Abstract
Introduction
Large datasets are required to ensure reliable non-invasive glioma assessment with radiomics-based machine learning methods. This can often only be achieved by pooling images from different centers. Moreover, trained models should perform with high accuracy when applied to data from different centers. In this study, the impact of reconstruction settings and segmentation methods on radiomic features derived from amino acid and TSPO PET images of glioma patients was examined. Additionally, the ability to model and thus reduce feature differences was investigated.
Methods
[18F]FET and [18F]GE-180 PET data were acquired from 19 glioma patients. For each acquisition, 10 reconstruction settings and 9 segmentation methods were included to emulate multicentric data. Statistical robustness measures were calculated before and after ComBat harmonization. Differences between features due to setting variations were assessed using Friedman test, coefficient of variation (CV) and inter-rater reliability measures, including intraclass and Spearman’s rank correlation coefficients and Fleiss’ Kappa.
Results
According to Friedman analyses, most features (>60%) showed significant differences. Yet, CV and inter-rater reliability measures indicated higher robustness. ComBat resulted in almost complete harmonization (>87%) according to Friedman test and little to no improvement according to CV and inter-rater reliability measures. [18F]GE-180 features were more sensitive to reconstruction settings than [18F]FET features.
Conclusions
According to Friedman test, feature distributions could be successfully aligned using ComBat. However, depending on settings, changes in patient ranks were observed for some features and could not be eliminated by harmonization. Thus, for clinical utilization it is recommended to exclude affected features.
Keywords: Radiomics, Robustness, Data Pooling, FET PET, TSPO PET, Glioma
1. Introduction
The most common type of primary malignant brain tumor is glioma with an overall incidence of approx. 6 per 100,000 persons. Glioblastoma, the most aggressive subtype of glioma, has a 5-year relative survival of only 7% and represents 49% of all malignant central nervous system tumors [1]. This dismal outcome underlines the urgent need for improved diagnosis, patient stratification and, consequently, improved treatment planning. Hence, many studies aim to improve the clinical performance of simple image statistics combined with clinical parameters by further including multi-modal and texture information and using machine or deep learning methods [2], [3], [4].
Several radiomic studies were performed using magnetic resonance imaging (MRI) data, which offer excellent spatial resolution and soft tissue contrast but lack specificity for tumor tissue. Therefore, positron emission tomography (PET) using amino acid radiotracers, which show increased uptake in neoplastic tissue, is now widely used [5], [6] and several related studies have shown the added value of radiomic analyses for patient survival [7], tumor classification [8], [9], [10], and identification of tumor recurrence and early tumor progression [11], [12]. Recently, the overexpression of the 18-kDa translocator protein (TSPO) in neoplastic tissue in addition to activated glial cells has also attracted attention as a novel imaging marker for assessing glioma microenvironment [13], [14].
To properly translate radiomic models into clinical routine, they should be validated on large datasets that preferably include data from multiple centers and thus improve reproducibility and generalizability of radiomics analyses. However, reports have shown that features are sensitive to variations of several factors, including image acquisition, image reconstruction, tumor segmentation, as well as test-retest imaging [15], [16], [17], [18], [19], [20], [21], [22]. Thus, it is essential to ensure the reproducibility and robustness of features in this regard. Several methods for removing unwanted variations have been introduced and tested. These so-called harmonization techniques aim to integrate data originating from different centers while preserving clinically relevant information [23]. The ComBat method outperformed other data adjustment methods [24] and was previously validated on radiomic features extracted from PET, MRI, and computed tomography (CT) images of cancer patients and phantoms [21], [25], [26], [27]. Several statistical measures have been used in previous publications to assess the robustness of features. Orlhac et al. [25], [26], [27] used Friedman test and the equivalent Wilcoxon test to validate the ComBat harmonization method. Differences between scanners or reconstruction algorithms and test-retest variability have been assessed using either coefficient of variation (CV) [15], [17], intra-class correlation coefficient (ICC) [16], [19], [20], [21], [22], or Spearman’s correlation coefficient [16], [22]. Since each of these measures reflects different properties of the data, their relevance may depend on the specific application. Thus, in this work, the robustness of radiomic features was analyzed by including all statistical measures applied in either of the aforementioned publications.
The main goal of this study was to assess whether radiomic feature harmonization is feasible for pooling amino acid or TSPO PET images of glioma patients. To achieve this, radiomic features were evaluated with respect to variations in image processing as encountered in multicentric studies, where data pooling is required for improved generalizability of clinical models. Furthermore, the effectiveness of ComBat feature harmonization was assessed for this specific application. Variations arising from multicentric data were emulated by reconstructing each patient dataset with different settings and applying multiple segmentation methods. To the best of our knowledge, these analyses have not been performed so far.
2. Methods
2.1. Patient data and imaging
PET images from a cohort of 19 patients diagnosed with glioma were included in this study. 10 patients were scanned at initial diagnosis before any treatment and 9 patients at tumor recurrence. Histological and molecular genetic classification according to the 2021 WHO guideline for brain tumors [28] revealed 13 glioblastomas, IDH wildtype; 4 astrocytoma IDH mutant without 1p/19q codeletion; 2 oligodendroglioma, IDH mutant, 1p/19q codeleted. All patients have given written informed consent to the data analysis. The study was approved by the local ethics committee (approval number 18-783).
The images were acquired on a Biograph 64 PET/CT scanner (Siemens Healthineers, Erlangen, Germany) at the Department of Nuclear Medicine of the University Hospital, LMU Munich. Immediately before each PET scan, low-dose CT was performed for attenuation correction. Each patient underwent one PET scan after administration of the radiolabeled TSPO ligand (S)-N,N-diethyl-9-(2-[18F]-fluoroethyl)-5-methoxy-2,3,4,9-tetrahydro-1H-carbazole-4-carboxamide ([18F]GE-180) and one PET scan after administration of the amino acid tracer O-(2-[18F]-fluoroethyl)-L-tyrosine ([18F]FET) on consecutive days.
