/protocols/totalseq-a-antibodies-and-cell-hashing-with-10x-single-cell-3-reagent-kit-v3-3-1-protocol). SPRI-purified HTO fractions were amplified using the SI-PCR primer and TruSeq D70×_LONG 8-bp library-indexing primers for 13–15 cycles. Products were purified again using SPRI purification to generate final libraries and quantified using Agilent TapeStation 4200 D1000 high-sensitivity screen tapes.
To sequence the libraries, each GEX and HTO library was diluted to 5 nM and pooled at a volume ratio of 4:1 for GEX:HTO, resulting in four times more GEX reads than HTO reads. Libraries were then sequenced on two lanes of an S4 version v1.5 flow cell on an Illumina NovaSeq6000 instrument to achieve an average sequencing depth of 50,000 reads per cell. The sequencing parameters used were 28:8:0:91 cycles (Read1:i7 index:i5 index:Read2).
scRNA-seq alignment, preprocessing, and normalization
Raw sequencing files were demultiplexed by GEX and HTO library i7 barcodes to generate FASTQ files for each GEX and HTO library. Reads from each library were aligned using cellranger count (CellRanger version 4.0.0 from 10X Genomics). More than 90% GEX reads aligned to the Ensembl 93 mm10–3.0.0 mouse reference genome containing the tdTomato gene (https://www.ensembl.org), and ∼65% of the total reads aligned to the transcriptome. HTO reads were aligned to a feature reference (using the argument --feature-ref) containing the TotalSeq-A barcodes used, and ∼60% of reads contained a valid HTO barcode. The unfiltered UMI count matrices generated by CellRanger were used for pre-processing of data.
scRNA-seq pre-processing
Quality control was performed using a multi-step procedure. For preliminary removal of cell barcodes containing only ambient mRNA or poor-quality transcriptomes, outlier barcodes that were more than three median average deviations below the median read count were removed. Cell barcodes with <250 mRNA features, <500 UMI, sequencing saturation <50%, and percent of mtRNA reads >20% were also considered to be poor-quality transcriptomes or dying cells and removed (Fig. S3 C).
HTO counts were normalized across cells using the centered log-ratio transformation and demultiplexed using the HTODemux() function with default settings in the Seurat v4.0.0 R package (Hao et al., 2021; Stoeckius et al., 2018; Fig. S3 D). A single HTO barcode could be assigned to 72.3% of cells, and HTO labels were later used to filter cell multiplets. Alignment of sequencing reads generated 2.7 billion total genome-mapped reads across and 951 million hashtag barcode-mapped reads (89.7% associated with gene expression cell barcodes) from 10 gene expression and antibody hashtag libraries.
For preliminary removal of multiplets by read count and number of features, barcodes with a read cutoff of 10 median average deviations above the median read number and >6,000 mRNA features were removed. Next, cell barcodes with two or more HTO barcodes identified by the HTO classifier were removed. Finally, the R package DoubletFinder (McGinnis et al., 2019) was applied to each 10X sample library to remove doublets not removed by either method. Briefly, artificial doublets containing averaged expression values from two cell barcodes were simulated and integrated into the dataset. Normalization, scaling, and dimension reduction using principal-components analysis were used to embed cell barcodes and artificial doublets. The proportion of artificial nearest neighbors (pANN) were then computed using the k nearest neighbors for every cell barcode in principle-component space, and the barcodes with top pANN values were classified as doublets. Overall, we observed a 77.7% concordance in predicted singlets between HTO labeling and DoubletFinder, with 15.6% of doublets detected by either HTODemux() or DoubletFinder (Fig. S3, E and F).
Cell cycle stage was calculated for each cell using the machine learning approach previously described (Scialdone et al., 2015). This approach trains a classifier on pairs of genes that change expression directionality across different cell cycle phases. When applied to our data, each cell’s phase could be predicted by observing the calculated gene expression difference for either G1, G2M, or S phases (Fig. S3 J).
All 10 libraries were normalized using the procedure outlined previoulsy (Ellwanger et al., 2021). In short, scaling factors for each library were first calculated. Then, similar cells were binned into pools containing at least 10% of the total number of cells per cluster and a minimum average gene expression of 1 using the quickCluster function in the R package scran (Lun et al., 2016). Pool-based size factors were calculated and deconvolved to yield cell-based size factors. The cell-based size factors were normalized between libraries based on the ratio of average UMI counts between libraries using the R package batchelor. The resulting UMI count matrix was log2-transformed and used for downstream analyses.
