Abstract
Salt and drought stresses are major factors limiting soybean (Glycine max [L.] Merr.) growth and development; thus, improving soybean stress tolerance is critical. In this study, both salt stress and drought stress induced mRNA levels of CONSTANS-like 1a (GmCOL1a) and stabilized the GmCOL1a protein. Transgenic 35S:GmCOL1a soybean plants exhibited enhanced salt and drought tolerance, with higher relative water content in leaves, greater proline content, lower malondialdehyde (MDA) content, and less reactive oxygen species (ROS) production compared with wild-type plants; the GmCOL1a knockout co-9 mutant showed opposite phenotypes. In addition, GmCOL1a promoted the expression of genes related to salt tolerance, effectively reducing the Na+/K+ ratio in soybean plants, especially in stems and leaves of 35S:GmCOL1a soybean. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis identified two potential direct targets of GmCOL1a, late embryogenesis abundant (GmLEA) and Δ1-pyrroline-5-carboxylate synthetase (GmP5CS) genes, which were verified by chromatin immunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR), electrophoretic mobility shift assay (EMSA), and transient transcriptional activation assays. GmCOL1a bound directly to the Myc(bHLH)-binding and Che-binding motifs of GmLEA and GmP5CS promoters to stimulate mRNA expression. Analysis of transgenic hairy-root GmP5CS:GmP5CS soybean plants in wild type, co-9, and 35S:GmCOL1a backgrounds further revealed that GmCOL1a enhances salt and drought tolerance by promoting GmP5CS protein accumulation in transgenic soybean hairy roots. Therefore, we demonstrate that GmCOL1a plays an important role in tolerance to abiotic stress in soybean.
The Glycine max CONSTANS-like 1a transcription factor positively regulates tolerance to salt and drought stresses by promoting the accumulation of Δ1-pyrroline-5-carboxylate synthetase.
Introduction
Soybean (Glycine max) plants are important economic oil crops that provide a major source of oil and protein. When plants are subjected to abiotic stressors, such as salt and drought, they affect seed germination, crop growth, and yield (Farooq et al., 2009; Zelm et al., 2020). Currently, saline soils cover more than 800 million hectares, accounting for about 6% of the world's total soil area (Rengasamy, 2002; Munns and Tester, 2008). Drought stress is one of the most severe constraints that inhibit plant growth and development and limits plant productivity (de Brito et al., 2021). Therefore, it is important to understand the genetic and regulatory mechanisms of salt and drought tolerance in soybean and breed soybean varieties with excellent salt and drought tolerance.
Salt stress is usually caused by high concentrations of sodium and chloride ions in the soil (Ismail et al., 2014). Salt stress includes three pathways: osmotic, ionic, and secondary stress. A high salt concentration disrupts the dynamic balance of water potential and ion distribution in plant cells, causing loss of greenery, dehydration, and cell necrosis and preventing normal plant growth (Munns and Tester, 2008; Xie et al., 2008). For plants, sodium ions (Na+), the major ion in saline soils, are absorbed from the root system and accumulate in photosynthetic tissues, resulting in ion imbalance and reduced productivity. In response to salt stress, plants have a range of salt tolerance mechanisms, including enhanced Na+ rejection, limited Na+ uptake, and regulation of cellular ion balance (especially Na+/K+ ratio; Zhu, 2002; Ishikawa and Shabala, 2019). When plants are stimulated by abiotic stress, the balance between the production and scavenging of reactive oxygen species (ROS) in vivo is disrupted, and ROS levels are elevated, resulting in oxidative damage in plants (Fahnenstich et al., 2008). In addition to the plant's ability to balance intracellular ROS through enzymatic degradation and the use of small molecules, such as ascorbic acid, mannitol, and flavonoids, for reduction (Lu et al., 2013; Yan et al., 2014), ROS accumulation can also be limited by enhancing the antioxidant system and regulating the expression of defense-related genes to protect plant cell membranes from damage (Ketehouli et al., 2021). The E2 knockout mutant e2CR not only has a shortened flowering time but also positively regulates the transcription level of ROS scavenging-related genes, which enhances the salt tolerance of soybean, laying a foundation for the selection of early maturing and salt-tolerant soybean varieties (Dong et al., 2022).
Transcription factors (TFs) can be activated in response to specific abiotic stressors (Song et al., 2016; Li et al., 2019) and have been widely used in transgenic plants to enhance plant tolerance under abiotic stress (Wang et al., 2016). NAC (NAM, ATAF, and CUC) proteins are one of the largest families of TFs involved in abiotic stress. GmNAC8 overexpression lines promote the production of proline content and superoxide dismutase (SOD) activity, and GmNAC8 improves tolerance under drought stress (Yang et al., 2020). Stress associated proteins 16 (GmSAP16) overexpression in transgenic soybean hairy roots enhances salt and drought tolerance in soybean seedlings and increases the transcript levels of the stress-related genes drought response element-binding factors (GmDREB1B; 1, and GmDREB2), 9-cis-epoxycarotenoid dioxygenase 3 (GmNCED3), responsive to dehydration protein 22 (GmRD22), a vacuolar Na+/H+ antiporter gene (GmNHX1), and Salt Overly Sensitive 1 (GmSOS1; Zhang et al., 2019). Therefore, the identification of key genes is essential to understand the complex molecular mechanisms of salt and drought tolerance in soybean.
Late embryogenesis abundant (LEA) proteins are involved in plant stress tolerance mechanisms and play a crucial role in adversity resistance (Abdul Aziz et al., 2021). In rice (Oryza sativa), five LEA genes (OsLEA1, OsLEA2, OsLEA3, OsLEA4, and OsLEA5) were substantially up-regulated under drought stress and provided better physiological adaptation to drought stress (Kamarudin et al., 2019). The GmLEA2-1 protein enhances salt and drought tolerance in Arabidopsis (Arabidopsis thaliana; Putterill et al., 1995; Robson et al., 2001). Notably, GmLEA is a soybean PM2 protein (Accession No. M80664), belonging to the LEA3 protein family (Hsing et al., 1992), that has been shown to enhance tolerance to high salt stress in Escherichia coli (E. coli) with some hydrophilic and thermal stability, protecting proteins and enzyme activities against freeze–thaw or high-temperature stress (Min et al., 2015; Song et al., 2018). In Arabidopsis, the gene encoding Δ1-pyrroline-5-carboxylate synthetase (P5CS) is induced by salt stress, drought stress, and abscisic acid (ABA; Liu et al., 2016). Proline biosynthesis is catalyzed by P5CS, and the resulting Δ1-pyrroline-5-carboxylate (P5C) is reduced by P5CR to proline (Hu et al., 1992). Overexpression of P5CS in transgenic Nicotiana benthamiana (N. benthamiana) plants increases proline content, improves tolerance to osmotic stress, and induces participation in the proline biosynthetic pathway in plants (Kishor et al., 1995).
The CONSTANS-like (COL) gene was first isolated in Arabidopsis, and the COL protein encoded by the COL gene contains two homologous domains: the B-box domain (zinc-finger domain) with a unique crystal structure (Dahal et al., 2022) and the CCT (CO, CO-like, and TOC1) domain. To date, only a few members of the CO family have been involved in abiotic stress responses. AtCOL4 and ZmCOLs in maize (Zea mays L.) might be dependent on ABA signaling to participate in salt and drought stress responses (Min et al., 2015; Song et al., 2018). In rice, a COL TF, Ghd7, is a negative regulator involved in developmental and drought-induced leaf senescence pathways (Liu et al., 2016). In Tamarix hispida, ThCOL2 overexpression enhances ROS clearance to reduce cell damage and death caused by salt stress (Lei et al., 2021). Zinc-finger protein B-BOX 7/CONSTANS-LIKE 9 (MdBBX7/MdCOL9), which formed a module with MdMIEL1, played an active role in drought resistance in apple (Malus domestica; Chen et al., 2022). In mango (Mangifera indica L.), MiCOL1A and MiCOL1B are involved in both the flowering pathway and enhanced drought tolerance in Arabidopsis (Guo et al., 2022). Therefore, we also aimed to identify whether the floral suppressor GmCOL1a was involved in the regulation of stress responses in soybean.
In this study, 35S:GmCOL1a transgenic soybeans had enhanced salt and drought tolerance by increasing water absorption, Na+/K+ ratio, and proline content, decreasing MDA content, and reducing ROS production, whereas the co-9 mutant showed the opposite phenotypes. GmCOL1a directly bound to Myc(bHLH)-binding and Che-binding motifs in the promoters of stress-related genes GmLEA and GmP5CS to promote mRNA expression and suppress ROS production, achieving salt and drought tolerance. These results provided important insights into the functional diversity of the GmCOL1a protein, a positive regulator involved in salt and drought tolerance pathways.
Results
Subcellular localization of the GmCOL1a protein and spatial expression patterns
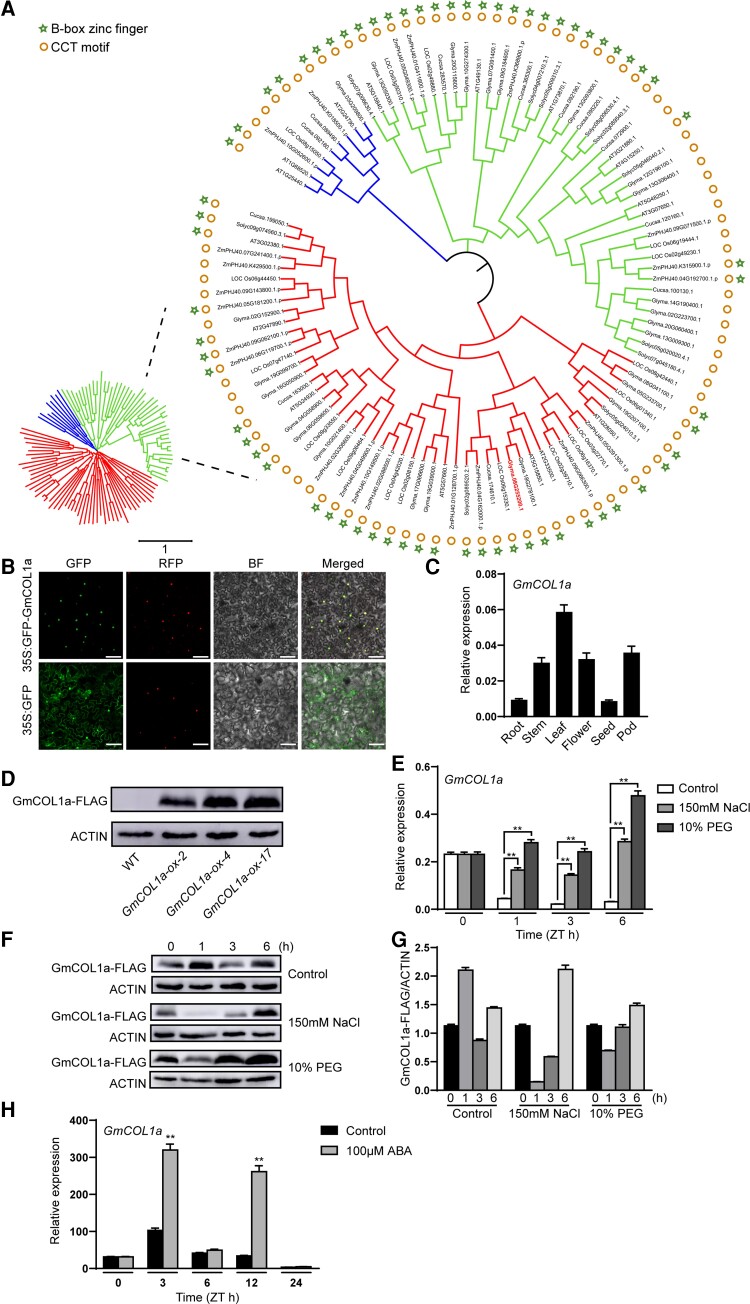
GmCOL1a, belonging to the BBX family of zinc-finger TFs, contained one or two B-box patterns and sometimes a CCT domain, which played important roles in plant growth and development, including the photoperiodic regulation of flowering and responses to biotic and abiotic stress (Putterill et al., 1995; Robson et al., 2001). To investigate the phylogenetic relationships among soybean GmCOL1a genes, 105 CO protein sequences from Glycine max, Arabidopsis thaliana, Zea mays, tomato (Solanum lycopersicum), cucumber (Cucumis sativus), and Oryza sativa were compared, and a phylogenetic tree was constructed using a neighbor-joining (NJ) algorithm, indicating that these neighboring genes may exhibit similar functions, with most genes containing B-box patterns and a CCT domain and a few genes containing only a CCT domain (Figure 1A).
