Abstract
Antigen tests to detect SARS-CoV-2 have emerged as a promising rapid diagnostic method for COVID-19, but they are unable to differentiate between variants of concern (VOCs). Here, we report a rapid point-of-care test (POC-T), termed CoVariant-SPOT, that uses a set of antibodies that are either tolerant or intolerant to spike protein mutations to identify the likely SARS-CoV-2 strain concurrent with COVID-19 diagnosis using antibodies targeting nucleocapsid protein. All reagents are incorporated into a portable, multiplexed, and sensitive diagnostic platform built upon a non-fouling polymer brush. To validate CoVariant-SPOT, we tested recombinant SARS-CoV-2 proteins, inactivated viruses, and nasopharyngeal swab samples from COVID-19 positive and negative individuals and showed that CoVariant-SPOT can readily distinguish between two VOCs: Delta and Omicron. We believe CoVariant-SPOT can serve as a valuable adjunct to next-generation sequencing to rapidly identify variants using a scalable and deployable POC-T, thereby enhancing community surveillance efforts worldwide and informing treatment selection.
Keywords: Point-of-care, multiplexed immunoassay, COVID-19, SARS-CoV-2 variants
Graphical Abstract
INTRODUCTION
Diagnostic tests that detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—the causative pathogen of coronavirus disease 2019 (COVID-19)—that are fast, sensitive, and user-friendly are urgently needed to combat the ongoing pandemic. This need is further exacerbated by the emergence of SARS-CoV-2 variants of concern (VOCs) that are more virulent, transmissible, and capable of partially evading both natural and vaccine-induced humoral immunity.1-3 New variants are currently identified using next generation sequencing (NGS) after a positive diagnosis. NGS uses an unbiased method to identify nucleic acid sequences without prior knowledge of the mutations, and thus is a vital tool for SARS-CoV-2 surveillance.4 It is through these surveillance systems that we have identified several VOCs, such as Alpha (B.1.1.7),5 Beta (B.1.351),6 Gamma (P.1),7 Delta (B.1.617.2),8 and most recently Omicron (B.1.529).9
During the COVID-19 pandemic, researchers have deposited sequences into databases such as the EpiCov run by the Global Initiative on Sharing Avian Influenza Data (GISAID).10 However, only a small fraction of the total cases have been sequenced and deposited into GISAID repositories, thus limiting the ability to effectively track these variants.10 In addition, NGS surveillance is often not available in low- and middle-income countries. For example, as of July 20, 2021, >2 million SARS-CoV-2 genomes have been submitted to GISAID, of which 94% from high income countries and only 6% are from low- and middle-income countries.11 Beyond the need for robust genomic epidemiological surveillance, determining the variant causing COVID-19 is important to inform clinical decision making, as some variants can evade commonly used monoclonal antibody therapies.12, 13 Therefore, there is an urgent need for a companion test that can detect circulating strains previously identified via NGS in a rapid and easy-to-use format, ideally concurrent with initial diagnosis.
To address this need, we report herein a rapid point-of-care test (POC-T), termed CoVariant-SPOT (Covid-19 Variant Spike Protein Observation Test) which uses a panel of monoclonal antibodies that are incorporated into a portable, multiplexed, and sensitive diagnostic platform (the D4) that is built upon a non-fouling polymer brush.14-17 We sought to exploit the ability of antibodies that are either tolerant or intolerant to spike (S) protein mutations to identify the likely SARS-CoV-2 strain concurrent with COVID-19 diagnosis using antibodies targeting nucleocapsid (N) protein. As proof-of-principle, we demonstrate the performance of CoVariant-SPOT for diagnosis of acute COVID-19 infection and differentiation between two VOCs: Delta and Omicron. To validate CoVariant-SPOT, we tested recombinant SARS-CoV-2 proteins, inactivated viruses, and nasopharyngeal swab samples from COVID-19 positive and negative individuals and found that we could readily distinguish between SARS-CoV-2 VOCs by examining the ratio of fluorescence intensity from multiple anti-S antibodies. Given its ease of use and rapid turnaround time, we believe CoVariant-SPOT can serve as a valuable adjunct to NGS to rapidly identify mutant variants using a scalable and readily deployable POC-T, thereby enhancing community-based SARS-CoV-2 surveillance efforts worldwide and informing treatment.
MATERIALS AND METHODS
CoVariant-SPOT testing procedure
CoVariant-SPOT employs the technology of the D4 assay, described previously.14 Additional experimental details regarding fabrication are described in the supporting information. CoVariant-SPOT chips were secured in a 96-well microarray hybridization cassette that separates the chip into 24 separate wells. To perform the assay, 60 μL of sample was added directly to an assay well, covered, and incubated at room temperature for 1 h. After incubation, samples were aspirated and chips were rinsed in wash buffer (0.1% Tween-20 in 1x PBS), dried, and then scanned with an Axon Genepix 4400 tabletop scanner (Molecular Devices LLC). The average fluorescence intensity at each capture spot was quantified using the Genepix Pro 7 analysis software. All fluorescence intensities were log transformed prior to analysis.
Analytical validation with recombinant SARS-CoV-2 antigens was performed by testing a 15-point dose-response curve in triplicate with SARS-CoV-2 N and S1 protein antigens (N: Acro Biosystems, catalog numbers: NUN-C5227, NUN-C52Hr, NUN-C52Ht; S1: Sino Biological, catalog numbers: 40591-V08H, 40591-V08H23, 40591-V08H41). N and S proteins of the same SARS-CoV-2 variant were mixed and diluted with extraction buffer (Acro Biosystems, catalog number: LY14) to a starting concentration of 300 ng/mL for each antigen. Antigens for S trimer and BA.2 S1 were purchased from Sino Biological. The LOD was calculated as described elsewhere.18 Log transformed values were used for calculating the cAb ratios shown in Figure 2C.
