CONSPECTUS:
After decades of extensive fundamental studies and clinical trials, lipid nanoparticles (LNPs) have demonstrated effective mRNA delivery such as the Moderna and Pfizer-BioNTech vaccines fighting against COVID-19. Moreover, researchers and clinicians have been investigating mRNA therapeutics for a variety of therapeutic indications including protein replacement therapy, genome editing, and cancer immunotherapy. To realize these therapeutics in the clinic, there are many formidable challenges. First, novel delivery systems such as LNPs with high delivery efficiency and low toxicity need to be developed for different cell types. Second, mRNA molecules need to be engineered for improved pharmaceutical properties. Lastly, the LNP–mRNA nanoparticle formulations need to match their therapeutic applications.
In this Account, we summarize our recent advances in the design and development of various classes of lipids and lipid derivatives, which can be formulated with multiple types of mRNA molecules to treat diverse diseases. For example, we conceived a series of ionizable lipid-like molecules based on the structures of a benzene core, an amide linker, and hydrophobic tails. We identified N1,N3,N5-tris(3-(didodecylamino)propyl)benzene-1,3,5-tricarboxamide (TT3) as a lead compound for mRNA delivery both in vitro and in vivo. Moreover, we tuned the biodegradability of these lipid-like molecules by introducing branched ester or linear ester chains. Meanwhile, inspired by biomimetic compounds, we synthesized vitamin-derived lipids, chemotherapeutic conjugated lipids, phospholipids, and glycolipids. These scaffolds greatly broaden the chemical space of ionizable lipids for mRNA delivery. In another section, we highlight our efforts on the research direction of mRNA engineering. We previously optimized mRNA chemistry using chemically-modified nucleotides to increase the protein expression, such as pseudouridine (ψ), 5-methoxyuridine (5moU), and N1-methylpseudouridine (me1ψ). Also, we engineered the sequences of mRNA 5′ untranslated regions (5′-UTRs) and 3′ untranslated regions (3′-UTRs), which dramatically enhanced protein expression. With the progress of LNP development and mRNA engineering, we consolidate these technologies and apply them to treat diseases such as genetic disorders, infectious diseases, and cancers. For instance, TT3 and its analog-derived lipid-like nanoparticles can effectively deliver factor IX or VIII mRNA and recover the clotting activity in hemophilia mouse models. Engineered mRNAs encoding SARS-CoV-2 antigens serve well as vaccine candidates against COVID-19. Vitamin-derived lipid nanoparticles loaded with antimicrobial peptide-cathepsin B mRNA enable adoptive macrophage transfer to treat multidrug resistant bacterial sepsis. Biomimetic lipids such as phospholipids formulated with mRNAs encoding costimulatory receptors lead to enhanced cancer immunotherapy.
Overall, lipid–mRNA nanoparticle formulations have considerably benefited public health in the COVID-19 pandemic. To expand their applications in clinical use, research work from many disciplines such as chemistry, engineering, materials, pharmaceutical sciences, and medicine need to be integrated. With these collaborative efforts, we believe that more and more lipid–mRNA nanoparticle formulations will enter the clinic in the near future and benefit human health.
Graphical Abstract
1. INTRODUCTION
Discovered in the 1960s, the mRNA molecules are translated to corresponding proteins via the protein synthesis machinery.5–8 This natural process makes mRNA-based therapeutics feasible for numerous biological and therapeutic applications: vaccines, protein replacement therapy, cancer immunotherapy, genome editing, cellular reprogramming, etc.6,9 On the basis of traditional concepts of druggability, mRNAs themselves are not favorable drug molecules. These molecules have large molecular weight and negative charges; they are unstable in physiological conditions, and they may induce immunogenicity. To address these issues and concerns, researchers have spent extensive efforts on developing mRNA delivery vehicles and engineering mRNA molecules.10
Since the 1970s, researchers have developed many types of delivery materials for mRNA, i.e., lipid-based nanoparticles, polymers, protein–mRNA complexes, and other biomaterials.10–17 Among them, lipid nanoparticles (LNPs) have been recently used as the delivery system for COVID-19 mRNA vaccines in the clinic.18 Naturally, lipids are major components of cell membranes and play essential roles in maintaining membrane structures and their biological functions.19,20 It has been recognized for a long time that lipid formulated liposomes can serve as delivery vehicles for many cargos, including mRNA molecules.21 In the 1980s, a cationic lipid named 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) was reported to deliver nucleic acids in vitro.22 Later on, many other cationic lipids were developed for gene delivery, such as 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), dimethyldioctadecylammonium (DDAB), and 3β-[N-(N′,N′-dimethylaminoethane)carbamoyl]cholesterol (DC-cholesterol).23 Recent results showed that cationic lipids with permanent positive charges may be toxic to cells.10,24 Meanwhile, researchers produced ionizable lipids, which generally possess three parts: amine heads, linkage regions, and lipid chains. With a pKa value less than 7.0, ionizable lipids are neutral at physiological pH and can be protonated at acidic conditions. It should be noted that the pKa of ionizable lipids may be different from that of the corresponding LNPs.25 In 2018, DLin-MC3-DMA (MC3) LNPs, an ionizable lipid-based formulation, was the first FDA approved LNP for an RNAi therapeutic, which delivered a double-stranded siRNA to liver hepatocytes for treating transthyretin (TTR) amyloidosis.23,26 In the late 2020, the COVID-19 mRNA vaccines, BNT162b2 and mRNA-1273, obtained emergency use authorization to fight against the COVID-19 pandemic, utilizing two different ionizable lipids, ALC-0315 and SM-102, respectively.27–29
On the other hand, it is also critical to understand and optimize the properties of mRNA, that is, to augment mRNA translation efficiency and reduce its undesired immunogenicity.30 Chemical modification of the nucleobases is a practical approach to engineer mRNA molecules since several naturally existing chemically-modified nucleotides were discovered in biological systems, such as inosine (I), dihydrouridine (D), and pseudouridine (ψ).31,32 Additionally, mRNA molecules consist of multiple components: 5′ caps, 5′ untranslated regions (5′-UTRs), coding sequences, 3′ untranslated regions (3′-UTRs), and poly(A) tails. Through sequence engineering, researchers can tune the properties of mRNA and its interactions with other cellular elements.6 Specifically, a functional 5′ cap is important for the initiation of mRNA translation, which can be added via enzymatic or synthetic methods. 5′-UTR and 3′-UTR may bind to sequence specific proteins or RNAs that regulate mRNA translation. Coding sequences can theoretically carry any proteins of interest. The length and sequence of the poly(A) tail is also a crucial factor for mRNA stability. Recently, a CleanCap technology was developed, which provides a chemical approach for the capping process of mRNA.33 These engineering strategies facilitate the translation of mRNA therapeutics from bench to bedside.
In the Account, we summarize our design and development of synthetic ionizable lipids such as lipid-like molecules, vitamin-derived lipids, chemotherapy drug-derived amino lipids, phospholipids, and glycolipids. We also describe our mRNA engineering approaches (chemical modifications of nucleobases and optimizations of untranslated regions) to improve the mRNA properties. We then overview these lipid–mRNA nanoparticle formulations for the treatment of genetic disorders, infectious diseases, and cancers. Finally, we provide a brief perspective on future directions and challenges of this field.
2. LIPIDS AND LIPID NANOPARTICLES
2.1. Synthetic Ionizable Lipids and Lipid-Like Nanoparticles
Traditionally, phospholipids are key components of eukaryotic cell membranes and composed of a single hydrophilic head and two hydrophobic chains. On the basis of these natural molecules, many lipids like DLin-MC3-DMA, ALC-0315, and SM-102 have been developed and applied for RNA delivery.24 To expand the lipid chemistry, the Anderson and Langer groups used a combinatorial approach to synthesize a large library of lipid derivatives termed lipid-like molecules or lipidoids, which consist of more than 1200 structures with a variety of amines and lipid chains. Among these structures, 98N12-5 LNPs silenced up to 85% of Apolipoprotein B expression in nonhuman primates at an siRNA dose of 6.25 mg/kg.34 Since then, a variety of these lipidoids have been reported such as C12-200, cKK-E12, 5A2-SC8, 306Oi10, BAMEA-O16B, etc.14,35–39 These lipidoids can be formulated into lipid-like nanoparticles (LLNs), which greatly enlarge the diversity of the lipid nanoparticles (LNPs). In practice, these LLNs have been applied to deliver siRNA, mRNA, and the CRISPR system.10
In 2015, we reported a series of 1,3,5-benzenetricarbonyl trichloride (TT)-derived lipid-like molecules consisting of three amino groups, three amide linkers, and a benzene ring. Scheme 1 shows the synthetic route. 1,3,5-Benzenetricarbonyl trichloride was condensed with mono-boc-protected diamines to obtain the compound (1). Deprotection of 1 and a subsequent reductive amination gave the products TT2–TT8 (Scheme 1A).1 Then, TT compounds were formulated with 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol (Chol), 1,2-dimyristoyl-sn-glycerolmethoxypolyethylene glycol (DMG-PEG2000), and firefly luciferase mRNA. The delivery efficiency of these resulting LLNs was evaluated by in vitro screenings. Among these ionizable lipids, the TT3 LLNs induced the highest luminescence intensity in the Hep3B cells. In order to further optimize the formulation of the TT3 LLNs, we utilized an orthogonal method by tuning the molar ratios of each component. After two rounds of optimization, the TT3 LLNs (molar ratio: TT3/DOPE/Chol/DMG-PEG2000 = 20/30/40/0.75) were identified as a lead formulation. Recently, we studied the long-term storage of these nanoparticles. We added different amounts of sucrose, trehalose, or mannitol to the TT3 LLNs under freeing or lyophilization conditions and then evaluated their stability. The results showed that the inclusion of 5% (w/v) trehalose or sucrose to these LLNs was able to maintain the mRNA delivery efficiency when stored in liquid nitrogen for at least 3 months.40 Next, we examined the biodegradability of lipid-like compounds by installing various ester chains, which may be degraded in the presence of esterases or lipases.41 We first synthesized a set of amino esters and compared their biodegradability with amino alcohols. The results showed that amino esters were biodegradable, and the lipid chemistry was able to tune the biodegradability rate. Subsequently, we incorporated these ester chains on the core structure of the TT compounds and prepared functionalized TT (FTT) molecules.2 With different lipid chain structures, we classified these FTT derivatives into three types: hydrocarbon chains, linear ester chains, and branched ester chains. FTT1 holds a polyunsaturated hydrocarbon chain; FTT2–FTT6 have a variety of branched esters, different from FTT2–FTT6, and FTT7–FTT10 bear several linear ester structures with less steric effects (Scheme 1B). Then, the mRNA delivery efficiency of FTT1–10 LLNs were tested in mice by intravenously injecting firefly luciferase (FLuc) mRNA-loaded FTT LLNs (0.5 mg/kg). After quantification of the bioluminescence intensity from major organs, 6 h after administration, FTT5 LLNs were found to increase the luminescent signal over 2-fold in the liver compared to that of TT3 LLNs.
