Abstract
Research on schizophrenia typically focuses on one paradigm for which clear-cut differences between patients and controls are established. Great efforts are made to understand the underlying genetical, neurophysiological, and cognitive mechanisms, which eventually may explain the clinical outcome. One tacit assumption of these “deep rooting” approaches is that paradigms tap into common and representative aspects of the disorder. Here, we analyzed the resting-state electroencephalogram (EEG) of 121 schizophrenia patients and 75 controls. Using multiple signal processing methods, we extracted 194 EEG features. Sixty-nine out of the 194 EEG features showed a significant difference between patients and controls, indicating that these features detect an important aspect of schizophrenia. Surprisingly, the correlations between these features were very low. We discuss several explanations to our results and propose that complementing “deep” with “shallow” rooting approaches might help in understanding the underlying mechanisms of the disorder.
Keywords: electroencephalography, schizophrenia, psychosis, neuroimaging, resting-state, psychiatry
Introduction
Schizophrenia patients show strong abnormalities in many domains, including personality, cognition, perception, and even immunology. In many experimental paradigms, the differences between patients and controls have large effect sizes, indicating that important aspects of the disease are detected. This provokes two questions: What do these abnormalities have in common, and how representative are they of the disease? For example, patients exhibit strong deficits in cognition, such as in working memory tasks (Meyer-Lindenberg et al. 2001), which are attributed to the abnormalities of cortico-cerebellar-thalamic-cortical circuits (Andreasen et al. 1998). Patients show also diminished skin flushing with the niacin skin test (Rybakowski and Weterle 1991), which is attributed to dysfunctional phospholipase A2 arachidonic acid signaling (Messamore 2012). How do the working memory deficits correspond to deficits in skin functioning? Very few studies have correlated deficits with each other (Toomey et al. 1998; Braff et al. 2006, 2007; Price et al. 2006; Dickinson et al. 2011; Seidman et al. 2015). The Consortium on the Genetics of Schizophrenia studied neurocognitive and neurophysiological abnormalities in schizophrenia patients with a battery of 15 paradigms (Seidman et al. 2015). They found that neurocognitive measures shared a significant amount of variance, while neurophysiological measures were almost entirely independent. Price et al. (2006) studied four candidate electrophysiological endophenotypes of schizophrenia (mismatch negativity, P50, P300, and antisaccades). Even though patients and their family members showed deficits in each of these endophenotypes, the features were largely uncorrelated.
Here, we took another road. Instead of comparing different paradigms, we analyzed the very same data of the very same patients and controls with different electroencephalogram (EEG) analysis methods, including many that have shown atypical patterns in patients (Kim et al. 2000; Boutros et al. 2008; Uhlhaas and Singer 2010; Nikulin et al. 2012; Sun et al. 2014; Andreou et al. 2015; Di Lorenzo et al. 2015; da Cruz et al. 2020a). Data were recorded from a 5-min resting-state session during which the participants did nothing else than relaxing. Many of the resting-state EEG features we extracted are thought to reflect brain mechanisms linked to important aspects of the disorder. For example, schizophrenia patients exhibit reduced long-range temporal correlations (LRTC) in the alpha and beta frequency bands (Nikulin et al. 2012) suggested to reflect excessive switching of neuronal states. Patients also have shown atypical patterns in the dynamics of the EEG microstates classes C and D (Rieger et al. 2016; da Cruz et al. 2020a), which were proposed to correspond to imbalances in attentional and information processing. Schizophrenia patients have shown increased power in the delta, theta, and beta frequency bands (Venables et al. 2009). Increased beta power was suggested to reflect cortical hyperexcitability, and increased power in the delta and theta bands were proposed to relate to atypical dopaminergic function, to name a few examples. All these results, individually, suggest that each EEG feature captures important aspects of schizophrenia. But how representative are these abnormalities of the disorder? Does a patient showing abnormal microstate dynamics also show deficits in LRTC or in other EEG features?
Aiming to shed light on this EEG “multiverse” of schizophrenia, we analyzed the resting-state EEG data of 121 schizophrenia patients and 75 healthy controls with multiple methods. We extracted 194 EEG features, such as time-domain features, frequency-domain, and connectivity features both in electrode and source space, and nonlinear dynamical features. Then, we correlated the features that showed significant group differences to evaluate how these abnormalities/deficits relate to each other. We also examined whether these EEG features show adequate predictive power to clinical scales measuring key symptoms of schizophrenia.
Materials and methods
Participants
Two groups of participants joined the experiment: schizophrenia patients (n = 121) and healthy controls (n = 75). All participants took part in a battery of tests comprising perceptual and cognitive tasks as well as EEG recordings. Data of 101 patients and 75 controls have already been published in different contexts (Favrod et al. 2018; da Cruz et al. 2020a, 2020b; Garobbio et al. 2021). Patients were recruited from the Tbilisi Mental Health Hospital or the psycho-social rehabilitation center. Patients were invited to participate in the study when they had recovered sufficiently from an acute psychotic episode. Thirty-five were inpatients and 86 were outpatients. Patients were diagnosed using the Diagnostic and Statistical Manual of Mental Disorders Fourth Edition (DSM-IV) by means of an interview based on the Structured Clinical Interview for DSM-IV, Clinical Version, information from staff, and study of patients’ records. Psychopathology of patients was assessed by an experienced psychiatrist using the Scale for the Assessment of Negative Symptoms (SANS) and the Scale for the Assessment of Positive Symptoms (SAPS). Out of the 121 patients, 106 were receiving neuroleptic medication. Chlorpromazine (CPZ) equivalents are indicated in Table 1. Controls were recruited from the general population in Tbilisi, aiming to match the patients’ demographics as closely as possible. All controls were free from psychiatric axis I disorders and had no family history of psychosis. General exclusion criteria were alcohol or drug abuse, severe neurological incidents or diagnoses, developmental disorders (autism spectrum disorder or intellectual disability), or other somatic mind-altering illnesses, which were assessed through interview by certified psychiatrists. All participants were no older than 55 years. Group characteristics are presented in Table 1. All participants signed informed consent and were informed that they could quit the experiment at any time. All procedures complied with the Declaration of Helsinki (except for preregistration) and were approved by the Ethical Committee of the Institute of Postgraduate Medical Education and Continuous Professional Development (Georgia); protocol number: 09/07; title: “Genetic polymorphisms and early information processing in schizophrenia.”
