Abstract
Background:
The most-common strategy for zebrafish Cre/lox-mediated lineage labeling experiments combines ubiquitously expressed, lox-based Switch reporter transgenes with tissue-specific Cre or 4-OH-Tamoxifen-inducible CreERT2 driver lines. Although numerous Cre driver lines have been produced, only a few broadly expressed Switch reporters exist in zebrafish and their generation by random transgene integration has been challenging due to position-effect sensitivity of the lox-flanked recombination cassettes. Here, we compare commonly used Switch reporter lines for their recombination efficiency and reporter expression pattern during zebrafish development.
Results:
Using different experimental setups, we show that ubi:Switch and hsp70l:Switch outperform current generations of the two additional Switch reporters actb2:BFP-DsRed and actb2:Stop-DsRed. Our comparisons also document preferential Cre-dependent recombination of ubi:Switch and hsp70l:Switch in distinct zebrafish tissues at early developmental stages. To investigate what genomic features may influence Cre accessibility and lox recombination efficiency in highly functional Switch lines, we mapped these transgenes and charted chromatin dynamics at their integration sites.
Conclusions:
Our data documents the heterogeneity among lox-based Switch transgenes towards informing suitable transgene selection for lineage labeling experiments. Our work further proposes that ubi:Switch and hsp70l:Switch define genomic integration sites suitable for universal transgene or switch reporter knock-in in zebrafish.
Keywords: Zebrafish, Cre/lox, CreERT2, Tamoxifen, lineage tracing, site-specific recombinases, transgenes
Introduction
Site-specific recombinase (SSR)-based techniques provide powerful versatility to transgenic models. Techniques based on Cre/lox, Flp/FRT, or phiC31 recombinase systems allow permanent excision or rearrangement of transgene cassettes to modify their activity and function 1–5. As a widely used recombinase system in vertebrate models, the Cre/lox system combines i) a Cre recombinase-providing transgene driven by a tissue-specific cis-regulatory element and ii) a recombination-competent reporter transgene. The Cre recombinase, originally derived from the P1 bacteriophage, directionally recombines DNA at specific 13 bp palindromic repeats called loxP sites in a variety of species including mice and zebrafish 2,4,6,7. A major application of the Cre/lox system in zebrafish involves the recombination of lox-flanked fluorophore cassettes that switch upon Cre activity to lineage-label cell populations of interest. This strategy has been successfully employed to reveal developmental lineage origins, to follow post-embryonic stem cells, and to track the lineage composition of regenerating organs 1,8–16. These experiments hinge upon combining a tissue- or cell type-specific Cre or 4-OHT-Tamoxifen (4-OHT)-inducible CreERT2 driver with temporal control and a broadly active or ubiquitous lox-based reporter transgenic 14,17–19. Particularly simple to control in developing zebrafish by 4-OHT addition to the embryo medium, CreERT2 transgenics have rapidly increased in number across the field and include Tol2-based transgenes driving CreERT2 with tissue-specific regulatory elements, gene trap collections, and first CRISPR-based knock-in lines 1,18,20–22. In contrast, the generation of suitable and reproducibly well-performing lox-based reporter transgenes has proven challenging.
The heat shock protein 70-like (hsp70l), beta-actin2 (actb2), and ubiquitin (ubi or ubb) promoter elements have been routinely used to drive quasi-ubiquitous transgene gene expression in zebrafish and have been successfully applied in several lox-based Switch reporters 1,7,14,15,23–26. hsp70l-based transgenes are a mainstay of the zebrafish’s transgenic tool kit, as heat shock induction around 37°C causes rapid ubiquitous reporter expression that is however not sustained long beyond the heat shock pulse 24. The actb2 promoter provides strong, rapid transgene expression during development and in individual tissues, yet its activity drastically diminishes or silences in select cell types such as erythrocytes 25–28. The ubi promoter drives widespread and persistent transgene expression in various independent transgenics including the Cre-sensitive loxP reporter ubi:loxP-GFP-loxP_mCherry (ubi:Switch) that has found widespread use 14. Nonetheless, ubi-based transgenics including ubi:Switch show slow reporter accumulation at early developmental stages 14,28,29; this property causes considerable latency between Cre-triggered lox cassette recombination and reporter detection, rendering short lineage labeling timeframes (under 24 hours) challenging to achieve.
The advantages and drawbacks of each regulatory element require careful characterization of individual transgene insertions over multiple generations, data that is rarely available to guide experimental design. Generating reproducibly functional, single-insertion lox-based Switch transgenics by randomly integrating Tol2 or ISce-I transgenesis is also notoriously screening-intensive due to position-effect sensitivity of lox cassette recombination 1,30,31. Consequently, together with the challenges of sharing transgenic zebrafish lines internationally, the majority of labs only have access to one or few Switch reporter lines. Means to efficiently generate Switch transgenics are therefore highly desirable, requiring the identification of suitable genomic loci for transgene knock-ins.
Here, we compared previously published and validated Switch reporter lines in combination with ubiquitous and tissue-specific CreERT2 drivers. Our results document the heterogeneity in recombination efficiency and preferential tissue expression of individual reporters. Together with genomic integration mapping of tested transgenes, our results define ubi:Switch and hsp70l:Switch as integrations in favorable loci for lox cassette recombination and transgene expression.
Results
Ubiquitous Switch reporters feature variable recombination efficiency and tissue activity
To gain insight into recombination efficiency and reporter expression across tissues by ubiquitous Switch reporters in development, we compared transgenic zebrafish reporters that have been previously used to follow lineage trajectories. We focused on Switch lines in our collection that have been documented to drive ubiquitous transgene expression: ubi:Switchcz1701, hsp70l:Switchzh701, Tg(bactin2:loxP-Stop-LoxP-DsRed-express)sd5 (shortened to actb2:Stop-DsRed), and Tg(bactin2:loxP-BFP-loxP-DsRed)sd27 (shortened to actb2:BFP-DsRed) 14,15,23,32. The ubi:creERT2 driver has been widely used for ubiquitous CreERT2 expression in early development and provides a first assay for Cre responsiveness of individual Switch lines 14. To establish switching efficiency under ubi:creERT2 and homogeneity versus heterogeneity of reporter expression, we treated ubi:creERT2;promoter:Switch embryos with 4-OHT at shield stage overnight, and imaged whole larvae at 3 dpf (Fig. 1). In all crosses, the ubi:creERT2-carrying parent was male to avoid maternal CreERT2 contribution.
Figure 1. Ubiquitous Switch reporter lines show variable recombination efficiency.
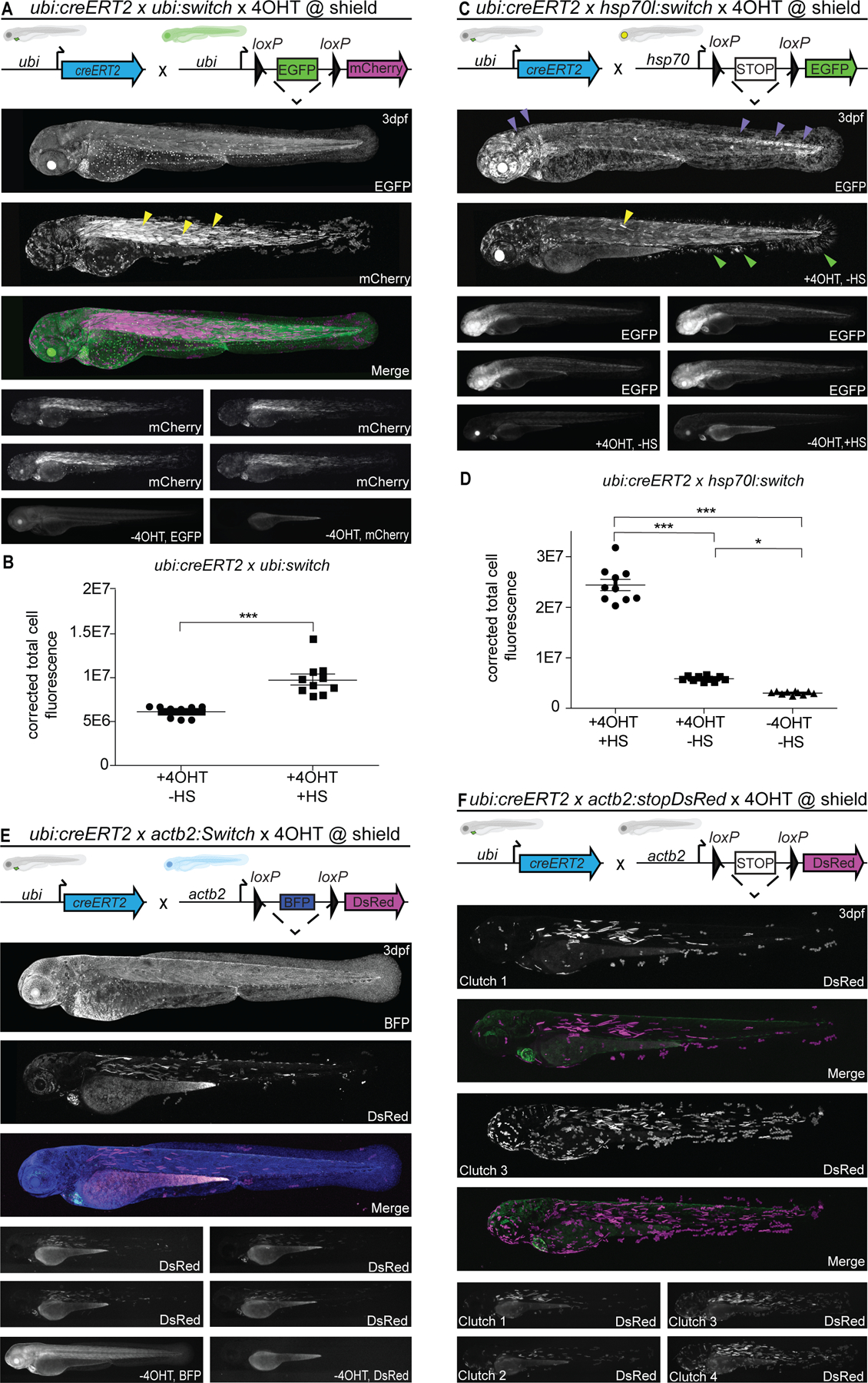
(A,B) ubi:Switch, (C,D) hsp70l:Switch, (E) actb2:BFP-DsRed and (F) actb2:Stop-DsRed crossed to ubi:creERT2, induced with 10 µM 4-OHT at shield stage, and imaged laterally at 3 dpf. Schematics of fluorophore cassettes for each Switch transgene are shown at the top of each panel and larval schematics represent secondary transgenic markers (A,B,E,F). One representative confocal image, and four representative stereo microscope images are presented per reporter. ubi:Switch (A) shows preferential recombination in somitic myofiber (yellow arrowheads), hsp70l:Switch (C) shows preferential switching in CNS (brain, neural tube) (purple arrowheads). Non heat-shocked (+4-OHT) controls (C, second panel) show faint EGFP expression in somitic myofiber (yellow arrowheads) and fin fibroblasts (green arrowheads). (B) Corrected total cell fluorescence (CTCF) measurements of 3 dpf ubi:creERT2 crossed to ubi:Switch, induced with 10 µM 4-OHT at shield, and heat-shocked 3 hours prior to lateral view imaging; fluorescence intensity was compared to non-heat-shocked sibling (n=10, 2 clutches). Note increased CTCF following heat-shock at 3 dpf (n=10, Mann-Whitney, P>0.0001). (D) CTCF ubi:creERT2 crossed to hsp70l:Switch, induced with 10 µM 4-OHT at shield. Non-heat shocked controls (center), and non-heat shocked, non-treated controls (right) are included (n=10, 1-way ANOVA, P<0.0001). ubi:Switch (A) and hsp70l:Switch (B) show more spatially complete recombination compared to both and actb2:BFP-DsRed (E) and actb2:Stop-dsRED (F) lines that only display sparse recombination.
