Abstract
Purpose:
Patients with resected localized clear-cell renal cell carcinoma (ccRCC) remain at variable risk of recurrence. Incorporation of biomarkers may refine risk prediction and inform adjuvant treatment decisions. We explored the role of tumor genomics in this setting, leveraging the largest cohort to date of localized ccRCC tissues subjected to targeted gene sequencing.
Experimental Design:
The somatic mutation status of 12 genes was determined in 943 ccRCC cases from a multinational cohort of patients, and associations to outcomes were examined in a Discovery (n = 469) and Validation (n = 474) framework.
Results:
Tumors containing a von-Hippel Lindau (VHL) mutation alone were associated with significantly improved outcomes in comparison with tumors containing a VHL plus additional mutations. Within the Discovery cohort, those with VHL+0, VHL+1, VHL+2, and VHL+≥3 tumors had disease-free survival (DFS) rates of 90.8%, 80.1%, 68.2%, and 50.7% respectively, at 5 years. This trend was replicated in the Validation cohort. Notably, these genomically defined groups were independent of tumor mutational burden. Amongst patients eligible for adjuvant therapy, those with a VHL+0 tumor (29%) had a 5-year DFS rate of 79.3% and could, therefore, potentially be spared further treatment. Conversely, patients with VHL+2 and VHL+≥3 tumors (32%) had equivalent DFS rates of 45.6% and 35.3%, respectively, and should be prioritized for adjuvant therapy.
Conclusions:
Genomic characterization of ccRCC identified biologically distinct groups of patients with divergent relapse rates. These groups account for the ∼80% of cases with VHL mutations and could be used to personalize adjuvant treatment discussions with patients as well as inform future adjuvant trial design.
Translational Relevance.
Determination of recurrence risk in patients with resected localized renal cell carcinoma (RCC) remains reliant on pathologic grounds alone. Despite extensive characterization of these tumors at the genomic level, the application of such information for patient benefit has not been fully realized. We undertook targeted DNA sequencing of tumor-normal sample pairs from a large multinational cohort of patients with localized clear-cell RCC and explored the impact of these data on patient outcome. Using a 12-gene classifier, we are able to classify almost 80% of patients into one of four groups with highly divergent risk of recurrence or death from RCC, independent of tumor stage and grade or patient age. Tumors found to contain a von-Hippel Lindau mutation alone were associated with most favorable outcomes and defines a group of patients who may potentially be spared adjuvant therapy. Conversely, patients at very high risk of recurrence are identified as those who should be managed aggressively.
Introduction
Renal cell carcinoma (RCC) presents a growing global health problem, with over 400,000 new cases each year and rising incidence rates worldwide (1, 2). The majority (70%–80%) of RCCs are clear-cell RCC (ccRCC), which are characterized by their highly variable clinical course. For clinicians, this heterogeneity poses significant challenges to the delivery of individual patient care.
Most patients (75%–80%) present with apparent localized disease and are offered curative intent treatment in the form of surgery or ablation, but 20% to 30% of these patients will subsequently relapse (3). The estimation of likely patient outcomes underpins further decision-making, guiding patient counselling, length and intensity of follow-up, and the selection of patients for adjuvant therapy.
Immune checkpoint inhibitors (ICI) have been explored for their efficacy in the adjuvant RCC setting within large randomized trials involving thousands of patients (4). Pembrolizumab, a programmed death 1—targeted agent, has recently received approval in both the US and Europe for use in patients with resected intermediate-high and high-risk RCC, based on results from the ongoing Phase III KEYNOTE-564 study. A significant disease-free survival (DFS) advantage in favor of 1 year of adjuvant pembrolizumab versus placebo was demonstrated in this trial (5, 6). Determination of risk was based on tumor–node–metastasis (TNM) stage and tumor grade, with the majority (86%) of recruited patients having either pT2 (high grade) or pT3 (any grade) tumors. However, 68% of patients in the placebo arm of this study remained free of recurrence at 2 years (5), indicating the limitations of current risk-estimation methods. Given the subsequent recent publication of three negative adjuvant RCC studies also employing ICIs (7–9), and the financial cost and potentially severe immune-mediated adverse events associated with these agents, careful consideration must be given to both the risks versus benefits of such therapy. Those at highest risk of recurrence should be prioritized whilst sparing those with biologically lower-risk tumors, highlighting an urgent need for easily implemented molecular tools to improve individual patient stratification (10, 11).
ccRCC has been extensively characterized at the genetic, epigenetic, and transcriptomic level (12, 13). Mutation or methylation of the von-Hippel Lindau (VHL) tumor suppressor gene occurs in the majority (≈80%) of sporadic cases (13). In addition, recurrent mutations in PBRM1 (≈40%), SETD2 (≈20%), and BAP1 (≈15%) are observed, along with a large number of lower frequency events. The application of genomics in ccRCC to inform clinical practice is yet to be realized. Clinical association studies to date have typically considered each gene in isolation (mutated versus non-mutated tumors), often using small cohorts and much of our current understanding comes from a single dataset, The Cancer Genome Atlas (TCGA) study (12, 14). Recently, distinct evolutionary subtypes of ccRCC have been proposed that appear biologically and clinically distinct, including subtypes that are VHL wild-type (WT), VHL monodrivers, and those that have multiple clonal drivers (15). This has advanced our understanding of how genomic alterations may impact on disease progression, and has defined a new paradigm in linking genomic signatures of tumors to clinical outcome.
Our recent study “Cancer Genomics of the Kidney (CAGEKID)”, as part of the International Cancer Genome Consortium, provided the first comprehensive multinational description of the molecular architecture of ccRCC (13). Using a larger multinational validation cohort with extensive associated demographic, clinical, and follow-up data, we have now further explored some of the more commonly observed genetic changes in ccRCC to enable their molecular classification and demonstrate how such information may be applied in the clinic for patient benefit.
Materials and Methods
Patients and samples
Patients undergoing nephrectomy for suspected renal cancer between March 1998 and February 2014 across 17 centers (detailed in Supplementary Methods) in the UK, Czech Republic, Romania, Russia, and Serbia donated blood and tissue samples for research following written informed consent, based on the Declaration of Helsinki principles. Ethical approvals were obtained from the Leeds (East) Local Research Ethics Committee, the International Agency for Research on Cancer Ethics Committee, as well as from local ethics committee for recruiting centers in Czech Republic, Romania, Russia, Serbia, and Bosnia & Herzegovina. Inclusion and exclusion criteria and sampling were as previously described (13). All samples were subject to panel pathology review. A single frozen tissue block was used for sequencing in the majority (86%) of cases, with a minimum cutoff of 70% (cohorts 1 and 2; C1 and C2 – combined to form the Discovery cohort) or 50% (cohort 3; C3 – the Validation cohort) viable tumor cells in sections flanking the analyzed tissue. For the remaining cases, formalin-fixed, paraffin-embedded (FFPE) tissue was used with three targeted 1-mm punches from a single block in each case.
Preparation of DNA
DNA was isolated from buffy coats and frozen tumor tissue using FlexiGene DNA Kit and DNeasy Blood & Tissue Kit (Qiagen, Toronto, Canada) respectively, following manufacturers’ instructions. Chemagic DNA Cell Kit Special (Perkin Elmer) was used to isolate DNA from FFPE tissues, and DNA samples were quantified using Quant-iT PicoGreen dsDNA Assay (Life Technologies, Burlington, Canada).
