Significance
Circular RNAs (circRNAs) play important roles in tumor metastasis. Our results identified circNEIL3 (hsa_circ_0001460) as a TGFβ-repressive circRNA and also down-regulated in metastatic tumor lesions and demonstrated that circNEIL3 potently represses tumor metastasis in vitro and in vivo. circNEIL3 promoted Nedd4L-mediated proteasomal degradation of YBX1 through its direct interaction with YBX1. Our study provides a reported association of circRNA, protein stability, and metastasis and sheds lights on the molecular mechanism of circRNA in metastasis.
Keywords: circRNA, metastasis, YBX1, Nedd4L
Abstract
Distant metastasis is a major contributor to cancer-related mortality. However, the role of circRNAs in this process remains unclear. Herein, we profiled the circRNA expression in a cohort of 68 colorectal carcinoma (CRC) primary tumors and their paired liver metastatic lesions. By overlapping with the TGFβ-responsive circRNAs, circNEIL3 (hsa_circ_0001460) was identified as a TGFβ-repressive and metastasis-related circRNA. Functionally, circNEIL3 effectively inhibited tumor metastasis in both and in vivo and in vivo models of various cancer types. Mechanistically, circNEIL3 exerts its metastasis-repressive function through its direct interaction with oncogenic protein, Y-box-binding protein 1 (YBX1), which consequently promotes the Nedd4L-mediated proteasomal degradation of YBX1. Importantly, circNEIL3 expression was negatively correlated to YBX1 protein level and metastatic tendency in CRC patient samples. Collectively, our findings indicate the YBX1-dependent antimetastatic function of circNEIL3 and highlight the potential of circNEIL3 as a biomarker and therapeutic option in cancer treatment.
Distant metastasis is a major contributor to cancer-related mortality and an insurmountable barrier to extend life expectancy (1). As a complex and multistep biological process, tumor metastasis consists of sequential steps, including local invasion, intravasation into blood, circulatory survival, extravasation, and colonization (2). Cancer cells acquire metastatic potential by reprogramming their gene expression spectrums in transcriptional and posttranscriptional manners (3). The complexity of this metastatic reprogramming is further evidenced by the recently published study where thousands of somatic mutations were identified to be correlated with tumor metastasis (4). Still, specific biomarkers and efficient therapeutics to handle tumor metastasis are in urgent needs. Therefore, a better understanding of tumor metastasis will improve the clinical outcomes of cancer patients.
Y-box-binding protein 1 (YBX1, also known as YB-1) is a pivotal factor to promote tumor metastasis (5–7). As an RNA-binding protein, YBX1 is up-regulated in various cancer types, including breast cancer (8), hepatocellular carcinoma (9), lung cancer (10), and acute myeloid leukemia (11). Moreover, YBX1 is tightly implicated in the metastatic reprogramming through regulating the transcription (8, 9), alternative splicing (12), stability (13), and translation (6, 7) of many oncogenic transcripts. Notably, the elevated YBX1 protein level is a more predominant event than the increased YBX1 mRNA level in advanced tumors (14), implying that the dysfunction in posttranslational regulation of YBX1 may be more common for its upregulation. In accordance, several E3 ligases were identified to target YBX1 for proteasomal degradation (15–17), and among which, Nedd4L serves as a tumor suppressor and is down-regulated in metastatic lung cancer (18). However, the detailed mechanism underlying YBX1 degradation during tumor metastasis is still obscure.
The degradation of several RNA-binding proteins was reported to be modulated by circular RNAs (circRNAs) (19–21). For example, circPABPC1 represses tumor metastasis in hepatocellular carcinoma by targeting proteasomal degradation of ITGB1, a key member of the integrin family to weaken cell adhesion and promote cell migration (19). As a class of covalently closed loop RNA molecules, circRNAs are generated from the back-splicing of primary transcripts to exert their functions at transcriptional and posttranscriptional levels (22, 23). During tumor metastasis, dysfunction of splicing machinery may cause abnormal expression of circRNAs (24). Although few circRNAs were identified to be dysregulated in metastatic tumors (25–27), the expression pattern and function of circRNAs during cancer progression remain largely unknown.
In this study, we analyzed the differentially expressed circRNAs between colorectal cancer (CRC) primary tumors and their paired metastatic lesions in livers, followed by overlapping with the TGFβ-responsive circRNAs. As a result, circNEIL3 (hsa_circ_0001460) was identified as a TGFβ-repressive circRNA which is also significantly down-regulated in the metastatic tumors. Functionally, circNEIL3 robustly represses tumor metastasis in various and in vivo and in vivo models. Mechanistically, circNEIL3 plays the antimetastatic role through its direct association with YBX1 and consequently targeting YBX1 for Nedd4L-mediated proteasomal degradation. Importantly, circNEIL3 expression was observed to be negatively correlated to YBX1 abundance and tumor metastasis in CRC samples. Our findings indicate that circNEIL3 is a crucial metastasis-repressive circRNA by promoting proteasomal degradation of YBX1.
Results
circNEIL3 Was Significantly Down-Regulated in Metastatic CRC Tumors.
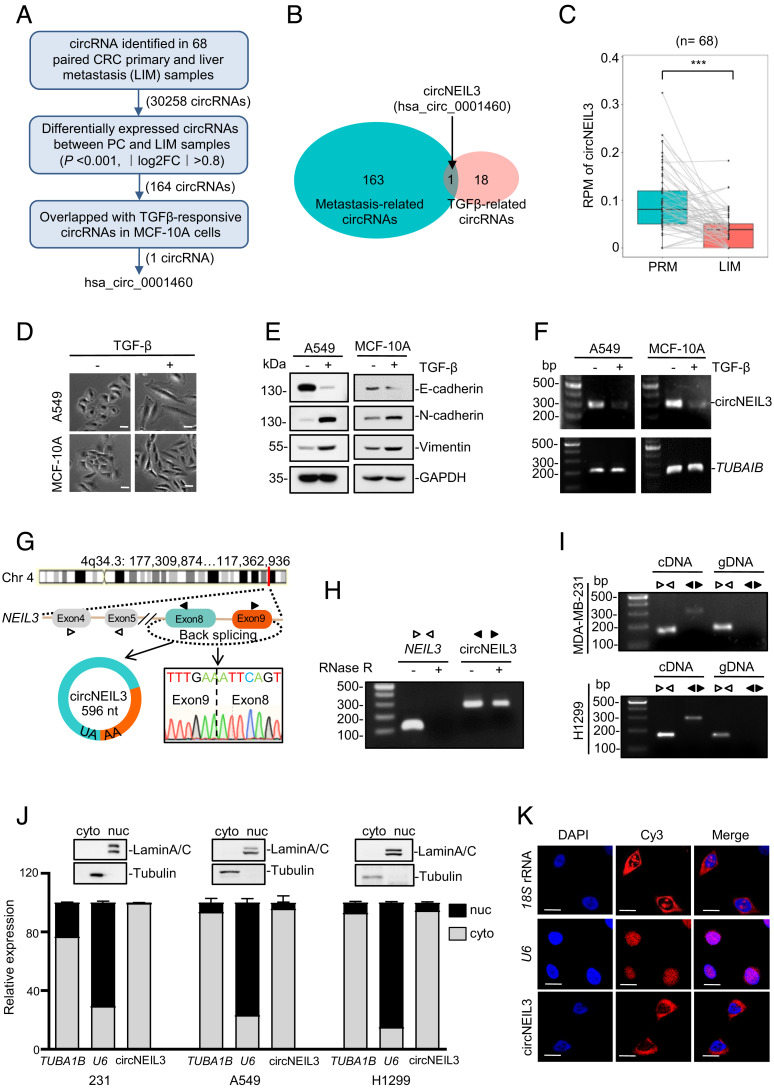
Distant metastasis is a lethal cause of CRC patients (1). To explore the implication of circRNAs in tumor metastasis, we analyzed expression profiles of circRNA in 68 paired CRC primary and liver metastatic (LIM) tissues. A total of 30,258 circRNAs were identified, among which 164 circRNAs demonstrated significantly differential expression (log2 |average normalized fold change| (FC) > 0.8 and P < 0.001) (Fig. 1A and SI Appendix, Fig. S1 A–D). Considering the pivotal role of TGFβ signaling in tumor metastasis and its implication in the CRC paired specimens (SI Appendix, Fig. S1E), these differentially expressed 164 circRNAs were overlapped with 19 TGFβ-responsive circRNAs in MCF10A cells with TGFβ treatment (GEO accession number: GSE165576). circNEIL3 (hsa_circ_0001460) was identified as the only metastasis-related and TGFβ-responsive circRNA (Fig. 1 A and B and SI Appendix, Tables S1 and S2), with dramatically decreased expression in metastatic samples compared to CRC primary tumors (Fig. 1C). To prove that circNEIL3 is regulated by TGFβ, the epithelial MCF10A and A549 cells were treated with TGFβ. Consistent with previous reports (28), TGFβ stimulation conferred on both cells a fibroblast-like shape (Fig. 1D), decreased expression of the epithelial marker E-cadherin, and increased expression of mesenchymal makers N-cadherin and vimentin (Fig. 1E), indicating the effective TGFβ treatment. Indeed, TGFβ stimulation significantly decreased circNEIL3 expression (Fig. 1F), with a decrease of circNEIL3 more striking in TGFβ-activated CRC pairs (SI Appendix, Fig. S1F). Collectively, these results demonstrate that circNEIL3 is significantly down-regulated in metastatic tumors and also upon TGFβ stimulation.