Table 1.
Group average statistics (± standard deviation).
| Patients | Controls | Statistics | |
|---|---|---|---|
| Gender (F/M) | 22/99 | 39/36 | χ2(1) = 24.702, P = 6.690e-7a |
| Age (years) | 35.8 ± 9.2 | 35.1 ± 7.7 | t(194) = 0.519, p = 0.604b |
| Education (years) | 13.3 ± 2.6 | 15.1 ± 2.9 | t(194) = −4.418, P = 1.657e-5b |
| Handedness (L/R) | 6/115 | 4/71 | χ2(1) = 0.013, p = 0.908a |
| Illness duration (years) | 10.8 ± 8.7 | ||
| SANS | 10.1 ± 5.2 | ||
| SAPS | 8.6 ± 3.2 | ||
| CPZ equivalentc | 561.1 ± 389.4 |
aPearson’s chi-squared test.
bTwo-sided independent samples t-test.
cAverage CPZ equivalents calculated over the 106 patients receiving neuroleptic medication.
EEG recording and data processing
Participants were sitting in a dim lit room. They were instructed to keep their eyes closed and to relax for 5 min. Resting-state EEG was recorded using a BioSemi Active Two Mk2 system (Biosemi B.V., The Netherlands) with 64 Ag-AgCl sintered active electrodes referenced to the common mode sense electrode. The recording sampling rate was 2,048 Hz. Offline data were downsampled to 256 Hz and were preprocessed using an automatic pipeline (da Cruz et al. 2018). Preprocessed EEG data were analyzed using multiple signal processing methods in the electrode and source space. In total, 194 EEG features were extracted (see Supplementary Table 1). Out of the 194 EEG features, 50 were obtained in the source space and 144 in the electrode space. For source space analysis, we defined 80 brain regions (40 per hemisphere) according to the AAL atlas (see Supplementary Table 2). See Supplementary Methods for a detailed description of the analysis methods.
Group comparisons
We compared patients’ and controls’ scores for each of the 194 EEG features. For each of the  variables (i.e. 64 electrodes, 80 brain regions, or 12 microstate parameters, depending on the number of variables of each EEG feature) of a given feature, we performed a two-way ANCOVA, with Group (patients and controls) and Gender (male and female) as factors and with Education as a covariate. The P-values for the effect of Group were corrected for
variables (i.e. 64 electrodes, 80 brain regions, or 12 microstate parameters, depending on the number of variables of each EEG feature) of a given feature, we performed a two-way ANCOVA, with Group (patients and controls) and Gender (male and female) as factors and with Education as a covariate. The P-values for the effect of Group were corrected for  comparisons using false discovery rate (FDR; with an error rate of 5%). Group effects’
comparisons using false discovery rate (FDR; with an error rate of 5%). Group effects’  were converted to Cohen’s d.
were converted to Cohen’s d.
Pearson, partial least squares, and distance correlations
First, for each EEG feature that contained at least one variable showing a significant difference between patients and controls (after correcting for multiple comparisons), we selected the variable (i.e. electrode, brain region, or microstate parameter) with the biggest effect size to be the representative variable for that feature. Then, for patients and controls separately, we computed pairwise Pearson correlations between the representative variables of each significant EEG feature. As a complementary analysis, we computed Pearson correlations between the first principal components of the EEG features showing significant group differences for patients and controls separately. Second, to quantify the overall relationship, i.e. the amount of shared information, between pairs of multivariate EEG features, we used partial least squares correlation (PLSC). PLSC generalizes correlations between two variables to two matrices (Tucker 1958; McIntosh et al. 1996). The shared information can be quantified as the inertia common to the 2 features (Krishnan et al. 2011). The statistical significance of the inertia was assessed using a permutation test (McIntosh et al. 2004; Abdi and Williams 2013). The inertia values were normalized. Hence, the normalized inertias (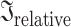 ) ranged from 0 (the two EEG features are completely unrelated) to 1 (the two EEG features contain the same information). PLSC analysis was done for patients and controls separately. Finally, for patients and controls separately, we quantified the relationship between pairs of multivariate EEG features using distance correlations (Székely and Rizzo 2013). Distance correlations are close to 0 if the multivariate features are unrelated and are close to 1 if features are strongly related. See Supplementary Methods for details.
) ranged from 0 (the two EEG features are completely unrelated) to 1 (the two EEG features contain the same information). PLSC analysis was done for patients and controls separately. Finally, for patients and controls separately, we quantified the relationship between pairs of multivariate EEG features using distance correlations (Székely and Rizzo 2013). Distance correlations are close to 0 if the multivariate features are unrelated and are close to 1 if features are strongly related. See Supplementary Methods for details.
