Abstract
Children’s cognitive functioning and educational performance are socially stratified. Social inequality, including classism and racism, may operate partly via epigenetic mechanisms that modulate neurocognitive development. Following preregistered analyses of data from 1,183 participants, ages 8 to 19 years, from the Texas Twin Project, we found that children growing up in more socioeconomically disadvantaged families and neighborhoods and children from marginalized racial/ethnic groups exhibit DNA methylation profiles that, in previous studies of adults, were indicative of higher chronic inflammation, lower cognitive functioning, and a faster pace of biological aging. Furthermore, children’s salivary DNA methylation profiles were associated with their performance on in-laboratory tests of cognitive and academic skills, including processing speed, general executive function, perceptual reasoning, verbal comprehension, reading, and math. Given that the DNA methylation measures that we examined were originally developed in adults, our results suggest that children show molecular signatures that reflect the early life social determinants of lifelong disparities in health and cognition.
Keywords: DNA methylation, epigenetics, cognition, children, socioeconomic status, racism, preregistered
Children’s cognitive function and educational performance are sensitive to environmental input, robustly predict their future social attainments and health, and consistently differ by major dimensions of social inequality, such as parental education, income, and race (Engelhardt et al., 2019; Korous et al., 2020). Socioeconomic and racial disparities in child cognitive development arise through various factors tied to classism and racism, including inequitable access to high-quality childcare, educational resources, health care, nutrition, and differences in exposure to toxicants, family stress, and neighborhood threat, among other factors (Anglin et al., 2021). For example, the social advantage of White identity, or White privilege, describes the generational legacy of social power experienced by White people through state-sanctioned social marginalization, which persistently shapes the disadvantaged context that Black and Latinx youths face in the United States. Because of the chronic nature of interpersonal and vicarious discrimination in the day-to-day lives of these youths, indicators of socioeconomic disadvantage do not fully capture the effects of racism on child development (Williams et al., 2019). Given that research in developmental psychology investigating the role of race, including racial disparities in adversity, is rare, scientific understanding of how racism manifests in children’s lives and affects their development remains limited (Roberts et al., 2020).
Epigenetic mechanisms that regulate the expression of genes are hypothesized to be involved in the biological embedding of environmental privilege and disadvantage (Danese & Lewis, 2017). With regard to cognition, a consistent finding from experimental manipulations of the social environment in nonhuman animals is that social adversity increases expression of genes linked to inflammation (Snyder-Mackler et al., 2020), which can modulate the continued development and function of the brain (Danese & Lewis, 2017). The central nervous system, and the cognitive functions that it supports, is susceptible to inflammation because the blood-brain barrier can be disrupted by peripheral inflammation via multiple pathways (Huang et al., 2021). Inflammatory insults on neurocognitive development may have lasting effects on late-life cognitive function by way of molecular processes originating in childhood and adolescence. In this way, epigenetic mechanisms actuated by classism and racism may, in part, contribute to social disparities in children’s cognitive function.
New advances of genomewide technology and “omic” approaches have now quantified molecular signatures of a host of exposures, biological processes, and phenotypes that can be used to investigate the etiology of social disparities in life-course development. For example, studies have identified patterns of DNA methylation across the epigenome in association with a peripheral proxy for systemic inflammation (Ligthart et al., 2016), multisystem biological aging processes (Horvath & Raj, 2018), and psychological phenotypes (McCartney et al., 2022). Results from such discovery studies can be used in prediction studies to construct epigenetic profiles in new samples that can then be examined in relation to a wide range of measured variables. Specifically, DNA methylation profiles may be useful for examining the early life etiology of social disparities in life-span development. In particular, if DNA methylation measures that were originally developed in adults are shown to be related to pediatric phenotypes and exposures, this result is evidence of a molecular continuity between childhood social inequalities and adulthood health disparities. However, little work has been conducted to date to examine whether DNA methylation measures known to be related to adult health disparities are sensitive to social inequality in childhood (Raffington et al., 2021) and associated with psychological and health-relevant phenotypes in children and adolescents.
DNA methylation is a dynamic process and can be tissue specific with, for example, different epigenetic signatures in brain, blood, and saliva (Bakulski et al., 2016). DNA methylation discovery studies most commonly analyze methylation from blood or other tissues, rather than methylation in salivary DNA, which comes from a mixture of buccal cells and leukocytes (https://ngdc.cncb.ac.cn/ewas/statistics). However, because DNA methylation profiling using saliva is amenable at large scale in pediatric samples, this method offers distinct opportunities for large-scale epidemiological and longitudinal studies. It is currently unclear whether DNA methylation profiles developed using blood samples in adults translate not just across development (from adults to youths) but also across tissue (from blood to saliva). Of particular interest in this study, then, is to use these molecular signatures to investigate the etiology of social disparities in life-course disparities in health and cognition. Following preregistered analyses (https://osf.io/x978n/), we examined whether salivary DNA methylation measures derived from adult discovery studies trained on inflammation, cognitive function, and the pace of biological aging are (a) stratified by major dimensions of social inequality and (b) associated with cognitive functions in children and adolescents.
Statement of Relevance.
A child’s cognitive function can be harmed if their environment is stressful—for example, if they regularly experience social inequalities due to things such as their social class or race/ethnicity. We can measure how much stress a person experiences by looking at an epigenetic profile—a score based on markers on the DNA that turn genes “on” or “off.” Using salivary DNA taken from a sample of adolescents from the Texas Twin Project, we created epigenetic profiles that were developed to predict chronic inflammation, lower cognitive function, and a faster pace of biological aging. We found that (a) the epigenetic profiles of children from disadvantaged backgrounds looked worse than other children and (b) children’s epigenetic profiles were associated with their performance on a range of cognitive tests. Epigenetic profiles are a promising tool and can help us better understand how social inequalities become embedded in the body and impact the mind.
Three salivary DNA methylation composite scores were of particular interest in the present study because their blood-derived composites have been associated with cognitive function or they have been found to be sensitive to socioeconomic inequality. First, we examined DNA methylation profiles of C-reactive protein (CRP; i.e., DNAm-CRP), which in blood samples have previously been found to be associated with cognitive functions in adults (Stevenson et al., 2020) and children (Barker et al., 2018). Second, we examined DNA methylation profiles of cognitive performance (i.e., Epigenetic-g) from a blood-based epigenome-wide association study of general cognitive functions (g) in adults, which accounted for 3.4% and 4.5% of the variance in general cognitive functioning in two external adult cohorts using methylation from blood samples (McCartney et al., 2022). Third, we examined Dunedin methylation pace of aging (i.e., DunedinPoAm), which was developed from an analysis of rate of longitudinal change in organ-system integrity occurring in middle adulthood in a cohort of individuals who were all the same chronological age (Belsky et al., 2020).
We previously reported (Raffington et al., 2021) that socioeconomic disadvantage and Latinx identity compared with White identity are associated with a faster pace of biological aging, as indicated by the DunedinPoAm, in an earlier data freeze of Texas Twin salivary DNA methylation data (n = 600). In contrast, epigenetic clocks and the mortality predictor “GrimAge” were not sensitive to socioeconomic inequality and therefore not considered in analyses reported here (profiles of inflammation and cognitive functioning were not previously examined). Participants in the current study were 1,183 (609 female) children and adolescents with at least one DNA methylation sample from the population-based Texas Twin Project, including 426 monozygotic twins and 757 dizygotic twins, ages 8 to 19 years (M = 13.6 years, SD = 3.05), from 611 unique families.
Method
Sample
The Texas Twin Project is an ongoing longitudinal study that includes the collection of saliva samples for DNA and DNA methylation extraction (Harden et al., 2013). Participants were recruited via public school rosters, support groups for parents of twins and other multiples, and word of mouth. Children with severe developmental and/or cognitive delays who would be unable to fully participate in the research study were excluded from participation. Participants in the current study were 1,183 (609 female) children and adolescents, including 426 monozygotic twins and 757 dizygotic twins (see zygosity measure), ages 8 to 19 years (M = 13.6 years, SD = 3.05), from 617 unique families, who had at least one DNA methylation sample. Within the analytic sample, 183 participants contributed two DNA methylation samples (time between repeated samples: M = 22 months, SD = 6.5, range = 3–38 months), and 15 samples were assayed in duplicate for reliability analyses. For descriptive statistics of the analytic sample, see Table 1. The University of Texas Institutional Review Board granted ethical approval.
Table 1.