Fig. 1.
Characterization of the metastasis-related circNEIL3. (A and B) Flowchart (A) and Venn diagram (B) of profiling circRNAs in CRC tumors and TGFβ-stressed cell samples. (C) circNEIL3 expression between CRC primary tumors and their paired liver metastatic lesions. ***P < 0.001 by a paired two-sided Wilcoxon signed-rank test. (D) Cell morphology under 5 ng/mL TGFβ for 72 h. (E) Immunoblot assays to examine EMT-related makers in A549 and MCF-10A cells stressed by TGFβ. (F) Semiquantitative RT–PCR of circNEIL3 expression in cells treated with or without TGFβ (5 ng/mL) for 72 h. (G) Schematic illustration showing the back-splicing of circNEIL3 from NEIL3 primary transcript, which was confirmed by RT–PCR and Sanger sequencing. The divergent and convergent primers for PCR are indicated by solid and hollow triangles, respectively. (H) RT–PCR assays to detect circNEIL3 and its linear transcript in MDA-MB-231 cells after RNase R digestion. NEIL3 mRNA was amplified with convergent primers targeting the coding sequences outside the circularized exon. (I) RT–PCR assays using cDNA or gDNA from MDA-MB-231 or H1299 cells as the template to detect circNEIL3 and NEIL3 mRNA. (J) Subcellular distribution of circNEIL3 determined in different cancer cells by RT–qPCR assays. TUBA1B and U6 transcripts were used as the cytoplasmic and nucleus indicators, respectively. Immunoblot assays with antitubulin and antilamin A/C confirmed good cytoplasmic/nucleus fractionation. Cyto, cytoplasmic; Nuc, nuclear. Data represent mean ± SD from three independent experiments. (K) RNA in situ hybridization analysis for circNEIL3 in A549 cells. (Scale bar, 10 µm.) 18S rRNA and U6 RNA were used as the cytoplasmic and nuclear markers, respectively. Nuclei were stained with 4,6-Diamidino-2-phenylindole dihydrochloride (DAPI).
Characteristics of circNEIL3.
According to the circBase (http://www.circbase.org), circNEIL3 arises from exons 8 to 9 of the NEIL3 gene with a length of 596 nucleotides, and its back-splicing site was confirmed by Sanger sequencing of RT–PCR products using specific divergent primers (Fig. 1G). Moreover, circNEIL3 is resistant to the digestion of RNase R exonuclease that degrades linear RNAs (Fig. 1H), supporting that circNEIL3 harbors a closed loop structure. To rule out the possibility that the head-to-tail splicing of circNEIL3 was produced by genomic rearrangements, RT–PCR was performed using convergent and divergent primers (marked in Fig. 1G), respectively. As expected, circNEIL3 was only amplified with divergent primers using cDNAs, rather than genomic DNA (gDNA), as a template from the breast cancer cell MDA-MB-231 and the lung cancer cell H1299 (Fig. 1I). Subsequently, the subcellular distribution of circNEIL3 was determined by RT–qPCR after cytoplasmic and nuclear fractionations. The results indicated that circNEIL3 mainly localized in the cytoplasm of MDA-MB-231, A549, and H1299 cells (Fig. 1J), which was further validated in A549 cells by fluorescence in situ hybridization assays (Fig. 1K). Collectively, these results demonstrate that circNEIL3 is a bona fide circRNA, predominately localizing in the cytoplasm.
circNEIL3 Inhibits Tumor Metastasis In Vitro and In Vivo.
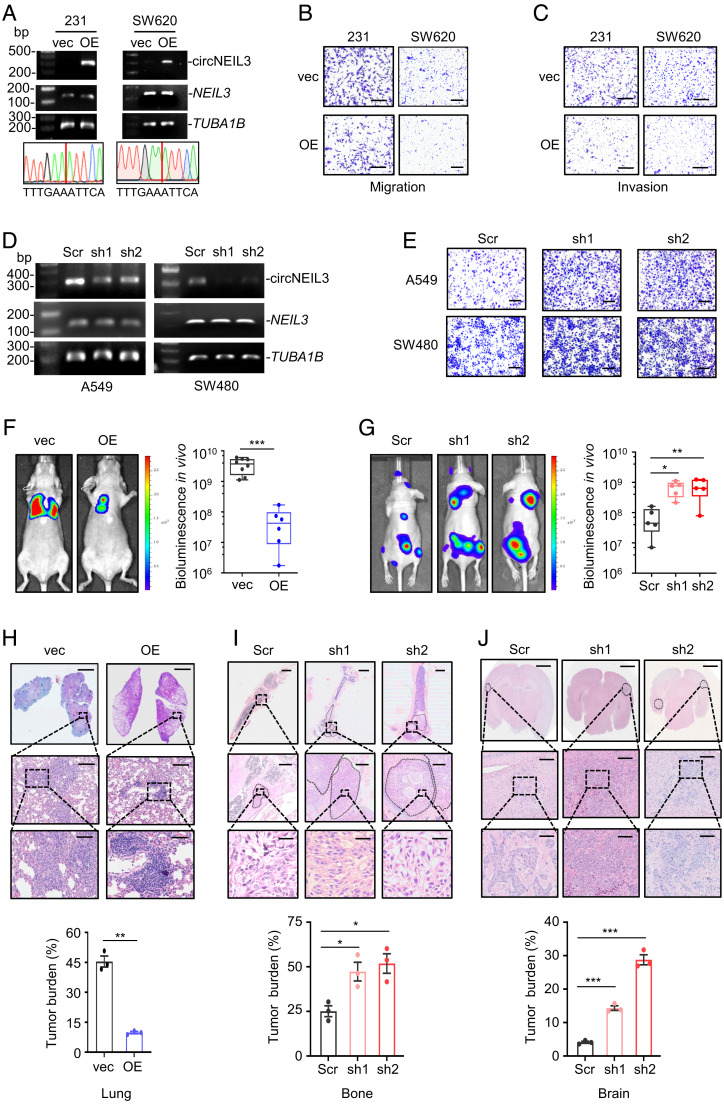
To investigate the role of circNEIL3 in tumor metastasis, we measured the endogenous expression of circNEIL3 in various cancer cells (SI Appendix, Fig. S2A). Based on the endogenous circNEIL3 expression, we generated circNEIL3 overexpression (OE) cancer cells (SW620, MDA-MB-231, BT549, and H1299 cells) and circNEIL3 knockdown A549 and SW480 cells, respectively. First, we confirmed both the expression efficiency and junction site sequence of enforced circNEIL3 in aforementioned cell lines (Fig. 2A and SI Appendix, Fig. S2B). Subsequently, Transwell assays showed that circNEIL3 overexpression significantly suppressed the migration and invasion capability of different cancer cells (Fig. 2 B and C and SI Appendix, Fig. S2 C–E). In accordance, circNEIL3 knockdown by two independent shRNAs strengthened cell migration in A549 and SW480 cells (Fig. 2 D and E and SI Appendix, Fig. S2F) without affecting the expression of its host transcript, NEIL3 mRNA (Fig. 2D). Besides, circNEIL3 overexpression or knockdown exhibited little impact on cell proliferation, as indicated by cell growth curve or colony formation (SI Appendix, Fig. S2 G and H), supporting the specific antimetastatic function of circNEIL3. Considering the well-documented role of Epithelial-Mesenchymal Transition (EMT) in metastasis (3), we also examine the impact of circNEIL3 on EMT markers by immunoblot assays. The minimal change of EMT markers upon circNEIL3 expression (SI Appendix, Fig. S2I) indicated that circNEIL3 may play its metastasis-repressive effect independent of the EMT process.
