Abstract
Background
We sought to use deep learning to extract anatomic features from postnatal kidney ultrasounds and evaluate their performance in predicting the risk and timing of chronic kidney disease (CKD) progression for boys with posterior urethral valves (PUV). We hypothesized that these features would predict CKD progression better than clinical characteristics such as nadir creatinine alone.
Methods
We performed a retrospective cohort study of boys with PUV treated at two pediatric health systems from 1990 to 2021. Features of kidneys were extracted from initial postnatal kidney ultrasound images using a deep learning model. Three time-to-event prediction models were built using random survival forests. The Imaging Model included deep learning imaging features, the Clinical Model included clinical data, and the Ensemble Model combined imaging features and clinical data. Separate models were built to include time-dependent clinical data that were available at 6 months, 1 year, 3 years, and 5 years.
Results
Two-hundred and twenty-five patients were included in the analysis. All models performed well with C-indices of 0.7 or greater. The Clinical Model outperformed the Imaging Model at all time points with nadir creatinine driving the performance of the Clinical Model. Combining the 6-month Imaging Model (C-index 0.7; 95% confidence interval [CI] 0.6, 0.79) with the 6-month Clinical Model (C-index 0.79; 95% CI 0.71, 0.86) resulted in a 6-month Ensemble Model that performed better (C-index 0.82; 95% CI 0.77, 0.88) than either model alone.
Conclusions
Deep learning imaging features extracted from initial postnatal kidney ultrasounds may improve early prediction of CKD progression among children with PUV.
Keywords: Posterior urethral valves, Chronic kidney disease, Machine learning, Kidney ultrasound
Graphical Abstract
Introduction
Posterior urethral valves (PUV) are a congenital obstruction of the urinary tract caused by leaflets of tissue in the posterior urethra of boys [1]. These valves cause urinary obstruction in utero, which can cause kidney failure, pulmonary hypoplasia, and mortality in infancy [1]. PUV are detected in approximately 1/1250 ultrasound screenings [2, 3].
In boys with PUV, kidney injury is often established at birth. Some children, however, maintain preserved kidney function into adulthood, while others progress to kidney failure in childhood [4-7]. Our ability to effectively implement therapies to slow chronic kidney disease (CKD) progression is limited by our lack of understanding of which patients are at greatest risk for CKD progression and therefore would be most likely to benefit from early intervention such as intermittent urethral catheterization and closer surveillance [4, 8, 9]. Additionally, this uncertainty hinders physicians from accurately counseling families about their child’s expected clinical course at the time of birth. Currently, predicting future risk of CKD progression relies heavily on nadir creatinine during the first year of life [10-12]. A serum creatinine of less than 0.8 mg/dL has been associated with minimal risk of CKD progression, and a value greater than 1.2 mg/dL at 1 year of age has been associated with a higher risk for developing kidney failure [11, 13]. However, the clinical utility of nadir creatinine is limited by its lack of availability at birth. Recently, kidney features obtained from ultrasound have been shown to be associated with CKD progression. Our group and others have reported that low kidney parenchymal area measured on postnatal ultrasound is associated with CKD progression among boys with PUV [14-17]. Limitations to kidney parenchymal area include that it is laborious to measure and does not use all the information contained in the ultrasound images.
Our goal was to use deep learning to automatically extract features of kidneys from ultrasound images and to determine the performance of these features in predicting CKD progression among children with PUV [18]. Our secondary objective was to compare our deep learning models to models built using clinical data alone. We hypothesized that these features would predict CKD progression better than clinical characteristics such as nadir creatinine alone.
Methods
Study design and population
Following institutional research board approval, we performed a retrospective cohort study at the Children’s Hospital of Philadelphia (CHOP) and the Hospital for Sick Children (HSC). The electronic medical records at both institutions were queried to identify all patients with a history of PUV treated at the respective institutions between 1990 and 2021. The ICD-9 code 753.6 and the ICD-10 code Q64.2 were used to identify potential patients with PUV. Manual review of each patient’s chart was performed to verify the diagnosis. All patients with a history of PUV who presented to our respective institutions in the first year of life and had their diagnosis verified via cystoscopy were included in the analysis. Data from both institutions were recorded in a single Research Electronic Data Capture (REDCap) database [19].
