Highlights
-
•
Fluorescently tagged 3DNA can be visualized in vivo non-invasively.
-
•
Folic acid or transferrin conjugated 3DNA increases delivery to pancreatic tumors.
-
•
Non-tumor-bearing organs have significantly less 3DNA detection.
-
•
Folic acid conjugated 3DNA effectively delivers siRNA with high durability.
Keywords: Pancreatic cancer, Nanotherapy, Sirna delivery, Active Targeting
Abstract
Introduction
Standard-of-care systemic chemotherapies for pancreatic ductal adenocarcinoma (PDAC) currently have limited clinical benefits, in addition to causing adverse side effects in many patients. One factor known to contribute to the poor chemotherapy response is the poor drug diffusion into PDAC tumors. Novel treatment methods are therefore drastically needed to improve targeted delivery of treatments. Here, we evaluated the efficacy of the 3DNA® Nanocarrier (3DNA) platform to direct delivery of therapeutics to PDAC tumors in vivo.
Materials and Methods
A panel of PDAC cell lines and a patient tissue microarray were screened for established tumor-specific proteins to identify targeting moieties for active targeting of the 3DNA. NRG mice with or without orthotopic MIA PaCa-2-luciferase PDAC tumors were treated intraperitoneally with 100 μl of fluorescently labeled 3DNA.
Results
Folic acid and transferrin receptors were significantly elevated in PDAC compared to normal pancreas. Accordingly, both folic acid- and transferrin-conjugated 3DNA treatments significantly increased delivery of 3DNA specifically to tumors in comparison to unconjugated 3DNA treatment. In the absence of tumors, there was an increased clearance of both folic acid-conjugated 3DNA and unconjugated 3DNA, compared to the clearance rate in tumor-bearing mice. Lastly, delivery of siLuciferase by folic acid-conjugated 3DNA in an orthotopic model of luciferase-expressing PDAC showed significant and prolonged suppression of luciferase protein expression and activity.
Conclusion
Our study progresses the 3DNA technology as a reliable and effective treatment delivery platform for targeted therapeutic approaches in PDAC.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer-related deaths with a five-year overall survival rate of roughly 11.5% [1,2]. Currently, the only curative therapy is surgical resection for patients with an early stage of disease, complemented with chemotherapy. However, for the majority of patients (>80%), tumors are unresectable, and many present with metastatic disease (50–60%) [3]. At this stage, patients are typically given a combination of systemic chemotherapies, which is the current standard-of-care (SOC). Unfortunately, systemic chemotherapies have been largely unsuccessful, with a median survival rate of less than one year for patients with metastatic disease [4], [5], [6]. Recent improvements with the approval of FOLFIRINOX and nab-paclitaxel plus gemcitabine have offered only modest survival benefits, and at a cost of significantly higher toxicities [4,7,8].
A major obstacle to PDAC treatment efficacy lies in the dense and heterogeneous tumor microenvironment [9], [10], [11]. Desmoplasia in PDAC tumors is linked to chemoresistance through creating a physical barrier and constricting blood vessels, subsequently leading to poor drug delivery [12], [13], [14]. Poor drug delivery is thought to be one reason treatments which show efficacy in experimental studies do not achieve similar potency in clinical trials [15]. Nanotherapeutics (i.e., submicron-sized drug delivery systems ranging from 1 to 100 nm) offer solutions to improve penetration of stroma-rich tumors that previous SOC could not address. Due to the leaky tumor vasculature and poor lymphatic drainage, nanotherapeutics exploit the “enhanced permeability and retention” effect to increase passive therapy delivery by optimizing size and charge of treatments [16,17]. Active targeting can also be achieved with nanotherapeutics through ligand-targeting, localizing treatments to tissues that overexpress the corresponding receptor or antigen [18]. Other benefits to nanotherapeutics include increased solubility, volume distribution and plasma clearance of therapies, while limiting their toxicity [19], [20], [21].
To address the need for improved delivery of treatments to PDAC tumors, we propose the use of the proprietary DNA-based nanotherapeutic, 3DNA® Nanocarrier (3DNA), designed by Code Biotherapeutics (Code Bio, Hatfield, PA). The 3DNA is an assembly of modified double-stranded DNA cross-linked at the center, with unique single-stranded arms that do not anneal due to non-complementary bases [22]. The 3DNA is assembled layer-by-layer to control for size and the number of single-stranded arms (e.g., 36 arms for the 2-layered 3DNA) [22], [23], [24]. These single-stranded arms can be linked to a variety of molecules (e.g., aptamers, peptides, or antibodies) which can be used for active targeting to specific tissues.
A fundamental advantage of the 3DNA platform is that with several single-stranded arms available for conjugations, there is an abundance of opportunities for unique composition as well as size of the payload. Using the 3DNA platform to deliver small molecules directly to tumor cells may increase tumor drug concentrations while minimizing deleterious off-target effects. In addition to small molecules, siRNAs and whole-gene constructs can be attached to the 3DNA platform (Supplemental Fig. 1a). This is significant because the ability to genetically alter tumor cells provides opportunities to target proteins and pathways which have been previously deemed “undruggable” or have had little success with small molecule approaches. The versatility of the 3DNA platform and its ability to deliver siRNAs has the potential to broaden the possible targets for personalized therapies, an avenue that in pancreatic cancer treatment has not had much success to date [5,[25], [26], [27], [28], [29], [30]]. Herein, we describe the optimization of 3DNA for direct delivery to PDAC tumors by targeting folic acid and transferrin receptors found to be significantly upregulated in PDAC. We demonstrate a significant increase and specific uptake of targeted 3DNA by PDAC tumors, in addition to robust and durable delivery of functional siRNA to PDAC tumors by folic acid-conjugated 3DNA.
Materials and methods
Tissue microarray
A tissue microarray of PDAC and normal pancreas samples were constructed using formalin-fixed, paraffin-embedded samples from patients at Thomas Jefferson University Hospital (Philadelphia, PA). Patients consented to the Institutional Review Board approved protocol. Samples were deidentified before authors’ acquisition and analysis. This tissue microarray was originally published in Brown, et al. 2022 [31].
Adjacent serial sections of tissue microarrays were stained for transferrin, folic acid, and epidermal growth factor receptors via immunohistochemistry and scored by pathologist WJ. Scoring was defined on a 2+ (strong staining) to 0 (low-no staining) scale.
Cell lines
HPNE, MIA PaCa-2, PANC-1, Hs766T, and KB cells were obtained from ATCC (Manassas, VA). Conditionally reprogramed PDAC cells (4671-T-CRC) were a gift from Dr. Rosalie Sears from Oregon Health & Science University. 4671-T cells were cultured in 3:1 DMEM: F12 medium supplemented with 5% FBS, 0.4 μg/mL hydrocortisal, 5 μg/mL insulin, 8.4 ng/mL cholera toxin, 10 ng/mL EGF, 24 μg/mL adenine, 0.25 μg/mL amphotericin B, 1X primocin, and 1X ROCK inhibitor. KB cells were cultured in RPMI folic acid-free media supplemented with 10% FBS and 1% penicillin-streptomycin. All other cell lines were cultured in DMEM medium supplemented with 10% FBS, 1% l-glutamine and 1% penicillin-streptomycin. All cells were incubated at 37 °C and 5% CO2, as recommended. Cells were STR authenticated via short tandem repeat analysis and mycoplasma-tested monthly using PCR based mycoplasma detection kit (# MP0035, Sigma Aldrich, St. Louis, MO). Cells were passaged twice after thawing and before experimental use.