Fig. 2.
circNEIL3 inhibits cell migration in vitro and tumor metastasis in vivo. (A) Agarose gel electrophoresis and Sanger sequencing analysis of RT–PCR products to confirm successful expression and correct circularization of exogenously expressed circNEIL3 in MDA-MB-231 and SW620 cells. (B and C) Effects of enforced circNEIL3 expression on cell migration (B) and invasion (C) in MDA-MB-231 and SW620 cells. (D) Semiquantitative RT–PCR analysis to confirm the effective knockdown of endogenous circNEIL3 in A549 and SW480 cells. (E) circNEIL3 knockdown in A549 and SW480 cells promotes cell migration. (F and G, Left) Representative bioluminescence images of mice after tail vein injection of MDA-MB-231 cells with (n = 8) or without (n = 6) circNEIL3 overexpression (F) or after left ventricular injection of A549 cells with or without circNEIL3 knockdown (n = 5/group). (G, Right) Bioluminescence signals were quantified, and significance of difference in Fig. 2F was determined by the Student t test, while significance of difference in Fig. 2G was determined by the one-way ANOVA test. Data were presented as means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001. (H–J) Representative H&E staining images of metastatic lesions in the lung from (F) experiments, and in the bone (I) and brain (J) from (G) experiments. (Scale bar, 5 mm [Upper], 1 mm [Medium] or 200 μm [Lower].) Percentage of tumor area in each slide was measured using inForm software (Akoya Biosciences). Data were presented as means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
Next, we sought to further evaluate the function of circNEIL3 with metastasis models in mice. To this end, mice accepting tail vein injection of MDA-MB-231 cells were employed to evaluate the effect of circNEIL3 on pulmonary metastasis, while mice accepting left ventricular injection of A549 cells were used to examine the effect of circNEIL3 on metastasis in the bone and brain. As expected, circNEIL3 overexpression markedly repressed the formation of lung metastasis (Fig. 2F), whereas circNEIL3 knockdown significantly promoted tumor metastasis in the brain and bone (Fig. 2G), which was further confirmed by hematoxylin and eosin (H&E) staining (Fig. 2 H–J). Taken together, our results demonstrated that circNEIL3 can potently inhibit tumor metastasis in vitro and in vivo.
circNEIL3, Rather Than Its Linear Transcript, Represses Tumor Metastasis.
Although circNEIL3 is the dominant product of wild-type circNEIL3-overexpressing plasmids, the circNEIL3-OE plasmids also introduce some linear transcripts (SI Appendix, Fig. S3A), so the observed metastasis-repressive effect of circNEIL3 may be caused by the linear transcripts. To exclude this possibility, we constructed the circNEIL3 circulation-dead (CD) plasmid by mutating the Alu elements flanking circNEIL3 (SI Appendix, Fig. S3B). As shown in SI Appendix, Fig. S3C, the circNEIL3-CD plasmids did not form circNEIL3 but expressed linear transcripts in both MDA-MB-231 and BT549 cells. As expected, Transwell experiments indicated that circNEIL3-CD failed to exhibit antimigration or anti-invasion effects compared to its wild-type (WT) counterparts (SI Appendix, Fig. S3 D and E). Moreover, mice vein injected with MDA-MB-231 cells stably expressing circNEIL3-CD exhibited comparable metastatic lesions in the lungs to those bearing the empty vector, whereas mice bearing circNEIL3-WT cells have few lung metastases (SI Appendix, Fig. S3 F and G). The failure of circNEIL3-CD to inhibit tumor metastasis excludes that the antimetastatic role of circNEIL3 is contributed by its linear transcripts. Therefore, circNEIL3 represses tumor metastasis independent of its linear transcripts.
circNEIL3 Interacts with YBX1 to Repress Tumor Metastasis.
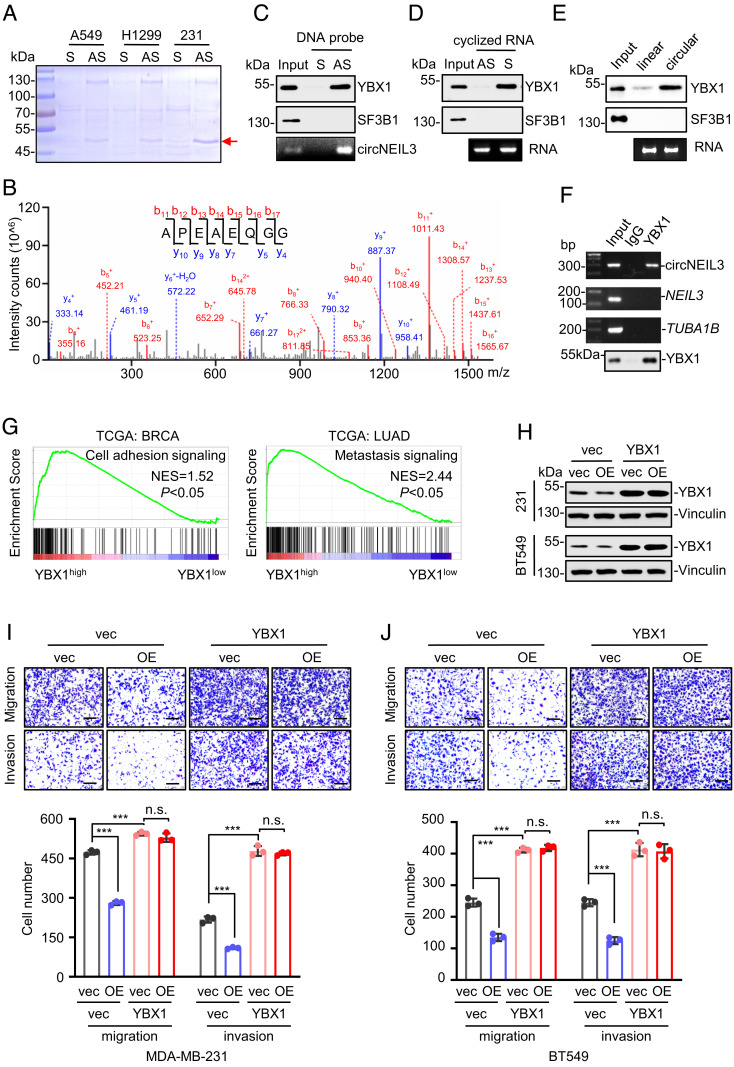
Accumulating evidence supports that circRNAs exert their biological functions commonly through their interaction with proteins to regulate gene expression (29). To identify the circNEIL3-interacting protein, we performed RNA pull-down assays after transfecting into cells with biotin-labeled sense (S) or antisense (AS) DNA oligomers crossing the junction site of circNEIL3, and the pull-down proteins were subjected to Sodium Dodecyl Sulfate PolyAcrylamide Gel Electrophoresis (SDS-PAGE) and Coomassie blue staining. As shown in Fig. 3A, two bands at ~130 kDa and ~45 kDa were pulled down by the AS probe consistently from three different cancer cell lines. Mass spectrometry analysis identified the ~45 kDa of band as the RNA-binding protein YBX1 because it exhibited the highest scores (Score Sequest HT = 272.24) and the top abundance (unique peptide = 21 and coverage = 62%) (Fig. 3B and SI Appendix, Table S3). Moreover, the association of circNEIL3 with YBX1 was confirmed by immunoblot assays after pull-down by the antisense probe (Fig. 3C) or the in vitro transcribed and circularized circNEIL3 (Fig. 3D). Notably, circularized RNA demonstrates much more striking association with YBX1 than its linear transcript (Fig. 3E), supporting the dominant role of circNEIL3 as a YBX1-associating factor. Besides, such endogenous circNEIL3–YBX1 association within cells was validated by RNA immunoprecipitation (RIP) assays using anti-YBX1 antibody (Fig. 3F). Notably, the linear transcripts of NEIL3 exhibited minimal enrichment in YBX1-precipitated fractions (Fig. 3F), indicating that YBX1 selectively associated with circNEIL3 rather than its host gene transcript.
Fig. 3.