Outcome
The outcome of interest, CKD progression, was defined as developing kidney failure (receiving dialysis or kidney transplant) or decline more than 50% in eGFR. eGFR was calculated using the bedside Schwartz equation. eGFR was only calculated in instances when both creatinine and height were recorded in the electronic medical record within 6 months of each other. In those instances, the height measurement in nearest temporal proximity to the creatinine value was used to calculate eGFR. Only outpatient creatinine values obtained when patients were at their baseline health status were used for eGFR calculations.
Exposures
Imaging features
The first available postnatal kidney ultrasound images were obtained for all patients. All ultrasounds were performed prior to any intervention (ablation or diversion). Kidney ultrasound images from HSC were transferred to CHOP, and imaging analysis was performed in CHOP’S secure bioinformatics environment. Only patients with images available prior to day 90 of life were included in the analysis. All ultrasound images were resized to have the same size of 224×224 with cropping, and each image’s intensity values were normalized to (0, 255). For each patient, transverse and sagittal kidney ultrasound images were identified using Python 3.7.0 Pytesseract optical character recognition to identify existing orientation labels, and results were manually verified for accuracy. Python 3.7.0 Pytesseract optical recognition was used to classify bladders as decompressed or full at the time of ultrasound. We included ultrasounds performed at our institutions as well as ultrasounds performed at outside institutions. As a result, the ultrasound images were obtained using a wide variety of ultrasound machines and transducers.
Clinical variables
Clinical data were aggregated longitudinally within distinct 6-month time periods beginning with each patient’s initial presentation to CHOP or HSC until they reached the outcome or their last date of follow-up. Age at time of valve ablation or urinary diversion, history of vesicostomy creation, history clean intermittent catheterization and/or catheterizable channel creation, anticholinergic use within each distinct 6-month window, and the presence of a concomitant genetic abnormality were obtained from the medical records. The initial postnatal voiding cystourethrogram was analyzed for the presence of vesicoureteral reflux. The lowest creatinine measurement in the first year of life was designated as their nadir creatinine. Nadir creatinine and age at time of ablation were treated as continuous variables, while the remaining variables were treated as binary. Patients were followed until the date they reached the outcome or were censored at their last known clinical encounter.
Machine learning analysis
The deep learning method was implemented using Python 3.7.0 and TensorFlow 2.34. Deep learning imaging features were extracted by a pretrained ResNet-50 deep learning model [20]. We applied the pretrained ResNet-50 model to all available transverse and sagittal kidney ultrasound images from each patient. Deep learning imaging features were extracted from the last layer (avg_pool) of this model. Three base time-to-event prediction models were built. The first model (Imaging Model) used random survival forests on the deep learning imaging features alone. The second model (Clinical Model) used random survival forests on clinical features alone. Clinical features included were history of vesicoureteral reflux, nadir creatinine at 1 year of life, need for intermittent catheterization via urethra or catheterizable channel, prior vesicostomy creation, and prior history of treatment with anticholinergics for at least 6 months at any point in the patient’s life. A time-to-event prediction model combining imaging features and clinical characteristics (Ensemble Model) was then generated by averaging the risk scores of the 2 models (Imaging Model and Clinical Model).
Four models using time-dependent covariates were created for each base model at 6 months, 1 year, 3 years, and 5 years. The time-dependent covariates included intermittent catheterization, anticholinergic use, vesicostomy creation, catheterizable channel creation, and nadir creatinine in the first year of life. For these time-dependent models, only data that were available at the particular time points after birth were included. For example, if a patient has started on intermittent catheterization at age 2 years, that information was only included in the year 3 and year 5 Clinical Model but not in the 6-month and 1-year models because this information would not be available to a clinician until the patient was 2 years old.