[18F]FET was synthesized in a 2-step process by [18F]-fluoroethylation of L- and D-tyrosine as described by Wester et al. [29] and [18F]GE-180 was synthesized using a FASTlab synthesizer with single-use cassettes (GE Healthcare, Chicago, Illinois, USA) [30]. Dynamic acquisitions were obtained in list mode and corrected for scattered and random coincidences, photon attenuation, radionuclide decay and detector dead time during image reconstruction. For both radiotracers, late tracer uptake was used for radiomics analysis. The respective late static images were derived by averaging motion corrected 10-minute time frames of the dynamic studies. Frame-wise motion correction to an early 0-3 min post-injection (p.i.) image was performed using the PVIEW tool of the PMOD software (version 3.502, PMOD Technologies, Zürich, Switzerland).
For each patient, a 90-minute scan was performed after intravenous bolus injection of 172 ± 11 MBq of [18F]GE-180. The aforementioned 10-minute frames were generated from 60-90 min p.i. acquisition data according to previous research [31], [32], [33]. On the following day, a 40-minute scan was carried out after bolus injection of 177 ± 9 MBq of [18F]FET. In this case, 20-40 min p.i. acquisition data were used for the static images following international practice guidelines for glioma imaging with amino acid tracers [6].
2.2. Image reconstruction
Each image was reconstructed 10 times with different settings. One setting was defined as the default and comprised reconstruction parameters that were optimized for clinical quantification of brain PET images at our department [32], [34]. For the remaining settings, the respective parameters were fixed to the default setting, while either the reconstruction algorithm, the matrix size, the number of subsets or the filter size were varied individually. The default reconstruction setting was OSEM3D algorithm with 4 iterations, 21 subsets, and 5 mm Gaussian post-reconstruction filter. The default matrix size of 336 × 336 × 109 with a zoom factor of 2 resulted in a voxel size of 1.018 × 1.018 × 2.027 mm3. An overview of all included settings is given in Table 1.
Table 1.
Image reconstruction settings for PET data of 19 glioma patients acquired on a Biograph 64 PET/CT system. The bold entries were selected as the default. FBP: filtered back-projection; OSEM: ordered subsets expectation maximization; FWHM: full width half maximum.
| Parameter | Values |
|---|---|
| Algorithm | OSEM3D; OSEM2D; TrueX; FBP with 4.9 mm Hann |
| Matrix | 128; 168; 336 |
| Subsets | 8; 16; 21 |
| Filter | 2; 4; 5 mm Gaussian |
2.3. Tumor segmentation
The background intensity was defined on the PET images as the mean intensity in a crescent shaped volume manually delineated in a non-affected brain region encompassing both white and grey matter, as recommended in the EANM/EANO/RANO/SNMMI joint practice guidelines for amino acid PET imaging [6] and described by Unterrainer et al. [35]. For comparison of reconstruction settings, volumes-of-interest (VOI) were segmented using the background intensity multiplied by a factor of 1.6 as a threshold for [18F]FET and 1.8 for [18F]GE-180 [34], [36]. Semiautomatic segmentation was performed inside of a manually defined confining volume, using initial seeds and the region growing algorithm provided by the simpleITK library (version 2.1.1, [37]) in Python 3.9.
For comparison of feature values derived using different segmentation methods, three different threshold-based segmentation methods were employed each with three different threshold values, resulting in nine segmentation methods (Table 2). These analyses were performed on patient images reconstructed with the default setting. The intensity threshold was either derived using background intensity (), maximal intensity (), or contrast (). The values of the threshold factors , , and defined in Table 2 were chosen using previous literature [34], [36], [38]. VOIs with less than 18 voxels were considered too small [39]. Thus, the data of 2/19 patients were excluded.
Table 2.
Image segmentation methods for PET data of 19 glioma patients acquired on a Biograph 64 PET/CT system.
| Method | Threshold | Empirical factors F |
|---|---|---|
| Background intensity | 1.4; 1.6; 1.8 | |
| Maximum intensity | 0.4; 0.45; 0.5 | |
| Contrast | 0.3; 0.35; 0.4 |
2.4. Radiomic feature extraction
Initially, PET images were normalized to the background signal by dividing all voxel intensities by to improve inter-patient comparability yielding tumor-to-background ratio (TBR) images. Feature extraction was performed with the Python package PyRadiomics (version 3.0.1, [40]). Voxels were resampled to 2 × 2 × 2 mm3 with a b-spline interpolator and TBR values were discretized using a fixed bin width as recommended by Leijenaar et al. [22] to preserve quantitative characteristics and improve inter- and intra-patient comparability of radiomic features. In accordance with previous publications, the bin width was set to the interquartile range of TBR values devided by 4, which yields 0.13 [41], [42]. Overall, 107 features from the following categories were extracted from each image: first order statistics (n = 18), 3D shape features (n = 14), and texture features (n = 75). Detailed feature definitions, most of which are compliant with the definitions published by the Image Biomarker Standardization Initiative (IBSI, [43]), can be found in the PyRadiomics documentation [40].
2.5. Feature harmonization
The ComBat method is an empirical Bayes framework that was proposed to harmonize data originating from different sites [44]. It assumes that the data are affected by site-specific additive and multiplicative effects. The neuroCombat package (version 0.2.12, [45]) was implemented in Python to apply ComBat for each feature separately with no adjustments for biological covariates assuming non-parametric variables. In this study, different radiomic feature distributions resulting from a variation of reconstruction settings and segmentation methods were aligned using ComBat by removing batch effects. ComBat was fitted and applied independently to assimilate the features derived from each of the following subgroups with the respective number of feature distributions given in brackets: all reconstructions (10), algorithm (4), matrix (3), subsets (3), filter (3), all segmentations (9), background (3), maximum (3), and contrast (3) (see Table 1, Table 2).
