Differences in the effects of LKB1 loss in human versus murine lung adenocarcinoma are attributed to differential TPI1 regulation that leads to divergent metabolic liabilities and has implications for therapeutic targeting in cancer.
Abstract
KRAS is the most frequently mutated oncogene in human lung adenocarcinomas (hLUAD), and activating mutations frequently co-occur with loss-of-function mutations in TP53 or STK11/LKB1. However, mutation of all three genes is rarely observed in hLUAD, even though engineered comutation is highly aggressive in mouse lung adenocarcinoma (mLUAD). Here, we provide a mechanistic explanation for this difference by uncovering an evolutionary divergence in the regulation of triosephosphate isomerase (TPI1). In hLUAD, TPI1 activity is regulated via phosphorylation at Ser21 by the salt inducible kinases (SIK) in an LKB1-dependent manner, modulating flux between the completion of glycolysis and production of glycerol lipids. In mice, Ser21 of TPI1 is a Cys residue that can be oxidized to alter TPI1 activity without a need for SIKs or LKB1. Our findings suggest this metabolic flexibility is critical in rapidly growing cells with KRAS and TP53 mutations, explaining why the loss of LKB1 creates a liability in these tumors.
Significance:
Utilizing phosphoproteomics and metabolomics in genetically engineered human cell lines and genetically engineered mouse models (GEMM), we uncover an evolutionary divergence in metabolic regulation within a clinically relevant genotype of human LUAD with therapeutic implications. Our data provide a cautionary example of the limits of GEMMs as tools to study human diseases such as cancers.
This article is highlighted in the In This Issue feature, p. 799
INTRODUCTION
Lung cancer remains the most common cause of cancer mortality in the United States and worldwide due to high incidence coupled with poor response to standard-of-care therapies in most patients (1). However, lung cancer is not a single disease but comprises a spectrum of distinct subtypes that arise from different cellular origins, governed by specific somatic mutations affecting different sets of proto-oncogenes and tumor suppressor genes (2–5).
Previous genomic research has defined the underlying genetic mutations that govern specific disease subtypes and have revealed genotypes that are favored or disfavored in the mutational landscape of numerous cancers, including lung adenocarcinoma (LUAD; refs. 6–10). In efforts to understand these combinations of mutations in cancers of the lung and other tissues, most attention has been accorded to the selective advantages of mutations that promote cell growth and other tumorigenic traits referred to as “hallmarks of cancer” (11, 12). But some combinations of mutations, such as mutations that activate both the EGFR and KRAS oncogenes, are detrimental to cancer cells and selected against (13, 14).
Metabolic reprogramming is a cancer hallmark, required to support tumorigenesis in diverse environments (15, 16). Despite improvements in our understanding of metabolic discrepancies between normal and oncogenic tissues, accurately modeling and exploiting these differences for therapeutic intervention have achieved only marginal success. The serine/threonine protein kinase liver kinase B1 (LKB1) has been defined as a conserved central regulator of metabolic processes under both physiologic and pathologic contexts (17, 18). LKB1 governs organismal and cellular response to energy stress by activation of members of a family of kinases, the AMPK-related kinases (AMPKR), with the role of AMP-activated protein kinase (AMPK) most extensively defined (19–21). Further research is needed to annotate the role of other AMPKRs in governing the metabolic stress response.
Inherent differences between humans and mice may also have significant effects on tumor development through divergent mechanisms of response to the oxidative environment and to metabolic determinants (22). The nature and extent of such differences are unknown, but their mechanisms may reveal new molecular targets for therapy.
In this report, we have examined why the three most common genetic mutations found in human LUAD—loss-of-function TP53 mutations, oncogenic KRAS activation, and loss-of-function LKB1 mutations—rarely co-occur, despite earlier observations that loss of LKB1 is associated with a more aggressive growth of LUADs in genetically engineered mouse models (GEMM) harboring Trp53 and Kras mutations (23–27). Therefore, we sought to identify differences between human and mouse lung adenocarcinoma (hLUAD and mLUAD) with the most common genotype, mutated KRAS and TP53 (KP-mutant), and to determine the effects of loss of the tumor suppressor LKB1 on metabolic regulation and the growth of such tumors. We have focused on two especially intriguing aspects of the problem: the role of LKB1 in metabolic reprogramming and the physiologic consequences conferred by species-specific differences in the sequence of a critical enzyme governing cell metabolism, triosephosphate isomerase (TPI1). Our findings have implications for the design of therapeutic strategies that affect cancer cell metabolism and for the interpretation of results with mouse models of human cancers.
RESULTS
Co-occurrence of KRAS, TP53, and LKB1 Mutations Differentially Affects Growth of Human and Mouse LUADs
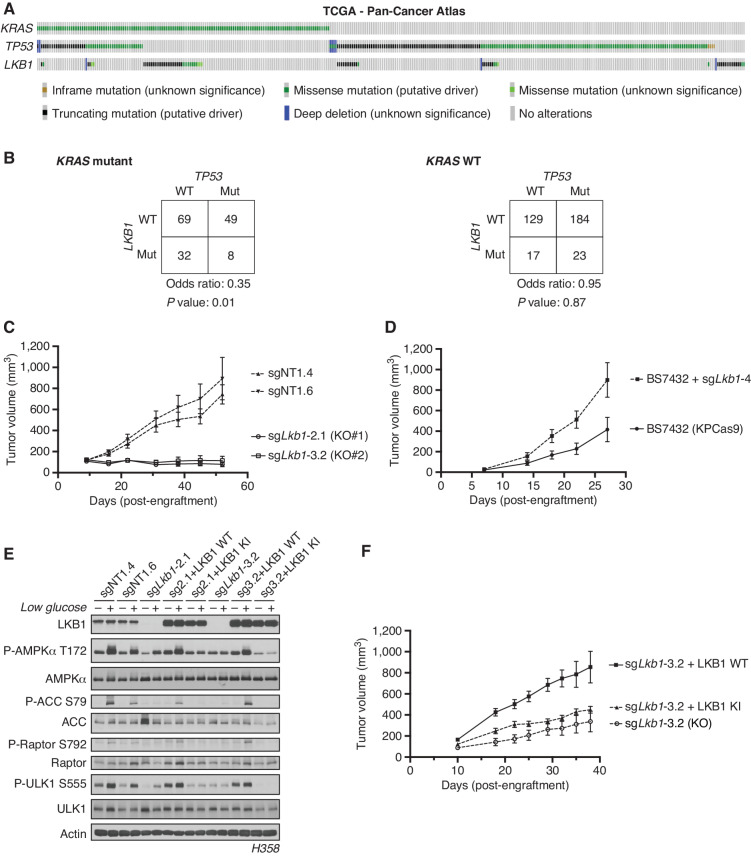
We used the The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas to determine the frequency of co-occurrence of mutations in the three most mutated genes in hLUAD—KRAS, TP53, and LKB1—and found that only 8 of 511 tumors carried mutations in all three genes (Fig. 1A). A Fisher exact test showed that the co-occurrence of LKB1 and TP53 mutations in hLUADs with a KRAS mutation was less frequent than expected by chance based on the overall frequency of mutations in these three genes, with an odds ratio of 0.35 and a P value of 0.01 (Fig. 1B). No similar reduction was observed in the comutation of TP53 and LKB1 in the absence of KRAS mutations (odds ratio = 0.95; P = 0.87; Fig. 1B). A second dataset from the Memorial Sloan Kettering Cancer Center derived from sequencing of tumor DNA from 1,357 patients with lung cancer revealed similar infrequency of triple mutations (Supplementary Fig. S1A and S1B; ref. 9).
Figure 1.
Co-occurrence of KRAS, TP53, and LKB1 mutations differentially affects the growth of human and mouse LUADs. A, The TCGA Pan-Cancer Atlas oncoprint of co-occurrence of KRAS, TP53, and LKB1 in hLUAD patients. B, Fisher exact test of the statistical likelihood of co-occurrence of LKB1 and TP53 mutations in a KRAS-mutant or wild-type (WT) background, respectively. C, Graph of mean (± SEM) tumor volumes of subcutaneous flank injections of H358 (KRAS;TP53) isogenic clones expressing Cas9 and a nontargeting (sgNT1.4 and sgNT1.6) or LKB1-specific (sgLkb1-2.1 and sgLkb1-3.2) guide RNA; 1 × 106 cells were implanted in the right hind flank (n = 10 per cohort). KO, knockout. D, Mean (± SEM) volumes of isogenic KPCas9 LUAD allograft tumors expressing a nontargeting (BS7432) or Lkb1-specific (sgLkb1-4) guide RNA; 0.25 × 106 cells were implanted in left and right flank of C57Bl/6J mice (n = 10 per cohort). E, Western blot analysis of H358 (KRAS;TP53) isogenic clones (KP: sgNT1.4 and sgNT1.6; KPL: sgLkb1-2.1 and sgLkb1-3.2) and KPL lines with additional transgenic expression of guide RNA resistant LKB1 WT (sgLkb1-2.1 + LKB1 WT and sgLkb1-3.2 + LKB1 WT) or LKB1 kinase-inactive (KI; sgLkb1-2.1 + LKB1 KI and sgLkb1-3.2 LKB1 KI) and treated with 11.1 mmol/L or 0.5 mmol/L glucose for 6 hours as indicated. Restoration of AMPK signaling in LKB1 WT lines in response to 0.5 mmol/L glucose validated by blotting for P-AMPK Thr172 and downstream substrates (P-ACC S79, P-ULK1 S555, and P-Raptor S792). Similar results were observed in three independent experiments and in an additional KRAS;TP53 cell line, H2009 (Supplementary Fig. S1E). F, Graph of mean (± SEM) tumor volumes of subcutaneous flank injections of H358 (KRAS;TP53) isogenic clones with transgenic expression of an empty vector (KO) or guide RNA resistant LKB1 WT or LKB1 KI; 1 × 106 cells were implanted in the right hind flank (n = 10 per cohort).
Although mutations in KRAS, TP53, and LKB1 together are rare in hLUAD, previous studies have shown that GEMMs harboring conditional mutations in all three genes develop mLUAD that is more aggressive and more likely to metastasize than with only two of these genes mutated (23–27). To investigate this discrepancy between LUAD in human patients and mouse models, we generated isogenic human and mouse KP cell lines with and without LKB1 and also compared them with existing GEMM-derived tumor lines with parallel genotypes (25). Human KP lines engrafted and formed tumors in vivo, whereas isogenic lines in which LKB1 was deleted (KPL) did not (Fig. 1C). Furthermore, human KP lines from two different human KP LUAD parental cell lines readily formed spheroids in organotypic culture, but KPL lines did not or were substantially regressed (Supplementary Fig. S1C and S1D). In contrast, both mouse isogenic cell lines formed tumors in vivo with subsequent Lkb1 inactivation accelerating observed tumor growth (Fig. 1D), whereas GEMM-derived KP and KPL lines both formed tumors in vivo and spheroids in vitro with equal frequency (Supplementary Fig. S1E and S1F). Additionally, treatment of human isogenic lines in organotypic culture with the KRASG12C inhibitor AMG-510 reduced the spheroid formation of a G12C-mutant KP line but partially rescued the spheroid growth of a KPL line (Supplementary Fig. S1G).
To determine whether these observations were attributable to LKB1 kinase activity, wild-type (WT) or kinase-inactive (KI; K78I) LKB1 was reexpressed in multiple isogenic KPL hLUAD lines. We first verified that wild-type LKB1 restored the activity of AMPK, a known substrate of LKB1, under conditions of energy stress. Glucose restriction caused LKB1-dependent phosphorylation of AMPK at Thr172 and of its downstream substrates [Acetyl CoA Carboxylase (ACC) at Ser79, Raptor at Ser792, and Unc-51 Like autophagy activating kinase 1 (ULK1) at Ser555; Fig. 1E; Supplementary Fig. S1H]. Expression of WT but not KI LKB1 rescued growth of xenografts in immunodeficient mice, suggesting that LKB1 kinase activity is required to support tumor formation by human KP LUAD cells (Fig. 1F).
Phosphorylation of Human TPI1 Is LKB1-Dependent
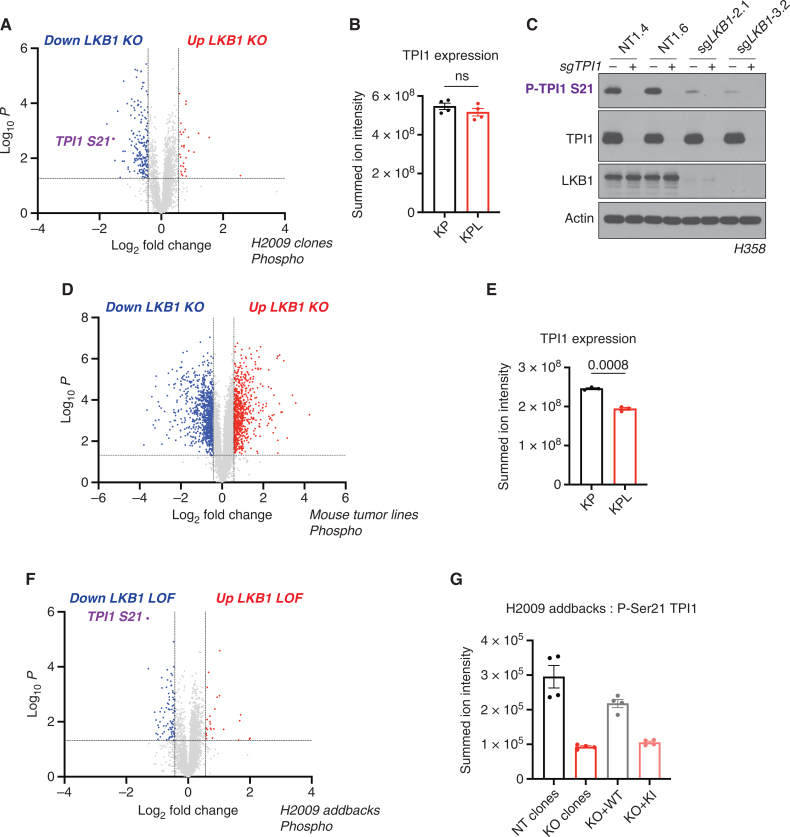
Because LKB1 phosphorylates and activates a family of AMPK-related Ser/Thr protein kinases (AMPKR) involved in regulating various metabolic and stress response pathways, we used comprehensive quantitative proteomics and phosphoproteomics under glucose-limited conditions to assess differences in protein phosphorylation between KP and KPL isogenic human lines. Phosphorylation of Ser21 on the glycolytic enzyme TPI1 was one of the most significantly downregulated phosphorylation events observed when comparing KPL with KP (Fig. 2A; Supplementary Fig. S2A and S2B), with peptide identification and phosphosite localization confirmed by manual inspection of the derived MS/MS spectra (Supplementary Fig. S2C). This effect was not due to differences in protein levels, as observed in our corresponding total proteomic analyses in multiple human KP and KPL isogenic lines (Fig. 2B; Supplementary Fig. S2D and S2E). Furthermore, utilizing a developed phosphospecific antibody against P-Ser21 of TPI1 confirmed phosphoproteomic results and validated this phosphorylation as an LKB1-dependent event (Fig. 2C). In contrast, using the same experimental design with tumor-derived mouse cell lines, we did not detect phosphorylation of Tpi1 in cells with either KP or KPL genotypes, although TPI1 protein levels were reduced in cells with a KPL genotype (Fig. 2D and E).
Figure 2.