The ubi:Switch transgenic permanently recombines from EGFP to mCherry following Cre activity (Fig. 1A), yet mCherry levels only reach detectable levels after 12–24 hours post-Cre activation 14. When combined with ubi:creERT2, we observed strongest mCherry activity in the somitic myofibers along the entire trunk and tail (Fig. 1A). Skin and fin epithelium showed more sparse switching, predominantly in the median fin and head (Fig. 1A). We observed strong mCherry signal in the heart, while other tissues including the eye lens and neural tube consistently displayed comparatively lower mCherry fluorescence (Fig. 1A). Promoters of ubiquitin genes in several species have been postulated to harbor heat shock-responsive elements, possibly to support protein degradation by increased Ubiquitin polypeptide production upon heat or other stress 33–38. To test if ubi:Switch as driven by the zebrafish ubb gene promoter reaches higher expression levels upon heat shock, we compared the levels of mCherry fluorescence at 3 dpf with and without a 1 hour-long, 37°C heat shock prior to imaging: compared to non-heat-shocked siblings, heat-shocked ubi:creERT2;ubi:Switch larvae showed increased fluorescent intensity 3 hours post-heat shock (n = 10 for quantification, 2 clutches) (Fig. 1B). These observations document that ubi:Switch expression can be further augmented by a brief heat shock prior to imaging.
Following Cre activity, the hsp70l:Switch transgenic line permanently switches from a non-fluorescent Stop cassette to EGFP 15. Heat shock for 1 hour at 37°C induces prominent EGFP expression within one hour post-heat shock treatment. To trigger reporter expression of hsp70l:Switch, we heat-shocked recombined embryos 2–3 hours prior to imaging (Fig. 1C). When crossed to ubi:creERT2 and treated with 4-OHT at shield stage, hsp70l:Switch showed strong EGFP signal across the entire embryo, indicating near complete switching, with the central nervous system (brain, neural tube) displaying higher EGFP signal (Fig. 1C). Notably, in non-heat-shocked controls that were 4-OHT-induced, we also detected faint EGFP expression (n = 10) in the skeletal myofibers and fin fibroblasts (Fig. 1C,D); these observations indicate that tissue-specific accessibility of the hsp70l element at the genomic location of hsp70l:Switch leads to transcriptional activity even at 28°C. Embryos that were left untreated with 4-OHT did not show any detectable EGFP expression with or without heat shock, confirming that the used Stop cassette 15,39 efficiently interrupts any EGFP transcription.
The actb2:Stop-DsRed transgene recombines from no fluorophore to DsRed and actb2:BFP-DsRed recombines from BFP to DsRed, enabling versatile combination with existing tissue-specific reporters based on EGFP 23,32. After recombination with ubi:creERT2 at shield stage, both actb2 Switch lines imaged at 3 dpf showed overall sporadic switching with the most prominent DsRed signal in the skeletal myofibers, fin epidermis, and the heart (Fig. 1E,F). Of note, actb2:Stop-DsRed showed two distinct switching efficiencies across clutches, potentially due to multiple integrations within the current or most-recent generation (Fig. 1F). Under these conditions, switching was absent from a large majority of tissues including, but not limited to, endothelial cells, skin, brain and neural tube, and fin fibroblasts (Fig. 1F). Ubiquitous BFP expression is observed with actb2:BFP-DsRed throughout the 4-OHT induction regime (Fig. 1E).
Taken together, our observations from triggering ubiquitous CreERT2 activity after onset of gastrulation reveal heterogeneous recombination efficiencies and reporter expression across four distinct, broadly expressed lox reporter lines. The results with ubi:Switch are in line with previously reported Cre-based lineage labeling experiments across various cell types and developmental stages. Nonetheless, hsp70l:Switch displayed distinctively more homogenous reporter expression than ubi:Switch; this effect is possibly in part due to the pulsed fluorescence activation following heat shock shortly before analysis in hsp70l:Switch experiments, while in ubi:Switch the mCherry reporter dynamically accumulates and degrades in various cell types following 4-OHT treatment. In contrast, the lower switching efficiency observed with the actb2-based Switch lines indicates low Cre accessibility of the insertions selected for these transgenics over the past decade 23,32.
hsp70l:Switch shows widespread and reproducible Cre sensitivity
The widespread Cre-induced recombination and reporter expression observed with ubi:Switch and hsp70l:Switch indicate favorable genomic integration sites for loxP cassette recombination. To further evaluate the switching efficiencies between ubi:Switch and hsp70l:Switch, we turned to two previously established, highly active, and tissue-specific CreERT2 driver lines: i) Tg(drl:creERT2;cryaa:Venus) (drl:creERT2 for short) that expresses in lateral plate mesoderm (LPM)-primed mesendoderm during gastrulation before refining to cardiovascular lineages by mid-somitogenesis 40,41 (Fig. 2A,B); and ii) Tg(tbx1:creERT2;cryaa:Venus) (tbx1:creERT2 for short) that starts expressing in late gastrulation and predominantly labels cardiopharyngeal progenitors together with tbx1-expressing ectodermal and endodermal lineages in the head 15 (Fig. 2C,D). After crossing, we treated double-transgenic embryos with 10 µM 4-OHT at shield stage and imaged the head region at 3 dpf laterally and ventrally to assess lineage labeling.
Figure 2. hsp70l:Switch high degrees of lineage labeling crossed to tissue-specific CreERT2 driver lines.
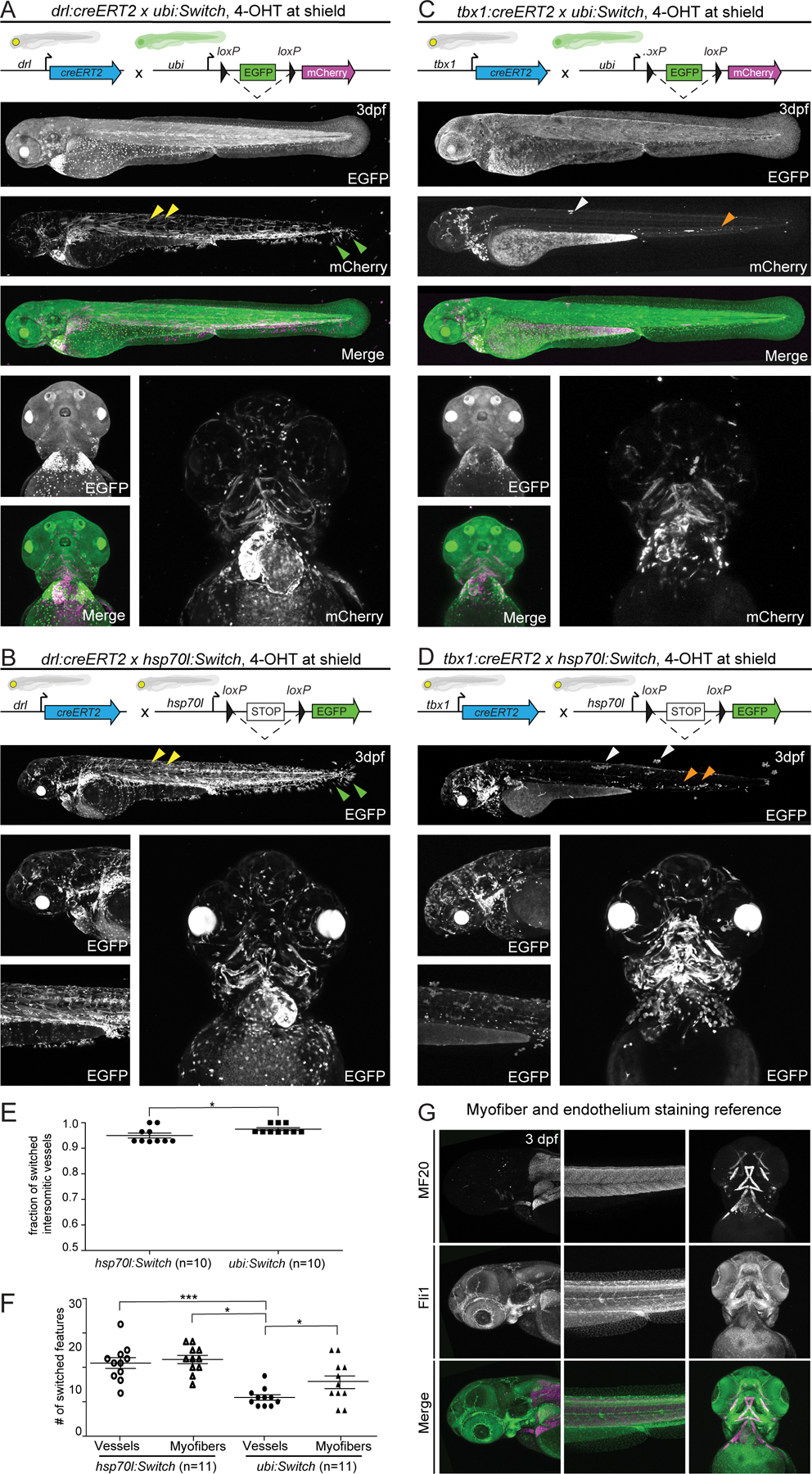
(A,B) drl:creERT2, and (C,D) tbx1:creERT2 crossed to ubi:Switch and hsp70l:Switch, respectively induced with 4-OHT at shield stage and imaged laterally and ventrally at 3 dpf. Quantifications depicted as bar diagrams with individual data points (E,F). Schematics of fluorophore cassettes for each Switch transgene are shown at the top of each panel and larval schematics represent secondary transgenic markers (A,B,C,D). drl:creERT2 yields more complete recombination when crossed to hsp70l:Switch (B) compared to ubi:Switch (A). The higher degree of recombination is detectable in the somitic myofiber (yellow arrowheads), and fin fibroblasts (green arrowhead) (A,B). A higher percentage of switched intersomitic vessels (ISV) is observed when combining drl:creERT2 with ubi:Switch than with hsp70l:Switch (n=10, unpaired t-test, p=0.0345) (E). tbx1:creERT2 yields less switching mosaicism when crossed to hsp70l:Switch (D), compared to ubi:Switch (C). Higher recombination efficiency is readily observed in trunk skin (white arrowhead) and hematopoietic cells (orange arrowheads) (C, D). When crossed to hsp70l:Switch, tbx1:creERT2 yields significantly more switched head vessels and myofibers compared to ubi:Switch (n=11, 1-way ANOVA p<0.0001) (F). Representative lateral and ventral images (A,B;D,E). (G) Max projections of z-stack confocal images of fli1a:EGFP transgenic zebrafish fixed at 3 dpf and stained for myofibers using an MF20 antibody to visualize vasculature and myofiber anatomy in the zebrafish head at 3 dpf.
