Here, Mitchell et al. investigated the molecular mechanisms critical for CSC population maintenance. They developed a spatially resolved loss-of-function screen in GBM patient-derived organoids to identify essential epigenetic regulators in the SOX2-enriched, therapy-resistant niche and identified WDR5 as indispensable for this population. Their findings highlight the role of WDR5 and the WRAD complex in maintaining the CSC state and provide a rationale for therapeutic development of WDR5 inhibitors for GBM and other advanced cancers.
Keywords: epigenetics, glioblastoma, stem cell
Abstract
Glioblastomas (GBMs) are heterogeneous, treatment-resistant tumors driven by populations of cancer stem cells (CSCs). However, few molecular mechanisms critical for CSC population maintenance have been exploited for therapeutic development. We developed a spatially resolved loss-of-function screen in GBM patient-derived organoids to identify essential epigenetic regulators in the SOX2-enriched, therapy-resistant niche and identified WDR5 as indispensable for this population. WDR5 is a component of the WRAD complex, which promotes SET1 family-mediated Lys4 methylation of histone H3 (H3K4me), associated with positive regulation of transcription. In GBM CSCs, WDR5 inhibitors blocked WRAD complex assembly and reduced H3K4 trimethylation and expression of genes involved in CSC-relevant oncogenic pathways. H3K4me3 peaks lost with WDR5 inhibitor treatment occurred disproportionally on POU transcription factor motifs, including the POU5F1(OCT4)::SOX2 motif. Use of a SOX2/OCT4 reporter demonstrated that WDR5 inhibitor treatment diminished cells with high reporter activity. Furthermore, WDR5 inhibitor treatment and WDR5 knockdown altered the stem cell state, disrupting CSC in vitro growth and self-renewal, as well as in vivo tumor growth. These findings highlight the role of WDR5 and the WRAD complex in maintaining the CSC state and provide a rationale for therapeutic development of WDR5 inhibitors for GBM and other advanced cancers.
Glioblastoma (GBM) is the most common primary malignant brain tumor and remains highly lethal despite an aggressive multimodal standard of care approach. GBM displays profound cellular heterogeneity, with tumor growth and therapeutic resistance driven by populations of cells with stem cell-like properties (cancer stem cells [CSCs]). Neurodevelopmental transcription factors such as SOX2, POU3F2, SALL2, and OLIG2 are expressed in subpopulations of GBM tumor cells, are necessary and sufficient for tumor propagation in vivo, and cooperate to maintain stem cell-like epigenetic landscapes (Suvà et al. 2014). Such epigenetic regulation controls the access of transcription factors to defined sets of genes and allows for the transition of cells between states (Huang 2013). Thus, transcriptional and epigenetic programs cooperatively promote maintenance of the CSC state in GBM.
GBM cell state plasticity in response to external stimuli is facilitated, at least in part, by chromatin reorganization. Specific DNA methylation, histone methylation/acetylation, and chromatin accessibility patterns are predictive of patient response to therapy in GBM. Accordingly, several epigenetic regulators are elevated in GBM CSCs, are necessary and sufficient for self-renewal, and likely generate permissive chromatin states that facilitate maintenance of the CSC state amid cell interactions and therapeutic pressures (for reviews, see Flavahan et al. 2017; Valor and Hervás-Corpión 2020; Yabo et al. 2022).
Primary patient-derived GBM organoids show regional heterogeneity with a proliferative, SOX2-enriched outer rim and a hypoxic core harboring quiescent CSCs and non-CSC tumor cells (Hubert et al. 2016). To elucidate epigenetic factors responsible for maintenance of the CSC state in GBM in the context of heterogeneous cell populations in each niche, we used an unbiased organoid-based screen targeting epigenetic regulators. Through this screen, we identified WDR5 as a key CSC regulator. Targeting WDR5 provides an alternative approach to directly inhibiting core “stemness” transcription factors (such as the transcriptional master regulator and GBM CSC marker SOX2) that regulate CSCs but are challenging to individually target due to their complex and varied interactions with proteins and DNA (Gangemi et al. 2009). Here we tested the hypothesis that targeting WDR5 could be a means to compromise the ability of GBM CSCs to maintain a favorable epigenetic state.
Results
Patient-derived GBM organoid specimens exhibit increased SOX2 expression within the highly proliferative rim region
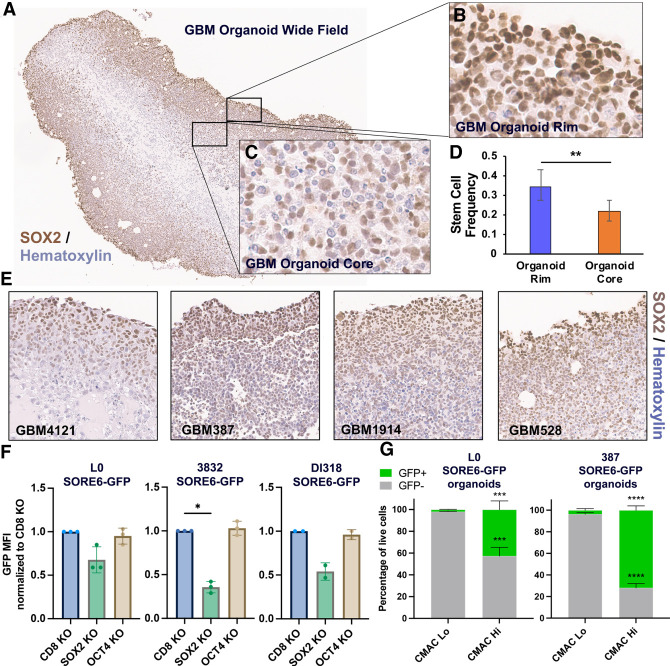
GBM CSCs reside in defined tumor niches and display complex interactions with their microenvironment and surrounding cell populations (for review, see Mitchell et al. 2021). To better model the cellular heterogeneity in GBM and capture the complex dependencies of CSCs, we leveraged 3D organoid models of GBM, which recapitulate cellular states observed in primary patient tumors, including niches of SOX2+ CSCs, and allow for interaction of cells in various states in an in vitro system. This GBM organoid outer rim is highly proliferative and enriched for SOX2+ cells, but SOX2+ cells within this region are resistant to standard of care and other clinically relevant therapeutics (Hubert et al. 2016; Sundar et al. 2022). A range of patient-derived GBM organoid specimens recapitulated enriched SOX2 expression within the rim region (Fig. 1A–C,E; Supplemental Fig. S1A,B). We isolated viable GBM populations from the rim and core regions by labeling with CellTracker blue CMAC fluorescent dye followed by FACS. We found the outer rim of GBM organoids is enriched for functionally self-renewing SOX2+ CSCs compared with the inner hypoxic core (Fig. 1D).
Figure 1.
SOX2 is enriched in the highly proliferative rim region of patient-derived GBM organoids. (A–C) SOX2 IHC shows enrichment of SOX2+ cells in the GBM organoid rim. (D) Stem cell behavior (sphere formation) shown by limiting dilution assay in cells isolated from the organoid rim and core. Error bars are 95% confidence intervals. P-value was determined by χ2 test. (E) SOX2 IHC of four different patient-derived GBM organoid specimens. Fields of view (20× magnification) for individual specimens are shown. (F) SOX2 and OCT4 were knocked out via CRISPR:Cas9 in SORE6-GFP transduced GBM CSCs, and SORE6-GFP reporter expression was measured by flow cytometry. Bars represent geometric mean fluorescence intensity (MFI) for GFP relative to CD8A control, ±SD; symbols are biological replicates. P-values were determined by two-tailed, paired t-tests. (G) GBM organoids derived from SORE6-GFP transduced GBM CSCs were regionally labeled with the CellTracker blue CMAC fluorescent dye, dissociated, and analyzed by flow cytometry to measure GFP expression in the CMAC+ and CMAC− niches. Error bars are SD; n = 3 biological replicates per line. P-values were determined by two-tailed, unpaired t-tests.
To measure the abundance of SOX2+ cells in the organoid rim, we turned to the lentiviral-based SOX2/OCT4 response element (SORE6) reporter system, where a destabilized copGFP is expressed in response to binding to tandem repeats of the SOX2::OCT4 motif (Supplemental Fig. S1C; Tang et al. 2015). The destabilized GFP allows for real-time monitoring of transcription factor activity. To interrogate functionality of the SORE6-GFP system in GBM CSCs, we knocked out SOX2 and OCT4 via CRISPR:Cas9-mediated genome editing and monitored the effect on SORE6-GFP expression (Fig. 1F; Supplemental Fig. S1D–G). In the three GBM CSC isolates tested, SOX2 KO, but not OCT4 KO, reduced GFP reporter activity, suggesting that SOX2 is the main driver of the SOX2::OCT4 motif in these cells, and this reporter can be used as a readout of SOX2 activity. To quantify SOX2-expressing cells in organoid regions, we generated and matured organoids from SORE6-GFP transduced GBM CSCs. Next, we regionally labeled GBM organoids with CMAC and assessed dissociated organoids by flow cytometry. The CMAC+ population contained the vast majority of GFP+ cells (Fig. 1G; Supplemental Fig. S1H). These data corroborate our IHC data demonstrating enrichment of SOX2 in the outer organoid niche.