The performance of each time-dependent survival model was assessed using concordance index (C-index) and receiver operating curves (ROC) with areas under the curve (AUC) measurements at 2, 5, and 10 years. In order to create ROCs at 2, 5, and 10 years, follow-up was stopped at those points, and predictions for the entire cohort up to those time points were analyzed. Each of the models was evaluated using a stratified 5 K-fold cross validation on the entire data set with an 80/20 training and test split. Each split was the same for all three models.
Based on each model’s prediction results (the risk scores assigned to each subject), a risk threshold was used to stratify patients into high risk (top 20%) and low risk (bottom 80%) groups. The high-risk group was those patients predicted to have CKD progression, while the low-risk group was patients predicted to not reach the outcome. The high-risk group included 20% of our patients because roughly 20% (22.6%) of our patients were observed to have CKD progression. Observed CKD progression over time was compared between these groups using Kaplan–Meier curves and log-rank tests.
Sensitivity analysis
We performed two sensitivity analyses. First, we redefined CKD progression as a decline of ≥ 35% in eGFR or receiving dialysis or kidney transplant. A 35% decline in GFR was chosen as an outcome measure to assess our model’s ability to identify milder CKD progression. Adult CKD studies have shown eGFR decline of 30–40% over a 2- to 3-year period as a reliable surrogate for development of dialysis-dependent kidney failure [21, 22].
Second, we used the chronic kidney disease in children (CKiD) U25 eGFR equation to calculate eGFR. The U25 CKiD GFR equation was analyzed to validate our model using this new GFR calculation that has been purported to be appropriate for our patient population [23, 24].
Results
We identified 537 patients with PUV who were treated at CHOP (n = 370) and HSC (n = 167) during the study period. We included in the analysis the 225 patients who had initial postnatal ultrasound images and a full clinical history including nadir creatinine within the first year of life available. Fifty-one patients (22.6%) had CKD progression (37 patients who developed kidney failure and 14 patients who had a decline in eGFR of 50%) over a median follow-up of 6.1 years (IQR 2.1, 11). Cohort characteristics are summarized in Table 1.
Table 1.
Cohort characteristics
| CHOP cohort | HSC cohort | Entire cohort | Analyzed cohort | |
|---|---|---|---|---|
| N = 370 | N = 167 | N = 537 | N = 225 | |
| Age at presentation, median (IQR), days | 88 (2,962) | 16 (3,142) | 31.5 (2,731.5) | 3 (1, 19) |
| Length of follow-up, median (IQR) years | 6.1 (1.8, 11.1) | 6.32 (2.96, 11.7) | 6.32 (2, 11.2) | 6.1 (2.1, 11) |
| Age at time of initial treatment (ablation or diversion), median (IQR) days | 26 (6,342) | 26.5 (10,120) | 26 (7,221) | 15 (6, 34.75) |
| Vesicoureteral reflux, n (%) | 127 (34.3%) | 88 (52.6%) | 215 (40%) | 118 (52.4%) |
| Vesicostomy creation (%) | 66 (17.8%) | 25 (15%) | 91 (16.9%) | 39 (17.3%) |
| Clean intermittent catheterization, n (%) | 50 (13.5%) | 36 (21.6%) | 86 (16%) | 28 (12.4%) |
| Catheterizable channel (%) | 26 (7%) | 22 (13.2%) | 48 (8.9%) | 23 (10.2%) |
| Anticholinergic use for at least 6 months at any point in follow-up | 120 (32.4%) | 40 (24%) | 160 (29.8%) | 78 (34.7%) |
| Genetic abnormality | 10 (2.7%) | 5 (3%) | 15 (2.8%) | 4 (1.8%) |
| Nadir creatinine available | 145 (39.2%) | 126 (75.4%) | 271 (50.5%) | 225 (100%) |
| Nadir creatinine, median (IQR) mg/dL | 0.3 (0.23, 0.5) | 0.33 (0.25, 0.42) | 0.31 (0.25, 0.5) | 0.33 (0.26, 0.5) |
| Initial postnatal ultrasound available | 267 (72.2%) | 114 (68.3%) | 381 (70.9%) | 225 (100%) |
| Age at time of initial postnatal ultrasound, median (IQR) days | 122 (3.5, 1594) | 15 (4,137) | 43 (4,1007.5) | 6 (1.26) |
| Transplant and/or dialysis | 54 (14.6%) | 24 (14.3%) | 78 (14.5%) | 37 (16.4%) |
| 50% decline in eGFR (calculated using bedside Schwartz equation) | 13 (3.5%) | 12 (7.2%) | 25 (4.6%) | 14 (6.2%) |
Primary analysis
From our cohort of 225 patients, a mean of 7.8 transverse images of the kidney (SD 5.2) and a mean of 6.4 sagittal images (SD 4.8) were available for each patient. All images were static. Forty-eight (21%) of the 225 patients had a fully decompressed bladder at the time of ultrasound.
