Abstract
Objectives
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become established in the human population, making the need to develop safe and effective treatments critical. We have developed the small-molecule antiviral ensitrelvir, which targets the 3C-like (3CL) protease of SARS-CoV-2. This study evaluated the in vitro and in vivo efficacy of ensitrelvir compared with that of another SARS-CoV-2 3CL PI, nirmatrelvir.
Methods
Cultured cells, BALB/cAJcl mice and Syrian hamsters were infected with various SARS-CoV-2 strains, including the ancestral strain WK-521, mouse-adapted SARS-CoV-2 (MA-P10) strain, Delta strain and Omicron strain. Ensitrelvir efficacy was compared with that of nirmatrelvir. Effective concentrations were determined in vitro based on virus-induced cytopathic effects, viral titres and RNA levels. Lung viral titres, nasal turbinate titres, body-weight changes, and animal survival were also monitored.
Results
Ensitrelvir and nirmatrelvir showed comparable antiviral activity in multiple cell lines. Both ensitrelvir and nirmatrelvir reduced virus levels in the lungs of mice and the nasal turbinates and lungs of hamsters. However, ensitrelvir demonstrated comparable or better in vivo efficacy than that of nirmatrelvir when present at similar or slightly lower unbound-drug plasma concentrations.
Conclusions
Direct in vitro and in vivo efficacy comparisons of 3CL PIs revealed that ensitrelvir demonstrated comparable in vitro efficacy to that of nirmatrelvir in cell culture and exhibited equal to or greater in vivo efficacy in terms of unbound-drug plasma concentration in both animal models evaluated. The results suggest that ensitrelvir may become an important resource for treating individuals infected with SARS-CoV-2.
Introduction
Coronavirus disease 2019 (COVID-19) has become a health challenge worldwide, resulting from the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The establishment of SARS-CoV-2 in the human population makes the need for the development of safe and effective treatments to prevent or limit COVID-19 critical. It is reasonable to assume that a combination of vaccination and oral antiviral medications will provide the most beneficial approach for managing SARS-CoV-2 infection and COVID-19.
SARS-CoV-2, a member of the family Coronaviridae, has a virion particle that contains four structural proteins, including the spike (S) protein.1 The S protein forms a trimeric structure that is anchored to the viral membrane and directly contacts the cellular receptor angiotensin-converting enzyme 2 (ACE2) on the host cell via its receptor-binding domain.2,3 The biological properties of S protein make it the primary target for vaccine development against COVID-19. However, since the beginning of the COVID-19 pandemic, multiple virus variants have emerged, including five variants of concern: Alpha, Beta, Gamma, Delta and Omicron variants. The variants have characteristic mutations that especially accumulate in the S protein.4 This variation and the risk of vaccines losing their efficacy as the viruses evolve increase the need for other treatment options for individuals who may become infected with SARS-CoV-2.
There are several antivirals targeting SARS-CoV-2 that have been approved or are under development. These can be categorized as monoclonal antibodies (mAbs) or small molecules that aim to interfere with virus replication.4 Currently, mAbs require administration in a hospital setting, and because they target the S protein, they may lose their clinical efficacy, as observed with the Omicron variants.4,5 Unlike vaccines and mAbs, oral antivirals are direct-acting and less susceptible to viral mutations and changes in the S protein. Oral therapeutic agents approved for use, or showing potential, target the viral RNA-dependent RNA polymerase (RdRp) or 3C-like (3CL) protease. Clinically developed agents include molnupiravir, targeting RdRp, and nirmatrelvir, targeting 3CL protease.6–8 Molnupiravir is the prodrug form of the antiviral nucleotide analogue, β-D-N4-hydroxycytidine (NHC). Another RdRp-targeting drug is remdesivir, a nucleotide analogue prodrug that was originally developed to treat individuals with Ebola virus infections.
The SARS-CoV-2 3CL protease is an essential viral protein involved in the processing of the SARS-CoV-2-encoded polyprotein and viral replication.9 Importantly, 3CL protease is conserved among coronaviruses, but no human host-cell proteases with the same substrate specificity have been identified.9 The characteristics and biological importance of 3CL protease makes it an ideal antiviral target in SARS-CoV-2.
We have developed the antiviral compound ensitrelvir fumaric acid (S-217622 fumaric acid, hereinafter ensitrelvir), which, like nirmatrelvir, targets the 3CL protease of SARS-CoV-2. Ensitrelvir exhibits anti-SARS-CoV-2 activity in non-clinical settings using cultured cells and mouse infection models,10 and significantly reduced viral titres in nasal lavage fluid in Phase 2a and Phase 2b studies.11,12
The current study evaluated the in vitro and in vivo efficacy of ensitrelvir compared with that of nirmatrelvir. The efficacies were evaluated in vitro using various cell lines and different SARS-CoV-2 variants and in vivo using mouse and hamster model systems.
Materials and methods
Ethics
The study design and execution were compliant with the Standards for the Reliability of Application Data (Article 43, Enforcement Regulations, Law for the Assurance of Quality, Efficacy, and Safety of Pharmaceuticals and Medical Devices). All animal protocols were approved by the Director of Shionogi Pharmaceutical Research Centre Institute (Shionogi & Co., Ltd., Toyonaka, Japan) based on the review by the Institutional Animal Care and Use Committee (Approval Nos. S21068D-0015, S21199D-0007 and S20121D-0113). The institutional animal facilities were Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International accredited.
