Here, Treen et al. investigated the precise timing of gene induction during the earliest phases of embryogenesis and used high-resolution in situ detection methods, along with genetic and experimental manipulations, to study the timing of zygotic activation in the simple model chordate Ciona intestinalis with minute-scale temporal precision. Their findings support a FGF timing mechanism that is driven by ERK-mediated derepression of the ERF repressor.
Keywords: Ciona, developmental biology, zygotic genome activation
Abstract
Zygotic genome activation has been extensively studied in a variety of systems including flies, frogs, and mammals. However, there is comparatively little known about the precise timing of gene induction during the earliest phases of embryogenesis. Here we used high-resolution in situ detection methods, along with genetic and experimental manipulations, to study the timing of zygotic activation in the simple model chordate Ciona with minute-scale temporal precision. We found that two Prdm1 homologs in Ciona are the earliest genes that respond to FGF signaling. We present evidence for a FGF timing mechanism that is driven by ERK-mediated derepression of the ERF repressor. Depletion of ERF results in ectopic activation of FGF target genes throughout the embryo. A highlight of this timer is the sharp transition in FGF responsiveness between the eight- and 16-cell stages of development. We propose that this timer is an innovation of chordates that is also used by vertebrates.
The earliest stages of embryonic development are remarkable, as the cells are not transcribing RNA and function purely through maternal contributions. This abruptly changes at precise points in development when the zygotic genome begins to transcribe RNA and maternal RNAs are depleted (Vastenhouw et al. 2019).
In the ascidian Ciona, zygotic genome activation occurs between the eight- and 32-cell stages (Lamy et al. 2006; Satou 2020). At the eight-cell stage, transcription is extremely limited, with Foxa.a being one of the only genes expressed (Lamy et al. 2006; Treen et al. 2018). At the 16-cell stage, a greater set of genes is expressed and there is some lineage specification (Oda-Ishii et al. 2016; Treen et al. 2018). By the 32-cell stage, there is full zygotic genome activation, with the germ layers being mostly specified (Satou 2020; Tokuoka et al. 2021). Additionally, at the 32-cell stage there is a sufficiently complicated embryonic geometry, as well as activating and inhibiting signals, for neural induction to occur through FGF/ERK signaling, resulting in localized expression of Otx in the a6.5 and b6.5 blastomeres (Bertrand et al. 2003; Hudson et al. 2003; Ohta and Satou 2013; Williaume et al. 2021).
Genes activated by cell signaling pathways are generally repressed in the absence of an inductive signal (Barolo and Posakony 2002; Affolter et al. 2008). For example, the Ets-class transcriptional repressor ERF is known to repress target genes in the absence of ERK phosphorylation. Activation of ERK by RTK signaling pathways such as FGF triggers ERF phosphorylation and derepression of its target genes (Sgouras et al. 1995; Le Gallic et al. 1999).
We previously determined that ERF undergoes phase separation in response to FGF signaling. When FGF is active, ERF is phosphorylated, causing condensates to dissolve, coinciding with derepression of target genes (Weaver et al. 2022). ERF was also shown to repress Otx in Ciona embryos at the 32-cell stage (Williaume et al. 2021). Interestingly, ERF condensates are able to dissolve and reform within a single interphase (Weaver et al. 2022), suggesting a dynamic response to endogenous FGF signals.
We identified Prdm1-r.a and Prdm1-r.b as the earliest target genes that are regulated by FGF signaling, at the 16-cell stage. However, unlike Otx, this activation does not depend on cell–cell contacts. We show that Prdm1-r.a, Prdm1-r.b, and Otx are activated in a precise temporal order from the eight- to 32-cell stages in the a5.3/a5.4 lineages. Sequential expression depends on derepression of ERF. ERF inhibition results in coactivation of all three genes at the onset of the 16-cell stage. We further show that localized expression of Prdm1-r.a/b depends on the ERF repressor and Foxa.a activator. Altogether, these results suggest that an FGF timer acts through derepression of ERF at the 16-cell stage. This timer is augmented by higher levels of FGF signals at the 32-cell stage. The consequence of these processes is the precise temporal activation of the earliest genes during zygotic genome activation.
Results and Discussion
ERF is present as a maternal mRNA in Ciona unfertilized eggs and early embryos (Imai et al. 2004; Treen et al. 2018). We hypothesized that it could be acting as a maternal repressor of the earliest zygotically expressed genes. To test this, we incubated embryos from the one-cell stage in high concentrations of FGF, or FGF and the MEK inhibitor U0126, and measured the expression of all known regulatory genes expressed at the 16-cell stage (Satou 2020) by qPCR (Supplemental Fig. S1). We measured samples at the eight-cell stage to see whether any genes are precociously expressed by FGF treatment. The only gene that responded to this treatment was Prdm1-r.a, which encodes a zinc finger transcriptional repressor in Ciona (Ikeda et al. 2013). It is linked to Prdm1-r.b, a paralogous gene that is reported to be expressed from the 32-cell stage and has overlapping expression and function with Prdm1-r.a (Ikeda et al. 2013). Prdm-1r.a was up-regulated by FGF treatment at the eight-cell stage; this up-regulation is abrogated by adding U0126 (Supplemental Fig. S1).