Scheme 1.
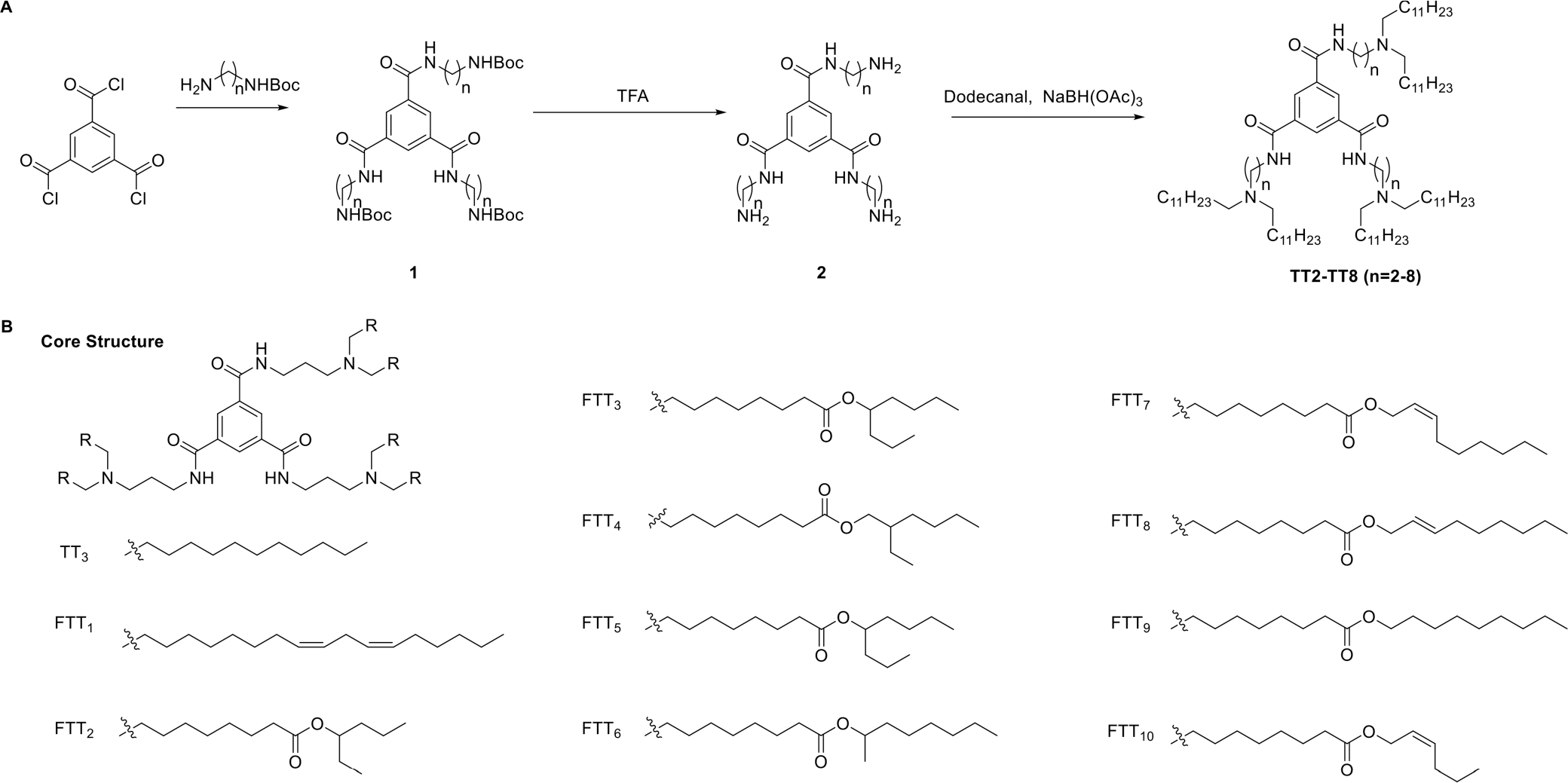
Synthesis of 1,3,5-Benzenetricarbonyl Trichloride (TT)-Derived Lipid-Like Molecules (A) and Functionalized TT (FTT) Molecules (B)1,2
2.2. Biomimetic Molecule-Derived Ionizable Lipid Nanoparticles
Lipids conjugated with bioactive molecules, such as oligonucleotides, oligopeptides/amino acids, sugars, or small molecule drugs, have been developed as formulation components for RNA delivery.21 In 2007, Desigaux et al. formulated a series of lipidic aminoglycoside molecules with DOPE to deliver siRNA in several human cancer cell lines, resulting in gene silencing.42 In 2016, siRNA loaded LNPs were reported to show significant tumor inhibition in vivo using lipids conjugated with a cell penetrating peptide oleoyloctaarginine.43 Recently, van der Meel et al. prepared LNPs from lipophilic taxane prodrugs for the delivery of siRNA against the androgen receptor (AR) to treat prostate cancer.44
In our group, we synthesized several series of bioactive molecule-derived ionizable lipids for mRNA delivery. For example, vitamins are indispensable nutrients for cell functions.45,46 We conjugated vitamins or their derivatives with an amino lipid bearing a carboxylic acid group through ester or amide bond formation to prepare vitamin-derived lipids (Scheme 2A). We then apply them to encapsulate an mRNA encoding a hybrid protein of antimicrobial peptides and cathepsin B.4 These vitamin-derived lipid nanoparticles (VLNPs) were screened and optimized in RAW264.7 cells for mRNA delivery. The results displayed that VcLNPs (molar ratio: Vc-lipid/DOPE/cholesterol = 30/30/40; mass ratio: Vc-Lipid/mRNA = 15/1) were over 70- and 300-fold more efficient for mRNA delivery in these cells than that of Lipofectamine 3000 and electroporation, respectively. In another study, we conjugated chemotherapy drugs, paclitaxel and camptothecin, with an amino lipid via an ester bond to explore the synergistic effects of anticancer agents and tumor suppressor genes (Scheme 2B). Paclitaxel-derived LNPs exhibited effective mRNA delivery. We then loaded these LNPs with mRNA encoding p53 as a potential therapy for triple-negative breast cancer.47
Scheme 2.