Figure 1.
Subcellular localization of GmCOL1a and spatial expression patterns. A, Phylogenetic tree was constructed using the NJ method (with a bootstrap of 1000) with CONSTANS. The DIR nomenclature is as follows: Glycine max (Glyma), Arabidopsis thaliana (At), Zea mays (Zm), Solanum lycopersicum (Solyc), Cucumis sativus (Cucsa), and Oryza sativa (Os). The 1 scales show substitution distance. B, Subcellular localization of the GmCOL1a protein. After infiltration, the Nicotiana benthamiana leaves were grown for 2 days, and the GFP signal was detected by fluorescence microscopy. A red nuclear marker plasmid (H2B-RFP) was used to confirm the location of the cell nucleus. GFP, green fluorescent protein; RFP, red fluorescent protein; BF, bright field; Merge, GFP, RFP, and bright-field images. Scale bars = 100 μm. C, Tissue-specific expression of GmCOL1a under LDs. Roots, stems, and leaves were collected from 20-day-old plants. Flowers were collected from 52-day-old plants. Seeds and pods were collected from 68-day-old plants. All data were normalized with the soybean GmActin4 gene as an internal reference. D, Immunoblot analysis of the 35S:GmCOL1a transgenic soybean. The GmCOL1a-FLAG protein was expressed in transgenic seedlings grown for 20 days under LDs. Actin was used as a control. E, Expression of GmCOL1a in leaves of WT under salt (150 mM) and PEG 6000 (10%) treatments determined by RT-qPCR. Asterisks indicated values significantly different from 150 mM NaCl or 10% PEG compared to control. F, Expression of GmCOL1a protein in GmCOL1a-ox-2 seedlings grown in LDs for 14 days and treated with 150 mM NaCl or 10% PEG 6000 for 0, 1, 3, and 6 h. Leaf samples were collected (ZT0 is 7:00 a.m.), western blots of protein extracts are shown. G, FLAG-tagged proteins were detected using an anti-FLAG antibody. Actin served as the loading control. The levels of the GmCOL1a protein were determined by normalization of the GmCOL1a signals to ACTIN signals and were presented as GmCOL1a/ACTIN. H, RT-qPCR analysis of the mRNA level of GmCOL1a in the leaves of 14-day-old WT plants under ABA (100 µM) treatment. Asterisks indicated values significantly different between the control and ABA in each group. For each experiment, three technical replicates were conducted. Data shown are the mean ± SD of three independent experiments (*, P < 0.05 and **, P < 0.01, Student's t test).
To determine the subcellular localization of GmCOL1a, its localization was analyzed by expressing a gene encoding a green fluorescent protein (GFP)-GmCOL1a fusion protein under the control of the 35S promoter. The fluorescence microscopic observations showed that GFP fluorescence was dispersed throughout the cells, bombarded with the control plasmid 35S:GFP. In contrast, the 35S:GFP-GmCOL1a fusion protein was localized exclusively to the nucleus of Nicotiana benthamiana (N. benthamiana) cells (Figure 1B).
To investigate the tissue expression patterns in soybean “DongNong 50” (DN50), the transcript abundance of the GmCOL1a gene in soybean root, stem, leaf, flower, pod, and seed tissues was also detected by reverse-transcription quantitative PCR (RT-qPCR). The expression of GmCOL1a was detected in all tissues, with the highest expression in the leaves (Figure 1C).
mRNA level of GmCOL1a was induced by salt, drought, and ABA
To further confirm the function of GmCOL1a in soybean salt and drought tolerance, we generated three T3 generation GmCOL1a-ox (overexpression) transgenic lines under the control of the CaMV35S promoter. The expression of the GmCOL1a-FLAG protein with a size of 58 kDa was detected by western-blot analysis in three GmCOL1a-ox transgenic soybean lines (Figure 1D). RT-qPCR showed that the transcript levels of GmCOL1a were induced in leaves at 1, 3, and 6 h after salt and drought treatment (Figure 1E). The expression of the GmCOL1a protein immediately decreased after salt and drought treatment and gradually stabilized with a longer treatment time, as shown by western-blot analysis of GmCOL1a-ox-2 transgenic leaves (Figure 1, F and G). In addition, the transcript level of GmCOL1a was especially induced after 3 and 12 h of ABA treatment (Figure 1H).
GmCOL1a promoted salt tolerance in soybean
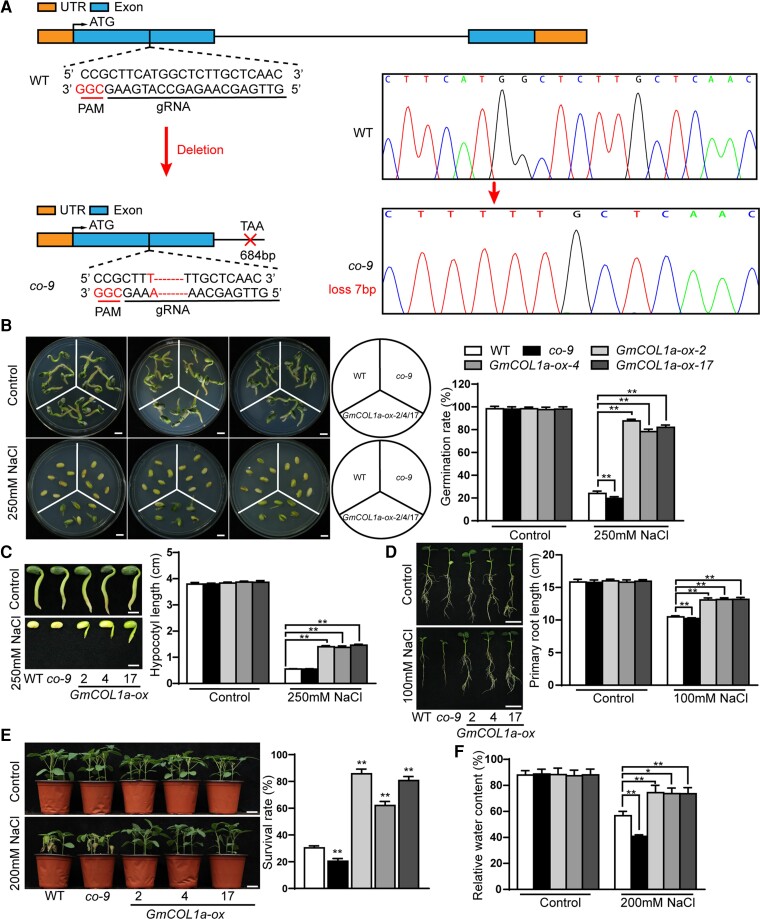
To clarify the biological functions of GmCOL1a in soybean, we generated an independent CRISPR/Cas9-mediated loss-of-function mutant (named co-9) in the DN50 background. DNA sequencing confirmed that seven bases (ATGGCTC) upstream of the PAM site were deleted in the co-9 mutant, which resulted in amino acid conversions that ultimately stopped translation at 684 bp (Figure 2A).
Figure 2.
GmCOL1a positively regulated salt tolerance in soybean. A, Gene structures of GmCOL1a with a CRISPR/Cas9 target site designed in the exon. Black lines, orange strips, and blue strips indicate introns, untranslated regions (UTRs), and exons, respectively. Nucleotide sequences indicate regions targeted by the gRNA designed in this study, and nucleotides in red indicate proto-spacer adjacent motifs (PAMs). B, Images of 6-day-old seedlings were taken, and germination rates were recorded (n ≥ 20). Scale bars = 1 cm. C, Images of whole seedlings were taken. Scale bars = 1 cm. Hypocotyl length of WT and transgenic lines under 0 (control) or 250 mM NaCl treatment. Each column represents the hypocotyl length (n ≥ 20). D, Primary root length of 4-day-old soybean seedlings treated with 0 (control) or 100 mM NaCl. Images were taken 21 days after stress treatment. Each column represents the primary root length (n ≥ 20). Root growth bags from seedlings grown in 10 plastic bags were collected and treated similarly. Each bag contained two seedlings from three GmCOL1a-ox transgenic lines, the co-9 mutant, and the WT soybeans. Scale bar = 5 cm. E, Phenotypes and survival rates of three GmCOL1a-ox transgenic lines, the co-9 mutant, and the WT soybeans after 200 mM salt treatment for 8 days. Scale bars = 5 cm. Survival rates were calculated (n ≥ 20). F, Relative leaf water content was measured from three GmCOL1a-ox transgenic lines, the co-9 mutant, and the WT soybeans (n ≥ 10) grown under various concentrations of NaCl for 8 days. Asterisks indicated values significantly different between the transgenic lines and the WT in each group. For each experiment, three technical replicates were conducted. Data shown are the mean ± SD of three independent experiments (*, P < 0.05 and **, P < 0.01, Student's t test).
For germination assays, the seeds of three GmCOL1a-ox transgenic lines, co-9, and the wild type (WT) were sown on germination medium [1/2 Murashige and Skoog basic nutrient salts with B5 vitamins (MSB) with 2% sucrose] with various concentrations of NaCl. There was no difference in germination rates on 0 mM NaCl medium; however, the germination of WT and co-9 seeds was inhibited to a greater extent than that of GmCOL1a-ox seeds in the medium containing 250 mM NaCl (Figure 2B). Furthermore, the hypocotyl length of the three GmCOL1a-ox transgenic lines was significantly longer than that of the WT and co-9 seedlings after 250 mM NaCl treatment, although salt stress also repressed the growth rate of GmCOL1a-ox transgenic lines to a lesser extent (Figure 2C). We further examined the effect of salt treatments on soybean root growth; seedlings grown under normal growth conditions in a bag showed no significant difference in primary roots. However, the WT and co-9 produced shorter primary roots, and the three GmCOL1a-ox transgenic lines showed longer roots and more and longer lateral roots under 100 mM NaCl treatment (Figure 2D). For seedlings grown in soil treated with salt, co-9 and WT plants began to wilt substantially 8 days after treatment with 200 mM NaCl. Although the three GmCOL1a-ox transgenic lines were shorter than the WT seedlings when not treated with salt, the three GmCOL1a-ox transgenic lines showed higher heights and healthier growth than the co-9 and WT plants. Statistical analysis of survival rates further showed that the three GmCOL1a-ox transgenic lines had 62%–85% survival rates, while the WT and co-9 plants had 30% and 20% survival rates, respectively (Figure 2E).
It is important to maintain the water balance in plants under stress conditions. To investigate the ability of transgenic soybean plants to regulate osmotic stress, the leaf relative water content (RWC) was assessed during salt treatment. After 8 days of salt treatment, RWCs decreased by 31% and 48% in WT and co-9 plants, respectively, but only decreased by 14%–15% in the three GmCOL1a-ox transgenic lines (Figure 2F).
Taken together, the overexpression of GmCOL1a increased salt tolerance, and the co-9 mutant increased salt sensitivity.
