Abstract
Brain metastases (BM) is one of the main reasons for lung cancer-related deaths but lack prediction methods. Many patients with BMs do not benefit from immunotherapy. A comprehensive genomic analysis of matched primary tumors (PT) and their BM lesions may provide new insight into the evolutionary and immune characteristics. To describe evolutionary features and immune characteristic differences, we analyzed whole-exome sequencing data for 28 paired PT and BM samples from 14 patients with non-small cell lung cancer. In addition, we used another 26 matched PT and BM samples as a validation cohort. We found that total mutational signatures were relatively consistent between paired primary and brain metastatic tumors. Nevertheless, the shared mutations of the two lesions were fewer than the mutations present in each of the lesions alone. In the process of BM, driver genes undergo evolutionary branches. Typical driver genes, including EGFR and TP53, appear relatively conserved throughout evolution; however, specific signals are enriched in BM lesions. We found several main characteristics of lung cancer BMs that were different from primary lung cancer, such as genomic instability, novel driver genes, tumor mutation burden, and BM lesion private neoantigens. In addition, the estimated timing of dissemination showed that BMs might occur early in lung cancer.
Implications:
Mechanistic insight from this study provides new insight into the biology of the metastatic brain process and a new beneficial approach for preventing and treating lung cancer BMs.
Introduction
Brain metastasis (BM) is one of the most common malignancies in the central nervous system, which is associated with poor survival outcomes and poses serious clinical challenges. Approximately 30%–50% of patients with non–small lung cancer (NSCLC), particularly those with lung adenocarcinoma, would develop BMs during disease progression (1).
A recent new understanding of the genomic etiology of NSCLC has contributed to developing personalized therapeutic options for patients. Unique genomic traits of BM of lung cancer have also been reported previously (2–8). Gene variations, chromosomal alterations, mutation spectrum, mutational signature, clonality, evolutionary pattern, metastasis pathways, and tumor mutation burden (TMB) may exhibit differences between primary lung lesions and BM of lung cancer. Clinically, these findings suggest that usual genetic detection of the primary tumor (PT), as in routine clinical practice, might overlook potential therapeutic targets in BMs. Moreover, despite their clinical importance, understanding of BMs of NSCLC is still limited.
To thoroughly investigate the evolutionary characteristics and immune divergence between primary and BM tumors, we analyzed whole-exome sequencing (WES) data for paired primary and metastatic brain samples from 14 patients with NSCLC. In addition, we used another series of WES data as a validation set (n = 13).
Materials and Methods
Patient samples and clinical characteristics
We collected 28 paired tumor specimens from 14 patients with NSCLC at the Cancer Hospital of the Chinese Academy of Medical Sciences and Peking Union Medical College who underwent surgical resection from 2009 to 2018. The study was approved by the ethics committee of the Cancer Hospital of the Chinese Academy of Medical Sciences (no. 21/288-2959). The validation dataset included 13 patients with NSCLC with paired tumor specimens: 12 patients from a previous study (5) and another patient from the same institution. The patient's clinical characteristics were collected from medical records, including sex, age, smoking history, and tumor subtype (according to International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society Classification system). WES was performed for all tumor samples (Shanghai Tongshu Biotechnology Co., Ltd).
DNA extraction, next-generation sequencing, variant calling, and annotation
DNA from formalin-fixed paraffin-embedded samples was extracted using DNeasy Blood and Tissue Kit (69504, QIAGEN). Tissue sections were examined by pathologists and were required to contain at least 50% tumor cell nuclei with <20% necrosis per The Cancer Genome Atlas protocol requirements. Sequencing libraries were constructed on the basis of the Illumina standard library construction instructions (Illumina, Inc.).
For WES, DNA-targeted capture pulldown and exon-wide libraries were generated from native genomic DNA using xGen Exome Research Panel (Integrated DNA Technologies) and TruePrep DNA Library Prep Kit V2 of Illumina (#TD501, Vazyme) according to the manufacturer's instructions. Paired-end sequencing data were generated using a NovaSeq6000, with an average sequencing depth of 200× for tumor tissues. Sequencing reads were mapped to the human reference genome (GRCh37) using Burrows-Wheeler Aligner (9), and PCR duplicates were sorted and removed with GATK4.0. Mutation calling and filtering were performed following protocols described previously (10).
CNA burden, weighted genome integrity index, intratumor heterogeneity, and clonal diversity
Copy number alternations (CNAs) were called from aligned WES data using CNVkit (11). The CNA burden was calculated as described previously (12). The weighted genome instability index (wGII) was calculated on the basis of the segment results from CNVkit (13). Intratumor heterogeneity (ITH) and clonal diversity (Shannon diversity index, SDI) were evaluated according to methods described previously (12). A driver gene set was constructed by combining the two driver gene lists defined in previous studies (14, 15). Mutations of driver genes were annotated using cosmic89_coding, oncoKB, and other information by ANNOVAR.
Spectrum and signatures
Non-negative matrix factorization and model selection were used to identify major mutational signatures (16). In the cohort, one sample was considered to have a strong association with a mutational signature if the proportion of the contribution was >20% with Mutational Patterns (version 1.10; ref. 17) and deconstructed Sigs (18).
Estimation of the timing of metastatic seeding
Chronologic estimates of seeding time relative to PT diagnosis and parameter selection were performed following the multicancer study by Hu and colleagues (19). The tumor expansion age, that is, the tumor size and doubling time at diagnosis, was determined with the Gompertzian model (20).
TMB, human leukocyte antigen type, and neoantigen burden
TMB was calculated by the number of nonsynonymous somatic mutations [single-nucleotide variants (SNV) and small insertions/deletions] per mega-base in coding regions. We used the NetMHCpan version 4.0 binding strength predictor and Pickpocket to predict potential tumor neoantigens (21). According to the recommended algorithm, HLA typing was performed in silico using HLAscan (22).