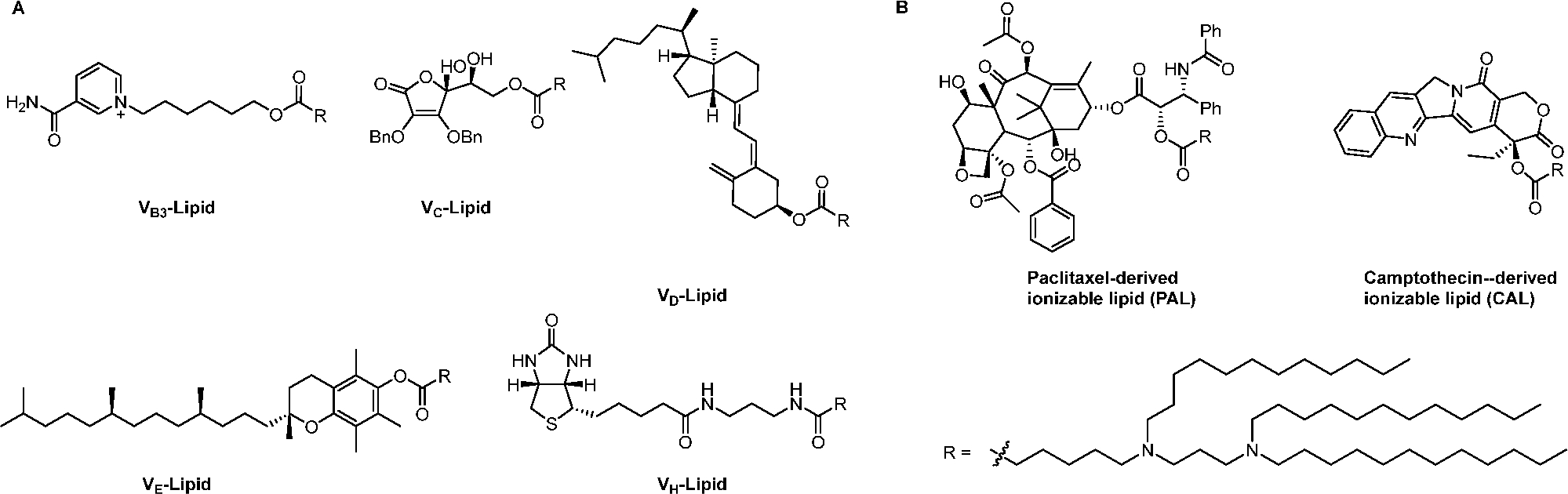
Chemical Structures of Vitamin-Derived Ionizable Lipids (A) and Chemotherapy Drug-Derived Ionizable Lipids (B)4,47
Phospholipids are essential components of the cell membrane and not only construct the membrane structure but also are responsible for cellular functions such as molecular transportation.19,20 Inspired by these natural lipids, we produced ionizable phospholipids (PLs). The synthetic routes started from phosphorylation of trimethylene bromohydrin, leading to a phosphate triester 4. Then, the substitution of bromine in compound 4 with boc-protected trimethylenediamine gave compound 5. Finally, the removal of the boc protecting group, followed by a reductive amination with aldehydes, yielded phospholipids (Scheme 3A).48 Additionally, glycolipids are an important class of lipids with carbohydrate moieties. They are critical for cell membrane structures and other cellular functions such as immune responses.49 Similar to the synthetic route to ionizable phospholipids, ionizable glycolipids (GLs) were synthesized (Scheme 3B). Several PLs and GLs were able to effectively deliver mRNA in E.G7 cells, a T cell line.50
Scheme 3.
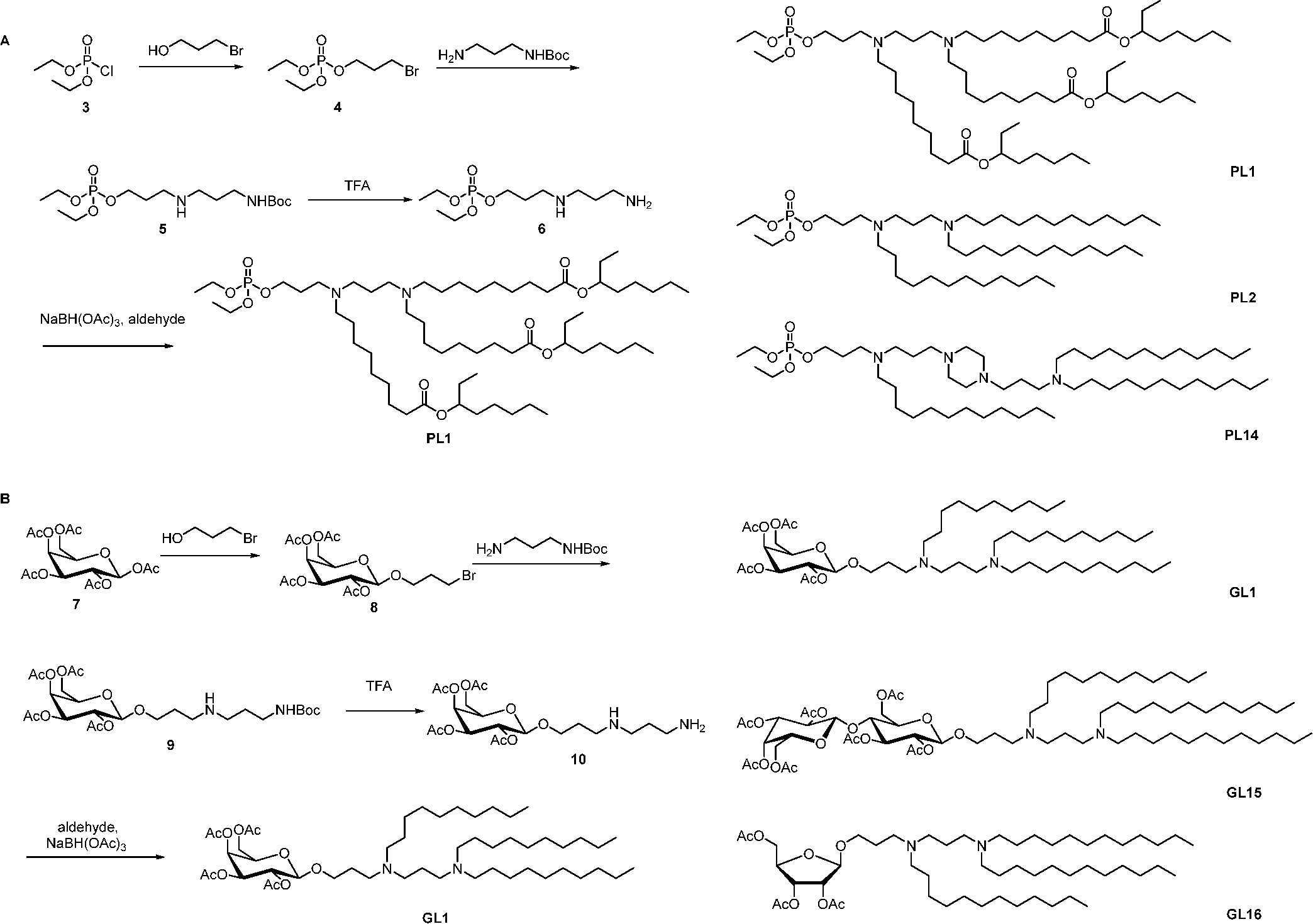
Synthesis of Ionizable Phospholipids (A) and Glycolipids (B)48
3. MRNA ENGINEERING
3.1. Chemical Modification of mRNA
Chemical modification, such as ribose, backbone, and nucleobase modifications of mRNA, has been shown to increase the protein expression and reduce undesired immunogenicity.30 In 2005, Karikò et al. reported that certain nucleoside-modified RNA was less immunogenetic than the unmodified RNA.51 Afterward, they found that pseudouridine-modified mRNA, which can be applied to produce multiple types of proteins such as erythropoietin, increased protein expression.52,53 Recently, phosphorothioate-modified mRNA was constructed, which increased the protein synthesis rate.54 To test the translation efficiency of different mRNAs, we prepared a library of chemically-modified mRNAs (Scheme 4), including 5-methylcytidine (5meC), 5-hydroxymethyl-cytidine (5hmC), 5-methyluridine (5meU), pseudouridine (ψ), N1-methylpseudouridine (me1ψ), N6-methyladenosine (me6A), etc. Then, we measured the protein expression level and compared the expression level under different translation conditions. Consequently, pseudouridine (ψ), 5-methoxyuridine (5moU), and N1-methylpseudouridine (me1ψ) were advantageous nucleotides that increased the production of proteins. Specifically, 5moU modification increased eGFP mRNA stability in Hep3B cancer cells.30
Scheme 4.