Spatially resolved organoid screening reveals WDR5 is essential for CSC survival
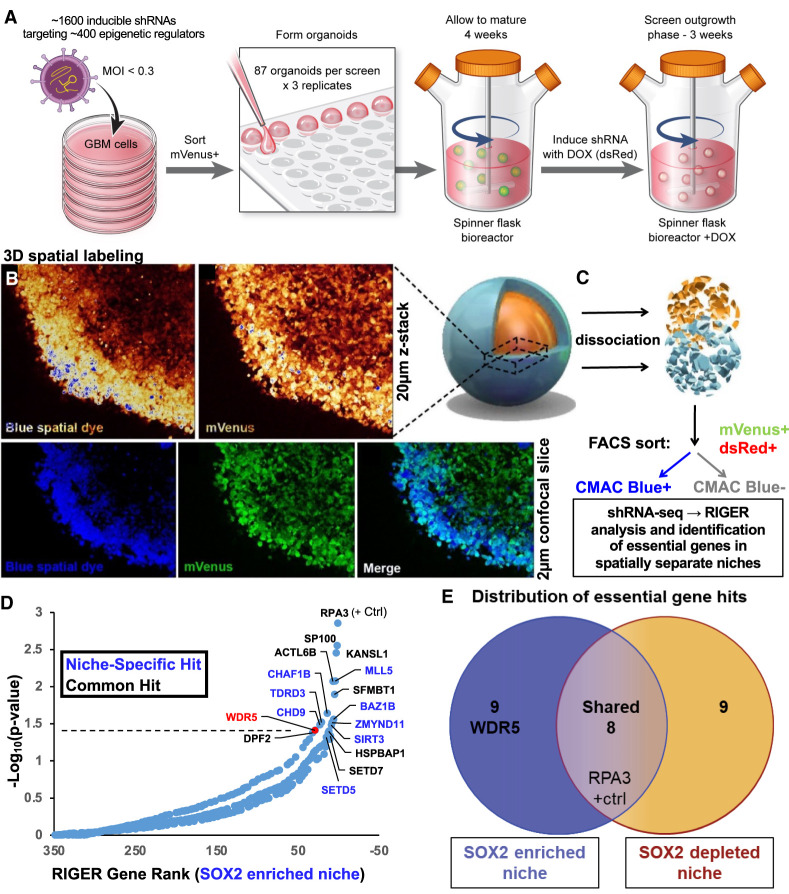
To elucidate epigenetic regulators responsible for maintenance of the SOX2+, therapy-resistant cellular state, we adapted our methods to enable high-throughput functional genomics screening in 3D organoid culture. We used a pooled inducible lentiviral shRNA library to target ∼400 epigenetic-modifying genes in GBM CSCs, FACS-sorted virus-infected (mVenus+) CSCs, seeded CSCs to generate several hundred organoids in parallel, and allowed the organoids to mature for 1 mo in spinning bioreactors prior to shRNA induction and outgrowth. We waited 1 mo prior to doxycyline-induced shRNA induction (dsRed+) to avoid affecting cells prior to stable microenvironment formation (Fig. 2A; Supplemental Fig. S2A). We validated viral integration (mVenus+) and shRNA induction (dsRed+) in the entire organoid by microscopy. We then spatially labeled GBM organoids with CMAC as described above and verified proper organoid regional labeling using live confocal imaging (Fig. 2B) prior to organoid dissociation. We FACS-sorted on successfully Dox-induced mVenus+dsRed+ cells in each region (CMAC+ or CMAC−), and DNA was isolated from sorted populations, tagged with unique molecular barcodes, and deconvolved by high-throughput sequencing of the integrated shRNA libraries as previously described (Fig. 2C; Miller et al. 2017). Greater than 500-fold representation of library complexity was maintained at every step in the screened populations, and full screens were performed in triplicate. Genes identified by RIGER analysis (Broad Institute) as essential were separated into overlapping or niche-specific targets (Fig. 2D,E; Supplemental Fig. S2B). RPA3 knockdown, known to be broadly cell-lethal, was found to be essential in both niches in our screen. Since knockdowns common to both organoid regions are likely enriched for universally required genes and therefore are less likely to have a therapeutic window upon translation to therapy, we focused on genes uniquely essential in the SOX2+-enriched niche (Fig. 2D). This included MLL5, a gene critical for maintaining CSC self-renewal in GBM (Gallo et al. 2015), underscoring the ability of our platform to identify biologically meaningful hits in GBM CSCs.
Figure 2.
Spatial functional genomics screening defines genes essential in cancer niches. (A–C) GBM CSCs were infected with an inducible shRNA library targeting epigenetic modifiers (mVenus reporter) and grown into organoids. (A) shRNAs were induced with doxycycline (leading to dsRed reporter expression), and after 3 wk, organoids were incubated with CellTracker blue CMAC fluorescent dye to label the entire outer rim region. Live confocal imaging of screened GBM528 organoids was used to verify proper spatial labeling prior to subsequent dissociation. (B) Z-stacks showing labeling intensity (pseudocolored) and individual image slices showing labeling overlap are shown using a 20× objective. (C) Subsequently, single cells were isolated from the organoids and separated into rim (CMAC+) or core (CMAC−) populations by FACS, and then DNA was isolated for barcode sequencing and analysis. (D) Rank-ordered list of genes targeted in the shRNA screen, ranked by depletion of the shRNA in the SOX2-enriched niche (CMAC+). The dotted line represents P = 0.05 as determined by RIGER analysis of all hairpin sequences and replicates. Niche-specific hits are in blue, and common hits (including RPA3-positive control) are shown in black. (E) Venn diagram of screen hits showing localization in SOX2-enriched or SOX2-depleted niches or common to both regions.
Our screen also identified the trithorax protein WD repeat domain 5 (WDR5) as an essential gene for growth within the SOX2-enriched niche of GBM organoids. WDR5 is best characterized as a core subunit of the WRAD complex, which also includes the proteins RBBP5, ASH2L, and DPY30, two of which were found to be important for the GBM CSC state, including in vivo (Alvarado et al. 2017; Miller et al. 2017; Dixit et al. 2022). The WRAD complex interacts with SET1/MLL methyltransferases to facilitate post-translational modifications on histone tails, including histone 3 lysine 4 (H3K4) monomethylation, dimethylation, and trimethylation, which are associated with transcriptionally permissive chromatin (Santos-Rosa et al. 2002). WDR5 mediates self-renewal in embryonic stem cells by regulating the OCT4–SOX2–NANOG pluripotency transcription factor network (Ang et al. 2011). WDR5 is functionally important in serum grown GBM cell lines (Dai et al. 2020; Wang et al. 2020), but its function has not been investigated in CSC or GBM organoid culture, and therapeutic inhibition of WDR5 has not been tested in GBM.
Our screen identified KANSL1 as essential in both organoid niches. WDR5 binds KANSL1 as an essential part of the nonspecific lethal H4 acetyltransferase complex (Dias et al. 2014). KANSL1 and MLL1 both bind WDR5, mutually exclusively, at its WIN site, an arginine binding cavity. While WDR5 WIN inhibition disrupted WDR5/MLL1 binding (Tian et al. 2020), WDR5/KANSL1 binding was not affected (Guarnaccia et al. 2021). As a tool to inhibit WDR5 without affecting the commonly essential KANSL1, we first treated organoids with a commercially available WDR5 WIN site peptide inhibitor, MM-102, previously shown to inhibit MLL1 histone methyltransferase (HMT) activity in vitro (Karatas et al. 2013). We quantified cell proliferation in each organoid region using immunohistochemistry (IHC) for a mitotic marker (phosphorylated histone H3 [pHH3]). MM-102 reduced pHH3+ cells in the SOX2-enriched niche, recapitulating the screen results (Supplemental Fig. S2C). We turned to conventional CSC-enriching culture conditions to expand cells and validate WDR5 function in CSCs more efficiently. We detected SOX2 expression in GBM models in CSC-enriching culture conditions, and this expression was reduced in serum conditions (Supplemental Fig. S2D). We observed a reduction in CSC number and proliferation upon treatment with MM-102 in a dose-dependent manner (Supplemental Fig. S2E,F). These data demonstrated that WDR5 inhibition is detrimental to GBM CSC growth in organoid and sphere culture and provided rationale for the further study of WDR5 in GBM.
WDR5 small molecule inhibition reduces the interaction between WDR5 and WRAD complex members and broadly diminishes histone 3 lysine 4 trimethylation (H3K4me3)
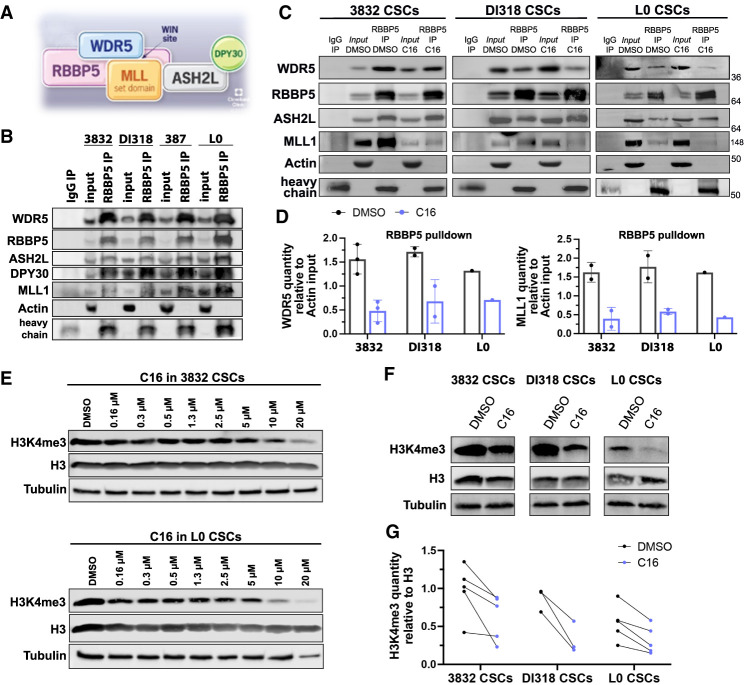
We next sought to detect the interaction between WDR5 and members of the WRAD complex in GBM CSCs. By immunoprecipitation of either RBBP5 or WDR5 and immunoblotting, we detected the association of WDR5 with RBBP5 and the other WRAD complex members (Fig. 3A,B; Supplemental Fig. S3A). MLL1 is functionally important in GBM CSCs (Heddleston et al. 2012; Gallo et al. 2013) and, unlike for other SET1 family methyltransferases (SET1A, SET1B, and MLL2–MLL4), is specifically dependent on WDR5 for its methyltransferase activity (Dou et al. 2006; Li et al. 2016). Thus, we immunoblotted for MLL1 and found that it was bound to WDR5 and RBBP5 (Fig. 3B). To assess WDR5 WIN site inhibition in the context of this complex, we turned to compound 16 (C16), a recently disclosed small molecule WDR5 WIN site inhibitor (Supplemental Fig. S3B; Tian et al. 2020). C16 was synthesized in a series of compounds that have improved on-target potency and drug-like properties compared with previously described WDR5 WIN site inhibitors. C16 was the most potent in this series, with picomolar binding affinity for WDR5 and low nanomolar inhibition of MLL1 HMT activity. In addition, an X-ray cocrystal structure of C16 bound to WDR5 has been solved (Tian et al. 2020). Using recombinant WDR5, C16 robustly displaced a fluorescently labeled MLL1-derived peptide using the time-resolved fluorescence energy transfer (TR-FRET) assay (Supplemental Fig. S3C). To gain further insight into the effects of C16 in GBM CSCs, we performed coimmunoprecipitation studies in GBM CSCs after C16 treatment with a focus on MLL1 and core WRAD complex members. WIN site inhibition specifically inhibits MLL1 and SET1A HMT activity (Patel et al. 2008; Alicea-Velázquez et al. 2016). As expected, MLL1 was displaced from WDR5 and RBBP5 upon C16 treatment. The interaction between WDR5 and RBBP5 was also reduced (Fig. 3C,D; Supplemental Fig. S3D,E). C16 targets the WDR5/MLL1 interaction site, which is distinct from the WDR5/RBBP5 interaction site, yet the RBBP5 interaction appears to be allosterically affected in GBM CSCs. In previous studies, WDR5 WIN site inhibitor OICR-9429 reduced the WDR5/RBBP5 interaction in HEK293 cells (Grebien et al. 2015), but similar results were not seen with WIN site inhibitor C6 in HEK293-WDR5-FLAG cells (Guarnaccia et al. 2021) or with C16 in AML cells (Tian et al. 2020). Thus, WDR5/WIN site inhibition may have varied effects in different tumor contexts.