Phosphorylation of human TPI1 is LKB1-dependent and regulates triose phosphate levels. A, Volcano plot of quantitative phosphoproteomic data of genetic sensitivity in H2009 clones (2 KP clones and 2 KPL clones); 2 biological replicates each, n = 4 per genotype. Cells were grown in 0.5 mmol/L glucose for 6 hours. Phosphopeptides that pass statistical criteria (P < 0.05) are highlighted in red and blue; peptides that do not satisfy this are colored gray. Phosphopeptides colored red satisfy a fold change > 1.5 and those colored blue satisfy a fold change < −1.5. The TPI1 P-Ser21 peptide labeled in purple text. KO, knockout. B, Bar graph of summed ion intensities for TPI1 protein expression in H2009 isogenic lines (KP: sgNT1.1 and sgNT1.2; KPL: sgLKB1-3.1 and sgLKB1-3.7). Cell lines were treated with 0.5 mmol/L glucose for 6 hours prior to collection. Data are representative of 4 independent biological experiments and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. ns, not significant. C, Western blot analysis of H358 (KRAS;TP53) isogenic cell (KP: sgNT1.4 and sgNT1.6; KPL: sgLKB1-2.1 and sgLKB1-3.2) and KPL lines to validate phosphospecific antibody. D, Volcano plot for comparison of quantitative phosphoproteomic data of genetic sensitivity in mLUAD cell lines, 634T (KP) and Lkb1-t2 (KPL), in biological triplicate for each condition. Analysis was conducted on cells treated with 0.5 mmol/L glucose for 6 hours in culture. Statistical criteria and color scheme are the same as for other volcano plots presented. E, Bar graph of summed ion intensities for TPI1 protein expression in mLUAD lines from companion unenriched total proteomic analysis. Data are representative of 3 independent biological experiments and reported as the mean (±SEM). Statistical significance was determined by a two-tailed paired t test. F, Volcano plot of quantitative phosphoproteomic data of genetic sensitivity in H2009 isogenic clones including clones with transgenic expression of guide RNA resistant WT or KI LKB1 in LKB1-specific KO (sgLKB1-3.1 and sgLKB1-3.7) from Supplementary Fig. S1E; 4 biological replicates each. The LKB1 loss-of-function (LOF) group consisted of merging LKB1 KO lines (KPL: sgLKB1-3.1 and sgLKB1-3.7) with lines expressing guide RNA resistant LKB1 KI (KPL + LKB1 KI: sgLKB1-3.1 + LKB1 KI and sgLKB1-3.7 + LKB1 KI), and was compared with H2009 lines containing nontargeting guide RNAs (KP: sgNT1.1 and sgNT1.2) merged with LKB1 KO lines expressing guide RNA resistant LKB1 WT (KPL + LKB1 WT: sgLKB1-3.1 + LKB1 WT and sgLKB1-3.7 + LKB1 WT) at the phosphopeptide level. Cells were grown in 0.5 mmol/L glucose for 6 hours. Statistical criteria and color scheme same as for A. The TPI1 P-Ser21 peptide is labeled in purple text. G, Summed ion intensity of the H2009 (KRAS;TP53) isogenic clones (KP: sgNT1.1 and sgNT1.2; KPL: sgLKB1-3.1 and sgLKB1-3.7) and lines with additional transgenic expression of guide RNA resistant LKB1 WT (sgLKB1-3.1 + LKB1 WT and sgLKB1-3.7 + LKB1 WT) or LKB1 KI (sgLKB1-3.1 + LKB1 KI and sgLKB1-3.7 LKB1 KI) for the phosphopeptide containing Ser21 of TPI1 from the experiments from which the volcano plot in Supplementary Fig. S2E was derived. Bar graph depicts each genotype individually and shows the restoration of TPI1 phosphorylation in KPL lines expressing transgenic WT LKB1 but not KI LKB1. Ion intensities were normalized to identify TPI1 protein expression from paired unenriched total proteomic analysis across conditions to control for protein expression; the relevant phosphopeptide was observed 3 times in each biological replicate.
To assess if the restoration of LKB1 kinase activity reestablished phosphorylation of TPI1 during metabolic stress, we used quantitative proteomics and phosphoproteomics to analyze human KPL cells expressing WT or KI LKB1 in parallel with KP and KPL cells under glucose-limited conditions. Again, Ser21 was one of the sites at which phosphorylation was most significantly reduced when KPL cells, and KPL cells expressing exogenous KI LKB1 were compared with KP cells and KPL cells expressing WT LKB1 (Fig. 2F). Quantification of phosphopeptide ion intensities within individual genotypes confirmed the restoration of P-Ser21 levels in human KPL lines expressing WT but not KI LKB1 (Fig. 2G), without significant variation in the abundance of TPI1 protein.
Phosphorylation of TPI1 Regulates Triose Phosphate Levels and Metabolic Flux
To examine the possibility that loss of regulation of TPI1 in hLUAD might explain selection against the KPL genotype, we studied the metabolic consequences of LKB1 deficiency. It is known that TPI1 controls interconversion of the triose phosphates dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP), both of which are generated by aldolase from the upstream glycolytic intermediate fructose-1,6-bisphosphate (F-1,6-BP). This conversion in carbon metabolism lies at a critical bifurcation point: One product, GAP, is used for glycolysis and energy homeostasis, whereas the other, DHAP, is used for lipid synthesis and cellular growth, and has recently been shown to activate mTOR (28). Additionally, previous studies have shown that increased oxidative burden due to KRAS and/or TP53 mutations causes metabolic flux to flow primarily through the oxidative pentose phosphate pathway (oxPPP) to increase the reductive potential and restore redox balance (29–31).
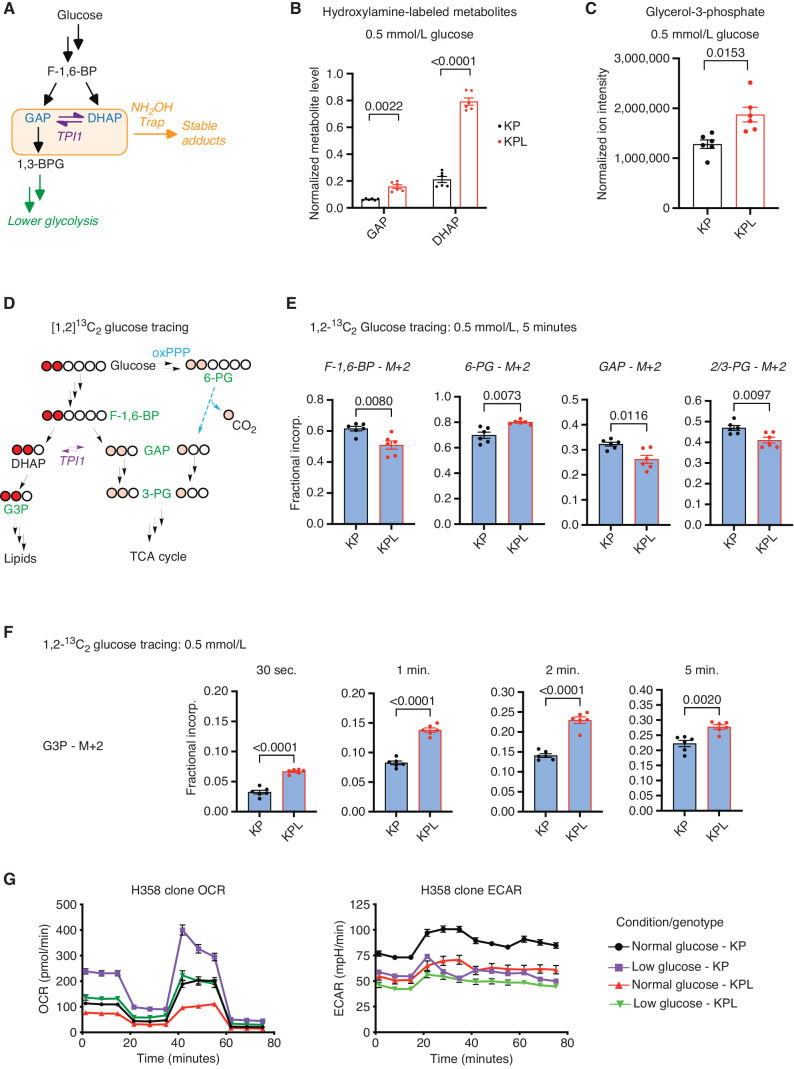
We assessed the influence of LKB1-dependent phosphorylation on TPI1 activity by measuring the pools of GAP and DHAP in KP and KPL hLUAD cell lines. Due to the inherent instability and complex chromatographic separation of the triose phosphates, we used in situ chemical-trapping metabolomics with hydroxylamine labeling of live cells under normal and glucose-limited conditions to create quantifiable, stable adducts (Fig. 3A; Supplementary Fig. S3A; ref. 32). These analyses confirmed the relative elevation of DHAP in KPL lines under normal and low glucose, further suggesting that TPI1 phosphorylation limited DHAP accumulation to maintain GAP for glycolysis (Fig. 3B; Supplementary Fig. S3B). Additionally, in a parallel analysis of KPL cell lines reexpressing WT and KI LKB1, levels of GAP and DHAP in LKB1 WT lines more accurately replicated endogenous levels in KP human cells, whereas DHAP remained elevated in LKB1 KI cells (Supplementary Fig. S3C). Furthermore, steady-state analysis revealed that human KPL cells had a significant increase in glycerol-3-phosphate (G3P), the next metabolic intermediate in lipid and triglyceride synthesis, in response to both normal and low glucose (Fig. 3C; Supplementary Fig. S3D). Collectively, the observed changes in metabolites and phosphorylation of human TPI1 suggested that LKB1 regulates the distribution of glycolytic metabolites via the triose phosphates through a regulatory phosphorylation site of TPI1.
Figure 3.
TPI1 phosphorylation regulates triose phosphate levels and metabolic flux. A, Schematic showing metabolites (shaded in the orange box) chemically labeled to create stable adducts. 1,3-BPG, 1,3-bisphosphoglycerate. B, Bar graph depicting in situ chemical-trapping metabolomics of hydroxylamine-labeled GAP and DHAP in H2009 clones (KP: sgNT1.1 and sgNT1.2; KPL: sgLKB1-3.1 and sgLKB1-3.7) treated in culture for 6 hours with 0.5 mmol/L. Data are representative of 3 independent biological experiments each containing 3 technical replicates and reported as the mean (±SEM). Cell number was normalized across models 12 hours prior to assay, and samples were normalized to an exogenous standard, d3-serine. Statistical significance was determined by a two-tailed paired t test. C, Normalized ion intensity of G3P from steady-state analysis of H2009 clones treated for 30 minutes with 0.5 mmol/L glucose. Analysis conducted in H2009 isogenic clones (KP: sgNT1.1 and sgNT1.2; KPL: sgLKB1-3.1 and sgLKB1-3.7) in biological triplicate and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. D, Schematic showing isotopic glucose tracing using positionally labeled 1,2-13C2 glucose, with circles representing carbons in each metabolite. Red circles indicate isotopic carbons and direct path to lipid synthesis through DHAP. Pink circles indicate readout of TPI1 conversion of DHAP to GAP and downstream glycolytic intermediates as well as alternate flux through the oxPPP. Green text indicates metabolites monitored and presented in histograms. E, Isotopic tracing results for M+2 isotopologues at 5-minute time point. Analysis conducted in H358 isogenic lines (KP: sgNT1.4 and sgNT1.6; KPL: sgLKB1-2.1 and sgLKB1-3.2) in biological triplicate (n = 6 per genotype) and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. F, Isotopic tracing results for M+2 isotopologue for G3P at 30 seconds and 1, 2, and 5 minutes. Analysis conducted in H358 isogenic lines (KP: sgNT1.4 and sgNT1.6; KPL: sgLKB1-2.1 and sgLKB1-3.2) in biological triplicate (n = 6 per genotype) and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. G, Mitochondrial stress test results; oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) plotted over the course of the assay. Analysis conducted in H358 isogenic lines (KP: sgNT1.4 and sgNT1.6; KPL: sgLKB1-2.1 and sgLKB1-3.2) in biological triplicate (n = 6 per genotype) and reported as the mean (± SEM) and treated as indicated with normal (11.1 mmol/L) or low (0.5 mmol/L) glucose for 6 hours prior.
To ascertain the extent to which glucose-derived central carbon flux was affected by the loss of LKB1 in human KP LUAD, we conducted metabolic flux analysis (MFA) using positionally labeled 1,2-13C2 glucose under low (0.5 mmol/L) glucose conditions at 0.5, 1, 2, and 5 minutes. The positional labeling of glucose allowed for accurate assessment of the flux of glucose-derived carbon through primary glycolysis, lipid synthesis, and oxPPP by monitoring differences in the isotopic incorporation of key metabolic intermediates in each pathway (Fig. 3D). More importantly, this tracer allows annotation of TPI1 activity more broadly within the context of central carbon metabolism, as the three carbons that produce GAP from F-1,6-BP would be unlabeled (M+0), and any observed incorporation of both labeled carbons (M+2) into GAP and lower glycolytic intermediates would require the conversion of DHAP to GAP by TPI1. If oxPPP is a route to generate GAP for lower glycolysis and the tricarboxylic acid (TCA) cycle, single carbon labeled (M+1) intermediates would be observed in significant amounts and can be readily determined by 3-phosphoglycerate (3-PG) isotopologue distribution (Fig. 3D; refs. 33–35).
We first looked at the differential incorporation of both carbons (M+2) into the glycolytic intermediates F-1,6-BP, GAP, and 3-PG, as well as the oxPPP intermediate 6-phosphogluconate (6-PG). We observed reduced incorporation of all glycolytic intermediates and an increase in 6-PG incorporation in the KPL versus KP hLUAD cells, with a linear increase of incorporation across the time course for F-1,6-BP and 6-PG (Fig. 3E; Supplementary Fig. S3E). The increase in 6-PG incorporation suggested that oxPPP flux may be higher in KPL. However, neither an appreciable accumulation of singly labeled carbon (M+1) incorporation into GAP and 3-PG nor a significant difference between KP and KPL hLUAD cells was observed. However, differences in unlabeled (M+0) GAP and 3-PG were observed at later time points, suggesting a difference in primary glycolytic flux (Supplementary Fig. S3F and S3G). In addition, when we measured the fractional incorporation of M+2 into the lipid and triglyceride synthesis pathway intermediate G3P, we observed a significant increase at all time points in KPL when compared with KP hLUAD cells (Fig. 3F).
Given the observed difference in metabolic flux after LKB1 was mutated in KP hLUAD cells, we next assessed mitochondrial function and metabolic rate in mouse and human LUAD lines with different genotypes. Human and mouse LUAD cells grown in normal or low glucose were analyzed for mitochondrial respiration and metabolic rate. Strikingly, maximal respiration was blunted in human KPL compared with KP lines, but no significant differences were found in the GEMM-derived mouse lines with KP and KPL genotypes (Fig. 3G; Supplementary Fig. S3H). Additionally, extracellular acidification rate (ECAR) values supported a pronounced defect in the metabolic rate selectively in human KPL cells. Under normal (11.1 mmol/L) glucose conditions, ECAR values for human KPL cells were similar to those in KP cells grown under low (0.5 mmol/L)-glucose conditions. This discordance was not observed in the mouse counterparts (Fig. 3G; Supplementary Fig. S3H). Collectively, the MFA and mitochondrial stress results indicate that the reduced central carbon flux observed in KPL hLUAD is diverted into lipid and triglyceride synthesis pathways, contributing to a metabolic defect, and compromised mitochondrial respiration not observed in mLUAD cells with a KPL genotype.
LKB1-Activated Members of the SIK Family Phosphorylate Human TPI1
We next sought to determine whether human TPI1 is phosphorylated directly by LKB1 or by one of the downstream LKB1-dependent Ser/Thr protein kinases of the AMPKR family; these kinases, all of which require phosphorylation by LKB1 for activity, are known to mediate responses to various metabolic stresses (Fig. 4A; refs. 18, 36, 37).
Figure 4.