Combined with drl:creERT2, both Switch lines displayed switching in the same tissues and cell types, yet hsp70l:Switch consistently resulted in more complete recombination (lower mosaicism). This feature was most evident in the prevalence of switched fin fibroblasts, skeletal myofiber cells, macrophages, and head vasculature (Fig. 2A,B). To compare recombination efficiency in a major cell type, we quantified switched versus unlabeled intersomitic vessels (ISV) 29: we observed slighty more switched ISVs with ubi:Switch than with hsp70l:Switch when recombined by drl:creERT2 (n = 10, p = 0.0454) (Fig. 2E). Of note, induction of drl:creERT2 at shield stage results in complete labeling of LPM-derived structures, but also sparse labeling of paraxial mesoderm-derived structures including somitic skeletal myofiber cells and median fin fold fibroblasts, reflecting the starting segregation of individual mesodermal territories at shield stage 40,41 (Fig. 2A,B).
When crossed to tbx1:creERT, both ubi:Switch and hsp70l:Switch displayed broad lineage labeling in the ventricular cardiomyocytes, pharyngeal arches, cranial vasculature, head myofibers and cartilage, and hatching gland, consistent with previous work 15 (Fig. 2C,D). Once more, ubi:Switch animals showed preferential switching in myofibers, versus hsp70l:Switch that displayed more complete switching across multiple cell types, especially head vasculature. We quantified head myofiber and endothelial switching of each Switch line, separately using myofiber (MF20 antibody)-stained fli1:EGFP embryos as reference for vessel/myofiber number and position (n = 11) (Fig. 2F,G). This analysis documented that tbx1:creERT2 had significantly higher recombination efficiency in the head vasculature and musculature combined with hsp70l:Switch than when combined with ubi:Switch (Fig. 2F). These results further underline the widespread, yet differential sensitivity to Cre activity of both ubi:Switch and hsp70l:Switch.
ubi:creERT2 shows decreased recombination activity at 48 hpf
The ubi:creERT2cz1702 transgenic line has been widely applied to provide broad 4-OHT-inducible Cre activity, as desirable for testing new Switch transgenes 1,14. Our data presented above further confirmed the reproducible activity of ubi:creERT2 at shield stage paired with responsive Switch lines. Nevertheless, anecdotal observations have suggested that ubi:creERT2 becomes less responsive to 4-OHT at later stages 14,29. To what extent this effect depends on choice of lox Switch line remains uncertain.
To test the recombination capacity after gastrulation stages, we crossed ubi:creERT2 to ubi:Switch and hsp70l:Switch, treated with 10 μM 4-OHT at 48 hpf, and performed lateral-whole embryo imaging at 3 dpf (Fig. 3). In contrast to shield stage induction, 4-OHT induction at 48 hpf showed markedly reduced switching at 3 dpf with both ubi:Switch and hsp70l:Switch (Fig. 3A,B). When crossed to hsp70l:Switch, 10 μM 4-OHT induction at 48 hpf of ubi:creERT2 resulted in switching predominantly in the skeletal myofibers and the fin fibroblasts (Fig. 3A). When crossed to ubi:Switch and induced with this regimen, ubi:creERT2 induced switching predominantly in the blood and heart (Fig. 3B). Due to the cardiac myl7:EGFP reporter incorporated in the ubi:creERT2 transgene, we could not evaluate switching efficiency in the heart using hsp70l:Switch. While 10 μM results in reproducible CreERT2 activity across transgenes with negligible impact on the treated embryos, previous work has indicated that 4-OHT reaches saturation for inducing CreERT2 activity at 25 μM 14,19,29. In line with this observation, increasing 4-OHT concentration from 10 μM to 30 μM at 48 hpf did not notably increase switching efficiency in ubi:creERT2;ubi:Switch embryos (Fig. 3C).
Figure 3. ubi:creERT2 shows decreased 4-OHT and recombination response at larval stages.
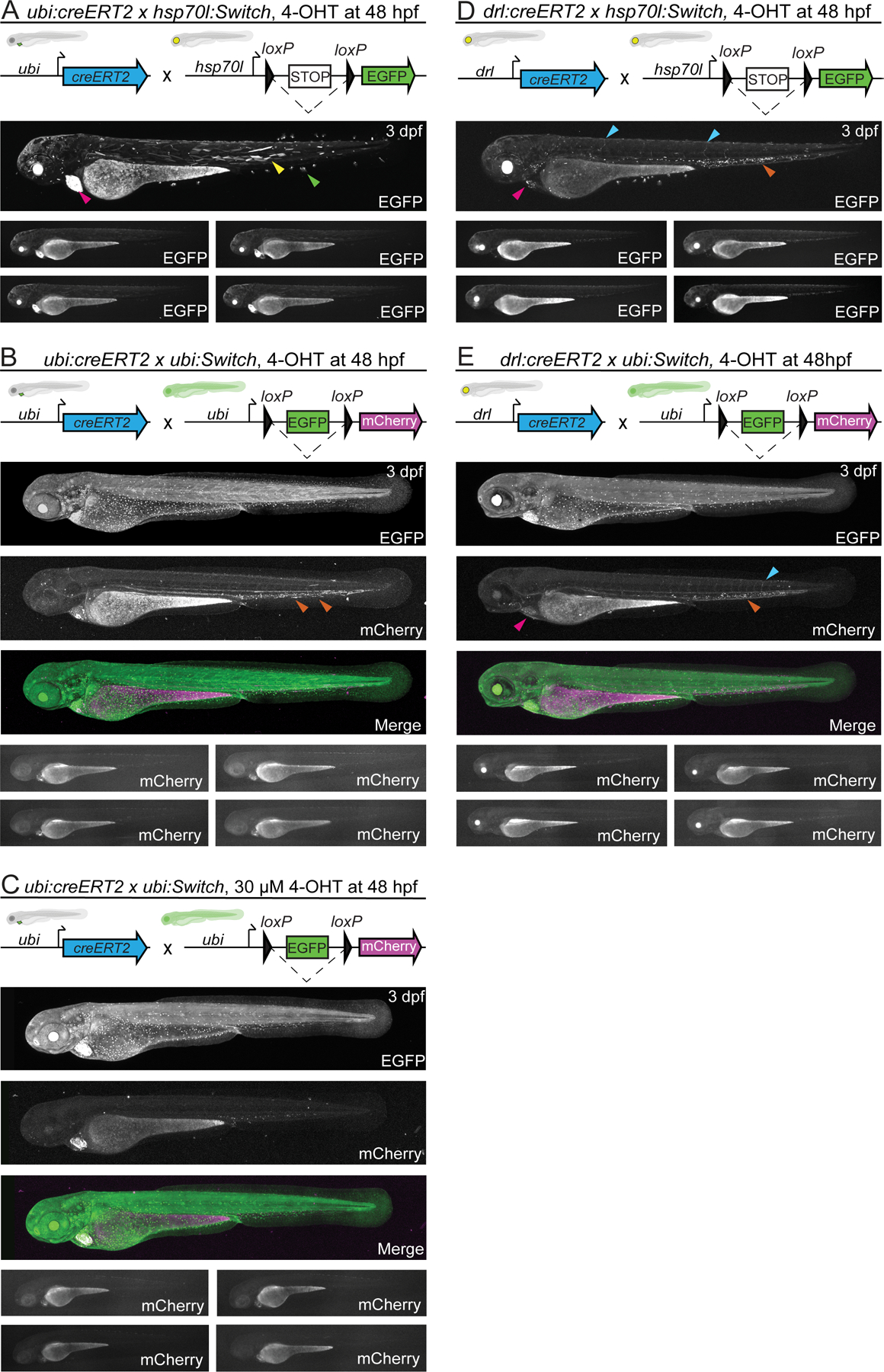
(A,B,C) ubi:creERT2 and (D, E) drl:creERT2 crossed to ubi:Switch, and hsp70l:Switch, induced with 4-OHT at 48hpf, and imaged laterally at 3 dpf. Schematics of fluorophore cassettes for each Switch transgene and larval schematics representing secondary transgenic markers are shown at the top of each panel; one representative confocal image, and four representative stereo microscope images are presented here (A-E). ubi:creERT2 shows sparse switching with both reporters when induced with 4-OHT at 48 hpf (A,B,C). Predominant switching occurs in somitic myofiber (yellow arrowheads), and fin fibroblasts (green arrowheads) when crossed to hsp70l:Switch (A), and hematopoietic cells (orange arrowhead) when crossed to ubi:Switch, induced with 10 μM (B) and 30 μM (C) 4-OHT. drl:creERT2 with 4-OHT induction at 48 hpf results in switching in the heart (pink arrowhead), vasculature (blue arrowhead), and hematopoietic cells (orange arrowhead) when crossed to both hsp70l:Switch (D), and ubi:Switch (E). Note for ubi:Switch stereo microscope images that are representative of routine laboratory imaging, extended exposure (30 seconds) was used to capture all traces of mCherry fluorescence (B,C,E).
To test whether the observed decrease in switching efficiency was specifically associated with the ubi:creERT2 line, we crossed drl:creERT2 to hsp70l:Switch and ubi:Switch, treated with 10 μM 4-OHT at 48 hpf, and performed lateral-whole embryo imaging at 3 dpf (Fig 3D,E). drl-based labeling is restricted predominantly to LPM-derived cardiovascular, hematopoietic, and fin fibroblast lineages by 48 hpf 40,41. Accordingly, when crossed to hsp70l:Switch, 48 hpf induction of drl:creERT2 resulted in robust switching in hematopoietic cells with sporadic labeling in the heart, endothelial cells, as well as fin fibroblasts (Fig. 3D). When crossed to ubi:Switch, 48 hpf induction of drl:creERT2 again resulted in robust switching in hematopoietic cells, with sporadic labeling in the heart, endothelial cells, and fin fold fibroblasts (Fig. 3E). Despite maximum exposure lengths, visualizing mCherry fluorescence at 24 hours post-4OHT induction is challenging with standard stereo microscopy due to the slow reporter accumulation in ubi:Switch (Fig. 3D,E). We did not observe any switched pectoral fin cells, further supporting our conclusion that hsp70l:Switch provides more thorough recombination reporting than ubi:Switch. Together, these results underscore that the original ubi:creERT2 transgenic (cz1702Tg, ZDB-ALT-110121–1 14) features diminished recombination potency at later developmental stages.