Statistical analysis
All tests were performed in the R environment version 3.6.0 (R core team). The comparison between paired lung tumors and BMs was based on the Wilcoxon sign-rank test. The nonparametric Wilcoxon rank-sum test was applied to compare wGII, SDI, and ITH between smoking and no smoking statuses. Associations with FDR-adjusted P values [P(FDR), also called q values] <0.05 were considered significant.
Data availability
Raw WES data of the discovery cohort (GSA: HRA003466) re-ported in this article have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation, Chinese Academy of Sciences that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. Any information required to reanalyze the data from the discovery cohort reported in this article is available from Puyuan Xing (xingpuyuan@cicams.ac.cn) upon request. Professor Deng approved the validation cohort, and most of the dataset had been published previously.
Results
Overview of Chinese clinical cohorts
We performed WES among 14 patients with pairwise primary lung tumor and BM. There were 7 males, and the median age was 52 years. WES data for another 13 patients with matched samples were used as a validation cohort. The clinical characteristics of the 27 patients in the case series are listed in Supplementary Table S1.
Genomic landscape in PTs and paired BMs
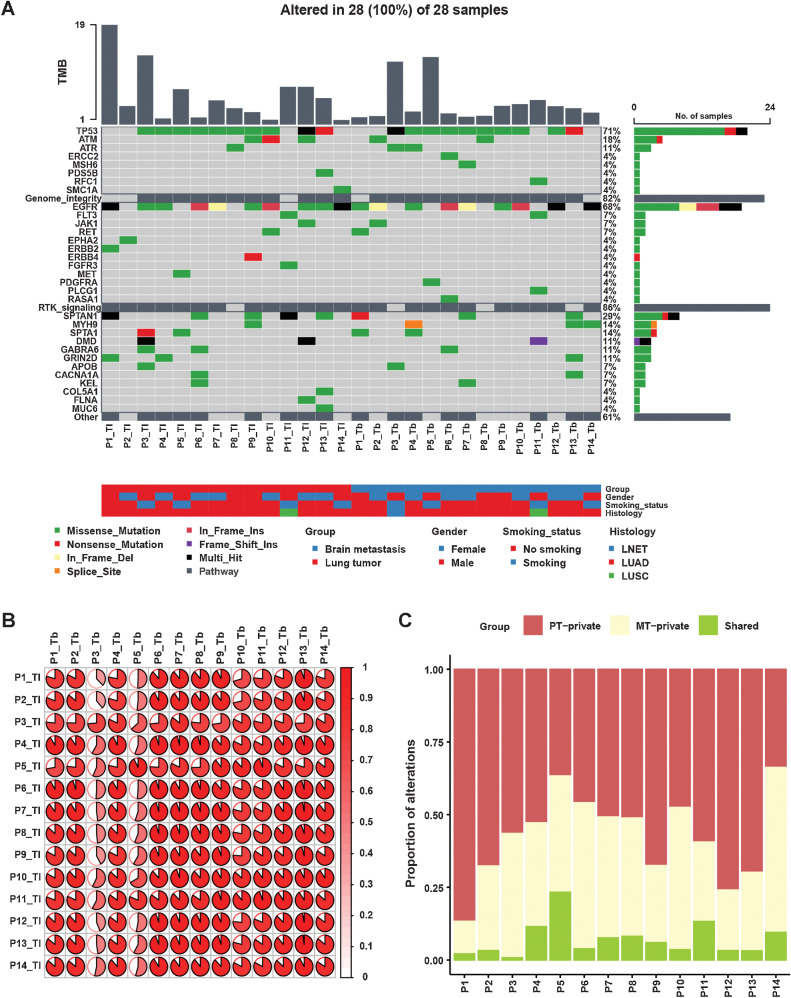
Divergence of BMs and primary lung tumors has been suggested by several studies, such as mutation landscape, mutation driver, pathway alterations, and TMB (2–6, 23). We compared the mutational discrepancy between PT and BM tumors in our discovery cohort. The most typically mutated genes were mainly enriched in two pathways: genome integrity and receptor tyrosine kinase (RTK) signaling (Fig. 1A). The most frequent driver alterations involving transcription factors were in the following genes: WT1 (14%), MYCN (11%), ZFHX3 (11%), and ZNF750 (11%). The most frequent alterations involving genome integrity were in the following genes: TP53 (71%), ATM (18%), and ATR (11%). In RTK signaling pathway, most driver alterations were in EGFR (68%), FLT3 (7%), JAK1 (7%), and RET (7%). We then calculated cosine similarity between mutational profiles in primary lesions and their matched BMs. As illustrated in Fig. 1B, we observed that almost all patients showed high concordance of mutations between primary and matched BM lesions. Interestingly, we also observed mutational similarity in paired samples among different patients, except for Patient 3. To investigate ITH, genetic mutations were classified as private mutations (present only in the primary or brain metastatic tumors) or shared mutations (present in both primary and brain metastatic tumors; ref. 24). We observed significantly higher heterogeneity between PTs and BMs (Fig. 1C). Shared genetic mutations, with a median proportion of 4.9% (range, 0.8%–23%), exhibited lower proportions than private mutations, which were present only in the primary or brain metastatic tumors [respectively, 54.6%, (range, 33.8%–86.7%); 37.9%, (range, 11.2%–56.7%)].
Figure 1.
Distinct mutational landscape of PTs and matched BMs. A, Comparison of the mutational landscape between primary lesions and matched BMs. The top panel represents the TMB of each sample. The middle panel represents the matrix of alterations in a selection of frequently mutated genes. Columns represent samples. Three clinicopathologic characteristics (gender, smoking history, and histology) are presented below. B, The cosine similarity of mutational profiles in primary lesions and their matched BMs was calculated. C, Proportions of private and shared alterations of primary and metastatic lesions of patients.