MIA PaCa-2-luciferase cells were transduced with firefly luciferase viral particles supplemented with 1 μg/mL polybrene (# TR-1003-G, Sigma Aldrich, St. Louis, MO). Viral particles were generously provided by Drs. Scott Waldman and Adam Snook from Thomas Jefferson University. Cells containing firefly luciferase were selected by puromycin (# P8833, Sigma Aldrich, St. Louis, MO) and validated for luciferase activity using ONE-Glo Luciferase Assay System (# E6110, Promega, Madison, WI).
Detection of binding and internalization of 3DNA to cells, in vitro
KB cells were plated at 5000 cells per well in a 96-well plate and used immediately for live cell staining. Fluorescent (Alexa-647) 3DNA reagents with and without FA-oligo conjugate hybridized were prepared in cell culture media with Hoechst dye for nuclei staining. 3DNA reagents were added to KB cells with and without 0.5 µM LysoSensor™ Green DND-189 (# L7535, Thermo Fisher Scientific, Waltham, MA) and incubated for 30 and 15 min at 37 °C. Next, the plate was centrifuged, staining reagents were aspirated, and the cells were washed with 1X phosphate buffered saline (PBS) and spun again. Cells were fixed in 2% paraformaldehyde, washed, and centrifuged again. Finally, cells were washed two more times with 1X PBS and then transferred to imaging 96-well plate for visualization with the BioTek Cytation 5 instrument.
Immunoblot analysis
Protein was extracted from cells via ice cold RIPA buffer (# sc-24948A, Santa Cruz Biotechnology Inc., Dallas, TX) supplemented with protease inhibitors (# 78,430, Life Technologies Corp, Carlsbad, CA). Lysates were immunoblotted and membranes were scanned with ChemiDoc Imaging System (# 17,001,402, BioRad, Hurcules, CA). Immunoblots were blocked with Intercept® (TBS) Blocking Buffer (# 927–60,003, LI-COR Biosciences, Lincoln, NE) for one hour. Primary antibodies used were epidermal growth factor receptor (1:1000, # 4267S, Cell Signaling Technology, Danvers, MA), transferrin receptor (1:1000, # 136,800, Life Technologies Corp, Carlsbad, CA), folate receptor α (1:1000, # PA5–27,465, Thermo Fisher Scientific, Waltham, MA), β-actin (1:5000, # AM4302, Thermo Fisher Scientific, Waltham, MA), followed by LI-COR IRDye secondary antibodies IRDye 800CW Goat anti-Rabbit IgG (1:10,000, # 926–32,211, LI-COR, Lincoln, NE) or IRDye 680RD Goat anti-Mouse IgG (1:20,000, # 926–68,070, LI-COR, Lincoln, NE) diluted in blocking buffer. Immunoblots were incubated with primary antibodies overnight and secondary antibodies for one hour.
Luciferase activity analysis, in vitro
MIA PaCa-2-luciferase cells were transfected with 30 nM of oligonucleotides and Lipofectamine 2000 (# 11,668,019, Thermo Fisher Scientific, Waltham, MA) according to manufacturer's instructions. The next day, cells were counted and 1000 cells/ siRNA treatment were plated in triplicate in white bottom plates with 100 μL of media. After 24 h, 100 μL of One-Glo Luciferin (# E6110, Promega, Madison, WI) was added for 3 min prior to the plate being measured for bioluminescence with the GloMax Plate Reader (# GM300, Promega, Madison, WI).
Animal studies
All mouse protocols were approved by the Thomas Jefferson University or the Oregon Health & Science University Institutional Animal Care Regulations and Use Committee.
Six-nine-week-old NRG mice (mixed male and female) were generously provided by Drs. Scott Waldman and Adam Snook from Thomas Jefferson University, with the exception of mice used for siRNA delivery assessment which were purchased from The Jackson Laboratory (# 007,799, JAX, Bar Harbor, ME). Mice were randomized into experimental arms. For all in vivo targeting experiments, three mice per experimental arm were used. For siRNA delivery in vivo experiments, five mice per experimental arm were used. During experimental use, mice were exposed to 3% isoflurane for anesthesia purposes.
Mice were housed five mice per cage in 70°F and 30–70% humidity. Light cycle was kept at 12 h on, 12 h off. Bedding and PicoLab Mouse Diet 20 (5058) were replaced every two weeks. Mice had access to water via automatic watering system.
Orthotopic PDAC mouse model
Using orthotopic survival surgery, 1E6 MIA PaCa-2-luciferase cells were injected directly into the tail of the pancreas of 6–9-week-old NRG mice (mixed male and female). Injection of cells were 50 µL in 1:1 cold 1X PBS: cold Matrigel. After injection, needle was exchanged with a cotton swab to eliminate leakage of cells. The peritoneal cavity was closed using absorbable vicryl rapide sutures (# VR834, Mckesson, Irving, TX), and the skin was closed using wound clips (# 12,022–09, Fine Science Tools, Foster City, CA). Mice were injected with 0.1 mg/kg buprenorphine for pain maintenance and monitored every day for a week for any signs of stress.
To monitor tumor volume, mice were injected behind the neck subcutaneously with 100 µL of d-luciferin (15 mg/mL; LUCK, Gold Biotechnology, St Louis, MO). Bioluminescence was assessed with the In Vivo Imaging System. Once bioluminescence signals reached ∼1E6 counts, mice were randomized into treatment groups.
3DNA Nanocarrier treatments
3DNA was manufactured by Code Biotherapeutics (Hatfield, PA) in a series of sequential DNA strand hybridization and crosslinking steps [24,32,33]. For targeting and internalization studies, fluorophore-labeled oligos were crosslinked to the 2-layer 3DNA structure, then targeting DNA conjugates were hybridized to the fluorescent 3DNA. Targeting DNA conjugates were prepared by Code Biotherapeutics using click-chemistry to attach dibenzocyclooctyne-modified oligo to Folate-TEG-azide (Berry & Associated, Dexter, MI) or by using LC-SMCC crosslinker to attach amine-oligo to TCEP-reduced antibody via a maleimide attachment to the free thiol group. For siRNA efficacy studies, the 2-layer 3DNA structure was hybridized with modified siRNA and either folic acid-oligo conjugate or antibody-oligo conjugate. The siRNA oligos were purchased from Avecia (Milford, MA) or IDT (Newark, NJ) with bases modified for stability and short DNA extension on sense strand to hybridize 3DNA. All 3DNA treatments were formulated in 1X PBS.
For in vivo treatments, mice received 1 mg/kg 3DNA treatments via 100 μL intraperitoneal injections. Mice were monitored for signs of distress and body weights were monitored to assess for possible toxicity.
Serum and organ collection
Using a glass capillary tube coated with K2EDTA, blood was collected retro-orbitally into an Eppendorf containing 10 μL of K2EDTA. Samples were immediately placed on ice, then centrifuged at 2000 g for 15 min at 4 °C. The top serum layer was transferred to a new Eppendorf.