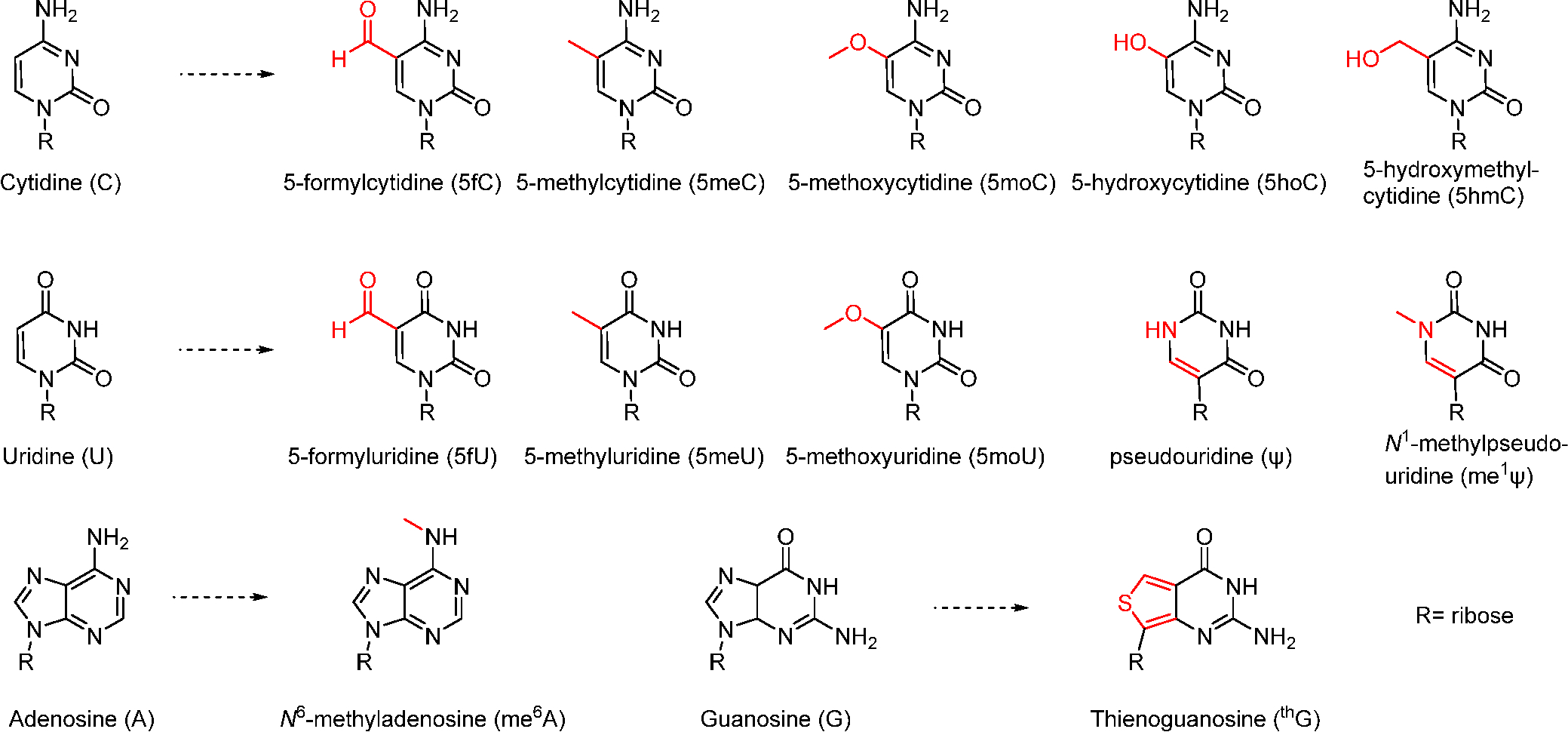
Examples of Chemically-Modified Nucleobases30
Subsequently, we applied this mRNA engineering strategy to the CRISPR-Cpf1 system, a class-II CRISPR system that enables effective gene editing in human cells.55 To increase Cpf1-mediated genome editing, we designed and assessed a library of engineered Cpf1 mRNA in human cell lines. ψ-modified AsCpf1 mRNA increased the efficiency of gene cutting by 177% compared to the AsCpf1 plasmid. Also, we incorporated chemically-modified CRISPR RNAs (crRNAs) in the system. From 42 engineered AsCpf1 crRNAs, we found that cr3′5F containing five 2′-fluoro ribose at the 3′ terminus increased the efficiency over 127% compared to that of wild-type crRNA. The combination of the optimal Cpf1 crRNA and mRNA enhanced gene-cutting efficiency over 3-fold compared with wild-type crRNA and plasmid-encoding AsCpf1. Meanwhile, these data were further validated by targeted deep-sequencing, which showed that cr3′5F together with ψ-modified mRNA significantly improved the gene cutting efficiency without increasing off-target effects.56,57
3.2. Sequence Engineering of mRNA
The mRNA molecules consist of several essential components, including 5′ cap, 5′ untranslated region (5′-UTR), encoding region, 3′ untranslated region (3′-UTR), and the polyadenylated (poly-A) tail. Within these components, the 5′ untranslated region (5′-UTR) and 3′ untranslated region (3′-UTR) played important regulating roles for protein expression.3 For example, when a β-globin in the 5′-UTR was replaced with an α-globin mRNA, the translation efficiencies of mRNA were increased.58 Thus, it is critical to design proper 5′-UTR and 3′-UTR to increase protein production. In our study, we first performed bioinformatic analysis of the mRNA UTR, from which the number of protein copies and amino acids were quantified for each mRNA molecule in the database. The murine Rps27a mRNA displayed the highest translation ability. Meanwhile, we conducted a de nova design of the 5′-UTR. Several criteria were utilized including minimal secondary structures, removal of miRNA binding sites, and inclusion of a Kozak sequence (GCCACC). The results indicated that sequence engineering greatly augmented protein production in both Hep3B and 293T cells and the optimal length of 5′-UTR was found to be 70 nt in these experiments. Besides length, the composition of A, U, G, and C is another important factor for the 5′-UTRs. We analyzed nucleotide compositions and created several 5′-UTRs, named, NCA-1 to NCA-8, with different nucleotide compositions. The results showed that NCA-7 and NCA-8 with relatively high U content (11 and 18 out of 70 nt, respectively) were preferred compositions. Then, we installed functional motifs after the S27a 3′-UTR, which resulted in repeated sequence element 3 and U-rich element (R3U) as a lead motif. Lastly, we chose NCA-7d as a 5′-UTR and S27a with R3U as a 3′-UTR, which was termed NASAR UTR. These UTR engineering methods enable us to understand the effects of UTR on protein expression and facilitate broader applications of mRNA.
4. THERAPEUTIC AREAS
4.1. Genetic Disorders
Genetic disorders are a large class of diseases caused by defective or mutated genome sequences.59 Hemophilia B is a typical rare genetic disease in which patients have insufficient blood clotting protein, named factor IX.60 In our work, we formulated TT3 LLNs to deliver human FIX (hFIX) mRNA. After in vitro and in vivo testing, the optimal TT3 LLN group showed a notably higher biodistribution signal in spleen and liver than that of the control groups of C12–200 LLNs. Then, the TT3 LLNs formulated with human factor IX mRNA were intravenously injected into both wild-type mice and hemophilia B mice with control groups of free hFIX mRNA. In the FIX-knockout mice groups, TT3-hFIX mRNA LLNs produced 608 and 1740 ng/mL hFIX at an mRNA dose of 0.55 and 1.1 mg/kg, respectively. In the meantime, the hFIX activities were quantified. Importantly, the TT3 hFIX LLNs were able to recover the hFIX level to the normal range in hemophilia B mice.
We later prepared functionalized TT (FTT) LLNs from which FTT5 LLNs were selected as a lead material after in vitro and in vivo screening. The biodegradability of FTT5 and FTT9 LLNs was evaluated by mass spectrometry after intravenous administration at different time points (Figure 1A,B). FTT9 was almost fully cleared within 24 h, while FTT5 demonstrated a slower clearance rate in mouse liver. These results were consistent with our design that branched ester chains with higher steric effects than linear ester chains can tune the rate of biodegradability. Next, we formulated FTT5 LLNs to deliver the hFVIII mRNA.60 FTT5-hFVIII LLNs were injected intravenously at a dose of 2 mg/kg to wild-type mice and hFVIII-knockout mice with control groups of free hFVIII mRNA. Similar to wild-type mice, hFVIII-knockout mice treated with FTT5-hFVIII LLNs produced over 200 ng/mL hFVIII protein 6 and 12 h post-administration. Meanwhile, more than 70% of the hFVIII activity was restored at the time points (Figure 1C,D).
Figure 1.
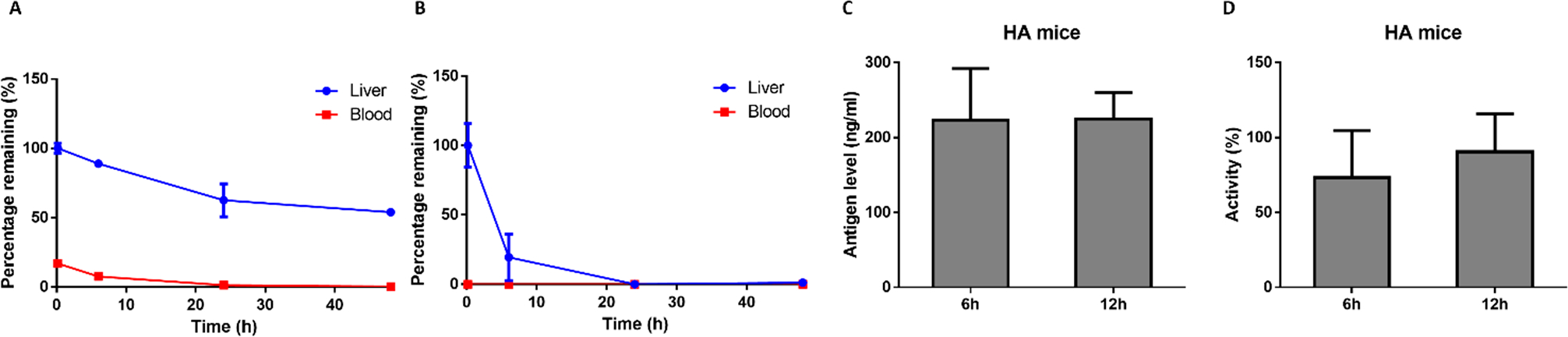
Biodegradability of (A) FTT5 and (B) FTT9 in vivo; (C) the level and (D) activity of hFIX in hemophilia A mice after i.v. injection of FTT5-hFIX mRNA.2 Reproduced with permission from ref 2. Copyright 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. Distributed under a Creative Commons Attribution License 4.0 (CC BY); https://creativecommons.org/licenses/by/4.0/.
Additionally, we applied FTT5 LLNs to deliver an adenine base editor mRNA (~5.5k nucleotides) and an sgRNA targeting proprotein convertase subtilisin/kexin type 9 (PCSK9). The PCSK9 protein binds to low-density lipoprotein receptors and plays a significant role in regulating blood cholesterol level.61 The base editing efficiency of PCSK9 was potent at low doses. These FTT lipid-like molecules further increased the delivery efficiency of TT3, which may lead to potential clinical applications.
