Abstract
The true benefits of large single-cell transcriptome and epigenome data sets can be realized only with the development of new approaches and search tools for annotating individual cells. Matching a single-cell epigenome profile to a large pool of reference cells remains a major challenge. Here, we present scEpiSearch, which enables searching, comparison, and independent classification of single-cell open-chromatin profiles against a large reference of single-cell expression and open-chromatin data sets. Across performance benchmarks, scEpiSearch outperformed multiple methods in accuracy of search and low-dimensional coembedding of single-cell profiles, irrespective of platforms and species. Here we also demonstrate the unconventional utilities of scEpiSearch by applying it on single-cell epigenome profiles of K562 cells and samples from patients with acute leukaemia to reveal different aspects of their heterogeneity, multipotent behavior, and dedifferentiated states. Applying scEpiSearch on our single-cell open-chromatin profiles from embryonic stem cells (ESCs), we identified ESC subpopulations with more activity and poising for endoplasmic reticulum stress and unfolded protein response. Thus, scEpiSearch solves the nontrivial problem of amalgamating information from a large pool of single cells to identify and study the regulatory states of cells using their single-cell epigenomes.
Single-cell epigenome profiling enables the identification of active and poised cis-regulatory sites and the underlying genome regulation across in vivo and in vitro cell types and tissues. Due to several advantages like slower degradation than RNA and a better understanding of heterogeneity in cellular responses, epigenome profiling for single cells is increasingly adapted for atlas-scale data sets and more accurate insights into underlying cell state regulation (Buenrostro et al. 2015; Corces et al. 2020). Hence, an important challenge is how to handle the problem of searching and meta-analysis of single-cell epigenome profiles. A search tool can handle such tasks, revealing various stages of dedifferentiation of cancer cells and predicting a cell's behavior in an unknown state. Such an approach can lead to better annotation and regulatory inference from new single-cell open-chromatin profiles as it is supported by a reference pool of cells in different cellular states. The challenges and opportunities of such an approach have been discussed as one of the 11 grand challenges in single-cell data science by (Lähnemann et al. 2020) under the topic of mapping a single cell to a reference atlas. They have also listed integrating single-cell data across samples and experiments as another grand challenge. There have been efforts from several groups (Srivastava et al. 2018; Cao et al. 2020) to build search engines for single-cell expression profiles. Some tools like scfind (Lee et al. 2021) help to identify cell type–specific and housekeeping genes. However, they do not resolve the challenges associated with large single-cell epigenome data sets. There have been a few studies on integrating single-cell epigenome with single-cell expression profiles (Jin et al. 2020; Wang et al. 2020; Danese et al. 2021; Wu et al. 2021), but they have not used the approach of searching a large pool of reference cells. Such tools also include Seurat (Stuart et al. 2019), LIGER (Liu et al. 2020), and Conos (Barkas et al. 2019), which have been proposed for the integration of single-cell open-chromatin profiles. However, a recent benchmarking study by Leucken et al. (2022) compared more than 30 such single-cell integration methods and revealed that the most integrative approaches performed poorly for batch correction while integrating scATAC-seq profiles. Leucken et al. revealed a fact about such integrative methods that only 27% of their integration outputs for scATAC-seq profiles performed better than the best-unintegrated results (Luecken et al. 2022). Therefore, despite the availability of large single-cell epigenome atlases (Cusanovich et al. 2018; Domcke et al. 2020) and methods for their low-dimensional visualization, there is a scarcity of a robust mapping method which can lead to the development of search engines meant to correctly match query single-cell epigenome profile to a large number of single-cell profiles irrespective of batch effect.
Most of the published approaches that utilize canonical correlation and principal component analysis focus on visualization and analysis of scATAC-seq profiles within a group and can miss rare cells (very few cells across the entire data set). On the other hand, a search engine approach that can help handle every single-cell independent of each other would provide the tremendous benefit of preserving the information of rare and unique cells in a study and utilizing the data sets from other studies. Multiple kinds of single-cell transcriptomes (UMI or non-UMI) could be mentioned as single-cell RNA sequencing (scRNA-seq) profile for brevity. In comparison to the scRNA-seq, single-cell open-chromatin profiles offer new obstacles. Currently, single-cell epigenome profiling mainly aims to capture open chromatin regions (preferentially at promoters, enhancers, etc.) using DNase-seq (DNase I hypersensitive site sequencing) (Jin et al. 2015), MNase-seq (Micrococcal Nuclease digestion with deep sequencing) (Lai et al. 2018) or ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) (Cusanovich et al. 2018). Besides having more noise and sparseness, read-count matrices for single-cell open-chromatin profiles have a higher number of genomic loci (peaks) as features than a similar matrix for a single-cell expression data set. Most often, genomic loci (peaks) in the read-count matrices of single-cell open chromatin profiles compiled by different research groups are not the same. Hence existing algorithms and search methods proposed for single-cell expression profiles cannot be used directly for single-cell open-chromatin profiles.
Here we describe scEpiSearch, consisting of novel computational methods to match queried single-cell open-chromatin profiles with a large pool of reference single-cell open-chromatin and single-cell expression data sets. scEpiSearch resolves the issue of handling nonsimilar peak lists of single-cell epigenome profiles from multiple scientific groups and solves the problem of calculating the statistical significance of the match of the query with single-cell expression and open-chromatin profiles. scEpiSearch uses a gene-enrichment (GE) score as a proxy for cell type specificity (instead of gene activity [Cusanovich et al. 2018]), while minimizing the bias and noise bias across reference cells (see Methods). scEpiSearch also resolves the problem of using reference cell atlases for highly efficient joint-embedding of query single-cell open-chromatin profiles irrespective of batch effect, species, and peak list. Here, we apply scEpiSearch to single-K562 and embryonic stem cell epigenomes and capture heterogeneity, lineage bias and stress response across single cells to better understand regulatory behaviors through their epigenomes.
Results
scEpiSearch first preprocesses the reference pool of single-cell epigenome and expression profiles (see Methods). In fact, it also has its own processed reference pool of single-cell transcriptomes and epigenomes (Fig. 1). The current reference pool of scEpiSearch has 4.3 million expression profiles from human and mouse cells (Supplemental Table S1), and approximately 800,000 single-cell epigenome profiles from human and mouse cells (Supplemental Fig. S1A). To handle such a large reference pool of single-cell expression and epigenome profiles, scEpiSearch keeps it in a clustered format so that it can be searched in a hierarchical manner (Supplemental Fig. S1B; Fig. 1).
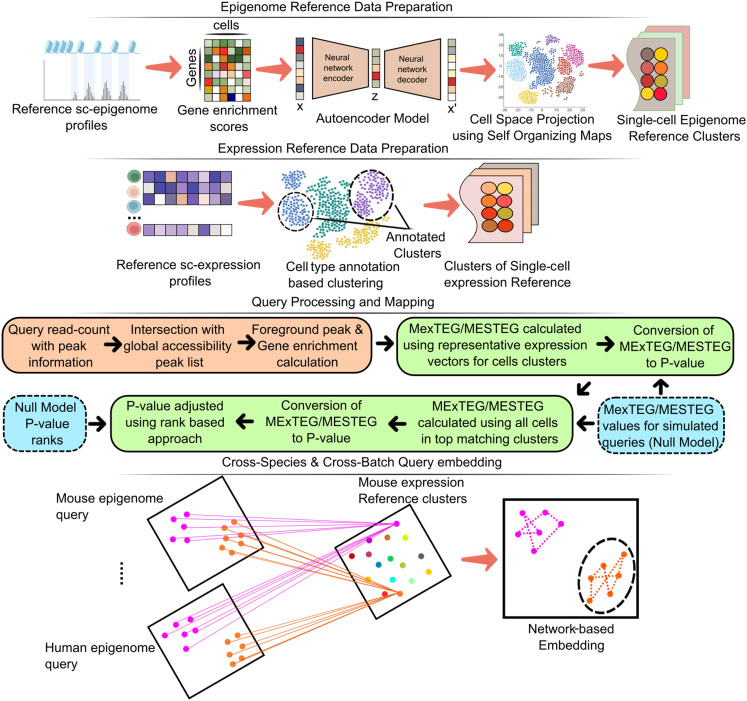
Figure 1.
A graphical outline describing the proposed approach and algorithms in scEpiSearch for annotation of new single-cell open-chromatin profiles and better inference of their regulatory states using the collection of available data sets of single-cell epigenomes and transcriptomes. It involves the steps named as: Epigenome and Expression Reference Data Preparation, Query Processing and Mapping, and Cross-Species and Cross-Batch Query embedding. The cross-species and cross-batch query embedding represents the coembedding of multiple open-chromatin profiles irrespective of differences in peak list in the read-count matrix, batch effect, and species using existing reference single-cell profiles.