The landscape of CNAs and increased chromosomal instability in human BMs
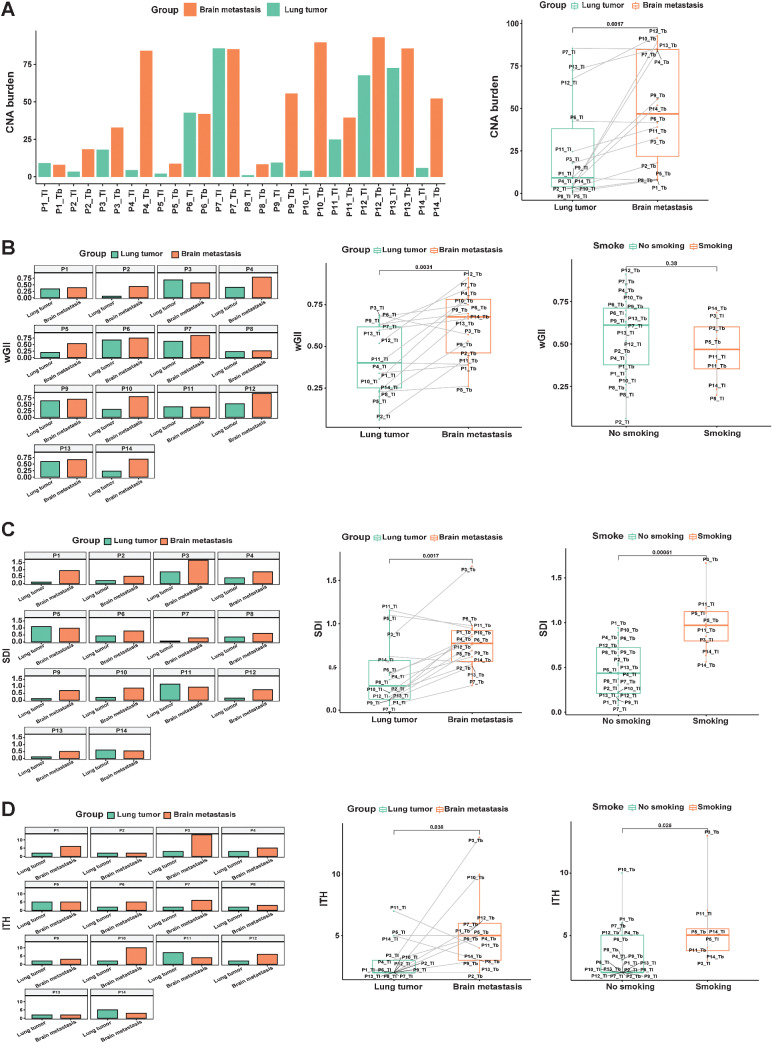
The CNA burden was similar between lesions from an individual index focus in P1, P6, P7, P11, and P13; the CNA burden of brain lesions was higher in P2, P3, P4, P5, P8, P9, P10, P12, and P14 (Fig. 2A). The median CNA burden (46.8) in BMs was significantly higher than that in PTs (9.1; P = 0.0017).
Figure 2.
Distinct CNA burden, wGII, SDI, ITH of PTs, and matched BMs. A, Comparison of the CNA burden between primary lesions and matched BMs. B, Comparison of wGII between primary lesions and matched BMs. C, SDI analysis of primary lesions and matched BMs. D, ITH analysis in primary lesions and matched BMs.
Chromosomal instability (CIN) is associated with tumor evolution and metastases (25). Matched PTs and BMs in our all-Chinese cohorts were compared, including levels of wGII, ITH index, and SDI. We applied wGII to evaluate the CIN difference among the matched samples. The wGII was similar between lesions for P1, P3, P6, P7, P8, P9, P11, and P13, whereas wGII differed between lesions for P2, P4, P5, P10, P12, and P14 (Fig. 2B). The median wGII (0.67) in BMs was significantly higher than that (0.4) in PTs (P = 0.0031; Fig. 2B). However, there was no significant difference in wGII between smoking and nonsmoking cases in the discovery cohort (Fig. 2B). SDI was similar between lesions for P4, P5, P6, P8, P11, and P14, but it differed between lesions for P1, P2, P3, P7, P9, P10, P12, and P13 (Fig. 2C). ITH was similar between lesions for P1, P2, P4, P5, P6, P7, P8, P9, P11, P12, P13, and P14, but it was different between lesions for P3 and P10 (Fig. 2D). SDI (P = 0.0017; Fig. 2C) and ITH (P = 0.036; Fig. 2D) of brain samples were higher than those of primary samples in the discovery cohort. In short, CIN might increase in BMs more than in PTs of Chinese patients with NSCLC.
The landscape of mutational signatures
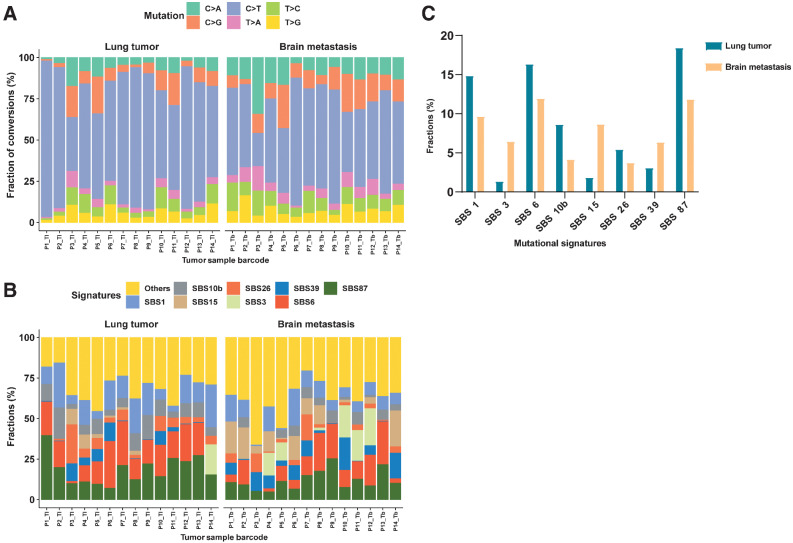
All samples mainly displayed the C>T transition (Fig. 3A). The median percentages of C>T, C>G, C>A, T>C, T>G, and T>A variants were 57.1%, 9.2%, 9.0%, 8.3%, 6.1%, and 3.6%, respectively. Discrepant variant spectra were observed between different lesions in all patients at the level of substitution composition, and the difference was statistically significant for P3 and P12 (Fig. 3A).