Genomic sequencing
Samples from C1 were analyzed using whole-genome sequencing (WGS) as previously reported (13). Libraries were generated using the Nextera Rapid Capture Enrichment library preparation Kit (Illumina; 42-gene panel, C2) and the Lucigen AmpFree library preparation kit with xGen Dual Index UMI adapters [Integrated DNA Technologies (IDT); 12-gene panel, C3] according to the manufacturers’ recommendations. Libraries were quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) and the Kapa Illumina GA with Revised Primers-SYBR Fast Universal kit (Kapa Biosystems). Average fragment size was determined using a LabChip GX (PerkinElmer) instrument. Captured libraries were sequenced on a HiSeq 2500 (2×100 cycles; C2) or on a NovaSeq 6000 (2×150 cycles; C3), and bcl2fastq (Illumina) was used to de-multiplex samples and generate fastq reads.
Bioinformatic analyses
Data analysis was performed using the GenPipes DNA-Seq High coverage pipeline (16) with the default parameters, and an added step for generating consensus reads (described below). Adapters and low quality reads were removed using Trimmomatic (RRID: SCR_011848), and reads were aligned to the human genome build GRCh37 using bwa-mem. Mapped reads were further refined using GATK (RRID: SCR_001876) and Picard (RRID: SCR_006525) for indel realignment and verifying mate-pairs. Deduplication (Cohorts 1 and 2) was performed with Picard. UMIs (Cohort 3) were processed using fgbio (17) according to the IDT analysis guidelines for xGen Dual Index UMI Adapters to generate consensus reads. The consensus reads were input back into the pipeline for refining with GATK and Picard. Somatic and germline calls were generated using VarScan2 (ref. 18; RRID: SCR_006849) and the identified indels as well as complex variants were re-called using VarDict (19) to ensure proper identification of complex variants that were not properly resolved by VarScan2. Calls were further processed with the addition of functional annotations using snpEff (20) and genomic annotation by Gemini (21) for prioritization of candidate mutations. We extracted somatic variants with predicted high or moderate functional impact and filtered the candidate somatic mutations for a minimum allele frequency of 5% in tumor samples. Among the resulting variants, those with a frequency less than 0.01% in the 1000 Genomes database were selected for manual validation in Integrative Genomics Viewer (IGV) to ensure the quality of the calls and the absence in patient-matched germline DNA.
Survival definitions
DFS was defined as time from date of surgery to local or regional recurrence, metastases, contralateral kidney cancer, or death, whichever occurred first. Any patients without disease recurrence were censored at the date they were last known to be recurrence free (for patients who died without recurrence this was date of death). RCC-specific survival was defined as time from date of surgery to death from RCC. Non-RCC–related survival was defined as time from date of surgery to death from causes other than RCC.
Statistical methods
Cox proportional hazards (PH) models were used to estimate the association between gene mutation frequency and the selected endpoints. The Kaplan–Meier method was used to estimate survival functions and multivariable survival models were constructed to assess the independence of associations between gene mutation status and each endpoint.
To validate the Cox PH model constructed for comparison of genomically defined groups, patients were divided into Discovery and Validation datasets. The Kaplan–Meier curves for each dataset were used to confirm good separation between risk groups, and the HRs between risk groups were well maintained between the Discovery and Validation datasets.
Cumulative incidence functions were estimated using the cmprsk package (22), and cause-specific hazard models were used to assess RCC-related death in the presence of competing risks. All statistical analyses were undertaken in the R environment for statistical computing.
Data availability
Clinical information and list of somatic mutations of the 12 genes are included in the supplementary data (Supplementary Tables S1 and S2). Raw sequence data are available in the European Genome-phenome Archive under accession codes EGAS00001000083 (C1 and C2) and EGAS00001007004 (C3).
Results
Patients
A total of 943 patients with ccRCC were included in this study. Patient characteristics are summarized in Table 1 (detailed in Supplementary Table S3). Most patients (806; 85%) had stage I–III disease. Median follow-up was 5.7 years [interquartile range (IQR), 3.8–7.3], with 160 (17%) recurrences and 192 (20%) cancer-specific deaths recorded.
Table 1.
Clinical and demographic characteristics in all patients (additional detail in Supplementary Table S1).
| Characteristic | All (n = 943) | Discovery (n = 469) | Validation (n = 474) | |
|---|---|---|---|---|
| Age at surgery (years) | Median (range) | 61 (23–86) | 62 (23–86) | 61 (26–85) |
| Sex | Female | 359 (38.1) | 193 (41.2) | 166 (35.0) |
| Male | 580 (61.5) | 276 (58.8) | 304 (64.1) | |
| Missing | 4 (0.4) | 0 (0) | 4 (0.8) | |
| Body mass index | Median (range) | 27.8 (14.9–49.6) | 27.8 (14.9–49.6) | 27.7 (16–49.2) |
| Country | Czech Republic | 342 (36.3) | 133 (28.4) | 209 (44.1) |
| UK | 291 (30.9) | 150 (32.0) | 141 (29.7) | |
| Russia | 197 (20.9) | 129 (27.5) | 68 (14.3) | |
| Romania | 75 (8.0) | 50 (10.7) | 25 (5.3) | |
| Serbia | 34 (3.6) | 7 (1.5) | 27 (5.7) | |
| Missing | 4 (0.4) | 0 (0) | 4 (0.8) | |
| Pathologic tumor size (mm) | Median (range) | 55 (13–220) | 58 (13–220) | 55 (12–225) |
| Pathologic tumor stage | 1a | 277 (29.4) | 128 (27.3) | 149 (31.4) |
| 1b | 215 (22.8) | 114 (24.3) | 101 (21.3) | |
| 2 | 107 (11.3) | 57 (12.2) | 50 (10.5) | |
| 3 | 288 (30.5) | 155 (33.0) | 133 (28.1) | |
| 4 | 10 (1.1) | 7 (1.5) | 3 (0.6) | |
| Missing | 46 (4.9) | 8 (1.7) | 38 (8.0) | |
| Overall stage (TNM) | I | 486 (51.5) | 235 (50.1) | 251 (53.0) |
| II | 95 (10.1) | 47 (10.0) | 48 (10.1) | |
| III | 225 (23.9) | 117 (24.9) | 108 (22.8) | |
| IV | 132 (14.0) | 70 (14.9) | 62 (13.1) | |
| Missing | 5 (0.5) | 0 (0) | 5 (1.1) | |
| FFPE Fuhrman gradea | 1 | 122 (12.9) | 66 (14.1) | 56 (11.8) |
| 2 | 405 (42.9) | 215 (45.8) | 190 (40.1) | |
| 3 | 279 (29.6) | 139 (29.6) | 140 (29.5) | |
| 4 | 91 (9.7) | 45 (9.6) | 46 (9.7) | |
| Missing | 46 (4.9) | 4 (0.9) | 42 (8.9) | |
| Follow-up (years) | Median (IQR) | 5.7 (3.8–7.3) | 6.0 (3.6–7.6) | 5.5 (4.1–6.8) |
Note: n (%) unless otherwise stated.
aAs per original reporting pathologist at each center.