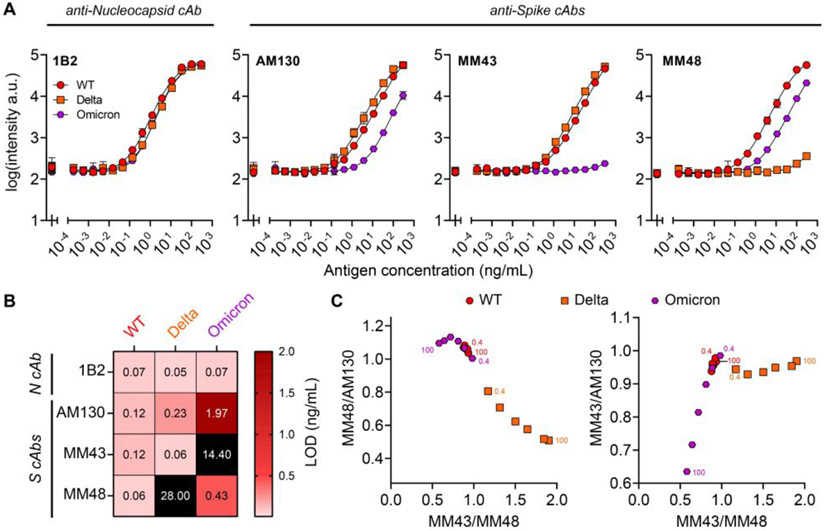
Figure 2. Analytical validation of CoVariant-SPOT using recombinant antigens.
(A) Dose-response curves partitioned by cAb for each SARS-CoV-2 strain spiked into lysis buffer. Each data point represents the average of three replicates with standard deviation (SD) shown. Furthest left point is a blank. (B) LOD summary for data presented in Figure 2A. (C) Anti-S cAb ratios to differentiate variants. Numbers in the graph represent the concentration of S1 in ng/mL. The accuracy of differentiation is improved at higher S1 concentrations.
Analytical validation with UV inactivated SARS-CoV-2 isolates was performed by testing a 15-point dose-response curve in triplicate. Three different isolates were procured, representing each variant tested: WT isolate USA-WA1/2020 (ZeptoMetrix, catalog number: 0810587UV), Delta (B.1.617.2) isolate USA/PHC658/2021 (ZeptoMetrix, catalog number: 0810624UV), and Omicron (B.1.1.529 BA.1) isolate USA/MD-HP20874/2021 (ZeptoMetrix, catalog number: 0810642UV). Isolates were diluted in extraction buffer to a starting TCID50/mL of 1 x 106 for WT and Delta, and 2 x 105 for Omicron. All fluorescence intensities were log transformed and then normalized by dividing each value by the cAb intensity of a blank sample.
Clinical testing
Clinical samples were either purchased commercially (Discovery Life Sciences) or from patients identified through the Duke University Health System or the Durham Veterans Affairs Health System and enrolled into the Molecular and Epidemiological Study of Suspected Infection (MESSI; Pro00100241) approved by the Duke Health Institutional Review Board (IRB). Samples were accessed via an exempt protocol (Pro00105331, PI Ashutosh Chilkoti). For the samples collected under the MESSI protocol, flocked nasopharyngeal swabs were added to 3 mL of VTM (Dasky Medical, catalog #: 88-221KC; VWR (BD), catalog #: 10769-896) and frozen at −80 °C until testing by CoVariant-SPOT. In addition, viral load was determined using the methods described in the supporting information. All samples are summarized in Table S1.
Point-of-care implementation of CoVariant-SPOT
Analytical validation of the microfluidic CoVariant-SPOT was performed by testing a 6-point dose-response curve in triplicate with SARS-CoV-2 N and S1 proteins and an additional blank (n=4). As before, N and S1 proteins of the same SARS-CoV-2 variant were mixed and diluted with extraction buffer to a starting concentration of 300 ng/mL for each antigen, from which a series of 1/4th dilutions was obtained. To run the assay, 72 μL of sample was added to the sample inlet. Immediately after, 200 μL of wash buffer was added to the wash buffer inlet, and the device was left to incubate in the vertical position. Once the reaction chamber had completely drained of fluid, the cassette was inserted into the D4Scope, manually aligned using the fiducial spots, imaged, and analyzed. Microfluidic cassettes imaged on the D4Scope were analyzed in the same way as described in “CoVariant-SPOT testing procedure”. The D4Scope has been described in greater detail in previous publications.15, 16 The only modification made to the D4Scope for this study was the introduction of a higher power excitation laser with a higher quality collimator lens. Additional details are outlined in the supporting information.