2.6. Statistical analysis
Each of the statistical measures described below was calculated using the radiomic features of the entire patient cohort before and after ComBat harmonization. The evaluation was performed separately for [18F]FET and [18F]GE-180 data. The different statistical measures allow for a separate quantification of differences between feature distributions, variability of feature values, and changes in patient ranks.
Friedman test was employed using the Python package SciPy (version 1.7.3, [46]) to compare the distributions of feature values with respect to patients, whereby each distribution originated from a different setting. Statistically significant differences between distributions were indicated by p-values less than 0.05, therefore percentages of robust features exhibiting p-values greater than 0.05 were reported.
Mean coefficients of variation (CV) over all patients were calculated in a feature-wise manner with SciPy to characterize the within-patient variance relative to the mean value from different settings. A threshold of 0.1 was used to identify robust features to provide a reference value for comparison with results from previous robustness studies [15], [17].
Intraclass correlation coefficients (ICC) were estimated in Python with the Pingouin package (version 0.5.0, [47]) to quantify the within-patient variance relative to the between-patient variance [48]. According to the guideline published by Koo and Li [49], the model for two-way mixed effects, consistency, single rater, and single measurement was selected. Following their recommendation for evaluating reliability without considering the 95% confidence interval of the ICC estimate, features were categorized as robust when their ICC exceeded a value of 0.9.
Differences between patient ranks were quantified by computing pair-wise Spearman’s rank correlation coefficients and Fleiss’ Kappa using the Python packages SciPy and statsmodels (version 0.14.0, [50]). Whereas Spearman’s rank correlation coefficient was calculated directly from the feature values, Fleiss’ Kappa was derived from patient ranks. Since the Spearman’s correlation coefficient can only be determined for two rankings at a time, the calculation was performed by averaging over all pair-wise coefficients. Thus, Fleiss’ Kappa was also computed directly on the patient ranks to include a measure that eliminates the need for averaging. Thresholds for defining robust features were set to 0.9 for Spearman’s rank correlation and to 0.4 for Fleiss’ Kappa based on previous studies and recommendations [16], [22], [51].
Furthermore, spaghetti plots were generated for an exemplary feature to visually inspect the influence of setting variations.
3. Results
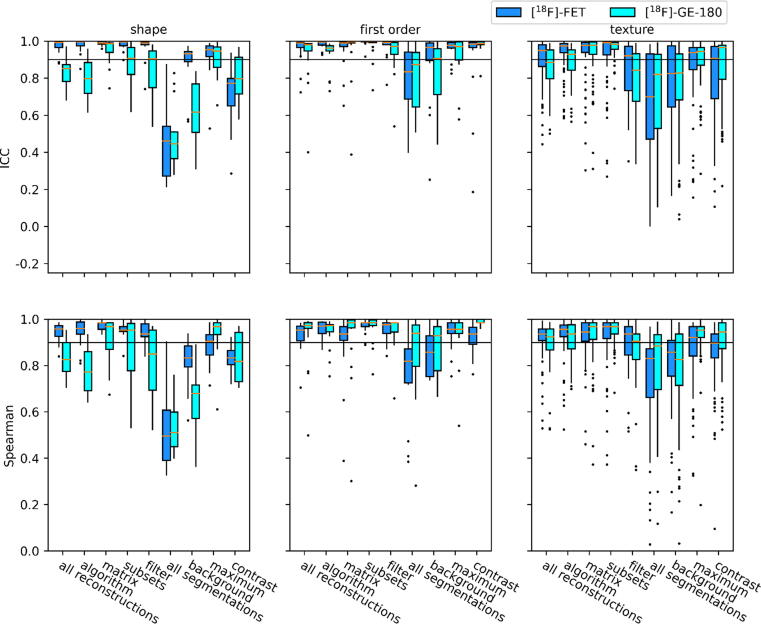
Fig. 1 shows boxplots of ICC and Spearman’s correlation coefficient, and supplementary Fig. S1 shows boxplots of CV and Fleiss’ Kappa. The percentages of features with p > 0.05 are listed in Table 3 for reconstruction settings and Table 4 for segmentation methods. Percentages for CV > 0.1, ICC < 0.9, Spearman’s correlation coefficient > 0.9, and Fleiss’ Kappa > 0.4 are listed in supplementary Tables S2a and S2b (Supplementary Material 2). All percentages reported in this section are for a variation of all settings and/or all feature classes. A complete list of individual values for every feature is provided in Tables S3a-S3d (Supplementary Material 3) for the variation of all settings. All tables contain results with and without ComBat harmonization.
Figure 1.
Boxplots showing intraclass correlation coefficients (ICC) and Spearman’s rank correlation coefficients of shape, first order and texture features. Statistics were calculated by comparing feature values between all reconstruction settings, all segmentation methods, and subgroups thereof. The horizontal lines indicate the robustness thresholds for the respective measure.
Table 3.
Percentages of features without significant differences between reconstruction settings before and after ComBat harmonization. Percentages are color-coded with shades of green ranging from white for low values to dark green for high values. CV: coefficient of variation; ICC: intraclass correlation coefficient.
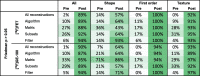 |
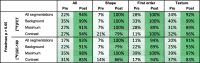
Table 4.