For both query and reference single-cell open-chromatin profiles, scEpiSearch first normalizes the read count of every peak with its global accessibility score to highlight potential enhancers (Fu et al. 2018). For both species, human and mouse, we used the global accessibility peak list compiled using several published open-chromatin profiles of bulk samples published by different groups and consortiums (The ENCODE Project Consortium 2012; Bujold et al. 2016). Normalization by global accessibility score for peaks removes the bias that could have come from other cells in the same query. Thus, every cell in the query is treated independently of the other.
scEpiSearch enables correct matching to reference cells irrespective of technical biases
scEpiSearch uses peaks with high normalized counts as foreground and others as the background set to calculate the GE score using the Fisher's exact test (hypergeometric test). In order to compare to a reference scRNA-seq profile scEpiSearch estimates the median of normalized expression values (MExTEG) (in same reference cell) for the top 1000 enriched genes for every query cell separately (see Methods). It uses the normalized expression (see Methods) values in the reference scRNA-seq profile to calculate a median expression for query cells’ top 1000 enriched genes (MExTEG). For a query, the MExTEG of a reference cell is converted to P-value using precalculated MExTEG values for cells in the null model (see Fig. 1; Supplemental Fig. S1B). To compare to a large pool of reference scRNA-seq profiles, scEpiSearch uses a hierarchical approach (Supplemental Fig. S1B; Supplemental Methods). scEpiSearch further calculates a new P-value based on the ranks of reference cells for a query to reduce bias in the data set and search procedures (see Methods, Supplemental Fig. S1B). Such bias can occur due to the presence of doublets of different cell types in reference data and even due to unseen artifacts in the null model. scEpiSearch makes a rank adjustment for hits using their precalculated ranks for the null model. First, we compared our method using a reference set of 10,100 mouse single-cell expression profiles from the mouse cell atlas (MCA). We compared it against three different approaches: (i) comparing gene expression to gene-activity of scATAC-seq calculated using Seurat (Stuart et al. 2019), (ii) correlating GE scores of scATAC-seq and expression profile of reference, (iii) calculating the correlation between the BABEL- (Wu et al. 2021) based predicted expression of query scATAC-seq profile to reference expression (Fig. 2A). We found that our MExTEG based approach is much superior to direct comparison (or correlation) of reference gene-expression values to gene activity, GE scores or predicted expression of query scATAC-seq profiles (Fig. 2A; Supplemental Fig. S2A,B).
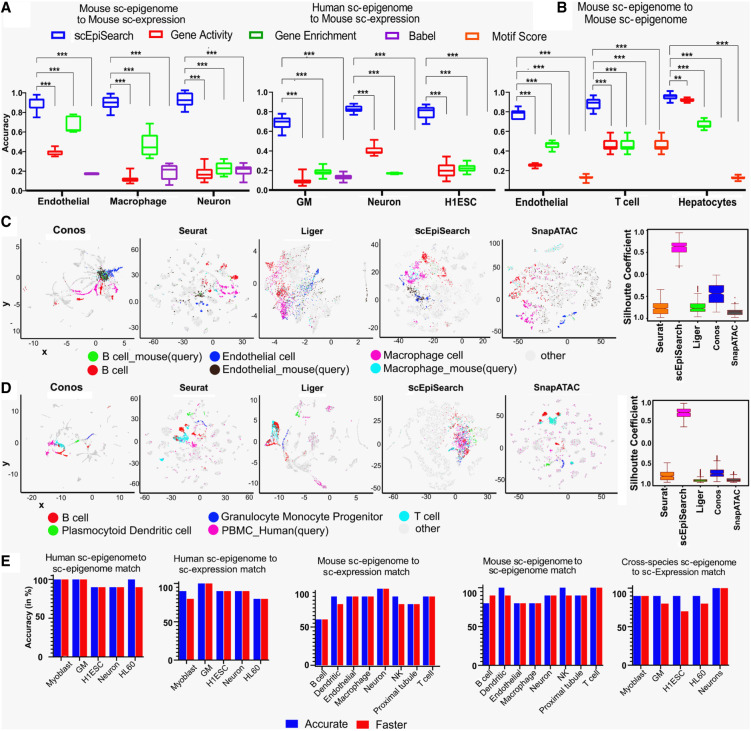
Figure 2.
Evaluation of the accuracy of scEpiSearch. (A) Comparison of scEpiSearch with three approaches based on correlation of gene scores (activity, enrichment and BABEL-based predicted expression) for matching single-cell open-chromatin profiles to a pool of reference single-cell transcriptome. Here the reference data set consisted of single-cell expression profiles of 10,100 cells chosen from the mouse cell atlas (MCA). Accuracy here shows the percentage of query cells which had the correct cell type among the top five matches. (B) Comparison of five methods for matching query single cell open-chromatin profile to reference sci-ATAC-seq profile of ∼81,000 mouse cells published by Cusanovich et al. (2018). (***) P-value <0.001; (**) P-value <0.01. (C) Comparison of scEpiSearch integrative method using reference single-cell expression profiles of 10,100 cells from MCA data set. Here query consisted of scATAC-seq profiles of three types of mouse cells, namely, B cells, macrophages, and endothelial. The silhouette index of query cells, for being in proximity to correct reference cell types, is shown for different methods, on the right panel. (D) Evaluation of cross-species search for integrative methods and approach of scEpiSearch using human PBMC scATAC-seq profiles as query and reference single-cell expression profiles from MCA. Silhouette coefficients for human PBMCs are also shown for different methods. Here immune cells in references and query cells were considered to belong to one class, whereas other cell types as second class for calculation of silhouette coefficients. (E) Accuracy achieved by scEpiSearch engine for matching query scATAC-seq read-count matrices to its own collection of reference single-cell profiles; shown from left to right as such: (i) query human single-cell epigenome (open-chromatin) profile to reference human single-cell epigenome; (ii) query human single-cell open-chromatin profile to reference human single-cell expression collection; (iii) query mouse single-cell epigenome to reference single-cell expression profiles; (iv) query mouse single-cell epigenome (open-chromatin) to reference mouse single-cell expression profile; (v) cross-species search, query human single-cell epigenome to reference mouse single-cell expression. The y-axis shows accuracy in the percentage of query cells for which correct annotation came among the top five hits. It shows accuracies as bar plots for faster and accurate modes of scEpiSearch.
For matching to a reference scATAC-seq profile, its median enrichment score for top enriched genes (MESTEG) of query cells is calculated. The MESTEG value is converted to P-value using precalculated MESTEG scores for cells (vectors of enriched genes) in the null model (Methods, Fig. 1). scEpiSearch also uses a hierarchical approach to find the most matching scATAC-seq profile in the reference data set (Supplemental Fig. S1B). After determining the rank of reference cells for a query cell, it calculates a new P-value using the precalculated rank of the same reference for the null model (see Methods). We compared the MESTEG based approach to the direct correlation between gene activity, GE score, and BABEL-based (Wu et al. 2021) predicted expression of query and reference open-chromatin profiles. In our evaluation, the query scATAC-seq profiles were not from the same data sets used to make a reference pool of single-cell epigenomes. The MESTEG-based approach was substantially better than other procedures in finding correct matching open-chromatin profiles (Fig. 2B; Supplemental Fig. S2C).
We also compared scEpiSearch to integrative methods, Seurat (Stuart et al. 2019), LIGER (Liu et al. 2020), Conos (Barkas et al. 2019), and SnapATAC (Fang et al. 2021) in terms of finding the closest matching single-cell expression profile for various types of scATAC-seq read-count matrices. Here we provided the same reference single-cell expression pool of 10,100 cells from the MCA data set published by Han et al. (Han et al. 2018) to Seurat (Stuart et al. 2019), LIGER (Liu et al. 2020), Conos (Barkas et al. 2019), SnapATAC (Fang et al. 2021), and scEpiSearch. For a two-dimensional (2D) visualization of scEpiSearch results, we used the average of coordinates (in the t-SNE plot) of the top five matching cells. We found that with homogeneous and smaller query sets of single-cell open-chromatin profiles, the coembedding results by integrative methods were unsatisfactory (Supplemental Fig. S3A,B). It could be due to the design of integrative methods like Seurat and LIGER to exploit heterogeneity in single-cell profiles to find anchors, resulting in the wrong grouping of homogenous query single-cell ATAC-seq profiles with various nonsimilar cells. In fact, Leucken et al. (Luecken et al. 2022) also revealed the poor performance of integrative methods for scATAC-seq data and mentioned that gene activity scores used by many integrative methods could be poorly suited to represent scATAC-seq data. Another reason for the revelation of such a result could be that, unlike previous studies, we did not calculate silhouette coefficients/index for reference single-cell expression data points in the coembedding plots, as it could have overwhelmed the corresponding values for query single-cell ATAC-seq profiles (as shown in Supplemental Fig. S3D). Our target was to evaluate the process of finding matching expression profiles for single-cell ATAC-seq data sets (like a search engine); hence we calculated silhouette coefficients only for query cells.
The coembedding plots improved when we increased heterogeneity in the query to include the single-cell ATAC-seq profiles of three types of mouse cells (macrophages, B cells and endothelial cells) (Fig. 2C). However, based on the measure of silhouette coefficients for query single-cell ATAC-seq profiles, the integrative methods (Seurat, LIGER, and Conos) were not comparable to scEpiSearch (Fig. 2C; Supplemental Fig. S3C). A similar trend was observed when scATAC-seq profiles of human cells were used as a query for the same reference expression data set consisting of 10,100 cells from MCA (Han et al. 2018). When scATAC-seq profiles of Human embryonic stem cells (hESC) were used as a query, scEpiSearch-based plots showed them closer to mouse ESC. Similarly, for the scATAC-seq profile of Human Neuron cells, results based on scEpiSearch showed their proximity to reference neuronal cells from MCA (Supplemental Fig. S4B). Seurat and LIGER had the same problem, such that homogenous query cells were spread and colocalized with multiple groups of nonsimilar reference expression profiles in coembedding plots (see Supplemental Fig. S4A,B). When we used a query consisting of scATAC-seq profiles of peripheral blood mononuclear cells (PBMCs) with higher heterogeneity, the results for LIGER and Seurat improved such that query cells appeared closer to immune cells in their coembedding plots (Fig. 2D; Supplemental Fig. S4C). However, when we calculated the silhouette coefficients for query cells to evaluate the efficacy of the integrative method as a search engine, their performance was not comparable to simple embedding based on the results of scEpiSearch (Supplemental Fig. S4A,B,D; Fig. 2D). Overall, our results indicate that integrative methods could not provide an efficient search for matched expression profiles for query scATAC-seq data sets like scEpiSearch.