Figure 3.
The distinct signature landscape of PTs and matched BMs. A, The fraction of six substitution subtypes SNV in each group. B, Fractions of SBS signatures in each group. C, The average fraction difference of the top eight signatures in each group.
Mutational signature spectra are presented in Fig. 3B. The composition of single base substitution (SBS) signatures showed extensive heterogeneity between patients and between different lesions of an individual patient. As shown, SBS87, 6, 1, 10b, 26, 39, 15, and 3 were dominant in lung samples, with average percentages of 18.4%, 16.4%, 14.9%, 8.6%, 5.4%, 3.1%, 1.8%, and 1.3%, respectively. SBS6, 87, 1, 15, 3, 39, 10b, and 26 were dominant in brain samples, with average percentages of 11.9%, 11.8%, 9.7%, 8.7%, 6.4%, 6.3%, 4.1%, and 3.7%, respectively (Fig. 3C). Among the top eight signatures, four showed significant differences between primary and metastatic tumors, including SBS 15, 87, 1, and 3.
Furthermore, we compared between these four signatures and previously defined COSMIC signatures, which helps to reveal clinical significance, such as molecular mechanisms and etiology. SBS15 and SBS3 were significantly increased in BM tumors (Fig. 3C). SBS15 is associated with defective DNA mismatch repair and microsatellite instability (MSI) and is often found with other MSI-associated signatures. SBS3 is strongly related to somatic and germline BRCA1/2 variants and BRCA1 promoter methylation in breast, ovarian, and pancreatic cancers. SBS3 is also associated with response to platinum therapy and homologous recombination repair defect therapy, which is thought to be a predictor of defective homologous recombination–based repair. Nonetheless, PTs increased significantly with SBS87 and SBS1. According to COSMIC, SBS87 may be associated with thiopurine chemotherapy treatment, which needs to be experimentally validated. SBS1 is observed in many cancer types, and SBS1 mutations over time differ markedly among cancer types. These differences correlate with estimated stem cell division rates in different tissues, and SBS1 may therefore be associated with cell division and/or mitosis.
The timing of dissemination and clonal evolution analysis between primary and BM tumors
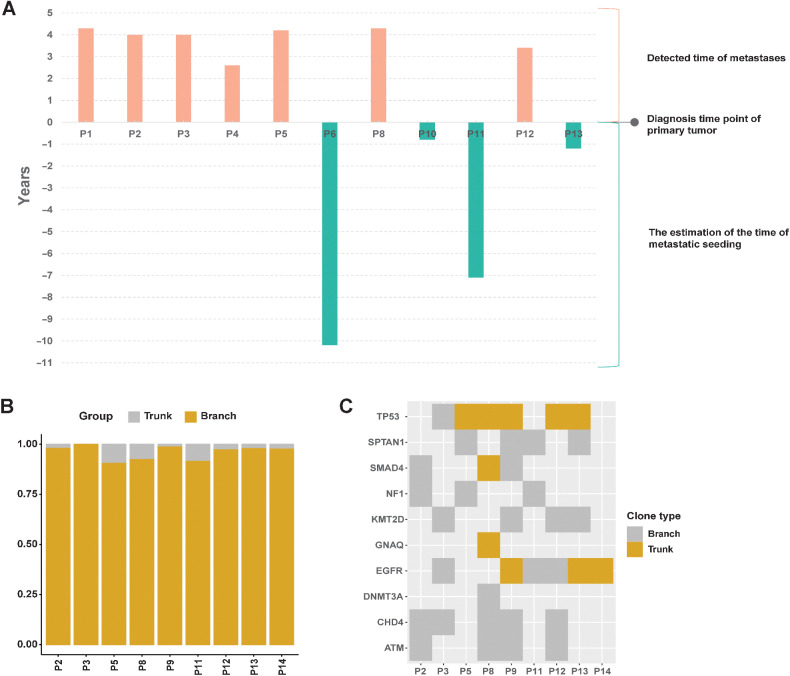
To infer the evolution of NSCLC with BM, we investigated clonality discrepancy and timing of dissemination between primary and metastatic tumors. According to a previous study, Ts is identified as the time (in years) from metastatic seeding to PT diagnosis (26). Ts was estimated for BMs (monoclonal metastases) in our Chinese cancer cohort (Fig. 4A). According to this method's average tumor expansion age (4.3 years), the seeding time estimate relative to the PT's diagnosis was 0.68 years. It is worth noting that 4 patients (P6, P10, P11, P13) may have had metastases before PT diagnosis. Nevertheless, our clinical data showed that 2 patients (P6 and P8) had BMs when they were initially diagnosed. This result indicates that BM may occur before the primary diagnosis and may even not be detected long after diagnosis.
Figure 4.
The timing of dissemination and clonal evolution analysis. A, Estimated time of metastatic seeding (Ts) for metastases in each cohort. B, Distribution of trunk and branch variants in each sample. Trunk variants encompassed those that occurred on the root node and its only child node; all others were classified to have occurred in the branch. C, Genes associated with lung cancer in each sample.
