Abstract
Background
Binary diagnosis of coronary artery disease (CAD) does not preserve the complexity of disease or quantify its severity or mortality risk; hence, a quantitative marker of CAD is warranted. We evaluated a quantitative marker of CAD derived from probabilities of a machine learning model.
Methods
A CAD-predictive machine learning model was developed and validated using 95,935 electronic health records, and its probabilities were assessed as in silico scores for CAD (ISCAD; range, 0 [lowest probability] to 1 [highest probability]) in participants in two longitudinal biobank cohorts. The relationship of ISCAD with coronary artery stenosis, obstructive CAD, multivessel CAD, all-cause mortality, and CAD sequela was measured.
Results
Among 95,935 participants, 35,749 were from BioMe Biobank (median [IQR] age, 61 [18] years; 13,290 [37%] male; 5,130 [14%] with diagnosed CAD) and 60,186 were from UK Biobank (median [IQR] age, 62 [15] years; 25,031 [42%] male; 8,128 [14%] with diagnosed CAD). The model predicted CAD with an area under the receiver-operating-characteristic curve of 0.95 and 0.93 in the BioMe validation and holdout sets, respectively, and 0.91 in the UK Biobank external test set. ISCAD captured CAD risk from known risk factors, pooled cohort equations, and polygenic risk scores. Coronary artery stenosis increased quantitatively with ascending ISCAD quartiles (12 percentage-point increase per quartile), including risk of obstructive CAD, multivessel CAD, and stenosis of major coronary arteries. Hazard ratio and prevalence of all-cause mortality increased stepwise over ISCAD deciles (1.0 [0.20%], 11 [3.1%], and 56 [11%] in bottom, middle, and top deciles, respectively). A similar trend was observed for recurrent myocardial infarction. Almost half of undiagnosed individuals with high ISCAD had clinical evidence of CAD according to clinical guidelines.
Interpretation
EHR-based machine learning was used to generate an in silico marker for CAD that non-invasively quantifies atherosclerosis and mortality risk on a continuous spectrum, and identifies underdiagnosed individuals.
INTRODUCTION
Detection of CAD enables initiation of preventive measures, including lifestyle modifications and lipid-lowering therapies, to prevent cardiovascular disease.1–3 However, CAD is a complex disease with many contributing factors and varied clinical manifestations.4,5 Quantitative differences in the amount of coronary stenosis and plaque composition result in gradations of risk for myocardial infarction and mortality.6,7 This phenotypic spectrum of CAD is missed with the binary classification of CAD as case versus control. Misclassification of CAD is also possible whereby individuals lacking a diagnosis of CAD have evidence of disease.8–10 Missed diagnosis of CAD may lead to myocardial infarction, stroke, and death.11–15
Risk factors can inform the screening and diagnosis of CAD, including the presence of hypertension, diabetes, smoking, and dyslipidemia.3,16 These variables are included in risk scores that predict CAD events, such as the Framingham Risk Score,17 SCORE2,18 and pooled cohort equations (PCE).19 However, these tools use a small number of predictors and discard large amounts of data contained in electronic health records (EHRs); for example, most vital signs, laboratory tests, medications, symptoms, and other clinical features are not used. Millions of these heterogenous clinical data points are accrued by patients longitudinally in EHR-based health systems, but are difficult to analyze or interpret without the use of machine learning.20–24
Machine learning models have been developed to accurately predict 5- or 10-year risk of CAD based on EHR data;25,26 we recently developed an EHR-based model that outperforms PCE and conventional risk factors in predicting one-year CAD status.27 However, these models are primarily tested as a classification tool to predict case-control status of disease (binary framework) and do not attempt to measure disease on a continuous scale (quantitative framework). Individuals occupy a spectrum of CAD, rather than rigid categories of case versus control, and evaluation of CAD in a quantitative manner may better represent this spectrum and improve personalized care.28–30 Here, we asked whether a quantitative in silico score for CAD (ISCAD) derived from a machine learning model has clinical utility as a marker in the detection, risk stratification, and prognostication of CAD. Conventionally, markers are molecules or anthropometrics measured in the body as an in vivo indicator of disease31; we sought to examine ISCAD, an amalgam of clinical data points in the EHR, as an in silico marker for CAD. We evaluated the relationship of ISCAD with clinical outcomes of CAD—atherosclerotic plaque burden, all-cause mortality, and CAD sequela including recurrent myocardial infarction—and identified underdiagnosed individuals who had high ISCAD and EHR evidence of disease, but lacked a corresponding diagnosis.
METHODS
Study design
We performed a study to train, validate, and externally test a CAD-predictive machine learning model using clinical features extracted from EHRs in two large biobanks. This model was adapted from a previous model27 for the short-term risk prediction of CAD in a binary framework based on EHR data. In the present study, probability scores from the model were instead evaluated as a quantitative CAD marker.
First, we trained and validated the machine learning model using 20,497 EHRs from the BioMe Biobank (BioMe), tested the model on a holdout set of 15,252 EHRs from BioMe, and externally tested the model on 60,186 EHRs from UK Biobank. Second, we assessed the relationship of ISCAD with clinical outcomes relevant to CAD in participants from both biobanks. Study protocols were approved by the Institutional Review Board at the Icahn School of Medicine at Mount Sinai (GCO#07–0529; STUDY-11–01139) and all participants provided informed consent. Use of data from UK Biobank was approved with the UK Biobank Resource under Application Number 16218.
Study participants
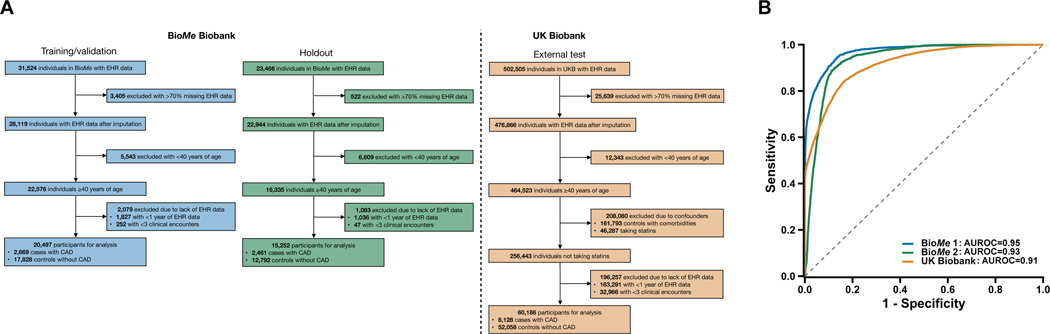
The study included participants from two EHR-linked biobanks in two countries (Supplementary Methods). The machine learning model was trained and validated, and ISCAD was evaluated, in BioMe. BioMe comprises >60,000 individuals of diverse ethnicities recruited from outpatient centers in the Mount Sinai Health System across New York City from 2007 onwards.32 All individuals consented to providing biological and DNA samples linked to de-identified EHRs. Participants at least 40 years old were selected to ensure adequate representation of CAD cases in an age group for whom PCE guides statin initiation27 (Fig. 1A). The model was externally tested in UK Biobank, a community-based cohort of >500,000 individuals chiefly of British self-reported ethnicity between 40–69 years old enrolled across the United Kingdom between 2006–2010.33 Participants were selected with the same criteria as in BioMe (Fig. 1A).