Organs were harvested and washed in 1X PBS twice before imaging. After imaging, organs were flash frozen in liquid nitrogen.
Serum folic acid detection
Serum was collected as described above. Folic acid levels were quantified using a Folic Acid ELISA Kit (# MET-5068, Cell Biolabs Inc, San Diego, CA), following the manufacturer's protocol.
qPCR detection of 3DNA nanocarrier
Organs were added to 2 mL screw-cap microtubes containing high impact zirconium beads (3.0 mm) and TL buffer (# PD061, Omega, Norcross, GA), then shaken at 4000 RPM for 1–2 min in a Bead Bug Microtube Homogenizer until completely homogenized. Tubes were spun down in a microcentrifuge and the liquid homogenate was transferred to clean microcentrifuge tubes. Plasma and organ homogenates from animals with no 3DNA treatment were spiked with the appropriate 3DNA treatment to prepare standard curves specific for each sample and treatment. qPCR detection of 3DNA in plasma and organ homogenates was completed using primers specific for 3DNA sequences and the Promega GoTaq® Probe qPCR Master Mix (# A6102, Promega, Madison, WI). Each sample was run in triplicate using the Bio-Rad CFX96 Touch Real-Time PCR Detection System instrument and analyzed with CFX Maestro software.
Immunofluorescence histochemistry detection of luciferase in tumors
Tumors were fixed in 4% paraformaldehyde (# NC9288315, Thermo Fisher Scientific, Waltham, MA) overnight, then transferred to 70% EtOH. Tissue blocks and slides were made by the Histopathology Shared Resource Core at OHSU. Two sections of each tumor were deparaffinized and rehydrated prior to antigen retrieval (# H-3300–250, Vector Laboratories, Burlingame, CA). Tissues were blocked with 5% BSA, 2% normal goat serum in TBS-T for 1 hour and then stained with luciferase primary antibody (1:200, # ab185924, Abcam, Cambridge, UK) in blocking solution overnight at 4 °C. After washing slides in TBST, tissues were incubated in goat anti-rabbit-Alexa 488 secondary antibody (1:500, # ab150077, Abcam, Cambridge, UK) in PBS for 1 hour. Tissues were then washed with PBS, after which autofluorescence was quenched using the TrueVIEW reagent (# SP-8400, Vector Laboratories, Burlingame, CA). Tissues were then stained with DAPI and mounted with Vectashield mounting medium provided in the TrueVIEW quenching kit. Slides were scanned by the Advanced Light Microscopy Core at OHSU. ZEN Digital Imaging for Light Microscopy (RRID:SCR_013672) was used to quantify the mean fluorescent intensity for each section. Values were normalized to mean fluorescent intensity of sections not stained with either luciferase primary antibody or goat anti-rabbit-Alexa 488 secondary antibody.
Statistical analysis
All analyses were performed using GraphPad Prism software (San Diego, CA). Specific test used is indicated in the legend of each figure. Briefly, statistical analysis of in vitro studies was calculated using either multiple Student's two-sample t-test or a one-way ANOVA. Relationship between fluorescence and concentration of Alexa-750–3DNA was assessed by Pearson correlation. Statistical analysis of in vivo animal studies was calculated using a Student's two-sample t-test to assess clearance, or a two-way ANOVA when assessing organ qPCR data. Tumor volumes were statistically assessed with a Student's two-sample t-test. Lastly, luciferase activity in tumor-bearing mice over time was assessed by linear regression followed by the extra sum-of-squares F test comparison model. All data are expressed as mean ± standard error of mean.
Results
Folic acid and transferrin receptors are highly expressed in pancreatic ductal adenocarcinoma
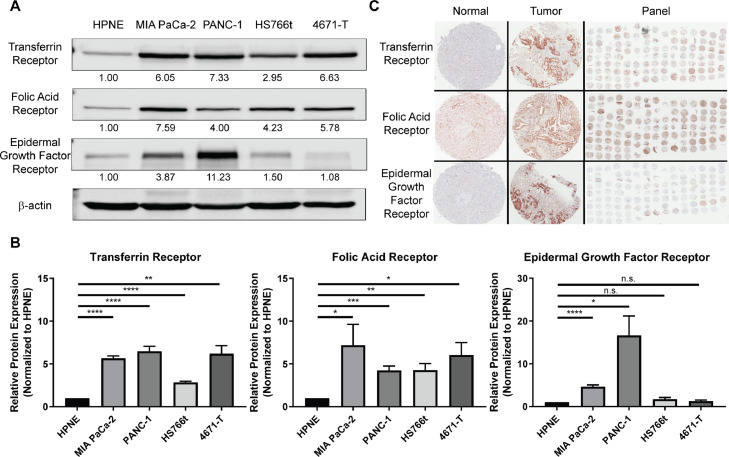
To improve therapeutic delivery, we sought to identify receptors upregulated in PDAC cells to exploit for active targeting. Since transferrin, folic acid, and epidermal growth factor receptors are common nanoparticle targets due to their high expression in a number of solid tumors, relative to normal tissues [20,[34], [35], [36]], we quantified and evaluated these receptors in a panel of PDAC cell lines. Expression of transferrin and folic acid receptors were significantly higher in all four PDAC cell lines analyzed, in comparison to the normal immortalized pancreatic ductal cell line HPNE (ranging from -3–7-fold, p<0.01–0.0001 and -4–8-fold, p<0.05–0.001, respectively). Epidermal growth factor receptor expression was significantly higher in only two PDAC cells lines in the panel (MIA PaCa-2: -4-fold, p<0.0001 in; PANC-1: -16-fold, p<0.05 in; Fig. 1a.b). Moreover, a tissue microarray of patient PDAC samples showed positive staining for transferrin receptor in 100% of PDAC samples (n = 77; 2+: 64%, 1+: 36%), positive staining for folic acid receptor in 100% of PDAC samples (n = 80; 2+: 54%, 1+: 46%), and positive staining for epidermal growth factor receptor in 42% of cases (n = 72; 2+: 4%, 1+: 38%; Fig. 1c, Supplemental Table 1). Importantly, in normal pancreas tissue, transferrin and epidermal growth factor receptor expression was not detected, and only low levels of folic acid receptor was detected (Supplemental Table 1). These results prompted further pursuit of transferrin and folic acid receptors for active targeting of 3DNA to PDAC tumors, by conjugating folic acid (FA) and transferrin (Tf) to the 3DNA.
Fig. 1.
Transferrin and folic acid receptors are highly expressed in pancreatic ductal adenocarcinoma. A) Representative Western blot analysis of transferrin receptor, folic acid receptor, and epidermal growth factor receptor from whole protein lysates of the normal immortalized pancreatic ductal cell line HPNE and a panel of PDAC cells (MIA PaCa-2, PANC-1, HS766t, 4671-T). Cell line 4671-T-CRC (4671-T) is a conditionally reprogrammed cell line from a primary PDAC tumor generated and gifted by Dr. Rosalie Sears from Oregon Health & Sciences University. Protein quantification depicted under each band was normalized to β-actin and relative to HPNE. B) Quantification of A. Average of relative protein expression ± standard error of the mean (n = 4). Statistical analysis was calculated using multiple Student's two-sample t-test. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, n.s. not significant. C) Representative immunohistochemistry staining for transferrin receptor, folic acid receptor, and epidermal growth factor receptor of patient tissues, both normal and PDAC, as well as the full panel of tissues.