Modifying EGFP in ubi:Switch does not alter recombination efficiency
CRISPR-Cas9-mediated genome editing has become a standard tool to generate zebrafish mutants. Cas9 can also be harnessed to target transgenic insertions to mutate regulatory elements and transgenic cargo such as fluorescent protein ORFs. ubi:Switch expresses EGFP by default, resulting in ubiquitous green fluorescence that prohibits combining the reporter with GFP-based transgenic reporters (Fig. 1A). Since we predict integration site is critical to recombination efficiency, creating new stable Tol2 transgenics with modified stop cassettes lacking EGFP would likely show differential switching capacity. Instead, to disrupt EGFP translation and consequently evaluate the impact of in situ modification of a Switch locus in zebrafish, we isolated a CRISPR-induced four base-pair frameshift mutation (Δ4) in the EGFP ORF, resulting in a premature stop codon (Fig. 4). We derived the ubi:Switch-based transgenic strain Tg(ubi:loxP-delta-EGFP-loxP_mCherry) (ubi:delta-EGFP for short) and compared this new transgene to the original ubi:Switch (Fig. 4).
Figure 4. Modifying EGFP in ubi:Switch in situ does not alter recombination efficiency.
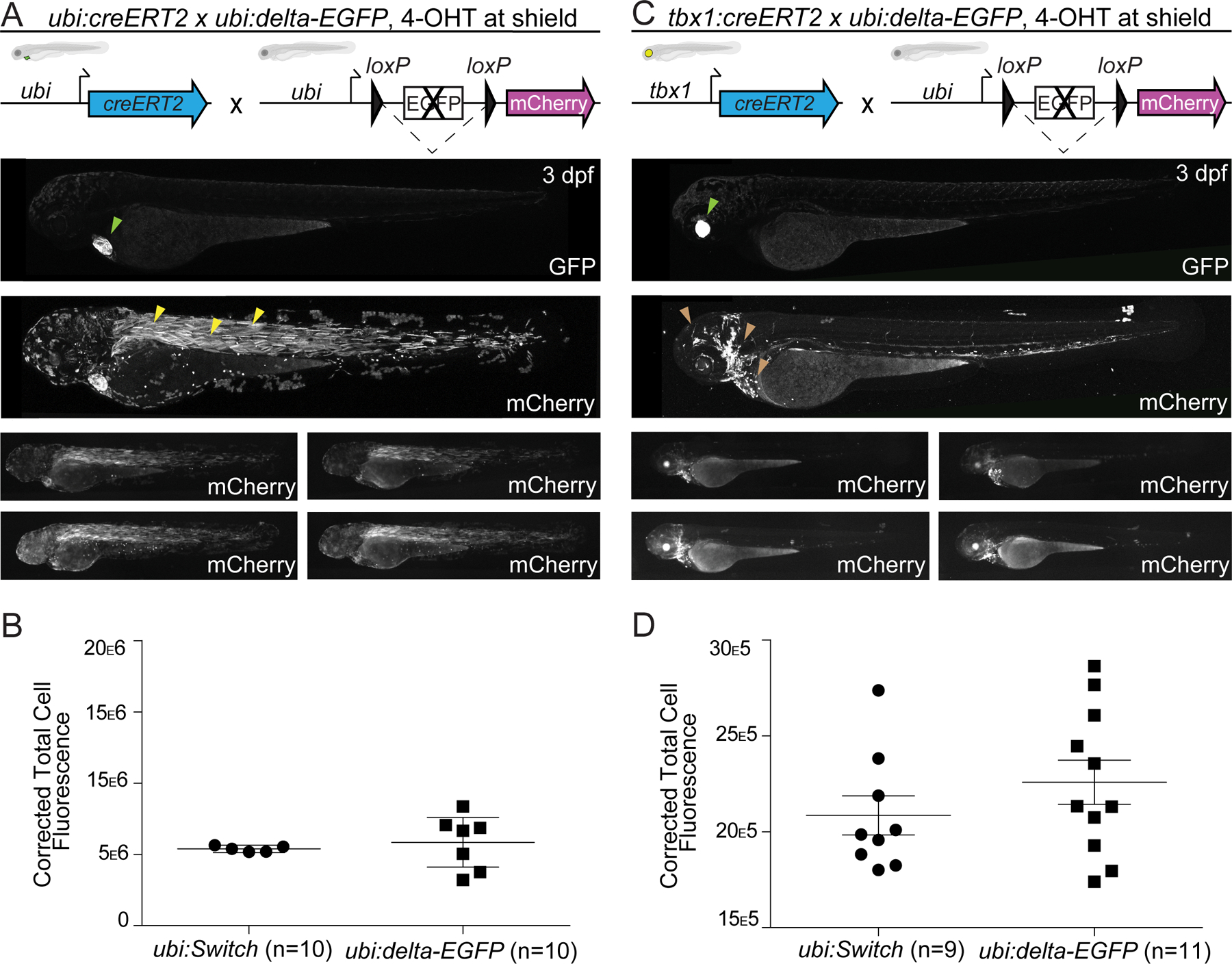
(A,B) ubi:creERT2 and (C,D) tbx1:creERT2 crossed to ubi:delta-EGFP that bases on ubi:Switch with a disrupted EGFP cassette, induced with 4-OHT at shield stage, and imaged laterally at 3 dpf plus corrected total cell fluorescence for quantification (B,D). Schematics of fluorophore cassettes for the Switch transgene are shown at the top of each panel and larval schematics represent secondary transgenic markers. One representative confocal image, and four representative stereoscopic images are presented here (A,C). ubi:creERT2 shows high recombination efficiency with preferential switching in somitic myofiber (yellow arrowheads) (A). No significant difference is observed when ubi:creERT2 is combined with either ubi:Switch or ubi:delta-EGFP (n=10, unpaired t-test, p>0.05) (B). tbx1:creERT2 switching in the ventricular cardiomyocytes, pharyngeal arches, cranial vasculature, head myofibers and cartilage, and hatching gland with ubi:delta-EGFP (brown arrowheads) (C). No significant difference in recombination efficiency is observed between tbx1:creERT2 crossed to ubi:Switch or ubi:delta-EGFP (n=9–11, unpaire t-test, p>0.05) (D). Note visibility of screening markers (myl7:EGFP for ubi:creERT2; cryaa:Venus for tbx1:creERT2) when EGFP ORF is removed in ubi:delta-EGFP (lime arrowhead) (A,C).
When crossed to ubi:creERT2 and treated with 4-OHT at shield stage, ubi:delta-EGFP showed preferential switching in the zebrafish skeletal myofibers at 3 dpf (Fig. 4A). To quantify the switching efficiencies of ubi:Switch and ubi:delta-EGFP, we performed whole-mount lateral view measurements of fluorescence intensity. These measurements did not reveal any significant differences in recombination pattern, mosaicism, or mCherry intensity at these experimental time points (n = 10, Fig. 4B). When crossed to tbx1:creERT2, ubi:delta-EGFP showed labeling in the ventricular cardiomyocytes, pharyngeal arches, cranial vasculature, head myofibers and cartilage, and hatching gland at 3 dpf (Fig. 4C), with no significant difference to tbx1:creERT2, ubi:Switch (n = 9–11, Fig. 4D). From this limited experimental paradigm, we conclude that ubi:delta-EGFP broadly preserves the recombination efficiency of ubi:Switch after modification of the fluorescent ORF.
Transgene mapping reveals genomic features at Switch reporter integrations
The rational selection of suitable locations for universal, inert, and permissive genomic integration spots for transgenes is highly desirable to minimize position effects. While the majority of zebrafish transgenesis has been performed with random integration methods including Tol2 and ISce-I, CRISPR-Cas9 now provides a first means for targeted transgene integrations. However, neither rational design principles for safe harbor sites or long term-validated loci for functional transgene integration, especially of functional lox-based Switch reporters, have been reported in zebrafish 1. Our data above established that the Tol2-based transgenes hsp70l:Switch and ubi:Switch integrated into loci that are highly permissive to loxP cassette recombination and can be maintained for over a dozen generations 14,15, possibly providing suitable genomic coordinates for universal transgene landing sites in the genome.
We used Thermal Asymmetric Interlaced (TAIL)-PCR to map the genomic integration loci of the ubi:Switch and hsp70l:Switch transgenes 42. In contrast to ligation-mediated (LM)-PCR that involves restriction enzyme digestion, relegation, and sequencing, TAIL-PCR uses a series of nested PCRs involving transgene-specific primers and established degenerate primers. We sequenced the resulting TAIL-PCR products, charted their genomic position by BLAST, and confirmed individual transgene integrations with independent primers. The results of our transgene mapping are depicted in Fig. 5A,B. The ubi:Switch transgene integrated at chr14:32826253, proximal and upstream of the gene inppl1b and distal and upstream of the gene arr3b (Fig. 5A); despite this proximity to gene bodies, homozygous ubi:Switch zebrafish are viable with no overt morphological phenotypes. The hsp70l:Switch transgene integrated at chr24:26386749, placing it 122 bp from the transcription start of an annotated transcript of the gene skilb, which itself is annotated to be within the gene body of the gene si:ch211–230g15.5 (Fig. 5B). While no loss-of-function alleles for skilb or si:ch211–230g15.5 have been reported to our knowledge, we found also hsp70l:Switch to be homozygous viable and fertile with no discernible phenotype.
Figure 5. Transgene mapping reveals genomic features at the ubi:Switch and hsp70l:Switch loci.