4.2. Infectious Diseases
Infectious diseases are severe threats of human health. mRNA-based vaccines and therapeutics have emerged in fighting multiple types of infectious diseases. For example, SARS-CoV-2 virus led to the worldwide COVID-19 pandemic. Pfizer-BioNTech and Moderna have successfully constructed ionizable lipid–mRNA formulations as COVID-19 vaccines. Similarly, we prepared TT3 LLNs loaded with NASAR mRNA encoding SARS-CoV-2 antigens (Figure 2A). After the in vitro screening, these TT3-NASAR LLNs were intramuscularly injected into mice and induced more than 70-fold higher luciferase intensity than that of DLin-MC3-DMA LNPs. Next, the mice were vaccinated with a prime at day 0 and a boost at day 14. The TT3-mRNA LLNs vaccine produced more than 300-fold higher anti-S1 antibodies compared to the DLin-MC3-DMA LNP vaccine on day 30 (Figure 2B). Also, over 5-fold antigen-specific antibodies were detected by intramuscular injection than by subcutaneous injection. Lastly, we observed dose-dependent vaccination effects at the doses from 0.012 to 12 μg (Figure 2C).
Figure 2.
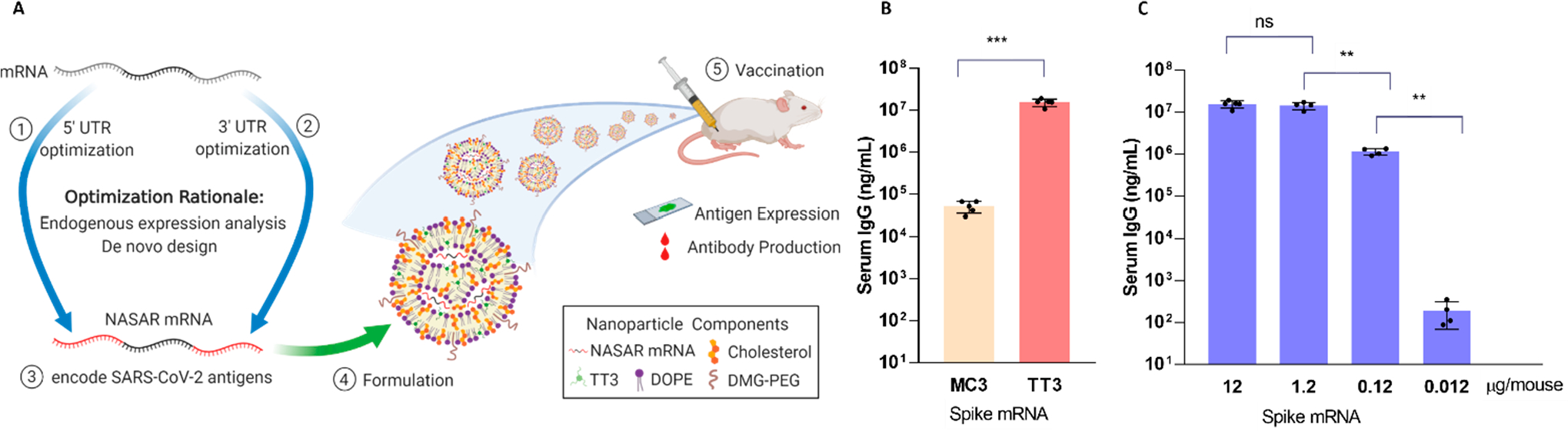
(A) Illustration of mRNA UTR engineering and its application to COVID-19 mRNA vaccines. (B) Serum IgG level in mouse sera post-intramuscular injection. (C) Dosage dependency effects of TT3-NASAR mRNA formulation as a COVID-19 mRNA vaccine.3 Reproduced with permission from ref 3. Copyright 2020 Wiley-VCH.
Lipid–mRNA nanoparticle formulations may also be applied to treat bacterial infections. For instance, sepsis can be caused by many different bacteria.62 To address this deadly disease, we developed mRNAs encoding antimicrobial peptide (AMP), cathepsin B (Cat B), and a Cat B sensitive linker. Then, we encapsulated this mRNA in vitamin C-derived LNPs to enable the adoptive transfer of macrophages, boost the immunocytes, and clear multidrug-resistant bacteria infections (Figure 3A).
Figure 3.
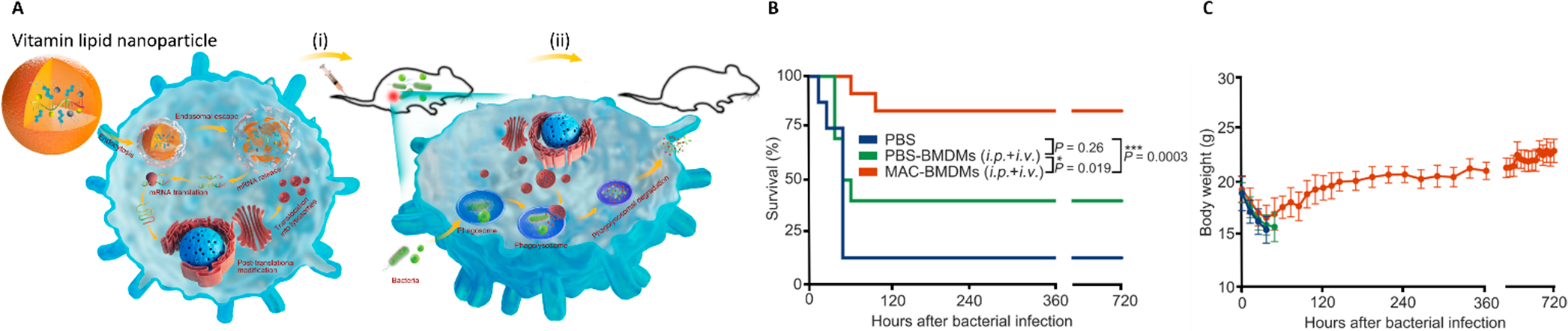
(A) Illustration of macrophages containing antimicrobial peptides linked to cathepsin B in the lysosomes (MACs) for the treatment of multidrug resistant bacterial sepsis: (i) adoptive macrophage transfer; (ii) recovery from multidrug resistant bacteria induced sepsis. (B) Survival rate and (C) body weights of septic mice after treatments.4 Adapted with permission from ref 4. Copyright 2020 Springer Nature.
To study the activity of these engineered macrophages, the intracellular survival of multidrug resistant Staphylococcus aureus in RAW264.7 cells was measured. The macrophages containing antimicrobial peptides linked to cathepsin B in the lysosomes (MAC-RAWs) induced by AMP-CatB VcLNPs showed the strongest bactericidal activity compared to the control groups of free mRNAs, empty VcLNPs, and AMP-CatB inhibitor VcLNPs. To investigate the therapeutic effects in vivo, MAC-RAWs were injected to MDRSA-induced sepsis mice. Colony forming units (CFUs) in mouse blood were reduced almost 2-fold by MAC-RAWs compared to control groups. Furthermore, primary bone marrow-derived macrophages (BMDMs) were used to evaluate the therapeutic activity. These macrophages (MAC-BMDMs) induced by AMP-CatB VcLNPs showed more effective delivery than other groups, which had similar results as in the RAW264.7 cells. For in vivo studies, MAC-BMDMs showed 58% of mice survival from sepsis compared to 10% survival in control groups (Figure 3B). After 30 days, the levels of the body weight (BW), white blood cells (WBCs), and lymphocytes (LYMs) of the seven surviving mice returned to normal and were fully restored (Figure 3C). There were no bacteria detected in major organs of these treated mice. To study the biodistribution of the macrophages, 6 h post-intraperitoneal and -intravenous injections of the FLuc-BMDMs, the biodistribution luminescence signals were measured and ranked in the order of peritoneal cavity, spleen, liver, lungs, kidneys, heart, and blood.
4.3. Cancers
Triple-negative breast cancer (TNBC) is a type of breast cancer with an insufficient level of estrogen receptors (ERs), progesterone receptors (PRs), and human epidermal growth factor receptors (HER2s).63 Prior studies reported that the alteration of tumor suppressor gene TP53 was at a high level in certain TNBC patients.64 Currently, chemotherapy drugs are one of the key approaches for TNBC treatment. Thus, we developed a new treatment approach for TNBC through the combination of chemotherapy drugs and tumor suppressor p53 mRNA. In this work, paclitaxel and camptothecin were coupled to ionizable amino lipids to synthesize PAL lipid and CAL lipid, respectively. These PAL and CAL lipids were formulated with p53 mRNA to be assembled into the lipid nanoparticles. After in vitro screening and orthogonal optimization, PAL-p53 LNPs were i.v. injected to nude mice with orthotopic TNBC; the PAL-p53 LNPs showed significantly higher antitumor effects compared to control groups of the free drug, free p53 mRNA, and PAL-control mRNA LNPs. The combination of chemotherapy and LNP–mRNA-mediated gene therapy provides a favorable synergistic effect for the treatment of TNBC.
Immunotherapy has significantly advanced the treatment against various cancer types, many of which are under preclinical and clinical studies.65 In our studies, we proposed to stimulate the interactions between costimulatory receptors and their ligands in tumor environments. For example, CD137 and OX40 are glycoproteins that can act as costimulatory receptors on T cells, which are involved in the activation and proliferation of CD8 and CD4 T cells.66 However, the level of these receptors are relatively low on tumor-infiltrating lymphocytes. To address this issue, we applied the phospholipids mentioned in Section 1 to encapsulate mRNAs encoding OX40. As PL1 was selected as the lead compound based on cell screening, we encapsulated mRNAs into PL1 lipid nanoparticles for in vivo studies. After intratumoral (i.t.) injection into mice with A20 tumors, PL1-OX40 LNPs were accumulated at tumor sites. Compared to the control groups, the tumor growth rate was dramatically inhibited using PL1-OX40 LNPs (Figure 4A). Six out of 10 mice showed a complete response to the treatment of PL1-OX40 LNPs (Figure 4B). More importantly, these mice that survived were completely resistant to the A20 tumor rechallenge (Figure 4C). Our results indicated that the integration of biomimetic lipid nanoparticles loaded with costimulatory receptor mRNA together with their corresponding agonistic antibodies can greatly enhance the immunotherapeutic effects in multiple cancer types.50
Figure 4.