Folic acid-conjugated 3DNA allows for targeting and internalization of 3DNA in folic acid receptor-positive cells
We then tested the ability of the 3DNA to specifically bind and internalize into tumor cells by means of folic acid receptor targeting. Unconjugated 3DNA (UN-3DNA) and folic acid-conjugated 3DNA (FA-3DNA) were incubated with folic acid receptor-positive KB cells in FA-depleted media, a model for testing surface binding and internalization of FA-based treatments [37], [38], [39], [40]. For visualization, both UN-3DNA and FA-3DNA were conjugated with Alexa-647 fluorophores. Imaging revealed that only FA-3DNA bound to the KB cell surface as indicated by the co-localization of Alexa-647 and Hoechst nuclear staining (Supplemental Fig. 2a). To visualize internalization of the 3DNA, this experiment was repeated in the presence of LysoSensor™ Green DND-189, a dye that fluoresces in acidic environments such as late endosomes and lysosomes. Co-localization of Alexa-647 and LysoSensor was observed in the majority of cells treated with FA-3DNA (Supplemental Fig. 2b). Few cells showed only membrane localization of Alexa-647, suggestive of binding but not internalization. These data indicate that the FA conjugation to the 3DNA platform facilitated binding and internalization of the 3DNA into the cancer cells.
3DNA targets primary tumors in an orthotopic model of PDAC
Given that conjugating FA to 3DNA allowed binding and internalization in vitro, we next aimed to determine whether targeting transferrin or folic acid receptors improves binding and internalization of 3DNA into PDAC tumors, in vivo. Utilizing an orthotopic in vivo model of PDAC, MIA PaCa-2-luciferase tumor-bearing mice were treated with Alexa-750-conjugated 3DNA treatments (Alexa-750–3DNA; Supplemental Fig. 1b). Importantly, we validated that there was no cross-reactivity between the fluorescence of the Alexa-750–3DNA and the luciferase bioluminescence of the tumor cells (Supplemental Fig. 3a). Moreover, we showed that fluorescence of the Alexa-750–3DNA detected by the In Vivo Imaging Software (IVIS) was correlated with the concentration of Alexa-750–3DNA (R2=0.9239, p = 0.0022; Supplemental Fig. 3b).
Since the high FA levels found in many rodent laboratory chows has been shown to outcompete and interfere with responses to FA-conjugated drugs by elevating FA serum levels well beyond the human range (i.e., 6–37 nM) [41,42], we assessed the FA serum levels of mice on our standard laboratory chow. We found mouse serum FA concentrations within the human range, therefore not requiring the use of a FA-free diet for testing FA-3DNA treatments in this model (Supplemental Fig. 3c).
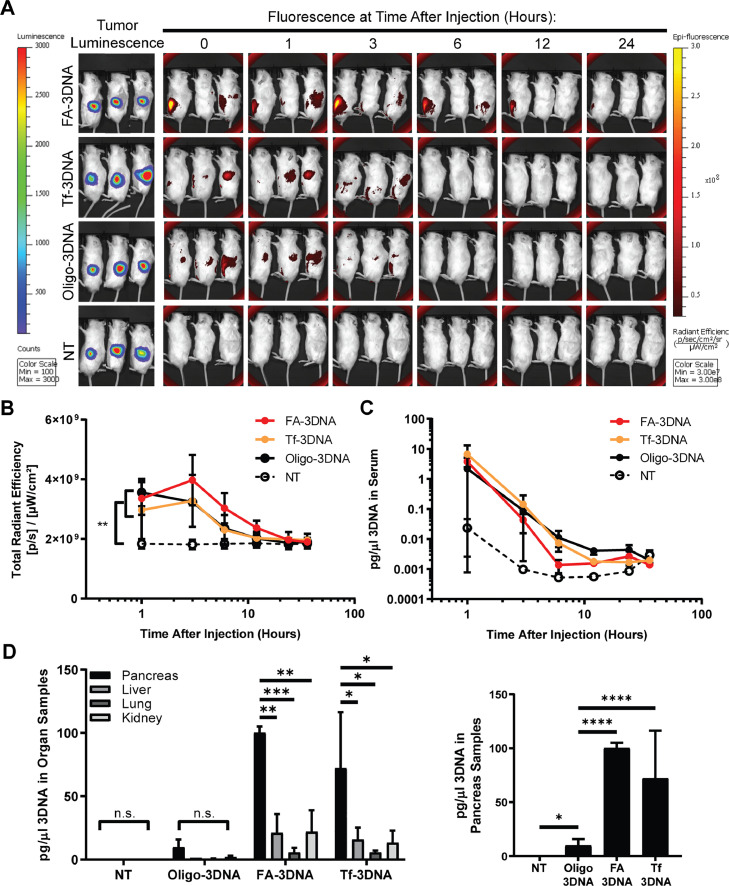
Orthotopic PDAC tumor-bearing mice were treated with Alexa-750–3DNA conjugated with either folic acid (FA-3DNA), transferrin (Tf-3DNA), or an oligonucleotide control (Oligo-3DNA) which contains only the single-stranded DNA extension used to attach targeting moieties. An additional group received no treatment (NT). Mice were imaged at several time points between 0 and 36 h after treatment for fluorescent detection of the 3DNA (Fig. 2a). Analysis of fluorescence levels during the time course showed no significant differences between the 3DNA treatment groups, but each had significant increase over the NT group (Fig. 2b). Using primers designed against the 3DNA, qPCR detection and precise quantification of 3DNA in serum showed no significant difference in the amount of 3DNA in circulation over time between the different treatment groups (Fig. 2c).
Fig. 2.
FA-3DNA and Tf-3DNA increase targeting of 3DNA to PDAC tumors, in vivo. A) Images of tumor-bearing mice over time after treatment with folic acid-conjugated 3DNA (FA-3DNA), transferrin-conjugated 3DNA (Tf-3DNA), oligo control 3DNA (Oligo-3DNA), or no treatment (NT). Images taken at 36 h are not shown. Bioluminescence of tumors are shown in the left panels. 3DNA was monitored over time by fluorescence, as seen in the right panels. Bioluminescence and fluorescence intensity for each image is to the same scale. B) Quantification of the fluorescence depicted in A. C) qPCR detection of 3DNA in vascular circulation at indicated time points. Serum was extracted from blood collected at time points and used for 3DNA qPCR quantification. Values were normalized to 18S. Statistical analysis of time course data presented in B and C were calculated using a Student's two-sample t-test of the area under the curve. **p<0.01. D) qPCR detection of 3DNA in homogenized organs 3 h after re-injection of the same treatment. Right graph depicts only pancreas samples. Values were normalized to 18S. Statistical analysis was calculated using two-way ANOVA. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, n.s. not significant. n = 3 mice for each experimental arm.