GmCOL1a enhanced salt tolerance by maintaining the Na+/K + ratio
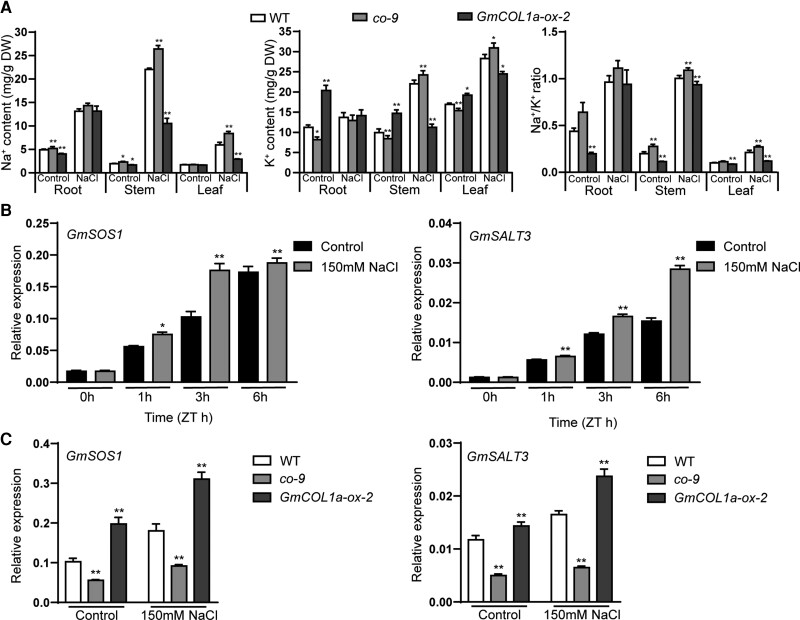
To assess the functional properties of GmCOL1a, we compared the accumulation of K+ and Na+ in the roots, stems, and leaves of different transgenic plants. Under normal growth conditions, GmCOL1a-ox-2 transgenic soybean showed a higher K+ content and lower Na+ content than WT and co-9 in roots, stems, and leaves. However, under salt stress, the Na+ content in the roots, stems, and leaves of WT, co-9, and GmCOL1a-ox-2 was obviously increased compared to each of the nontreated plants, which led to a substantial increase in the Na+/K+ ratio. Although there were no significant differences in the Na+/K+ ratio in the roots of different transgenic plants under salt stress conditions, Na+/K+ ratios in the stems and leaves of GmCOL1a-ox-2 transgenic soybean were significantly lower than those of the WT and co-9 under salt stress (Figure 3A).
Figure 3.
Salt sensitivity of different GmCOL1a-ox plants. A, Concentrations of Na+ and K+ and Na+/K+ ratio of roots, stems, and leaves in WT, co-9, and GmCOL1a-ox soybean materials grown under 0 mM NaCl or 150 mM NaCl treatments for 8 days, respectively. Asterisks indicated values significantly different between the transgenic lines and the WT in each group. B, Transcripts of GmSOS1 and GmSALT3 in the WT under control and salt (150 mM NaCl) conditions. Asterisks indicated values significantly different between the control and 150 mM NaCl in each group. C, RT-qPCR analysis of the mRNA levels of GmSOS1 and GmSALT3 in 14-day-old leaves from GmCOL1a-ox and co-9 plants under 150 mM NaCl treatments for 3 h, determined by RT-qPCR. Asterisks indicated values significantly different between the transgenic lines and the WT in each group. For each experiment, three technical replicates were conducted. Data shown are the mean ± SD of three independent experiments (*, P < 0.05 and **, P < 0.01, Student's t test).
To further understand whether GmCOL1a transgenic plants maintained ion homeostasis by regulating the acquisition and distribution of K+ and Na+ under salt stress, the expression levels of GmSOS1 in Na+ efflux transporters (Zhang et al., 2022) and GmSALT3 (salt tolerance-associated gene on chromosome 3; Guan et al., 2014) were analyzed. Both GmSOS1 and GmSALT3 were induced at 0, 1, 3, and 6 h after salt treatment in the WT plants (Figure 3B). We found that the expression of GmSOS1 and GmSALT3 was more up-regulated in different transgenic soybean plants than the control under 150 mM NaCl treatment for 3 h. Therefore, the transcript levels of GmSOS1 and GmSALT3 were studied in GmCOL1a transgenic plants under salt stress for 3 h. The transcript levels of GmSOS1 and GmSALT3 were higher in GmCOL1a-ox-2 transgenic soybean plants than those in WT. The opposite result was observed in co-9 transgenic soybean plants (Figure 3C). GmCOL1a increased the expression of GmSOS1 and GmSALT3 by maintaining the Na +/K+ ratio to improve salt tolerance.
GmCOL1a promoted drought tolerance in soybean
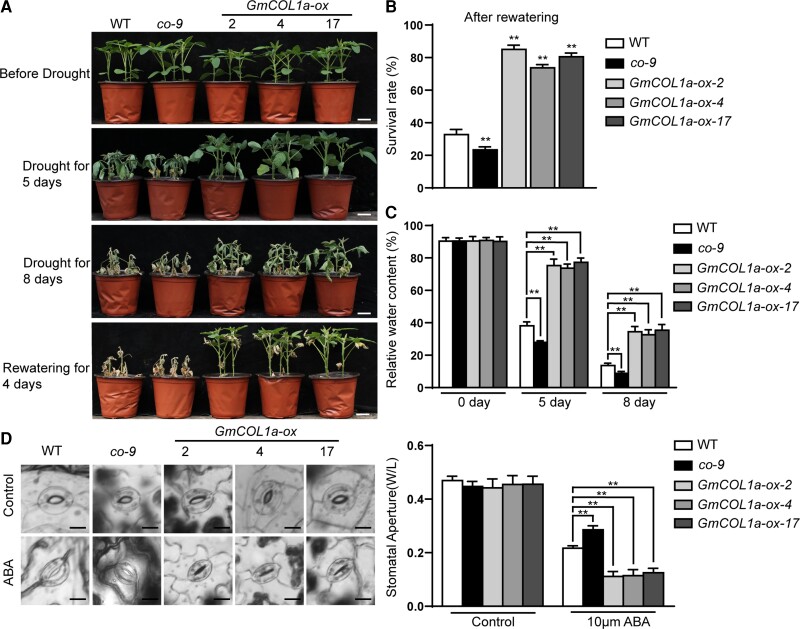
In the drought treatment, the green coloration of the three overexpression transgenic lines persisted longer than that in the WT and co-9 plants. After re-watering for 4 days, most WT and co-9 plants failed to recover (33% and 23% survival, respectively), whereas the three overexpression transgenic lines showed higher recovery (74%–85% survival; Figure 4, A and B). Additionally, RT-qPCR assays showed that the transcript levels of GmCOL1a were induced in leaves after 1, 3, and 6 h of drought treatment (Figure 1E). The RWC was then measured during the water deficit treatment. The RWCs decreased by 52% and 63% in WT and co-9 plants, respectively, but the RWCs of the three overexpression transgenic lines decreased by only 13%–17% on day 5 of the water deficit treatment. In contrast, the RWCs of WT and co-9 soybeans decreased by 77% and 82%, respectively, while those of the three overexpression transgenic lines decreased by only 55%–58% on day 8 of the drought treatment (Figure 4C).
Figure 4.
Analyses of drought stress phenotypes in different soybean plants. A, B, Phenotypes and survival rates of three GmCOL1a-ox transgenic lines, co-9 mutant, and WT soybeans (n ≥ 20) after dehydration for 5 days, 8 days, and recovery for 4 days. The control group was watered with half-strength Hoagland solution every 3 days when the first trifoliate leaves of the plants were fully expanded. Scale bars = 5 cm. C, Relative leaf water content of three GmCOL1a-ox transgenic lines, co-9 mutant, and WT soybean seedlings (n = 10) after drought treatment. The water content was measured at 0, 5, and 8 days after drought treatment. D, Left panel: Images of epidermal stomata after 10 µm ABA treatment observed with a bright field microscope. Scale bars = 10 µm. Right panel: Stomatal aperture of the second trifoliate leaf at the top after ABA treatment; width/length of the stomatal aperture was measured using ImageJ (n = 10). Asterisks indicated values significantly different between the transgenic lines and the WT in each group. Data are the means ± SD of three independent experiments (*, P < 0.05 and **, P < 0.01, Student's t test).
ABA plays an important role in regulating stomatal movement and is closely related to water loss under drought stress (Verslues et al., 2006; Tai et al., 2014). We further verified that ABA treatment resulted in reduced stomatal opening in the three overexpression transgenic lines, and co-9 showed a larger stomatal aperture than the WT (Figure 4D). These results suggest that GmCOL1a overexpression increases drought tolerance by ABA-mediated stomatal movement.
Physiological index analysis under salt and drought stress
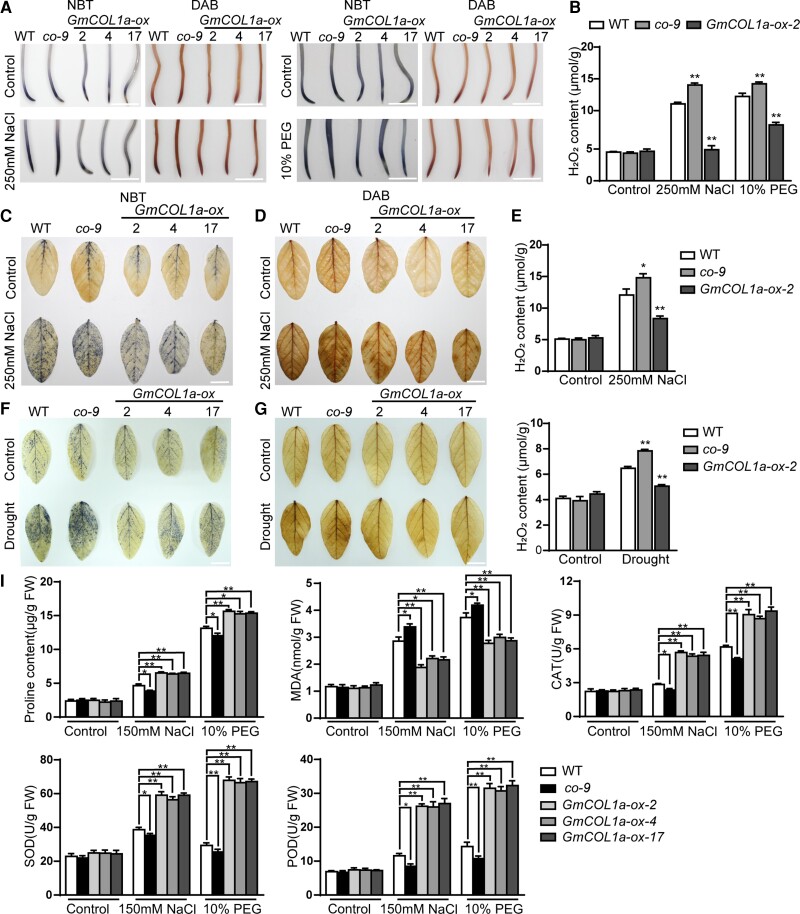
To further investigate the role of GmCOL1a in regulating salt stress and drought tolerance, we measured the proline, MDA, and H2O2 content, as well as catalase (CAT), SOD, and peroxidase (POD) activities, to reflect the response ability of plants for resilience. First, we stained soybean root tips and leaves with nitro blue tetrazolium (NBT) and 3,3-diaminobenzidine (DAB) to detect H2O2 content under normal or treatment conditions in three GmCOL1a-ox transgenic lines, co-9, and WT plants. No substantial difference was observed under normal conditions with NBT and DAB staining; however, the color depth of WT and co-9 plants was substantially higher than that of the three GmCOL1a-ox transgenic lines under water deficit or 250 mM NaCl treatment (Figure 5A). In contrast, the leaf color depth of the three GmCOL1a-ox transgenic lines was substantially lower than that of the co-9 and WT plants (Figure 5, C, D, F, G). Finally, we selected the best phenotypic GmCOL1a-ox-2 line for H2O2 content measurement and found that the H2O2 content was the highest in both root tips and leaves of the co-9 mutant and the lowest in the GmCOL1a-ox-2 soybean (Figure 5, B, E, H).
Figure 5.
