Abstract
Rationale
Prostate cancer treatment response may be automatically quantified using a molecular imaging analysis platform targeting prostate-specific membrane antigen (PSMA).
Methods
A retrospective analysis of patients with castration-sensitive prostate cancer who underwent PSMA-targeted molecular imaging prior to and 3 months or more after treatment was conducted. Disease burden was analyzed with aPROMISE, an artificial intelligence imaging platform that automatically quantifies PSMA-positive lesions. The calculated PSMA scores for prostate/bed, nodal, and osseous disease sites were compared with prostate-specific antigen (PSA) values.
Results
Of 30 eligible patients, the median decline in prostate/bed, nodal, and osseous disease PSMA scores were 100% (range 52–100%), 100% (range − 87–100%), and 100% (range − 21–100%), respectively. PSMA score decline was significantly associated with PSA decline.
Conclusion
Changes in aPROMISE PSMA scores are associated with changes in PSA and may quantify treatment response.
Keywords: PSMA, Automated, Artificial intelligence
Introduction
High-risk localized, recurrent, or metastatic CSPC patients receive androgen receptor (AR) axis inhibition and local therapy to disease sites. PSA changes traditionally gauge treatment response, but cannot discern lesion-level response (Scher et al. 2016). Currently employed imaging biomarkers allow noninvasive disease measurement using standard imaging modalities, but suffer limitations despite anatomy-specific quantitative techniques (Schwartz et al. 1990; van Persijn van Meerten 2010; Ulmert et al. 2012; Mitsui et al. 2012; Anand et al. 2016; Anand et al. 2020).
Disease-specific molecular imaging with PSMA-targeted radiotracers allows quantitative spatial resolution of individual tumors with positron emission tomography (PET) when combined with computed tomography (CT). PSMA PET-CTs are increasingly used for prostate cancer staging and can quantify treatment response (Fanti et al. 2020; Gafita et al. 2022). [18F]DCFPyL is a PSMA-targeted radiotracer with exceptional sensitivity and specificity for lesion detection (Morris et al. 2021; Pienta et al. 2021). Prostate cancer molecular imaging standardized evaluation (PROMISE) criteria provides a standardized framework for classifying and quantifying PSMA tracer-avid disease (Eiber et al. 2018). Automated PROMISE (aPROMISE) is an AI platform that enhances PROMISE’s approach by auto-segmenting organs, quantifying radiotracer uptake in reference organs, identifying pathologic lesions, and quantifying lesion uptake to facilitate calculation of PSMA scores that take into account both lesion volume and standardized uptake values (SUV) in specific anatomic regions (Nickols et al. 2022; Johnsson et al. 2022).
Materials and methods
Clinical data for CSPC patients treated from 2018 onwards were reviewed. Patients who had both baseline and follow-up PSMA PET-CTs at least 3 months after treatment initiation were identified. PSMA PET-CTs were read by a nuclear medicine physician for baseline and follow-up disease assessment.
aPROMISE was used to identify, quantify, and calculate changes in PSMA tracer-avid disease (Fig. 1) (Calais et al. 2022). Patient anatomy was auto-segmented from the low-dose CT portion of the PSMA PET-CT using a convolutional neural network (CNN) trained on CTs with expertly defined anatomy. Auto-segmented liver and aorta structures were overlayed on the PET dataset to calculate reference standard uptake values (SUVsref) from the liver and aorta SUVsmean, which were then used to identify other normal tissues. Putative lesion identification was divided into hot spot detection and segmentation phases. Hot spot detection utilizes a second CNN trained on expertly annotated PSMA PETs. The segmentation phase uses an adaptive threshold based on contextual information from the two deployed CNNs (Fig. 2). The hot spot detection algorithm has high sensitivity to minimize the number of lesions that miss detection and require manual contouring. A nuclear medicine physician assists the process by reviewing auto-segmented lesions to identify true versus fals positives. A PSMA score for the selected set of lesions was calculated as the product of the individual lesion volume and SUVmean, normalized by the SUVmean of reference tissue. An anatomic compartment PSMA score for prostate/bed, nodal, or osseous disease was calculated as the sum of individual lesion PSMA scores in the compartment (e.g.,). An additional composite score was calculated as a sum of the compartment PSMA scores. A Pearson’s R test was performed for total PSMA scores, as well as for the prostate/bed, nodal, and osseous compartment PSMA scores. Significance was determined by a 2-tailed T test.