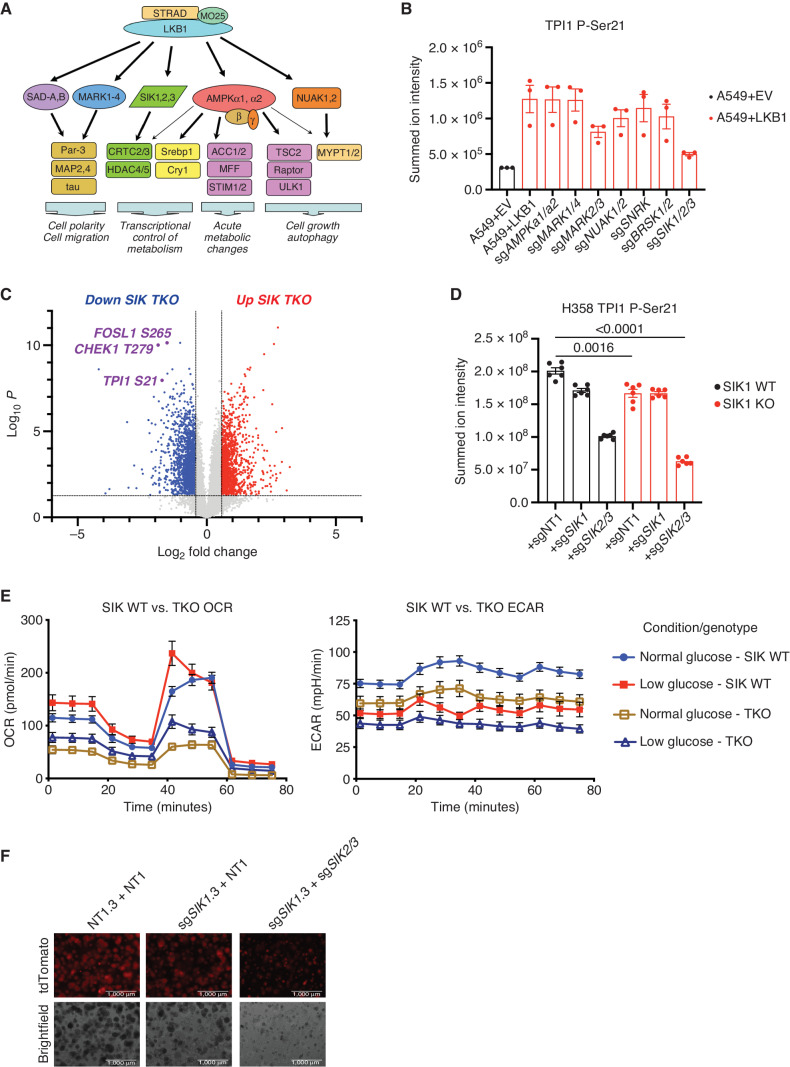
SIKs phosphorylate human TPI1 in KP hLUAD cell lines. A, Cartoon depicting regulation of the AMPKR kinase family members by LKB1 and their downstream substrates. B, Bar graph of summed ion intensity for the TPI1-derived Ser21 phosphopeptide from extracts of A549 cell lines infected with an empty vector (EV) or a vector expressing WT LKB1; the indicated guide RNAs were used to inactivate members of the AMPKR subfamilies in LKB1 transgenic expressing cells. Cell lines were cultured in 11.1 mmol/L glucose prior to analysis. Ion intensities were normalized to identified nonphosphorylated peptides across conditions to control for protein expression and reported as the mean (±SEM). C, Volcano plot of quantitative phosphoproteomic data used to compare phosphorylation in H358 clones (2 KP clones and 2 KP SIK TKO clones, with 3 biological replicates of each). Cells were cultured in 0.5 mmol/L glucose for 6 hours before lysis. Phosphopeptides that pass statistical criteria (P < 0.05) are highlighted in red and blue; those that do not satisfy this criterion are colored gray. Proteins highlighted in red satisfy the fold change threshold of >1.5 after triple deletion of SIK1,2,3. Phosphopeptides highlighted in blue satisfy the fold change threshold of < −1.5 for a decrease after SIK1,2,3 triple deletion. D, Bar graph of summed ion intensities for the TPI1-derived Ser21 phosphopeptide in extracts of isogenic H358 cell lines containing a nontargeting control (sgNT1.3 and sgNT1.4) or SIK1-specific (sgSIK1.3 and sgSIK1.4) guide RNA and additional control (NT1), SIK1 (sgSIK1), or dual SIK2 and SIK3 (sgSIK2/3) guide RNAs in a polyclonal population. Ion intensities were normalized against identified nonphosphorylated variant across conditions. Cell lines were cultured in 0.5 mmol/L glucose prior to lysis, analyzed in biological triplicate per clone (n = 6 per genotype), and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. KO, knockout. E, Mitochondrial stress test results; oxygen consumption rate (OCR) and ECAR plotted over the course of the assay, respectively. Analysis conducted in H358 isogenic lines used for phosphoproteomic analysis in D in biological triplicate (n = 6 per genotype) and reported as the mean (± SEM) and treated as indicated with normal (11.1 mmol/L) or low (0.5 mmol/L) glucose for 6 hours prior. F, 3D spheroid growth in Matrigel of isogenic clones of the H358 cell line labeled with a tdTomato fluorescent reporter and expressing Cas9 and nontargeting controls (sgNT1.3) or SIK1-specific (sgSIK1-2.3) guide RNA and additional control (NT1) or dual SIK2 and SIK3 (sgSIK2/3) guide RNAs in a polyclonal population. Five thousand cells were seeded into Matrigel and grown for 14 days, and the media were changed every 24 hours. Images were taken on an EVOS fluorescence microscope under 4× magnification and filtered to resolve tdTomato signal intensity and brightfield.
To determine whether AMPKR kinases are directly responsible for TPI1 phosphorylation downstream of LKB1, we monitored the phosphorylation of Ser21 in TPI1 in a panel of human KRAS;LKB1-mutant LUAD cell lines in which subfamilies of the AMPKR kinases have been mutated, after restoring the stable expression of WT LKB1 from a transgene (Fig. 4B; ref. 24). Restoration of LKB1 increased the phosphorylation of Ser21 in TPI1, consistent with results in Fig. 2F and G, and inactivation of the SIK subfamily significantly reduced Ser21 phosphorylation (Fig. 4B). Mutational inactivation of other AMPKR superfamily members [the microtubule affinity regulating kinases, the NUAK family kinases (NUAK), the brain-specific serine/threonine protein kinases, the catalytic subunits of AMPK, and SNF-related kinase] did not have significant effects on the phosphorylation of TPI1 at Ser21. Furthermore, by analyzing specific combinations of SIK family member mutants, we found that SIK1 and SIK3 together made the greatest contribution to phosphorylation of Ser21 in TPI1 (Supplementary Fig. S4A).
The phosphorylation of Ser551 in SIK3, known to regulate activity by altering molecular association, was one of the most significantly downregulated phosphorylation sites in the LKB1-inactivated, KP-mutant hLUAD cell lines, supporting the concept that the SIK subfamily of protein kinases drives the phosphorylation of Ser21 in TPI1 (Supplementary Fig. S2B; refs. 38, 39). Additionally, the phosphorylation of Ser62, Ser329, and Ser370 in CREB regulated transcription coactivator 3 (CRTC3), all SIK family substrate sites, was also reduced in KPL versus KP human lines (Supplementary Fig. S2A and S2B). Furthermore, we found that the amount of SIK1 mRNA was significantly increased upon inactivation of LKB1 in multiple human KP lines, suggesting that a signaling network might increase transcription of SIK1 via a feedback mechanism to recover SIK activity after the loss of LKB1 (Supplementary Fig. S4B).
We next asked if SIK family kinases were responsible for the phosphorylation of TPI1 in human KP lines that express LKB1 from the endogenous LKB1 locus. We generated a series of cell lines deficient in members of the SIK gene family, including two SIK1/2/3 triple-knockout lines (SIK TKO). Analysis of this series of cell lines by Western blot confirmed the deletion of all SIK family members in the SIK TKO lines (Supplementary Fig. S4C). A quantitative total proteomic analysis of KP hLUAD cells under low-glucose conditions with various combinations of deletions of SIK1, SIK2, and SIK3 in KP hLUAD revealed numerous proteomic changes (Supplementary Fig. S4D). However, the amount of TPI1 protein was not significantly altered (Supplementary Fig. S4E). Quantitative phosphoproteomic analyses on the same cells under low glucose revealed that Ser21 in TPI1 was among the phosphorylation events with the greatest reduction when SIK TKO cells were compared with SIK WT cells, mirroring LKB1 loss. In addition, numerous proteins and phosphosites known to regulate the cell cycle, including CHEK1 and FOSL1, were affected; the latter findings were validated by Western blot analysis without a notable difference in phospho-ERK1/2 (Fig. 4C; Supplementary Fig. S4F). Additionally, measurements of the TPI1 P-Ser21 tryptic peptide from these cells indicated that each of the SIK kinases contributes to the phosphorylation of TPI1 at Ser21 in KP hLUAD cells downstream of LKB1 (Fig. 4D). Furthermore, SIK TKO in KP hLUAD cells phenocopied LKB1 loss in compromised mitochondrial respiration and extracellular acidification under normal and low-glucose conditions and reduced spheroid formation in organotypic culture (Fig. 4E and F).
These results indicate that in the examined KP hLUAD cell lines, the SIK family of LKB1-regulated protein kinases appears to dominate the phosphorylation of Ser21 in TPI1. However, because deletion of all three SIK family members did not eliminate phosphorylation at this site, other LKB1-regulated protein kinases may also contribute to TPI1 phosphorylation or compensate for the loss of the SIK kinases.
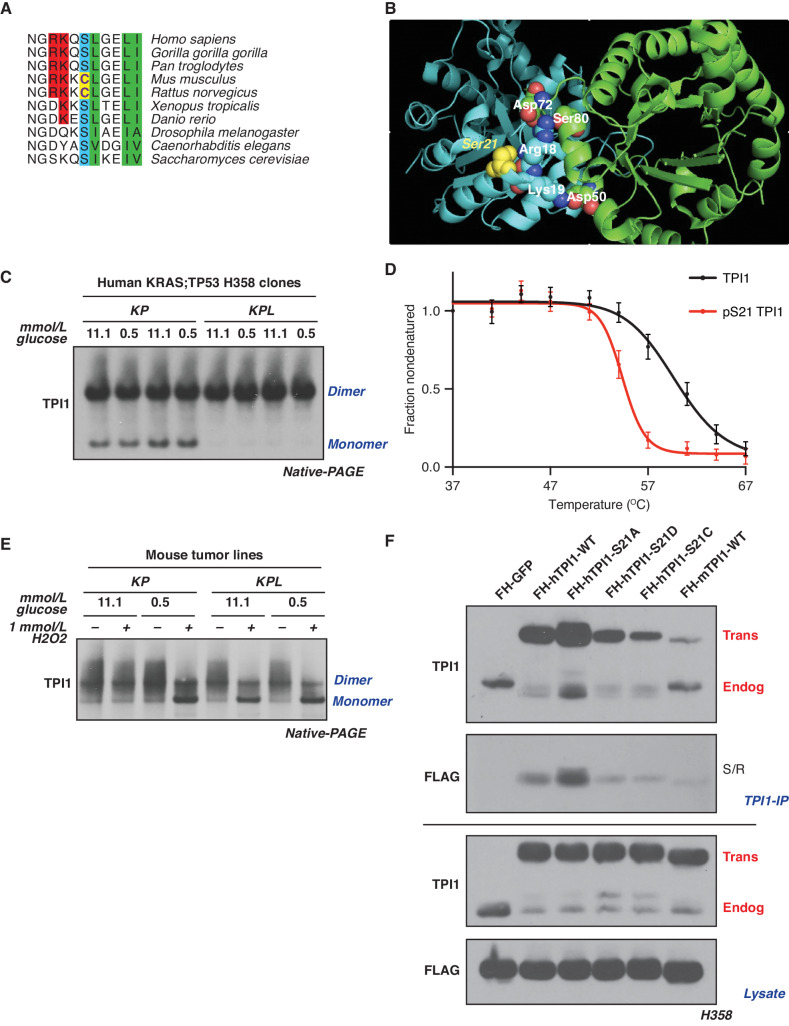
The Nonconserved Amino Acid Sequence of TPI1 Requires LKB1 to Regulate Its Multimeric State in Human but Not Mouse LUAD
To determine whether differences between LKB1 loss in human and mouse LUAD cells could be explained by differences in the regulation of human and mouse TPI1, we explored the evolutionary conservation of the primary amino acid sequence surrounding position 21 of this enzyme. Ser21 and the surrounding residues are conserved in most mammals and other metazoans, including yeast. However, in mouse and rat TPI1, Ser21 has been replaced by cysteine (Fig. 5A). In a published crystal structure of human TPI1, the hydroxyl moiety of Ser21 is located at a region of subunit–subunit interactions, presumably stabilizing the homodimer. Notably, the nearby residues Arg18 and Lys19 form intersubunit electrostatic interactions predicted to further stabilize the highly active homodimer state. This structure raises the possibility that phosphorylation of Ser21 could produce intrasubunit charge interactions with Arg18 and/or Lys19, interfering with the ability of these amino acids to confer stability to the dimer, thereby altering enzymatic activity (Fig. 5B; ref. 40). Because the sulfur atom of Cys21 in rodent TPI1 could be oxidized to sulfinic or sulfonic acid, mimicking phosphorylation of Ser21, it is possible that rodents have a mechanistic alternative to phosphodependent regulation of TPI1 activity. This could explain differences in the response to loss of LKB1 in mouse and human KP tumors, circumventing the requirement for LKB1 activity in murine tumors in response to stress.
Figure 5.
LKB1 regulates the multimeric state of hTPI1 but not mTPI1 due to an amino acid difference at position 21. A, Sequence alignment of TPI1 amino acid residues 16 to 26 across species, showing conservation of Ser21 from H. sapiens to S. cerevisiae, with cysteine at position 21 in mouse and rat TPI1. B, Crystal structure of TPI1 homodimer (cyan and green, respectively), with critical residues highlighted in space-filling atoms. Ser21 on the cyan monomer is highlighted in yellow. C, Western blot analysis of (BN-PAGE) of human isogenic clones derived from KP H358 hLUAD cell line. Cells were grown under normal (11.1 mmol/L) or low (0.5 mmol/L)-glucose conditions for 6 hours prior to collection. D, Melting curve plot from thermal profiling of unmodified and Ser21-phosphorylated TPI1. Analysis conducted in H2009 and H358 isogenic clones expressing Cas9 and a nontargeting (sgNT1.1 and sgNT1.2 or sgNT1.4 and sgNT1.6, respectively) guide RNA. Data presented are from 7 biological replicates and reported as the mean (±SEM). E, Western blot (Blue Native PAGE) of extracts from mLUAD cell lines. Cells were cultured in either 11.1 mmol/L or 0.5 mmol/L glucose for 6 hours and then treated with 1 mmol/L H2O2 for 15 minutes. F, Western blot of proteins coimmunoprecipitated from extracts of H358 cells expressing Cas9 and a nontargeting (FH-GFP cell line) or TPI1-specific (all other cell lines) guide RNA and transgenic expression of FLAG-HA tagged GFP or guide RNA resistant TPI1 allelic variants using a polyclonal antibody against full-length TPI1. Cells were cultured in 0.5 mmol/L glucose for 6 hours prior to collection. IP, immunoprecipitation.
Based on the structural features of human and mouse TPI1, we asked whether the loss of LKB1 kinase activity and reduced phosphorylation of human TPI1 at Ser21 would differentially affect the dimerization and activity of this enzyme in cells from the two species. We used native gel electrophoresis (BN-PAGE) and Western blot to determine the proportions of monomeric and dimeric TPI1 in extracts of two human KP cell lines, in the presence and absence of LKB1, when cells are grown under normal glucose conditions (Supplementary Fig. S5A). Loss of LKB1 promoted the dimeric (more slowly migrating) form of the human TPI1 protein; conversely, cell lines expressing LKB1 had increased monomeric TPI1 (Supplementary Fig. S5A). Furthermore, analysis of one set of isogenic hLUAD lines under normal and low-glucose conditions revealed that LKB1 loss dictated the biophysical distribution of dimeric and monomeric TPI1 under both growth conditions and that the availability of environmental glucose had no observable effects (Fig. 5C). Thermal proteome profiling was also used to measure the thermal stability of TPI1 proteoforms (41, 42). The ΔTm (measured at 0.5 fraction nondenatured) of the phosphorylated variant was 5.8°C lower than that of unmodified TPI1, further supporting the prediction that phosphorylation of Ser21 limits dimerization of TPI1 (Fig. 5D).