RESULTS AND DISCUSSION
Fabrication of CoVariant-SPOT
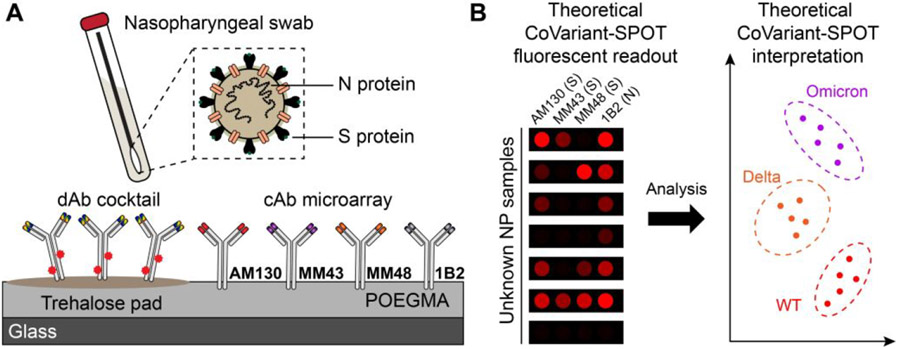
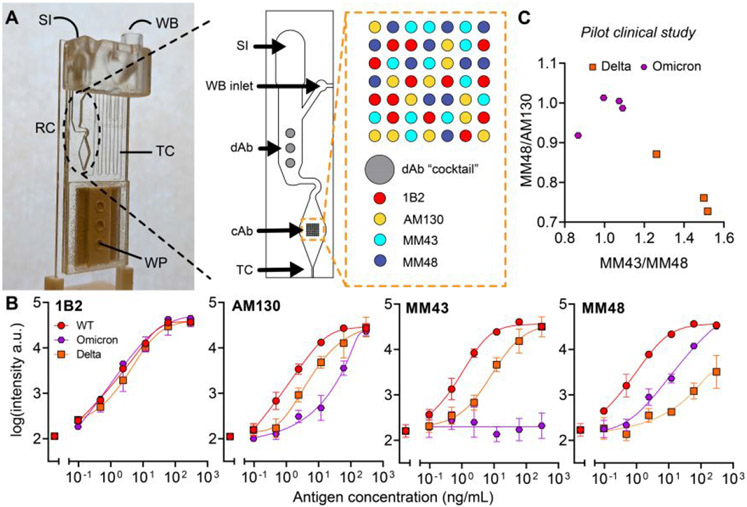
CoVariant-SPOT is a multiplexed sandwich immunoassay where all the biomolecular reagents needed to complete the assay are stored stably on a poly(oligoethylene glycol methyl ether methacrylate) (POEGMA) surface coating (Figure 1A), as we have described in more detail elsewhere.14, 17, 19 Multiplexing is accomplished using inkjet printing of spatially discrete immobilized capture antibodies (cAbs) that exhibit differential levels of binding to the S proteins of each variant. Nearby, a fluorescently labeled detection antibody (dAb) which binds similarly to the S proteins of all variants is co-printed with an excipient (trehalose), making the dAbs “dissolvable” upon addition of biological fluid. In addition, we incorporated an antibody pair for N protein into the assay, as it is expressed more abundantly than S protein and is more conserved across SARS-CoV-2 variants.20, 21 After incubation with a sample, variation in the fluorescence intensity at the cAb spots allows diagnosis of COVID-19 infection (based on N protein) and identification of the likely SARS-CoV-2 variant causing infection (Figure 1B).
Figure 1. CoVariant-SPOT schematic.
(A) Schematic of CoVariant-SPOT, which consists of 4 cAbs and a dAb cocktail of 2 dAbs printed on a trehalose pad. The primary targets of CoVariant-SPOT are SARS-CoV-2 N and S proteins from nasopharyngeal swabs. (B) Cartoon schematic of CoVariant-SPOT readout, demonstrating the ability to differentiate variants depending on ratio of the fluorescence intensity at different cAb spots.
Analytical performance of CoVariant-SPOT
To identify antibodies for CoVariant-SPOT, we conducted high throughput screens to determine optimal cAb/dAb pairs (Table S2) that bound to SARS-CoV-2 variants differentially, as described in the supporting information. The final version of CoVariant-SPOT featured four cAbs (three targeting S protein and one targeting N protein) and a dAb cocktail consisting of one dAb targeting S protein and one dAb targeting N protein. To investigate the analytical performance of CoVariant-SPOT, we first evaluated the response to WT, Delta, and Omicron recombinant S1 and N proteins at various dilutions. Experiments were performed in a commercially available extraction/lysis buffer (Acro Biosystems). To build dose-response curves, S1 and N proteins for WT, Delta, and Omicron were added to CoVariant-SPOT at 14 dilutions starting at 300 ng/mL as the highest concentration. The results are shown in Figure 2A. For this experiment, we used a 1 h incubation; however, in a separate set of experiments we found the time can be lowered to 15 min with only a modest impact on analytical sensitivity (Figure S3). Dose-response curves were fit using a 5-parameter logistic regression22 and the limit-of-detection (LOD) was calculated, as described elsewhere.18 A summary of the LODs is shown in Figure 2B.