Figure 3.
WDR5 inhibition reduces the interaction between WDR5 and WRAD complex members and diminishes the H3K4me3 mark. (A) Model of the WRAD complex indicating points of protein–protein interactions, based on structures solved in Xue et al. (2019). (B) Immunoprecipitation of RBBP5 in GBM CSCs. Immunoblotting was performed for WRAD complex members and MLL1. Inputs are 10%. Representative experiments are shown. (C) Immunoprecipitation of RBBP5 after C16 inhibitor treatment (5 µM for 24 h) in GBM CSC models. Immunoblotting was performed for WRAD complex and MLL1. Inputs are 10%. Representative experiments are shown. (D) Related to C. Quantification of WDR5 and MLL1 immunoprecipitated by RBBP5, relative to actin quantity from each sample's input. Circles are biological replicates. (E) Western blots showing H3K4me3 levels in whole-cell lysates after C16 treatment for 72 h at different doses in CSC models. Representative experiments are shown. (F) Western blots showing H3K4me3 levels in whole-cell lysates after C16 treatment (5 µM for 72 h). Representative experiments are shown. (G) Quantification of H3K4me3, relative to H3 quantity after C16 treatment (5 µM for 72 h). Circles are biological replicates; lines connect DMSO- and C16-treated specimens from the same experiment.
We next aimed to assess MLL1 function in response to C16 treatment by measuring H3K4me3 levels. We observed a global reduction in H3K4me3 by Western blot in a dose-dependent manner (Fig. 3E–G). These data corroborate previous findings that WDR5 depletion or inhibition reduces genome-wide H3K4me3 (Benayoun et al. 2014; Zhang et al. 2018; Siladi et al. 2022).
WDR5 inhibition leads to H3K4me3 loss at essential CSC genes
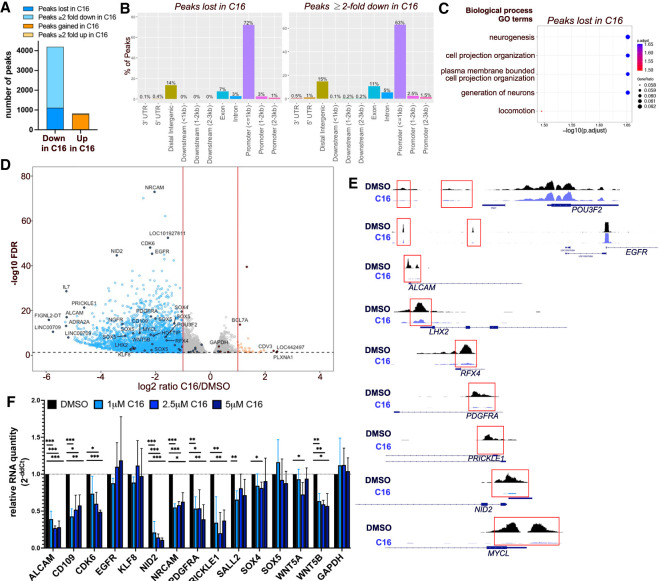
Based on changes in H3K4me3 by Western blot, we used the cleavage under targets and tagmentation (CUT&Tag) approach (Kaya-Okur et al. 2019) to identify genome-wide changes in H3K4me3 localization and abundance after C16 treatment. We treated DI318, a newly derived CSC model from a GBM patient specimen, with its IC50 dose of C16 (3 μM) for 72 h and performed CUT&Tag for H3K4me3. We used MACS2 for peak calling and found that replicates from each group clustered closely together based on similarities of peak sequences (Supplemental Fig. S4A). In DMSO-treated cells, we identified an average of 21,224 H3K4me3 peaks across replicates, while in C16-treated cells, we identified an average of 19,502 peaks across replicates. We first sought to identify peaks that disappear or appear following C16 treatment and are therefore unique to each group. After excluding peaks <50 bp, we identified 1110 H3K4me3 peaks unique to the DMSO group (i.e., lost in the C16 treatment group; corresponding to 995 genes) and 783 peaks unique to the C16 group (corresponding to 758 genes) (Fig. 4A; Supplemental Table S1). Peaks lost with C16 treatment were generally larger than peaks gained with C16 treatment (Supplemental Fig. S4B). H3K4me3 is enriched immediately downstream from TSSs (Guenther et al. 2005); thus, as expected, the majority (72%) of peaks lost with C16 treatment were contained within 1 kb of transcription start sites (TSSs). Peaks in exons and distal intergenic regions made up 20% of peaks lost (Fig. 4B). Genes with peaks lost with C16 treatment were enriched for biological processes including neurogenesis and cell projection (Fig. 4C), while genes with peaks gained with C16 were enriched for catabolic processes, tube formation, intracellular protein transport, and RNA metabolic processes (Supplemental Fig. S4C), suggestive of compensatory metabolic and developmental pathways being up-regulated in response to WDR5 inhibition.
Figure 4.
WDR5 inhibition leads to H3K4me3 loss at essential CSC genes. (A) Number of H3K4me3 CUT&Tag peaks lost, gained, decreased, or increased (log2FC ≤ −1 or log2FC ≥ 1) after C16 treatment (3 μM for 72 h) of DI318 cells. (B) Bar plot showing distribution of peaks in gene regions that were lost in the C16 group (unique to the DMSO group) or reduced twofold or greater in the C16 treatment group. (C) MSigDB gene set annotations enriched among CUT&Tag peaks lost in the C16 treatment group. (D) Volcano plot of differential H3K4me3 peaks detected by CUT&Tag in DI318 CSCs. Blue dots represent H3K4me3 peaks reduced twofold or greater (log2FC ≤ −1) with C16 treatment, and orange circles are H3K4me3 peaks increased twofold or greater (log2FC ≥ 1) with C16 treatment. Multiple peaks (circles) may map to the same gene. (E) Representative H3K4me3 peaks from CUT&Tag at the indicated genes (from Integrative Genomics Viewer). (F) qPCR for specified genes in DI318 CSCs treated with the indicated doses of C16 for 72 h. Bars represent mean expression of n = 3 biological replicates, normalized to ACTB levels by ddCt method, ±SD.
We next identified consensus peaks between the DMSO and C16 treatment groups to determine whether there was differential enrichment of these peaks between groups (Supplemental Table S2). Seventy-seven percent of consensus peaks were contained within 1 kb of TSSs (Supplemental Fig. S4D). Of all consensus peaks with FDR of <0.05 (7728 peaks), 3085 were diminished at least twofold in the C16 group compared with the DMSO group (Fig. 4A,D). These 3085 peaks corresponded to 2454 genes, 599 of which were the same genes that lost a peak with C16 treatment (Supplemental Fig. S4E). Only 39 peaks were increased twofold or greater in the C16 treatment group (corresponding to 37 genes) (Supplemental Fig. S4E). Sixty-three percent of peaks diminished twofold or greater in C16 were contained within 1 kb of TSSs (Fig. 4B). Loss of H3K4me3 in C16-treated CSCs occurred on genes with previously described roles in GBM, such as ALCAM, CD109, EGFR, KLF8, NRCAM, PDGFRA, and SOX4 (Sehgal et al. 1999; Jackson et al. 2006; Kijima et al. 2012; Schnell et al. 2012; Zhang et al. 2014; Furnari et al. 2015; Filppu et al. 2021), and stem cell master transcription factors LHX2, MYCL, RFX4, CITED1, HEY2, SOX5, POU3F2, and SALL2 (Fig. 4D,E), of which the latter two are essential for GBM propagation (Suvà et al. 2014). Genes with H3K4me3 peaks diminished with C16 treatment were enriched for brain developmental and differentiation pathways; synaptic signaling; small GTPase signal transduction; oncogenic signatures including KRAS, E2F3 (involved in G1/S transition), EGFR, LEF1, and WNT/Frizzled binding; and the WNT signaling pathway (Supplemental Fig. S5A–D). WDR5 has been previously implicated in regulating β-catenin transcription and activating the Wnt/β-catenin signaling pathway via interaction with the long noncoding RNA HOTTIP (Liu et al. 2020), whose corresponding gene also displayed loss of H3K4me3 in C16-treated CSCs (Fig. 4D; Supplemental Table S2). We measured expression of several of these targets at the mRNA level and found that some, but not all, CSC-relevant genes with H3K4me3 loss also had reduced expression with C16 treatment (Fig. 4F; Supplemental Fig. S4F). For some genes (EGFR, KLF8, SOX4, and SOX5), there was minimal or no reduction in expression despite reduction in H3K4me3. It is likely that stringent regulation of these key factors requires coordination of multiple chromatin modifications to significantly alter gene expression.