To verify our qPCR results, we developed a four-color hybridization chain reaction (HCR) in situ detection method. We used probes for both Prdm1-r.a and Prdm1-r.b, the known FGF-responsive gene Otx (Bertrand et al. 2003), and the earliest expressed zygotic gene Foxa.a (Fig. 1A). For each gene, two large spots could be observed within the nucleus that are likely to correspond to sites of active transcription. This observation was supported by the colocalization of the large spots for Prdm1-r.a and Prdm1-r.b, as the transcriptional start sites for these genes are only ∼18 kb apart (Satou et al. 2019). Smaller spots could also be detected within the nucleus, as well as in the cytoplasm, that are presumably clusters of RNAs or possibly individual transcripts. Four-color HCR in situ was able to recapitulate previously described gene expression patterns from the eight- to 32-cell stages (Fig. 1; Supplemental Figs. S2, S3, Lamy et al. 2006; Ikeda et al. 2013; Williaume et al. 2021) as well as reliably distinguish between cells that are actively transcribing genes and cells where the HCR in situ signal is exclusively cytoplasmic due to expression at earlier stages.
Figure 1.
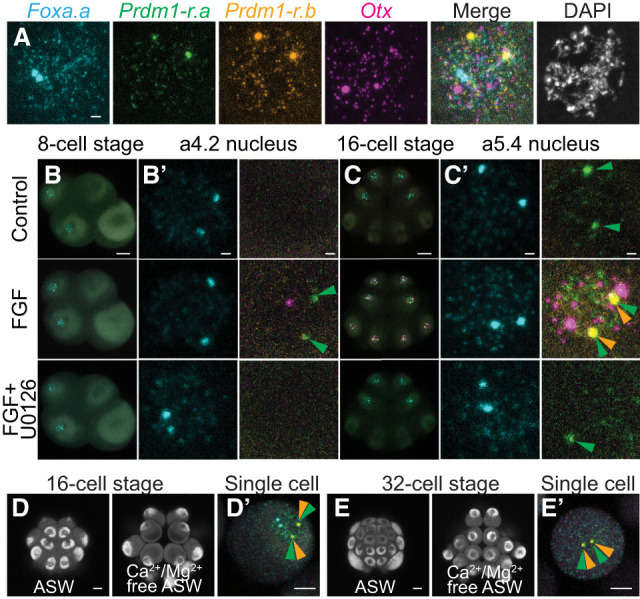
FGF/ERK signaling can induce Prdm1-r.a/b and Otx expression at the eight-/16-cell stages. (A) Four-color multiplex HCR fluorescence in situ hybridization can detect the expression of Foxa.a (cyan), Prdm1-r.a (green), Prdm1-r.b (orange), and Otx (magenta). The image shown is a single nucleus from a 16-cell stage Ciona embryo treated with FGF to induce the expression of FGF-responsive genes. DNA is stained with DAPI (white). (B,C) In situ hybridization at the eight-cell stage (B) and 16-cell stage (C) for the genes in A. Embryos were treated with FGF or with FGF and U0126. Whole-embryo and zoomed-in views of an a4.2 (B′) and a5.4 (C′) nucleus are shown. (D,E) Autofluorescence of an intact 16-cell stage (D) and 32-cell stage (E) Ciona embryo cultured in artificial sea water (ASW) or in calcium/magnesium-free artificial sea water (Ca2+/Mg2+-free ASW) to dissociate the blastomeres. (D′) In situ hybridization of a single dissociated blastomere from a 16-cell stage embryo that was treated with Ca2+/Mg2+-free ASW from the eight-cell stage. Nuclear expression of Foxa.a and Prdm1-r.a can be seen. (E′) In situ hybridization of a single dissociated blastomere from a 32-cell stage embryo that was treated with Ca2+/Mg2+-free ASW from the 16-cell stage. Cytoplasmic expression of Foxa.a and nuclear expression of Prdm1-r.a/b can be seen, but no expression of Otx is detected. Eight-cell stage embryos are oriented lateral view, anterior left, and animal hemisphere up. Sixteen-cell embryos are oriented animal view and anterior up. Arrowheads indicate sites of active Prdm1-r.a (green) and Prdm1-r.b (orange) transcription. Scale bars: for whole embryo, 20 µm; for single nucleus, 2 µm; for dissociated cell views, 10 µm.