Regression and classification analyses
To evaluate whether EEG features predict the psychopathology scores (SAPS and SANS) adequately, we used elastic net regression models (Zou and Hastie 2005). Elastic nets can handle regression problems where the number of predictors is relatively large compared to the number of samples as well as multicollinearity (i.e. the predictors are not linearly independent) by combining the 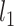 and
and  penalties to achieve regularization. For each of the 194 EEG features (with all its variables), we built 2 regression models, one to predict SAPS scores and one to predict SANS scores. We performed 20 repetitions of a 3-fold nested cross-validation procedure. First, one third of the data (1-fold) was left out for validation (test set), while the remaining data (2-folds; train set) were used to find the optimal parameters, namely the amount of penalization and the compromise between
penalties to achieve regularization. For each of the 194 EEG features (with all its variables), we built 2 regression models, one to predict SAPS scores and one to predict SANS scores. We performed 20 repetitions of a 3-fold nested cross-validation procedure. First, one third of the data (1-fold) was left out for validation (test set), while the remaining data (2-folds; train set) were used to find the optimal parameters, namely the amount of penalization and the compromise between  and
and  penalties, using 3-fold cross-validation. The model with the parameters leading to best performance in the train set was tested on the left-out data (test set). The entire procedure was repeated 20 times, with different allocations of the patients in the train and test sets. Using the same crossvalidation procedure, i.e. 20 repetitions of a 3-fold cross-validation, we also evaluated predictive performance using a nonlinear random forest regression model, setting the maximum depth of the tree to 10 and the number of trees to 100. Random forests are meta estimators that average several decision trees trained on subsets of the dataset to improve accuracy and to avoid overfitting. Prediction performance was calculated using the coefficient of determination (R2) and the root-mean-squared error (RMSE). The distribution of the prediction performance values was obtained from the 60 aggregated RMSE and R2 across repetitions of the procedure. Further, we evaluated the classification performance of the EEG features, i.e. we aimed to discriminate between patients and controls using penalized logistic regression. Accuracy (ACC) and area under the curve (AUC) were obtained using a training procedure consisting of 100 repetitions of a 3-fold cross-validation method. First, 33% of the data were separated as the testing set, and the remaining 67%, i.e. training set, were used to estimate the amount of penalization (
penalties, using 3-fold cross-validation. The model with the parameters leading to best performance in the train set was tested on the left-out data (test set). The entire procedure was repeated 20 times, with different allocations of the patients in the train and test sets. Using the same crossvalidation procedure, i.e. 20 repetitions of a 3-fold cross-validation, we also evaluated predictive performance using a nonlinear random forest regression model, setting the maximum depth of the tree to 10 and the number of trees to 100. Random forests are meta estimators that average several decision trees trained on subsets of the dataset to improve accuracy and to avoid overfitting. Prediction performance was calculated using the coefficient of determination (R2) and the root-mean-squared error (RMSE). The distribution of the prediction performance values was obtained from the 60 aggregated RMSE and R2 across repetitions of the procedure. Further, we evaluated the classification performance of the EEG features, i.e. we aimed to discriminate between patients and controls using penalized logistic regression. Accuracy (ACC) and area under the curve (AUC) were obtained using a training procedure consisting of 100 repetitions of a 3-fold cross-validation method. First, 33% of the data were separated as the testing set, and the remaining 67%, i.e. training set, were used to estimate the amount of penalization ( norm, 10 values between
norm, 10 values between 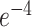 and
and  ) using 3-fold cross validation. The model giving the best fit on the training set was tested on the left out 33% of the data and the classification ACC and AUC were estimated. The entire procedure was repeated for 100 times, allocating the participants differently at each iteration, and the values of ACC and AUC were aggregated. The mean ACC and AUC were obtained for each EEG feature. To identify the features that classified patients and controls significantly, we repeated the above-mentioned procedure for 1,000 times and aggregated the ACC and AUC values. We assigned different EEG feature values to different participants at each repetition (random label permutation). The mean AUCs obtained in the previous step were compared to the null distribution of 1,000 AUC values and a P-value was obtained. The P-value indicated the probability of a value of AUC obtained from random label permutation to be larger than that obtained from the original data. We declare that the features were significant if the value was <5%.
) using 3-fold cross validation. The model giving the best fit on the training set was tested on the left out 33% of the data and the classification ACC and AUC were estimated. The entire procedure was repeated for 100 times, allocating the participants differently at each iteration, and the values of ACC and AUC were aggregated. The mean ACC and AUC were obtained for each EEG feature. To identify the features that classified patients and controls significantly, we repeated the above-mentioned procedure for 1,000 times and aggregated the ACC and AUC values. We assigned different EEG feature values to different participants at each repetition (random label permutation). The mean AUCs obtained in the previous step were compared to the null distribution of 1,000 AUC values and a P-value was obtained. The P-value indicated the probability of a value of AUC obtained from random label permutation to be larger than that obtained from the original data. We declare that the features were significant if the value was <5%.
Results
Multiple EEG features reveal significant group effects and classification performance
For 121 patients (22 females, 35.8 ± 9.2 years old, 13.3 ± 2.6 years of education) and 75 age-matched healthy controls (39 females, 35.1 ± 7.7 years old, 15.1 ± 2.9 years of education; Table 1), we extracted, in total, 194 features from the resting-state EEG recordings, including time-domain, frequency-domain, connectivity, and nonlinear dynamical features (Supplementary Table 1). Among the 194 EEG features, 69 (35.57%) showed significant differences between patients and controls with medium to large effect sizes (Cohen’s d varied from 0.463 to 1.037, Fig. 1). Patients showed significantly reduced values in 24 out of the 69 EEG features, revealing significant group differences (illustrated as negative effect size in Fig. 1). Patients exhibited significantly higher values than controls in 45 EEG features.
Fig. 1.

Effect size (Cohen’s d) of the group differences between patients and controls for each of the 194 EEG features. We took the values of the electrode, brain region, or microstate parameter, with the largest effect size according to Cohen’s d ( values were converted to Cohen’s d) to be the representative variable for each feature. Significant group differences, after correction for multiple comparisons (using FDR), are depicted in red, with dotted red horizontal lines serving as a guide to their labels. > and < were added to the feature labels to indicate if patients had significantly higher or lower values than controls, respectively. The non-significant effects are shown in blue. Error bars represent 95% confidence intervals. A list with the abbreviations and the corresponding name of each feature is presented in Supplementary Table 1.
values were converted to Cohen’s d) to be the representative variable for each feature. Significant group differences, after correction for multiple comparisons (using FDR), are depicted in red, with dotted red horizontal lines serving as a guide to their labels. > and < were added to the feature labels to indicate if patients had significantly higher or lower values than controls, respectively. The non-significant effects are shown in blue. Error bars represent 95% confidence intervals. A list with the abbreviations and the corresponding name of each feature is presented in Supplementary Table 1.
Using cross validated classification analysis, we found 91 EEG features with a significant AUC performance compared to the null models. The AUC values of the EEG features with significant classification performance ranged between 0.610 and 0.848 for the training sets and between 0.523 and 0.715 for the testing sets. The classification accuracies of the significant EEG features ranged between 0.691 and 0.873 for the training sets and between 0.590 and 0.736 for the testing sets. Out of the 69 EEG features, which showed a significant effect in the group comparison using ANCOVA, 57 features also showed a significant classification performance (Supplementary Table 3).