In contrast, we observed no changes in the ratio of the monomeric and dimeric forms of mouse TPI1 in KP versus KPL cell lines grown in media with normal or low-glucose concentrations (Fig. 5E). However, acute treatment of mouse lines with the oxidant peroxide caused a dramatic shift toward the monomeric form of TPI1 in low-glucose medium, independent of LKB1 status (Fig. 5E). Additionally, peroxide treatment caused a similar shift toward monomeric TPI1 under normal (11.1 mmol/L) glucose conditions in KPL but not in KP mouse cells (Fig. 5E), consistent with earlier reports that loss of LKB1 increases basal oxidative stress (43–46).
To further explore the functional significance of the amino acid difference at position 21 of human and mouse TPI1 and its effect on homodimer formation, we created a series of tagged TPI1 variants expressed as transgenes in human KP cells following the CRISPR/Cas9-mediated depletion of endogenous TPI1. Replacement of serine with alanine (S21A) increased recovery of both the transgenic and remaining endogenous TPI1 by immunoprecipitation under competitive, antibody-limited conditions and low-glucose treatment, implying that an inability to phosphorylate position 21 of TPI1 stabilized the TPI1 dimer. In contrast, the phosphomimetic S21D-mutant form of TPI1 or a mutant in which Ser21 is replaced by an oxidizable cysteine (S21C) significantly reduced coimmunoprecipitation of the remaining endogenous TPI1 under low or normal glucose conditions, with no observed effect from acute oxidant treatment with diamide in the latter (Fig. 5F; Supplementary Fig. S5B). These findings were further confirmed when TPI1 variants were analyzed by BN-PAGE. WT transgenic human TPI1 was found in both the dimeric and monomeric states, but the S21A mutant was detected solely in the dimeric state, and the S21C variant was mostly monomeric (Supplementary Fig. S5C). Collectively, these findings support the conclusion that phosphorylation of human TPI1 or oxidation of murine TPI1 destabilizes its dimeric form, providing a structural mechanism by which TPI1 activity can be regulated in response to LKB1-dependent phosphorylation or by oxidative stress.
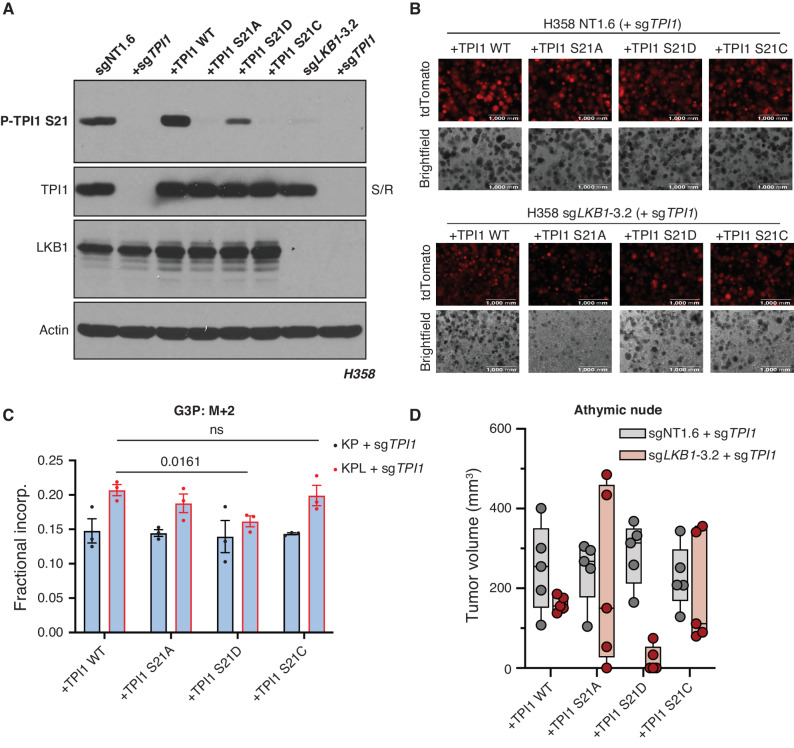
To gauge the phenotypic effects of single amino acid substitutions at position 21 of TPI1, we created an untagged allelic panel of TPI1 variants following CRISPR/Cas9-mediated depletion of endogenous TPI1 and verified loss of phosphorylation (Fig. 6A). First, we tested TPI1 allelic variants in both human KP and KPL isogenic lines for spheroid growth in organotypic culture (Fig. 6B). As seen previously, all TPI1 variants in the KPL isogenic cells were substantially reduced in spheroid growth, in both size and number, relative to TPI1 allelic variants in isogenic KP cells. Strikingly, we also observed near-complete ablation of observable spheroid formation of the S21A variant in the KPL cells, further suggesting that regulation of this site is critical upon LKB1 loss in KP hLUAD (Fig. 6B). To verify our previous metabolic flux analyses, we next conducted isotopic tracing in our human TPI1 allelic variants in both KP and KPL isogenic cells using positionally labeled 1,2-13C2 glucose under low (0.5 mmol/L)-glucose conditions at 1 minute. When we measured the fractional incorporation of M+2 into the lipid and triglyceride synthesis pathway intermediate G3P, we again observed a statistically significant increase in all TPI1 variants in the KPL genotype when compared with KP hLUAD cells except for TPI1 S21D, which was compelling (Fig. 6C). Additionally, we observed a similar effect when we looked at differential incorporation of both carbons (M+2) into GAP, where again only TPI1 S21D in the KPL cells showed a reduction in incorporation when compared with TPI1 S21D–expressing KP cells, with no significant difference observed for any other TPI1 variant in KP versus KPL genotypes (Supplementary Fig. S6A). Furthermore, no change was observed when comparing TPI1 variants in KP versus KPL cells for the glycolytic intermediates F-1,6-BP and 3-PG (Supplementary Fig. S6B and S6C), as well as the oxPPP intermediate 6-PG (Supplementary Fig. S6D). Lastly, we engrafted all TPI1 variant cell lines into the flanks of athymic nude mice to look for changes in tumor growth rates. We observed the most pronounced reduction in tumor growth with the TPI1 S21D variant expressed in human KPL cells (Fig. 6D).
Figure 6.
TPI1 amino acid differences at position 21 affect growth in organotypic culture, in vivo, and alter metabolic flux. A, Western blot analysis of human H358 (KRAS;TP53) isogenic KP and KPL cells (KP: sgNT1.6; KPL: sgLKB1-3.2) and derived untagged TPI1 allelic panel with phosphospecific antibody. B, 3D spheroid growth in Matrigel of isogenic clones of the H358 cell line labeled with a tdTomato fluorescent reporter and expressing Cas9- and TPI1-specific (sgTPI1-3) guide RNA and transgenic expression of guide RNA resistant TPI1 allelic variants. 5,000 cells were seeded into Matrigel and grown for 14 days, and the media were changed every 24 hours. Images were taken on an EVOS fluorescence microscope under 4× magnification and filter to resolve tdTomato signal intensity and brightfield. C, Isotopic tracing results for M+2 isotopologue for G3P at 1 minute. Analysis conducted in H358 isogenic lines (KP: sgNT1.6; KPL: sgLKB1-3.2) with additional expression of sgTPI1 guide RNA and guide RNA resistant TPI1 allelic variants in biological triplicate (n = 3 per genotype) and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. ns, not significant. D, Subcutaneous tumor growth of human H358 isogenic lines (KP: sgNT1.6; KPL: sgLKB1-3.2) with additional expression of sgTPI1 guide RNA and guide RNA resistant TPI1 allelic variants. Cells were first infected with control or LKB1-targeting guide RNA to produce isogenic KP and KPL, respectively. Derived lines were then infected with lentiviruses encoding TPI1 single-guide RNA with a subsequent lentiviral expression of transgenic guide RNA resistant hTPI1 WT, S21A, S21D, or S21C and measured 30 days following engraftment. Injections in athymic nude mice (n = 5/group).
These unexpected observations with the TPI1 S21D variant suggested the possibility of an unusual mechanistic event. During the revision of our manuscript, a study reported the use of native mass spectrometry and in vitro reconstitution experiments to deduce that TPI1 Ser21 phosphorylation existed in a hemiphosphorylated state in the TPI1 dimer (i.e., only one of two monomers is phosphorylated; ref. 47). Furthermore, this phosphorylation increased enzymatic activity by 4-fold over the unmodified dimer. The authors found no evidence of a doubly phosphorylated TPI1 dimer in both cell lysate and in vitro reconstitution studies, which collectively supports our biochemical, metabolomic, and allelic xenograft data, suggesting that TPI1 S21D would be the most deleterious allele for tumor growth because TPI1 would be entirely monomeric.
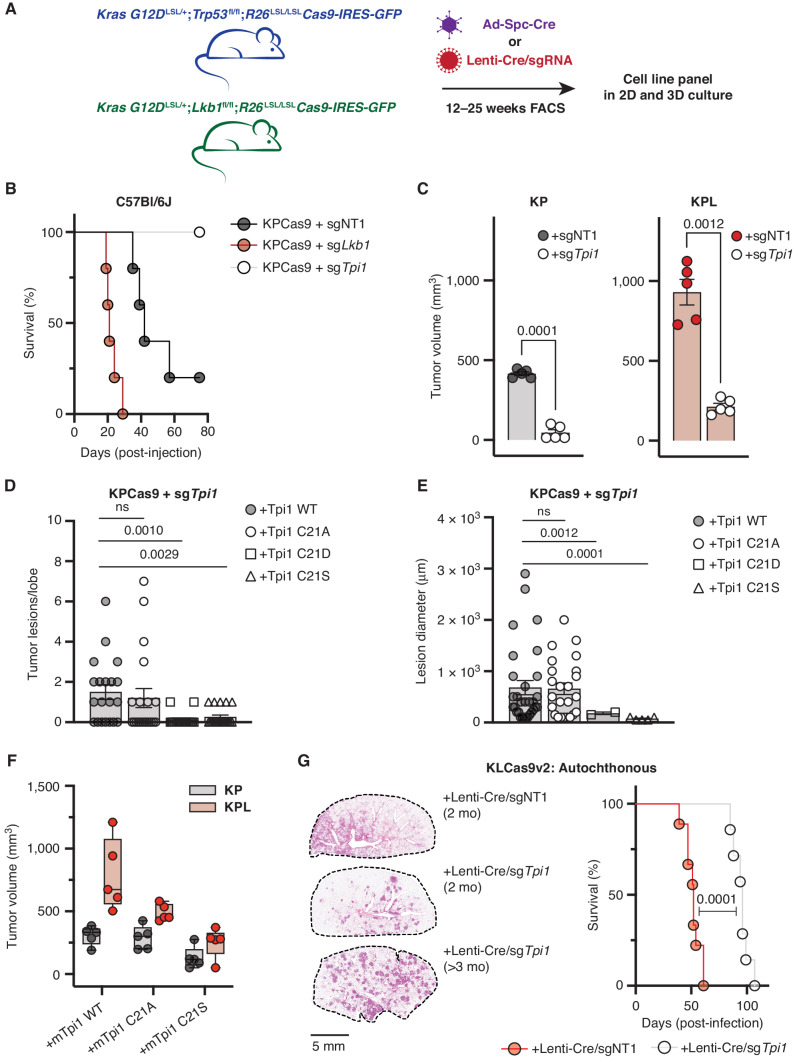
To assess the effects of TPI1 alterations on tumor growth and to explore the therapeutic relevance of TPI1 as a target, we generated conditional KPCas9 and two different KLCas9 (v1 and v2) mouse models that can inactivate genes targeted by CRISPR-mediated mutagenesis (KRAS G12DLSL/+;Trp53fl/fl;R26LSL/LSLCas9-IRES-GFP, KRAS G12DLSL/+;Lkb1fl/fl;R26LSL/LSLCas9-IRES-GFP and KRAS G12DLSL/+;Lkb1fl/fl;H11LSL/LSLCas9;R26LSL/LSLtdTomato; see Methods; Fig. 7A). Tumor-derived mLUAD cell lines were established from these mice and used to characterize the functional consequences of genetic changes by injection into immune-competent, syngeneic hosts. CRISPR/Cas9-mediated mutation of Lkb1 in KPCas9 tumor cells (now KPLCas9 cells) delivered by tail-vein injection reduced the median survival of mice by approximately 50% compared with control cells, with increased lung colonization and tumor burden (Fig. 7B; Supplementary Fig. S7A). Conversely, mutational inactivation of Tpi1 permitted 100% survival of mice receiving tail-vein injections, with blunted lung colonization and absence of clearly defined tumor lesions in the experimental time sampled (Fig. 7B; Supplementary Fig. S7A). We next compared Tpi1 loss by paired flank injections in syngeneic hosts of isogenic KP versus KPL cells and observed that loss of Tpi1 in both KP and KPL significantly reduced tumor volume 1 month following engraftment (Fig. 7C).
Figure 7.
TPI1 is required for tumor growth in KP and KPL LUAD models, and humanizing TPI1 regresses tumor growth and burden. A, Cartoon schematic depicting generated conditional genetic mouse models and subsequent derived tumor-derived cell lines. B, C57Bl/6J mouse survival following tail-vein injection of the KPCas9 cell line (BS7341) infected with lentiviruses encoding control or targeting sgRNAs (n = 5/group). C, Subcutaneous tumor growth of KP- or KPL-derived tumor cell lines (BS7432) infected with lentiviruses encoding control or targeting sgRNAs 30 days following engraftment in C57Bl/6J mice (n = 5/group); paired t test provided. D, Histogram quantifying tumor lesions per lobe in tail-vein-injected KPCas9 (BS7431) Tpi1 allelic variants in C57Bl/6J mice (n = 5/group) and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. ns, not significant. E, Histogram quantifying tumor lesion diameter in tail-vein-injected KPCas9 (BS7431) Tpi1 allelic variants in C57Bl/6J mice (n = 5/group) and reported as the mean (± SEM). Statistical significance was determined by a two-tailed paired t test. F, Subcutaneous tumor growth of KPCas9-derived tumor cell line (BS7432) first infected with control or Lkb1-targeting guide RNA to produce isogenic KP and KPL, respectively. Derived lines were then infected with lentiviruses encoding Tpi1 sgRNA with subsequent lentiviral expression of transgenic guide RNA resistant mTpi1 WT, C21A, or C21S and measured 30 days following engraftment. Paired flank injections in C57Bl/6J mice were conducted (KP/KPL for each transgene) per mouse (n = 5/group). G, KLCas9v2 autochthonous tumor model survival following intratracheal administration of lentiviruses encoding Cre-recombinase and control (sgNT) or targeting (sgTpi1) sgRNAs (n = 12/group). Representative lung tumor burden in groups of mice at 8 weeks (sgNT and sgTpi1) and 14.5 weeks (sgTpi1) following intubation; histologic appearance of tumor lesions.
We next assessed the effects on tumor growth and lung colonization of single amino acid variations at position 21 in mouse TPI1 in our tumor-derived cell lines. As with our human TPI1 allelic panels, CRISPR-based mutation of endogenous TPI1 was followed by expression of transgenes encoding single-guide RNA (sgRNA) resistant WT, C21A, C21D, C21E, or C21S versions of TPI1, and we tested for evidence of phosphorylation using the previously derived phosphospecific antibody. Results verified that the C21S variant is phosphorylated in the mouse tumor-derived KPCas9 cells (Supplementary Fig. S7B). Furthermore, deletion of Lkb1 in culture or de novo generation of tumor-derived KPLCas9 cells by the administration of sgTrp53 guide RNA (gRNA) with Cre-recombinase in our KLCas9v1 model and generation of additional TPI1 allelic variants revealed reduced phosphorylation of TPI1 C21S by both routes (Supplementary Fig. S7C). Next, tail-vein injection of the established KPCas9 TPI1 allelic panel was conducted to assess lung colonization and tumor burden in response to a single amino acid substitution. No difference was observed in tumor lesions per lobe or lesion diameter between reexpression WT and C21A; however, both C21D and C21S resulted in a significant reduction in both tumor burden and tumor size in the lung (Fig. 7D and E). Additionally, histologic staining revealed reduced colonization of the lung and absence of clearly defined tumor lesions using cells with TPI1 C21D and C21S, with the same time course previously for cells in which TPI1 had been mutationally inactivated (Fig. 7B; Supplementary Fig. S7A), phenocopying those observations (Supplementary Fig. S7D).