Descriptive Statistics of the Analytic Sample
| Variable | Frequency |
|---|---|
| Zygosity | |
| Monozygotic | 426 (36.0%) |
| Dizygotic | 757 (64.0%) |
| Agea | M = 13.6 years (SD = 3.05) |
| Sex | |
| Female | 609 (51.5%) |
| Male | 574 (48.5%) |
| Race/ethnicity | |
| White only | 731 (61.8%) |
| Latinx only | 144 (12.2%) |
| Latinx & White | 94 (7.9%) |
| Black+ | 116 (9.8%) |
| Asian+ | 89 (7.5%) |
| Other | 9 (0.8%) |
Note: N = 1,183 twins.
This includes repeated observations from 183 individuals for whom methylation data were extracted on two occasions.
Measures
DNA methylation preprocessing
Saliva samples were collected during a laboratory visit using Oragene kits (DNA Genotek, Ottawa, ON, Canada). DNA extraction and methylation profiling were conducted by the Edinburgh Clinical Research Facility (UK). The Infinium MethylationEPIC BeadChip kit (Illumina, Inc., San Diego, CA) was used to assess methylation levels at 850,000 methylation sites. DNA methylation preprocessing was primarily conducted with the “minfi” package (Aryee et al., 2014) in the R programming environment (Version 4.0.4; R Core Team, 2021). Within-array normalization was performed to address array background correction, red/green dye bias, and probe type I/II correction, and it has been noted that at least part of the probe type bias is a combination of the first two factors (Dedeurwaerder et al., 2014). Noob preprocessing as implemented by minfi’s “preprocessNoob” (Triche et al., 2013) is a background correction and dye-bias equalization method that has similar within-array normalization effects on the data as probe type correction methods such as BMIQ (Teschendorff et al., 2013).
In line with our preregistered preprocessing plan, CpG probes with detection p greater than .01 and fewer than three beads in more than 1% of the samples and probes in cross-reactive regions were excluded (Pidsley et al., 2016). None of these failed probes overlapped with the probes used for DNA methylation scores. Forty-four samples were excluded because (a) they showed low intensity probes as indicated by the log of average methylation of less than 9 and their detection p was greater than .01 in greater than 10% of their probes, (b) their self-reported and methylation-estimated sex was mismatched, and/or (c) their self-reported and DNA-estimated sex was mismatched. Cell composition of immune and epithelial cell types (i.e., CD4+ T-cell, natural killer cells, neutrophils, eosinophils, B cells, monocytes, CD8+ T-cell, and granulocytes) was estimated using a newly developed child saliva reference panel implemented in the R package “BeadSorted.Saliva.EPIC” within “ewastools” (Middleton et al., 2022). Surrogate variable analysis was used to correct methylation values for batch effects using the “combat” function in the “SVA” package (Johnson et al., 2007).
DNA methylation profiles
DNAm-CRP
Profiles of inflammation were computed on the basis of an epigenome-wide association study of CRP (Ligthart et al., 2016). Using the summary statistics of the associations between CpG sites and adult CRP, we created one methylation score per person by summing the product of the weight and the individual beta estimate for each individual at each of the 218 CpG sites significantly associated (p < 1.15 × 10−7) with CRP.
DunedinPoAm
DunedinPoAm was developed from DNA methylation analysis of pace of aging in the Dunedin Study birth cohort. Pace of aging is a composite phenotype derived from analysis of longitudinal change in 18 biomarkers of organ-system integrity measured when Dunedin Study members were all 26, 32, and 38 years of age (Belsky et al., 2015). In contrast, so-called epigenetic clocks are trained on chronological age (Raffington & Belsky, 2022). Elastic-net regression machine learning analysis was used to fit pace of aging to Illumina 450k DNA methylation data generated from blood samples collected when participants were 38 years old. The elastic-net regression produced a 46-CpG algorithm. Increments of DunedinPoAm correspond to “years” of physiological change occurring per 12 months of chronological time. The Dunedin Study mean was 1, that is, the typical pace of aging among 38-year-olds in that birth cohort. Thus, 0.01 increment of DunedinPoAm corresponds to a percentage point increase or decrease in an individual’s pace of aging relative to the Dunedin birth cohort at midlife. DunedinPoAm was calculated on the basis of the published algorithm (Belsky et al., 2020) using code available at https://github.com/danbelsky/DunedinPoAm38.
DNA methylation profiles of cognitive functioning
Salivary DNA methylation profiles of cognitive functioning, or Epigenetic-g, were computed on the basis of weights from a blood-based epigenome-wide association study of general cognitive functions (g) in adults using BayesR+ (McCartney et al., 2022). General cognitive ability was derived from the first unrotated principal component of logical memory, verbal fluency and digit symbol tests, and vocabulary. We calculated DNA methylation profiles of cognitive functioning on the basis of the algorithm available at https://gitlab.com/danielmccartney/ewas_of_cognitive_function. Prior to computation, methylation values were scaled within each CpG site (M = 0, SD = 1). All DNA methylation scores were residualized for array, slide, batch, and cell composition and then standardized to ease interpretation.
Cognitive function
A battery of cognitive tasks was administered on the same day as the saliva samples were taken.
Processing speed
Three tasks were used to construct a latent measure of processing speed and were available to participants in Grades 3 through 8: Symbol Search (Wechsler, 2003), Pattern Comparison, and Letter Comparison (Salthouse & Babcock, 1991). Each task assessed how quickly and accurately participants identified similarities between symbols, patterns, or letters.
Executive functions
The current study included 15 tasks assessing four executive function domains that were available to participants in Grades 3 through 8: inhibition, switching, working memory, and updating. Tasks were administered orally, on the computer, or on paper. Inhibition was assessed with four tasks: Animal Stroop (Wright et al., 2003), Mickey (Lee et al., 2013), and Stop Signal. The study originally used an auditory Stop Signal task (Logan et al., 1997), which was replaced with a visual Stop Signal task (Verbruggen et al., 2008) after the 3rd year of data collection to accommodate the needs of administering executive function tasks in the MRI scanner. Switching was assessed using four tasks: Trail Making (Salthouse, 2011), Local-Global, Plus-Minus (Miyake et al., 2000), and a computerized Cognitive Flexibility task (Baym et al., 2008). Cognitive Flexibility replaced the Plus-Minus task, again to accommodate MRI task administration after the 3rd year of data collection. Working memory was assessed using three tasks: Symmetry Span (Kane et al., 2004), Digit Span Backward (Wechsler, 2003), and Listening Recall (Daneman & Carpenter, 1980). These tasks tap spatial, verbal, and auditory working memory, respectively. Updating was assessed with four tasks: Keeping Track (Miyake et al., 2000), Running Memory for Letters (Broadway & Engle, 2010), Two-Back task (Jaeggi et al., 2010), and, as a replacement for the Two-Back task after the 3rd year of data collection, a One- and Two-Back task (Jaeggi et al., 2010). More comprehensive task descriptions can be found in the work by Engelhardt et al. (2015).
Previous research in this sample (Engelhardt et al., 2015; Sabhlok et al., 2022) demonstrated that variation in executive function is best captured by a hierarchical factor model, with individual executive function tasks loading onto one of four latent factors representing each executive function domain and each of these loading onto a common executive function factor (for graphical depiction, see Sabhlok et al., 2022). We controlled for age-related differences in performance by regressing first-order latent executive function factors onto age in all models. This same hierarchical model was adopted in all the analyses presented in the current research to examine general executive function.
Verbal comprehension and perceptual reasoning
We administered the Wechsler Abbreviated Scale of Intelligence (Wechsler, 2011) to all participants to assess perceptual reasoning, also called nonverbal fluid intelligence, and verbal comprehension, also called verbal crystallized intelligence. Perceptual reasoning is the sum of the age-normed t scores on the Block Design and Matrix Reasoning subtests. Verbal comprehension is the sum of the age-normed t scores on the Vocabulary and Similarities subtests.
Math and reading
To assess more specific reading comprehension and mathematics skills, we had participants in Grades 3 through 8 complete the Passage Comprehension and Calculation subtests, respectively, of the Woodcock-Johnson III Tests of Academic Achievement (Woodcock et al., 2001). The dependent variable for the reading and math subtests is the total number of items correct.
Socioeconomic context
Family-level socioeconomic disadvantage
The family-level measure was computed from parent reports of household income, parental education, occupation, history of financial problems, food insecurity (based on the U.S. Household Food Security Survey Module, 2012), father absence, residential instability (changes in home address), and family receipt of public assistance. These were aggregated to form a composite measure of household-level cumulative socioeconomic disadvantage described by Engelhardt et al. (2019) and coded so that higher scores reflect greater disadvantage.