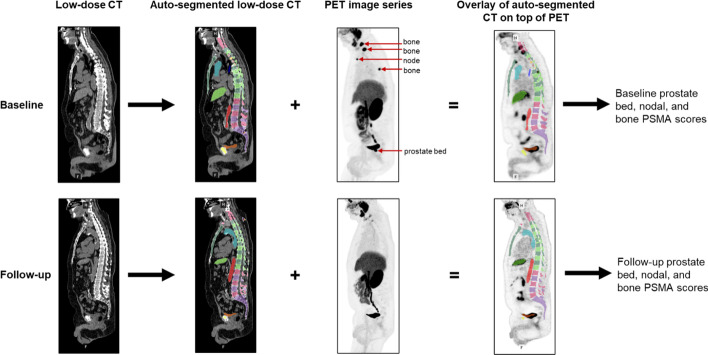
Fig. 1.
aPROMISE workflow. Anatomy is auto-segmented, radiotracer avid lesions are identified and auto-segmented, and then, the CT and PET image sets are overlaid to obtain annotated and quantifiable lesions. Sagittal PSMA PET-CT slices before and after therapy for a patient are shown
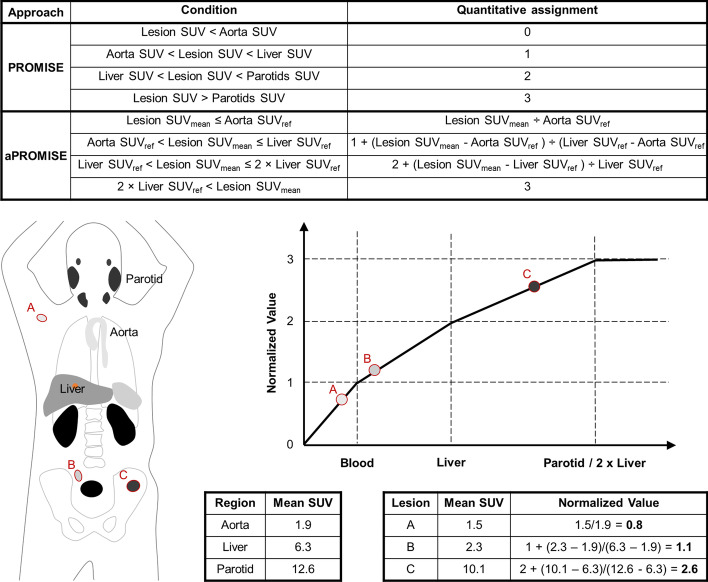
Fig. 2.
PROMISE and aPROMISE uptake value normalization comparison with schematic example of aPROMISE normalized SUV calculation
Results
Thirty patients were eligible for analysis. All patients demonstrated a decline in PSA before the second PSMA PET-CT. The median interval between PSMA PET-CTs was 8 months (range: 3–30). Baseline prostate/bed, regional nodal, non-regional nodal, and osseous disease was present in 27, 25, 9, and 18 patients, respectively. Treatment details were available for 29 patients. Of these, 4 patients had prior prostatectomy with 1 patient experiencing bed recurrence. Patient clinicopathologic data, treatment course, and biochemical outcome information are shown in Table 1.
Table 1.