Overview of gene sequencing results
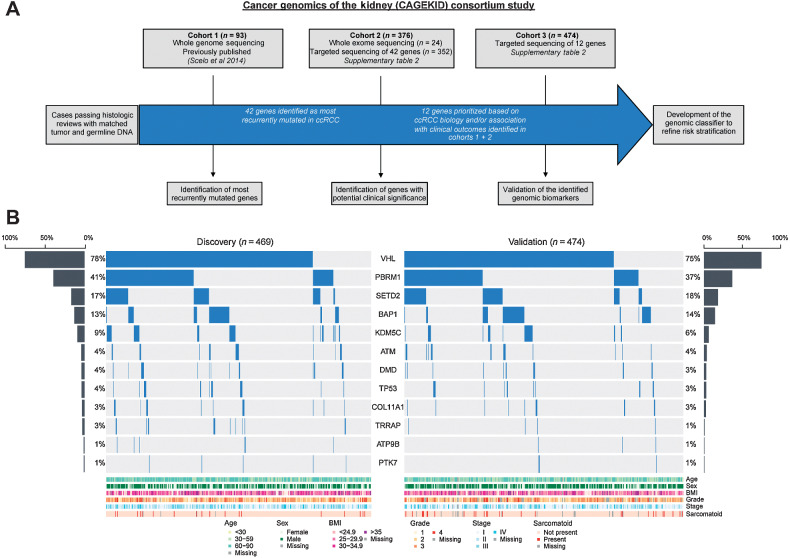
Patient samples were sequenced in three cohorts, as summarized in Fig. 1A. C1 consisted of 93 patient sample pairs (tumor and matched germline DNA) subjected to WGS, previously reported in a descriptive study (13). C2 consisted of sample pairs from an additional 376 patients, of which 24 were analyzed by exome sequencing and 352 underwent targeted sequencing of 42 genes, identified as being most frequently mutated in C1 or other large-scale genomic studies (refs. 12, 23; Supplementary Table S4). C3 consisted of sample pairs from an additional 474 patients, which were analyzed by targeted sequencing of 12 genes (ATM, ATP9B, BAP1, COL11A1, DMD, KDM5C, PBRM1, PTK7, SETD2, TP53, TRRAP, and VHL), included in an RCC-focused gene panel (24). These genes were selected on the basis of their known role in ccRCC biology/previously reported clinical associations (BAP1, KDM5C, PBRM1, SETD2, TP53, VHL), and/or our preliminary observed associations with outcome or other clinical parameters in C1 and C2 (ATM, ATP9B, COL11A1, DMD, PTK7, TRRAP).
Figure 1.
Study summary (A) and mutational profiling of the Discovery and Validation cohorts (B).
For the main analysis presented here, we focus on the final selected 12 genes only, irrespective of sequencing approach/panel. Amongst all cases (n = 943), the most frequently mutated genes were VHL (76%), PBRM1 (39%), SETD2 (18%), BAP1 (14%), and KDM5C (8%). Our VHL mutation detection rate is notably higher than that reported in previous studies (14, 25–27) and is likely to reflect the high depth of sequencing coverage in our targeted sequencing approach (average 296X and 1475X in C2 and C3, respectively), sensitivity for detection of indel mutations, and the exclusion of non-ccRCCs. Furthermore, the similar rates of gene mutations between cohorts and irrespective of whether frozen or FFPE tissue, confirm the sensitivity of our screen, and consistency of genomic results across examined cohorts (Supplementary Table S4). Therefore, to identify reproducible and robust associations, we combined cases from C1 and C2 to form a Discovery cohort (n = 469) and considered C3 as a Validation cohort (n = 474). The Discovery and Validation cohorts showed no significant differences in mutation rates for each gene, and for relevant clinical features such as tumor stage, grade, and patient age (Fig. 1B and Table 1).
Association of gene mutation status with survival
In multivariable models stratified for TNM stage, and incorporating patient age, mutations in BAP1 were found to be significantly associated with DFS in the Discovery cohort (q = 0.02). However, no genes retained significance in both cohorts once adjusted for multiple testing (Supplementary Table S5). When conducting a competing risks analysis using cause-specific hazards models, no genes retained significant associations with RCC-specific survival in either cohort (Supplementary Tables S6 and S7).
Genomically defined subgroups in ccRCC
Following the recently proposed evolutionary trajectory of RCC (15), we next explored the classification of tumors based on the number of identified mutated genes to create genomically defined groups. Specifically, we focused on cases with a VHL mutation (n = 720) detected in isolation (VHL+0; n = 245; 26%) versus those with a VHL mutation plus other driver events; VHL+1 (n = 284; 30%), VHL+2 (n = 148; 16%), and VHL+≥3 (n = 43; 5%). VHL WT cases (n = 218; 23%) were excluded from the analysis due to the potential differences in biological drivers and associated clinical behavior that are suggested to exist between VHL-mutated and VHL WT ccRCCs, including the prevalence of sarcomatoid features and copy-number aberrations in tumors that are VHL WT (15).
The characteristics of tumors in each group are presented in Table 2, illustrating the ability to identify clinically distinct subgroups of ccRCC. Tumors with a VHL mutation alone (VHL+0) were predominantly composed of stage I and II (n = 175; 72%), low-grade (grade 1 or 2; n = 154; 63%) cancers, with none (0/245) reported as containing sarcomatoid and/or rhabdoid change. By comparison, half of tumors containing a VHL mutation plus 2 or more other mutations (VHL+2 and VHL+≥3) were stage III or IV or high-grade cancers (n = 96; 50%) and 9.9% (19/191) contained sarcomatoid and/or rhabdoid change. Tumors containing a VHL mutation plus only one additional mutation were a mix of low-stage (n = 177; 63%) and high-stage cancers (n = 88; 31%), with 4.2% (12/284) reported as containing sarcomatoid and/or rhabdoid change.
Table 2.
Clinical characteristics of cases by gene group.
| Genomically defined groups | ||||||
|---|---|---|---|---|---|---|
| Characteristic | VHL+0 (n = 245) | VHL+1 (n = 284) | VHL+2 (n = 148) | VHL+≥3 (n = 43) | P b | |
| Age at surgery (years) | Median (range) | 60 (23–86) | 62 (26–85) | 63 (38–83) | 64 (43–83) | <0.001 |
| Sex | Female | 86 (35.1) | 118 (41.5) | 46 (31.1) | 16 (35.2) | 0.157 |
| Male | 158 (64.5) | 165 (58.1) | 102 (68.9) | 27 (64.8) | ||
| Missing | 1 (0.4) | 1 (0.4) | 0 (0) | 0 (0) | ||
| Pathologic tumor size (mm) | Median (range) | 50 (15–160) | 55 (12–170) | 60 (22–225) | 65 (18–165) | 0.002 |
| Fuhrman grade | 1 | 30 (12.2) | 46 (16.2) | 11 (7.4) | 3 (7.0) | <0.001 |
| 2 | 124 (50.6) | 116 (40.8) | 61 (41.2) | 12 (27.9) | ||
| 3 | 75 (30.6) | 80 (28.2) | 42 (28.4) | 18 (41.9) | ||
| 4 | 8 (3.3) | 25 (8.8) | 27 (18.2) | 9 (20.9) | ||
| Missing | 8 (3.3) | 17 (6.0) | 7 (4.7) | 1 (2.3) | ||
| Overall stage (TNM) | I | 147 (60.0) | 148 (52.1) | 66 (44.6) | 16 (37.2) | <0.001 |
| II | 28 (11.4) | 24 (8.5) | 11 (7.4) | 2 (4.7) | ||
| III | 43 (17.6) | 71 (25.0) | 43 (29.1) | 17 (39.5) | ||
| IV | 26 (10.6) | 39 (13.7) | 28 (18.9) | 8 (18.6) | ||
| Missing | 1 (0.4) | 2 (0.7) | 0 (0) | 0 (0) | ||
| Sarcomatoid and/or rhabdoid changea | Present | 0 (0) | 12 (4.2) | 15 (10.1) | 4 (9.3) | <0.001 |
| Absent | 238 (97.1) | 264 (93.0) | 130 (87.8) | 39 (90.7) | ||
| Missing | 7 (2.9) | 8 (2.8) | 3 (2.0) | 0 (0) | ||
Note: n (%) unless otherwise stated.
aSeven tumors containing sarcomatoid and/or rhabdoid change were reported as grade 2 or grade 3 by original diagnostic pathologist.
bBased on comparisons using Kruskal–Wallis test.