Fig. 1.
Performance of the machine learning model for the detection of coronary artery disease (CAD) in the validation, holdout, and external test sets.
The machine learning model was trained/validated in the BioMe Biobank (BioMe 1), assessed in a holdout set in BioMe (BioMe 2), and externally tested in the UK Biobank. a, Electronic health records (EHRs) of study participants contained both categorical data (i.e., diagnosis codes and medications) and continuous data (i.e., laboratory readings and vital measurements). Only EHR data prior to the earliest date of coronary artery disease (CAD) diagnosis, procedure (e.g., angioplasty), or medication (e.g., statins) prescription were used for CAD cases. In UK Biobank, date of statins prescription is unavailable and individuals with statins were excluded; controls with an Elixhauser comorbidity index of zero were retained. Participants with >70% missing data in the EHR were removed, and the EHR data of the remaining individuals underwent imputation with a random forest-based algorithm. We restricted to participants at least 40 years of age as the target population for which CAD is prevalent and the pooled cohort equations (PCE) is designed to guide statin initiation. Age was defined by the last considered clinical feature entry. Participants with at least one year of EHR data and three recorded clinical encounters were retained. b, The machine learning model discriminated CAD controls from cases with area under the receiver-operating-characteristic curves (AUROCs) of 0.95 (95% CI, 0.94–0.95), 0.93 (95% CI, 0.92–0.93), and 0.91 (95% CI, 0.91–0.91) for the validation, holdout, and external test datasets, respectively.
CAD and clinical outcomes
In BioMe, cases of CAD were identified by the presence of CAD diagnosis codes27 while controls had the absence of all CAD diagnosis codes (Supplementary Methods). In UK Biobank, CAD cases were identified by the presence of CAD diagnosis and procedure codes27; controls were identified by the absence of all CAD diagnosis/procedure codes.
Clinical outcomes relevant to CAD were obtained for 35,749 participants in BioMe. A total of 3,858 participants had undergone cardiac catheterization, with coronary angiography data available for 2,131 participants (Supplementary Methods). These reported the procedure date, coronary vessel, and segment; a subset of 905 also noted stenosis extent (graded as 7 categories of percent stenosis), SYNTAX score,34 and long (>20mm) or heavily calcified (>270°) lesions. Obstructive CAD,6 multivessel CAD,35 and left main, proximal left anterior descending, left circumflex, and right coronary artery stenosis were evaluated. All-cause mortality information available for 35,242 (99%) participants reported whether the participant had died and year of death. CAD sequela comprised recurrent myocardial infarction (MI), defined by an episode of MI >28 days after first MI,36 arrhythmia, and heart failure after CAD diagnosis.
Clinical features obtained from the EHR
In BioMe, both categorical and continuous data from the EHR were obtained as clinical features. Only clinical feature data before the first instance of CAD diagnosis, procedure (e.g., angioplasty, coronary artery bypass graft), or statin use were considered for CAD cases. Categorical features comprised 14,695 unique diagnosis codes and 27,802 medications. Continuous features included 105 laboratory measurements and 9 vital traits. Stringent quality control was performed to filter missing and correlated data (Supplementary Methods) yielding 88 diagnosis codes, 104 medications, 81 laboratory results, and 9 vital traits to train the machine learning model (Supplementary Table 1).
In UK Biobank, we externally tested the machine learning models with EHR data before the first instance of CAD diagnosis or procedure for CAD cases (medication dates are unavailable; individuals using statins in UK Biobank were therefore removed). Continuous features and participants with >70% missing values were removed, and the remaining values were imputed using random forest-based algorithms.
Development of the machine learning model
Our model comprised a random forest-based machine learning system27,37 to predict CAD using clinical features from the EHR (Supplementary Methods). The workflow used a random sample of 90% of cases and an equal number of controls for training, and the remaining 10% of cases and an equal number of controls for validation, iterated 100 times to minimize sampling bias. Feature selection was performed on the training dataset and applied to the validation dataset to reduce model complexity38 and increase clinical interpretability.20 A 10-fold cross-validation was used to optimize the model’s hyperparameters. The resulting model predicted CAD status in the validation dataset; performance metrics were reported for the mean and 95% CI across 100 iterations. This workflow was repeated for an external test dataset from UK Biobank using BioMe Biobank features present in UK Biobank to train 100 new models (Supplementary Methods). A series of sensitivity analyses using variations of the model and its features was performed to improve validity and portability (Supplementary Methods).
ISCAD derivation
Probability scores from the machine learning model were obtained for each participant as their ISCAD. ISCAD ranged from 0 (lowest CAD probability) to 1 (highest CAD probability), and was subsequently evaluated as a quantitative marker for CAD.
Pooled cohort equations, polygenic risk score, and baseline risk
We computed PCE and polygenic risk scores (PRS) for all participants in BioMe (Supplementary Methods; Supplementary Figure 2). PCE represents 10-year atherosclerotic cardiovascular disease risk19 and PRS measures the aggregated effects of genetic variants on CAD risk39. We also considered a baseline of known CAD risk factors19 comprising age, sex, ethnicity, total cholesterol, high-density lipoprotein cholesterol, systolic blood pressure, treatment for blood pressure, type 2 diabetes, smoking status, and body mass index (BMI).
Underdiagnosed CAD identified by ISCAD
The utility of ISCAD in identifying underdiagnosed cases of CAD was assessed. A subset of 26 individuals with high ISCAD ≥0.9 lacked a prior CAD diagnosis. Manual EHR review was performed for these undiagnosed individuals and a propensity score-matched set of 26 undiagnosed participants with low ISCAD ≤0.1 (nearest neighbor matching on age, sex, statin use, and Elixhauser comorbidity index) while blinded to ISCAD to identify those with evidence of CAD according to clinical guidelines16.
Statistical analysis
Differences in categorical and continuous variables were assessed with 2-sided unpaired Fisher’s exact tests and t-tests, respectively. Models to predict CAD were evaluated with area under the receiver-operating characteristic curve (AUROC) and area under the precision-recall curve (AUPRC) using the pROC package (version 1.16.2),40 and sensitivity, specificity, accuracy, positive predictive value (PPV), and negative predictive value (NPV). Linear, logistic, and Cox proportional hazards regression tested the association of ISCAD with continuous, categorical, and temporal outcomes, respectively. Regression models were adjusted for age, sex, BMI, smoking status, and self-reported ethnicity, unless otherwise stated. Significance level was set at 0.05. Associations of ISCAD with atherosclerosis and survival were assessed in pre-specified subgroups of males and females. In each case, interaction between sex and ISCAD was tested, and heterogeneity of sex-specific results was evaluated with Cochran’s Q test. All statistical tests and plots were generated with R (version 3.5.3).
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
RESULTS
Study population
The study population included 95,935 participants from two biobanks with EHR data to train, validate, and externally test the machine learning model (Fig. 1A). The model was trained and validated on EHR data for 20,497 participants from BioMe (median age, 61 years [interquartile range (IQR), 18]; 6,718 [38%] male; 5,887 [29%] European ethnicity) with 2,669 (13%) cases of CAD (Table 1). A holdout EHR dataset comprised 15,252 participants from BioMe (median age, 61 years [IQR, 18]; 6,572 [43%] male; 5,201 [34%] European ethnicity) with 2,461 (16%) cases of CAD (Supplementary Table 3). An EHR dataset for 60,186 participants from UK Biobank (median age, 62 years [IQR, 15]; 25,031 [42%] male; 56,986 [95%] European ethnicity) was used for external testing, including 8,128 (14%) cases (Supplementary Table 4). The association of ISCAD with clinical outcomes of CAD—atherosclerotic burden, all-cause mortality, and CAD sequela such as recurrent MI—was evaluated in the BioMe cohort.