Macrophages were distinguished from monocytes and dendritic cells based on established combinatorial markers (e.g., ItgaxlowNr4a1highLy6c2high for monocytes and Flt3highLyz2low for dendritic cells). Alveolar macrophages could be separated from interstitial macrophages based on marker genes such as Ear2, Siglecf, and Lpl.
Variance modeling, spectral embedding, and cell projection
To quantitatively find the subset of genes that have significantly high variance between cells, we used the approach implemented in (Ellwanger et al., 2021). Briefly, the variance of each gene was fitted as a function of mean expression. Genes were tested for significance using the R package scran by modeling the residuals with an F-distribution, and genes with a P value <0.05 were considered significant. To retain tissue and FACS-specific variance, the union of all significant variable genes in all sample libraries was used.
Data dimensionality was reduced using principal component analysis (PCA) on the highly variable genes (R package irlba). Library- and animal-specific variations in cellular expression were detected and determined to be the main confounding variables to mask tumor and tdTomato-based variation using the plotExplanatoryVariables function in the R package scater (version 1.0.4). Therefore, Harmony was used to integrate PCA embeddings across library and animal (Korsunsky et al., 2019). After integration, we aimed to increase the signal-to-noise ratio of the spectral embedding. A factor analysis using a scree plot determined the optimal number of components to use for computing a two-dimensional uniform manifold approximation and projection of single cells.
Clustering and cell classification
To cluster cells for cell type and subset identification, a shared nearest neighbor graph was first constructed using the top 40 integrated principal components. The Louvain algorithm in the R package Seurat was used to cluster cells. The optimal resolution parameter for unsupervised clustering was empirically determined using a systematic assessment of clustering robustness provided in the R package chooseR (Patterson-Cross et al., 2021). Briefly, cells were subsampled and iteratively clustered across 20 different resolution parameters, ranging from 0.2 to 12. For each resolution, 100 bootstrap replicates were performed. Next, co-clustering frequencies for each cell were summed at each resolution and used to calculate mean per-cluster silhouette scores. From the distribution of Silhouette scores, a resolution of 1.6 was selected. Clustering resulted in the detection of 33 cell clusters, which could be combined into 16 major cell types.
Differential expression using the Wilcoxon rank sum test with a cutoff of FDR-adjusted P value <0.05 and log2FC >0.25 (FindAllMarkers function in Seurat) was utilized to identify marker genes for each cell. Cells were assigned cell types by matching cluster marker genes to previously reported cell marker genes and by expert annotation.
To manually confirm that minor populations were not missed at this clustering resolution, cells were clustered at resolutions of 1.8 and 2.0 and FindAllMarkers() was applied to corroborate that additional clusters identified did not express unique marker genes.
Differential expression and analysis of functional scores
To calculate differential gene expression between any two populations of cells, we applied MAST (Finak et al., 2015) to evaluate the significance of each gene, using the Bonferroni correction for multiple hypothesis correction. Genes with an adjusted P value < 0.05 and log2-fold change (log2FC) > |0.25| were considered to be differentially expressed.
To score cells using gene sets pertaining to shared cell function, we used the approach reported in Tirosh et al. (2016). We used the log-normalized expression of signature genes to calculate functional scores using the AddModuleScore function in Seurat. Differential expression of functional scores was tested using the Wilcoxon rank sum test in R.
Trajectory inference
For determination of alveolar macrophage, monocyte-to-interstitial macrophage, and classical dendritic cell development and differentiation trajectories, relevant classified cell types were first subsetted. Variable features were selected using the F-distribution (see Variance modeling). Dimensionality reduction and cell embedding were calculated using diffusion maps (Coifman and Lafon, 2006), which resolves both linear and non-linear substructures across cell states in a deterministic manner. Diffusion components were calculated from a cell-by-cell expression matrix of the distance function representing each cell’s relative dissimilarity was first generated. Then diffusion components were calculated using multiple local sigmas in the diffusion kernel as performed in Haghverdi et al. (2015). Correction of batch effects across libraries and animals was performed using Harmony. The number of components for lower-dimensional embedding was selected using a scree plot analysis, and the resulting manifold was visualized to approximate the relationship between cells from the same lineage. Pseudotime values and relative ordering of cell subsets were then assigned to cells using the getLineages function in the R package slingshot (Street et al., 2018).