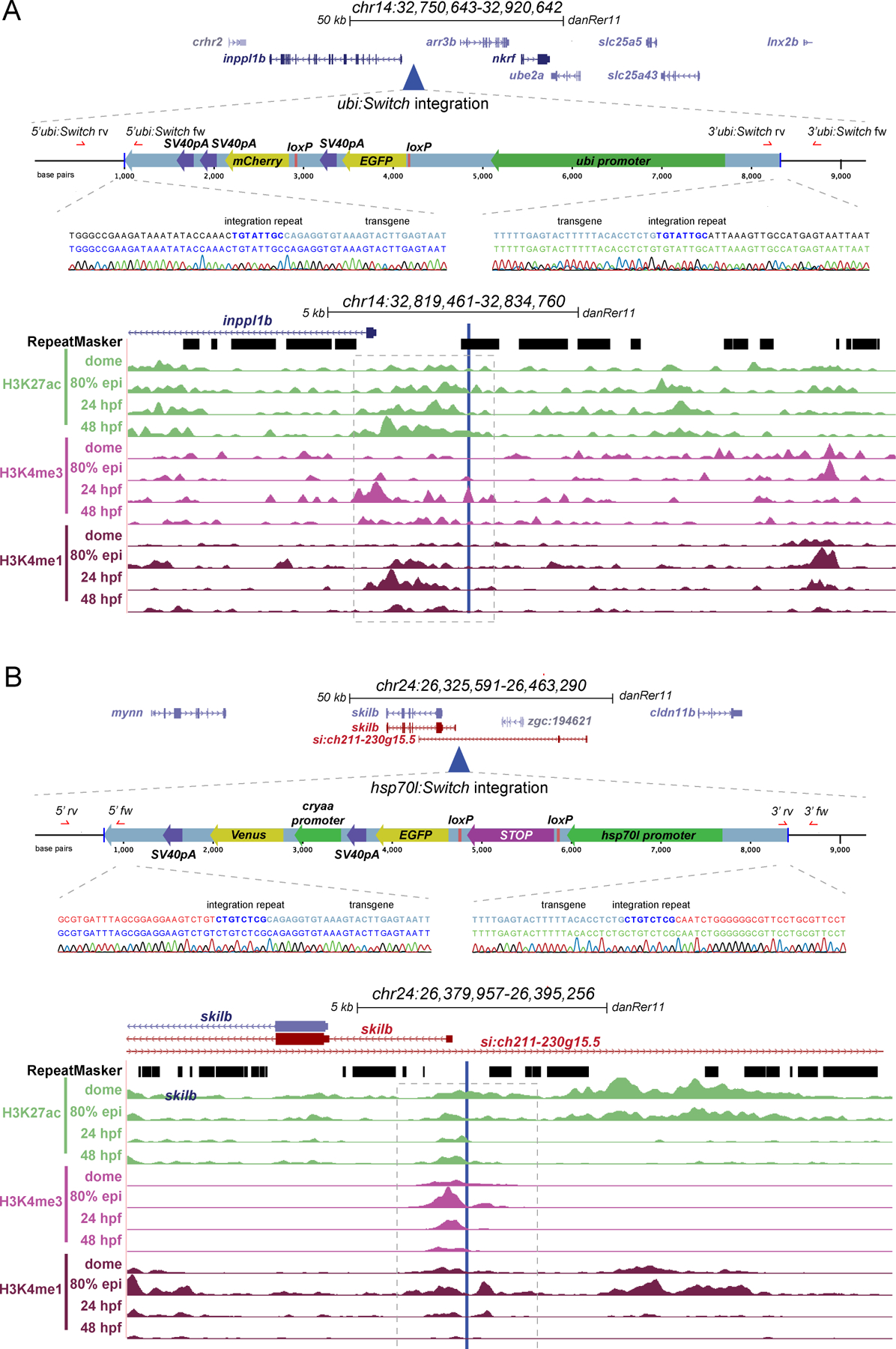
(A,B) Genomic integration sites of ubi:Switch and hsp70l:Switch determined by Thermal asymmetric Interlaced (TAIL)-PCR. For each transgene, the integration site is displayed in UCSC genome browser, a schematic of the transgene is depicted, the integration site is aligned to RepeatMasker track, and ChIP-seq tracks for active chromatin marks at key developmental stages. Sequencing reads for 5’ and 3’ transgene/genome boundaries are provided for ubi:Switch and hsp70l:Switch integration sites. ubi:Switch and hsp70l:Switch integration site loci contain peaks for all three chromatin modification signatures (H3K27ac, H3K4me3, H3K4me1, dashed boxes) (A,B).
In contrast to the ubi:Switch and hsp70l:Switch transgenic lines, we observed significantly less recombination with both the actb2:Stop-DsRed and the actb2:BFP-DsRed transgenic lines under the same experimental conditions (Fig. 1A–D). We again used TAIL-PCR to map the genomic integration loci of the actb2:Stop-DsRed and actb2:BFP-DsRed transgenes (Fig. 6A,B). The actb2:Stop-DsRed transgene integrated at chr7:5941511, within an intron of two distinct annotated genes: dusp19b and si:dkey-23a13.11 (Fig. 6A). The actb2:BFP-DsRed transgene integrated at chr19:25353744, within the intron of the gene glcci1a (Fig. 6B). We were unable to detect any additional integrations of actb2:Stop-DsRed using TAIL-PCR.
Figure 6. Transgene mapping reveals genomic features of the tested actb2-based Switch transgenes.
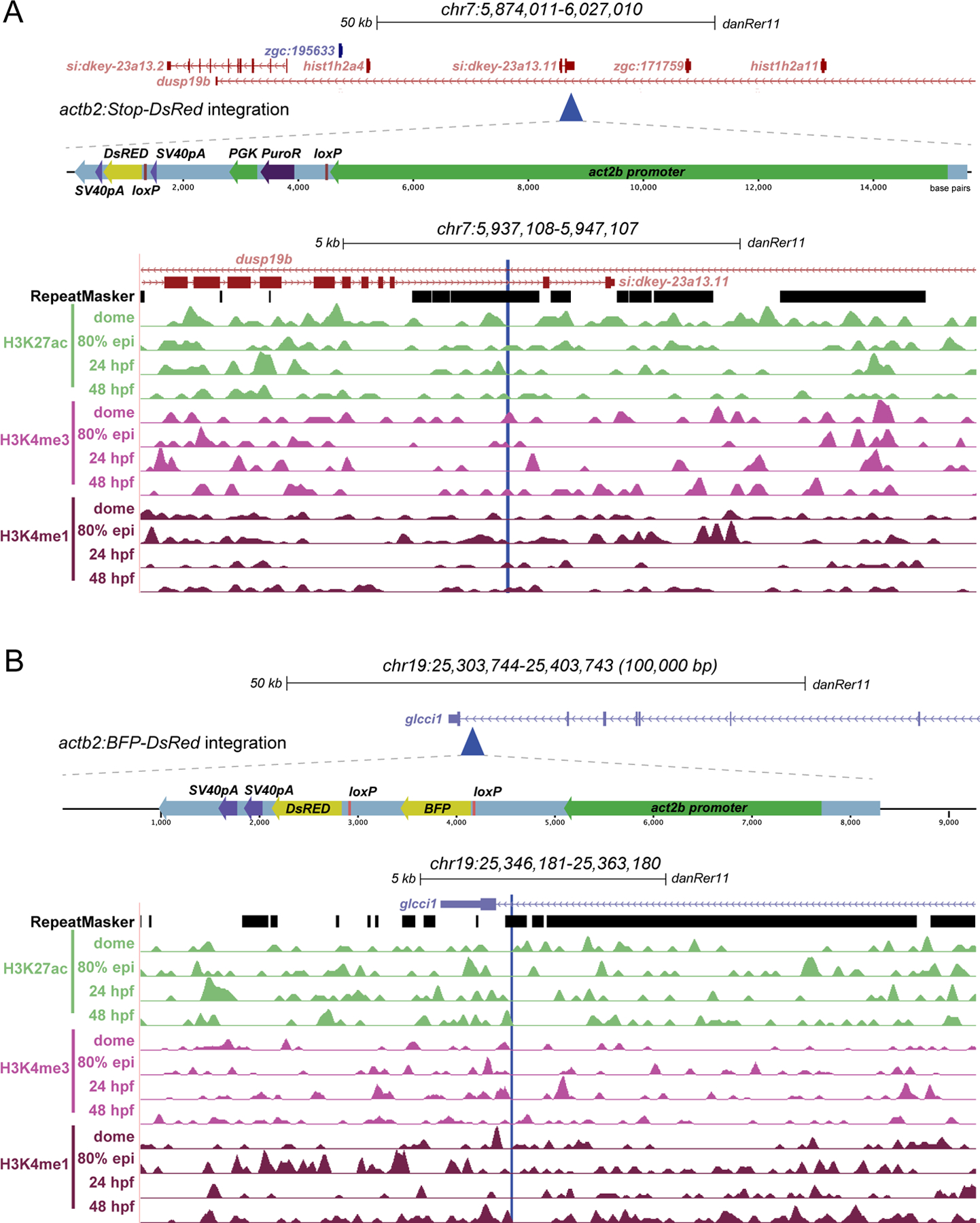
(A,B) Genomic integration sites of actb2-Stop-DsRed and actb2:BFP-DsRed determined by Thermal Asymmetric Interlaced (TAIL)-PCR. For each transgene, the integration site is displayed in UCSC genome browser, a schematic of the transgene is depicted, the integration site is aligned to RepeatMasker track, and ChIP-seq tracks for active chromatin marks at key developmental stages (A,B). Note the intronic integration sites for both transgenes and the absence of concise active chromatin marks compared to ubi:Switch and hsp70l:Switch.
To gain insight into what genomic features might influence Switch transgene recombination efficiency, we compared the integration sites of the mapped transgenes. All four transgenes mapped within or near protein-coding loci, with ubi:Switch, actb2:Stop-DsRed, and actb2:BFP-DsRed landing in highly repetitive regions (Fig. 5A,6A,6B). While ubi:Switch and hsp70l:Switch mapped to intergenic regions (hsp70l:Switch may lie within one possible gene body annotation) (Fig. 5A,B), both actb2 Switch transgenes mapped within introns of annotated and studied genes (dusp19b and glcc1a) (Fig. 6A,B). Using publicly available ChIP-seq datasets 43, we explored whether distinct histone marks at the integration loci of these individual Switch transgenic reporters may associate with high recombination. Distinct from surrounding regions, the ubi:Switch integration site showed a series of peaks for H3K27ac, H3K4me3, and H3K4me1 that represent open, active chromatin between 80% epiboly and 48 hpf (Fig. 5A). The immediate vicinity of the hsp70l:Switch integration locus featured signal for all histone marks between dome stage and 48 hpf (Fig. 5B). In contrast, the integration sites of the actb2-based Switch lines did not show active histone marks at any developmental stage we plotted (Fig. 6A,B). Based on a small sample size of four transgene insertions of different recombination qualities, these observations suggest that higher recombination efficiency might correspond with transgene integration in native areas containing active chromatin that may be primed for transcription. In addition, our mapping provides the genomic coordinates of the ubi:Switch and hsp70l:Switch reporters for potential future modifications and knock-ins of alternative reporters or lox-based Switch transgenes at these validated loci.
Discussion
Transgenic experiments in zebrafish depend on reliable, reproducible reagents including single-copy insertion transgenes with predictable expression patterns in space and time. Cre/lox experiments in particular depend on efficient lox-based Switch reporters, the generation of which has so far been serendipitous due to favorable Tol2-based random transgene integrations. Here, we have documented the Cre-dependent recombination or switching efficiencies of four established lox-based Switch reporters with both ubiquitous and tissue-specific CreERT2 driver lines at two different developmental time points. Further, our results suggest two long-term validated genomic loci as potential safe harbor sites suitable for lox-based Switch reporters based on their reproducible performance over generations and across labs.
While seemingly straight-forward given access to suitable transgenic lines, Cre/lox-based lineage tracing studies require careful consideration to ensure successful experimental outcomes 1. The specific combination of fluorophores in the lox-based reporter may impede visualization if used in parallel with other desired transgenic reporter lines, such as when seeking to define switched cells with a tissue-specific reporter in parallel. Our conversion of ubi:Switch to ubi:delta-EGFP shows that modulating functional Switch transgenics in situ by small edits is feasible without drastically perturbing their properties (Fig. 4). Disruption of the EGFP ORF in ubi:delta-Switch facilitates the visualization of screening markers (myl7:EGFP, cryaa:Venus, etc.) and enables the use of this line in combination with commonly used EGFP/GFP-based reporter lines.