Importantly, WT, Delta, and Omicron can be differentiated by the relative fluorescence of the anti-S cAb spots, likely due to some cAbs having weaker binding affinities to VOC S1 proteins because of mutations relative to WT. Figure 2C shows the ratio of MM43/MM48 plotted against MM48/AM130 (left) and MM43/AM130 (right). On both plots, Delta can be clearly differentiated from WT and Omicron across S1 concentrations ranging from 100 ng/mL to 0.4 ng/mL if MM43/MM48 is greater than ~1.0, and/or MM48/AM130 is less than ~1.0. Conversely, Omicron and WT can only be differentiated at higher concentrations of Omicron S1 if MM43/MM48 is less than ~0.75 or MM43/AM130 is less than ~0.8, suggesting that MM43 does not bind as well to Omicron S1. The MM43 cAb also binds poorly to the BA.2 sub lineage of Omicron (Figure S4). Of note, antibodies that bind to WT but do not bind—or bind weakly—to S proteins from certain variants has been observed with other antibodies and is potentially one of the drivers of SARS-CoV-2 variant escape from natural or vaccine induced humoral immunity.2, 12, 13, 23-26
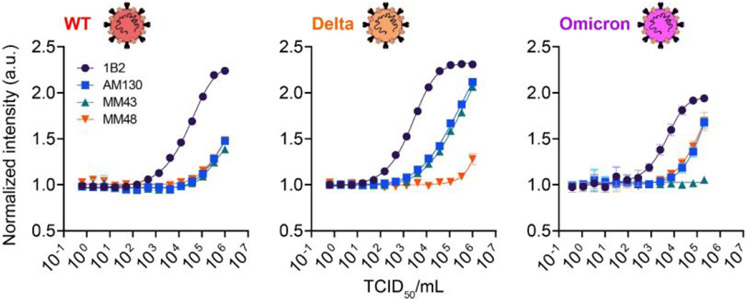
Next, we investigated the performance of CoVariant-SPOT to detect SARS-CoV-2 virus samples that had been propagated in cultured cells and inactivated using ultraviolet (UV) irradiation. Samples for WT, Delta, and Omicron viruses were added to lysis buffer, and then added to CoVariant-SPOT assays at various dilutions. The resulting dose-response curves are shown in Figure 3, which plot the normalized intensity against median tissue culture infectious dose per milliliter (TCID50/mL). As expected, the analytical sensitivity in terms of TCID50/mL for N protein is superior to that of S1 protein, likely because N protein is expressed more abundantly than S.20, 27, 28 We also observed similar trends in terms of cAb specificity for each variant. Notably, MM48 cAb does not bind as efficiently to Delta S protein and MM43 cAb does not bind efficiently to Omicron S protein. By examining the ratio of different anti-S cAb, similar patterns exist compared to the recombinant samples (Figure S5), further supporting our hypothesis that CoVariant-SPOT can differentiate between variants, especially at high viral loads. In a separate experiment, we found that the Acro Biosystems lysis/extraction buffer performed better in terms of analytical sensitivity compared to standard viral transport media (VTM) for N protein, while it had no impact on the detection of S protein—and thus does not appear to make a difference in VOC differentiation—presumably due to its location on the viral membrane (Figure S6). Overall, these experiments strongly suggest that CoVariant-SPOT could be useful to diagnose COVID-19 and differentiate between specific SARS-CoV-2 variants based on the fluorescence output of the assay.
Figure 3. Analytical validation of CoVariant-SPOT using UV inactivated virus.
Dose-response curves for each SARS-CoV-2 variant shown as a function of normalized intensity (y-axis) versus TCID50/mL (x-axis). Isolates were spiked into lysis buffer and incubated for 1 h. Each data point represents the average of three replicates, with SD shown.
Assessment of CoVariant-SPOT against clinical specimens
As proof-of-principle, we next sought to apply CoVariant-SPOT to diagnose COVID-19 infection and differentiate between Delta and Omicron variants in clinical specimens, as these strains were circulating concurrently at the time these experiments were performed. To demonstrate the clinical performance of CoVariant-SPOT, we tested biobanked nasopharyngeal swab samples from 32 COVID-19 negative and 76 positive individuals (Table S1). All samples were collected in VTM or universal transport media (UTM) and confirmed as COVID-19 positive or negative via reverse transcriptase polymerase chain reaction (RT-PCR). For a subset of the samples, viral load was quantified using quantitative RT-PCR (see Methods section). Of the 76 COVID-19 samples, 62 were sequenced using Illumina NextSeq500, of which ~32.3% were Omicron and ~24.2% were Delta (a full breakdown is shown in Table S1). Although the remaining 14 positive samples were not sequenced due to sample volume limitations, the probability of infection being from the predicted variant is high based on surveillance data collected by GISAID.10 An unavoidable limitation of this study is the use of biobanked samples as: (1) samples were collected in VTM/UTM rather than a lysis buffer that helps extract N protein, (2) the samples were collected in a large volume (~3 mL) which causes significant protein dilution relative to ideal collection methods (~150 μL), and (3) the samples were stored at −80 °C rather than tested fresh.
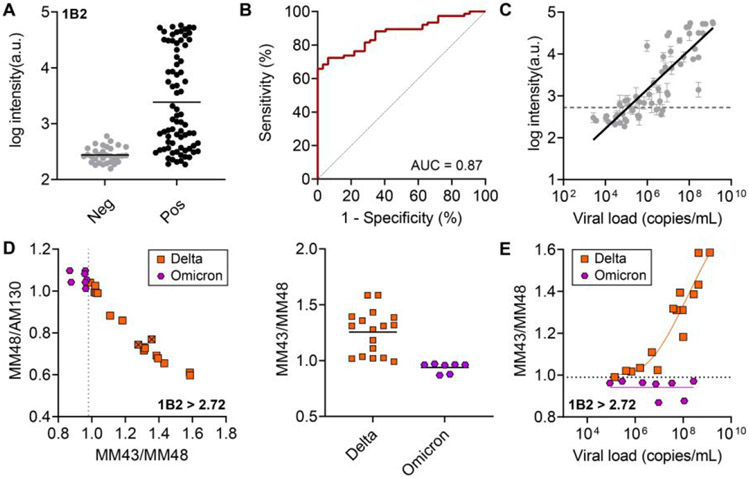
For clinical validation, each sample was tested in duplicate on CoVariant-SPOT. We first examined the ability of CoVariant-SPOT to diagnose COVID-19 via detection of N protein. The aggregate data for all samples is shown in Figure 4A. We found a statistically significant difference between the mean intensity for COVID-19–positive and –negative samples (P < 0.001), as determined by a two-tailed unpaired t-test. Sensitivity and specificity were determined by receiver operator curve (ROC) analysis (Figure 4B) which yielded an area under the ROC curve (AUC) of 0.87. At the optimal cut point of 2.72 arbitrary units for N, the sensitivity is 68.4% (95% CI: 57.3% – 77.8%) and the specificity is 96.9% (95% CI: 84.3% – 99.8%), which is similar to the performance metrics of other rapid antigen tests.29 For a subset of the positive samples, viral load was quantified using quantitative RT-PCR. We found that the D4 intensity for the N protein antibody pair was highly correlated with viral load (R2 = 0.72) (Figure 4C), which is consistent with other studies in the literature.30
Figure 4. Clinical validation of CoVariant-SPOT.