RNA velocity analysis
RNA velocity was determined to understand the directionality and differentiation potential for cells within lineages. Spliced and unspliced mRNA UMI counts were tallied for the 10X scRNA-seq CellRanger output from each sample library using the python package velocyto (version 0.6). Spliced and unspliced count matrices were then merged across libraries and normalized by library size factors (see Normalization section under scRNA-seq pre-processing). Genes were filtered to select genes expressed in only 3% or more cells for each lineage in both count matrices. The python package scvelo (version 0.2.3) was used to derive the velocity kinetics. First, principal components analysis was performed on the filtered data, and a k-nearest neighbor graph (k = 30) was built using the top 30 principal components to connect similar cells. For each cell, first- and second-order means and uncentered gene variances (moments) were calculated using scvelo.pp.moments, and these moments were passed to scvelo.tl.recover_dynamics to derive splicing kinetics of selected genes. Dynamical modeling of velocity was calculated using scvelo.tl.velocity (mode = “dynamical”), which was then used in scvelo.tl.velocity_graph to construct a velocity graph based the cosine similarities between potential state transitions and velocities. Last of all, velocity streams were visualized from the velocity graph using scvelo.pl.velocity_embedding_stream and embedded onto DiffusionMap cell projections.
TCGA analysis of clinical outcomes
Clinical phenotypes and expression data from the TCGA Clinical Data Resource (Liu et al., 2018) were downloaded using the Diseaseland Explorer (from Qiagen OmicSoft Studio V11.5). To test for the correlation between the PHAT-related gene expression and overall survival (OS) in lung adenocarcinoma (LUAD) and head and neck squamous cell carcinoma (HNSC), a signature score was calculated using the mean mRNA expression values for PHAT genes shared between monocytes, macrophages, and dendritic cells. The mean expression was then divided by the expression of PTPRC to adjust for CD45+ cell levels for each sample. Samples were then grouped into PHAThigh or PHATlow categories based on expression above the 60th (PHAThigh) or below the 40th (PHATlow) percentiles. The Cox proportional hazards model was used in GraphPad Prism 9.3.1 to test for significance between the selected genes and OS after correcting for the effect of gender, race, age, and pathologic stage.
Meta-analyses with published scRNA-seq datasets
For the Sanin et al. (2022) datasets, cluster-defining genes were extracted from supplemental data files 1 and 2. Ribosomal proteins were excluded from the author’s gene lists and were subsequently removed from the interstitial macrophage signature gene list. For the Jaitin et al. (2019) datasets, differentially expressed gene lists provided in dataset S5 and dataset S6 were filtered to remove genes with P value≥0.05. Gene enrichment in the Mac3/LAMs or Mac1/Macs clusters was determined by further filtering by log2 fold change ≥0.25 or log2 fold change ≤0.25, respectively. For both Sanin et al. (2022) and Jaitin et al. (2019) datasets, overlap of the interstitial macrophage signature genes and the cluster-defining genes was determined from the Jaccard index using GeneOverlap (version 1.34.0).
For the Lantz et al. (2020) dataset, normalized and pre-processed single cell data were imported from BioTuring as a Seurat object (Hao et al., 2021). Cells were scored for expression of the PHAT signature using UCell (version 2.2; Andreatta and Carmona, 2021). Statistical significance of signatures scores was determined using the unpaired Wilcoxon test.
PHAT gene signature analysis of human TAMs
Tumor samples from patients with non-small cell lung cancer, with a primary diagnosis of lung adenocarcinoma or lung squamous carcinoma were collected and analyzed as described in Combes et al. (2022). Briefly, myeloid cells were sorted from single-cell suspension of freshly harvested tumors or normal adjacent tissue using the expression of the following protein markers: negative for viability dye; CD45+ CD3− CD19− CD56− HLADR+ CD11b+/−; CD90+ CD44+. Cells were sorted into lysis buffer and mRNA was isolated. Libraries were created and sequencing was performed as described. Median expression of genes comprising PHAT signature; M2 or M1 CIBERSORT signature (Newman et al., 2015) was calculated for each myeloid sorted sample. Statistical analysis used was a one-tailed t test with two-sample unequal variance, tumor vs. normal.
Online supplemental material
Fig. S1 shows the frequency and phenotype of tdTompos macrophages in syngeneic, autochthonous, and in vitro tumor models. Fig. S2 shows the frequency and phenotype of tdTompos macrophages in Adeno-SPCcre autochthonous lung tumors. Fig. S3 shows a summary of quality control metrics for scRNA-seq, including tissue dissections, cell sorting gating strategy, library sample preparation, doublet discrimination, heat map of cell clusters and cell cycle scores. Fig. S4 shows the identification, frequency, and gene expression trajectory of AM and IM subsets in naive lung or dissected autochthonous tumors at the single cell level. Fig. S5 shows the mitochondrial levels in tdTompos or tdTomneg immune cells from AdenoSPcre or syngeneic tumors and the metabolic phenotypes of AMs, IMs, and DCs from AdenoSPCcre autochthonous tumors. Table S1 shows summary of differentially expressed genes between tumor tdTom-pos vs. tumor tdTom-neg cells (Fig. 4 A). Table S2 shows human orthologs of PHAT signature genes. Table S3 shows differential expression results for tumor tdTom-pos alveolar macrophages vs. tumor tdTom-neg alveolar macrophages. Table S4 shows differential expression results for tumor tdTom-pos interstitial macrophages vs. tumor tdTom-neg interstitial macrophages. Table S5 shows differential expression results for tumor tdTom-pos monocytes vs. tumor tdTom-neg monocytes.