Cell lines and SARS-CoV-2 variants
Transmembrane serine protease 2 (TMPRSS-2)-expressing VeroE6 (VeroE6/TMPRSS2) cells13 were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (Osaka, Japan). HEK293T/ACE2-TMPRSS2 cells were obtained from GeneCopoeia (Rockville, MD, USA). The VeroE6/TMPRSS2 and HEK293T/ACE2-TMPRSS2 cells were maintained in culture medium of DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% heat-inactivated FBS (Sigma–Aldrich Co. Ltd., St. Louis, MO, USA) and 1% penicillin-streptomycin (Thermo Fisher Scientific). Human nasal cavity MucilAir™ cells and MucilAir™ medium were purchased from Epithelix (Plan-les-Ouates, Switzerland). The National Institute of Infectious Diseases (NIID) of Japan provided the hCoV-19/Japan/TY/WK-521 strain (WK-521, Pango Lineage A), which was originally isolated from a traveller returning from Wuhan at the end of January 2020.13 The mouse-adapted SARS-CoV-2 (MA-P10) strain (mouse-adapted hCoV-19/Japan/TY/WK-521/2020, Pango lineage A)10 was obtained from the Division of Molecular Pathobiology, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan. The SARS-CoV-2 strains hCoV-19/Japan/TY11-927/2021 (Delta, Pango Lineage B.1.617.2), hCoV-19/Japan/TY38-873/2021 (Omicron, Pango Lineage BA.1) and hCoV-19/Japan/TY40-385/2022 (Omicron, Pango Lineage BA.2) were provided by NIID, Japan. All the SARS-CoV-2 viruses were propagated on VeroE6/TMPRSS2 cells. Virus stocks were prepared and titred using TCID50 assays with VeroE6/TMPRSS2 cells.
Anti-SARS-CoV-2 test compounds
Ensitrelvir (Shionogi & Co. Ltd., Osaka, Japan) was synthesized as previously described.14 Nirmatrelvir was also synthesized at Shionogi & Co., Ltd. Remdesivir and NHC, the active ingredients of the prodrug molnupiravir, were obtained from MedChemExpress (Monmouth Junction, NJ, USA). The permeability glycoprotein (P-gp) inhibitor CP-100356 was purchased from Sigma–Aldrich Co. Ltd.
Virus replication inhibition assay using cell lines
Ensitrelvir, nirmatrelvir, remdesivir and NHC were diluted with DMSO (Nacalai Tesque, Kyoto City, Japan). Antiviral activity of the agents against SARS-CoV-2 was assessed in the VeroE6/TMPRSS2 cells by monitoring viral RNA levels in the infected cells using quantitative RT–PCR (qRT–PCR) assays. The VeroE6/TMPRSS2 cells were suspended in culture medium consisting of DMEM supplemented with 10% FBS and 1% penicillin-streptomycin, seeded into 96-well plates (1.5 × 104 cells/well) and incubated at 37°C with 5% CO2 overnight. The next day, the cells were infected with hCoV-19/Japan/TY/WK-521 strain at 100 TCID50/well in viral assay medium of MEM (Thermo Fisher Scientific), containing 2% FBS and 1% penicillin-streptomycin and incubated at 37°C with 5% CO2 for 1 h. Post-adsorption, the inoculum was removed and viral assay medium containing the diluted compounds was added. The infected cells were incubated at 37°C with 5% CO2 for 1 day, followed by extraction of the viral RNA from the cell lysate using TRIzol LS (Thermo Fisher Scientific) and a Direct-zol-96 RNA Kit (Zymo Research, Irvine, CA, USA). The viral RNA was quantified by real-time PCR using an Applied Biosystems QuantStudio 5 system (Thermo Fisher Scientific) with SARS-CoV-2-specific primers and probes.10
Antiviral activity against SARS-CoV-2 in HEK293T/ACE2-TMPRSS2 cells was assessed by monitoring cell viability based on induced cytopathic effects (CPEs). The HEK293T/ACE2-TMPRSS2 cells were suspended in the viral assay medium and seeded with the diluted compounds into 96-well plates (1.5 × 104 cells/well). The cells were then infected with hCoV-19/Japan/TY/WK-521 strain at 3000 TCID50/well and cultured for 3 days at 37°C with 5% CO2. Cell viability was assessed using a CellTiter-Glo 2.0 assay (Promega, Madison, WI, USA). The luminescent signal (Relative Light Unit) was measured using an EnSpire multiplate reader (PerkinElmer Japan Co., Ltd.), and the percent inhibition of CPE induced by SARS-CoV-2 was calculated. Cell-control wells were not infected nor treated with any test or reference substance. Virus-control wells were infected with virus but not treated with a test or reference substance. The concentrations of substances achieving 50% inhibition (EC50) or 90% inhibition (EC90) against SARS-CoV-2 replication were calculated using software XLfit 5.3.1.3 (fit model: 205). The mean and SD values were calculated based on three independent experiments.
Virus replication inhibition assay using human airway epithelial cells
Human nasal epithelial cells (MucilAir™; Epithelix Sàrl, Switzerland) were seeded at approximately 5.0 × 105 cells/well into a 24-well Transwell plate and then infected with the SARS-CoV-2 Delta strain (hCoV-19/Japan/TY11-927/2021) or SARS-CoV-2 Omicron BA.1 strain (hCoV-19/Japan/TY38-873/2021) at approximately 5000 TCID50/well. The cells were incubated at 37°C in a 5% CO2 incubator for 2 h. After incubation, the cells were washed with MucilAir™ culture medium to remove unabsorbed viruses and transferred to a 24-well Transwell plate containing serially diluted compound solutions prepared in 700 μL of MucilAir™ culture medium. The infected cells were incubated at 37°C in 5% CO2. The cell culture fluids were collected at 2 and 3 days post infection (p.i.) for the Delta strain and 1 and 2 days p.i. for the Omicron strain. The collected samples were stored at −80°C until use. The samples were subjected to viral titration using VeroE6/TMPRSS2 cells.10 The concentrations achieving EC90 against SARS-CoV-2 replication were calculated using the two-point method. The mean and SD values were calculated based on three independent experiments.