Table 1.
Characteristics of participants in the training/validation set.
| Characteristic | All participants (N=20,497) | CAD controls (N=17,828) | CAD cases (n=2,669) |
|---|---|---|---|
| Median age (IQR) — years | 62 (19) | 61 (18) | 72 (16) |
| Sex — no. (%) | |||
| Female | 12,470 (61) | 11,110 (62) | 1,360 (51) |
| Male | 8,027 (39) | 6,718 (38) | 1,309 (49) |
| Ethnicity — no. (%) | |||
| African | 5,480 (27) | 4,755 (27) | 725 (27) |
| European | 5,887 (29) | 5,276 (30) | 611 (23) |
| Hispanic | 7,424 (36) | 6,276 (35) | 1,148 (43) |
| Other | 1,704 (8.3) | 1,519 (8.5) | 185 (6.9) |
| Ever smoked — no. (%) | 3,830 (19) | 3,308 (19) | 522 (20) |
| Median vitals (IQR) | |||
| Weight — lbs | 172 (53) | 170 (55) | 180 (37) |
| Height — inches | 65 (5.1) | 65 (5.0) | 66 (5.3) |
| Systolic blood pressure — mmHg | 128 (17) | 127 (18) | 133 (9.7) |
| Diastolic blood pressure — mmHg | 73 (10) | 74 (11) | 72 (6) |
| Pulse — sec−1 | 77 (12) | 77 (12) | 75 (11) |
| Oxygen saturation — % on room air | 98 (1.0) | 98 (1.5) | 97 (0.72) |
| Respirations — min−1 | 18 (1.0) | 18 (1.1) | 18 (0.52) |
| Temperature — °F | 98 (0.55) | 98 (0.60) | 98 (0.21) |
| Median laboratory measurement (IQR) | |||
| Low-density lipoprotein cholesterol — mg/dL | 100 (35) | 101 (34) | 90 (31) |
| High-density lipoprotein cholesterol — mg/dL | 53 (19) | 54 (19) | 48 (17) |
| Triglycerides — mg/dL | 115 (67) | 114 (67) | 124 (65) |
| Glucose — mg/dL | 92 (22) | 91 (20) | 103 (43) |
| Hemoglobin A1c — % | 5.7 (0.80) | 5.7 (0.73) | 5.1 (1.5) |
| Troponin-I — ng/dL | 0.011 (0.040) | 0.010 (0.035) | 0.023 (0.13) |
| Lactate dehydrogenase — U/L | 213 (44) | 211 (42) | 224 (53) |
| Erythrocyte sedimentation rate — mm/hr | 22 (24) | 21 (23) | 33 (27) |
| Comorbidities — no. (%) | |||
| Hypercholesterolemia | 10,748 (52) | 8,506 (48) | 2,207 (83) |
| Hypertension | 11,762 (57) | 9,376 (53) | 2,365 (89) |
| Type 2 diabetes | 6,395 (31) | 4,834 (27) | 1,529 (57) |
IQR, interquartile range; no., number; ethnicity, self-reported ethnicity; other, other miscellaneous ethnicities besides the ones listed.
Model performance in validation, holdout, and external testing datasets
In the validation dataset, the model predicted CAD with an AUROC of 0.95 (95% CI, 0.94–0.95), sensitivity of 0.94 (95% CI, 0.94–0.95), and specificity of 0.82 (95% CI, 0.81–0.83) (Table 2 and Fig. 1). The prevalence of CAD was 13% in the validation dataset, with a NPV of 0.93 (95% CI, 0.93–0.93) and PPV of 0.84 (95% CI, 0.83–0.95). In the holdout dataset, the model predicted CAD with an AUROC of 0.93 (95% CI, 0.92–0.93), sensitivity of 0.90 (95% CI, 0.89–0.90), and specificity of 0.88 (95% CI, 0.87–0.88). The prevalence of CAD was 16% in the holdout dataset, with a NPV of 0.89 (95% CI, 0.89–0.89) and PPV of 0.88 (95% CI, 0.88–0.88). In the external test dataset, the model predicted CAD with an AUROC of 0.91 (95% CI, 0.91–0.91), sensitivity of 0.84 (95% CI 0.83–0.84), and specificity of 0.83 (95% CI, 0.82–0.83). The prevalence of CAD was 14% in the external test dataset, with a NPV of 0.84 (95% CI, 0.83–0.84) and PPV of 0.83 (95% CI, 0.82–0.83). The models were calibrated with Brier scores of 0.048, 0.053, and 0.056 in the validation, holdout, and external test datasets, respectively (Supplementary Figure 3).
Table 2.
Classification performance and predictive values of the machine learning model for the detection of coronary artery disease (CAD) in the validation, holdout, and external test datasets.
| Dataset | Total no. | CAD control, no. (%) | CAD case, no. (%) | AUROC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) | NPV (95% CI) | PPV (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| Validation dataset | 20,497 | 17,828 (87) | 2,669 (13) | 0.95 (0.94 – 0.95) | 0.94 (0.94 – 0.95) | 0.82 (0.81 – 0.83) | 0.88 (0.87 – 0.89) | 0.93 (0.93 – 0.94) | 0.84 (0.83 – 0.85) |
| Holdout dataset | 15,252 | 12,791 (84) | 2,461 (16) | 0.93 (0.92 – 0.93) | 0.90 (0.89 –0.90) | 0.88 (0.87 – 0.88) | 0.89 (0.89 – 0.89) | 0.89 (0.89 –0.89) | 0.88 (0.88 –0.88) |
| External test dataset | 60,186 | 52,058 (86) | 8,128 (14) | 0.91 (0.91 – 0.91) | 0.84 (0.83 – 0.84) | 0.83 (0.82 – 0.83) | 0.85 (0.85 – 0.85) | 0.84 (0.83 – 0.84) | 0.83 (0.82 – 0.83) |
No., number; AUROC, area under the receiver-operating-characteristic curve; NPV, negative predictive value; PPV, positive predictive value.
Sensitivity analyses of ISCAD
Sensitivity analyses showed similar performance of a portable model (with shared features in validation and external test datasets) in the external test dataset with no retraining (Supplementary Figure 5); a portable model derived from the external test dataset and assessed in the training/validation dataset with no retraining (Supplementary Figure 6; Supplementary Table 7); and a streamlined model solely based on routine laboratory measurements, vitals, and demographics (Supplementary Figure 7; Supplementary Table 8). Removal of heart failure-related features such as echocardiography measurements, electrocardiogram data, cardiac enzymes, and heart failure medications had minimal effect on the performance of the model (Supplementary Figure 8). In contrast, an ascertainment model testing indicators of the presence or absence of features instead of the values of features performed poorly (Supplementary Figure 9).