Identification of YBX1 as the circNEIL3-associated protein. (A) Pull-down assays and Coomassie blue staining of circNEIL3-interacting proteins after transfecting DNA probes into A549, H1299, or MDA-MB-231 cells. S and AS represent the DNA probe identical and complementary to circNEIL3 sequence crossing back-splicing junction site, respectively. (B) A representative YBX1 peptide identified by LC-MS/MS mass spectrometry. (C–E) Immunoblot assays for YBX1 pulled down from MDA-MB-231 cell lysates by DNA probes (C), in vitro circularized circNEIL3 (D), or its linear transcribed RNA (E). SF3B1 served as a negative control. Enrichment efficiency of circNEIL3 by pull-down assay was determined by measuring its abundance in the precipitation (C); enrichment efficiency of circularized (D) or linear RNA (E) was detected by electrophoresis. (F) RIP and semiquantitative RT–PCR assays showed the association of YBX1 with circNEIL3 in MDA-MB-231 cells using anti-YBX1 antibody or the control IgG. TUBA1B served as negative controls. (G) Gene Set Enrichment Analysis showing the correlation of YBX1 protein abundance with cellular adhesion and metastatic pathways in breast cancer and lung adenocarcinoma samples. (H) Immunoblot analyses of YBX1 in different stable cell lines. (I and J) The decreased cell migration and invasion by circNEIL3 overexpression (OE) are attenuated by YBX1 overexpression in MDA-MB-231 (I) and BT549 cells (J). Data are presented as means ± SD. n.s., not significant; ***P < 0.001.
Given the reported roles of YBX1 in tumor metastasis (5–7), we wonder whether YBX1 acts as an important effector of circNEIL3. To this end, we first explored the correlation between YBX1 expression and tumor progression. Our bioinformatics analyses showed that higher YBX1 expressions in breast cancer and lung adenocarcinoma samples were significantly enriched in cell adhesion and tumor metastasis pathways, respectively (Fig. 3G). Moreover, the metastasis-promoting role of YBX1 is confirmed by Transwell assays as enforced YBX1 expression promoted cell migration and invasion (SI Appendix, Fig. S4 A and B), while YBX1 knockdown exhibited opposite effects (SI Appendix, Fig. S4 C and D). Importantly, cell migration and invasion assays showed that the antimetastatic effects of circNEIL3 were abolished by enforced YBX1 expression (Fig. 3 H–J), indicating that the function of circNEIL3 depends on YBX1. Therefore, YBX1 may be a critical circNEIL3-associating factor to affect cell migration and invasion.
Two YBX1 Recognition Consensus Motifs within circNEIL3 Mediate the Direct Interaction of circNEIL3 with YBX1.
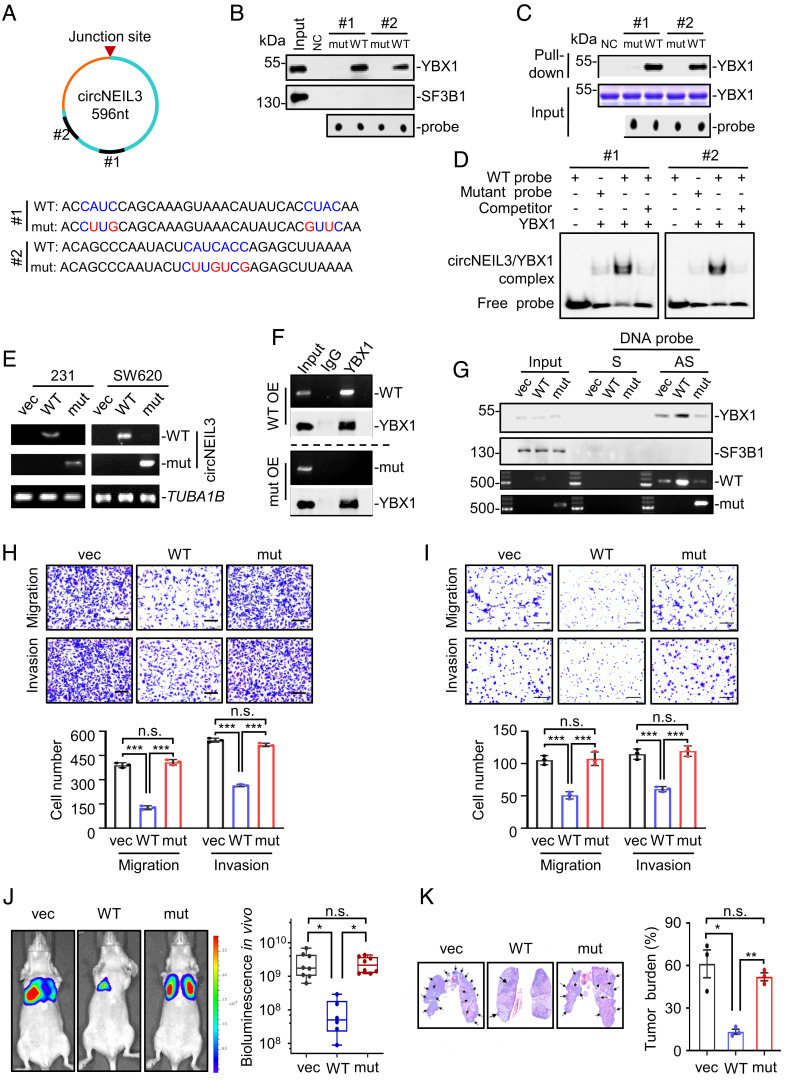
Previous studies indicated that YBX1 preferentially bound to the “CAUC” or “CACC” consensus motifs (30). To dissect the molecular basis underlying the association of circNEIL3 with YBX1, we identified two potential YBX1-recognized regions (containing either “CAUC” or “CAUCACC”) within circNEIL3. First, two biotin-labeled RNA probes (#1-WT and #2-WT) and their mutant probes (“CUUG” #1-mut and “CUUGUCG” #2-mut) were synthesized for RNA pull-down assays (Fig. 4A). As shown in Fig. 4B, comparable enrichments of endogenous YBX1 were observed in the precipitates of two wild-type probes, whereas such YBX1 enrichments were diminished by the replacement of the critical nucleotides within the putative YBX1-recognized motifs. These findings indicated that both “CAUC” and “CAUCACC” consensus motifs are responsible for their association with YBX1. Next, recombinant YBX1 proteins were incubated with these RNA probes to examine whether their interaction is direct or indirect. The results from both RNA pull-down assays (Fig. 4C) and RNA-EMSA assays (Fig. 4D) supported direct interaction of YBX1 with the probes harboring YBX1-recognized consensus motifs. Then, we wondered whether these YBX1-recognized consensus motifs mediate circNEIL3–YBX1 interaction. To this end, we generated the circNEIL3 mutant (circNEIL3-mut) by introducing the aforementioned mutations into wild-type circNEIL3 overexpression plasmids. As expected, the interaction of YBX1 with wild-type circNEIL3, rather than circNEIL3-mut, was validated by both RIP experiments (Fig. 4 E and F) and pull-down assays (Fig. 4G), thus reinforcing reliance of direct circNEIL3–YBX1 interaction on these two YBX1 recognition consensus motifs.
Fig. 4.
The metastasis inhibitory effect of circNEIL3 depends on its direct interaction with YBX1. (A) Schematic illustration showing the location of two predicted YBX1-binding motifs within circNEIL3. The 5’-end labeled wild-type (WT) and mutated RNA probes (mut) were synthesized. (B) YBX1 was pulled down by WT rather than mut RNA probes from MDA-MB-231 cell extracts. SF3B1 serves as a negative control. (C and D) RNA pull-down assays (C) and RNA-EMSA assays (D) showing the direct interaction between YBX1 and wild-type probes. (E) RT–PCR analysis to detect the expression of WT or mutated circNEIL3 in stable cells using specific primers complementary to the wild-type or mutant sequence, respectively. (F) RIP assays using anti-YBX1 antibody showing the association of YBX1 with WT circNEIL3. (G) RNA pull-down using DNA probes to show the association of circNEIL3 with YBX1 in MDA-MB-231 stable cell lines. (H and I) Cell migration and invasion assays for different MDA-MB-231 (H) and SW620 stable cell lines (I). Data are presented as means ± SD. n.s., not significant; ***P < 0.001. (J, Left) Representative bioluminescence images of mice (n = 8 for vector, n = 6 for WT, and n = 8 for mutant groups) after tail vein injection of different MDA-MB-231 stable cells. (Right) Bioluminescence signals were quantified, and significance of difference was determined by the one-way ANOVA test. Data are presented as means ± SEM. n.s., not significant; *P < 0.05. (K) Representative H&E staining images of the metastatic lesions in the lung. Tumor burdens were reflected by the percentages of tumor area in each slide. Data are presented as means ± SEM. n.s., not significant; *P <0.05 and **P < 0.01.