H2O2 levels and analysis of physiological parameters of different soybean plants under salt and drought treatment. A, Hairy roots of 6-day-old seedling plants of three GmCOL1a-ox transgenic lines, co-9, and WT plants at 3 h after 250 mM NaCl or 10% PEG 6000 treatment were stained by NBT or DAB, and half-strength Hoagland nutrient solution was used as a control. Scale bars = 0.5 cm. B, Determination of H2O2 content in the roots of GmCOL1a-ox-2, co-9, and WT plants under 250 mM NaCl or 10% PEG 6000 treatment for 3 h. C, NBT and D, DAB staining of the leaves of 14-day-old three GmCOL1a-ox transgenic lines, co-9, and WT plants exposed to 250 mM NaCl for 6 h. F, NBT and G, DAB staining of the leaves of 14-day-old three GmCOL1a-ox transgenic lines, co-9, and WT plants subjected to drought (no irrigation) treatment for 4 days. Scale bars = 0.5 cm. H2O2 content of 14-day-old seedlings of GmCOL1a-ox-2, co-9, and WT plants under E, salt or H, drought treatment. I, Proline and MDA content and CAT, POD, and SOD activities of 2-week-old seedlings of three GmCOL1a-ox transgenic lines, co-9, and WT plants. All values were measured after 6 h of exposure to salt or drought stress (150 mM NaCl or 10% PEG 6000). Asterisks indicated values significantly different between the transgenic lines and the WT in each group. For each experiment, three technical replicates were conducted. Data shown are the means ± SD (*, P < 0.05 and **, P < 0.01, Student's t test).
Proline accumulation enhances plant stress tolerance (Ashraf and Foolad, 2007). The proline accumulation rate among the different transgenic soybean plants showed no significant differences under control conditions; however, under salt or drought stress, the proline content was significantly higher in the three GmCOL1a-ox transgenic lines. The MDA content is used to indicate lipid peroxidation products and to reflect the extent of plant damage caused by adversity (Cao et al., 2007). Compared with the co-9 and WT plants, the MDA content in GmCOL1a-ox transgenic plants significantly decreased after salt and drought stress. The enzymatic activities of SOD, POD, and CAT scavenge intracellular ROS and reduce H2O2 production, thereby enhancing salt and drought tolerance (Han et al., 2020). The SOD, POD, and CAT activities of the co-9 and WT plants were significantly lower than those of the three GmCOL1a-ox transgenic lines (Figure 5I). These results suggest that GmCOL1a overexpression in soybeans enhanced salt and drought tolerance by increasing the proline content, reducing the MDA content, and increasing the enzyme activities of SOD, POD, and CAT to reduce ROS production.
GmCOL1a directly bound to the promoters of GmLEA and GmP5CS
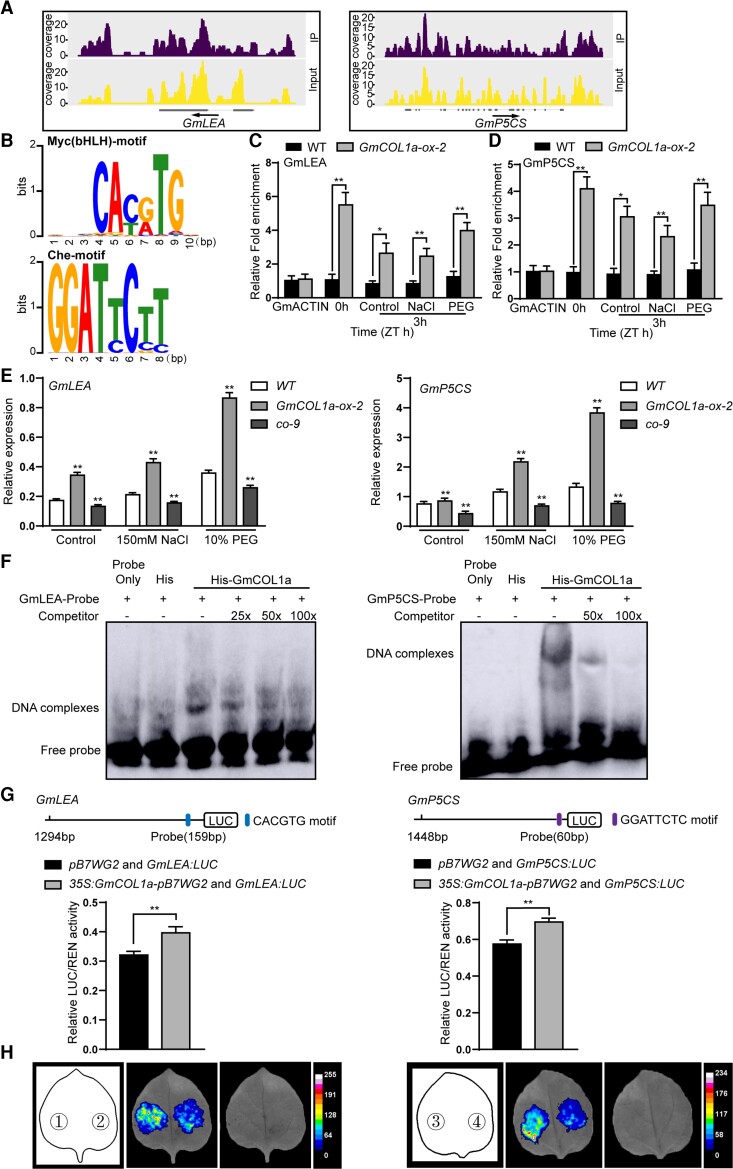
To identify the DNA-binding sites and the downstream target genes of GmCOL1a in soybean and further elucidate the potential mechanism by which GmCOL1a promotes salt and drought tolerance, we performed ChIP-seq using GmCOL1a-ox-2 soybean leaves. Based on the sequencing data, two promoter regions of the GmCOL1a target genes, GmLEA and GmP5CS, were putatively directly bound by GmCOL1a (Figure 6A; Supplemental Table S2).
Figure 6.
Binding of GmCOL1a protein to the promoters of GmLEA and GmP5CS. A, Peak graphs showing the ChIP-seq raw reads at the indicated gene loci in Integrative Genomics Viewer. The arrow indicated the direction of transcription, and the gray bars indicated the transcripts of the gene. B, Motif analysis of GmCOL1a-binding sequences. The distribution of a single pattern in the sequences, the parameter was set to zero or one per sequence. ChIP-qPCR assay of GmCOL1a binding to the GmLEA promoter and GmP5CS promoter using the soybean leaf samples treated with 150 mM NaCl and 10% PEG 6000 for 0 or 3 h. 0 h sample collection time was the same in C and D. The GmActin4 locus was used as a negative control. The asterisk indicated a significant difference between the WT and GmCOL1a-ox-2. Data shown are the means ± SD of three independent experiments (*, P < 0.05 and **, P < 0.01, Student's t test). E, The soybean seedlings were treated with 150 mM NaCl and 10% PEG 6000 for 3 h to collect samples. GmLEA and GmP5CS expressions in 21-day-old leaves from WT, GmCOL1a-ox-2 and co-9 plants. Values were means ± SD (n = 3). Asterisks indicated a significant change compared with WT. (*, P < 0.05 and **, P < 0.01, Student's t test). F, EMSA assays showing the specific binding of GmCOL1a to the GmLEA and GmP5CS promoter fragments containing CACGTG and GGATTCTC motifs, respectively. The unlabeled competitive probe is 25-, 50-, 100-fold excess of the labeled probe. HIS served as a negative control. G, The effect of GmCOL1a protein on the promoter activities of GmLEA and GmP5CS. Relative luciferase activity of co-transfected effector and reporter genes in Nicotiana benthamiana leaves was detected in LDs ZT 12 h. The activities of firefly LUC were normalized by the activities of 35S:Renilla LUC. Upper panel: physical locations of fragments harboring putative motifs were shown in the schematic diagram. Asterisks indicated values significantly different the line indicated. Results represented means ± SD of eight independent samples (*, P < 0.05 and **, P < 0.01, Student's t test). H, Luciferase activities of GmLEA and GmP5CS under LDs. Panel: ①: 35S:GmCOL1a-pB7WG2 + GmLEA:LUC ②: pB7WG2 + GmLEA:LUC ③: 35S:GmCOL1a-pB7WG2 + GmP5CS:LUC ④:pB7WG2 + GmP5CS:LUC. The two leaf images were digitally extracted for comparison under the same imaging conditions.
Then, Myc(bHLH)-binding, and Che-binding motifs that bind to GmCOL1a were identified (Figure 6B). We further performed ChIP-qPCR assays to verify the potential enrichment of GmLEA and GmP5CS promoters by GmCOL1a binding using GmCOL1a-ox-2 transgenic soybean plants expressing a GmCOL1a-FLAG fusion protein under salt and drought treatments with a WT sample as a negative control. The promoter fragments of GmLEA and GmP5CS carrying the identified GmCOL1a-binding sites were highly enriched in DNA chromatin immunoprecipitation (ChIPed) at 0 and 3 h of salt and drought stress, indicating that GmLEA and GmP5CS promoters containing Myc(bHLH)-binding and Che-binding motifs, respectively, were bound by GmCOL1a, despite the decreasing enrichment after 3 h of salt or drought treatment compared with the nontreated (0 h; Figure 6, C and D). We then examined the transcript levels of GmLEA and GmP5CS in different transgenic soybean plants under salt and drought stress. RT-qPCR showed that the transcript levels of GmLEA and GmP5CS were significantly elevated in GmCOL1a-ox-2 and decreased in co-9 soybean plants at 3 h of salt and drought treatment (Figure 6E).
To determine whether GmCOL1a could directly bind to the GmLEA promoter Myc(bHLH) motif (CACGTG) and the GmP5CS promoter Che motif (GGATTCTC), in vitro binding was analyzed using electrophoretic mobility shift assay (EMSA). HIS-GmCOL1a dramatically reduced the migration of the 40-bp and 32-bp probes, indicating that GmCOL1a directly bound to both CACGTG and GGATTCTC motifs (Figure 6F). Reciprocal competitive EMSA showed strong and specific binding of GmCOL1a to the two motifs.
Moreover, the reporters GmLEA:LUC and GmP5CS:LUC were constructed with GmLEA and GmP5CS promoters containing GmCOL1a-binding sites, the CACGTG motif and the GGATTCTC motif, respectively, driving the LUC reporter gene (Figure 6G). When co-infiltrating Agrobacterium expressing 35S:GmCOL1a-pB7WG2 effectors together with GmLEA:LUC and GmP5CS:LUC reporters into the leaves of N. benthamiana, LUC activity was elevated by GmCOL1a protein expression, demonstrating that GmCOL1a could promote the transcriptional activation activities of both GmLEA and GmP5CS (Figure 6H).
Overall, the results suggest that the GmCOL1a protein directly promotes the expression of GmLEA and GmP5CS by binding directly to their promoters.