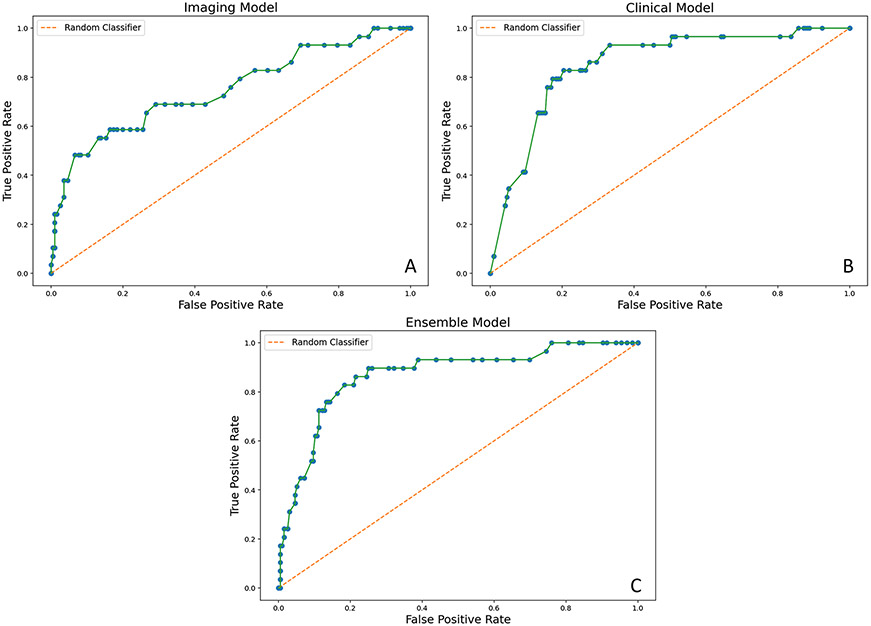
Table 2 reports the C-indices of our 6-month models and AUCs assessing outcomes at 2, 5, and 10 years of follow-up. C-indices and AUCs of our other models can be found in Supplemental Table 1. All models had C-indices of 0.7 or higher. The Clinical Models consistently outperformed the Imaging Models at all time points. Permutation scoring identified nadir creatinine as the strongest contributor to the Clinical Models’ performance. However, the Clinical Model created using only time-dependent covariates that were available at 6 months of life (6-month Clinical Model) performed worse than the Clinical Model built on clinical information available at 1 year and beyond. Combining the 6-month Imaging Model (C-index 0.7; 95% confidence interval [CI] 0.6, 0.79) with the 6-month Clinical Model (C-index 0.79; 95% CI 0.71, 0.86) resulted in a 6-month Ensemble Model that performed better (C-index 0.82; 95% CI 0.77, 0.88) than either model alone. Figure 1 demonstrates the ROC curves of all three models built upon information available at 6 months of life. ROC curves for all other models are available in the supplement.