Mice were then re-injected with the same treatment 24 h after 3DNA clearance, as determined by fluorescence signal equaling that of NT mice. For optimal 3DNA detection, organs were collected at the time point corresponding to when fluorescence of the 3DNA began to wane in Fig. 2a–c (i.e., 3 h post-treatment). 3DNA detection in organs via qPCR showed a significant increase in 3DNA presence in tumor-bearing pancreas when mice were treated with either FA-3DNA or Tf-3DNA, in comparison to Oligo-3DNA treatment (p<0.0001; Fig. 2d). This strongly indicated that conjugation of a targeting moiety significantly increased delivery of the 3DNA to tumors. Passive delivery of Oligo-3DNA was seen, as indicated by increased 3DNA detection in tumor-bearing pancreas of mice treated with Oligo-3DNA, in comparison to NT mice. Moreover, FA-3DNA and Tf-3DNA detection in the liver, lung, and kidneys were significantly lower than in the tumor-bearing pancreas, while the Oligo-3DNA treatment showed no significant changes in detection across any organs in that treatment group.
3DNA clears quickly without the presence of a tumor
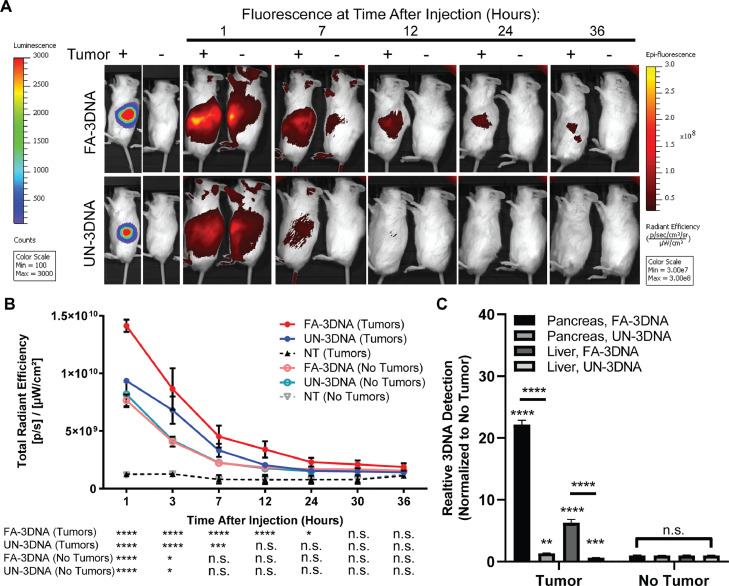
To assess specific delivery to tumor cells in the pancreas, both tumor-bearing and non-tumor-bearing mice were treated with Alexa-750 unconjugated 3DNA (UN-3DNA), FA-3DNA, or NT (images not shown). Mice were imaged at several time points after treatment (Fig. 3a). Defining 3DNA clearance as a fluorescent signal equaling to that of NT mice, we found that in tumor-bearing mice, UN-3DNA cleared in half the time it took for FA-3DNA to clear (p<0.05; Fig. 3b). In mice without tumors (non-tumor-bearing), FA-3DNA and UN-3DNA treatments cleared faster compared to their clearance rate in mice with tumors.
Fig. 3.
3DNA clears quickly without the presence of a tumor. A) Representative images of both tumor- and non-tumor-bearing mice over time after treatment with folic acid-conjugated 3DNA (FA-3DNA) or unconjugated 3DNA (UN-3DNA). Mice not treated (NT) are not pictured. Tumors are shown by bioluminescence in the left panels. 3DNA was monitored over time by fluorescence, as seen in the right panels. B) Quantification of the fluorescence depicted in A. Statistical analysis was calculated using a Student's two-sample t-test for each treatment at each time point in comparison to NT mice. Significance is indicated at the bottom of the graph. C) qPCR detection of 3DNA in homogenized organs 7 h after re-injection of the same treatment. Values were normalized to 18S. Statistical analysis was calculated using a two-way ANOVA. Brackets mark significance between organs and treatments. Bars mark significance between treatments of the same organ. Asterisks directly above of bars indicate significance between tumor-bearing organs and non-tumor-bearing organs. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, n.s. not significant. n = 3 mice for each experimental arm. Bioluminescence and fluorescence intensity for each image is to the same scale.
After 24 h post-clearance, mice were re-injected with the same 3DNA treatment, and organs were collected at the time point corresponding to the approximate half-life of 3DNA in Fig. 3a,b (i.e., 7 h post-treatment). In tumor-bearing mice, there was a higher accumulation of FA-3DNA in both pancreas and liver samples, in comparison to UN-3DNA samples, as shown by qPCR quantification (Fig. 3c). Additionally, both FA-3DNA and UN-3DNA treatments showed significantly higher quantification of 3DNA detected in the pancreas and livers of tumor-bearing mice, in comparison to the non-tumor-bearing mice (Fig. 3c). Altogether, these data validate that the addition of FA targeting moieties allows for accumulation in tumor-bearing pancreas, and that a tumor is required for prolonged presence of the 3DNA in the pancreas and other organs.
Targeted 3DNA successfully delivers siRNA to tumors
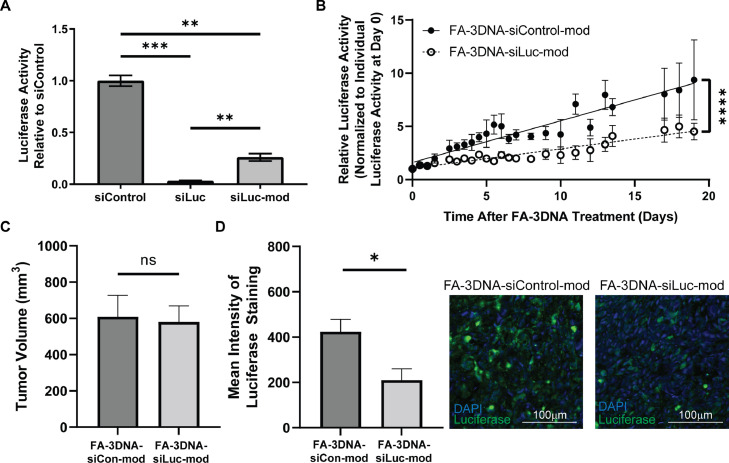
Next, we determined whether 3DNA can efficiently deliver functional siRNA payloads to tumors. Because our in vivo model utilizes PDAC cells exogenously expressing luciferase, we sought to test the efficacy of 3DNA-siLuciferase delivery. To ensure stability of siRNA when in circulation in vivo, the standard ribonucleotide bases were swapped for proprietary chemically modified versions which protect the sugar backbone from ribonuclease attack. We confirmed that the modified siLuciferase was able to significantly decrease luciferase activity of MIA PaCa-2-luciferase cells in comparison to siControl (p = 0.0003), although with a slight loss in potency as compared to unmodified siLuciferase (Fig. 4a).
Fig. 4.
