Abstract
Despite the approval of vaccines, monoclonal antibodies and restrictions during the pandemic, the demand for new efficacious and safe antivirals is compelling to boost the therapeutic arsenal against the COVID-19. The viral 3-chymotrypsin-like protease (3CLpro) is an essential enzyme for replication with high homology in the active site across CoVs and variants showing an almost unique specificity for Leu-Gln as P2–P1 residues, allowing the development of broad-spectrum inhibitors.
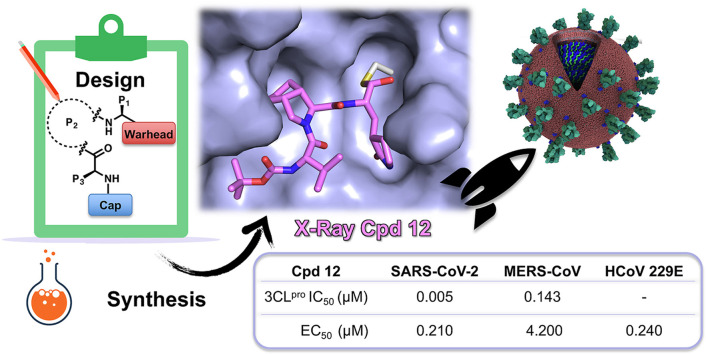
The design, synthesis, biological activity, and cocrystal structural information of newly conceived peptidomimetic covalent reversible inhibitors are herein described. The inhibitors display an aldehyde warhead, a Gln mimetic at P1 and modified P2–P3 residues. Particularly, functionalized proline residues were inserted at P2 to stabilize the β-turn like bioactive conformation, modulating the affinity. The most potent compounds displayed low/sub-nM potency against the 3CLpro of SARS-CoV-2 and MERS-CoV and inhibited viral replication of three human CoVs, i.e. SARS-CoV-2, MERS-CoV, and HCoV 229 in different cell lines. Particularly, derivative 12 exhibited nM-low μM antiviral activity depending on the virus, and the highest selectivity index. Some compounds were co-crystallized with SARS-CoV-2 3CLpro validating our design. Altogether, these results foster future work toward broad-spectrum 3CLpro inhibitors to challenge CoVs related pandemics.
Keywords: SARS-CoV-2, 3C-like protease inhibitors, Peptidomimetics, Reversible covalent inhibitors, COVID-19, Crystal structure
Graphical abstract
1. Introduction
Since the early 2020, the pandemic Coronavirus Disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) poses a global threat due to its ability to rapidly spread person-to-person via respiratory droplets and its capacity to suppress human immune surveillance [1]. SARS-CoV-2 is a member of the Coronaviridae family, specifically belonging to the β group, which includes other highly pathogenic coronaviruses (CoVs) such as Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV), that caused the 2003 and the 2011 epidemics, respectively [2]. Other known human CoVs, such as HCoV-229E and HCoV-NL63, belong to the α group and are associated to the seasonal common cold diseases [3,4]. Unfortunately, SARS-CoV-2 spread has been much more extensive in comparison to the latter two family members, exceeding the 676 million reported cases and the 6.8 million deaths [5], representing the worst pandemic in the last 100 years. Fear for forthcoming CoV outbreaks beyond COVID-19 is justified by the increasing processes of anthropization, deforestation, and human mobility that may accelerate virus transmission from wildlife to humans and human-to-human spread.
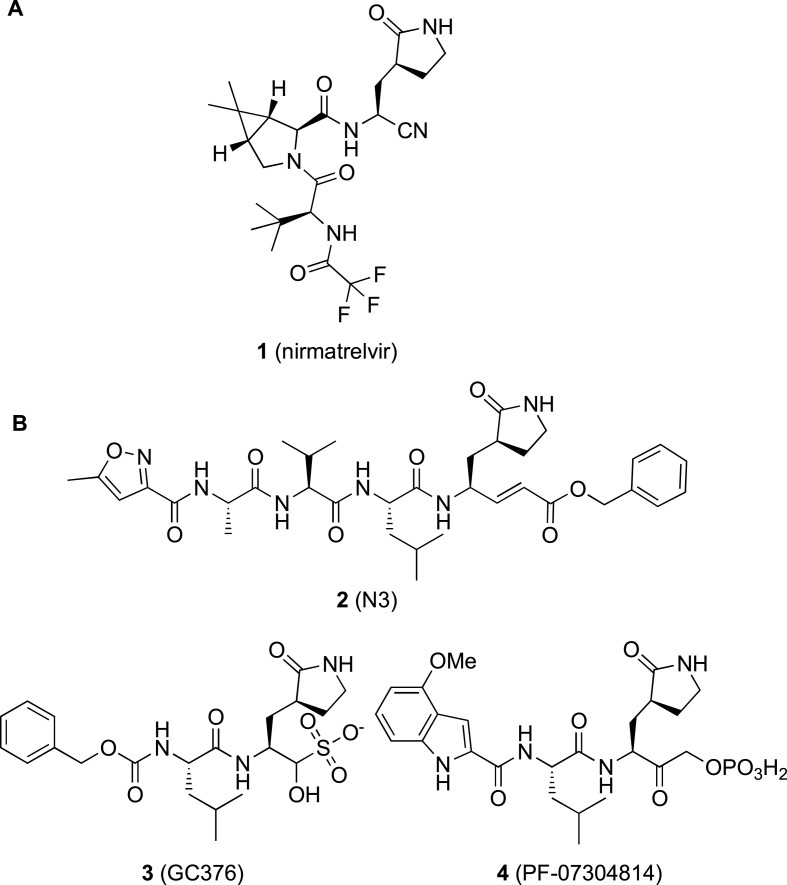
Throughout the last two years the scientific community has been ramping-up efforts to identify COVID-19 treatments and preventives at an unprecedented speed. Vaccines had a huge impact on the pandemic control, effectively protecting from serious symptoms but without blocking the virus transmission [6]. To contain outbreaks by emerging viral variants, like omicron, the last entry in 2021, a third and in some cases a fourth booster dose of vaccine were necessary [6]. Nevertheless, fully vaccinated people and COVID-19 experienced patients can be re-infected. Major benefits will come from the availability of direct-acting antivirals (DAAs) specifically targeting essential CoV enzymes for replication [7]. Most importantly, DAAs are the only approach to treat immunocompromised patients, and their manufacturing, distribution, and administration are easier than for vaccines, thus likely reaching also developing countries where vaccination rate is still very low. Despite the unprecedented effort of repurposing campaigns and focused medicinal chemistry programs, only three DAAs have been approved so far [7]. Specifically, remdesivir (intravenous injection) [8] and molnupiravir (oral administration) [9] are repurposed nucleotide inhibitors of the viral RNA-dependent RNA polymerase with moderate efficacy [7]. Random drug repurposing, however, failed to identify highly effective COVID-19 therapies, thus the development of specific DAAs meets a pressing medical need. The discovery of nirmatrelvir [10] (1 in Fig. 1 A) represents a step forward, being the first-in-class approved inhibitor of the 3-chymotrypsin-like-cysteine-protease, called also Main protease (3CLpro or Mpro, respectively). Compound 1 was designed as peptidomimetic reversible covalent inhibitor of SARS-CoV-2 3CLpro, but due to its high metabolic degradation it is co-dosed in combination with ritonavir (a cytochrome inhibitor) to allow an acceptable oral dosing regimen. Paxlovid® is indeed a combination of nirmatrelvir and ritonavir, administered in two high doses a day. Different research groups have reported additional peptidomimetic [11] and non-peptidic [12] inhibitors of 3CLpro that are currently in different phases of clinical investigation for COVID-19.
Fig. 1.
A) First-in-class 3CLpro inhibitor nirmatrelvir 1 approved as active component of the oral drug Paxlovid® for COVID-19 treatment; B) representative examples of peptidomimetic inhibitors (2–4) of 3CLpro from different CoVs repurposed on SARS-CoV-2.
The SARS-CoV-2 genome is a large (∼30 kb) single strand of 5′-capped (+)-RNA which contains two open reading frames (ORF1a and ORF1ab) and encodes multiple structural and nonstructural proteins [13]. Translation of ORF1a and ORF1ab into polypeptides pp1a and pp1ab is followed by proteolytic processing by the viral 3CLpro and a papain-like cysteine protease, resulting in 16 mature nonstructural proteins which are involved in the replication−transcription complex. The two proteases are essential for viral replication, making them attractive targets for therapeutic intervention.
SARS-CoV-2 3CLpro is a 33.8 kDa homodimer with a catalytic Cys−His dyad (Cys145−His41) and an extended substrate binding site, with an almost unique specificity, not present in mammalian proteases, for Leu-Gln-Ser (Ala, Gly) as preferred P2–P1–P1’ sequence [14]. Since the pandemic started, several peptidomimetics have been investigated as SARS-CoV-2 3CLpro inhibitors and many crystal structures of such compounds in complex with the enzyme have been solved [14]. Several compounds targeting the 3CLpro of different CoVs have been successfully repurposed as anti-SARS-CoV-2 agents, alongside novel chemical series of peptidomimetics have been reported, for the most structurally related to compounds 2–4 (Fig. 1B) [14]. These compounds are covalent reversible/irreversible inhibitors functionalized with an electrophilic warhead acting as cysteine trap (i.e., the catalytic Cys145), and with P1–P4 groups mimicking the sequence of the natural substrate to gain affinity and binding specificity. Indeed, a similar design strategy had previously afforded the α-ketoamide inhibitors of Hepatitis C Virus (HCV) NS3/4A serine protease, telaprevir and boceprevir, approved by FDA as the first HCV DAAs. In addition, considering that the length of COVID-19 treatment should be relatively short, this approach appears as a privileged route to discovery drugs targeting SARS-CoV-2 3CLpro with limited side effects. The discovery of nirmatrelvir also relied on this approach: this molecule is indeed a hybrid obtained by merging the P2–P3 residues present in the drug boceprevir, and the Gln mimetic group shared as P1 by most of the known potent 3CLpro inhibitors.
In this contribution we reported on a new series of tripeptides as covalent reversible inhibitors of SARS-CoV-2 3CLpro, mainly investigating the effect of differently functionalized proline residues in the P2 position, describing the rational design, synthesis, biological evaluation in biochemical and antiviral assays, and X-ray structures of representative compounds in complex with the molecular target. The most promising inhibitors were also investigated as broad-spectrum 3CLpro inhibitors against MERS and HCoV 229E, as representative members of the β- and a α-CoV, respectively. Indeed, MERS and HCoV 229E 3CLpros are structurally similar to SARS-CoV-2 homologue enzyme, respectively sharing 77% and 69% similarity in the active site [15], thus potent 3CLpro inhibitors could retain activity against a panel of CoVs.
2. Results and discussion
2.1. Rational design and molecular modelling
In the first released X-ray structure, the SARS-CoV-2 3CLpro was in complex with the repurposed tetrapeptide 2 [16] (PDB 6LU7, superseded by 7BQY; Fig. 1, Fig. 3A), an irreversible inhibitor endowed with a vinyl carboxyl ester, acting as the Michael acceptor warhead, while sharing with other peptidomimetic inhibitors a Gln mimetic γ-lactam and a Leu residue in P1 and P2 positions, respectively. Previously, 2 was shown to inhibit SARS- and MERS-CoVs 3CLpros in enzymatic assays with K i = 9 μM [17] and IC50 = 0.3 μM [18] respectively, and was then repurposed also against SARS-CoV-2, showing EC50 = 16.77 μM in a phenotypic cell-based assay. However, no data in biochemical assays were reported against SARS-CoV-2 3CLpro [16].
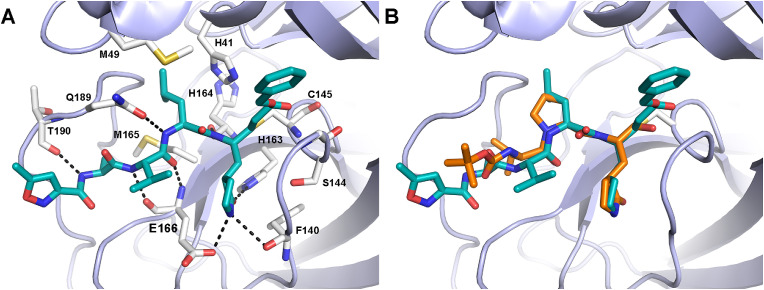
Fig. 3.
A) Crystallographic pose of compound 2 (cyan sticks) covalently bound to the active site of SARS-CoV-2 3CLpro (PDB: 7BQY). B) Docking pose of the designed tripeptide aldehyde 6 (orange sticks) overlaid with the X-ray conformation of 2. The enzyme is represented as light-blue cartoons. Residues important for ligand binding are displayed as white sticks. H-bonds are depicted as dashed black lines. Hydrogens are omitted for clarity.
Starting from this inhibitor, we designed a truncated analogue by: i) removing the P4 Ala residue, in order to reduce the molecular size while retaining the natural substrate sequence, and ii) replacing the vinyl ester with an aldehyde to obtain a more suitable covalent reversible inhibitor. The resulting tripeptide 5 (Fig. 2 ; MW = 468.6 vs 680.8 of 2) inhibits the SARS-CoV-2 3CLpro with sub-nM potency (IC50 = 0.7 nM; Table 1 ), similar to the most potent inhibitors reported in literature and to compound 3 (Fig. 1B and Table 1; in house data IC50 = 0.22 nM; reported IC50 = 30 nM [19], a commercial broad-spectrum inhibitor commonly included as reference compound for biological assays (Table 1). The difference between the IC50 values is due to different assay conditions, as already described in our previous work [20] (in house data: experiment carried out with buffer at pH = 7.3, 5 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1% bovine serum albumin (BSA), and the proteins were preincubated for 30 min at 37 °C; lit. data [19]: experiment carried out with buffer at pH = 6.5, without TCEP and BSA, and pre-incubation of 30 min at 30 °C).
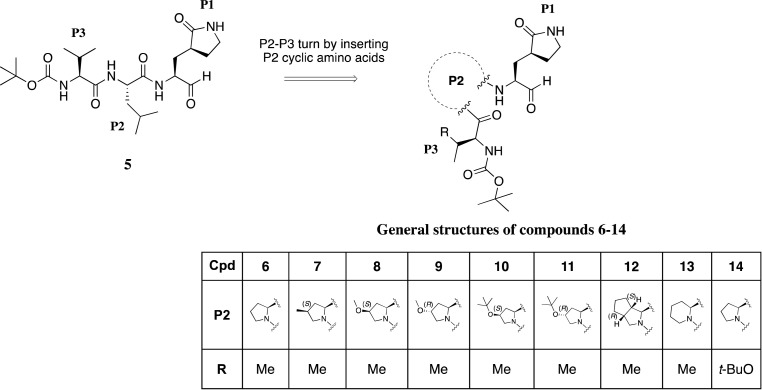
Fig. 2.
Chemical structures of designed and synthetized aldehyde tripeptides as SARS-CoV-2 3CLpro inhibitors (5-14).
Table 1.
Biological activities of target compounds 5–14 and reference compound 3.
| Cpd | 3CLpro IC50 (nM)a (n. of replicates) | EC50 (μM)b (SI)c | CC50 (μM)d | ||||
| SARS-CoV-2 | MERS | SARS-CoV-2e | SARS-CoV-2f | MERSf | HCoV229Eg | BEAS2B | |
| 3 | 0.22 ± 0.05 (10) | 0.24 ± 0.06 (10) | 3.55 ± 1.4 (>28) | 0.40 ± 0.12 (>250) | 0.75 ± 0.21 (>133) | 0.12 ± 0.03 (>828) | >100 |
| 5 | 0.70 ± 0.02 (4) | 0.02 ± 0.003 (4) | 40.1 ± 4.4 (>2.5) | 3.6 ± 0.5 (>27.7) | 1.58 ± 0.91 (>63) | 2.3 ± 1.4 (>30.1) | 70.5 ± 11.7 |
| 6 | 6.23 ± 0.70 (3) | 0.02 ± 0.0007 (3) | 22.5 ± 8.0 (>4.4) | 16.0 ± 5.2 (>6.2) | 4.56 ± 1.39 (>22) | >17.9 | 17.9 ± 4.3 |
| 7 | 0.47 ± 0.10 (3) | 2.33 ± 0.50 (3) | 47.2 ± 11.2 (>2) | 16.9 ± 3.9 (>5.9) | NTh | NTh | NTh |
| 8 | 0.24 ± 0.02 (5) | 0.007 ± 0.0005 (5) | 23.6 ± 10.8 (>4.2) | 8.7 ± 0.1 (>11.5) | 0.58 ± 0.34 (>172) | >3.6 | 3.6 ± 0.7 |
| 9 | 4196 ± 944 (7) | NTh | 19.6 ± 3.2 (>5.1) | 2.8 ± 0.7 (>36) | NTh | NTh | NTh |
| 10 | 20290 ± 7000 (4) | NTh | >100 | >100 | NTh | NTh | NTh |
| 11 | 2.5 ± 0.46 (4) | 26.0 ± 2.0 (4) | 33.4 ± 5.2 > 3 | 3.4 ± 1.2 (>29.4) | 33.56 ± 6.8 (>3) | >48.2 | 48.2 ± 15.6 |
| 12 | 5.0 ± 0.25 (4) | 143.4 ± 12.79 (4) | 5.3 ± 1.7 (>19) | 0.21 ± 0.13 (>476) | 4.2 ± 1.3 (>24) | 0.24 ± 0.19 (116) | 28.4 ± 2.7 |
| 13 | 3350 ± 280 (7) | NTh | >100 | 5.83 ± 0.81 (>17) | NTh | NTh | NTh |
| 14 | 5.3 ± 0.73 (5) | 0.022 ± 0.002 (5) | 22.9 ± 8.18 (>4.3) | 1.86 ± 0.96 (>54) | NTh | NTh | NTh |
Compound concentration required to reduce the 3CLpro activity by 50%.
Compound concentration required to reduce the viral cytopathic effect in cell by 50%.
SI is the selectivity index calculated as the CC50/EC50 ratio.
Compound concentration required to reduce BEAS2B cell viability by 50%.