Table 2.
Performance results of each model
| Model | C-index (95% CI) | AUC 2 years | AUC 5 years | AUC 10 years |
|---|---|---|---|---|
| 6-month Clinical Model | 0.79 (0.71, 0.86) | 0.77 (0.72, 0.87) | 0.82 (0.74, 0.89) | 0.82 (0.73, 0.89) |
| 6-month Imaging Model | 0.70 (0.6, 0.79) | 0.65 (0.58, 0.7) | 0.75 (0.68, 0.84) | 0.65 (0.59, 0.73) |
| 6-month Ensemble Model | 0.82 (0.77, 0.88) | 0.78 (0.73, 0.88) | 0.85 (0.76, 0.9) | 0.82 (0.74, 0.89) |
Fig. 1.
ROC curves illustrating the performance of the 6-month models. A AUC of Imaging Model at 5 years = 0.75 (0.68, 0.84). B AUC of Clinical Model at 5 years = 0.82 (0.74, 0.89). C AUC of Ensemble Model at 5 years = 0.85 (0.76, 0.9)
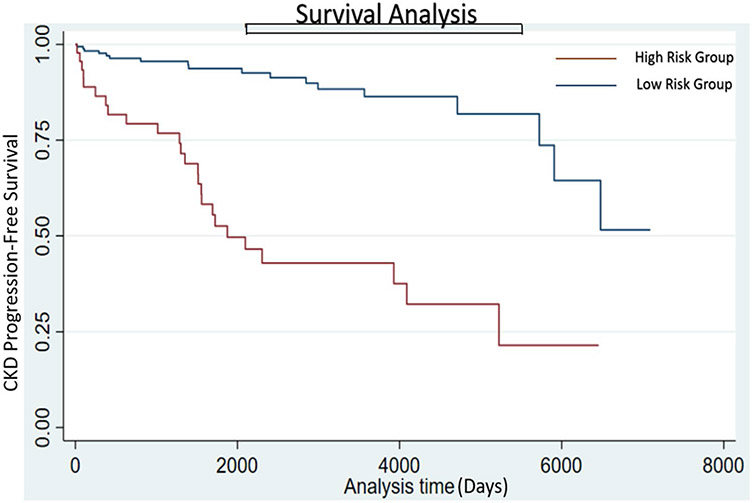
Twenty-seven of 51 (52.9%) patients classified into the high-risk group based on their risk score provided by our 6-month Ensemble Model developed CKD progression, while 19 of the 174 (10.9%) patients in the low-risk group reached the outcome (p-value<0.01) (Fig. 2). Kaplan–Meier curves for risk groups derived from each model can be found in supplemental figures.
Fig. 2.
Kaplan–Meier plots depicting difference in CKD progression in patients classified by the 6-month Ensemble Model as low or high risk of CKD progression. Progression-free survival was longer in the low-risk group compared to the high-risk group. Median time to CKD progression was 1875 (1512, 4089) days in the high-risk group
Sensitivity analyses
We identified 32 patients who had a decline in eGFR of 35%, which increased the number of patients who developed CKD progression from 51 (22.6%) to 69 (30.7%). Using the CKiD U25 equation, we identified 11 patients with a 50% decline in eGFR. Similar to the primary results, the Clinical Model outperformed the Imaging Model at all time points, and combining the 6-month models resulted in an Ensemble Model that outperformed each individual model. Both changing the threshold of eGFR decline to 35%, and using the CKiD U25 equation to calculate eGFR resulted in a decline in the C-index of all 3 models. See complete results in the supplemental table.