GmP5CS improved salt and drought stress tolerance in transgenic soybean hairy roots
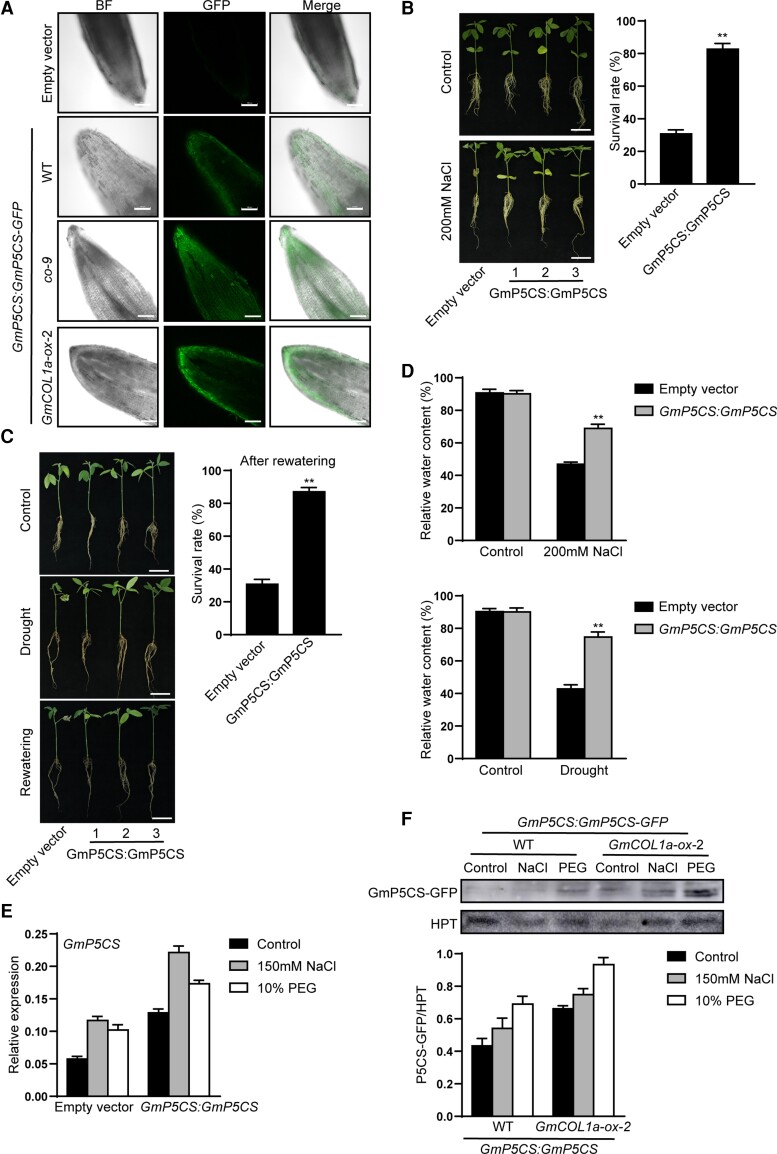
Notably, GmCOL1a bound directly to the promoters of GmLEA and GmP5CS. GmLEA has been shown to be one of the major genes regulating salt stress (Liu and Zheng, 2005; Liu et al., 2010). However, the function of GmP5CS under salt or drought stress has not been reported. To explore the molecular mechanism of GmP5CS involved in salt and drought tolerance in soybean, transgenic soybean plants expressing GmP5CS:GmP5CS-GFP were generated by Agrobacterium rhizogene (A. rhizogene)-mediated hairy-root transformation in the WT, co-9, and GmCOL1a-ox-2 backgrounds, forming “composite” transgenic plants. The plasmid carrying pCAMBIA1302-no35S (control) produced by A. rhizogenis strain K599 was used as a control hairy root for the soybean plants. Transgenic hairy roots were identified and selected using GFP under a confocal microscope (Figure 7A), and the remaining non-GFP hairy roots and main roots were removed.
Figure 7.
Assessment of the effects of salt and drought stress on GmP5CS transgenic soybean hairy roots. Empty vector indicated soybean hairy roots carrying the pCAMBIA1302-no35S vector and produced by A, rhizogenis strain K599. GmP5CS:GmP5CS indicated soybean plants with transgenic hairy roots expressing GmP5CS. GFP fluorescence was detected in hairy roots infected by A. rhizogenis strain K599 harboring pCAMBIA1302-no35S (control) and GmP5CS:GmP5CS-GFP-pCAMBIA1302 plasmids, in WT, co-9, and GmCOL1a-ox-2 background, respectively. Scale bar = 100 µm. B, Phenotypes and survival rates of “composite” plants with GmP5CS:GmP5CS transgenic hairy roots in WT background after 200 mM NaCl treatment for 6 days. Scale bar = 5 cm. Survival rates were quantified (n ≥ 20). Asterisks indicated a significant change compared with the empty vector. Data shown are means ± SD (*, P < 0.05 and **, P < 0.01, Student's t test). C, Phenotypes and survival rates of “composite” plants with GmP5CS:GmP5CS transgenic hairy roots in WT background after dehydration for 5 days and recovery for 2 days. Scale bar = 5 cm. Survival rates were quantified (n ≥ 20). Asterisks indicated a significant change compared with the empty vector. Data shown are means ± SD (*, P < 0.05 and **, P < 0.01, Student's t test). D, RWCs of leaves were measured from “composite” plants with GmP5CS:GmP5CS transgenic hairy roots in WT background after 200 mM NaCl treatment for 6 days or dehydration for 5 days. RWCs of leaves were calculated (n ≥ 10). Asterisks indicated a significant change compared with the empty vector. Data shown are means ± SD (*, P < 0.05 and **, P < 0.01, Student's t test). E, The transcript level of GmP5CS in transgenic hairy roots in WT background. Data shown are means ± SD of three independent experiments. F, Immunoblotting showing GmP5CS protein levels of GmP5CS:GmP5CS-GFP using GFP-Tag antibodies, Anti-HPT served as the loading control. Protein expression profiles of GmP5CS-GFP exposed to 150 mM NaCl or 10% PEG treatment for 6 h. Roots of GmP5CS-GFP protein calculated by normalization of the GmP5CS-GFP signals to Anti-HPT signals and presented as GmP5CS-GFP/HPT. Data shown are means ± SD of three independent experiments.
After salt treatment, the “composite” transgenic soybean plants with GmP5CS hairy roots in the WT background showed healthier leaves and larger root elongation than the control plants. GmP5CS also increased the survival rates of “composite” plants with GmP5CS hairy roots under salt stress (Figure 7B). In addition, after drought treatment, the GmP5CS hairy roots retained life processes, unlike the control plants. After rewatering for 2 days, the majority of GmP5CS transgenic soybean plants were rejuvenated (87% survival), but most of the control plants were not (31% survival; Figure 7C). Then, we measured the RWC of GmP5CS “composite” transgenic soybean plants after salt and drought treatment. Under salt and drought treatment, the RWC of transgenic GmP5CS “composite” soybean plants decreased significantly compared to that of control plants (Figure 7D). GmP5CS enhanced tolerance to both salt and drought stress in the GmP5CS:GmP5CS-GFP “composite” transgenic soybean with A. rhizogene-mediated hairy roots.
We also measured the proline content in hairy roots after salt and drought stress. GmP5CS transgenic soybean hairy roots contained higher levels of proline compared to soybean control hairy roots, and the proline content of GmP5CS transgenic soybean hairy roots increased under salt and drought conditions (Supplemental Figure S1, A). To determine whether GmP5CS:GmP5CS-GFP “composite” transgenic plants regulated salt and drought stress in relation to ROS, we measured CAT, SOD, and POD activities. The CAT, SOD, and POD activities of GmP5CS:GmP5CS-GFP “composite” transgenic plants were significantly higher than those of empty vector plants (Supplemental Table S1, B–D). The results suggest that the GmP5CS:GmP5CS-GFP “composite” transgenic soybean plants enhanced salt and drought tolerance by increasing SOD, POD, and CAT enzyme activities to reduce ROS production.
The mRNA levels of GmP5CS were higher in GmP5CS:GmP5CS-GFP transgenic hairy roots than in empty vectors under salt and drought conditions (Figure 7E). GmP5CS:GmP5CS was transformed into WT and GmCOL1a-ox-2 soybean backgrounds. After 6 h of treatment with 150 mM NaCl or 10% PEG 6000, the protein levels of GmP5CS-GFP in transgenic seedlings were observed. As expected, the GmP5CS-GFP protein was induced in roots by salt and drought stress in both the WT and GmCOL1a-ox-2 backgrounds, and GmP5CS-GFP protein expression was higher in the GmCOL1a-ox-2 background than in the WT background (Figure 7F), which further validated that GmCOL1a promoted GmP5CS expression.
Thus, the evidence above strongly suggests that GmP5CS was a downstream gene of GmCOL1a, which was required to promote the stress-responsive GmP5CS protein in soybean and improved salt tolerance and drought resistance by increasing the expression of GmP5CS to increase the RWC, proline content, and SOD, POD, and CAT enzyme activities.
Overexpression of GmP5CS rescued the salt- and drought-sensitive phenotypes of co-9 plants
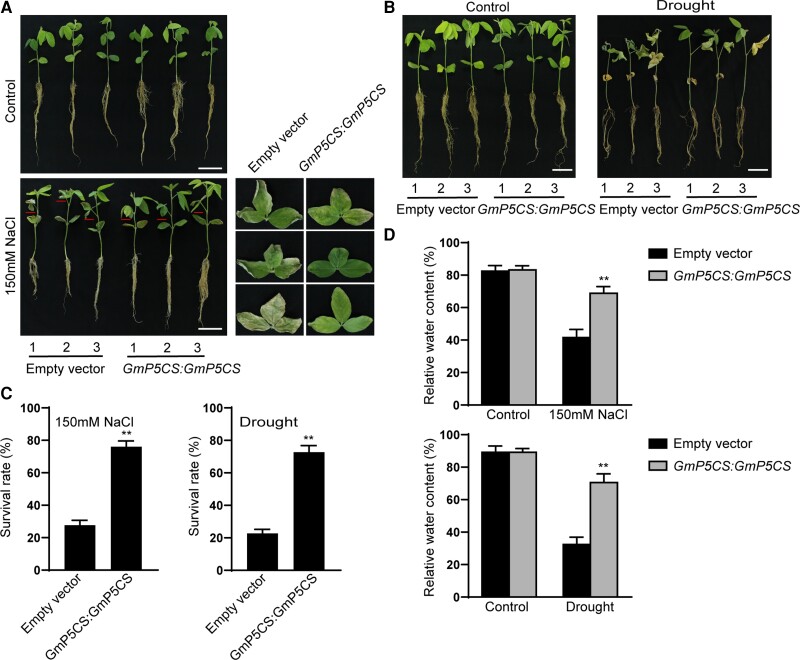
Since GmP5CS has been proven to increase salt and drought tolerance, we further investigated whether the transformation of GmP5CS:GmP5CS-GFP into the co-9 mutant background by A. rhizogene-mediated hairy-root transformation could rescue the salt- and drought-sensitive phenotype of co-9 plants (Figure 8, A and B). Under salt and drought stress, the survival rates of “composite” plants with GmP5CS hairy roots increased by 48.33% and 50%, respectively (Figure 8C). Similarly, the RWCs of GmP5CS “composite” transgenic soybean plants increased by 27.66% and 38.18% after salt and drought treatment, respectively (Figure 8D).
Figure 8.
Overexpression of GmP5CS in transgenic soybean hairy roots rescued the salt and drought-sensitive phenotype of the co-9 plants. A, Phenotypes of “composite” plants with GmP5CS:GmP5CS transgenic hairy roots in co-9 background after 150 mM NaCl treatment for 5 days. Scale bar = 5 cm. B, Phenotypes of “composite” plants with GmP5CS:GmP5CS transgenic hairy roots in co-9 background after dehydration for 6 days. Scale bar = 5 cm. C, Survival rates of “composite” plants with GmP5CS:GmP5CS transgenic hairy roots in co-9 background under salt and drought stress. Survival rates were quantified (n ≥ 20). D, RWCs of leaves were measured from “composite” plants with GmP5CS:GmP5CS transgenic hairy roots in co-9 background under salt and drought stress. RWCs of leaves were calculated (n ≥ 10). Asterisks indicated a significant change compared with the empty vector. Data shown are means ± SD of three independent experiments (*, P < 0.05 and **, P < 0.01, Student's t test).