(A) Raw aggregate data for 1B2 for COVID-19 negative and positive samples. Each data point represents the average intensity of a unique sample, run in duplicate. (B) ROC analysis for 1B2 in diagnosing COVID-19. At the optimal cut point of 2.72 arbitrary units for N, the sensitivity is 68.4% (95% CI: 57.3% – 77.8%) and the specificity is 96.9% (95% CI: 84.3% – 99.8%). The AUC achieved is 0.87. (C) Correlation of 1B2 intensity compared to viral load, as quantified by RT-PCR. (D) Anti-S cAb ratios to differentiate between Delta and Omicron variants for all positive COVID-19 samples with 1B2 intensity greater than 2.72 arbitrary units. At a MM43/MM48 cut point of 0.99, all Delta and Omicron samples are perfectly discriminated (right). Samples with an “x” have not been sequenced but are presumed to be a given variant based on sample collection date. (E) MM43/MM48 plotted against viral load for Omicron and Delta samples with 1B2 > 2.72. As viral load increases, discrimination improves. The horizontal dashed line represents the optimal MM43/MM48 cut point.
Next, we examined the ability of CoVariant-SPOT to differentiate between two VOCs—Delta and Omicron—via detection of S protein. To test how well the two VOCs can be discriminated, we plotted MM43/MM48 against MM48/AM130 for all samples where the 1B2—the cAb for N—intensity is greater than 2.72 arbitrary units (i.e., tested positive), as shown in Figure 4D. We found that all Delta and Omicron samples are perfectly discriminated if MM43/MM48 is greater than 0.99 (Figure 4D). While this cut point was chosen based on the given dataset, future studies should validate the performance across an independent clinical test dataset. Consistent with results from recombinant samples and UV inactivated viruses, discrimination between Delta and Omicron improves with increasing viral load, as shown in Figure 4E. In line with Figure 2C, differentiation between Omicron and WT is less evident (Figure S7), suggesting additional antibodies should be incorporated into future iterations of CoVariant-SPOT to improve variant identification. Overall, these results suggest that the discriminatory power of CoVariant-SPOT is best for samples with high viral load and that the overall performance would likely improve in a prospective study where samples are collected in a small volume with extraction buffer.
Integration into a point-of-care format
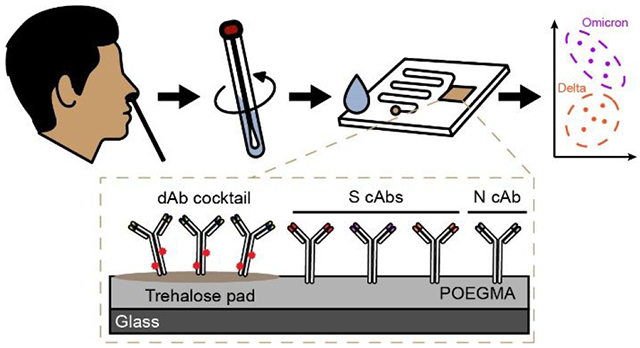
Finally, we sought to demonstrate the deployability of our test at the POC, de-coupled from laboratory or clinical infrastructure. To do this, we integrated CoVariant-SPOT into a microfluidic cassette (Figure 5A) that we developed recently.16 The microfluidic cassette automates CoVariant-SPOT passively with capillary and gravity driven flow and requires users to only add sample and wash buffer at the time of testing. Further, the microfluidic cassettes can be imaged with a portable, low-cost, and easy-to-use fluorescent detector—the D4Scope—which we have previously used to detect biomarkers of Ebola and antibodies against SARS-CoV-2.15, 16 The D4Scope costs ~$1,000 and can be operated using either battery or wall power. Combined, the microfluidic cassette and D4Scope could allow for sample testing and variant discrimination to occur simply by swabbing the nose, adding the swab to extraction buffer, adding a few drops to the microfluidic cassette, and imaging on the D4Scope after an incubation period (Figure S8).
Figure 5. Microfluidic implementation of CoVariant-SPOT.
(A) Photograph of the microfluidic cassette. Sample is added at the sample inlet (SI) followed by addition of wash buffer to the wash buffer inlet (WB). CoVariant-SPOT reagents are printed in the reaction chamber (RC) where all binding occurs. Inset: detailed view of the RC and zoom in of the cAb array. The incubation time is governed by the length of the timing channel (TC) which ends at the wicking pad (WP) that pulls sample and wash buffer through the channel, leaving a dry and clean RC for imaging. (B) Dose-response curves for recombinant N and S1 proteins spiked into lysis buffer and added to the microfluidic CoVariant-SPOT. Each data point represents the average of three replicates, with SD shown as error bars. (C) Proof of concept study testing a subset of clinical samples from Figure 4 on the microfluidic CoVariant-SPOT. For all positive Delta and Omicron COVID-19 samples with 1B2—the N protein cAb—intensity greater than 2.72 arbitrary units, anti-S cAb ratios are plotted to visually discriminate the two VOCs.
To validate the performance of the microfluidic CoVariant-SPOT, we tested WT, Delta, and Omicron recombinant S1 and N proteins at various dilutions. The resulting dose-response curves are shown in Figure 5B. The sensitivity is comparable to the results presented in Figure 2 (Table S3) and demonstrates the same intensity attenuation at MM48 cAb spots for Delta S1 protein and at MM43 cAb spots for Omicron S1 protein, suggesting that the microfluidic CoVariant-SPOT can distinguish between Delta and Omicron. As a proof-of-principle, we tested a subset of clinical samples using the microfluidic CoVariant-SPOT and found that we could reliably distinguish between Delta and Omicron by examining the anti-S cAb ratios, as done previously (Figure 5C).