Then, we further investigated the evolution of driver variants. All 14 patient samples were assessed; results for driving gene mutation branching were obtained for 9 patients, whereas the other 5 patients did not show the driving gene branching result. The median percentages of trunk and branch variants in each sample were 2.4% and 97.6%, respectively. Branch variants were dominant in all detected samples (Fig. 4B). As shown in Fig. 4C, four driver genes were trunk variants, including two common driver genes, EGFR and TP53.
The immune landscape in PTs and their metastases
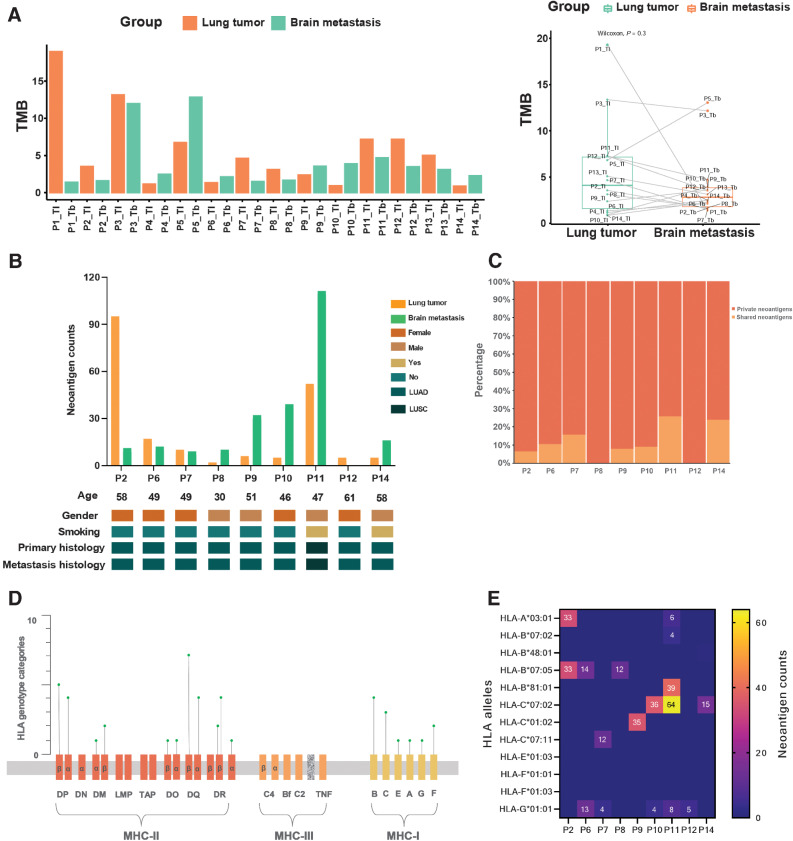
Genetic mutations were analyzed to describe TMB, HLA genotypes, and tumor neoantigens to investigate the immune characteristics of primary and BM lesions. We first calculated each tumor specimen's TMB. Heterogeneity existed in every patient's pairwise samples (Fig. 5A), even though nearly all histology was lung adenocarcinoma. The TMB was compared between the primary and matched BM lesions (Fig. 5A). Moreover, the TMB of primary lesions was higher than that of BM lesions (P = 0.3).
Figure 5.
The immune landscape in paired PTs and their metastases. A, The TMB distribution of all 28 primary and metastatic samples. The total TMB (defined as the number of nonsynonymous mutations) in each group. B, Baseline comparison and characteristics of neoantigen counts in PTs and BMs. TNB, tumor neoantigen burden; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma. C, Percentage of shared and private neoantigens. D, The distribution of identified HLA genotype categories in the HLA complex. E, Corresponding HLA genotypes of clonal neoantigens. HLA, human leukocyte antigen.
Next, we examined HLA genotypes and neoantigens of 28 samples from 14 cases with primary and brain lesions. Neoantigens were predicted according to the identified somatic mutations in all tumors. Among them (Fig. 5B), 9 (64%) patients showed neoantigens, 4 (29%) patients were male, and 2 (14%) had a smoking history. There were 8 patients with primary lung adenocarcinoma and 1 with primary lung squamous cell carcinoma. All patients displayed consistent pathology between primary and metastatic tumors. A wide range of tumor neoantigen burden (TNB) was identified in both PTs (median 6, range 2–95) and BMs (median 12, range 0–111; Fig. 5B). The TNB of PTs was similar to that of matched BMs (P = 0.406). Moreover, sex and smoking history had no impact on TNB of primary lesions or metastases, which might be due to the limited sample number. To investigate the characteristic discrepancy of PTs and BMs, neoantigens were classified as private (present only in PTs or BMs) and shared (present in both), as in previous studies (24, 27). We observed a relatively low percentage of shared neoantigens, with a median of 9.1% (range, 0%–25.8%; Fig. 5C). These results are consistent with those of previous studies (28).
As neoantigens are commonly considered clinically significant in personalized vaccine therapy, we further examined the distribution of HLA genotypes and shared neoantigens between PTs and BMs. All 14 patients were included in the analysis. Neoantigen-relevant results were not obtained for 5 patients. In total, 44 genotypes identified, including 12 MHC class I (MHC I) molecules and 32 MHC class II (MHC II) molecules (Fig. 5D). Most of them belong to HLA-B and HLA-C. Of note, more neoantigens were related to HLA-C*07:02 and HLA-B*07:05 alleles, especially the HLA-C*07:02 allele, which predicted 115 neoantigens (Fig. 5E). Intriguingly, the HLA-G*01:01 allele was found in five cases (55.5%); it is predicted to 34 neoantigens (Fig. 5E). In addition, several HLA-A alleles were identified, which could predicted neoantigens (Fig. 5E).
