Abstract
Phenylketonuria is an inborn error of phenylalanine metabolism caused by a phenylalanine hydroxylase deficiency. To prevent the occurrence of neurological symptoms and maternal complications resulting from phenylketonuria, patients must adhere to a strict diet therapy, tetrahydrobiopterin supplementation, or pegvaliase injection to maintain blood phenylalanine levels within a recommended range throughout their lives. Therefore, monitoring blood phenylalanine levels is necessary to determine the recent metabolic status of phenylalanine in patients with PKU; however, there are no available instruments for individuals to monitor their own blood phenylalanine levels using whole fingertip blood. We developed a phenylalanine monitoring system (designated as PheCheck) that included a pre-existing portable ammonia detection device and phenylalanine ammonia-lyase, which converts phenylalanine to trans-cinnamic acid and ammonia. This system was able to remove 86.7% ± 0.03% of the ammonia contained in fingertip blood and successfully reduce background ammonia levels. A good correlation was found between the estimated plasma phenylalanine levels detected by PheCheck and plasma phenylalanine levels detected by high-performance liquid chromatography (R2 0.97). The entire PheCheck process for measuring blood phenylalanine takes only 20 min. PheCheck can lay the foundation for home phenylalanine monitoring with high feasibility because all the components are easily accessible. Further studies with a more user-friendly PheCheck optimized for practice are needed to improve blood phenylalanine control, reduce the burden on patients and/or caregivers, and prevent the sequelae associated with phenylketonuria.
Keywords: Phenylketonuria, Phenylalanine, Point-of-care testing, Phenylalanine self-monitoring, Phenylalanine ammonia-lyase
Highlights
-
•
Blood Phe monitoring is essential to manage treatment for patients with phenylketonuria.
-
•
Instruments are not available for individuals to monitor their own blood Phe levels using whole fingertip blood.
-
•
A portable ammonia detection device and phenylalanine ammonia-lyase can estimate plasma Phe levels using fingertip blood.
-
•
The point-of-care system (designated as PheCheck) can lay the foundation for home Phe monitoring with high feasibility.
1. Introduction
Phenylketonuria (PKU, MIM# 261600) is an inborn error of phenylalanine metabolism in which phenylalanine hydroxylase (EC 1.14.16.1) is defective. This results in increased phenylalanine (Phe) levels in the body [1]. The sustained, elevated Phe levels following birth causes white matter disruption and neurological sequelae include severe developmental delay and epilepsy [2,3]. A newborn screening program identified individuals with PKU to prevent irreversible complications [4]. Even after the critical period of brain development, an improper diet in adult PKU patients may induce neurological symptoms, such as dementia, visual abnormalities, ataxia, tremor, and/or sensory manifestations [5]. Maternal PKU should also be monitored to avoid fetal complications during the pregnancy in female PKU patients [6]. The well-established surrogate biomarker blood Phe is used for the management of PKU [[6], [7], [8], [9]]. Along with monitoring blood Phe, diet conditioning, which consists of both phenylalanine restriction and other amino acid supplementation, can prevent the devastating complications of PKU, except in patients with the effective therapeutic effects of tetrahydrobiopterin or pegvaliase [10,11]. Monitoring blood Phe levels and adjusting diet requires frequent blood sampling throughout a patient's life, and hospital visits are expensive and time-consuming [12]. Sending dried blood samples collected at home could remedy this inconvenience; however, the time lag between sampling and receiving blood Phe data can occur. Therefore, a convenient, immediate, and reliable point-of-care testing (POCT) system to measure blood Phe levels is hoped for the management of PKU. Many studies [[13], [14], [15], [16], [17], [18], [19], [20]] have successfully established quantitative techniques for measuring blood Phe levels; however, practical POCT instruments for self-monitoring do not exist [17]. For instance, the developmental cost, which is inevitable for new equipment, remains an obstacle.