The standalone and web server versions have their database of reference expression and GE scores of scATAC-seq profiles. Both versions of scEpiSearch have scalable visualization, where results of more than 1000 query scATAC-seq profiles can be visualized interactively. Evaluation using several scATAC-seq profiles with known cell-type annotation revealed that for most of the queries, the accuracy of the search of scEpiSearch is about 80%–100% in highlighting the correct cell type among the top five matches (Fig. 2E; Supplemental Fig. S5A,B; Supplemental Tables S2, S3). The results from several query scATAC-seq profiles also demonstrated that our approach of rank-based P-value adjustment improves the accuracy of search in many cases (Supplemental Fig. S5C) and pseudocount value of 1 for normalization by global accessibility score is better than smaller values (Supplemental Fig. S5D). In the faster version of scEpiSearch, a predefined peak list is used to determine the proximal genes directly. For a query with more than 200,000 peaks, an intersection with a predefined peak list with global accessibility speeds up the process of determining proximal genes by 1000 times (Supplemental Fig. S5E), with almost 75%–90% peaks covered most of the time (Supplemental Fig. S5F). Our approach of using proximal genes enrichment and expression profiles also allows matching query scATAC-seq profiles from human cells to mouse reference scRNA-seq profiles with high accuracy (see Fig. 2D,E; Supplemental Table S4).
To ensure that scEpiSearch does not report false matches in the absence of the correct cell type in its reference data set, we performed a test with negative controls. From the reference single-cell expression pool of 10,100 cells from the MCA data set (used for Fig. 2A,B), we first removed the correct matching cell types for query scATAC-seq profiles. We found that in the absence of the suitable relevant cell types from the reference, most often, scEpiSearch reports insignificant P-values (more than 0.05) for top matches (Supplemental Fig. S6A,B). A similar pattern was observed while using reference scATAC-seq profiles without correct cell types for query (Supplemental Fig. S6C). Such negative control based evaluations hint about the reliability of scEpiSearch.
There are several possible applications of scEpiSearch, including (i) finding cell types for scATAC-seq profiles from unannotated cells from in vivo samples, (ii) studying heterogeneity and tracking divergences in the state of cells and their potency, for example, one cell line showing the differentiation potential toward several lineages, such as K562 cells (Tetteroo et al. 1984), (iii) finding a matching mouse model for a human cell which is not characterized well, (iv) highlighting marker genes representing query cells, (v) embedding and clustering multiple scATAC-seq profiles irrespective of their sources, technique, and species. Using scEpiSearch search-engine for unannotated cells in single-cell indexed ATAC-seq (sciATAC-seq) data set published by Cusanovich et al. (2018) and single-cell epigenome profiles for human cells provided very relevant hits and important genes (see Supplemental Results; Supplemental Figs. S7A,B, S8A–D, S9A–D, S10A,B). We further applied scEpiSearch for different types of case studies to demonstrate its scientific utility.
Determining lineage of cancer cells and understanding their multipotent behavior using scEpiSearch
Recently several groups have started profiling scATAC-seq profiles of frozen nuclei derived from tumor samples known to have heterogeneity and cells with unreported intermediate cellular states. Reprogramming and dedifferentiation in cancer cells are often associated with drug resistance and unexpected lineage switching (Slany 2009; Jacoby et al. 2016). Thus, it becomes crucial to compare the scATAC-seq profile of cancer cells to the existing pool of cells to find their lineage and potency to better understand tumor pathogenesis. Hence as proof of concept, we first evaluated the performance of scEpiSearch in identifying lineage using scATAC-seq profiles of HL60 and K562 cell lines. HL-60 cells derived from myeloid leukaemia patients show neutrophilic promyelocytic morphology (Gallagher et al. 1979). For scATAC-seq read-count matrices of HL60 cells, scEpiSearch found that top-matching expression profiles were from myeloid lineage cells (dendritic and Langerhans cells). For 18% of the HL60 cells, scEpiSearch also reported monocytes among the top five matching cells (Fig. 3A; Supplemental Table S5). Such results could be due to the differentiation potential of HL60 toward monocyte, which is well known (Imaizumi et al. 1987). The most frequently enriched genes included LYST, ALOX5, RASSF4, AOAH, RXRA, MEF2A, PRAM1, and AKAP13 (Supplemental Fig. S11A), which have been cataloged in gene sets for myeloid lineage in the Human_Gene_Atlas listed in Enrichr (Kuleshov et al. 2016) (https://maayanlab.cloud/Enrichr/#stats).
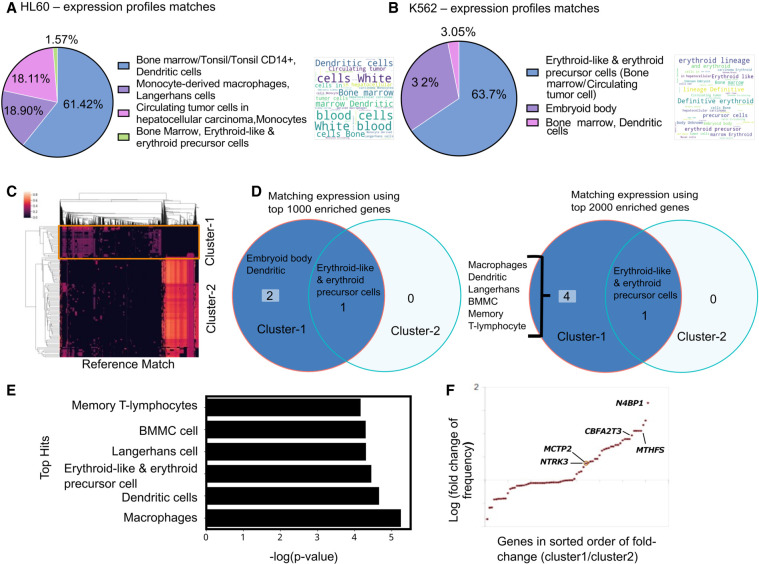
Figure 3.
Case study of using scEpiSearch to reveal lineage and underlying multipotency of cancer cells. (A) The pie chart shows the proportion of cell types for the top five matching single-cell expression profiles for scATAC-seq read-count matrix for HL60 cells. A word-cloud of annotations of matching single-cell expression profiles is also shown on the right side of the pie chart. (B) The proportions of cell types for the top five matching single-cell expression profiles for scATAC-seq read-count matrix for K562 cells. The corresponding word-cloud is shown on the right. (C) Heatmap of match scores achieved with top matching reference epigenome profiles for scATAC-seq read-count matrices of K562 cells. Columns in the heatmap show matching epigenome profiles from reference, and every row represents a Query cell (K562 cell). Two clusters of query cells are also shown. (D) The annotations of cells for the top five match expression profiles matching to a scATAC-seq data set of K562 cells in cluster-1 and cluster-2. The result has been shown when top-1000 and top-2000 enriched genes are used. (E) Average P-values for matching (while using top 2000 enriched genes) to cluster-1 K562 cells with different cell types. (F) Fold change in frequency of being in the top 50 enriched genes is shown for query K562 cells from two clusters. Each dot represents a gene. It is plotted only for those genes which have a frequency of at least 10% in either class and have a fold change above 1.05. Genes whose names are displayed in the plot are known to be markers for dendritic lineage.
The K562 leukaemia cell line has been broadly utilized in consideration of erythroid differentiation. K562 cells serve as an experimental model to study the early steps of megakaryoblast and macrophage commitment and differentiation (Tetteroo et al. 1984; Sutherland et al. 1986). Using the default mode of using the top 1000 enriched genes in scEpiSearch, the top five hits for all K562 cells consisted mainly of erythroid-like and erythroid precursor cells (Fig. 3B). In the top five matching expression profiles, we also found a few cells from the embryoid body whose lineage was not annotated. Genes with a higher frequency of being among the top 50 enriched genes for query scATAC-seq profiles of K562 included KSR1 and PRKCB (Supplemental Fig. S11B), which have been reported to have higher expression in erythroid cells in the mouse cell atlas scRNA-seq data set by Han et al. (2018). At the same time, frequent top enrichment of genes involved in early erythropoiesis like NR2F2 and SOCS1 (Sarna et al. 2003; Fugazza et al. 2021) hints at the dedifferentiated state of K562 and their similarity with erythrocyte progenitors. Overall, scEpiSearch can predict the major lineage of cancer cells and is useful in highlighting relevant genes.