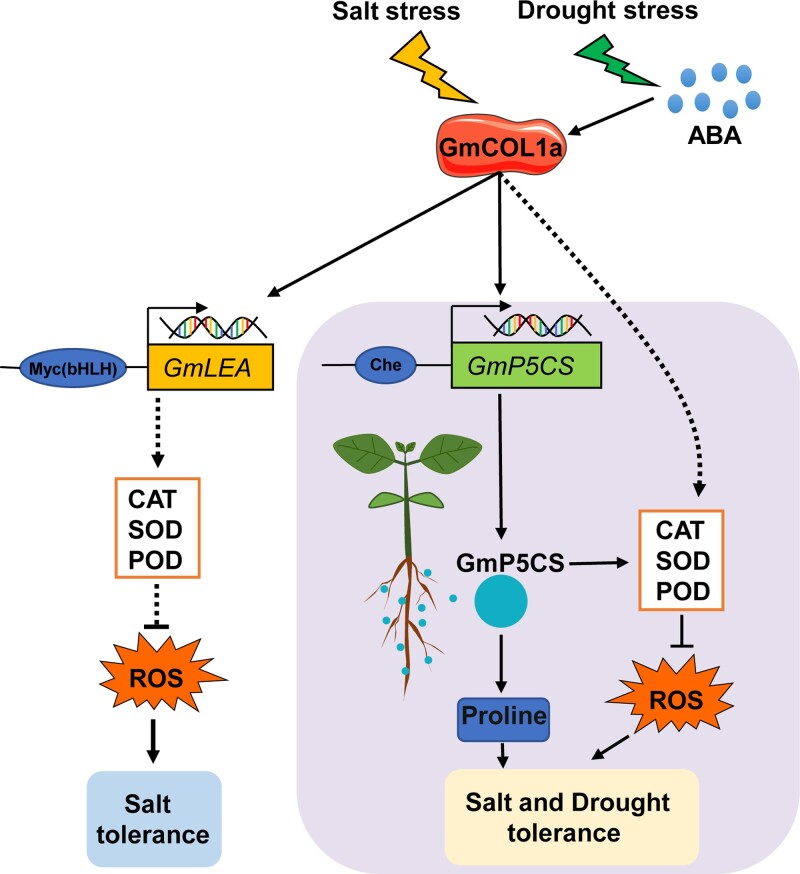
Based on all experimental evidence, we proposed that GmCOL1a was a nuclear-localized protein that regulated salt and drought tolerance in soybean. GmCOL1a promoted drought tolerance dependent on the ABA signaling pathway. Our proposed model clarified that GmCOL1a regulates salt and drought stress responses by directly binding to Myc(bHLH) and Che motifs in the promoters of the stress resistance-related genes GmLEA and GmP5CS, respectively, to up-regulate their transcription. GmCOL1a enhanced salt and drought tolerance by promoting GmP5CS accumulation, which increased the proline content and reduced ROS production to protect soybean plants from salt and drought stress damage. Meanwhile, GmCOL1a might also directly inhibit ROS production independent of downstream GmP5CS (Figure 9).
Figure 9.
Proposed model depicting the function of GmCOL1a in the regulation of salt and drought stress tolerance. The solid arrows indicate activation of transcription, dotted-lines denote a relationship that has not been established as direct, and lines ending with a dash denote repression of transcription. GmCOL1a is induced by salt and drought tolerance. GmCOL1a is mediated by the abscisic acid (ABA), which enhances drought resistance. GmCOL1a promoted the transcriptional level of GmLEA, probably by decreasing the accumulation of ROS, thus improving salt tolerance in GmCOL1a-overexpressing transgenic plants. In addition, GmCOL1a not only increased the transcript level of GmP5CS but also promoted GmP5CS protein accumulation, which increased proline content and reduced ROS production, thereby improving stress tolerance in soybean plants.
Discussion
GmCOL1a was involved in the stress resistance of soybean plants
Currently, the molecular mechanisms involved in the photoperiodic pathway of CO in plants are relatively well established. For example, the TF CO is a crucial gene affecting the circadian rhythm and flowering time in plants, regulated by the circadian and biological clock, which promotes flowering by affecting the expression of the flowering hormone FLOWERING LOCUS T (FT; Suárez-López et al., 2001; Hayama et al., 2003). The four cis-elements (CORE1, CORE2, P1, and P2) of the FT promoter region are direct targets of CO, forming a CO-NF-Y complex that binds to the FT promoter with high overall binding specificity and affinity, further revealing the importance of CO in the photoperiodic pathway (Lv et al., 2021). A few studies have shown the involvement of the CO family in abiotic stress responses in Arabidopsis, maize, rice, apple, mango, and Tamarix hispida plants. However, the analysis of CO gene expression in soybean under abiotic stress has not been reported. In this study, we demonstrated that GmCOL1a overexpression in transgenic soybean plants improved salt tolerance and drought resistance, and knockout mutant co-9 transgenic soybean plants showed opposite phenotypes (Figures 2E and 4A). The GmCOL1a gene contains two B-box domains and a CCT domain, which are typical B-box proteins (BBXs; Putterill et al., 1995; Gangappa and Botto, 2014). Previous studies have demonstrated that GbBBX25 improves salt acclimation in transgenic Populus (Huang et al., 2021). The homolog of GmCOL1a was AtCOL4 (BBX5), with a sequence similarity of 35.62%, which also contained two B-box domains and a CCT domain. This enhances plant tolerance to abiotic stress by participating in an ABA-dependent signaling pathway (Min et al., 2015). MdBBX7/MdCOL9 overexpression enhanced drought tolerance in transgenic apples and regulated the effect of the MdMIEL1-MdBBX7 module on the drought stress response (Chen et al., 2022). Therefore, we hypothesized that GmCOL1a was involved in the abiotic stress response in soybean plants, probably because of the roles of the B-Box domain and CCT domain.
Transgenic GmCOL1a-ox-2 soybean had a reduced Na+/K+ ratio, improving salt tolerance
Reducing the Na+/K+ ratio was a key factor in improving salt tolerance in plants by regulating the ion dynamic balance. Salt induction affects the circulation and transport of K+ in specific tissues, balancing excess Na+ and thus conferring salt tolerance to the plant (Álvarez-Aragón et al., 2016). In the present study, we initially investigated the effect of the GmCOL1a gene on the Na+/K+ ratio under salt stress conditions. We observed that the Na+/K+ ratio of GmCOL1a-ox-2 transgenic soybean was significantly lower in stems and leaves but not in the roots compared to those in the WT and co-9 after salt stress (Figure 3A). We speculated that GmCOL1a reduced Na+ ion toxicity by reducing its transport from the roots to the stems and leaves and especially maintained the lower dynamic balance of Na+/K+ in leaves. Most NHX transgenic plants have improved salt stress adaptation by reducing the Na+ content in the cytoplasm or by maintaining a higher K+/Na+ ratio (Wu et al., 2016; Sun et al., 2021). PgTIP1-trasgenic soybean reduced the Na+/K+ ratio in stems and leaves but not in roots (An et al., 2017), which was similar to the results of the present study. Although the K+ content of GmCOL1a-ox-2 transgenic soybean stems and leaves was reduced under salt stress conditions, the Na+/K+ ratio remained relatively low, improving plant salt tolerance.
GmP5CS increased the proline content
The GmP5CS protein regulated abiotic stress responses in the WT and GmCOL1a-overexpressing transgenic soybeans. Previous research has established that the gene encoding AtP5CS was induced by salt stress, drought stress, and ABA in Arabidopsis (Yoshiba et al., 1995). The two homologs of GmP5CS were Glycine max (gmp5cs), with a sequence similarity of 28.69%, and Lactuca sativa (lsp5cs), with a sequence similarity of 29.86%, which were reported to be up-regulated only under drought stress (Porcel et al., 2004). However, a functional analysis of the P5CS gene in soybean plants under salt stress has not been performed. Here, we reported the facilitation of abiotic stress responses by the GmP5CS gene in soybean because GmP5CS enhanced the ability of soybean to resist high salt and drought conditions (Figure 7, B and C). Furthermore, P5CS plays a key role in plant proline biosynthesis (Hu et al., 1992). This was consistent with the results that GmP5CS transgenic hairy roots were able to accumulate more proline content, further increasing the proline content under salt and drought treatments (Supplemental Figure S1, A). In addition, we described a previously uncharacterized mechanism whereby GmCOL1a not only increased the transcript level of GmP5CS but also promoted GmP5CS protein accumulation, resulting in an increased proline content and reduced ROS production, thereby improving stress resistance in soybean plants.
Materials and methods
Identification and phylogenetic relationship analysis of GmCOL1a genes
Multiple full-length GmCOL1a amino acid sequences of soybean (Glycine max) and other plants were aligned using the ClustalW program (http://www.clustal.org/). An NJ tree was constructed using MEGA7.0 and polished using EvolView with 1,000 bootstrap trials.
Plant materials and growth conditions
For the germination assay, three T3 generation transgenic soybean lines GmCOL1a-ox-2, GmCOL1a-ox-4, and GmCOL1a-ox-17, T3 generation co-9 Crispr/Cas9 mutant, and WT (DN50) seeds were chlorine-sterilized and placed on 1/2 MSB medium (1/2 Murashige and Skoog basal nutrient salts, B5 vitamins, and 2% sucrose, pH 5.7) supplemented with 0 and 250 mM NaCl and germinated at 25°C under LDs (16 h/8 h light/dark). Images were taken, and germination rates and hypocotyl lengths were recorded at the end of the experiment.
Three GmCOL1a-ox transgenic lines, the co-9 mutant, and WT soybeans were also used for phenotype analysis. Soybean seeds were germinated for 4 days in grass charcoal soil and vermiculite (1:1 ratio), and uniformly growing seedlings were transferred to LDs at 25°C and 250 µmol m−2 s−1 with white light and 60% relative humidity and grown in plastic root growth bags containing a half-strength Hoagland solution. In the root growth phenotype assay, the movement of roots was minimized, and five parallel bags were used for each treatment, with all genotypes contained in the same number of bags. Survival rate and root length were photographed and recorded by adding 100 mM NaCl to the hydroponic solution and changing the solution every 3 days for 21 days of treatment.
N. benthamiana was grown in a greenhouse at 22°C under LDs.
RNA extraction and RT-qPCR analysis
To analyze the expression pattern of GmCOL1a (Glyma.08G255200), GmLEA (Glyma.13G149000), and GmP5CS (Glyma.18G188000) genes under salt and drought stress, the soybean cultivar “DN50” was grown under LDs at 25°C and 250 µmol m−2 s−1 with white light. When the first trifoliate leaves were fully expanded, soybean seedlings were treated with control (a half-strength Hoagland solution), 10% [w/v] PEG 6000, and 150 mM NaCl. Samples were collected after 0, 1, 3, and 6 h, immediately frozen in liquid nitrogen, and stored at −80°C.
Total RNA was extracted from soybean plants using Trizol reagent (Invitrogen), as described previously (Zhao et al., 2013). The resulting cDNA was used as a template using an ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA). GmActin4 (GenBank accession number AF049106) was used as an internal control. Three biological replicates and three technical replicates were applied to the assays. The primers used for RT-qPCR are listed in Supplemental Table S1.
Plasmid construction and generation of transgenic soybean
Total RNA was reverse transcribed into first-strand cDNA. A 1047 bp GmCOL1a gene fragment was cloned from the DN50 using the GmCOL1a-3F6H-F and GmCOL1a-3F6H-R primers (Supplemental Table S1). The GmCOL1a gene fragment was reassembled with the pENTRY-3F6H vector (named 35S:GmCOL1a-3F6H-pENTRY) using the In-Fusion Cloning System (Clontech, USA). The recombinant plasmid 35S:GmCOL1a-3F6H-pENTRY was synthesized into the pB7WG2 vector by the LR reaction (named 35S:GmCOL1a-3F6H-pB7WG2). The construct was then transferred into Agrobacterium tumefaciens (EHA105). According to the method described previously (Zhao et al., 2018), transgenic soybean DN50 expressing 35S:GmCOL1a-3F6H-pB7WG2 was obtained.
The design of the target sequence adapters for the GmCOL1a gene was performed using the online tool CRISPR-P (http://cbi.hzau.edu.cn/crispr/). The DNA oligonucleotide pairs (GmCOL1a-cas9-F and GmCOL1a-cas9-R) of the synthesized sgRNA were annealed and generated into a dimer bound to the pGES-201 vector (Bai et al., 2020). The recombinant vector was introduced into the Agrobacterium tumefaciens strain, EHA105, and then transformed into soybean DN50 (Zhao et al., 2018). Transgenic soybean plants were screened using PCR. One representative homozygous line (co-9) was selected for further study.
For the cloning of GmCOL1a into the pDEST17 vector to express the protein in E. coli, the full-length coding region of GmCOL1a was amplified by PCR using GmCOL1a-TOPO-F and GmCOL1a-TOPO-R primers (Supplemental Table S1), cloned into pENTR/D-TOPO (Thermo Fisher Scientific, USA), and transferred to the expression vector pDEST17 through an LR reaction to generate the HIS-GmCOL1a fusion vector. The construct was transformed into the E. coli competent cell line BL21 (DE3; Transgene, Beijing, China) to produce recombinant proteins. For subcellular localization, the GmCOL1a-TOPO plasmid was also transferred to the expression vector pGWB506 through an LR reaction to generate the 35S:GFP-GmCOL1a fusion vector.