FA-3DNA can deliver functional siRNA to PDAC tumors. A) Relative luciferase activity measured with the One-Glo Luciferase Assay. Averages of bioluminescence counts ± standard error of the mean (n = 3). Statistical analysis was calculated using a one-way ANOVA. B) Luciferase activity overtime observed in mice treated with either FA-3DNA-siControl-mod or FA-3DNA-siLuc-mod, normalized to luciferase activity for each mouse at day 0. Data represents mean relative luciferase activity ± standard error of the mean (n = 5). Statistical analysis was calculated using linear regression model. C) Tumor volumes at the end of the experiment in B (i.e., day 19). Statistical analysis was calculated using a Student's two-sample t-test. D) Averages of mean intensity of luciferase expression ± standard error of the mean, measured by fluorescent immunohistochemistry of tumors 5.5 days after treatment with either FA-3DNA-siControl-mod or FA-3DNA-siLuc-mod. Two sections of the same tumor were used for analysis. Representative images are found to the right of the corresponding bar graph. Statistical analysis was calculated using a Student's two-sample t-test. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, n.s. not significant.
To test the capability of the 3DNA system to effectively deliver the modified siLuciferase to PDAC tumors, orthotopic tumor-bearing mice were treated with either FA-3DNA or Tf-3DNA conjugated with modified siLuciferase (FA-3DNA-siLuc-mod; Tf-3DNA-siLuc-mod) or control siRNA with similar modifications (FA-3DNA-siControl-mod; Tf-3DNA-siControl-mod). Linear regression analysis of luciferase activity over time indicated that FA-3DNA-siLuc-mod treatment significantly reduced luciferase activity compared to FA-3DNA-siControl-mod treatment (p<0.0001, Fig. 4b). Tf-3DNA-siLuc-mod showed decreased luciferase activity in comparison to Tf-3DNA-siControl-mod, although not reaching statistical significance (Supplemental Fig. 4a). Importantly, at the termination of these experiments, there was no difference in tumor volumes between the siControl and siLuciferase groups, indicating that the difference in luciferase activity is a direct consequence of the siRNA efficacy and not differences in tumor growth (Fig. 4c, Supplemental Fig. 4b). This indicates that FA-3DNA delivery of siRNA is highly effective and durable, showing significant decrease in luciferase activity up to day 13. Moreover, 3DNA treatments were well tolerated in mice, as indicated by no significant changes in body weights (Supplemental Fig. 5).
For the FA-3DNA delivery system, the most significant and largest difference in luciferase activity in FA-3DNA-siLuc-mod treated versus FA-3DNA-siControl-mod treated mice was observed at 5.5 days post treatment. Therefore, we repeated this experiment and tumors were assessed at 5.5 days post treatment for luciferase expression via fluorescent immunohistochemistry. We found that FA-3DNA-siLuc-mod treated mice had significantly less luciferase protein expression compared to FA-3DNA-siControl-mod treated tumors (p = 0.0266, Fig. 4d). Taken together, these data show that the FA-3DNA is capable of delivering functional siRNA to PDAC tumors with high durability after a single injection.
Discussion
Nanoparticle therapies provide the potential to increase treatment accumulation in tumors while simultaneously reducing adverse effects [16], [17], [18], [19], [20], [21]. Here, we provide evidence that the 3DNA® Nanocarrier developed by Code Biotherapeutics is capable of selectively targeting PDAC tumors for functional siRNA delivery; thus, holding the possibility of addressing the need for improved treatment delivery and efficacy.
We were able to identify that the expression of transferrin and folic acid receptors were significantly elevated in PDAC compared to normal pancreas (Fig. 1), and that this could be exploited for targeting of the 3DNA to tumors. Both FA-3DNA and Tf-3DNA increased 3DNA accumulation in PDAC tumors in comparison to an Oligo-3DNA control (Fig. 2), demonstrating the ability to improve delivery of 3DNA to PDAC tumors by targeting overexpressed receptors on the PDAC cell surface. We also showed that FA-3DNA accumulated in tumor-bearing pancreas more than normal pancreas and other organ sites (e.g., liver, lung, kidney; Fig. 3), supporting previous work in which investigators assessing 3DNA biodistribution found 3DNA did not accumulate in the liver [32]. Moreover, 3DNA treatments were well tolerated in our model, as indicated by no significant changes in behavior or body weight. This supplements existing 3DNA safety studies where mouse and rabbit models showed no toxicity associated with chronic treatment of various 3DNA formulations as measured by behavior, body weight, and cytokine activation [24,33,43]. Additionally, investigators showed no integration of 3DNA into the host genome, eliminating concerns of unintentional 3DNA genome integration [43]. These data suggest that actively targeting PDAC tumors with FA-3DNA is not only safe, but increases delivery of the therapy to tumors while minimizing non-specific effects, due to little 3DNA accumulation in non-tumor-bearing organs. Assessment of additional non-tumor-bearing organs (e.g., brain, heart, intestines, etc.), as well as elucidating the mechanism of uptake and the uptake rate would be beneficial for fully characterizing and understanding this delivery system.
We demonstrated for the first time that the FA-3DNA can deliver functional siRNA to PDAC tumors, supplementing published work showing FA-3DNA delivery of siRNA against Human Antigen R (HuR, ELAVL1) to ovarian tumors [24]. Utilizing the FA-3DNA for siRNA delivery, we were able to reduce the activity of the exogenously expressed luciferase in MIA PaCa-2-luciferase tumors (Fig. 4). A single treatment of FA-3DNA-siLuc-mod was capable of decreasing luciferase activity for up to 13 days, demonstrating robust durability of the treatment. In all, these data suggest that 3DNA is a promising platform to deliver therapeutics directly to PDAC tumors.
A limitation of these studies is that they were performed in an immunocompromised model. Due to this, we were unable to address the influence of the immune system on 3DNA delivery and vice versa. Of note, others have determined 3DNA treatments in immunocompetent models do not induce immunogenicity [33,43], yet they did not assess whether there were immune responses to the siRNAs being delivered by the 3DNA platform. Performing these studies in immunocompetent models would provide insight on whether the modified siRNAs are capable of inducing an immune reaction, in addition to determining if the targeting ability of the 3DNA is impacted by an intact immune system.
Delivery of siRNA via FA-3DNA holds potential for inhibiting promising targets for which small molecule inhibitors either have had minimal clinical success or do not yet exist. The Know Your Tumor initiative and others have demonstrated that up to 50% of patients with PDAC have actionable mutations (e.g., AKT, MEK/ERK) [44], [45], [46]. Small molecule approaches of targeting these activated pathways in PDAC have shown limited therapeutic success [5,[25], [26], [27], [28], [29], [30]]. Use of FA-3DNA to deliver either siRNA against oncogenic targets (e.g., KRAS) or whole genes to overcome loss of tumor suppressors (e.g., TP53) could expand therapeutic possibilities and target “undruggable” proteins through use of genetic manipulation. Additionally, this platform could be utilized to enhance and personalize immuno-oncologic therapies, such as delivery of immune checkpoint inhibitors or neoantigens [47]. With the 2-layer 3DNA having 36 single-stranded arms available for conjugations, multiple payloads targeting different oncogenic pathways can be delivered. The clinical relevance of delivering various payloads is supported by the Know Your Tumor initiative, which data shows on average, patient tumors have four different oncogenic driver mutations. Thus, using the 3DNA platform, treatments can be tailored to a patient's tumor genomic profile by conjugating multiple siRNAs or whole genes that would target the mutational landscape, opposed to a singular pathway. Inhibiting multiple oncogenic pathways may help increase effectiveness while minimizing the chance of compensation or resistance by the tumor [48], [49], [50], [51], [52], ultimately providing better therapeutic outcomes.