Further for detecting multipotency and heterogeneity among K562 cells, we used the clustering result provided by scEpiSearch, which is based on their match with the reference epigenome profile. Clustering of K562 scATAC-seq profiles revealed two major clusters (Fig. 3C). We compared results of matching scRNA-seq profiles found by scEpiSearch using the top 1000 and top 2000 enriched genes for the query K562 cells. Cells in cluster-2 always had the top five matching scRNA-seq profiles from erythroid-like or erythroid precursor cells with both parameter settings (top-1000 and top-2000 enriched genes) (Fig. 3C,D). However, cells in cluster-1 also had other types of matching expression profiles in addition to erythroid-like cells. Using top-2000 enriched genes of query cells, other matching expression profiles for cluster-1 cells were Macrophages, dendritic cells (including Langerhans) and a few bone marrow mononuclear cells (BMMC) (Fig. 3D). We took an average of P-values for top-five expression matches provided by scEpiSearch for K562 cells from cluster-1. We found that expression profiles of macrophages had the most significant P-values of the match with cluster-1 cells (Fig. 3E). For dendritic and erythroid lineage cells, average P-values for the match to cluster-1 cells were also significant but comparable to each other. We also found a few genes labeled as markers of dendritic cells (available in Enrichr [Kuleshov et al. 2016]) like NTRK3, MCTP2, MTFHS, N4BP1, CBFA2T3, which had a higher frequency of appearance in the top 50 genes for cluster-1 cells in comparison to cluster-2 (Fig. 3F; Supplemental Fig. S11C). The differentiation potential of K562 cells toward macrophage lineage is well known (Sutherland et al. 1986). Several groups have also reported the potency of K562 cells to differentiate toward dendritic lineage (Zhao et al. 2005). Our analysis revealed that a minor population of K562 cells had slightly more significant similarity with macrophages than erythroid lineage. It also hints toward a potential application of scEpiSearch in studying heterogeneity in the underlying poised state of cancer cells for oncology studies.
scEpiSearch enables joint embedding and visualization of single-cell epigenome profiles across batches and species
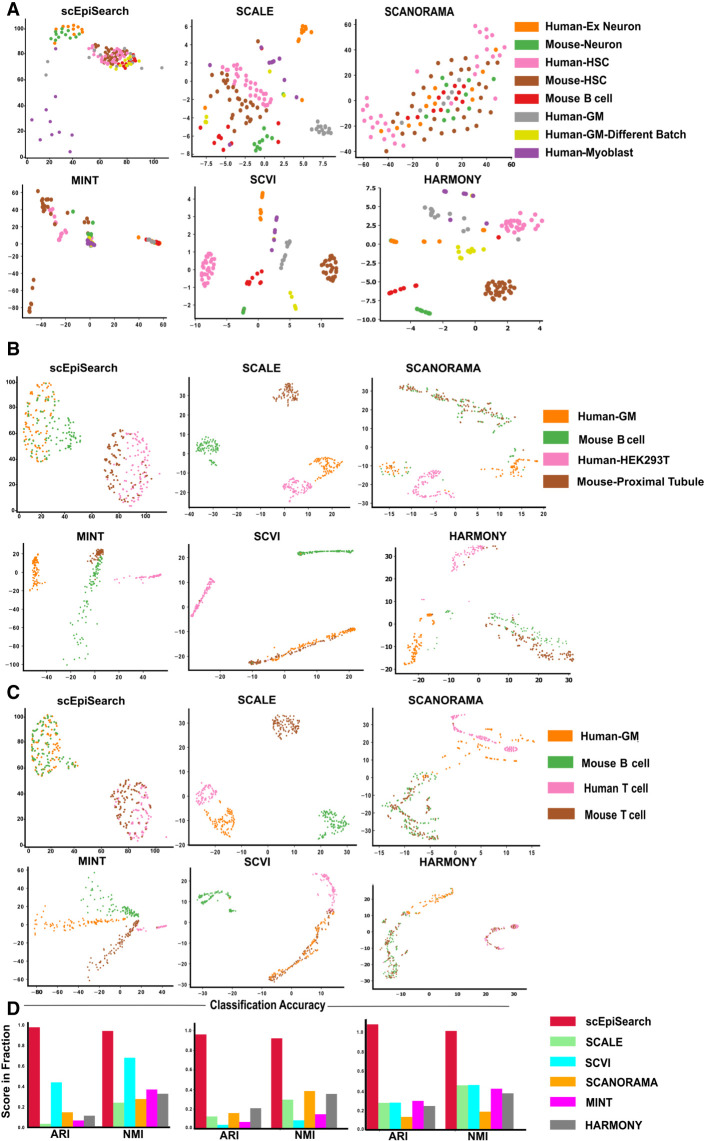
Even though scEpiSearch finds matching transcriptome and open-chromatin profiles for single query cell epigenome, it is often necessary to visualize and cluster cellular profiles from multiple sources to get an insight into discrete or mixed cellular states. Therefore, scEpiSearch is also designed to embed and provide an integrated visualization of multiple, scATAC-seq profiles with different peak list and batch-effect irrespective of their species of origin. scEpiSearch calculates distances among query cells based on the similarity of top-matching mouse reference expression profiles. Here, mouse expression profiles are called similar if they belong to the same cluster in the processed reference scRNA-seq data set of scEpiSearch. We compared the performance of scEpiSearch with five methods meant for embedding (SCANORAMA, MINT, SCVI, SCALE, HARMONY) (Rohart et al. 2017; Lopez et al. 2018; Hie et al. 2019; Korsunsky et al. 2019; Xiong et al. 2019) using four different collections of scATAC-seq read-count matrices. SCANORMA, MINT, and SCVI use genes as features hence GE scores from multiple scATAC-seq profiles (query cells) were provided to them for 2D embedding. Whereas for SCALE, its latent space representation of scATAC-seq read-count matrices was used with t-SNE to perform 2D embedding. As shown in Fig. 4A–D and Supplemental Fig. S12A,B, the 2D embedding plot made by scEpiSearch for scATAC-seq profiles has almost correct colocalization of similar cell types irrespective of the species and laboratory of origin. Other available embedding (SCANORAMA, MINT, SCVI SCALE, and HARMONY) provided the wrong grouping of cells (Fig. 4A–D; Supplemental Fig. S12A). For further confirmation, we estimated clustering purity after density-based spatial clustering (using DBSCAN [Ester et al. 1996]) of the embedding results using cell type labels as true clusters. The clustering purity-based adjusted Rand index (ARI) and normalized mutual information (NMI) scores showed the superiority of scEpiSearch in the embedding of open-chromatin profiles in an unbiased manner (Fig. 4A–D; Supplemental Fig. S12B). The silhouette coefficients-based comparison is shown in Supplemental Figure S12B. For further evaluation of coembedding by scEpiSearch, we tested it for cases where as negative control there were cell types which did not have any matching cell group (Supplemental Fig. S13). In such cases scEpiSearch did not force colocalization of nonmatching cell types.
Figure 4.
Evaluation of embedding of query sets of single-cell open-chromatin profiles irrespective of batch effect, species, differences in peak list and their source (scientific group). (A) For this case study, queries consisted of separate read-count matrices for scATAC-seq profiles of human-neuron, mouse-neuron, human-HSC (hematopoietic stem cells), mouse-HSC, human-myoblast, human-GM12878 (GM) cells from two batches, and mouse B cells. Here the x-axis shows dimension-1 whereas the y-axis shows dimension-2 of low dimensional embedding. The peak lists of query read-count matrices were different from each other. Embedding plots from five other methods are also shown here. While SCANORAMA mixed the location of all cells, MINT could not group cells of the same type together like scEpiSearch. (B) The plot of embedding results shows the alignment of the same cells from different species and batches together. Queries are made for human-GM12878 (GM) cell, mouse B cell, human-HEK293T, and mouse-proximal tubule. The embedding plot from scEpiSearch derived from projections onto mouse expression profiles. (C) The plots show the 2D embedding of cells from different species and batches. Queries were made for human-GM12878 (GM), mouse B cell, human T cell, and mouse T cell. (D) The purity of density-based spatial clustering (using DBSCAN) with embedded coordinates is also shown here in terms of ARI and NMI scores. The silhouette coefficients calculated without DBSCAN-based clustering are shown in Supplemental Figure S12B.