CUT&Tag reveals loss of H3K4me3 at specific POU domain DNA binding motifs
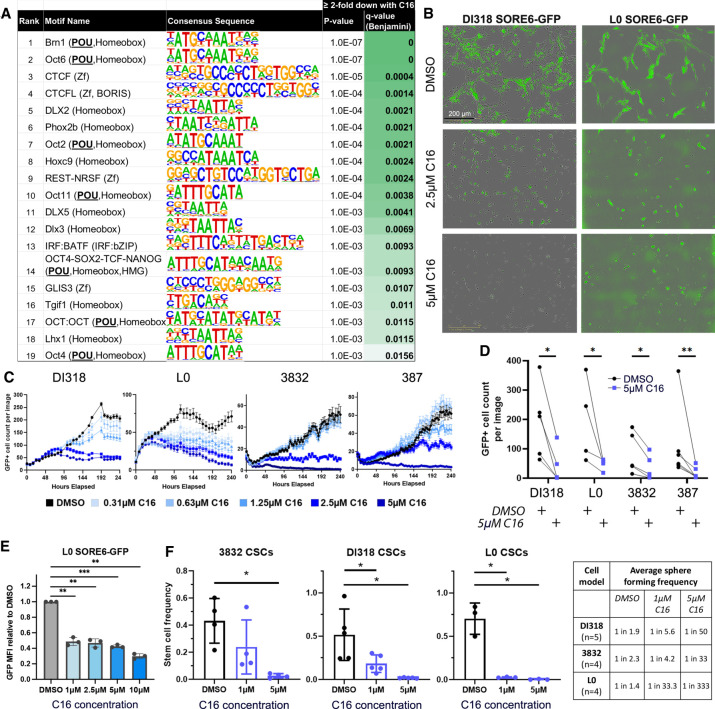
To ask mechanistically which gene regulatory programs are most affected by WDR5 perturbation in CSCs, we identified enriched DNA binding motifs within H3K4me3 peaks reduced twofold or greater in the C16 treatment group via HOMER analysis. The most significantly changed motifs (Q-value < 0.02) correspond to transcription factors important in GBM maintenance, including OCT4 (POU5F1), SOX2, BRN1 (POU3F3), CTCF, and REST, and were enriched [p(X ≥ 7) < 5.7 × 10−7] by hypergeometric test for members of the POU domain transcription factor family (Fig. 5A). In embryonic stem cells, WDR5 and the POU transcription factor OCT4 interact, and DNA specificity conferred by OCT4 directs WDR5 to specific loci; namely, those driving self-renewal (Ang et al. 2011). As it is currently unknown how WDR5 is specified to histone targets in GBM, these data provide insight into factors that may cooperate with WDR5 in promoting activation of target gene programs.
Figure 5.
WDR5 inhibition leads to H3K4me3 loss preferentially at POU domain DNA binding motifs, diminishes SORE6 reporter+ cells, and compromises GBM CSC self-renewal. (A) Top DNA binding motifs enriched within H3K4me3 peaks reduced (log2FC ≤ −1) with C16 treatment. Enrichment P-values were computed using the HOMER motif analysis tool set with all significant peaks from DiffBind analysis as background. (B–D) GBM CSC models transduced with the SORE6-GFP reporter were treated with C16. (B) Representative images of day 7 after treatment. (C) GFP+ cell numbers were quantified over 10 d using IncuCyte live-cell imaging. One representative time course experiment is shown for each CSC model. Average GFP+ cell count per image is plotted at each time point, ±SEM per image. Multiple images were taken per well with n = 3 technical replicates (wells). (D) GFP+ cell numbers at day 7 after treatment with 5 µM C16. Each line is a biological replicate. P-values were determined by two-tailed, paired t-tests. (E) GFP intensity of live L0 SORE6-GFP cells after C16 treatment for 3 d. (F) In vitro limiting dilution analysis of CSCs in the presence of C16. Bars represent mean sphere formation frequency, ±SD; symbols are biological replicates. Sphere formation frequency from multiple independent replicates is shown in the table. For E and F, P-values were determined by one-way ANOVA and post-hoc Dunnett's multiple comparisons test.
Among the top DNA binding motifs enriched within H3K4me3 peaks reduced with C16 treatment was the OCT4–SOX2–TCF–NANOG (POU, homeobox, and HMG) motif, which matches the POU5F1(OCT4)::SOX2 motif. Considering our original identification of WDR5 as critical in the SOX2-enriched GBM niche, we again used the SORE6 reporter system (Supplemental Fig. S1C) here to test whether WDR5 inhibition affects activity of loci regulated by the core stem cell transcription factors SOX2 and OCT4. While this system has been used as a tool in a variety of tumors, it has not yet been used to inform the GBM CSC phenotype. The SORE6 reporter responds to SOX2 and OCT4 individually (Tang et al. 2015), and our CRISPR experiments (Fig. 1) demonstrate that SOX2 mainly drives SORE6 reporter expression in GBM CSCs. Live-cell imaging of SORE6 reporter transduced CSCs revealed that C16 treatment decreased the number of GFP+ cells over time, indicating that WDR5 inhibition diminished reporter activity and/or inhibited growth of cells with high reporter activity (Fig. 5B–D; Supplemental Fig. S6A). SORE6-GFPhigh cells gave rise to both GFPhigh and GFPlow cells, with GFPhigh cells having a higher self-renewal frequency (Supplemental Fig. S6B). Mean GFP intensity of live SORE6-GFP cells was reduced after C16 treatment, suggestive of inhibited transcription of SORE6-regulated loci (Fig. 5E). These data mechanistically validate the SORE6 motif as a WDR5-regulated sequence. To further interrogate the CSC state in response to WDR5 inhibition, we performed limiting dilution sphere formation assays and found that C16 treatment reduced CSC self-renewal in a concentration-dependent manner (Fig. 5F; Supplemental Fig. S6C). Taken together, these data provide proof of concept that targeting of WDR5 suppresses the CSC phenotype and disrupts epigenetic programs that maintain the CSC transcriptional state.
Reduction of CSC growth and viability via WDR5 inhibition
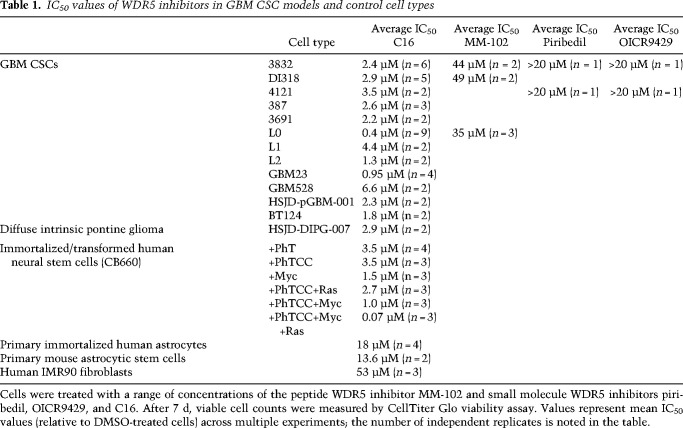
Given our initial functional and mechanistic assessments of the WDR5 inhibitors MM-102 and C16, we aimed to further assess the effects of these and other WDR5 inhibitors in CSC-enriched cultures. As with MM-102, C16 led to a reduction in CSC number in a dose-dependent manner (Supplemental Fig. S7A). For C16, half-maximal inhibitory concentration (IC50) values ranged from 0.4 to 6.6 µM across CSCs from more than eight PDX models (Table 1). We also obtained two previously described small molecule WDR5 inhibitors: piribedil and OICR-9429. These two compounds displayed IC50 values similar to those observed with MM-102. With WDR5 binding affinity confirmed for C16 (Supplemental Fig. S3C), and since GBM CSC models were more sensitive to C16 compared with the other WDR5 inhibitors tested (Table 1), we moved forward with further testing of C16.
Table 1.
IC50 values of WDR5 inhibitors in GBM CSC models and control cell types
WDR5 is differentially expressed in human GBM patient tumors compared with normal brain tissue
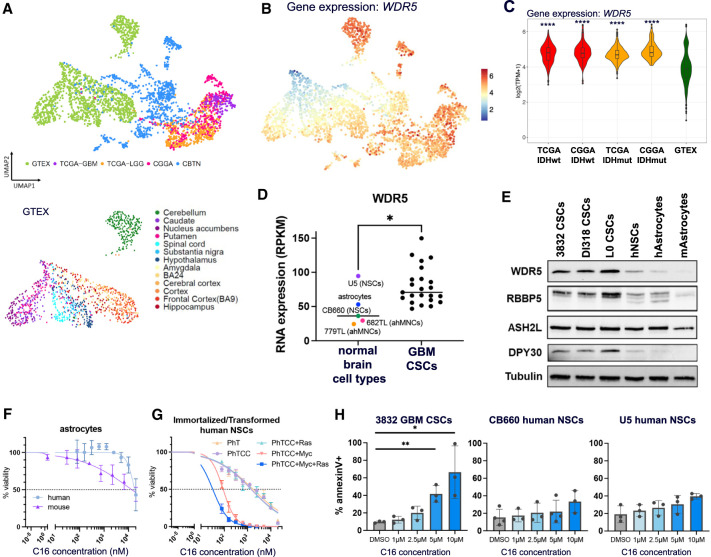
To investigate the relevance of WDR5 in the context of human GBM patients, we interrogated publicly available gene expression data from patients’ tumors (Bowman et al. 2017). Importantly, WDR5 expression is elevated in GBM compared with normal brain tissue (Supplemental Fig. S7B). To examine WDR5 expression in more detail, we queried WDR5 and WRAD complex member expression in RNA sequencing data through a recently reported “Brain-UMAP” (Arora et al. 2023). Batch-corrected log2-normalized gene expression data from different uniformly processed pipelines, including 702 adult glioma samples from The Cancer Genome Atlas (TCGA), 270 adult glioma samples from the Chinese Glioma Genome Atlas (CGGA), 1409 healthy normal brain samples from the Genotype–Tissue Expression Project (GTEx) across 12 GTEx-defined brain regions, and 802 pediatric tumor samples from the Children's Brain Tumor Network (CBTN), were used to generate the Brain-UMAP. Coloring the UMAP projections for gene expression of the WRAD complex members revealed enriched WDR5 and WRAD complex member expression in glioma samples from TCGA and the CGGA compared with normal brain tissues from the GTEx (Fig. 6A–C; Supplemental Fig. S7C,D). WDR5 expression in patients’ tumors also correlates with increased SOX2 expression (Supplemental Fig. S7E). We also interrogated WDR5 and WRAD complex expression in RNA sequencing data from a panel of normal brain cell types and GBM CSC lines (Toledo et al. 2015). These data show significantly increased expression of WDR5, RBBP5, and DPY30 in GBM compared with normal brain cells (Fig. 6D; Supplemental Fig. S7F). Next, we compared protein expression of WDR5 and other WRAD complex members in GBM CSCs, NSCs, and astrocytes and found diminished expression of WDR5, RBBP5, and DPY30 in normal brain cell types compared with GBM (Fig. 6E). Finally, increased WDR5 expression in patients’ tumors trends with poorer overall survival in GBM (Supplemental Fig. S7G).