EC50 in VERO E6-GFP cell in absence of P-gp inhibitor (CP-100356).
EC50 in Vero E6-GFP cell in presence of P-gp inhibitor (CP-100356) at the fixed concentration of 2 μM.
EC50 in BEAS2B cell.
NT = not tested. Values represent the mean ± SDs of two independent experiments in triplicate Note: all compounds showed CC50 > 100 μM in Vero E6-GFP cells.
Compound 5 was thus considered as a starting hit for further modifications in the peptide sequence. At this regard, we observed that the 3CLpro-bound conformation of compound 2 is characterized by a β turn-like motif around P1–P3 residues (φi+1 ≅ −75° and ψi+1 ≅ 120°; distance between Cαi and pseudo Cαi+3 ≅ 6.5 Å). Therefore, we sought to introduce a proline residue at the P2 position, with the aim to stabilize the ligand's bioactive conformation, and we designed the tripeptide 6 (Fig. 2) by replacing the P2 leucine of the starting hit 5 with a proline.
Covalent docking calculations suggested that the newly designed compound 6 could effectively bind at the 3CLpro catalytic site (Fig. 3B and S1), establishing all the primary interactions observed for the co-crystallized derivative 2. In detail, docking predicted that essential hydrogen bonds can be established between: i) the thiohemiacetal moiety resulting by the nucleophilic attack of the reactive cysteine over the aldehyde warhead and the backbone of Gly143 and Cy145; ii) the Gln-mimetic γ-lactam and the side chains of His163 and Glu166 in the S1 pocket; iii) the ligand's P3 backbone atoms and the main chain of Glu166. Additionally, according to docking, proline could allow the P3 lipophilic Val to establish van der Waals contacts with P1 γ-lactam, thus further stabilizing the ligand's binding conformation. Remarkably, in the course of our studies, the crystal structure of the proline-based repurposed HCV protease inhibitor boceprevir in complex with the SARS-CoV-2 3CLpro was released [21], which further corroborated our design strategy. Moreover, a work related to the focus of the present research has been published while our studies were ongoing, reporting on a series of dipeptide aldehydes having in P2 only two different proline residues [22].
The introduction of a rigid and hydrophobic proline in the peptidomimetic sequence is however a widespread approach adopted for all oral HCV protease inhibitors approved as drugs to improve the pharmacokinetic properties [23]. Moreover, in such proline-based compounds, the peptide bond between P2–P3 can only act as an H-bond acceptor, hindering the formation of ordered secondary structures and avoiding aggregations. Furthermore, the presence of a proline residue in the peptide sequence increases the proteolytic stability.
According to the docking of 6, unlike the natural leucine, a proline residue partially occupied the S2 subpocket of SARS-CoV-2 3CLpro. Therefore, we first explored a series of modifications of this amino acid, while keeping constant the N-Boc-Val in P3, the Gln mimetic γ-lactam in P1, and the aldehyde warhead (Fig. 2). Accordingly, also considering the synthetic accessibility, in this set we included compounds endowed either with a proline functionalized at the 4 position with groups of variable size and lipophilicity, such as Me (7), MeO (8), and tBuO (9) in S configuration, R MeO (10) and tBuO (11), or a bicycloproline (i.e. (1S,3aR,6aS)-octahydrocyclopenta[c]pyrrole-1-formamide) (12), which could suitably occupy the S2 subpocket of the 3CLpro. Moreover, we designed two additional tripeptides, by either replacing the proline with a 6-term piperidine analogue (13), in order to get further insights into the optimal ring size of the cyclic P2 amino acid, or interrogating the outcome of a P3 tBuO-threonine moiety as in derivative 14 used instead of the valine residue present in all the other compounds, with the aim to better explore the S3/S4 sites and to modulate the compound lipophilicity.
2.2. Chemistry
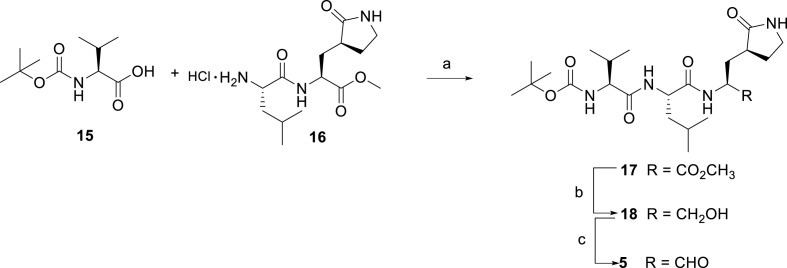
A tert-butyloxycarbonyl peptide solution synthesis was applied for the synthesis of tripeptides 5–14. Compound 5 was prepared by a linear synthetic approach, starting from a coupling reaction between the commercially available N-Boc-valine 15 and the intermediate 16, synthesized as reported in literature [24], to afford compound 17. The ester derivative 17 was first reduced with lithium borohydride to the primary alcohol 18, and subsequentially oxidized into the desired aldehyde 5 under Parikh-Doering conditions, using DMSO as oxidant activated by the sulfur trioxide pyridine complex in the presence of diisopropylethylamine as base (Scheme 1 ).
Scheme 1.
Synthesis of compound 5a.
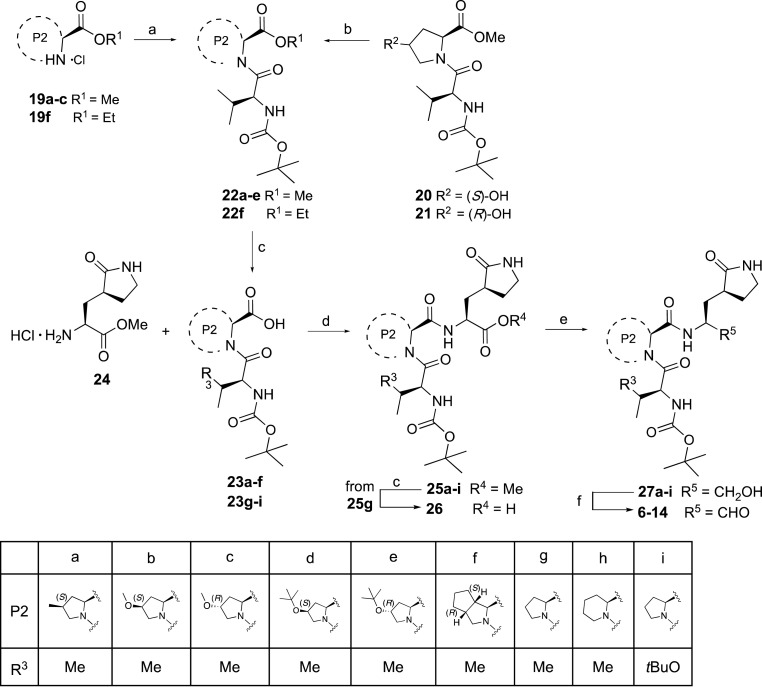
The synthesis of compounds 6–14 was performed with a convergent synthetic procedure, as shown in Scheme 2 . A condensation reaction, using HBTU in presence of DIPEA, between the commercial proline analogues 19a-c, 19f and N-Boc-valine 15 provided the dipeptide intermediates 22a-c, and 22f. The dipeptide intermediates 20 and 21 were functionalized at the C-4 hydroxyl group of the P2 proline residue by reaction with di-tert-butyl dicarbonate under Lewis acid promoted conditions in presence of ytterbium (III) trifluoromethanesulfonate, as reported in literature for the synthesis of other tert-butyl ethers [25], in order to afford the corresponding tert-butyl ether intermediates 22d,e with around 45% yield. Alkaline hydrolysis of the ester derivatives 22a-f gave the correspondent carboxylic acids 23a-f in quantitative yields, which together with the dipeptide carboxylic acids 23g-i [26] were reacted under coupling conditions, using EDCI in presence of HOBt and DIPEA, with the commercial P1 amine 24 to afford the tripeptides 25a-i in yields ranging from 37 to 71%. Then, the ester derivatives 25a-i were reduced into the alcohol intermediates 27a-i, which were finally oxidized by Parikh-Doering reaction to obtain the final aldehydes 6–14, while hydrolysis of the ester intermediate 25g afforded the corresponding acid 26.
Scheme 2.
Synthesis of compounds 6-14.a.
2.3. Enzymatic inhibition against SARS-CoV-2 and MERS 3CLpros
All the synthesized tripeptide aldehydes 6–14 were tested in a Fluorescence Resonance Energy Transfer (FRET) biochemical assay in order to evaluate their inhibitory activity against the isolated SARS-CoV-2 3CLpro processing a fluorescent substrate. Moreover, also tripeptides 25g, 26, and 27g, namely the corresponding C-terminal ester, acid, and alcohol analogues (structures in Scheme 2), respectively, of aldehyde 6, were tested in order to indirectly assess the key role of the electrophilic warhead as a requirement for the inhibitory activity. The commercial compound 3 was included as reference inhibitor in the biochemical assays.
All the compounds were tested at fixed 30 μM concentration and only active compounds (≥70% enzyme inhibition) were tested for dose-effect curves, to obtain the IC50 values. All aldehydes effectively inhibited the SARS-CoV-2 3CLpro (>70%) and almost all showed IC50 values in the sub-to low-nM range (0.24–6.23 nM) (Table 1), while derivatives 9, 10, and 13 resulted in a loss of potency with IC50 values in the μM range (4.2, 20.43, and 3.3 μM, respectively). Interestingly, compound 6 having a non-functionalized proline in P2 showed an IC50 = 6.23 nM, which is comparable to that of the close analogue 5 and of reference 3, which are both characterized by a P2 leucine. This result validated our rational design strategy of replacing the P2 Leu with Pro maintaining the same level of intrinsic potency on the enzyme. On the contrary, compounds 25g, 26, and 27g (compound structures are reported in Scheme 2), that only differ from 6 for the replacement of the aldehyde with similar but not reactive C-terminal groups, resulted inactive at 30 μM (data not reported in Table 1), thereby confirming that the electrophilic warhead is essential to obtain enzyme inhibition with peptidomimetics.
Furthermore, the introduction of (S)-pipecolinic acid as P2 turned out to be very detrimental, with the corresponding analogue 13 (IC50 = 3.35 μM) resulting around 500-fold less potent than the proline-based analogue 6. This suggested that a larger 6-membered ring could either be less tolerated by the S2 pocket or not allow the P1 and P3 residues to properly accommodate in the enzyme active site. More interesting are the effects of the substituents on the proline in P2. Here, the introduction at the C4 position of small substituents such as a methyl (7) and a methoxy (8) with S configuration provided the most potent inhibitors with IC50 < 1 nM, while a bulkier t-butoxy (9) was very detrimental (IC50 = 4.2 μM). The results are reversed in the presence of the corresponding R epimers, with the R-MeO derivative 10 resulting in low inhibition (IC50 = 20.43 μM) of the protease while the R-tBuO analogue 11 displaying nM potency (IC50 = 2.5 nM). The introduction of a bulkier bicycloproline analogue as P2 residue (12) also provided a potent compound with IC50 = 5.3 nM, in line with the potency reported for the nirmatrelvir 1 that is endowed with a similar bicycloproline. All these analyses confirmed that the P2 proline residue is a key region to modulate the potency. Finally, compound 14, having a tBu-threonine in place of a valine as P3 residue, demonstrated a potency (IC50 = 5.3 nM) similar to that of 6 suggesting a marginal or neutral contribution to the binding of this larger substituent to the protein.
Almost all tripeptide aldehydes resulted potent inhibitors of SARS-CoV-2 3CLpro by the FRET-based enzymatic assay, albeit with different degree of potencies, prompting their evaluation as broad-spectrum CoV 3CLpro inhibitors. Thus, for compounds showing IC50 < 1 μM on SARS-CoV-2 3CLpro, the screening was extended also to MERS-CoV 3CLpro, taken as representative homologue protein in order to evaluate the potential broad inhibition within the same virus family (Table 1). The selected compounds (5–8, 11, 12, and 14) resulted in a broad-spectrum inhibition of CoVs 3CLpro being active also against the MERS homologue. Actually, the compounds were generally found more potent against this protease in comparison to the SARS-CoV-2 homologue, with IC50 spanning from 0.007 to 143 nM (Table 1). Derivative 8 showed an IC50 = 0.007 nM, resulting to our knowledge the most potent MERS-CoV 3CLpro inhibitor so far reported. Conversely, derivative 12 exhibited a lower but still significant inhibitory activity on MERS 3CLpro (IC50 = 143 nM), if compared to the data obtained on SARS-CoV-2 (IC50 = 5 nM). In order to rationalize these results, we performed in silico studies to compare co-crystallographic structures obtained for some of our compounds in complex with the SARS-CoV-2 3CLpro and data reported in literature for the MERS homologue protein, as described in the next sections.
2.4. Phenotypic assays against SARS-CoV-2, MERS-CoV and HCoV 229E
All the active compounds, i.e. all the aldehydes, were first tested in a phenotypic assay against SARS-CoV-2 replicating in Vero E6-Green Fluorescent Protein (GFP) stably expressing a GFP protein under the control of cytomegalovirus promoter. Results were expressed as EC50 and determined by using the cytopathic effect (CPE) method. The host cell toxicity was also evaluated quantifying the viability as proportional to the GFP signal of untreated cells and expressed as CC50. Indeed, active compounds causing cytotoxicity, for instance non-specific cellular protease inhibitors, may show antiviral activity coupled to reduced viability, resulting in a low selectivity index (SI = CC50/EC50). Active compounds with low or no cytotoxicity exert a genuine antiviral effect resulting in suitable SI (≥10), which is a very important feature for developable antiviral agents. In the assay, compound 3 was again included as positive control.
All the compounds were tested at 100 μM and those able to reduce >95% viral replication were tested in concentration-effect curves for EC50 values determination (Table 1). With the only exceptions of derivatives 13 and 10, which showed modest or null inhibition also in the biochemical assay against SARS-CoV-2 3CLpro, the other compounds displayed EC50s ranging from 5 to 47 μM, comparable to the positive control 3 (EC50 = 3.55 μM), and no significant cell toxicity (CC50 > 100 μM) (Table 1). However, a significant drop in the compounds’ potency from the biochemical to the cell-based assays was observed. Considering that Vero E6 cells express high level of active membrane transporters such as the P-glycoprotein (P-gp), we speculated that drug efflux could contribute to the high antiviral EC50/enzyme IC50 ratio, as already observed for other peptidomimetic inhibitors of CoVs 3CLpro (i.e., compounds 1 [10]). The underestimation of the antiviral potency may lead to discard compounds likely able to potently inhibit SARS-CoV-2 replication in human lung cells, the relevant tissue for human CoVs and COVID-19. Thus, according to recent literature, all the compounds were co-dosed with a commercial non-toxic P-gp inhibitor (CP-100356) at the fixed concentration of 2 μM [10], resulting in a shift for the antiviral activity without any detectable impact on cell viability/toxicity (CC50s > 100 μM) (Table 1). This effect was considerable especially for derivative 12, which showed a potent antiviral activity with an EC50 = 0.21 μM (w/o CP-100356: EC50 = 5.3 μM), a value closer to its IC50 and 2-fold lower than reference 3 (w/CP-100356: EC50 = 0.4 μM). Consequently, also the SI values of all the compounds were significantly improved, with 12 having the highest SI (>476). Excluding derivatives 6–8, the remaining compounds (5, 9, 11, 13, and 14) gained more than 3-fold improvement in their antiviral potency displaying EC50s in the low μM range and SI values > 11.5 (Table 1). Although derivates 6 and 14 showed a comparable in vitro enzymatic potency, 14 showed an almost 10-fold increase in antiviral activity (w/CP-100356: EC50 = 1.86 μM) compared to 6 (w/CP-100356: EC50 = 16.0 μM), indicating that the replacement in P3 of valine with tBuO-Thr was favorable to enhance the cell activity. Indeed, the impact of tBuO-Thr at the P3 position as chemical moiety to enhance the cellular antiviral potency was also observed in another recent study on peptidomimetic inhibitors of CoV 3CLpro [27]. In summary, derivative 12 resulted the most active inhibitor in cellular assay, showing EC50 and SI values comparable to the most potent SARS-CoV-2 3CLpro inhibitors reported in literature when evaluated in the same cellular system, including the oral inhibitor nirmatrelvir 1 [10].
To further investigate the broad-spectrum activity of this series, five representative compounds (5, 6, 8, 11 and 12) were selected for in vitro cellular evaluation against the β-CoV MERS and α-HCoV 229E. Selected compounds were tested against MERS-CoV in Vero E6-GFP cells by CPE assay, in presence of the P-gp inhibitor (CP-100356 at 2 μM), and GC-376 was the reference compound as described above for SARS-CoV-2. All compounds showed to be active also against MERS, with EC50 values spanning from 0.58 to 33.6 μM, without cellular toxicity up to 100 μM (Table 1). Noteworthy, the antiviral activities were consistent with the results obtained in the enzymatic assays against SARS-CoV-2 and MERS 3CLpros. Indeed, derivatives 5, 6 and 8 inhibited MERS replication in cells in a larger extent respect to SARS-CoV-2 (w/CP-100356: EC50 = 1.58 μM, EC50 = 4.56 μM, EC50 = 0.58 μM, respectively), while the opposite profile was observed for compounds 11 and 12 (w/CP-100356: EC50 = 33.56 μM, EC50 = 4.20 μM, respectively). In particular, compound 8 was the most potent of the series on MERS, showing a 15-fold increase in potency on MERS respect to SARS-CoV-2 in the CPE cellular assay (Table 1). Indeed, this result agreed well with the data in the enzymatic assays, where derivative 8 resulted in the most potent inhibition with IC50 value in the picomolar range (Table 1). Derivative 12, which was the most active against SARS-CoV-2 in cell-based assay, exhibited a 20-fold lower activity against MERS compared to SARS-CoV-2, with an EC50 = 4.2 μM, as expected from the enzyme inhibition data on the respective 3CLpros (Table 1).