CAD risk captured by ISCAD
We next used CAD probabilities from the model to generate ISCAD for all 20,497 participants in BioMe and examined its characteristics. Mean ISCAD was greater in CAD cases compared to controls by 0.55 (95% CI, 0.54–0.55; P<0.0001) (Supplementary Figure 10) and CAD onset occurred 1.8 years earlier per quartile increase in ISCAD (95% CI, 1.5–2.3; P<0.0001). Known risk factors for CAD were associated with ISCAD (Supplementary Figure 11); for example, ISCAD increased monotonically by 0.085 per decade of age (95% CI, 0.082–0.087; P<0.0001) and was greater for individuals with dyslipidemia (0.076; 95% CI, 0.069–0.083; P<0.0001) and type 2 diabetes (0.13; 95% CI, 0.12–0.14; P<0.0001). In addition, ISCAD captured axes of CAD risk from PCE and PRS (Supplementary Figure 11): ISCAD increased by 0.080 per 10-point increase in PCE (95% CI, 0.076–0.084; P<0.0001) and by 0.023 per standard deviation (SD) increase in PRS (95% CI, 0.018–0.027; P<0.0001). Non-traditional clinical features in the EHR were also important in the model, including acetaminophen, ejection fraction, T axis, hemoglobin A1c, and glucose (Supplementary Table 5). Analysis of the model’s interpretability with SHAP values showed that features that are CAD risk factors contributed to the model predictions in the direction expected with their biological effects (Supplementary Figures 12–13).
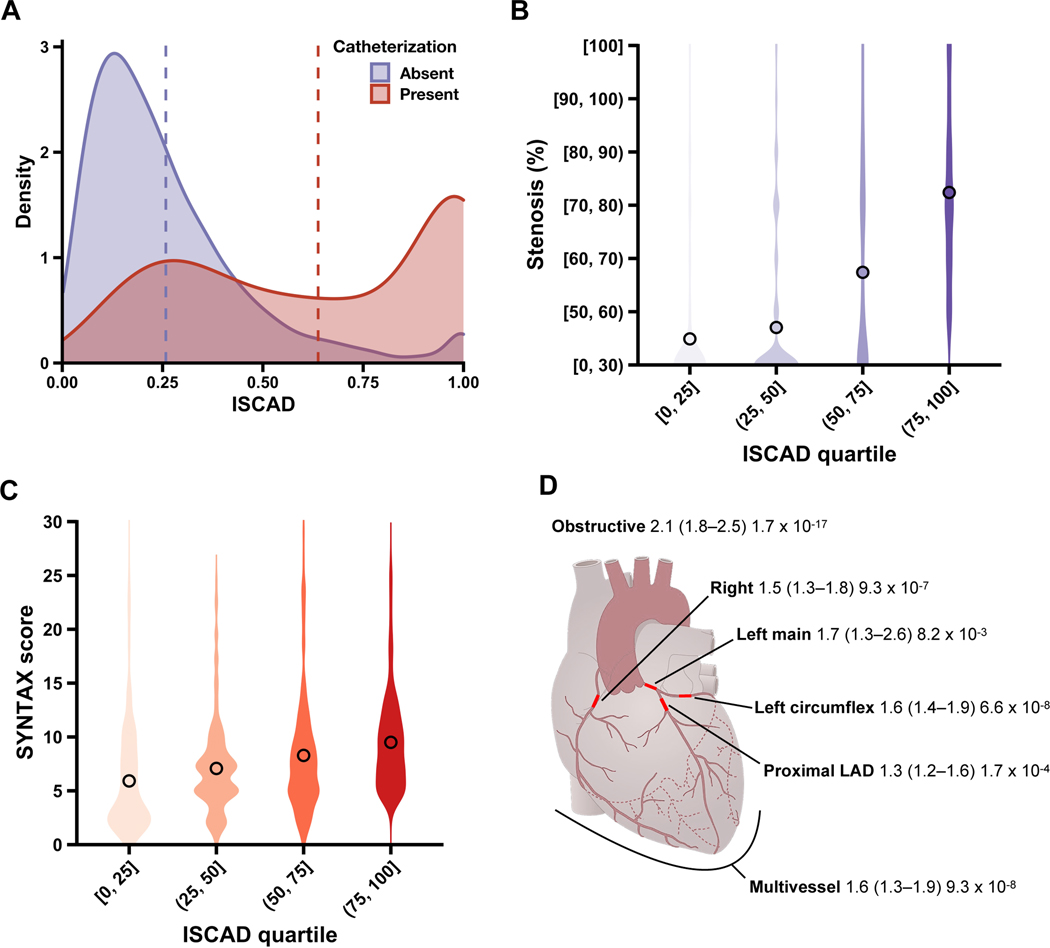
Evaluation of atherosclerosis with ISCAD
After establishing the CAD risk captured by ISCAD, we assessed if ISCAD can represent a quantitative marker for coronary atherosclerosis. Mean ISCAD was greater in participants who underwent cardiac catheterization than those who had not by 0.38 (95% CI, 0.37–0.39; P<0.0001) (Fig. 2A). Stenosis increased quantitatively by 12-percentage points per ISCAD quartile (95% CI, 10–13 percentage points; P<0.0001) (Fig. 2B). Complexity of atherosclerosis, measured by SYNTAX score, also increased monotonically by 1.2 per ISCAD quartile (95% CI, 0.93–1.54; P<0.0001) (Fig. 2C). We also evaluated clinically significant angiographic outcomes: there was an adjusted odds ratio (OR) of 2.1 for obstructive CAD (95% CI, 1.8–2.5; P<0.0001), 1.6 for multivessel CAD (95% CI, 1.3–1.9; P<0.0001), 2.8 for long atherosclerotic lesions (95% CI, 1.7–4.6; P<0.0001), and 2.3 for heavily calcified lesions (95% CI, 1.4–4.1; P=0.002) per ISCAD quartile. These relationships of ISCAD with atherosclerotic burden were consistent in pre-specified sex subgroups (Supplementary Figure 14); for both stenosis and SYNTAX, no significant heterogeneity between sexes (P=0.070, P=0.16) or interaction between sex and ISCAD (P=0.21, P=0.69) was observed. Stenosis of major anatomical vessels and coronary vasculature regions were further examined with ISCAD (Fig. 2D). Adjusted OR for stenosis increased quantitatively with ascending ISCAD quartiles for all coronary sites tested, including left main (1.7; 95% CI, 1.3–2.6; P=0.008) and proximal left anterior descending coronary arteries (1.3; 95% CI, 1.2–1.6; P<0.0001).
Fig. 2.
Relationship of in silico score for CAD (ISCAD) with coronary stenosis and atherosclerosis complexity on cardiac catheterization.
Cardiac catheterization data were examined for association with ISCAD. This comprised percent coronary stenosis, recorded as 7 strata ranging from [0, 30), less than 30%, to [100], 100%, and SYNTAX score ranging from 0, low complexity, to 30, high complexity. ISCAD were stratified by quartiles. a, Individuals who underwent cardiac catheterization (red) had higher mean ISCAD (dashed line) than those who had not underwent cardiac catheterization (purple). b, Violin plots show the distribution of samples across coronary stenosis values along with the mean value overlaid as a point for each ISCAD quartile. c, Violin plots show the distribution of samples across SYNTAX score values along with the mean value overlaid as a point for each ISCAD quartile. d, Schematic of coronary arteries depicts the association of ISCAD with obstructive CAD (≥50% stenosis in the left main coronary artery, ≥70% stenosis in any other coronary artery, or both), multivessel CAD (≥70% stenosis in at least two coronary arteries, or ≥50% stenosis in left main coronary artery and ≥70% stenosis in another coronary artery), left main stenosis (≥50%), proximal left anterior descending (LAD) stenosis (≥70%), left circumflex stenosis (≥70%), and right coronary artery stenosis (≥70%). Results are reported as adjusted odds ratio (95% CI) P value per increase in ISCAD quartile.