A case study of embedding: understanding multiple phenotype acute leukemia
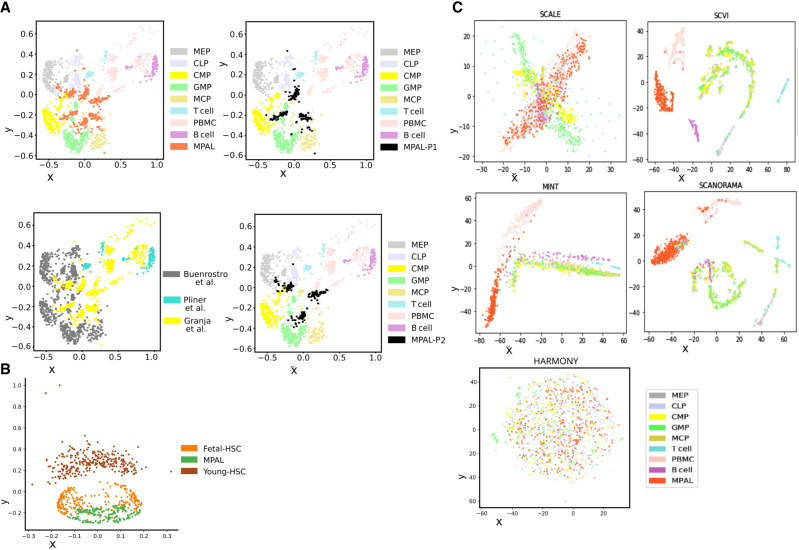
To get further insights from the joint embedding of single-cell epigenome profiles and underlying cell states, we analyzed scATAC-seq profiles from patient blood cells with mixed-phenotype acute leukaemia (MPAL) (Granja et al. 2019). An initial analysis of MPAL cells and PBMCs from healthy patients in the same study revealed a change in the fraction of cell types. Such that for single-cell epigenomes of PBMCs from healthy individuals, the matching scRNA-seq profiles were from similar fractions of blood cell types as reported by others (Supplemental Fig. S10A; Kleiveland 2015). However, for MPAL cells from two patients, we found an increase in the fraction of cell types of dendritic, monocyte and erythrocyte lineage (Supplemental Fig. S14A). It is not trivial to find whether it represented true fractions or it was due to sampling bias during scATAC-seq profiling. Nevertheless, we performed embedding of scATAC-seq profiles of MPAL cells from two patients, PBMCs from healthy individuals and progenitors of cells in the blood (progenitors of hematopoietic cells) (Buenrostro et al. 2018), T cells, and B cells (Supplemental Methods; Pliner et al. 2018). For our analysis, we included the scATAC-seq profiles of progenitors of hematopoietic cells (Buenrostro et al. 2018) isolated from human bone marrow, namely megakaryocytic-erythroid progenitor (MEP), common myeloid progenitor (CMP), common lymphoid progenitor (CLP), granulocyte-monocyte progenitor (GMP), and mast cell progenitor (MCP). In the 2D embedding results from scEpiSearch many MPAL cells overlapped with different types of hematopoietic progenitor cells. Multiple PBMCs colocalized with T cells, B cells and a few with MPAL cells (Fig. 5A). In our embedding results, PBMCs rarely overlapped with hematopoietic progenitor cells. PBMCs colocalizing with B cells and T cells in coembedding plot also had a top matching hit as a transcriptome from B and T cells (Supplemental Fig. S14B). A few PBMCs with a top-matching hit as a transcriptome (Supplemental Fig. S14B) from dendritic or monocytic cell were close to MPAL cells from patient-2 (Fig. 5A). Such results hint about the levels and types of undifferentiated states of MPAL cells. The undifferentiated states of MPAL cells could explain their plasticity and lineage-switching capability (Slany 2009). Further detailed analysis revealed that only a few MPAL cells for two patients showed overlap (Fig. 5A). The majority of cells from two patients with MPAL did not overlap with each other and showed closeness with hematopoietic progenitor cells (Fig. 5A). We further used scEpiSearch to perform coembedding of the scATAC-seq profile of MPAL cells and hematopoietic stem cells (HSC) from young and fetal mice. In our result, fetal mice HSC were closer to human MPAL cells than HSC from young mice (Fig. 5B). Such results hint that MPAL cells and fetal mice HSC could share common features such as fast cell-cycling and possibly some similarity in potency. We also applied SCANORAMA, MINT, SCVI, SCALE, and HARMONY on the same set of read-count matrices (Supplemental Methods) and found that they either mixed the location of different types of blood cell progenitors or showed no colocalization of PBMCs with B cell or T cell (Fig. 5C). Given the fact that B cells and T cells are frequently present among PBMCs, it became quite evident that using other methods could not lead to the result achieved by scEpiSearch (Fig. 5C). We also performed another case study of coembedding of the single-cell open-chromatin profile of renal cancer cells (Wang et al. 2022) and adult and human fetal kidney (Supplemental Fig. S15) cells. The renal cancer cells colocalizing with fetal human kidney cells also had top-5 matching hits from fetal mice kidneys while searching for their match in the reference mouse transcriptome (Supplemental Fig. S15). In summary, scEpiSearch can display closeness and differences among subpopulations of cells irrespective of source and batch effect and highlight undifferentiated states and plasticity of cells derived from patient samples.
Figure 5.
Using scEpiSearch for 2D embedding to track the dedifferentiated state of leukemia cells from blood-cancer patients. (A) scEpiSearch-based 2D embedding of single-cell open chromatin profiles of three types of cells: blood cells collected from patients with mixed MPAL, PBMC, from healthy (normal) individuals, and progenitors of blood cells (derived from bone marrow). The scATAC-seq profiles of MPAL and PBMCs were published by Granja et al. (2019), and progenitor cell epigenome profiles are from a different study. In the embedding plot by scEpiSearch, most of the PBMCs are far from progenitor cells and closer to B cells. MPAL cells are closer to progenitor cells. Some MPAL cells overlap with blood cell progenitors, highlighting their highly undifferentiated state. The same 2D embedding plot from scEpiSearch is shown with a different color for cells according to the source of data and patient. (B) scEpiSearch based 2D embedding of single-cell open chromatin profile of MPAL cells with HSC from fetal and young mice. (C) Results from other tools for the 2D embedding of the single-cell open-chromatin profile of three types of cells: blood cells collected from patients with MPAL, PBMCs from healthy individuals, and progenitors of blood cells. Other methods either mixed up the locations of different types of hematopoietic progenitor cells or could not colocalize B and T cells with PBMCs.
Application in highlighting unique regulatory patterns in a subpopulation of stem cells
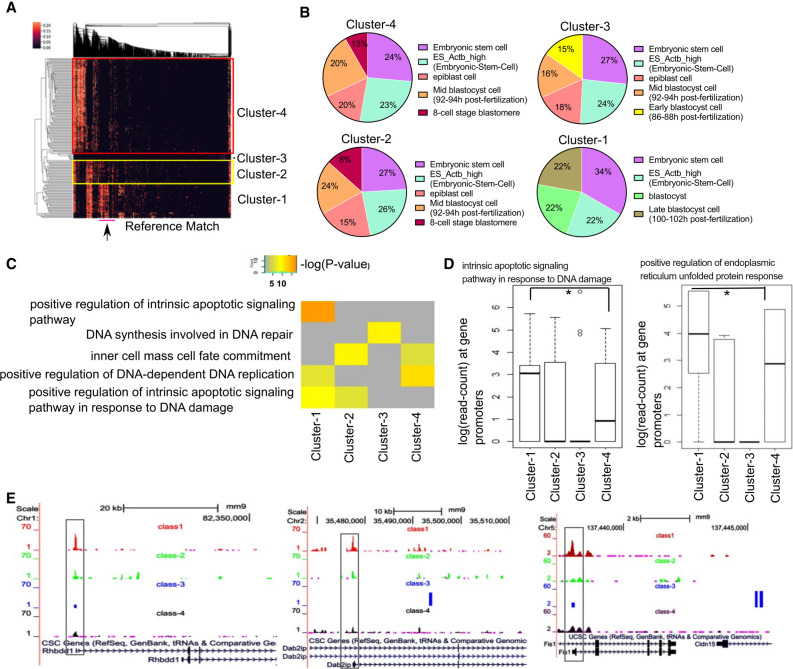
Heterogeneity within a population of stem cells has been widely identified in single-cell genomics studies. We hypothesized that clustering scATAC-seq profiles of embryonic stem cells based on matched scores with reference data sets could highlight cells with differential peak enrichment across features. We generated and reanalyzed plate-based scATAC-seq data from mouse embryonic stem cells (mESC) in Serum conditions (Chen et al. 2018), applied scEpiSearch combining scATAC-seq profiles and calculated read counts per cell for clustering of open-chromatin patterns. The hierarchical clustering of our queried scATAC-seq profiles using match score with reference data sets (using top 2000 enriched genes) captured four major clusters of mESC cells. The cells in four clusters had high matching scores with reference cells belonging to published embryonic stem cells, epiblast cells, and different blastocyst stage cells (Fig. 6A). The cluster-1 cells matched closely to late-blastocyst cells, whereas cells in clusters 2 and 4 matched with mid- and early-blastocyst cells (Fig. 6B). Using the top 10,000 peaks per cluster with the highest normalized read counts, we performed Gene Ontology (GO) enrichment using GREAT (McLean et al. 2010). We found cluster-1 cells were enriched for negative regulation of the G2/M phase, apoptosis, cellular response to unfolded protein, H4K5 and H4K8 acetylation and DNA damage terms (Fig. 6C; Supplemental Table S6). To have a systematic overview, we selected a few terms appearing among top-enriched from each of the four clusters of cells (Supplemental Table S6). Then for the selected terms, we curated enrichment scores (P-values) calculated by GREAT for each cluster (see Fig. 6C). We found that terms like positive regulation of intrinsic apoptotic signaling pathway were specifically enriched for cluster-1 cells (Fig. 6C).
Figure 6.
Studying single-cell epigenome profile of stem cells using scEpiSearch. (A) Heatmap obtained from scEpiSearch showing biclustering of match scores of query scATAC-seq profiles of mouse embryonic cells (rows) with top matching reference single-cell epigenome profile (columns). The columns, highlighted with the pink bar below (and arrow), belong to reference cells from the late blastocyst, which show high similarity with the mESC of clusters-1. (B) The pie charts show cell types of the top five matching expression profiles to query scATAC-seq profiles of mESCs belonging to different clusters. It is based on a top-2000 enriched genes-based search. (C) The result of gene-set enrichment for selected biological functions using genes proximal to the top 10,000 peaks specific to mESC cells belongs to different clusters. The gene-set enrichment was performed using the GREAT Gene Ontology enrichment tool using default parameters. (D) The read counts at the promoter of genes belonging to the gene set for biological function terms: “intrinsic apoptotic signaling pathway in response to DNA damage” and “positive regulation of endoplasmic reticulum unfolded protein response.” Here a matrix consisting of a read count for four clusters is quantile normalized to avoid bias. The star (*) shows a significant P-value (<0.03) calculated using Wilcoxon rank-sum test. (E) The snapshot of the UCSC Genome Browser showing the difference in the activity level of the promoter of two genes (Rhbdd1, Dab2ip) associated with unfolded protein response and endoplasmic reticulum stress and one gene (Fis1) associated with apoptosis, cell cycle, and mitochondrion fission.