In parallel, the dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was used to measure the HCoV 229E induced CPE and cytotoxicity in human bronchial epithelial BEAS-2B cells, in presence of GC-376 as positive reference. With the only exception of derivative 11, all compounds displayed antiviral activity against HCoV 229E with EC50 values comparable to SARS-CoV-2 cellular assay, ranging from 0.24 to 22.9 μM (Table 1). Derivative 12 showed the most potent activity (EC50 = 0.24 μM) and the higher SI (116), thus comparable to the anti-HCoV 229E activity of the reference GC-376 (EC50 = 0.12 μM) and the approved nirmatrelvir (EC50 = 0.19 μM [10]). Compound 5 showed to inhibit the HCoV 229E replication with a suitable SI (>30) and an EC50 value in low micromolar range (EC50 = 2.3 μM), although 10-fold less potent respect to derivative 12. Unfortunately, in this cellular type a more significant cytotoxicity was observed for some derivatives (6, 8 and 11), with a sub-optimal SI < 10 (Table 1).
Considering the overall results from biochemical and antiviral cell-based assays, compound 12 resulted the best inhibitor in this series and thus its antiviral activity was further investigated in a more physio-pathological relevant cell line for COVID-19 research. Viral load reduction assays were thus performed in human epithelial lung adenocarcinoma Calu-3 cells, a cell type closer to the airway epithelium, that is more relevant for a respiratory virus such as SARS-CoV-2, and with low expression of efflux pumps to eliminate the artifacts of drug extrusion on compound activity. The extracellular viral load was determined by qPCR, titrating S gene copy number in the RNA extracted from the supernatant 48 h post infection. Results showed that 12 effectively reduced viral titers, causing a 3 Log decrease of viral titer in the cell supernatant at 20 μM and 4 μM, and showing an EC50 = 0.449 μM, without sign of cytotoxicity (Figure S4). Therefore, these results confirmed the potent antiviral activity observed in the Vero cell assays in presence of the P-gp inhibitor.
2.5. Binding and kinetic assays on compound 12
Compound 12 was further characterized by additional experiments. In particular, a differential scanning fluorimetry assay was carried out to validate the specific binding of this compound to the SARS-CoV-2 3CLpro. Compounds 12 and reference 3 increased the thermal stability of the target protein with considerable thermal shifts. Derivative 12 caused the highest increase in the protein melting point (+11 °C), thus suggesting that bicycloproline could better fill the S2 pocket of this enzyme, while the GC376 produced a less pronounced effect (only +4 °C). In order to elucidate the mechanism of action of 12, we performed a kinetics study by reading the signal generated by the proteolytic cleavage of the FRET substrate for 60 min. The results for 12 (Figure S5) showed a biphasic enzymatic progression curve in the presence but not in absence of inhibitors, which is characteristic behavior of a slow covalent binding inhibitor, more evident in the first 30 min of reaction. The same profile is reported for reference compound 3. Moreover, binding affinity of derivative 12 for SARS-CoV-2 3CLpro was estimated, resulting in an apparent K i = 3.2 ± 0.96 nM.
2.6. Structural biology studies
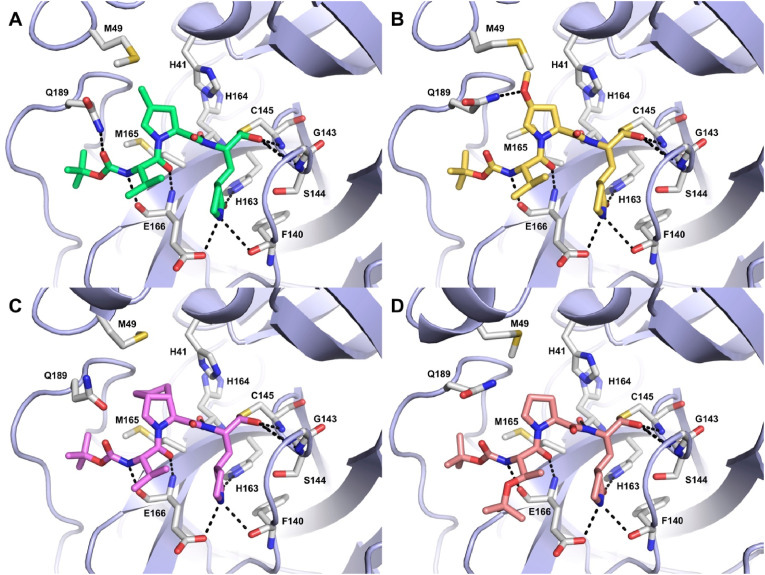
Single crystal X-ray crystallography was exploited to solve the structures of the most potent compound 8 and some other selected inhibitors (7, 12, and 14) bound in the catalytic site of SARS-CoV-2 3CLpro. Crystallization experiments were carried out through seeding in the Morpheus screening (Molecular Dimensions) as previously described [28]. For the four compounds, well diffracting crystals grew in several conditions in space group P212121 reaching resolutions in the range of 1.35–1.66 Å (7: 1.63 Å, 8: 1.5 Å, 12: 1.66 Å, 14: 1.35 Å). In all the resulting crystal structures, the well-known heart-shape dimer of SARS-CoV-2 3CLpro is present in the asymmetric unit [28,29]. Refinement was carried out alternating manual model building cycles in coot [30] and automated refinement cycles in Phenix [31]. Accordingly, each inhibitor was unambiguously modelled covalently bound to the catalytic Cys145 in both protomers (Fig. 4 and Figure S6). Data collection and refinement statistics, including the final Rwork/Rfree values are reported in Table S1.
Fig. 4.
Crystal structures of compounds 7 (A, green), 8 (B, yellow), 12 (C, violet), and 14 (D, salmon) into SARS-CoV-2 3CLpro active site represented as light-blue cartoons. Residues important for ligand binding are displayed as white sticks and labeled. H-bonds discussed in the text are depicted as dashed black lines.
The four solved crystal structures show that, following the attack of the catalytic Cys145 to the aldehyde group of the inhibitors, a new chiral center is formed and that the resulting thiohemiacetal is in the S configuration, confirming that this reaction is typically enantioselective, as previously reported [21].
In the covalent adducts, the thiohemiacetal occupies the oxyanion hole formed by the backbone amide groups of Gly143, Ser144, and Cys145, where it forms hydrogen bonds with the amide groups of Gly143 and Cys145. According to our initial design hypothesis, the four inhibitors adopt a β-turn like motif around P1–P2 groups and extend along the S1–S4 binding subsites interacting with active site residues through several hydrogen bonds and hydrophobic contacts. The S1 cavity is occupied by the P1 γ-lactam ring that forms hydrogen bonds with the backbone of Phe140 and with the side chains of His163 and Glu166. The proline ring (14) occupies the S2 pocket where, as expected, also substituted residues can be hosted. Indeed, both the prolines functionalized at the 4 position, with either Me (7) or MeO (8) and the bicycloproline (i.e., (1S,3aR,6aS)-octahydrocyclopenta[c]pyrrole-1-formamide) (12), fit in this cleft establishing, albeit to a different extent, hydrophobic interactions with the side chains of residues such as His41, Met49, Met165 and Gln189. On the other hand, these residues could partly hinder an optimal accommodation of the bulkiest groups, explaining the lower affinity of compounds 12 and especially 9 compared to 7 and 8. It is also interesting to remark that: i) the MeO oxygen of 8 forms an additional H-bond with the Gln189 side chain, which might explain why this compound is the most potent within the series, and ii) the S configuration of the proline γ-carbon is required to ensure the optimal positioning of the substituent within the S2 pocket, as demonstrated by the very low affinity displayed by 10.
In the S3–S4 pockets, each ligand forms two hydrogen bonds with the backbone of Glu166 through the backbone of its P3 group, while 7 forms an additional H-bond with the Gln189 side chain through its terminal N-Boc cap. Finally, the side chain of the P3 residue (Val in 7, 8 and 12, tBu-threonine in 14) can form intramolecular hydrophobic contacts with the P1 γ-lactam that contribute to stabilize the ligand binding conformation. Altogether, the four solved X-ray structures confirm the predictions from the docking studies and the validity of our rational design approach.
2.7. Molecular modelling on MERS-CoV 3CLpro
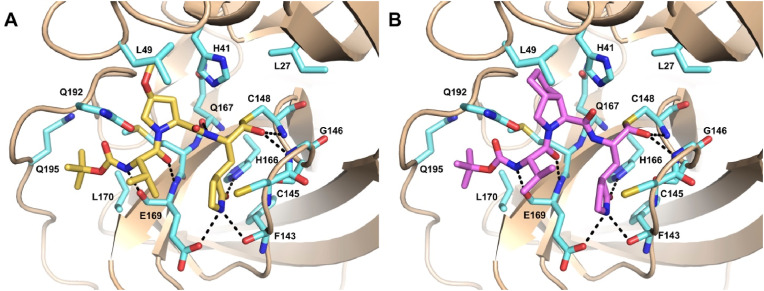
CoVs 3CLpro possess a well-conserved active site, with highly similar substrate recognition profiles. Particularly, SARS-CoV-2 and MERS-CoV 3CLpro share overall 50% identity. Indeed, the overlay of their X-ray structures indicates a high degree of structural similarity and conservation, with very few mutations occurring at the substrate binding site. Herein, the S1’, S1 and S2 subsites show mostly conservative substitutions, whereas the main differences involve the residues lining the S3–S4 region and the lid covering the S2 (Figure S2). These mutations result in slight differences in the overall active site architecture. In order to gain insights into the possible binding mode of the most potent compound 8 into MERS 3CLpro active site and to help interpretation of SAR data, docking studies were performed through the same covalent docking procedure used for the SARS-CoV-2 homologue (see Experimental Section for details). According to docking, the binding mode of 8 observed in the crystal structure of SARS-CoV-2 3CLpro is essentially maintained also in MERS-CoV 3CLpro active site. The docked pose of 8 displays the thiohemiacetal adduct in the S configuration, with the hydroxy group accepting two H-bonds from the backbone amide groups of the oxyanion hole residues Gly146 and Cys148 (Fig. 5 A). The P1 γ-lactam moiety engages the canonical H-bonds with His166 and Glu169 side chains and with Phe143 main chain within the S1 pocket, whereas the ligand backbone interacted with Gln167 and Glu169 main chain. The P2 fragment is deeply inserted into the S2 pocket, with the Leu49 side chain clamping the MeO substituent through tight hydrophobic interactions. This latter, in turn, could potentially accept a H-bond from the side chain of Gln192, likely accounting for the higher potency of 8 also towards MERS 3CLpro compared to compounds such as 6, bearing an unsubstituted proline, and 5, characterized by the P2 leucine. Finally, the Boc cap can establish further hydrophobic interactions with Leu170 and Gln195.
Fig. 5.
Predicted binding mode of 8 (A, yellow sticks) and 12 (B, violet sticks) into MERS 3CLpro (PDB ID: 4RSP) [32] active site represented as wheat ribbon model. Residues important for ligand binding are displayed as aquamarine sticks and labeled. H-bonds discussed in the text are depicted as dashed black lines.
At variance with 8, compound 12, bearing a bicycloproline as P2 fragment, displayed a significant drop in potency towards MERS 3CLpro (Fig. 5B). To rationalize these data, we also performed covalent docking calculations of this compound in the active site of the MERS enzyme. Notably, the presence of the bicycloproline caused unfavorable close contacts with Leu49 and His41 (Figure S3). Thus, the replacement of Met49 in SARS-CoV-2 with Leu in MERS exerts a certain impact on the shape and size of the S2 subsite, thereby influencing the inhibitory activity of functionalized P2 proline fragments. Indeed, also the lower potency of 7, bearing a 4-methyl group on the proline ring could be due to the potential clashes that this substituent can form with the side chain of Leu49. On the other hand, the presence of the oxygen atom at position 4 of the P2 proline rescues activity towards MERS, allowing the ligand i) to better orient the methyl (8) to form profitable hydrophobic interactions with Leu49; ii) to potentially establish a H-bond with Gln192. Importantly, the S configuration ensures the optimal positioning of the substituents within the S2 pocket.
3. Conclusions
The destructive impact of COVID-19 pandemic rapidly boosted the research on this disease, leading to the approval of vaccines and DAAs within a short time frame, if compared to any other infections. However, only three DAAs are available to date, which show limitations in efficacy and/or PK properties. An elective target to identify DAAs against SARS-CoV-2 is the 3CLpro; indeed, different research groups reported peptidomimetic and non-peptidic inhibitors of this enzyme. These efforts culminated in the development of nirmatrelvir 1, the first-in-class approved DAA targeting 3CLpro. Compound 1 was designed as a proline-based peptidomimetic reversible covalent inhibitor, similar to the previously approved first-in-class α-ketoamide inhibitors of HCV NS3/4A serine protease, telaprevir and boceprevir.
Here, we designed and synthesized a novel series of proline-based tripeptides as potential covalent reversible inhibitors of SARS-CoV-2 3CLpro bearing an aldehyde warhead as Cysteine trap and a cyclic analogue of Gln in P1, similarly to nirmatrelvir and other potent reported inhibitors. In addition, we replaced the Leucine in P2 of the natural substrate with a proline and we particularly investigated the effect of different substitution patterns at 4 position.
Notably, most of our newly synthesized compounds showed excellent inhibition of the SARS-CoV-2 3CLpro with some displaying IC50 values in the sub-nM range. Nonetheless, the inhibitory potency was influenced by the size, the nature and the chirality of the substituents at the P2 position, indicating that: i) a 5-membered cyclic amino acid (i.e. proline) is preferred over a 6-membered residue (i.e. pipecolinic acid), and that ii) small substituents at the 4 position of the proline ring are preferred in S configuration, while bigger moieties are better tolerated in R configuration. The compounds active in the biochemical assays effectively inhibited SARS-CoV-2 replication in Vero cells, however showing a large shift in the antiviral potency without a clear correlation with the enzymatic activity. To be noted, this behavior is common to other peptidomimetic inhibitors reported in this field, including nirmatrelvir 1, being such molecules substrates of cellular efflux pumps like P-gp. Indeed, compounds’ potencies in cell-based assays improved when co-dosed with a P-gp inhibitor, with derivative 12 displaying the highest antiviral activity, in the nM range. These results were further confirmed in human epithelial lung adenocarcinoma Calu-3 cells, a cell type not only more relevant to evaluate the infection of a respiratory virus such as SARS-CoV-2, but also characterized by low expression of efflux pumps. In addition to the 3CLpro of SARS-CoV-2, the majority of the compounds displayed high affinity also towards the MERS homologue (IC50 values ranged from mid-to sub-nM). In light of the high sequence and structural similarity of the 3CLpros within the Coronoviridae family, the activity of compounds was also investigated in additional phenotypic assays against MERS and another human CoV of the alfa group, i.e. confirming the potential anti-coronavirus activity of this series. In particular, compound 12 displayed a potent antiviral activity on MERS and HCoV 229E. Moreover, our derivatives, particularly compound 12, demonstrated a promising in-cell safety index in different cell line, thus overcoming the toxicity warning related to the reactivity of aldehydes. In this perspective, the exploiting of molecules bearing an aldehyde warhead can be advantageous in terms of target engagement kinetics for the formation of the reversible covalent adduct.
These results will prompt us to run chemical optimization in order to obtain novel derivatives with improved PK properties as suitable candidates for future in vivo studies, in the challenge against COVID-19 and other coronaviruses-related diseases.
4. Experimental section
4.1. Molecular docking
For docking calculations, we selected the crystal structures of SARS-CoV-2 3CLpro (PDB ID: 7BQY) [16] and MERS-CoV 3CLpro (PDB ID: 4RSP) [32] in complex with compound 2 and a peptide inhibitor, respectively. Prior to docking, the protein structure was prepared with the Protein Preparation Wizard tool within Maestro. In particular, bond orders were assigned, and missing hydrogens added. A prediction of the receptor side chains ionization and tautomeric states was performed using Epik. Then, an optimization of the hydrogen-bonding network was carried out and the positions of the hydrogen atoms were minimized using the OPLS3e force field. Finally, both the co-crystallized water molecules and inhibitor were removed. Compounds' 3D structures were generated using the graphical interface of Maestro and prepared for docking using LigPrep. Calculations were performed using the Covalent Docking procedure implemented in Maestro in the Pose Prediction (thorough) mode. A grid box of 30 Å × 30 Å × 30 Å was centered on the reactive Cys145 in SARS-CoV-2 3CLpro and Cys148 in MERS-CoV 3CLpro. The ‘nucleophilic addition to a double bond’ reaction type was selected to account for the aldehyde warhead. For the refinement phase, the maximum number of poses to retain was set to 250, while the cutoff for minimization was set to 5.0 Å. Macrocycles sampling and MM-GBSA scoring were disabled. Otherwise, default parameters were applied. For each ligand, 5 poses were finally retained and visually inspected.
4.2. Experimental synthetic procedures
General methods. Reagents and solvents were purchased from commercial sources and used without further purification. Reactions were carried out at ambient temperature, unless otherwise specified. Moisture-sensitive reactions were performed under a positive pressure of dry nitrogen in oven-dried glassware. Analytical thin-layer chromatography (TLC) on silica gel 60 F254 plates (250 μm thickness) was performed to monitor the reaction progress, using UV and KMNO4 as revelation method. Flash chromatography on silica gel (70–230 mesh) was performed for purification. All products were characterized by their NMR and MS spectra. (ESI)-MS spectra were performed on a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) by infusion into the ESI source using MeOH as solvent. 1H and 13C NMR spectra were recorded in CDCl3 or DMSO‑d 6 at 25 °C on Bruker Avance NEO 400 MHz and 700 MHz instruments equipped with a RT-DR-BF/1H-5 mm-OZ SmartProbe. Chemical shifts (δ) are reported in part per million (ppm) downfield from tetramethylsilane, using residual solvent signal as the internal reference.
The final compounds were characterized by HPLC-MS/MS, using a Dionex ULTIMATE 3000 (Thermo Fisher Scientific) HPLC module and a LTQ XL mass spectrometer with electrospray ionization in positive mode and an Ion-Trap detector. Separation was performed with a Kinetex column C18 Polar column (250 mm × 4.6 mm; particle size 5 μm, Phenomenex, Torrance, CA, USA) at 30 °C, using a 17 min gradient, 5% [0.1% TFA/CH3CN]/95% [0.1% TFA/H2O] to 95% [0.1% TFA/CH3CN]. High resolution mass spectra were obtained using an Orbitrap Q-Exactive (Thermo Fisher Scientific) for the final compounds, by infusion into the ESI source using DMSO as solvent.