Correlations between EEG features
To evaluate to what extent features that showed significant group differences are sensitive to the same aspects of the disorder, we computed Pearson’s correlations between pairs of features (Fig. 2). As the representative variable for each feature, we took the values of the electrode, brain region, or microstate parameter which showed the largest group difference according to Cohen’s d (Fig. 1). Surprisingly, we found that, in the patients group, only 36.49% of the pairwise correlations were significant at a level of 0.05 (without correcting for multiple comparisons). For the control group, only 26.73% of the correlations were significant. Since significance depends on the sample size, here, we focus on the magnitude of the correlation coefficients (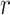 -values). In general, the magnitudes of the
-values). In general, the magnitudes of the 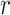 -values were very low in both patients (0.055, 0.122, and 0.251 for the 25th, 50th, and 75th percentiles, respectively) and controls (0.059, 0.129, and 0.242 for the 25th, 50th, and 75th percentiles, respectively; Fig. 2). Strong correlations were found mainly for pairs of very closely related features (Supplementary Tables 4 and 5), such as between waiting-time statistics of gamma bursts (“waiting time gamma”) and life-time statistics of gamma bursts (“life time gamma”;
-values were very low in both patients (0.055, 0.122, and 0.251 for the 25th, 50th, and 75th percentiles, respectively) and controls (0.059, 0.129, and 0.242 for the 25th, 50th, and 75th percentiles, respectively; Fig. 2). Strong correlations were found mainly for pairs of very closely related features (Supplementary Tables 4 and 5), such as between waiting-time statistics of gamma bursts (“waiting time gamma”) and life-time statistics of gamma bursts (“life time gamma”; 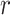 = 0.836 and
= 0.836 and 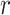 = 0.926 in patients and controls, respectively). Similar results were found when, instead of the variable showing the largest group difference, we selected the first principal component as the representative variable of each EEG feature showing a significant group difference between patients and controls. The
= 0.926 in patients and controls, respectively). Similar results were found when, instead of the variable showing the largest group difference, we selected the first principal component as the representative variable of each EEG feature showing a significant group difference between patients and controls. The 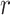 -values were low in both patients (0.060, 0.152, and 0.313 for the 25th, 50th, and 75th percentiles, respectively) and controls (0.059, 0.135, and 0.264 for the 25th, 50th, and 75th percentiles, respectively). Similar results were found using disattenuated
-values were low in both patients (0.060, 0.152, and 0.313 for the 25th, 50th, and 75th percentiles, respectively) and controls (0.059, 0.135, and 0.264 for the 25th, 50th, and 75th percentiles, respectively). Similar results were found using disattenuated 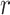 -values (see Supplementary Results). Interestingly, when we put together all variables from all EEG features, 13,112 variables in total, and we corrected for multiple comparisons using Holm method, we found 272 variables from 16 EEG features which showed significant differences (see Supplementary Table 6). When we correlated these 16 EEG features, selecting the variable showing the largest effect as the representative variable, we found that correlations were stronger in patients (0.163, 0.317, and 0.454 for the 25th, 50th, and 75th percentiles, respectively) than in controls (0.088, 0.164, and 0.302 for the 25th, 50th, and 75th percentiles, respectively). Potentially, these features might be interesting for future investigations.
-values (see Supplementary Results). Interestingly, when we put together all variables from all EEG features, 13,112 variables in total, and we corrected for multiple comparisons using Holm method, we found 272 variables from 16 EEG features which showed significant differences (see Supplementary Table 6). When we correlated these 16 EEG features, selecting the variable showing the largest effect as the representative variable, we found that correlations were stronger in patients (0.163, 0.317, and 0.454 for the 25th, 50th, and 75th percentiles, respectively) than in controls (0.088, 0.164, and 0.302 for the 25th, 50th, and 75th percentiles, respectively). Potentially, these features might be interesting for future investigations.
Fig. 2.
Pairwise correlations between the 69 EEG features which showed significant group differences between patients and controls. Patients’ 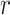 -values are presented in the upper triangle and controls’
-values are presented in the upper triangle and controls’  -values are shown in the lower triangle. Strong negative and positive
-values are shown in the lower triangle. Strong negative and positive 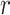 -values are depicted in red and blue, respectively, and
-values are depicted in red and blue, respectively, and 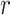 -values around 0 in yellow. For each feature, we used the values of the electrode, brain region, or microstate parameter which showed the largest effect size as the representative variable for the correlations. A list with the abbreviations and corresponding name of each feature is shown in Supplementary Table 1.
-values around 0 in yellow. For each feature, we used the values of the electrode, brain region, or microstate parameter which showed the largest effect size as the representative variable for the correlations. A list with the abbreviations and corresponding name of each feature is shown in Supplementary Table 1.
To quantify the overall shared information between pairs of EEG features, which showed significant group differences, by taking not only variables with the largest effect size into account but all variables of the features, we used PLSC and distance correlations. For the patients, 55.92% of the pairwise inertias were significant (without correcting for multiple comparisons) and for controls, 40.28%. In general, relative inertias were not very high in both patients (0.254, 0.329, and 0.409 for the 25th, 50th, and 75th percentiles, respectively) and controls (0.305, 0.387, and 0.472 for the 25th, 50th, and 75th percentiles, respectively; Fig. 3). As in the Pearson’s correlation results, features that showed strong associations were mainly similar features, such as the same network statistics for different connectivity measures in the theta band, for example, at the electrode level: clustering coefficient connectivity estimated with the phase locking value (“clust coeff e-plv theta”) and with the imaginary part of coherence (“clust coeff e-icoh theta”; 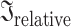 = 0.804 and
= 0.804 and 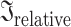 = 0.826, in patients and controls, respectively). Distance correlations show similar results. The distance correlation values were low in both patients (0.096, 0.189, and 0.329 for the 25th, 50th, and 75th percentiles, respectively) and controls (0.102, 0.168, and 0.303 for the 25th, 50th, and 75th percentiles, respectively). For the patients, 61.59% of the pairwise distance correlations were significant and 47.02% of the pairwise distance correlations were significant for controls (without correction for multiple comparisons). Disattenuated values were stronger for relative inertias, whereas for distance correlations, the values were not strong (see Supplementary Results).