Survival outcomes amongst genomically defined groups
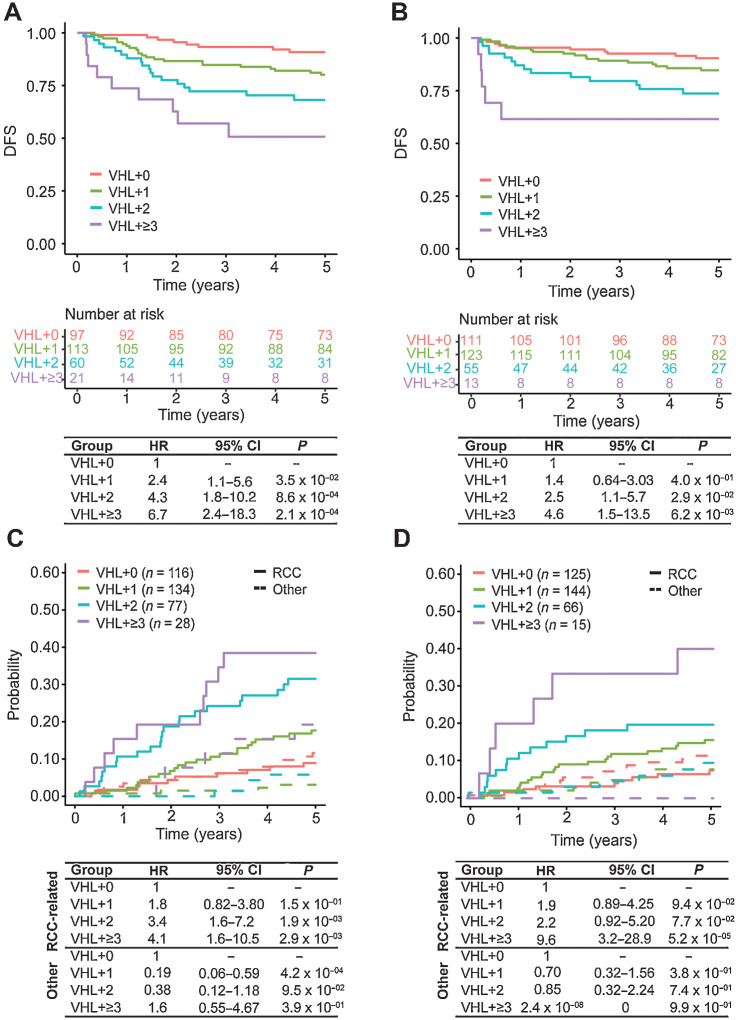
We explored whether consideration of these gene groups would allow stratification of patients by outcome. Within both the Discovery and Validation cohorts, we observed that with an increasing number of mutations in driver genes, the risk of disease recurrence increases. In the Discovery cohort, the 5-year DFS rate was 50.7% [95% confidence interval (CI), 32–80%] for patients with VHL+≥3 tumors, 68.2% (95% CI, 57%–82%) for patients with VHL+2 tumors, and 80.1% (95% CI, 73%–88%) for patients with VHL+1 tumors, compared with 90.8% (95% CI, 85%–97%) for patients with only mutations in VHL (Fig. 2A). A similar trend was observed within the Validation cohort, with 5-year DFS rates of 61.5%, 73.7%, 84.7%, and 90.4% for patients with VHL+≥3, VHL+2, VHL+1, and VHL+0 tumors, respectively (Fig. 2B). Furthermore, this association remained independently significant when accounting for stage and patient age. We observed within both cohorts increasing risk of disease recurrence from VHL+1 to VHL+≥3 (Figs. 2A and B), and association with disease recurrence was significant for VHL+2 [Discovery HR = 4.3 (1.8–10.2), P = 0.000862; Validation HR = 2.5 (1.1–5.7), P = 0.02883] and VHL+≥3 [Discovery HR = 6.7 (2.4–18.3), P = 0.000212; Validation HR = 4.6 (1.5–13.5), P = 0.00615] groups (Figs. 2A and B). These observations were independently replicated amongst the 247 VHL-mutated ccRCCs from the TCGA dataset (ref. 12; Supplementary Fig. S1).
Figure 2.
DFS outcomes and Competing Risks Analysis for RCC-related death amongst patients with VHL mutations stratified into genomically defined groups. Kaplan–Meier survival curves comparing DFS amongst VHL+0, VHL+1, VHL+2, and VHL+≥3 groups within the (A) Discovery and (B) Validation cohorts. Cox PH models estimating association between genomically defined groups and 5-year DFS within the Discovery (left) and Validation (right) cohorts. Cumulative incidence functions amongst VHL+0, VHL+1, VHL+2, and VHL+≥3 groups comparing risk of death caused by RCC (solid line) compared with other causes (dotted line) within the (C) Discovery and (D) Validation cohorts. Cox PH models estimating association between genomically defined groups and 5-year RCC-related death compared with death from other causes within the Discovery (left) and Validation (right) cohorts.
We observed a similar association between these genomically defined groups and risk of RCC-related death. A competing-risks analysis showed that risk of RCC-related death increases with the number of mutated genes, whereas the cumulative incidence curves for patients with VHL+0 tumors were almost identical for RCC-related death and death from other causes (Fig. 2C). This pattern was also demonstrated in the Validation dataset (Fig. 2D). When stratifying for tumor stage, and considering patient age as a covariate, a cause-specific hazards model showed increasing risk with additional driver mutations and a significant association with RCC-related death for VHL+2 [HR = 3.4 (1.6–7.2), P = 0.00190] and VHL+≥3 groups [HR = 4.1 (1.6–10.5), P = 0.00286], which was also seen in the Validation cohort (Figs. 2C and D). This trend was not observed for non-RCC–related death.
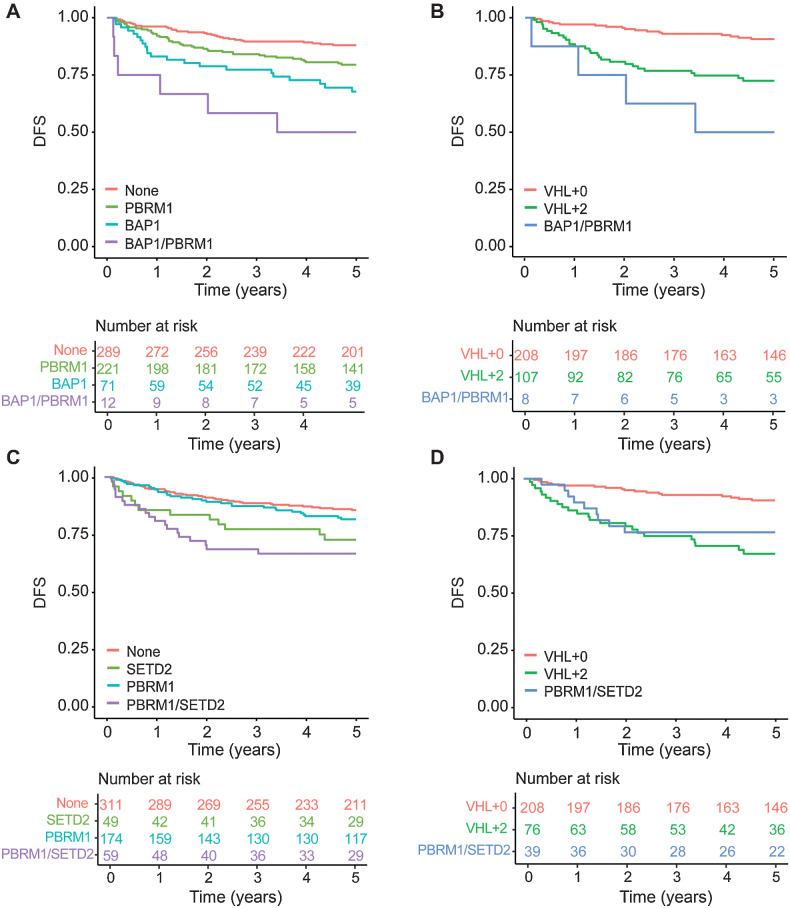
Because tumors containing both a BAP1 and PBRM1 mutation are known to be associated with poor outcomes (28, 29), we examined the effects of mutations in PBRM1, BAP1, and in both genes on DFS in patients with somatic VHL mutations in our cohort. In line with previous reports, we observed poorer DFS when tumors harbored PBRM1 [HR 1.72 (95% CI, 1.09–2.72); P = 0.20] or BAP1 mutations [HR 2.59 (95% CI, 1.51–4.44); P = 0.0006], compared with those without these mutations, whereas patients with co-occurrence of BAP1 and PBRM1 mutations were associated with the poorest survival [HR 7.29 (95% CI, 3.03–17.54); P < 0.0001; Fig. 3A]. To assess the added value of our classifier to these known associations, we divided patients with VHL+2 tumors into two groups: those with PBRM1 and BAP1 mutations and those whose tumors harbor mutations in any two other genes from our 12-gene classifier, except both PBRM1 and BAP1. We then compared DFS outcomes of these groups with that of patients with VHL+0 status. We observed that whilst BAP1/PBRM1–mutated tumors are associated with a higher risk of recurrence [HR 12.08 (95% CI, 3.95–136.95); P < 0.0001], remaining VHL+2 tumors continue to be associated with significantly poorer outcomes [HR 2.74 (95% CI, 1.49–5.01); P = 0.001; Fig. 3B; Supplementary Table S8]. Given that VHL+2 tumors account for approximately 20% of all classifiable tumors in our cohort, and PBRM1/BAP1 co-occurrence accounts for just 7% (8/115) of these cases, our genomic classifier allows for the stratification of a greater proportion of patients by outcome. We performed similar analyses for tumors containing both a PBRM1 and SETD2 mutation (30) and observed comparable results (Fig. 3C and D; Supplementary Table S9).