Analytical HPLC was performed on Shimatzu-1100 HPLC using a Kinetex C18 column (4.6 mm × 150 mm, 5 μm, 100 Å) with an acetonitrile (0.1%TFA) − water (0.1% TFA) custom gradient. The purities of the final compounds were all >95%, as determined by HPLC (UV λ = 220 and 254 nm).
A Jasco P-2000 digital polarimeter with a sodium lamp at 589 nm and a photomultiplier tube detector was used for the measurement of specific optical rotation ([α]D), using a 100 mm cell, sample concentration 1 mg/1 mL in MeOH, at 25 °C.
4.2.1. Methyl (6S,9S,12S)-9-isobutyl-6-isopropyl-2,2-dimethyl-4,7,10-trioxo-12-(((S)-2-oxopyrrolidin-3-yl)methyl)-3-oxa-5,8,11-triazatridecan-13-oate (17)
To a solution of Boc-l-Val-OH 15 (189 mg, 0.870 mmol) in anhydrous DMF (2 mL) in a three-necked oven-dried flask, EDCI (208.5 mg, 1.09 mmol, 1.2 eq.) and HOBt (167 mg, 1.09 mmol, 1.2 eq.) were added under a positive anhydrous nitrogen pressure, and the mixture was left stirring for 30 min. In a separate flask, a solution of amine hydrochloride intermediate 16 [24] (292 mg, 0.870 mmol, 1 eq.) in anhydrous DMF (3 mL) was cooled to 0–5 °C and DIPEA (0.61 mL, 3.48 mmol, 4 eq.) was added dropwise. After 30 min, this solution was added to the reaction mixture containing activated intermediate of 15 and was left stirring for 16 h at room temperature. The reaction mixture was diluted with brine and extracted with EtOAc (25 mL x 4). The collected organic layers were washed with brine (15 mL x 10), dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The reaction crude was purified by column chromatography (CH2Cl2/MeOH, 97:3 v/v) to afford the pure product 17 (138 mg, 40%) as a white solid. 1H NMR (400 MHz, CDCl3): δ 7.79 (d, J = 6.3 Hz, 1H), 6.94 (d, J = 8.1 Hz, 1H), 6.46 (s, 1H), 5.03 (d, J = 8.7 Hz, 1H), 4.64 (td, J = 9.1, 4.7 Hz, 1H), 4.52 (s, 1H), 3.90–3.81 (m, 1H), 3.72 (s, 3H), 3.33 (dd, J = 13.3, 7.7 Hz, 2H), 2.39 (dd, J = 12.1, 5.5 Hz, 2H), 2.25–2.05 (m, 2H), 1.94–1.78 (m, 2H), 1.75–1.61 (m, 2H), 1.55–1.46 (m, 1H), 1.43 (s, 9H), 1.00–0.90 (m, 12H). MS (ESI) m/z: [M+H]+ calcd for C24H43N4O7 + 499.3, found 499.2.
4.2.2. tert-Butyl ((S)-1-(((S)-1-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)-4-methyl-1-oxopentan-2-yl)amino)-3-methyl-1-oxobutan-2-yl)carbamate (18)
In oven-dried three-necked round bottom flask, the tripeptide methyl ester 17 (138 mg, 0.277 mmol) was dissolved in anhydrous THF (1.6 mL) under a positive anhydrous nitrogen pressure, then a 2 M solution of LiBH4 (18 mg, 0.830 mmol, 3 eq.) in anhydrous THF (0.415 mL) was added dropwise at 0 °C. The reaction mixture was stirred at room temperature for 5 h, then quenched by acidifying with a saturated citric acid solution to pH 2 and extracted with EtOAc (25 mL x 3). The collected organic layers were washed with brine (15 mL x 10), dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The reaction crude was purified by column chromatography (CH2Cl2/MeOH, 98:2 to 95:5 v/v) to afford the desired alcohol 18 (82 mg, 63%) as a white solid. 1H NMR (400 MHz, CDCl3): δ 7.65 (d, J = 7.0 Hz, 1H), 7.13 (d, J = 7.7 Hz, 1H), 6.43 (s, 1H), 5.12 (d, J = 8.4 Hz, 1H), 4.56 (td, J = 9.0, 4.7 Hz, 1H), 4.01 (s, 1H), 3.92 (t, J = 7.4 Hz, 1H), 3.62 (d, J = 15.7 Hz, 2H), 3.45–3.24 (m, 3H), 2.38 (d, J = 5.7 Hz, 2H), 2.07 (dt, J = 23.8, 8.6 Hz, 2H), 1.90–1.73 (m, 3H), 1.71–1.57 (m, 3H), 1.55–1.49 (m, 1H), 1.43 (s, 9H), 0.99–0.86 (m, 12H). MS (ESI) m/z: [M+H]+ calcd for C23H43N4O6 + 471.3, found 471.3.
4.2.3. tert-Butyl ((S)-3-methyl-1-(((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)amino)-1-oxobutan-2-yl)carbamate (5)
Compound 18 (82 mg, 0.174 mmol) was dissolved in anhydrous DMSO (0.06 mL, 0.870 mmol, 5 eq.), anhydrous CH2Cl2 (0.43 mL) under a positive anhydrous nitrogen pressure, and DIPEA (0.106 mL, 0.609 mmol, 3.5 eq.) was added at −5 °C. In another flask, pyridine sulfur trioxide complex (55.4 mg, 0.348 mmol, 3 eq.) and pyridine (0.028 mL, 0.384 mmol, 3 eq.) were suspended in anhydrous DMSO (0.06 mL, 0.870 mmol, 7.5 eq.) at room temperature under stirring for 10 min, and then the resulting mixture was added to the previously formed 18/DMSO mixture at −5 °C. After 24 h, the reaction mixture was poured into ice/water (1:1) and extracted with CH2Cl2 (25 mL x 3). The collected organic layers were washed with a saturated citric acid solution (15 mL x 1), water (15 mL x 1), saturated solution of NaHCO3 (15 mL x 1) and brine (15 mL x 1), dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The reaction crude was purified by column chromatography (CH2Cl2/MeOH, 95:5 v/v) to afford the target aldehyde 5 (17 mg, 21%) as a white solid. [α]20 D −50.0 (c = 1.0, MeOH). 1H NMR (700 MHz, CDCl3): δ 9.50 (s, 1H), 8.22 (s, 1H), 6.77 (d, J = 7.3 Hz, 1H), 6.32 (s, 1H), 5.05 (d, J = 8.6 Hz, 1H), 4.63 (td, J = 8.9, 4.9 Hz, 1H), 4.35 (s, 1H), 3.93–3.86 (m, 1H), 3.44–3.24 (m, 2H), 2.51–2.33 (m, 2H), 2.13 (dt, J = 13.3, 6.7 Hz, 1H), 2.00 (ddd, J = 14.5, 10.5, 6.6 Hz, 1H), 1.92 (ddd, J = 14.4, 6.6, 4.4 Hz, 1H), 1.87–1.81 (m, 1H), 1.69 (ddd, J = 16.3, 10.0, 5.9 Hz, 2H), 1.58–1.54 (m, 1H), 1.43 (s, 9H), 1.01–0.88 (m, 12H).13C NMR (176 MHz, CDCl3): δ 199.7, 180.1, 173.3, 171.9, 156.2, 129.9, 80.3, 60.4, 57.7, 51.8, 42.0, 40.7, 38.2, 30.7, 29.9, 28.6, 24.9, 23.1, 22.0, 19.4, 18.0. HRMS (ESI) m/z: [M+H]+ calcd for C23H41N4O6 + 469.3026, found 469.3534. LC-MS (ESI) m/z: [M+H]+ calcd for C23H41N4O6 + 469.3, found 469.2 (rt: 7.07).
4.2.4. General procedure for the synthesis of dipeptide ester intermediates 22a-c, 22f (Method A)
In a three-necked oven-dried round-bottom flask, N-Boc-Valine 15 (1 eq) was dissolved in anhydrous CH2Cl2 (0.3 M) under a positive anhydrous nitrogen pressure, then HBTU (1.1 eq.), the appropriate amine hydrochloride (1.3 eq.) and DIPEA (2 eq.) were added at 0 °C. The reaction mixture was stirred for 4 h at room temperature. A saturated aqueous solution of NaHCO3 was added to the reaction mixture and the resulting aqueous mixture was extracted with CH2Cl2 (25 mL x 3), the collected organic layers were washed with a saturated citric acid solution (15 mL x 1), brine (15 mL x 1), dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The reaction crude was purified by flash chromatography to yield the desired product as a colorless oil.
4.2.4.1. Methyl (2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methylpyrrolidine-2-carboxylate (22a)
Compound 22a was prepared following the general procedure Method A and using methyl (2S,4S)-4-methylpyrrolidine-2-carboxylate hydrochloride 19a (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 149 mg, 45%). 1H NMR (400 MHz, CDCl3): δ 5.22 (d, J = 9.1 Hz, 1H), 4.41 (dd, J = 9.7, 7.6 Hz, 1H), 4.29 (dd, J = 9.3, 6.0 Hz, 1H), 4.03–3.89 (m, 1H), 3.71 (d, J = 3.7 Hz, 3H), 3.12 (dd, J = 16.6, 6.7 Hz, 1H), 2.41 (dd, J = 12.2, 6.6 Hz, 1H), 2.33 (d, J = 6.6 Hz, 1H), 2.02 (dd, J = 13.4, 6.4 Hz, 1H), 1.56–1.48 (m, 1H), 1.43 (s, 9H), 1.09 (d, J = 6.4 Hz, 3H), 1.02 (d, J = 6.8 Hz, 3H), 0.96–0.89 (m, 3H). MS (ESI) m/z: [M+H]+ calcd for C17H31N2O5 + 343.2, found 343.1.
4.2.4.2. Methyl (2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxylate (22b)
Compound 22b was prepared following the general procedure Method A and using methyl (2S,4S)-4-methoxypyrrolidine-2-carboxylate hydrochloride 19b (purification method: flash chromatography eluting CH2Cl2/MeOH 98:2 v/v; yield: 1.45 g, 70%). 1H NMR (400 MHz, CDCl3): δ 5.20 (d, J = 9.4 Hz, 1H), 4.70 (dd, J = 8.8, 3.8 Hz, 1H), 4.22 (dd, J = 9.3, 6.3 Hz, 1H), 3.99 (ddd, J = 16.1, 9.3, 4.3 Hz, 1H), 3.69 (s, 3H), 3.59 (dd, J = 10.6, 2.8 Hz, 1H), 3.28 (s, 3H), 2.37–2.28 (m, 1H), 2.25–2.14 (m, 2H), 2.08–1.98 (m, 1H), 1.42 (s, 9H), 1.05 (d, J = 6.8 Hz, 3H), 0.96 (d, J = 6.7, 2.4 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C17H31N2O6 + 359.2, found 359.1.
4.2.4.3. Methyl (2S,4R)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxylate (22c)
Compound 22c was prepared following the general procedure Method A and using methyl (2S,4R)-4-methoxypyrrolidine-2-carboxylate hydrochloride 19c (purification method: flash chromatography eluting CH2Cl2/MeOH 98:2 v/v; yield: 986 mg, 74%). 1H NMR (400 MHz, CDCl3): δ 5.18 (d, J = 9.4 Hz, 1H), 4.69 (dd, J = 8.8, 3.8 Hz, 1H), 4.21 (dd, J = 9.3, 6.3 Hz, 1H), 4.01 (ddd, J = 16.1, 9.3, 4.3 Hz, 1H), 3.68 (s, 3H), 3.60 (dd, J = 10.6, 2.8 Hz, 1H), 3.28 (s, 3H), 2.36–2.27 (m, 1H), 2.25–2.14 (m, 2H), 2.09–1.98 (m, 1H), 1.42 (d, J = 5.3 Hz, 9H), 1.06 (d, J = 6.8 Hz, 3H), 0.96 (dd, J = 6.7, 2.4 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C17H31N2O6 + 359.2, found 359.1.
4.2.4.4. Ethyl (1S,3aR,6aS)-2-((tert-butoxycarbonyl)-l-valyl)octahydrocyclopenta[c]pyrrole-1-carboxylate (22f)
Compound 22f was prepared following the general procedure Method A and using ethyl (1S,3aR,6aS)-octahydrocyclopenta[c]pyrrole-1-carboxylate hydrochloride 19f (purification method: flash chromatography eluting CH2Cl2/MeOH 97:3 v/v; yield: 580 mg, 87%). 1H NMR (400 MHz, CDCl3): δ 5.21 (d, J = 9.3 Hz, 1H), 4.35 (d, J = 3.9 Hz, 1H), 4.27 (dd, J = 9.3, 6.2 Hz, 1H), 4.17 (q, J = 7.1 Hz, 2H), 3.81 (dd, J = 10.3, 7.7 Hz, 1H), 3.66 (dd, J = 10.4, 3.5 Hz, 1H), 2.74 (dd, J = 7.7, 3.8 Hz, 1H), 2.65 (dd, J = 8.0, 3.9 Hz, 1H), 2.09–1.81 (m, 3H), 1.75 (dd, J = 12.2, 6.1 Hz, 1H), 1.68–1.56 (m, 3H), 1.39 (s, 9H), 1.26 (t, J = 7.1 Hz, 3H), 1.01 (d, J = 6.8 Hz, 3H), 0.91 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C20H35N2O5 + 383.2, found 383.1.
4.2.5. General procedure for the synthesis of dipeptide ester intermediates 22d and 22e (Method B)
In an oven-dried three-necked round-bottom flask, the dipeptidyl alcohol (1 eq.) and Yb(OTf)3 (0.2 eq.) were left stirring in dry CH2Cl2 (0.1 M) under anhydrous nitrogen atmosphere until complete dissolution. Subsequently, di-tertbutyl carbonate (3.3 eq.) was added and the reaction was stirred at reflux for 24 h. The mixture reaction was quenched with distilled water, filtered over Celite, and the filtrate was extracted with CH2Cl2 (25 mL x 3). The collected organic layers were washed with brine (15 mL x 1), dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The reaction crude was purified by flash chromatography to yield the desired product as a white solid.
4.2.5.1. Methyl-(2S,4S)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxylate (22d)
Compound 22d was prepared following the general procedure Method B and using methyl-(2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-hydroxypyrrolidine-2-carboxylate 20 (purification method: flash chromatography eluting CHCl3/MeOH 99:1 v/v; yield: 384 mg, 40%). 1H NMR (400 MHz, CDCl3): δ 5.26–5.06 (m, 1H), 4.74 (dd, J = 8.7, 3.9 Hz, 1H), 4.49 (t, J = 7.8 Hz,1H), 4.12 (dd, J = 11.5, 5.7 Hz, 1H), 3.95 (dd, J = 10.0, 6.5 Hz, 1H), 3.71 (s, 3H), 3.36 (dd, J = 10.1, 6.4 Hz, 1H), 2.47–2.31 (m, 2H), 2.09–1.91 (m, 1H), 1.45 (s, 9H), 1.17 (s, 9H), 0.95 (d, J = 6.8 Hz, 3H), 0.91 (d, J = 6.7 Hz, 3H). MS(ESI) m/z: [M + H]+ calcd for C20H37N2O6 + 401.3, found 401.2.
4.2.5.2. Methyl-(2S,4R)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxylate (22e)
Compound 22e was prepared following the general procedure Method B and using methyl-(2S,4R)-1-((tert-butoxycarbonyl)-l-valyl)-4-hydroxypyrrolidine-2-carboxylate 21 (purification method: flash chromatography eluting CHCl3/MeOH 99:1 v/v; yield: 516 mg, 46%). 1H NMR (400 MHz, CDCl3): δ 5.28–5.04 (m, 1H), 4.73 (dd, J = 8.7, 3.9 Hz, 1H), 4.50 (t, J = 7.8 Hz,1H), 4.11 (dd, J = 11.5, 5.7 Hz, 1H), 3.93 (dd, J = 10.0, 6.5 Hz, 1H), 3.69 (s, 3H), 3.35 (dd, J = 10.1, 6.4 Hz, 1H), 2.48–2.31 (m, 2H), 2.10–1.92 (m, 1H), 1.44 (s, 9H), 1.15 (s, 9H), 0.95 (d, J = 6.8 Hz, 3H), 0.92 (d, J = 6.7 Hz, 3H). MS(ESI) m/z: [M + H]+ calcd for C20H37N2O6 + 401.3, found 401.2.
4.2.6. General procedure for the synthesis of acid derivatives 23a-f, and 26 (Method C)
A solution of 1 N aq. LiOH (2 eq.) was added dropwise to a solution of ester intermediate (1 eq.) in THF (0.2 M) at 0 °C, and the resulting mixture (1:3) was stirred for 3 h at room temperature. The reaction mixture was acidified to pH 3 with a saturated solution of citric acid and extracted with EtOAc (25 mL x 4). The collected organic layers were dried over anhydrous Na2SO4, filtered and concentrated under vacuum to quantitatively yield the product as a white solid, used in the next reaction step without further purification.
4.2.6.1. (2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methylpyrrolidine-2-carboxylic acid (23a)
Compound 23a was prepared following the general procedure Method C and using ester intermediate 22a (yield: 130 mg, 97%). 1H NMR (400 MHz, CDCl3): δ 5.23 (d, J = 8.9 Hz, 1H), 4.53 (t, J = 8.5 Hz, 1H), 4.31 (dd, J = 9.0, 6.4 Hz, 1H), 4.07–3.97 (m, 1H), 3.08 (dd, J = 13.4, 6.2 Hz, 1H), 2.43–2.32 (m, 2H), 1.99 (dd, J = 13.4, 6.7 Hz, 1H), 1.90 (dd, J = 21.3, 9.8 Hz, 1H), 1.41 (s, 9H), 1.11 (d, J = 6.3 Hz, 3H), 0.98 (d, J = 6.6 Hz, 3H), 0.94 (s, 3H). MS (ESI) m/z: [M]- calcd for C16H27N2O5 − 327.2, found 327.1.