To resolve the hurdles, we focused on phenylalanine ammonia-lyase (PAL, EC 4.3.1.24), which exists in plants, fungi, and bacteria, does not require a cofactor, and catalyzes the conversion of Phe to trans-cinnamic acid and ammonia (Fig. 1A) [21]. The ammonia produced from blood Phe by PAL can be detected with an ammonia detecting device. Because ammonia and Phe are equivalent in molar amounts, Phe levels in blood may be quantified. Although this concept is not a novel idea and previous studies successfully demonstrated this proof-of-principle [22], high levels of intrinsic ammonia in fingertip blood would influence the results and have prevented the achievement of Phe home monitoring system utilizing PAL until now. In this study, we describe the development of a fundamental system capable of realizing POCT, by which fingertip blood Phe levels can be easily measured using a PAL solution and a portable ammonia detecting system with a marked reduction of blood ammonia.
Fig. 1.
Recombinant PAL solution to measure blood Phe levels. (A) The reaction of PAL. (B) Coomassie brilliant blue staining of recombinant PAL. Arrow indicates the expected position of PAL. (C) Michaelis–Menten curve for recombinant PAL. (D) The relationship between phenylalanine concentration in whole blood and the ammonia produced in three healthy volunteers. (E) PAL enzyme activity pre- or post-freeze-drying. PAL, phenylalanine ammonia-lyase.
2. Materials and methods
2.1. Recombinant PAL preparation
Recombinant PAL protein was expressed and purified as previously described with minor modifications [23]. A plasmid containing pal cDNA was obtained (78,286, Addgene, Cambridge, MA, USA) and the cDNA was transferred into the pET21d(+) vector (69,743, Novagen Merck Millipore, Darmstadt, Germany) using an In-Fusion HD Cloning kit (TaKaRa, Shiga, Japan). A His6 tag was added to the C-terminus as previous studies showed decreased enzyme activity of N-terminal His-tagged PAL [24]. Sanger sequencing identified six variants in the construct including 149A > C (p.Glu50Asp), 465C > A (p.Ser155Ser), and 1158G > T (p.Pro386Pro) in the coding sequencing. One T to C substitution at the 10th base from the last base of the F1 ori, one C deletion at the next base from the last base of the F1 ori, and a TAT deletion in the BOM site. The PAL protein was expressed in the HMS174(DE3) E. coli strain (69,453, Novagen Merck Millipore, Darmstadt, Germany). The cells were cultured in LB medium containing 100 μg/mL ampicillin and 40 μg/mL rifampicin. The flask was shaken at 37 °C and 250 rpm until the OD600 reached 0.6–1.0. PAL protein expression was induced by the addition of 1 mM isopropyl thio-α-D-galactopyranoside (cat# 9030, TaKaRa, Shiga, Japan). Secondary culture was done for 15–20 h at 25 °C and 250 rpm. The pellet was collected by centrifugation at 10,000 rpm for 10 min and eluted with BugBuster protein extraction reagent (cat# 70584, Novagen Merck Millipore, Darmstadt, Germany). The soluble fraction included the tagged PAL protein, which was collected using a HisSpinTrap (cat# 28401353, GE Healthcare, Buckinghamshire, UK) and dialyzed overnight in 50 mM Tris-HCl, pH 8.8. The dialyzed solution was concentrated with an Amicon Ultra filtration unit (UFC503096, Merck Millipore, Billerica, MA, USA).
2.2. PAL solution
PAL solution consisted of PAL enzyme protein (1–3 mg/mL), 50 mM Tris-HCl, 154 mM Na, and 0.1% (w/v) azide. The 5-fold concentrated solution was stored at −20 °C and was diluted before using. The enzymatic activity was determined based on the maximum amount of Phe per assay, which was 30 nmol in 15 μL of whole blood containing 2 mM Phe, and the minimum amount of enzyme was 3 mU per 5 μL after a 10-min incubation. The molecular weight of PAL to calculate Kcat was 79,999. The PAL solution was combined with 10 mg/mL trehalose and lyophilized using an FDU-2200 (Eyela, Tokyo, Japan) according to the manufacturer's instructions overnight at −80 °C. Lyophilized PAL was stored at room temperature and was reconstituted with 50 mM Tris-HCl each time immediately before performing the assay.