= 0.826, in patients and controls, respectively). Distance correlations show similar results. The distance correlation values were low in both patients (0.096, 0.189, and 0.329 for the 25th, 50th, and 75th percentiles, respectively) and controls (0.102, 0.168, and 0.303 for the 25th, 50th, and 75th percentiles, respectively). For the patients, 61.59% of the pairwise distance correlations were significant and 47.02% of the pairwise distance correlations were significant for controls (without correction for multiple comparisons). Disattenuated values were stronger for relative inertias, whereas for distance correlations, the values were not strong (see Supplementary Results).
Fig. 3.
Shared information between the 69 EEG features which showed significant group differences, as measured by the relative inertia (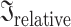 ) computed with PLSC. The relative inertia ranges from 0 (the two features are completely unrelated) to 1 (the two features’ values move together by the exact same percentage). Patients’ relative inertias are presented in the upper triangle, and controls’ relative inertias are shown in the lower triangle. A list with the abbreviations and corresponding name of each feature is shown in Supplementary Table 1.
) computed with PLSC. The relative inertia ranges from 0 (the two features are completely unrelated) to 1 (the two features’ values move together by the exact same percentage). Patients’ relative inertias are presented in the upper triangle, and controls’ relative inertias are shown in the lower triangle. A list with the abbreviations and corresponding name of each feature is shown in Supplementary Table 1.
Prediction of psychopathology scores
We evaluated whether EEG features were adequate predictors of psychopathology scores determined by the Scale for the Assessment of Positive Symptoms (SAPS) and the Scale for the Assessment of Negative Symptoms (SANS), which target positive (hallucinations, delusions, bizarre behavior, and positive formal thought disorder) and negative (affective flattening, alogia, apathy, anhedonia, and attention) symptoms, respectively. All 194 EEG features exhibited very weak out-of-sample predictive ability to both the SANS and SAPS scores. Results were very similar for both the linear (i.e. elastic net) and nonlinear (i.e. random forest) models. See Supplementary Tables 7 and 8 for details.
Discussion
Traditionally, most studies in schizophrenia research focus on a single experimental paradigm and analysis method, which shows significant differences between patients and controls. Extensive research with the paradigm tries to derive the underlying genetic and neurophysiological causes of the disorder. This approach has been quite successful in the formulation of hypotheses, such as the dopamine hypothesis (Howes and Kapur 2009), the social brain hypothesis (Burns 2006), the glutamate hypothesis (Hu et al. 2015), or the dysconnection hypothesis (Friston et al. 2016), just to name a few.
Here, we took a different road and examined to what extent abnormalities, quantified by different EEG features, correlate with each other. Many of the investigated features were previously linked to different abnormalities of brain processes in schizophrenia. Here, we reproduced many of these results, such as imbalance in microstates dynamics (Rieger et al. 2016; da Cruz et al. 2020a), decreased LRTC in the alpha and beta bands (Nikulin et al. 2012), decreased life time and waiting time in the beta band (Sun et al. 2014), increased spectral amplitude in the theta band (Boutros et al. 2008), increased connectivity in the theta band at the source level (Andreou et al. 2015; Di Lorenzo et al. 2015), and decreased Lyapunov exponent (Kim et al. 2000), among others. With our systematic analysis, we also found abnormalities in EEG features, which, to the best of our knowledge, have not been reported yet, namely, delta-phase gamma-amplitude coupling, range EEG coefficient of variation and asymmetry in the theta and alpha bands, etc. In some way, deeper analysis of each feature may have warranted an in-depth study and a potential publication. However, we did not want to elaborate on these methods individually because we wanted to understand how all EEG features relate to each other in their entirety.
The surprising insight from our analysis is that, even though we are probing the same signals from the same participants, we found only weak correlations between the 69 significant features. The only strong correlations we found were between features that are similar from the outset, thereby resembling test-retests. This suggests that, even though each EEG feature reveals clear-cut and reproducible differences between patients and controls, none of the features is truly representative for the disease. Hence, the traditional approach of focusing on a single experimental paradigm and analysis method has its limitations. These results remind us that schizophrenia is indeed a very heterogeneous disease, a well-known fact, which is however not always taken seriously enough because, as mentioned above, most research tries to find the one or a few causes of schizophrenia within one well-described paradigm by digging as deep as possible into the underlying neurophysiological and genetic mechanisms. In analogy to botany, one may call these approaches “deep rooting” approaches.
There can be several reasons why we did not find strong correlations between EEG features even though they show clear-cut group effects. First, test re-test reliability may be low. However, similar EEG features showed strong correlations. Second, EEG features show clear-cut group differences, but variance in the patients and controls is low, leading to low correlations, the well-known reliability paradox (Hedge et al. 2018). However, variance is high, particularly, in the schizophrenia patients. Third, it may be that the linear and nonlinear methods we used are blind to more complex structures. Fourth, EEG features pick up disease-related and, to a substantial amount, also disease-unrelated aspects. When different EEG features tap into different of these disease-unrelated mechanisms, correlations may be low. For example, one EEG feature may strongly depend on the level of fatigue and another one on cardiac functions, which may be both intact in the patients. In this case, variance may be high in both populations but correlations may be low. We cannot determine to what extent this scenario holds true in our study. Fifth, schizophrenia is a heterogeneous disease and different EEG features tap into different aspects of the disease.
Particularly the fifth scenario suggests to complement “deep rooting” approaches with “shallow rooting” approaches, representing schizophrenia within a high-dimensional space, where many tests and analysis outcomes are used instead of one. In this respect, low correlations between tests are a wanted feature because different aspects of the disease are targeted—as long as the tests do not measure mainly disease-unrelated aspects. Tests should ideally have large effect sizes, low mutual correlations, and a “flat” factor structure. Whether this is possible is an open question and depends very much on the underlying causes of schizophrenia.