Patient clinicopathologic data and treatment course
| Patient number | Disease timing | miTNM at diagnosis | Prostate/bed disease | Regional nodal disease | Non-regional Nodal diseaseˤ | Osseous disease | Imaging interval (months) | Interval treatment | % reduction in PSA |
|---|---|---|---|---|---|---|---|---|---|
| 1 | De novo | T3bN1bM0 | Yes | Yes | No | No | 30 | ADT + radiotherapy | 100 |
| 2 | De novo | T2mN0M1b | Yes | No | No | Yes | 14 | Surgery + ADT + radiotherapy | 100 |
| 3 | De novo | T2uN0M0 | Yes | No | No | No | 13 | Surgery | 84 |
| 4 | Recurrence | T2mN1bM0 | Yes | Yes | No | No | 10 | ADT + radiotherapy | 99 |
| 5 | De novo | T2mN1bM1b | Yes | Yes | No | Yes | 21 | ADT + radiotherapy | 100 |
| 6 | Recurrence | T2mN0M1b | Yes | No | No | Yes | 3 | ADT + radiotherapy | 100 |
| 7 | De novo | T2mN1aM0 | Yes | Yes | No | No | 3 | Surgery | 92 |
| 8 | De novo | T3bN1aM1a | Yes | Yes | Yes | No | 20 | Surgery + ADT + radiotherapy | 95 |
| 9 | De novo | T3bN1bM1a | Yes | Yes | Yes | No | 21 | Surgery + ADT | 92 |
| 10 | De novo | T3aN1aM1b | Yes | Yes | No | Yes | 11 | ADT + radiotherapy | 100 |
| 11˘ | Recurrence | T0N0M1b | No | No | Yes | Yes | 12 | ADT + radiotherapy | 100 |
| 12˜ | Recurrence | T0N1aM0 | No | Yes | No | No | 12 | Unknown˜ | Unknown˜ |
| 13 | De novo | T3bN1bM1b | Yes | Yes | Yes | Yes | 4 | ADT | 69 |
| 14 ˠ | De novo | T3bN1bM1b | Yes | Yes | No | Yes | 5 | ADT + radiotherapy | 100 |
| 15 | De novo | T3bN0M1b | Yes | Yes | Yes | Yes | 7 | ADT + radiotherapy | 83 |
| 16 | De novo | T3bN1bM1b | Yes | Yes | No | Yes | 7 | ADT + radiotherapy | 98 |
| 17˘ | Recurrence | TrN1bM1b | Yes | Yes | No | Yes | 15 | ADT + radiotherapy | 100 |
| 18 | De novo | T2uN1aM1b | Yes | Yes | No | Yes | 26 | Surgery + ADT + radiotherapy | 100 |
| 19 | De novo | T3bN1bM1b | Yes | Yes | No | Yes | 7 | ADT + radiotherapy | 100 |
| 20 | De novo | T2mN1aM1b | Yes | Yes | No | Yes | 7 | ADT + radiotherapy | 100 |
| 21 | De novo | T3bN1aM1b | Yes | Yes | No | Yes | 7 | ADT + radiotherapy | 100 |
| 22 | De novo | T2mN1aM0 | Yes | Yes | No | No | 7 | ADT + radiotherapy | 100 |
| 23 | De novo | T2mN1bM1a | Yes | Yes | Yes | No | 7 | ADT + radiotherapy | 100 |
| 24 | De novo | T3bN1bM1b | Yes | Yes | No | Yes | 18 | ADT + radiotherapy | 100 |
| 25 | De novo | T2mN0M1b | Yes | No | No | Yes | 9 | ADT + radiotherapy | 100 |
| 26 | De novo | T2mN1aM0 | Yes | Yes | No | Yes | 6 | ADT + radiotherapy | 99 |
| 27 | De novo | T2mN0M1b | Yes | No | No | Yes | 7 | ADT + radiotherapy | 100 |
| 28˘ | Recurrence | T0N1bM1a | No | Yes | Yes | No | 7 | ADT | 100 |
| 29 | De novo | T2mN1bM1a | Yes | Yes | Yes | No | 8 | ADT + radiotherapy | 100 |
| 30 | De novo | T2mN1bM1a | Yes | Yes | Yes | No | 7 | ADT + radiotherapy | 97 |
˘Patients with recurrent disease who had undergone prior prostatectomy
˜Patient 12’s treatment course and PSA response was not available for review
ˠPatient 14 had concurrent lung metastases at initial PSMA PET-CT
ˤNon-regional nodal disease was any nodal disease above the bifurcation of the common iliac arteries
Changes in composite and compartment-specific PSMA scores are shown in Table 2. Median prostate/bed, nodal, osseous, and composite PSMA scores at baseline were 21.6 (range: 0.9–150.5), 5.3 (range: 0.1–105.6), 2.2 (0.1–96.2), and 9.7 (0.2–106.8), respectively. Median PSMA scores for prostate/bed, nodal, osseous, and composite at follow-up PSMA PET-CT were 0 (0–6.2), 0 (0–91.1), 0 (0–36.2), and 0 (0–91.6), respectively. Baseline prostate/bed PSMA scores were significantly correlated with baseline PSA values (p < 0.001); however, neither nodal (p = 0.53) nor osseous (p = 0.65) baseline PSMA scores were correlated with baseline PSA values.
Table 2.