To investigate whether direct interaction of YBX1 with circNEIL3 mediate the antimetastatic effect of circNEIL3, cells overexpressing circNEIL3-WT or circNEIL3-mut variants were used to perform in vitro and in vivo experiments. The results showed that overexpression of circNEIL3-mut failed to inhibit cell migration and invasion, whereas circNEIL3-WT exhibited a potent antimigration and anti-invasion effect (Fig. 4 H and I). Moreover, wild-type circNEIL3, rather than its YBX1 interaction-deficient variants, dramatically relieved metastatic tumor burden in the mouse tail vein metastasis model (Fig. 4 J and K). Collectively, the direct interaction of circNEIL3 with YBX1 contributes to repressing tumor metastasis.
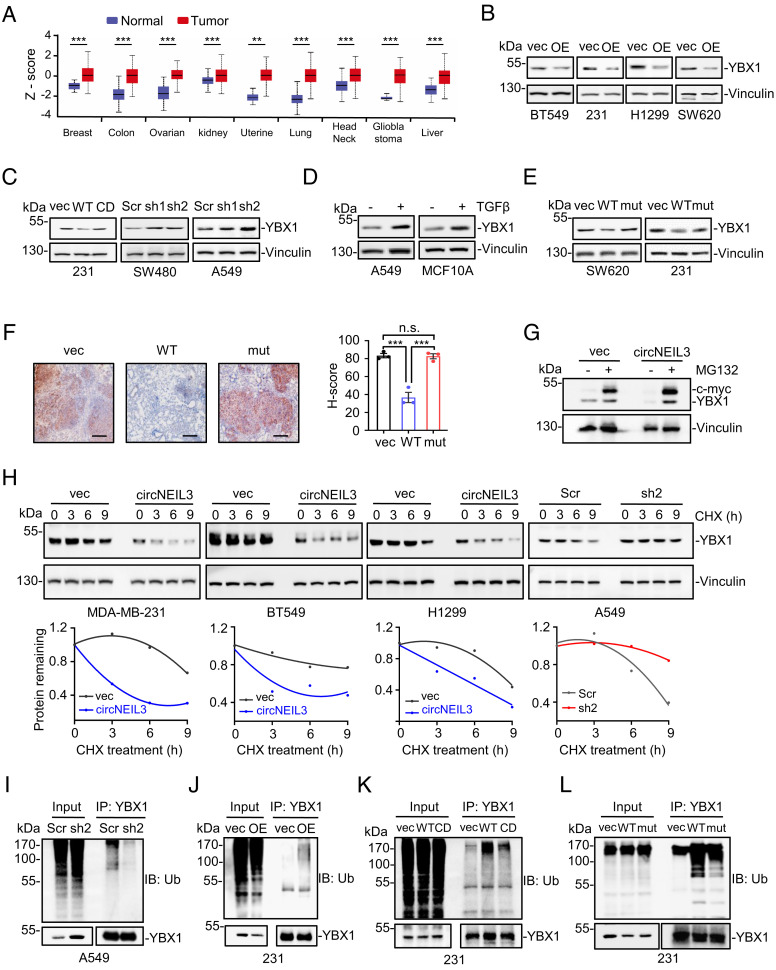
circNEIL3 Promotes YBX1 Degradation through Ubiquitination/Proteasome Pathway.
Since circNEIL3 was identified to directly interact with YBX1, we sought to elucidate the molecular consequences of such interaction. Considering the pivotal role of YBX1 in promoting cell metastasis (5–7), we first analyzed YBX1 protein levels in the Clinical Proteomic Tumor Analysis Consortium (CPTAC) database (http://ualcan.path.uab.edu) and found a consistent upregulation of YBX1 in various cancer types (Fig. 5A), supporting that dysregulation of YBX1 protein may act as an important step during cancer progression. Thus, we wonder whether circNEIL3 could affect YBX1 expression. To this end, YBX1 protein levels were measured in the aforementioned stable cells. As expected, enforced circNEIL3 expression robustly decreased YBX1 protein level in various cancer cells (Fig. 5B), whereas enforced expression of the linear form of circNEIL3 (circNEIL3-CD) showed a minimal effect on YBX1 expression (Fig. 5 C, Left), indicating that the decrease of YBX1 protein by circNEIL3 depends on the mature form of circNEIL3. Consistently, circNEIL3 knockdown increased YBX1 expression in both SW480 and A549 cells (Fig. 5 C, Middle and Right), thus reinforcing the conclusion that circNEIL3 down-regulates YBX1 protein levels. Furthermore, YBX1 protein level was elevated upon TGFβ stimulation in both A549 and MCF10A cells (Fig. 5D) as a consequence of decreased circNEIL3 expression (Fig. 1F). Substantially, the effect of circNEIL3 on YBX1 protein levels was further confirmed to depend on their direct interaction as overexpressing the YBX1-binding-deficient circNEIL3 variant failed to affect YBX1 expression in two cancer cell lines (Fig. 5E) and pulmonary metastatic tissues (Fig. 5F). Collectively, these findings demonstrate that circNEIL3 represses YBX1 protein level through their direct interaction.
Fig. 5.
circNEIL3 promotes YBX1 degradation through the ubiquitin/proteasome pathway. (A) Bioinformatics analysis of YBX1 protein levels in various cancers from the CPTAC database. (B) circNEIL3 overexpression decreased YBX1 expression in different cancer cells. (C) Immunoblot assays to measure YBX1 expression in cells stably expressing the empty vector, WT circNEIL3, circularization-dead (CD) mutant, or circNEIL3 shRNAs. (D) Immunoblotting assays for YBX1 expression in A549 and MCF10A cells treated with 5ng/mL TGFβ for 72h. (E) Immunoblot assays to measure YBX1 expression in cells stably expressing the empty vector, WT circNEIL3, or YBX1-binding-deficient mutants (mut). (F) Representative Immunohistochemistry (IHC) staining images using an anti-YBX1 antibody of the metastatic lesions in the lung from mice tail vein injected with MDA-MB-231 stable cells. Histochemistry score (H score) is presented as mean scores ± SEM of three mice in each group. n.s., not significant; ***P < 0.001. (G) Effects of MG132 treatment on protein levels of YBX1 and c-myc in MDA-MB-231 cells with or without circNEIL3 overexpression. (H) Effect of circNEIL3 on YBX1 protein stability. Different cancer cells with circNEIL3 overexpression or knockdown were treated with CHX for indicated time and then subjected to immunoblot assays. (I–L) Polyubiquitination status of YBX1 immunoprecipitated from different cancer cells stably expressing circNEIL3 shRNA (I), WT circNEIL3 (J), circularized-dead (CD) circNEIL3 (K), or its YBX1-binding-deficient mutants (mut) (L).
Given that overexpression of circNEIL3 has little effect on YBX1 mRNA level (SI Appendix, Fig. S5A), we wonder whether circNEIL3 could affect YBX1 protein stability. To this end, cells were treated with the proteasome inhibitor MG132 to effectively block protein degradation, as indicated by the increased c-Myc expression after treatment (Fig. 5G). Interestingly, circNEIL3-induced YBX1 decline was fully prevented by MG132 treatment (Fig. 5G), indicating that circNEIL3 promotes proteasomal degradation of YBX1. To further confirm this, cells were treated with cycloheximide (CHX) to block protein synthesis for indicated times and then subjected to immunoblot assays. As expected, circNEIL3 overexpression dramatically shortened the half-life of YBX1 proteins in different cancer cells, while circNEIL3 depletion exhibited the opposite effect (Fig. 5H and SI Appendix, Fig. S5 B and C). Considering that polyubiquitination modification plays a central role in protein degradation (31), we next examined the effect of circNEIL3 on ubiquitination status of YBX1. The results showed that circNEIL3 knockdown significantly decreased the polyubiquitination levels of YBX1 (Fig. 5I), while circNEIL3 overexpression had the opposite effect (Fig. 5 J and K). Notably, the expression of the YBX1-binding-deficient circNEIL3 variant demonstrated less upregulation of YBX1 polyubiquitination compared to its wide-type counterpart (Fig. 5L). Taken together, our results indicate that circNEIL3 promotes YBX1 degradation through the ubiquitination/proteasome pathway.