Current machine learning approaches are well within this spirit (Yang et al. 2010; Mothi et al. 2019; Phang et al. 2020; Morgan et al. 2021). For example, Clementz et al. (2016) analyzed 9 variables, including evoked EEG variables, with k-means clustering. Three clusters were found, which, however, did not correspond to DSM psychosis categories. Using sparse canonical correlation analysis, a bundle of neuroimaging features showed strong links to lifestyle and demographic variables in schizophrenia and bipolar disorder patients (Moser et al. 2018). Future research will tell what we gain from “shallow rooting” approaches. The gain will strongly depend on the complexity of the disease.
Within a multifactorial framework, there are several possible scenarios of complexity. Our results show that there cannot be one cause. However, on the lowest complexity level, there may be a few independent causes, which were not found yet. Given the heterogeneity of the disease, including abnormalities in the cognitive (Andreasen et al. 1998), but also the skin functioning domain (Messamore 2012), the causes need to be on a rather general level, likely subcellular, present in all human functioning. On a medium complexity level, schizophrenia may be an approximatively “additive” disease, where many small abnormalities add up to severe symptoms. For example, the many single-nucleotide polymorphisms (SNPs) involved in schizophrenia may each contribute a little (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014). In an even more complex scenario, schizophrenia is a disease where many causes act in a truly combinatorial manner, i.e. focusing on a single or a few causes is of no avail. One needs always to take all causes into account, which may be impossible because such approaches require impossible sample sizes. For example, only certain combinations of redundant functions, each coming with at least two variants, cause the disease. If one function is upregulated and another one is downregulated in an individual, there are no abnormalities. Deficits manifest only when all or most functions are either up- or down-regulated. In such a combinatorial scenario, it would be difficult to find the underlying causes since each variant itself does not lead to a deficit; only certain combinations do.
Our study has several limitations. There are demographic differences between patients and controls, which might affect our group comparisons. However, we attempted to minimize these demographic effects by using education as a covariate and gender as factor in the analyses. Similarly, we cannot exclude effects of medication in our results. Nonetheless, we find similar patterns of correlations between EEG features, i.e. weak associations, in both patients and controls, suggesting that if there is an effect of medication, it is small. Further, our sample size is relatively small for achieving reliable estimates of predictive power (Schnack and Kahn 2016; Varoquaux 2018; Poldrack et al. 2020). Importantly, during resting-state EEG recordings, participants might be differently engaged into different aspects of cognitive processing. However, the group effects revealed by the 69 EEG features indicate that there is abnormal processing even if the patients would engage differently into different aspects of cognition. Moreover, task-based EEG features also do not correlate strongly (Braff et al. 2006; Price et al. 2006; Seidman et al. 2015). In the healthy control group, the low correlations are only partly surprising since we do not know to what extent different EEG features tap into similar mechanisms, which is contrary to the patient group for which we know that the features are related to processing abnormalities. Still, it is surprising that so few features correlate in the control group as well and how similar the correlations look in patients and controls.
Our results and the complexity of the disease may explain a deep mystery in schizophrenia research. Schizophrenia has an estimated heritability of 70%–85% (Burmeister et al. 2008). For example, the chance to also suffer from schizophrenia for monozygotic twins is about 33% when the partner twin has the disease (Hilker et al. 2018). Furthermore, about 0.25%–0.75% people of a population suffer from schizophrenia and related psychotic disorders (Kessler et al. 2005; Saha et al. 2005; Moreno-Küstner et al. 2018). These values are rather stable across cultures (Simeone et al. 2015). Given that schizophrenia patients have less offspring (Bassett et al. 1996; Avila et al. 2001; Keller and Miller 2006; MacCabe et al. 2009), this provokes the question why schizophrenia has not been extinguished during the course of evolution (Keller and Miller 2006; Liu et al. 2019). In the above-mentioned combinatorial scenario with many redundant functions, this may simply happen because evolution operates on the individual SNP level and not on the combinatorial one. As long as most of the population shows average functioning, there will be no change of the allele distributions. In the additive scenario, evolution may extinct harmful alleles, of which each constitutes only a little risk, very slowly and these may be replaced by harmful de novo mutations (Keller and Miller 2006). To what extent such considerations hold true will be shown by “shallow rooting” approaches using a plethora of paradigms and a multiverse of analysis methods.
Supplementary Material
Acknowledgements
We would like to thank Marc Repnow for his comments and Ben Lönnqvist for proofreading the manuscript. M.H.H., E.C., A.B., and M.R. designed the research; M.R. and E.C. performed the research; J.R.d.C., D.G., W.-H.L., and O.F. analyzed the data; J.R.d.C., D.G., O.F. A.B., P.F., and M.H.H. wrote the paper. The codes that support the findings of this study are available upon request. The authors declare no competing interests.
Contributor Information
Dario Gordillo, Laboratory of Psychophysics, Brain Mind Institute, School of Life Sciences, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland.
Janir Ramos da Cruz, Laboratory of Psychophysics, Brain Mind Institute, School of Life Sciences, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland; Institute for Systems and Robotics – Lisboa, Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa,1049-001 Lisbon, Portugal; Wyss Center for Bio and Neuroengineering, CH-1202 Geneva, Switzerland.
Eka Chkonia, Department of Psychiatry, Tbilisi State Medical University (TSMU), 0186 Tbilisi, Georgia; Institute of Cognitive Neurosciences, Free University of Tbilisi, 0159 Tbilisi, Georgia.
Wei-Hsiang Lin, Laboratory of Psychophysics, Brain Mind Institute, School of Life Sciences, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland.
Ophélie Favrod, Laboratory of Psychophysics, Brain Mind Institute, School of Life Sciences, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland.
Andreas Brand, Laboratory of Psychophysics, Brain Mind Institute, School of Life Sciences, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland.
Patrícia Figueiredo, Institute for Systems and Robotics – Lisboa, Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, 1049-001 Lisbon, Portugal.
Maya Roinishvili, Institute of Cognitive Neurosciences, Free University of Tbilisi, 0159 Tbilisi, Georgia; Laboratory of Vision Physiology, Ivane Beritashvili Centre of Experimental Biomedicine, 0160 Tbilisi, Georgia.
Michael H Herzog, Laboratory of Psychophysics, Brain Mind Institute, School of Life Sciences, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland.
Funding
This work was partially funded by the Fundação para a Ciência e a Tecnologia under grant FCT PD/BD/105785/2014 and the National Centre of Competence in Research (NCCR) Synapsy financed by the Swiss National Science Foundation under grant 51NF40-185897.
