The recent mpox (monkeypox) outbreak has caused >86,000 infections and 96 deaths (https://ourworldindata.org/monkeypox) and was declared a global health emergency by the WHO (1, 2). The disease is caused by mpox virus (MPXV), a virus endemic in an animal reservoir in the sub-Saharan Africa (Fig. 1). MPXV transmits occasionally to humans, and it spread in a few occasions outside the African continent, causing limited outbreaks. The recent 2022 international outbreak has caused an unprecedented transmission of the virus mainly in men that have sex with men. The extent of the outbreak has decreased dramatically after a few months, with 13 to 37 daily cases worldwide in February 2023. We still do not understand the reasons why the MPXV clade circulating in 2022 was so transmissible in humans. In PNAS, Americo et al. (3) report that the MPXV clade causing the 2022 international outbreak is less virulent in a mouse model than the MPXV clades previously identified and suggest that the virus may be adapting to other species.
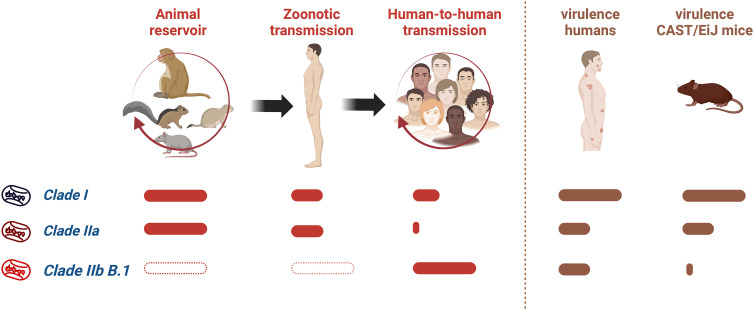
Fig. 1.
Transmission properties of different clades of MPXV and their virulence in humans and mice. The finding of MPXV clades in an animal reservoir, their ability to spread to humans from animals, and their efficiency of human-to-human transmission are indicated. The virulence of MPXV clades in humans and CAST/EiJ mice, as described by Americo et al. (3), is shown. Solid bars show a relative quantification of the properties. Empty bars indicate that no information is available.
MPXV is an orthopoxvirus closely related to variola virus (VARV), a human virus that caused smallpox and was eradicated in 1980 (4). The possibility that an orthopoxvirus such as MPXV, causing a less severe smallpox-like disease, could be introduced into the human population and fill the niche occupied by VARV has been a concern.
MPXV is a zoonotic virus present in a natural animal reservoir in rodents in Africa, including squirrels, Gambian rats, and dormice, and can be transmitted to monkeys and humans. Two clades of MPXV have been identified (Fig. 1). Clade I (Central Africa or Congo Basin) causes high mortality (10%) and long chains of human-to-human transmission. By contrast, clade IIa (West Africa) shows low mortality (1%) and is less transmissible between humans. The MPXV isolate-causing infections outside Africa are related to clade IIa and were designated as clade IIb. The MPXV genome consists of a double-stranded DNA molecule of ~200,000 bp encoding 190 proteins. Differences between clades I and IIa map to the genes involved in virus–host interaction located at the ends of the genome. Changes in clade IIa include the fragmentation or deletion of genes involved in immune evasion, such as the interleukin-1β receptor, an apoptosis regulator, and a complement inhibitor.
The genomic sequences of >1,200 MPXV isolates circulating during the 2022 outbreak (www.nextstrain.org/monkeypox) are related to clade IIb and were defined as lineage B.1 (5). Lineage B.1 has accumulated 46 nucleotide changes compared to a clade IIb MPXV isolated in the United Kingdom in 2018. The number of mutations accumulated in B.1 was 6 to 12 higher than that estimated for a highly stable orthopoxvirus DNA genome. Many mutations were consistent with the action of host APOBEC3 cytosine deaminase activity, which may have contributed to an accelerated evolution after human-to-human transmission during the 2022 outbreak. Genomic sequencing of MPXV isolates from Nigeria in 2019 and 2020 identified variants circulating in humans before the emergence of lineage B.1 (6) and including the lineage A.2 introduced independently into the United States in 2022 (7).
The contributions of the clade IIb B.1 mutations to MPXV pathogenicity are unknown, and an animal model is of interest. Nonhuman primates, ground squirrels, African dormice, and prairie dogs have been experimentally infected with MPXV, but these models have limitations due to complex husbandry requirements and availability of immunoreagents (8). A survey of 38 inbred mouse strains identified three susceptible wild-derived strains, and the CAST/EiJ mice were developed as an mpox model (9). Deficient IFNγ production is associated with the susceptibility of CAST/EiJ mice to MPXV (10).