2.3. Measurement of PAL enzyme kinetic parameters
PAL enzyme activity was determined based on a previous report [25]. First, 96 μL of 30 mM Phe solution was added to a 96-well plate and incubated at 30 °C for 6 min. Next, 4 μL of PAL enzyme solution was added and the absorbance at 275 nm was immediately measured every 5 s at 30 °C. Specific activity was calculated based on the O.D. at 275 nm for 80 s using a SpectraMax M2e (Molecular Devices, CA, USA). The path length was set at 0.28 cm. Molar absorption coefficients for trans-cinnamic acid and p-hydroxycinnamic acid were 17.212 and 13.426 mM−1 cm−1, respectively.
2.4. Measurement of blood phenylalanine levels
A total of 18 patients (14 classical PKU and 4 BH4-responsive hyperphenylalaninemia) were included. Blood samples from patients and healthy volunteers were collected with informed consent. Plasma Phe levels were determined using high-performance liquid chromatography (HPLC) at BML, Inc. (Tokyo, Japan). The plasma was deproteinized, separated by HPLC, and incubated with ninhydrin reagent using an L-8900 automated amino acid analyzer (Hitachi High-Technologies, Tokyo, Japan). Dried blood spot (DBS) Phe levels were measured by tandem mass spectrometry (TQD, Waters, Manchester, UK). The precursor ion (m/z), fragment ion (m/z), Dwell (s), Cone (V), and Collision (V) of Phe were set to 166.1, 120.1, 0.050, 20, 15, respectively. Ammonia concentrations were measured with a PocketChem Ammonia Checker II (Arkray Inc., Kyoto, Japan) and a PocketChem BA PA-4140 (Arkray Inc., Kyoto, Japan).
2.5. Data analysis and visualization
Enzyme kinetics, statistical analysis, and visualization were done using Prism 8 for MacOS (Dotmatics, San Diego, CA, USA). Chemical structures were generated using MarvinSketch 20.11 (ChemAxon Ltd., Budapest, Hungary).
3. Results
3.1. Combining a recombinant PAL solution with a portable ammonia detection system to measure blood Phe levels
Recombinant PAL enzyme was produced using the pET system (Fig. 1B). The C-terminal His-tagged PAL exhibited Phe deamination activity (Fig. 1C). The kinetic parameters of the recombinant PAL for Phe were as follows: Km 184.1 μM (95%CI 136.0–248.2), Vmax 1.543 μmol/min*mg (95%CI 1.443–1.649), Kcat 1.23*105 /s, and Kcat/Km 670.5 /μM*s. PAL enzymatic activity for tyrosine may be ignored as the lyase activity for tyrosine was faint (Km 3947 μM (95%CI 1462–infinity), Vmax 0.2467 μmol/min*mg (95%CI 0.1301–infinity). To confirm that blood Phe levels can be quantified, venous whole blood from three healthy volunteers was serially diluted with Phe and assayed using a portable ammonia detection system with a PAL solution. The ammonia levels and added Phe concentration exhibited good linearity (R2 were 0.99, 0.97, and 0.99 from left to right in Fig. 1D). Furthermore, to create a PAL solution with a long shelf life, freeze-drying was performed, because freezing PAL solutions often, but not always, caused aggregation of PAL-containing elements and inactivation of the enzyme. Lyophilization retained 63.2% of PAL enzyme activity after 6 weeks compared with the pre-freeze-dried sample (Fig. 1E).