4.2.6.2. (2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxylic acid (23b)
Compound 23b was prepared following the general procedure Method C and using ester intermediate 22b (yield: 1.36 g, 98%). 1H NMR (400 MHz, CDCl3): δ 5.22 (d, J = 9.2 Hz, 1H), 4.71 (dd, J = 9.2, 2.8 Hz, 1H), 4.22 (dd, J = 9.0, 6.9 Hz, 1H), 3.96 (dd, J = 10.8, 4.9 Hz, 1H), 3.64 (d, J = 10.4 Hz, 1H), 3.33 (d, J = 7.7 Hz, 3H), 2.60 (d, J = 13.7 Hz, 1H), 2.15 (ddd, J = 13.8, 9.3, 4.6 Hz, 1H), 2.09–1.97 (m, 2H), 1.41 (s, 9H), 1.02 (d, J = 6.8 Hz, 3H), 0.96 (d, J = 6.8 Hz, 3H). MS (ESI) m/z: [M]- calcd for C16H27N2O6 − 343.2, found 343.3.
4.2.6.3. (2S,4R)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxylic acid (23c)
Compound 23c was prepared following the general procedure Method C and using ester intermediate 22c (yield: 1.36 g, 99%). 1H NMR (400 MHz, CDCl3): δ 5.25 (d, J = 9.2 Hz, 1H), 4.69 (dd, J = 9.2, 2.8 Hz, 1H), 4.25 (dd, J = 9.0, 6.9 Hz, 1H), 3.94 (dd, J = 10.8, 4.9 Hz, 1H), 3.65 (d, J = 10.4 Hz, 1H), 3.34 (d, J = 7.7 Hz, 3H), 2.58 (d, J = 13.7 Hz, 1H), 2.16 (ddd, J = 13.8, 9.3, 4.6 Hz, 1H), 2.11–1.95 (m, 2H), 1.40 (s, 9H), 1.02 (d, J = 6.8 Hz, 3H), 0.95 (d, J = 6.8 Hz, 3H). MS (ESI) m/z: [M]- calcd for C16H27N2O6 − 343.2, found 343.3.
4.2.6.4. (2S,4S)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxylic acid (23d)
Compound 23d was prepared following the general procedure Method C and using ester intermediate 22d (yield: 300 mg, 100%). 1H NMR (700 MHz, CDCl3): δ 5.30–5.23 (m, 1H), 4.53 (dd, J = 8.4, 7.1 Hz, 1H), 4.29–4.21 (m, 1H), 3.97 (dd, J = 10.2, 6.1 Hz, 1H), 3.41 (dd, J = 10.2, 6.0 Hz, 1H), 2.39 (ddd, J = 14.1, 10.7, 6.3 Hz, 2H), 2.14 (dt, J = 13.1, 6.6 Hz, 1H), 2.04–1.99 (m, 1H), 1.42 (s, 9H), 1.20 (s, 9H), 1.00 (d, J = 6.6 Hz, 3H), 0.92 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M]- calcd for C19H33N2O6 − 385.2, found 385.3.
4.2.6.5. (2S,4R)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxylic acid (23e)
Compound 23e was prepared following the general procedure Method C and using ester intermediate 22e (yield: 497 mg, 97%). 1H NMR (700 MHz, CDCl3): δ 5.31–5.22 (m, 1H), 4.54 (dd, J = 8.4, 7.1 Hz, 1H), 4.30–4.19 (m, 1H), 3.98 (dd, J = 10.2, 6.1 Hz, 1H), 3.39 (dd, J = 10.2, 6.0 Hz, 1H), 2.35 (ddd, J = 14.1, 10.7, 6.3 Hz, 2H), 2.16 (dt, J = 13.1, 6.6 Hz, 1H), 2.06–1.97 (m, 1H), 1.41 (s, 9H), 1.22 (s, 9H), 1.01 (d, J = 6.6 Hz, 3H), 0.94 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M]- calcd for C19H33N2O6 − 385.2, found 385.3.
4.2.6.6. (1S,3aR,6aS)-2-((tert-butoxycarbonyl)-l-valyl)octahydrocyclopenta[c]pyrrole-1-carboxylic acid (23f)
Compound 23f was prepared following the general procedure Method C and using ester intermediate 22f (yield: 531 mg, 100%). 1H NMR (400 MHz, CDCl3): δ 5.25 (d, J = 9.5 Hz, 1H), 4.41 (d, J = 3.5 Hz, 1H), 4.26 (dd, J = 9.1, 7.0 Hz, 1H), 3.82–3.69 (m, 1H), 2.92–2.84 (m, 1H), 2.79 (dd, J = 7.5, 4.0 Hz, 1H), 2.05–1.95 (m, 2H), 1.92–1.82 (m, 1H), 1.81–1.71 (m, 1H), 1.68–1.47 (m, 4H), 1.41 (s, 9H), 0.97 (d, J = 6.7 Hz, 3H), 0.92 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M calcd]- for C18H29N2O5 − 353.2, found 353.3.
4.2.6.7. (S)-2-((S)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoic acid (26)
Compound 26 was prepared following the general procedure Method C and using ester intermediate 25g (yield: 180 mg, 100%). 1H NMR (400 MHz, DMSO‑d 6): δ 8.19 (s, 1H), 7.54 (s, 1H), 6.79 (d, J = 8.1 Hz, 1H), 4.30 (d, J = 28.8 Hz, 1H), 3.99 (d, J = 8.1 Hz, 1H), 3.70 (s, 1H), 3.56–3.52 (m, 1H), 3.12–3.04 (m, 2H), 2.10–1.73 (m, 8H), 1.55 (d, J = 11.2 Hz, 2H), 1.36 (s, 9H), 0.89 (d, J = 6.2 Hz, 3H), 0.84 (d, J = 6.1 Hz, 3H). MS (ESI) m/z: [M]- calcd for C22H35N4O7 − 467.2, found 467.2.
4.2.7. General procedure for the convergent synthesis of tripeptide intermediates 25a-i (Method D)
To a solution of the appropriate dipeptide acid intermediate (1 eq.) in anhydrous DMF (0.4 M) in an oven-dried three-necked round-bottom flask, EDCI (1.2 eq.) and HOBt (1.2 eq.) were added under a positive anhydrous nitrogen pressure, and the reaction mixture was left under stirring at room temperature for 30 min. In a separate flask, a solution of amine hydrochloride 24 [33] (1 eq.) in anhydrous DMF (0.3 M) was cooled to 0–5 °C and DIPEA (4 eq.) was added dropwise. After 30 min, this solution was added to the reaction mixture containing the activated acid, and the resulting mixture was left under stirring at room temperature for 16 h. The resulting mixture was washed with brine and extracted with EtOAc (25 mL x 4). The collected organic layers were washed with brine (15 mL x 10), dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The reaction crude was purified by flash chromatography to yield the desired product as a white solid.
4.2.7.1. Methyl (S)-2-((2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methylpyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25a)
Compound 25a was prepared following the general procedure Method D and using (2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methylpyrrolidine-2- carboxylic acid 23a (purification method: flash chromatography eluting CH2Cl2/MeOH 97:3 v/v; yield: 72 mg, 39%). 1H NMR (400 MHz, CDCl3): δ 7.51 (d, J = 7.4 Hz, 1H), 5.64 (s, 1H), 5.27 (d, J = 9.2 Hz, 1H), 4.55 (ddd, J = 11.3, 7.3, 4.0 Hz, 1H), 4.37–4.25 (m, 2H), 3.98–3.90 (m, 1H), 3.72 (s, 3H), 3.31 (dd, J = 8.8, 6.2 Hz, 2H), 3.12 (t, J = 10.1 Hz, 1H), 2.60 (d, J = 8.1 Hz, 1H), 2.49–2.32 (m, 2H), 2.27 (d, J = 6.6 Hz, 1H), 2.14 (ddd, J = 14.2, 11.4, 5.7 Hz, 1H), 2.00 (dd, J = 13.0, 6.5 Hz, 1H), 1.92–1.78 (m, 2H), 1.71 (dd, J = 16.8, 6.6 Hz, 1H), 1.41 (s, 9H), 1.09 (d, J = 6.4 Hz, 3H), 0.99 (d, J = 6.8 Hz, 3H), 0.88 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C24H41N4O7 + 497.3, found 497.3.
4.2.7.2. Methyl (S)-2-((2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25b)
Compound 25b was prepared following the general procedure Method D and using (2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2- carboxylic acid 23b (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 890 mg, 45%).1H NMR (400 MHz, CDCl3): δ 7.29 (brs, 1H), 5.61 (s, 1H), 5.23 (d, J = 9.2 Hz, 1H), 4.62 (ddd, J = 9.3, 7.2, 3.5 Hz, 2H), 4.27 (dd, J = 9.3, 5.8 Hz, 1H), 4.01 (d, J = 2.6 Hz, 1H), 3.92 (dd, J = 10.7, 5.0 Hz, 1H), 3.71 (s, 3H), 3.63 (d, J = 10.6 Hz, 1H), 3.35–3.27 (m, 5H), 2.59–2.39 (m, 3H), 2.20–2.00 (m, 2H), 1.84 (ddd, J = 14.0, 8.1, 3.9 Hz, 2H), 1.41 (s, 9H), 1.05 (d, J = 6.8 Hz, 3H), 0.96 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C24H41N4O8 + 513.3, found 513.3.
4.2.7.3. Methyl (S)-2-((2S,4R)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25c)
Compound 25c was prepared following the general procedure Method D and using (2S,4R)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2- carboxylic acid 23c (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 366 mg, 52%).1H NMR (400 MHz, CDCl3): δ 7.28 (brs, 1H), 5.59 (s, 1H), 5.25 (d, J = 9.2 Hz, 1H), 4.61 (ddd, J = 9.3, 7.2, 3.5 Hz, 2H), 4.28 (dd, J = 9.3, 5.8 Hz, 1H), 4.0 (d, J = 2.6 Hz, 1H), 3.92 (dd, J = 10.7, 5.0 Hz, 1H), 3.73 (s, 3H), 3.61 (d, J = 10.6 Hz, 1H), 3.33–3.29 (m, 5H), 2.57–2.36 (m, 3H), 2.19–1.99 (m, 2H), 1.82 (ddd, J = 14.0, 8.1, 3.9 Hz, 2H), 1.41 (s, 9H), 1.03 (d, J = 6.8 Hz, 3H), 0.94 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C24H41N4O8 + 513.3, found 513.3.
4.2.7.4. Methyl (S)-2-((2S,4S)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25d)
Compound 25d was prepared following the general procedure Method D and using (2S,4S)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxylic acid 23d (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 147 mg, 37%).1H NMR (700 MHz, CDCl3): δ 7.39 (d, J = 7.2 Hz, 1H), 5.58 (d, J = 20.1 Hz, 1H), 4.58 (ddd, J = 11.4, 7.6, 3.9 Hz, 1H), 4.36 (t, J = 8.0 Hz, 1H), 4.28 (dd, J = 9.1, 5.5 Hz, 1H), 4.22–4.18 (m, 1H), 3.92 (dd, J = 10.0, 6.3 Hz, 1H), 3.71 (s, 3H), 3.40 (dd, J = 10.0, 6.8 Hz, 1H), 3.31 (s, 3H), 2.53 (dd, J = 9.8, 5.1 Hz, 1H), 2.49–2.42 (m, 2H), 2.41–2.36 (m, 1H), 2.17–2.02 (m, 2H), 1.89–1.82 (m, 2H), 1.42 (s, 9H), 1.20 (s, 9H), 1.01 (d, J = 6.8 Hz, 3H), 0.89 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C27H47N4O8 + 555.3, found 555.3.
4.2.7.5. Methyl (S)-2-((2S,4R)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25e)
Compound 25e was prepared following the general procedure Method D and using (2S,4R)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxylic acid 23e (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 600 mg, 83%).1H NMR (700 MHz, CDCl3): δ 7.41 (d, J = 7.2 Hz, 1H), 5.59 (d, J = 20.1 Hz, 1H), 4.56 (ddd, J = 11.4, 7.6, 3.9 Hz, 1H), 4.38 (t, J = 8.0 Hz, 1H), 4.27 (dd, J = 9.1, 5.5 Hz, 1H), 4.21–4.17 (m, 1H), 3.91 (dd, J = 10.0, 6.3 Hz, 1H), 3.73 (s, 3H), 3.41 (dd, J = 10.0, 6.8 Hz, 1H), 3.33 (s, 3H), 2.54 (dd, J = 9.8, 5.1 Hz, 1H), 2.51–2.42 (m, 2H), 2.39–2.35 (m, 1H), 2.15–2.04 (m, 2H), 1.89–1.83 (m, 2H), 1.43 (s, 9H), 1.21 (s, 9H), 1.03 (d, J = 6.8 Hz, 3H), 0.89 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C27H47N4O8 + 555.3, found 555.3.
4.2.7.6. Methyl-(S)-2-((1S,3aR,6aS)-2-((tert-butoxycarbonyl)-l-valyl)octahydrocyclopenta[c]pyrrole-1-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25f)
Compound 25f was prepared following the general procedure Method D and using (1S,3aR,6aS)-2-((tert-butoxycarbonyl)-l-valyl)octahydrocyclopenta[c]pyrrole-1-carboxylic acid 23f (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 427 mg, 62%).1H NMR (400 MHz, CDCl3): δ 7.54 (d, J = 7.1 Hz, 1H), 5.61 (s, 1H), 5.22 (d, J = 9.3 Hz, 1H), 4.52 (ddd, J = 11.2, 7.2, 4.2 Hz, 1H), 4.31–4.23 (m, 2H), 3.82 (dd, J = 10.2, 7.4 Hz, 1H), 3.72 (s, 3H), 3.67–3.60 (m, 1H), 3.37–3.25 (m, 2H), 2.80 (s, 3H), 2.55–2.35 (m, 2H), 2.13 (ddd, J = 14.3, 10.4, 5.0 Hz, 1H), 2.06–1.94 (m, 2H), 1.93–1.70 (m, 5H), 1.50–1.41 (m, 10H), 0.98 (d, J = 6.8 Hz, 3H), 0.89 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C26H43N4O7 + 523.3, found 523.4.
4.2.7.7. Methyl (S)-2-((S)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25 g)
Compound 25g was prepared following the general procedure Method D and using (tert-butoxycarbonyl)-l-valyl-l-proline 23g [26] (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 180 mg, 65%). 1H NMR (400 MHz, DMSO‑d 6): δ 8.43 (d, J = 8.4 Hz, 1H), 7.59 (s, 1H), 6.80 (d, J = 8.4 Hz, 1H), 4.43–4.29 (m, 2H), 3.97 (t, J = 8.0 Hz, 1H), 3.75–3.66 (m, 1H), 3.62 (s, 3H), 3.55 (d, J = 8.6 Hz, 1H), 3.14–3.02 (m, 2H), 2.16–1.70 (m, 8H), 1.63–1.50 (m, 2H), 1.36 (s, 9H), 0.88 (d, J = 6.6 Hz, 3H), 0.84 (d, J = 6.4 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C23H39N4O7 + 483.3, found 483.3.
4.2.7.8. Methyl (S)-2-((S)-1-((tert-butoxycarbonyl)-l-valyl)piperidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25h)
Compound 25h was prepared following the general procedure Method D and using (S)-1-((tert-butoxycarbonyl)-l-valyl)piperidine-2-carboxylic acid 23h [34] (purification method: flash chromatography eluting CH2Cl2/MeOH 98:2 to 95:5 v/v; yield: 397 mg, 49%).1H NMR (400 MHz, CDCl3): δ 7.62 (d, J = 6.3 Hz, 1H), 5.81 (t, J = 23.4 Hz, 1H), 5.57 (d, J = 7.9 Hz, 1H), 5.32 (dd, J = 35.4, 12.9 Hz, 1H), 4.67–4.55 (m, 1H), 4.53–4.45 (m, 1H), 4.41–4.31 (m, 1H), 3.86 (t, J = 12.5 Hz, 1H), 3.71 (s, 3H), 3.38–3.25 (m, 2H), 2.42 (t, J = 13.2 Hz, 4H), 2.00 (dddd, J = 26.7, 22.7, 16.5, 8.9 Hz, 8H), 1.43 (s, 9H), 0.99 (d, J = 5.4 Hz, 3H), 0.86 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C24H41N4O7 + 497.3, found 497.1.
4.2.7.9. Methyl-(S)-2-((S)-1-(N-(tert-butoxycarbonyl)-O-(tert-butyl)-l-threonyl)pyrrolidin-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate (25i)
Compound 25i was prepared following the general procedure Method D and using N-(tert-butoxycarbonyl)-O-(tert-butyl)-l-threonyl-l-proline 23i (purification method: flash chromatography eluting CH2Cl2/MeOH 97:3 v/v; yield: 296 mg, 41%). 1H NMR (400 MHz, CDCl3): δ 7.43 (d, J = 7.8 Hz, 1H), 5.76 (s, 1H), 5.40 (d, J = 8.1 Hz, 1H), 4.66–4.51 (m, 2H), 4.46 (dd, J = 8.1, 5.3 Hz, 1H), 4.00–3.84 (m, 2H), 3.71 (s, 3H), 3.32 (dd, J = 18.7, 9.1 Hz, 2H), 2.42 (ddd, J = 12.4, 9.0, 3.4 Hz, 2H), 2.26–1.76 (m, 8H), 1.41 (s, 9H), 1.23 (s, 9H), 1.15 (d, J = 5.2 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C26H45N4O8 + 541.3, found 541.3.
4.2.8. General procedure for the synthesis of tripeptide alcohols 27a-i (Method E)
In oven-dried three-necked round-bottom flask, the tripeptide methyl ester (1 eq.) was dissolved in anhydrous THF (0.2 M) under a positive anhydrous nitrogen pressure, then a 2 M solution of LiBH4 (3 eq.) in anhydrous THF was added dropwise at 0 °C, and the reaction mixture was stirred at room temperature for 5 h. The reaction was quenched by acidifying with a saturated citric acid solution to pH 2 and extracted with EtOAc (25 mL x 3). The organic phase was washed with brine (15 mL x 10), dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The reaction crude was purified by flash chromatography to yield the desired product as a white solid.