Figure 6.
WDR5 is more highly expressed in human GBM patient tumors than normal brains, and WDR5 inhibition is preferentially detrimental to malignant cells. (A) UMAP projection showing normal brain tissue and brain tumor sample source. (GTEx) Normal brain tissue samples from the Genotype–Tissue Expression Project, (TCGA) The Cancer Genome Atlas, (CGGA) the Chinese Glioma Genome Atlas, (CBTN) the Children's Brain Tumor Network. (B) UMAP projection of gene expression for WDR5 on Brain-UMAP. Scale bar is log2(TPM + 1). (C) RNA expression of WDR5 in selected tumor and normal groups. P-values of each group compared with GTEx were determined by unpaired t-tests with correction for multiple comparisons. (D) WDR5 expression from RNA sequencing of normal brain cell types and GBM lines. Each point represents average expression from multiple sequencing replicates. (E) WRAD complex member expression in CSCs, human NSCs, and human (h) and mouse (m) astrocytes. Mouse cells are primary astrocytic stem cells. Human astrocytes are h-TERT immortalized. (F,G) Astrocytes and immortalized/transformed CB660 human NSCs were treated with a range of concentrations of WDR5 inhibitor C16. After 7 d, viable cell counts were measured by CellTiter Glo viability assay. Values represent mean luminescence values normalized to DMSO-treated cells. One representative curve per cell model is shown. Average IC50 values and number of replicates are shown in Table 1. Key for immortalized/transformed NSC-CB660 cells: (PhT) addition of dominant-negative p53DD and hTERT; (PhTCC) dominant-negative p53DD, hTERT, CyclinD1, and CDK4R24C; (Myc) c-Myc; and (Ras) H-RasV12. (H) Human GBM CSCs or NSCs were treated with WDR5 inhibitor C16 for 4 d and assessed for apoptosis by annexinV and DAPI staining by flow cytometry. Error bars are ±SD; circles are biological replicates. P-values were determined by one-way ANOVA and post-hoc Dunnett's multiple comparisons test.
WDR5 inhibition preferentially reduces growth and viability of GBM CSCs relative to normal brain cell types
We next tested the effect of WDR5 inhibition in normal brain cell types. We treated astrocytes, NSCs, and fibroblasts with C16 in 7-d IC50 assays to assess toxicity on these relevant normal cell populations. Compared with GBM CSCs, human and mouse astrocytes had, on average, a fivefold and sevenfold increased IC50 dose, human immortalized NSCs had a 1.3-fold increased IC50 dose, and human fibroblasts had a 20-fold increased IC50 dose (Fig. 6F; Table 1). To directly compare the effect of C16 in nonmalignant and malignant cells, we used a series of immortalized and/or transformed human NSCs from the same background (CB660 line) (Hubert et al. 2013). These cells were modified via the following combinations of loss of tumor suppressors and addition of oncogenes: dominant-negative p53DD and hTERT (PhT), CyclinD1 and CDK4R24C (CC), c-Myc, and H-RasV12. Sensitivity to C16 dramatically increased as a result of exogenous c-Myc expression, and most acutely (50-fold decrease in IC50) in c-Myc and H-RasV12 transformed cells, which are malignant (Fig. 6G; Table 1). These data directly demonstrate that malignant transformation sensitizes cells to WDR5 inhibition. Additionally, WDR5 and RBBP5 protein expression increases modestly in normal NSCs with the overexpression of c-Myc (Supplemental Fig. S7H). Due to similar IC50 values between NSCs and GBM CSCs, we further investigated the effects of C16 on normal NSCs. Observation of C16-treated cells under the microscope suggested that NSCs were undergoing a slowing of growth rather than cell death upon WDR5 inhibitor treatment, while cell death was evident in C16-treated GBM CSCs. To test this, we measured apoptosis in human NSCs in parallel with GBM CSCs treated with WDR5 inhibitor C16 via annexinV staining by flow cytometry. We observed a significant increase in apoptosis in GBM CSCs but not in NSCs (Fig. 6H). Similarly, we did not observe a significant increase in apoptosis in primary astrocytes after C16 treatment (Supplemental Fig. S7I).
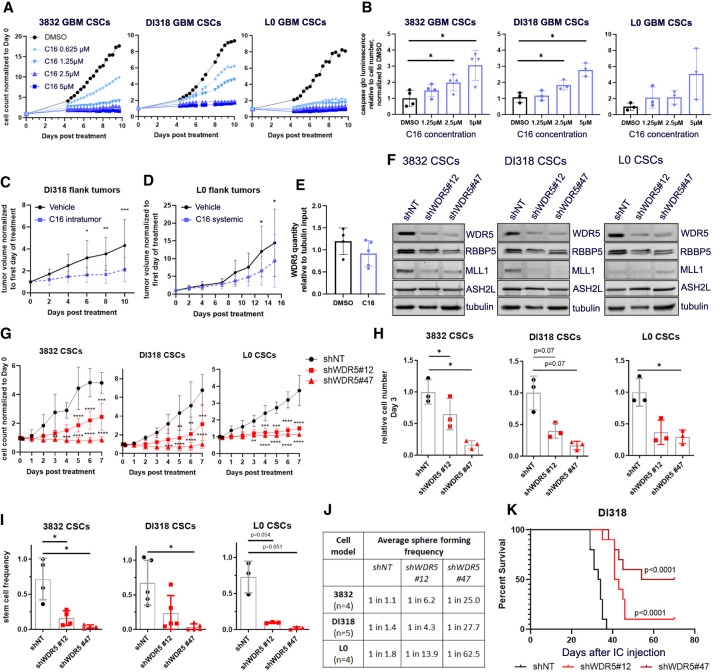
As the proliferation rate of immortalized NSCs increased with exogenous H-RasV12 and MYC expression, we investigated whether increased sensitivity to C16 could be attributed to an increased proliferation rate of cells. We found no correlation between doubling times and C16 IC50s in a panel of CSCs and immortalized/transformed NSCs (Supplemental Fig. S8A), suggesting that sensitivity to WDR5 inhibition is not solely due to proliferation rate. Based on these observations, we further tested the effects of C16 in a subset of CSC models (3832, DI318, and L0) and found that C16 reduced cell number over time and increased apoptosis in a concentration-dependent manner (Fig. 7A,B; Supplemental Fig. S8B,C). C16 also reduced CSC proliferation in patient-derived GBM organoids (Supplemental Fig. S8D). Importantly, loss of proliferation specifically occurred in SOX2+ cells in organoids with C16 treatment (Supplemental Fig. S8E).
Figure 7.
Pharmacologic and genetic inhibition of WDR5 reduces GBM CSC growth, self-renewal, and in vivo tumor growth. (A) Proliferation of WDR5 inhibitor C16-treated CSCs over 10 d, measured by IncuCyte live-cell imaging. Values represent mean fold change in cell count relative to day 0, ±SD; n = 3 technical replicates; one representative experiment is shown per CSC model. (B) GBM CSCs were treated with a range of concentrations of C16 and subjected to caspase 3/7 Glo luminescence assay after 4 d to measure caspase 3/7 activity. Bars represent fold change in caspase 3/7 activity per cell relative to the average for DMSO-treated cells, ±SD; circles are biological replicates. P-values were determined by one-way ANOVA and post-hoc Dunnett's multiple comparisons test. (C) Five-hundred-thousand DI318 CSCs were implanted into the flanks of mice, and once tumors developed, 3 mg/kg C16 was injected into the tumors daily. n = 10 per group. (D) Five-hundred-thousand L0 CSCs were implanted into the flanks of mice, and once tumors developed, 10 mg/kg C16 was injected intraperitoneally daily. n = 9 (DMSO); n = 10 (C16). For C and D, tumor volume over time normalized to tumor size at day 0 is shown. P-values were determined by two-tailed, unpaired t-test comparing means per group at each time point. (E) Immunoprecipitation of RBBP5 and immunoblot for WDR5 were performed on flank tumor lysates isolated from treated mice in D. Symbols are individual mice. WDR5 quantity in RBBP5 pull-down relative to tubulin quantity in input (10%) is plotted. (F) Short hairpin RNA-mediated targeting of WDR5 with two nonoverlapping short hairpins in CSC models. Western blots show the level of WDR5 protein in CSCs infected with a nontargeting (shNT) control virus or WDR5 knockdown (KD) viruses. (G) Proliferation of WDR5 KD and shNT control CSCs over 7 d, determined by IncuCyte live-cell imaging. Values represent mean fold change in cell count relative to day 0, ±SD; n = 3 biological replicates, P-values were determined by two-tailed, unpaired t-tests comparing means per group at each time point. (H) Equal numbers of WDR5 KD and shNT control CSCs were plated, and viable cell counts were measured by CellTiter Glo luminescence viability assay after 72 h. Bars represent mean luminescence values relative to the average for shNT control cells, ±SD; circles are biological replicates, (I) In vitro limiting dilution analysis was performed on WDR5 KD and shNT control CSCs. Bars represent mean sphere formation frequency, ±SD; symbols are biological replicates. For H and I, P-values were determined by one-way ANOVA and post-hoc Dunnett's multiple comparisons test. (J) Related to I, mean sphere formation frequency for each group is shown. (K) Kaplan–Meier survival plot of mice intracranially implanted with WDR5 KD or shNT control CSCs. P-values indicate comparisons between shNT and shWDR5 and were determined by log rank analysis. n = 10 per group; median survival shNT: 33.5 d; shWDR5 #12: 42 d; shWDR5 #47: 62 d.