3.2. Simple manipulation to remove most ammonia from blood and a POCT for Phe using a portable ammonia detection system and a PAL solution
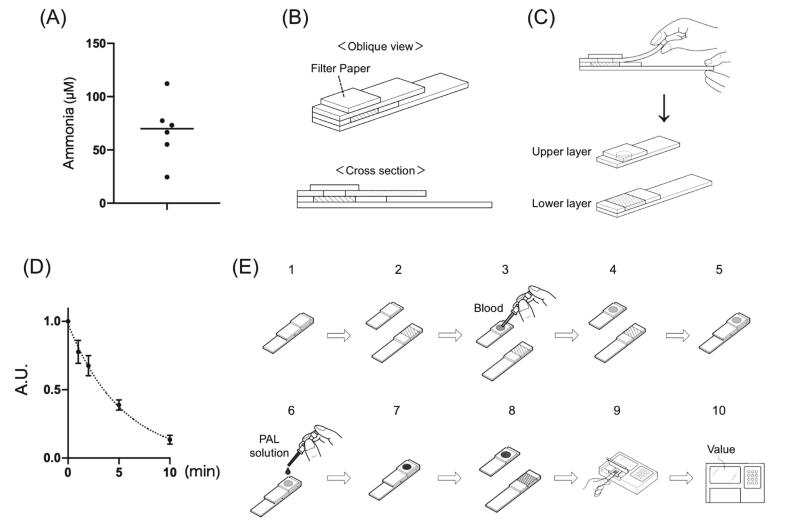
Next, we quantified blood Phe levels using a PAL solution and a portable ammonia detection system. Fingertip blood is suitable for home Phe detection in PKU patients; however, as previously reported [26], high ammonia levels are present in the fingertip capillary blood of healthy volunteers (Fig. 2A). This blood ammonia must be easily removed and as much as possible to facilitate accurate Phe measurements. To resolve this problem, we focused on the components of the ammonia detection kit (Fig. 2B). This kit involves the following steps: blood sampling, applying the blood to a piece of filter paper, incubating for 180 s, tearing off the upper layer, and detecting changes in the indicators marked as diagonal lines (Fig. 2C). The filter paper used for sample retention contains boric acid to evaporate blood ammonium ions as ammonia gas. To exclude intrinsic ammonia from the blood, we applied blood samples onto the filter, which was already separated in the upper and lower layers. A 10-min incubation after spotting removed 86.7 ± 0.03% of the ammonia from the blood (Fig. 2D). This method for reducing ammonia levels in blood samples satisfied the background requirements and was convenient. The resultant test process was established as follows (Fig. 2E). The upper and lower layers of the test strip were ripped off (2E-1, 2), 15 μL of fingertip blood was applied to the filter (2E-3) and incubated for 10 min (2E-4), the layers were overlapped as before (2E-5), and 5 μL of PAL solution was applied to the filter (2E-6) and incubated for 10 min (2E-7). Finally, the upper layer of the test strip was removed (2E-8) and placed on the ammonia meter (2E-9, 10). We named this system “PheCheck.”
Fig. 2.
PheCheck system for fingertip blood Phe levels. (A) Ammonia concentration in the fingertip blood of volunteers. (B) Diagram of the test strip from the Ammonia Test Kit II. (C) The ripping procedure in the test strip composed of an upper and lower layer. The filter paper in the upper layer has a hole for ammonia gas to pass through the upper layer and react with the indicator marked as diagonal lines on the lower layer. (D) Ammonia changes in the filter paper after blood spotting and incubation over time. Each bar represents the mean ± SD. (E) Step-by-step procedure of the PheCheck system.
3.3. PheCheck accurately quantifies Phe in fingertip capillary blood
To quantify blood Phe levels using PheCheck, a standard curve was generated. It was difficult to collect a large number of plasma samples representing several Phe concentrations; thus, a two-step approach was performed. First, the relationship between Phe in the plasma and fingertip blood of PKU patients was assessed. Similar to a previous report [27], an excellent linear correlation was observed between plasma Phe levels determined via HPLC and fingertip blood Phe levels with tandem mass spectrometry (Fig. 3A, R2 0.99). A positive relationship between Phe levels detected by tandem mass spectrometry and those detected via PheCheck using fingertip blood was observed (Fig. 3B, Y = 0.5007*X + 50.48). The combination of these data resulted in the following equation to estimate plasma Phe levels: (plasma Phe levels) = 2.2875 × (PheCheck values) − 100.87. Next, we estimated plasma Phe levels in PKU patients using PheCheck. The results demonstrated that the plasma Phe levels estimated by PheCheck was accurately correlated with plasma Phe levels determined via HPLC (85.6–460.6 μM) (Fig. 3C, R2 0.97). Considering that the LOD of the ammonia meter was 235 μM, the LOD of the PheCheck system can be estimated to be 437 μM in plasma Phe, which encompasses the recommended plasma Phe levels for almost all PKU patients, including maternal PKU.