Supplementary Material
shows a summary of differentially expressed genes between tumor tdTom-pos vs. tumor tdTom-neg cells (Fig. 4 A).
shows human orthologs of PHAT signature genes.
shows differential expression results for tumor tdTom-pos alveolar macrophages vs. tumor tdTom-neg alveolar macrophages.
shows differential expression results for tumor tdTom-pos interstitial macrophages vs. tumor tdTom-neg interstitial macrophages.
shows differential expression results for tumor tdTom-pos monocytes vs. tumor tdTom-neg monocytes.
Acknowledgments
We thank Stanford Veterinary Animal Care Staff, the Amgen In Vivo Pharmacology department, and the Amgen Flow Cytometry Services department. We thank Tyler Jacks and Tushar Desai for providing mouse lines. We also thank Sarah Pierce for generating the KPT cell lines in the Winslow lab.
M. Yousefi was supported by a National Institutes of Health (NIH) Ruth L. Kirschstein National Research Service Award (NIH F32-CA236311). C.W. Murray was supported by the NSF Graduate Research Fellowship Program and an Anne T. and Robert M. Bass Stanford Graduate Fellowship. E.L. Ashkin was supported by Howard Hughes Medical Institute Gilliam Fellowship for Advanced Study (GT14928). This work was supported in part by NIH R01-CA231253 (to M.M. Winslow) and NIH R01-CA234349 (to M.M. Winslow).
Author contributions: M.A. Gonzalez and D.R. Lu contributed equally to the completion of the manuscript. M.A. Gonzalez and D.R. Lu performed data curation, formal analysis, investigation, methodology, visualization, and writing. M.A. Gonzalez performed all flow cytometry analyses. H. Zhou, V. Arias, and D.R. Lu performed sample preparation for Nanostring and scRNA-seq cell capture and downstream analysis. M Yousefi contributed to methodology and development of autochthonous lung tumor mouse models, data generation and writing. A Kroll contributed analysis to compare signatures across datasets. C.W. Murray, E.L. Ashkin, M.K. Tsai provided technical assistance and editing. C.G. Briseño performed scRNA-seq analysis of DC phenotypes and contributed to editing. J.E.V. Watson performed data curation and analysis for Immunoprofiler dataset and contributed to writing. S. Novitskiy performed methodology and visualization of immunofluorescence and contributed to writing. A.P. Stapper performed QC and data curation for the Immunoprofiler dataset. C.M. Li provided project administration and resources. K.V. Tarbell and M.M. Winslow contributed through study conceptualization, methodology, writing, project administration, and supervision.
Data and availability
scRNA-seq metadata and sequencing files can be found in the Gene Expression Omnibus (accession ID: GSE217368).
References
- A-Gonzalez, N., Bensinger S.J., Hong C., Beceiro S., Bradley M.N., Zelcer N., Deniz J., Ramirez C., Díaz M., Gallardo G., et al. 2009. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 31:245–258. 10.1016/j.immuni.2009.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- A-Gonzalez, N., Quintana J.A., García-Silva S., Mazariegos M., González de la Aleja A., Nicolás-Ávila J.A., Walter W., Adrover J.M., Crainiciuc G., Kuchroo V.K., et al. 2017. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214:1281–1296. 10.1084/jem.20161375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahl, P.J., Hopkins R.A., Xiang W.W., Au B., Kaliaperumal N., Fairhurst A.M., and Connolly J.E.. 2020. Met-Flow, a strategy for single-cell metabolic analysis highlights dynamic changes in immune subpopulations. Commun. Biol. 3:305. 10.1038/s42003-020-1027-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard, B., Panariti A., and Martin J.G.. 2018. Alveolar macrophages in the resolution of inflammation, tissue repair, and tolerance to infection. Front. Immunol. 9:1777. 10.3389/fimmu.2018.01777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta, M., and Carmona S.J.. 2021. UCell: Robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19:3796–3798. 10.1016/j.csbj.2021.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala, M., Gupta P., Gupta S., Dua A., Injeti E., and Mittal A.. 2021. Efficient and modified 2-NBDG assay to measure glucose uptake in cultured myotubes. J. Pharmacol. Toxicol. Methods. 109:107069. 10.1016/j.vascn.2021.107069 [DOI] [PubMed] [Google Scholar]
- Casanova-Acebes, M., Dalla E., Leader A.M., LeBerichel J., Nikolic J., Morales B.M., Brown M., Chang C., Troncoso L., Chen S.T., et al. 2021. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature. 595:578–584. 10.1038/s41586-021-03651-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassetta, L., and Pollard J.W.. 2018. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17:887–904. 10.1038/nrd.2018.169 [DOI] [PubMed] [Google Scholar]