4.2.8.1. tert-Butyl ((S)-1-((2S,4S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)-4-methylpyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27a)
Compound 27a was prepared following the general procedure Method E and using methyl (S)-2-((2S,4S)-1-((tert-butoxycarbonyl)- l-valyl)-4-methylpyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25a (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 30 mg, 54%). 1H NMR (400 MHz, CDCl3): δ 7.50 (d, J = 7.2 Hz, 1H), 5.58 (s, 1H), 5.27 (d, J = 9.2 Hz, 1H), 4.56 (ddd, J = 11.3, 7.3, 4.0 Hz, 1H), 4.35–4.25 (m, 2H), 3.96–3.90 (m, 1H), 3.31–3.20 (m, 4H), 3.10 (t, J = 10.1 Hz, 1H), 2.61 (d, J = 8.1 Hz, 1H), 2.47–2.30 (m, 2H), 2.25 (d, J = 6.6 Hz, 1H), 2.12 (ddd, J = 14.2, 11.4, 5.7 Hz, 1H), 2.00 (dd, J = 13.0, 6.5 Hz, 1H), 1.92–1.79 (m, 2H), 1.71 (dd, J = 16.8, 6.6 Hz, 2H), 1.39 (s, 9H), 1.08 (d, J = 6.4 Hz, 3H), 0.98 (d, J = 6.8 Hz, 3H), 0.86 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C23H41N4O6 + 469.3, found 469.3.
4.2.8.2. tert-Butyl ((S)-1-((2S,4S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)-4-methoxypyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27b)
Compound 27b was prepared following the general procedure Method E and using methyl (S)-2-((2S,4S)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25b (purification method: flash chromatography eluting CH2Cl2/MeOH 93:7 v/v; yield: 624 mg, 74%). 1H NMR (700 MHz, CDCl3) δ 7.29 (d, J = 8.0 Hz, 1H), 7.04 (d, J = 6.5 Hz, 1H), 5.94 (s, 1H), 5.24 (d, J = 9.0 Hz, 1H), 4.53 (dd, J = 9.1, 3.7 Hz, 1H), 4.15 (dd, J = 8.8, 5.5 Hz, 1H), 3.91 (dd, J = 9.8, 4.9 Hz, 2H), 3.80 (dd, J = 11.9, 2.4 Hz, 1H), 3.72 (dd, J = 13.7, 4.9 Hz, 1H), 3.51–3.45 (m, 2H), 3.33–3.29 (m, 5H), 2.40 (dd, J = 17.0, 7.1 Hz, 2H), 2.07 (dd, J = 11.8, 6.7 Hz, 2H), 2.00–1.94 (m, 2H), 1.81 (dd, J = 21.0, 9.4 Hz, 2H), 1.42 (s, 9H), 1.03 (d, J = 6.6 Hz, 3H), 0.97 (d, J = 6.8 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C23H41N4O7 + 485.3, found 485.3.
4.2.8.3. tert-Butyl ((S)-1-((2S,4R)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)-4-methoxypyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27c)
Compound 27c was prepared following the general procedure Method E and using methyl (S)-2-((2S,4R)-1-((tert-butoxycarbonyl)-l-valyl)-4-methoxypyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25c (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 160 mg, 47%). 1H NMR (700 MHz, CDCl3) δ 7.28 (d, J = 8.0 Hz, 1H), 7.02 (d, J = 6.5 Hz, 1H), 5.95 (s, 1H), 5.26 (d, J = 9.0 Hz, 1H), 4.54 (dd, J = 9.1, 3.7 Hz, 1H), 4.17 (dd, J = 8.8, 5.5 Hz, 1H), 3.92 (dd, J = 9.8, 4.9 Hz, 2H), 3.79 (dd, J = 11.9, 2.4 Hz, 1H), 3.71 (dd, J = 13.7, 4.9 Hz, 1H), 3.52–3.44 (m, 2H), 3.35–3.31 (m, 5H), 2.42 (dd, J = 17.0, 7.1 Hz, 2H), 2.09 (dd, J = 11.8, 6.7 Hz, 2H), 1.99–1.92 (m, 2H), 1.82 (dd, J = 21.0, 9.4 Hz, 2H), 1.41 (s, 9H), 1.05 (d, J = 6.6 Hz, 3H), 0.99 (d, J = 6.8 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C23H41N4O7 + 485.3, found 485.3.
4.2.8.4. tert-Butyl ((S)-1-((2S,4S)-4-(tert-butoxy)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27d)
Compound 27d was prepared following the general procedure Method E and using methyl (S)-2-((2S,4S)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25d (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 53 mg, 40%). 1H NMR (400 MHz, CDCl3): δ 7.31 (d, J = 7.6 Hz, 1H), 5.93 (s, 1H), 5.33–5.25 (m, 1H), 4.29 (dt, J = 9.1, 7.0 Hz, 2H), 4.22–4.14 (m, 1H), 3.99 (d, J = 3.4 Hz, 1H), 3.95–3.85 (m, 1H), 3.65 (s, 1H), 3.52–3.39 (m, 3H), 3.31 (dd, J = 9.0, 4.4 Hz, 2H), 2.57–2.47 (m, 1H), 2.46–2.35 (m, 2H), 2.13–1.93 (m, 3H), 1.82 (dd, J = 27.7, 17.3 Hz, 2H), 1.42 (s, 9H), 1.19 (s, 9H), 1.01 (d, J = 6.7 Hz, 3H), 0.90 (d, J = 6.6 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C26H47N4O7 + 527.3, found 527.3.
4.2.8.5. tert-Butyl ((S)-1-((2S,4R)-4-(tert-butoxy)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27e)
Compound 27e was prepared following the general procedure Method E and using methyl (S)-2-((2S,4R)-4-(tert-butoxy)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidin-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25e (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 225 mg, 75%). 1H NMR (400 MHz, CDCl3): δ 7.31 (d, J = 7.6 Hz, 1H), 5.95 (s, 1H), 5.34–5.23 (m, 1H), 4.27 (dt, J = 9.1, 7.0 Hz, 2H), 4.24–4.15 (m, 1H), 4.01 (d, J = 3.4 Hz, 1H), 3.94–3.87 (m, 1H), 3.67 (s, 1H), 3.53–3.41 (m, 3H), 3.32 (dd, J = 9.0, 4.4 Hz, 2H), 2.59–2.45 (m, 1H), 2.48–2.36 (m, 2H), 2.14–1.92 (m, 3H), 1.81 (dd, J = 27.7, 17.3 Hz, 2H), 1.43 (s, 9H), 1.17 (s, 9H), 1.02 (d, J = 6.7 Hz, 3H), 0.89 (d, J = 6.6 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C26H47N4O7 + 527.3, found 527.3.
4.2.8.6. tert-Butyl ((S)-1-((1S,3aR,6aS)-1-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)hexahydrocyclopenta[c]pyrrol-2(1H)-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27f)
Compound 27f was prepared following the general procedure Method E and using methyl-(S)-2-((1S,3aR,6aS)-2-((tert-butoxycarbonyl)-l-valyl)octahydrocyclopenta[c]pyrrole-1-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25f (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 to 93:7 v/v; yield: 250 mg, 62%). 1H NMR (700 MHz, CDCl3): δ 7.64–7.60 (m, 1H), 5.81 (d, J = 14.7 Hz, 1H), 5.30 (dd, J = 11.7, 6.1 Hz, 1H), 4.30–4.24 (m, 1H), 4.21 (d, J = 3.7 Hz, 1H), 4.06 (dd, J = 9.0, 4.6 Hz, 1H), 3.96 (dt, J = 10.6, 5.6 Hz, 1H), 3.86 (dd, J = 10.0, 8.1 Hz, 1H), 3.68 (d, J = 10.0 Hz, 1H), 3.64–3.55 (m, 2H), 3.51 (ddd, J = 32.2, 15.7, 9.0 Hz, 1H), 3.32 (dd, J = 9.0, 4.5 Hz, 2H), 2.80 (dd, J = 7.6, 3.9 Hz, 1H), 2.76–2.72 (m, 1H), 2.55–2.48 (m, 1H), 2.40 (ddd, J = 12.4, 8.6, 4.3 Hz, 1H), 2.02–1.96 (m, 1H), 1.95–1.90 (m, 1H), 1.84 (ddd, J = 26.6, 17.7, 9.8 Hz, 2H), 1.65–1.58 (m, 3H), 1.47 (dt, J = 11.8, 5.5 Hz, 2H), 1.42 (d, J = 5.8 Hz, 9H), 0.98 (t, J = 6.0 Hz, 3H), 0.89 (d, J = 6.7 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C25H43N4O6 + 495.3, found 495.2.
4.2.8.7. tert-Butyl ((S)-1-((S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27 g)
Compound 27g was prepared following the general procedure Method E and using methyl (S)-2-((S)-1-((tert-butoxycarbonyl)-l-valyl)pyrrolidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25g (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v; yield: 90 mg, 50%).1H NMR (400 MHz, DMSO‑d 6): δ 7.66 (d, J = 8.9 Hz, 1H), 7.45 (s, 1H), 6.80 (d, J = 8.4 Hz, 1H), 4.64 (d, J = 5.7 Hz, 1H), 4.23 (d, J = 7.8 Hz, 1H), 3.98 (s, 1H), 3.77 (s, 1H), 3.69 (s, 2H), 3.13–3.04 (m, 2H), 3.03–2.95 (m, 2H), 2.16 (d, J = 14.8, 2H), 2.05–1.90 (m, 6H), 1.50 (d, J = 11.4 Hz, 2H), 1.36 (s, 9H), 0.88 (d, J = 6.7 Hz, 3H), 0.84 (d, J = 6.6 Hz, 3H). MS (ESI) m/z: [M+H]+ calcd for C22H39N4O6 + 455.3, found 455.2.
4.2.8.8. tert-Butyl ((S)-1-((S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)piperidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (27h)
Compound 27h was prepared following the general procedure Method E and using methyl (S)-2-((S)-1-((tert-butoxycarbonyl)-l-valyl)piperidine-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25h (purification method: flash chromatography eluting CH2Cl2/MeOH 97:3 v/v; yield: 274 mg, 73%). 1H NMR (400 MHz, CDCl3): δ 7.87 (d, J = 7.9 Hz, 1H), 5.80 (m, 1H), 5.23 (m, 1H), 4.57 (m, 1H), 4.49 (d, J = 5.2 Hz, 1H), 4.25–4.15 (m, 1H), 4.03–3.71 (m, 1H), 3.59 (dd, J = 22.9, 12.1 Hz, 2H), 3.61–3.53 (m, 2H), 3.40–3.32 (m, 3H), 2.57–2.30 (m, 3H), 1.93–1.77 (m, 8H), 1.63 (s, 9H), 1.01 (d, J = 3.8 Hz, 3H), 0.89 (d, J = 7.6 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C23H41N4O6 + 469.3, found 469.2.
4.2.8.9. tert-Butyl ((2S,3R)-3-(tert-butoxy)-1-((S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-1-oxobutan-2-yl)carbamate (27i)
Compound 27i was prepared following the general procedure Method E and using methyl-(S)-2-((S)-1-(N-(tert-butoxycarbonyl)-O-(tert-butyl)-l-threonyl)pyrrolidin-2-carboxamido)-3-((S)-2-oxopyrrolidin-3-yl)propanoate 25i (purification method: flash chromatography eluting CH2Cl2/MeOH 98:2 to 95:5 v/v; yield: 190 mg, 62%). 1H NMR (400 MHz, CDCl3): δ 6.84 (d, J = 8.3 Hz, 1H), 5.71 (s, 1H), 5.38 (d, J = 7.6 Hz, 1H), 4.59–4.44 (m, 2H), 4.04 (ddd, J = 23.9, 18.6, 5.5 Hz, 2H), 3.90 (dt, J = 12.6, 6.4 Hz, 1H), 3.85–3.71 (m, 1H), 3.70–3.61 (m, 1H), 3.59–3.41 (m, 1H), 3.37–3.29 (m, 2H), 3.18 (s, 1H), 2.54–2.29 (m, 2H), 2.27–1.92 (m, 5H), 1.88–1.76 (m, 2H), 1.43 (s, 9H), 1.26 (s, 9H), 1.18 (d, J = 9.0 Hz, 3H). MS (ESI) m/z: [M + H]+ calcd for C25H45N4O7 + 513.3, found 513.3.
4.2.9. General procedure for the synthesis of target aldehydes 6–14 (Method F)
The tripeptide alcohol intermediate (1 eq.) was dissolved in anhydrous DMSO (5 eq.) and anhydrous CH2Cl2 (0.4 M) under anhydrous nitrogen atmosphere, and DIPEA (3.5 eq.) was added at −5 °C. In another flask, pyridine sulfur trioxide complex (3 eq.) and anhydrous pyridine (3 eq.) were suspended in anhydrous DMSO (7.5 eq.) at room temperature. The suspension was left under stirring for 10 min and then added to the previously formed tripeptide alcohol/DMSO mixture at −5 °C. After 24 h, the reaction mixture was poured into ice/water (1:1) and extracted with CH2Cl2 (25 mL x 3). The collected organic layers were washed with a saturated citric acid solution (15 mL x 1), water (15 mL x 1), saturated solution of NaHCO3 (15 mL x 1), brine (15 mL x 1), dried over anhydrous Na2SO4, filtered, and concentrated under vacuum. The reaction crude was purified by flash chromatography to yield the desired product as a white solid.
4.2.9.1. tert-Butyl ((S)-3-methyl-1-oxo-1-((S)-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)butan-2-yl)carbamate (6)
Compound 6 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate 27g (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 to 93:7 v/v; yield: 22 mg, 25%). [α]20 D −84.5 (c = 1.0, MeOH). 1H NMR (400 MHz, DMSO‑d 6): δ 9.42 (s, 1H), 8.44 (d, J = 7.9 Hz, 1H), 7.60 (s, 1H), 6.80 (t, J = 11.1 Hz, 1H), 4.32 (s, 1H), 4.26 (d, J = 7.6 Hz, 1H), 4.00 (t, J = 7.8 Hz, 1H), 3.71 (s, 1H), 3.59 (d, J = 23.9 Hz, 1H), 3.18–3.03 (m, 2H), 2.20–2.06 (m, 2H), 1.94 (d, J = 17.2 Hz, 2H), 1.86 (d, J = 12.3 Hz, 4H), 1.67–1.54 (m, 2H), 1.36 (s, 9H), 0.88 (d, J = 6.5 Hz, 3H), 0.84 (d, J = 6.5 Hz, 3H).13C NMR (176 MHz, CDCl3): δ 199.9, 180.1, 172.8, 172.0, 156.0, 131.2, 79.7, 60.4, 57.9, 57.0, 47.8, 40.6, 37.9, 31.4, 30.0, 29.1, 28.9, 25.4, 19.6, 17.4. HRMS (ESI) m/z: [M+H]+ calcd for C22H37N4O6 + 453.2713, found 453.1683. LC-MS (ESI) m/z: [M+H]+ calcd for C22H37N4O6 + 453.3, found 453.2 (rt: 6.65).
4.2.9.2. tert-Butyl ((S)-3-methyl-1-((2S,4S)-4-methyl-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-1-oxobutan-2-yl)carbamate (7)
Compound 7 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((2S,4S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)-4-methylpyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate 27a (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5; yield: 18 mg, 60%). [α]20 D −38.9 (c = 1.0, MeOH). 1H NMR (400 MHz, CDCl3): δ 9.50 (s, 1H), 8.16–8.07 (m, 1H), 6.00 (d, J = 27.5 Hz, 1H), 5.37–5.28 (m, 1H), 4.43–4.35 (m, 1H), 4.30 (dd, J = 9.0, 6.0 Hz, 1H), 3.99–3.89 (m, 1H), 3.37–3.29 (m, 2H), 3.15 (t, J = 10.2 Hz, 1H), 2.71–2.54 (m, 1H), 2.48–2.35 (m, 2H), 2.28 (td, J = 12.9, 6.5 Hz, 1H), 2.10–1.75 (m, 6H), 1.42 (s, 9H), 1.09 (t, J = 6.5 Hz, 3H), 0.99 (t, J = 5.9 Hz, 3H), 0.90–0.83 (m, 3H). 13C NMR (176 MHz, CDCl3): δ 199.4, 177.1, 172.9, 144.8, 79.6, 77.3, 77.2, 77.0, 70.4, 65.3, 63.5, 61.3, 58.0, 57.0, 54.9, 40.7, 37.6, 34.3, 31.4, 29.8, 28.5, 19.5, 17.3, 16.6. HRMS (ESI) m/z: [M+H]+ calcd for C23H39N4O6 + 467.2870, found 467.2862. LC-MS (ESI) m/z: [M+H]+ calcd for C23H39N4O6 + 467.3, found 467.2 (rt: 6.96).
4.2.9.3. tert-Butyl ((S)-1-((2S,4S)-4-methoxy-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (8)
Compound 8 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((2S,4S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)-4-methoxypyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate 27b (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v, yield: 76 mg, 35%). [α]20 D −49.6 (c = 1.0, MeOH). 1H NMR (400 MHz, DMSO‑d 6): δ 9.40 (s, 1H), 8.30 (d, J = 7.7 Hz, 1H), 7.61 (s, 1H), 6.89–6.73 (m, 1H), 4.38–4.27 (m, 1H), 4.22 (s, 1H), 4.16–4.04 (m, 1H), 4.05–3.93 (m, 2H), 3.67 (d, J = 8.2 Hz, 1H), 3.24 (s, 3H), 3.20–2.99 (m, 3H), 2.35 (ddd, J = 27.0, 15.4, 6.9 Hz, 2H), 2.17 (dd, J = 12.4, 6.9 Hz, 1H), 1.89 (ddd, J = 29.8, 13.6, 6.3 Hz, 3H), 1.65–1.56 (m, 1H), 1.37 (s, 9H), 0.90 (d, J = 6.6 Hz, 3H), 0.84 (d, J = 6.4 Hz, 3H).13C NMR (176 MHz, CDCl3): δ 199.5, 180.0, 172.9, 172.1, 156.1, 129.9, 79.8, 79.2, 59.5, 57.4, 57.0, 52.9, 40.5, 37.5, 33.2, 31.2, 30.4, 28.6, 28.5, 19.8, 17.4. HRMS (ESI) m/z: [M+H]+ calcd for C23H39N4O7 + 483.2819, found 483.2814. LC-MS (ESI) m/z: [M+H]+ calcd for C23H39N4O7 + 483.3, found 483.2 (rt: 6.60).