Percentages of features without significant differences between segmentation methods before and after ComBat harmonization. Percentages are color-coded with shades of green ranging from white for low values to dark green for high values. CV: coefficient of variation; ICC: intraclass correlation coefficient.
 |
3.1. Robustness of radiomic features
The percentages of robust features according to Friedman test were low for the variation of reconstruction settings ([18F]FET: 2%; [18F]GE-180: 1%) with highest robustness for shape features (14%; 7%) and for changes in matrix size (27%; 33%) and number of subsets (26%; 29%). Less than 1% of first order and texture features were robust to a variation of all reconstruction settings. In contrast, CV and inter-rater reliability measures indicated a moderate to high robustness for variable reconstruction settings (Fig. 1 and S1), whereby texture features presented with lower percentages compared to shape and first order features. Furthermore, lower robustness was observed for [18F]GE-180 compared to [18F]FET. All statistical measures implied a high sensitivity to the choice of post-reconstruction filter, especially for texture features.
For variation of segmentation methods, the fraction of robust features according to Friedman test was slightly increased but still low ([18F]FET: 22%; [18F]GE-180: 17%), whereby shape features were least robust (7%; 7%). ICC and Spearman’s correlation indicated a moderate to low robustness (Fig. 1), with first order features being the most robust and shape features being the least robust. In this case, both measures indicated a lower feature robustness for [18F]GE-180.
3.2. Effect of ComBat harmonization
ComBat feature harmonization enabled an almost perfect assimilation of features as assessed using Friedman test for the variation of both reconstruction settings and segmentation methods ([18F]FET: >89%; [18F]GE-180: >90%). The only exception was a residual sensitivity of several shape features to the variation of reconstruction settings. According to CV and ICC, ComBat caused an overall improvement in robustness, whereas the settings which already presented with very high percentages of features with CV < 0.1 and ICC > 0.9 showed little to no increase. For a variation of reconstruction settings, the percentage of robust features after ComBat harmonization according to CV and ICC remained lowest for texture features. Similarly, the percentages of shape features with CV < 0.1 and ICC > 0.9 remained lowest for variable segmentation methods. Spearman’s rank correlation and Fleiss’ Kappa were not affected by the ComBat method (see Tables S2a-S2b, Supplementary Material 2).
4. Discussion
A large number of publications report the clinical relevance of radiomic features derived from PET images of glioma patients [3]. Hence, pooling data and applying trained models to data from different centers becomes essential to improve generalizability of models and ultimately enable translation into clinical routine. Therefore, in this study, sensitivity of radiomic features derived from [18F]FET and [18F]GE-180 PET images of glioma patients was quantified with respect to variations in image reconstruction settings and tumor segmentation methods. Since feature robustness has previously been evaluated by different statistical measures, we compared their results and critically assessed their usefulness in judging the success of harmonization.
In previous studies, Friedman test was applied for evaluation of ComBat performance [25], [26], [27], whereas CV and ICC were frequently applied to assess feature variance and inter-rater reliability and have been complemented by Spearman’s rank correlation coefficient [15], [16], [17], [19], [20], [21], [22]. Friedman test quantifies significant differences in feature distributions considering the paired nature of feature values of each patient. Since feature harmonization using ComBat allows to improve the correspondence between feature value distributions, this property can be directly evaluated using Friedman test [25], [26], [27]. CV and inter-rater reliability measures describe substantially different aspects of feature robustness. CV quantifies the within-patient variance relative to the mean feature value of the patient and ICC relative to the between-patient variance [48]. Spearman’s rank correlation coefficient and Fleiss’ Kappa quantify whether patients are ranked differently within each setting, which could adversely affect e.g. classification tasks. Since changes in patient ranks cannot be compensated using feature harmonization, variations leading to a low rank correlation need to be avoided.
Overall results showed that PET radiomic features were highly sensitive to the choice of image segmentation methods and, in accordance with the literature for [18F]FDG PET, reconstruction settings [15], [17], [52]. Rank-based measures implied that a variation of segmentation methods is more likely to change patient ranks with respect to feature values than a variation of reconstruction settings. As this variability cannot be diminished, it is important to first carefully select a clinically meaningful segmentation method and then consistently apply the chosen segmentation method to all patient data.
The high impact of different post-reconstruction filters is most likely explained by the strong effect of smoothing on object boundaries, image texture, as well as voxel intensities in general. This finding contradicts results of a previous study using [18F]FDG for the assessment of lung lesions [15], where the choice of matrix size had the strongest impact on radiomic features as assessed by CV. However, the impact of matrix size might be reduced in this study as the applied radiomics pipeline included resampling to the same voxel size before feature extraction.
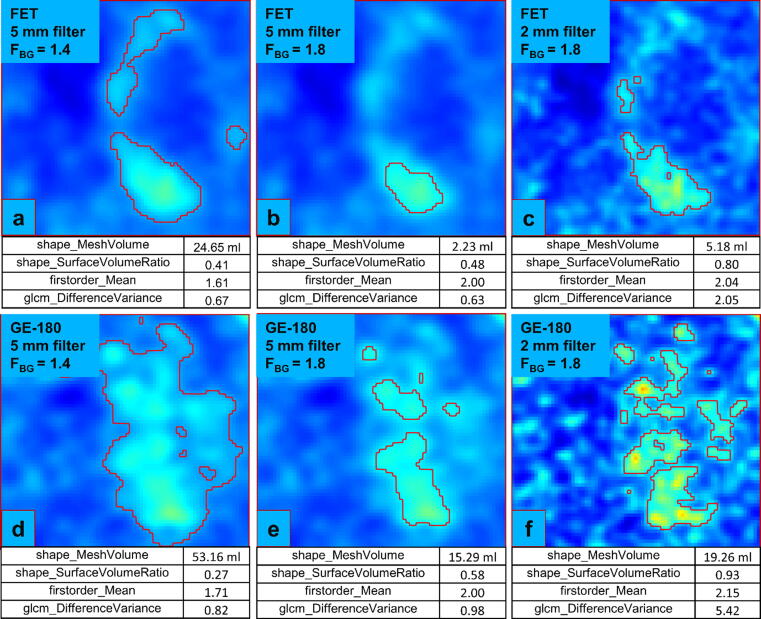
The sensitivity difference of shape features between reconstruction and segmentation was expected, as image segmentation directly relates to the shape of a VOI, whereas reconstruction rather affects voxel intensities and their interrelations. Especially in lesions with a more spread-out tracer uptake, VOIs that were generated with different segmentation methods showed significant differences in shape features, as exemplarily seen in Fig. 2.