The gene-set enrichment performed by GREAT (McLean et al. 2010) often uses genes lying far away from peaks; hence, some more evidence is needed to support its estimation. Therefore, we calculated the read count at promoters of all RefSeq genes for all four clusters of mESC and performed their quantile normalization. Except for cluster-3, which had a lower number of cells, we got decent normalized read counts at promoters for other clusters. It revealed that genes associated with intrinsic_apoptotic_signalling_pathway_in_response_to_DNA_damage and ER_unfolded_protein_response had higher open-chromatin accessibility at their promoters in cluster-1 cells (see Fig. 6D). For comparison, we also made box plots for read counts on promoters of other control gene-sets (see Supplemental Fig. S16A). We believe that cells in cluster-1 might have higher chromatin plasticity poised for cellular responses like apoptosis and ER stress compared to cluster-4 with post-replicated single-cells and a defined chromatin state.
We visualized the differences in peak accessibility at single-gene promoters (UCSC Genome Browser) across clusters. Whereas pluripotency factors (Oct4/Pou5f1 and Sox2) had no substantial differences in peak accessibility at promoters (Supplemental Fig. S16B), we observed higher accessibility in cluster-1 cells for Fis1 related to apoptosis and Rhbdd1 (Lastun et al. 2016) and Dab2ip (Fig. 6E; Bellazzo et al. 2017) associated with ER stress and unfolded protein response. Our results are consistent with earlier studies describing ER stress due to unfolded proteins in stem cells (Yang et al. 2016). It also demonstrates a better understanding and interpretation of single-cell open-chromatin peaks can provide novel insights into the heterogeneous stem cell chromatin landscape and underlying regulation.
Discussion
A major challenge in single-cell genomics is to have an accurate and robust projection of single-cell epigenome profiles to reference epigenome and expression atlases and meaningful interpretation of matching cells. This challenge is confounded by batch and technical biases, including cell-to-cell variability both in signal (peak accessibility) and noise, differential read depths, protocols, platforms, and laboratories. Here, we proposed our approach and showed how such challenges could be handled to enable the cross matching of single-cell epigenomic profiles. Notably, our method does not use distance-based measures (correlation or cosine distance) or hashing, and latent-feature extraction approaches but leverages median expression and enrichment of top genes and peaks (MExTEG and MESTEG) to mitigate batch and technical biases.
The uniqueness of scEpiSearch also lies in its statistical approach to reducing bias during the search for matching transcriptome and open-chromatin profiles. It is resourceful in terms of search using a large pool of reference cell profiles and various facilities it provides, such as low-dimensional embedding robust to batch effect, summary word-cloud for overall notion about query scATAC-seq profiles and enrichment scores of genes to highlight possible markers for cell types. Thus, scEpiSearch can also be useful for cross validation of rare cellular states discovered using single-cell open chromatin profiles, especially from patient samples. The standalone version of scEpiSearch also has an in-built preprocessed reference and can be used securely and locally to maintain the confidentiality of sensitive and clinical data.
Our analysis revealed the benefits of mapping single-cell epigenomes to reference-cell profiles, as observed in the coembedding of scATAC-seq. scEpiSearch-based results were substantially better than integrative methods like Seurat, LIGER Conos, and SnapATAC. Most integrative methods depend on the dimensionality reduction approach (such as canonical correlation, principal component analysis, and linear matrix factorization), which may not efficiently capture the nonlinear relationships among the modality of different types of single-cell profiles. Our results indicate that integration of single-cell profiles of two different modalities could be improved if regulatory link between them is exploited with proper approach to avoid batch effect such as scEpiSearch relies on P-value for MExTEG values to avoid reference specific artifacts.
Further, runtime benchmarking using a single CPU core revealed that with preprocessed reference data set, scEpiSearch needed the lowest amount of memory for the same size of reference and query cells in comparison to integrative methods evaluated here (Supplemental Table S7). The initial preprocessing of reference data set by scEpiSearch can also be done with limited memory and time (Supplemental Table S7). Our analysis using scEpiSearch also highlighted a conceptual advancement that using reference cell profiles for feature extraction and calculating distances among cells can achieve better embedding of open-chromatin profiles than other latent feature extraction methods like SCALE, MINT, and SCANORAMA.
In addition, clustering based on similarity with reference cells could highlight new features in minor populations of cells that could have been overwhelmed by properties of cell-states in the majority due to the reference-free feature extraction method. While cells exist in a continuum of states across tumors and cell cultures and show heterogeneity in activity response to the environment, scEpiSearch was able to group these K562 and mESCs into discrete clusters based on single-cell accessibility peaks and similarity to reference data sets. We found a subset of K562 cells increasingly poised toward macrophages and dendritic lineage, indicative of regulatory rewiring. We believe that the scEpiSearch-based clustering and comparison can help create a new hypothesis and better understand cellular heterogeneity. For example, we observe four mESC clusters based on chromatin accessibility after matching with reference data sets, where cluster-1 cells are similar to late blastocyst cells and likely with high cellular plasticity for response to stress. Some of the top enriched terms for mESC in cluster-1, like, unfolded protein response with stress in the endoplasmic reticulum (ER) (Lin et al. 2019), apoptosis, H4K5 and H4K8 acetylation, and DNA damage, are known to co-occur (Dhar et al. 2017; Bolland et al. 2021). Such results hint that heterogeneity in poising toward ER stress and unfolded protein response in multiple types of stem cells and preimplantation embryos (Yang et al. 2016) can also be studied using their scATAC-seq profile with scEpiSearch. Various reports have also linked DNA damage, apoptosis, and ER stress in the development of diabetes, cancer, and other disorders (Yoshida 2007; Urra et al. 2016). Hence analysis of their single-cell open chromatin profiles using a pool of large reference cells with scEpiSearch could help better elucidate the cause or effect of such disorders.
Overall, we have shown a few capabilities of scEpiSearch, such as (i) correct matching of query single-cell open chromatin profile to a large pool of single-cell profiles, (ii) cross-species search for query single-cell open-chromatin profile, (iii) correct coembedding of single-cell open-chromatin profiles from two species (iv) highlighting footprints of poising for stress-response and apoptotic behavior in a subpopulation of embryonic stem cells. The current version of scEpiSearch has a limitation that it can efficiently handle queries of only single-cell open-chromatin profiles. Currently, scEpiSearch cannot be used for single-cell DNA methylation profiles or a few kinds of histone modification profiles. In the future, we anticipate that scEpiSearch can incorporate single-cell histone modification data sets (Supplemental Fig. S10B; Rotem et al. 2015) and provide an increasingly efficient search engine for major epigenetic regulators.
Methods
Preprocessing of single-cell ATAC-seq reference data
For each cell in the reference data set of scATAC-seq, the read count on every peak is normalized by its global accessibility score to enumerate its cell type specificity. In other words, for every cell, peaks with cell type–specific activity (possible enhancers) are highlighted by normalization with its global accessibility score (see Supplemental Methods). The normalized read count tij of a peak i in a single-cell j is calculated as
| (1) |
Here, the read count rij of the peak i in the single-cell j is normalized by its global accessibility score ai added with a pseudocount ɛ. Here, we kept the value of pseudocount as 1 after testing various values for accuracy of scEpiSearch (Supplemental Fig. S5D). Considering proximal (nearest) genes to all peaks as background, and genes nearest to the top 10,000 enriched peaks are taken in the foreground set for every cell to calculate the P-value (of gene enrichment) using Fisher's exact test (based on hypergeometric distribution). Thus, the equation of calculation of the P-value of enrichment of genes can be written as
| (2) |
Here Km represents the number of times a gene m appears in the background set, and N is the number of the appearance of all genes in the background (includes foreground). Whereas Km is the number of times, the gene m appears in the foreground. At the same time, n is the total number of all genes in the foreground. For single-cell epigenome profiles, we selected 10,000 peaks after analyzing multiple data sets as a robust cutoff. However, for data sets with lower than 10,000 peaks, we utilize all peaks (with nonzero read count) toward analysis. Notice that it is the gene enrichment calculation, and it is not the same as gene-set enrichment performed by different tools (McLean et al. 2010; Chawla et al. 2021). Using our approach, we processed each single-cell open-chromatin profile data set separately to make a large pool of reference in scEpiSearch.
Preprocessing reference single-cell expression
Data sets from different studies for human cells are assembled together using the same set of genes. Similarly, expression profiles of single-cells from mouse samples were assembled together. The single-cell RNA-seq based expression (FPKM) profiles were quantile normalized (Cole et al. 2019) with respect to one of the reference data sets. For every reference cell, the expression of a gene is normalized by its mean expression across all the cells to achieve its cell-specific expression. After the calculation of cell-specific-expression values of genes, each reference expression data set was processed separately. Further details are provided in the Supplemental Methods.
Query preprocessing
For query scATAC-seq profiles, scEpiSearch highlights cell-specific peaks by normalization with a global accessibility score and calculate the GE score. For this purpose, it also optimizes proximal gene finding as explained in detail in the Supplemental Methods.
Finding a match in the scRNA-seq reference data set
scEpiSearch computes MExTEG (Median expression of top enriched genes) for query cells in representative expression vectors for clusters or reference cells, such that for query cell q the MExTEG value in reference cell (or cluster) m is
| (3) |
Thus, a MExTEG value represents the median of cell type–specific (or normalized) expression (in a single reference cell) of the top 1000 enriched genes of a query scATAC-seq profile. If the sequencing depth of queried scATAC-seq is low, it uses only those genes which have nonzero enrichment value. Additionally, the user has the choice to utilize the top 2000 enriched genes, for a more detailed analysis of poised states or regulatory epigenomic signatures. It calculates P-value corresponding to a MExTEG value by comparison with null model to reference specific bias such variable sparseness and noise. Further, the search for matching single-cell transcriptome for query scATAC-seq profile is done hierarchically (Supplemental Methods; Supplemental Fig. S1B). As a final refinement score, a new P-value is calculated based on rank using MExTEG based P-value ranks (Supplemental Methods).