Neighborhood-level socioeconomic disadvantage
The neighborhood-level measure was composed from tract-level U.S. Census data according to the method described by Engelhardt et al. (2019). Briefly, participant addresses were linked to tract-level data from the U.S. Census Bureau American Community Survey averaged over 5 years (https://www.census.gov/programs-surveys/acs). A composite score of neighborhood-level socioeconomic disadvantage was computed from tract-level proportions of residents reported as unemployed, living below the federal poverty threshold, having fewer than 12 years of education, not being employed in a management position, and single mothers. These were aggregated using principal component analysis to form a neighborhood-level socioeconomic disadvantage composite measure described by Engelhardt et al. (2019) and coded so that higher scores reflect greater disadvantage.
Neighborhood opportunity
The neighborhood opportunity measure indexed the intergenerational economic mobility of children of low-income parents. It examines average annual household income in 2014–2015 of offspring (born between 1978 and 1983, who are now in their 30s and 40s) of low-income parents (defined as mean pretax income at the household level across 5 years—1994, 1995, and 1998–2000—at the 25th percentile of the national income distribution, or $27,000/year) within each census tract. Household income was obtained from federal tax return records between 1989 and 2015, the 2000 and 2010 Decennial Census (United States Census Bureau / American FactFinder, 2000, 2010; https://data2.nhgis.org/main), and 2005–2015 American Community Surveys (https://www.census.gov/programs-surveys/acs). Census tracts reflect where the child resided through the age of 23 years. These data were compiled by and obtained from the Opportunity Atlas (https://opportunityatlas.org; Chetty et al., 2018).
Developmental covariates
Body mass index (BMI)
BMI is socially patterned with high BMI being more common in children from families and neighborhoods of low socioeconomic status (Datar & Chung, 2015). BMI is also associated with differential DNA methylation patterns across the genome (Wahl et al., 2017). We therefore considered BMI in our analysis. We measured BMI from in-laboratory measurements of height and weight transformed to gender- and age-normed z scores according to the method published by the U.S. Centers for Disease Control and Prevention (https://www.cdc.gov/growthcharts/percentile_data_files.htm).
Pubertal development
The onset of puberty is sometimes reported at younger ages in children growing up in conditions of socioeconomic disadvantage (Colich et al., 2020). Puberty is also associated with a range of DNA methylation changes (Almstrup et al., 2016; Aylwin et al., 2019). We therefore considered children’s pubertal development in our analysis. Pubertal development was measured using children’s self-reports on the Pubertal Development Scale (Petersen et al., 1988). The scale assesses the extent of development across five sex-specific domains (for both sexes: height, body hair growth, skin changes; for girls: onset of menses, breast development; for boys: growth in body hair, deepening of voice). A total pubertal status score was computed as the average response (1 = not yet begun to 4 = has finished changing) across all items. Pubertal development was residualized for age, gender, and an age-by-gender interaction.
Tobacco exposure
Smoking is a socially patterned health behavior to which children from families and neighborhoods of low socioeconomic status are disproportionately exposed. It is also associated with differential DNA methylation patterns across the genome (Joehanes et al., 2016; Joubert et al., 2016). We therefore considered tobacco exposure in our analysis. We measured tobacco exposure using a DNA methylation smoking (DNAm-smoke) score created by summing the product of the weight and the individual beta estimate for each individual at each CpG site significantly associated with smoking in the discovery epigenome-wide association study (EWAS; Joehanes et al., 2016). Excluding self-reported tobacco users (n = 53) did not significantly alter results. For descriptive statistics of all study variables, see Table S1 in the Supplemental Material available online.
Statistical analyses
Following the preregistered analysis plan (https://osf.io/x978n/), we conducted the analysis in three main steps. Supplemental analyses (not preregistered) were also conducted and are clearly indicated below. A preliminary analysis examined the reliability of and interrelatedness between the methylation measures used in the current study. After this preliminary analysis, the first step in the analysis explored the association between DNA methylation profiles and measures capturing different levels/aspects of socioeconomic factors.
The second step shifted focus to race/ethnicity and examined disparities in DNA methylation across groups. A supplemental analysis (not preregistered) additionally examined whether racial/ethnic disparities observed in levels of DNA methylation were robust to adjustment for socioeconomic factors.
The third step in the analysis examined associations between DNA methylation profiles and six cognitive outcomes, including (a) processing speed, (b) general executive function, (c) perceptual reasoning, (d) verbal comprehension, (e) reading, and (f) math. Supplemental analyses of the third step (not preregistered) included sensitivity analyses relating to the sample selection process and adjustment for genetic and environmental factors using a biometric (twin) model. A final supplemental analysis was conducted to examine the potential of genetic confounding by examining the association between genetic profiles for CRP and cognitive outcomes in a subsample of European ancestry participants.
The analyses that we performed were multilevel, multivariate regression models fitted with FIML in Mplus (Version 8.2; Muthén & Muthén, 2017). To account for nesting of repeated measures within individuals, and multiple twin pairs within families, we used the Mplus sandwich correction to estimate cluster-robust standard errors. All models included a random intercept, representing the family-level intercept of the dependent variable, to correct for nonindependence of twins. All models included age, gender, and an age-by-gender interaction as covariates. All effect sizes are reported as standardized regression coefficients that are interpretable as Pearson’s r. We controlled for multiple testing using the Benjamini-Hochberg false-discovery-rate method (Benjamini & Hochberg, 1995).
Results
Salivary DNA methylation profiles in children are reliably measured and show expected patterns of association with covariates
DNA methylation profiles of inflammation (DNAm-CRP), cognitive functioning (Epigenetic-g), and DunedinPoAm measured from salivary DNA were approximately normally distributed (for descriptive statistics before correction for the cell composition of saliva samples, see Table S1). Analyses of 15 technical replicates suggested moderate-to-good reliability of DNA methylation profiles residualized for technical artifacts and cell composition (intraclass correlation coefficient for DNAm-CRP = .73, Epigenetic-g = .80, DunedinPoAm = .84). Biometric models using the twin family structure, where the similarity between twins due to both additive genetic factors (A) and environmental factors shared by twins living in the same home (C) represents a lower bound estimate of reliability, also suggested good reliability of DNA methylation profiles (A + C variation for DNAm-CRP = 60.7%, Epigenetic-g = 55.3%, DunedinPoAm = 54.2%, accounting for age and gender).
Higher profiles of inflammation (DNAm-CRP) were strongly correlated with profiles of a faster DunedinPoAm (r = .89, 95% confidence interval, or CI = [.81, .96], p < .001, accounting for age and gender) and moderately correlated with lower profiles of cognitive performance (Epigenetic-g, r = −.31, 95% CI = [−.42, −.19], p < .001). This result is unsurprising because CRP levels were one of the 18 biomarkers that the DunedinPoAm measure was trained on (Belsky et al., 2020). Lower DNA methylation profiles of cognitive performance were weakly correlated with a faster pace of aging (r = −.17, 95% CI = [−.29, −.04], p = .011).
Older children had DNA methylation profiles of higher inflammation (DNAm-CRP: r = .35, 95% CI = [.26, .44], p < .001), higher cognitive performance (Epigenetic-g: r = .64, 95% CI = [.56, .72], p < .001), and a faster pace of aging (DunedinPoAm: r = .13, 95% CI = [.02, .23], p = .018). Boys had profiles of lower inflammation (d = −0.26, 95% CI = [−.34, −.18], p < .001) and a slower pace of aging (d = −0.18, 95% CI = [−.27, −.10], p < .001) but not different profiles of cognitive performance (d = 0.06, 95% CI = [−.02, .14], p = .143). All models included age, gender, and an age-by-gender interaction as covariates.