Figure 3.
DFS outcomes by BAP1/PBRM1 and PBRM1/SETD2 mutation status. Kaplan–Meier survival curves based on (A) BAP1 and PBRM1 mutation status and (B) VHL+0 tumors, VHL+2 tumors containing both a BAP1 and PBRM1 mutation, and remaining VHL+2 tumors (i.e., those not containing both a BAP1 and PBRM1 mutation). Kaplan–Meier survival curves based on (C) PBRM1 and SETD2 mutation status and (D) VHL+0 tumors, VHL+2 tumors containing both a PBRM1 and SETD2 mutation, and remaining VHL+2 tumors (i.e., those not containing both a PBRM1 and SETD2 mutation).
Risk stratification among patients eligible for adjuvant therapy
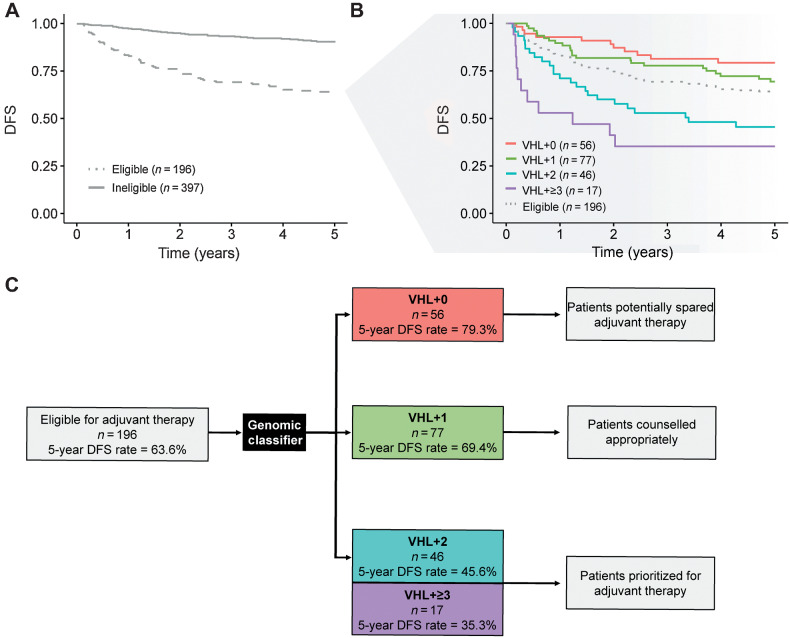
We investigated the potential utility of the genomic classifier for selection of patients for adjuvant therapies (clinically eligible defined as pT2 grade 3–4; pT3 or pT4 (any grade); any pT, any grade, N+). DFS rate at 5 years amongst the 397 patients not considered eligible for adjuvant therapy was 90.6% (95% CI, 88%–94%) versus 63.6% (95% CI, 57%–71%) for the 196 patients eligible for adjuvant therapy (Fig. 4A). Patients defined as being eligible for adjuvant therapy could be further stratified by risk of relapse based on the genomic classification of their tumors. Five-year DFS rates were 79.3% (95% CI, 69%–91%) amongst the 56 (29%) patients with VHL+0 tumors, 69.4% (95% CI, 60%–81%) amongst the 77 (39%) patients with VHL+1 tumors, 45.6% (95% CI, 33%–63%) amongst the 46 (23%) patients with VHL+2 tumors, and 35.3% (95% CI, 19%–67%) amongst the 17 (9%) patients with VHL+≥3 tumors (Fig. 4B).
Figure 4.
Patients eligible for adjuvant therapy stratified by the genomic classifier. A, DFS amongst patients considered eligible [pT2 grade 3–4; pT3 or pT4 (any grade); any pT, any grade, N+] versus ineligible [pT1 (any grade); pT2 grade 1–2] for adjuvant therapy. B, DFS by VHL+0, VHL+1, VHL+2, and VHL+≥3 groups amongst patients considered eligible for adjuvant therapy. C, Flow diagram demonstrating potential clinical application. Application of the genomic classifier to patients typically considered eligible for adjuvant therapy allows sub-stratification of patients into groups with highly divergent risk of relapse. This information could usefully inform individual patient discussions around the benefit versus risks of adjuvant therapy.
Notably, the VHL+2 and VHL+≥3 groups had significantly poorer survival compared with the VHL+0 group (P = 0.00055 and P < 0.0001, respectively). The potential clinical application of these findings, to inform individual patient counselling and decision making, is depicted in Fig. 4C. Groups of patients who may be both spared (VHL+0) versus prioritized (VHL+2 and VHL+≥3) for adjuvant treatment, based on risk of recurrence, are identified.
We also explored the added utility of our genomic classifier when first considering the Leibovich score, a validated prognostic nomogram that incorporates tumor stage, grade, size, necrosis, and lymph node status (31), available in 181/196 (92%) patients. As expected, patients with high-risk disease had significantly poorer DFS rates in comparison with those with intermediate-risk disease (P < 0.001; Supplementary Fig. S2A; Supplementary Table S10). We found that our genomic classifier retains the ability to meaningfully substratify within these groups. For example, amongst patients with Leibovich score defined intermediate-risk disease, VHL+0 patients had a 5-year DFS rate of 89% (95% CI, 80%–99%) compared with 66% (95% CI, 47%–92%) for patients with VHL+2 tumors (P = 0.02) and 25% (95% CI, 5%–100%) for VHL+≥3 tumors (P < 0.0001), although it should be noted there were only 4 patients in the latter group (Supplementary Fig. S2B).
Finally, we examined outcomes by our classifier amongst patients with stage I disease, who are not usually considered for adjuvant therapy. Patients with stage I VHL+0 tumors (140/365; 38%) were associated with excellent outcomes, with a 5-year DFS rate of 96% (95% CI, 93%–99%). By contrast, patients with a stage I VHL+≥3 tumor had relatively poorer outcomes, with a 5-year DFS of 78% (95% CI, 59%–100%; P = 0.01), although the wide CIs reflect the small number of patients in this group (16/365; 4%) and should be considered an exploratory finding. Outcomes by tumor stage stratified by our genomic classifier are shown in Supplementary Fig. S3 and Supplementary Table S11.