Validation in another Chinese cohort
The WES data were validated in another cohort. We compared the genomic characteristics of primary and BM tumors in this Chinese cohort. In accordance with the results of the discovery cohort, we observed consistency of primary and metastatic tumors (Fig. 6). The most frequent variants and associated pathway variants are presented in Fig. 6A. The frequent mutations were mainly enriched in three pathways: RTK signaling, genome integrity, chromatin and transcription factors pathways. The genome integrity and RTK signaling pathway-related genes were similar to those in the discovery cohort. Relatively uniform mutations between primary and matched BM lesions were also observed in this cohort (Fig. 6A). The BM tumor wGII was also higher than that of lung cancer samples (P = 0.11; Fig. 6B). In the validation cohort, we further found that the ITH (Fig. 6C) and SDI (Fig. 6D) of the primary samples were lower than those of the BM samples (P = 0.17 and P = 0.13, respectively). Similar to the discovery cohort, high concordance of mutations between primary and matched BM lesions was observed for most patients (Fig. 6E and F). There was no significant difference in wGII and ITH between smoking and nonsmoking cases in the validation cohort (P = 0.84 and P = 0.47, respectively), but SDI in smoking cases was significantly higher than that in nonsmoking cases (P = 0.023). The CNA burden in BMs was also higher than that in PTs (P = 0.056; Fig. 6G and H). These trends of CNA burden, wGII, ITH, and SDI between primary and matched BM lesions were consistent with the discovery cohort results.
Figure 6.
Distinct genomic landscape of PTs and matched BMs. A, Comparison of the genomic landscape between primary lesions and their matched BMs. B, Comparison of wGII between primary lesions and their matched BMs. C, ITH analysis in primary lesions and matched BMs. D, SDI analysis in primary lesions and matched BMs. E, The cosine similarity of mutational profiles in primary lesions and matched BMs was calculated. F, Proportions of alterations private to the primary or metastatic lesion or shared by primary and metastatic lesions in each tumor specimen. G–H, Comparison of CNA burden between primary lesions and matched BMs.
In addition, we investigated immune-relevant biomarkers in the validation cohort. Unlike the discovery cohort results, TMB in BM samples was higher than that in PT samples in this cohort (P = 0.15; Supplementary Fig. S1).
Discussion
Despite recent therapeutic progress in lung cancer, metastasis remains a major challenge for treatment failure, with a dismal long-term survival. In our discovery cohort, after surgery, 4 (28.6%, 4/14) patients were treated with EGFR-directed therapy, 4 (28.6%, 4/14) were treated with radiotherapy, and 6 (42.9%, 6/14) were treated with chemotherapy. Ten patients were treated with different kinds of adjuvant therapies, yet all 10 experienced BM. Increasing evidence shows that metastases are, to some extent, a product of the evolution of PTs (29). Tang and colleagues further suggested that metastatic seeding occurs approximately 2.74 years before clinical detection, with the PTs mainly seeding most nonlymph node metastases (30). Metastasis-competent clones may arise at the early and late stages of PT progression, even before primary diagnosis. Thus, a better understanding of the brain metastatic process may identify novel strategies to prevent and control distant metastases. Furthermore, elucidating the immune landscape divergence between paired samples might shed light on the development of a new approach for clinical therapeutics. Therefore, it is necessary to research a well-defined cohort of paired PTs and BMs and perform comparative sequencing analyses to explore these metastasis events. Nevertheless, most patients with BM of lung cancer are in late stages at diagnosis and are usually treated with palliative approaches instead of neurosurgical resection. Thus, researchers rarely have the opportunity to investigate pairwise primary-metastatic tumors in terms of genomic and immune characteristics on a large scale (31).
This study collected matched lung lesion and BM samples to investigate the mutational signature landscape, evolutionary characteristics, and immune characteristic divergence in NSCLC. Recent reports have found some novel metastasis-related variants that are potential biomarkers for targeted therapeutics and prognosis (3, 5). However, genomic landscape results revealed relative consistency between PTs and BMs in our cohorts. In addition to the typical driving genes and relevant variants of NSCLC, some uncommon variants were found. The mutant gene mainly showed enrichment in chromosome repair, genome stability, RTK signaling, and transcription factor–related signaling pathways, which might be involved in BM. Despite few identical variations in PTs and BMs, the overall mutation characteristics were consistent with previous reports (3). However, it is worth noting that we observed mutational differences in the PT and BM of Patient 3, who had lung neuroendocrine tumors (LNETs) in BM lesions. Recently, a single-cell RNA-sequencing study found that most of the same individual lung cancer cells simultaneously express classical marker genes of more than two histologic subtypes (adenocarcinoma; squamous cell carcinoma; neuroendocrine, NET) of NSCLC (32). Hence, we infer possible higher heterogeneity in the PT for Patient 3 than in the other patients, with more NET histologic subtype cells in the PT. In addition, we explored genomic stability–related indicators of paired primary and BM tumors and validated them with another dataset. The CNA burden, wGII, ITH, and SDI in BM lesions were similar to those of a previous study (6) and higher than in PTs, suggesting more increased genomic instability of BM tumors. Interestingly, in Fig. 2C and D, it can be seen that the SDI and ITH of Patient 3 have the largest changes from PT to BM, and the ITH and SDI in BM of Patient 3 were higher than those of the other patients in the discovery cohort. Previous study reported that the clonal divergence of small cell lung cance ancestors from the NSCLC cells occurred before the first EGFR tyrosine kinase inhibitors treatments, and the heterogeneity may be related to apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC)-induced hypermutation (33). We therefore speculate that Patient 3 most likely also underwent this process, resulting in the generation of additional subclones, some of which progressed to form neuroendocrine tumors.
We further analyzed signature differences between the two lesions to explore the causes of differences in genomic instability. The main base-substitution mutation types in both lesions were similar, but the SBS type with the highest proportion differed slightly. This result suggests that the difference in SBS might be related to increased genomic instability of brain lesions. The signature results provide insight into why genetic mutations associated with genome integrity are prevalent in BMs. These results suggest that the BM branch develops along a more heterogeneous and genomic instability path. In general, higher CIN may be present in BMs. In conclusion, clones with a preference for increased heterogeneity are more likely to form metastases, whereas clones with a preference for proliferation remain in the PT.