The percent decrease in each PSMA score was calculated from the quantified pre-treatment and post-treatment PSMA PET-CT for the total composite and each anatomic compartment
| Patient number | Lymph node % change | Osseous disease % change | Primary (prostate or prostate bed) % change | Total composite % change | % decrease PSA |
|---|---|---|---|---|---|
| 1 | − 100% | N/A | − 100% | − 100% | − 100 |
| 2 | N/A | 14% | − 100% | − 97% | − 100 |
| 3 | N/A | N/A | − 100% | − 100% | − 84 |
| 4 | − 66% | N/A | − 100% | − 70% | − 99 |
| 5 | − 100% | − 47% | − 100% | − 100% | − 100 |
| 6 | N/A | − 87% | − 100% | − 89% | − 100 |
| 7 | N/A | N/A | − 100% | − 100% | − 92 |
| 8 | − 100% | N/A | − 100% | − 100% | − 95 |
| 9 | − 62% | N/A | − 100% | − 94% | − 92 |
| 10 | − 100% | − 100% | − 100% | − 100% | − 100 |
| 11 | − 100% | − 75% | N/A | − 76% | − 100 |
| 12˜ | − 20% | N/A | N/A | − 20% | Unknown |
| 13 | 87% | − 45% | − 100% | 31% | − 69 |
| 14ˠ | − 96% | − 62% | − 100% | − 70% | − 100 |
| 15 | N/A | − 35% | − 100% | − 98% | − 83 |
| 16 | − 100% | − 80% | − 41% | − 49% | − 98 |
| 17 | − 100% | − 100% | N/A | − 100% | − 100 |
| 18 | N/A | − 100% | − 100% | − 100% | − 100 |
| 19 | − 100% | − 100% | − 100% | − 100% | − 100 |
| 20 | N/A | N/A | − 100% | − 100% | − 100 |
| 21 | − 100% | − 64% | − 100% | − 98% | − 100 |
| 22 | − 100% | − 100% | − 100% | − 100% | − 100 |
| 23 | − 100% | − 100% | − 100% | − 100% | − 100 |
| 24 | − 100% | − 100% | − 100% | − 100% | − 100 |
| 25 | N/A | − 100% | − 100% | − 100% | − 100 |
| 26 | − 100% | − 100% | − 95% | − 98% | − 99 |
| 27 | N/A | − 100% | − 95% | − 95% | − 100 |
| 28 | − 59% | N/A | N/A | − 59% | − 100 |
| 29 | − 100% | N/A | − 100% | − 100% | − 100 |
| 30 | − 100% | N/A | − 52% | − 56% | − 97 |
| Pearson R | 0.95 | 0.38 | − 0.06 | 0.61 | |
| p value | 4 E-10 | 0.09 | Not calculated | 5 E-4 |
A Pearson R correlation coefficient was calculated between the percent decrease in the PSMA score, and PSA was calculated and is shown. “N/A” indicates that the patient did not have relevant disease that could be scored for that anatomic compartment
˜Patient 12’s treatment course and PSA response was not available for review
ˠPatient 14 had concurrent lung metastases at initial PSMA PET-CT
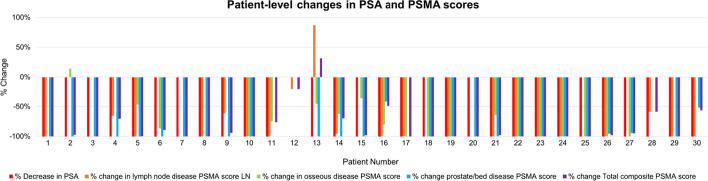
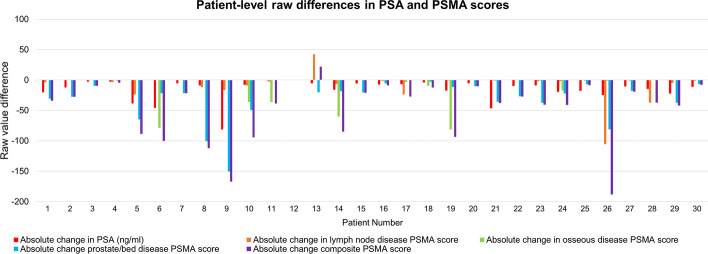
The median PSA decrease was 100% (range: 68–100%). Changes in PSMA scores were significantly correlated with corresponding decreases in PSA for composite and nodal disease, but not for prostate/bed or osseous disease (Table 2). Patient-level changes in PSMA scores and PSA are shown in Fig. 3. Patient 2 and Patient 13 had an increase in osseous, and nodal and composite PSMA scores, respectively, despite PSA decline after treatment. The osseous and prostate PSMA scores before/after treatment for Patient 2 were 0.62/0.70 and 27.5/0, respectively; thus, the before/after composite PSMA score of 28.1/0.7 was only minimally affected by increased osseous disease uptake. The osseous, lymph node, and prostate PSMA scores before/after treatment for Patient 13 were 0.99/0.48, 48.71/91.06, and 20.12/0, respectively; thus, the before/after composite PSMA score of 69.72/91.55 was heavily weighted by increased nodal disease uptake. Absolute changes in PSMA scores are shown in Fig. 4.