Conflict of interest statement: None declared.
References
- Abdi H, Williams LJ. Partial least squares methods: Partial least squares correlation and partial least square regression. In: Reisfeld B, Mayeno AN, editors. Computational toxicology. Methods in molecular biology. Totowa (NJ): Humana Press; 2013. pp. 549–579 [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Paradiso S, O’Leary DS. “Cognitive dysmetria” as an integrative theory of schizophrenia: A dysfunction in cortical-subcortical-cerebellar circuitry? Schizophr Bull. 1998:24:203–218. [DOI] [PubMed] [Google Scholar]
- Andreou C, Leicht G, Nolte G, Polomac N, Moritz S, Karow A, Hanganu-Opatz IL, Engel AK, Mulert C. Resting-state theta-band connectivity and verbal memory in schizophrenia and in the high-risk state. Schizophr Res. 2015:161:299–307. [DOI] [PubMed] [Google Scholar]
- Avila M, Thaker G, Adami H. Genetic epidemiology and schizophrenia: a study of reproductive fitness. Schizophr Res. 2001:47:233–241. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Bury A, Hodgkinson KA, Honer WG. Reproductive fitness in familial schizophrenia. Schizophr Res. 1996:21:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros NN, Arfken C, Galderisi S, Warrick J, Pratt G, Iacono W. The status of spectral EEG abnormality as a diagnostic test for schizophrenia. Schizophr Res. 2008:99:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL, Freedman R, Schork NJ, Gottesman II. Deconstructing schizophrenia: an overview of the use of endophenotypes in order to understand a complex disorder. Schizophr Bull. 2006:33:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL, Light GA, Swerdlow NR. Prepulse inhibition and P50 suppression are both deficient but not correlated in schizophrenia patients. Biol Psychiatry. 2007:61:1204–1207. [DOI] [PubMed] [Google Scholar]
- Burmeister M, McInnis MG, Zöllner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008:9:527–540. [DOI] [PubMed] [Google Scholar]
- Burns J. The social brain hypothesis of schizophrenia. World Psychiatry Off J World Psychiatr Assoc WPA. 2006:5:77–81. [PMC free article] [PubMed] [Google Scholar]
- Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD, Keshavan MS, Tamminga CA. Identification of distinct psychosis biotypes using brain-based biomarkers. Am J Psychiatry. 2016:173:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz JR, Chicherov V, Herzog MH, Figueiredo P. An automatic pre-processing pipeline for EEG analysis (APP) based on robust statistics. Clin Neurophysiol. 2018:129:1427–1437. [DOI] [PubMed] [Google Scholar]
- da Cruz JR, Favrod O, Roinishvili M, Chkonia E, Brand A, Mohr C, Figueiredo P, Herzog MH. EEG microstates are a candidate endophenotype for schizophrenia. Nat Commun. 2020a:11:3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz JR, Shaqiri A, Roinishvili M, Favrod O, Chkonia E, Brand A, Figueiredo P, Herzog MH. Neural compensation mechanisms of siblings of schizophrenia patients as revealed by high-density EEG. Schizophr Bull. 2020b:46:1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo G, Daverio A, Ferrentino F, Santarnecchi E, Ciabattini F, Monaco L, Lisi G, Barone Y, Di Lorenzo C, Niolu C, et al. Altered resting-state EEG source functional connectivity in schizophrenia: the effect of illness duration. Front Hum Neurosci. 2015:9:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D, Goldberg TE, Gold JM, Elvevag B, Weinberger DR. Cognitive factor structure and invariance in people with schizophrenia, their unaffected siblings, and controls. Schizophr Bull. 2011:37:1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favrod O, Roinishvili M, da Cruz JR, Brand A, Okruashvili M, Gamkrelidze T, Figueiredo P, Herzog MH, Chkonia E, Shaqiri A. Electrophysiological correlates of visual backward masking in patients with first episode psychosis. Psychiatry Res Neuroimaging. 2018:282:64–72. [DOI] [PubMed] [Google Scholar]
- Friston K, Brown HR, Siemerkus J, Stephan KE. The dysconnection hypothesis (2016). Schizophr Res. 2016:176:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garobbio S, Roinishvili M, Favrod O, da Cruz JR, Chkonia E, Brand A, Herzog MH. Electrophysiological correlates of visual backward masking in patients with bipolar disorder. Psychiatry Res Neuroimaging. 2021:307:111206. [DOI] [PubMed] [Google Scholar]
- Hedge C, Powell G, Sumner P. The reliability paradox: why robust cognitive tasks do not produce reliable individual differences. Behav Res Methods. 2018:50:1166–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, Nordentoft M, Glenthøj B. Heritability of schizophrenia and schizophrenia spectrum based on the nationwide Danish twin register. Biol Psychiatry. 2018:83:492–498. [DOI] [PubMed] [Google Scholar]
- Howes OD, Kapur S. The dopamine hypothesis of schizophrenia: version III—the final common pathway. Schizophr Bull. 2009:35:549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, MacDonald ML, Elswick DE, Sweet RA. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies: glutamate system and schizophrenia. Ann N Y Acad Sci. 2015:1338:38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: Which evolutionary genetic models work best? Behav Brain Sci. 2006:29:385–404. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Birnbaum H, Demler O, Falloon IRH, Gagnon E, Guyer M, Howes MJ, Kendler KS, Shi L, Walters E, et al. The prevalence and correlates of nonaffective psychosis in the National Comorbidity Survey Replication (NCS-R). Biol Psychiatry. 2005:58:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-J, Jeong J, Chae J-H, Park S, Yong Kim S, Jin Go H, Paik I-H, Kim K-S, Choi B. An estimation of the first positive Lyapunov exponent of the EEG in patients with schizophrenia. Psychiatry Res Neuroimaging. 2000:98:177–189. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Williams LJ, McIntosh AR, Abdi H. Partial least squares (PLS) methods for neuroimaging: a tutorial and review. NeuroImage. 2011:56:455–475. [DOI] [PubMed] [Google Scholar]