Animals and in vivo models
Female BALB/cAJcl mice were purchased from CLEA Japan, Inc. (Tokyo, Japan). Male Syrian hamsters were purchased from Japan SLC, Inc. (Shizuoka, Japan). All the mice and hamsters were maintained in an environment controlled at 20°C–26°C, 30%–70% relative humidity, and a 12 h light/dark cycle. The mice were housed 4–5/cage, and the hamsters were housed 2/cage. The animals received a standard chow diet of CE-2 (CLEA Japan, Inc.) and were provided water ad libitum.
The mice (20–22 weeks of age) were intranasally infected as previously described with 3.0 × 103 TCID50 of the MA-P10 strain.10,15 The mice were then orally treated twice a day for 5 days starting 1 day p.i. Dosing included 16 or 32 mg/kg ensitrelvir in 0.5% methylcellulose (MC), 300 or 1000 mg/kg nirmatrelvir in 0.5% MC with 2% Tween 80,15 or vehicle (0.5% MC). The pharmacokinetic (PK) analysis of nirmatrelvir in mice is described in the Supplementary Methods (available as Supplementary data at JAC Online), and the findings are shown in Figure S1. The PK data for ensitrelvir in mice were previously reported.14 Mice were monitored daily for weight and survival through 14 days p.i. (n = 5/group). Mice were euthanized on Days 2, 3 and 4 p.i., and their lungs dissected and homogenized as previously described (n = 4–5/group).10
The hamsters (7–9 weeks of age) were intranasally infected as previously described with 1.0 × 104 TCID50 of SARS-CoV-2 Omicron BA.2.16–18 The hamsters were then treated twice a day at an 8/16 h interval for 5 days starting 1 day p.i. Dosing included oral ensitrelvir (12.5 or 50 mg/kg) in 0.5% MC, oral vehicle (0.5% MC), subcutaneous nirmatrelvir [250 or 750/250 mg/kg (initial dose 750 mg/kg; subsequent doses 250 mg/kg)] in 0.5% MC, or subcutaneous vehicle (0.5% MC). The PK analysis of nirmatrelvir in hamsters is described in the Supplementary Methods, and the findings are shown in Figure S1. The PK data of ensitrelvir in hamsters were previously reported.18,19 As we found the bioavailability of the crystalline form of nirmatrelvir to be low following oral administration, it was administered subcutaneously in the current study in order to produce long-lasting drug exposure. The hamsters were monitored daily for weight and survival through 14 days p.i. (n = 3–4/group). Hamsters were euthanized at 2 and 4 days p.i., their lungs and nasal turbinates were dissected, and lung homogenates and nasal turbinate homogenates prepared (n = 6–8/group).18 Viral titres of lung and nasal turbinate homogenates were determined using TCID50 assays with VeroE6/TMPRSS2 cells as previously described.10
Evaluation of ensitrelvir and nirmatrelvir protein binding in vitro
Blood was collected from female BALB/cAJcl mice and male Syrian hamsters (5–6 weeks, non-fasted) and centrifuged to obtain the serum. Pooled serum from female BALB/cAJcl mice (8 weeks, fasted) was purchased from CLEA Japan, Inc. for the protein-binding study of ensitrelvir in mice.
Radiolabelled ensitrelvir, [14C]-ensitrelvir, was synthesized at Shionogi & Co., Ltd. (Osaka, Japan). After preparation of the serum samples containing 5 or 50 µg/mL [14C]-ensitrelvir (n = 3 each), 0.8 mL of the samples were transferred to a Centrifree Ultrafiltration device (Merck Millipore Corporation, Burlington, MA, USA) and then centrifuged at 37°C for 3 min. A portion of the serum samples (50 µL) and all the filtrate were weighed in counting vials. The radioactivity levels were determined using a TRI-CARB 3100TR liquid scintillation counter (PerkinElmer Co., Ltd., Waltham, MA, USA). The protein binding ratio was then calculated using the following equation:
where Cfiltrate and Cserum sample represent the concentration of radioactivity in the filtrate and the concentration of radioactivity in the serum samples, respectively.
In the evaluation of nirmatrelvir, protein binding ratios were evaluated using equilibrium dialysis assays. Serum samples containing 4 µM nirmatrelvir (n = 2) were loaded into an equilibrium dialysis device and dialysed against PBS (pH 7.4). After incubation at 37°C for 24 h, the peak areas of nirmatrelvir in the serum and PBS samples were determined by LC MS. The protein binding ratio was calculated using the following equation:
where Peak AreaPBS sample and Peak Areaserum sample represent the peak area of ensitrelvir in the PBS samples and serum samples, respectively.
Plasma concentration analysis
To evaluate the plasma concentration of ensitrelvir and nirmatrelvir in the mice and hamsters, blood samples were collected 24, 48 and 72 h after the first dosing (n = 4–8/group per timepoint) and centrifuged to obtain the plasma. The plasma concentrations of ensitrelvir and nirmatrelvir were determined by LC/MS/MS analysis using a SCIEX API5000 or Triple Quad 6500+ system (Sciex, Framingham, MA, USA) after protein precipitation with acetonitrile (MeCN).
Statistical analysis
Statistical analysis was performed using SAS Version 9.4 software (SAS Institute, Cary, NC, USA). The significance level was set at P = 0.05 for two-sided tests. The uniformity of mean body weight was confirmed among the different experimental groups at the time of group assignment using one-way analysis of variance. Endpoint viral titres in the lungs and nasal turbinates were determined using a logarithmic scale. Tukey’s method was applied to assess the effects of ensitrelvir and nirmatrelvir on viral titres in the lungs and nasal turbinates. Survival time comparisons between each ensitrelvir-treated or nirmatrelvir-treated group and the vehicle-treated group were analysed using a log-rank test. The proportion of current body weight to initial body weight was imputed at 80% for all timepoints post-death for mice that died during the experimental period. The multiplicity of statistical tests for survival-time analyses were adjusted using the fixed-sequence procedure.