Fig. 3.
Percent change in PSA and prostate/bed, nodal, and osseous disease PSMA scores for each patient
Fig. 4.
Absolute raw changes in PSMA scores and PSA after treatment
Conclusion
Changes in prostate cancer disease burden as identified by PSMA-targeting radiotracers correlate with PSA response, but few studies have shown correlation between PSA changes and qualitative PET-based disease response (Hope et al. 2017; Schmidkonz et al. 2018; Zacho and Petersen 2018; Afshar-Oromieh et al. 2018; Aggarwal et al. 2018; Ettala et al. 2020; Emmett et al. 2019; Shagera et al. 2022). For instance, RECIP 1.0 assesses response for patients with castration resistant prostate cancer treated with targeted radionuclide therapy (Gafita et al. 2022). Here we present a method to automatically annotate and measure lesion changes for patients with CSPC using a previously validated tool for PSMA radiotracer uptake quantification. While there are no prospective data to demonstrate utility of PSMA-targeted PET-CTs for response assessment, the quantifiable PSMA score changes in this study were significantly correlated with PSA decline (Fanti et al. 2021).
PSMA expression is down-regulated by AR axis activation and up-regulated by its suppression, which may complicate response assessment of immediate post-treatment PSMA PET-CTs (Hope et al. 2017; Lückerath et al. 2018). Initial responses to AR-directed therapy as assessed on PSMA PET-CT were previously evaluated in small studies with variable time courses (Hope et al. 2017; Zacho and Petersen 2018; Aggarwal et al. 2018; Ettala et al. 2020; Emmett et al. 2019; Wondergem et al. 2020; Plouznikoff et al. 2019). These studies report inter-patient, intra-patient, and intra-patient inter-lesion radiotracer uptake heterogeneity after AR-directed therapy, which further complicates global interpretation of treatment efficacy. Discordance between changes in anatomically stratified PSMA scores and PSA were also observed here. Patients 2 and 13 had PSMA score increases. While the percent change in osseous PSMA score for Patient 2 was + 14%, this represented a raw change from 0.6 to 0.7. By contrast, Patient 13 had an increase of the nodal PSMA score of 87%, which represented a raw change from 48.7 to 91.1. This impacted the composite PSMA score and was qualitatively appreciated on imaging. Thus, utilization of PSMA score percent change may only be relevant above a certain raw score threshold. These cases also demonstrate how PSA changes may not reflect disease response at the individual lesion level, thus underscoring the need for imaging biomarkers that assess response at the lesion level.
It remains unclear whether aPROMISE may serve as an imaging biomarker and how these data affect clinical outcomes or provider decision-making. Additionally, as our approach was geared toward high sensitivity for lesion detection, clinical guidance to determine true versus false positive disease is required. The putative concordance between PSA and PSMA imaging response justifies PSMA PET-CT inclusion into prospective studies as a method to stratify therapeutic approaches. Given the few discordances between PSA and PSMA score changes here, additional work to track and compare changes at the lesion-specific level to assess intra-patient, inter-lesion heterogeneity is warranted.
Acknowledgements
Not applicable.