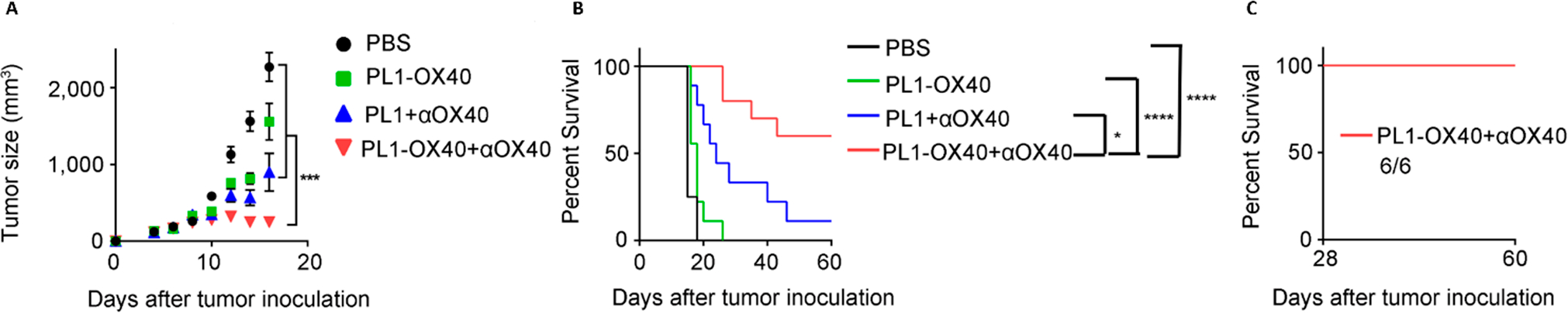
Therapeutic effects in an A20 mouse tumor model: (A) tumor size; (B) percent survival; (C) percent survival of rechallenged mice.
5. CONCLUSIONS AND PERSPECTIVES
Lipid nanoparticle–mRNA formulations have been administered in hundreds of millions of people worldwide as COVID-19 vaccines, which paves the way for developing more and more lipid–mRNA-based therapeutics. By employing diverse synthetic strategies, we prepared many classes of ionizable lipids including lipid-like molecules, biodegradable lipids, chemotherapy drug-derived amino lipids, biomimetic phospholipids and glycolipids, and vitamin-derived ionizable lipids that can efficiently deliver mRNAs in vitro and in vivo when formulated into lipid nanoparticles. Meanwhile, the chemical modification and the sequence engineering of mRNA significantly improve the mRNA stability and translation. In a variety of diseases models, TT3 LLNs were able to effectively deliver hFIX mRNA to treat hemophillia B and mediate the delivery of mRNA encoding SARS-CoV-2 antigens as an mRNA vaccine against COVID-19. FTT5 LLNs induced potent base editing activity at low doses in mice. Vitamin C-derived LNPs loaded with antimicrobial mRNA can engineer macrophages to treat multidrug resistant bacteria-induced sepsis. Biomimetic phospholipid nanoparticles encapsulating chemically-modified OX40 mRNAs together with corresponding antibodies were demonstrated as a promising strategy for cancer immunotherapy. With the advances of lipid structures and mRNA engineering, lipid nanoparticle formulated mRNA can be specifically delivered to many organ and cell types to treat various diseases. Several aspects of lipid nanoparticle–mRNA formulations can be further investigated in future studies. (i) Lipids and lipid derivatives with novel structures can be conceived on the basis of diverse design strategies. For example, we have incorporated biomimetic, bioinspired, and bioactive components into the molecule design.24 (ii) Biodegradability and biocompatibility are critical aspects for the translation of LNPs to the clinic. Lipids with proper biodegradability can minimize possible side effects and improve the safety of LNPs.67,68 (iii) The formulation of LNPs can be carefully tuned to include certain components or targeting ligands, which may enhance delivery efficiency or increase targeting specificity. Also, biomaterials other than LNPs have been developed for mRNA delivery, such as polymers and extracellular vesicles, which can be greatly expanded.69–74 (iv) The chemistry, sequence, and structure of mRNAs need to be systematically examined to improve the pharmaceutical properties of mRNAs. Prior studies found that many types of chemical modifications of mRNA affected the mRNA translation, and the related mechanisms are not fully understood.10 (v) The lipid–mRNA formulations have entered clinical trials for preventing or treating infectious diseases, genetic disorders, and cancer. Many other indications may also benefit from this new class of medicine.18 With the collaborative efforts from academia and industry, lipid nanoparticle–mRNA formulations will make incredible contributions to human health in the future.
ACKNOWLEDGMENTS
Y.D. acknowledges the support of the Early Career Investigator Award from the Bayer Hemophilia Awards Program, Research Awards from the National PKU Alliance, New Investigator Grant from the AAPS Foundation, Maximizing Investigators’ Research Award R35GM119679 from the National Institute of General Medical Sciences, and the R01HL136652 grant from the National Heart, Lung, and Blood Institute as well as the start-up fund from the College of Pharmacy at The Ohio State University.
Biographies
Chang Wang is a postdoctoral associate in the College of Pharmacy at The Ohio State University. He received his B.Sc. in Pharmacy from Tianjin University, China, and his Ph.D. in Chemistry from Lehigh University, PA, where he investigated fundamental and translational studies of lipids and membranes. Currently, under the mentorship of Dr. Yizhou Dong at The Ohio State University, he is devoted to developing multifunctional and biodegradable lipid-like molecules for mRNA delivery and genome editing.
Yuebao Zhang received his B.S. degree in pharmaceutical science from Zhengzhou University in 2011. In 2017, he received his Ph.D. degree in medicinal chemistry from Sichuan University under the supervision of Professor Zhenlei Song. He is currently a postdoctoral research associate in the lab of Dr. Yizhou Dong at The Ohio State University. His current research focuses on the development of novel ionizable lipids and evaluation of lipids for their application in mRNA delivery.
Yizhou Dong is an Associate Professor in the Division of Pharmaceutics and Pharmacology, College of Pharmacy at The Ohio State University. He received his B.S. in pharmaceutical sciences from Peking University, Health Science Center, and M.S. in organic chemistry from Shanghai Institute of Organic Chemistry. In 2009, he received his Ph.D. degree in pharmaceutical sciences from the University of North Carolina at Chapel Hill (UNC-CH) under the supervision of Professor K.-H. Lee. From 2010 to 2014, he was a postdoctoral fellow in the laboratory of Professors Robert Langer and Daniel Anderson at Harvard Medical School and Massachusetts Institute of Technology. His research focuses on the design and development of nanomaterial and biotechnology platforms for the treatment of genetic disorders, infectious diseases, and cancers. Several of his inventions have been licensed and are currently under development as drug candidates for clinical trials. He serves as a board member of the Gene Delivery and Editing (GDE) Focus Group of the Controlled Release Society and an editorial board member for Bioactive Materials. He is a scientific advisory board member of Oncorus, Inc.
Footnotes
The authors declare the following competing financial interest(s): C.W. and Y.Z. declare no competing interests. Y.D. is a scientific advisory board member of Oncorus, Inc.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.accounts.1c00550
Contributor Information
Chang Wang, Division of Pharmaceutics & Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States.
Yuebao Zhang, Division of Pharmaceutics & Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States.
Yizhou Dong, Division of Pharmaceutics & Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States; Department of Biomedical Engineering, The Center for Clinical and Translational Science, The Comprehensive Cancer Center, Dorothy M. Davis Heart & Lung Research Institute, Department of Radiation Oncology, The Ohio State University, Columbus, Ohio 43210, United States.
REFERENCES
- (1). Li B; Luo X; Deng B; Wang J; McComb DW; Shi Y; Gaensler KML; Tan X; Dunn AL; Kerlin BA; Dong Y An Orthogonal Array Optimization of Lipid-Like Nanoparticles for mRNA Delivery in Vivo. Nano Lett. 2015, 15, 8099–8107. This paper reported the development of TT3 lipid-like nanoparticles for mRNA delivery in a hemophilia B mouse model.
- (2). Zhang X; Zhao W; Nguyen GN; Zhang C; Zeng C; Yan J; Du S; Hou X; Li W; Jiang J; Deng B; McComb DW; Dorkin R; Shah A; Barrera L; Gregoire F; Singh M; Chen D; Sabatino DE; Dong Y Functionalized Lipid-Like Nanoparticles for in Vivo mRNA Delivery and Base Editing. Sci. Adv. 2020, 6, No. eabc2315. This paper reported the chemical modifications of lipid-like molecules. The lead material FTT5 LLNs effectively delivered human factor VIII mRNA and base editing components in mouse models.