Assessment of all-cause mortality by ISCAD
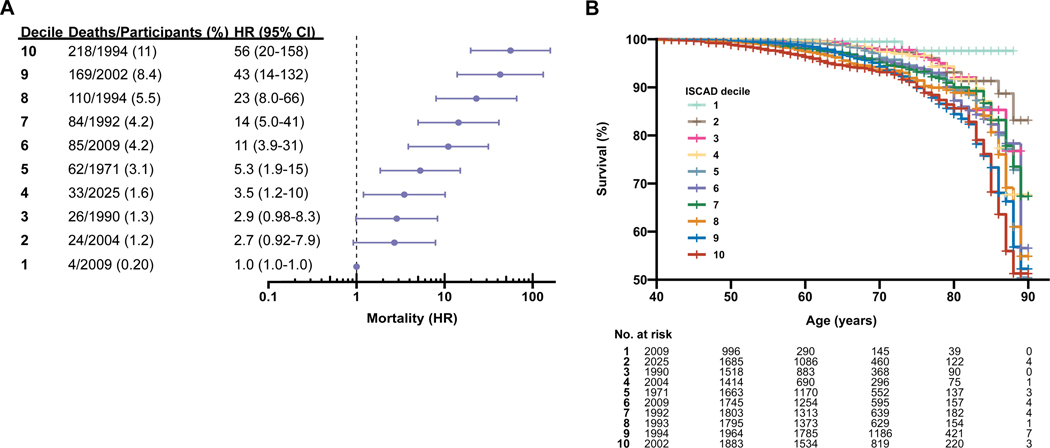
We then evaluated the relationship of ISCAD with all-cause mortality using survival analyses. A total of 815 (4.1%) participants were deceased at the time of analysis (median age of death [IQR], 68 [17] years). ISCAD was associated with a quantitative increase in risk of death with an adjusted hazard ratio [HR] of 1.5 (95% CI, 1.4–1.6; P<0.0001) per SD increase in ISCAD. Prevalence and risk of death increased stepwise over ascending ISCAD deciles: 4 of 2009 (0.20%) deceased participants in the bottom decile, 62 of 1971 (3.1%) in the middle decile (adjusted HR, 5.3; 95% CI, 1.9–15; P=0.002), and 218 of 1994 (11%) in the top decile (adjusted HR, 56; 95% CI, 20–158; P<0.0001) (Fig. 3). Results were similar when stratified by ISCAD quartiles (Supplementary Figure 15) and sex (Supplementary Figure 16). No heterogeneity between sexes (P=0.80) or interaction between ISCAD and sex (P=0.35) was observed.
Fig. 3.
All-cause mortality stratified by in silico score for CAD (ISCAD).
All-cause mortality was stratified by ISCAD deciles and adjusted hazard ratios (HR) were compared to the lowest decile. a, Percent mortality and adjusted HR for mortality increased monotonically over ascending ISCAD deciles. b, Kaplan-Meier survival curves relate age in increments of one year on the X-axis to cumulative survival for each age on the Y-axis and differed by ISCAD decile. Higher ISCAD deciles had lower survival over increasing ages compared to lower ISCAD deciles.
CAD sequela tracked by ISCAD
We examined if ISCAD is also associated with CAD sequela, including recurrent myocardial infarction, arrhythmia, and heart failure. Among 2,481 CAD cases without reinfarction within 28 days of incident myocardial infarction, 505 (20%) had recurrent myocardial infarction. There was an adjusted OR of 1.5 (95% CI, 1.3–1.7; P<0.0001) for recurrent myocardial infarction per SD increase in ISCAD. Prevalence and odds of recurrent myocardial infarction increased quantitatively with higher ISCAD quartiles from 79 of 666 individuals (12%) in the lowest quartile to 155 of 666 (23%) in the highest quartile (adjusted OR, 2.2; 95% CI, 1.6–3.1; P<0.0001) (Supplementary Figure 17). Among 2,620 CAD cases without an arrhythmia prior to CAD, 402 (15%) had post-CAD arrhythmia. A SD increase in ISCAD was accompanied by an adjusted OR of 1.5 (95% CI, 1.3–1.8; P<0.0001) for arrhythmia, and higher ISCAD quartiles had increasing prevalence and odds of arrhythmia from 42 of 655 individuals (6.4%) in the lowest quartile to 132 of 655 (20%) in the highest quartile (adjusted OR, 3.6; 95% CI, 2.5–5.2; P<0.0001) (Supplementary Figure 17). Out of 2,540 CAD cases without heart failure prior to CAD, 842 (33%) had post-CAD heart failure. An adjusted OR of 1.8 (95% CI 1.6–2.0; P<0.0001) for heart failure was associated with a SD increase in ISCAD. The prevalence and odds of heart failure rose with higher ISCAD quartiles from 97 of 635 individuals (15%) in the lowest quartile to 272 of 635 (43%) in the highest quartile (adjusted OR, 3.7; 95% CI, 2.8–5.0; P<0.0001) (Supplementary Figure 17).
ISCAD compared to PCE and PRS
We evaluated the performance of ISCAD in tracking atherosclerosis, mortality, and CAD sequela compared to that of PCE and PRS. Across all outcomes, ISCAD demonstrated greater associations compared to PCE or PRS (Supplementary Figures 18–22; Supplementary Table 9). For example, stenosis increased by 12-percentage points per ISCAD quartile (95% CI, 10–13; P<0.0001), 2.8-percentage points per PCE quartile (95% CI, 0.61–5.0; P=0.012), and 1.9-percentage points per PRS quartile (95% CI, 0.18–3.6; P=0.031). Furthermore, we examined the performance of ISCAD in tracking outcomes after accounting for PCE, PRS, and a baseline of known CAD risk factors19 (Supplementary Table 10). The magnitude and statistical significance of associations of outcomes with ISCAD adjusted for a combination of PCE, PRS, and baseline risk were similar to associations of outcomes with ISCAD alone in the primary analysis.