WDR5 inhibitor C16 is tolerated in vivo and reduces tumor growth in a flank tumor model
To understand the potential toxicity of chronic administration of C16 in vivo, we treated a small cohort of healthy, non-tumor-bearing mice daily for 32 d with vehicle or 10 mg/kg C16 intraperitoneally (i.p.). Mice were weighed twice weekly and monitored for signs of distress or inhibitor-related decline. We did not observe significant reduction in weight or other phenotypic signs of potential toxicity over the 32-d period (Supplemental Fig. S9A). Following the conclusion of the experimental time frame, whole blood was collected, and separated plasma was used to evaluate a basic metabolic panel. Mice showed no significant changes in ALT, AST, creatinine, chloride, sodium, or glucose, indicative of unimpaired liver and kidney function (Supplemental Fig. S9B). Recently published work testing C16-related WDR5 small molecule inhibitors in mice also found no systemic toxicity and did not note any neurologic defects, observing a desirable oral pharmacokinetic profile with manageable intravenous clearance and high oral bioavailability (Teuscher et al. 2022). Mouse and human WDR5 are identical (Guarnaccia and Tansey 2018); therefore, C16 is predicted to bind mouse WDR5, and, in this sense, toxicity studies with WDR5 inhibitors in mice are translatable to humans. However, assessment of brain penetration via a snapshot brain-to-plasma time-course concentration profile after a single i.p. bolus dose of 10 mg/kg C16 in mice revealed it has limited brain penetration, with an area under the curve brain to plasma ratio of <10% (AUCbrain/AUCplasma ratio < 0.1) (Supplemental Fig. S9C). Blood–brain barrier (BBB) penetration potential was also assessed using MDR1–MDCK cell monolayers, a routine methodology used to predict the likelihood of passive CNS penetration. An average A–B passive permeability (Papp) of 0.715 × 10−6 cm/sec was measured for C16, a value that is indicative of low brain penetration potential. Typical CNS therapeutics maintain moderate to high passive permeability (Papp > 10 × 10−6 cm/sec) and lack active transport/efflux from transporters expressed at the BBB (Mahar Doan et al. 2002). Despite the expected low brain penetrance of C16, we tested C16 in vivo in the context of orthotopic brain tumors. NSG mice were intracranially implanted with DI318 CSCs, and daily treatment of 10 mg/kg C16 commenced 10 d after implantation. As expected, we did not observe a difference in survival. Of note, the mice tolerated daily i.p. treatment of C16 well for almost 3 wk in this experiment, supporting that C16 is tolerable and not overtly toxic to mice (Supplemental Fig. S9D). To evaluate the effect of C16 on tumors in vivo, we implanted GBM CSCs as flank tumors. Both direct tumoral injection and systemic i.p. injection of C16 modestly reduced tumor volume (Fig. 7C,D). Mice were treated until vehicle group mice reached a humane end point based on flank tumor size. Finally, to demonstrate that the tumor-suppressive effect is related to WDR5, we prepared lysates from end point tumors (from Fig. 7D) and performed coimmunoprecipitations to measure protein–protein interactions between WDR5 and its binding partner, RBBP5. We saw a trend of reduction in WDR5 bound to RBBP5 (Fig. 7E), supporting a WDR5-specific effect in tumors after systemic i.p. injection of C16.
WDR5 knockdown reduces CSC growth, self-renewal, and tumor initiation
We finally sought to validate our inhibitor studies with WDR5 genetic loss of function. We silenced WDR5 using short hairpin RNA (shRNA)-mediated interference (Fig. 7F) and found that WDR5 loss led to reduced expression of RBBP5 (and MLL1 in two out of three CSC models), indicating that WDR5 inhibition may destabilize the WRAD complex. We next tested how WDR5 loss affected stem cell behavior in the 3832, DI318, and L0 CSC models. Depletion of WDR5 resulted in reduced CSC growth (Fig. 7G,H) and attenuated self-renewal (Fig. 7I,J) compared with nontargeting (NT) controls. As CSCs are functionally defined in part by their capacity for tumor initiation, we intracranially implanted CSCs expressing control or WDR5 targeted shRNAs and found that WDR5 knockdown increased tumor latency (Fig. 7K). Half of the mice implanted with shWDR5 #47 CSCs, which produce greater knockdown of WDR5, did not develop tumors within the time frame of the study, implying that below a certain threshold of WDR5 expression, tumors are not viable. Future studies will further examine mechanisms of tumor growth slowing and increased tumor latency in response to genetic and pharmacologic WDR5 inhibition. Taken together, these knockdown data more broadly validate our initial organoid screening results, demonstrate that WDR5 is essential for the CSC phenotype, and provide a rationale for the pharmacological targeting of WDR5 and the WRAD complex to impair CSC population viability in GBM (Supplemental Fig. S10).
Discussion
In this study, we provide proof of concept for WDR5 as a viable therapeutic target in GBM based on an unbiased screen; the use of a WDR5-directed tool compound, C16; and genetic loss-of-function studies. Our findings demonstrate that the GBM CSC phenotype is reliant on WDR5 to a greater extent than nonmalignant neural populations. We demonstrate a role of WDR5 in regulating epigenetic maintenance of the GBM CSC state and validate its functional necessity for CSC self-renewal and tumor initiation.
Identifying mechanisms to attenuate the CSC state remains an immediate priority for malignant cancers, including GBM, and it is well established that self-renewal is driven by the coordinate action of transcription factors and epigenetic programs. There have been recent promising efforts to target individual transcription factors, such as with the OLIG2 inhibitor CT-129 (Oasa et al. 2020), but DNA/protein interactions have been historically difficult to target (Bushweller 2019). Therefore, understanding the upstream molecular network of self-renewal transcriptional programs may provide more rational therapeutic targets. Here, we link WDR5 WIN site inhibition in GBM to reduced H3K4 methyltransferase activity facilitated by the WRAD complex and diminished H3K4me3 on genes involved in pathways previously shown to promote CSC function in GBM, including the WNT and EGFR pathways (Kim et al. 2013; Furnari et al. 2015). These effects represent a vulnerability to tumor populations as compared with nonmalignant control cells; however, it remains unclear exactly how C16 leads to cell death. This constitutes an immediate future direction, as it may also provide insight into putative therapeutic resistance mechanisms.
Our findings align with previous observations of the importance of MLL1 in GBM CSCs (Heddleston et al. 2012; Gallo et al. 2013). Notably, MI-2-2, a menin-MLL inhibitor, repressed temozolomide-resistant GBM clones and reduced subcutaneous GBM growth in vivo (Lan et al. 2017). Optimal HMT activity of the MLL1 complex depends on WDR5 (and specifically the WIN site), while the other SET1 family methyltransferases can be fully activated by just RBBP5 and ASH2L (Dou et al. 2006; Patel et al. 2008; Alicea-Velázquez et al. 2016; Li et al. 2016). Therefore, in theory, WDR5 WIN site inhibition should preferentially affect MLL1 function compared with other SET1 family members. As one possibility, our observation of global diminishment of H3K4me3 after C16 treatment may suggest an effect broader than MLL1, since MLL1 has been previously demonstrated to have proportionally fewer targets than other methyltransferases in the family (Sze et al. 2020). As another possibility, since the loci targeted by MLL1 in GBM have not been previously examined, MLL1 may methylate a broader/different array of targets in GBM than in previously examined cell types. For instance, MLL1 is known to regulate HOX genes, yet we did not observe down-regulation of H3K4me3 on HOX genes in our GBM models. Furthermore, while H3K4me3 levels generally positively correlate with gene expression, it remains unclear how exactly H3K4me3 loss affects transcription. In previous studies, loss of H3K4me3 resulted in minimal global transcriptional changes, and H3K4me3 was dispensable for maintenance of transcription. Experimental evidence suggests different roles for this mark, including regulation of alternative splicing, regulation of miRNA genes, transcriptional memory, and stabilization of transcriptional noise (Guenther et al. 2005; Howe et al. 2017; Murray et al. 2019). Regardless of the specific role of H3K4me3, our studies show that WDR5 inhibitor-mediated H3K4me3 loss accompanies compromised CSC survival.
Additionally, given that C16 led to disruption of the non-WIN site-mediated interaction between WDR5 and RBBP5, it is likely that WDR5 functions beyond those mediated through MLL1 are affected by this inhibitor. Along these lines, other WRAD complex members were not hits in our screen, indicating that WDR5 likely has additional, WRAD-independent roles in GBM CSCs. Due to the nature of screens where efficiency of knockdown from gene to gene can vary, it is also possible that other WRAD members and MLL1 were false negative hits in our screen. However, we propose that WDR5's known myriad functions result in a stronger phenotype upon WDR5 knockdown than knockdown of individual components of the WRAD complex.
Data from the DepMap portal (Broad Institute) classify WDR5 as “common essential” in large, pan-cancer screens, indicating that WDR5 inhibitors could be broadly applicable in multiple tumor types. In fact, a variety of cancers show dependence on WDR5 for survival (Guarnaccia and Tansey 2018; Guarnaccia et al. 2021). Data suggest context-specific and tumor type-specific roles of WDR5, likely underlain by differences in expression and function of binding partners, available chromatin binding sites, and downstream signaling networks. Therefore, a more focused assessment of the role of WDR5 in each cancer, including GBM, is warranted, as is the discovery of biomarkers associated with increased sensitivity to WDR5 inhibition.
A current limitation is the restricted brain penetration of C16, which reveals a clear need for medicinal chemistry efforts to surmount challenges within the current existing scaffolds (such as poor permeability and potential transporter efflux) and turns attention to inhibitor modifications that will lead to overall improved brain penetration. Our experiments in immortalized/transformed NSC models and the elevated expression of WDR5 in GBM compared with normal brain tissue suggest the existence of a therapeutic window to target WDR5 in CSCs without compromising normal neural function, and future drug developments must take care to maintain this window. Overall, our findings highlight a key role for WDR5 in the epigenetic maintenance of the CSC state and provide proof of concept that WDR5 inhibitors can neutralize CSC populations in GBM.