Fig. 3.
Quantification of blood Phe levels with PheCheck. Association of (A) Phe levels between plasma and fingertip capillary blood, (B) Phe levels in fingertip capillary blood and PheCheck levels, and (C) plasma Phe levels and estimated plasma Phe levels based on the regression equation in (A) and (B). Spots in each figure were quantified with simultaneously sampled blood. Phe, phenylalanine.
4. Discussion
This study showed that the PheCheck, which combines a PAL solution and a pre-existing portable ammonia detection system, can precisely estimate plasma Phe levels in fingertip blood. Although intrinsic ammonia in fingertip capillary blood hampered the direct use of the ammonia detection system and PAL, a step was introduced to apply blood and incubate on a separated layer for removal of this background ammonia. PheCheck has the potential to provide PKU patients with a home Phe monitoring system that allows them to check their own blood Phe levels and adjust their diet on their own.
A POCT to monitor one's own blood Phe at home has been needed for a long time. [[28], [29], [30]] One major issue in the development of the POCT is cost, which is an inevitable problem for the development of novel medical devices. The developmental cost of equipment and consumables may not be recouped, especially for rare diseases [31]. The PheCheck system did not require a significant development cost. It also has several advantages, such as the pocket-size of the instrument, easy to handle and operate, and the short time required for sample preparation and measurement (approximately 20 min); however, the more user-friendly type of PheCheck should be desired because the current PheCheck system has some difficulties regarding the operation for those unfamiliar with it. A more convenient and easier-to-use version of PheCheck should be developed so that patients with PKU or their families can conveniently perform the test at home.
PheCheck may help manage the dietary needs of PKU patients. Currently, the primary method to obtain blood Phe levels involves DBS sampling at home and mailing the samples to a lab. Although some steps may reduce the time lag [32], a few days are still needed to transfer and process samples for measuring Phe levels. However, the interval between sampling and obtaining results may affect a patient's judgment to increase or decrease dietary protein amounts. The PheCheck system provides blood Phe levels in only 20 min. This rapid test not only enables dietary adjustments but also allows one to avoid any excessive dietary restrictions. Thus, the ideal diet for a PKU patient can be implemented regardless of location or timing. Furthermore, quality of life of PKU patients and their families will improve because of the reduced number of visits to clinics or hospitals [29]. The psychological stress will also be reduced by avoiding venipuncture. Clinical studies are needed to demonstrate the abovementioned advantages and predicted outcomes with an improved version of PheCheck.
There are some limitations to the PheCheck. First, the PAL solution for PheCheck may not be available in commercial packages. Second, a more suitable mixture or condition should be evaluated to extend the shelf life of the lyophilized PAL or PAL solution. Third, the measurable range of blood Phe should be extended. In this study, the PheCheck system can detect up to 437 μM in plasma Phe levels, which does not fully cover the levels presented by older or uncontrolled PKU patients [33,34]. Although blood sample dilution could improve the measurement sensitivity according to the dilution factor (data not shown), the added manipulation would reduce its usability. Other ingenuity should be required to extend the dynamic range. Fourth, home Phe monitoring should be covered by health insurance for daily use of PheCheck. Lastly, it is unknown whether the PheCheck system can reflect the correct Phe levels in the blood of patients treated with pegvaliase, which is a recombinant PEGylated PAL and a drug for enzyme substitution therapy for PKU approved in the Food and Drug Administration and the European Medicines Agency [[35], [36], [37]]. Blood pegvaliase may not significantly affect the Phe levels in the PheCheck system because the enzyme activity of the blood pegvaliase is estimated using data from previous reports [38,39] may be equivalent to 1–3% of PAL contained in the solution of PheCheck. The influence of blood pegvaliase should be assessed with clinical samples from patients treated with pegvaliase.