Given these collective observations, we next explored the effects on tumor growth of single amino acid variations at position 21 in mouse TPI1 in our derived cysteine to alanine or serine in mouse TPI1 isogenic KP and KPL cells by injection into the flanks of syngeneic hosts. Paired engraftment into mice demonstrated that transgenic reexpression of TPI1 WT showed increased tumor volume in KPL versus KP as previously observed (Fig. 7F). However, KPL cells with C21A TPI1 produced smaller tumors, whereas little or no effect on tumor size was observed when C21A TPI1 was produced in KP cells, further supporting the necessity of regulating TPI1 at position 21 upon Lkb1 loss and confirming our lung colonization data (Fig. 7B, D, and E). KP and KPL tumor cells producing C21S TPI1 both showed reduced tumor volumes, and previous Western blot analysis (Supplementary Fig. S7C) suggests that residual phosphorylation is still present in the LKB1-inactivated lines.
Lastly, we asked if the inactivation of TPI1 in an autochthonous lung cancer model was disadvantageous. Utilizing our KLCas9v2 GEMM, we intratracheally administered equivalent titers of lentiviral particles encoding Cre-recombinase and gRNAs targeting Tpi1 (sgTpi1) or a nontargeting control (sgNT). The median survival of mice receiving Lenti-Cre/sgNT (KLN) was 52 days, in line with previous reports on the latency of KL lung tumorigenesis (24, 48–51). However, mice administered Lenti-Cre/sgTpi1 (KLT) had a median survival of 96 days (Fig. 7G). Comparative histology when KLN mice exhibited labored breathing (2 months) revealed tumor burden that was multifocal/multinodular in both KLN and KLT mice, but KLN mice had larger, more diffuse tumors. In contrast, lesions in KLT mice were smaller and less diffuse (Fig. 7G). KLT mice had normal-appearing lung tissue in between the tumor nodules, while in KLN mice there was diffuse spreading of tumor cells along the alveolar septa (i.e., lepidic growth; Supplementary Fig. S8A). Additionally, histology in moribund KLT mice on study greater than 90 days revealed sustained, well-demarcated lesions with no observable lepidic growth, suggesting distinct pathology and tumor progression upon TPI1 inactivation (Fig. 7G; Supplementary Fig. S8A). Finally, to confirm whether observed lesions fully inactivated TPI1, we performed immunofluorescence on temporally matched, tumor-bearing lungs. Although TPI1 is expressed in all cells, and highly expressed in infiltrating immune/proliferative cells, the smaller lesions from KLT mice were less intense in their staining for TPI1 (Supplementary Fig. S8B). However, all KLT mice died from lung tumor burden, and examination of these late lesions showed an increase in TPI1 signal as compared with early-stage tumors (Supplementary Fig. S8B). Therefore, we cannot rule out that TPI1 was inactivated in every tumor cell and a significant amount of tumor mass observed may be attributed to tumor cells that escaped TPI1 inactivation. Taken together, these results suggest that TPI1 may be a relevant therapeutic target in KP hLUAD and validate the identified regulatory axis as a contributing factor in the selection against the KPL genotype in human LUAD.
DISCUSSION
Although much progress has been made using GEMMs to decipher mechanisms of tumor initiation and progression, comparisons of human and mouse tumors often lead to conflicting observations (52). In particular, accurately recapitulating the tumor metabolic environment remains a significant challenge, but an important one, as discrepancies between mouse and human tumors are likely to have implications in the development of novel therapeutic agents (53). Here we provide a mechanistic explanation for why the loss of LKB1 in hLUADs driven by KRAS and TP53 mutations is a rare event that appears to be selected against in human tumors, but not in mouse tumors, where the loss of Lkb1 enhances tumorigenesis and metastasis.
Several recent reports have implicated the SIKs as effectors of LKB1-mediated tumor suppression in Kras- and Kras;Tp53-mutant mLUAD; however, similar findings have not been reported in hLUADs (24, 27). Here, we find that this discrepancy may be due, in part, to a single amino acid difference between rodent and other metazoan versions of the glycolytic enzyme TPI1. In turn, that difference can influence subsequent metabolic events, which determine the flow of glucose-derived tricarbon substrates into pathways for glycolysis or lipid synthesis. In humans, the abundance of the products of TPI1 is governed by the LKB1–SIK–TPI1 signaling axis that we have elucidated in this article. In rodents, the substitution of an oxidizable cysteine for a phosphorylatable serine at residue 21 of Tpi1 enables direct redox regulation, circumventing the requirement for regulation by LKB1–SIK-mediated phosphorylation.
Our biochemical, proteomic, and metabolomic data support the conclusion that phosphorylation of TPI1 in hLUAD regulates the biophysical distribution of monomeric and dimeric forms, altering enzymatic activity and in turn triose phosphate pools. This reduces the conversion of GAP to DHAP, an energetically downhill reaction, and thereby shifts the balance away from glycerol lipid production and toward alternate metabolic pathways, including glycolysis and the TCA cycle. Regulation of metabolites at this central point in the glycolytic pathway could help to overcome metabolic stresses experienced during tumorigenesis and to improve the efficiency of energy production. In addition, this regulation allows rapidly growing cells to balance pathways for lipid synthesis versus serine/glycine synthesis. Collectively, these metabolic differences could strongly influence a wide range of protumorigenic processes and have significant effects on tumor cell phenotype, all of which warrant future study.
Additional features of these phenomena—such as how specific KRAS and TP53 mutations influence this phenotype and their contributions to the response to metabolic and oxidative stresses—have yet to be deciphered. Previous research suggests that TP53 can upregulate the enzyme TIGAR in response to oxidative and metabolic stresses, which suggests a critical role for p53 expression in response to LKB1 loss with anticipated increased oxidative stress (30, 54–56). Yet further work is needed to decipher whether TIGAR expression is differentially affected in p53-null versus p53-mutant cells. In fact, our preliminary data suggest that homozygous deleted TP53 KP hLUAD cells (NCI-H358) respond more poorly to acute LKB1 loss than TP53-mutant KP hLUAD cells (NCI-H2009) in organotypic culture and warrants further investigation (Supplementary Fig. S1C and S1D).
Knowledge of the differences in human and mouse TPI1 may not only explain the different consequences of loss of LKB1 in human and mouse LUADs but also help to design next-generation mouse models in which the mechanisms of metabolic regulation of human cancers are more accurately replicated. Furthermore, the findings reported here suggest that selective inhibitors of LKB1 or SIK family protein kinases could be effective in treating human KRAS/TP53-mutant lung cancers or other cancers with KRAS/TP53 mutations. Our work also raises the cautionary note that preclinical trials with such inhibitors would likely fail in currently available GEMMs with Kras and Trp53 mutations.
Finally, the observations reported here also reveal new ways for LKB1 to regulate metabolism beyond its known capacity to respond to cellular energy levels through activation of AMPK (20). Although enzymes such as hexokinase, pyruvate kinase, and phosphofructokinase have been intensely studied in regard to phosphorylation-dependent regulation in cancers, TPI1 has not been considered a likely factor in cancer-dependent regulation of metabolic flux. Additional research is needed to understand whether regulation of TPI1 by LKB1 is important in other types of human cancers and whether this knowledge provides a basis for seeking new cancer therapies across multiple organ types and mutational backgrounds.
METHODS
No statistical methods were used to predetermine sample size. Data were visualized and statistical analyses were performed using Prism 9 software (GraphPad) or R statistical package. P < 0.05 was considered statistically significant. P values for paired comparisons between two groups with comparable variance were calculated by a two-tailed Student t test.
Reagents
Media and sera were purchased from Life Technologies and R&D Systems, respectively. AMG-510 (sotorasib) was acquired from Selleck Chemicals (catalog no. S8830). All other reagents were from Sigma-Aldrich unless otherwise noted.
Cell Lines
All cell lines (A549, H358, H441, H2009, 634T, and HEK293T) were purchased from ATCC or kindly provided by Kwok-Kin Wong at NYU Langone Medical Center (634T and Lkb1-t2). Cells were maintained in RPMI 1640 medium (Life Technologies, 11879020) supplemented with glucose concentrations as indicated (Life Technologies, A2494001) except for HEK293T cells, which were propagated in DMEM with sodium pyruvate and L-glutamine (Corning). All media were supplemented with 10% FBS, 100 units/mL of penicillin, and 100 μg/mL streptomycin and grown at 37°C in a 5% CO2, humidified incubator. All cell lines were confirmed to be Mycoplasma-free using the MycoAlert Mycoplasma detection kit (Lonza, LT07-218).
CRISPR/Cas9 Reagents and Plasmids
The control and LKB1 knockout (KO) lines were generated by infecting the cell lines with lentivirus generated from the LentiCRISPRv2 plasmid (Addgene, 52961). The control and TPI1 KO or SIK KO lines were generated by infecting the Cas9-expressing lines (LentiCRISPRv2) with lentivirus generated from the LRT2B plasmid (Addgene, 110854). The sgRNA sequences were as follows: sgNT1, CCAATACGGACCGGATTGCT; sgLKB1-2, TGTATAACACATCCACCAGC; sgLKB1-3, TGCACAAGGACATCAAGCCG; sgSAFE, GGTTGGATAAGGCTTAGAAA; sgTPI1-3, GAAGTACACGAGAAGCTCCG; sgTPI1-4, GGAAGCCATCCACATCAGGC; sgSIK1, ATGGTCGTGACAGTACTCCA; sgSIK2, GCACCGGATCACCAAGACGG; sgSIK3, GTGCTTGCAGATCTGCTCCA; and sgTpi1, TGAAGGTCAGTACAAACGCA. The TPI1 alleles were synthesized by Twist Biosciences and cloned into pHAGE-CMV-N-Flag-HA-IRES-Puro-DEST or pLenti-PGK-Hygro via Gateway cloning or Gibson Assembly using NEB HiFi DNA Assembly Master Mix.
Lentivirus Production, Transduction, and Single-Cell Cloning
Lentivirus was generated by transfecting the target plasmid with the packaging plasmids pMD2.G (Addgene, 12259) and psPAX2 (Addgene, 12260) into 293T cells using Lipofectamine 3000 (Invitrogen, L3000015). Media were changed 6 hours after transfection, and then the viral supernatant was collected at 24 and 48 hours after transfection. Transduction was conducted in a 6-well format on 1 × 105 cells, and cells were plated in suspension into viral supernatant containing 8 μg/mL polybrene (Santa Cruz Biotechnology, SC-134220) and incubated overnight (16 hours). Viral supernatant was aspirated, and fresh culturing media were added to transduced cells for recovery for 24 hours. Puromycin (Life Technologies, A1113803) was supplemented into media 48 hours after transduction for relevant plasmids (LentiCRISPRv2 and pHAGE-CMV-N-Flag-HA-IRES-PURO) at a concentration of 2 μg/mL, and selection was conducted for 72 hours. Blasticidin (InvivoGen, ANT-BL-1) was supplemented into media 48 hours after transduction for relevant plasmid (LRT2B) at a concentration of 10 μg/mL, and selection was conducted for 5 days. Following selection, single-cell cloning was conducted by serial dilution and plating into a 96-well plate, and cells were maintained under relevant selection criteria during the cloning process. Clones that grew out from single cells were expanded, and validation of KO was conducted by Western blot or qPCR as indicated.
LUAD Clinical Dataset Analysis
hLUAD datasets (TCGA and Memorial Sloan Kettering Cancer Center) were downloaded from cBioPortal, and KRAS, TP53, and LKB1 mutational and copy-number status was assessed. Samples were divided into KRAS-mutant and KRAS WT cohorts for further analysis. Using the R statistical software package, a Fisher exact test was performed on each cohort to determine the odds of TP53 and LKB1 mutations co-occurring.
Athymic Nude Mice and Xenografts
Animal procedures were performed with the approval of the Weill Cornell Medicine Institutional Animal Care and Use Committee (IACUC). Tumor volume was not allowed to exceed 1,000 mm3 in any experiment. Prior to implantation, cells were resuspended in PBS and mixed 1:1 with Matrigel (Corning, 356231). Cells were then injected subcutaneously into single flanks of 6-week-old male and female athymic mice (Envigo). Caliper measurements were performed weekly to monitor tumor growth. For the H358 LKB1 KO clones, 1 × 106 cells were injected per flank; for the murine lung tumor lines (634T and Lkb1-t2), 1 × 104 cells were injected per flank.
Western Blotting
Protein lysates were prepared in CST lysis buffer (Cell Signaling Technology, 9803) supplemented with cOmplete mini EDTA-free protease inhibitor (Roche, 04693159001) and quantified using the BCA protein assay (Thermo Scientific, 23225). Lysates were prepared at a concentration of 1 mg/mL and supplemented with 4x Laemmli Sample Buffer (Bio-Rad, 1610747) supplemented with fresh 2-mercaptoethanol (Sigma, M3148). Proteins were separated on self-cast Tris-glycine polyacrylamide gels, transferred to Polyscreen PVDF membranes (PerkinElmer, NEF1002), and probed with Cell Signaling Technology antibodies used at 1:1,000 in 5% BSA (Sigma, A4503) in TBS-T: P-ACC Ser79 (#3661), ACC (#3662), P-Raptor Ser792 (#2083), Raptor (#2280), P-AMPKα Thr172 (#2535), AMPKα1/2 (#2532), LKB1 (#3047), P-ULK1 Ser555 (#5869), ULK1 (#8054), SIK2 (#6919), P-FRA1 (FOSL1) Ser265 (#5841), and phospho-p44/42 MAPK (Erk1/2) Thr202/Tyr204 (#4370). Antibodies from Abcam were used at concentrations indicated in 5% BSA in TBS-T against β-actin (ab6276, 1:20,000), TPI1 (ab96696, 1:3,000), and P-SIK1 Thr182 + P-SIK2 Thr175 + P-SIK3 Thr163 (ab199474, 1:1,000). Antibody from Novus Biologicals was used at 1:20,000 in 5% BSA in TBS-T against SIK3 (NBP2-47278). Antibodies from Sigma-Aldrich against FLAG epitope tag (F7425, 1:5,000 and F3165, 1:1,000) were used at indicated concentrations in 5% BSA in TBS-T. Secondary antibodies from Millipore against Rabbit (AP132PMI) and Mouse (AP124PMI) primary antibodies were resuspended per the manufacturer's instructions and used at 1:10,000 in 5% nonfat dried milk in TBS-T. Western blots were then developed in the dark room on an autoradiograph following incubation with homemade ECL.