In Cre/lox-based lineage labeling, the onset of fluorescent reporter expression following loxP recombination dictates the earliest possible analysis timepoints. As documented in previous work, ubi-driven transgenes feature a latency in reporter expression, restricting detectable visualization and imaging to 24 hours or more after Cre-based recombination (Fig. 3C,D) 1,14,28,29. We note that, even with extended exposure length for imaging, visualizing mCherry fluorescence 24 hours post-4-OHT induction, remains challenging using standard stereo microscopy (Fig. 3C,D). Consequently, ubi:Switch may not be the preferred choice of lox reporter for short-term trace experiments (i.e. analysis within hours following first Cre activity), unless in situ hybridization for mCherry mRNA is used as a readout, rather than mCherry fluorescence. However, we here document that a simple heat shock prior to imaging can boost the reporter activity of ubi:Switch (Fig. 1B); if this pragmatic way to facilitate reporter detection with this transgene is based in heat shock elements in the zebrafish ubi promoter or based on other effects warrants further analyses. In contrast, the hsp70l:Switch transgenic line bypasses long latency in fluorescent reporter expression, yielding bright EGFP expression within 2 hours post heat-shock (Fig. 1B, Fig. 2A,B). As a potential caveat, heat-shock efficiency has not been extensively tested in adult hsp70l:Switch zebrafish; nonetheless the heat-shock system has been successfully used in adults with Tg(hsp70:loxP-DsRed-Stop-loxP-EGFP)tud107, as well as other hsp70l-based transgenic lines 15,44–49.
ubi:creERT2 provides a versatile tool to test new lox-based Switch lines and to establish 4-OHT regimens 1,14,29. In our assays, inducing CreERT2 activity with 4-OHT during gastrulation leads to ubiquitous recombination in all tested Switch reporters (Fig. 1). When combined with ubi:Switch, we noted increased mCherry signal in the heart relative to other tissues, possibly a consequence of the heart-specific myl7:EGFP transgenesis marker of ubi:creERT2 that could influence creERT2 expression 50,51. Nonetheless, CreERT2 activity from ubi:creERT2 is drastically reduced by 48 hpf despite substantial evidence that the ubi regulatory element remains active throughout all stages of zebrafish development 14. Our data here adds to the notion that this diminished activity is due to the particular ubi:creERT2 transgene insertion and not due to issues with 4-OHT uptake of the zebrafish embryo and larva: we have documented efficient switching with drl:creERT2 when induced at 48 hpf, confirming 10 μM 4-OHT penetration in superficial tissues such as fin fibroblasts and blood (Fig. 3C,D). Efficient loxP recombination has also been documented through labeling of oligodendrocytes at 6 dpf following a 5 μM 4-OHT induction at 5 dpf Tg(mbpa:mCherry-T2A-CreERT2), and in hepatocytes in 1–2 year old zebrafish livers following 2 μM 4-OHT induction from 5–7 dpf and 10–11 dpf (tp1:creERT2) 47,52. Labeled liver hepatocytes were observed at 10 weeks of age using TRE:creERT2 following 1 μM 4-OHT induction at 5 weeks for 3 consecutive days, however no description of 4-OHT delivery was described 12. The maintained integration of ubi:creERT2 is therefore best applied for basic tests and at early developmental stages to recombine lox-based Switch reporters.
Our observations here present the case that, ideally, multiple independent Switch lines should be used with individual Cre drivers to confirm findings and avoid biasing lineage information as not all Switch lines are created equal (Table 1). lox-based Switch reporters are highly sensitive to position effects, possibly arising from differential chromatin accessibility across independent transgenic insertions 1. Our transgene mapping provides first details of the chromatin environment and dynamics at the integration sites of the four transgenes tested here. From this limited sample size, the well-recombining hsp70l:Switch stands out as having integrated adjacent to a region with native H3K4me1, K3K4me3, and H3K27Ac marks indicative of open, active chromatin from early developmental stages 43 (Fig. 5). The ubi:Switch integration locus showed H3K27Ac marks across developmental timepoints, yet lacks H3K4 methylation marks. In contrast, the two less Cre-responsive actb2-based Switch lines that integrated into introns show no consistent histone marks associated with open chromatin (Fig. 6). Nonetheless, all observed correlation with open histone marks is based on native genome context without inserted transgene, and how the Tol2-based integrations themselves affect the chromatin context remains unknown. Our mapping further uncovered that while hsp70l:Switch did not integrate within a repetitive region, ubi:Switch, actb2:BFP-DsRed, and actb2:Stop-DsRed integrated within genomic repeat sequences of different classes (Fig. 5,6). These first insights should encourage future mapping of Tol2 transgene insertions towards identifying features that support consistent transgene expression also beyond Switch lines.
Table 1. Summary of the advantages and disadvantages of Switch transgenes.
Comparison of the relative switching efficiencies and logistical considerations for experimental design using the different Switch transgene lines described.
| Line | Advantages | Disadvantages |
|---|---|---|
| ubi:Switch | Second highest recombination efficiency | Cannot be combined with other EGFP lines |
| No need for heat-shock | Latency in reporter expression | |
| ubi:delta-EGFP | Can be combined with other EGFP lines | No screening marker |
| Same recombination efficiency as ubi:Switch | Latency in reporter expression | |
| hsp70l:Switch | Highest recombination efficiency | Need to heat-shock |
| Easy screening marker cryaa:Venus | ||
| Rapid transgene expression after heat-shock | ||
| actb2:BFP-DsRed | Can be combined with other EGFP lines | Low recombination efficiency |
| actb2:Stop-DsRed | Can be combined with other EGFP lines | Low recombination efficiency |
| No screening marker | ||
| Multiple integrations in current generation |
So-called safe harbor sites to integrate transgenes provide a key tool for reproducible transgene deployment in any given model. Previously generated transgene integrations for repeated insertion based on the phiC31 recombinase system have either shown weak to medium expression levels, have not been validated for lox-based recombination, or have not been maintained beyond proof-of-principle 1,53–58. Consequently, versatile and validated safe harbor sites are currently missing in zebrafish, rendering Tol2-or ISce-I-based random integration and subsequent screening for functional single-copy transgenes labor-intensive. Recent work has reported the Tol2-based generation of a phiC31-targetable attB integration site called SHH-SBE2 that is suitable for gene-regulatory element analysis through reproducible transgenesis into the same locus 57; while promising, if SHH-SBE2 shows long-term stability and is amenable to lox-based Switch reporters warrants further investigation. Retained switching efficiency over generations is a highly desirable feature of lox-based Switch reporter transgenics. CRISPR-Cas9-based knockin would provide means to target transgene insertion into suitable loci 21,59–62; nonetheless, little predictive information is available as to what genomic loci provide safe harbor sites. In mammalian systems, the serendipitously discovered Hipp11 (H11) locus is widely used for reproducible transgenesis 63–65. Our data argues that, in zebrafish, functional and stable Tol2 insertions of lox-based Switch transgenics indicate potential safe harbor sites that are also suitable for recombinase-sensitive cassettes. ubi:Switch and hsp70l:Switch have been maintained as functional lines in numerous laboratories since their isolation in 2009 14 and 2016 15, respectively. Considering the high recombination efficiency, and retained activity over generations of both ubi:Switch and hsp70l:Switch transgenes, we propose these loci as potential safe harbor sites for future transgenic work. A possible approach involves CRISPR-Cas9-based removal of either Switch transgene and integration of a phiC31-targeted landing site, such as by targeting the Tol2 transposon arms 5’ and 3’ of the respective transgene and screening for lost reporter expression. Altogether, our observations argue that mapping of well-working Tol2 transgene insertions, especially of lox-based Switch reporters, should become more widespread in the zebrafish community to inform about suitable transgene insertion sites with validated activity.
Experimental Design
Zebrafish Husbandry and Procedures
Animal care and procedures were carried out in accordance with the veterinary office of the IACUC of the University of Colorado School of Medicine (protocol #00979), Aurora, CO, USA. Females from the included Switch lines were crossed with male [regulatory-element]:creERT2 transgenic zebrafish using dividers and embryos were incubated at 28.5° C in E3 medium.
Drug Administration and Heat shock Protocol
Activity of CreERT2 was induced with 10 μM final concentration of (Z)-4-Hydroxytamoxifen (Sigma Aldrich, St. Louis, MO, USA, H7904, abbreviated as 4-OHT) in E3. 4-OHT stock is stored at −20°C in the dark as 10mM single-use aliquots dissolved in DMSO and used within 2 months of dissolving. Prior to administration, the 4-OHT aliquots were incubated at 65°C for 10 minutes and vortexed. For shield stage treatment, 4-OHT was administered overnight and then replaced with N-Phenylthiourea (Sigma Aldrich, P7629, abbreviated as PTU) at a final concentration of 200 μM in DMSO embryo medium each morning to inhibit melanogenesis. For embryos treated with 4-OHT at 2 dpf, PTU was administered at 24 hpf, 4-OHT/PTU was administered at 48hpf, replaced with PTU at 56 hpf and refreshed the next morning prior to imaging.
Tg(hsp70l:Switch) embryos were heat-shocked for 1 hour at 37°C in a water bath. Prior to heat shock embryos were dechorionated and transferred to a glass vial with E3 medium. Embryos were imaged 2 hours (stereo microscope) and 3 hours (confocal microscope) later.
The ubi:delta-EGFP transgenic line was made using the following sgRNA to target the EGFP ORF in ubi:Switch: 5’-GAGCTGGACGGCGACGTAAA-3’. The mutant allele harbors a 4 bp deletion resulting in a frameshift mutation beginning at Y40A, and premature stop at Stop42K. ubi:delta-EGFP was generated by injecting in vitro-transcribed sg RNA and recombinant Cas9 protein (PNA Bio) into ubi:Switch zygotes 66–68. Resulting F0s were screened for mosaic EGFP expression and a single, highly mosaic individual was selected to establish the line. Mutagenesis was confirmed by Sanger sequencing the PCR product amplified with Fw 5’-TTTAACATGGGAGAAGTGCAAAA-3’; Rev 5’-GTCGTCCTTGAAGAAGATGGTG-3’.
Imaging
Embryos were anesthetized at 3 dpf with 0.016% Tricaine-S (MS-222, Pentair Aquatic Ecosystems, Apopka, FL, USA, NC0342409) in E3 embryo medium. Basic fluorescence imaging was performed on a Leica M205FA with a DFC450 C camera. Laser scanning confocal microscopy was performed on a Zeiss LSM880 following embedding in E3 with 1% low-melting-point agarose (Sigma Aldrich, A9045) on glass bottom culture dishes (Greiner Bio-One, Kremsmunster, Austria, 627861). Images were collected with a x10/0.8 air-objective lens with all channels captured sequentially with maximum speed in bidirectional mode, with the range of detection adjusted to avoid overlap between channels. Maximum projections of acquired Z-stacks were made using ImageJ/Fiji 69 and cropped and rotated using Adobe Photoshop 2021.