Genomic classifier independence from tumor mutational burden
To investigate whether the increasing number of driver mutations in RCC-focused genes act as a classifier independent of tumor mutational burden (TMB), a known prognostic marker in some cancers, we compared TMB values between the genomically defined groups. TMB differed only between the VHL+0 and other groups within the WGS samples (Cohort 1). Notably, TMB was not significantly different between VHL+1, VHL+2, and VHL+≥3 tumors (Supplementary Table S12). This analysis was replicated using the publically available TCGA dataset, which showed only marginally significant difference between the VHL+0 and VHL+2 groups, and no differences between the VHL+1, VHL+2, and VHL+≥3 tumors (Supplementary Table S12). In addition, whereas TMB alone did not show a significant association to DFS, when incorporating TMB as a covariate, VHL+2 and VHL+≥3 groups showed significantly poorer DFS in both the C1 cohort (P = 0.025 and P = 0.043 for VHL+2 and VHL+≥3, respectively) and TCGA (P = 0.002 and P = 0.046 for VHL+2 and VHL+≥3, respectively) cohorts (Supplementary Table S13). These results demonstrate that the performance of the classifier is independent from TMB.
Discussion
The prospect of offering effective adjuvant therapy to patients with RCC represents a major step forward in the management of this disease. With this, however, comes the challenge of deciding who should or should not be treated, based on risk of cancer recurrence. In this multinational study, comprising final data from a total of 943 patients, we have examined the role of targeted gene sequencing to improve individual risk stratification. We show that by considering a panel of 12 RCC-focused genes, clinically and biologically distinct groups can be identified, accounting for 76% of cases in our cohort, that can be used to refine individual risk-estimates with potential immediate application for selection of patients for adjuvant therapy. Importantly, these findings were observed in both the Discovery and Validation cohorts, and were independent from patient age, tumor stage, and TMB.
Studies examining the clinical impact of somatic mutations in localized ccRCC have consistently revealed the association of mutations in specific genes, such as BAP1 and SETD2, with poorer outcomes (28, 32). However, few studies have examined their independent prognostic value. In a pooled analysis of 1,049 patients with ccRCC, incorporating four independent cohorts (including patients from C1 of the current study), only mutations in SETD2 remained marginally significant for recurrence-free survival in a model including TNM stage and patient age (33). We found mutations in BAP1, but not SETD2, to retain significance on multivariate testing when considering DFS. However, this finding failed to replicate between our Discovery and Validation cohorts. Given these inconsistencies, the value of considering the mutation status of any single gene to inform clinical practice remains uncertain.
The existence of seven distinct, genomically defined, evolutionary subtypes of ccRCC was proposed in a study employing multi-region sampling of primary tumors, elegantly described amongst 63 cases (15). These included tumors containing multiple clonal drivers (n = 12; defined as tumors with mutations in 2 or more of BAP1, PBRM1, SETD2, or PTEN clonal mutations), demonstrating limited intra-tumor heterogeneity (ITH) and associated exclusively with higher-stage disease. Another 11 tumors were defined as VHL monodrivers, again demonstrating limited subclonality and predominantly composed of stage I cancers. A third group (n = 6), defined as VHL WT, were associated with increased somatic copy-number alterations and enriched for tumors containing sarcomatoid differentiation (15).
These intriguing exploratory observations led us to examine our data using a similar principle, in our much larger sample set, but subject to single region sequencing. Most patients within the CAGEKID cohort demonstrated a mutation in the VHL gene (n = 718; 76%) and could be classified as those with tumors containing a VHL mutation alone (VHL+0), or a VHL mutation plus additional driver events (VHL+1, VHL+2, VHL+≥3), within the sequenced region and equate to the ‘VHL monodriver’ and ‘multiple clonal driver’ subtypes proposed above. These genomic groups showed striking divergence in terms of their clinical behavior. Importantly, whilst a preponderance of either early stage, low grade, or high stage, high grade cancers were found in VHL+0 and VHL+2/VHL+≥3 tumors, respectively, no exclusivity was observed and all groups were represented across the disease spectrum.
Determination of recurrence risk in localized RCC currently remains reliant on clinicopathologic criteria alone. TNM stage and tumor grade provide useful broad stratification of patients into low-, intermediate-, and high-risk groups, but ultimately fail to adequately account for individual variance in tumor biology and outcomes. Efforts to improve risk prediction has led to the development of several prognostic nomograms (31, 34, 35), incorporating additional tumor or clinical characteristics, which have also been widely adopted. However, their predictive accuracy appears, at best, only marginally better than TNM alone and their performance has declined over time (36–38).
These deficiencies lead to a significant risk that many patients may be overtreated in the adjuvant setting. The ability of the current genomic classifier to identify over a quarter of such patients (VHL+0) with a 5-year DFS rate approaching 80% is therefore important. Potential avoidance of adjuvant therapy carries significant benefits to both patients and healthcare systems, given the costs and toxicity of ICI-based therapies. Conversely, patients with tumors containing multiple driver mutations appear to be at extremely high risk of recurrence (5-year DFS rate 35%–46%), representing a group who may benefit most from adjuvant treatment. Our findings also carry important implications for the design, costs, and success of future adjuvant RCC trials.
This study adds to a growing literature in ccRCC demonstrating the ability to infer tumor biology from genomic data derived from a single tumor region (33, 39–42). The impact of spatial ITH, well characterized in ccRCC (43), on clinical association studies such as ours remains poorly understood. Whilst multi-region sequencing would almost certainly increase mutation detection (15), in all but the VHL gene, the clinical impact of a small subclonal driver event within the context of an otherwise largely clonal tumor, for example, remains unknown (44).
Our gene panel is not definitive and it is likely that further refinement is possible. For example, other than COL11A1, we did not consider genes within the PI3K/AKT/mTOR pathway. Furthermore, the significance, at an individual level, of the lower frequency events included in our panel could not be robustly established despite the size of our cohort and will require even larger studies to achieve this. Analysis of other known genomic features, such as copy-number alterations, were not undertaken and may allow further refinement of genomic groups. Whilst VHL+2 and VHL+≥3 tumors are associated with the highest risk of disease recurrence, the benefit of adjuvant ICI in these patients remains unknown and warrants investigation. The TCGA sample set represents the next largest available cohort of genetically defined ccRCC and was useful in validating our findings and in investigating associations to TMB, but ideally a larger cohort, with a similarly high VHL mutation rate to our study (notably the TCGA cohort reported a rate of VHL mutation of 52.3%; ref. 14), would have been employed.
In conclusion, this study establishes the ability to define biologically distinct molecular subgroups of ccRCC that could be used to better inform patients and their physicians regarding individual risk of tumor recurrence following nephrectomy. These genetic groups can be defined based on the mutation status of a small panel of genes captured within a single tumor region and, therefore, readily applied to the clinic. Further prospective evaluation of these findings is warranted.
Study Centers
Patients were recruited from the following centers (with lead investigator(s) at each site): St James's University Hospital, Leeds, Dr. Naveen Vasudev; Newcastle Upon Tyne Hospitals NHS Foundation Trust, Professor Naeem Soomro; Stockport NHS Foundation Trust, Mr. Adebanji Adeyoju; Nottingham University Hospitals NHS Trust, Professor Poulam Patel; NHS Lothian, Professor Grant Stewart; Charing Cross Hospital, Mr. David Hrouda; Oxford University Hospitals NHS Foundation Trust, Mr. Mark Sullivan; Northwick Park Hospital, Mr. Jeff Webster; University Hospital Motol, Dr. Antonin Brisuda; General University Hospital, Dr. Roman Sobotka; Masaryk Memorial Cancer Institute, Dr. Lenka Foretova; Palacký University Hospital, Dr. Vladimir Janout; České Budějovice Regional Hospital, Dr. Vladimir Janout; Th. Burghele Hospital, Bucharest, Dr. Viorel Jinga; N.N. Blokhin Cancer Research Center, Dr. David Zaridze; Clinical Center of Serbia (KCS), Dr. Ljiljana Bogdanovic; Military Medical Academy, Dr. Bozidar Kovacevic.