ISCAD application to holdout and external test datasets
We assessed ISCAD in the holdout and external test datasets for association with CAD-related outcomes. In the holdout dataset, ISCAD was associated with clinically significant CAD (e.g., OR of 2.4 for obstructive CAD [95% CI, 2.0–2.9; P<0.0001] per quartile) and stenosis of major vessels (e.g., OR of 2.5 for proximal left anterior descending CAD [95% CI, 2.0–3.1; P<0.0001]); all-cause mortality (HR=2.7 per SD increase [95% CI, 2.5–2.9; P<0.0001]); and recurrent myocardial infarction (OR=1.2 per SD increase [95% CI, 1.1–1.3; P<0.0001]), arrhythmia (OR=1.2 per SD increase [95% CI, 1.1–1.2; P<0.0001]), and heart failure (OR=1.2 per SD increase [95% CI, 1.1–1.3; P<0.0001]) (Supplementary Table 6; Supplementary Figure 4). In the external test dataset, ISCAD was associated with all-cause mortality (HR=2.7 per SD increase [95% CI, 2.5–2.9; P<0.0001]), recurrent myocardial infarction (OR=1.2 per SD increase [95% CI, 1.1–1.3; P<0.0001]), arrhythmia (OR=1.2 per SD increase [95% CI, 1.1–1.2; P<0.0001]), and heart failure (OR=1.2 per SD increase [95% CI, 1.1–1.3; P<0.0001]) (Supplementary Figure 5). We further examined in the training/validation dataset (BioMe) outcomes tracked by ISCAD derived from the external test dataset (UK Biobank) and observed similar associations with atherosclerosis, mortality, and CAD sequela (Supplementary Table 7; Supplementary Figure 6).
Underdiagnosed CAD
We asked whether ISCAD can identify individuals who have clinical evidence of CAD but lack a diagnosis. Twenty-six participants with high ISCAD ≥0.9 and no prior CAD diagnosis were identified, along with a propensity-matched set of 26 participants with low ISCAD ≤0.1 and no prior CAD diagnosis (median age, 73 years [IQR, 20]; 31 [60%] female; 26 [50%] European ethnicity) (Supplementary Figure 23). Twelve individuals (46%) in the high ISCAD group were found to have clinical evidence of CAD according to 2014 American College of Cardiology/American Heart Association Task Force Guidelines16 upon clinician-conducted manual EHR review while blinded to ISCAD group.
We then evaluated ISCAD as a marker for CAD risk among 17,828 participants without a diagnosis of CAD. In these undiagnosed individuals, ISCAD was associated with PCE score (0.063 increase in ISCAD per 10-point increase in PCE score; 95% CI, 0.060–0.067; P<0.0001) and PRS (0.0031 increase in ISCAD per SD increase in PRS; 95% CI, 0.00063–0.0055; P=0.014) (Supplementary Figure 24). Stenosis and SYNTAX score were generally low in 120 undiagnosed individuals with coronary angiography data (median [IQR], 14% [0] and 5.1 [3.2], respectively); yet, even in this group with subclinical atherosclerosis, ISCAD still tracked plaque burden with a 2.8-percentage point increase in stenosis (95% CI, 0.28–5.1 percentage points; P=0.031) and 0.58 increase in SYNTAX score (95% CI, 0.046–1.1; P=0.036) per ISCAD quartile (Supplementary Figure 25). All-cause mortality was also associated with ISCAD among 17,524 undiagnosed individuals with mortality data. A SD increase in ISCAD was accompanied by an adjusted HR of 8.8 (95% CI, 5.6–14; P<0.0001) for death. Both the prevalence and risk of mortality increased continuously with ascending ISCAD deciles from 4 of 1,789 individuals (0.22%) in the lowest decile to 43 of 1,761 (2.4%) in the middle decile (adjusted HR, 3.7; 95% CI, 1.3–11; P=0.017) to 149 of 1,733 (8.6%) in the highest decile (adjusted HR, 62; 95% CI, 22–178; P<0.0001) (Supplementary Figure 26).
DISCUSSION
Here, we sought to evaluate the performance of a novel in silico quantitative marker for CAD, generated from a machine learning model trained on EHR data in two large biobanks, to capture CAD risk, atherosclerosis, and mortality in a diverse population. The primary finding was that an artificial intelligence-derived marker could capture the clinical risk of PCE and genetic risk of PRS for CAD, and non-invasively quantify plaque burden and mortality risk. The marker illustrates the phenotypic spectrum of CAD, revealing distinct gradations of disease risk, atherosclerosis, and survival that would otherwise be missed with binary case-versus-control schemas. The breadth and richness of EHR data contained in biobanks unselected for a specific disease—for example, a median of 10 years (IQR, 6.3) of longitudinal EHR data and millions of unique diagnosis codes, laboratory tests, and medications in BioMe—opens an avenue to apply machine learning analyses of disease spectrums for a wide array of conditions.
Machine learning of EHR data can predict CAD with high accuracy,25,26 expanding the pool of informative features beyond traditional risk factors to include lifestyle, biomarker, and genetic features that improve predictive ability.41–44 These multimodal models, including the ones we used recently27 and in the present study, demonstrate good performance in classifying individuals as a CAD case or control. However, it was unknown whether probability scores from a machine learning model could be used as a quantitative marker for CAD. Prior studies indicate that CAD exists as a continuous phenotype and that small changes in the stenosis of coronary arteries, even subclinical, confer different degrees of risk for myocardial infarction and death.6,7 The ISCAD marker recapitulated this continuum and showed a quantitative increase in levels of atherosclerosis, all-cause mortality, and CAD sequela (Figs. 2 and 3). This is analogous to coronary artery calcium scoring, which is often used as a clinical marker of CAD, but requires specialized cardiac computed tomography and may miss early atherosclerotic lesions or non-calcified risk features of lesions.45,46 Common diseases, including CAD, lie on a spectrum and assigning case versus control categories discretizes this spectrum.28 Indeed, we found that almost half of the 26 individuals with high ISCAD who lacked a case label had clinical evidence of CAD. Disease assessment in a quantitative manner more faithfully represents its phenotype and has the potential to improve risk stratification and diagnosis of individuals.29,30 The potential of machine learning to create a digital biomarker extends beyond CAD to other diseases that may also be evaluated on continuums of severity, such as Parkinson’s disease47, carpal tunnel syndrome48, and lung cancer49. As discussed previously27, application of machine learning for precision medicine more broadly necessitates a consistent set of features across different health systems, access to EHRs available for research and development, and infrastructure and resources for evaluation and deployment in clinical settings. Transparent and explainable models, as exemplified with SHAP and feature importance analyses, are needed for adoption by health systems and clinicians.
There were study limitations. First, CAD case status was obtained using diagnosis codes, which may lead to misclassification. However, we used codes from previous studies27,39,50 that recapitulated established genetic associations with CAD. Second, low sample size may predispose machine learning models to overfitting and worsen generalizability.51 This is likely not the case for our model as it demonstrated consistent performance in validation, holdout, and external test datasets. Third, the study was retrospective, examining EHR data over decades from two biobanks. This led to imbalanced counts of cases and controls. We mitigated bias due to imbalance by selecting equal numbers of cases and controls in the training and testing of the model. Fourth, only a subset of participants had coronary angiography data available for analysis. Larger, prospective studies are needed to further validate the utility of ISCAD in the evaluation of atherosclerotic plaque burden. Fifth, clinical outcomes of ISCAD were assessed in a health system-based biobank and may not reflect other clinical practices or the general population. Sixth, only all-cause mortality information was available and death attributed to specific causes was unknown.
We leveraged a machine learning model trained on EHR data to synthesize an in silico quantitative marker for CAD. Atherosclerotic plaque burden, mortality, and CAD complications increased on a continuous spectrum with ISCAD. The marker identified individuals with a potentially missed diagnosis of CAD. Further research in prospective studies is required to assess the relationship of in silico markers with incident CAD events and death, and to examine its efficacy in other populations.
Supplementary Material
RESEARCH IN CONTEXT.