When eight-cell embryos were treated with FGF, we could detect extremely weak nuclear HCR in situ signals for Prdm1-r.a in a minority of embryos (Fig. 1B). A single example of precocious activation of Otx at the eight-cell stage was also observed. Expression of Prdm1-r.a could not be detected when embryos were treated with both FGF and U0126, consistent with our qPCR results. Overall, the response of the eight-cell embryo to FGF treatment was extremely modest. This is in dramatic contrast to the 16-cell stage. Substantial up-regulation of Prdm1-r.a could be seen, as well as ectopic and precocious activation of Prdm1-r.a, Prdm1-r.b, and Otx in all cells of 16-cell stage embryos except the transcriptionally silent B5.2 cells (Fig. 1C; Supplemental Figs. S2, S3; Shirae-Kurabayashi et al. 2011). These effects were overcome when embryos were treated with both FGF and U0126. Under these conditions, Prdm1-r.a nuclear signals could still be detected in a5.3/a5.4 blastomeres; however, the signal appeared weaker than normal (Fig. 1C′).
Early embryos tend to be made up of a small number of large cells, making the establishment of diffusible morphogen gradients difficult. In Ciona, it has been shown that Otx expression is dependent instead on juxtracrine signaling using extensive cell–cell contacts (Ohta and Satou 2013; Williaume et al. 2021), and it is likely that most signaling events in the embryo use this mechanism (Guignard et al. 2020). We tested whether Prdm1-r.a expression was dependent on cell–cell contacts by transferring eight-cell embryos to Ca2+/Mg2+-free artificial sea water and allowing them to develop until the late 16-cell stage. This treatment eliminated cell–cell contacts and partially or completely dissociated the embryos (Fig. 1D). HCR in situs were performed on these dissociated embryos, and Prdm1-r.a was expressed in similar proportions of cells to those of intact embryos (Supplemental Fig. S4A). In contrast, as expected, Otx expression was lost from cells that were dissociated from the 16- to 32-cell stages (Fig. 1E; Supplemental Fig. S4B). We conclude that, unlike for Otx, cell–cell contacts are not required for Prdm1-r.a expression.
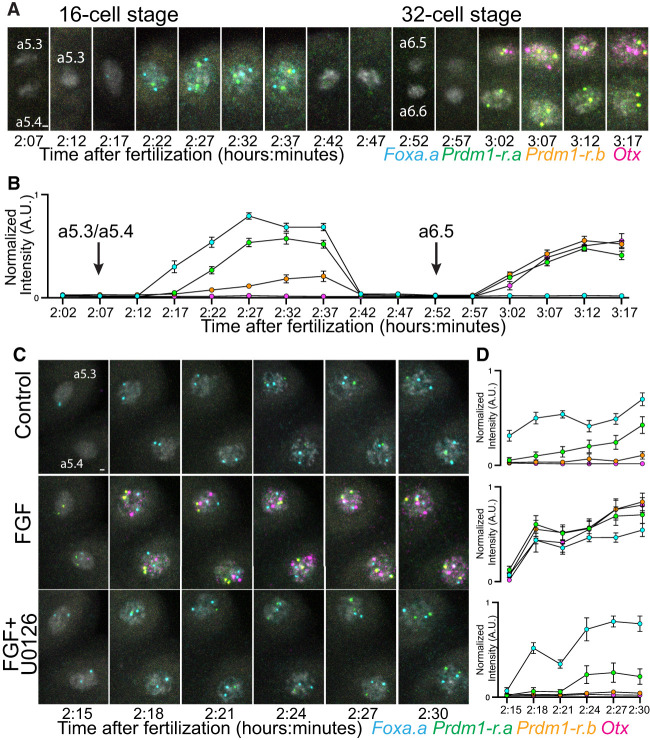
Using Ciona embryos, we can precisely control the timing of fertilization and synchronize development. We performed an in-depth analysis of gene expression from the end of the eight-cell stage (a4.2 cells) to the 32-cell stage (a6.5 and a6.6 cells) (Fig. 2A; Supplemental Fig. S5) using samples taken in 5-min intervals. This revealed a staged onset of transcription within the 16-cell stage. First, Foxa.a is expressed immediately after mitosis, followed by Prdm1-r.a. Unexpectedly, we detected transcription of Prdm1-r.b at low levels late in the 16-cell stage. This expression was probably too low to be detected with less sensitive methods (Ikeda et al. 2013). At the 32-cell stage, Prdm1-r.a, Prdm1-r.b, and Otx all came on simultaneously in the a6.5 cells (Fig. 2B). In the a6.6 cells, the same temporal expression of Prdm1-r.a and Prdm1-r.b could be seen, but there was no expression of Otx (Supplemental Fig. S5A).
Figure 2.