“In PNAS, Americo et al. report that the MPXV clade causing the 2022 international outbreak is less virulent in a mouse model than the MPXV clades previously identified and suggest that the virus may be adapting to other species.”
Americo et al. publish in PNAS investigations on the virulence of the new MPXV clade IIb B.1 in the CAST/EiJ mouse model they developed (3). The CAST/EiJ mice recapitulate the pathogenicity of MPXV clades in humans, with mice being more susceptible to clade I than clade IIa by both intranasal and intraperitoneal routes of infection (9) (Fig. 1). This difference is more pronounced in the systemic, peritoneal infection with an LD50 <1 plaque-forming unit (PFU) for clade I and an LD50 1,000 PFU for clade IIa. Interestingly, the new clade IIb B.1 is highly attenuated in CAST/EiJ mice administered by either route of infection, with no lethality observed at high viral doses (105 PFU) and low virus replication in different organs. The authors propose that increased adaptation of this MPXV lineage to humans, as a result of the recent human-to-human transmission events, may have led to a major change in host specificity and reduced ability to infect rodents. The CAST/EiJ mouse model may help to narrow down the mutations responsible for this dramatic change in host specificity by constructing clade IIa viruses bearing specific mutations identified in clade IIb B.1. This will point at the immune response pathways involved in this resistance.
Identifying which of the 46 mutations found in clade IIb B.1, including 24 nonsynonymous, 18 synonymous, and four intergenic, may favor MPXV adaptation to humans remains a research challenge. The nonsynonymous changes affect immunomodulatory proteins (TNF decoy receptor, CC-chemokine binding protein), proteins of the entry–fusion complex, transcription factors, proteins involved in virus replication and morphogenesis, and the major antigenic B21 surface protein. The possibility that a few amino acid mutations may lead to relevant changes in the function of these proteins will need to be addressed experimentally. Mutations in intergenic regions may affect the expression level of some genes. Lastly, genomic rearrangements that consist of translocations of DNA between terminal sequences in the genome may cause the duplication, inactivation, and deletion of genes. One example of this gene rearrangement has been identified in MPXV isolates from the 2022 outbreak, and previously in clade I virus isolates in Africa (11, 12).
Gene loss events are common in orthopoxvirus evolution during adaptation to new host species (13), but the impact on virulence is unpredictable. The loss of genes that target specific immune pathways or promote virus replication normally causes viral attenuation. But, gene loss may increase virulence, as demonstrated for the vaccinia virus interleukin-1β receptor that inhibits fever and systemic responses in the infected host (14). This is counterintuitive, but illustrates that viral proteins may reduce the immunopathology caused by an excessive immune response induced after infection.
The establishment of MPXV in the human population as a new disease replacing smallpox is uncertain. The evolution of smallpox, a human-specific virus, may shed light into potential scenarios of mpox evolution in humans. VARV lost 29 genes involved in immune evasion and host range as compared to an orthopoxvirus ancestor, likely similar to cowpox virus, which causes limited pathology and infects a broad range of species. Recent studies identified VARV in archaeological human remains from the Viking Age (600 to 1050 CE) that still retained 14 immune modulatory and host range genes that were lost in modern VARV (15). One possibility is that smallpox was initially a zoonotic disease causing a mild infection in humans and VARV progressively lost genes and increased its virulence as it became more restricted to humans (16). Maybe the mpox infections we are seeing represent the beginning of a similar evolutionary pattern and MPXV will mutate or suffer genomic rearrangements, leading to a better adaption to humans. The 2022 international outbreak is providing MPXV a good opportunity to accelerate its molecular evolution generating more transmissible and better adapted new clades. Whether they will evolve toward increased virulence, as we saw with smallpox, or attenuation is at present unpredictable.
Transmission of MPXV occurs through direct contact with skin lesions, body fluids, respiratory droplets, and fomites (2). Epidemiological data of the current outbreak suggest that contact with skin lesions is the dominant route of transmission (17). Recent studies on mpox patients have identified high virus loads in the respiratory tract (throat and nasopharyngeal swabs), saliva, and air samples (17, 18), but transmission through respiratory droplets or at longer distances through aerosols does not appear to play a role in the current outbreak. The possibility that MPXV may mutate and increase viral loads in droplets and aerosols, or its efficiency to infect human cells in the respiratory mucosa, will enhance its respiratory transmission, and this should be monitored in the future. Evidence of airborne transmission of smallpox was reported (19), and mpox may evolve in the same direction.