Abbreviations
- PSMA
Prostate-specific membrane antigen
- CSPC
Castration-sensitive prostate cancer
- PSA
Prostate-specific antigen
- PROMISE
Prostate cancer molecular imaging standardized evaluation
- AI
Artificial intelligence
- PET
Positron emission tomography
- CT
Computed tomography
- SUV
Standardized uptake value
- CNN
Convolutional neural network
Author contributions
SD and NN performed data analysis, acquisition, and statistical analysis. SD was responsible for writing this manuscript. GB and MR performed clinical interpretation of patient data, with MR responsible for disease-/cancer-related data, and GB responsible for clinical PET/CT interpretation. ST was responsible for patient recruitment and imaging coordination. All authors read and approved the final manuscript.
Funding
This work was supported through a cooperative research and development agreement with Lantheus.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to their nature as protected PHI of VA patients, but are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The Veteran Administration Greater Los Angeles Healthcare System Research and Development IRB committee approved this project under reference number 1615935-5. The study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments. All patients underwent informed consent for participation and use of their clinical data in this work.
Consent for publication
All patients underwent informed consent for participation and use of their clinical data in this work.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Afshar-Oromieh A, et al. Impact of long-term androgen deprivation therapy on PSMA ligand PET/CT in patients with castration-sensitive prostate cancer. Eur J Nucl Med Mol Imaging. 2018;45:2045–2054. doi: 10.1007/s00259-018-4079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal R, et al. Heterogeneous flare in prostate-specific membrane antigen positron emission tomography tracer uptake with initiation of androgen pathway blockade in metastatic prostate cancer. Eur Urol Oncol. 2018;1:78–82. doi: 10.1016/j.euo.2018.03.010. [DOI] [PubMed] [Google Scholar]
- Anand A, et al. Analytic validation of the automated bone scan index as an imaging biomarker to standardize quantitative changes in bone scans of patients with metastatic prostate cancer. J Nucl Med off Publ Soc Nucl Med. 2016;57:41–45. doi: 10.2967/jnumed.115.160085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, et al. Assessing radiographic response to 223Ra with an automated bone scan index in metastatic castration-resistant prostate cancer patients. J Nucl Med off Publ Soc Nucl Med. 2020;61:671–675. doi: 10.2967/jnumed.119.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calais J, et al. Prospectively planned and Independent validation of aPROMISE in a phase III CONDOR study for rapid lesion detection and standardized quantitative evaluation for 18F-DCFPyL (PSMA) imaging in prostate cancer. J Nucl Med. 2022;63:2496. [Google Scholar]
- Eiber M, et al. Prostate cancer molecular imaging standardized evaluation (PROMISE): proposed miTNM classification for the interpretation of PSMA-ligand PET/CT. J Nucl Med off Publ Soc Nucl Med. 2018;59:469–478. doi: 10.2967/jnumed.117.198119. [DOI] [PubMed] [Google Scholar]
- Emmett L, et al. Rapid modulation of PSMA expression by androgen deprivation: serial 68Ga-PSMA-11 PET in men with hormone-sensitive and castrate-resistant prostate cancer commencing androgen blockade. J Nucl Med off Publ Soc Nucl Med. 2019;60:950–954. doi: 10.2967/jnumed.118.223099. [DOI] [PubMed] [Google Scholar]
- Ettala O, et al. Prospective study on the effect of short-term androgen deprivation therapy on PSMA uptake evaluated with 68Ga-PSMA-11 PET/MRI in men with treatment-naïve prostate cancer. Eur J Nucl Med Mol Imaging. 2020;47:665–673. doi: 10.1007/s00259-019-04635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti S, Hadaschik B, Herrmann K. Proposal for systemic-therapy response-assessment criteria at the time of PSMA PET/CT imaging: the PSMA PET progression criteria. J Nucl Med off Publ Soc Nucl Med. 2020;61:678–682. doi: 10.2967/jnumed.119.233817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti S, et al. Consensus statements on PSMA PET/CT response assessment criteria in prostate cancer. Eur J Nucl Med Mol Imaging. 