Materials and methods
Organoid screen
Our study used a previously described inducible RNAi screening system (Miller et al. 2017). This shRNA library contained 1586 shRNAs targeting 406 known chromatin and transcriptional regulator genes (two to four shRNAs per gene), with positive and negative control shRNAs. GBM528 patient-derived CSCs were transduced with the shRNA library pool at low MOI to ensure single viral integration, and cells with genomic integration of shRNAs, as monitored by expression of a constitutive mVenus fluorescent reporter, were isolated using FACS. Cells were then allowed to recover and expand for three passages. Organoids for shRNA screening were formed by seeding 30,000 positively infected cells per 20-μL Matrigel pearl using custom 96-well format parafilm molds and multichannel pipettes. Organoids were allowed to grow and mature uninduced for 1 mo in 500-mL spinner flasks in 250 mL of media at 37°C with 5% CO2, prior to the addition of 1 µg/mL doxycycline (Dox) to induce gene knockdown. Organoids were maintained on Dox for 21 d. Each of three organoid screen cohorts, consisting of 55 independent organoids each, was processed and analyzed separately. This represents an ∼1000-fold library coverage for each screen replicate. Day 0 controls were also collected, stored frozen, and processed in parallel for comparison. At the end of the screen, organoids were regionally labeled with CellTracker blue CMAC for 2 h as previously described (Shakya et al. 2021), and single organoids from each screen cohort were spot-checked by confocal microscopy to ensure proper CMAC labeling. Labeled organoids were dissociated and separated by FACS, marked by positivity for constitutive mVenus expression and Dox-induced dsRed expression, and sorted into regional populations based on retention of CMAC regional blue dye. Genomic DNA was isolated from each screened population and sequenced as described (Miller et al. 2017). Briefly, deep-sequencing libraries were generated by PCR amplification of shRNA guide strands using barcoded primers that tag the product with standard Illumina adapters (see Supplemental Table S3). Libraries were sequenced on the HiSeq 2500 platform at the Cleveland Clinic Genomics Core Facility using a primer that reads in reverse into the guide strand. Sequence processing was performed using two custom workflows at https://usegalaxy.org. Raw read counts were converted to reads per million (RPM) to control for variations in total shRNA reads in each sample. shRNAs were scored using RIGER and extension of the GENE-E package (Broad Institute). Median RPM value for each replicate was used for analysis. The signal to noise ratio of replicates was used to calculate individual shRNA score based on their ability to deplete cells in the induced cohorts compared with the control inputs, and second-best shRNA score was used to rank genes. Expressed genes with a total RIGER P-value score of <0.05 for depletion compared with controls were considered hits.
CUT&Tag
DI318 CSCs were treated for 72 h with 3 μM C16. Cells were harvested and counted, and the CUT&Tag-IT assay and library preparation were performed on 500,000 cells per replicate, according to the manufacturer's manual (Active Motif) using rabbit antihuman trimethyl-histone H3 (Lys4; C42D8; CST 9751) antibody. Size distribution and concentration of libraries were assessed using an Agilent 4200 TapeStation with D1000 reagents and Qubit assay. Barcoded libraries were mixed to achieve equal representation, and paired-end Illumina sequencing was performed on the barcoded libraries on a NovaSeq SP100 with the following parameters: read1:i7:i5:read2 = 28:10:10:90.
SORE6-GFP reporter experiments
For SORE6-GFP reporter experiments, SORE6-dsCopGFP lentiviral particles were generated by transfection of 293T cells (Fugene transfection reagent) with pPACKH1 vectors and SORE6-dsCopGFP plasmid DNA (kindly provided by Wakefield Laboratory, National Institutes of Health) according to the manufacturer's protocol (System Biosciences). Viral supernatant was collected at 48 h, and virus was concentrated with PEG-it virus precipitation solution (System Biosciences). SORE6-GFP virus was added to CSCs plated on Geltrex. Forty-eight hours after infection, 2–3 µg/mL puromycin was added to cells. After puromycin selection, cells were collected, and GFPhigh (10%–20% brightest) or GFPnegative cells were isolated by FACS. GFPhigh and GFPnegative cells were subjected to limiting dilution analysis as described in the Supplemental Material, or protein was isolated for Western blot. GFPhigh cells were cultured further and used for inhibitor treatment experiments, CRISPR experiments, and organoid experiments. Fluorescence images were taken with the IncuCyte live-cell analysis system (Sartorius).
Supplementary Material
Acknowledgments
We thank members of the Lathia, Hubert, Stauffer, Paddison, and Patel laboratories for critical feedback, and Ms. Amanda Mendelsohn for illustration assistance. This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) T32 CA059366 and NIH/National Institute of Neurological Disorders and Stroke F32 NS116109 to K.M.; NIH/NCI K00 CA253768 to S.A.S.; NIH/NCI F30CA250254 to A.L.; NIH/National Center for Advancing Translational Sciences Clinical and Translational Science Award KL2 TR002547, Clinical Translational Science Collaborative of Cleveland grant UL1 TR002548, American Brain Tumor Association Discovery grant DG1800016 in memory of Dr. Joseph Weiss, support from the American Cancer Society Institutional Research Grant award IRG-16-186-21 to the Case Comprehensive Cancer Center grant RES513876, and support from the VeloSano Cancer Research Fund to C.G.H.; R35 CA197718 to J.N.R.; and the Lerner Research Institute (LRI) and Case Comprehensive Cancer Center to J.D.L. Work in the Lathia laboratory is also supported by NIH grants P01 CA245705 and R35 NS127083. S.R.S. thanks the Lerner Research Institute for laboratory startup seed support. We thank the Vanderbilt University Medical Center Metabolic Physiology Shared Resource Core and the Diabetes Research and Training Center for in-life brain plasma studies using compound C16. We thank Alan Pratt and the Prevention Research Laboratory and Lab Diagnostic Core at Cleveland Clinic Foundation for blood chemistry studies using C16. The Metabolic Physiology Shared Resource is supported by NIH grant P30 DK020593.
Author contributions: K.M., C.G.H., J.D.L., S.R.S., J.N.R., and J.A. designed the study and experiments. K.M., S.S., S.A.S., D.J.S., C.M.G., L.W., G.R., R.E.S., K.K., C.G.H., A.L., J.D.K., A.K., S.J., and J.A. performed experiments and analyzed data. T.E.M. provided guidance on the organoid screen. A.P.P. and P.J.P. provided guidance on CUT&Tag experiments and data analysis. S.A., K.M., and J.B. performed statistical analyses. K.M., C.G.H., J.D.L., S.R.S., and E.E.M.-H. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349803.122.
Competing interest statement
The authors declare no competing interests.
References
- Alicea-Velázquez NL, Shinsky SA, Loh DM, Lee JH, Skalnik DG, Cosgrove MS. 2016. Targeted disruption of the interaction between WD-40 repeat protein 5 (WDR5) and mixed lineage leukemia (MLL)/SET1 family proteins specifically inhibits MLL1 and SETd1A methyltransferase complexes. J Biol Chem 291: 22357–22372. 10.1074/jbc.M116.752626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado AG, Thiagarajan PS, Mulkearns-Hubert EE, Silver DJ, Hale JS, Alban TJ, Turaga SM, Jarrar A, Reizes O, Longworth MS, et al. 2017. Glioblastoma cancer stem cells evade innate immune suppression of self-renewal through reduced TLR4 expression. Cell Stem Cell 20: 450–461.e4. 10.1016/j.stem.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197. 10.1016/j.cell.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Szulzewsky F, Jensen M, Nuechterlein N, Pattwell SS, Holland EC. 2023. An RNA seq-based reference landscape of human normal and neoplastic brain. bioRxiv. 10.1101/2023.01.03.522658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. 2014. H3k4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158: 673–688. 10.1016/j.cell.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RL, Wang Q, Carro A, Verhaak RGW, Squatrito M. 2017. Gliovis data portal for visualization and analysis of brain tumor expression datasets. Neuro-Oncology 19: 139–141. 10.1093/neuonc/now247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushweller JH. 2019. Targeting transcription factors in cancer—from undruggable to reality. Nat Rev Cancer 19: 611–624. 10.1038/s41568-019-0196-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Xiao Z, Zhu G, Mao B, Huang H, Guan F, Lin Z, Peng W, Liang X, Zhang B, et al. 2020. WD repeat domain 5 promotes invasion, metastasis and tumor growth in glioma through up-regulated zinc finger E-box binding homeobox 1 expression. Cancer Manag Res 12: 3223–3235. 10.2147/CMAR.S237582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J, Van Nguyen N, Georgiev P, Gaub A, Brettschneider J, Cusack S, Kadlec J, Akhtar A. 2014. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev 28: 929–942. 10.1101/gad.240200.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit D, Prager BC, Gimple RC, Miller TE, Wu Q, Yomtoubian S, Kidwell RL, Lv D, Zhao L, Qiu Z, et al. 2022. Glioblastoma stem cells reprogram chromatin in vivo to generate selective therapeutic dependencies on DPY30 and phosphodiesterases. Sci Transl Med 14: eabf3917. 10.1126/scitranslmed.abf3917 [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13: 713–719. 10.1038/nsmb1128 [DOI] [PubMed] [Google Scholar]
- Filppu P, Tanjore Ramanathan J, Granberg KJ, Gucciardo E, Haapasalo H, Lehti K, Nykter M, Le Joncour V, Laakkonen P. 2021. CD109-GP130 interaction drives glioblastoma stem cell plasticity and chemoresistance through STAT3 activity. JCI Insight 6: e141486. 10.1172/jci.insight.141486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: eaal2380. 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. 2015. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer 15: 302–310. 10.1038/nrc3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M, Ho J, Coutinho FJ, Vanner R, Lee L, Head R, Ling EK, Clarke ID, Dirks PB. 2013. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res 73: 417–427. 10.1158/0008-5472.CAN-12-1881 [DOI] [PubMed] [Google Scholar]