- (3). Zeng C; Hou X; Yan J; Zhang C; Li W; Zhao W; Du S; Dong Y Leveraging mRNA Sequences and Nanoparticles to Deliver SARS-CoV-2 Antigens in Vivo. Adv. Mater. 2020, 32, 2004452. This paper reported the approaches for engineering mRNA UTR and applied the lead UTR for constructing COVID-19 mRNA vaccine candidates.
- (4). Hou X; Zhang X; Zhao W; Zeng C; Deng B; McComb DW; Du S; Zhang C; Li W; Dong Y Vitamin Lipid Nanoparticles Enable Adoptive Macrophage Transfer for the Treatment of Multidrug-Resistant Bacterial Sepsis. Nat. Nanotechnol. 2020, 15, 41–46. This paper reported the development of vitamin C lipid nanoparticle (VcLNP) and applied the VcLNP–mRNA formulation to engineer macrophages for the treatment of multidrug-resistant bacterial sepsis.
- (5).Brenner S; Jacob F; Meselson M An Unstable Intermediate Carrying Information from Genes to Ribosomes for Protein Synthesis. Nature 1961, 190, 576–581. [DOI] [PubMed] [Google Scholar]
- (6).Sahin U; Karikó K; Türeci Ö mRNA-based Therapeutics — Developing a New Class of Drugs. Nat. Rev. Drug Discovery 2014, 13, 759–780. [DOI] [PubMed] [Google Scholar]
- (7).Weissman D; Karikó K mRNA: Fulfilling the Promise of Gene Therapy. Mol. Ther. 2015, 23, 1416–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gros F; Hiatt H; Gilbert W; Kurland CG; Risebrough RW; Watson JD Unstable Ribonucleic Acid Revealed by Pulse Labelling of Escherichia Coli. Nature 1961, 190, 581–585. [DOI] [PubMed] [Google Scholar]
- (9).Russick J; Delignat S; Milanov P; Christophe O; Boros G; Denis CV; Lenting PJ; Kaveri SV; Lacroix-Demazes S Correction of bleeding in experimental severe hemophilia A by systemic delivery of factor VIII-encoding mRNA. Haematologica 2020, 105, 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hajj KA; Whitehead KA Tools for Translation: Non-Viral Materials for Therapeutic mRNA Delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar]
- (11).Whitehead KA; Langer R; Anderson DG Knocking Down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discovery 2009, 8, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Singha K; Namgung R; Kim WJ Polymers in Small-interfering RNA Delivery. Nucleic Acid Ther. 2011, 21, 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Li J; Wu C; Wang W; He Y; Elkayam E; Joshua-Tor L; Hammond PT Structurally Modulated Codelivery of siRNA and Argonaute 2 for Enhanced RNA Interference. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E2696–E2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Love KT; Mahon KP; Levins CG; Whitehead KA; Querbes W; Dorkin JR; Qin J; Cantley W; Qin LL; Racie T; Frank-Kamenetsky M; Yip KN; Alvarez R; Sah DWY; de Fougerolles A; Fitzgerald K; Koteliansky V; Akinc A; Langer R; Anderson DG Lipid-like Materials for Low-dose, in Vivo Gene Silencing. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kaczmarek JC; Patel AK; Rhym LH; Palmiero UC; Bhat B; Heartlein MW; DeRosa F; Anderson DG Systemic Delivery of mRNA and DNA to the Lung Using Polymer-lipid Nanoparticles. Biomaterials 2021, 275, 120966. [DOI] [PubMed] [Google Scholar]
- (16).Zhao W; Zhang C; Li B; Zhang X; Luo X; Zeng C; Li W; Gao M; Dong Y Lipid Polymer Hybrid Nanomaterials for mRNA Delivery. Cell. Mol. Bioeng. 2018, 11, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Islam MA; Xu Y; Tao W; Ubellacker JM; Lim M; Aum D; Lee GY; Zhou K; Zope H; Yu M; Cao W; Oswald JT; Dinarvand M; Mahmoudi M; Langer R; Kantoff PW; Farokhzad OC; Zetter BR; Shi J Restoration of Tumour-Growth Suppression in Vivo via Systemic Nanoparticle-Mediated Delivery of PTEN mRNA. Nat. Biomed. Eng. 2018, 2, 850–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hou X; Zaks T; Langer R; Dong Y Lipid Nanoparticles for mRNA Delivery. Nat. Rev. Mater. 2021, 3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang C; Krause MR; Regen SL Push and Pull Forces in Lipid Raft Formation: The Push Can Be as Important as the Pull. J. Am. Chem. Soc. 2015, 137, 664–666. [DOI] [PubMed] [Google Scholar]
- (20).van Meer G; Voelker DR; Feigenson GW Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pattni BS; Chupin VV; Torchilin VP New Developments in Liposomal Drug Delivery. Chem. Rev. 2015, 115, 10938–10966. [DOI] [PubMed] [Google Scholar]
- (22).Felgner PL; Gadek TR; Holm M; Roman R; Chan HW; Wenz M; Northrop JP; Ringold GM; Danielsen M Lipofection: A Highly Efficient, Lipid-Mediated DNA-Transfection Procedure. Proc. Natl. Acad. Sci. U. S. A. 1987, 84, 7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cullis PR; Hope MJ Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhang Y; Sun C; Wang C; Jankovic KE; Dong Y Lipids and Lipid Derivatives for RNA Delivery. Chem. Rev. 2021, 121, 12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Carrasco MJ; Alishetty S; Alameh M-G; Said H; Wright L; Paige M; Soliman O; Weissman D; Cleveland TE; Grishaev A; Buschmann MD Ionization and Structural Properties of mRNA Lipid Nanoparticles Influence Expression in Intramuscular and Intravascular Administration. Commun. Biol. 2021, 4, 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Garber K Alnylam Launches Era of RNAi Drugs. Nat. Biotechnol. 2018, 36, 777–778. [DOI] [PubMed] [Google Scholar]
- (27).Polack FP; Thomas SJ; Kitchin N; Absalon J; Gurtman A; Lockhart S; Perez JL; Pérez Marc G; Moreira ED; Zerbini C; Bailey R; Swanson KA; Roychoudhury S; Koury K; Li P; Kalina WV; Cooper D; Frenck RW; Hammitt LL; Türeci Ö; Nell H; Schaefer A; Ünal S; Tresnan DB; Mather S; Dormitzer PR; Şahin U; Jansen KU; Gruber WC Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Baden LR; El Sahly HM; Essink B; Kotloff K; Frey S; Novak R; Diemert D; Spector SA; Rouphael N; Creech CB; McGettigan J; Khetan S; Segall N; Solis J; Brosz A; Fierro C; Schwartz H; Neuzil K; Corey L; Gilbert P; Janes H; Follmann D; Marovich M; Mascola J; Polakowski L; Ledgerwood J; Graham BS; Bennett H; Pajon R; Knightly C; Leav B; Deng W; Zhou H; Han S; Ivarsson M; Miller J; Zaks T Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zeng C; Hou X; Bohmer M; Dong Y Advances of Nanomaterials-based Strategies for Fighting Against COVID-19. VIEW 2021, 2, 20200180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Li B; Luo X; Dong Y Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjugate Chem. 2016, 27, 849–853. [DOI] [PubMed] [Google Scholar]
- (31).McCown PJ; Ruszkowska A; Kunkler CN; Breger K; Hulewicz JP; Wang MC; Springer NA; Brown JA Naturally Occurring Modified Ribonucleosides. Wiley Interdiscip. Rev.: RNA 2020, 11, No. e1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Corey DR Chemical Modification: the Key to Clinical Application of RNA Interference? J. Clin. Invest. 2007, 117, 3615–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wadhwa A; Aljabbari A; Lokras A; Foged C; Thakur A Opportunities and Challenges in the Delivery of mRNA-Based Vaccines. Pharmaceutics 2020, 12, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Akinc A; Zumbuehl A; Goldberg M; Leshchiner ES; Busini V; Hossain N; Bacallado SA; Nguyen DN; Fuller J; Alvarez R; Borodovsky A; Borland T; Constien R; de Fougerolles A; Dorkin JR; Narayanannair Jayaprakash K; Jayaraman M; John M; Koteliansky V; Manoharan M; Nechev L; Qin J; Racie T; Raitcheva D; Rajeev KG; Sah DWY; Soutschek J; Toudjarska I; Vornlocher H-P; Zimmermann TS; Langer R; Anderson DG A Combinatorial Library of Lipid-like Materials for Delivery of RNAi Therapeutics. Nat. Biotechnol. 2008, 26, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Dong Y; Love KT; Dorkin JR; Sirirungruang S; Zhang Y; Chen D; Bogorad RL; Yin H; Chen Y; Vegas AJ; Alabi CA; Sahay G; Olejnik KT; Wang W; Schroeder A; Lytton-Jean AKR; Siegwart DJ; Akinc A; Barnes C; Barros SA; Carioto M; Fitzgerald K; Hettinger J; Kumar V; Novobrantseva TI; Qin J; Querbes W; Koteliansky V; Langer R; Anderson DG Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cheng Q; Wei T; Jia Y; Farbiak L; Zhou K; Zhang S; Wei Y; Zhu H; Siegwart DJ Dendrimer-Based Lipid Nanoparticles Deliver Therapeutic FAH mRNA to Normalize Liver Function and Extend Survival in a Mouse Model of Hepatorenal Tyrosinemia Type I. Adv. Mater. 2018, 30, 1805308. [DOI] [PubMed] [Google Scholar]