4.2.9.4. tert-Butyl ((S)-1-((2S,4S)-4-(tert-butoxy)-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (9)
Compound 9 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((2S,4S)-4-(tert-butoxy)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl) carbamate 27d (purification method: flash chromatography eluting CH3Cl/MeOH 93:7 v/v, yield: 13 mg, 27%). [α]20 D −12.1 (c = 1.0, MeOH). 1H NMR (400 MHz, CDCl3): δ 9.50 (s, 1H), 7.98 (d, J = 5.3 Hz, 1H), 5.67 (s, 1H), 5.24 (d, J = 8.3 Hz, 1H), 4.48–4.34 (m, 2H), 4.33–4.25 (m, 1H), 4.22 (dd, J = 13.8, 6.9 Hz, 1H), 3.95 (dd, J = 10.0, 6.3 Hz, 1H), 3.43 (dd, J = 10.0, 6.9 Hz, 1H), 3.34 (dd, J = 8.6, 6.3 Hz, 2H), 2.61–2.50 (m, 1H), 2.47–2.38 (m, 2H), 2.12–2.03 (m, 2H), 1.94 (t, J = 6.7 Hz, 2H), 1.85 (dd, J = 12.3, 9.8 Hz, 1H), 1.43 (s, 9H), 1.19 (d, J = 7.2 Hz, 9H), 1.01 (d, J = 6.7 Hz, 3H), 0.89 (d, J = 6.7 Hz, 3H). 13C NMR (176 MHz, CDCl3): δ 199.9, 179.9, 172.4, 171.9, 156.1, 129.9, 79.7, 74.5, 69.7, 59.2, 57.8, 57.0, 53.8, 40.6, 38.0, 37.0, 31.2, 30.2, 28.9, 28.4, 19.7, 17.2. HRMS (ESI) m/z: [M+H]+ calcd for C26H45N4O7 + 525.3288, found 525.3279. LC-MS (ESI) m/z: [M+H]+ calcd for C26H45N4O7 + 525.3, found 525.3 (rt: 7.45).
4.2.9.5. tert-Butyl ((S)-1-((2S,4R)-4-methoxy-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (10)
Compound 10 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((2S,4R)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)-4-methoxypyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate 27c (purification method: flash chromatography eluting CH2Cl2/MeOH 95:5 v/v, yield: 10 mg, 17%). [α]20 D −54.2 (c = 1.0, MeOH). 1H NMR (700 MHz, CDCl3): δ 9.50 (s, 1H), 8.16 (d, J = 6.0 Hz, 1H), 5.91 (s, 1H), 5.30 (dd, J = 17.2, 9.1 Hz, 1H), 4.54 (t, J = 7.9 Hz, 1H), 4.40–4.33 (m, 1H), 4.27 (dd, J = 9.1, 6.6 Hz, 1H), 4.08 (d, J = 2.2 Hz, 1H), 3.95 (d, J = 11.0 Hz, 1H), 3.70 (dd, J = 11.0, 4.2 Hz, 1H), 3.32 (s, 3H), 2.64–2.59 (m, 1H), 2.42–2.37 (m, 1H), 2.35–2.30 (m, 1H), 2.22–2.18 (m, 1H), 2.05–1.89 (m, 4H), 1.86–1.79 (m, 2H), 1.42 (s, 9H), 0.97–0.95 (m, 3H), 0.88 (d, J = 6.7 Hz, 3H). 13C NMR (176 MHz, CDCl3) δ 190.6, 180.6, 179.1, 178.7, 155.8, 130.8, 79.2, 79.1, 72.8, 65.6, 60.0, 56.7, 47.5, 46.5, 35.7, 34.8, 31.3, 29.8, 28.5, 19.6, 19.2. HRMS (ESI) m/z: [M+H]+ calcd for C23H39N4O7 + 483.2819, found 483.2815. LC-MS (ESI) m/z: [M+H]+ calcd for C23H39N4O7 + 483.3, found 483.2 (rt: 6.43).
4.2.9.6. tert-Butyl ((S)-1-((2S,4R)-4-(tert-butoxy)-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate (11)
Compound 11 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((2S,4R)-4-(tert-butoxy)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl) carbamate 27e (purification method: flash chromatography eluting CH3Cl/MeOH 93:7 v/v, yield: 50 mg, 25%). [α]20 D −36.7 (c = 1.0, MeOH). 1H NMR (700 MHz, CDCl3): δ 9.50 (s, 1H), 8.07 (dd, J = 22.8, 8.9 Hz, 1H), 6.05 (t, J = 43.5 Hz, 1H), 5.41–5.21 (m, 1H), 4.68–4.53 (m, 1H), 4.42 (dd, J = 28.9, 24.1 Hz, 1H), 4.28–4.21 (m, 1H), 3.83–3.74 (m, 1H), 3.64–3.51 (m, 1H), 3.36–3.27 (m, 2H), 2.64–2.54 (m, 1H), 2.43–2.30 (m, 2H), 2.01–1.89 (m, 3H), 1.87–1.77 (m, 3H), 1.42 (s, 9H), 1.18 (d, J = 5.3 Hz, 9H), 0.96 (d, J = 5.6 Hz, 3H), 0.88 (d, J = 6.4 Hz, 3H). 13C NMR (176 MHz, DMSO‑d 6): δ 201.1, 178.6, 172.3, 170.5, 155.6, 149.6, 77.9, 73.5, 69.6, 65.0, 58.5, 57.4, 56.0, 54.5, 46.0, 40.0, 37.0, 28.2, 28.0, 27.4, 19.0, 18.4. HRMS (ESI) m/z: [M+H]+ calcd for C26H45N4O7 + 525.3288, found 525.3282. LC-MS (ESI) m/z: [M+H]+ calcd for C26H45N4O7 + 525.3, found 525.3 (rt: 7.13).
4.2.9.7. tert-Butyl ((S)-3-methyl-1-oxo-1-((1S,3aR,6aS)-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)hexahydrocyclopenta[c]pyrrol-2(1H)-yl)butan-2-yl)carbamate (12)
Compound 12 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((1S,3aR,6aS)-1-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)hexahydrocyclopenta[c]pyrrol-2(1H)-yl)-3-methyl-1-oxobutan-2-yl)carbamate 27f (purification method: flash chromatography eluting CH2Cl2/MeOH 93:7 v/v, yield: 44 mg, 36%). [α]20 D −40.6 (c = 1.0, MeOH). 1H NMR (700 MHz, CDCl3): δ 9.50 (s, 1H), 8.15–8.06 (m, 1H), 6.08 (s, 1H), 5.27 (d, J = 9.3 Hz, 1H), 4.36 (dd, J = 10.1, 5.0 Hz, 1H), 4.32 (d, J = 3.6 Hz, 1H), 4.27 (dd, J = 9.2, 6.3 Hz, 1H), 3.86 (dd, J = 10.2, 7.8 Hz, 1H), 3.66 (dd, J = 10.3, 3.6 Hz, 1H), 3.38–3.32 (m, 2H), 2.87–2.73 (m, 2H), 2.55 (s, 1H), 2.42–2.36 (m, 1H), 2.02 (dd, J = 13.8, 7.2 Hz, 1H), 1.98–1.81 (m, 6H), 1.78–1.73 (m, 1H), 1.62 (ddd, J = 14.5, 11.5, 6.1 Hz, 2H), 1.42 (s, 9H), 0.99–0.95 (m, 3H), 0.89 (dd, J = 13.2, 6.8 Hz, 3H).13C NMR (176 MHz, CDCl3): δ 199.9, 180.3, 173.0, 171.8, 156.0, 129.9, 79.6, 66.8, 57.8, 57.0, 53.6, 47.5, 43.5, 40.7, 38.0, 32.5, 31.7, 31.1, 30.0, 28.9, 25.4, 19.3, 17.4. HRMS (ESI) m/z: [M+H]+ calcd for C25H41N4O6 + 493.3026, found 493.3011. LC-MS (ESI) m/z: [M+H]+ calcd for C25H41N4O6 + 493.3, found 493.3 (rt: 7.26).
4.2.9.8. tert-Butyl ((S)-3-methyl-1-oxo-1-((S)-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)piperidin-1-yl)butan-2-yl)carbamate (13)
Compound 13 was prepared following the general procedure Method F and using tert-butyl ((S)-1-((S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)piperidin-1-yl)-3-methyl-1-oxobutan-2-yl)carbamate 27h (purification method: flash chromatography eluting CH3Cl/MeOH 96:4 v/v, yield: 74 mg, 60%). [α]20 D −9.3 (c = 1.0, MeOH). 1H NMR (400 MHz, CDCl3): δ 9.51 (d, J = 1.1 Hz, 1H), 8.84 (s, 1H), 8.84 (s, 1H), 5.97 (s, 1H), 5.54 (d, J = 7.8 Hz, 1H), 5.31 (d, J = 17.6 Hz, 1H), 4.68–4.55 (m, 1H), 4.20 (dd, J = 9.8, 4.8 Hz, 1H), 3.90 (d, J = 14.1 Hz, 1H), 3.36 (ddd, J = 26.9, 10.7, 6.4 Hz, 3H), 2.40 (dd, J = 13.7, 5.2 Hz, 3H), 1.98–1.68 (m, 8H), 1.43 (d, J = 5.0 Hz, 9H), 0.99 (t, J = 7.1 Hz, 3H), 0.87 (t, J = 6.0 Hz, 3H). 13C NMR (176 MHz, CDCl3): δ 199.6, 180.2, 172.2, 171.5, 156.0, 79.9, 58.5, 55.7, 53.1, 44.1, 40.7, 38.6, 31.3, 29.8, 29.5, 29.0, 28.5, 25.7, 20.9, 20.0, 17.0. HRMS (ESI) m/z: [M+H]+ calcd for C23H39N4O6 + 467.2870, found 467.2864. LC-MS (ESI) m/z: [M+H]+ calcd for C23H39N4O6 + 467.3, found 467.1 (rt: 7.10).
4.2.9.9. tert-Butyl ((2S,3S)-3-(tert-butoxy)-1-oxo-1-((S)-2-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)butan-2-yl)carbamate (14)
Compound 14 was prepared following the general procedure Method F and using tert-butyl ((2S,3R)-3-(tert-butoxy)-1-((S)-2-(((S)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)carbamoyl)pyrrolidin-1-yl)-1-oxobutan-2-yl)carbamate 27i (purification method: flash chromatography eluting CH2Cl2/MeOH 98:2 to 95:5 v/v, yield: 47 mg, 25%). [α]20 D −62.8 (c = 1.0, MeOH). 1H NMR (400 MHz, DMSO‑d 6): δ 9.41 (s, 1H), 7.44 (t, J = 8.5 Hz, 1H), 6.39 (dd, J = 15.2, 8.9 Hz, 1H), 5.69 (dd, J = 19.6, 5.8 Hz, 1H), 4.32 (d, J = 7.6 Hz, 1H), 4.23–4.16 (m, 1H), 3.73 (dd, J = 12.7, 6.3 Hz, 2H), 3.12 (dd, J = 18.3, 9.0 Hz, 2H), 2.42–2.29 (m, 1H), 2.13 (dd, J = 23.9, 15.4 Hz, 2H), 1.94 (dd, J = 14.5, 8.3 Hz, 2H), 1.88–1.76 (m, 3H), 1.67–1.57 (m, 1H), 1.54–1.43 (m, 1H), 1.36 (s, 9H), 1.12 (s, 9H), 1.03 (dd, J = 5.7, 2.9 Hz, 3H). 13C NMR (176 MHz, CDCl3): δ 199.6, 179.7, 172.8, 170.2, 155.4, 79.9, 75.2, 68.9, 60.4, 57.2, 56.4, 48.5, 40.4, 37.7, 30.6, 29.3, 28.5, 28.5, 28.3, 25.1, 18.6. HRMS (ESI) m/z: [M + H]+ calcd for C25H43N4O7 + 511.3132, found 511.3126. LC-MS (ESI) m/z: [M + H]+ calcd for C25H43N4O7 + 511.3, found 511.2 (rt: 7.09).
4.3. Biological studies
4.3.1. Protein purification
The SARS-CoV-2 and MERS-CoV 3CLpro proteins for the biochemical assay were expressed using a pET vector in E. coli cells BL21 (DE3) and the expression pellets were clarified by ultracentrifugation. The proteins were purified as previously described [35]. Eluted fractions containing the 3CLpro SARS-2 were pooled and subjected to buffer exchange (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.8) by using Amicon Ultra 15 centrifugal filters at 4000×g, at 4 °C. 3CLpro MERS-CoV protein was purified using a Ni-Sepharose column and by HiTrap Q HP column. Eluted fractions containing the target protein were pooled and subjected to buffer exchange (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.8) by using Amicon Ultra 15 centrifugal filters at 4000×g, at 4 °C. Proteins purity was checked by SDS-PAGE analysis. The SARS-CoV02 3CLpro for crystallographic studies was expressed in BL21 (DE) E. coli grown in YT medium using the pGEX vector provided by the Hilgenfeld's group and described in literature [29]. Protein was purified using the protocol reported in literature [28].
4.3.2. SARS-CoV-2 and MERS-CoV 3CLpro biochemical assays
The 3CLpro SARS-CoV-2 and MERS-CoV enzymes inhibition assays was performed with the substrate DABCYL-KTSAVLQ↓SGFRKM-EDANS (Bachem) in an assay buffer containing 20 mM Tris (pH 7.3), 100 mM NaCl, 5 mM TCEP, 0.1% BSA and 1 mM EDTA [10,28]. The proteins were preincubated for 30 min at 37 °C with different concentrations of compounds before the addition of the substrate. The signal was monitored after 15 min and 30 min of incubation for 3CLpro SARS-CoV-2 and 3CLpro MERS, respectively. Following enzymatic cleavage, generation of the fluorescent product was monitored (Ex 340 nm, Em 490 nm). Compound GC376 was used as positive control.
In the Kinetics assay for the determination of the MOA and Ki of compound 12 the fluorescence signal was followed for 60 min and acquired every 60 s. The apparent Ki was determined with the single-point equation as reported [36] in triplicate and averaged. Initial enzymatic velocities in presence and absence of the inhibitor were determined by linear regression in the first 15 min of the generated curves.
4.3.3. SARS-CoV-2 replication assay
The African green monkey kidney cell line was previously engineered to constitutively express GFP (Vero E6-GFP) and were kindly provided by Janssen Pharmaceutical. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% v/v fetal beef serum (FBS; Gibco), 0.075% Sodium Bicarbonate (7.5% solution, Gibco) and 1x Pen-strep (Euroclone) and kept under 5% CO2 on 37 °C. SARS-CoV-2 strain BetaCov/Belgium/GHB-03021/2020 was provided by KU Leuven. All virus-related work was carried out in certified, high-containment biosafety level-3 facilities at the University of Cagliari. Cells were seeded at 10000 cells/well in 96-well black cell-treated plates. The following day, cells were incubated with the control compounds at different concentrations and the virus at MOI 0.01. GC376 compound was used as positive control [19], in presence of 2 μM P-gp inhibitor CP-100356 [10]. 72 h post infection the media was removed and total well GFP fluorescence was measured with a Victor 3 with 485/535 nm excitation wavelength. The inhibition of viral replication was calculated as percentage of viral induced cytopathic effect on infected untreated controls, minus blanks (empty wells). EC50 value was calculated with Prism 9. Version 9.1.2 via non-linear regression. The experiments represent average and standard deviation of at least two independent experiments in triplicate.
For quantification of viral copy number, by RT-PCR human epithelial lung adenocarcinoma cells Calu-3 cells were plated in 96 well plates (20000 cells/well). The next day, drugs were added to cells and cells were infected with SARS-CoV-2 (MOI = 0.3) for 1 h and subsequently, the virus inoculum was removed. The cells were placed into fresh media with the indicated drugs. At 48 h post-infection, viral RNA was extracted from the supernatant with the QIAamp Viral RNA Mini Kit (Qiagen) following manufacturer instructions.
One-step RT-qPCR was performed in 20 mL to detect SARS-CoV-2 S-gene copy number using the primers: forward _GTGTTTATTTTGCTTCCACTG; reverse_GGCTGAGAGACATATTCAAAA with Luna Universal One-Step RT-qPCR Kit (New England Biolabs) according to manufacturer's instructions in a CFX-96 RT-PCR (Biorad). Results report the mean and standard deviation of two independent replicates.
4.3.4. MERS replication assay
Middle East respiratory syndrome (MERS) Coronavirus, strain IP/COV/MERS/Hu/France/FRA2 (Ref-SKU: 014V-02970) was provided by EVA-GLOBAL Jessica VANHOMWEGEN laboratory, Pasteur Institute. All virus-related work was carried out in certified, high-containment biosafety level-3 facilities at the University of Cagliari. Cells were seeded at 10000 cells/well in 96-well black cell-treated plates. The following day, cells were incubated with the control compounds at different concentrations and the virus at MOI 0.005. GC376 compound (Kuzikov et al., 2021) was used as positive control, in presence of 2 mM Pgp inhibitor CP-100356 (Hu et al., 2021). 4 days post infection the media was removed and total well GFP fluorescence was measured with a Victor 3 with 485/535 nm excitation wavelength. The inhibition of viral replication was calculated as percentage of viral induced cytopathic effect on infected untreated controls, minus blanks (empty wells). EC50 value was calculated with Prism 9. Version 9.1.2 via non-linear regression. The experiments represent average and standard deviation of at least two independent experiments in triplicate.