Immunoprecipitation
Following indicated treatments, cells were rinsed in ice-cold PBS collected and lysed in ice-cold CST lysis buffer (Cell Signaling Technology, 9803) supplemented with cOmplete mini EDTA-free protease inhibitor (Roche, 04693159001) and rotated end-over-end for 30 minutes at 4°C. The soluble fraction of each lysate was generated by centrifugation at 13,200 RPM for 15 minutes at 4°C, and the supernatant was transferred to clean microcentrifuge tubes. Generated lysates were then precleared with protein A dynabeads (Invitrogen, 10001D) preequilibrated in CST lysis buffer for 1 hour with gentle rocking at 4°C. Precleared lysates were then centrifuged at 500 × g and placed on a magnet for 2 minutes, and the supernatant was moved to clean microcentrifuge tubes. Clarified soluble protein lysate was then quantified by the micro-BCA assay (Thermo Scientific, 23225). Following quantification, 1 mg of each lysate was then incubated with 1.2 μg of TPI1 polyclonal antibody (Proteintech, 10713-1-AP) for 16 hours at 4°C. Following 16-hour incubation, samples were supplemented with 10 μL preequilibrated protein A dynabeads and allowed to incubate at 4°C for the additional hour rotating end-over-end. Immunoprecipitated proteins bound to the resin were then washed 3 times in cold lysis buffer, and proteins were eluted from beads by pulse boiling in Laemmli sample buffer (Bio-Rad) for 5 minutes at 90°C. Following boiling, the mixture was centrifuged at 500 × g at room temperature for 5 minutes and then placed on a magnet for 2 minutes to separate beads, and the supernatants were loaded onto the gel for Western blot analysis.
Sn-Glycerol-3-Phosphate Steady-State Analysis Metabolite Extraction
Cells were cultured in a medium reconstituted from glucose-free RPMI 1640 medium (Life Technologies, 11879020) supplemented with 11.1 or 0.5 mmol/L glucose and 10% dialyzed FBS. The day prior to treatment and collection, cells were lifted and counted, and 2 × 106 cells were plated in a 10-cm culture dish. Cells were rinsed twice with PBS before the addition of normal or low-glucose medium. Metabolites were extracted 30 minutes after addition as indicated in the text.
MFA
Cells were cultured in medium reconstituted from glucose-free RPMI 1640 medium (Life Technologies, 11879020) supplemented with 0.5 mmol/L 1,2-13C2 glucose (Cambridge Isotope Laboratories, CLM-504-PK) and 10% dialyzed FBS. The day prior to treatment and collection, cells were lifted and counted, and 2 × 106 cells were plated in a 10-cm culture dish in RPMI 1640 medium supplemented with 11.1 mmol/L glucose and 10% FBS. Cells were given a medium change 1 hour before the addition of tracing medium, and glucose conditions were lowered to 0.5 mmol/L glucose at that time. Cells were rinsed twice with PBS before the addition of the tracing medium. The time of addition of tracer medium was designated time 0. Metabolites were extracted at 0.5, 1, 2, and 5 minutes after addition as indicated in the text (n = 6 per genotype and time point), and PBS washes were not implemented at the collection time point to ensure accurate collection times.
Metabolite Extraction and LC-MS/MS Analysis
Cells were washed twice with PBS and LC-MS–grade H2O (except for tracing analyses). Five hundred microliters of 80% methanol at −80°C was added to quench metabolic reactions, and the cells were collected by scraping. The lysate was then transferred to a fresh 2.0-mL Eppendorf tube prechilled on dry ice, and an additional 500 μL of 80% methanol was added to the original plate and scraped again. The second lysate was added to the first and incubated on dry ice for 20 minutes with intermittent vortexing then centrifuged at 16,000 × g for 10 minutes to allow cellular debris to be pelleted. The aqueous volume was then transferred to a clean, fresh prechilled 2.0 mL Eppendorf tube and dried under vacuum in a speedvac and stored at −80°C. Dried sample pellets were resuspended in HPLC-grade water (20 μL) and centrifuged at 20,000 × g for 5 minutes to remove insoluble material. Following centrifugation, 16 μL of supernatant was transferred to virgin polypropylene autosampler vials, capped, and placed on dry ice. Supernatants (5 μL) were injected and analyzed using a hybrid 6500 QTRAP triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) via selected reaction monitoring (MRM). ESI voltage was +4,950 V in positive ion mode and –4,900 V in negative ion mode with polarity switching. A dwell time of 3 ms per SRM transition was used. Approximately 10 to 12 datapoints were acquired per detected metabolite. Q1/Q3 SRM transitions were developed to target both the unlabeled and labeled isotopomer forms of each tested metabolite in either positive or negative mode. Samples were delivered to the mass spectrometer via hydrophilic interaction chromatography (HILIC) using a 4.6 mm i.d. × 10 cm Amide XBridge column (Waters) at 400 μL/minute using a chilled autosampler. Gradients were run starting from 85% buffer B (HPLC-grade acetonitrile) to 42% B from 0 to 5 minutes; 42% B to 0% B from 5 to 16 minutes; 0% B was held from 16 to 24 minutes; 0% B to 85% B from 24 to 25 minutes; and 85% B was held for 7 minutes to reequilibrate the column. Buffer A was comprised of 20 mmol/L ammonium hydroxide/20 mmol/L ammonium acetate (pH = 9.0) in 95:5 water:acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v3.0 software (AB/SCIEX). Tubes containing cellular debris were retained to determine protein concentration for data normalization. Briefly, pellet was resuspended by the addition of 600 μL of sodium hydroxide and boiled at 90°C for 30 minutes with intermittent vortexing. Resolubilized pellets were allowed to come to room temperature, and protein was quantified using the DC protein assay (Bio-Rad, 5000111). Derived metabolite data were normalized to protein concentration and median ion intensity per injection across the dataset.
In Situ Hydroxylamine Trapping in Live Cells
Two 15-cm dishes per condition were plated with 9 × 106 cells 24 hours prior to treatment. Plated cells were washed twice with PBS and then grown in RPMI 1640 media containing 11.1 mmol/L or 0.5 mmol/L glucose as indicated for 6 hours. Cells were then washed twice with PBS, and 3 mL of PBS containing protease inhibitors was added to the plate and cells were scraped. Cell homogenate was transferred to a 15-mL conical tube and centrifuged at 1,400 × g for 3 minutes to pellet cells. Cellular pellets were resuspended in 300 μL ice-cold 80% methanol and transferred to a 1.5-mL Eppendorf tube. Chemical labeling of live cells was achieved by adding 10 μL of hydroxylamine solution (Sigma, 467804, ∼15 mol/L solution) and incubated for 10 minutes with gentle vortexing intermittently. Following a 10-minute incubation, the suspended cells were lysed with a probe sonicator set to 30% amperage pulse (1:1 pulse:pause 16 seconds total). Lysed cellular homogenates were then centrifuged at 20,000 × g for 10 minutes at 4°C. Clarified supernatant was transferred to a fresh 1.5-mL Eppendorf tube and dried under nitrogen gas flow until all solvent was evaporated. Dried pellets were then stored at −80°C until ready for analysis. Dried metabolites were resuspended in 100 μL of an 80:20 mixture of MeOH/H2O, and an internal deuterated standard, 10 nmol d3-serine, was added to the dried metabolome solution for quantification and sample normalization.
Targeted LC-MS/MS for Hydroxylamine Trapping
Resuspended metabolites were separated by HILIC with a Gemini reverse-phase C18 column (50 mm × 4.6 mm with 5 μm diameter particles) from Phenomenex together with precolumn (C18, 3.5 mm, 2 mm × 20 mm). Mobile phase A was composed of 100% H2O (10 mmol/L tributylamine aqueous solution, adjusted to pH 4.95 with 15 mmol/L acetic acid), and mobile phase B was composed of 100% methanol. We used a multistep gradient with buffer A and B as follows: from 0 to 5 minutes, 95% buffer A; from 5 to 15 minutes, linear change from 95% to 90% buffer A; from 15 to 22 minutes, linear change from 90% to 85% buffer A; from 22 to 26 minutes, 10% buffer A; and from 30 to 33 minutes, 95% buffer A, and maintained for 7 minutes. The flow rate was 0.2 mL/minute for 0 to 15 minutes and 30 to 40 minutes, and 0.3 mL/minute for 15 to 30 minutes. Targeted MS/MS analysis was performed on an Agilent triple quadrupole LC-MS/MS instrument (Agilent Technologies 6460 QQQ). The capillary voltage was set to 4.0 kV. The drying gas temperature was 350°C, the drying gas flow rate was 10 L/minute, and the nebulizer pressure was 45 psi. Relative metabolite abundance was quantified by integrated peak area for the given MRM transition. Data presented are representative of 3 independent biological experiments each containing 3 technical replicates for a given condition.
Proteomics and Phosphoproteomic Sample Preparation
Protein lysates were prepared in CST lysis buffer (Cell Signaling Technology, 9803) supplemented with cOmplete mini EDTA-free protease inhibitor (Roche, 04693159001) and quantified using the BCA protein assay (Thermo Scientific, 23225). Following quantification, 100 μg of each protein lysate was moved into a clean 1.5-mL tube. Following the distribution of protein, each tube was brought to a final volume of 300 μL by the addition of PBS followed by precipitation with trichloroacetic acid (Sigma) to a final concentration of 25%, vigorously vortexed, and incubated on ice overnight. Trichloroacetic acid precipitates were centrifuged at 21,130 × g for 30 minutes at 4°C, washed twice in 500 μL of ice-cold acetone, and centrifuged at 21,130 × g for 10 minutes after each wash. Following precipitation and washes, pellets were allowed to completely dry at room temperature. Dry pellets were resuspended in 100 μL of 100 mmol/L TEAB with 0.5% SDS and reduced with 9.5 mmol/L tris-carboxyethyl phosphine (TCEP) for 60 minutes at 55°C. Following reduction of disulfide bonds with TCEP, the denatured protein mix was centrifuged at 21,130 × g for 5 minutes and then alkylated with 4.5 mmol/L iodoacetamide (IA) for 30 minutes in the dark at room temperature. After reduction and alkylation of disulfide bonds, the denatured protein mixture was precipitated out of solution by the addition of 600 μL of ice-cold acetone and placed in the −20°C freezer overnight. The following day, precipitated proteins were centrifuged at 8,000 × g for 10 minutes to pellet precipitated protein. Following centrifugation, the supernatant was decanted off and pellets were allowed to air-dry at room temperature. Once dry, protein pellets were reconstituted in 100 μL 100 mmol/L TEAB and CaCl2 was supplemented to a final concentration of 1 mmol/L, 2 μg of sequencing-grade Trypsin (Promega) was added, and reactions were placed in the dark on a thermal mixer (Eppendorf) set to 37°C and shaking at 850 RPM for 16 hours.
Thermal Proteomic Profiling
Cells were lifted using TrypLE Express (Thermo Fisher Scientific–Gibco) and neutralized following 5-minute incubation using complete media (RPMI + 10% FBS penicillin/streptomycin) and centrifuged at 1,100 RPM. for 4 minutes. The cell pellet was reconstituted in 10 mL PBS containing protease and phosphatase inhibitors (Roche) and centrifuged again at 1,100 RPM for 4 minutes. Following centrifugation, the cell pellet was resuspended in 1 mL PBS with inhibitors and distributed into thin-wall PCR tubes at 100 μL of cell suspension in each tube. Thermal denaturation was performed as previously described, the resulting cellular suspension was transferred to clean 1.5-mL microcentrifuge tubes, and PCR tubes were additionally rinsed with 30 μL of PBS with inhibitors to ensure complete transfer of cellular suspension. Cellular suspension was next snap-frozen in liquid nitrogen for 1 minute, followed by thawing and reequilibration back to room temperature. This freeze–thaw cycle was repeated 2 additional times, and the soluble fraction of each lysate was generated by centrifugation at 21,130 × g for 30 minutes at 4°C. Supernatants were transferred to clean 1.5-mL microcentrifuge tubes, and protein was quantified in the supernatant for temperatures 37°C and 41°C by micro-BCA assay (Thermo Fisher Scientific–Pierce). Following quantification, the average of the two lowest temperatures was taken and the volume equivalent to 30 μg of protein in the lowest temperature was moved from each temperature fraction into a clean 1.5-mL tube. Following the distribution of protein, each tube was brought to a final volume of 300 μL by the addition of PBS with inhibitors, followed by precipitation with trichloroacetic acid (Sigma) to a final concentration of 25%, vigorously vortexed, and incubated on ice overnight. Trichloroacetic acid precipitates were centrifuged at 21,130 × g for 30 minutes at 4°C, washed twice in 500 μL of ice-cold acetone, and centrifuged at 21,130 × g for 10 minutes after each wash. Following precipitation and washes, pellets were allowed to completely dry at room temperature. Dry pellets were resuspended in 100 μL of 100 mmol/L TEAB with 0.5% SDS and reduced with 9.5 mmol/L TCEP for 60 minutes at 55°C. Following the reduction of disulfide bonds with TCEP, the denatured protein mix was centrifuged at 21,130 × g for 5 minutes and then alkylated with 4.5 mmol/L IA for 30 minutes in the dark at room temperature. After reduction and alkylation of disulfide bonds, the denatured protein mixture was precipitated out of solution by the addition of 600 μL of ice-cold acetone and placed in the −20°C freezer overnight. The following day, precipitated proteins were centrifuged at 8,000 × g for 10 minutes to pellet precipitated protein. Following centrifugation, the supernatant was decanted off and pellets were allowed to air-dry at room temperature. Once dry, protein pellets were reconstituted in 100 μL 100 mmol/L TEAB and CaCl2 was supplemented to a final concentration of 1 mmol/L, 1 μg of sequencing-grade Trypsin (Promega) was added, and reactions were placed in the dark on a thermal mixer (Eppendorf) set to 37°C and shaking at 850 RPM for 16 hours. The next day, digested samples were centrifuged at 21,130 × g for 10 minutes and proceeded to TMT labeling of digested samples.
TMT Labeling, Fractionation, and Phosphopeptide Enrichment
TMT labeling was performed generally as per the manufacturer's protocol. Briefly, each TMT tag was resuspended in 164 μL anhydrous acetonitrile with intermittent vortexing for 10 minutes. Following resuspension, 41 μL was added to corresponding temperatures (TMT-126 = 37°C; four separate aliquots of each temperature for subsequent desalting and fractionation) and labeling reaction was allowed to proceed for 1 hour at room temperature. Reactions were quenched by the addition of 8 μL of 5% hydroxylamine in 100 mmol/L TEAB and incubated for 15 minutes. Labeled temperature fractions were pooled and then desalted on 1cc/50 mg C18 SepPAK columns (Waters, WAT054955) on a vacuum manifold, and desalted peptides were dried down in a SpeedVac. Dried peptides were reconstituted in 300 μL of 0.1% TFA in H2O, high-pH reverse-phase spin-columns (Thermo Fisher Scientific–Pierce) were equilibrated, and samples were fractionated per the manufacturer's instructions into 8 fractions, 2 washes, and a flow-through fraction (11 total). Separate samples from the same fractions were then combined and dried. Peptide fractions were reconstituted in 200 μL of 5% acetonitrile with 0.1% TFA in water, and 10 μL was removed for bulk HTP analysis. The remaining fractionated labeled peptides were dried and redissolved in 40% acetonitrile, 6% TFA in water before phosphopeptide enrichment with Titansphere 5-μm TiO2 beads (GL Sciences). Titansphere TiO2 beads (GL Sciences) were reconstituted in a buffer containing 80% acetonitrile, 6% TFA, and 2,5-dihydroxybenzoic acid (20 mg/mL) and rotated for 15 minutes at 25°C. An equal amount of bead slurry (∼5:1 bead-to-peptide ratio based on the concentration of peptides in 37°C aliquot) was added to each temperature aliquot of reconstituted peptides and rotated for 20 minutes 25°C. Beads were then washed twice with a higher percentage of acetonitrile (10% and 40%) in 6% TFA, and the supernatant was removed by centrifugation at 500 × g for 2 minutes. Washed beads were then added to a self-packed stage tip with C8 SPE (Sigma-Aldrich) and washed once more with 60% acetonitrile in 6% TFA. Phosphopeptides were first eluted with 5% NH4OH, then 10% NH4OH, and 25% acetonitrile, and dried with a SpeedVac. Dried phosphopeptides were reconstituted in 5% acetonitrile, 1% TFA, desalted with a self-packed stage tip with C18 SPE (Sigma-Aldrich), and dried with a SpeedVac once more. The final processed phosphopeptides were reconstituted in 5% acetonitrile, 0.1% TFA in water for LC-MS3 analysis.