Statistical approach to calculate the significance of match. A null model was prepared by randomly selecting a few normalized scATAC-seq profiles using global accessibility scores of peaks. For the randomly selected 500 cells, the top 1000 genes with the highest GE score were extracted. Random pairs were made from selected 500 cells, and the top enriched gene lists of two cells in a pair were merged. Among the merged list of 2000 genes for every pair of cells, 1000 genes were randomly selected. Thus, we made 1000 query vectors that served as false queries (cells), each having a thousand genes. For every cell in the set of false queries, MExTEG is calculated using cell-specific expression profiles of the reference cell to get a matrix of size 1000 × No of reference single cells. For a reference cell, we calculate the P-value of similarity as the fraction of null model cells (false queries), which have higher values of MExTEG than for the query cell. Thus, the significance (P-value) of the match between query q and the reference expression profile of cell m is calculated as
| (4) |
Finding a match among a huge set of reference single-cell epigenome profiles
Similar to the expression matching procedure, MESTEG (median gene enrichment scores of top enriched genes) for query cells q is calculated using its representative GE vectors of reference cell n is
| (5) |
Further, the conversion of the MESTEG value for the query to the P-value is done using the null model for representative GE vectors for clusters. After finding the top matching clusters using P-value for MESTEG, the matching is done at the single-cell level following the hierarchical approach of search (Supplemental Fig. S1B). Again, these MESTEG values for the query are converted to P-values to find the significance of match with each reference cell in top matching clusters. Further using the rank of matched reference cells, the new P-value is calculated to reduce such bias in the search (as explained below).
Statistical approach to calculate the significance of match with reference open-chromatin profile. Initially, a random selection of 500 normalized ATAC-seq cells GE scores is made from various profiles. Then, random pairs of cells were taken, and for every pair of cells, the mean normalized read count was calculated. Thus, a total of 1000 false queries is made whose MESTEG is calculated for all scATAC-seq reference profiles. For every reference cell, the P-value of the match with the query is calculated as the fraction of null model cells (false queries), which have higher values of MESTEG than the query cell. Thus P-value for the match between query q and reference cell n,
| (6) |
After converting MESTEG to P-value of the match, the rank of a reference cell for a query cell is used to calculate the new P-value. The new P-value of the match between a reference cell and a query cell is calculated as the fraction of cells in the null model for which the same reference cell has a better rank than for the query cell.
Embedding of multiple query scATAC-seq profile
We devised a novel method included in scEpiSearch to handle multiple query read count matrices with a different peak list from both human and mouse cells and perform their coembedding. For this purpose, scEpiSearch computes GE scores for cells in each batch of query scATAC-seq profiles with different peaks/species separately. Further it integrates GE scores for different read count matrices into the same matrix. However, scEpiSearch does not use GE directly for embedding as it is influenced by the batch effect. Once GE scores from multiple read count matrices are assembled together, matches for queries are found in mouse single-cell expression profiles using the procedure described above. For mouse query cells, the null model for the mouse is used. For human query cells, scEpiSearch uses a null model made using human cells, just as explained above for cross-species search. It is to be noted that because we use P-value based on MExTEG to find the top matching reference cluster or cell, there is less chance of artifacts in the result due to batch effect in the reference expression profile. After finding matching mouse cells for all query cells, scEpiSearch builds a network with every node representing a query cell. It connects two query cells (nodes) with an edge with a weight equal to the number of top-matching reference cells belonging to the same cluster. For example, consider a query cell A for which four out of the top ten matching mouse expression profiles belong to a subcluster X. If another query cell B has five out of the top ten matching expression profiles from the same subcluster X, then the weight of an edge between query cells A and B would be 4.
After calculating all edge weights scEpiSearch builds a KNN-based graph with nodes as query cells. Further, it uses the Fruchterman and Reingold algorithm (Supplemental Methods) to calculate the 2D coordinate of nodes. (Fruchterman and Reingold 1991). After determining the 2D coordinate of nodes (representing query cells), it uses the networkX library (draw_network_nodes function) to plot the network for visualization (Hagberg et al. 2008). To assist users, it also performs spectral clustering (Xiang and Gong 2008) using the KNN-based network of queries to find clusters of cells.
The description about other functionalities of scEpiSearch is provided in Supplemental Methods and Results.
Mouse embryonic stem cell culture
E14 mESC were cultured in 6-well dishes, precoated with 0.1% gelatin, and in Serum + LIF media containing DMEM knockout (Gibco 10829), 15% FBS (Gibco 10270), 1 × Pen-Strep- Glutamine (Gibco 10378), 1 × MEM (Gibco 11140), 1 × B-ME (Gibco 21985), and 1000 U/mL LIF (Merck ESG1107).
Single-cell chromatin accessibility
We generated and reanalyzed additional single-cell chromatin accessibility profiles from mESCs (Chen et al. 2018). Briefly, mESCs were trypsinized, washed in 1 × DPBS, and counted, and 50,000 cells were used for plate-based scATAC-seq analysis, as previously described (Xu et al. 2021).
Data access
The raw and processed scATAC-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE192644. The standalone version of scEpiSearch, with a user-friendly graphical user interface and functionality of embedding, can be downloaded from http://reggen.iiitd.edu.in:1207/episearch/index.php?view=download and from GitHub (https:// github.com/reggenlab/scEpiSearch). The code of scEpiSearch is also available as Supplemental Material (Supplemental Code and Data 1 and Supplemental Code and Data 2). The webserver version of scEpiSearch without the functionality of embedding is available at http://www.scepisearch.com/ or http://reggen.iiitd.edu.in:1207/episearch/index.php. The codes and data used for the evaluation of different methods and figure generation are also available at the GitHub link and at http://reggen.iiitd.edu.in:1207/episearch/index.php?view=download.
Supplementary Material
Acknowledgments
We are grateful to the Indraprastha Institute of Information Technology - Delhi (IIIT-D) for supporting computational facilities and fellowship of PhD students. The research in the KNN lab is supported by the Villum Young Investigator Grant (VYI#00025397), Novo Nordisk Grants (#NNF18OC0052874 and #NNF19OC0056962) and TIH Anubhuti–IIIT Delhi (IHUB Anubhuti/Project Grant/06).
Author contributions: S.M. implemented the code and tested it and contributed to the writing of the manuscript. S.C., O.C., N.P., and D.S. helped in the curation of the reference data set and I.P.J., N.P., and M.S. tested other tools for comparison. K.N.N. performed mESC experiments and scATAC-seq profiling used in this study. V.K. and K.N.N. wrote the manuscript. V.K. also conceptualized the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277015.122.
Competing interest statement
The authors declare no competing interests.
References
- Barkas N, Petukhov V, Nikolaeva D, Lozinsky Y, Demharter S, Khodosevich K, Kharchenko PV. 2019. Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat Methods 16: 695–698. 10.1038/s41592-019-0466-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellazzo A, Di Minin G, Collavin L. 2017. Block one, unleash a hundred. Mechanisms of DAB2IP inactivation in cancer. Cell Death Differ 24: 15–25. 10.1038/cdd.2016.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland H, Ma TS, Ramlee S, Ramadan K, Hammond EM. 2021. Links between the unfolded protein response and the DNA damage response in hypoxia: a systematic review. Biochem Soc Trans 49: 1251–1263. 10.1042/BST20200861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523: 486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, Majeti R, Chang HY, Greenleaf WJ. 2018. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173: 1535–1548.e16. 10.1016/j.cell.2018.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujold D, Morais DAL, Gauthier C, Côté C, Caron M, Kwan T, Chen KC, Laperle J, Markovits AN, Pastinen T, et al. 2016. The International Human Epigenome Consortium Data Portal. Cell Syst 3: 496–499.e2. 10.1016/j.cels.2016.10.019 [DOI] [PubMed] [Google Scholar]
- Cao ZJ, Wei L, Lu S, Yang DC, Gao G. 2020. Searching large-scale scRNA-seq databases via unbiased cell embedding with cell BLAST. Nat Commun 11: 3458. 10.1038/s41467-020-17281-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla S, Samydurai S, Kong SL, Wu Z, Wang Z, Tam WL, Sengupta D, Kumar V. 2021. UniPath: a uniform approach for pathway and gene-set based analysis of heterogeneity in single-cell epigenome and transcriptome profiles. Nucleic Acids Res 49: e13. 10.1093/nar/gkaa1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Miragaia RJ, Natarajan KN, Teichmann SA. 2018. A rapid and robust method for single cell chromatin accessibility profiling. Nat Commun 9: 5345. 10.1038/s41467-018-07771-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MB, Risso D, Wagner A, DeTomaso D, Ngai J, Purdom E, Dudoit S, Yosef N. 2019. Performance assessment and selection of normalization procedures for single-cell RNA-seq. Cell Syst 8: 315–328.e8. 10.1016/j.cels.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Shcherbina A, Kundu S, Gloudemans MJ, Frésard L, Granja JM, Louie BH, Eulalio T, Shams S, Bagdatli ST, et al. 2020. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer's and Parkinson's diseases. Nat Genet 52: 1158–1168. 10.1038/s41588-020-00721-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174: 1309–1324.e18. 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese A, Richter ML, Chaichoompu K, Fischer DS, Theis FJ, Colomé-Tatché M. 2021. EpiScanpy: integrated single-cell epigenomic analysis. Nat Commun 12: 5228. 10.1038/s41467-021-25131-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Gursoy-Yuzugullu O, Parasuram R, Price BD. 2017. The tale of a tail: histone H4 acetylation and the repair of DNA breaks. Philos Trans R Soc Lond B Biol Sci 372: 20160284. 10.1098/rstb.2016.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Hill AJ, Daza RM, Cao J, O'Day DR, Pliner HA, Aldinger KA, Pokholok D, Zhang F, Milbank JH, et al. 2020. A human cell atlas of fetal chromatin accessibility. Science 370: eaba7612. 10.1126/science.aba7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, Xu X. 1996. A density-based algorithm for discovering clusters in large spatial databases with noise. In KDD'96: Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Vol. 96, pp. 226–231. AAAI Press, Portland, OR. [Google Scholar]