CoVariant-SPOT has several limitations. First, as was reinforced by validation with UV inactivated virus and clinical samples, N protein is the superior diagnostic target for COVID-19 due to its abundance in the SARS-CoV-2 virion. This presents a challenge for CoVariant-SPOT as variant differentiation may only be possible for samples with high viral loads. As a result, this may limit the ability to use CoVariant-SPOT as a tool for identifying patients for relevant monoclonal antibody therapies. From an epidemiological standpoint, however, it is not critical to obtain strain information from every positive test, so long as an informative number of samples that do reach the S protein detection threshold are collected within a community. Second, due to limitations in sample availability and quantity, we were unable to perform sequencing on all the clinical samples used. This precludes us from making a definitive statement about the ability to differentiate variants in all clinical samples, although the probability is high that samples collected during the various time frames are likely from the given strain that was most prevalent at that time. In addition, we were able to sequence >80% of the samples tested giving us confidence in the results. Third, given the retrospective nature of this study, we were not able to test samples under ideal conditions, and instead tested samples collected in VTM or UTM using a large volume. Ideally, samples would be tested prospectively and collected using a small volume (~150 μL) of extraction buffer. We hope to implement such a study in the future and to compare the performance of our test against emergency use authorization approved options. In addition, we plan to implement pattern recognition software into the D4Scope to automatically determine the likely SARS-CoV-2 variant, which will be important as we plan to further multiplex the assay with additional antibodies. Finally, Omicron and Delta are no longer concurrently circulating, which limits the applicability of this version of CoVariant-SPOT. However, this work demonstrates our ability to rapidly adapt the platform to target emerging VOCs. Despite these limitations, we believe CoVariant-SPOT is a promising tool for SARS-CoV-2 variant surveillance, particularly in locations where sequencing infrastructure does not currently exist.
CONCLUSIONS
As the COVID-19 pandemic has progressed, epidemiological surveillance of emerging SARS-CoV-2 VOCs has proven essential in guiding the public health response. Our assay, CoVariant-SPOT, helps addresses this need by enabling simultaneous diagnosis of COVID-19 and differentiation between SARS-CoV-2 strains in an easy-to-use POC platform. Although the platform can be easily expanded to detect other variants, as proof-of-principle, we chose to limit our target strains in our validation studies to the WT, Delta, and Omicron variants given that Delta and Omicron were the two most globally dominant strains at the time we performed our experiments. A major strength of our assay platform is the highly multiplexed nature of the D4 microarray format that has two important attributes for VOC identifcation.23 First, the microarray format of CoVariant-SPOT can also be used as a high-throughput antibody screening platform. To identify antibodies useful for VOC identification by CoVariant-SPOT, we printed a panel of 29 commercially available and in-house antibodies as cAb’s in a D4 microarray and compared the binding of each antibody to VOC specific antigens, that enabled us to identify a subset of antibodies with differing sensitivity against WT, Delta, and Omicron. This approach will continue to be useful to enable differentiation of new VOCs as they emerge. Second as described in detail in this paper, the microarray format of CoV-SPOT enables discrimination between VOC’s.
We believe that CoVariant-SPOT could have a transformative effect on COVID-19 surveillance, and, more broadly, other infectious diseases caused by pathogens that shed measurable amount of antigen. CoVariant-SPOT opens the possibility of a new diagnostic paradigm where COVID-19 infection and mutant variant identification can occur simultaneously using a single sample. SARS-CoV-2 variant surveillance in the US (and worldwide) has been lackluster due to the difficulty in implementing NGS within the current clinical workflow. This is also true in many low- and middle-income countries where access to facilities that can conduct NGS is cost prohibitive or non-existent. We envision CoVariant-SPOT as a valuable adjunct to NGS that would significantly enhance surveillance capabilities in low- and middle-income countries. While NGS will remain a critical tool for identifying new variants, recognizing recurring SNPs, and tracking sequence evolution, we believe our assay is far better equipped to readily determine strain dominance down to the community level, regardless of available resources. Moreover, identifying COVID-19 strain can help personalize treatment. For instance, strain identification is clinically relevant because currently available therapeutic monoclonal antibodies have less therapeutic efficacy against certain variants (e.g., Omicron). Beyond the utility of this platform as a tool for SARS-CoV-2 management, multiplexed platforms such as CoVariant-SPOT could be useful for a variety of other indications, making this technology broadly applicable to the field of clinical diagnostics.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Duke University School of Medicine for the use of the Sequencing and Genomic Technologies Shared Resource, which provided SARS-CoV-2 variant sequencing and analysis services.
Funding:
This work was supported by National Institutes of Health (NIAID) grant R01 AI159992 (A.C. and C.W.W.). We would also like to acknowledge the Gilhuly Accelerator Fund who helped support this work.
Footnotes
SUPPORTING INFORMATION
Additional CoVariant-SPOT experimental methods, results for recombinant proteins, isolates, and clinical samples, and additional information regarding antibodies / clinical samples.