Consent
Tissue microarray of PDAC and normal pancreas samples from patients at Thomas Jefferson University Hospital (Philadelphia, PA) was created with patients consent to the Institutional Review Board approved protocol. Samples were deidentified prior to authors’ acquisition and analysis. No identifying details are reported in this manuscript. This tissue microarray was originally published in Brown, et al. 2022.
Sources of funding
This work was supported by Code Biotherapeutics. Additionally, this work was supported by the National Institutes of Health (R21 CA263996 to J.R.B.; R01 CA212600 to J.R.B.; U01 CA224012 to J.R.B.; R37 CA227865 to J.R.B.; 15–90–25-BROD to J.R.B.) as well as the AACR-PanCAN RAN Grant to J.R.B.. Additional funding was supported by grants from the Brenden-Colson Center for Pancreatic Care and Knight Cancer Institute at Oregon Health & Science University, the Hirshberg Foundation, and the Sarah Parvin Foundation.
CRediT authorship contribution statement
Grace A. McCarthy: Conceptualization, Investigation, Validation, Formal analysis, Resources, Data curation, Writing – original draft, Writing – review & editing. Aditi Jain: Resources, Data curation, Writing – original draft, Investigation. Roberto Di Niro: Investigation, Resources, Writing – original draft. Christopher W. Schultz: Investigation, Resources, Writing – original draft. Wei Jiang: Investigation, Resources, Formal analysis. Charles J Yeo: Investigation, Resources, Writing – original draft. Jessica Bowers: Resources, Writing – original draft, Methodology, Formal analysis, Investigation. Jennifer Finan: Investigation, Resources, Writing – original draft. Kelly Rhodes: Methodology, Formal analysis, Investigation, Resources, Writing – original draft. Lou Casta: Methodology, Formal analysis, Investigation, Resources, Writing – original draft. Vivi Hou: Investigation, Resources, Writing – original draft. Anthony Stefanoni: Methodology, Formal analysis, Investigation, Resources, Writing – original draft. Samantha Z. Brown: Investigation, Resources, Writing – original draft. Avinoam Nevler: Investigation, Resources, Writing – original draft. Lebaron C. Agostini: Investigation, Resources, Writing – original draft. Lori Getts: Methodology, Formal analysis, Investigation, Resources, Writing – original draft. Robert Getts: Conceptualization, Investigation, Resources, Funding acquisition, Supervision, Writing – original draft. Jonathan R. Brody: Conceptualization, Investigation, Resources, Funding acquisition, Supervision, Writing – original draft.
Declaration of Competing Interest
All other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. This work and G.A.M. were supported Code Biotherapeutics through a Sponsored Research Agreement. Materials for 3DNA were supplied by Code Biotherapeutics. A.S., S.Z.B., L.G., and R.G. are all employees of Code Biotherapeutics.
Acknowledgments
We are grateful for the technical help and support from the Advanced Light Microscopy Shared Resource at Oregon Health & Science University. Also, thank you to Dr. Rosalie Sears from Oregon Health & Science University for generously providing the patient derived, conditionally reprogrammed cell line 4671-T-CRC.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101662.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N., Noone A.M., Krapcho M., Miller D., Brest A., Yu M., Ruhl J, Tatalovich Z., Mariotto A., Lewis D.R., Chen H.S., Feuer E.J. 2021. SEER Cancer Statistics Review, 1975-2018. [Google Scholar]
- 3.Howlader N N.A., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., Chen H.S., Feuer E.J., Cronin K.A. SEER Cancer Stat. Rev. 2019:1975–2016. based on November 2018 SEER data submission, posted to the SEER web site, AprilNational Cancer Institute. Bethesda, MD. [Google Scholar]
- 4.Conroy T., et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 2018;379(25):2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 5.Deplanque G., et al. A randomized, placebo-controlled phase III trial of masitinib plus gemcitabine in the treatment of advanced pancreatic cancer. Ann. Oncol. 2015;26(6):1194–1200. doi: 10.1093/annonc/mdv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore M.J., et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the national cancer institute of canada clinical trials group. J. Clin. Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 7.Von Hoff D.D., et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conroy T., et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364(19):1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 9.Waghray M., et al. Deciphering the role of stroma in pancreatic cancer. Curr. Opin. Gastroenterol. 2013;29(5):537–543. doi: 10.1097/MOG.0b013e328363affe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang C., et al. Complex roles of the stroma in the intrinsic resistance to gemcitabine in pancreatic cancer: where we are and where we are going. Exp. Mol. Med. 2017;49(12):e406. doi: 10.1038/emm.2017.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahadevan D., Von Hoff D.D. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol. Cancer Ther. 2007;6(4):1186–1197. doi: 10.1158/1535-7163.MCT-06-0686. [DOI] [PubMed] [Google Scholar]
- 12.Schober M., et al. Desmoplasia and chemoresistance in pancreatic cancer. Cancers. 2014;6(4):2137–2154. doi: 10.3390/cancers6042137. Basel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minchinton A.I., Tannock I.F. Drug penetration in solid tumours. Nat. Rev. Cancer. 2006;6(8):583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 14.Olive K.P., et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali E.S., et al. Targeting cancer cells with nanotherapeutics and nanodiagnostics: current status and future perspectives. Semin. Cancer Biol. 2021;69:52–68. doi: 10.1016/j.semcancer.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Cabral H., et al. Accumulation of sub-100nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 2011;6(12):815–823. doi: 10.1038/nnano.2011.166. [DOI] [PubMed] [Google Scholar]
- 17.Lieleg O., Baumgartel R.M., Bausch A.R. Selective filtering of particles by the extracellular matrix: an electrostatic bandpass. Biophys. J. 2009;97(6):1569–1577. doi: 10.1016/j.bpj.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peer D., et al. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007;2(12):751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 19.Awasthi R., et al. Nanoparticles in Cancer Treatment: opportunities and Obstacles. Curr. Drug Targets. 2018;19(14):1696–1709. doi: 10.2174/1389450119666180326122831. [DOI] [PubMed] [Google Scholar]
- 20.Dawidczyk C.M., et al. State-of-the-art in design rules for drug delivery platforms: lessons learned from FDA-approved nanomedicines. J Control Release. 2014;187:133–144. doi: 10.1016/j.jconrel.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sparreboom A., et al. Comparative preclinical and clinical pharmacokinetics of a cremophor-free, nanoparticle albumin-bound paclitaxel (ABI-007) and paclitaxel formulated in Cremophor (Taxol) Clin. Cancer Res. 2005;11(11):4136–4143. doi: 10.1158/1078-0432.CCR-04-2291. [DOI] [PubMed] [Google Scholar]