The temporal onset of early zygotic genes. (A) In situ hybridization from the eight- to 32-cell stage, tracing the a4.2 lineage. DNA is stained with DAPI and shown in white. (B) Quantifications of active transcription levels from samples shown in A. Each data point indicates the mean (n = 24 from three embryos), and error bars indicate 95% confidence intervals. (C) In situ hybridization from the early to late 16-cell stage. The a5.3 and a5.4 nuclei from one half embryo is shown. DNA is stained with DAPI and shown in white. Embryos are treated with FGF or FGF and U0126. (D) Quantifications of active transcription levels from samples shown in C. Each data point indicates the mean (n = 24 from three embryos), and error bars indicate 95% confidence intervals. Active transcription levels are calculated by measuring the normalized fluorescence intensity of individual sites of active transcription. Scale bars, 2 µm.
We next investigated how this precise onset of transcription was affected by perturbing FGF signaling. We augmented temporal resolution by using samples taken every 3 min. Gene expression was measured during the 16-cell stage in normal development and upon perturbing FGF signaling (Fig. 2C,D). When embryos were treated with FGF, we observed Prdm1-r.a, Prdm1-r.b, and Otx all transcribing simultaneously, immediately after mitosis, at the same time as Foxa.a. This precocious transcription was eliminated when embryos were treated with both FGF and U0126. The kinetics of Prdm1-r.a, Prdm1-r.b, and Otx in response to FGF treatment at the 16-cell stage were remarkably similar to what was seen in the a6.5 cell in untreated embryos (Fig. 2B,D).
To confirm that our observations are due to repression by maternal ERF, we knocked down translation using morpholino oligonucleotides (MOs). In ERF MO-injected embryos, weak transcription of Prdm1-r.a (but not Prdm1-r.b or Otx) could be detected at the eight-cell stage (Supplemental Fig. S6). At the 16-cell stage, ERF knockdown resulted in up-regulation of Prdm1-r.a, Prdm1-r.b, and Otx in all transcriptionally active cells (Fig. 3; Supplemental Fig. S7). This demonstrates that ERF is a repressor of Prdm1-r.a, Prdm1-r.b, and Otx at the 16-cell stage.
Figure 3.
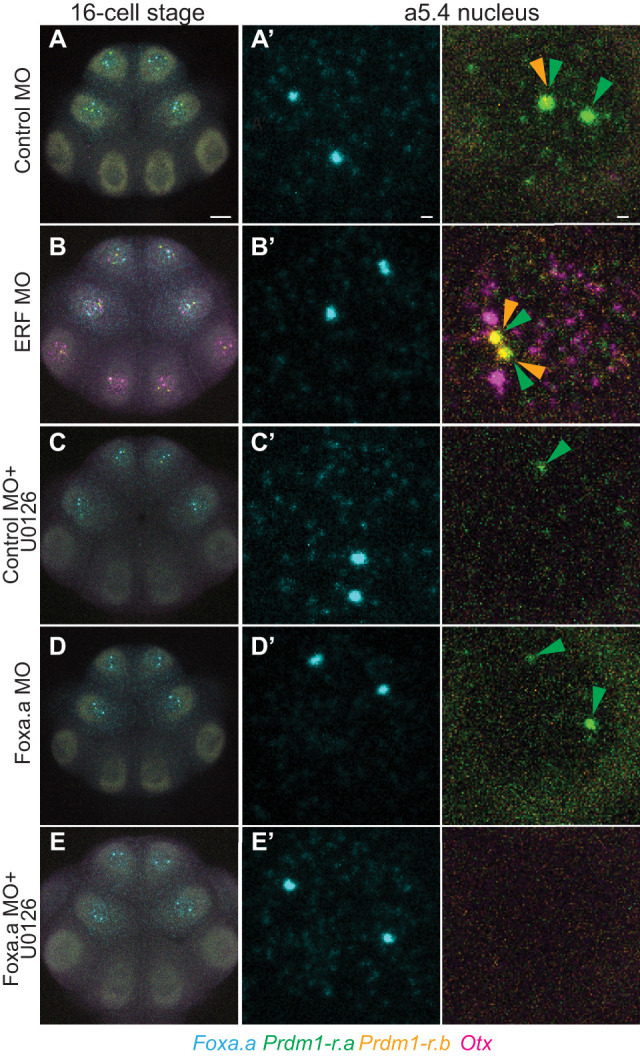
Regulation of Prdm1-r.a/b and Otx by ERF and Foxa.a. (A) In situ hybridizations at the 16-cell stage for embryos injected with a control MO. (A′–E′) Zoomed-in views of the a5.4 nucleus. (B) Same as A, but embryos are injected with an ERF MO. (C) Same as A, but embryos are also treated with U0126. (D) Same as A, but embryos are injected with a Foxa.a MO. (E) Same as D, but embryos are also treated with U0126. Arrowheads indicate sites of active Prdm1-r.a (green) and Prdm1-r.b (orange) transcription. Scale bars: for whole embryo, 20 µm; for single-nucleus views, 2 µm.