- Coifman, R.R., and Lafon S.. 2006. Diffusion maps. Appl. Comput. Harmon. Anal. 21:5–30. 10.1016/j.acha.2006.04.006 [DOI] [Google Scholar]
- Combes, A.J., Samad B., Tsui J., Chew N.W., Yan P., Reeder G.C., Kushnoor D., Shen A., Davidson B., Barczak A.J., and Immunoprofiler Consortium. et al. 2022. Discovering dominant tumor immune archetypes in a pan-cancer census. Cell. 185:184–203.e19. 10.1016/j.cell.2021.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim, T.Q., Lee E., Merriott D.J., Goulbourne C.N., Cheng J., Cheng A., Gonen A., Allen R.M., Palladino E.N.D., Ford D.A., et al. 2017. ABCG1 regulates pulmonary surfactant metabolism in mice and men. J. Lipid Res. 58:941–954. 10.1194/jlr.M075101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadon, M., Torzilli G., Cortese N., Soldani C., Di Tommaso L., Franceschini B., Carriero R., Barbagallo M., Rigamonti A., Anselmo A., et al. 2020. Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis. J. Exp. Med. 217:e20191847. 10.1084/jem.20191847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwanger, D.C., Wang S., Brioschi S., Shao Z., Green L., Case R., Yoo D., Weishuhn D., Rathanaswami P., Bradley J., et al. 2021. Prior activation state shapes the microglia response to antihuman TREM2 in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 118:e2017742118. 10.1073/pnas.2017742118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming, S.A., Murray P.J., and Pearce E.J.. 2021. Metabolic orchestration of the wound healing response. Cell Metab. 33:1726–1743. 10.1016/j.cmet.2021.07.017 [DOI] [PubMed] [Google Scholar]
- Finak, G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. 2015. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16:278. 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessain, G., Blériot C., and Ginhoux F.. 2020. Non-genetic heterogeneity of macrophages in diseases-A medical perspective. Front. Cell Dev. Biol. 8:613116. 10.3389/fcell.2020.613116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S.R., Maute R.L., Dulken B.W., Hutter G., George B.M., McCracken M.N., Gupta R., Tsai J.M., Sinha R., Corey D., et al. 2017. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 545:495–499. 10.1038/nature22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams, M., Thierry G.R., Bonnardel J., and Bajenoff M.. 2020. Establishment and maintenance of the macrophage niche. Immunity. 52:434–451. 10.1016/j.immuni.2020.02.015 [DOI] [PubMed] [Google Scholar]
- Gundra, U.M., Girgis N.M., Gonzalez M.A., San Tang M., Van Der Zande H.J.P., Lin J.D., Ouimet M., Ma L.J., Poles J., Vozhilla N., et al. 2017. Vitamin A mediates conversion of monocyte-derived macrophages into tissue-resident macrophages during alternative activation. Nat. Immunol. 18:642–653. 10.1038/ni.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi, L., Buettner F., and Theis F.J.. 2015. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 31:2989–2998. 10.1093/bioinformatics/btv325 [DOI] [PubMed] [Google Scholar]
- Hanayama, R., Tanaka M., Miyasaka K., Aozasa K., Koike M., Uchiyama Y., and Nagata S.. 2004. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 304:1147–1150. 10.1126/science.1094359 [DOI] [PubMed] [Google Scholar]
- Hao, Y., Hao S., Andersen-Nissen E., Mauck W.M. III, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184:3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herber, D.L., Cao W., Nefedova Y., Novitskiy S.V., Nagaraj S., Tyurin V.A., Corzo A., Cho H.I., Celis E., Lennox B., et al. 2010. Lipid accumulation and dendritic cell dysfunction in cancer. Nat. Med. 16:880–886. 10.1038/nm.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S.C., Everts B., Ivanova Y., O’Sullivan D., Nascimento M., Smith A.M., Beatty W., Love-Gregory L., Lam W.Y., O’Neill C.M., et al. 2014. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15:846–855. 10.1038/ni.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, E.L., Olive K.P., Tuveson D.A., Bronson R., Crowley D., Brown M., and Jacks T.. 2005. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65:10280–10288. 10.1158/0008-5472.CAN-05-2193 [DOI] [PubMed] [Google Scholar]