- Gallo M, Coutinho FJ, Vanner RJ, Gayden T, Mack SC, Murison A, Remke M, Li R, Takayama N, Desai K, et al. 2015. MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H3.3 and globally reorganizing chromatin. Cancer Cell 28: 715–729. 10.1016/j.ccell.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. 2009. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells 27: 40–48. 10.1634/stemcells.2008-0493 [DOI] [PubMed] [Google Scholar]
- Grebien F, Vedadi M, Getlik M, Giambruno R, Grover A, Avellino R, Skucha A, Vittori S, Kuznetsova E, Smil D, et al. 2015. Pharmacological targeting of the Wdr5–MLL interaction in C/EBPα N-terminal leukemia. Nat Chem Biol 11: 571–578. 10.1038/nchembio.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia AD, Tansey WP. 2018. Moonlighting with WDR5: a cellular multitasker. J Clin Med 7: 21. 10.3390/jcm7020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia AD, Rose KL, Wang J, Zhao B, Popay TM, Wang CE, Guerrazzi K, Hill S, Woodley CM, Hansen TJ, et al. 2021. Impact of WIN site inhibitor on the WDR5 interactome. Cell Rep 34: 108636. 10.1016/j.celrep.2020.108636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. 2005. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci 102: 8603–8608. 10.1073/pnas.0503072102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston JM, Wu Q, Rivera M, Minhas S, Lathia JD, Sloan AE, Iliopoulos O, Hjelmeland AB, Rich JN. 2012. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ 19: 428–439. 10.1038/cdd.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe FS, Fischl H, Murray SC, Mellor J. 2017. Is H3K4me3 instructive for transcription activation? Bioessays 39: 1–12. 10.1002/bies.201600095 [DOI] [PubMed] [Google Scholar]
- Huang S. 2013. Genetic and non-genetic instability in tumor progression: link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev 32: 423–448. 10.1007/s10555-013-9435-7 [DOI] [PubMed] [Google Scholar]
- Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Girard EJ, Davison J, Berndt J, Corrin P, et al. 2013. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev 27: 1032–1045. 10.1101/gad.212548.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, Rich JN. 2016. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res 76: 2465–2477. 10.1158/0008-5472.CAN-15-2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. 2006. PDGFRα-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron 51: 187–199. 10.1016/j.neuron.2006.06.012 [DOI] [PubMed] [Google Scholar]
- Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L, Lei M, Dou Y, Wang S. 2013. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc 135: 669–682. 10.1021/ja306028q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10: 1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima N, Hosen N, Kagawa N, Hashimoto N, Nakano A, Fujimoto Y, Kinoshita M, Sugiyama H, Yoshimine T. 2012. CD166/activated leukocyte cell adhesion molecule is expressed on glioblastoma progenitor cells and involved in the regulation of tumor cell invasion. Neuro Oncol 14: 1254–1264. 10.1093/neuonc/nor202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Seol HJ, Kim EH, Rheey J, Jin HJ, Lee Y, Joo KM, Lee J, Nam DH. 2013. Wnt/β-catenin signaling is a key downstream mediator of MET signaling in glioblastoma stem cells. Neuro Oncol 15: 161–171. 10.1093/neuonc/nos299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Jörg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, et al. 2017. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549: 227–232. 10.1038/nature23666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Han J, Zhang Y, Cao F, Liu Z, Li S, Wu J, Hu C, Wang Y, Shuai J, et al. 2016. Structural basis for activity regulation of MLL family methyltransferases. Nature 530: 447–452. 10.1038/nature16952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Li Z, Song E, Hu P, Yang Q, Hu Y, Liu H, Jin A. 2020. LncRNA HOTTIP enhances human osteogenic BMSCs differentiation via interaction with WDR5 and activation of Wnt/β-catenin signalling pathway. Biochem Biophys Res Commun 524: 1037–1043. 10.1016/j.bbrc.2020.02.034 [DOI] [PubMed] [Google Scholar]
- Mahar Doan KM, Humphreys JE, Webster LO, Wring SA, Shampine LJ, Serabjit-Singh CJ, Adkison KK, Polli JW. 2002. Passive permeability and P-glycoprotein-mediated efflux differentiate central nervous system (CNS) and non-CNS marketed drugs. J Pharmacol Exp Ther 303: 1029–1037. 10.1124/jpet.102.039255 [DOI] [PubMed] [Google Scholar]
- Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, Factor DC, Kim LJY, Morrow JJ, Wu Q, et al. 2017. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature 547: 355–359. 10.1038/nature23000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K, Troike K, Silver DJ, Lathia JD. 2021. The evolution of the cancer stem cell state in glioblastoma: emerging insights into the next generation of functional interactions. Neuro Oncol 23: 199–213. 10.1093/neuonc/noaa259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SC, Lorenz P, Howe FS, Wouters M, Brown T, Xi S, Fischl H, Khushaim W, Rayappu JR, Angel A, et al. 2019. H3k4me3 is neither instructive for, nor informed by, transcription. bioRxiv 10.1101/709014 [DOI] [Google Scholar]
- Oasa S, Vukojević V, Rigler R, Tsigelny IF, Changeux JP, Terenius L. 2020. A strategy for designing allosteric modulators of transcription factor dimerization. Proc Natl Acad Sci 117: 2683–2686. 10.1073/pnas.1915531117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V, Cosgrove MS. 2008. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem 283: 32162–32175. 10.1074/jbc.M806317200 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411. 10.1038/nature01080 [DOI] [PubMed] [Google Scholar]
- Schnell O, Romagna A, Jaehnert I, Albrecht V, Eigenbrod S, Juerchott K, Kretzschmar H, Tonn JC, Schichor C. 2012. Krüppel-like factor 8 (KLF8) is expressed in gliomas of different WHO grades and is essential for tumor cell proliferation. PLoS One 7: e30429. 10.1371/journal.pone.0030429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Ricks S, Warrick J, Boynton AL, Murphy GP. 1999. Antisense human neuroglia related cell adhesion molecule hNr-CAM, reduces the tumorigenic properties of human glioblastoma cells. Anticancer Res 19: 4947–4953. [PubMed] [Google Scholar]
- Shakya S, Gromovsky AD, Hale JS, Knudsen AM, Prager B, Wallace LC, Penalva LOF, Brown HA, Kristensen BW, Rich JN, et al. 2021. Altered lipid metabolism marks glioblastoma stem and non-stem cells in separate tumor niches. Acta Neuropathol Commun 9: 101. 10.1186/s40478-021-01205-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siladi AJ, Wang J, Florian AC, Thomas LR, Creighton JH, Matlock BK, Flaherty DK, Lorey SL, Howard GC, Fesik SW, et al. 2022. WIN site inhibition disrupts a subset of WDR5 function. Sci Rep 12: 1848. 10.1038/s41598-022-05947-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar SJ, Shakya S, Barnett A, Wallace LC, Jeon H, Sloan A, Recinos V, Hubert CG. 2022. Three-dimensional organoid culture unveils resistance to clinical therapies in adult and pediatric glioblastoma. Transl Oncol 15: 101251. 10.1016/j.tranon.2021.101251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et al. 2014. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157: 580–594. 10.1016/j.cell.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CC, Ozark PA, Cao K, Ugarenko M, Das S, Wang L, Marshall SA, Rendleman EJ, Ryan CA, Zha D, et al. 2020. Coordinated regulation of cellular identity-associated H3K4me3 breadth by the COMPASS family. Sci Adv 6: eaaz4764. 10.1126/sciadv.aaz4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Raviv A, Esposito D, Flanders KC, Daniel C, Nghiem BT, Garfield S, Lim L, Mannan P, Robles AI, et al. 2015. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports 4: 155–169. 10.1016/j.stemcr.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher KB, Meyers KM, Wei Q, Mills JJ, Tian J, Alvarado J, Sai J, Van Meveren M, South TM, Rietz TA, et al. 2022. Discovery of potent orally bioavailable WD repeat domain 5 (WDR5) inhibitors using a pharmacophore-based optimization. J Med Chem 65: 6287–6312. 10.1021/acs.jmedchem.2c00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Teuscher KB, Aho ER, Alvarado JR, Mills JJ, Meyers KM, Gogliotti RD, Han C, Macdonald JD, Sai J, et al. 2020. Discovery and structure-based optimization of potent and selective WD repeat domain 5 (WDR5) inhibitors containing a dihydroisoquinolinone bicyclic core. J Med Chem 63: 656–675. 10.1021/acs.jmedchem.9b01608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo CM, Ding Y, Hoellerbauer P, Davis RJ, Basom R, Girard EJ, Lee E, Corrin P, Hart T, Bolouri H, et al. 2015. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep 13: 2425–2439. 10.1016/j.celrep.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valor LM, Hervás-Corpión I. 2020. The epigenetics of glioma stem cells: a brief overview. Front Oncol 10: 602378. 10.3389/fonc.2020.602378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang J, Ke X, Peng W, Zhao G, Peng S, Xu J, Xu B, Cui H. 2020. WDR5-Myc axis promotes the progression of glioblastoma and neuroblastoma by transcriptional activating CARM1. Biochem Biophys Res Commun 523: 699–706. 10.1016/j.bbrc.2019.12.101 [DOI] [PubMed] [Google Scholar]
- Xue H, Yao T, Cao M, Zhu G, Li Y, Yuan G, Chen Y, Lei M, Huang J. 2019. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature 573: 445–449. 10.1038/s41586-019-1528-1 [DOI] [PubMed] [Google Scholar]
- Yabo YA, Niclou SP, Golebiewska A. 2022. Cancer cell heterogeneity and plasticity: a paradigm shift in glioblastoma. Neuro Oncol 24: 669–682. 10.1093/neuonc/noab269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jiang H, Shao J, Mao R, Liu J, Ma Y, Fang X, Zhao N, Zheng S, Lin B. 2014. SOX4 inhibits GBM cell growth and induces G0/G1 cell cycle arrest through Akt–p53 axis. BMC Neurol 14: 207. 10.1186/s12883-014-0207-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zheng X, Yang H, Yan J, Fu X, Wei R, Xu X, Zhang Z, Yu A, Zhou K, et al. 2018. Piribedil disrupts the MLL1–WDR5 interaction and sensitizes MLL-rearranged acute myeloid leukemia (AML) to doxorubicin-induced apoptosis. Cancer Lett 431: 150–160. 10.1016/j.canlet.2018.05.034 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.