Restricted expression of Prdm1-r.a/b throughout the anterior animal portion of the embryo suggests that Foxa.a might serve as their activator at the 16-cell stage. To test this, we knocked down Foxa.a by MO injection. Prdm1-r.a/b expression is quantitatively reduced in morphants, although weak spots of active transcription could still be reliably detected (Fig. 3; Supplemental Fig. S7). Only when we knocked down Foxa.a and treated embryos with U0126 could we completely abolish Prdm1-r.a expression at the 16-cell stage (Fig. 3; Supplemental Fig. S7), similar to observations at the 32-cell stage (Ikeda and Satou 2017).
It is well appreciated that maternally expressed transcriptional repressors are important for inhibiting zygotic transcription and that these repressors must be deactivated so that ZGA can proceed (Vastenhouw et al. 2019). However, we still have a poor understanding of how these repressors are being inactivated at precise time points in embryogenesis. The sudden acquisition of FGF competence provides one such mechanism.
Over the past few years, single-cell sequencing atlases have provided comprehensive gene expression profiles during zygotic genome activation (Xue et al. 2013; Tintori et al. 2016; Treen et al. 2018; Alda-Catalinas et al. 2020; Asami et al. 2022). We also have a general understanding of how early signals can influence what genes are expressed in specific embryonic territories (Stathopoulos et al. 2002; Ohta and Satou 2013; Gentsch et al. 2019). However, breaking down the precise timing of expression with minute-to-minute accuracy is currently beyond the sensitivity of single-cell RNA-seq. In the present study, we used a combination of four-color HCR in situ detection methods along with genetic and experimental perturbation methods to obtain a complete understanding of how a set of signaling-responsive genes is activated in early Ciona development.
Our results suggest that the embryo is rapidly changing from the eight- to 32-cell stages. At the eight-cell stage, when the embryo is first able to perform zygotic transcription, there appears to be an almost total incompetence to respond to FGF signaling. This can be partially overcome with FGF treatment or ERF knockdown; nevertheless, the effects are minimal. At the 16-cell stage, only ∼30 min later, the embryo is fully able to respond to FGF treatment or ERF knockdown by ectopically and precociously expressing FGF-responsive genes. The difference between responses at the eight- and 16-cell stages suggests the occurrence of an important transition. While the nature of this switch is unknown, it is easy to imagine that it involves the synthesis or activation of FGF receptors. In Drosophila, the repressor Capicua can be rapidly inactivated by ERK phosphorylation in response to EGFR signals in a switch-like manner (Keenan et al. 2020; Patel et al. 2021). We propose that levels of ERK phosphorylation that can inactivate ERF are controlled through the FGF pathway through a timing mechanism that begins to operate at the 16-cell stage. Because Prdm1-r.a is still expressed in dissociated embryos, the mechanism does not depend on cell–cell contacts. Instead, autocrine signaling appears to act as an internal timer at the 16-cell stage to delay the onset of Prdm1-r.a/b expression after mitosis (Fig. 4A). Although the identity of this signaling molecule is unknown, one candidate is an FGF4/5/6 ligand that is maternally expressed in Ciona eggs and early embryos (Imai et al. 2004).
Figure 4.
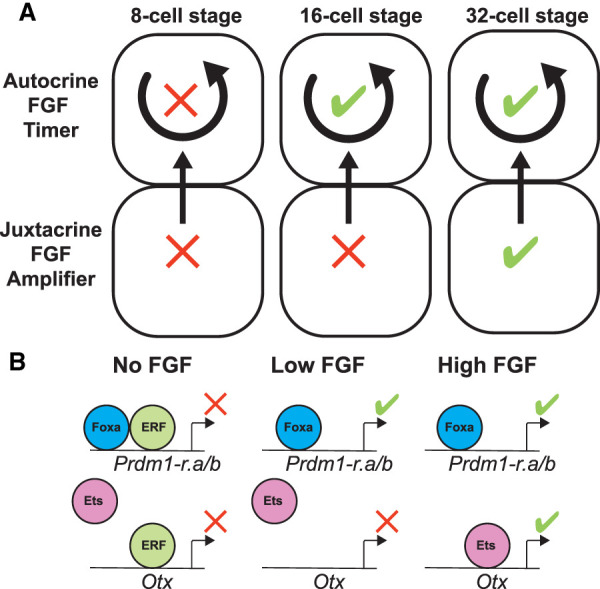
An FGF timer for zygotic genome activation. (A) Schematic depicting an absolute block on FGF signaling at the eight-cell stage that is relieved during the 16-cell stage. At the 32-cell stage, zygotic expression of FGF, as well as compaction of the embryonic cells, has reached a level where it can amplify the signal level. (B) Schematic depicting how the timing mechanisms shown in A is interpreted within the nucleus, resulting in the repression or activation of Prdm1-r.a/b and Otx at specific developmental stages in response to no, low, or high FGF signals.