- Jackson, E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., and Tuveson D.A.. 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15:3243–3248. 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin, D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H., Lundgren P., Bleriot C., Liu Z., Deczkowska A., et al. 2019. Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. Cell. 178:686–698.e14. 10.1016/j.cell.2019.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, A.K., Huang S.C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K.M., Ashall J., Everts B., et al. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 42:419–430. 10.1016/j.immuni.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Wei W., Yang M., Du Y., and Wan Y.. 2014. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 20:483–498. 10.1016/j.cmet.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere, A.I., Smeitink J.A., and Rodenburg R.J.. 2012. Mitochondrial ATP synthase: Architecture, function and pathology. J. Inherit. Metab. Dis. 35:211–225. 10.1007/s10545-011-9382-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers, J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., and Berns A.. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 29:418–425. 10.1038/ng747 [DOI] [PubMed] [Google Scholar]
- Katzenelenbogen, Y., Sheban F., Yalin A., Yofe I., Svetlichnyy D., Jaitin D.A., Bornstein C., Moshe A., Keren-Shaul H., Cohen M., et al. 2020. Coupled scRNA-seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell. 182:872–885.e19. 10.1016/j.cell.2020.06.032 [DOI] [PubMed] [Google Scholar]
- Keren-Shaul, H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Koo, S.J., and Garg N.J.. 2019. Metabolic programming of macrophage functions and pathogens control. Redox Biol. 24:101198. 10.1016/j.redox.2019.101198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky, I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., and Raychaudhuri S.. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 16:1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczanowsky, F., Raker V., Graulich E., Domogalla M.P., and Steinbrink K.. 2016. IL-10-modulated human dendritic cells for clinical use: Identification of a stable and migratory subset with improved tolerogenic activity. J. Immunol. 197:3607–3617. 10.4049/jimmunol.1501769 [DOI] [PubMed] [Google Scholar]
- Lantz, C., Radmanesh B., Liu E., Thorp E.B., and Lin J.. 2020. Single-cell RNA sequencing uncovers heterogenous transcriptional signatures in macrophages during efferocytosis. Sci. Rep. 10:14333. 10.1038/s41598-020-70353-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionova, I., Tuguzbaeva G., Ponomaryova A., Stakheyeva M., Cherdyntseva N., Pavlov V., Choinzonov E., and Kzhyshkowska J.. 2020. Tumor-associated macrophages in human breast, colorectal, lung, ovarian and prostate cancers. Front. Oncol. 10:566511. 10.3389/fonc.2020.566511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin, Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., and Amit I.. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 159:1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Yu J., Huber A., Kryczek I., Wang Z., Jiang L., Li X., Du W., Li G., Wei S., et al. 2022. Metabolism drives macrophage heterogeneity in the tumor microenvironment. Cell Rep. 39:110609. 10.1016/j.celrep.2022.110609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V.. et al. 2018. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 173:400–416.e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke, P., and Lin J.D.. 2022. Redefining inflammatory macrophage phenotypes across stages and tissues by single-cell transcriptomics. Sci. Immunol. 7:eabo4652. 10.1126/sciimmunol.abo4652 [DOI] [PubMed] [Google Scholar]
- Lun, A.T., Bach K., and Marioni J.C.. 2016. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17:75. 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Garner A.R., Shimaoka D., Chuong A.S., Klapoetke N.C., Li L., van der Bourg A., Niino Y., Egolf L., Monetti C., et al. 2015. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 85:942–958. 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, B., Leader A.M., Chen S.T., Tung N., Chang C., LeBerichel J., Chudnovskiy A., Maskey S., Walker L., Finnigan J.P., et al. 2020. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 580:257–262. 10.1038/s41586-020-2134-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, S., Vooijs M., van Der Gulden H., Jonkers J., and Berns A.. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14:994–1004. 10.1101/gad.14.8.994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, C.S., Murrow L.M., and Gartner Z.J.. 2019. DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8:329–337.e4. 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, M.M., Weinberg S.E., and Chandel N.S.. 2017. Mitochondrial control of immunity: Beyond ATP. Nat. Rev. Immunol. 17:608–620. 10.1038/nri.2017.66 [DOI] [PubMed] [Google Scholar]
- Metchnikoff, E.J.B.Z. 1883. Untersuchungen über die mesodermalen Phagocyten einiger Wirbeltiere. Biol. Zentralblatt. 3:560–565. [Google Scholar]