To clone GmP5CS into the pCAMBIA1302-no35S plasmid, pCAMBIA1302 was first digested with Hind III and Nco I to remove the 35S fragment, blunted by T4 DNA polymerase, and ligated by T4 DNA ligase to produce the circular plasmid pCAMBIA1302-no35S. The full-length sequences consisting of 1427-bp promoter regions were amplified from the genomic DNA of DN50 using GmP5CS-P-F and GmP5CS-P-R, and 2163-bp coding sequences (CDSs) of GmP5CS were amplified from the cDNA of DN50 by overlapping PCR with pairs of GmP5CS-CDS-F and GmP5CS-CDS-R. PCR was then performed using GmP5CS-P-F and GmP5CS-CDS-R primers to obtain the GmP5CS:GmP5CS-GFP PCR product (Supplemental Table S1; Yang et al., 2021). The PCR products were purified and cloned into the pCAMBIA1302-no35S vector containing the GFP sequence in the C-terminus of the cloning site linearized by Bgl II using the In-Fusion cloning system to construct the recombinant GmP5CS:GmP5CS-GFP-pCAMBIA1302 fusion expression vector. The recombinant vector (GmP5CS:GmP5CS-GFP-pCAMBIA1302) was transferred into the Agrobacterium rhizogene (A. rhizogenes) strain K599.
Identification of phenotypes under salt and drought stress
Three transgenic GmCOL1a-ox lines, the co-9 mutant, and WT soybeans were planted in 10 pots, each with three seedlings. For the salt stress tests, the plants were immersed in a 200 mM NaCl solution when the first trifoliate leaves were fully expanded. After 8 days of salt treatment, the RWC was determined in the first pair of unifoliolate leaves of soybean plants, as previously described (Bao et al., 2014). According to a previous report, measurements of Na+ and K+ concentration were performed using atomic absorption spectrophotometry (AAS; Hitachi Z-2000; Xu et al., 2006; Sun et al., 2021). The analysis was repeated in triplicate. For dehydration experiments, the water supply was stopped at the first untreated growth stage to ensure consistent watering in each pot, and the control group was watered with half-strength Hoagland solution every 3 days when the first trifoliate leaves of the plants were fully expanded.
Stomatal aperture measurement
Three-week-old soybean leaves were placed in stomatal opening solution (30 mM KCl and 10 mM MES-KOH, pH 6.15) for 2 h and then treated with 10 µM ABA for 3 h (Li et al., 2013). Stomatal observation was performed with a confocal microscope (TCS SP5; Leica Microsystems). The experimental setup followed by: bit depth was eight bit, depth of focus was 1.72 µm, digital offset was 0, digital gain was 1 (Martin et al., 2009).
Histochemical and physiological index analysis
For 3,3-DAB staining, root tips or leaf samples were immersed in 50 mM DAB solution (Solarbio, China) for 12 or 24 h and then decolorized in 95% [v/v] ethanol until the color turned white. For NBT staining, root tips or leaf samples were immersed in 50 mM NBT solution (Creek Huizhi, China) for 15 min or 16 h and then decolorized in 95% [v/v] ethanol until the color turned white (Wang et al., 2017). Images were taken using a Canon 50D (Canon, Japan) camera. After treatment, 100 mg of root or leaf samples was subjected to H2O2 content measurement (Solarbio, China).
For physiological parameter measurements, 2-week-old seedlings of three GmCOL1a-ox transgenic lines, the co-9 mutant, and WT soybeans were exposed to 150 mM NaCl or 10% [w/v] PEG 6000 for 6, and 100 mg of leaf tissues was used to measure the free proline (Bates et al., 1973) and MDA content (Gao et al., 2011) and catalase (CAT; Abei, 1984), SOD (He et al., 2009), and POD activity (Kong et al., 2005). A half-strength Hoagland nutrient solution was used as a control.
Immunoblot analysis
To measure the protein expression of the GmCOL1a gene driven by the 35S promoter in transgenic soybean, the 35S:GmCOL1a-3F6H protein was extracted in extraction buffer for the extraction of whole proteins from soybean leaves (Wang et al., 2021). The HRP-coupled anti-FLAG antibody (A8592, Sigma) was used to detect the 35S:GmCOL1a-3F6H protein. A mouse β-actin monoclonal antibody (HRP-60008, Proteintech) was used as a control to detect actin proteins.
The GmP5CS:GmP5CS protein in transgenic hairy roots of in vitro-generated “composite” plants was extracted in the same extraction buffer. The protein extracts were detected with a GFP-Tag antibody (AE011, Abclonal), followed by an HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H + L) Secondary Antibody (SA00001–2, Proteintech). Anti-HPT (AbM59707–5-PU, BPI), followed by HRP-conjugated goat anti-mouse IgG (H + L) Secondary Antibody (G-21040, Thermo Fisher Scientific), was used as a control to detect HPT proteins.
Super Signal West Pico chemiluminescent substrate kits (Thermo Fisher Scientific, USA) were used to detect the signals by chemiluminescent imaging (Amersham Imager 600). All experiments were performed at least three times with independent biological replicates.
ChIP-qPCR
After 14 days of growth, 35S:GmCOL1a-3F6H transgenic soybean seedlings and WT seedlings were transferred to 150 mM NaCl or 10% [w/v] PEG 6000 solution for 0 or 3 h at LDs. Half-strength Hoagland solution was used as a control, and the tissues were fixed with a 1% [v/v] formaldehyde crosslinking method for 15 min. ChIP-qPCR assays were performed as previously described (Wang et al., 2021). Immunoprecipitation reactions were performed using an anti-FLAG antibody (mouse-produced Monoclonal ANTI-FLAG M2 antibody, F1804, Sigma-Aldrich) and an anti-IgG antibody (Normal Rabbit IgG, 2729, Cell Signaling Technology) control. Complexes of chromatin antibodies were captured with protein G beads (Invitrogen), and DNA was purified using a QIAquick PCR purification kit (QIAGEN). Enriched DNA fragments were identified by RT-qPCR analysis, and three biological replicates and three technical replicates were performed for each sequence segment. The primers used in ChIP-qPCR are listed in Supplemental Table S1.
EMSA
Binding of the GmCOL1a protein to the Myc(bHLH) motif and Che motif in the promoter regions of GmLEA and GmP5CS, respectively, was examined by EMSA. Oligonucleotide probes containing the Myc(bHLH) motif (CACGTG) and the Che motif (GGATTCTC) were synthesized and labeled with biotin at the 5′ end. Probe sequences are shown in Supplemental Table S1. For competition with the unlabeled probe, an unlabeled probe was added to the reactions. BL21(DE3) containing the HIS-GmCOL1a fusion vector was induced using 0.5 mM isopropyl-b-ß-1-thiogalactopyranoside at 18°C for 12 h. The empty vector (pET-28a) was used as the negative control. The recombinant protein was purified using a 6×HIS-Tagged Protein Purification Kit (Soluble Protein; CWBIO, China) according to the manual provided by the manufacturer. The HIS-GmCOL1a fusion protein was incubated with the labeled probe and unlabeled DNA fragments (100-fold excess of the labeled probe). According to the manufacturer's protocol, the biotin-labeled probes were visualized using a Light Shift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, USA). The protein–DNA complexes were separated with 6% [w/v] native polyacrylamide gels and visualized on nylon membranes by chemiluminescence imaging (Amersham Imager 600).
Transient expression assays in N. benthamiana leaves
GmLEA and GmP5CS promoters were amplified from the genomic DNA of DN50 using the specific primers listed in Supplemental Table S1. The pGreenII-0800-GmLEA and pGreenII-0800-GmP5CS recombinant vectors were constructed using the In-Fusion Cloning System (Clontech, USA). The recombinant plasmid vectors were introduced into Agrobacterium tumefacien GV3101 by electroporation and transformed into N. benthamiana, as previously described (Sheikh et al., 2014). Firefly and Renilla luciferase activities were detected and analyzed, as described previously (Yang et al., 2021).
Transformation and detection of soybean hairy roots
Seven-day-old soybean DN50 seedlings were transformed by A. rhizogenes to form hairy roots using hypocotyl inoculation. Transgenic hairy roots were generated as described previously (Kereszt et al., 2007; Wang et al., 2015). The “composite” transgenic soybean plants were treated with 200 mM NaCl or subjected to drought for phenotypic observations, and positive “composite” transgenic soybean plants were used to determine the RWC, proline content, and CAT, SOD, and POD activities.
The use of A. rhizogenes-mediated cotyledonary transformation to obtain an in vitro-generated plant followed the inoculation method (Li et al., 2010; Cao et al., 2011). When hairy roots emerged, they were screened with a confocal microscope (TCS SP5; Leica Microsystems) to visualize GFP expression. The experimental setup followed by: excitation wavelength and acquisition bandwidth of laser at 488/490–540 nm for GFP, bit depth was eight bit, depth of focus was 1.72 µm, digital offset was 0, digital gain was 1(Martin et al., 2009). Expression of the GmP5CS gene in in vitro-generated plants with transgenic cotyledon hairy roots was tested using GFP fluorescence, RT-qPCR, and western blot.
Subcellular localization of GmCOL1a
The recombinant 35S:GFP-GmCOL1a construct was introduced into Agrobacterium GV3101 and subsequently transformed into N. benthamiana (Sheikh et al., 2014). A red nuclear marker plasmid (H2B-RFP) was used to confirm the location of the cell nucleus (Goodin et al., 2002). Two days after transfection, the leaves of N. benthamiana were imaged using a confocal microscope (TCS SP5; Leica Microsystems). The experimental setup followed by: excitation wavelength and acquisition bandwidth of laser at 561/580–630 nm for RFP, 488/490–540 nm for GFP, respectively, bit depth was eight bit, depth of focus was 1.72 µm, digital offset was 0, digital gain was 1(Martin et al., 2009).
Accession numbers
Sequence data from this article can be found in the Phytozome data libraries under accession numbers: GmCOL1a (Glyma.08G255200), GmLEA (Glyma.13G149000), GmP5CS (Glyma.18G188000), GmACTIN (Glyma.12G063400).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Physiological assays of GmP5CS:GmP5CS transgenic hairy root “composite” plants under salt and drought stress.
Supplemental Table S1 . Primer sequences used in this study.
Supplemental Table S2 . Peak sequences.
Supplementary Material
Acknowledgments
We thank Dr. Yuefeng Guan (Fujian Agriculture and Forestry University) for sharing plasmid materials.
Contributor Information
Chongjing Xu, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Jinming Shan, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Tianmeng Liu, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Qi Wang, Institute of Crop Cultivation and Tillage, Heilongjiang Academy of Agricultural Sciences, Harbin 150028, China.
Yujia Ji, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Yuntong Zhang, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Mengyuan Wang, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Ning Xia, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Lin Zhao, Key Laboratory of Soybean Biology of Ministry of Education China, Northeast Agricultural University, Harbin 150030, China.
Funding
This study was financially supported by the National Key Research and Development Program of China (2021YFF1001203), National Natural Science Foundation of China (32072086), Heilongjiang Province Natural Science Foundation (ZD2020C002).