Our results suggest that FGF-mediated derepression of ERF allows Foxa.a to activate Prdm1-r.a and Prdm1-r.b during the 16-cell stage. However, these low levels of FGF signaling are not sufficient to activate Otx, which might depend on both derepression of ERF and induction of additional activators such as Ets transcriptional activators. We propose that temporal precision is tuned during zygotic genome activation by this interplay of localized activators and repressors.
Zebrafish embryos have maternal ERF mRNA at high levels (White et al. 2017), suggesting that ERF repression of early zygotic genes could have been used by vertebrate ancestors. In mouse embryos, ERF does not appear to be maternally expressed but can be detected in early epiblast stages (Vega-Sendino et al. 2021), suggesting that mammalian early development has deviated from the ancestral state. We propose that mechanisms like those we observed in Ciona are used by a range of vertebrate embryos to control the precise timings of gene expression during zygotic genome activation and early embryogenesis.
Materials and methods
Animals
Adult Ciona intestinalis type A (Pacific populations, also referred to as Ciona robusta) were sourced commercially from M-REP. Live adults and embryos were handled at 18°C.
Quantitative PCR
Three batches of embryos from different adult individuals were developed until the eight-cell stage. Total RNA purification, cDNA synthesis, and qPCR amplifications and quantifications were done as previously described (Treen et al. 2018). Primers used for qPCR are listed in Supplemental Table S1.
Treatments
U0126 (Sigma-Aldrich U120) stock solutions were dissolved in DMSO, and embryo treatment concentrations were 10 μM. FGF treatments were done using recombinant Fibroblast Growth Factor-Basic (Sigma-Aldrich F3685), and concentrations were 300 ng/mL. Control samples were treated with 0.1% DMSO. All treatments were done from the one-cell stage ∼30 min after fertilization.
Microinjections
Antisense morpholino oligonucleotides (MOs) were commercially synthesized by GeneTools. The MO sequences were ERF (5′-CACATACGAGCAGTGCATGATTAAG 3′) and Foxa.a (5′-GAGACGACAACATCATTTTTGTAC-3′). Control MOs used the GeneTools standard control oligo (5′-CCTCTTACCTCAGTTACAATTTATA 3′). MO injection concentrations were 0.5 mM. Microinjections were performed using unfertilized Ciona eggs as previously described (Treen et al. 2018).
Hybridization chain reaction probe design
Probes sets for HCR in situ hybridization were produced commercially by Molecular Instruments based on the following gene models: KY21.Chr11.1129.v1.SL1-1 (Foxa.a), KY21.Chr12.997.v1.SL1-1 (Prdm1-r.a), KY21.Chr12.994.v1.SL1-1 (Prdm1-r.b), and KY21.Chr4.720.v2.SL2-1 (Otx). To ensure no cross-hybridization between Prdm1-r.a and Prdm1-r.b probe sets, the probe sets were designed to target nucleotides 1–844 (Prdm1-r.a) and 1–877 (Prdm1-r.b) of the predicted transcripts. Probes were designed to be compatible with the following HCR amplifiers: Foxa.a: B2, Prdm1-r.a: B1, Prdm1-r.b: B5, and Otx: B3. Twenty split initiator pairs were used for each gene.
Hybridization chain reaction in situ hybridization
Embryos were fixed in 100 mM HEPES, 500 mM NaCl, 2 mM MgSO4, 2 mM EGS [ethylene glycol bis(succinimidyl succinate)], and 1% formaldehyde for 5 min with constant agitation and then for a further 55 min without agitation at room temperature. Samples were then washed four times in phosphate-buffered saline and 0.1% Tween 20 (PBST). Samples were then dehydrated by replacing the PBST with 50% ethanol twice and then with 80% ethanol twice. Samples were stored in 80% ethanol for between 1 and 7 d at −20°C before use.
The HCR protocol was based on a previously published protocol for sea urchin embryos (Choi et al. 2018) with some modifications. Dehydrated embryos were rehydrated from 80% ethanol by gradually adding 5× sodium chloride/sodium citrate and 0.1% Tween 20 (5× SSCT) until the remaining ethanol concentration was ∼20%. Samples were then washed three times in 5× SSCT. For a control experiment, we performed an RNase A treatment before hybridization. Embryos were treated with 20 μg/mL RNase A diluted in 5× SSCT for 30 min at 37°C and then washed three times with 5× SSCT. Rehydrated samples were prehybridized in a prehybridization buffer (5× sodium chloride/sodium citrate, 50% formamide, 5× Denhardt's solution, 100 μg/mL yeast tRNA, 100 μg/mL salmon sperm DNA) for 2 h at 37°C. We found that shorter prehybridization times of 30 min to 1 h, as well as the absence of yeast tRNA and salmon sperm DNA in the prehybridization solution, resulted in nonspecific fluorescence signals at cell surfaces. The prehybridization buffer was then replaced with 0.8 pmol of each HCR probe set diluted in fresh prehybridization buffer and incubated for 16–18 h at 37°C. Samples were washed in HCR wash buffer (Molecular Instruments) twice for 5 min and then twice for 30 min at 37°C. Samples were washed twice in 5× SSCT at room temperature. The amplification solution was prepared by heating and snap cooling HCR hairpins (Alexa 488: B1, Alexa 647: B2, Alexa 514: B5, and Alexa 546: B3) as previously described (Choi et al. 2018). Six picomoles of each HCR hairpin was diluted in HCR amplification buffer (Molecular Instruments). Samples were incubated with HCR hairpins diluted in HCR amplification buffer for 3 h at room temperature in the dark. Samples were washed twice in 5× SSCT for 5 min and then twice for 30 min. In the penultimate wash, DAPI was added to stain DNA. The 5× SSCT was replaced with PBST, and samples were kept in the dark at 4°C until they were imaged.