Evidence before this study
On July 2, 2022, we searched PubMed without language or date restrictions for studies reporting the development and validation of machine learning-based markers for coronary artery disease, including atherosclerosis, mortality, and myocardial infarction. The following terms and related terms were used when searching: (“machine learning”, “artificial intelligence”, or “random forest”) and (“coronary artery disease”, “atherosclerosis”, “plaque”, or “myocardial infarction”). We identified several machine learning models in the past decade that predict coronary artery disease. However, these studies used machine learning models as a classification tool to simply predict case-control status of coronary artery disease (binary framework of disease) and none use models to capture coronary artery disease on a spectrum of disease probabilities (quantitative framework of disease). Many of the prior studies are based on a limited set of features or predetermined risk factors. Hence, assessments of the clinical utility of coronary artery disease-predictive machine learning models are limited. Therefore, we investigated probabilities generated by a machine learning model as an in silico marker for coronary artery disease. Its clinical utility to quantify atherosclerotic plaque burden, survival, and risk of myocardial infarction on a continuum was assessed in a longitudinal multi-ethnic cohort, while underdiagnosed individuals with CAD were identified as an example of its intervenability. Our multimodal model analyzes millions of diverse clinical datapoints of diagnoses, laboratory test results, medications, and vitals contained in the electronic health records of participants.
Added value of this study
This marks the first study to our knowledge that constructs a quantitative marker for coronary artery disease risk, severity, and prognosis from a machine learning model trained on clinical data from electronic health records. Individuals with common diseases occupy a spectrum of disease that represents an individual’s mix of risk factors and pathogenic processes; quantitative differences in coronary stenosis, for example, result in gradations of mortality risk. Quantifying where an individual falls on the disease spectrum is needed for clinical screening and management. We developed and externally tested a coronary artery disease-predictive machine learning model using 95,935 electronic health records in the multi-ethnic BioMe Biobank and UK Biobank, and from it generated an in silico score for coronary artery disease (ISCAD). We found that coronary stenosis from angiography data increased quantitatively with ascending ISCAD, including risk of obstructive CAD, multivessel CAD, and stenosis of each major coronary artery such as the left main and proximal left anterior descending arteries. All-cause mortality increased stepwise over ascending ISCAD and sequela such as recurrent myocardial infarction rose in gradations with ISCAD. ISCAD showed greater associations with these CAD outcomes than did conventional risk scores of pooled cohort equations and polygenic risk scores. We identified participants with high ISCAD who had no prior CAD diagnosis and found that almost 50% of them had clinical evidence of underdiagnosed CAD upon manual chart review.
Implications of all the available evidence
Our study demonstrates a reconceptualization of coronary artery disease—including atherosclerosis, mortality, and sequela—as a spectrum of disease that is quantifiable with artificial intelligence trained on clinical data. This in silico marker derived from machine learning captured CAD pathophysiology and clinical outcomes on a continuum. The model is holistic in drawing on a wide array of clinical information from population-based biobanks, inclusive in representing diverse populations, and faithful in preserving the complexity of disease. The implementation of machine learning-based quantitative markers for CAD may help define the disease state and clinical outcomes in patients, while optimizing the detection of disease and reducing underdiagnosis.
Acknowledgements:
Bruce D. Gelb, MD, Sander Houten, PhD, Paz Polak, PhD, Stuart Scott, PhD, and Ethylin Jabs, MD, all of whom are on the thesis advisory committee of ISF, provided critical feedback and expertise.
Funding/Support:
Mr. Forrest is supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) (T32-GM007280). Dr. Do is supported by the National Institute of General Medical Sciences of the NIH (R35-GM124836) and the National Heart, Lung, and Blood Institute of the NIH (R01-HL139865 and R01-HL155915).
Footnotes
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data Sharing: The dataset from UK Biobank analyzed in the study is available via application to the Access Management System at https://bbams.ndph.ox.ac.uk/ams/. Further information regarding the BioMe Biobank and its dataset is available at https://icahn.mssm.edu/research/ipm/programs/biome-biobank. Code used for the analyses is available at https://data.mendeley.com/datasets/ncmpmvv5tm/draft?a=a7dbf30b-17c9-401b-9cbd-4be2dc067dca.
Declaration of Interests: Dr. Do reported receiving grants from AstraZeneca, grants and nonfinancial support from Goldfinch Bio, being a scientific co-founder, consultant and equity holder for Pensieve Health, and being a consultant for Variant Bio, all not related to this work. Dr. Nadkarni reported being a scientific co-founder, consultant, advisory board member, and equity owner of Renalytix AI, a scientific co-founder and equity holder for Pensieve Health, a consultant for Variant Bio, and receiving grants from Goldfinch Bio and personal fees from Renalytix AI, BioVie, Reata, AstraZeneca, and GLG Consulting, all not related to this work. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith SC, Benjamin EJ, Bonow RO, et al. AHA/ACCF secondary prevention and risk reduction therapy for patients with coronary and other atherosclerotic vascular disease: 2011 update: A guideline from the American Heart Association and American College of Cardiology Foundation. Circulation 2011; 124: 2458–73. [DOI] [PubMed] [Google Scholar]
- 2.Sidney C. Smith J, Allen J, Blair SN, et al. AHA/ACC Guidelines for Secondary Prevention for Patients With Coronary and Other Atherosclerotic Vascular Disease: 2006 Update. Circulation 2006; 16: 60–2. [Google Scholar]
- 3.Knuuti J, Wijns W, Saraste A, et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromesThe Task Force for the diagnosis and management of chronic coronary syndromes of the European Society of Cardiology (ESC). Eur Heart J 2020; 41: 407–77. [DOI] [PubMed] [Google Scholar]
- 4.Kitsios GD, Dahabreh IJ, Trikalinos TA, Schmid CH, Huggins GS, Kent DM. Heterogeneity of the Phenotypic Definition of Coronary Artery Disease and Its Impact on Genetic Association Studies. Circ Cardiovasc Genet 2011; 4: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox KAA, Metra M, Morais J, Atar D. The myth of ‘stable’ coronary artery disease. Nat Rev Cardiol 2020; 17: 9–21. [DOI] [PubMed] [Google Scholar]
- 6.Maddox TM, Stanislawski MA, Grunwald GK, et al. Nonobstructive coronary artery disease and risk of myocardial infarction. JAMA 2014; 312: 1754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park D-W, Clare RM, Schulte PJ, et al. Extent, Location, and Clinical Significance of Non–Infarct-Related Coronary Artery Disease Among Patients With ST-Elevation Myocardial Infarction. JAMA 2014; 312: 2019–27. [DOI] [PubMed] [Google Scholar]
- 8.Sequist TD, Marshall R, Lampert S, Buechler EJ, Lee TH. Missed Opportunities in the Primary Care Management of Early Acute Ischemic Heart Disease. Arch Intern Med 2006; 166: 2237–43. [DOI] [PubMed] [Google Scholar]
- 9.Turkay M, Senol Y, Alimoglu MK, Aktekin MR, Deger N. Missed Opportunities for Coronary Heart Disease: Primary Care Experience. Croat Med J 2007; 48: 362. [PMC free article] [PubMed] [Google Scholar]