- Fang R, Preissl S, Li Y, Hou X, Lucero J, Wang X, Motamedi A, Shiau AK, Zhou X, Xie F, et al. 2021. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat Commun 12: 1337. 10.1038/s41467-021-21583-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruchterman TM, Reingold EM. 1991. Graph drawing by force-directed placement. Softw Pract Exp 21: 1129–1164. 10.1002/spe.4380211102 [DOI] [Google Scholar]
- Fu S, Wang Q, Moore JE, Purcaro MJ, Pratt HE, Fan K, Gu C, Jiang C, Zhu R, Kundaje A, et al. 2018. Differential analysis of chromatin accessibility and histone modifications for predicting mouse developmental enhancers. Nucleic Acids Res 46: 11184–11201. 10.1093/nar/gky753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugazza C, Barbarani G, Elangovan S, Marini MG, Giolitto S, Font-Monclus I, Marongiu MF, Manunza L, Strouboulis J, Cantù C, et al. 2021. The Coup-TFII orphan nuclear receptor is an activator of the γ-globin gene. Haematologica 106: 474–482. 10.3324/haematol.2019.241224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher R, Collins S, Trujillo J, McCredie K, Ahearn M, Tsai S, Metzgar R, Aulakh G, Ting R, Ruscetti F, et al. 1979. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood 54: 713–733. 10.1182/blood.V54.3.713.713 [DOI] [PubMed] [Google Scholar]
- Granja JM, Klemm S, McGinnis LM, Kathiria AS, Mezger A, Corces MR, Parks B, Gars E, Liedtke M, Zheng GXY, et al. 2019. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol 37: 1458–1465. 10.1038/s41587-019-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg AA, Schult DA, Swart PJ. 2008. Exploring network structure, dynamics, and function using NetworkX. In Proceedings of the 7th Python in Science conference (SciPy 2008), Pasadena, CA (ed. Varoquaux G, et al. ), Vol. 2008, pp. 11–15. [Google Scholar]
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. 2018. Mapping the mouse cell atlas by Microwell-seq. Cell 173: 1307. 10.1016/j.cell.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Hie B, Bryson B, Berger B. 2019. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat Biotechnol 37: 685–691. 10.1038/s41587-019-0113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi M, Uozumi J, Breitman TR. 1987. Retinoic acid-induced monocytic differentiation of HL60/MRI, a cell line derived from a transplantable HL60 tumor. Cancer Res 47: 1434–1440. [PubMed] [Google Scholar]
- Jacoby E, Nguyen SM, Fountaine TJ, Welp K, Gryder B, Qin H, Yang Y, Chien CD, Seif AE, Lei H, et al. 2016. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat Commun 7: 12320. 10.1038/ncomms12320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, et al. 2015. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528: 142–146. 10.1038/nature15740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zhang L, Nie Q. 2020. scAI: an unsupervised approach for the integrative analysis of parallel single-cell transcriptomic and epigenomic profiles. Genome Biol 21: 25. 10.1186/s13059-020-1932-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiveland CR. 2015. Peripheral blood mononuclear cells. In The impact of food bioactives on health: in vitro and ex vivo models (ed. Verhoeckx K, et al. ), pp. 161–167. Springer, Cham (CH). [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, Raychaudhuri S. 2019. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods 16: 1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lähnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, Vallejos CA, Campbell KR, Beerenwinkel N, Mahfouz A, et al. 2020. Eleven grand challenges in single-cell data science. Genome Biol 21: 31. 10.1186/s13059-020-1926-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Gao W, Cui K, Xie W, Tang Q, Jin W, Hu G, Ni B, Zhao K. 2018. Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562: 281–285. 10.1038/s41586-018-0567-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastun VL, Grieve AG, Freeman M. 2016. Substrates and physiological functions of secretase rhomboid proteases. Semin Cell Dev Biol 60: 10–18. 10.1016/j.semcdb.2016.07.033 [DOI] [PubMed] [Google Scholar]
- Lee JTH, Patikas N, Kiselev VY, Hemberg M. 2021. Fast searches of large collections of single-cell data using scfind. Nat Methods 18: 262–271. 10.1038/s41592-021-01076-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Lee JE, Kang JW, Shin HY, Lee JB, Jin DI. 2019. Endoplasmic reticulum (ER) stress and unfolded protein response (UPR) in mammalian oocyte maturation and preimplantation embryo development. Int J Mol Sci 20: 409. 10.3390/ijms20020409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gao C, Sodicoff J, Kozareva V, Macosko EZ, Welch JD. 2020. Jointly defining cell types from multiple single-cell datasets using LIGER. Nat Protoc 15: 3632–3662. 10.1038/s41596-020-0391-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. 2018. Deep generative modeling for single-cell transcriptomics. Nat Methods 15: 1053–1058. 10.1038/s41592-018-0229-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken MD, Büttner M, Chaichoompu K, Danese A, Interlandi M, Mueller MF, Strobl DC, Zappia L, Dugas M, Colomé-Tatché M, et al. 2022. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods 19: 41–50. 10.1038/s41592-021-01336-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. 2018. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol Cell 71: 858–871.e8. 10.1016/j.molcel.2018.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F, Eslami A, Matigian N, Bougeard S, Lê Cao KA. 2017. MINT: a multivariate integrative method to identify reproducible molecular signatures across independent experiments and platforms. BMC Bioinformatics 18: 128. 10.1186/s12859-017-1553-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. 2015. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 33: 1165–1172. 10.1038/nbt.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarna MK, Ingley E, Busfield SJ, Cull VS, Lepere W, McCarthy DJ, Wright MJ, Palmer GA, Chappell D, Sayer MS, et al. 2003. Differential regulation of SOCS genes in normal and transformed erythroid cells. Oncogene 22: 3221–3230. 10.1038/sj.onc.1206381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slany RK. 2009. The molecular biology of mixed lineage leukemia. Haematologica 94: 984–993. 10.3324/haematol.2008.002436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Iyer A, Kumar V, Sengupta D. 2018. CellAtlasSearch: a scalable search engine for single cells. Nucleic Acids Res 46: W141–W147. 10.1093/nar/gky421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland JA, Turner AR, Mannoni P, McGann LE, Turc JM. 1986. Differentiation of K562 leukemia cells along erythroid, macrophage, and megakaryocyte lineages. J Biol Response Mod 5: 250–262. [PubMed] [Google Scholar]
- Tetteroo PA, Massaro F, Mulder A, Schreuder-van Gelder R, von dem Borne AE. 1984. Megakaryoblastic differentiation of proerythroblastic K562 cell-line cells. Leuk Res 8: 197–206. 10.1016/0145-2126(84)90143-7 [DOI] [PubMed] [Google Scholar]
- Urra H, Dufey E, Avril T, Chevet E, Hetz C. 2016. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer 2: 252–262. 10.1016/j.trecan.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Wang C, Sun D, Huang X, Wan C, Li Z, Han Y, Qin Q, Fan J, Qiu X, Xie Y. 2020. Integrative analyses of single-cell transcriptome and regulome using MAESTRO. Genome Biol 21: 198. 10.1186/s13059-020-02116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Zhang B, Fu Y, Zhao X, Zhang J, Zuo K, Xing Y, Jiang S, Qin Z, et al. 2022. Single-cell chromatin accessibility landscape in kidney identifies additional cell-of-origin in heterogenous papillary renal cell carcinoma. Nat Commun 13: 31. 10.1038/s41467-021-27660-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KE, Yost KE, Chang HY, Zou J. 2021. BABEL enables cross-modality translation between multiomic profiles at single-cell resolution. Proc Natl Acad Sci 118: e2023070118. 10.1073/pnas.2023070118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Gong S. 2008. Spectral clustering with eigenvector selection. Pattern Recognit 41: 1012–1029. 10.1016/j.patcog.2007.07.023 [DOI] [Google Scholar]
- Xiong L, Xu K, Tian K, Shao Y, Tang L, Gao G, Zhang M, Jiang T, Zhang QC. 2019. SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nat Commun 10: 4576. 10.1038/s41467-019-12630-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Wen Y, Liang Y, Xu Q, Wang X, Jin W, Chen X. 2021. A plate-based single-cell ATAC-seq workflow for fast and robust profiling of chromatin accessibility. Nat Protoc 16: 4084–4107. 10.1038/s41596-021-00583-5 [DOI] [PubMed] [Google Scholar]
- Yang Y, Cheung HH, Tu J, Miu KK, Chan WY. 2016. New insights into the unfolded protein response in stem cells. Oncotarget 7: 54010–54027. 10.18632/oncotarget.9833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. 2007. ER stress and diseases. FEBS J 274: 630–658. 10.1111/j.1742-4658.2007.05639.x [DOI] [PubMed] [Google Scholar]
- Zhao C, Wang B, Meng D, Cao Y, Yang J, Zhao X, Chen B. 2005. Study on rapid generation of dendritic cells from K562 cell line induced by A23187 alone. Zhonghua Xue Ye Xue Za zhi 26: 408–412. 10.3760/cma.j.issn.0253-2727.2005.07.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.