Results
In vitro efficacy of ensitrelvir compared with that of nirmatrelvir, remdesivir and NHC
The anti-SARS-CoV-2 efficacy of ensitrelvir was initially evaluated against the SARS-CoV-2 ancestral strain (WK-521) in VeroE6/TMPRSS2 cells with and without the P-gp inhibitor CP-100356 (0.75 µmol/L) and in HEK293T/ACE2-TMPRSS2 cells and compared with that of nirmatrelvir, remdesivir and NHC (Table 1). Nirmatrelvir efficacy is known to be affected by P-gp.18 Because nirmatrelvir and remdesivir serve as major substrates for the plasma membrane multidrug transporter P-gp, these antivirals require a P-gp inhibitor, such as CP-100356, to inhibit the efflux of the antivirals from Vero cells, which express high levels of P-gp.20 Ensitrelvir and nirmatrelvir showed comparable antiviral activity using VeroE6/TMPRSS2+ P-gp inhibitor and HEK293T/ACE2-TMPRSS2 cells. The antiviral activities of ensitrelvir, nirmatrelvir and remdesivir improved 2.74-, 61.4- and 51.8-fold, respectively, with the addition of the P-gp inhibitor, CP-100356, as indicated by the decreased EC90 values (Table 1).
Table 1.
In vitro efficacy of ensitrelvir in cell lines compared with that of nirmatrelvir, remdesivir and NHC
| Cells | SARS-CoV-2 | Assay | Ensitrelvir | Nirmatrelvir | Remdesivir | NHC |
|---|---|---|---|---|---|---|
| VeroE6/TMPRSS2 | WK-521 | qRT–PCR EC90 (nmol/L) |
91.1 ± 26.4 | 1320 ± 150 | 906 ± 243 | 606 ± 128 |
| VeroE6/TMPRSS2 + P-gp inhibitor | WK-521 | qRT-PCR EC90 (nmol/L) |
33.3 ± 2.7 | 21.5 ± 4.0 | 17.5 ± 3.2 | 1450 ± 130 |
| HEK293T/ACE2-TMPRSS2 | WK-521 | CPE EC50 (nmol/L) |
27 ± 2 | 41 ± 13 | 12 ± 2 | 4000 ± 3500 |
Results are presented as mean ± SD calculated from three independent experiments.
+P-gp inhibitor, 0.75 µmol/L CP-100356.
In vitro efficacy of ensitrelvir against SARS-CoV-2 Delta and Omicron BA.1 variants in MucilAir™ cells
The in vitro efficacy of ensitrelvir was evaluated against SARS-CoV-2 Delta and Omicron BA.1 variants in human airway epithelial (MucilAir™) cells and compared with that of nirmatrelvir, remdesivir and NHC (Table 2). Ensitrelvir and nirmatrelvir showed comparable antiviral activity in the human primary airway epithelial cells infected with the SARS-CoV-2 variant strains (Table 2).
Table 2.
In vitro efficacy of ensitrelvir in human airway epithelial cells compared with that of nirmatrelvir, remdesivir and NHC
| Cells | SARS-CoV-2 | Assay | Day (p.i.) | Ensitrelvir | Nirmatrelvir | Remdesivir | NHC |
|---|---|---|---|---|---|---|---|
| MucilAir (nasal) | Delta | Viral titre | 2 | 51.4 ± 18.2 | 32.9 ± 2.9 | 35.9 ± 9.7 | 3830 ± 890 |
| EC90 (nmol/L) | 3 | 117 ± 26 | 46.8 ± 10.2 | 35.9 ± 14.0 | 6720 ± 3910 | ||
| MucilAir (nasal) | Omicron(BA.1) | Viral titre | 1 | 66.0 ± 12.5 | 37.6 ± 17.2 | 8.97 ± 6.64 | 2350 ± 890 |
| EC90 (nmol/L) | 2 | 160 ± 57 | 69.2 ± 14.1 | 21.1 ± 9.0 | 3660 ± 920 |
Results are presented as mean ± SD calculated from three independent experiments.