5. Conclusions
We developed the PheCheck system with a PAL solution and a pre-existing portable ammonia detection system, along with a little ingenuity. Although PheCheck has some obstacles to overcome, this study provides an outline of POCTs to determine blood Phe levels at a low cost. A revised PheCheck system with improved convenience and measurement range would realize a home Phe monitoring system in the future. The development of PheCheck warrants further research to investigate the clinical usefulness and effectiveness of Phe self-monitoring.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or non-for-profit sectors.
Author contributions
Conceptualization: YW, SK. Methodology: YW, SK. Validation: YW, ET, YM-S, SK. Formal analysis: YW, ET, YM-S. Investigation: YW, ET, YM-S. Resources : YW, ET, YM-S, AK, TM, SK. Writing - Original Draft: YW, ET. Writing - Review & Editing: YW, ET, YM-S, AK, TM, SK. Supervision: YW, AK, TM, SK. Project administration: YW, AK, TM, SK.
Ethics approval
This study was approved by the Ethics Committee of Tohoku University School of Medicine (approval number: 2020–1-362).
Patient consent statement
Written informed consent was obtained from all parents of the children enrolled in this study.
Acknowledgments
We thank all patients, their family members, and volunteers. We also thank Ms. Shino Miyakawa, Ms. Tomoe Kanno, Ms. Ai Kurihara, and Ms. Yuko Sato for quantification of DBS Phe at the Public Health Society of Miyagi Prefecture. We also acknowledge the support of the Biomedical Research Core of the Tohoku University Graduate School of Medicine and the Biomedical Research Unit of Tohoku University Hospital.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.van Spronsen F.J., Blau N., Harding C., Burlina A., Longo N., Bosch A.M. Phenylketonuria. Nat. Rev. Dis. Prim. 2021;7(1):36. doi: 10.1038/s41572-021-00267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson A.J., Youl B.D., Kendall B., et al. Neurological deterioration in young adults with phenylketonuria. Lancet. 1990;336(8715):602–605. doi: 10.1016/0140-6736(90)93401-a. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira B.K., Rodrigues M.T., Streck E.L., Ferreira G.C., Schuck P.F. White matter disturbances in phenylketonuria: possible underlying mechanisms. J. Neurosci. Res. 2021;99(1):349–360. doi: 10.1002/jnr.24598. [DOI] [PubMed] [Google Scholar]
- 4.Levy H.L. Robert guthrie and the trials and tribulations of newborn screening. Int. J. Neonatal. Screen. 2021;7(1):5. doi: 10.3390/ijns7010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaulent P., Charriere S., Feillet F., Douillard C., Fouilhoux A., Thobois S. Neurological manifestations in adults with phenylketonuria: new cases and review of the literature. J. Neurol. 2020;267(2):531–542. doi: 10.1007/s00415-019-09608-2. [DOI] [PubMed] [Google Scholar]
- 6.van Spronsen F.J., van Wegberg A.M., Ahring K., et al. Key European guidelines for the diagnosis and management of patients with phenylketonuria. Lancet Diabetes Endocrinol. 2017;5(9):743–756. doi: 10.1016/S2213-8587(16)30320-5. [DOI] [PubMed] [Google Scholar]
- 7.Vockley J., Andersson H.C., Antshel K.M., et al. Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet. Med. 2014;16(2):188–200. doi: 10.1038/gim.2013.157. [DOI] [PubMed] [Google Scholar]
- 8.Singh R.H., Rohr F., Frazier D., et al. Recommendations for the nutrition management of phenylalanine hydroxylase deficiency. Genet. Med. 2014;16(2):121–131. doi: 10.1038/gim.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shintaku H., Ohura T., Takayanagi M., et al. Guide for diagnosis and treatment of hyperphenylalaninemia. Pediatr. Int. 2021;63(1):8–12. doi: 10.1111/ped.14399. [DOI] [PubMed] [Google Scholar]