References
- Abei H (1984) Catalase in vitro. Methods Enzymol 105(C): 121–126 [DOI] [PubMed] [Google Scholar]
- Álvarez-Aragón R, Haro R, Benito B, Rodríguez-Navarro A (2016) Salt intolerance in Arabidopsis: shoot and root sodium toxicity, and inhibition by sodium-plus-potassium overaccumulation. Planta 243(1): 97–114 [DOI] [PubMed] [Google Scholar]
- An J, Hu Z, Che B, Chen H, Yu B, Cai W (2017) Heterologous expression of Panax ginseng PgTIP1 confers enhanced salt tolerance of soybean cotyledon hairy roots, composite, and whole plants. Front Plant Sci 8: 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59(2): 206–216 [Google Scholar]
- Aziz MA, Sabeem M, Mullath SK, Brini F, Masmoudi K (2021) Plant group II LEA proteins: intrinsically disordered structure for multiple functions in response to environmental stresses. Biomolecules 11(11): 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Yuan J, Kuang H, Gong P, Li S, Zhang Z, Liu B, Sun J, Yang M, Yang L, (2020) Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol J 18(3): 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao AK, Wang YW, Xi JJ, Liu C, Zhang JL, Wang SM (2014) Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 enhances salt and drought tolerance in transgenic Lotus corniculatus by increasing cations accumulation. Funct Plant Biol 41(2): 203–214 [DOI] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39(1): 205–207 [Google Scholar]
- Cao D, Hou W, Liu W, Yao W, Wu C, Liu X, Han T (2011) Overexpression of TaNHX2 enhances salt tolerance of ‘composite’ and whole transgenic soybean plants. Plant Cell Tissue Organ Cult 107(3): 541–552 [Google Scholar]
- Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143(2): 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhi F, Li X, Shen W, Yan M, He J, Bao C, Fan T, Zhou S, Ma F, (2022) Zinc-finger protein MdBBX7/MdCOL9, a target of MdMIEL1 E3 ligase, confers drought tolerance in apple. Plant Physiol 188(1): 540–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal P, Kwon E, Pathak D, Kim DY (2022) Crystal structure of a tandem B-box domain from Arabidopsis CONSTANS. Biochem Biophys Res Commun 599: 38–42 [DOI] [PubMed] [Google Scholar]
- de Brito YMA, Rufino IAA, Braga CFC, Mulligan K (2021) The Brazilian drought monitoring in a multi-annual perspective. Environ Monit Assess 193(1): 31. [DOI] [PubMed] [Google Scholar]
- Dong L, Hou Z, Li H, Li Z, Fang C, Kong L, Li Y, Du H, Li T, Wang L, (2022) Agronomical selection on loss-of-function of GIGANTEA simultaneously facilitates soybean salt tolerance and early maturity. J Integr Plant Biol 64(10): 1866–1882 [DOI] [PubMed] [Google Scholar]
- Fahnenstich H, Scarpeci TE, Valle EM, Flügge UI, Maurino VG (2008) Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol 148(2): 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M, Wahid A, Kobayashi N, Fujita D, Basra S (2009) Plant drought stress: effects, mechanisms and management. Agron Sustain Dev 29(1): 185–212 [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19(7): 460–470 [DOI] [PubMed] [Google Scholar]
- Gao S, Yuan L, Zhai H, Liu C, He S, Liu Q (2011) Transgenic sweetpotato plants expressing an LOS5 gene are tolerant to salt stress. Plant Cell Tissue Organ Cult 107(2): 205–213 [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31(3): 375–383 [DOI] [PubMed] [Google Scholar]
- Guan R, Qu Y, Guo Y, Yu L, Liu Y, Jiang J, Chen J, Ren Y, Liu G, Tian L, (2014) Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J 80(6): 937–950 [DOI] [PubMed] [Google Scholar]
- Guo Y, Luo C, Liu Y, Liang R, Yu H, Lu X, Mo X, Chen S, He X (2022) Isolation and functional analysis of two CONSTANS-like 1 genes from mango. Plant Physiol Biochem 172: 125–135 [DOI] [PubMed] [Google Scholar]
- Han D, Du M, Zhou Z, Wang S, Li T, Han J, Xu T, Yang G (2020) Overexpression of a Malus baccata NAC transcription factor gene MbNAC25 increases cold and salinity tolerance in Arabidopsis. Int J Mol Sci 21(4): 1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422(6933): 719–722 [DOI] [PubMed] [Google Scholar]
- He S, Han Y, Wang Y, Hong Z, Liu Q (2009) In vitro selection and identification of sweetpotato (Ipomoea batatas (L. Lam.) plants tolerant to NaCl. Plant Cell Tissue Organ Cult 96(1): 69–74 [Google Scholar]
- Hsing Y, Chen Z, Chow T (1992) Nucleotide sequences of a soybean complementary DNA encoding a 50-kilodalton late embryogenesis abundant protein. Plant Physiol 99(1): 354–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CA, Delauney AJ, Verma DP (1992) A bifunctional enzyme (Δ1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci U S A 89(19): 9354–9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Chen C, Xu M, Wang G, Xu L, Wu Y (2021) Overexpression of Ginkgo BBX25 enhances salt tolerance in transgenic Populus. Plant Physiol Biochem 167: 946–954 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Shabala S (2019) Control of xylem Na+ loading and transport to the shoot in rice and barley as a determinant of differential salinity stress tolerance. Physiol Plant 165(3): 619–631 [DOI] [PubMed] [Google Scholar]
- Ismail A, Takeda S, Nick P (2014) Life and death under salt stress: same players, different timing? J Exp Bot 65(12): 2963–2979 [DOI] [PubMed] [Google Scholar]
- Kamarudin ZS, Yusop MR, Ismail MR, Tengku Muda Mohamed M, Harun AR, Yusuff O, Magaji U, Fatai A (2019) LEA Gene expression assessment in advanced mutant rice genotypes under drought stress. Int J Genomics 2019(Pt.2): 8406036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, Gresshoff PM (2007) Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nature Protoc 2(4): 948–952 [DOI] [PubMed] [Google Scholar]
- Ketehouli T, Zhou YG, Dai SY, Carther KFI, Sun DQ, Li Y, Nguyen QVH, Xu H, Wang FW, Liu WC, (2021) A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J Plant Physiol 256(4): 153331. [DOI] [PubMed] [Google Scholar]
- Kishor P, Hong Z, Miao GH, Hu C, Verma D (1995) Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108(4): 1387–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Wang M, Bi D (2005) Selenium modulates the activities of antioxidant enzymes, osmotic homeostasis and promotes the growth of sorrel seedlings under salt stress. Plant Growth Regul 45(2): 155–163 [Google Scholar]
- Lei X, Tan B, Liu Z, Wu J, Lv J, Gao C (2021) ThCOL2 improves the salt stress tolerance of Tamarix hispida. Front Plant Sci 12: 653791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET (2013) Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol 200(2): 457–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lin YJ, Wang P, Zhang B, Li M, Chen S, Shi R, Tunlaya-Anukit S, Liu X, Wang Z, (2019) The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell 31(3): 663–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Todd TC, Trick HN (2010) Rapid in planta evaluation of root expressed transgenes in chimeric soybean plants. Plant Cell Rep 29(2): 113–123 [DOI] [PubMed] [Google Scholar]
- Liu J, Shen J, Xu Y, Li X, Xiao J, Xiong L (2016) Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J Exp Bot 67(19): 5785–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li W (2014) bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 201(4): 1192–1204 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng Y (2005) PM2, A group 3 LEA protein from soybean, and its 22-mer repeating region confer salt tolerance in Escherichia coli. Biochem Biophys Res Commun 331(1): 325–332 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng Y, Zhang Y, Wang W, Li R (2010) Soybean PM2 protein (LEA3) confers the tolerance of Escherichia coli and stabilization of enzyme activity under diverse stresses. Currt Microbiol 60(5): 373–378 [DOI] [PubMed] [Google Scholar]
- Lu Y, Lam H, Pi E, Zhan Q, Tsai S, Wang C, Kwan Y, Ngai S (2013) Comparative metabolomics in Glycine max and Glycine soja under salt stress to reveal the phenotypes of their offspring. J Agric Food Chem 61(36): 8711–8721 [DOI] [PubMed] [Google Scholar]
- Lv X, Zeng X, Hu H, Chen L, Zhang F, Liu R, Liu Y, Zhou X, Wang C, Wu Z, (2021) Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex. Plant cell 33(4): 1182–1195 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59(1): 150–162 [DOI] [PubMed] [Google Scholar]
- Min JH, Chung JS, Lee KH, Kim CS (2015) The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J Integr Plant Biol 57(3): 313–324 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59(1): 651–681 [DOI] [PubMed] [Google Scholar]
- Porcel R, Azcón R, Ruiz-Lozano JM (2004) Evaluation of the role of genes encoding for Δ1-pyrroline-5-carboxylate synthetase (P5CS) during drought stress in arbuscular mycorrhizal Glycine max and Lactuca sativa plants. Physiol Mol Plant Pathol 65(4): 211–221 [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80(6): 847–857 [DOI] [PubMed] [Google Scholar]
- Rengasamy P (2002) Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: an overview. Austral J Exp Agricul 42(3): 351–361 [Google Scholar]
- Robson F, Costa MM, Hepworth SR, Vizir I, Piñeiro M, Reeves PH, Putterill J, Coupland G (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28(6): 619–631 [DOI] [PubMed] [Google Scholar]
- Sheikh AH, Raghuram B, Eschen-Lippold L, Scheel D, Lee J, Sinha AK (2014) Agroinfiltration by cytokinin-producing Agrobacterium sp. Strain GV3101 primes defense responses in Nicotiana tabacum. Mol Plant Microbe Interact 27(11): 1175–1185 [DOI] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354(6312): aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Xu Z, Wang J, Qin Q, Jiang H, Si W, Li X (2018) Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment. Gene 673: 1–11 [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410(6832): 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sun T, Ma N, Wang C, Fan H, Wang M, Zhang J, Cao J, Wang D (2021) A Golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front Plant Sci 12: 638340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45(4): 523–539 [DOI] [PubMed] [Google Scholar]
- Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, Ma B, Bi YD, Lai YC, Liu XL, (2015) GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J 83(2): 224–236 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang H, Shao H, Tang X (2016) Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front Plant Sci 7(248): 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu C, Sun J, Dong L, Li M, Liu Y, Wang J, Zhang X, Li D, Sun J, (2021) GmRAV confers ecological adaptation through photoperiod control of flowering time and maturity in soybean. Plant Physiol 187(1): 361–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zhang W, Qin M, Li S, Qiao M, Liu Z, Xiang F (2017) Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol 58(10): 1764–1776 [DOI] [PubMed] [Google Scholar]
- Wu XX, Li J, Wu XD, Liu Q, Wang ZK, Liu SS, Li SN, Ma YL, Sun J, Zhao L, (2016) Ectopic expression of Arabidopsis thaliana Na+(K+)/H+ antiporter gene, AtNHX5, enhances soybean salt tolerance. Genet Mol Res 15(2): gmr.15027483. [DOI] [PubMed] [Google Scholar]
- Xie Z, Duan L, Tian X, Wang B, Eneji AE, Li Z (2008) Coronatine alleviates salinity stress in cotton by improving the antioxidative defense system and radical-scavenging activity. J Plant Physiol 165(4): 375–384 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125(7): 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang B, Jiang Y, Cheng L, Wu T (2014) GmFNSII-controlled soybean flavone metabolism responds to abiotic stresses and regulates plant salt tolerance. Plant Cell Physiol 55(1): 74–86 [DOI] [PubMed] [Google Scholar]
- Yang C, Huang Y, Lv W, Zhang Y, Akhter Bhat J, Kong J, Xing H, Zhao J, Zhao T (2020) GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci 293: 110442. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang Y, Shan J, Sun J, Li D, Zhang X, Li W, Zhao L (2021) GmIDD is induced by short days in soybean and may accelerate flowering when overexpressed in Arabidopsis via inhibiting AGAMOUS-LIKE 18. Front Plant Sci 12: 629069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K (1995) Correlation between the induction of a gene for Δ1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 7(5): 751–760 [DOI] [PubMed] [Google Scholar]
- Zelm E, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Ann RevPlant Biol 71(1): 403–433 [DOI] [PubMed] [Google Scholar]
- Zhang M, Cao J, Zhang T, Xu T, Yang L, Li X, Ji F, Gao Y, Ali S, Zhang Q, (2022) A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in Glycine max. Front Plant Sci 13: 870695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zheng W, Cao X, Cui X, Zhao S, Yu T, Chen J, Zhou Y, Chen M, Chai S, (2019) Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front Plant Sci 10: 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Li M, Xu C, Xue Y, Li D, Zhao X, Wang K, Li Y, Zhang X, Liu L, (2018) Natural variation in GmGBP1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J 96(1): 147–162 [DOI] [PubMed] [Google Scholar]
- Zhao L, Wang Z, Lu Q, Wang P, Li Y, Lv Q, Song X, Li D, Gu Y, Liu L, (2013) Overexpression of a GmGBP1 ortholog of soybean enhances the responses to flowering, stem elongation and heat tolerance in transgenic tobaccos. Plant Mol Biol 82(3): 279–299 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Ann Rev Plant Biol 53(1): 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.