2021;48:469–476. doi: 10.1007/s00259-020-04934-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafita A, et al. Novel framework for treatment response evaluation using PSMA-PET/CT in patients with metastatic castration-resistant prostate cancer (RECIP 10): an international multicenter study. J Nucl Med off Publ Soc Nucl Med. 2022 doi: 10.2967/jnumed.121.263072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope TA, et al. 68Ga-PSMA-11 PET imaging of response to androgen receptor inhibition: first human experience. J Nucl Med off Publ Soc Nucl Med. 2017;58:81–84. doi: 10.2967/jnumed.116.181800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson K, et al. Analytical performance of aPROMISE: automated anatomic contextualization, detection, and quantification of [18F]DCFPyL (PSMA) imaging for standardized reporting. Eur J Nucl Med Mol Imaging. 2022;49:1041–1051. doi: 10.1007/s00259-021-05497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lückerath K, et al. Preclinical evaluation of PSMA expression in response to androgen receptor blockade for theranostics in prostate cancer. EJNMMI Res. 2018;8:96. doi: 10.1186/s13550-018-0451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui Y, et al. Prediction of survival benefit using an automated bone scan index in patients with castration-resistant prostate cancer. BJU Int. 2012;110:E628–634. doi: 10.1111/j.1464-410X.2012.11355.x. [DOI] [PubMed] [Google Scholar]
- Morris MJ, et al. Diagnostic performance of 18F-DCFPyL-PET/CT in men with biochemically recurrent prostate cancer: results from the CONDOR phase III, multicenter study. Clin Cancer Res off J Am Assoc Cancer Res. 2021;27:3674–3682. doi: 10.1158/1078-0432.CCR-20-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols N, et al. aPROMISE: a novel automated PROMISE platform to standardize evaluation of tumor Burden in 18F-DCFPyL images of veterans with prostate cancer. J Nucl Med off Publ Soc Nucl Med. 2022;63:233–239. doi: 10.2967/jnumed.120.261863. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, et al. A phase 2/3 prospective multicenter study of the diagnostic accuracy of prostate specific membrane antigen PET/CT with 18F-DCFPyL in prostate cancer patients (OSPREY) J Urol. 2021;206:52–61. doi: 10.1097/JU.0000000000001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouznikoff N, et al. Evaluation of PSMA expression changes on PET/CT before and after initiation of novel antiandrogen drugs (enzalutamide or abiraterone) in metastatic castration-resistant prostate cancer patients. Ann Nucl Med. 2019;33:945–954. doi: 10.1007/s12149-019-01404-2. [DOI] [PubMed] [Google Scholar]
- Scher HI, et al. Trial design and objectives for castration-resistant prostate cancer: updated recommendations from the prostate cancer clinical trials working group 3. J Clin Oncol off J Am Soc Clin Oncol. 2016;34:1402–1418. doi: 10.1200/JCO.2015.64.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidkonz C, et al. 68Ga-PSMA-11 PET/CT-derived metabolic parameters for determination of whole-body tumor burden and treatment response in prostate cancer. Eur J Nucl Med Mol Imaging. 2018;45:1862–1872. doi: 10.1007/s00259-018-4042-z. [DOI] [PubMed] [Google Scholar]
- Schwartz LH, et al. RECIST 1.1—update and clarification: from the RECIST committee. Eur J Cancer Oxf Engl 1990. 2016;62:132–137. doi: 10.1016/j.ejca.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagera QA, et al. 68Ga-PSMA PET/CT for response assessment and outcome prediction in metastatic prostate cancer patients treated with taxane-based chemotherapy. J Nucl Med off Publ Soc Nucl Med. 2022;63:1191–1198. doi: 10.2967/jnumed.121.263006. [DOI] [PubMed] [Google Scholar]
- Ulmert D, et al. A novel automated platform for quantifying the extent of skeletal tumour involvement in prostate cancer patients using the Bone Scan Index. Eur Urol. 2012;62:78–84. doi: 10.1016/j.eururo.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Persijn van Meerten EL, Gelderblom H, Bloem JL. RECIST revised: implications for the radiologist. A review article on the modified RECIST guideline. Eur Radiol. 2010;20:1456–1467. doi: 10.1007/s00330-009-1685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondergem M, van der Zant FM, Broos WAM, Knol RJJ. Clinical impact of PSMA PET in biochemically recurrent prostate cancer; a review of the literature. Tijdschr Voor Urol. 2020;10:109–121. doi: 10.1007/s13629-020-00296-6. [DOI] [Google Scholar]
- Zacho HD, Petersen LJ. Bone flare to androgen deprivation therapy in metastatic, hormone-sensitive prostate cancer on 68Ga-prostate-specific membrane antigen PET/CT. Clin Nucl Med. 2018;43:e404–e406. doi: 10.1097/RLU.0000000000002273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to their nature as protected PHI of VA patients, but are available from the corresponding author on reasonable request.