- Mosser, D.M., and Edwards J.P.. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958–969. 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan, L., Odegaard J.I., Morel C.R., Heredia J.E., Mwangi J.W., Ricardo-Gonzalez R.R., Goh Y.P., Eagle A.R., Dunn S.E., Awakuni J.U., et al. 2009. PPAR-δ senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 15:1266–1272. 10.1038/nm.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder, K., Patel, A.A., Kong, W.T., Piot, C., Halitzki, E., Dunsmore, G., et al. Ginhoux, F.. 2021. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. 54:1883–1900 e1885. 10.1016/j.immuni.2021.07.007 [DOI] [PubMed] [Google Scholar]
- Murphy, T.L., Grajales-Reyes G.E., Wu X., Tussiwand R., Briseño C.G., Iwata A., Kretzer N.M., Durai V., and Murphy K.M.. 2016. Transcriptional control of dendritic cell development. Annu. Rev. Immunol. 34:93–119. 10.1146/annurev-immunol-032713-120204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness, S., Lin S., and Gordon J.R.. 2021. Regulatory dendritic cells, T cell tolerance, and dendritic cell therapy for immunologic disease. Front. Immunol. 12:633436. 10.3389/fimmu.2021.633436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., and Alizadeh A.A.. 2015. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 12:453–457. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson-Cross, R.B., Levine A.J., and Menon V.. 2021. Selecting single cell clustering parameter values using subsampling-based robustness metrics. BMC Bioinformatics. 22:39. 10.1186/s12859-021-03957-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., and Hogan B.L.. 2011. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA. 108:E1475–E1483. 10.1073/pnas.1117988108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszer, T., Menéndez-Gutiérrez M.P., Lefterova M.I., Alameda D., Núñez V., Lazar M.A., Fischer T., and Ricote M.. 2011. Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator-activated receptor γ or retinoid X receptor α deficiency. J. Immunol. 186:621–631. 10.4049/jimmunol.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanin, D.E., Ge Y., Marinkovic E., Kabat A.M., Castoldi A., Caputa G., Grzes K.M., Curtis J.D., Thompson E.A., Willenborg S., et al. 2022. A common framework of monocyte-derived macrophage activation. Sci. Immunol. 7:eabl7482. 10.1126/sciimmunol.abl7482 [DOI] [PubMed] [Google Scholar]
- Schulz, D., Severin Y., Zanotelli V.R.T., and Bodenmiller B.. 2019. In-depth characterization of monocyte-derived macrophages using a mass cytometry-based phagocytosis assay. Sci. Rep. 9:1925. 10.1038/s41598-018-38127-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone, A., Natarajan K.N., Saraiva L.R., Proserpio V., Teichmann S.A., Stegle O., Marioni J.C., and Buettner F.. 2015. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 85:54–61. 10.1016/j.ymeth.2015.06.021 [DOI] [PubMed] [Google Scholar]
- Scott, R.S., McMahon E.J., Pop S.M., Reap E.A., Caricchio R., Cohen P.L., Earp H.S., and Matsushima G.K.. 2001. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 411:207–211. 10.1038/35075603 [DOI] [PubMed] [Google Scholar]
- Singh, M., Murriel C.L., and Johnson L.. 2012. Genetically engineered mouse models: Closing the gap between preclinical data and trial outcomes. Cancer Res. 72:2695–2700. 10.1158/0008-5472.CAN-11-2786 [DOI] [PubMed] [Google Scholar]
- Stoeckius, M., Zheng S., Houck-Loomis B., Hao S., Yeung B.Z., Mauck W.M. III, Smibert P., and Satija R.. 2018. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19:224. 10.1186/s13059-018-1603-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street, K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., and Dudoit S.. 2018. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 19:477. 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, K.D., Song J.Y., Kwon M.C., Proost N., Zevenhoven J., and Berns A.. 2014. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc. Natl. Acad. Sci. USA. 111:4952–4957. 10.1073/pnas.1319963111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas, I., and Bornfeldt K.E.. 2016. Macrophage phenotype and function in different stages of atherosclerosis. Circ. Res. 118:653–667. 10.1161/CIRCRESAHA.115.306256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippimanchai, D.D., Nolan K., Poczobutt J., Verzosa G., Li H., Scarborough H., Huang J., Young C., DeGregori J., Nemenoff R.A., and Malkoski S.P.. 2018. Adenoviral vectors transduce alveolar macrophages in lung cancer models. OncoImmunology. 7:e1438105. 10.1080/2162402X.2018.1438105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh, I., Izar B., Prakadan S.M., Wadsworth M.H. II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352:189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tur, J., Vico T., Lloberas J., Zorzano A., and Celada A.. 2017. Macrophages and mitochondria: A critical interplay between metabolism, signaling, and the functional activity. Adv. Immunol. 133:1–36. 10.1016/bs.ai.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Tuveson, D.A., Shaw A.T., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., et al. 2004. Endogenous oncogenic K-rasG12D stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 5:375–387. 10.1016/s1535-6108(04)00085-6 [DOI] [PubMed] [Google Scholar]