In addition to exploring the genomic characteristics of tumors in different lesions, we noted that patients with lung cancer BM had different prognoses after surgery. Previous studies have revealed that PTs likely seed most distant metastases in colorectal cancer but that pancreatic cancer exhibits highly diversified spread routes (34, 35). Therefore, we estimated the timing of dissemination of metastasis transmission and analyzed the evolution of driver gene branches. The results showed that the predicted metastatic results for 1 patient (P6) was consistent with our existing clinical follow-up. Because current clinical information is limited, more detailed clinical follow-up information is needed to confirm the predicted results of several other cases (P10, P11, and P13). These Ts results may predict the relatively poor prognosis for these patients in advance. The time of BM is estimated to be earlier, and the prognosis of patients was necessary to pay more attention. Although BMs and PTs derive from a common ancestor, they may develop in two different directions early in the evolution of tumor cells. Hence, it is possible to prevent lung cancer metastases by early cancer detection or periodic physical examination. Some novel molecular technologies, such as circulating cell-free DNA screening (36, 37), may be more sensitive than low-dose CT for detecting cancer. Sensitive cancer diagnostic methods could be significant in preventing lung cancer metastases as early as possible. The typical driver genes in NSCLC may appear in either the trunk or the branch in branching evolution, which suggests that these driver genes are relatively conserved in branching evolution. At the same time, we found some new driver genes present in the branch. These genes reflect the genetic mutation differences between metastatic and primary lesions and provide a new possible molecular basis for therapeutic strategies for patients with NSCLC with BM.
Furthermore, we explored the immunologic characteristics of primary lesions and BMs. In TMB analysis, our two datasets led to different results, which may be related to differences in mutation characteristics of the patients in the two datasets. Neoantigen analysis showed that the number of neoantigens in BMs was slightly higher than that in PTs, suggesting differences in immune characteristics between the two lesions. These immune results should be validated in larger sample sizes and in multiomics analysis research. The results of the current study show that patients with lung cancer with BM NSCLC respond to immunotherapy (38–41).
Increased genomic instability of BM tumors may induce higher TMB or TNB. Nevertheless, the TMB of BMs was lower than that of PTs in the discovery cohort, and the shared mutation proportion was also lower than that of the private mutation of both BMs and PTs. Hence, BM may have a more diverse immune microenvironment, which is a better basic condition for effective immunotherapy. In addition, although the mutation numbers of BMs are lower than those of PTs, the heterogeneity of BMs is higher. This indicates that BM lesions may have organ-specific antigens, which may support novel predictors for lung cancer with BM. A more detailed study of these differences might further improve the response rate to immunotherapy in patients with NSCLC with BM. We also explored HLA typing characteristics to provide theoretical references for developing new immune drugs.
There are also several limitations to this study. First, this was a retrospective study, which may lead to some bias. Second, because paired primary and metastatic samples are not readily available in the clinic, we further need more large sample size to validate these results. Third, multiomics analysis is needed to reveal and verify more details about tumor immune microenvironment divergence of lung and BM lesions in NSCLC.
In this study, we identified characteristics of lung cancer BMs that were different from primary lung cancer in four dimensions: genomic differences, evolutionary features, immune characteristics, and timing of dissemination, such as higher genomic instability, novel driver genes, BM lesion private neoantigens, and a more diverse immune microenvironment. These findings provide new insight into the biology of the metastatic brain process. Moreover, despite the conservation and similarity of driver mutations, specific signals are still enriched in BM lesions, providing valuable information for developing novel strategies to prevent and target BMs. In addition, research on the heterogeneity of PTs in depth may help to distinguish the histologic transformation of metastases early, determine prognosis and provide a theoretical basis for individualized treatment.
Supplementary Material
The TMB distribution of all primary and metastases samples. The total TMB (defined as the number of nonsynonymous mutations) in each group.
The basic clinical information of all patients.
Supplementary Figure 1. The TMB distribution of all primary and metastases samples. The total TMB (defined as the number of nonsynonymous mutations) in each group.Supplementary Table 1. The basic clinical information of all patients.
Acknowledgments
We want to thank the participants and their relatives for their cooperation and contribution to this work. Support for this work was provided by the CAMS Innovation Fund for Medical Sciences (CIFMS; no. 2020-I2M-C&T-A-016). The authors thank Shanghai Tongshu Biotechnology Co., Ltd. for technical support. We also thank MS Taiyan Guo from Shanghai Tongshu for her excellent assistance.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 291
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
T. Xie: Software, formal analysis, visualization, methodology, writing–original draft, project administration, writing–review and editing. Z. Liu: Data curation, visualization, methodology, writing–original draft, project administration, writing–review and editing. Y. Li: Data curation, validation, investigation. S. Wang: Supervision, validation, visualization, methodology, writing–review and editing. Y. Zhai: Data curation, writing–review and editing. F. Teng: Data curation, software. X. Hao: Data curation, validation, methodology. Y. Wang: Resources, data curation, methodology. H. Wang: Resources, data curation, validation. X. Zhang: Resources, data curation. X. Wu: Resources, data curation. J. Ying: Conceptualization, resources, data curation, supervision, validation, investigation, methodology. J. Li: Conceptualization, resources, data curation, supervision, investigation. Y. Zhang: Resources, funding acquisition, methodology. Y. Deng: Conceptualization, resources, data curation, software, formal analysis, supervision, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. P. Xing: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Cagney DN, Martin AM, Catalano PJ, Redig AJ, Lin NU, Lee EQ, et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: a population-based study. Neuro Oncol 2017;19:1511–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tomasini P, Barlesi F, Gilles S, Nanni-Metellus I, Soffietti R, Denicolai E, et al. Comparative genomic analysis of primary tumors and paired brain metastases in lung cancer patients by whole exome sequencing: a pilot study. Oncotarget 2020;11:4648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang T, Fang Z, Tang S, Cheng R, Li Y, Ren S, et al. Mutational landscape and evolutionary pattern of liver and brain metastasis in lung adenocarcinoma. J Thorac Oncol 2021;16:237–49. [DOI] [PubMed] [Google Scholar]