Supplementary Material
Clinical information and 12 gene mutation status for cohorts C1-C3
Somatic mutations detected in 12 interrogated genes for cohorts C1-C3
Supplementary Tables S3-S13
Disease-Free Survival outcomes amongst genomic groups when applied to the 247 VHL mutated ccRCCs from the TCGA dataset
DFS amongst Leibovich risk groups stratified by genomic classifier
DFS by tumor stage stratified by genomic classifier
Supplementary Methods
Acknowledgments
We thank all the staff at each of the recruiting centers, including the technical support provided by staff of genomic facilities of McGill University and Genome Québec Innovation Centre. We thank Drs. Patricia Harnden and Morag Seywright (both now retired) for their substantial contributions to panel pathology review. We are grateful to the patients for consenting to take part in this study.
This work was supported by the EU FP7 under grant agreement number 241669 (the CAGEKID project, http://www.cng.fr/cagekid) and grants from Génome Québec, le Ministère de l'Enseignement supérieur, de la Recherche, de la Science et de la Technologie (MESRST) Québec, Cancer Research Society (CRS 22592), Kidney Foundation of Canada (2020KHRG-673291), and McGill University. Y. Riazalhosseini is a research scholar of the Fonds de recherche du Québec – Santé (FRQS). The study in Brno, Czech Republic, was supported by MH CZ - DRO (MMCI, 00209805). A.Y. Warren is supported by the NIHR Cambridge Biomedical Research Centre. We also acknowledge the support of Cancer Research UK, Experimental Cancer Medicine Centre and National Institute for Health Research (NIHR) Clinical Research Facility infrastructure funding in Leeds, the Leeds Multidisciplinary and NIHR Research Tissue Banks and are grateful to the sample processing, urology, pathology, and oncology clinical teams at St James's University Hospital, Leeds.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 1161
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
N.S. Vasudev reports grants, personal fees, and nonfinancial support from Bristol Myers Squibb; personal fees and nonfinancial support from EUSA Pharma and Ipsen; and personal fees from Eisai Ltd, Merck Serono, Pfizer, and 4D Pharma outside the submitted work. M.B. Wozniak reports personal fees from Novartis Ireland Ltd outside the submitted work. J. Tost reports grants from European Union during the conduct of the study, as well as grants from European Union, Agence National de Recherche (ANR), Fondation pour la Recherche Medicale (FRM), DBV Technologies, MSD Avenir, and Agence Nationale Sécurité Sanitaire Alimentaire Nationale (Anses) outside the submitted work. Y. Riazalhosseini reports grants from Cancer Research Society, Kidney Foundation of Canada, and Fonds de recherche du Québec – Santé (FRQS) during the conduct of the study. R.E. Banks reports grants from EU and NIHR during the conduct of the study. No disclosures were reported by the other authors.
Disclaimer
The views and opinions expressed by the authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, the UK Department of Health, or the International Agency for Research on Cancer/World Health Organization.
Authors' Contributions
N.S. Vasudev: Resources, data curation, investigation, methodology, writing–original draft, writing–review and editing. G. Scelo: Conceptualization, resources, data curation, formal analysis, investigation, methodology, project administration, writing–review and editing. K.I. Glennon: Formal analysis, investigation, methodology, writing–review and editing. M. Wilson: Formal analysis, investigation, writing–review and editing. L. Letourneau: Formal analysis, writing–review and editing. R. Eveleigh: Formal analysis, writing–review and editing. N. Nourbehesht: Resources, investigation, writing–review and editing. M. Arseneault: Resources, investigation, writing–review and editing. A. Paccard: Resources, investigation, writing–review and editing. L. Egevad: Data curation, formal analysis, investigation, writing–review and editing. J. Viksna: Software, writing–review and editing. E. Celms: Software, writing–review and editing. S.M. Jackson: Data curation, writing–review and editing. B. Abedi-Ardekani: Formal analysis, investigation, writing–review and editing. A.Y. Warren: Formal analysis, investigation, writing–review and editing. P.J. Selby: Conceptualization, writing–review and editing. S. Trainor: Data curation, writing–review and editing. M. Kimuli: Resources, writing–review and editing. J. Cartledge: Resources, writing–review and editing. N. Soomro: Resources, writing–review and editing. A. Adeyoju: Resources, writing–review and editing. P.M. Patel: Resources, writing–review and editing. M.B. Wozniak: Data curation, formal analysis, writing–review and editing. I. Holcatova: Resources, writing–review and editing. A. Brisuda: Resources, writing–review and editing. V. Janout: Resources, writing–review and editing. E. Chanudet: Data curation, project administration, writing–review and editing. D. Zaridze: Resources, writing–review and editing. A. Moukeria: Resources, writing–review and editing. O. Shangina: Resources, writing–review and editing. L. Foretova: Resources, writing–review and editing. M. Navratilova: Resources, writing–review and editing. D. Mates: Resources, writing–review and editing. V. Jinga: Resources, writing–review and editing. L. Bogdanovic: Resources, writing–review and editing. B. Kovacevic: Resources, writing–review and editing. A. Cambon-Thomsen: Supervision, methodology, writing–review and editing. G. Bourque: Formal analysis, writing–review and editing. A. Brazma: Conceptualization, investigation, writing–review and editing. J. Tost: Conceptualization, investigation, writing–review and editing. P. Brennan: Conceptualization, supervision, writing–review and editing. M. Lathrop: Conceptualization, supervision, funding acquisition, writing–review and editing. Y. Riazalhosseini: Conceptualization, formal analysis, supervision, investigation, methodology, writing–original draft, writing–review and editing. R.E. Banks: Conceptualization, data curation, supervision, project administration, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol 2015;67:519–30. [DOI] [PubMed] [Google Scholar]
- 3. Dabestani S, Beisland C, Stewart GD, Bensalah K, Gudmundsson E, Lam TB, et al. Long-term outcomes of follow-up for initially localized clear- cell renal cell carcinoma: RECUR database analysis. Eur Urol Focus 2019;5:857–66. [DOI] [PubMed] [Google Scholar]
- 4. Larroquette M, Peyraud F, Domblides C, Lefort F, Bernhard JC, Ravaud A, et al. Adjuvant therapy in renal cell carcinoma: current knowledge and future perspectives. Cancer Treat Rev 2021;97:102207. [DOI] [PubMed] [Google Scholar]
- 5. Choueiri TK, Tomczak P, Park SH, Venugopal B, Ferguson T, Chang YH, et al. Adjuvant pembrolizumab after nephrectomy in renal cell carcinoma. N Engl J Med 2021;385:683–94. [DOI] [PubMed] [Google Scholar]
- 6. Powles T, Tomczak P, Park SH, Venugopal B, Ferguson T, Symeonides SN, et al. Pembrolizumab versus placebo as post-nephrectomy adjuvant therapy for clear-cell renal cell carcinoma (KEYNOTE-564): 30-month follow-up analysis of a multicenter, randomized, double-blind, placebo-controlled, phase III trial. Lancet Oncol 2022;23:1133–44. [DOI] [PubMed] [Google Scholar]
- 7. Pal SK, Uzzo R, Karam JA, Master VA, Donskov F, Suarez C, et al. Adjuvant atezolizumab versus placebo for patients with renal cell carcinoma at increased risk of recurrence following resection (IMmotion010): a multicenter, randomized, double-blind, phase III trial. Lancet 2022;400:1103–16. [DOI] [PubMed] [Google Scholar]
- 8. Motzer RJ, Russo P, Gruenwald V, Tomita Y, Zurawski B, Parikh OA, et al. Adjuvant nivolumab plus ipilimumab (NIVO+IPI) vs placebo (PBO) for localized renal cell carcinoma (RCC) at high risk of relapse after nephrectomy: results from the randomized, Phase III CheckMate 914 trial. Ann Oncol 2022;33:S808–S69. [Google Scholar]
- 9. Allaf M, Kim SE, Harshman LC, McDermott DF, Master VA, Signoretti S, et al. Phase III randomized study comparing perioperative nivolumab (nivo) versus observation in patients (Pts) with renal cell carcinoma (RCC) undergoing nephrectomy (PROSPER, ECOG-ACRIN EA8143), a national clinical trials network trial. Ann Oncol 2022;33:S808–S69. [Google Scholar]
- 10. Figlin RA, Leibovich BC, Stewart GD, Negrier S. Adjuvant therapy in renal cell carcinoma: does higher risk for recurrence improve the chance for success? Ann Oncol 2018;29:324–31. [DOI] [PubMed] [Google Scholar]
- 11. Rossi SH, Blick C, Handforth C, Brown JE, Stewart GD. Renal Cancer Gap Analysis Collaborative. Essential research priorities in renal cancer: a modified Delphi consensus statement. Eur Urol Focus 2020;6:991–98. [DOI] [PubMed] [Google Scholar]
- 12. Ricketts CJ, De Cubas AA, Fan H, Smith CC, Lang M, Reznik E, et al. The Cancer Genome Atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep 2018;23:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scelo G, Riazalhosseini Y, Greger L, Letourneau L, Gonzalez-Porta M, Wozniak MB, et al. Variation in genomic landscape of clear- cell renal cell carcinoma across Europe. Nat Commun 2014;5:5135. [DOI] [PubMed] [Google Scholar]
- 14. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear- cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 2018;173:595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bourgey M, Dali R, Eveleigh R, Chen KC, Letourneau L, Fillon J, et al. GenPipes: an open-source framework for distributed and scalable genomic analyses. Gigascience 2019;8:giz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. FulcrumGenomics. 1/09/2020. fgbio. Available from:https://github.com/fulcrumgenomics/fgbio.