In vivo efficacy of ensitrelvir against mouse-adapted SARS-CoV-2 strain infection in mice compared with that of nirmatrelvir
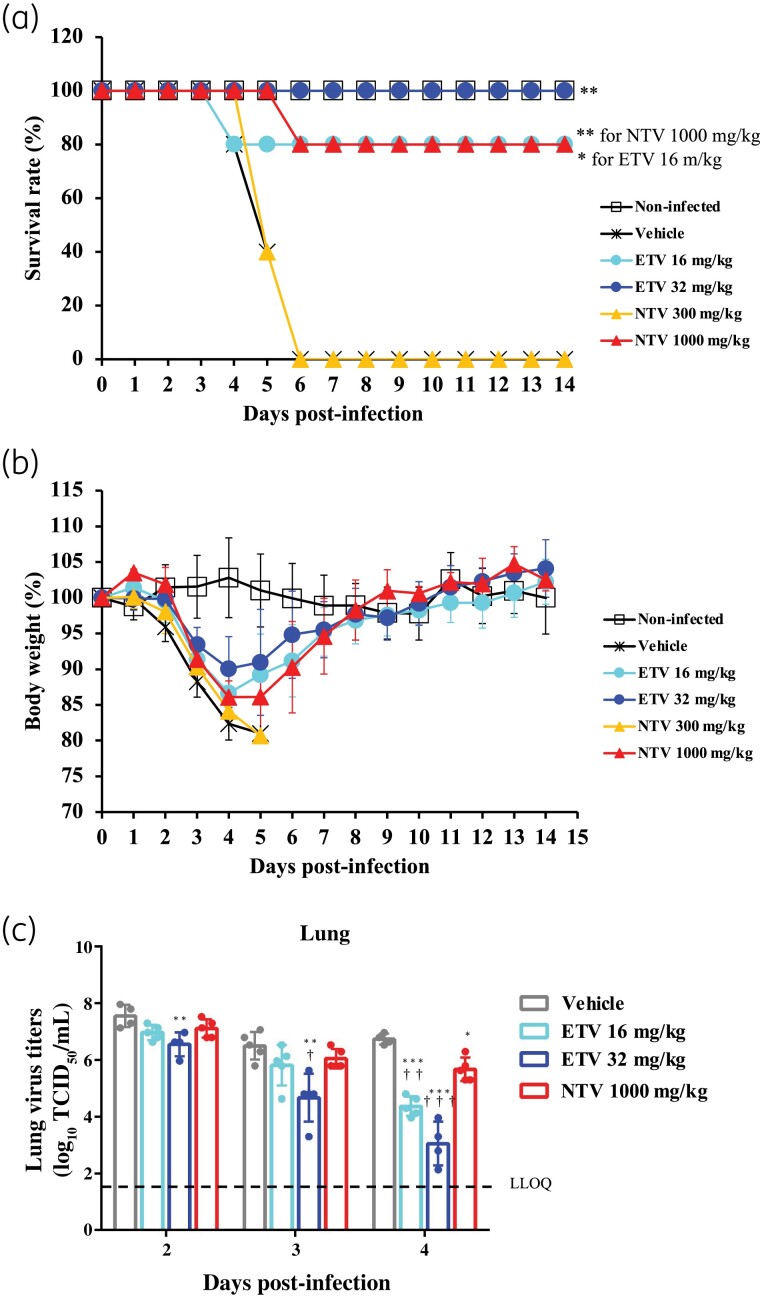
The in vivo efficacy of ensitrelvir against the MA-P10 strain in infected mice was compared with that of nirmatrelvir. Ensitrelvir at 16 and 32 mg/kg, and nirmatrelvir at 1000 mg/kg resulted in improved survival rates in the mice [Figure 1(a)]. However, 300 mg/kg nirmatrelvir failed to demonstrate any improvement in the survival rates. Furthermore, treatment of MA-P10 strain-infected mice with 16 and 32 mg/kg ensitrelvir and 1000 mg/kg nirmatrelvir improved the body-weight loss induced by SARS-CoV-2 infection [Figure 1(b)]. Ensitrelvir at 16 and 32 mg/kg and nirmatrelvir at 1000 mg/kg also demonstrated antiviral activity in the lungs of the mice at 4 days p.i. [Figure 1(c)]. The antiviral activity at 32 mg/kg ensitrelvir was greater than that of 1000 mg/kg nirmatrelvir, while 16 mg/kg ensitrelvir was equal to or greater than that of 1000 mg/kg nirmatrelvir. Ensitrelvir had higher protein binding rates in mice, at 97.6%, compared with that of nirmatrelvir, at 30.1% (Table 3). The unbound concentration of 32 mg/kg ensitrelvir in the plasma of mice at 24, 48 and 72 h post-treatment was comparable to that of 1000 mg/kg nirmatrelvir (Table 4).
Figure 1.
In vivo efficacy of ensitrelvir compared with that of nirmatrelvir in a mouse lethality model. Female BALB/cAJcl mice were nasally infected with the mouse-adapted SARS-CoV-2 (MA-P10) strain and then orally administered various doses of ensitrelvir (ETV), nirmatrelvir (NTV) or vehicle (0.5% MC solution) every 12 h (twice daily) for 5 days. The first administration was performed 1 day p.i. (a) Mouse survival was monitored daily. n = 5 mice/group. P values were calculated using log-rank test versus vehicle. *P < 0.05 and **P < 0.001. (b) Body-weight values are presented as a percentage of initial body weight through to Day 14 p.i. The graph bars represent the mean ± SD of 5 mice/group. (c) Lung viral titres were determined using TCID50 assays on the days p.i. indicated. Each point represents the mean ± SD of n = 4–5 mice. The dashed line represents the lower limit of quantification (LLOQ) of 1.80 log10 TCID50/mL. P values were calculated using Tukey’s test. *P < 0.05 versus vehicle; **P < 0.01 versus vehicle; ***P < 0.001 versus vehicle; †P < 0.05 versus 1000 mg/kg NTV; ††P < 0.01 versus 1000 mg/kg NTV; †††P < 0.001 versus 1000 mg/kg NTV. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Table 3.
Protein binding rate of ensitrelvir and nirmatrelvir in mice and hamsters
| Animal | Protein binding rate (%) | |
|---|---|---|
| Ensitrelvir | Nirmatrelvir | |
| Mouse | 97.6 | 33.1 |
| Hamster | 98.3 | 40.4 |
Table 4.