Discussion
We found that clinical characteristics and kidney features extracted from initial postnatal ultrasounds were strong predictors of CKD progression for boys with PUV. Deep learning imaging features extracted from initial postnatal ultrasounds performed well on their own (C-index 0.7), but time-varying clinical features alone consistently outperformed the imaging features, with nadir creatinine at 1 year of life the most important feature. At the 6-month time point, we found combining imaging features from first postnatal ultrasound with clinical data had better predictive performance than either clinical characteristics or imaging features alone. These results suggest that features extracted from routine kidney ultrasounds can improve prediction of CKD progression in the immediate postnatal period when additional clinical information is not yet available, thus allowing earlier prediction and months later by increasing the predictive performance of nadir creatinine.
Kidney features obtained from ultrasounds have previously been shown to be associated with future CKD progression. Our group and others have reported that low kidney parenchymal area measured on postnatal ultrasound is associated with higher odds of CKD progression among boys with PUV [17, 25]. Here, we extend upon the evidence base of imaging biomarkers of CKD progression by using deep learning to automatically extract features from postnatal kidney ultrasounds of infants with PUV to predict CKD progression. These deep learning imaging features could serve as novel biomarkers that are easily reproducible, do not require invasive testing, and are potentially available as early as the first day of life. Deep neural network learning (aka deep learning) avoids the need to identify data features of interest prior to analysis. Instead of adopting empirically derived image features (i.e., what clinicians think about from prior experience), deep learning-based methods detect informative features (i.e., see patterns we have not considered). This approach is automated and free of the need for any type of human selection of regions of interest in order to eliminate bias and increase clinical uptake. In addition, this approach makes feature extraction more robust to noise. As a result, the features extracted by our model cannot be equated to familiar clinical parameters such as parenchymal thickness or echogenicity.
We are not the first to report the strong predictive capabilities of nadir creatinine in this population [8, 10, 12]. A serum creatinine of less than 0.4 to 0.8 mg/dL has been associated with a low risk of CKD progression, and value greater than 0.8 to 1.2 mg/dL at 1 year of age has been associated with a higher risk of future kidney failure [11, 13]. Additionally, Coleman et al. reported that a nadir creatinine greater than 0.85 mg/dL at 1 month of life is associated with a higher risk of kidney failure, and values of less than 0.4 mg/dL are associated with a lower risk [26, 27]. Clinical prediction models have recently been published that rely heavily on nadir creatinine. Our group previously reported that CKD progression among patients with PUV could be predicted with fairly high accuracy (C-index 0.77) using clinical characteristics [28]. Vasconcelos et al. similarly reported that the probability of CKD ≥ stage 3 at 10 years of age was estimated at 6%, 40%, and 70% for patients assigned to the low-risk, medium-risk, and high-risk groups, respectively (P < 0.001) [29].
A limitation of nadir creatinine is that it is not available until the patient is 1 year old. Ultrasounds, on the other hand, are available as early as the first day of life. Should these results be confirmed by others and in prospective cohorts, imaging features could help guide family counseling immediately after birth and improve clinical decision-making in the first year of life, such as considering aggressive management (e.g., vesicostomy) for children predicted to have a high risk of CKD progression as predictive performance increases when nadir creatinine becomes available.
Our machine learning model allows clinicians to identify PUV children at highest risk of CKD earlier than any existing tools. Patients can be risk stratified within days to weeks after birth as opposed to months to years. The potential clinical ramifications of having this information available at such an early stage in life cannot be overstated. Currently, our ability to make informed treatment decisions for these patients immediately after birth is compromised by our inability to effectively risk stratify patients in the newborn period. Our machine learning model can augment a physician’s clinical decision-making process in the first month of life.
Our models were less accurate in predicting CKD progression defined as a 35% decline in eGFR. However, the models still performed well and suggest that machine learning of ultrasound images can predict even more subtle changes in eGFR with additional clinical information. In our second sensitivity analysis, using the U25 CKiD equation resulted in 3 patients, who were classified as having a 50% eGFR decline by the bedside Schwartz equation, being reclassified as not having a 50% decline in eGFR. Reclassification of these 3 patients resulted in models that were slightly less accurate at predicting CKD progression. Studies with greater power would be needed to better understand this change in model performance.