The recent mpox outbreak has been largely reduced, but a basal level of virus transmission is likely to continue, including asymptomatic infections. The increase in recent years of mpox cases in Africa, where new MPXV clades appear to be evolving (6), may reflect waning of protective immunity against poxviruses after the smallpox vaccination ceased. A major concern is the possibility that clade I, more virulent and transmissible than clade II, may become established in humans and cause new epidemics. It is likely that new mpox outbreaks will occur in the near future.
Genomic surveillance, particularly in African countries, is required to monitor the evolution of MPXV and its human adaptation. Further investigations on the role of MPXV proteins involved in immune evasion and host interaction are needed to identify factors that contribute to transmission and pathogenesis. The development of animal models, like the CAST/EiJ model described by Americo et al. in PNAS (3), is necessary to allow these investigations. Lastly, as we have learned during the COVID-19 pandemic, rapid diagnostic tests, antivirals, and vaccines are urgently needed to control future mpox outbreaks.
Acknowledgments
The author’s research is supported by the Spanish Ministry of Science and Innovation and European Union (European Regional Development’s Funds) (grant PID2021-128580OB-I00), Nextgeneration European Union (EU), and Consejo Superior de Investigaciones Científicas (Plataforma Temática Interdisciplinar Salud Global).
Author contributions
A.A. wrote the paper.
Competing interests
The author declares no competing interest.
Footnotes
See companion article, “Virulence differences of mpox (monkeypox) virus clades I, IIa, and IIb.1 in a small animal model,” 10.1073/pnas.2220415120.
References
- 1.Gessain A., Nakoune E., Yazdanpanah Y., Monkeypox. N. Engl. J. Med. 387, 1783–1793 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Reynolds M. G., Damon I. K., Outbreaks of human monkeypox after cessation of smallpox vaccination. Trends Microbiol. 20, 80–87 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Americo J. L., Earl P. L., Moss B., Virulence differences of mpox (monkeypox) virus clades I, IIa and IIb.1 in a small animal model. Proc. Natl. Acad. Sci. U.S.A. 120, e2220415120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith G. L., McFadden G., Smallpox: Anything to declare? Nat. Rev. Immunol. 2, 521–527 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Isidro J., et al. , Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 28, 1569–1572 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ndodo A., et al. , Different coexisting mpox lineages were continuously circulating in humans prior to 2022. bioRxiv [Preprint] (2023). 10.1101/2023.01.03.522633 (Accessed 15 February 2023). [DOI]
- 7.Gigante C. M., et al. , Multiple lineages of monkeypox virus detected in the United States, 2021–2022. Science 378, 560–565 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker S., Buller R. M., A review of experimental and natural infections of animals with monkeypox virus between 1958 and 2012. Future Virol. 8, 129–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Americo J. L., Moss B., Earl P. L., Identification of wild-derived inbred mouse strains highly susceptible to monkeypox virus infection for use as small animal models. J. Virol. 84, 8172–8180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earl P. L., Americo J. L., Moss B., Lethal monkeypox virus infection of CAST/EiJ mice is associated with a deficient gamma interferon response. J. Virol. 86, 9105–9112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones T., et al. , Genetic variability, including gene duplication and deletion, in early sequences from the 2022 European monkeypox outbreak. bioRxiv [Preprint] (2022), 10.1101/2022.07.23.501239 (Accessed 15 February 2023). [DOI]
- 12.Kugelman J. R., et al. , Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo. Emerg Infect Dis. 20, 232–239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson R. C., Wang C., Hatcher E. L., Lefkowitz E. J., Orthopoxvirus genome evolution: The role of gene loss. Viruses 2, 1933–1967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alcamí A., Smith G. L., A mechanism for the inhibition of fever by a virus. Proc. Natl. Acad. Sci. U.S.A. 93, 11029–11034 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mühlemann B., et al. , Diverse variola virus (smallpox) strains were widespread in northern Europe in the Viking Age. Science 369, eaaw8977 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Alcamí A., Was smallpox a widespread mild disease? Science 369, 376–377 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Mitjà O., et al. , Monkeypox. Lancet 401, 60–74 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernaez B., et al. , Monitoring monkeypox virus in saliva and air samples in Spain: A cross-sectional study. Lancet Microbe. 4, e21–e28 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milton D. K., What was the primary mode of smallpox transmission? Implications for biodefense. Front Cell Infect. Microbiol. 2, 150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]