Plasma concentrations of ensitrelvir and nirmatrelvir in mice and hamsters
| Plasma concentration | ||||
|---|---|---|---|---|
| Mouse | ETV 16 mg/kg | ETV 32 mg/kg | NTV 1000 mg/kg | |
| t-C24h (ng/mL) | 4820 ± 2500 | 20 700 ± 10 400 | 1010 ± 910 | |
| t-C48h (ng/mL) | 22 400 ± 7500 | 37 700 ± 16 200 | 2050 ± 610 | |
| t-C72h (ng/mL) | 26 100 ± 12 200 | 56 300 ± 22 700 | 2230 ± 650 | |
| f-C24h (ng/mL) | 116 ± 60 | 497 ± 250 | 676 ± 609 | |
| f-C48h (ng/mL) | 538 ± 180 | 905 ± 389 | 1370 ± 410 | |
| f-C72h (ng/mL) | 626 ± 293 | 1350 ± 540 | 1490 ± 430 |
| Hamster | ETV 12.5 mg/kg | ETV 50 mg/kg | NTV 250 mg/kg | NTV 750/250 mg/kg |
| t-C24h (ng/mL) | 587 ± 183 | 7770 ± 4660 | 635 ± 375 | 1370 ± 190 |
| t-C48h (ng/mL) | 4490 ± 1310 | 29 300 ± 5100 | 1290 ± 60 | 1710 ± 470 |
| t-C72h (ng/mL) | 7530 ± 3960 | 16 700 ± 4000 | 1920 ± 600 | 1900 ± 660 |
| f-C24h (ng/mL) | 9.98 ± 3.11 | 132 ± 79 | 378 ± 224 | 817 ± 113 |
| f-C48h (ng/mL) | 76.3 ± 22.3 | 498 ± 87 | 769 ± 36 | 1020 ± 280 |
| f-C72h (ng/mL) | 128 ± 67 | 284 ± 68 | 1140 ± 360 | 1130 ± 390 |
ETV, ensitrelvir; NTV, nirmatrelvir; t-C, total concentration; f-C, unbound drug concentration; Cxxh, plasma concentration at xx h after the first administration of antiviral agent; f-Cxxh = t-Cxxh × (100%−protein binding rate).
In vivo efficacy of ensitrelvir against Omicron BA.2 infection in Syrian hamsters compared with that of nirmatrelvir
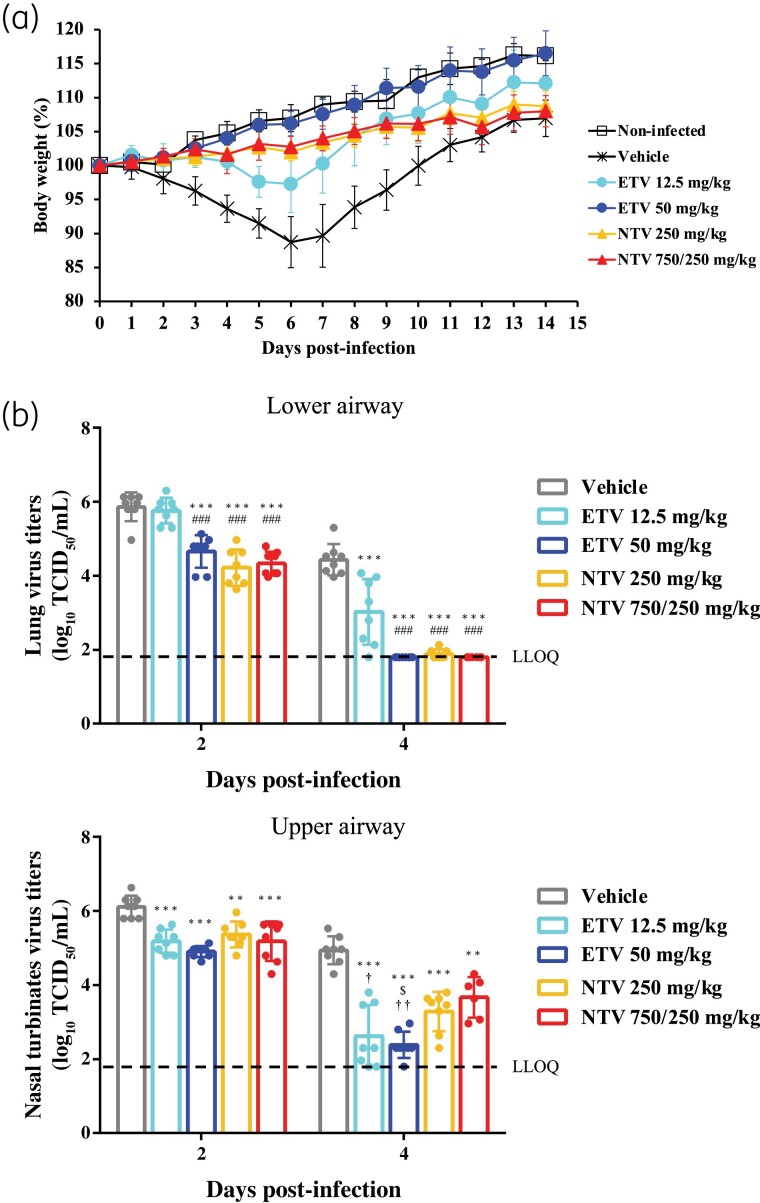
The in vivo efficacy of ensitrelvir was compared with that of nirmatrelvir in Syrian hamsters infected with the Omicron BA.2 strain. Ensitrelvir at 12.5 and 50 mg/kg, and nirmatrelvir at 250 and 750/250 mg/kg improved body-weight loss induced by SARS-CoV-2 infection [Figure 2(a)]. At 2 days p.i., 50 mg/kg ensitrelvir, and 250 and 750/250 mg/kg nirmatrelvir demonstrated antiviral activity in the lungs of hamsters, but 12.5 mg/kg ensitrelvir failed to show antiviral activity [Figure 2(b)]. Meanwhile, all the compound dosing groups showed antiviral activity in hamster lungs at 4 days p.i. In comparison, all compound dosing groups showed antiviral activity in the nasal turbinates of the hamsters at 2 and 4 days p.i. [Figure 2(b)]. In particular, at 4 days p.i., 12.5 and 50 mg/kg ensitrelvir showed greater antiviral activity in the nasal turbinates compared with that of nirmatrelvir. Ensitrelvir also had higher protein binding rates, at 98.3%, in hamsters compared with that of nirmatrelvir, at 40.4% (Table 3). The unbound concentration of 50 mg/kg ensitrelvir in the plasma of hamsters at 24, 48 and 72 h post-treatment was lower than that of 250 and 750/250 mg/kg nirmatrelvir.
Figure 2.