- Uribe-Querol, E., and Rosales C.. 2020. Phagocytosis: Our current understanding of a universal biological process. Front. Immunol. 11:1066. 10.3389/fimmu.2020.01066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola, A., Munari F., Sánchez-Rodríguez R., Scolaro T., and Castegna A.. 2019. The metabolic signature of macrophage responses. Front. Immunol. 10:1462. 10.3389/fimmu.2019.01462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wang Y., Chu Y., Li Z., Yu X., Huang Z., Xu J., and Zheng L.. 2021. Tumor-derived adenosine promotes macrophage proliferation in human hepatocellular carcinoma. J. Hepatol. 74:627–637. 10.1016/j.jhep.2020.10.021 [DOI] [PubMed] [Google Scholar]
- Watanabe, S., Alexander M., Misharin A.V., and Budinger G.R.S.. 2019. The role of macrophages in the resolution of inflammation. J. Clin. Invest. 129:2619–2628. 10.1172/JCI124615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wculek, S.K., Dunphy G., Heras-Murillo I., Mastrangelo A., and Sancho D.. 2022. Metabolism of tissue macrophages in homeostasis and pathology. Cell. Mol. Immunol. 19:384–408. 10.1038/s41423-021-00791-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt, J. and Porse B.. (2008). Bone marrow-derived macrophages (BMM): Isolation and applications. CSH protoc. 2008:prot5080. 10.1101/pdb.prot5080 [DOI] [PubMed] [Google Scholar]
- Xia, H., Li S., Li X., Wang W., Bian Y., Wei S., Grove S., Wang W., Vatan L., Liu J.R., et al. 2020. Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. JCI Insight. 5:e141115. 10.1172/jci.insight.141115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J., and Horng T.. 2020. Lipid metabolism in regulation of macrophage functions. Trends Cell Biol. 30:979–989. 10.1016/j.tcb.2020.09.006 [DOI] [PubMed] [Google Scholar]
- Yin, C., and Heit B.. 2021. Cellular responses to the efferocytosis of apoptotic cells. Front. Immunol. 12:631714. 10.3389/fimmu.2021.631714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona, S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, N.P., Crowley D., and Jacks T.. 2011. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 71:4040–4047. 10.1158/0008-5472.CAN-10-4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.R., O’Koren E.G., Hotten D.F., Kan M.J., Kopin D., Nelson E.R., Que L., and Gunn M.D.. 2016. A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PLoS One. 11:e0150606. 10.1371/journal.pone.0150606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurdagul, A., Jr. 2021. Metabolic consequences of efferocytosis and its impact on atherosclerosis. Immunometabolism. 3:e210017. 10.20900/immunometab20210017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Weinberg S., DeBerge M., Gainullina A., Schipma M., Kinchen J.M., Ben-Sahra I., Gius D.R., Yvan-Charvet L., Chandel N.S., et al. 2019. Efferocytosis fuels requirements of fatty acid oxidation and the electron transport chain to polarize macrophages for tissue repair. Cell Metab. 29:443–456.e5. 10.1016/j.cmet.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F., Xiao C., Evans K.S., Theivanthiran T., DeVito N., Holtzhausen A., Liu J., Liu X., Boczkowski D., Nair S., et al. 2018. Paracrine Wnt5a-β-Catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity. 48:147–160.e7. 10.1016/j.immuni.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Herndon J.M., Sojka D.K., Kim K.W., Knolhoff B.L., Zuo C., Cullinan D.R., Luo J., Bearden A.R., Lavine K.J., et al. 2017. Tissue-resident macrophages in pancreatic Ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity. 47:323–338.e6. 10.1016/j.immuni.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows a summary of differentially expressed genes between tumor tdTom-pos vs. tumor tdTom-neg cells (Fig. 4 A).
shows human orthologs of PHAT signature genes.
shows differential expression results for tumor tdTom-pos alveolar macrophages vs. tumor tdTom-neg alveolar macrophages.
shows differential expression results for tumor tdTom-pos interstitial macrophages vs. tumor tdTom-neg interstitial macrophages.
shows differential expression results for tumor tdTom-pos monocytes vs. tumor tdTom-neg monocytes.
Data Availability Statement
scRNA-seq metadata and sequencing files can be found in the Gene Expression Omnibus (accession ID: GSE217368).