Salivary DNA methylation profiles are socially stratified in children
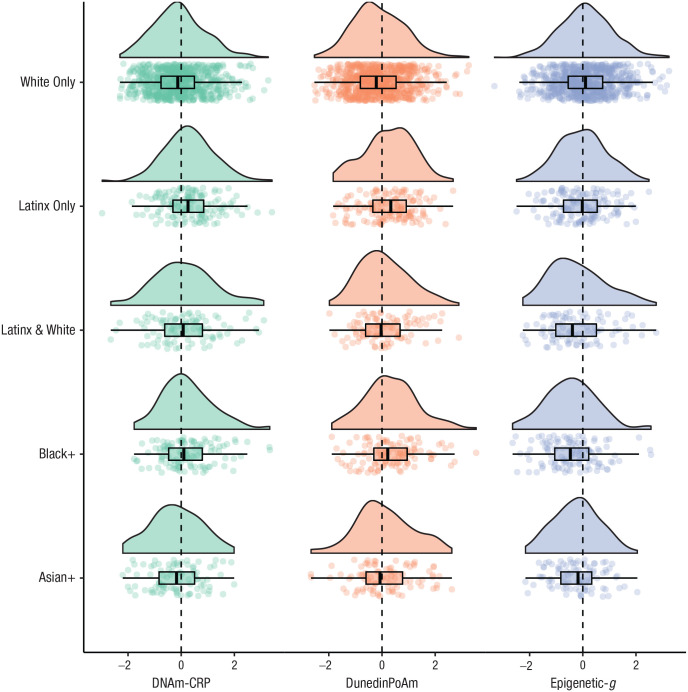
Salivary DNA methylation profiles in children were socially stratified. Children from socioeconomically disadvantaged families, socioeconomically disadvantaged neighborhoods, and neighborhoods with less intergenerational economic mobility (i.e., neighborhood opportunity) demonstrated higher levels of DNA methylation profiles associated with poor health and lower levels of DNA methylation profiles associated with cognition (see Fig. 1; see Table S2 in the Supplemental Material). Children reporting Latinx-only or Black+ identity relative to White-only identity exhibited DNA methylation profiles associated with higher chronic inflammation, a faster pace of biological aging, and lower cognitive functioning (see Fig. 2; see Table S2). Children reporting Black+ and Latinx-only identities lived in the most socioeconomically disadvantaged families and neighborhoods compared with children reporting White-only identity (see Table S3 in the Supplemental Material). Family-level socioeconomic disadvantage accounted for racial/ethnic disparities in DNA methylation profiles of inflammation, but not cognitive functioning. Family-level socioeconomic disadvantage accounted for the difference in DunedinPoAm between Black+, but not Latinx-only, and White-only identity (see Table S2). For effect size estimates between socioeconomic inequality and DNA methylation profiles reported separately for each racial/ethnic group, see Table S4 in the Supplemental Material (this analysis was not preregistered).
Fig. 1.
Associations between three measures of socioeconomic factors and three DNA methylation profiles in children and adolescents. DNA methylation profiles and socioeconomic disadvantage values are in standard deviation units. For standardized regression coefficients with and without covariate controls for body mass index and puberty, see Table S2 in the Supplemental Material available online. DNAm-CRP = DNA methylation profiles of C-reactive protein; DunedinPoAm = Dunedin methylation pace of aging; Epigenetic-g = DNA methylation profiles of cognitive performance.
Fig. 2.
Raincloud plot of associations between self-identified racial/ethnic identity and three DNA methylation profiles in children and adolescents. DNA methylation profile values are in standard deviation units. The racial/ethnic identity boxplots display group DNA methylation differences in the mean (black midline), standard errors of the mean (error bars), the first and third quartiles (lower and upper hinges), and the mean across groups (black dashed line). Participants self-identified as White only (62%), Latinx only (12.2%), Latinx and White (8.1%), Black and potentially another race/ethnicity (10%), Asian and potentially another race/ethnicity but not Latinx or Black (7.5%), and Indigenous American, Pacific Islander or other, but not Latinx, Black, or Asian (0.6%, not shown because of small sample size). For standardized regression coefficients with and without covariate controls for body mass index, puberty, and socioeconomic inequality, see Table S2 in the Supplemental Material available online. DNAm-CRP = DNA methylation profiles of C-reactive protein; DunedinPoAm = Dunedin methylation pace of aging; Epigenetic-g = DNA methylation profiles of cognitive performance.
Comparing White-only identifying children with all other groups (this comparison was not preregistered) indicated that the advantage, or privilege, of White identity compared with other racial/ethnic categories was evident in all three DNA methylation profiles (DNAm-CRP: r = −.14, 95% CI = [−.22, −.06], p < .001; Epigenetic-g: r = .23, 95% CI = [.16, .31], p < .001; DunedinPoAm: r = −.25, 95% CI = [−.34, −.16], p < .001). White identity remained evident in DNA methylation profiles of cognitive functioning (Epigenetic-g: r = .21, 95% CI = [.13, .29], p < .001) and DunedinPoAm (r = −.19, 95% CI = [−.29, −.09], p < .001) but not inflammation (DNAm-CRP: r = −.08, 95% CI = [−.17, .01], p = .067), after accounting for the lower rates of family-level disadvantage experienced by White children (r = −.29, 95% CI = [−.38, −.19], p < .001). Effects of White identity were reduced but also still remained evident in DNA methylation profiles of cognitive functioning (r = .17, 95% CI = [.09, .25], p < .001) and DunedinPoAm (r = −.16, 95% CI = [−.26, −.05], p = .003) but not inflammation (r = −.04, 95% CI = [−.13, .05], p = .373), after accounting for the lower rates of both family-level (r = −.30, 95% CI = [−.39, −.21], p < .001) and neighborhood-level (r = −.34, 95% CI = [−.42, −.26], p < .001) socioeconomic disadvantage experienced by White children.
We next examined the role of BMI, pubertal stage, and DNA methylation profiles related to smoking (DNAm-smoke) in associations of social inequality and DNA methylation profiles of interest. Socioeconomic and racial/ethnic inequalities in DNA methylation largely remained after we included these covariates, with the exception that correlations with DNA methylation profiles of inflammation were attenuated by including BMI (see Table S2).
Salivary DNA methylation profiles are associated with cognitive functions
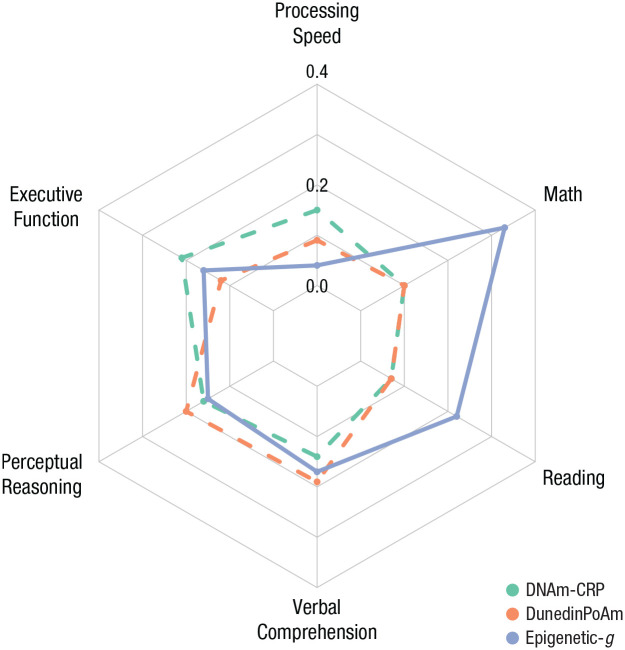
Salivary DNA methylation profiles were associated with performance on multiple in-laboratory tests of cognitive functioning (see Fig. 3). Higher DNA methylation profiles of inflammation were associated with worse performance on tests of processing speed, general executive function, perceptual reasoning, and verbal comprehension. Lower DNA methylation profiles of cognitive performance were associated with lower scores on tests of perceptual reasoning, verbal comprehension, reading, and math. Finally, a faster pace of aging was associated with lower scores on tests of verbal comprehension and perceptual reasoning (see Table 2). Notably, the largest effect size was observed for DNA methylation profiles of cognitive performance and math, where DNA methylation profiles of cognitive performance explained an R2 of 11.1% of the variation in math performance. For effect size estimates between DNA methylation profiles and cognition reported separately for each racial/ethnic group, see Table S5 in the Supplemental Material (this analysis was not preregistered).
Fig. 3.
Radar plot of effect sizes (Pearson’s r) for three DNA methylation profiles across six measures of cognition in children and adolescents. The plot depicts the standardized regression coefficients (rs) calculated by regressing cognitive functions on DNA methylation measures separately. Note that effect sizes are presented as absolute values to facilitate plotting; the polarity of original effect sizes is indicated by line type, with solid lines indicating positive values and dashed lines representing negative values. All models included covariate adjustment for child’s age, gender, and technical covariates. For standardized regression coefficients with and without covariate controls for body mass index, puberty, and socioeconomic inequality, see Table 2. DNAm-CRP = DNA methylation profiles of C-reactive protein; DunedinPoAm = Dunedin methylation pace of aging; Epigenetic-g = DNA methylation profiles of cognitive performance.