- 4. Dono A, Takayasu T, Yan Y, Bundrant BE, Arevalo O, Lopez-Garcia CA, et al. Differences in genomic alterations between brain metastases and primary tumors. Neurosurgery 2021;88:592–602. [DOI] [PubMed] [Google Scholar]
- 5. Liu Z, Zheng M, Lei B, Zhou Z, Huang Y, Li W, et al. Whole-exome sequencing identifies somatic mutations associated with lung cancer metastasis to the brain. Ann Transl Med 2021;9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shih DJH, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E, et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet 2020;52:371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H, Ou Q, Li D, Qin T, Bao H, Hou X, et al. Genes associated with increased brain metastasis risk in non-small cell lung cancer: Comprehensive genomic profiling of 61 resected brain metastases versus primary non-small cell lung cancer (Guangdong Association Study of Thoracic Oncology 1036). Cancer 2019;125:3535–44. [DOI] [PubMed] [Google Scholar]
- 8. Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov 2015;5:1164–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kimura K, Koike A. Ultrafast SNP analysis using the burrows-wheeler transform of short-read data. Bioinformatics 2015;31:1577–83. [DOI] [PubMed] [Google Scholar]
- 10. Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res 2018;24:3097–107. [DOI] [PubMed] [Google Scholar]
- 11. Talevich E, Shain AH, Botton T, BBC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 2019;179:219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013;494:492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 2020;20:555–72. [DOI] [PubMed] [Google Scholar]
- 15. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;174:1034–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Scott Shaffer J, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 2019;51:1113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benzekry S, Lamont C, Beheshti A, Tracz A, Ebos JM, Hlatky L, et al. Classical mathematical models for description and prediction of experimental tumor growth. PLoS Comput Biol 2014;10:e1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med 2016;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ka S, Lee S, Hong J, Cho Y, Sung J, Kim HN, et al. HLAscan: genotyping of the HLA region using next-generation sequencing data. BMC Bioinformatics 2017;18:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng Z, Cui L, Li P, Ren N, Zhong Z, Tang Z, et al. Genomic comparison between cerebrospinal fluid and primary tumor revealed the genetic events associated with brain metastasis in lung adenocarcinoma. Cell Death Dis 2021;12:935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109–21. [DOI] [PubMed] [Google Scholar]
- 25. Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553:467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu Z, Li Z, Ma Z, Curtis C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat Genet 2020;52:701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang T, Cheng R, Pan Y, Zhang H, He Y, Su C, et al. Heterogeneity of neoantigen landscape between primary lesions and their matched metastases in lung cancer. Transl Lung Cancer Res 2020;9:246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turajlic S, Swanton C. Metastasis as an evolutionary process. Science 2016;352:169–75. [DOI] [PubMed] [Google Scholar]
- 30. Tang WF, Wu M, Bao H, Xu Y, Lin JS, Liang Y, et al. Timing and origins of local and distant metastases in lung cancer. J Thorac Oncol 2021;16:1136–48. [DOI] [PubMed] [Google Scholar]
- 31. Preusser M, Berghoff AS, Koller R, Zielinski CC, Hainfellner JA, Liebmann-Reindl S, et al. Spectrum of gene mutations detected by next generation exome sequencing in brain metastases of lung adenocarcinoma. Eur J Cancer 2015;51:1803–11. [DOI] [PubMed] [Google Scholar]
- 32. Li Q, Wang R, Yang Z, Li W, Yang J, Wang Z, et al. Molecular profiling of human non-small cell lung cancer by single-cell RNA-seq. Genome Med 2022;14:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee JK, Lee J, Kim S, Kim S, Youk J, Park S, et al. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol 2017;35:3065–74. [DOI] [PubMed] [Google Scholar]
- 34. Naxerova K, Reiter JG, Brachtel E, Lennerz JK, van de Wetering M, Rowan A, et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 2017;357:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakamoto H, Attiyeh MA, Gerold JM, Makohon-Moore AP, Hayashi A, Hong J, et al. The evolutionary origins of recurrent pancreatic cancer. Cancer Discov 2020;10:792–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fiala C, Diamandis EP. New approaches for detecting cancer with circulating cell-free DNA. BMC Med 2019;17:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dudley JC, Schroers-Martin J, Lazzareschi DV, Shi WY, Chen SB, Esfahani MS, et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov 2019;9:500–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou Q, Chen M, Jiang O, Pan Y, Hu D, Lin Q, et al. Sugemalimab versus placebo after concurrent or sequential chemoradiotherapy in patients with locally advanced, unresectable, stage III non-small-cell lung cancer in China (GEMSTONE-301): interim results of a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol 2022;23:209–19. [DOI] [PubMed] [Google Scholar]
- 39. Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018–28. [DOI] [PubMed] [Google Scholar]
- 40. Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. [DOI] [PubMed] [Google Scholar]
- 41. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The TMB distribution of all primary and metastases samples. The total TMB (defined as the number of nonsynonymous mutations) in each group.
The basic clinical information of all patients.
Supplementary Figure 1. The TMB distribution of all primary and metastases samples. The total TMB (defined as the number of nonsynonymous mutations) in each group.Supplementary Table 1. The basic clinical information of all patients.
Data Availability Statement
Raw WES data of the discovery cohort (GSA: HRA003466) re-ported in this article have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation, Chinese Academy of Sciences that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. Any information required to reanalyze the data from the discovery cohort reported in this article is available from Puyuan Xing (xingpuyuan@cicams.ac.cn) upon request. Professor Deng approved the validation cohort, and most of the dataset had been published previously.