LC-MS3 Analysis and Data Acquisition
High-pH reverse-phase fractions were run on a 4-hour instrument method with an effective linear gradient of 180 minutes from 5% to 25% mobile phase B with the following mobile phases: A: 0.1% formic acid in H2O, and B: 80% acetonitrile/0.1% formic acid in water on a 50-cm Acclaim PepMap RSLC C18 column (Thermo Fisher Scientific, #164942) operated by a Dionex ultimate 3000 RSLC nano pump with column heating at 50°C connected to an Orbitrap Fusion Lumos. Briefly, the instrument method was a data-dependent analysis, and the cycle time was set to 3 seconds total. Each cycle consisted of one full-scan mass spectrum (400–1,500 m/z) at a resolution of 120,000, RF Lens: 60%, maximum injection time of 100 ms followed by data-dependent MS/MS spectra with precursor selection determined by the following parameters: AGC target of 4.0e5, maximum injection time of 100 ms, monoisotopic peak determination: peptide, charge state inclusion: 2–7, dynamic exclusion 10 seconds with an intensity threshold filter: 5.0e3. Data-dependent MS/MS spectra were generated by isolating in the quadrupole with an isolation window of 0.4 m/z with CID activation and corresponding collision energy of 35%, CID activation time of 10 ms, activation Q of 0.25, detector type Ion Trap in Turbo mode, AGC target of 1.0e4, and maximum injection time of 120 ms. Data-dependent multinotch MS3 was done in synchronous precursor selection mode (SPS, multinotch MS3) with the following settings: precursor selection range; mass range 400 to 1,200, precursor ion exclusion properties m/z low: 18 high: 5, isobaric tag loss exclusion properties: TMT. The number of SPS precursors was set to 10, and data-dependent MS3 was detected in the Orbitrap (60,000 resolution; scan range, 120–500) with an isolation window of 2 m/z HCD activation type with a collision energy of 55%, AGC target of 1.2e5, and a maximum injection time of 150 ms. Raw files were parsed into MS1, MS2, and MS3 spectra using RawConverter.
Proteomic, Phosphoproteomic, and Thermal Profiling Data Analysis
Data generated were searched using the ProLuCID algorithm in the Integrated Proteomics Pipeline (IP2) software platform. Human and mouse proteome data were searched using concatenated target/decoy UniProt databases. Basic searches were performed with the following search parameters: HCD fragmentation method; monoisotopic precursor ions; high resolution mode (3 isotopic peaks); precursor mass range, 600 to 6,000 and initial fragment tolerance at 600 p.p.m.; enzyme cleavage specificity at C-terminal lysine and arginine residues with 3 missed cleavage sites permitted; static modification of +57.02146 on cysteine (carboxyamidomethylation), +229.1629 on N-terminal and lysine for TMT-10-plex tag; 4 total differential modification sites per peptide, including oxidized methionine (+15.9949), and phosphorylation (+79.9663) on serine, threonine, and tyrosine (only for phosphoenriched samples); primary scoring type by XCorr and secondary by Z-score; and minimum peptide length of 6 residues with a candidate peptide threshold of 500. A minimum of one peptide per protein and half-tryptic peptide specificity were required. Starting statistics were performed with a Δmass cutoff = 10 p.p.m. with modstat, and trypstat settings. FDRs of protein (pfp) were set to 1% (for unenriched datasets) or peptide (sfp) set to 1% (for phosphoproteomics datasets). TMT quantification was performed using the isobaric labeling 10-plex labeling algorithm, with a mass tolerance of 5.0 p.p.m. or less. Reporter ions 126.127726, 127.124761, 127.131081, 128.128116, 128.134436, 129.131417, 129.13779, 130.134825, 130.141145, and 131.13838 were used for relative quantification.
Seahorse Analysis
Oxygen consumption rate (OCR) and ECAR measurements were performed using a Seahorse XFe96 analyzer (Seahorse Biosciences). Seahorse XFe96 plates were coated with 0.56 μg Cell-Tak (Corning) in 24 mL coating buffer (8 mL H2O + 16 mL). In vitro–cultured cells were collected and plated in 96-well plates at 40,000 cells per well 16 hours prior to analysis in Seahorse RPMI media pH 7.4 (Agilent, 103576-100) supplemented with 11.1 mmol/L glucose (Agilent) and 10% FBS. To measure the OCR and ECAR of normal and low glucose–treated cells, Seahorse RPMI media pH 7.4 was supplemented with 11.1 mmol/L or 0.5 mmol/L glucose (Agilent), and previously plated cells were washed 2× with PBS and then media were added as indicated with 6 representative wells per cell line and glucose concentration. Respiratory rates were measured in RPMI Seahorse media pH 7.4 (Agilent) supplemented with 11.1 or 0.5 mmol/L glucose (Agilent) and 2 mmol/L glutamine (Agilent) in response to sequential injections of oligomycin (1 mmol/L), FCCP (0.5 mmol/L), and antimycin/rotenone (1 mmol/L; all Agilent, 103015-100). After analysis, media were aspirated from analyzed cells, and 50 μL of RIPA lysis buffer was directly added and micro-BCA (Pierce) conducted to normalize results to protein concentration.
KRAS G12DLSL/+;TP53fl/fl;R26LSL/LSLCas9-IRES-GFP (KPCas9) Mouse Model
All transgenic animals were bred and maintained in the animal facility of Weill Cornell Medicine with the approval of the university IACUC panel (protocol #:2013-0116). KRAS G12DLSL/+ mice were obtained from The Jackson Laboratory (JAX; B6.129S4-Krastm4Tyj/J; Strain #:008179) and maintained on a C57BL/6J background (JAX; C57BL/6J; Strain #:000664). TP53fl/fl mice were obtained from The Jackson Laboratory (JAX; B6.129P2-Trp53tm1Brn/J; Strain #:008462). R26LSL/LSLCas9-IRES-GFP mice were obtained from The Jackson Laboratory [JAX; B6J.129(B6N)-Gt(ROSA)26Sortm1(CAG-cas9*,-EGFP)Fezh/J; Strain #:026175]. TP53fl/fl mice were crossed to R26LSL/LSLCas9-IRES-GFP mice, and then heterozygous progeny were backcrossed. Homozygous mice were then backcrossed for three generations to generate TP53fl/fl; R26LSL/LSLCas9-IRES-GFP mice. Concurrently, KRAS G12DLSL/+ mice were crossed to R26LSL/LSLCas9-IRES-GFP mice. Confirmed progeny were then crossed to R26LSL/LSLCas9-IRES-GFP mice. Confirmed progeny were then backcrossed for three generations to R26LSL/LSLCas9-IRES-GFP mice. Next, F3 generation TP53fl/fl;R26LSL/LSLCas9-IRES-GFP mice were crossed and backcrossed to F3 KRAS G12DLSL/+;R26LSL/LSL Cas9-IRES-GFP mice to generate KRAS G12DLSL/+;TP53fl/fl;R26LSL/LSL Cas9-IRES-GFP progeny. Confirmed mice were maintained by continual breeding to previously generated F3 TP53fl/fl;R26LSL/LSLCas9-IRES-GFP mice for four generations prior to use.
KRAS G12DLSL/+;LKB1fl/fl;R26LSL/LSLCas9-IRES-GFP (KLCas9v1) Mouse Model
All transgenic animals were bred and maintained in the animal facility of Weill Cornell Medicine with the approval of the university IACUC panel (protocol #:2013-0116). KRAS G12DLSL/+ mice were obtained from The Jackson Laboratory (JAX; B6.129S4-Krastm4Tyj/J; Strain #:008179) and maintained on a C57BL/6J background (JAX; C57BL/6J; Strain #:000664). LKB1fl/fl mice were obtained from The Jackson Laboratory (JAX; STOCK Stk11tm1.1Sjm/J; Strain #:014143). R26LSL/LSLCas9-IRES-GFP mice were obtained from The Jackson Laboratory [JAX; B6J.129(B6N)-Gt(ROSA)26Sortm1(CAG-cas9*,-EGFP)Fezh/J; Strain #:026175]. LKB1fl/fl mice were crossed to R26LSL/LSLCas9-IRES-GFP mice, and then heterozygous progeny were backcrossed. Homozygous mice were then backcrossed for three generations to generate LKB1fl/fl;R26LSL/LSLCas9-IRES-GFP mice. Concurrently KRAS G12DLSL/+ mice were crossed to R26LSL/LSLCas9-IRES-GFP mice. Confirmed progeny were then crossed to R26LSL/LSLCas9-IRES-GFP mice. Confirmed progeny were then backcrossed for three generations to R26LSL/LSLCas9-IRES-GFP mice. Next, F3 generation LKB1fl/fl;R26LSL/LSLCas9-IRES-GFP mice were crossed and backcrossed to F3 KRAS G12DLSL/+;R26LSL/LSLCas9-IRES-GFP mice to generate KRAS G12DLSL/+;LKB1fl/fl;R26LSL/LSLCas9-IRES-GFP progeny. Confirmed mice were maintained by continual breeding to previously generated F3 LKB1fl/fl;R26LSL/LSLCas9-IRES-GFP mice for four generations prior to use.
KRAS G12DLSL/+;LKB1fl/fl;H11LSL/LSLCas9;R26LSL/LSLtdTomato (KLCas9v2) Mouse Model
All transgenic animals were bred and maintained in the animal facility of Weill Cornell Medicine with the approval of the university IACUC panel (protocol #:2013-0116). KRAS G12DLSL/+ mice were obtained from The Jackson Laboratory (JAX; B6.129S4-Krastm4Tyj/J; Strain #:008179) and maintained on a C57BL/6J background (JAX; C57BL/6J; Strain #:000664). LKB1fl/fl mice were obtained from The Jackson Laboratory (JAX; STOCK Stk11tm1.1Sjm/J; Strain #:014143). H11LSL/LSLCas9 mice were obtained from The Jackson Laboratory (JAX; B6.129-Igs2tm1(CAG-cas9*)Mmw/J; Strain #:027632). R26LSL/LSLtdTomato mice were obtained from The Jackson Laboratory (JAX; B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J; Strain #:007914). R26LSL/LSLtdTomato mice were crossed to H11LSL/LSLCas9 mice, and then heterozygous progeny were backcrossed. Homozygous mice were then backcrossed for three generations to generate H11LSL/LSLCas9;R26LSL/LSLtdTomato mice. Next, LKB1fl/fl mice were crossed and backcrossed to derive H11LSL/LSLCas9;R26LSL/LSLtdTomato mice. Concurrently KRAS G12DLSL/+ mice were crossed to H11LSL/LSLCas9;R26LSL/LSLtdTomato mice. Confirmed progeny were then backcrossed for three generations to H11LSL/LSLCas9;R26LSL/LSLtdTomato mice. Next, F3 generation LKB1fl/fl;H11LSL/LSLCas9;R26LSL/LSLtdTomato mice were crossed and backcrossed to F3 KRAS G12DLSL/+; H11LSL/LSLCas9; R26LSL/LSLtdTomato mice to generate KRAS G12DLSL/+; LKB1fl/fl; H11LSL/LSLCas9;R26LSL/LSLtdTomato progeny. Confirmed mice were maintained by continual breeding to previously generated F3 LKB1fl/fl;H11LSL/LSLCas9;R26LSL/LSLtdTomato mice for 4 generations prior to use.
Intratracheal Infection
Mice were anesthetized using inhaled 2.5% isoflurane and infected with 107 pfu Ad5.Spc-Cre (Iowa Viral Vector Core Facility) via an intranasal or intratracheal route. Infections were performed in a total volume of 50 μL, where adenoviral concentrates were gently mixed with serum-free EMEM and CaCl2 and allowed to salt out at room temperature for 20 minutes before placing on ice and using within the hour (57). To knock out specific genes in the mouse airway on top of a tumorigenic genetic background (“KPCas9”), guide RNAs were cloned into the Lenti-sgNT/Cre plasmid (gift of M. Winslow; Addgene, 66895) using PCR mutagenesis. Crude lentiviral supernatants were generated in HEK293T cells, packaged with psPAX2 (gift of D. Trono; Addgene, 12260) and pseudotyped with pMD2.G (gift of D. Trono; Addgene, 12259), as above. After transfection, media were changed at 6 and 72 hours, and supernatants were collected and concentrated using ultracentrifugation above a 30% sucrose cushion for 2 hours at 25,000 RPM (Beckman, SW32 Ti). Supernatants were then discarded, and pelleted lentiviruses were resuspended overnight at 4°C in 200 μL of PBS. Lentiviral titers were determined on early passage tail tip fibroblasts generated from R26R-tdTomato mice (JAX 007908). A titer of ∼106 virions per mouse was used for indicated experiments and prepared similar to adenoviral concentrates, as detailed above.
Tail Tip Fibroblast Generation
Briefly, mice were euthanized with inhaled carbon dioxide, and the entire tail was excised and sliced lengthwise. The outer epidermis was sheathed, and then the remaining tail was sectioned into 1-cm segments and washed 3× with PBS containing Primocin (InvivoGen). Tail sections were placed into culture on plates coated with 0.1% gelatin (Stem Cell Technologies) in DMEM/F12 (Corning) with 10% FBS and 100 μg/mL penicillin/streptomycin (Corning). Three days following fibroblast migration out of tail sections, tail tips were removed, and culture media were replaced every 3 to 4 days. Primary tail tip fibroblasts were expanded for 2 to 3 passages before use in lentiviral titer determination.
Lung Digestion and Generation of Lung Tumor Cell Lines
When mice showed labored breathing from advanced disease, animals were euthanized as before, lungs perfused with PBS and inflated through the trachea using 2 to 3 mL of a mixture containing collagenase type IV (Worthington, LS004188), elastase (Worthington, LS002292), Liberase (Sigma, 05401119001), and DNAse I (Worthington, LS002139) in serum-free EMEM supplemented with 0.5% BSA. Enzyme stock solutions were prepared at 100× working concentrations in water or PBS and stored at −80°C prior to use. A hemostat was used to clamp the trachea, ensuring the digestion cocktail was retained in the lung, and then lobes were grossly dissected away from the heart, thymus, and trachea and transferred into a Miltenyi C tube. Lungs were digested for 45 minutes on a preset program using a gentleMACS Octo Dissociator. Crude single-cell suspensions were then poured over a 70-μm mesh filter (Corning) and washed 3× using PBS containing 0.5% BSA and 1 mmol/L EDTA. Red blood cells were lysed for 3 minutes using 5 mL of hypotonic ACK buffer (Gibco), and then cells were washed 2 more times before resuspension in wash buffer containing DAPI and DNAse I. Viable EGFP+ or tdTomato+ single cells were sorted on a BD Influx, BD Aria II, or Sony MA900 at the Weill Cornell Flow Cytometry Core Facility. Approximately 105 sorted cells were plated per well of a 6-well dish in MTEC+ media (58), and media were changed every 3 to 4 days until cultures were expanding and required splitting. Once cell lines had adapted to 2D growth in MTEC+ media, they were easily cultured in RPMI 1640 + 10% FBS for further studies. Tumor line genotypes were confirmed by PCR and/or Western blot.
Tail Vein in Syngeneic Hosts
C57Bl/6J mice were purchased from The Jackson Laboratory and bred for no more than four generations. Both male and female mice, 8 to 12 weeks of age, were used for these studies. Briefly, mice were anesthetized using inhaled 2.5% isoflurane and vasodilated for 3 minutes with a heat lamp. Tumor cells (5 × 105) were injected into the right lateral tail vein in a total volume of 100 μL of PBS. After recovery, mice were monitored biweekly for 3 months. Survival events were recorded when animals were found dead or moribund from respiratory distress (e.g., gasping).
Allograft in Syngeneic Hosts
For allograft studies, the fur of C57Bl/6J mice was trimmed and then a depilatory cream was applied to both flanks to denude the skin, permitting accurate monitoring of tumor growth. Tumor cells (106) were subcutaneously injected in a total volume of 100 μL in a 1:1 mixture of PBS and growth factor reduced Matrigel (Corning). Both male and female mice were used, and in most cases, both hind flanks of mice were used for engraftments. Mice were monitored biweekly; tumor volumes were monitored by caliper measurements and calculated using a modified ellipsoid formula. Tumor volumes exceeding 1 cm3 were outside of protocol limits and required termination of experiments when reached.