Competing interests:
A.C., J.T.H., R.J.B. and D.S.K. are inventors on a provisional patent filled by Duke University (Application number: 63/429,316) on 12/1/2022; entitled “methods for concurrent sars-cov-2 strain identification and covid-19 diagnosis and treatment.” (A.C., D.S.K., C.M.F., A.M.H., J.L., and J.T.H. are inventors on the patent filed by Duke University [PCT/US2021/046833, filed (20 August 2021) entitled “Microfluidic assay device” that describes the D4 microfluidic cassette in this work. A.C. and J.L. are inventors on a patent related to this work filed by Duke University [no. WO/2020/223713, filed (2 May 2020), published (5 May 2020)]. The patent is entitled “Devices and methods for imaging microarray chips” that describes innovations used for the D4Scope. Immucor Inc. has acquired the rights to the D4 assay on POEGMA brushes for in vitro diagnostics from Sentilus Inc. (cofounded by A.C. and A.M.H.). All other authors declare that they have no competing interests.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
References
- 1.Abdool Karim SS; de Oliveira T, New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N Engl J Med 2021, 384 (19), 1866–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mlcochova P; Kemp SA; Dhar MS; Papa G; Meng B; Ferreira I; Datir R; Collier DA; Albecka A; Singh S; Pandey R; Brown J; Zhou J; Goonawardane N; Mishra S; Whittaker C; Mellan T; Marwal R; Datta M; Sengupta S; Ponnusamy K; Radhakrishnan VS; Abdullahi A; Charles O; Chattopadhyay P; Devi P; Caputo D; Peacock T; Wattal C; Goel N; Satwik A; Vaishya R; Agarwal M; Indian S-C-GC; Genotype to Phenotype Japan, C.; Collaboration, C.-N. B. C.-.; Mavousian A; Lee JH; Bassi J; Silacci-Fegni C; Saliba C; Pinto D; Irie T; Yoshida I; Hamilton WL; Sato K; Bhatt S; Flaxman S; James LC; Corti D; Piccoli L; Barclay WS; Rakshit P; Agrawal A; Gupta RK, SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599 (7883), 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCallum M; Czudnochowski N; Rosen LE; Zepeda SK; Bowen JE; Walls AC; Hauser K; Joshi A; Stewart C; Dillen JR; Powell AE; Croll TI; Nix J; Virgin HW; Corti D; Snell G; Veesler D, Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 2022, 375 (6583), 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiara M; D'Erchia AM; Gissi C; Manzari C; Parisi A; Resta N; Zambelli F; Picardi E; Pavesi G; Horner DS; Pesole G, Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Brief Bioinform 2021, 22 (2), 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.du Plessis L; McCrone JT; Zarebski AE; Hill V; Ruis C; Gutierrez B; Raghwani J; Ashworth J; Colquhoun R; Connor TR; Faria NR; Jackson B; Loman NJ; O'Toole A; Nicholls SM; Parag KV; Scher E; Vasylyeva TI; Volz EM; Watts A; Bogoch II; Khan K; Consortium, C.-G. U.; Aanensen DM; Kraemer MUG; Rambaut A; Pybus OG, Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 2021, 371 (6530), 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegally H; Wilkinson E; Giovanetti M; Iranzadeh A; Fonseca V; Giandhari J; Doolabh D; Pillay S; San EJ; Msomi N; Mlisana K; von Gottberg A; Walaza S; Allam M; Ismail A; Mohale T; Glass AJ; Engelbrecht S; Van Zyl G; Preiser W; Petruccione F; Sigal A; Hardie D; Marais G; Hsiao M; Korsman S; Davies M-A; Tyers L; Mudau I; York D; Maslo C; Goedhals D; Abrahams S; Laguda-Akingba O; Alisoltani-Dehkordi A; Godzik A; Wibmer CK; Sewell BT; Lourenço J; Alcantara LCJ; Pond SLK; Weaver S; Martin D; Lessells RJ; Bhiman JN; Williamson C; de Oliveira T, Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020, 2020.12.21.20248640. [Google Scholar]
- 7.Tao K; Tzou PL; Nouhin J; Gupta RK; de Oliveira T; Kosakovsky Pond SL; Fera D; Shafer RW, The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 2021, 22 (12), 757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherian S; Potdar V; Jadhav S; Yadav P; Gupta N; Das M; Rakshit P; Singh S; Abraham P; Panda S; Team N, SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena SK; Kumar S; Ansari S; Paweska JT; Maurya VK; Tripathi AK; Abdel-Moneim AS, Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol 2022, 94 (4), 1738–1744. [DOI] [PubMed] [Google Scholar]
- 10.Shu Y; McCauley J, GISAID: Global initiative on sharing all influenza data – from vision to reality. Eurosurveillance 2017, 22 (13), 30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brito AF; Semenova E; Dudas G; Hassler GW; Kalinich CC; Kraemer MUG; Ho J; Tegally H; Githinji G; Agoti CN; Matkin LE; Whittaker C; Danish Covid-19 Genome, C.; Project, C.-I.; Network for Genomic Surveillance in South, A.; team, G. c. c.; Howden BP; Sintchenko V; Zuckerman NS; Mor O; Blankenship HM; Oliveira T. d.; Lin RTP; Siqueira MM; Resende PC; Vasconcelos ATR; Spilki FR; Aguiar RS; Alexiev I; Ivanov IN; Philipova I; Carrington CVF; Sahadeo NSD; Gurry C; Maurer-Stroh S; Naidoo D; von Eije KJ; Perkins MD; Kerkhove M. v.; Hill SC; Sabino EC; Pybus OG; Dye C; Bhatt S; Flaxman S; Suchard MA; Grubaugh ND; Baele G; Faria NR, Global disparities in SARS-CoV-2 genomic surveillance. medRxiv 2021, 2021.08.21.21262393. [Google Scholar]
- 12.Liu L; Iketani S; Guo Y; Chan JF; Wang M; Liu L; Luo Y; Chu H; Huang Y; Nair MS; Yu J; Chik KK; Yuen TT; Yoon C; To KK; Chen H; Yin MT; Sobieszczyk ME; Huang Y; Wang HH; Sheng Z; Yuen KY; Ho DD, Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022, 602 (7898), 676–681. [DOI] [PubMed] [Google Scholar]