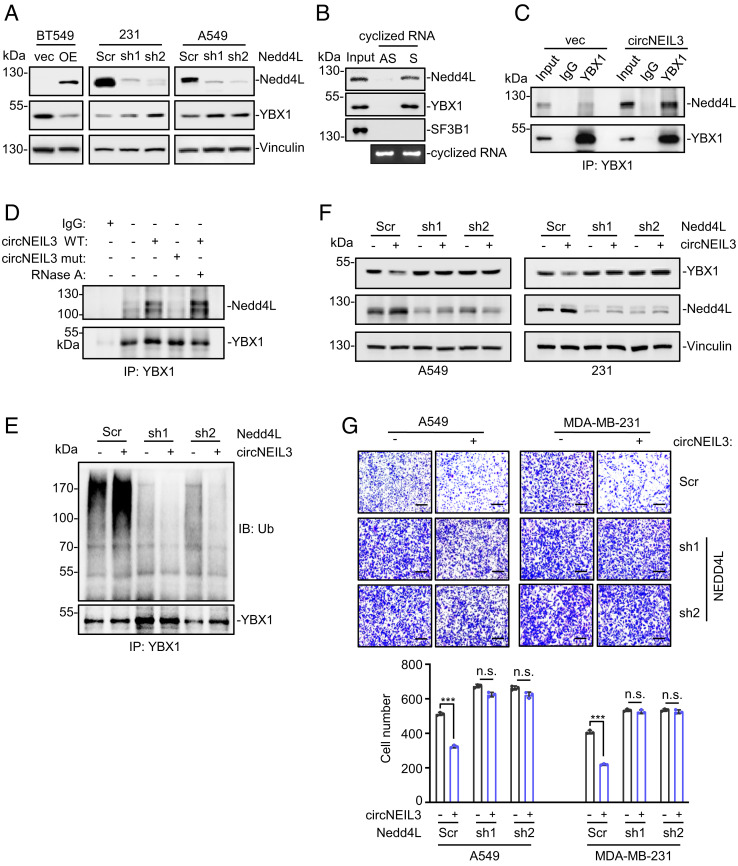
circNEIL3 Recruits the E3 Ligase Nedd4L to Promote YBX1 Degradation.
Previous studies reported that Nedd4L is the specific E3 ligase to trigger YBX1 degradation (15), which was experimentally confirmed in our cell systems after Nedd4L overexpression or knockdown (Fig. 6A). So we speculated that circNEIL3 could recruit Nedd4L and YBX1 together, thus destabilizing YBX1 protein. To test this assumption, the in vitro transcribed and circularized circNEIL3 was incubated with cell extracts. The results showed that both YBX1 and Nedd4L were captured by circNEIL3 (Fig. 6B), indicating the formation of circNEIL3–Nedd4L–YBX1 complex. Moreover, circNEIL3 overexpression significantly increased the association of Nedd4L with YBX1 (Fig. 6C). Besides, such increased Nedd4L–YBX1 association required the direct interaction between YBX1 and circNEIL3, as enforced expression of the YBX1-binding-deficient circNEIL3 variant failed to strengthen Nedd4L–YBX1 association (Fig. 6D). The circNEIL3-increased Nedd4L–YBX1 association is resistant to the exonuclease RNase A, pronouncing the requirement of mature circNEIL3, rather than its linear transcripts, for their association (Fig. 6D). Notably, the circNEIL3-mediated YBX1 degradation relied on Nedd4L because the increase of YBX1 polyubiquitination (Fig. 6E) and the decrease of YBX1 protein (Fig. 6F) by circNEIL3 were attenuated after Nedd4L knockdown with two independent shRNAs. In accordance to these findings, the antimetastatic effects of circNEIL3 were effectively weakened by Nedd4L knockdown (Fig. 6G). Collectively, these results support that circNEIL3 recruits Nedd4L and YBX1 together, leading to polyubiquitination and degradation of YBX1.
Fig. 6.
circNEIL3 recruits the E3 ligase Nedd4L to promote YBX1 degradation. (A) Effects of Nedd4L overexpression (WT) or knockdown by two independent shRNAs on YBX1 expression. (B) RNA pull-down assays showing the association of the in vitro circularized circNEIL3 with YBX1 and Nedd4L. (C) circNEIL3 overexpression in MDA-MB-231 cells increased the association of Nedd4L with YBX1. (D) The increased Nedd4L–YBX1 association by circNEIL3 depends on the direct interaction between YBX1 and circNEIL3. (E) Polyubiquitination status of YBX1 immunoprecipitated from MDA-MB-231 cells stably expressing circNEIL3 or Nedd4L shRNA. (F) Immunoblot assays to measure the expression of YBX1 and Nedd4L proteins in A549 and MDA-MB-231 stable cell lines expressing circNEIL3 or Nedd4L shRNA. (G) Cell migration assays of different stable cells expressing circNEIL3 or Nedd4L shRNA. Data are presented as means ± SD. n.s., not significant; ***P < 0.001.
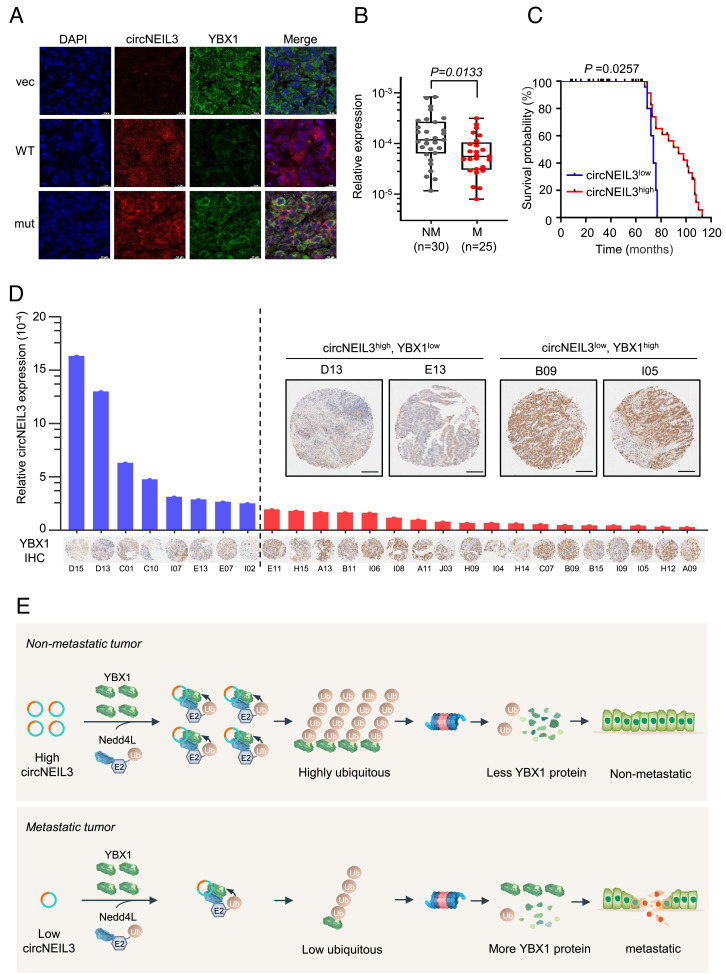
circNEIL3 Expression Is Negatively Correlated to YBX1 Abundance and Tumor Metastasis in CRC Samples.