In vivo efficacy of ensitrelvir compared with that of nirmatrelvir against Omicron BA.2 in a hamster infection model. Syrian hamsters were intranasally infected with 1.0 × 104 TCID50 of SARS-CoV-2 Omicron BA.2. The hamsters were treated twice a day at an 8 h/16 h interval for 5 days starting 1 day p.i. with various doses of oral ensitrelvir (ETV), subcutaneous nirmatrelvir (NTV) or vehicle (0.5% MC solution). Hamsters were monitored daily. (a) Body-weight values are presented as a percentage of initial body weight through to Day 14 p.i. The graph bars represent the mean ± SD. n = 4/infected group; n = 3/non-infected group. (b) Hamster lungs (lower airway) and nasal turbinates (upper airway) were harvested and prepared 2 and 4 days p.i. Viral titres were determined using TCID50 assays. Each point represents the mean ± SD of n = 6–8/group. The dashed line represents the LLOQ of 1.80 log10 TCID50/mL. P values were calculated using Tukey’s test. **P < 0.01 versus vehicle; ***P < 0.001 versus vehicle; ###P < 0.01 versus 12.5 mg/kg ETV; §P < 0.05 versus 250 mg/kg NTV; †P < 0.05 versus 750/250 mg/kg NTV; ††P<0.01 versus 750/250 mg/kg NTV. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
The need for safe and effective treatments for individuals infected with SARS-CoV-2 is critical, given the global health concern caused by COVID-19. Ensitrelvir is a newly developed antiviral that targets the 3CL protease of SARS-CoV-2. Ensitrelvir and nirmatrelvir had comparable in vitro antiviral activity in the cell lines evaluated, with the antiviral efficacy of ensitrelvir in human airway epithelial cells being comparable to that of nirmatrelvir and greater than that of NHC. Meanwhile, nirmatrelvir and remdesivir were more susceptible to the effects of P-gp than that of ensitrelvir. In vivo, ensitrelvir improved survivability of SARS-CoV-2-infected mice and exhibited antiviral activity at 16 and 32 mg/kg, which was equal to or greater than that of 1000 mg/kg nirmatrelvir. Ensitrelvir and nirmatrelvir each improved body-weight changes induced by SARS-CoV-2 infection in both mice and hamsters, including the Omicron BA.2 strain. Similar to nirmatrelvir, ensitrelvir exhibited antiviral activity in the lungs and nasal turbinates of hamsters.
Ensitrelvir and nirmatrelvir both exhibited in vitro and in vivo efficacy; however, when ensitrelvir and nirmatrelvir were at equivalent or slightly lower plasma concentrations of unbound drug, ensitrelvir showed better in vivo efficacy. Generally, antiviral compounds in plasma that are free from protein binding are distributed to tissues21 and likely to exhibit antiviral effects. As the in vitro efficacy for ensitrelvir and nirmatrelvir was comparable, it is possible that drug efflux transporters, such as P-gp, affected overall in vivo efficacy. Furthermore, in the hamster infection model, 12.5 mg/kg ensitrelvir was less effective in lowering lung viral titres than 750/250 mg/kg nirmatrelvir but showed greater antiviral activity against virus present in the nasal turbinates. Previous reports have also shown that ensitrelvir tends to lower virus titres in nasal turbinates.19 Although the reason why ensitrelvir is so effective in the nasal turbinates is unknown, its ability to easily suppress virus excreted from the nose may contribute clinically to the suppression of virus transmission.
The use of small-animal models that recapitulate SARS-CoV-2 infection is critical when it comes to investigating COVID-19 and evaluating potential therapeutic agents. The employment of mice and mouse-adapted SARS-CoV-2 strains as model systems has been previously described,10,15,22,23 as has the use of hamsters.18,24 Our current findings further demonstrate the utility of mice and hamsters as small-animal models for SARS-CoV-2 replication and infection studies, confirming them as an important resource for evaluating the efficacy of antivirals targeting SARS-CoV-2.
This study involved the direct comparison of the in vitro and in vivo efficacy of ensitrelvir under clinical development with that of nirmatrelvir, which also targets SARS-CoV-2 3CL protease. Identifying and developing safe and effective antiviral therapies is essential to expanding the arsenal available for combatting the worldwide health threat of COVID-19. Because epidemic strains of SARS-CoV-2 mutate and change over time, it is often difficult to compare therapeutic drug candidates that are evaluated in different clinical trials. Therefore, the comparison of non-clinical therapeutic effects is important for considering the potential of each clinical therapeutic effect.
The current findings indicate that ensitrelvir has the potential to suppress virus replication and prevent clinical disease caused by SARS-CoV-2 at levels equal to or better than that of nirmatrelvir. This suggests that ensitrelvir may become an important resource for individuals who become infected with SARS-CoV-2.
Supplementary Material
Acknowledgements
Portions of the reported findings were previously presented as posters at OPTIONS-XI Belfast (abstract nos. AOXI0134 and AOXI0168, 26–29 September 2022). We acknowledge the National Institute of Infectious Diseases (NIID) for providing the SARS-CoV-2 Delta strain (hCoV-19/Japan/TY11-927/2021) and Omicron strains (hCoV-19/Japan/TY38-873/2021 and hCoV-19/Japan/TY40-385/2022). We also acknowledge Prof. Hirofumi Sawa (Hokkaido University, Sapporo, Japan) for providing the SARS-CoV-2 (MA-P10) strain. We are grateful to all our colleagues who participated in the COVID-19 antiviral program at Shionogi and thank Shionogi TechnoAdvance Research Co., Ltd. for technical support in the pharmacological studies. Editorial support, in the form of medical writing, assembling tables and creating high-resolution images based on authors’ detailed directions, collating author comments, copyediting, fact checking and referencing, was provided by Editage, Cactus Communications, and funded by Shionogi & Co., Ltd.