Histology and Tissue Immunofluorescence
Mice were euthanized by carbon dioxide and dissected, and lungs were perfused with warm PBS through the left ventricle of the heart. Low melt agarose (2% in PBS) was instilled through the trachea, the trachea was clamped, and then lungs were quickly excised and plunged into freshly prepared, cold 4% paraformaldehyde. Following overnight fixation at 4°C, lungs were washed for 30 minutes in PBS at room temperature and then transferred into 70% ethanol for at least 24 hours. Lobes of lungs were grossly dissected and arranged for embedding to maximize sampled surface areas. Allograft tumor lesions were treated in a similar fashion, omitting cardiac perfusion. Embedding, cutting (4- to 5-μm sections), and routine histologic staining (hematoxylin and eosin) were contracted through Histowiz. Tissue sections were dewaxed in Histoclear and then rehydrated using decreasing alcohol washes and unmasked in a basic retrieval buffer (1× TBS + 0.05% Tween-20 + 2 mmol/L EDTA, pH 9.0) for 10 minutes in a pressure cooker. Slides were cooled in a water bath for 10 minutes and then blocked using 0.5% BSA + 1X TBS + 0.01% Triton X-100 supplemented with mouse-on-mouse blocking reagent (Vector Biolabs MKB-2213-1) for 1 hour at room temperature. Primary antibody incubations were performed overnight at 4°C in a humidified chamber (1:500 rabbit anti-RFP, Rockland, #600-401-379 or 1:500 rabbit anti-Tpi1, Proteintech, 10713-1-AP). Slides were then washed extensively using TBS + 0.5% Tween-20 and secondary antibody staining was performed at room temperature in the dark for 1 hour [1:1,000 DAPI counterstain + 1:1000 donkey anti-rabbit IgG (H+L) Alexa Fluor Plus 594; Thermo Fisher A32754]. Slides were again washed extensively, mounted (ProLong Diamond antifade; Thermo Fisher, P36961), and allowed to cure overnight before imaging. Slides were scanned on an Axio Scan 7 (ZEISS), and images were qualitatively analyzed using Zen 3.5 software. Hematoxylin and eosin comparisons were reviewed by a pathologist at Weill Cornell Medicine.
Data Availability
All derived UHPLC-MS3 proteomic data corresponding to data presented in the main figures have been deposited to the ProteomeXchange Consortium through MassIVE (https://massive.ucsd.edu) with the following identifiers:
Phosphoproteomic data in Fig. 2A: PXD039619 and MSV 000091122.
Total proteomic data in Fig. 2B: PXD039623 and MSV 000091125.
Phosphoproteomic data in Fig. 2D: PXD039639 and MSV 000091133.
Total proteomic data in Fig. 2E: PXD039638 and MSV 000091131.
Phosphoproteomic data in Fig. 2F and G: PXD039640, PXD039641 and MSV000091135, MSV000091136.
Phosphoproteomic data in Fig. 4C and D: PXD039650, PXD 039659, PXD039677, PXD039678 and MSV000091138, MSV 000091140, MSV000091151, MSV000091152
Due to size limitations, additional proteomic data corresponding to supplementary figures are available upon request (bds2005@med.cornell.edu).
Supplementary Material
Figure S1. Differential co-occurrence and effects of KRAS, TP53 and LKB1 mutations on human and mouse LUADs.
Figure S2. Phosphorylation of human TPI1 is LKB1-dependent.
Figure S3. TPI1 phosphorylation regulates triose phosphate levels and metabolic flux.
Figure S4. Members of the salt Inducible kinase family phosphorylate TPI1.
Figure S5. LKB1 regulates the multimeric state of hTPI1 but not mTpi1 and substitution of Cys in hTPI1 causes formation of a covalent TPI1 dimer.
Figure S6. TPI1 allelic variants effects central carbon metabolic flux.
Figure S7. Tpi1 is required for tumor growth in KP and KPL LUAD models.
Figure S8. Tpi1 inactivation in KL autochthonous tumor model alters histology and pathology.
Acknowledgments
We thank Kwok-Kin Wong for kindly providing the GEMM-derived LUAD cell lines, 634T and Lkb1-t2. We thank Jianxin Xie and Cell Signaling Technology for generating and kindly providing the Phospho-Ser21 TPI1 antibodies for this study. This work was supported, in part, by NIH grants P01CA120964 (to L.C. Cantley, R.J. Shaw, and J.M. Asara), R35CA197588 (to L.C. Cantley), R35-CA220538 (to R.J. Shaw), R01-DK080425 (to R.J. Shaw), R01AR076029 (to Q. Chen) and R21ES032347 (to Q. Chen); NSF-CAREER CHE-1945442 (to R.E. Moellering); and the Alfred P. Sloan Foundation (FG-2020–12839; to R.E. Moellering). H. Varmus is the Lewis Thomas University Professor at Cornell University. J.R. Ferrarone is the Lee Cooperman Physician-Scientist of the Damon Runyon Cancer Research Foundation (DRG 18-18). E.E. Gardner is the Kenneth G. and Elaine A. Langone Fellow of the Damon Runyon Cancer Research Foundation (DRG-2343-18). B. Ngo is supported by the NCI of the NIH under an F99/K00 Career Transition Fellowship (F99CA234950).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
R.E. Moellering reports grants from the NIH, the National Science Foundation, and the Sloan Foundation during the conduct of the study, as well as personal fees and nonfinancial support from ReAx Biotechnologies, and nonfinancial support from Anastasis Bio and Ama Therapeutics outside the submitted work. H. Varmus reports personal fees from Dragonfly Therapeutics, Surrozen, and Volastra outside the submitted work. L.C. Cantley reports grants from the NCI during the conduct of the study, as well as other support from Novartis and personal fees from Petra, Volastra, Larkspur, Faeth, Agios, and Loxo/Lilly outside the submitted work. No disclosures were reported by the other authors.
One of the Editors-in-Chief is an author on this article. In keeping with the AACR's editorial policy, the peer review of this submission was managed by a member of Cancer Discovery's Board of Scientific Editors, who rendered the final decision concerning acceptability.
Authors’ Contributions
B.D. Stein: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing, guided and performed experiments, performed all proteomics and biochemical experiments, metabolomics experiments, computational analyses, generated the KPCas9 and KLCas9 mouse models. J.R. Ferrarone: Resources, writing–review and editing, provided reagents and analyzed clinical data. E.E. Gardner: Investigation, visualization, methodology, writing–review and editing, performed some 3D Matrigel experiments, all in vivo tail vein and allograft studies. J.W. Chang: Investigation, methodology, writing–review and editing, performed and analyzed chemical-trapping metabolomics data. D. Wu: Investigation, writing–review and editing, assisted in xenograft studies. P.E. Hollstein: Writing–review and editing, provided cell lines and lysates utilized for A549 proteomics. R.J. Liang: Writing–review and editing, performed Seahorse experiments. M. Yuan: Writing–review and editing, performed metabolomics analyses. Q. Chen: Writing–review and editing, analyzed metabolomics experiments. J.S. Coukos: Writing–review and editing, performed and analyzed chemical-trapping metabolomics data. M. Sindelar: Writing–review and editing, analyzed metabolomics experiments. B. Ngo: Writing–review and editing, assisted in xenograft studies. S.S. Gross: Writing–review and editing, analyzed metabolomics experiments. R.J. Shaw: Writing–review and editing, provided cell lines and lysates utilized for A549 proteomics. C. Zhang: Formal analysis, validation. J.M. Asara: Methodology, writing–review and editing, performed metabolomics analyses. R.E. Moellering: Methodology, writing–review and editing, performed and analyzed chemical-trapping metabolomics data. H. Varmus: Conceptualization, supervision, writing–review and editing. L.C. Cantley: Conceptualization, supervision, funding acquisition, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang K, Creighton CJ, Davis C, Donehower L, Drummond J, Wheeler D, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, et al. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- 12. Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 13. Unni AM, Harbourne B, Oh MH, Wild S, Ferrarone JR, Lockwood WW, et al. Hyperactivation of ERK by multiple mechanisms is toxic to RTK-RAS mutation-driven lung adenocarcinoma cells. eLife 2018;7:e33718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Unni AM, Lockwood WW, Zejnullahu K, Lee-Lin SQ, Varmus H. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. eLife 2015;4:e06907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen PH, Cai L, Huffman K, Yang C, Kim J, Faubert B, et al. Metabolic diversity in human non-small cell lung cancer cells. Mol Cell 2019;76:838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem 2006;75:137–63. [DOI] [PubMed] [Google Scholar]
- 18. Shackelford DB, Shaw RJ. The LKB1–AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 2009;9:563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stein BD, Calzolari D, Hellberg K, Hu YS, He L, Hung CM, et al. Quantitative in vivo proteomics of metformin response in liver reveals AMPK-dependent and -independent signaling networks. Cell Rep 2019;29:3331–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell 2017;66:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 2018;19:121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demetrius L. Of mice and men. When it comes to studying ageing and the means to slow it down, mice are not just small humans. EMBO Rep 2005;6 Spec No:S39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eichner LJ, Brun SN, Herzig S, Young NP, Curtis SD, Shackelford DB, et al. Genetic analysis reveals AMPK is required to support tumor growth in murine kras-dependent lung cancer models. Cell Metab 2019;29:285–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hollstein PE, Eichner LJ, Brun SN, Kamireddy A, Svensson RU, Vera LI, et al. The AMPK-related kinases SIK1 and SIK3 mediate key tumor-suppressive effects of LKB1 in NSCLC. Cancer Discov 2019;9:1606–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov 2013;3:870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers ZN, McFarland CD, Winters IP, Seoane JA, Brady JJ, Yoon S, et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat Genet 2018;50:483–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murray CW, Brady JJ, Tsai MK, Li C, Winters IP, Tang R, et al. An LKB1-SIK axis suppresses lung tumor growth and controls differentiation. Cancer Discov 2019;9:1590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orozco JM, Krawczyk PA, Scaria SM, Cangelosi AL, Chan SH, Kunchok T, et al. Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat Metab 2020;2:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anastasiou D, Poulogiannis G, Asara JK, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011;334:1278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheung EC, DeNicola GM, Nixon C, Blyth K, Labuschagne CF, Tuveson DA, et al. Dynamic ROS control by TIGAR regulates the initiation and progression of pancreatic cancer. Cancer Cell 2020;37:168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011;475:106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang JW, Lee G, Coukos JS, Moellering RE. Profiling reactive metabolites via chemical trapping and targeted mass spectrometry. Anal Chem 2016;88:6658–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antoniewicz MR. A guide to (13)C metabolic flux analysis for the cancer biologist. Exp Mol Med 2018;50:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jang C, Chen L, Rabinowitz JD. Metabolomics and isotope tracing. Cell 2018;173:822–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Metallo CM, Walther JL, Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol 2009;144:167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Z, Jiang Q, Li J, Guo J. The potent roles of salt-inducible kinases (SIKs) in metabolic homeostasis and tumorigenesis. Signal Transduct Target Ther 2020;5:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakamoto K, Bultot L, Göransson O. The salt-inducible kinases: emerging metabolic regulators. Trends Endocrinol Metab 2018;29:827–40. [DOI] [PubMed] [Google Scholar]
- 38. Berggreen C, Henriksson E, Jones HA, Morrice N, Göransson O. cAMP-elevation mediated by β-adrenergic stimulation inhibits salt-inducible kinase (SIK) 3 activity in adipocytes. Cell Signal 2012;24:1863–71. [DOI] [PubMed] [Google Scholar]
- 39. Itoh Y, Sanosaka M, Fuchino H, Yahara Y, Kumagai A, Takemoto D, et al. Salt-inducible kinase 3 signaling is important for the gluconeogenic programs in mouse hepatocytes. J Biol Chem 2015;290:17879–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roland BP, Amrich CG, Kammerer CJ, Stuchul KA, Larsen SB, Rode S, et al. Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency. Biochim Biophys Acta 2015;1852:61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Savitski MM, Reinhard FB, Franken H, Werner T, Savitski MF, Eberhard D, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 2014;346:1255784. [DOI] [PubMed] [Google Scholar]
- 42. Huang JX, Lee G, Cavanaugh KE, Chang JW, Gardel ML, Moellering RE. High throughput discovery of functional protein modifications by hotspot thermal profiling. Nat Methods 2019;16:894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1α. Proc Natl Acad Sci U S A 2014;111:2554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu HG, Zhai YX, Chen J, Lu Y, Wang JW, Quan CS, et al. LKB1 reduces ROS-mediated cell damage via activation of p38. Oncogene 2015;34:3848–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li F, Han X, Li F, Wang R, Wang H, Gao Y, et al. LKB1 inactivation elicits a redox imbalance to modulate non-small cell lung cancer plasticity and therapeutic response. Cancer Cell 2015;27:698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B, et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 2017;546:168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schachner LF, Soye BD, Ro S, Kenney GE, Ives AN, Su T, et al. Revving an engine of human metabolism: activity enhancement of triosephosphate isomerase via hemi-phosphorylation. ACS Chem Biol 2022;17:2769–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007;448:807–10. [DOI] [PubMed] [Google Scholar]
- 49. Shackelford David B, Abt E, Gerken L, Vasquez Debbie S, Seki A, Leblanc M, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 2013;23:143–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med 2016;22:1108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deng J, Thennavan A, Dolgalev I, Chen T, Li J, Marzio A, et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1-mutant lung cancer. Nature Cancer 2021;2:503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 2012;482:405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ngo B, Kim E, Osorio-Vasquez V, Doll S, Bustraan S, Liang RJ, et al. Limited environmental serine and glycine confer brain metastasis sensitivity to PHGDH inhibition. Cancer Discov 2020;10:1352–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheung EC, Ludwig RL, Vousden KH. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc Natl Acad Sci U S A 2012;109:20491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J 2009;28:3015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee P, Vousden KH, Cheung EC. TIGAR, TIGAR, burning bright. Cancer Metab 2014;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 2009;4:1064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rock JR, Randell SH, Hogan BLM. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 2010;3:545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Differential co-occurrence and effects of KRAS, TP53 and LKB1 mutations on human and mouse LUADs.
Figure S2. Phosphorylation of human TPI1 is LKB1-dependent.
Figure S3. TPI1 phosphorylation regulates triose phosphate levels and metabolic flux.
Figure S4. Members of the salt Inducible kinase family phosphorylate TPI1.
Figure S5. LKB1 regulates the multimeric state of hTPI1 but not mTpi1 and substitution of Cys in hTPI1 causes formation of a covalent TPI1 dimer.
Figure S6. TPI1 allelic variants effects central carbon metabolic flux.
Figure S7. Tpi1 is required for tumor growth in KP and KPL LUAD models.
Figure S8. Tpi1 inactivation in KL autochthonous tumor model alters histology and pathology.
Data Availability Statement
All derived UHPLC-MS3 proteomic data corresponding to data presented in the main figures have been deposited to the ProteomeXchange Consortium through MassIVE (https://massive.ucsd.edu) with the following identifiers:
Phosphoproteomic data in Fig. 2A: PXD039619 and MSV 000091122.
Total proteomic data in Fig. 2B: PXD039623 and MSV 000091125.
Phosphoproteomic data in Fig. 2D: PXD039639 and MSV 000091133.
Total proteomic data in Fig. 2E: PXD039638 and MSV 000091131.
Phosphoproteomic data in Fig. 2F and G: PXD039640, PXD039641 and MSV000091135, MSV000091136.
Phosphoproteomic data in Fig. 4C and D: PXD039650, PXD 039659, PXD039677, PXD039678 and MSV000091138, MSV 000091140, MSV000091151, MSV000091152
Due to size limitations, additional proteomic data corresponding to supplementary figures are available upon request (bds2005@med.cornell.edu).