Imaging
HCR in situ samples were imaged using a Zeiss LSM 880 confocal microscope (Carl Zeiss) using a 20×/0.8 NA plan-apochromat objective. Z-sections of whole embryos were taken with a pixel size of 0.149 μm2 and 1-μm Z-stack steps. In order to minimize cross-talk, each fluorophore was imaged separately (except DAPI and Alexa 647, which were imaged simultaneously) using the following conditions: Alexa 488, 488-nm laser, 488- to 533-nm emission filters; Alexa 514, 514-nm laser, 544- to 554-nm emission filters; Alexa 546, 561-nm laser, 597- to 633-nm emission filters; and DAPI/Alexa 647, 405- and 633-nm lasers, 415- to 480-nm and 649- to 733-nm emission filters.
Quantification of HCR in situ signals
For each gene, the mean fluorescence intensity of a 2-μm diameter circular region was measured for the two brightest spots within the nucleus (assumed to be the sites of active transcription) at the Z-section that had the highest signal. If these signals were absent, the background signal of a random region within the nucleus was measured. Fluorescence intensity was normalized for each individual gene within a data set by dividing each signal level by the highest measurement for that gene, giving a data range of 0–1.
Supplementary Material
Acknowledgments
We thank members of the Levine laboratory for helpful discussions. This work was supported by grants from the National Institutes of Health (NS076542) and Princeton Catalysis Initiative.
Author contributions: N.T. and M.L. designed the experiments. N.T. and E.C. performed the experiments. C.J.W. and C.P.B. contributed essential reagents. N.T. and M.L. interpreted the results and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350164.122.
Competing interest statement
The authors declare no competing interests.
References
- Affolter M, Pyrowolakis G, Weiss A, Basler K. 2008. Signal-induced repression: the exception or the rule in developmental signaling? Dev Cell 15: 11–22. 10.1016/j.devcel.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Alda-Catalinas C, Bredikhin D, Hernando-Herraez I, Santos F, Kubinyecz O, Eckersley-Maslin MA, Stegle O, Reik W. 2020. A single-cell transcriptomics CRISPR-activation screen identifies epigenetic regulators of the zygotic genome activation program. Cell Syst 11: 25–41.e9. 10.1016/j.cels.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami M, Lam BYH, Ma MK, Rainbow K, Braun S, VerMilyea MD, Yeo GSH, Perry ACF. 2022. Human embryonic genome activation initiates at the one-cell stage. Cell Stem Cell 29: 209–216.e4. 10.1016/j.stem.2021.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Posakony JW. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev 16: 1167–1181. 10.1101/gad.976502 [DOI] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. 2003. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell 115: 615–627. 10.1016/S0092-8674(03)00928-0 [DOI] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, Pierce NA. 2018. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch GE, Owens NDL, Smith JC. 2019. The spatiotemporal control of zygotic genome activation. iScience 16: 485–498. 10.1016/j.isci.2019.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignard L, Fiúza UM, Leggio B, Laussu J, Faure E, Michelin G, Biasuz K, Hufnagel L, Malandain G, Godin C, et al. 2020. Contact area-dependent cell communication and the morphological invariance of ascidian embryogenesis. Science 369: eaar5663. 10.1126/science.aar5663 [DOI] [PubMed] [Google Scholar]
- Hudson C, Darras S, Caillol D, Yasuo H, Lemaire P. 2003. A conserved role for the MEK signalling pathway in neural tissue specification and posteriorisation in the invertebrate chordate, the ascidian Ciona intestinalis. Development 130: 147–159. 10.1242/dev.00200 [DOI] [PubMed] [Google Scholar]
- Ikeda T, Satou Y. 2017. Differential temporal control of Foxa.a and Zic-r.b specifies brain versus notochord fate in the ascidian embryo. Development 144: 38–43. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Matsuoka T, Satou Y. 2013. A time delay gene circuit is required for palp formation in the ascidian embryo. Development 140: 4703–4708. 10.1242/dev.100339 [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. 2004. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131: 4047–4058. 10.1242/dev.01270 [DOI] [PubMed] [Google Scholar]