- 10.Kure S., Hou D.-C., Ohura T., et al. Tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. J. Pediatr. 1999;135(3):375–378. doi: 10.1016/s0022-3476(99)70138-1. [DOI] [PubMed] [Google Scholar]
- 11.Thomas J., Levy H., Amato S., et al. Pegvaliase for the treatment of phenylketonuria: results of a long-term phase 3 clinical trial program (PRISM) Mol. Genet. Metab. 2018;124(1):27–38. doi: 10.1016/j.ymgme.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Rose A.M., Grosse S.D., Garcia S.P., et al. The financial and time burden associated with phenylketonuria treatment in the United States. Mol. Genet. Metabol. Rep. 2019;21 doi: 10.1016/j.ymgmr.2019.100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng L., Hu R., Chen J., et al. An enzyme cascade fluorescence-based assay for the quantification of phenylalanine in serum. Analyst. 2022;147(4):671–676. doi: 10.1039/d1an02038b. [DOI] [PubMed] [Google Scholar]
- 14.Sarı T., Dede S., Yusufoğlu B., Karakuş E. Determination of L-phenylalanine in human plasma samples with new Fluorometric method. Appl. Biochem. Biotechnol. 2022;194(3):1259–1270. doi: 10.1007/s12010-021-03694-7. [DOI] [PubMed] [Google Scholar]
- 15.Parrilla M., Vanhooydonck A., Watts R., Wael K.D. Wearable wristband-based electrochemical sensor for the detection of phenylalanine in biofluids. Biosens. Bioelectron. 2022;197 doi: 10.1016/j.bios.2021.113764. [DOI] [PubMed] [Google Scholar]
- 16.Jafari P., Beigi S.M., Yousefi F., et al. Colorimetric biosensor for phenylalanine detection based on a paper using gold nanoparticles for phenylketonuria diagnosis. Microchem. J. 2021;163 [Google Scholar]
- 17.Dinu A., Apetrei C. A review on electrochemical sensors and biosensors used in phenylalanine electroanalysis. Sens. Basel Switz. 2020;20(9):2496. doi: 10.3390/s20092496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dashtian K., Hajati S., Ghaedi M. L-phenylalanine-imprinted polydopamine-coated CdS/CdSe n-n type II heterojunction as an ultrasensitive photoelectrochemical biosensor for the PKU monitoring. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112346. [DOI] [PubMed] [Google Scholar]
- 19.Dinu A., Apetrei C. Voltammetric determination of phenylalanine using chemically modified screen-printed based sensors. Chemosens. 2020;8(4):113. [Google Scholar]
- 20.Idili A., Parolo C., Ortega G., Plaxco K.W. Calibration-free measurement of phenylalanine levels in the blood using an electrochemical aptamer-based sensor suitable for point-of-care applications. Acs Sens. 2019;4(12):3227–3233. doi: 10.1021/acssensors.9b01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barros J., Dixon R.A. Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci. 2020;25(1):66–79. doi: 10.1016/j.tplants.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Peterson K., Slover R., Gass S., Seltzer W.K., McCabe L.L., McCabe E.R.B. Blood phenylalanine estimation for the patient with phenylketonuria using a portable device. Biochem. Med. Metab. B. 1988;39(1):98–104. doi: 10.1016/0885-4505(88)90063-1. [DOI] [PubMed] [Google Scholar]
- 23.Wada Y., Kikuchi A., Arai-Ichinoi N., et al. Biallelic GALM pathogenic variants cause a novel type of galactosemia. Genet. Med. 2019;21(6):1286–1294. doi: 10.1038/s41436-018-0340-x. [DOI] [PubMed] [Google Scholar]
- 24.Cochrane F.C., Davin L.B., Lewis N.G. The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry. 2004;65(11):1557–1564. doi: 10.1016/j.phytochem.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Dreßen A., Hilberath T., Mackfeld U., Billmeier A., Rudat J., Pohl M. Phenylalanine ammonia lyase from Arabidopsis thaliana (AtPAL2): a potent MIO-enzyme for the synthesis of non-canonical aromatic alpha-amino acids part I: comparative characterization to the enzymes from Petroselinum crispum (PcPAL1) and Rhodosporidium toruloides (RtPAL) J. Biotechnol. 2017;258:148–157. doi: 10.1016/j.jbiotec.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Huizenga J.R., Gips C.H., Conn H.O., Jansen P.L.M. Determination of ammonia in ear-lobe capillary blood is an alternative to arterial blood ammonia. Clin. Chim. Acta. 1995;239(1):65–70. doi: 10.1016/0009-8981(95)06101-i. [DOI] [PubMed] [Google Scholar]