There are several limitations to our study. Our study is vulnerable to selection bias and confounding innate to any retrospective study. As a result of our inclusion criteria to only include ultrasounds performed within the first 90 days of life, we have excluded patients with PUV who present later in life. The results of our study cannot be extrapolated to this cohort of patients as their clinical course can be markedly different. In addition, overfitting and generalizability are two known limitations of machine learning. Our relatively large sample size that was drawn from two separate institutions and cross-validation method helped mitigate this risk. Additionally, we included ultrasounds performed at other imaging sites, which should increase generalizability. Nonetheless, continued calibration and validation of these models with additional patients at more institutions will be critical moving forward. Incorporating longitudinal images could potentially improve the performance of our model in the future as only the first postnatal ultrasound was included in the current study. Training and testing our model on ultrasound images performed prenatally as early as the 20th week of gestation will further increase its clinical utility. Future prospective trials are needed to validate our model.
Conclusion
Our study suggests that deep learning imaging features extracted from initial postnatal ultrasounds can improve the early prediction of CKD progression of children with PUV.
Funding
R21DK117297 NIH/NIDDK Anatomic Biomarkers of Chronic Kidney Disease, National Center for Advancing Translational Sciences of the National Institutes of Health under award number TL1TR001880 and 2UL1TR001878-06.
Footnotes
Supplementary information The online version contains supplementary material available at https://doi.org/10.1007/s00467-022-05677-0.
Conflict of interest The authors declare no competing interests.
Disclosures None.
References
- 1.Krishnan A, de Souza A, Konijeti R, Baski LS (2006) The anatomy and embryology of posterior urethral valves. J Urol 175:1214–1220 [DOI] [PubMed] [Google Scholar]
- 2.Gunn TR, Mora JD, Pease P (1995) Antenatal diagnosis of urinary tract abnormalities by ultrasonography after 28 weeks’ gestation: incidence and outcome. Am J Obstet Gynecol 172:479–486 [DOI] [PubMed] [Google Scholar]
- 3.Heikkila J, Holmberg C, Kyllonen L, Rintala R et al. (2011) Long-term risk of end stage renal disease in patients with posterior urethral valves. J Urol 186:2392–2396 [DOI] [PubMed] [Google Scholar]
- 4.Neild GH (2009) What do we know about chronic renal failure in young adults? II. Adult outcome of pediatric renal disease. Pediatr Nephrol 24:1921–1928 [DOI] [PubMed] [Google Scholar]
- 5.Groothoff J, Gruppen M, de Groot E, Offringa M (2005) Cardiovascular disease as a late complication of end-stage renal disease in children. Perit Dial Int 25:S123–S126 [PubMed] [Google Scholar]
- 6.Groothoff JW (2005) Long-term outcomes of children with end-stage renal disease. Pediatr Nephrol 20:849–853 [DOI] [PubMed] [Google Scholar]
- 7.Groothoff JW, Offringa M, Van Eck-Smit BL, Gruppen MP et al. (2003) Severe bone disease and low bone mineral density after juvenile renal failure. Kidney Int 63:266–275 [DOI] [PubMed] [Google Scholar]
- 8.Dodson JL, Jerry-Fluker JV, Ng DK, Moxey-Mims M et al. (2011) Urological disorders in chronic kidney disease in children cohort: clinical characteristics and estimation of glomerular filtration rate. J Urol 186:1460–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neild GH (2009) What do we know about chronic renal failure in young adults? I Primary renal disease Pediatr Nephrol 24:1913–1919 [DOI] [PubMed] [Google Scholar]
- 10.Heikkilä J, Holmberg C, Kyllonen L, Rintala R et al. (2011) Long-term risk of end stage renal disease in patients with posterior urethral valves. J Urol 186:2392–2396 [DOI] [PubMed] [Google Scholar]
- 11.Drozdz D, Drozdz M, Gretz N, Mohring K et al. (1998) Progression to end-stage renal disease in children with posterior urethral valves. Pediatr Nephrol 12:630–636 [DOI] [PubMed] [Google Scholar]