Quantifications
For CTCF (corrected total cell fluorescence) calculations, lateral images were taking using the stereo microscope and then processed using ImageJ 29,69. Whole embryos (ubi:creERT2) or anterior half (tbx1:creERT2) were traced using the magic wand tool and then measured. For every picture, the CTCF (corrected total cell fluorescence) was calculated by the formula ‘Integrated density whole–(area whole embryo x mean fluorescence background)’. This formula is loosely based on a method described for calculating cell-fluorescence 70.
For ISV quantification lateral view confocal z-stacks were taken and then max projections were generated using imageJ. %ISV (Intersomitic vessels) were manually counted and calculated by dividing total switched ISV/total ISV 29.
For head vessel and myofiber quantification ventral view confocal z-stacks were taken and then max projections were generated using imageJ. To obtain the total number of head myofibers and vessels, these structures were manually counted using fli1a:EGFP embryos stained with the MF20 antibody as a vessel/myofiber control reference, as well as published data that quantified these features 71,72. Lineage-traced embryos were then imaged and processed in the same manner, and structures were again manually counted.
Statistics
Unpaired non-parametric (Mann-Whitney) two-tailed t-test was done to compare the scores between two groups. For analyses with more than two groups, 1-Way ANOVA was performed to compare the scores between the groups. Adjusted p-values after multiple tests correction are reported and significance was set at p < 0.05.
MF20 immunostaining
fli1a:EGFP zebrafish 73 were dechorionated and fixed at 3 dpf with 4% formaldehyde, 0.1% Triton-X100 in PEM (0.1 M PIPES, 2 mM MgSO4, and 1 mM EDTA) overnight at 4°C. The next day, embryos were washed in 1x PBS with 0.1% Triton-X100 and 0.1% BSA, permeabilized for 1 hour at RT with ProtK diluted in PBS to a final concentration of 5ug/ul and next for 30 min in 1x PBS with 0.5% Triton-X100 for 30 min. After washing, embryos were blocked in PBS + 1% goat serum and 0.1% Triton-X100 for 2 hours at room temperature and incubated overnight at 4°C with primary antibody MF20 (DSHB, antibody ID# AB_2147781) 1:50 diluted in blocking buffer. The 3rd day, embryos were washed 3 times 10 minutes in 1x PBS with 0.1% Triton-X100 and 0.1% BSA and incubated overnight at 4°C with the secondary antibody goat-anti-mouse A568 1:500 (Abcam, antibody ID# ab175473). On the 4th day, embryos were washed 3× 30 min with 1x PBS with 0.1% Triton-X100 and 0.1% BSA and kept at 4° C in Vectashield with DAPI diluted 1:5 in the same wash buffer until imaging.
TAIL-PCR
Protocol based on previous work 42,53:
Extract DNA from fin clips or single embryos.
Prepare primer mixtures: Combine 1.5 μM of each Tol2-specific primer with 10 μM of each AD primer.
Pipet primary PCR reaction: 1 μl Genomic DNA, 4 μl Primer mix (primary + random), 2 μl 10x Polymerase Buffer, 2 μl dNTPs (2 mM), 0.2 μl Polymerase (Expand™ High Fidelity PCR System, Roche), 10.8 μl ddH2O.
Perform primary PCR step (See PCR conditions below).
Dilute 2 μl of the primary PCR product in 25 μl ddH2O and add to the secondary reaction: 2 μl Diluted PCR product, 4 μl Primer mix (secondary + random), 2 μl 10x Polymerase Buffer, 2 μl dNTPs (2 mM), 0.2 μl Polymerase, 9.8 μl ddH2O.
Perform secondary PCR reaction (See PCR conditions below).
Dilute 2 μl of the secondary PCR product in 25 μl ddH2O and add to tertiary reaction. Pipette reaction analogous to the secondary reaction with tertiary primer and secondary product.
Perform tertiary PCR reaction (See PCR conditions below).
Compare secondary and tertiary PCR product on a 1.5% (w/v) agarose gel. If the amplification of the genomic region was successful, a slight band shift should occur between the secondary and the shorter tertiary product.
Excise bands and purify with QIAquick gel extraction kit (#NA1111–1KT)
Sequence purified PCR products.
Use BLAST (http://blast.ncbi.nlm.nih.gov/) to align to the zebrafish genome and identify 5’ and 3’ flanking regions.
The following gene-specific primers and random primers (AD3, 5, 6, 11) were used for TAIL-PCR:
Tol2 5’1 forward GGGAAAATAGAATGAAGTGATCTCC Tol2 TIR 5’
Tol2 5’2 forward GACTGTAAATAAAATTGTAAGGAG Tol2 TIR 5’
Tol2 5’3 forward CCCCAAAAATAATACTTAAGTACAG Tol2 TIR 5’
Tol2 3’1 reverse CTCAAGTACAATTTTAATGGAGTAC Tol2 TIR 3’
Tol2 3’2 reverse ACTCAAGTAAGATTCTAGCCAGA Tol2 TIR 3’
Tol2 3’3 reverse CCTAAGTACTTGTACTTTCACTTG Tol2 TIR 3’
AD-3 WGTGNAGNANCANAGA Random
AD-5 WCAGNTGWTNGTNCTG Random
AD-6 STTGNTASTNCTNTGC Random
AD-11 NCASGAWAGNCSWCAA Random
The primers needed for the mapping of Tol2 integrations as described previously 74. W: Weak base (A ort T), S: Strong base (C or G), N: Any base (A, C, G, T).
The following PCR conditions were used to TAIL-PCR:
Primary PCR reaction:
2 min 94° C
0.5 min 94° C
1 min 62° C
2.5 min 72° C
Repetition of steps 2–4 (5x)
0.5 min 72° C
3 min 25° C
Ramping 0.3°C/s to 72° C
2.5 min 72° C
10 s 94° C
1 min 61° C
2.5 min 72° C
10 s 94° C
Secondary PCR reaction:
10 s 94° C
1 min 61° C
2.5 min 72° C
10 s 94° C
1 min 61° C
2.5 min 72° C
10 s 94° C
1 min 44° C
Ramping 1.5° C/s to 72° C
2.5 min 72° C
Repetition of steps 1–10 (15x)
5 min 72° C
Tertiary PCR reaction:
0.25 min 94° C
1 min 44° C
Ramping 1.5° C/s to 72° C
2.5 min 72° C
Repetition of steps 1–4 (30x)
5 min 72° C
Transgene mapping confirmation
The following locus-specific primers were used to confirm the integration sites of ubi:Switch and hsp70l:Switch:
5’ ubi:Switch Fw GGAGCATTCAGAGGTACC
5’ ubi:Switch Rev GACTGTAAATAAAATTGTAAGGAG
3’ ubi:Switch Fw CTCAAGTACAATTTTAATGGAGTAC
3’ ubi:Switch Rev GCTGTGAGACGATCAGGC
5’ hsp70l:Switch Fw GCATGACACGGCTAACCAAC
5’ hsp70l:Switch Rev GACTGTAAATAAAATTGTAAGGAG
3’ hsp70l:Switch Fw CCTAAGTACTTGTACTTTCACTTG
3’ hsp70l:Switch Rev TATCAGCACACACCTTTATCGC
We were unable to fully confirm the integration sites for the actb2 Switch transgenes with independent locus-specific primers. We confirmed the 5’ genomic border of the actb2:BFP-DsRed transgene, however the 3’ genomic border and both 5’ and 3’ genomic borders of acbt2:Stop-DsRed could not be confirmed.
Acknowledgements
We thank Christine Archer and Molly Waters for zebrafish husbandry support, Dr. Oscar Ruiz for support with TAIL-PCR reagents, Dr. Caleb Doll for input on imaging, and all members of the Mosimann lab for input on the manuscript.
Funding statement
This work was supported by the University of Colorado School of Medicine, Department of Pediatrics and Section of Developmental Biology to C.M. and A.B., the Children’s Hospital Colorado Foundation to C.M.; the Swiss Bridge Foundation to C.M. and A.B.; a Swiss National Science Foundation (SNSF) postdoc fellowship to J.K.R.; NIGMS GDDR T32 1T32GM141742–01 to H.R.M.
Footnotes
Conflict of interest disclosure
The authors declare no competing interests.
Ethics approval statement
Animal care and procedures were carried out in accordance with the veterinary office of the IACUC of the University of Colorado School of Medicine (protocol #00979), Aurora, CO, USA.
Patient consent statement:
Not applicable
Permission to reproduce material from other sources:
Not applicable
Data availability statement
All data and reagents are contained within manuscript, raw data of feature counts for quantifications available upon request.