- (37).Hajj KA; Ball RL; Deluty SB; Singh SR; Strelkova D; Knapp CM; Whitehead KA Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH. Small 2019, 15, 1805097. [DOI] [PubMed] [Google Scholar]
- (38).Liu J; Chang J; Jiang Y; Meng X; Sun T; Mao L; Xu Q; Wang M Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv. Mater. 2019, 31, 1902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Reichmuth AM; Oberli MA; Jaklenec A; Langer R; Blankschtein D mRNA Vaccine Delivery Using Lipid Nanoparticles. Ther. Delivery 2016, 7, 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhao P; Hou X; Yan J; Du S; Xue Y; Li W; Xiang G; Dong Y Long-term Storage of Lipid-like Nanoparticles for mRNA Delivery. Bioact. Mater. 2020, 5, 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Zhang X; Li B; Luo X; Zhao W; Jiang J; Zhang C; Gao M; Chen X; Dong Y Biodegradable Amino-Ester Nanomaterials for Cas9 mRNA Delivery in Vitro and in Vivo. ACS Appl. Mater. Interfaces 2017, 9, 25481–25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Desigaux L; Sainlos M; Lambert O; Chevre R; Letrou-Bonneval E; Vigneron J-P; Lehn P; Lehn J-M; Pitard B Self-Assembled Lamellar Complexes of siRNA with Lipidic Aminoglycoside Derivatives Promote Efficient siRNA Delivery and Interference. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 16534–16539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Li Y; Lee RJ; Yu K; Bi Y; Qi Y; Sun Y; Li Y; Xie J; Teng L Delivery of siRNA Using lipid Nanoparticles Modified with Cell Penetrating Peptide. ACS Appl. Mater. Interfaces 2016, 8, 26613–26621. [DOI] [PubMed] [Google Scholar]
- (44).van der Meel R; Chen S; Zaifman J; Kulkarni JA; Zhang XRS; Tam YK; Bally MB; Schiffelers RM; Ciufolini MA; Cullis PR; Tam YYC Modular Lipid Nanoparticle Platform Technology for siRNA and Lipophilic Prodrug Delivery. Small 2021, 17, 2103025. [DOI] [PubMed] [Google Scholar]
- (45).Semba RD The Discovery of the Vitamins. Int. J. Vitam. Nutr. Res. 2012, 82, 310–315. [DOI] [PubMed] [Google Scholar]
- (46).Wang Y-Q; Ji M-Y; Wang C Endoplasmic Reticulum Targeted Glutathione and pH Dual Responsive Vitamin Lipid Nanovesicles for Tocopheryl DM1 Delivery and Cancer Therapy. Int. J. Pharm. 2020, 582, 119331. [DOI] [PubMed] [Google Scholar]
- (47).Zhang C; Zhang X; Zhao W; Zeng C; Li W; Li B; Luo X; Li J; Jiang J; Deng B; McComb DW; Dong Y Chemotherapy Drugs Derived Nanoparticles Encapsulating mRNA Encoding Tumor Suppressor Proteins to Treat Triple-Negative Breast Cancer. Nano Res. 2019, 12, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Dong Y; Zhang X Biomimetic Nanomaterials and Uses Thereof. WO 2019027999, July 2, 2019. [Google Scholar]
- (49).Ariga T; McDonald MP; Yu RK Role of Ganglioside Metabolism in the Pathogenesis of Alzheimer’s Disease. J. Lipid Res. 2008, 49, 1157–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Dong Y; Li W; Zhang C Combination Immunoregulation and Uses Thereof. WO 2020198338, October 1, 2020. [Google Scholar]
- (51).Karikó K; Buckstein M; Ni H; Weissman D Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [DOI] [PubMed] [Google Scholar]
- (52).Karikó K; Muramatsu H; Welsh FA; Ludwig J; Kato H; Akira S; Weissman D Incorporation of Pseudouridine into mRNA Yields Superior Nonimmunogenic Vector with Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Karikó K; Muramatsu H; Keller JM; Weissman D Increased Erythropoiesis in Mice Injected With Submicrogram Quantities of Pseudouridine-containing mRNA Encoding Erythropoietin. Mol. Ther. 2012, 20, 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kawaguchi D; Kodama A; Abe N; Takebuchi K; Hashiya F; Tomoike F; Nakamoto K; Kimura Y; Shimizu Y; Abe H Phosphorothioate Modification of mRNA Accelerates the Rate of Translation Initiation to Provide More Efficient Protein Synthesis. Angew. Chem., Int. Ed. 2020, 59, 17403–17407. [DOI] [PubMed] [Google Scholar]
- (55).Makarova KS; Haft DH; Barrangou R; Brouns SJJ; Charpentier E; Horvath P; Moineau S; Mojica FJM; Wolf YI; Yakunin AF; van der Oost J; Koonin EV Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Li B; Zhao W; Luo X; Zhang X; Li C; Zeng C; Dong Y Engineering CRISPR-Cpf1 crRNAs and mRNAs to Maximize Genome Editing Efficiency. Nat. Biomed. Eng. 2017, 1, 0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Li B; Zeng C; Dong Y Design and Assessment of Engineered CRISPR-Cpf1 and its Use for Genome Editing. Nat. Protoc. 2018, 13, 899–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Babendure JR; Babendure JL; Ding JH; Tsien RY Control of Mammalian Translation by mRNA Structure Near Caps. RNA 2006, 12, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Zhao W; Hou X; Vick OG; Dong Y RNA Delivery Biomaterials for the Treatment of Genetic and Rare Diseases. Biomaterials 2019, 217, 119291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Coppola A; Di Capua M; Di Minno MND; Di Palo M; Marrone E; Ierano P; Arturo C; Tufano A; Cerbone AM Treatment of Hemophilia: A Review of Current Advances and Ongoing Issues. J. Blood Med. 2010, 1, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Horton JD; Cohen JC; Hobbs HH Molecular Biology of PCSK9: its Role In LdL Metabolism. Trends Biochem. Sci. 2007, 32, 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Reinhart K; Daniels R; Kissoon N; Machado FR; Schachter RD; Finfer S Recognizing Sepsis as a Global Health Priority — A WHO Resolution. N. Engl. J. Med. 2017, 377, 414–417. [DOI] [PubMed] [Google Scholar]
- (63).Denkert C; Liedtke C; Tutt A; von Minckwitz G Molecular Alterations in Triple-negative Breast Cancer-the Road to New Treatment Strategies. Lancet 2017, 389, 2430–2442. [DOI] [PubMed] [Google Scholar]
- (64).The Cancer Genome Atlas Network. Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).O’Donnell JS; Teng MWL; Smyth MJ Cancer Immunoediting and Resistance to T Cell-based Immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [DOI] [PubMed] [Google Scholar]
- (66).Mahoney KM; Rennert PD; Freeman GJ Combination Cancer Immunotherapy and New Immunomodulatory Targets. Nat. Rev. Drug Discovery 2015, 14, 561–584. [DOI] [PubMed] [Google Scholar]
- (67).Miao L; Lin J; Huang Y; Li L; Delcassian D; Ge Y; Shi Y; Anderson DG Synergistic Lipid Compositions for Albumin Receptor Mediated Delivery of mRNA to the Liver. Nat. Commun. 2020, 11, 2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Miao L; Zhang Y; Huang L mRNA Vaccine for Cancer Immunotherapy. Mol. Cancer 2021, 20, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Cheng Q; Wei T; Farbiak L; Johnson LT; Dilliard SA; Siegwart DJ Selective Organ Targeting (SORT) Nanoparticles for Tissue-specific mRNA Delivery and CRISPR-Cas Gene Editing. Nat. Nanotechnol. 2020, 15, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Riley RS; Kashyap MV; Billingsley MM; White B; Alameh M-G; Bose SK; Zoltick PW; Li H; Zhang R; Cheng AY; Weissman D; Peranteau WH; Mitchell MJ Ionizable Lipid Nanoparticles for in Utero mRNA Delivery. Sci. Adv. 2021, 7, No. eaba1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ma F; Yang L; Sun Z; Chen J; Rui X; Glass Z; Xu Q Neurotransmitter-derived Lipidoids (NT-Lipidoids) for Enhanced Brain Delivery Through Intravenous Injection. Sci. Adv. 2020, 6, No. eabb4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Lokugamage MP; Vanover D; Beyersdorf J; Hatit MZC; Rotolo L; Echeverri ES; Peck HE; Ni H; Yoon J-K; Kim Y; Santangelo PJ; Dahlman JE Optimization of Lipid Nanoparticles for the Delivery of Nebulized Therapeutic mRNA to the Lungs. Nat. Biomed. Eng. 2021, 5, 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Patel S; Ashwanikumar N; Robinson E; Xia Y; Mihai C; Griffith JP; Hou S; Esposito AA; Ketova T; Welsher K; Joyal JL; Almarsson Ö; Sahay G Naturally-Occurring Cholesterol Analogues in Lipid Nanoparticles Induce Polymorphic Shape and Enhance Intracellular Delivery of mRNA. Nat. Commun. 2020, 11, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Islam MA; Rice J; Reesor E; Zope H; Tao W; Lim M; Ding J; Chen Y; Aduluso D; Zetter BR; Farokhzad OC; Shi J Adjuvant-Pulsed mRNA Vaccine Nanoparticle for Immunoprophylactic and Therapeutic Tumor Suppression in Mice. Biomaterials 2021, 266, 120431. [DOI] [PMC free article] [PubMed] [Google Scholar]