To further confirm the negative correlation between circNEIL3 and YBX1, we stressed SW480 cells with hypoxia or oncostatin M (OSM, a JAK2/STAT3 activator), both of which are implicated in cancer metastasis. Both hypoxia and OSM decreased circNEIL3 levels (SI Appendix, Fig. S5D) while increased YBX1 protein levels (SI Appendix, Fig. S5E), thus supporting a wide existence of circNEIL3–YBX1 axis in cancer metastasis. Besides, circNEIL3 is decreased more strikingly in hypoxia-active CRC pairs (SI Appendix, Fig. S5F). Moreover, the strikingly higher YBX1 protein level, not its mRNA level, in metastatic SW620 CRC cells than its primary counterpart SW480 may be attributed to the distinguished circNEIL3 expression in these two cell lines (SI Appendix, Fig. S5 G and H). Next, we simultaneously examined circNEIL3 expression by FISH assay and YBX1 expression by immunofluorescence assay in the metastatic lesions within the lung of mice tail vein injected with MDA-MB-231 cells stably expressing empty vector or indicated circNEIL3 variants (Fig. 4 J and K). As shown in Fig. 7A, overexpression of wild-type circNEIL3 significantly decreased YBX1 protein levels, while the YBX1-binding-deficient circNEIL3 mutant failed to repress YBX1 expression. Thus, circNEIL3 negatively regulates YBX1 expression in vivo. Although circNEIL3 is up-regulated in primary CRC tissues than their adjacent tissues (SI Appendix, Fig. S1B), higher YBX1 proteins may be attributed to both the increased YBX1 mRNA levels (SI Appendix, Fig. S5I) and the decreased E3 ligase NEDD4L (SI Appendix, Fig. S5J). Given that circNEIL3 represses tumor metastasis (Fig. 2 and SI Appendix, Fig. S3) through promoting polyubiquitination and degradation of YBX1 (Fig. 5–6), we sought to explore their clinical relevance in tumor samples with metastasis information. To this end, the cDNA array containing a cohort of 80 CRC samples was employed to measure circNEIL3 abundance by RT–qPCR. After removing 25 samples because of inconvincible amplification curves, the remaining 55 samples were divided into two groups according to the presence or absence of lymph node metastatic status. The results showed that the CRC samples with lymph node metastasis exhibited lower circNEIL3 expression than those without metastasis (Fig. 7B and SI Appendix, Table S4), indicating that lower circNEIL3 expression may predict a metastatic tendency of CRC. In accordance with this finding, CRC patients with lower circNEIL3 expression exhibit shorter life spans compared to those with a higher circNEIL3 level (Fig. 7C and SI Appendix, Table S5), implying a negative correlation between circNEIL3 expression and the survival of CRC patients. Moreover, YBX1 protein levels were also examined by immunohistochemistry staining in another CRC tissue array. After omitting the invalid samples with less than 30% intact tissue area, the remaining 26 tumors that obtained from the same patients as the aforementioned cDNA array were used to analyze the correlation between YBX1 protein level and the circNEIL3 expression. As shown in Fig. 7D and SI Appendix, Fig. S5K, CRC tumors with higher circNEIL3 expression exhibited a lower YBX1 protein level, thus reinforcing a negative correlation between circNEIL3 expression and YBX1 protein levels. Therefore, circNEIL3 expression is negatively correlated to YBX1 abundance and tumor metastasis in CRC.
Fig. 7.
circNEIL3 expression is negatively correlated to YBX1 abundance and tumor metastasis in patients with CRC. (A) FISH assays to detect circNEIL3 expression and immunofluorescence assays to measure YBX1 expression in the metastatic lesions within the lung of mice tail vein injected with MDA-MB-231 cells stably expressing the empty vector or indicated circNEIL3 variants. (B) Comparison of circNEIL3 expression measured by RT–qPCR between nonmetastatic CRC tumors and metastatic samples. (C) The Kaplan–Meier curve for the overall survival of CRC patients, who were divided into two groups according to the mean expression of circNEIL3 variants. The P value was calculated by the two-sided log-rank test. (D) circNEIL3 expression measured by RT–qPCR and YBX1 expression by IHC staining in the tested CRC tumors. (E) Proposed working model for inhibitory effects of circNEIL3 on tumor metastasis. In nonmetastatic tumors, circNEIL3 recruits the E3 ligase Nedd4L and YBX1 together, leading to polyubiquitination and subsequent proteasomal degradation of YBX1. In metastatic tumors, circNEIL3 expression was down-regulated under certain stress such as TGFβ stimulation, causing increased YBX1 protein level to promote tumor metastasis.
Discussion
Accumulating evidence supports the implication of circRNA in tumor metastasis, but the detailed mechanism underlying this biological process remains elusive. In this study, a TGFβ-repressive circRNA, circNEIL3 (hsa_circ_0001460), was identified to be dramatically down-regulated in the liver metastatic samples of CRC patients (Fig. 1). As expected, circNEIL3 potently repressed tumor metastasis in various cancer models (Fig. 2 and SI Appendix, Fig. S3). Mechanistically, circNEIL3 promotes proteasomal degradation of YBX1, an oncogenic RBP, by recruiting YBX1 and its E3 ligase Nedd4L together to enhance its polyubiquitination (Figs. 4–6). Notably, decreased circNEIL3 expression is significantly correlated to elevated YBX1 protein level, serving as a negative indicator for metastasis and a predictor for worse CRC patient outcome (Fig. 7). In conclusion, circNEIL3 functions as a potent antimetastatic factor by directly targeting YBX1 for Nedd4L-mediated proteasomal degradation (Fig. 7E).
Although some circRNAs were identified to affect tumor metastasis (25–27), only few circRNAs were proven to be effective among different cancers (32). As a dysregulated circRNA identified in CRC, circNEIL3 demonstrated consistent antimigration and anti-invasion effects in colorectal cancer cells, breast cancer cells, and lung cancer cells (Fig. 2 A–E and SI Appendix, Fig. S2 B–F), supporting the pan-cancerous role of circNEIL3. Moreover, the antimetastatic effect of circNEIL3 was further validated in in vivo mouse models established with breast cancer cells and lung cancer cells (Fig. 2 F–J). Notably, such antimetastatic effects were proven to be contributed by the circularized form rather than the linear transcript of circNEIL3 (SI Appendix, Fig. S3). Importantly, the repressive effect of circNEIL3 on tumor metastasis depends on its direct interaction with the oncogenic RBP YBX1 as either restoration of YBX1 expression (Fig. 3 H–J) or destruction of the circNEIL3–YBX1 interaction (Fig. 4 H–K) attenuated the antimetastatic effect of circNEIL3. Collectively, these results strongly support that circNEIL3 functions as a potent antimetastatic factor through its direct interaction with YBX1.
Recent studies reported that circNEIL3 (hsa_circ_0001460) is up-regulated in glioma (33) and pancreatic ductal adenocarcinoma (34), which seems to be inconsistent to our finding that circNEIL3 is down-regulated in metastatic CRC samples compared to primary tumors (Fig. 1C). These discordances may be attributed to the change of circNEIL3 expression during different stages of cancer progression. Indeed, we also found that circNEIL3 demonstrated a higher expression level in the primary CRC samples than their adjacent colon tissues (SI Appendix, Fig. S1 B and C). However, circNEIL3 exhibited significantly decreased expression in metastatic lesions of liver in patients with CRC (Fig. 1C and SI Appendix, Fig. S1D). This may reflect that circNEIL3 plays an oncogenic role in tumor initiation and functions as a tumor suppressor for tumor metastasis. Such phenomena are also observed in the scenario of TGFβ signaling which represses cancer initiation at the early stage of tumorigenesis but promotes distant metastasis for advanced tumors (35). Therefore, exploring the function of a candidate should be focused on specific condition.
circRNAs were demonstrated as pivotal scaffolds to facilitate the assembly of protein complex (19, 36). For example, circACC1 was reported to form a ternary complex with the regulatory β and γ subunits of AMPK in response to serum deprivation, thus promoting the enzymatic activity of AMPK holoenzyme (36). Herein, we found that circNEIL3 strengthened the association between YBX1 and its E3 ligase Nedd4L to promote polyubiquitination and degradation of YBX1 (Fig. 6 C–E). Interestingly, we also observed that circNEIL3 enhanced Nedd4L expression (Fig. 6F), but the underlying molecular mechanism has not yet been elucidated. Combining the fact that Nedd4L plays a tumor-suppressive role in constraining TGFβ signaling by triggering the proteasomal degradation of SMAD2, SMAD3, and SMAD7 (37), we speculate that circNEIL3 mediates the formation of a positive feedback loop between Nedd4L and the TGFβ signaling pathway. Activated TGFβ signaling within tumor microenvironment (38) leads to the reduction of circNEIL3 expression, which in turn down-regulates Nedd4L expression to increase SMAD protein levels, thereby promoting TGFβ-mediated metastasis. This positive feedback loop in cancer cells provides insight into the regulatory mechanism in tumor metastasis.
In conclusion, our results demonstrate that the TGFβ-repressed circNEIL3 inhibits tumor metastasis by enhancing proteasomal degradation of YBX1. Considering that circRNAs possess unique features of higher stability and less immunogenicity, circNEIL3-based cancer therapy could be a potential therapeutic strategy.
Materials and Methods
Cell Lines and Tissues.
BT549 and MDA-MB-231 (human breast cancer cell lines), A549 and H1299 (human lung cancer cell lines), and SW480 and SW620 (human colorectal cancer cell lines) were cultured at 37 °C in an incubator with 5% CO2. MDA-MB-231, BT549, and H1299 were maintained in RPMI-1640 (Gibco) containing 10% fetal bovine serum (FBS) (PAN-Biotech, #ST30-3302). A549, SW480, and SW620 were cultured in DMEM (Gibco) supplemented with 10% FBS. cDNA array (# cDNA-HColA095Su02) and CRC tissue array (# HColA160Su02) were purchased from ShangHai OUTDO Biotech. In total, 68 CRC primary tumors, their paired liver metastatic tumor tissues, and adjacent normal tissues were collected from patients who underwent surgery at West China Hospital, Sichuan University. This study was approved by the Ethics Committee of West China Hospital, Sichuan University (2018(280); 2019(338)), and informed consent was obtained from patients or their families, as appropriate. All paired samples were confirmed by two pathologists independently and stored at −80 °C until use. Briefly, the patients’ tissues were dissected into small pieces and grounded into powder in the liquid nitrogen. Subsequently, cell lysates were obtained by adding RNAiso Plus reagent (TaKaRa, #9108). The samples were subjected to Ribo-Zero RNA sequencing.