Figure 2.
[18F]FET (a-c) and [18F]GE-180 (d-f) PET images showing the same axial slice through a lesion (TBR intensity window: 0-5). Tumor VOIs, marked by red contours, were generated after applying different smoothing filters and segmentation thresholds FBG. The values of four radiomic features are shown for each image.
The lower robustness to a variation of reconstruction settings of features from the [18F]GE-180 data compared to the [18F]FET data might be explained by the potential contribution of low inflammation-related PET signal in [18F]GE-180 images and by the lower activity concentration in healthy background resulting in an increased noise contribution especially when combined with a narrow post-reconstruction filter. This is visualized in Fig. 2 for an example glioma, where in case of a 2 mm Gaussian filter, the tumor volume is rather compact for [18F]FET, while it is broad and patchy for [18F]GE-180. Evidently, the feature extraction process is therefore also dependent on the interplay between reconstruction and segmentation. Hence, the robustness of radiomic features can be influenced by tracer-specific uptake patterns especially when a solely PET-based radiomics workflow including tumor segmentation is used. This implies that the distribution of suspiciously increased biological signal, which is driven by tracer characteristics, may affect the sensitivity of radiomic features to a variation of reconstruction settings or segmentation methods.
As assessed using Friedman test, ComBat harmonization successfully assimilated most radiomic features, which is in line with previous publications validating the ComBat method [25], [26], [27]. However, CV and ICC showed only little to no improvements and rank correlation measures were unchanged. Similarly, only little improvement of ICC was observed after Combat harmonization of CT based features as reported by Ligero et al. [21].
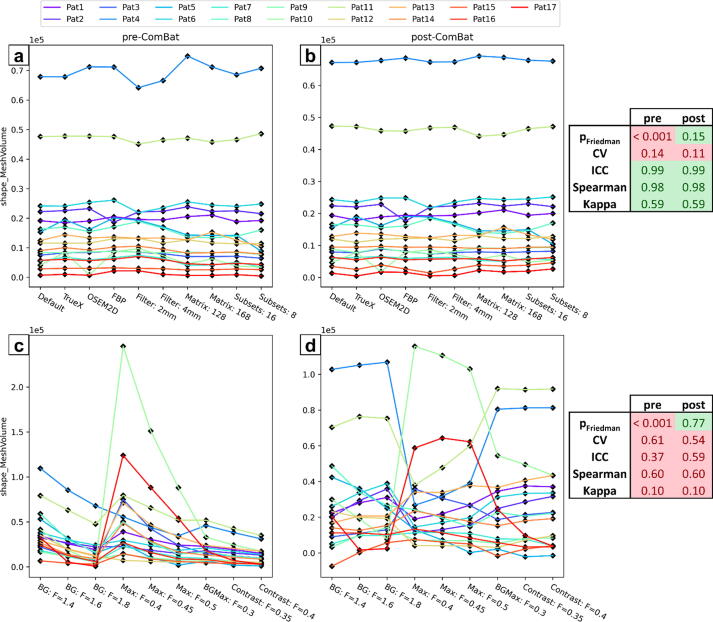
The different aspects quantified by statistical measures can be visualized using spaghetti plots as presented in Fig. 3 for the shape feature mesh volume derived from [18F]GE-180 PET data. Feature values of one patient for different settings are connected by lines. For zero variance, a straight horizontal line reflects perfect agreement of feature values from different settings, whereas a jagged line reflects increased variance. Fig. 3 depicts spaghetti plots before and after ComBat harmonization for a variation of reconstruction settings (Fig. 3a, b) and for a variation of segmentation methods (Fig. 3c, d). The mesh volume presents with only few intersecting lines and is therefore less sensitive to a variation of reconstruction settings according to rank correlation measures. Yet, for instance the upper-most blue line in Fig. 3a is slightly jagged and transformed to a straight line after harmonization (Fig. 3b), leading to a slightly decreased CV and satisfactory alignment of distributions according to Friedman test. In contrast, for a variation of segmentation methods, a large number of lines are jagged and intersecting before harmonization, which is reflected by all included statistical measures. Although feature distributions of each segmentation method were well aligned after harmonization, the feature values of each individual patient showed a high variability between settings as quantified using CV and ICC. Furthermore, mesh volume ranks of different patients remained the same as visualized by persisting intersecting lines and quantified using Spearman’s rank correlation and Fleiss’ Kappa. These observations are in line with the above-mentioned increased sensitivity of shape features to tumor segmentation.
Figure 3.
Spaghetti plots showing the values of the shape feature mesh volume after extraction from the [18F]GE-180 data before and after ComBat harmonization. a & b: distributions over all patients for every reconstruction setting. c & d: distributions over all patients for every segmentation method. Results of the statistical analyses are highlighted in green or red depending on the respective robustness threshold (pFriedman: 0.05; CV: 0.1; ICC: 0.9; Spearman: 0.9; Kappa: 0.4).
A limitation of this study is the small sample size, which was restricted due to the large number of included reconstruction protocols (20 per patient for both radiotracers, 380 overall). Results were derived from a mixed patient cohort, which comprises gliomas at initial diagnosis as well as recurrent tumors. To explicitly exclude features which cannot be harmonized using ComBat, the presented analyses should be reperformed with a larger sample size for each specific clinical task and patient group of interest.