- 10.Araújo C, Laszczyńska O, Viana M, et al. Missed Opportunities in Symptomatic Patients before a First Acute Coronary Syndrome: The EPIHeart Cohort Study. Cardiology 2018; 139: 71–82. [DOI] [PubMed] [Google Scholar]
- 11.Sanchis-Gomar F, Perez-Quilis C, Leischik R, Lucia A. Epidemiology of coronary heart disease and acute coronary syndrome. Ann Transl Med 2016; 4. DOI: 10.21037/ATM.2016.06.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Özcan C, Deleskog A, Schjerning Olsen A-M, Nordahl Christensen H, Lock Hansen M, Hilmar Gislason G. Coronary artery disease severity and long-term cardiovascular risk in patients with myocardial infarction: a Danish nationwide register-based cohort study. Eur Hear J - Cardiovasc Pharmacother 2018; 4: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jernberg T, Hasvold P, Henriksson M, Hjelm H, Thuresson M, Janzon M. Cardiovascular risk in post-myocardial infarction patients: Nationwide real world data demonstrate the importance of a long-term perspective. Eur Heart J 2015; 36: 1163–1170a. [DOI] [PubMed] [Google Scholar]
- 14.Zeitouni M, Clare RM, Chiswell K, et al. Risk Factor Burden and Long‐ Term Prognosis of Patients With Premature Coronary Artery Disease. J Am Heart Assoc 2020; 9. DOI: 10.1161/JAHA.120.017712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myerburg RJ, Junttila MJ. Sudden Cardiac Death Caused by Coronary Heart Disease. Circulation 2012; 125: 1043–52. [DOI] [PubMed] [Google Scholar]
- 16.Fihn SD, Blankenship JC, Alexander KP, et al. 2014 ACC/AHA/AATS/PCNA/SCAI/STS focused update of the guideline for the diagnosis and management of patients with stable ischemic heart disease: A report of the American College of Cardiology/American Heart Association Task Force on practice guidelines, a. J Am Coll Cardiol 2014; 64: 1929–49. [DOI] [PubMed] [Google Scholar]
- 17.Wilson PWF, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of Coronary Heart Disease Using Risk Factor Categories. Circulation 1998; 97: 1837–47. [DOI] [PubMed] [Google Scholar]
- 18.Hageman S, Pennells L, Ojeda F, et al. SCORE2 risk prediction algorithms: New models to estimate 10-year risk of cardiovascular disease in Europe. Eur Heart J 2021; 42: 2439–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goff DC, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: A report of the American college of cardiology/American heart association task force on practice guidelines. J Am Coll Cardiol 2014; 63: 2935–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med 2019; 380: 1347–58. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Cheng WY, Glicksberg BS, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med 2015; 7. DOI: 10.1126/SCITRANSLMED.AAA9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obermeyer Z, Lee TH. Lost in Thought — The Limits of the Human Mind and the Future of Medicine. N Engl J Med 2017; 377: 1209–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. npj Digit Med 2018. 11 2018; 1: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019. 251 2019; 25: 44–56. [DOI] [PubMed] [Google Scholar]
- 25.Agrawal S, Klarqvist MDR, Emdin C, et al. Selection of 51 predictors from 13,782 candidate multimodal features using machine learning improves coronary artery disease prediction. Patterns 2021; 2: 100364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward A, Sarraju A, Chung S, et al. Machine learning and atherosclerotic cardiovascular disease risk prediction in a multi-ethnic population. npj Digit Med 2020; 3. DOI: 10.1038/s41746-020-00331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrazzini BO, Chaudhary K, Márquez-Luna C, et al. Coronary Risk Estimation Based on Clinical Data in Electronic Health Records. J Am Coll Cardiol 2022; 79: 1155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy M, Birney E. Personalized profiles for disease risk must capture all facets of health. Nature 2021; 597: 175–7. [DOI] [PubMed] [Google Scholar]
- 29.Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nat Rev Genet 2009; 10: 872–8. [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Wang C, Khan A, et al. Quantitative disease risk scores from EHR with applications to clinical risk stratification and genetic studies. npj Digit Med 2021; 4: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS 2010; 5: 463–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tayo BO, Teil M, Tong L, et al. Genetic background of patients from a university medical center in Manhattan: Implications for personalized medicine. PLoS One 2011; 6. DOI: 10.1371/journal.pone.0019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018; 562: 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birim Ö, van Gameren M, Bogers AJJC, Serruys PW, Mohr FW, Kappetein AP. Complexity of Coronary Vasculature Predicts Outcome of Surgery for Left Main Disease. Ann Thorac Surg 2009; 87: 1097–105. [DOI] [PubMed] [Google Scholar]
- 35.Neumann FJ, Sousa-Uva M, Ahlsson A, et al. 2018 ESC/EACTS Guidelines on myocardial revascularization. Eur Heart J 2019; 40: 87–165. [DOI] [PubMed] [Google Scholar]
- 36.Thygesen K, Alpert JS, Jaffe AS, et al. Fourth Universal Definition of Myocardial Infarction (2018). Circulation 2018; 138: e618–51. [DOI] [PubMed] [Google Scholar]
- 37.Liaw A, Wiener M. Classification and Regression by randomForest. R News 2002; 2: 18–22. [Google Scholar]
- 38.Kursa MB, Rudnicki WR. Feature Selection with the Boruta Package. J Stat Softw 2010; 36: 1–13. [Google Scholar]
- 39.Khera A V, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018; 50: 1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011; 12: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimopoulos AC, Nikolaidou M, Caballero FF, et al. Machine learning methodologies versus cardiovascular risk scores, in predicting disease risk. BMC Med Res Methodol 2018; 18: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mozaffarian D, Wilson PWF, Kannel WB. Beyond established and novel risk factors lifestyle risk factors for cardiovascular disease. Circulation 2008; 117: 3031–8. [DOI] [PubMed] [Google Scholar]
- 43.Domínguez F, Fuster V, Fernández-Alvira JM, et al. Association of Sleep Duration and Quality With Subclinical Atherosclerosis. J Am Coll Cardiol 2019; 73: 134–44. [DOI] [PubMed] [Google Scholar]
- 44.Matsushita K, Coresh J, Sang Y, et al. Estimated glomerular filtration rate and albuminuria for prediction of cardiovascular outcomes: A collaborative meta-analysis of individual participant data. Lancet Diabetes Endocrinol 2015; 3: 514–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortensen MB, Gaur S, Frimmer A, et al. Association of Age with the Diagnostic Value of Coronary Artery Calcium Score for Ruling out Coronary Stenosis in Symptomatic Patients. JAMA Cardiol 2021; 7: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan SS, Navar AM. The Potential and Pitfalls of Coronary Artery Calcium Scoring. JAMA Cardiol 2021; 7: 11–2. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Yuan Y, Zhang G, et al. Artificial intelligence-enabled detection and assessment of Parkinson’s disease using nocturnal breathing signals. Nat Med 2022; : 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park D, Kim BH, Lee SE, et al. Machine learning-based approach for disease severity classification of carpal tunnel syndrome. Sci Rep 2021; 11: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan Q, Cai T, Hong C, et al. Performance of a Machine Learning Algorithm Using Electronic Health Record Data to Identify and Estimate Survival in a Longitudinal Cohort of Patients with Lung Cancer. JAMA Netw Open 2021; 4: e2114723–e2114723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inouye M, Abraham G, Nelson CP, et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J Am Coll Cardiol 2018; 72: 1883–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern Med 2018; 178: 1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.