References
- 1.Carney TJ, Mosimann C. Switch and Trace: Recombinase Genetics in Zebrafish. Trends Genet 2018;34(5):362–378. doi: 10.1016/j.tig.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 2.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell 2004;6(1):7–28. doi: 10.1016/S1534-5807(03)00399-X [DOI] [PubMed] [Google Scholar]
- 3.Guillou F Mammalian genome targeting using site-specific recombinases. Published online 2006. doi: 10.2741/1867 [DOI] [PubMed]
- 4.Rossant J, Nagy A. Genome engineering: the new mouse genetics. Nat Med 1995;1(6):592–594. Accessed February 21, 2018. http://www.ncbi.nlm.nih.gov/pubmed/7585128 [DOI] [PubMed] [Google Scholar]
- 5.McLellan MA, Rosenthal NA, Pinto AR. Cre-loxP-Mediated Recombination: General Principles and Experimental Considerations. Curr Protoc Mouse Biol 2017;7(1):1–12. doi: 10.1002/cpmo.22 [DOI] [PubMed] [Google Scholar]
- 6.Sauer B Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol 1987;7(6):2087–2096. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3037344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2005;102(17):6068–6073. doi: 10.1073/pnas.0408708102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta V, Gemberling M, Karra R, Rosenfeld GE, Evans T, Poss KD. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol 2013;23(13):1221–1227. doi: 10.1016/j.cub.2013.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dirian L, Galant S, Coolen M, et al. Spatial Regionalization and Heterochrony in the Formation of Adult Pallial Neural Stem Cells. Dev Cell 2014;30(2):123–136. doi: 10.1016/J.DEVCEL.2014.05.012 [DOI] [PubMed] [Google Scholar]
- 10.Kaufman CK, Mosimann C, Fan ZP, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 2016;351(6272):aad2197. doi: 10.1126/science.aad2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knopf F, Hammond C, Chekuru A, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell 2011;20(5):713–724. Accessed January 5, 2022. http://www.cell.com/article/S1534580711001651/fulltext [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Agrawal I, Gong Z, Y L, I A, Z G. Reversion of tumor hepatocytes to normal hepatocytes during liver tumor regression in an oncogene-expressing transgenic zebrafish model. Dis Model Mech 2019;12(10). doi: 10.1242/DMM.039578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi TY, Ninov N, Stainier DYR, et al. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 2014;146(3):776–788. doi: 10.1053/J.GASTRO.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 2011;138(1):169–177. doi: 10.1242/dev.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felker A, Prummel KD, Merks AM, et al. Continuous addition of progenitors forms the cardiac ventricle in zebrafish. Nat Commun 2018;9(1). doi: 10.1038/s41467-018-04402-6 [DOI] [PMC free article] [PubMed]
- 16.Le X, Langenau DM, Keefe MD, Kutok JL, Neuberg DS, Zon LI. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci U S A 2007;104(22):9410–9415. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17517602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 1997;237(3):752–757. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9299439 [DOI] [PubMed] [Google Scholar]
- 18.Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS One 2009;4(2):e4640. doi: 10.1371/journal.pone.0004640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hans S, Freudenreich D, Geffarth M, Kaslin J, Machate A, Brand M. Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev Dyn 2011;240(1):108–115. doi: 10.1002/dvdy.22497 [DOI] [PubMed] [Google Scholar]
- 20.Jungke P, Hans S, Brand M. The zebrafish crezoo: An easy-to-handle database for novel CreER T2-driver lines. Zebrafish 2013;10(3):259–263. doi: 10.1089/ZEB.2012.0834 [DOI] [PubMed] [Google Scholar]
- 21.Kesavan G, Hammer J, Hans S, Brand M. Targeted knock-in of CreER T2 in zebrafish using CRISPR/Cas9. Cell Tissue Res 2018;372(1):41–50. doi: 10.1007/S00441-018-2798-X [DOI] [PubMed] [Google Scholar]
- 22.Jungke P, Hammer J, Hans S, Brand M. Isolation of Novel CreERT2-Driver Lines in Zebrafish Using an Unbiased Gene Trap Approach. PLoS One 2015;10(6). doi: 10.1371/JOURNAL.PONE.0129072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi I, Kobayashi-Sun J, Kim AD, et al. Jam1a–Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nat 2014 5127514 2014;512(7514):319–323. doi: 10.1038/nature13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blechinger SR, Evans TG, Tang PT, Kuwada JY, Warren JT, Krone PH. The heat-inducible zebrafish hsp70 gene is expressed during normal lens development under non-stress conditions. Mech Dev 2002;112(1–2):213–215. doi: 10.1016/S0925-4773(01)00652-9 [DOI] [PubMed] [Google Scholar]
- 25.Burket CT, Montgomery JE, Thummel R, et al. Generation and characterization of transgenic zebrafish lines using different ubiquitous promoters. Transgenic Res 2008;17(2):265–279. doi: 10.1007/S11248-007-9152-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashijima S ichi, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol 1997;192(2):289–299. doi: 10.1006/DBIO.1997.8779 [DOI] [PubMed] [Google Scholar]
- 27.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol 2003;4(12):1238–1246. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14608381 [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Xia L, Bruchas MR, Solnica-Krezel L. Imaging early embryonic calcium activity with GCaMP6s transgenic zebrafish. Dev Biol Published online 2017. doi: 10.1016/j.ydbio.2017.03.010 [DOI] [PMC free article] [PubMed]
- 29.Felker A, Nieuwenhuize S, Dolbois A, et al. In Vivo Performance and Properties of Tamoxifen Metabolites for CreERT2 Control. PLoS One 2016;11(4):e0152989. doi: 10.1371/journal.pone.0152989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell 2004;7(1):133–144. doi: 10.1016/j.devcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 31.Kikuta H, Kawakami K. Transient and stable transgenesis using tol2 transposon vectors. Methods Mol Biol 2009;546:69–84. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19378098 [DOI] [PubMed] [Google Scholar]
- 32.Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010;464(7285):108–111. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20154733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HS, Simon JA, Lis JT. Structure and expression of ubiquitin genes of Drosophila melanogaster. Mol Cell Biol 1988;8(11):4727–4735. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=2463465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimuro M, Nishiya T, Nomura Y, Yokosawa H. Involvement of polyubiquitin chains via specific chain linkages in stress response in mammalian cells. Biol Pharm Bull 2005;28(12):2315–2318. doi: 10.1248/BPB.28.2315 [DOI] [PubMed] [Google Scholar]
- 35.Bond U, Schlesinger MJ. Ubiquitin is a heat shock protein in chicken embryo fibroblasts. Mol Cell Biol 1985;5(5):949–956. doi: 10.1128/MCB.5.5.949-956.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fornace AJ, Alamo I, Hollander MC, Lamoreaux E. Ubiquitin mRNA is a major stress-induced transcript in mammalian cells. Nucleic Acids Res 1989;17(3):1215–1230. doi: 10.1093/NAR/17.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 1992;18(4):675–689. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1313711 [DOI] [PubMed] [Google Scholar]
- 38.Nenoi M, Mita K, Ichimura S, et al. Heterogeneous structure of the polyubiquitin gene UbC of HeLa S3 cells. Gene 1996;175(1–2):179–185. doi: 10.1016/0378-1119(96)00145-X [DOI] [PubMed] [Google Scholar]
- 39.Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A 2009;106(35):14896–14901. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19706417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosimann C, Panáková D, Werdich AA, et al. Chamber identity programs drive early functional partitioning of the heart. Nat Commun 2015;6. doi: 10.1038/ncomms9146 [DOI] [PMC free article] [PubMed]
- 41.Prummel KD, Hess C, Nieuwenhuize S, et al. A conserved regulatory program initiates lateral plate mesoderm emergence across chordates. Nat Commun 2019;10(1):3857. doi: 10.1038/s41467-019-11561-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 1995;25(3):674–681. http://www.ncbi.nlm.nih.gov/pubmed/7759102 [DOI] [PubMed] [Google Scholar]
- 43.Bogdanovic O, Fernandez-Miñán A, Tena JJ, et al. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 2012;22(10):2043–2053. doi: 10.1101/gr.134833.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labonty M, Pray N, Yelick PC. A Zebrafish Model of Human Fibrodysplasia Ossificans Progressiva. Zebrafish 2017;14(4):293–304. doi: 10.1089/zeb.2016.1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duszynski RJ, Topczewski J, LeClair EE. Simple, Economical Heat-Shock Devices for Zebrafish Housing Racks. Zebrafish 2011;8(4):211. doi: 10.1089/ZEB.2011.0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duszynski RJ, Topczewski J, LeClair EE. Divergent requirements for fibroblast growth factor signaling in zebrafish maxillary barbel and caudal fin regeneration. Dev Growth Differ 2013;55(2):282–300. doi: 10.1111/DGD.12035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinzon-Olejua A, Welte C, Chekuru A, et al. Cre-inducible site-specific recombination in zebrafish oligodendrocytes. Dev Dyn 2017;246(1):41–49. doi: 10.1002/DVDY.24458 [DOI] [PubMed] [Google Scholar]
- 48.Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development 2011;138(22):4831–4841. doi: 10.1242/DEV.072587 [DOI] [PubMed] [Google Scholar]
- 49.Trompouki E, Bowman TV, Lawton LN, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 2011;147(3):577–589. doi: 10.1016/j.cell.2011.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwan KM, Fujimoto E, Grabher C, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- 51.Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn 2003;228(1):30–40. doi: 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- 52.Zhang W, Chen J, Ni R, Yang Q, Luo L, He J. Contributions of biliary epithelial cells to hepatocyte homeostasis and regeneration in zebrafish. iScience 2021;24(3):102142. doi: 10.1016/J.ISCI.2021.102142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosimann C, Puller AC, Lawson KL, Tschopp P, Amsterdam A, Zon LI. Site-directed zebrafish transgenesis into single landing sites with the phiC31 integrase system. Dev Dyn 2013;242(8):949–963. doi: 10.1002/dvdy.23989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu G, Goll MG, Fisher S. PhiC31 Integrase Mediates Efficient Cassette Exchange in the Zebrafish Germline. Dev Dyn 2011;240(9):2101–2107. doi: 10.1002/dvdy.22699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts JA, Miguel-Escalada I, Slovik KJ, et al. Targeted transgene integration overcomes variability of position effects in zebrafish. Development 2014;141(3):715–724. doi: 10.1242/dev.100347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Maddison LA, Chen W. PhiC31 integrase induces efficient site-specific excision in zebrafish. Transgenic Res 2011;20(1):183–189. doi: 10.1007/s11248-010-9394-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhatia S, Kleinjan DJ, Uttley K, Mann A, Dellepiane N, Bickmore WA. Quantitative spatial and temporal assessment of regulatory element activity in zebrafish. Elife 2021;10. doi: 10.7554/ELIFE.65601 [DOI] [PMC free article] [PubMed]
- 58.Lister JA. Transgene excision in zebrafish using the phiC31 integrase. Genesis 2010;48(2):137–143. doi: 10.1002/dvg.20595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Auer TO, Duroure K, De Cian A, Concordet J-PP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 2014;24(1):142–153. doi: 10.1101/gr.161638.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prykhozhij SV, Fuller C, Steele SL, et al. Optimized knock-in of point mutations in zebrafish using CRISPR/Cas9. Nucleic Acids Res 2018;46(17):e102–e102. doi: 10.1093/nar/gky512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 2014;4:6545. doi: 10.1038/srep06545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin J, Chen J, Solnica-Krezel L. Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development 2014;141(19):3807–3818. doi: 10.1242/dev.108019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hippenmeyer S, Youn YH, Moon HM, et al. Genetic Mosaic Dissection of Lis1 and Ndel1 in Neuronal Migration. Neuron 2010;68(4):695. doi: 10.1016/J.NEURON.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tasic B, Hippenmeyer S, Wang C, et al. Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc Natl Acad Sci U S A 2011;108(19):7902–7907. doi: 10.1073/pnas.1019507108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu F, Gamboa M, Farruggio AP, et al. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res 2014;42(5):e34. doi: 10.1093/NAR/GKT1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 2015;12(6):535–540. doi: 10.1038/nmeth.3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hwang WYY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013;31(3):227–229. doi: 10.1038/nbt.2501nbt.2501 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burger A, Lindsay H, Felker A, et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 2016;143(11):2025–2037. doi: 10.1242/dev.134809 [DOI] [PubMed] [Google Scholar]
- 69.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9(7):676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burgess A, Vigneron S, Brioudes E, Labbé J-C, Lorca T, Castro A. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci U S A 2010;107(28):12564–12569. doi: 10.1073/pnas.0914191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development 1997;124(15):2945–2960. doi: 10.1242/DEV.124.15.2945 [DOI] [PubMed] [Google Scholar]
- 72.Isogai S, Horiguchi M, Weinstein BM. The Vascular Anatomy of the Developing Zebrafish: An Atlas of Embryonic and Early Larval Development. Dev Biol 2001;230(2):278–301. doi: 10.1006/DBIO.2000.9995 [DOI] [PubMed] [Google Scholar]
- 73.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 2002;248(2):307–318. Accessed March 29, 2016. http://www.ncbi.nlm.nih.gov/pubmed/12167406 [DOI] [PubMed] [Google Scholar]
- 74.Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn 2004;231(2):449–459. doi: 10.1002/dvdy.20157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and reagents are contained within manuscript, raw data of feature counts for quantifications available upon request.