One potential caveat concerning ComBat is the occurrence of negative feature values after harmonization for features that can only assume positive values per definition, which was for example observed for the feature mesh volume for one patient (Fig. 3d). Thus, the biological meaning of the harmonized values is uncertain in these cases. In general, it is not clear how well biological variations are retained by ComBat, as ground truth data are usually unavailable to correlate radiomic feature values to the underlying biology. Furthermore, it remains to be evaluated, whether ComBat model fitted to a small patient cohort can be meaningfully applied on data from of a larger or even different patient population. Yet, the striking improvement of feature similarity among different settings in terms of overlapping feature distributions will increase the performance of clinically relevant features unless they are susceptible to patient rank variability. The clinical benefit of feature harmonization for glioma classification, survival prediction, or identification of tumor recurrence will be assessed in a separate study.
5. Conclusions
In this study, radiomic feature robustness and the applicability of ComBat feature harmonization was assessed with regard to variations that are typically encountered when data from multiple centers are pooled. From the findings it can be concluded that radiomic features derived from [18F]FET or [18F]GE-180 data of glioma patients are highly susceptible to setting variations, whereby [18F]GE-180 features display higher sensitivity to a variation of reconstruction settings compared to [18F]FET. This implies that feature robustness is tracer dependent. Although feature value distributions can be assimilated using ComBat, variable patient ranks cannot be compensated. However, poor agreement between patient ranks may have a significant impact on the biological interpretability and clinical applicability of radiomic features. Hence, multicentric data can be successfully pooled for selected clinically relevant features when ComBat harmonization is employed and rank variability is considered.
Ethics approval and consent to participate
The study was authorized by the local ethics committee (18-783) in accordance with the ICH Guideline for Good Clinical Practice (GCP) and the Declaration of Helsinki. Written informed consent was obtained from all individual patients included in this study.
Availability of data and material
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical restrictions.
Funding
This project was partly funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) (FOR 2858 project number 421887978 and Research Training Group GRK 2274). M.B. was funded by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy – ID 390857198).
Author contributions
AJZ, NLA, SZ, LK: Conceptualization; AJZ, SZ and LK: Methodology; AJZ and LK: Formal analysis and investigation; AJZ, AH and LK: Data curation; AJZ and LK: Software; AH, MU, JBL, SL, AB, GB, MB, LvB: Resources; AJZ and LK: Writing – original draft; NLA and SZ: Writing – review and editing; NLA, RR, JCT, PB, SZ: Funding acquisition; NLA, SZ and LK: Supervision. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: N.L.A. is member of the EANM Neuroimaging Committee (NIC). M.B. received speaker honoraria from Roche, GE healthcare and Life Molecular Imaging and is an advisor of Life Molecular Imaging and a member of the EANM NIC. All other authors declare that they have no conflict of interest.
Acknowledgements
The authors would like to thank PD Dr. rer. biol. hum. Michael Lauseker (Institute for Medical Information Processing, Biometry, and Epidemiology, LMU Munich) for kindly providing his expertise in the field of statistical analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.zemedi.2022.12.005.
Contributor Information
Adrian Jun Zounek, Email: adrian.zounek@med.uni-muenchen.de.
Nathalie Lisa Albert, Email: nathalie.albert@med.uni-muenchen.de.
Adrien Holzgreve, Email: adrien.holzgreve@med.uni-muenchen.de.
Marcus Unterrainer, Email: marcus.unterrainer@med.uni-muenchen.de.
Julia Brosch-Lenz, Email: julia.brosch-lenz@med.uni-muenchen.de.
Simon Lindner, Email: simon.lindner@med.uni-muenchen.de.
Andreas Bollenbacher, Email: andreas.bollenbacher@med.uni-muenchen.de.
Guido Boening, Email: guido.boening@med.uni-muenchen.de.
Rainer Rupprecht, Email: rainer.rupprecht@medbo.de.
Matthias Brendel, Email: matthias.brendel@med.uni-muenchen.de.
Louisa von Baumgarten, Email: louisa.vonbaumgarten@med.uni-muenchen.de.
Joerg-Christian Tonn, Email: Joerg.Christian.Tonn@med.uni-muenchen.de.
Peter Bartenstein, Email: peter.bartenstein@med.uni-muenchen.de.
Sibylle Ziegler, Email: sibylle.ziegler@med.uni-muenchen.de.