Table 2.
Associations Between DNA Methylation and Genetic Profiles With Cognitive Functions
| Processing speed | Executive functions | Perceptual reasoning | Verbal comprehension | Reading | Math | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | r | 95% CI | p | r | 95% CI | p | r | 95% CI | p | r | 95% CI | p | r | 95% CI | p | r | 95% CI | p |
| No further covariatesa | ||||||||||||||||||
| CRP | −.15 | [−.24, −.05] | .003 | −.21 | [−.36, −.07] | .004 | −.16 | [−.26, −.07] | .001 | −.14 | [−.25, −.03] | .013 | −.07 | [−.20, .06] | .310 | −.10 | [−.27, .06] | .218 |
| Epigenetic-g | .04 | [−.11, .17] | .657 | .16 | [−.05, .37] | .126 | .15 | [.07, .24] | < .001 | .17 | [.07, .27] | .001 | .22 | [.09, .42] | .001 | .33 | [.18, .49] | < .001 |
| PoAm | −.09 | [−.20, .01] | .082 | −.12 | [−.27, .02] | .090 | −.20 | [−.30, −.10] | < .001 | −.19 | [−.29, −.08] | < .001 | −.07 | [−.20, .08] | .366 | −.10 | [−.27, .06] | .218 |
| Adjusting for BMI | ||||||||||||||||||
| BMI | −.10 | [−.18, −.02] | .017 | −.24 | [−.35, −.12] | < .001 | −.18 | [−.27, −.10] | < .001 | −.19 | [−.29, −.08] | .001 | −.19 | [−.31, −.06] | .003 | −.24 | [−.36, −.12] | < .001 |
| CRP | −.09 | [−.20, .02] | .115 | −.09 | [−.26, .07] | .260 | −.10 | [−.20, .00] | .053 | −.07 | [−.20, .05] | .257 | .03 | [−.12, .17] | .737 | −.01 | [−.17, .15] | .918 |
| Epigenetic-g | −.01 | [−.21, .14] | .956 | .10 | [−.12, .32] | .351 | .14 | [.06, .22] | .001 | .15 | [.06, .25] | .001 | .20 | [.06, .34] | .005 | .30 | [.14, .45] | < .001 |
| PoAm | −.04 | [−.16, .08] | .499 | −.02 | [−.18, .14] | .825 | −.14 | [−.25, −.04] | .008 | −.12 | [−.24, −.01] | .030 | .02 | [−.13, .17] | .815 | .01 | [−.18, .19] | .975 |
| Adjusting for pubertal development | ||||||||||||||||||
| Puberty | −.12 | [−.21, −.02] | .019 | −.14 | [−.28, −.03] | .043 | .08 | [−.03, .18] | .159 | .04 | [−.09, .17] | .547 | −.11 | [−.25, .03] | .112 | −.07 | [−.20, .06] | .311 |
| CRP | −.12 | [−.22, −.02] | .014 | −.19 | [−.34, −.01] | .017 | −.17 | [−.26, −.07] | < .001 | −.14 | [−.26, −.03] | .013 | −.05 | [−.18, .09] | .523 | −.11 | [−.26, .04] | .134 |
| Epigenetic-g | .03 | [−.11, .16] | .680 | .16 | [−.08, .49] | .142 | .15 | [.07, .23] | < .001 | .17 | [.07, .26] | .001 | .22 | [.09, .36] | .001 | .33 | [.18, .49] | < .001 |
| PoAm | −.08 | [−.18, .01] | .125 | −.11 | [−.25, .04] | .137 | −.20 | [−.30, −.10] | < .001 | −.19 | [−.29, −.08] | < .001 | −.06 | [−.20, .09] | .446 | −.10 | [−.27, .07] | .250 |
| Adjusting for tobacco smoke exposure | ||||||||||||||||||
| DNAm-smoke | .08 | [−.06, .22] | .28 | .08 | [−.14, .30] | .480 | −.11 | [−.22, .01] | .081 | −.10 | [−.22, .03] | .119 | −.17 | [−.37, .04] | .114 | −.14 | [−.37, .08] | .205 |
| CRP | −.20 | [−.33, −.07] | .003 | −.25 | [−.44, −.06] | .012 | −.13 | [−.23, −.03] | .011 | −.12 | [−.23, −.01] | .045 | −.01 | [−.16, .16] | .972 | −.06 | [−.25, .13] | .541 |
| Epigenetic-g | .03 | [−.14, .20] | .737 | .22 | [−.03, .48] | .089 | .15 | [.07, .23] | < .001 | .16 | [.06, .26] | .002 | .23 | [.09, .37] | .001 | .35 | [.19, .50] | < .001 |
| PoAm | −.10 | [−.22, .01] | .070 | −.13 | [−.28, .03] | .121 | −.18 | [−.28, −.08] | < .001 | −.17 | [−.28, −.07] | .001 | −.03 | [−.18, .11] | .646 | −.07 | [−.25, .11] | .46 |
| Adjusting for family-level disadvantage | ||||||||||||||||||
| Family disadvantage | −.07 | [−.24, .04] | .170 | −.22 | [−.36, −.08] | .002 | −.20 | [−.29, −.11] | < .001 | −.29 | [−.40, −.19] | < .001 | −.17 | [−.31, −.03] | .019 | −.26 | [−.41, −.11] | .001 |
| CRP | −.12 | [−.39, −.04] | .013 | −.15 | [−.30, −.01] | .046 | −.11 | [−.21, −.02] | .020 | −.07 | [−.18, .05] | .237 | −.04 | [−.17, .10] | .590 | −.08 | [−.22, .07] | .292 |
| Epigenetic-g | .01 | [−.13, .15] | .877 | .12 | [−.10, .33] | .301 | .15 | [.07, .23] | < .001 | .16 | [.06, .25] | .001 | .21 | [.07, .35] | .003 | .31 | [.15, .47] | < .001 |
| PoAm | −.07 | [−.18, .04] | .210 | −.06 | [−.22, .09] | .426 | −.15 | [−.25, −.05] | .004 | −.11 | [−.22, −.01] | .041 | −.03 | [−.17, .11] | .673 | −.05 | [−.21, .12] | .564 |
Note: Standardized regression coefficients (rs) and 95% confidence intervals (CIs) and uncorrected p values calculated by regressing cognitive functions on DNA methylation measures with and without controlling for normed body mass index (BMI) z scores, puberty (residualized for age within each sex), DNA methylation profiles of smoking (DNAm-smoke), and family-level disadvantage separately. p values, where false-discovery-rate-corrected p < .05, are marked in bold. CRP = C-reactive protein; Epigenetic-g = DNA methylation profiles of cognitive performance; PoAm = Dunedin methylation pace of aging.
All models included covariate adjustment for age, gender, and an age-by-gender interaction. Methylation scores were residualized for technical covariates (i.e., array, slide, batch, and cell composition).
As all three DNA methylation profiles were associated with perceptual reasoning and verbal comprehension, we performed commonality analyses to examine the proportion of overlapping and unique variation explained. DNA methylation profiles of inflammation (DNAm-CRP) and DunedinPoAm explained largely overlapping variation in perceptual reasoning (DNAm-CRP alone: 2.6%, DunedinPoAm alone: 4%, combined: 3.8%) and verbal comprehension (DNAm-CRP alone: 2%, DunedinPoAm alone: 3.4%, combined: 3.8%). In contrast, DNA methylation profiles of cognitive functioning (Epigenetic-g) explained largely unique variation in perceptual reasoning (Epigenetic-g alone: 2.4%) and verbal comprehension (Epigenetic-g alone: 2.9%) relative to both inflammation (perceptual reasoning combined: 5.9%, verbal comprehension combined: 5.5%) and pace of aging (perceptual reasoning combined: 6.5%, verbal comprehension combined: 6.3%).
We next examined the role of BMI, puberty, DNAm-smoke, and family-level disadvantage in associations of DNA methylation measures with cognitive test performance. Associations were largely unaffected by controlling for BMI, puberty, and DNAm-smoke, with the exception that associations of DNA methylation profiles of inflammation with cognition were mostly accounted for by controlling for BMI. Associations of DNA methylation profiles of inflammation and DunedinPoAm with cognition were largely accounted for by controlling for family-level disadvantage, except for perceptual reasoning. In contrast, associations of DNA methylation profiles of cognitive functioning with cognitive test performance were unaffected by controlling for family-level disadvantage (see Table S2).