- 12.Lai R, Bhatnagar V, Mitra DK (1999) Long-term prognosis of renal function in boys treated for posterior urethral valves. Eur J Pediatr Surg 9:307–311 [DOI] [PubMed] [Google Scholar]
- 13.DeFoor W, Clark C, Jackson E, Reddy P et al. (2008) Risk factors for end stage renal disease in children with posterior urethral valves. J Urol 180:1705–1708 [DOI] [PubMed] [Google Scholar]
- 14.Yin S, Peng Q, Li H, Zhang Z et al. (2020) Multi-instance deep learning of ultrasound imaging data for pattern classification of congenital abnormalities of the kidney and urinary tract in children. Urology 142:183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin S, Peng Q, Li H, Zhang Z et al. (2020) Automatic kidney segmentation in ultrasound images using subsequent boundary distance regression and pixelwise classification networks. Med Image Anal 60:101602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Q, Furth SL, Tasian GE, Fan Y (2019) Computer-aided diagnosis of congenital abnormalities of the kidney and urinary tract in children based on ultrasound imaging data by integrating texture image features and deep transfer learning image features. J Pediatr Urol 15:75.e1–75.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Q, Tasian G, Fan Y (2018) Transfer learning for diagnosis of congenital abnormalities of the kidney and urinary tract in children based on ultrasound imaging data. Proc IEEE Int Symp Biomed Imaging 1487–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Wu Y, Song G, Li Z et al. (2018) A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal 43:98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris PA, Taylor R, Thielke R, Payne J et al. (2009) Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770–778. https://ieeexplore.ieee.org/document/7780459 [Google Scholar]
- 21.Coresh J, Turin TC, Matsushita K, Sang Y et al. (2014) Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. JAMA 311:2518–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levey AS, Inker LA, Matsushita K, Greene T et al. (2014) GFR decline as an end point for clinical trials in CKD: a scientific workshop sponsored by the National Kidney Foundation and the US Food and Drug Administration. Am J Kidney Dis 64:821–835 [DOI] [PubMed] [Google Scholar]
- 23.Warady BA, Abraham AG, Schwartz GJ, Wong CS et al. (2015) Predictors of rapid progression of glomerular and nonglomerular kidney disease in children and adolescents: the chronic kidney disease in children (CKiD) cohort. Am J Kidney Dis 65:878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierce CB, Munoz A, Ng DK, Warady BA et al. (2021) Age- and sex-dependent clinical equations to estimate glomerular filtration rates in children and young adults with chronic kidney disease. Kidney Int 99:948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viteri B, Elsingergy M, Roem J, Ng D et al. (2021) Ultrasound-based renal parenchymal area and kidney function decline in infants with congenital anomalies of the kidney and urinary tract. Semin Nephrol 41:427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman R, King T, Nicoara C, Bader M et al. (2015) Combined creatinine velocity and nadir creatinine: a reliable predictor of renal outcome in neonatally diagnosed posterior urethral valves. J Pediatr Urol 11:214.e1–e3 [DOI] [PubMed] [Google Scholar]
- 27.Coleman R, King T, Nicoara CD, Bader M et al. (2015) Nadir creatinine in posterior urethral valves: how high is low enough? J Pediatr Urol 11:356.e1–e5 [DOI] [PubMed] [Google Scholar]
- 28.Kwong JC, Khondker A, Kim JK, Chua M et al. (2022) Posterior urethral valves outcomes prediction (PUVOP): a machine learning tool to predict clinically relevant outcomes in boys with posterior urethral valves. Pediatr Nephrol 37:1067–1074 [DOI] [PubMed] [Google Scholar]
- 29.Vasconcelos MA, Silva AC, Gomes IR, Carvalho RA et al. (2019) A clinical predictive model of chronic kidney disease in children with posterior urethral valves. Pediatr Nephrol 34:283–294 [DOI] [PubMed] [Google Scholar]