Lena Kaiser, Email: Lena.Kaiser@med.uni-muenchen.de.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Low J.T., Ostrom Q.T., Cioffi G., Neff C., Waite K.A., Kruchko C., et al. Primary brain and other central nervous system tumors in the United States (2014–2018): A summary of the CBTRUS statistical report for clinicians. Neuro-Oncol Pract. 2022;9:165–182. doi: 10.1093/nop/npac015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galldiks N., Zadeh G., Lohmann P. Artificial Intelligence, Radiomics, and Deep Learning in Neuro-Oncology. Neurooncol Adv. 2020;2:iv1 -iv2. doi: 10.1093/noajnl/vdaa179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohmann P., Meissner A.K., Kocher M., Bauer E.K., Werner J.M., Fink G.R., et al. Feature-based PET/MRI radiomics in patients with brain tumors. Neurooncol Adv. 2020;2:iv15 -iv21. doi: 10.1093/noajnl/vdaa118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh G., Manjila S., Sakla N., True A., Wardeh A.H., Beig N., et al. Radiomics and radiogenomics in gliomas: a contemporary update. Br J Cancer. 2021;125:641–657. doi: 10.1038/s41416-021-01387-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albert N.L., Weller M., Suchorska B., Galldiks N., Soffietti R., Kim M.M., et al. Response Assessment in Neuro-Oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol. 2016;18:1199–1208. doi: 10.1093/neuonc/now058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law I., Albert N.L., Arbizu J., Boellaard R., Drzezga A., Galldiks N., et al. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [18F]FDG: version 1.0. Eur J Nucl Med Mol Imaging. 2019;46:540–557. doi: 10.1007/s00259-018-4207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyka T., Gempt J., Hiob D., Ringel F., Schlegel J., Bette S., et al. Textural analysis of pre-therapeutic [18F]-FET-PET and its correlation with tumor grade and patient survival in high-grade gliomas. Eur J Nucl Med Mol Imaging. 2016;43:133–141. doi: 10.1007/s00259-015-3140-4. [DOI] [PubMed] [Google Scholar]
- 8.Lohmann P., Lerche C., Bauer E.K., Steger J., Stoffels G., Blau T., et al. Predicting IDH genotype in gliomas using FET PET radiomics. Sci Rep. 2018;8:13328. doi: 10.1038/s41598-018-31806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haubold J., Demircioglu A., Gratz M., Glas M., Wrede K., Sure U., et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric 18F-FET PET-MRI and MR Fingerprinting. Eur J Nucl Med Mol Imaging. 2020;47:1435–1445. doi: 10.1007/s00259-019-04602-2. [DOI] [PubMed] [Google Scholar]
- 10.Li Z., Kaiser L., Holzgreve A., Ruf V.C., Suchorska B., Wenter V., et al. Prediction of TERTp-mutation status in IDH-wildtype high-grade gliomas using pre-treatment dynamic [18F]FET PET radiomics. Eur J Nucl Med Mol Imaging. 2021;48:4415–4425. doi: 10.1007/s00259-021-05526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann P., Kocher M., Ceccon G., Bauer E.K., Stoffels G., Viswanathan S., et al. Combined FET PET/MRI radiomics differentiates radiation injury from recurrent brain metastasis. Neuroimage Clin. 2018;20:537–542. doi: 10.1016/j.nicl.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohmann P., Elahmadawy M.A., Gutsche R., Werner J.-M., Bauer E.K., Ceccon G., et al. FET PET Radiomics for Differentiating Pseudoprogression from Early Tumor Progression in Glioma Patients Post-Chemoradiation. Cancers. 2020;12:3835. doi: 10.3390/cancers12123835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinnhardt B., Roncaroli F., Foray C., Agushi E., Osrah B., Hugon G., et al. Imaging of the glioma microenvironment by TSPO PET. Eur J Nucl Med Mol Imaging. 2021;49:174–185. doi: 10.1007/s00259-021-05276-5. [DOI] [PubMed] [Google Scholar]
- 14.Galldiks N., Langen K.-J., Albert N.L., Law I., Kim M.M., Villanueva-Meyer J.E., et al. Investigational PET tracers in neuro-oncology—What’s on the horizon? A report of the PET/RANO group. Neuro Oncol. 2022 doi: 10.1093/neuonc/noac131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J., Chu-Shern J.L., Loi H.Y., Khor L.K., Sinha A.K., Quek S.T., et al. Impact of Image Reconstruction Settings on Texture Features in 18F-FDG PET. J Nucl Med. 2015;56:1667–1673. doi: 10.2967/jnumed.115.156927. [DOI] [PubMed] [Google Scholar]
- 16.Whybra P., Parkinson C., Foley K., Staffurth J., Spezi E. Assessing radiomic feature robustness to interpolation in 18F-FDG PET imaging. Sci Rep. 2019;9:9649. doi: 10.1038/s41598-019-46030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papp L., Rausch I., Grahovac M., Hacker M., Beyer T. Optimized Feature Extraction for Radiomics Analysis of (18)F-FDG PET Imaging. J Nucl Med. 2019;60:864–872. doi: 10.2967/jnumed.118.217612. [DOI] [PubMed] [Google Scholar]
- 18.Park J.E., Park S.Y., Kim H.J., Kim H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J Radiol. 2019;20:1124. doi: 10.3348/kjr.2018.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barry N., Rowshanfarzad P., Francis R.J., Nowak A.K., Ebert M.A. Repeatability of image features extracted from FET PET in application to post-surgical glioblastoma assessment. Phys Eng Sci Med. 2021 doi: 10.1007/s13246-021-01049-4. [DOI] [PubMed] [Google Scholar]
- 20.Gutsche R., Scheins J., Kocher M., Bousabarah K., Fink G.R., Shah N.J., et al. Evaluation of FET PET Radiomics Feature Repeatability in Glioma Patients. Cancers. 2021;13:647. doi: 10.3390/cancers13040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ligero M., Jordi-Ollero O., Bernatowicz K., Garcia-Ruiz A., Delgado-Muñoz E., Leiva D., et al. Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis. Eur Radiol. 2021;31:1460–1470. doi: 10.1007/s00330-020-07174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leijenaar R.T., Nalbantov G., Carvalho S., van Elmpt W.J., Troost E.G., Boellaard R., et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Da-Ano R., Visvikis D., Hatt M. Harmonization strategies for multicenter radiomics investigations. Phys Med Biol. 2020;65:24TR02. doi: 10.1088/1361-6560/aba798. [DOI] [PubMed] [Google Scholar]