- 13.Planas D; Saunders N; Maes P; Guivel-Benhassine F; Planchais C; Buchrieser J; Bolland WH; Porrot F; Staropoli I; Lemoine F; Pere H; Veyer D; Puech J; Rodary J; Baele G; Dellicour S; Raymenants J; Gorissen S; Geenen C; Vanmechelen B; Wawina-Bokalanga T; Marti-Carreras J; Cuypers L; Seve A; Hocqueloux L; Prazuck T; Rey FA; Simon-Loriere E; Bruel T; Mouquet H; Andre E; Schwartz O, Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602 (7898), 671–675. [DOI] [PubMed] [Google Scholar]
- 14.Joh DY; Hucknall AM; Wei Q; Mason KA; Lund ML; Fontes CM; Hill RT; Blair R; Zimmers Z; Achar RK; Tseng D; Gordan R; Freemark M; Ozcan A; Chilkoti A, Inkjet-printed point-of-care immunoassay on a nanoscale polymer brush enables subpicomolar detection of analytes in blood. Proc Natl Acad Sci U S A 2017, 114 (34), E7054–E7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontes CM; Lipes BD; Liu J; Agans KN; Yan A; Shi P; Cruz DF; Kelly G; Luginbuhl KM; Joh DY; Foster SL; Heggestad J; Hucknall A; Mikkelsen MH; Pieper CF; Horstmeyer RW; Geisbert TW; Gunn MD; Chilkoti A, Ultrasensitive point-of-care immunoassay for secreted glycoprotein detects Ebola infection earlier than PCR. Sci Transl Med 2021, 13 (588). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heggestad JT; Kinnamon DS; Olson LB; Liu J; Kelly G; Wall SA; Oshabaheebwa S; Quinn Z; Fontes CM; Joh DY; Hucknall AM; Pieper C; Anderson JG; Naqvi IA; Chen L; Que LG; Oguin T 3rd; Nair SK; Sullenger BA; Woods CW; Burke TW; Sempowski GD; Kraft BD; Chilkoti A, Multiplexed, quantitative serological profiling of COVID-19 from blood by a point-of-care test. Sci Adv 2021, 7 (26). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heggestad JT; Fontes CM; Joh DY; Hucknall AM; Chilkoti A, In Pursuit of Zero 2.0: Recent Developments in Nonfouling Polymer Brushes for Immunoassays. Adv Mater 2020, 32 (2), e1903285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armbruster DA; Pry T, Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 2008, 29 Suppl 1, S49–52. [PMC free article] [PubMed] [Google Scholar]
- 19.Hucknall A; Kim D-H; Rangarajan S; Hill RT; Reichert WM; Chilkoti A, Simple Fabrication of Antibody Microarrays on Nonfouling Polymer Brushes with Femtomolar Sensitivity for Protein Analytes in Serum and Blood. Advanced Materials 2009, 21 (19), 1968–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-On YM; Flamholz A; Phillips R; Milo R, SARS-CoV-2 (COVID-19) by the numbers. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammad T; Choudhury A; Habib I; Asrani P; Mathur Y; Umair M; Anjum F; Shafie A; Yadav DK; Hassan MI, Genomic Variations in the Structural Proteins of SARS-CoV-2 and Their Deleterious Impact on Pathogenesis: A Comparative Genomics Approach. Front Cell Infect Microbiol 2021, 11, 765039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschalk PG; Dunn JR, The five-parameter logistic: a characterization and comparison with the four-parameter logistic. Anal Biochem 2005, 343 (1), 54–65. [DOI] [PubMed] [Google Scholar]
- 23.Heggestad JT; Britton RJ; Kinnamon DS; Wall SA; Joh DY; Hucknall AM; Olson LB; Anderson JG; Mazur A; Wolfe CR; Oguin TH 3rd; Sullenger BA; Burke TW; Kraft BD; Sempowski GD; Woods CW; Chilkoti A, Rapid test to assess the escape of SARS-CoV-2 variants of concern. Sci Adv 2021, 7 (49), eabl7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iketani S; Liu L; Guo Y; Liu L; Chan JF; Huang Y; Wang M; Luo Y; Yu J; Chu H; Chik KK; Yuen TT; Yin MT; Sobieszczyk ME; Huang Y; Yuen KY; Wang HH; Sheng Z; Ho DD, Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 2022, 604 (7906), 553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prevost J; Finzi A, The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe 2021, 29 (3), 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCallum M; Walls AC; Sprouse KR; Bowen JE; Rosen LE; Dang HV; De Marco A; Franko N; Tilles SW; Logue J; Miranda MC; Ahlrichs M; Carter L; Snell G; Pizzuto MS; Chu HY; Van Voorhis WC; Corti D; Veesler D, Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 2021, 374 (6575), 1621–1626. [DOI] [PubMed] [Google Scholar]
- 27.Ke Z; Oton J; Qu K; Cortese M; Zila V; McKeane L; Nakane T; Zivanov J; Neufeldt CJ; Cerikan B; Lu JM; Peukes J; Xiong X; Krausslich HG; Scheres SHW; Bartenschlager R; Briggs JAG, Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588 (7838), 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao H; Song Y; Chen Y; Wu N; Xu J; Sun C; Zhang J; Weng T; Zhang Z; Wu Z; Cheng L; Shi D; Lu X; Lei J; Crispin M; Shi Y; Li L; Li S, Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183 (3), 730–738 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brummer LE; Katzenschlager S; Gaeddert M; Erdmann C; Schmitz S; Bota M; Grilli M; Larmann J; Weigand MA; Pollock NR; Mace A; Carmona S; Ongarello S; Sacks JA; Denkinger CM, Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: A living systematic review and meta-analysis. PLoS Med 2021, 18 (8), e1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollock NR; Savage TJ; Wardell H; Lee RA; Mathew A; Stengelin M; Sigal GB, Correlation of SARS-CoV-2 Nucleocapsid Antigen and RNA Concentrations in Nasopharyngeal Samples from Children and Adults Using an Ultrasensitive and Quantitative Antigen Assay. J Clin Microbiol 2021, 59 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.