Animal Experiments.
All mouse experiment procedures performed in this study were approved by the Institutional Animal Care and Use Committee of West China Hospital, Sichuan University. The 4-wk-old female BALB/c nude mice were purchased from Beijing Hua Fu Kang (HFK) Bioscience. For the lung metastasis model, 1 × 106 luciferase-labeled MDA-MB-231 cells with enforced expression of circNEIL3 variants were injected into the tail veins of 4-wk-old nude mice. Nine weeks after cell injection, tumor metastasis was analyzed. For the systemic tumor metastasis model, 1 × 105 luciferase-labeled A549 cells with circNEIL3 knockdown by independent shRNAs were injected into the left cardiac ventricle of 4-wk-old nude mice. Five weeks later, mice were used for analysis of the systemic spread of tumor. Bioluminescence images of tumor-bearing mice were acquired with IVIS Spectrum (PerkinElmer) at 10 min after intraperitoneal injection of D-luciferin (15 mg/mL, Promega) and analyzed using Living Image 3.2 software package (Caliper Life Sciences). Next, the lungs and brains were obtained and fixed with 4% formalin, while the bones were fixed with 10% formaldehyde after decalcification in 4% hydrochloric acid/formic acid working solution for at least 1 wk. The numbers of metastatic nodules in the lungs, bones, and brains were carefully examined by H&E staining.
Statistical Analyses.
Statistical analyses were performed using SPSS 26.0 statistical software package International Business Machines Corporation (IBM) or GraphPad Prism 8.0. For statistical evaluation, data were analyzed by two-tailed Student’s t test for comparisons of two samples and one-way ANOVA for univariate comparisons. Significance of differences was determined by the paired two-sided Wilcoxon signed-rank test for circNEIL3 expressions in primary CRC sample and its corresponding liver metastasis or its adjacent colon tissues. Overall survival was assessed with the Kaplan–Meier analysis. Results are presented as mean ± SD in cellular experiments and ±SEM in the animal models. P values < 0.05 were considered statistically significant (*P < 0.05, **P < 0.01, and ***P < 0.001); P > 0.05, not significant (n.s.).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We appreciate all patients and control subjects who participated in this study. This work was supported by National Key R&D Program of China (2022YFA1303200 and 2018YFC2000305), the Fundamental Research Funds for the Central Universities (SCU2022D025), National Natural Science Foundation of China (82002570 and 82102982), Science and Technology Foundation of Sichuan Province, China (2022YFS0046, 2022NSFSC1461, and 2022NSFSC1296), and the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYGD20008 and ZYYC20007).
Author contributions
H.X. and Y.P. designed research; S.C., K.L., J.G., H.-N.C., Y.M., J.C., S.S., and Z.-G.Z. performed research; F.X., Z.Y., H.M., J.-K.Z., and Z.L. contributed new reagents/analytic tools; S.C., K.L., J.G., Y.J., T.Z., Y.Y., W.L., and L.D. analyzed data; and S.C., K.L., J.G., H.X., and Y.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Heng Xu, Email: xuheng81916@scu.edu.cn.
Yong Peng, Email: yongpeng@scu.edu.cn.
Data, Materials, and Software Availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or SI Appendix. RNA-seq data of TGFβ-treated MCF-10A cells have been deposited in the Gene Expression Omnibus database with accession no. GSE165576. The mass spectrum data have been uploaded to Proteomics Identification Database with project accession no. PXD039394. The datasets describing 68 CRC patient samples have been submitted to the Genome Sequence Archive database, China National Center for Bioinformation under accession No.HRA003989.
Supporting Information
References
- 1.Weiss F., Lauffenburger D., Friedl P., Towards targeting of shared mechanisms of cancer metastasis and therapy resistance. Nat. Rev. Cancer 22, 157–173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Lambert A. W., Pattabiraman D. R., Weinberg R. A., Emerging biological principles of metastasis. Cell 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Priestley P., et al. , Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan H., Li S., Bao L., Zhang X., Enhanced YB1/EphA2 axis signaling promotes acquired resistance to sunitinib and metastatic potential in renal cell carcinoma. Oncogene 39, 6113–6128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Naggar A. M., et al. , Translational activation of HIF1alpha by YB-1 promotes sarcoma metastasis. Cancer Cell 27, 682–697 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Evdokimova V., et al. , Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15, 402–415 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Jiang D., et al. , YB-1 is a positive regulator of KLF5 transcription factor in basal-like breast cancer. Cell Death Differ. 29, 1283–1295 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao Z., et al. , Targeting the YB-1/PD-L1 axis to enhance chemotherapy and antitumor immunity. Cancer Immunol. Res. 7, 1135–1147 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Shibahara K., et al. , Nuclear expression of the Y-box binding protein, YB-1, as a novel marker of disease progression in non-small cell lung cancer. Clin. Cancer Res. 7, 3151–3155 (2001). [PubMed] [Google Scholar]
- 11.Perner F., et al. , YBX1 mediates translation of oncogenic transcripts to control cell competition in AML. Leukemia 36, 426–437 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayavelu A. K., et al. , Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 588, 157–163 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Feng M., et al. , YBX1 is required for maintaining myeloid leukemia cell survival by regulating BCL2 stability in an m6A-dependent manner. Blood 138, 71–85 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gluz O., et al. , Y-box-binding protein YB-1 identifies high-risk patients with primary breast cancer benefiting from rapidly cycled tandem high-dose adjuvant chemotherapy. J. Clin. Oncol. 27, 6144–6151 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Huang S., et al. , Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 139, 2857–2876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutz M., Wempe F., Bahr I., Zopf D., von Melchner H., Proteasomal degradation of the multifunctional regulator YB-1 is mediated by an F-Box protein induced during programmed cell death. FEBS Lett. 580, 3921–3930 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Xu J., et al. , CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target. Ther. 5, 298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X., et al. , Decreased expression of NEDD4L contributes to NSCLC progression and metastasis. Biochem. Biophys. Res. Commun. 513, 398–404 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Shi L., et al. , A tumor-suppressive circular RNA mediates uncanonical integrin degradation by the proteasome in liver cancer. Sci. Adv. 7, eabe5043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Z. X., et al. , A novel NF-kappaB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol. Cancer 20, 103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B., et al. , circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 12, 295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L. L., The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21, 475–490 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Misir S., Wu N., Yang B. B., Specific expression and functions of circular RNAs. Cell Death Differ. 29, 481–491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conn S. J., et al. , The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Chen R. X., et al. , N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 10, 4695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanniford D., et al. , Epigenetic Silencing of CDR1as Drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell 37, 55–70.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., et al. , CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl. Acad. Sci. U.S.A. 118, e2012881118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peixoto P., et al. , EMT is associated with an epigenetic signature of ECM remodeling genes. Cell Death Dis. 10, 205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Sun D., Pu W., Wang J., Peng Y., Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 6, 319–336 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Wei W. J., et al. , YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 40, 8622–8636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swatek K. N., Komander D., Ubiquitin modifications. Cell Res. 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang C., et al. , The emerging picture of the roles of circRNA-CDR1as in cancer. Front. Cell Dev. Biol. 8, 590478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan Z., et al. , EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 21, 16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen P., et al. , CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing. Mol. Cancer 20, 51 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David C. J., Massague J., Contextual determinants of TGFbeta action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q., et al. , CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 30, 157–173 e157 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Gao S., et al. , Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol. Cell 36, 457–468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derynck R., Turley S. J., Akhurst R. J., TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 18, 9–34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or SI Appendix. RNA-seq data of TGFβ-treated MCF-10A cells have been deposited in the Gene Expression Omnibus database with accession no. GSE165576. The mass spectrum data have been uploaded to Proteomics Identification Database with project accession no. PXD039394. The datasets describing 68 CRC patient samples have been submitted to the Genome Sequence Archive database, China National Center for Bioinformation under accession No.HRA003989.