- 24.Chen C., Grennan K., Badner J., Zhang D., Gershon E., Jin L., et al. Removing Batch Effects in Analysis of Expression Microarray Data: An Evaluation of Six Batch Adjustment Methods. PLoS One. 2011;6:e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orlhac F., Boughdad S., Philippe C., Stalla-Bourdillon H., Nioche C., Champion L., et al. A Postreconstruction Harmonization Method for Multicenter Radiomic Studies in PET. J Nucl Med. 2018;59:1321–1328. doi: 10.2967/jnumed.117.199935. [DOI] [PubMed] [Google Scholar]
- 26.Orlhac F., Frouin F., Nioche C., Ayache N., Buvat I. Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiology. 2019;291:53–59. doi: 10.1148/radiol.2019182023. [DOI] [PubMed] [Google Scholar]
- 27.Orlhac F., Lecler A., Savatovski J., Goya-Outi J., Nioche C., Charbonneau F., et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur Radiol. 2021;31:2272–2280. doi: 10.1007/s00330-020-07284-9. [DOI] [PubMed] [Google Scholar]
- 28.Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wester H.J., Herz M., Weber W., Heiss P., Senekowitsch-Schmidtke R., Schwaiger M., et al. Synthesis and Radiopharmacology of <em>O</em>-(2-[<sup>18</sup>F]fluoroethyl)-<span class=“sc”>L</span>-Tyrosine for Tumor Imaging. J Nucl Med. 1999;40:205–212. [PubMed] [Google Scholar]
- 30.Wickstrøm T., Clarke A., Gausemel I., Horn E., Jørgensen K., Khan I., et al. The development of an automated and GMP compliant FASTlab™ Synthesis of [18F]GE-180; a radiotracer for imaging translocator protein (TSPO) J Label Compd Radiopharm. 2014;57:42–48. doi: 10.1002/jlcr.3112. [DOI] [PubMed] [Google Scholar]
- 31.Feeney C., Scott G., Raffel J., Roberts S., Coello C., Jolly A., et al. Kinetic analysis of the translocator protein positron emission tomography ligand [18F]GE-180 in the human brain. Eur J Nucl Med Mol Imaging. 2016;43:2201–2210. doi: 10.1007/s00259-016-3444-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unterrainer M., Fleischmann D.F., Vettermann F., Ruf V., Kaiser L., Nelwan D., et al. TSPO PET, tumour grading and molecular genetics in histologically verified glioma: a correlative 18F-GE-180 PET study. Eur J Nucl Med Mol Imaging. 2020;47:1368–1380. doi: 10.1007/s00259-019-04491-5. [DOI] [PubMed] [Google Scholar]
- 33.Vomacka L., Albert N.L., Lindner S., Unterrainer M., Mahler C., Brendel M., et al. TSPO imaging using the novel PET ligand [18F]GE-180: quantification approaches in patients with multiple sclerosis. EJNMMI Res. 2017;7:89. doi: 10.1186/s13550-017-0340-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaiser L., Holzgreve A., Quach S., Ingrisch M., Unterrainer M., Dekorsy F.J., et al. Differential Spatial Distribution of TSPO or Amino Acid PET Signal and MRI Contrast Enhancement in Gliomas. Cancers (Basel) 2021;14:53. doi: 10.3390/cancers14010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unterrainer M., Vettermann F., Brendel M., Holzgreve A., Lifschitz M., Zahringer M., et al. Towards standardization of (18)F-FET PET imaging: do we need a consistent method of background activity assessment? EJNMMI Res. 2017;7:48. doi: 10.1186/s13550-017-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauleit D., Floeth F., Hamacher K., Riemenschneider M.J., Reifenberger G., Muller H.W., et al. O-(2-[18F]fluoroethyl)-L-tyrosine PET combined with MRI improves the diagnostic assessment of cerebral gliomas. Brain. 2005;128:678–687. doi: 10.1093/brain/awh399. [DOI] [PubMed] [Google Scholar]
- 37.Lowekamp B., Chen D., Ibanez L., Blezek D. The Design of SimpleITK. Frontiers. Neuroinformatics. 2013;7 doi: 10.3389/fninf.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser L. Non-invasive quantification of CNS pathology with dynamic PET information: Investigation of advanced methods for the characterisation of multiple sclerosis and glioma lesions [Dissertation] LMU Munich. 2019 [Google Scholar]
- 39.Vomacka L., Unterrainer M., Holzgreve A., Mille E., Gosewisch A., Brosch J., et al. Voxel-wise analysis of dynamic 18F-FET PET: a novel approach for non-invasive glioma characterisation. EJNMMI Res. 2018;8:91. doi: 10.1186/s13550-018-0444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.can-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z., Kaiser L., Holzgreve A., Ruf V.C., Suchorska B., Wenter V., et al. Prediction of TERTp-mutation status in IDH-wildtype high-grade gliomas using pre-treatment dynamic [(18)F]FET PET radiomics. Eur J Nucl Med Mol Imaging. 2021;48:4415–4425. doi: 10.1007/s00259-021-05526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Holzgreve A., Unterrainer L.M., Ruf V.C., Quach S., Bartos L.M., et al. Combination of pre-treatment dynamic [(18)F]FET PET radiomics and conventional clinical parameters for the survival stratification in patients with IDH-wildtype glioblastoma. Eur J Nucl Med Mol Imaging. 2022 doi: 10.1007/s00259-022-05988-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwanenburg A., Vallières M., Abdalah M.A., Aerts H.J.W.L., Andrearczyk V., Apte A., et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2006;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 45.Fortin J.-P., Cullen N., Sheline Y.I., Taylor W.D., Aselcioglu I., Cook P.A., et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage. 2018;167:104–120. doi: 10.1016/j.neuroimage.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallat R. Pingouin: statistics in Python. J Open Source Softw. 2018;3:1026. doi: 10.21105/joss.01026. [DOI] [Google Scholar]
- 48.Shrout P., Fleiss J. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 49.Koo T.K., Li M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seabold S., Statsmodels P.J. Proceedings of the Python in Science Conference: SciPy. 2010. Econometric and Statistical Modeling with Python. [Google Scholar]
- 51.Fleiss J.L., Chilton N.W. The measurement of interexaminer agreement on periodontal disease. J Periodontal Res. 1983;18:601–606. doi: 10.1111/j.1600-0765.1983.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 52.Galavis P.E., Hollensen C., Jallow N., Paliwal B., Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010;49:1012–1016. doi: 10.3109/0284186x.2010.498437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical restrictions.