- Liu C, Everall I, Pantelis C, Bousman C. Interrogating the evolutionary paradox of schizophrenia: a novel framework and evidence supporting recent negative selection of schizophrenia risk alleles. Front Genet. 2019:10:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCabe JH, Koupil I, Leon DA. Lifetime reproductive output over two generations in patients with psychosis and their unaffected siblings: the Uppsala 1915–1929 Birth Cohort Multigenerational study. Psychol Med. 2009:39:1667. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Bookstein FL, Haxby JV, Grady CL. Spatial pattern analysis of functional brain images using partial least squares. NeuroImage. 1996:3:143–157. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Chau WK, Protzner AB. Spatiotemporal analysis of event-related fMRI data using partial least squares. NeuroImage. 2004:23:764–775. [DOI] [PubMed] [Google Scholar]
- Messamore E. Niacin subsensitivity is associated with functional impairment in schizophrenia. Schizophr Res. 2012:137:180–184. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Poline J-B, Kohn PD, Holt JL, Egan MF, Weinberger DR, Berman KF. Evidence for abnormal cortical functional connectivity during working memory in schizophrenia. Am J Psychiatry. 2001:158:1809–1817. [DOI] [PubMed] [Google Scholar]
- Moreno-Küstner B, Martín C, Pastor L. Prevalence of psychotic disorders and its association with methodological issues. A systematic review and meta-analyses. PLoS One. 2018:13:e0195687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SE, Young J, Patel AX, Whitaker KJ, Scarpazza C, van Amelsvoort T, Marcelis M, van Os J, Donohoe G, Mothersill D, et al. Functional magnetic resonance imaging connectivity accurately distinguishes cases with psychotic disorders from healthy controls, based on cortical features associated with brain network development. Biol Psychiatry Cogn Neurosci Neuroimaging. 2021:6:1125–1134. [DOI] [PubMed] [Google Scholar]
- Moser DA, Doucet GE, Lee WH, Rasgon A, Krinsky H, Leibu E, Ing A, Schumann G, Rasgon N, Frangou S. Multivariate associations among behavioral, clinical, and multimodal imaging phenotypes in patients with psychosis. JAMA Psychiatry. 2018:75:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothi SS, Sudarshan M, Tandon N, Tamminga C, Pearlson G, Sweeney J, Clementz B, Keshavan MS. Machine learning improved classification of psychoses using clinical and biological stratification: update from the bipolar-schizophrenia network for intermediate phenotypes (B-SNIP). Schizophr Res. 2019:214:60–69. [DOI] [PubMed] [Google Scholar]
- Nikulin VV, Jönsson EG, Brismar T. Attenuation of long-range temporal correlations in the amplitude dynamics of alpha and beta neuronal oscillations in patients with schizophrenia. NeuroImage. 2012:61:162–169. [DOI] [PubMed] [Google Scholar]
- Phang C-R, Noman F, Hussain H, Ting C-M, Ombao H. A multi-domain connectome convolutional neural network for identifying schizophrenia from EEG connectivity patterns. IEEE J Biomed Health Inform. 2020:24:1333–1343. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Huckins G, Varoquaux G. Establishment of best practices for evidence for prediction: a review. JAMA Psychiatry. 2020:77:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GW, Michie PT, Johnston J, Innes-Brown H, Kent A, Clissa P, Jablensky AV. A multivariate electrophysiological endophenotype, from a unitary cohort, shows greater research utility than any single feature in the Western Australian family study of schizophrenia. Biol Psychiatry. 2006:60:1–10. [DOI] [PubMed] [Google Scholar]
- Rieger K, Diaz Hernandez L, Baenninger A, Koenig T. 15 years of microstate research in schizophrenia—Where are we? A meta-analysis. Front Psychiatry. 2016:7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakowski J, Weterle R. Niacin test in schizophrenia and affective illness. Biol Psychiatry. 1991:29:834–836. [DOI] [PubMed] [Google Scholar]
- Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005:2:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium . Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014:511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack HG, Kahn RS. Detecting neuroimaging biomarkers for psychiatric disorders: sample size matters. Front Psychiatry. 2016:7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman LJ, Hellemann G, Nuechterlein KH, Greenwood TA, Braff DL, Cadenhead KS, Calkins ME, Freedman R, Gur RE, Gur RC, et al. Factor structure and heritability of endophenotypes in schizophrenia: findings from the Consortium On The Genetics of Schizophrenia (COGS-1). Schizophr Res. 2015:163:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone JC, Ward AJ, Rotella P, Collins J, Windisch R. An evaluation of variation in published estimates of schizophrenia prevalence from 1990–2013: a systematic literature review. BMC Psychiatry. 2015:15:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Tang Y, Lim KO, Wang J, Tong S, Li H, He B. Abnormal dynamics of EEG oscillations in schizophrenia patients on multiple time scales. IEEE Trans Biomed Eng. 2014:61:1756–1764. [DOI] [PubMed] [Google Scholar]
- Székely GJ, Rizzo ML. The distance correlation t -test of independence in high dimension. J Multivar Anal. 2013:117:193–213. [Google Scholar]
- Toomey R, Faraone SV, Seidman LJ, Kremen WS, Pepple JR, Tsuang MT. Association of neuropsychological vulnerability markers in relatives of schizophrenic patients. Schizophr Res. 1998:31:89–98. [DOI] [PubMed] [Google Scholar]
- Tucker LR. An inter-battery method of factor analysis. Psychometrika. 1958:23:111–136. [Google Scholar]
- Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010:11:100–113. [DOI] [PubMed] [Google Scholar]
- Varoquaux G. Cross-validation failure: small sample sizes lead to large error bars. NeuroImage. 2018:180:68–77. [DOI] [PubMed] [Google Scholar]
- Venables NC, Bernat EM, Sponheim SR. Genetic and disorder-specific aspects of resting state EEG abnormalities in schizophrenia. Schizophr Bull. 2009:35:826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu J, Sui J, Pearlson G, Calhoun VD. A hybrid machine learning method for fusing fMRI and genetic data: combining both improves classification of schizophrenia. Front Hum Neurosci. 2010:4:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B Stat Methodol. 2005:67:301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