4.3.5. HCoV229E replication assay
hCoV229E (ATCC® VR-740™) was in propagated in MRC-5 cells (ATCC:CCL-171™) maintained in MEM (Gibco) supplemented with 10% v/v fetal beef serum (FBS HI; Gibco), 1 mM Na Pyruvate (Euroclone), 1 mM Non Essential Amino Acids (Euroclone) and 1x Pen-strep (Euroclone) and kept under 5% CO2 on 37 °C. BEAS-2B cells were kindly provided by Pierre-Olivier Vidalain were maintained in DMEM/F-12 (Gibco), 5% FBS HI (Gibco), 1% Kanamycin (Thermo-Fisher Scientifics), at 37 °C with 5% CO2. 2*104 cells per well were seeded in transparent 96 well plate and incubated over night in order to reach 90% confluency. 24 h later cells were infected with hCoV-229E m. o.i. 0.06 in DMEM/F-12 in presence of compound or 0,1% DMSO (untreated controls). The cells were incubated for 2 h at 35 °C with 5% CO2, then the virus was removed, replaced with complete medium with or without compound, and incubated at 35 °C with 5% CO2. After 72 h, 20 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (Sigma-Aldrich) dissolved in PBS at 7,5 mg/mL were added to each well and incubated at 37 °C with 5% CO2 for 1 h. Then the supernatant was removed and cells were lysed with 100 μL/well of: 10% 2-Propanol, 0,004% Triton-X-100 (Sigma-Aldrich), 0,0004% HCl, then the absorbance was read at 570 nm with a plate reader Victor Nivo5 PerkinElmer.
4.3.6. Evaluation of cytotoxicity
Vero E6-GFP cells were seeded at 10,000 cells/well in 96-well black cell-treated plates, the following day, cells were incubated with the compounds and virus MOI of 0.002, 72 h post-infection the media was removed and total well GFP fluorescence was measured with a Victor 3 with 485/535 nm excitation wavelength. Cytotoxicity was calculated as a percentage of fluorescence of untreated controls, minus blanks (empty wells). CC50 value was calculated with Prism 9. Version 9.1.2 (225) via non-linear regression. The experiments represent average and standard deviation of at least two independent experiments in triplicate.
4.3.7. Differential scanning fluorimetry assay
The differential scanning fluorimetry (DSF) assay was performed in a buffer containing 20 mM HEPES pH 7.8 and 120 mM NaCl and 2 μM of 3CLpro protein was pre-incubated with 40 μM of compounds for of 30 min. 5 × SYPRO Orange dye was added to probe the thermal denaturation from 20 °C to 95 °C at a scan rate of 1.5 °C/min. The melt temperature (Tm) and the thermal shift (ΔTm) was calculated as described [22]. All experiments were performed in triplicate, and the values are presented as mean ± SD.
4.4. Protein crystallization and structure determination
4.4.1. Crystallization
Crystallization of SARS-CoV-2 3CLpro in complex with compounds was carried out as previously described [28]. Briefly, 3CLpro at a concentration of 5 mg/mL in 20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.8, 1 mM DTT was incubated for 1 h at RT with compounds at a final concentration of 5 mM. Crystallization experiments were then set up by seeding in sitting drops using the Morpheus® kit (Molecular Dimensions) with a Mosquito robot (STPlabtech Ltd., Melbourn Hertfordshire, UK). Crystals were flash frozen in liquid nitrogen after a few days of growth.
For compounds 7 and 8 the best diffracting crystals grew in condition G8: 0.1 M sodium formate, 0.1 M ammonium acetate, 0.1 M sodium citrate tribasic dihydrate, 0.1 M potassium sodium tartrate tetrahydrate, 0.1 M sodium oxamate, 0.1 M Hepes/Mops, pH 7.5, 12.5% v/v MPD; 12.5% PEG 1000, 12.5% w/v PEG 3350.
For compound 12 the best diffracting crystals grew in condition B8: 0.09 M sodium fluoride, 0.09 M sodium bromide, 0.09 M sodium iodide, 0.1 M Hepes/MOPS, pH 7.5, 0.1 M Hepes/Mops pH 7.5, 12.5% v/v MPD; 12.5% PEG 1000, 12.5% w/v PEG 3350.
For compound 14 the best diffracting crystals grew in condition D12: 0.12 M 1,6-Hexanediol, 0.12 M 1-butanol, 0.12 M 1,2-propanediol, 0.12 M 2-propanol, 0.12 M 1,4-butanediol, 0.12 M 1,3-propanediol, 0.1 M Tris/BICINE, pH 8.5, 12.5% v/v MPD; 12.5% PEG 1000, 12.5% w/v PEG 3350.
4.4.2. Data collection, data reduction, structure determination, refinement and final model analysis
Diffraction data were collected at the XRD2 beamline of the Elettra synchrotron (Trieste, Italy) [37] using a 1.000 Å wavelength, at 100 K. The collected datasets were processed with XDS [38] and Aimless [39] from the CCP4 suite [40].
Structures were solved by molecular replacement with Phaser [41] using the PDB structure 7BB2 [28] as search model. The initial models were refined alternating cycles of manual model building in COOT [30] and automatic refinement using Phenix (version 1.19.2–4085) [31]. Data collection and refinement statistics are reported in Table S1.
Figures were prepared using PyMOL [The PyMOL Molecular Graphics System, Version 2.1 Schrödinger, LLC., New York, NY, USA].
4.4.3. Data availability
Coordinates and structure factors were deposited into the Protein Data Bank with accession numbers 8OKK (3CLpro in complex with 7), 8OKL (3CLpro in complex with 8), 8OKM (3CLpro in complex with compound 12), 8OKN (3CLpro in complex with 14).
Funding sources
This research was supported by the project “EXaSCale smArt pLatform Against paThogEns for Corona Virus– Exscalate4CoV” founded by the EU's H2020-SC1-PHE-CORONAVIRUS-2020 call, grant N. 101003551, and by EU funding within the MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project N. PE00000007, INF-ACT). The publication was supported by the project “European Virus Archive GLOBAL (EVA-GLOBAL)”, founded by EU's H2020 research, grant. N. 871029.
CRediT authorship contribution statement
Irina Stefanelli: Methodology, and, Formal analysis, Writing – review & editing. Angela Corona: Formal analysis, Investigation, Data curation. Carmen Cerchia: computational studies, Investigation, Data curation, writing. Emilia Cassese: Formal analysis. Salvatore Improta: Formal analysis. Elisa Costanzi: X-Ray studies, Formal analysis, Investigation. Sveva Pelliccia: Formal analysis, Investigation. Stefano Morasso: protein production in X-Ray studies, Data curation. Francesca Esposito: Formal analysis, Investigation, Data curation. Annalaura Paulis: Formal analysis, Investigation, Data curation. Sante Scognamiglio: Formal analysis, Investigation, Data curation. Francesco Saverio Di Leva: computational studies, Investigation, Data curation, writing. Paola Storici: Formal analysis, Validation, Data curation, Supervision, Writing – review & editing. Margherita Brindisi: Writing – review & editing. Enzo Tramontano: Resources, Supervision. Rolando Cannalire: Conceptualization, Investigation, Supervision, Writing – original draft, Writing – review & editing. Vincenzo Summa: Conceptualization, Funding acquisition, Project administration, Supervision, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgements
Authors thank: Nicola Demitri (XRD2 beamline, Elettra - Sincrotrone Trieste S.C.p.A., SS 14 - km 163, 5 in AREA Science Park, Trieste, Basovizza, 34149, Italy) for helping in data collection and x-ray structural studies; Janssen Pharmaceutical for providing the African green monkey kidney cell line engineered to constitutively express GFP (Vero E6-GFP); Pieter Leyssen laboratory of KU Leuven for providing the SARS-CoV-2 strain BetaCov/Belgium/GHB-03021/2020 and Pierre-Olivier Vidalain of the Centre International de Recherche en Infectiologie (Université Lyon, Inserm, U1111, Université Claude Bernard Lyon 1, CNRS, Lyon, France) for providing BEAS-2B cell line. C.C. thanks MUR (Ministero dell’Universit a e della Ricerca), PON R&I 2014-2020-Asse IV “Istruzione e Ricerca per il recupero-REACT-EU” Azione IV.4- Contratti di Ricerca su tematiche dell'innovazione. R.C. thanks MIUR-Ministero dell’Istruzione, dell’Universita e della Ricerca (Italian Ministry of Education, University and Research), PON R&I 2014-2020- AIM (Attraction and International Mobility), project AIM1873131 Num. Attivita 2 Linea 2.1. S.P. acknowledges MUR (Ministero dell’Universit'a e della Ricerca), PON R&I 2014-2020- Asse IV “Istruzione e Ricerca per il recupero-REACT-EU”, Azione IV.6 ″Contratti di Ricerca su tematiche Green”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2023.115311.
Abbreviations
- 3CLpro
3-chymotrypsin-like protease
- COVID-19
coronavirus disease 2019
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- CoV
coronavirus
- SARS-CoV
severe acute respiratory syndrome coronavirus
- MERS-CoV
middle east respiratory syndrome coronavirus
- DAAs
direct-acting antivirals
- HCV
hepatitis C virus
- TCEP
tris(2-carboxyethyl)phosphine;
- BSA
bovine serum albumin
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- CPE
cytopathic effect
- P-gp
P-glycoprotein
- MTT
dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-NCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzo G. SARS-CoV-2 and other human coronavirus show genome patterns previously associated to reduced viral recognition and altered immune response. Sci. Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-90278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poutanen S.M. Elsevier Inc.; 2005. Human Coronaviruses. THIRD EDIT. [DOI] [Google Scholar]
- 5.Brier, J.; lia dwi jayanti. https://coronavirus.jhu.edu/map.html. https://coronavirus.jhu.edu/map.html, (n.d.)..
- 6.Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin. Microbiol. Infect. 2022;28(2):202–221. doi: 10.1016/j.cmi.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannalire R., Tramontano E., V. S. SARS-CoVs) 1 and 2; 2022. A Focus on Severe Acute Respiratory Syndrome (SARS) Coronavirus; pp. 449–485. [Google Scholar]
- 8.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zarenezhad E., Marzi M. Review on molnupiravir as a promising oral drug for the treatment of COVID-19. Med. Chem. Res. 2022;31(2):232–243. doi: 10.1007/s00044-021-02841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R. An oral SARS-CoV-2 mproinhibitor clinical candidatefor the treatment of COVID-19. Science. 2021;3(December):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 11.www.pardesbio.com/pipeline/#our-lead-program..
- 12.Unoh Y., Uehara S., Nakahara K., Nobori H., Yamatsu Y., Yamamoto S., Maruyama Y., Taoda Y., Kasamatsu K., Suto T., Kouki K., Nakahashi A., Kawashima S., Sanaki T., Toba S., Uemura K., Mizutare T., Ando S., Sasaki M., Orba Y., Sawa H., Sato A., Sato T., Kato T. Discovery of S - 217622, a noncovalent oral SARS-CoV - 2 3CL protease inhibitor clinical candidate for treating COVID-19. J. Med. Chem. 2022;65:6499–6512. doi: 10.1021/acs.jmedchem.2c00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adnan M., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection : emergence , transmission , and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannalire R., Cerchia C., Beccari A.R., Saverio F., Leva D. Targeting SARS-CoV - 2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem. 2022;65:2716–2746. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J.T., Yang Q., Gribenko A., Perrin B.S., Zhu Y., Cardin R., Liberator P.A., Anderson A.S., Hao L. Genetic surveillance of SARS-CoV-2 Mpro reveals high sequence and structural conservation prior to the introduction of protease inhibitor Paxlovid. mBio. 2022;13(4):1–15. doi: 10.1128/mbio.00869-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 17.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z., Irol J.V. Structures of two coronavirus main proteases : implications for substrate binding and antiviral drug design. J. Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10) doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir. GC-376 , and calpain inhibitors II , XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020 doi: 10.1038/s41422-020-0356-z. No. June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelliccia S., Cerchia C., Esposito F., Cannalire R., Corona A., Costanzi E., Kuzikov M., Gribbon P., Zaliani A., Brindisi M., Storici P., Tramontano E., Summa V. Easy access to α -ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route. Eur. J. Med. Chem. 2022;244(October) doi: 10.1016/j.ejmech.2022.114853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:1–8. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao J., Li Y., Zeng R., Liu F., Luo R., Huang C., Wang Y., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z., Zou Q., Zhang H., Qu W., Long Y., Li M., Yang R., Liu X., You J., Zhou Y., Yao R., Li W., Liu J., Chen P., Liu Y., Lin G., Yang X., Zou J., Li L., Hu Y., Lu G., Li W., Wei Y., Zheng Y., Lei J., Yang S. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;1378(March):1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leuw P. de, Stephan C. Protease inhibitors for the treatment of hepatitis C virus infection 2013 improved SVR rates, the amount of severe side effects was high. GMS Infect. Dis. 2017;5:1–14. doi: 10.3205/id000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prior A.M., Kim Y., Weerasekara S., Moroze M., Alliston K.R., Uy R.A.Z., Groutas W.C., Chang K.O., Hua D.H. Design, synthesis, and bioevaluation of viral 3C and 3C-like protease inhibitors. Bioorg. Med. Chem. Lett. 2013;23(23):6317–6320. doi: 10.1016/j.bmcl.2013.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartoli G., Bosco M., Locatelli M., Marcantoni E., Melchiorre P., Sambri L. Unusual and unexpected reactivity of T-butyl dicarbonate (Boc2O) with alcohols in the presence of magnesium perchlorate. A new and general route to t-butyl ethers. Org. Lett. 2005;7(3):427–430. doi: 10.1021/ol047704o. [DOI] [PubMed] [Google Scholar]
- 26.Agbowuro A.A., Mazraani R., Mccaughey L.C., Huston W.M., Gamble A.B., Tyndall J.D.A. Stereochemical basis for the anti-chlamydial activity of the phosphonate protease inhibitor JO146. Tetrahedron. 2018;74(12):1184–1190. doi: 10.1016/j.tet.2017.10.031. [DOI] [Google Scholar]
- 27.Ma Y., Yang K.S., Geng Z.Z., Alugubelli Y.R., Shaabani N., Vatansever E.C., Ma X.R., Cho C.C., Khatua K., Xiao J., Blankenship L.R., Yu G., Sankaran B., Li P., Allen R., Ji H., Xu S., Liu W.R. A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals. Eur. J. Med. Chem. 2022;240(June) doi: 10.1016/j.ejmech.2022.114570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costanzi E., Kuzikov M., Esposito F., Albani S., Demitri N., Giabbai B., Camasta M., Tramontano E., Rossetti G., Zaliani A., Storici P. Structural and biochemical analysis of the dual inhibition of Mg-132 against sars-cov-2 main protease (Mpro/3clpro) and human cathepsin-L. Int. J. Mol. Sci. 2021;22(21) doi: 10.3390/ijms222111779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P., Cowtan K. Coot: model-building tools for molecular Graphics. Biol. Crystallogr. 2004;60(12 I):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J., Richardson J.S., Richardson D.C., Sammito M.D., Sobolev O.V., Stockwell D.H., Terwilliger T.C., Urzhumtsev A.G., Videau L.L., Williams C.J., Adams P.D. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomar S., Johnston M.L., John S.E.S., Osswald H.L., Nyalapatla P.R., Paul L.N., Ghosh A.K., Denison M.R., Mesecar A.D. Ligand-induced dimerization of Middle East respiratory syndrome (MERS) coronavirus Nsp5 protease (3CLpro): implications for Nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015;290(32):19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandadapu S.R., Weerawarna P.M., Gunnam M.R., Alliston K.R., Lushington G.H., Kim Y., Chang K.O., Groutas W.C. Potent inhibition of norovirus 3CL protease by peptidyl α-Ketoamides and α-Ketoheterocycles. Bioorg. Med. Chem. Lett. 2012;22(14):4820–4826. doi: 10.1016/j.bmcl.2012.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cregge R.J., Curran T.T., Metz W.A. A convenient synthesis of peptidyl perfluoroalkyl Ketones. J. Fluor. Chem. 1998;88(1):71–77. doi: 10.1016/S0022-1139(97)00154-1. [DOI] [Google Scholar]
- 35.Kuzikov M., Costanzi E., Reinshagen J., Esposito F., Vangeel L., Wolf M., Ellinger B., Claussen C., Geisslinger G., Corona A., Iaconis D., Talarico C., Manelfi C., Cannalire R., Rossetti G., Gossen J., Albani S., Musiani F., Herzog K., Ye Y., Giabbai B., Demitri N., Jochmans D., Jonghe S. De, Rymenants J., Summa V., Tramontano E., Beccari A.R., Leyssen P., Storici P., Neyts J., Gribbon P., Zaliani A. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol. Transl. Sci. 2021;4(3):1096–1110. doi: 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzmič P., Sideris S., Cregar L.M., Elrod K.C., Rice K.D., Janc J.W. High-throughput screening of enzyme inhibitors: automatic determination of tight-binding inhibition constants. Anal. Biochem. 2000;281(1):62–67. doi: 10.1006/abio.2000.4501. [DOI] [PubMed] [Google Scholar]
- 37.Lausi A., Polentarutti M., Onesti S., Plaisier J.R., Busetto E., Bais G., Barba L., Cassetta A., Campi G., Lamba D., Pifferi A., Mande S.C., Sarma D.D., Sharma S.M., Paolucci G. Status of the crystallography beamlines at Elettra. Eur. Phys. J. Plus. 2015;130(3) doi: 10.1140/epjp/i2015-15043-3. [DOI] [Google Scholar]
- 38.Glusker J.P.X.D.S. Biol. Crystallogr. 1993;49(1) doi: 10.1107/s090744499201028x. 1–1. [DOI] [Google Scholar]
- 39.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69(7):1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., Read R.J., Vagin A., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67(4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40(4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors were deposited into the Protein Data Bank with accession numbers 8OKK (3CLpro in complex with 7), 8OKL (3CLpro in complex with 8), 8OKM (3CLpro in complex with compound 12), 8OKN (3CLpro in complex with 14).
Data will be made available on request.