- 22.Nilsen T.W., Grayzel J., Prensky W. Dendritic nucleic acid structures. J. Theor. Biol. 1997;187(2):273–284. doi: 10.1006/jtbi.1997.0446. [DOI] [PubMed] [Google Scholar]
- 23.Gerhart J., et al. Antibody-Conjugated, DNA-Based Nanocarriers Intercalated with Doxorubicin Eliminate Myofibroblasts in Explants of Human Lens Tissue. J. Pharmacol. Exp. Ther. 2017;361(1):60–67. doi: 10.1124/jpet.116.239079. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y.H., et al. Delivery of therapeutics targeting the mRNA-binding protein HuR Using 3DNA nanocarriers suppresses ovarian tumor growth. Cancer Res. 2016;76(6):1549–1559. doi: 10.1158/0008-5472.CAN-15-2073. [DOI] [PubMed] [Google Scholar]
- 25.Fong C.Y.K., et al. Up-to-date tailored systemic treatment in pancreatic ductal adenocarcinoma. Gastroenterol Res Pract. 2019;2019 doi: 10.1155/2019/7135437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottaiano A., et al. Gemcitabine mono-therapy versus gemcitabine plus targeted therapy in advanced pancreatic cancer: a meta-analysis of randomized phase III trials. Acta Oncol. 2017;56(3):377–383. doi: 10.1080/0284186X.2017.1288922. [DOI] [PubMed] [Google Scholar]
- 27.Infante J.R., et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur. J. Cancer. 2014;50(12):2072–2081. doi: 10.1016/j.ejca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 28.Chung V., et al. Effect of selumetinib and MK-2206vs oxaliplatin and fluorouracil in patients with metastatic pancreatic cancer after prior therapy: SWOG S1115 Study randomized clinical trial. JAMA Oncol. 2017;3(4):516–522. doi: 10.1001/jamaoncol.2016.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodoky G., et al. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest. New Drugs. 2012;30(3):1216–1223. doi: 10.1007/s10637-011-9687-4. [DOI] [PubMed] [Google Scholar]
- 30.Di Federico A., et al. Hacking Pancreatic Cancer: present and Future of Personalized Medicine. Pharmaceuticals. 2021;14(7) doi: 10.3390/ph14070677. Basel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown S.Z., et al. The RNA-Binding Protein HuR Posttranscriptionally Regulates the Protumorigenic Activator YAP1 in Pancreatic Ductal Adenocarcinoma. Mol. Cell. Biol. 2022 doi: 10.1128/mcb.00018-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roki N., et al. Unprecedently high targeting specificity toward lung ICAM-1 using 3DNA nanocarriers. J Control Release. 2019;305:41–49. doi: 10.1016/j.jconrel.2019.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerhart J., et al. Depletion of Myo/Nog Cells in the Lens mitigates posterior capsule opacification in rabbits. Invest. Ophthalmol. Vis. Sci. 2019;60(6):1813–1823. doi: 10.1167/iovs.19-26713. [DOI] [PubMed] [Google Scholar]
- 34.Scaranti M., et al. Exploiting the folate receptor alpha in oncology. Nat. Rev. Clin. Oncol. 2020;17(6):349–359. doi: 10.1038/s41571-020-0339-5. [DOI] [PubMed] [Google Scholar]
- 35.Daniels T.R., et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta. 2012;1820(3):291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mamot C., et al. Epidermal growth factor receptor (EGFR)-targeted immunoliposomes mediate specific and efficient drug delivery to EGFR- and EGFRvIII-overexpressing tumor cells. Cancer Res. 2003;63(12):3154–3161. [PubMed] [Google Scholar]
- 37.Siwowska K., et al. Folate receptor-positive gynecological cancer cells: in vitro and in vivo characterization. Pharmaceuticals. 2017;10(3) doi: 10.3390/ph10030072. Basel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathias C.J., et al. Indium-111-DTPA-folate as a potential folate-receptor-targeted radiopharmaceutical. J. Nucl. Med. 1998;39(9):1579–1585. [PubMed] [Google Scholar]
- 39.Lee R.J., Low P.S. Delivery of liposomes into cultured KB cells via folate receptor-mediated endocytosis. J. Biol. Chem. 1994;269(5):3198–3204. [PubMed] [Google Scholar]
- 40.Ab O., et al. IMGN853, a folate receptor-alpha (fralpha)-targeting antibody-drug conjugate, exhibits potent targeted antitumor activity against fralpha-expressing tumors. Mol. Cancer Ther. 2015;14(7):1605–1613. doi: 10.1158/1535-7163.MCT-14-1095. [DOI] [PubMed] [Google Scholar]
- 41.Mathias C.J., et al. Tumor-selective radiopharmaceutical targeting via receptor-mediated endocytosis of gallium-67-deferoxamine-folate. J. Nucl. Med. 1996;37(6):1003–1008. [PubMed] [Google Scholar]
- 42.Leamon C.P., et al. Impact of high and low folate diets on tissue folate receptor levels and antitumor responses toward folate-drug conjugates. J. Pharmacol. Exp. Ther. 2008;327(3):918–925. doi: 10.1124/jpet.108.143206. [DOI] [PubMed] [Google Scholar]
- 43.Robert C., Getts J.B. Platform technology – the 3DNA® platform for targeted drug delivery. Drug Dev. Deliv. 2016 November/December 2016. [Google Scholar]
- 44.Pishvaian M.J., et al. Molecular profiling of patients with pancreatic cancer: initial results from the know your tumor initiative. Clin. Cancer Res. 2018;24(20):5018–5027. doi: 10.1158/1078-0432.CCR-18-0531. [DOI] [PubMed] [Google Scholar]
- 45.Aung K.L., et al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the compass trial. Clin. Cancer Res. 2018;24(6):1344–1354. doi: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chantrill L.A., et al. Precision medicine for advanced pancreas cancer: the individualized molecular pancreatic cancer therapy (IMPaCT) trial. Clin. Cancer Res. 2015;21(9):2029–2037. doi: 10.1158/1078-0432.CCR-15-0426. [DOI] [PubMed] [Google Scholar]
- 47.Di Federico A., et al. Immunotherapy in pancreatic cancer: why do we keep failing? a focus on tumor immune microenvironment, predictive biomarkers and treatment outcomes. Cancers. 2022;14(10) doi: 10.3390/cancers14102429. Basel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhir T., et al. Abemaciclib is effective against pancreatic cancer cells and synergizes with HuR and YAP1 inhibition. Mol. Cancer Res. 2019;17(10):2029–2041. doi: 10.1158/1541-7786.MCR-19-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gout J., et al. Synergistic targeting and resistance to PARP inhibition in DNA damage repair-deficient pancreatic cancer. Gut. 2021;70(4):743–760. doi: 10.1136/gutjnl-2019-319970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agostini L.C., et al. Combined targeting of PARG and wee1 causes decreased cell survival and DNA damage in an S-phase-dependent manner. Mol. Cancer Res. 2021;19(2):207–214. doi: 10.1158/1541-7786.MCR-20-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madamsetty V.S., et al. Development of multi-drug loaded PEGylated nanodiamonds to inhibit tumor growth and metastasis in genetically engineered mouse models of pancreatic cancer. Nanoscale. 2019;11(45):22006–22018. doi: 10.1039/c9nr05478b. [DOI] [PubMed] [Google Scholar]
- 52.Blagosklonny M.V. Targeting cancer cells by exploiting their resistance. Trends Mol. Med. 2003;9(7):307–312. doi: 10.1016/s1471-4914(03)00111-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.