We assessed potential effects of differing sample sizes of cognitive measures on effect size estimates (this analysis was not preregistered). Effect size estimates based on models using listwise deletion were largely similar to reported results, suggesting that differing sample sizes across measures did not substantially affect effect sizes (see Fig. S1 in the Supplemental Material).
We further examined the extent to which DNA methylation associations with cognition are robust to complete genetic and family-level environmental control in a bivariate biometric model that used the twin family structure of the Texas Twin Project (see Table S6 in the Supplemental Material). Consistent with the hypothesis that DNA methylation associations with cognitive function represent (partially unmeasured) effects of family-level stratification, we found no evidence to suggest that identical twins who differ from their cotwins in DNA methylation show corresponding differences in their cognitive functioning. In other words, associations between interindividual variation in DNA methylation and cognitive functions entirely arose at the level of variation occurring across, rather than within, families. The DNA methylation indices examined here are therefore likely to index aspects of family-to-family differences, such as socioeconomic and school and neighborhood factors, underlying inequalities in cognitive development. Finally, we explored the possibility of confounding in our results due to genetic profiles specifically associated with inflammation. Using a polygenic score for CRP (see Supplemental Methods in the Supplemental Material) and a subsample of European ancestry dizygotic twins, we found that the polygenic score for CRP was not associated with any of the cognitive measures (see Table S7 in the Supplemental Material).
Discussion
We analyzed salivary DNA methylation data from 1,183 children and adolescents participating in the Texas Twin Project to investigate the etiology of social disparities in life-course disparities in health and cognition. We examined whether salivary DNA methylation measures of inflammation, cognitive function, and the pace of aging are (a) stratified by major dimensions of social inequality and (b) associated with performance on tests of cognitive function in childhood. We found that children and adolescents growing up in more socioeconomically disadvantaged families and neighborhoods and children from marginalized racial/ethnic groups compared with their more privileged peers exhibit DNA methylation profiles associated with higher chronic inflammation, lower cognitive functioning, and a faster pace of biological aging. Children reporting Black+ or Latinx-only identity lived, on average, in substantially more socioeconomically disadvantaged families and neighborhoods compared with children reporting White-only identity. Socioeconomic disadvantage statistically accounted for some, but not all, of the differences between racial/ethnic groups in DNA methylation profiles. For example, the social advantage of White identity, or White privilege, remained evident in DNA methylation profiles after accounting for the lower rates of both family-level and neighborhood-level disadvantage experienced by White families. Thus, our findings are consistent with observations that racial and ethnic disparities leave biological traces in the first two decades of life and reflect multiple dimensions of social inequality (Anglin et al., 2021).
Moreover, these socially stratified DNA methylation profiles were related to performance on multiple in-laboratory cognitive tests with nonnegligible effect sizes, including tests of processing speed, general executive function, perceptual reasoning, verbal comprehension, reading, and math. After we corrected for multiple comparisons, DNA methylation profiles of higher inflammation were associated with lower in-laboratory processing speed, general executive function, perceptual reasoning, and verbal comprehension. Lower DNA methylation profiles of cognitive performance (Epigenetic-g) were associated with lower perceptual reasoning, verbal comprehension, reading, and math performance. A faster pace of biological aging was correlated with lower verbal comprehension and perceptual reasoning. Notably, DNA methylation profiles of cognitive performance explained 11.1% of the variation in math performance. Associations of DNA methylation measures of inflammation and the pace of aging with cognition were largely accounted for by controlling for family-level socioeconomic disadvantage.
Given that the DNA methylation measures that we examined were originally developed in adults, our results suggest that social inequalities in childhood are reflected in molecular signatures that, when observed in adults, are associated with chronic inflammation, advanced aging, and reduced cognitive function. Our findings indicate that salivary DNA methylation measures may be useful for indexing social inequality and risk for disparities in cognitive function in childhood and adolescence. They also support the notion that adult cognitive function, morbidity, and mortality are partially driven by molecular processes that begin in childhood and adolescence, sensitive developmental periods in which cognitive functions, and the physiological processes that support them, are susceptible to environmental inputs.
DNA methylation is a dynamic process and can be tissue specific with, for example, different epigenetic signatures in brain, blood, and saliva. Whereas we measured methylation in salivary DNA, the original estimates on which our profiles were based were measured from DNA methylation in blood. We caution that there is still considerable uncertainty about what DNA methylation measures from peripheral tissues such as saliva reflect. Increasing confidence in our findings with salivary DNA methylation, epigenetic clocks were highly correlated with chronological age in a subsample of the present sample (Raffington et al., 2021). We have also observed good correspondence of pace of aging measurements across blood and saliva in adults (Raffington et al., 2021), although this type of analysis should be repeated in blood and saliva samples from children. Moreover, recent research suggests that salivary DNA methylation collected with Oragene kits (as was done here) in children is enriched for immune cells rather than epithelial cells (Middleton et al., 2022). It may therefore be particularly sensitive to inflammatory processes, which contribute to DNA methylation profiles of inflammation (DNAm-CRP) and pace of aging (DunedinPoAm). In contrast, genetic profiles related to inflammation (i.e., polygenic scores of CRP) were not associated with cognitive functioning.
Our findings linking DNA methylation profiles of inflammation with pace of aging and cognitive functioning are in line with experimental animal studies reporting that social adversity increases expression of genes linked to inflammation, which may be critically involved in multisystem aging processes (i.e., “inflammaging”) and can modulate the development of the brain (Danese & Lewis, 2017; Snyder-Mackler et al., 2020). Moreover, our results showing that socioeconomic and racial/ethnic inequalities in DNA methylation profiles of inflammation were attenuated by including BMI fits with conceptualizations of obesity as an inflammatory disease reflected in increased circulating levels of pro-inflammatory proteins in adults, adolescents, and children (de Heredia et al., 2012). Yet the measurements that we studied are molecular derivatives of unobserved inflammatory processes and not direct observations of chronic inflammation. Accordingly, this type of omics research is not well-suited to identifying precise biological processes. However, it may prove to be a powerful tool to elucidate social and developmental mechanisms.
We acknowledge several limitations. First, our cross-sectional, observational design did not allow us to examine whether policy changes mitigating socioeconomic inequality (e.g., increases in minimum wage, child tax credits) and structural racism (e.g., eliminating the legacy of redlining, police reforms) affect children’s DNA methylation profiles. Longitudinal samples including earlier childhood and experimental manipulations of social inequality, for example through income subsidies (https://www.childtrends.org/publications/lessons-from-a-historic-decline-in-child-poverty), are needed to examine potential pathways of epigenetic mediation and (bi-)directionality of effects between epigenetic mechanisms and cognition. Second, we used data from twins, and the conditions of twin pregnancy could alter DNA methylation patterns relative to singleton pregnancies. Although this may affect mean levels of DNA methylation, it is unlikely to lead to differential relationships with social inequality or cognitive performance.
Third, family and neighborhood indicators of socioeconomic disadvantage, privilege, and intergenerational mobility capture relevant but limited aspects of the effects of racism on child development (Williams et al., 2019). Additional factors that are often neglected, such as the impact of race-based discrimination in education and health care systems and chronic exposure to interpersonal and vicarious discrimination in daily life, may explain further variance in the effects of racial/ethnic marginalization (Goosby et al., 2018).
Our findings suggest that salivary DNA methylation profiles are promising candidate biomarkers of major dimensions of social inequality experienced in real-time during childhood. Because saliva can easily be collected in large-scale pediatric epidemiological studies, salivary DNA methylation profiles might be useful as surrogate endpoints in evaluation of ontogenetic theories and social programs that address the childhood social determinants of lifelong cognitive disparities.
Supplemental Material
Supplemental material, sj-docx-1-pss-10.1177_09567976221122760 for Socially Stratified Epigenetic Profiles Are Associated With Cognitive Functioning in Children and Adolescents by Laurel Raffington, Peter T. Tanksley, Aditi Sabhlok, Liza Vinnik, Travis Mallard, Lucy S. King, Bridget Goosby, K. Paige Harden and Elliot M. Tucker-Drob in Psychological Science
Acknowledgments
We gratefully acknowledge all participants of the Texas Twin Project. The University of Texas at Austin Institutional Review Board granted ethical approval. Informed consent to participate in the study was obtained from all participants and their parent or legal guardian. Because of the high potential for deductive identification in this special population from a geographically circumscribed area as well as the sensitive nature of information collected, data from the Texas Twin Project are not shared with individuals outside of the research team.