- 27.Moat S.J., Schulenburg-Brand D., Lemonde H., et al. Performance of laboratory tests used to measure blood phenylalanine for the monitoring of patients with phenylketonuria. J. Inherit. Metab. Dis. 2020;43(2):179–188. doi: 10.1002/jimd.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendel U., Langenbeck U. Towards self-monitoring and self-treatment in phenylketonuria — a way to better diet compliance. Eur. J. Pediatr. 1996;155(Suppl. 1):S105–S107. doi: 10.1007/pl00014224. [DOI] [PubMed] [Google Scholar]
- 29.Bilginsoy C., Waitzman N., Leonard C.O., Ernst S.L. Living with phenylketonuria: perspectives of patients and their families. J. Inherit. Metab. Dis. 2005;28(5):639–649. doi: 10.1007/s10545-005-4478-8. [DOI] [PubMed] [Google Scholar]
- 30.Feillet F., MacDonald A., Perron D.H., Burton B. Outcomes beyond phenylalanine: an international perspective. Mol. Genet. Metab. 2010;99:S79–S85. doi: 10.1016/j.ymgme.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 31.Iqbal C.W., Wall J., Harrison M.R. Challenges and climate of business environment and resources to support pediatric device development. Semin. Pediatr. Surg. 2015;24(3):107–111. doi: 10.1053/j.sempedsurg.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 32.ten Hoedt A.E., Hollak C.E., Boelen C.C., et al. “MY PKU”: increasing self-management in patients with phenylketonuria. A randomized controlled trial. Orphanet. J. Rare Dis. 2011;6(1) doi: 10.1186/1750-1172-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trefz F.K., van Spronsen F.J., MacDonald A., et al. Management of adult patients with phenylketonuria: survey results from 24 countries. Eur. J. Pediatr. 2015;174(1):119–127. doi: 10.1007/s00431-014-2458-4. [DOI] [PubMed] [Google Scholar]
- 34.Jurecki E.R., Cederbaum S., Kopesky J., et al. Adherence to clinic recommendations among patients with phenylketonuria in the United States. Mol. Genet. Metab. 2017;120(3):190–197. doi: 10.1016/j.ymgme.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Longo N., Harding C.O., Burton B.K., et al. Single-dose, subcutaneous recombinant phenylalanine ammonia lyase conjugated with polyethylene glycol in adult patients with phenylketonuria: an open-label, multicentre, phase 1 dose-escalation trial. Lancet. 2014;384(9937):37–44. doi: 10.1016/S0140-6736(13)61841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longo N., Dimmock D., Levy H., et al. Evidence- and consensus-based recommendations for the use of pegvaliase in adults with phenylketonuria. Genet. Med. 2019;21(8):1851–1867. doi: 10.1038/s41436-018-0403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha J.C., Bausell H., Bélanger-Quintana A., et al. Development of a practical dietitian road map for the nutritional management of phenylketonuria (PKU) patients on pegvaliase. Mol. Genet. Metabol. Rep. 2021;28 doi: 10.1016/j.ymgmr.2021.100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell S.M., Wendt D.J., Zhang Y., et al. Formulation and PEGylation optimization of the therapeutic PEGylated phenylalanine ammonia lyase for the treatment of phenylketonuria. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Y., Patel G., Henshaw J., et al. Pharmacokinetic, pharmacodynamic, and immunogenic rationale for optimal dosing of pegvaliase, a PEGylated bacterial enzyme, in adult patients with phenylketonuria. Clin. Transl. Sci. 2021;14(5):1894–1905. doi: 10.1111/cts.13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.