- 18. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy-number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016;44:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single-nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol 2013;9:e1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gray RJ. cmprsk: subdistribution analysis of competing risks 2010.
- 23. Riazalhosseini Y, Lathrop M. Precision medicine from the renal cancer genome. Nat Rev Nephrol 2016;12:655–66. [DOI] [PubMed] [Google Scholar]
- 24. Glennon KI, Maralani M, Abdian N, Paccard A, Montermini L, Nam AJ, et al. Rational development of liquid biopsy analysis in renal cell carcinoma. Cancers 2021;13:5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010;463:360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear- cell renal cell carcinoma. Nat Genet 2012;44:17–9. [DOI] [PubMed] [Google Scholar]
- 27. Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013;45:860–7. [DOI] [PubMed] [Google Scholar]
- 28. Kapur P, Pena-Llopis S, Christie A, Zhrebker L, Pavia-Jimenez A, Rathmell WK, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol 2013;14:159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joseph RW, Kapur P, Serie DJ, Parasramka M, Ho TH, Cheville JC, et al. Clear- cell renal cell carcinoma subtypes identified by BAP1 and PBRM1 expression. J Urol 2016;195:180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho TH, Kapur P, Joseph RW, Serie DJ, Eckel-Passow JE, Tong P, et al. Loss of histone H3 lysine 36 trimethylation is associated with an increased risk of renal cell carcinoma-specific death. Mod Pathol 2016;29:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leibovich BC, Blute ML, Cheville JC, Lohse CM, Frank I, Kwon ED, et al. Prediction of progression after radical nephrectomy for patients with clear- cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer 2003;97:1663–71. [DOI] [PubMed] [Google Scholar]
- 32. Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen YB, Gonen M, et al. Adverse outcomes in clear- cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 2013;19:3259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manley BJ, Zabor EC, Casuscelli J, Tennenbaum DM, Redzematovic A, Becerra MF, et al. Integration of recurrent somatic mutations with clinical outcomes: a pooled analysis of 1,049 patients with clear- cell renal cell carcinoma. Eur Urol Focus 2017;3:421–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear- cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade, and necrosis: the SSIGN score. J Urol 2002;168:2395–400. [DOI] [PubMed] [Google Scholar]
- 35. Zisman A, Pantuck AJ, Wieder J, Chao DH, Dorey F, Said JW, et al. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J Clin Oncol 2002;20:4559–66. [DOI] [PubMed] [Google Scholar]
- 36. Correa AF, Jegede O, Haas NB, Flaherty KT, Pins MR, Messing EM, et al. Predicting renal cancer recurrence: defining limitations of existing prognostic models with prospective trial-based validation. J Clin Oncol 2019;37:2062–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oza B, Eisen T, Frangou E, Stewart GD, Bex A, Ritchie AWS, et al. External validation of the 2003 leibovich prognostic score in patients randomly assigned to SORCE, an international phase III trial of adjuvant sorafenib in renal cell cancer. J Clin Oncol 2022;40:1772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vasudev NS, Hutchinson M, Trainor S, Ferguson R, Bhattarai S, Adeyoju A, et al. UK multicenter prospective evaluation of the Leibovich score in localized renal cell carcinoma: performance has altered over time. Urology 2020;136:162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rini B, Goddard A, Knezevic D, Maddala T, Zhou M, Aydin H, et al. A 16-gene assay to predict recurrence after surgery in localized renal cell carcinoma: development and validation studies. Lancet Oncol 2015;16:676–85. [DOI] [PubMed] [Google Scholar]
- 40. Rini BI, Escudier B, Martini JF, Magheli A, Svedman C, Lopatin M, et al. Validation of the 16-gene recurrence score in patients with locoregional, high-risk renal cell carcinoma from a phase III trial of adjuvant sunitinib. Clin Cancer Res 2018;24:4407–15. [DOI] [PubMed] [Google Scholar]
- 41. Tosoian JJ, Feldman AS, Abbott MR, Mehra R, Tiemeny P, Wolf JS Jr, et al. Biopsy cell-cycle proliferation score predicts adverse surgical pathology in localized renal cell carcinoma. Eur Urol 2020;78:657–60. [DOI] [PubMed] [Google Scholar]
- 42. Motzer RJ, Banchereau R, Hamidi H, Powles T, McDermott D, Atkins MB, et al. Molecular subsets in renal cancer determine outcome to checkpoint and angiogenesis blockade. Cancer Cell 2020;38:803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear- cell renal cell carcinomas defined by multi-region sequencing. Nat Genet 2014;46:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gulati S, Martinez P, Joshi T, Birkbak NJ, Santos CR, Rowan AJ, et al. Systematic evaluation of the prognostic impact and intra-tumor heterogeneity of clear- cell renal cell carcinoma biomarkers. Eur Urol 2014;66:936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical information and 12 gene mutation status for cohorts C1-C3
Somatic mutations detected in 12 interrogated genes for cohorts C1-C3
Supplementary Tables S3-S13
Disease-Free Survival outcomes amongst genomic groups when applied to the 247 VHL mutated ccRCCs from the TCGA dataset
DFS amongst Leibovich risk groups stratified by genomic classifier
DFS by tumor stage stratified by genomic classifier
Supplementary Methods
Data Availability Statement
Clinical information and list of somatic mutations of the 12 genes are included in the supplementary data (Supplementary Tables S1 and S2). Raw sequence data are available in the European Genome-phenome Archive under accession codes EGAS00001000083 (C1 and C2) and EGAS00001007004 (C3).