Footnotes
ORCID iD: Laurel Raffington  https://orcid.org/0000-0002-0144-5605
https://orcid.org/0000-0002-0144-5605
Supplemental Material: Additional supporting information can be found at http://journals.sagepub.com/doi/suppl/10.1177/09567976221122760
Transparency
Action Editor: Karen Rodrigue
Editor: Patricia J. Bauer
Author Contribution(s)
K. P. Harden and E. M. Tucker-Drob contributed equally to this study.
Laurel Raffington: Conceptualization; Formal analysis; Writing – original draft; Writing – review & editing.
Peter T. Tanksley: Formal analysis; Writing – review & editing.
Aditi Sabhlok: Formal analysis; Writing – review & editing.
Liza Vinnik: Data curation; Writing – review & editing.
Travis Mallard: Formal analysis; Writing – review & editing.
Lucy S. King: Writing – review & editing.
Bridget Goosby: Writing – review & editing.
K. Paige Harden: Conceptualization; Funding acquisition; Supervision; Writing – review & editing.
Elliot M. Tucker-Drob: Conceptualization; Funding acquisition; Supervision; Writing – review & editing.
The author(s) declared that there were no conflicts of interest with respect to the authorship or the publication of this article.
Funding: This research was supported by National Institutes of Health (NIH) Grants R01HD083613 and R01HD092548. L. Raffington was supported by the German Research Foundation. B. Goosby, K. P. Harden, and E. M. Tucker-Drob are faculty research associates of the Population Research Center at The University of Texas at Austin, which was supported by NIH Grant P2CHD042849. B. Goosby and E. M. Tucker-Drob are members of the Center on Aging and Population Sciences at The University of Texas at Austin, which was supported by NIH Grant P30AG066614. K. P. Harden and E. M. Tucker-Drob were also supported by Jacobs Foundation Research Fellowships. L. S. King was supported by NIH Fellowship F32HD105285-01.
Open Practices: This study was preregistered via the Open Science Framework and can be accessed at osf.io/x978n. This article has received the badge for Preregistration. More information about the Open Practices badges can be found at http://www.psychologicalscience.org/publications/badges. The study data include sensitive information about minors and are not publicly available. Code is available from the first author upon request.

References
- Almstrup K., Johansen M. L., Busch A. S., Hagen C. P., Nielsen J. E., Petersen J. H., Juul A. (2016). Pubertal development in healthy children is mirrored by DNA methylation patterns in peripheral blood. Scientific Reports, 6(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglin D. M., Ereshefsky S., Klaunig M. J., Bridgwater M. A., Niendam T. A., Ellman L. M., DeVylder J., Thayer G., Bolden K., Musket C. W., Grattan R. E., Lincoln S. H., Schiffman J., Lipner E., Bachman P., Corcoran C. M., Mota N. B., van der Ven E. (2021). From womb to neighborhood: A racial analysis of social determinants of psychosis in the United States. American Journal of Psychiatry, 178(7), 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., Irizarry R. A. (2014). Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30(10), 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylwin C. F., Toro C. A., Shirtcliff E., Lomniczi A. (2019). Emerging genetic and epigenetic mechanisms underlying pubertal maturation in adolescence. Journal of Research on Adolescence, 29(1), 54–79. [DOI] [PubMed] [Google Scholar]
- Bakulski K. M., Halladay A., Hu V. W., Mill J., Fallin M. D. (2016). Epigenetic research in neuropsychiatric disorders: The “tissue issue.” Current Behavioral Neuroscience Reports, 3(3), 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker E. D., Cecil C. A., Walton E., Houtepen L. C., O’Connor T. G., Danese A., Jaffee S. R., Jensen S. K. G., Pariante C., McArdle W., Gaunt T. R., Relton C. L., Roberts S. (2018). Inflammation-related epigenetic risk and child and adolescent mental health: A prospective study from pregnancy to middle adolescence. Development and Psychopathology, 30(3), 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym C., Corbett B., Wright S., Bunge S. (2008). Neural correlates of tic severity and cognitive control in children with Tourette syndrome. Brain, 131(1), 165–179. [DOI] [PubMed] [Google Scholar]
- Belsky D. W., Caspi A., Arseneault L., Baccarelli A., Corcoran D. L., Gao X., Hannon E., Harrington H. L., Rasmussen L. J., Houts R., Huffman K., Kraus W. E., Kwon D., Mill J., Pieper C. F., Prinz J. A., Poulton R., Schwartz J., Sugden K., . . . Moffitt T. E. (2020). Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. eLife, 9, Article 54870. 10.7554/eLife.54870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky D. W., Caspi A., Houts R., Cohen H. J., Corcoran D. L., Danese A., Harrington H., Israel S., Levine M. E., Schaefer J. D., Sugden K., Williams B., Yashin A. I., Poulton R., Moffitt T. E. (2015). Quantification of biological aging in young adults. Proceedings of the National Academy of Sciences, 112(30), E4104–E4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1), 289–300. [Google Scholar]
- Broadway J. M., Engle R. W. (2010). Validating running memory span: Measurement of working memory capacity and links with fluid intelligence. Behavior Research Methods, 42(2), 563–570. [DOI] [PubMed] [Google Scholar]
- Chetty R., Friedman J. N., Hendren N., Jones M. R., Porter S. R. (2018, October). The opportunity atlas: Mapping the childhood roots of social mobility (Working Paper No. 25147). National Bureau of Economic Research. https://www.nber.org/papers/w25147 [Google Scholar]
- Colich N. L., Rosen M. L., Williams E. S., McLaughlin K. A. (2020). Biological aging in childhood and adolescence following experiences of threat and deprivation: A systematic review and meta-analysis. Psychological Bulletin, 146(9), 721–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman M., Carpenter P. A. (1980). Individual differences in working memory and reading. Journal of Verbal Learning and Verbal Behavior, 19(4), 450–466. [Google Scholar]
- Danese A., Lewis S. J. (2017). Psychoneuroimmunology of early-life stress: The hidden wounds of childhood trauma? Neuropsychopharmacology, 42(1), 99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar A., Chung P. J. (2015). Changes in socioeconomic, racial/ethnic, and sex disparities in childhood obesity at school entry in the United States. JAMA Pediatrics, 169(7), 696–697. [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder S., Defrance M., Bizet M., Calonne E., Bontempi G., Fuks F. (2014). A comprehensive overview of Infinium HumanMethylation450 data processing. Briefings in Bioinformatics, 15(6), 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Heredia F. P., Gómez-Martínez S., Marcos A. (2012). Obesity, inflammation and the immune system. Proceedings of the Nutrition Society, 71(2), 332–338. [DOI] [PubMed] [Google Scholar]
- Engelhardt L. E., Briley D. A., Mann F. D., Harden K. P., Tucker-Drob E. M. (2015). Genes unite executive functions in childhood. Psychological Science, 26(8), 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt L. E., Church J. A., Harden K. P., Tucker-Drob E. M. (2019). Accounting for the shared environment in cognitive abilities and academic achievement with measured socioecological contexts. Developmental Science, 22(1), Article e12699. 10.1111/desc.12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosby B. J., Cheadle J. E., Mitchell C. (2018). Stress-related biosocial mechanisms of discrimination and African American health inequities. Annual Review of Sociology, 44, 319–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden K. P., Tucker-Drob E. M., Tackett J. L. (2013). The Texas Twin Project. Twin Research and Human Genetics, 16(1), 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., Raj K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature Reviews Genetics, 19(6), 371–384. [DOI] [PubMed] [Google Scholar]
- Huang X., Hussain B., Chang J. (2021). Peripheral inflammation and blood–brain barrier disruption: Effects and mechanisms. CNS Neuroscience & Therapeutics, 27(1), 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeggi S. M., Studer-Luethi B., Buschkuehl M., Su Y.-F., Jonides J., Perrig W. J. (2010). The relationship between n-back performance and matrix reasoning—implications for training and transfer. Intelligence, 38(6), 625–635. [Google Scholar]