- Keenan SE, Blythe SA, Marmion RA, Djabrayan NJV, Wieschaus EF, Shvartsman SY. 2020. Rapid dynamics of signal-dependent transcriptional repression by capicua. Dev Cell 52: 794–801.e4. 10.1016/j.devcel.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy C, Rothbächer U, Caillol D, Lemaire P. 2006. Ci-FoxA-a is the earliest zygotic determinant of the ascidian anterior ectoderm and directly activates Ci-sFRP1/5. Development 133: 2835–2844. 10.1242/dev.02448 [DOI] [PubMed] [Google Scholar]
- Le Gallic L, Sgouras D, Beal G, Mavrothalassitis G. 1999. Transcriptional repressor ERF Is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol Cell Biol 19: 4121–4133. 10.1128/MCB.19.6.4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Ishii I, Kubo A, Kari W, Suzuki N, Rothbächer U, Satou Y. 2016. A maternal system initiating the zygotic developmental program through combinatorial repression in the ascidian embryo. PLoS Genet 12: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N, Satou Y. 2013. Multiple signaling pathways coordinate to induce a threshold response in a chordate embryo. PLoS Genet 9: e1003818. 10.1371/journal.pgen.1003818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AL, Zhang L, Keenan SE, Rushlow CA, Fradin C, Shvartsman SY. 2021. Capicua is a fast-acting transcriptional brake. Curr Biol 31: 3639–3647.e5. 10.1016/j.cub.2021.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y. 2020. A gene regulatory network for cell fate specification in Ciona embryos. Curr Top Dev Biol 139: 1–33. 10.1016/bs.ctdb.2020.01.001 [DOI] [PubMed] [Google Scholar]
- Satou Y, Nakamura R, Yu D, Yoshida R, Hamada M, Fujie M, Hisata K, Takeda H, Satoh N. 2019. A nearly complete genome of Ciona intestinalis type A (C. robusta) reveals the contribution of inversion to chromosomal evolution in the genus Ciona. Genome Biol Evol 11: 3144–3157. 10.1093/gbe/evz228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouras DN, Athanasiou MA, Beal GJ, Fisher RJ, Blair DG, Mavrothalassitis GJ. 1995. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J 14: 4781–4793. 10.1002/j.1460-2075.1995.tb00160.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirae-Kurabayashi M, Matsuda K, Nakamura A. 2011. Ci-Pem-1 localizes to the nucleus and represses somatic gene transcription in the germline of Ciona intestinalis embryos. Development 138: 2871–2881. 10.1242/dev.058131 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. 2002. Whole-genome analysis of dorsal–ventral patterning in the Drosophila embryo. Cell 111: 687–701. 10.1016/S0092-8674(02)01087-5 [DOI] [PubMed] [Google Scholar]
- Tintori SC, Osborne Nishimura E, Golden P, Lieb JD, Goldstein B. 2016. A transcriptional lineage of the early C. elegans embryo. Dev Cell 38: 430–444. 10.1016/j.devcel.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuoka M, Maeda K, Kobayashi K, Mochizuki A, Satou Y. 2021. The gene regulatory system for specifying germ layers in early embryos of the simple chordate. Sci Adv 7: eabf8210. 10.1126/sciadv.abf8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treen N, Heist T, Wang W, Levine M. 2018. Depletion of maternal cyclin B3 contributes to zygotic genome activation in the Ciona embryo. Curr Biol 28: 1150–1156.e4. 10.1016/j.cub.2018.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL M, Cao WX, Lipshitz HD. 2019. The maternal-to-zygotic transition revisited. Development 146: 1–20. 10.1242/dev.161471 [DOI] [PubMed] [Google Scholar]
- Vega-Sendino M, Olbrich T, Tillo D, Tran AD, Domingo CN, Franco M, FitzGerald PC, Kruhlak MJ, Ruiz S. 2021. The ETS transcription factor ERF controls the exit from the naïve pluripotent state in a MAPK-dependent manner. Sci Adv 7: eabg8306. 10.1126/sciadv.abg8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CJ, Patel AL, Shvartsman SY, Levine MS, Treen N. 2022. ERK signaling dissolves ERF repression condensates in living embryos. Proc Natl Acad Sci 119: 1–6. 10.1073/pnas.2119187119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Collins JE, Sealy IM, Wali N, Dooley CM, Digby Z, Stemple DL, Murphy DN, Billis K, Hourlier T, et al. 2017. A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 6: 1–32. 10.7554/eLife.30860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williaume G, de Buyl S, Sirour C, Haupaix N, Bettoni R, Imai KS, Satou Y, Dupont G, Hudson C, Yasuo H. 2021. Cell geometry, signal dampening, and a bimodal transcriptional response underlie the spatial precision of an ERK-mediated embryonic induction. Dev Cell 56: 2966–2979.e10. 10.1016/j.devcel.2021.09.025 [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. 2013. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500: 593–597. 10.1038/nature12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.