Contributor Information
Takayuki Kuroda, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Haruaki Nobori, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Keita Fukao, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Kaoru Baba, Research Area for Drug Candidate Generation II, Shionogi TechnoAdvance Research Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Kazumi Matsumoto, Research Area for Drug Candidate Generation II, Shionogi TechnoAdvance Research Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Shinpei Yoshida, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Yukari Tanaka, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Ryosuke Watari, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Ryoko Oka, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Yasuyuki Kasai, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Kae Inoue, Research Area for Drug Candidate Generation II, Shionogi TechnoAdvance Research Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Sho Kawashima, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Alice Shimba, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Yoko Hayasaki-Kajiwara, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Miki Tanimura, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Qianhui Zhang, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Yuki Tachibana, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Teruhisa Kato, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Takao Shishido, Pharmaceutical Research Division, Shionogi & Co., Ltd., 1-1, Futaba-cho 3-chome, Toyonaka, Osaka 561-0825, Japan.
Funding
This work was supported by internal funding from Shionogi & Co., Ltd.
Transparency declarations
The authors declare the following competing financial interest(s): Shionogi has applied for a patent on ensitrelvir. T.K., H.N., K.F., K.B., K.M., S.Y., Y.T., R.W., R.O., Y.K., K.I., S.K., A.Y., Y.H.K., M.T., Q.Z., Y.T., T.K., and T.S. are employees of Shionogi & Co., Ltd., and its subsidiary company.
Supplementary data
Supplementary Methods and Figure S1 are available as Supplementary data at JAC Online.
References
- 1. Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol 2020; 41: 1100–15. 10.1016/j.it.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haynes BF, Corey L, Fernandes Pet al. Prospects for a safe COVID-19 vaccine. Sci Transl Med 2020; 12: eabe0948. 10.1126/scitranslmed.abe0948 [DOI] [PubMed] [Google Scholar]
- 3. Zhang J, Xiao T, Cai Yet al. Structure of SARS-CoV-2 spike protein. Curr Opin Virol 2021; 50: 173–82. 10.1016/j.coviro.2021.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vangeel L, Chiu W, De Jonghe Set al. Remdesivir, molnupiravir and nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res 2022; 198: 105252. 10.1016/j.antiviral.2022.105252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hammond J, Leister-Tebbe H, Gardner Aet al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med 2022; 386: 1397–408. 10.1056/NEJMoa2118542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imran M, Arora MK, Asdaq SMBet al. Discovery, development, and patent trends on molnupiravir: a prospective oral treatment for COVID-19. Molecules 2021; 26: 5795. 10.3390/molecules26195795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamb YN. Nirmatrelvir plus ritonavir: first approval. Drugs 2022; 82: 585–91. 10.1007/s40265-022-01692-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sendi P, Razonable RR, Nelson SBet al. First-generation oral antivirals against SARS-CoV-2. Clin Microbiol Infect 2022; 28: 1230–5. 10.1016/j.cmi.2022.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ullrich S, Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett 2020; 30: 127377. 10.1016/j.bmcl.2020.127377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uemura K, Nobori H, Sato Aet al. 2-Thiouridine is a broad-spectrum antiviral nucleoside analogue against positive-strand RNA viruses. BioRxiv 2022. 10.1101/2022.12.14.520006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mukae H, Yotsuyanagi H, Ohmagari Net al. A randomized phase 2/3 study of ensitrelvir, a novel oral SARS-CoV-2 3C-like protease inhibitor, in Japanese patients with mild-to-moderate COVID-19 or asymptomatic SARS-CoV-2 infection: results of the phase 2a part. Antimicrob Agents Chemother 2022; 66: e0069722. 10.1128/aac.00697-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mukae H, Yotsuyanagi H, Ohmagari Net al. Efficacy and safety of ensitrelvir in patients with mild-to-moderate COVID-19: the phase 2b part of a randomized, placebo-controlled, phase 2/3 study. Clin Infect Dis 2022; ciac933. 10.1093/cid/ciac933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsuyama S, Nao N, Shirato Ket al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 2020; 117: 7001–3. 10.1073/pnas.2002589117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Unoh Y, Uehara S, Nakahara Ket al. Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J Med Chem 2022; 65: 6499–512. 10.1021/acs.jmedchem.2c00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Owen DR, Allerton CMN, Anderson ASet al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021; 374: 1586–93. 10.1126/science.abl4784 [DOI] [PubMed] [Google Scholar]
- 16. Sia SF, Yan L-M, Chin AWHet al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020; 583: 834–8. 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imai M, Iwatsuki-Horimoto K, Hatta Met al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Nat Acad Sci U S A 2020; 117: 16587–95. 10.1073/pnas.2009799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sasaki M, Tabata K, Kishimoto Met al. Oral administration of S-217622, a SARS-CoV-2 main protease inhibitor, decreases viral load and accelerates recovery from clinical aspects of COVID-19. bioRxiv 2022. 10.1101/2022.02.14.480338 [DOI] [Google Scholar]
- 19. Uraki R, Kiso M, Iida Set al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature 2022; 607: 119–27. 10.1038/s41586-022-04856-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boras B, Jones RM, Anson BJet al. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat Commun 2021; 12: 6055. 10.1038/s41467-021-26239-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith DA, Di L, Kerns EH. The effect of plasma protein binding on in vivo efficacy: misconceptions in drug discovery. Nat Rev Drug Discov 2010; 9: 929–39. 10.1038/nrd3287 [DOI] [PubMed] [Google Scholar]
- 22. Gu H, Chen Q, Yang Get al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020; 369: 1603–7. 10.1126/science.abc4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun S, Gu H, Cao Let al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat Comm 2021; 12: 5654. 10.1038/s41467-021-25903-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abdelnabi R, Foo CS, Jochmans Det al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection with SARS-CoV-2 variants of concern. Nat Commun 2022; 13: 719. 10.1038/s41467-022-28354-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.