- Joehanes R., Just A. C., Marioni R. E., Pilling L. C., Reynolds L. M., Mandaviya P. R., Guan W., Xu T., Elks C. E., Aslibekyan S., Moreno-Macias H., Smith J. A., Brody J. A., Dhingra R., Yousefi P., Pankow J. S., Kunze S., Shah S. H., McRae A. F., . . . London S. J. (2016). Epigenetic signatures of cigarette smoking. Circulation: Cardiovascular Genetics, 9(5), 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E., Li C., Rabinovic A. (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8(1), 118–127. [DOI] [PubMed] [Google Scholar]
- Joubert B. R., Felix J. F., Yousefi P., Bakulski K. M., Just A. C., Breton C., Reese S. E., Markunas C. A., Richmond R. C., Xu C.-J., Küpers L. K., Oh S. S., Hoyo C., Gruzieva O., Söderhäll C., Salas L. A., Baïz N., Zhang H., Lepeule J., . . . London S. J. (2016). DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. The American Journal of Human Genetics, 98(4), 680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M. J., Hambrick D. Z., Tuholski S. W., Wilhelm O., Payne T. W., Engle R. W. (2004). The generality of working memory capacity: A latent-variable approach to verbal and visuospatial memory span and reasoning. Journal of Experimental Psychology: General, 133(2), 189–217. [DOI] [PubMed] [Google Scholar]
- Korous K. M., Causadias J. M., Bradley R. H., Luthar S. S., Levy R. (2020). A systematic overview of meta-analyses on socioeconomic status, cognitive ability, and achievement: The need to focus on specific pathways. Psychological Reports, 125(1), 55–97. [DOI] [PubMed] [Google Scholar]
- Lee K., Bull R., Ho R. M. (2013). Developmental changes in executive functioning. Child Development, 84(6), 1933–1953. [DOI] [PubMed] [Google Scholar]
- Ligthart S., Marzi C., Aslibekyan S., Mendelson M. M., Conneely K. N., Tanaka T., Colicino E., Waite L. L., Joehanes R., Guan W., Brody J. A., Elks C., Marioni R., Jhun M. A., Agha G., Bressler J., Ward-Caviness C. K., Chen B. H., Huan T., . . . Dehghan A. (2016). DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biology, 17(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan G. D., Schachar R. J., Tannock R. (1997). Impulsivity and inhibitory control. Psychological Science, 8(1), 60–64. [Google Scholar]
- McCartney D. L., Hillary R. F., Conole E. L. S., Trejo Banos D., Gadd D. A., Walker R. M., Nangle C., Flaig R., Campbell A., Murray A. D., Muñoz Maniega S., del C., Valdés-Hernández M., Harris M. A., Bastin M. E., Wardlaw J. M., Harris S. E., Porteous D. J., Tucker-Drob E. M., McIntosh A. M., . . . Marioni R. E. (2022). Blood-based epigenome-wide analyses of cognitive abilities. Genome Biology, 23(1), Article 26. 10.1101/2021.05.24.21257698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton L. Y. M., Dou J., Fisher J., Heiss J. A., Nguyen V. K., Just A. C., Faul J., Ware E. B., Mitchell C., Colacino J. A., Bakulski K. M. (2022). Saliva cell type DNA methylation reference panel for epidemiological studies in children. Epigenetics, 17(2), 161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake A., Friedman N. P., Emerson M. J., Witzki A. H., Howerter A., Wager T. D. (2000). The unity and diversity of executive functions and their contributions to complex “frontal lobe” tasks: A latent variable analysis. Cognitive Psychology, 41(1), 49–100. [DOI] [PubMed] [Google Scholar]
- Muthén L., Muthén B. (2017). Mplus user’s guide (8th ed.). [Google Scholar]
- Petersen A. C., Crockett L., Richards M., Boxer A. (1988). A self-report measure of pubertal status: Reliability, validity, and initial norms. Journal of Youth and Adolescence, 17(2), 117–133. [DOI] [PubMed] [Google Scholar]
- Pidsley R., Zotenko E., Peters T. J., Lawrence M. G., Risbridger G. P., Molloy P., Van Djik S., Muhlhausler B., Stirzaker C., Clark S. J. (2016). Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biology, 17(1), Article 208. 10.1186/s13059-016-1066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffington L., Belsky D. W. (2022). Integrating DNA methylation measures of biological aging into social determinants of health research. Current Environmental Health Reports, 9(2), 196–210. [DOI] [PubMed] [Google Scholar]
- Raffington L., Belsky D. W., Kothari M., Malanchini M., Tucker-Drob E. M., Harden K. P. (2021). Socioeconomic disadvantage and the pace of biological aging in children. Pediatrics, 147(6), Article e2020024406. 10.1542/peds.2020-024406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2021). R: A language and environment for statistical computing (Version 4.0.4) [Computer software]. R Foundation for Statistical Computing. http://www.R-project.org [Google Scholar]
- Roberts S. O., Bareket-Shavit C., Dollins F. A., Goldie P. D., Mortenson E. (2020). Racial inequality in psychological research: Trends of the past and recommendations for the future. Perspectives on Psychological Science, 15(6), 1295–1309. [DOI] [PubMed] [Google Scholar]
- Sabhlok A., Malanchini M., Engelhardt L. E., Madole J., Tucker-Drob E. M., Harden K. P. (2022). The relationship between executive function, processing speed, and attention-deficit hyperactivity disorder in middle childhood. Developmental Science, 25(2), e13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salthouse T. A. (2011). What cognitive abilities are involved in trail-making performance? Intelligence, 39(4), 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salthouse T. A., Babcock R. L. (1991). Decomposing adult age differences in working memory. Developmental Psychology, 27(5), 763–776. [Google Scholar]
- Snyder-Mackler N., Burger J. R., Gaydosh L., Belsky D. W., Noppert G. A., Campos F. A., Bartolomucci A., Yang Y. C., Aiello A. E., O’Rand A., Harris K. M., Shively C. A., Alberts S. C., Tung J. (2020). Social determinants of health and survival in humans and other animals. Science, 368(6493), Article eaax9553. 10.1126/science.aax9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson A. J., McCartney D. L., Hillary R. F., Campbell A., Morris S. W., Bermingham M. L., Walker R. M., Evans K. L., Boutin T. S., Hayward C., McRae A. F., McColl B. W., Spires-Jones T. L., McIntosh A. M., Deary I. J., Marioni R. E. (2020). Characterisation of an inflammation-related epigenetic score and its association with cognitive ability. Clinical Epigenetics, 12(1), Article 113. 10.1186/s13148-020-00903-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. (2013). A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics, 29(2), 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triche T. J., Jr., Weisenberger D. J., Van Den Berg D., Laird P. W., Siegmund K. D. (2013). Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Research, 41(7), Article e90. 10.1093/nar/gkt090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Census Bureau / American FactFinder. (2000). 2000 Census. U.S. Census Bureau. [Google Scholar]
- United States Census Bureau / American FactFinder. (2010). 2010 Census. U.S. Census Bureau. [Google Scholar]
- U.S. Household Food Security Survey Module. (2012). Economic research service.
- Verbruggen F., Logan G. D., Stevens M. A. (2008). STOP-IT: Windows executable software for the stop-signal paradigm. Behavior Research Methods, 40(2), 479–483. [DOI] [PubMed] [Google Scholar]
- Wahl S., Drong A., Lehne B., Loh M., Scott W. R., Kunze S., Tsai P.-C., Ried J. S., Zhang W., Yang Y., Tan S., Fiorito G., Franke L., Guarrera S., Kasela S., Kriebel J., Richmond R. C., Adamo M., Afzal U., . . . Chambers J. C. (2017). Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature, 541(7635), 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. (2003). Wechsler Intelligence Scale for Children–Fourth Edition (WISC-IV). The Psychological Corporation. [Google Scholar]
- Wechsler D. (2011). WASI-II: Wechsler Abbreviated Scale of Intelligence. PsychCorp. [Google Scholar]
- Williams D. R., Priest N., Anderson N. (2019). Understanding associations between race, socioeconomic status, and health: Patterns and prospects. In Oberlander J., Buchbinder M., Churchill L. R., Estroff S. E., King N. M. P., Saunders B. F., Strauss R. P., Walker R. L. (Eds.), The social medicine reader (Vol. II, 3rd ed., pp. 258–267). Duke University Press. [Google Scholar]
- Woodcock R. W., Mather N., McGrew K. S. (2001). Woodcock-Johnson III tests of cognitive abilities. Riverside Publishing Company. [Google Scholar]
- Wright I., Waterman M., Prescott H., Murdoch-Eaton D. (2003). A new Stroop-like measure of inhibitory function development: Typical developmental trends. Journal of Child Psychology and Psychiatry, 44(4), 561–575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-pss-10.1177_09567976221122760 for Socially Stratified Epigenetic Profiles Are Associated With Cognitive Functioning in Children and Adolescents by Laurel Raffington, Peter T. Tanksley, Aditi Sabhlok, Liza Vinnik, Travis Mallard, Lucy S. King, Bridget Goosby, K. Paige Harden and Elliot M. Tucker-Drob in Psychological Science